Significance
We delineate the downstream pathologic consequences underlying the known mitochondrial protein import defect caused by mutant huntingtin (mHTT). We show direct high-affinity mHTT interaction with the inner mitochondrial membrane protein importing complex subunit translocase of mitochondrial inner membrane 23 (TIM23) and show that mHTT more strongly associates with TIM23 than wild-type huntingtin (wtHTT). We find that endogenous full-length wtHTT and mHTT localize in the mitochondrial intermembrane space. We also find that reduction of TIM23-imported mitochondrial matrix proteins is likely due to mHTT binding to TIM23. Thus, the interaction between mHTT and TIM23 results in an altered mitochondrial proteome. Our findings explain a cause of the mitochondrial pathology in Huntington’s disease and provide insight into the mechanistic consequences of mitochondrial mHTT interactions.
Keywords: mutant huntingtin, TIM23, mitochondria, proteostasis, Huntington’s disease
Abstract
Mutant huntingtin (mHTT), the causative protein in Huntington’s disease (HD), associates with the translocase of mitochondrial inner membrane 23 (TIM23) complex, resulting in inhibition of synaptic mitochondrial protein import first detected in presymptomatic HD mice. The early timing of this event suggests that it is a relevant and direct pathophysiologic consequence of mHTT expression. We show that, of the 4 TIM23 complex proteins, mHTT specifically binds to the TIM23 subunit and that full-length wild-type huntingtin (wtHTT) and mHTT reside in the mitochondrial intermembrane space. We investigated differences in mitochondrial proteome between wtHTT and mHTT cells and found numerous proteomic disparities between mHTT and wtHTT mitochondria. We validated these data by quantitative immunoblotting in striatal cell lines and human HD brain tissue. The level of soluble matrix mitochondrial proteins imported through the TIM23 complex is lower in mHTT-expressing cell lines and brain tissues of HD patients compared with controls. In mHTT-expressing cell lines, membrane-bound TIM23-imported proteins have lower intramitochondrial levels, whereas inner membrane multispan proteins that are imported via the TIM22 pathway and proteins integrated into the outer membrane generally remain unchanged. In summary, we show that, in mitochondria, huntingtin is located in the intermembrane space, that mHTT binds with high-affinity to TIM23, and that mitochondria from mHTT-expressing cells and brain tissues of HD patients have reduced levels of nuclearly encoded proteins imported through TIM23. These data demonstrate the mechanism and biological significance of mHTT-mediated inhibition of mitochondrial protein import, a mechanism likely broadly relevant to other neurodegenerative diseases.
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disorder caused by expression of huntingtin (HTT) with a pathologically expanded polyglutamine (polyQ) stretch. In patients with the disease, the HTT CAG repeat region is expanded beyond 35 repeats on the coding region 5′ end (1). There is no effective treatment for HD, which affects 30,000 people in the United States, with ∼200,000 at risk (2). Although the mechanism by which mutant huntingtin (mHTT) mediates pathology is not fully understood, mitochondrial dysfunction plays a critical role in HD pathogenesis, and full-length and fragment mHTT directly associate with mitochondria (3–7). mHTT expression results in mitochondrial cytochrome c release (8, 9), caspase activation (10, 11), calcium dysregulation (7, 12, 13), decreased energetic function (14), impaired mitochondrial trafficking (15, 16), and disrupted mitochondrial dynamics (17, 18).
mHTT fragments bind with the translocase of inner mitochondrial membrane 23 (TIM23) complex and inhibit ornithine transcarbamylase (OTC) import into the mitochondria (19). In R6/2 mice, mHTT-induced mitochondrial protein import inhibition occurs in presymptomatic mice and is prominently manifested in synaptic mitochondria (19). Synaptic mitochondria are more vulnerable to cellular stress than somal mitochondria, a defect exacerbated by mHTT (20). The timing of this abnormality and the direct interaction between mHTT and the TIM23 complex suggest that this is a pathophysiologically important mechanism in HD. Since 99% of mitochondrial proteins are nuclearly encoded and imported (21, 22), we hypothesize that the mHTT–TIM23 complex interaction impairs mitochondrial protein import, altering the mitochondrial proteome. mHTT-mediated changes in the mitochondrial proteome may explain the profound mitochondrial dysfunction documented in HD.
To test this hypothesis, we used ST-Hdh-Q7/Q7 (Q7) and ST-Hdh-Q111/Q111 (Q111) knock-in mouse striatal cell lines that express full-length wild-type (polyQ7) and mutant (polyQ111) HTT. Q111 is a well-established cell line model of HD derived from an HTT knock-in murine embryo (23). Mitochondria isolated from Q111 cells demonstrate reduced OTC import compared with mitochondria from Q7 cells (19).
Proteomic studies explored mitochondrial proteome disturbances using the 2-dimensional difference gel electrophoretic (2D-DIGE) method (24–26). None of these studies directly evaluated mitochondrial proteome alterations in HD. Knowledge regarding mitochondrial protein changes in HD comes from studies performed on Q7 and Q111 cells whole-cell extracts (27), murine HD total brain lysates (28–31), and postmortem HD patient brain tissue (32). However, proteomic analysis of complex biological mixtures cannot delineate specific neuronal mitochondrial proteome alterations due to the presence of nonneuronal mitochondria in tissues and even in cell lines, due to the presence of mitochondrial proteins that may accumulate in the cytosol when import is impaired.
To quantify the impact of mHTT-mediated TIM23 complex activity inhibition on the mitochondrial proteome, we performed 2D-DIGE analysis followed by liquid chromatography-mass spectrometry on mitochondrial protein lysates from Q7 vs. Q111 cells and found that imported matrix proteins levels were decreased in mitochondria from mHTT-expressing cells. To confirm the human disease relevance of the data, we quantified the levels of specific proteins identified in the 2D-DIGE assay from human HD brain mitochondria. These data confirm that the in vitro data are relevant to changes that occur in human HD brain mitochondria.
Results
mHTT Fragment Binds TIM23.
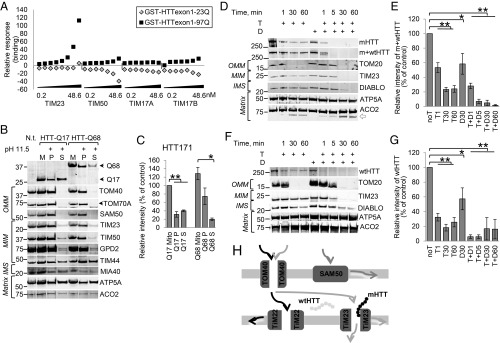
The mHTT fragment binds to the mitochondrial inner membrane TIM23 complex, which consists of TIM17A, TIM17B, TIM23, and TIM50 (19). The wild-type huntingtin (wtHTT) fragment also binds the TIM23 complex but with significantly less affinity. These experiments were performed using an HTT immunoprecipitation pull-down assay with whole-forebrain mitochondria followed by immunoblotting for multiple complex subunits and did not identify the specific complex member that interacts with HTT (19). To determine specificity and affinity of HTT binding to TIM23 complex proteins, we performed affinity binding assays using purified recombinant wtHTT (exon1-23Q) and mHTT (exon1-97Q) fragments and individual TIM23 complex subunits using a surface plasmon resonance (SPR) Biacore analysis platform (SI Appendix, Fig. S1). Data show that mHTT exon1 binds to TIM23 with high affinity (equilibrium dissociation constant [KD] = 5.05 × 10−13) (Fig. 1A and SI Appendix, Table S1), but wtHTT exon1 does not bind to TIM23. Neither wtHTT nor mHTT exon1 bound to any other subunits, including TIM50, TIM17A, and TIM17B. Therefore, mHTT directly binds with high affinity to the TIM23 subunit of the import complex.
