Significance
One of the long-standing questions in origins-of-life research centers on how the proteinaceous side chains and the protein backbone were selected during the earliest phases of evolution. Here we have studied oligomerization reactions of a group of positively charged amino acids, both proteinaceous and nonproteinaceous. Amino acids spontaneously oligomerized without the use of enzymes or activating agents, under mild, hydroxy acid-catalyzed, dry-down conditions. We observed that the proteinaceous amino acids oligomerized more extensively and with greater preference for reactivity through their α-amine compared with nonproteinaceous amino acids, forming predominantly linear, protein-like backbone topologies. These findings provide a purely chemical basis for selection of the positively charged amino acids found in today’s proteins.
Keywords: condensation dehydration, peptide evolution, chemical evolution, depsipeptides, prebiotic chemistry
Abstract
Numerous long-standing questions in origins-of-life research center on the history of biopolymers. For example, how and why did nature select the polypeptide backbone and proteinaceous side chains? Depsipeptides, containing both ester and amide linkages, have been proposed as ancestors of polypeptides. In this paper, we investigate cationic depsipeptides that form under mild dry-down reactions. We compare the oligomerization of various cationic amino acids, including the cationic proteinaceous amino acids (lysine, Lys; arginine, Arg; and histidine, His), along with nonproteinaceous analogs of Lys harboring fewer methylene groups in their side chains. These analogs, which have been discussed as potential prebiotic alternatives to Lys, are ornithine, 2,4-diaminobutyric acid, and 2,3-diaminopropionic acid (Orn, Dab, and Dpr). We observe that the proteinaceous amino acids condense more extensively than these nonproteinaceous amino acids. Orn and Dab readily cyclize into lactams, while Dab and Dpr condense less efficiently. Furthermore, the proteinaceous amino acids exhibit more selective oligomerization through their α-amines relative to their side-chain groups. This selectivity results in predominantly linear depsipeptides in which the amino acids are α-amine−linked, analogous to today’s proteins. These results suggest a chemical basis for the selection of Lys, Arg, and His over other cationic amino acids for incorporation into proto-proteins on the early Earth. Given that electrostatics are key elements of protein−RNA and protein−DNA interactions in extant life, we hypothesize that cationic side chains incorporated into proto-peptides, as reported in this study, served in a variety of functions with ancestral nucleic acid polymers in the early stages of life.
Coded protein is a linear polymer of 20 diverse amino acids, with a cohesive backbone that self-assembles via complementarity of H-bond acceptors and donors of the polypeptide backbone (1, 2). Proteins can fold spontaneously into “native” states with low configurational entropy and precise positioning of functional groups in 3D space. While sequences of proteins have been tuned by eons of evolution that is ongoing (3), it has been proposed that the polypeptide backbone is the product of prior chemical evolution (4). In these models, polypeptide is descended from ancestral proto-peptides that were heterogeneous both in backbone architecture and chemical structure and were composed of monomers that condensed readily under prebiotic conditions. Alex Rich, based, in part, on observations that the ribosome can synthesize polyester in vitro, proposed polyester (Fig. 1A) as ancestor of peptide. It has been recently demonstrated that depsipeptides, which contain mixtures of ester and amide linkages (Fig. 1A), form spontaneously under mild conditions during dry-down of mixtures of hydroxy acids and amino acids (5–9). The dry-down process concentrates reactants and reduces water activity and thus thermodynamically drives condensation reactions (10). Ester linkages lower activation energies for the formation of amide bonds through ester−amide exchange (Fig. 1D). In addition, facile ring opening of cyclic hydroxy acid dimers (dilactones, i.e., lactide and glycolide) and cyclic amino acid−hydroxy acid heterodimers (morpholinediones) promotes polymerization; in contrast, formation of cyclic amino acid dimers (diketopiperazines, DKPs) inhibits polymerization (11, 12). Depsipeptides are prebiotically plausible; hydroxy acids and amino acids are produced in model prebiotic reactions, are found together in some meteorites (5, 13–15), and can combine to form depsipeptides of up to 20 residues in mild dry-down conditions (5, 7).