Fig. 1.
mHTT localizes in the mitochondrial intermembrane space and binds TIM23. (A) Representative sensogram (n = 3) showing binding affinity of TIM23 complex subunits to HTT exon1-Q23 and -Q97 proteins. Each subunit (TIM23, TIM50, TIM17A, and TIM17B) was separately tested at the concentration range 0.2 to 48.6 nM. Raw sensograms were analyzed to extract relative response values for each subunit (SI Appendix, Table S1). Representative immunoblot (B) and quantification (C) demonstrate that mHTT fragments associate with mitochondria. Mitochondria from HEK293t cells transfected with 171-amino acid-long fragments of wtHTT (HTT-Q17) and mHTT (HTT-Q68) were subjected to alkaline treatment (pH 11.5); then, they were spun to separate mitochondria pellet (P) from supernatant (S) and subjected to immunoblot probing for HTT and for comparison with OMM proteins TOM40, TOM70A, SAM50, and MIM proteins TIM23, TIM50, GPD2, TIM44, soluble mitochondrial intermembrane space (IMS) protein MIA40, and matrix proteins ATP5A and ACO2. Black arrowheads indicate specific bands on blots with more than 1 band. Immunoblot relative fluorescence was normalized to untreated HTT171-Q17 (n = 4, data shown as mean ± SEM). M, untreated mitochondria; N.t., negative transfection sample. *P < 0.05 (t test); **P < 0.001 (t test). Mitochondrial full-length wtHTT and mHTT from cortex of HD4 patients are partially protected from trypsin degradation. Representative immunoblots (D) and quantification (E) of nonsynaptosomal mitochondria subjected to trypsin (T) or trypsin in combination with digitonin (D) treatment for different lengths of time and immunoblotted for mHTT and both wtHTT and mHTT with anti-polyQ (MAB1574) and anti-HTT (MAB2166) antibodies. Blots were also probed with TOM20 for OMM, TIM23 for MIM, DIABLO for IMS, and ATP5A and ACO2 for matrix for mitochondrial subcompartment comparisons. The white arrow indicates cleaved protein product. Immunoblots were normalized to the untreated HTT band (n = 5, data shown as mean ± SEM). *P < 0.05 (t test); **P < 0.001 (t test). Representative immunoblot (F) and quantification (G) demonstrating localization of full-length wtHTT probed with MAB2166 antibody in nonsynaptosomal mitochondria from surgically resected fresh human temporal lobe. Sample setup and quantification as described for D and E (n = 4, data shown as mean ± SEM). *P < 0.05 (t test); **P < 0.001 (t test). (H) Schematic drawing of mitochondrial protein import and localization of wtHTT and mHTT in the intermembrane space of mitochondria. The abbreviations of protein names in all figures are explained in SI Appendix, Table S3.
To validate the in vitro mHTT binding affinity with the TIM23 protein in cells, we performed an alkaline extraction on mitochondria purified from HEK293t cells expressing 171-amino acid-long HTT fragments with wild-type (Q17) and mutant (Q68) polyQ lengths (HTT171-Q17 and HTT171-Q68). High pH disrupts protein interactions, releasing proteins weakly associated with mitochondrial membranes (33). We examined whether HTT is released from the mitochondrial fraction after alkaline wash. We compared our data with well-studied integral mitochondrial membranes proteins (TOM40, TOM70A, SAM50, TIM23, TIM50, GPD2), soluble proteins in the intermembrane space (MIA40) or the matrix (ACO2), and proteins associated with the inner membrane multiprotein complexes (TIM44, ATP5A). Treatment with high pH (11.5) resulted in release of soluble proteins and proteins weakly associated with membranes (MIA40, TIM44, ATP5A, ACO2) into the supernatant, while outer mitochondrial membrane (OMM)-integrated proteins (TOM40, TOM70A, SAM50) were retained in the mitochondrial pellet (Fig. 1B). TIM23 as well as other mitochondrial inner membrane (MIM)-integrated proteins (TIM50, GPD2) were mostly retained in the mitochondrial pellet, with a small fraction released into the supernatant (34), suggesting that the inner membrane proteins are sometimes released by high pH, despite their transmembrane domains. In comparison with these controls, the HTT171-Q68 fragment is mostly retained in the mitochondrial fraction on alkaline extraction, with a small amount released to the supernatant (Fig. 1 B and C), a pattern similar to TIM23. In contrast, a greater proportion of HTT171-Q17 is released from the mitochondria into the supernatant at high pH similar to the soluble or weakly membrane-associated proteins, such as ATP5A, resulting in pellet to supernatant ratios of 4:1 (mHTT) and 1:1 (wtHTT). These data provide additional confirmation of the high-affinity association between mHTT and mitochondria (19).
Full-Length mHTT and wtHTT Reside in the Mitochondrial Intermembrane Space.
Full-length HTT and HTT fragments reside in mitochondria (4, 6, 35), but the intramitochondrial compartment containing HTT is unknown. Therefore, we investigated the mHTT mitochondrial localization. We utilized purified nonsynaptosomal mitochondria from frozen Huntington’s disease grade 4 (HD4) patient cortices. Mitochondria isolated from human brain were treated with trypsin and/or digitonin to assess HTT’s localization within mitochondrial subcompartments. HTT was detected with the MAB2166 antibody that binds both wtHTT and mHTT as well as the MAB1574 antibody that detects only mHTT. Considering the average small difference in polyQ length (about 20 to 25 glutamines, ∼4 kDa) between wtHTT and mHTT in human patients (SI Appendix, Table S2), the 2 different HTT lengths were not separated in our sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) conditions and thus, were analyzed as a single immunoblot band. Trypsin treatment digests proteins associated with the cytoplasmic face of the outer mitochondrial membrane (e.g., TOM20). Since trypsin cannot cross an intact outer mitochondrial membrane, it does not digest proteins inside the mitochondria. Mild digitonin treatment permeabilizes the outer mitochondrial membrane and in combination with trypsin, digests proteins in the mitochondrial intermembrane space and on the outer surface of the inner mitochondrial membrane (e.g., DIABLO, TIM23). Proteins in the matrix (e.g., ACO2) or in the inner membrane oriented toward the matrix are resistant to trypsin/digitonin treatment (e.g., ATP5A). We demonstrate that full-length mHTT and wtHTT are partially digested with trypsin alone and fully digested with the combination of trypsin and digitonin in nonsynaptosomal (Fig. 1 D and E) mitochondria, indicating the intermembrane localization of full-length HTT. About 20% of HTT resists 1-h digestion with trypsin alone. The HTT digestion pattern is similar to that of DIABLO and TIM23. DIABLO is an unbound protein in the mitochondrial intermembrane space, while TIM23 is a transmembrane protein embedded in the inner mitochondrial membrane with a large domain protruding into the intermembrane space. A similar digestion pattern was observed for full-length HTT in mitochondria from surgically resected fresh human temporal lobes, where HTT was completely digested only with the combination of trypsin and digitonin (Fig. 1 F and G). We observed some HTT reduction in digitonin only-treated samples, which may be due to endogenous cellular protease activity. In summary, wtHTT and mHTT reside in the mitochondrial intermembrane space where mHTT binds to the TIM23 complex, inhibiting mitochondrial protein import (Fig. 1H) (19).
Mitochondria from Q7 and Q111 Cell Lines Demonstrate Proteome Differences.
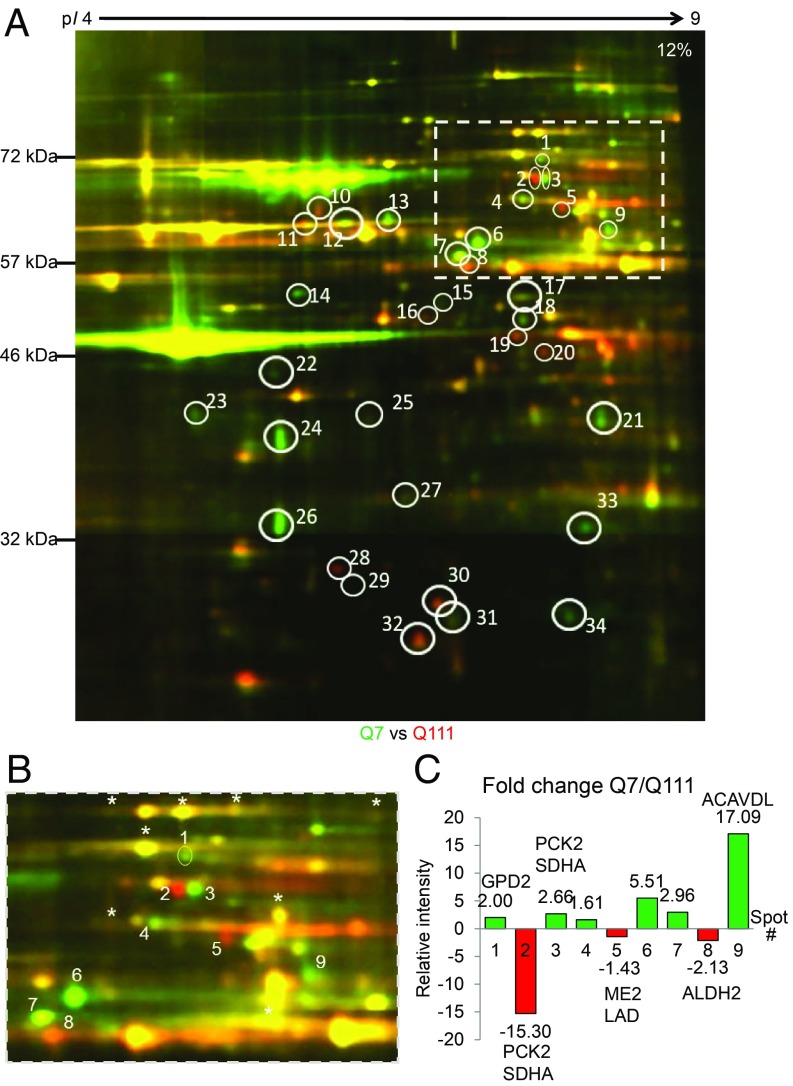
To assess the downstream impact of the mHTT–TIM23 association, which results in the previously demonstrated mitochondrial protein import inhibition (19), we performed 2D-DIGE comparing mitochondrial protein lysates from Q7 and Q111 cells labeled with fluorophores Cy3 and Cy5, respectively. After electrophoresis, SDS polyacrylamide gels were imaged to detect Cy3 and Cy5 signals separately (36). A representative 2D-DIGE image of Q7 and Q111 mitochondrial lysates is shown in Fig. 2A, where green spots denote proteins that are more abundant in the Q7 sample, red spots indicate proteins that are more prevalent in the Q111 sample, and yellow spots indicate proteins that are equal in abundance in both samples. The 2D-DIGE image shows numerous differences between Q7 and Q111 mitochondrial proteomes representing either variations in protein abundance or posttranslational modifications that alter electrophoretic mobility.
Fig. 2.
The mitochondrial proteome is altered in Q111 compared with Q7 striatal cell lines. (A) Representative 2D-DIGE image of Q7 and Q111 mitochondrial fractions. Green indicates Cy3-labeled Q7 sample; red indicates Cy5-labeled Q111 sample. n = 6 with reciprocally dye-labeled technical replicates for each n. The full 2D-DIGE gel image shown is a composite of 16 field-of-view camera shots (36). (B) Enlarged version of the cropped rectangle from A. Reproducible difference proteins are highlighted in white ovals. Area within the rectangle was analyzed in B to estimate fold changes of the difference proteins in Q7 and Q111 samples. *Proteins that do not change between Q7 and Q111. (C) The intensity of difference proteins (spots #1 to #9) was quantified and normalized using intensities of unchanging proteins. Fold change for each difference protein was calculated using the Cy3- and Cy5-normalized fluorescence intensities. Cy3/Cy5 ratios <1 were converted to negative fold change.
The excised gel plugs were processed and analyzed with liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) to identify proteins present in a specific spot. In total, 141 spots were successfully analyzed of 168 picked, yielding 34 identified highly reproducible proteins (SI Appendix, Table S3), of which 20 were mitochondrial proteins. In total, 116 mitochondrial or mitochondria-associated proteins, including isoforms, were identified (SI Appendix, Table S4). The vast majority of the identified mitochondrial proteins are soluble matrix proteins (68%), and about 12% constitute proteins of inner mitochondrial membrane and intermembrane space. Outer mitochondrial membrane proteins comprise only 5% of the identified species. The small proportion of outer mitochondrial membrane proteins observed could either be because TIM23, the translocase inhibited by mHTT, is not involved in recruitment of outer mitochondrial proteins or because hydrophobic multipass transmembrane proteins do not resolve well in isoelectric focusing, the first dimension of DIGE. The remaining identified proteins (15%) are noncanonical mitochondrial proteins known to regulate mitochondrial function.
We deduced 4 classes of protein differences (Fig. 2A) from the 2D-DIGE experiments.
1) Green spots (e.g., spot #1). Green color indicates that this protein is enriched in Q7 mitochondria compared with Q111 mitochondria. GPD2 is one such protein (SI Appendix, Table S3), an enzyme anchored in the inner mitochondrial membrane with import that is TIM23 dependent. GPD2 reduction in Q111 mitochondria is in line with our hypothesis of impaired mitochondrial protein import in mHTT-expressing cells.
2) Horizontal duo shifts (e.g., red spot #2 and green spot #3). Spots #2 and #3 contain mitochondrial PCK2 and SDHA (SI Appendix, Table S3). The horizontal shift is likely due to a change in the protein isoelectric point resulting from a posttranslational modification. SDHA has numerous acetylation and succinylation points, and PCK2 contains 3 phosphorylation sites. The differences in posttranslational modification could be an indirect consequence of mHTT toxicity.
3) Red spots (e.g., spot #8). Enhanced red color indicates increased abundance in Q111 mitochondria. This could be a consequence of gene overexpression, reduced protein degradation, or overall adaptation to mHTT expression. Spot #8 is mitochondrial ALDH2 (SI Appendix, Table S3), a matrix enzyme that converts toxic aldehyde to carboxylate in the ethanol degradation pathway. The overabundance of ALDH2 in Q111 cells is potentially a manifestation of a protective mechanism against the aldehyde buildup that occurs in neurodegeneration diseases and ischemic stroke (37).
4) Diagonal shifts from right to left (e.g., spots #15 and #16). This represents a change in isoelectric point and reduction in molecular weight. LC-MS/MS identified mitochondrial OAT in both spots (SI Appendix, Table S3). OAT is soluble mitochondrial matrix enzyme that is imported through TOM40 and TIM23. The observed changes could be a consequence of N-terminal cleavage of the premature OAT mitochondrial targeting sequence or could represent a different isoform of the protein.
We quantified 9 reproducible difference spot intensities from the selected area (Fig. 2B is an enlargement of cropped rectangle from Fig. 2A) to obtain a relative quantification of the difference proteins in Q7 and Q111 samples. Of 9 difference spots, 6 contained mitochondrial proteins (GPD2, PCK2, SHDA, ALDH2, ME2, LAD, ACADVL), of which 3 spots contained 2 proteins each (spots #2, #3, and #5) (Fig. 2C). The other 3 spots (spots #4, #6, and #7) were nonmitochondrial, likely contaminant proteins (not labeled on Fig. 2C). We found GPD2 and ACADVL (green spots #1 and #9) as well as ME2, LAD, and ALDH2 in red spots (#5 and #8). In horizontal duo shift spots (#2 and #3), PCK2 and SHDA were identified. We calculated the fold change for each difference protein found in spots #1 to #9 using the Cy3- and Cy5-normalized fluorescence intensities (Fig. 2C and SI Appendix, Table S5). We found that intensity increased in Q7 abundance spots, and we found intensity reduction in 2 of 3 Q111 spots, suggesting lower overall abundance of proteins in Q111 samples. Thus, mHTT expression in murine striatal cells results in mitochondrial proteome alterations affecting both protein amounts and posttranslational modifications.
Protein Changes Validated in Q7 and Q111 Mitochondria.
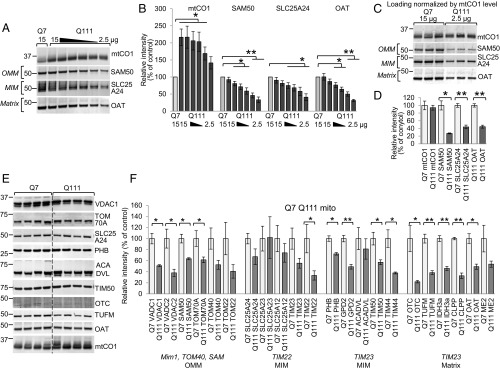
Since ∼99% of mitochondrial proteins are imported (38–40), we evaluated the impact of mHTT on the relative mitochondrial protein levels. Using total protein concentration in mitochondrial lysate as a loading parameter could erroneously correct for the biological change in the levels of imported mitochondrial proteins due to the import defect previously described (19), which would manifest as an artificial overabundance of nonimported mitochondria-encoded proteins. We demonstrated this effect using immunoblotting, where equal total protein loading per lane led to detection of increased levels of a nonimported mitochondrial protein, such as cytochrome c oxidase 1 (mtCO1), in Q111 mitochondria compared with Q7 samples (first 2 lanes of Fig. 3A). To avoid this technical issue, we validated mtCO1 as the loading control. This protein was chosen, because it is encoded by mitochondrial DNA and synthesized in the mitochondrial matrix, omitting the mitochondrial import step, and no major changes in mtCO1 RNA expression level were shown in mHTT-expressing cells (18, 41). We designed a serial dilution experiment (15 to 2.5 µg protein loaded per lane) to find conditions that equalize mtCO1 in Q111 and Q7 mitochondrial samples (Fig. 3A). These data demonstrated that, to obtain approximately equal mtCO1 levels in Q7 and Q111 samples, we need to load 2.5 µg of Q111 mitochondria and 15 µg of Q7 mitochondria per immunoblot lane (Fig. 3B). To monitor the effect of serial dilution on nuclearly encoded imported proteins, the same membrane was immunoblotted for representative proteins of mitochondrial subcompartments: outer membrane protein SAM50, inner membrane protein SLC25A24, and matrix protein OAT. Comparing these 3 nuclearly encoded mitochondrial proteins at an equivalent mtCO1 band intensity (i.e., 15 µg for Q7 cells and 2.5 µg for Q111 cells), there is significantly less protein in the Q111 compared with Q7 mitochondria (Fig. 3A). Interestingly, when we compare the mitochondrial proteins with 15 µg loaded for both samples, the band intensities are essentially equivalent, in sharp contrast to the mtCO1 band. To confirm the level of mtCO1 in equal numbers of Q7 and Q111 mitochondria, we used fluorescence-activated cell sorting (FACS) to obtain mito-eGFP–tagged mitochondria. The mtCO1 protein level is equivalent (P > 0.5) between equal numbers of mitochondria originating from Q7 and Q111 cells (SI Appendix, Fig. S2 A and B, Left, bars Q7 and Q111 4 MM Mito mito-eGFP).