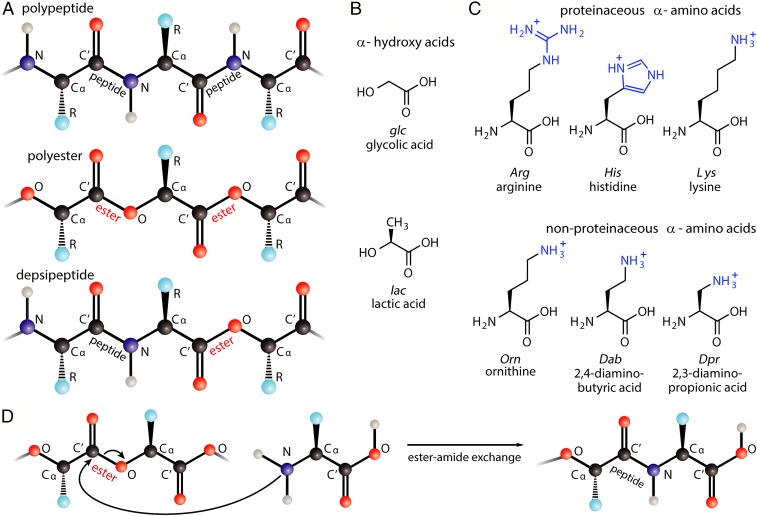
Fig. 1.
Chemical structures of monomers and oligomers investigated in this study. Cationic depsipeptides were generated by drying mixtures containing an α-hydroxy acid along with a cationic α-amino acid. (A) Chemical structures of polypeptide, polyester, and depsipeptide. Depsipeptides are copolymers of hydroxy acids and amino acids, linked by a heterogeneous backbone containing both amide and ester bonds. Depsipeptides have been hypothesized as ancestors of polypeptides. (B) The 2 hydroxy acids used here in dry-down reactions. (C) The 6 cationic amino acids used here in dry-down reactions. Cationic side-chain functional groups are blue. Amino acids are designated by uppercase 3-letter codes, and hydroxy acids are lowercase 3-letter codes. (D) The mechanism describing formation of depsipeptides via exchange of an ester for an amide, in a process called ester−amide exchange (i.e., ester aminolysis).
In the RNA World hypothesis (16–20) and in coevolutionary RNA−protein models for the origins of life (1, 21–23), proto-protein interacted extensively with negatively charged RNA (or proto-RNA) (24) and perhaps directly catalyzed the oligomerization/ligation of RNA (21, 25). These models require cationic proto-peptides, i.e., oligomers that are positively charged, which would enable noncovalent assemblies based on stabilizing electrostatic interactions between protomers of peptide and RNA. Two previous studies have reported depsipeptide formation in dry-down mixtures containing cationic amino acids with hydroxy acids (8, 9). However, only proteinaceous amino acids were included, and these studies did not characterize the linear versus branched topology of the products or demonstrate the presence of free cationic moieties. Cationic side-chain amino groups might compete with α-amine groups in chain extension reactions, giving rise to noncationic and/or branched nonbiological backbone architectures.
Here, we compare the spontaneous oligomerization of proteinaceous (lysine, Lys; arginine, Arg; and histidine, His) and nonproteinaceous (ornithine, Orn; 2,4-diaminobutyric acid, Dab; and 2,3-diaminopropionic acid, Dpr) cationic amino acids (Fig. 1). Proteinaceous amino acids are considered as those incorporated into proteins during translation. Amino acids with shorter cationic side chains, such as Orn and Dab, are thought to have been abundant on the prebiotic Earth (26–30). Lys, Arg, and His are considered less prebiotically plausible, even though Lys has been found in meteorites (31, 32) and prebiotic routes have been proposed for production of both Arg and His (33, 34). We reasoned that a comparative investigation of α-hydroxy acid-promoted oligomerizations of cationic amino acids could provide insights into the chemistry of cationic oligomers. We observe that proteinaceous amino acids condense more extensively, giving greater yields of higher molecular weight oligomers than this nonproteinaceous subset. The nonproteinaceous Orn and Dab cyclize to form lactams, while Dab and Dpr condense less efficiently. All 3 proteinaceous amino acids regioselectively link via their α-amines rather than through their side-chain amines, giving rise to oligomers that are predominantly linear and cationic. The results of this study provide a possible chemical rationale for the early selection of Lys, Arg, and His over other cationic candidates as proteinaceous amino acids.
Results
Depsipeptides Form via Dry-Down Reactions of Hydroxy Acids and Cationic Amino Acids.