Fig. 3.
Nuclearly encoded mitochondrial protein levels are reduced in the Q111 striatal cell line. (A–D) mtCO1 validated as a normalization control in mitochondria of Q7 and Q111 striatal cell lines. Representative immunoblot (A) and quantification (B) comparing mtCO1 level in Q7 samples (15 µg) and Q111 serial dilution mitochondrial samples (15 to 2.5 µg). The same membrane was immunoblotted for representative proteins of mitochondrial subcompartments: SAM50 for OMM, SLC25A24 for MIM, and OAT for matrix. Each protein was normalized to the Q7 sample (n = 3, data shown as mean ± SEM). *P < 0.05 (t test); **P < 0.001 (t test). Representative immunoblot (C) and quantification (D) of mtCO1 in 3 Q7 and 3 Q111 independently isolated mitochondrial fractions with differential protein loading to equalize the mtCO1 content (15 µg Q7 mitochondrial lysate and 2.5 µg Q111). The same membrane was immunoblotted for SAM50, SLC25A24, and OAT. For quantification, Q111 samples were normalized to Q7 samples for each protein (n = 3, data shown as mean ± SEM). *P < 0.05 (t test); **P < 0.001 (t test). Representative immunoblot (E) and quantification (F) of mitochondrial proteins from Q7 and Q111 cells. Mim1, TOM40, SAM, TIM22, and TIM23 represent import pathway of mitochondrial proteins. The immunoblot images for proteins that were analyzed but not shown in E are included in SI Appendix, Fig. S4. Band intensities were controlled for loading by dividing by mtCO1 intensity (F) and then normalized to the average Q7 band intensity (n = 4, data shown as mean ± SEM). *P < 0.05 (t test); **P < 0.001 (t test).
Thus, imported mitochondrial proteins are significantly underrepresented compared with mitochondrially encoded proteins. This underrepresentation would be overlooked if an equal amount of protein is loaded and analyzed.
Based on these findings, for subsequent experiments, we loaded 15 µg Q7 and 2.5 µg Q111 mitochondrial lysates to visually correct for the mtCO1 band intensity; all quantification was done by normalizing to the mtCO1 band (Fig. 3C). The representative nuclearly encoded mitochondrial proteins (SAM50, SLC25A24, and OAT) were compared in Q111 samples vs. Q7 (Fig. 3D). OAT, which is a matrix-targeted protein imported through the TIM23 pore, was reduced by 55% in Q111 mitochondria. Outer and inner membrane proteins SAM50 and SLC25A24 were also significantly reduced (SAM50 73% and SLC25A24 56%), likely due to a broad import dysfunction in Q111 cells (19). Therefore, mitochondrially encoded mtCO1 normalizes protein loading for mitochondrial content to prevent overrepresentation of nuclearly encoded mitochondrial proteins.
Nuclearly Encoded Mitochondrial Protein Levels Are Reduced in Q111 Compared with Q7 Cells.
Based on the DIGE data, we selected several soluble matrix proteins that are imported through TIM23 (TUFM, IDH3A, CLPP, OAT, ME2) for validation using immunoblotting (SI Appendix, Table S3). OTC was also analyzed based on previous mitochondrial protein import assay data from mHTT-expressing cells and R6/2 mice brains, where we have shown reduction of OTC import (19). We also included proteins anchored in the inner mitochondrial membrane (GPD2, ACADVL, PHB) for additional validation. Two inner membrane-associated subunits (TIM50, TIM44) were added (SI Appendix, Table S4) to supplement a group of TIM23-imported membrane-bound proteins. To get broader information about protein changes and test whether proteins imported by the TIM23 pathway demonstrate a greater reduction, we included in the analysis multipass transmembrane proteins localized in the mitochondrial outer membrane (VDAC1, VDAC2, SAM50, TOM70A) (SI Appendix, Table S4) that use the TOM40 and SAM50 pathways to integrate into the mitochondrial membrane (Fig. 1H) and multipass inner membrane proteins (SLC25A24, SLC25A23) (SI Appendix, Tables S3 and S4) that follow the TIM22 pathway for import (SI Appendix, Table S6). To make the number of TIM22-imported proteins equal to the number of TIM23-imported proteins, for statistical purposes, we added 5 canonical mitochondrial proteins (TOM40, TOM22, TIM23, TIM22, SLC25A12) (SI Appendix, Table S6). In total, we analyzed 22 nuclear-encoded mitochondrial proteins in mitochondrial fractions purified from Q7 and Q111 cell lines. Proteins identified in horizontal duo shift spots were excluded from the analysis, since this isoform change is likely caused by a posttranslational modification that may not be recognized using immunoblotting. Mitochondrial proteins with multiple known intracellular localization sites (HSP60, GCAT, PRDX3) were not included due to their potential presence in contaminating organelles that may confound our conclusions regarding mHTT mitochondrial effects. Mitochondrial fraction enrichment was verified by immunoblotting for mitochondrial markers VDAC1, ATP5A, ACO2, and mtCO1 and common contaminants, such as calreticulin (CALR) for endoplasmic reticulum, LAMP1 for lysosomes, RCAS1 for Golgi apparatus, and TUBA for cytosol (SI Appendix, Fig. S3A). Every biological repeat of Q7 and Q111 mitochondrial sample for analysis was obtained from a separate passage and independent isolation. To quantify the protein amount, each specific band signal intensity was normalized by the mtCO1signal on the same polyvinylidene difluoride (PVDF) membrane.
All mitochondrial proteins were reduced in Q111 samples (Fig. 3 E and F and SI Appendix, Fig. S4), except for SLC25A23, the inner membrane integral protein. The average reduction was the greatest (51%) for the group of TIM23-imported proteins (multivariate analysis of variance [MANOVA] P = 0.008 includes as dependent variables matrix soluble proteins CLPP, ACADVL, OTC, ME2) (SI Appendix, Table S7). Reduction in TIM22-imported proteins was 42% in Q111 samples compared with Q7 (SI Appendix, Table S7). Therefore, full-length mHTT expression results in global mitochondrial protein dysregulation in Q111 cells, most severely impacting soluble matrix proteins.
Mitochondrial Proteome Disturbance in HD Patients’ Striatum and Cortex.
HD is a complex disease, and cellular models cannot fully replicate pathogenesis. Even though mHTT is expressed in all tissues, medium spiny striatal neurons are the earliest and most impacted by mHTT, and their degeneration is followed by cortical neuron degeneration (42, 43). We, therefore, analyzed mitochondria-enriched fractions originating from Huntington’s disease grade 2 (HD2) (44) human striatal tissue and cortical tissues from HD4 patients. We attempted to isolate mitochondria from HD4 striatal tissue, but we could not analyze mitochondria due to extremely low yield after isolation procedures. This was likely due to the low number of surviving neurons in HD4 striatum as well as the inability to purify damaged mitochondria in these postmortem samples. Mitochondrial fractions were also isolated from control striatum and cortex for comparison. Mitochondrial fraction enrichment was assessed by probing isolated nonsynaptosomal and synaptosomal mitochondria for mitochondrial membrane and mitochondrial matrix protein markers (VDAC1, ATP5A, ACO2, mtCO1) (SI Appendix, Fig. S3B). The presence of other cellular compartments was assessed by probing for intracellular contaminant markers (TUBA, CALR, LAMP1, RCAS1, CAV2) in nonsynaptosomal and synaptosomal mitochondrial fractions alongside whole-brain homogenate and cytosolic fractions.
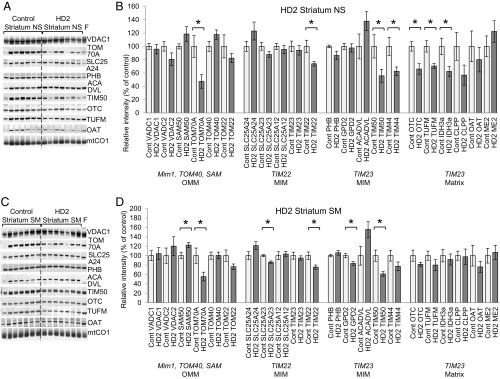
We tested the mitochondrial proteins as described for the cellular HD model in nonsynaptosomal and synaptosomal mitochondrial fractions purified from frozen HD2 striatum and HD4 cortex (SI Appendix, Table S2). The set of 7 HD2 and HD4 samples with 7 corresponding control samples was accompanied by mitochondrial fractions prepared from fresh surgically resected human temporal lobe tissue (last lanes in Figs. 4 A and C and 5 A and C and SI Appendix, Figs. S5 and S6) as an indicator of protein degradation in frozen tissue. We quantified the levels of individual mitochondrial proteins in HD2 striatum (Fig. 4 B and D and SI Appendix, Table S8) and HD4 cortex (Fig. 5 B and D and SI Appendix, Table S9). Given the expected variability in human postmortem brain specimens, we found few statistically significant individual protein changes. To increase the power of our analysis, we applied MANOVA for proteins grouped by import pathway categorized as TIM23 and non-TIM23 imported (SI Appendix, Table S6). TIM23-imported protein includes matrix and inner membrane with import that we expect to be affected by mHTT. Non-TIM23 imported proteins include outer mitochondrial membrane and inner membrane TIM22-imported proteins. In striatal nonsynaptosomal HD2 mitochondria, which include the neuronal soma as well as nonneuronal brain cells, the TIM23 imported proteins were reduced by 18% compared with control, and 7 proteins of 11 (PHB, TIM44, GPD2, ACADVL, CLPP, OAT, ME2) were dependent variables (MANOVA P = 0.003) (SI Appendix, Table S8). In the same mitochondria, outer membrane proteins and inner membrane non-TIM23–imported proteins were 8% lower than in controls (MANOVA P = 0.019; includes as dependent variables VDAC2, SAM50, TOM70A, TOM40, SLC25A12, SLC25A23, TIM22, TIM23). However, group analysis of HD2 synaptosomal mitochondria, a fraction that is restricted to only neurons, found that level of proteins did not decrease similarly (SI Appendix, Table S8). This may be due to the known widespread loss of vulnerable neurons in the HD striatum. Our additional data support this explanation, because we found that HD striatal tissue has only one-half as much (51.5% decrease) synaptosomal mitochondria compared with controls (SI Appendix, Fig. S7A). The decreased yield of synaptosomal, but not nonsynaptosomal, mitochondria could be a consequence of synaptic degradation in the striatum. This hypothesis is supported by the 2-fold decrease in the striatal neuronal marker GAD1 (SI Appendix, Fig. S7 B and C) (42), a 67-kDa form of glutamic acid decarboxylase that catalyzes the production of GABA, in HD2 striatal tissue homogenate, confirming degeneration of striatal neurons. The decrease in viable striatal neurons from which to obtain mitochondria may explain the absence of protein changes, because the vulnerable synapses were already degenerated.
Fig. 4.
Mitochondrial protein levels are dysregulated in the striatum of HD2 patients. Immunoblots and quantification of mitochondrial proteins in nonsynaptosomal (NS; A and B) and synaptosomal (SM; C and D) mitochondria. Samples prepared from the striatum of non-HD patients were used as a control, and mitochondrial lysates from fresh (F) surgically resected cortex tissue of non-HD patients were included. Each lane represents the mitochondrial fraction from an independent tissue block. Mim1, TOM40, SAM, TIM22, and TIM23 represent import pathway of mitochondrial proteins. The immunoblot images for additional analyzed proteins are shown in SI Appendix, Fig. S5. For quantification, the intensity of each nuclearly encoded protein band was normalized to mtCO1 level in the same sample and normalized to the average control sample band intensity. The normalized values in control samples were taken as 100% and used to recalculate values for HD2 samples (n = 7, data shown as mean ± SEM). *P < 0.05 (t test); **P < 0.001 (t test).
Fig. 5.
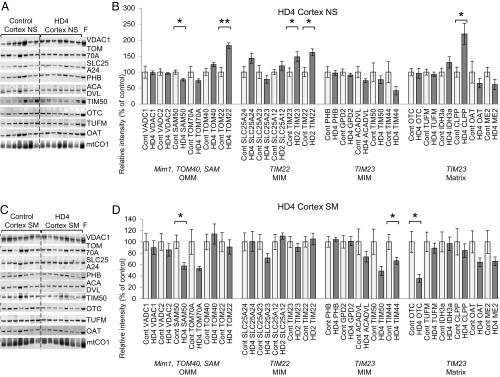
Protein dysregulation in cortical mitochondria of HD4 patients. Immunoblots and quantification of mitochondrial proteins in nonsynaptosomal (NS; A and B) and synaptosomal (SM; C and D) mitochondria. Samples prepared from cortex of non-HD patients were used as a control, and mitochondrial lysates from fresh (F) surgically resected cortex tissue of non-HD patients were included. Each lane represents the mitochondrial fraction from independent tissue block. Mim1, TOM40, SAM, TIM22, and TIM23 represent import pathway of mitochondrial proteins. The immunoblot images for additional analyzed proteins are shown in SI Appendix, Fig. S6. For quantification, the intensity of each nuclearly encoded protein band was normalized to mtCO1 level in the same sample and normalized to the average control sample band intensity. The normalized values in control samples were taken as 100% and used to recalculate values for HD4 samples (n = 7, data shown as mean ± SEM). *P < 0.05 (t test); **P < 0.001 (t test).
In synaptosomal mitochondria from HD4 cortex, group analysis showed a reduction of TIM23-imported proteins (24%, MANOVA P = 0.003 includes as dependent variables PHB, TIM44, ACADVL, OTC, CLPP, ME2) (Fig. 5D and SI Appendix, Table S9). This is the same import pathway demonstrated to be directly inhibited by mHTT expression (19). In nonsynaptosomal mitochondria, the same group of proteins did not change in abundance compared with control (Fig. 5B and SI Appendix, Table S9). This finding correlates with our previous study demonstrating inhibition of protein import specifically in synaptosomal mitochondria of R6/2 mice, a murine model of HD (19). Protein import inhibition was also demonstrated in R6/2 nonsynaptosomal mitochondria but to a lesser degree and at a later stage of disease progression, pointing at more significant impact in synaptic mitochondria. These data are consistent with our hypothesis that mHTT impairs TIM23-mediated protein import, resulting in reduced mitochondrial protein content in HD tissues.
Discussion
We demonstrate that endogenous full-length mHTT and wtHTT localize within the intermembrane space of neuronal mitochondria. HTT is a predominantly cytosolic protein that can be transported into the nucleus (35) and colocalize with the endoplasmic reticulum and Golgi apparatus (45). Our finding demonstrates an additional site of intracellular HTT localization. While the role of wtHTT inside mitochondria remains to be investigated, mHTT binds the TIM23 subunit of the inner membrane translocase complex and remains associated with membranes after high-pH treatment. mHTT interacts with TIM23, leading to a specific pattern of the mitochondrial proteome dysregulation in HD. Several comprehensive proteomic studies performed on human and mouse cell models of HD (27, 46) and brain tissues of HD patients and mHTT knock-in mouse (32, 47) documented only isolated alteration of some mitochondrial proteins. Considering that those studies were performed on whole-tissue or cell homogenates and were aimed to find general proteomic changes in mHTT-expressing models, accurate investigation of mitochondrial proteome was not feasible. To understand the mitochondrial protein import defect consequences, we applied a targeted strategy. First, we utilized isolated mitochondria to compare proteomic profiles of mHTT-expressing cells. Second, to assess the effect TIM23 impairment on mitochondrial protein level, we normalized nuclearly encoded protein to the mitochondrial-encoded mtCO1 protein. Third, we analyzed proteins by groups based on their localization and mitochondrial import pathway.
Mitochondria play an important role in the pathogenesis of neurodegenerative diseases, including HD. Most mitochondrial proteins are encoded in the nucleus and imported into mitochondria through pore complexes of translocases of mitochondrial membranes (TOM40, SAM50, TIM23, TIM22) (38–40). Alterations of mitochondrial protein abundance were first revealed in “global” proteomic studies of mouse ST-Hdh-Q111/Q111 cells (27) and HD-affected human embryonic stem cells (46), indicating proteome disturbance. Utilizing 2D-DIGE analysis of mitochondrial fractions isolated from Q111 and Q7 cells, we detected multiple protein differences between these 2 fractions. We picked for additional investigation only the abundance differences that may represent protein import dysregulation. We also observed changes suggesting posttranslational mHTT-driven dysregulation in mitochondrial proteome. Of 34 highly reproducible difference proteins, we found that 20 were mitochondrial proteins (SI Appendix, Table S3). Nonmitochondrial proteins could be introduced by impurities of mitochondria isolation techniques and potential association of these proteins with mitochondrial surface. Assembling groups of validated proteins for analysis based on import pathway and utilizing endogenous mtCO1 as normalization parameter were essential to test our hypothesis. Indeed, semiquantitative study of protein level and the analysis by the import pathway group revealed that mitochondria were deficient in TIM23-imported proteins in Q111 cells. Outer and inner membrane proteins were also reduced, likely due to a broad import dysfunction in Q111 cells resulting from TIM23 dysfunction. Although we do not directly measure mitochondrial function in this study, compromised function was documented previously in HD (4, 7, 8, 10) and could be a consequence of proteome alterations documented here.
Proteomic analysis of brain samples of HD patients (32) and HdhQ150 and HdhQ92 knock-in mouse models (47) showed increased oxidative stress, activation of antioxidant defense, and pronounced changes in protein abundance in the caudate region vs. the cortex. We tested the same mitochondrial proteins identified in 2D-DIGE with Q7 and Q111 samples on mitochondrial fractions originating from neurons and glia (nonsynaptosomal) and synaptic processes (synaptosomal) of striatum and cortex of HD patients. Interestingly, TIM23-imported proteins were reduced in brain mitochondria of HD2 and HD4 patients, indicating an identical pathological effect of mHTT on protein import in cell line models and HD. Interestingly, a group of TIM23-imported proteins in nonsynaptosomal mitochondria of HD4 patients did not show a decrease as did the same group in synaptosomal mitochondria, which correlates with decreased import activity in synaptosomal mitochondria of R6/2 mice (19). The most significant reduction of matrix proteins was observed in striatal nonsynaptosomal mitochondria of HD2 patients. This could be a consequence of the significant loss of neuronal processes at this stage as confirmed by a spiny neuron marker GAD1 and deterioration of synapses and synaptic mitochondria (48). Notably, not every protein change was consistent between the cell line data and the patient’s samples. We believe that this is not unexpected given the biologic variability among patient samples compared with the relative consistency of a monoclonal ST-Hdh-Q111/Q111 cell line when data were obtained from mitochondria of 4 independent cell passages, whereas data from human brain mitochondria represent an average of 7 individual patients in each group with different CAG repeats (CAG40-54 for mHTT allele), age (39 to 89 y), a mixture of genders, and a wide range of postmortem interval (5 to 85 h) (SI Appendix, Table S2).
In summary, our findings demonstrate alterations in the mitochondrial proteome that likely result in the known mitochondrial dysfunction in HD (49). Given the early interaction of mHTT with the TIM23 complex and inhibition of mitochondrial protein import, this work provides evidence of a direct and primary mechanism of mitochondrial pathogenesis in HD, which is in addition to and independent from transcriptional dysregulation and overall disturbance of protein degradation by mHTT. Obstruction of mitochondrial protein import may exacerbate other pathway and mitochondrial functions, like mtUPR, since major mtUPR regulators (CLPP, HSP60) are matrix-imported proteins.
Since mitochondrial protein import defects were demonstrated in Parkinson’s disease (50) and amyotrophic lateral sclerosis models (25), the mitochondrial proteome alterations and mitochondrial dysfunction documented here may be broadly applicable to other neurodegenerative diseases. Thus, improving mitochondrial function may be an important therapeutic approach in HD and other neurodegenerative disorders.
Materials and Methods
Additional detailed materials and methods are included in SI Appendix.
Plasmids and recombinant proteins preparation are described in details in SI Appendix.
Cell Culture.
ST-Hdh-Q111/Q111 (Q111) and ST-Hdh-Q7/Q7 (Q7) cell lines were provided by Marcy MacDonald, Massachusetts General Hospital, Harvard Medical School, Charlestown, MA (23). Cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 5% fetal bovine serum (FBS) and 1% sodium pyruvate at 33 °C in the presence of 5% CO2. For each 2D-DIGE gel, 8 10-cm2 tissue culture dishes of 80% confluent Q7 cells and 12 80% confluent dishes of Q111 cells were cultured for harvesting.
HEK293t cells were cultured in 5% FBS-supplemented DMEM at 37 °C and 5% CO2. The overexpression of HTT171 fragments was achieved using Lipofectamine2000 Transfection Reagent (Invitrogen) following the manufacturer’s recommendations for 10-cm2 tissue culture dishes volume. For each vector, 3 tissue dishes were transfected and incubated for 48 h before cell harvesting for mitochondria isolation.
SPR Biacore Analysis.
All Biacore experiments were performed using a Biacore 1000 instrument (GE Healthcare) and Hepes (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) saline running buffer supplemented with ethylenediaminetetraacetic acid (EDTA) and surfactant P20 (HBS-EP) (10 mM Hepes, 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P20, pH 7.4). Anti-GST antibody (30 μg/mL) included in the GST Capture kit (GE Healthcare) was coupled to the surface of Series S CM5 sensor chips (GE Healthcare) by an amine-coupling procedure as recommended by the manufacturer with a mixture containing N-ethyl-N′-(dimethylaminopropyl)-carbodiimide and N-hydroxysuccinimide. Typically, between 100 and 150 resonance units of GST antibody was immobilized, and unoccupied high-affinity binding sites were blocked with GST proteins. One of the sensor chips, a reference surface, was prepared with activation and deactivation treatments but not the adjacent protein-coupled surface. The examined GST-HTT exon1 proteins, each an immobilized ligand, were injected (5 µg/mL) for 2 min at a flow rate of 20 µL/min to enable binding with GST antibody. Purified TIM23, TIM50, TIM17A, and TIM17B proteins at different concentrations (0.2, 0.6, 1.8, 5.4, 16.2, and 48.6 nM) were injected into the immobilized ligand surface of the sensor chip to obtain SPR sensograms. The baseline-corrected sensograms (with the buffer blank run further subtracted) were globally fitted to a predefined binding model using BIAevaluation software (version 2.0.4). Between experiments, the surfaces were rigorously regenerated with multiple pulses of 2 M NaCl and 1.5 M glycine-HCl, pH 2.5, followed by an extensive washing with HBS-EP running buffer.
Alkaline and Trypsin Treatment.
Mitochondria isolated from HEK293t cells were treated with alkaline buffer (225 nM sucrose, 75 mM D-mannitol, 0.1 M Na2CO3, pH 11.5). Twenty micrograms of mitochondria resuspended in 20 µL of alkaline buffer and supplemented with protease inhibitor mixture Set III (Millipore Sigma) were incubated on ice for 15 min and centrifuged for 10 min at 13,000 × g to pellet mitochondria. The supernatant fraction was transferred to a separate tube. All samples were lysed with radioimmunoprecipitation assay (RIPA) buffer and analyzed in SDS/PAGE. Nonsynaptosomal mitochondria (40 µg) from cortex of HD4 patients and from surgically resected fresh human temporal lobes were subjected to trypsin (25 µg/mL) or trypsin and digitonin (0.25%) treatment at different time points. The reaction was stopped by adding protease inhibitors mixture. Samples were subjected to SDS/PAGE, and PVDF membranes were immunoblotted with anti-HTT (MAB2166, clone 1HU-4C8) or anti-polyQ (MAB1574, clone 5TF1-1C2) antibodies followed by striping and reprobing with antibodies detecting proteins of different mitochondrial compartments (OMM, MIM, and matrix).
Mitochondria Fractionation from Cell Lines.
Mitochondria from HEK293t, ST-Hdh-Q7/Q7, and ST-Hdh-Q111/Q111 cell lines were isolated using the Mitochondria Isolation MACS Kit (Miltenyi Biotec) according to the manufacturer’s protocol (the detailed procedure id described in SI Appendix). Mitochondria were eluted from MACS column with isolation buffer (225 mM sucrose, 75 mM mannitol, 10 mM Hepes, pH 8 adjusted with NaOH) on column removal from the magnetic field and pelleted at 13,000 × g for 4 min. For DIGE, mitochondrial samples were prepared using DIGE-lysis buffer (7 M urea, 2 M thiourea, 10 mM 1,4-dithiothreitol [DTT], 4% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate [CHAPS], 10 mM Hepes, pH 8 adjusted with NaOH, high performance liquid chromatography [HPLC] H2O) supplemented with protease inhibitor mixture Set III. Protein lysate that appeared viscose was sonicated for 30 s and spun at 20,000 × g to pellet and remove insoluble cell debris and iron beads from the final sample. Mitochondrial fractions utilized for immunoprobing with specific antibodies were eluted from the column with isolation buffer (225 mM sucrose, 75 mM mannitol, 5 mM Hepes-Tris). The mitochondrial pellet (13,000 × g, 4 min) was lysed in Pierce RIPA buffer (25 mM Tris⋅HCl, pH 7.6, 150 mM NaCl, 1% Nonidet P-40, 1% Na deoxycholate, 0.1% SDS; Thermo Scientific) supplemented with protease inhibitor mixture Set III (Calbiochem) and spun at 13,000 × g for 5 min to separate insoluble debris from protein lysate. Protein concentration was determined using Bradford reagent (BioRad) using a bovine serum albumin (BSA) standard made in DIGE-lysis buffer.
For alkaline treatment before mitochondria isolation, HEK293t cells were trypsinized and washed in phosphate-buffered saline (PBS buffer). Mitochondrial fractions were eluted from MACS column with isolation media II buffer (225 mM sucrose, 75 mM D-mannitol, 5 mM Hepes, pH 7.4 adjusted with Tris) and resuspended to measure protein concentration and aliquot for assay.
2D-DIGE and Spots Quantification.
One hundred micrograms of protein of the mitochondrial fraction obtained from Q7 cells was labeled with Cy3-NHS monoreactive dye (GE Healthcare), 100 µg from Q111 was labeled with Cy5-NHS (GE Healthcare) as described (51), and the 2 were combined. Seven biological replicates of Q7 vs. Q111 mitochondrial samples were run, and each biological replicate was performed with a reciprocally dye-labeled replicate. For isoelectric focusing (first dimension separation), mixed samples were applied to 18-cm IPG strips pH 3 to 10NL (GE Healthcare). IPG strips were transferred onto 12% SDS/PAGE (second dimension separation) using in-gel equilibration (52). Fluorescence images were acquired using a custom-built imager (53). Spots with relatively different Cy3 or Cy5 fluorescence intensity were excised on the fluorescence gel imager/spot picker platform (53, 54) and submitted for protein identification using LC-MS/MS. We picked on average 14 difference protein spots per gel, each with a reciprocal repeat. Each of the identified proteins was verified by matching the placement on the gel with the predicted molecular weight and isoelectric point. To estimate fold changes of the difference protein between Q7 and Q111 samples, the intensity of spots in the Cy3 and Cy5 channels was quantified using an open-source astronomy software package SExtractor (Source Extractor) (55). Cy3 and Cy5 channels were played in a video loop to identify “Guide star” proteins (marked with asterisk) that do not change between the 2 channels. Difference proteins were normalized using the intensities of Guide star proteins as previously described (56) (SI Appendix, Table S5).
LC-MS/MS.
Gel spots were reductively alkylated with DTT/3-indolacetic acid (IAA) and digested with trypsin according to standard protocol (57). Tryptic peptides were analyzed by nanoreverse-phase HPLC interfaced with an LTQ XL linear ion trap mass spectrometer (Thermo Fisher Scientific). The tandem mass spectra were analyzed using the MASCOT (Matrix Science) search engine. The identified peptides and proteins were further analyzed with the Scaffold software. Only proteins with the following high-confidence identifications were considered: 1) protein identification probability of 99% or above, 2) peptide identification probability of 95% or above, and 3) a minimum of 2 peptides. The molecular weight and isoelectric point of identified proteins were then cross-referenced against their position in 2D-DIGE gel images for confirmation.
Mitochondria Isolation and FACS Sorting.
For the single mitochondria sorting experiment, 8 dishes of Q7 and 12 dishes of Q111 were utilized. Forty-eight hours posttransfection, cells were harvested and subjected to mitochondria isolation as described above. The final mitochondrial fraction was eluted from the column with 2 mL of sorting buffer (125 mM KCl, 1% BSA) and separated based on fluorescent (eGFP) signal with a BD FACS Aria II sorter at The Unified Flow Core at the University of Pittsburgh.
Mitochondria Isolation from Human Brain Tissue.
Cortex samples of HD4 patients, striatum samples of HD2 patients, and control patients’ samples were obtained from the New York Brain Bank at Columbia University. Use of postmortem samples was approved by the University of Pittsburgh Committee for Oversight of Research and Clinical Training Involving Decedents (CORID). Clinical information of the patients and subjects is indicated in SI Appendix, Table S2. Fresh cortex tissue was surgically resected from human temporal lobes, placed in ice-cold isolation media, and used for mitochondria isolation immediately. Donation of residual patient tissue after surgery was approved by the University of Pittsburgh Institutional Review Board (PRO11080392). Fractions of nonsynaptosomal and synaptosomal mitochondria were isolated from tissues as described (58). Mitochondrial lysates were prepared in RIPA buffer for further immunoblotting (as described above).
Immunoblotting.
Proteins immunoblotted with primary and secondary IRDye infrared fluorescent dye-labeled antibody (SI Appendix, Table S10) were detected with the Odyssey CLx near-infrared fluorescence imager (LI-COR Biosciences). Detailed procedures are included in SI Appendix.
Statistical Analysis.
Statistical significance of differences in protein levels between the HD patients and controls in immunoblotting were compared by 2-tailed t tests. Proteins were further grouped according to import pathway. MANOVA was applied to assess the difference between HD patients and controls among the grouped proteins. P < 0.05 was used as the significance threshold.
Supplementary Material
Acknowledgments
We thank flow cytometry facility manager Dewayne Falkner from the Department of Immunology of the University of Pittsburgh for establishing the protocol for live mitochondria FACS sorting and R. Mark Richardson for providing fresh surgically resected human temporal lobe tissues. Human brain tissues were obtained from the New York Brain Bank at Columbia University/Taub Institute, supported by NIH Grant P50AG008702. For protein identification, this project used the Hillman Cancer Proteomics Facility that is supported in part by NIH Award P30CA047904. This work was supported by Walter L. Copeland Fund of The Pittsburgh Foundation Grant MR2014-67795 (to S.Y.) and NIH Grant R01NS039324 (to R.M.F.).
Footnotes
Conflict of interest statement: R.M.F. is on the board of NeuBase Therapeutics. No funding for this work was received from NeuBase Therapeutics.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904101116/-/DCSupplemental.
References
- 1.Gusella J. F., MacDonald M. E., Ambrose C. M., Duyao M. P., Molecular genetics of Huntington’s disease. Arch. Neurol. 50, 1157–1163 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Pringsheim T., et al. , The incidence and prevalence of Huntington's disease: A systematic review and meta-analysis. Mov. Disord. 27, 1083–1091 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Yu Z. X., et al. , Mutant huntingtin causes context-dependent neurodegeneration in mice with Huntington’s disease. J. Neurosci. 23, 2193–2202 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orr A. L., et al. , N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J. Neurosci. 28, 2783–2792 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ratovitski T., et al. , Huntingtin protein interactions altered by polyglutamine expansion as determined by quantitative proteomic analysis. Cell Cycle 11, 2006–2021 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo X., et al. , VCP recruitment to mitochondria causes mitophagy impairment and neurodegeneration in models of Huntington’s disease. Nat. Commun. 7, 12646 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choo Y. S., Johnson G. V., MacDonald M., Detloff P. J., Lesort M., Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum. Mol. Genet. 13, 1407–1420 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Wang X., et al. , Inhibitors of cytochrome c release with therapeutic potential for Huntington’s disease. J. Neurosci. 28, 9473–9485 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiechle T., et al. , Cytochrome C and caspase-9 expression in Huntington’s disease. Neuromolecular Med. 1, 183–195 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y., et al. , Huntingtin inhibits caspase-3 activation. EMBO J. 25, 5896–5906 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen M., et al. , Minocycline inhibits caspase-1 and caspase-3 expression and delays mortality in a transgenic mouse model of Huntington disease. Nat. Med. 6, 797–801 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Milakovic T., Quintanilla R. A., Johnson G. V., Mutant huntingtin expression induces mitochondrial calcium handling defects in clonal striatal cells: Functional consequences. J. Biol. Chem. 281, 34785–34795 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Giacomello M., Oliveros J. C., Naranjo J. R., Carafoli E., Neuronal Ca(2+) dyshomeostasis in Huntington disease. Prion 7, 76–84 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bossy-Wetzel E., Petrilli A., Knott A. B., Mutant huntingtin and mitochondrial dysfunction. Trends Neurosci. 31, 609–616 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trushina E., et al. , Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol. Cell. Biol. 24, 8195–8209 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang D. T., Rintoul G. L., Pandipati S., Reynolds I. J., Mutant huntingtin aggregates impair mitochondrial movement and trafficking in cortical neurons. Neurobiol. Dis. 22, 388–400 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Shirendeb U. P., et al. , Mutant huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’s disease. Hum. Mol. Genet. 21, 406–420 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manczak M., Reddy P. H., Mitochondrial division inhibitor 1 protects against mutant huntingtin-induced abnormal mitochondrial dynamics and neuronal damage in Huntington’s disease. Hum. Mol. Genet. 24, 7308–7325 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yano H., et al. , Inhibition of mitochondrial protein import by mutant huntingtin. Nat. Neurosci. 17, 822–831 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baranov S. V., et al. , Mitochondria modulate programmed neuritic retraction. Proc. Natl. Acad. Sci. U.S.A. 116, 650–659 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor S. W., et al. , Characterization of the human heart mitochondrial proteome. Nat. Biotechnol. 21, 281–286 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Sickmann A., et al. , The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. U.S.A. 100, 13207–13212 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trettel F., et al. , Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum. Mol. Genet. 9, 2799–2809 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Xie J., et al. , A two-dimensional electrophoretic map of human mitochondrial proteins from immortalized lymphoblastoid cell lines: A prerequisite to study mitochondrial disorders in patients. Proteomics 5, 2981–2999 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Li Q., et al. , ALS-linked mutant superoxide dismutase 1 (SOD1) alters mitochondrial protein composition and decreases protein import. Proc. Natl. Acad. Sci. U.S.A. 107, 21146–21151 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiorini A., et al. , Lack of p53 affects the expression of several brain mitochondrial proteins: Insights from proteomics into important pathways regulated by p53. PLoS One 7, e49846 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choudhury K. R., Das S., Bhattacharyya N. P., Differential proteomic and genomic profiling of mouse striatal cell model of Huntington’s disease and control; probable implications to the disease biology. J. Proteomics 132, 155–166 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Zabel C., et al. , A large number of protein expression changes occur early in life and precede phenotype onset in a mouse model for huntington disease. Mol. Cell. Proteomics 8, 720–734 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perluigi M., et al. , Proteomic analysis of protein expression and oxidative modification in r6/2 transgenic mice: A model of Huntington disease. Mol. Cell. Proteomics 4, 1849–1861 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Liu X., Miller B. R., Rebec G. V., Clemmer D. E., Protein expression in the striatum and cortex regions of the brain for a mouse model of Huntington’s disease. J. Proteome Res. 6, 3134–3142 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hosp F., et al. , Spatiotemporal proteomic profiling of Huntington’s disease inclusions reveals widespread loss of protein function. Cell Rep. 21, 2291–2303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorolla M. A., et al. , Proteomic and oxidative stress analysis in human brain samples of Huntington disease. Free Radic. Biol. Med. 45, 667–678 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Ferecatu I., et al. , Evidence for a mitochondrial localization of the retinoblastoma protein. BMC Cell Biol. 10, 50 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura Y., et al. , Identification of Tam41 maintaining integrity of the TIM23 protein translocator complex in mitochondria. J. Cell Biol. 174, 631–637 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rockabrand E., et al. , The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum. Mol. Genet. 16, 61–77 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Van P. T., Bass V., Shiwarski D., Lanni F., Minden J., High dynamic range proteome imaging with the structured illumination gel imager. Electrophoresis 35, 2642–2655 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Chen C. H., Joshi A. U., Mochly-Rosen D., The role of mitochondrial aldehyde dehydrogenase 2 (ALDH2) in neuropathology and neurodegeneration. Acta Neurol. Taiwan. 25, 111–123 (2016). [PMC free article] [PubMed] [Google Scholar]
- 38.Schatz G., Dobberstein B., Common principles of protein translocation across membranes. Science 271, 1519–1526 (1996). [DOI] [PubMed] [Google Scholar]
- 39.Pfanner N., Geissler A., Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell Biol. 2, 339–349 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Chacinska A., Koehler C. M., Milenkovic D., Lithgow T., Pfanner N., Importing mitochondrial proteins: Machineries and mechanisms. Cell 138, 628–644 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solans A., Zambrano A., Rodriguez M., Barrientos A., Cytotoxicity of a mutant huntingtin fragment in yeast involves early alterations in mitochondrial OXPHOS complexes II and III. Hum. Mol. Genet. 15, 3063–3081 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Reiner A., Deng Y. P., Disrupted striatal neuron inputs and outputs in Huntington’s disease. CNS Neurosci. Ther. 24, 250–280 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rangel-Barajas C., Rebec G. V., Dysregulation of corticostriatal connectivity in Huntington’s disease: A role for dopamine modulation. J. Huntingtons Dis. 5, 303–331 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vonsattel J. P., et al. , Neuropathological classification of Huntington’s disease. J. Neuropathol. Exp. Neurol. 44, 559–577 (1985). [DOI] [PubMed] [Google Scholar]
- 45.Atwal R. S., et al. , Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum. Mol. Genet. 16, 2600–2615 (2007). [DOI] [PubMed] [Google Scholar]
- 46.McQuade L. R., et al. , Proteomics of Huntington’s disease-affected human embryonic stem cells reveals an evolving pathology involving mitochondrial dysfunction and metabolic disturbances. J. Proteome Res. 13, 5648–5659 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Deschepper M., Hoogendoorn B., Brooks S., Dunnett S. B., Jones L., Proteomic changes in the brains of Huntington’s disease mouse models reflect pathology and implicate mitochondrial changes. Brain Res. Bull. 88, 210–222 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Li H., Li S. H., Yu Z. X., Shelbourne P., Li X. J., Huntingtin aggregate-associated axonal degeneration is an early pathological event in Huntington’s disease mice. J. Neurosci. 21, 8473–8481 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liot G., Valette J., Pepin J., Flament J., Brouillet E., Energy defects in Huntington’s disease: Why “in vivo” evidence matters. Biochem. Biophys. Res. Commun. 483, 1084–1095 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Di Maio R., et al. , Alpha-synuclein binds to TOM20 and inhibits mitochondrial protein import in Parkinson’s disease. Sci. Transl. Med. 8, 342ra378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Unlu M., Morgan M. E., Minden J. S., Difference gel electrophoresis: A single gel method for detecting changes in protein extracts. Electrophoresis 18, 2071–2077 (1997). [DOI] [PubMed] [Google Scholar]
- 52.Van P. T., et al. , In-gel equilibration for improved protein retention in 2DE-based proteomic workflows. Electrophoresis 35, 3012–3017 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Sellers K. F., Miecznikowski J., Viswanathan S., Minden J. S., Eddy W. F., Lights, Camera, Action! systematic variation in 2-D difference gel electrophoresis images. Electrophoresis 28, 3324–3332 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Minden J. S., Two-dimensional difference gel electrophoresis. Methods Mol. Biol. 869, 287–304 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Bertin E., Arnouts S., SExtractor: Software for source extraction. Astron. Astrophys. Suppl. Ser. 117, 393–404 (1996). [Google Scholar]
- 56.Gong L., et al. , Drosophila ventral furrow morphogenesis: A proteomic analysis. Development 131, 643–656 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Speicher K. D., Kolbas O., Harper S., Speicher D. W., Systematic analysis of peptide recoveries from in-gel digestions for protein identifications in proteome studies. J. Biomol. Tech. 11, 74–86 (2000). [PMC free article] [PubMed] [Google Scholar]
- 58.Khattar N. K., et al. , Isolation of functionally active and highly purified neuronal mitochondria from human cortex. J. Neurosci. Methods 263, 1–6 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.