We employed 6 cationic amino acids (Fig. 1C), including the 3 cationic proteinaceous amino acids (Lys, Arg, and His), along with 3 analogs of Lys having fewer methylene groups in the side chain (Orn, Dab, and Dpr). These homologated analogs of Lys are generally considered to have been more abundant than the proteinogenic cationic amino acids on the prebiotic Earth (13, 26–30). We employed the uncharged α-hydroxy acid monomers glycolic acid (glc, the hydroxy acid analog of glycine) and lactic acid (lac, the hydroxy acid analog of alanine). Depsipeptides were generated by drying down mixtures containing a single amino acid (unless otherwise noted) with a single hydroxy acid for 1 wk at 85 °C under unbuffered, mildly acidic conditions (pH ≈ 3). Preliminary studies indicated that product oligomers were generally longer when the starting mixture contained an excess of the hydroxy acid over the amino acid; we therefore employed a 5:1 hydroxy acid/amino acid molar ratio (unless otherwise noted). As detailed below, the product mixtures for each reaction were characterized by a variety of analytical methods including liquid chromatography−mass spectrometry (LC-MS), NMR, Fourier transform infrared (FTIR), enzyme assays, chemical probing, and high-performance LC (HPLC) with both hydrophobic and hydrophilic stationary phases.
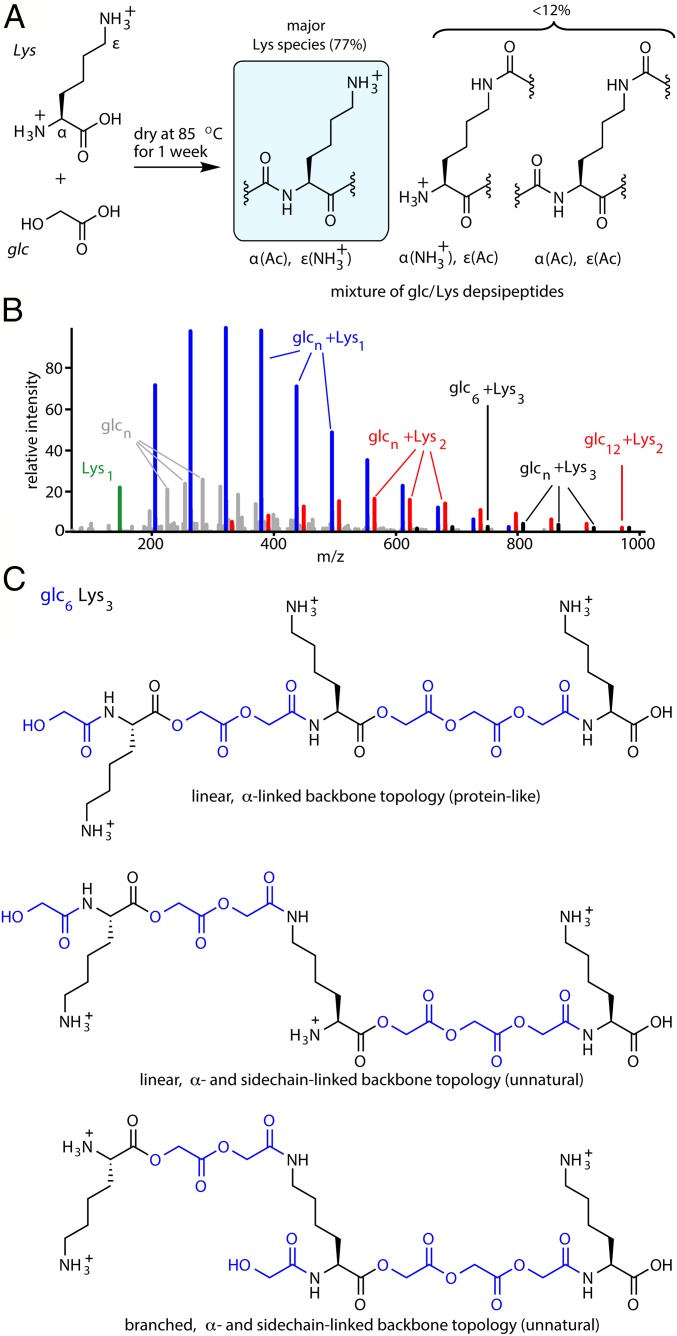
LC-MS analyses indicated that the dry-down reactions formed copolymers of hydroxy acids and amino acids (Fig. 2 A and B and SI Appendix, Figs. S1–S12). The mixture of glc and Lys produced oligomers up to 14-mers, with varying lengths and compositions of glc and Lys (Fig. 2B). Examples of observed depsipeptide species include 6gly3Lys and 12glc2Lys, with each compositional species observed by LC-MS representing multiple possible sequences (Fig. 2C). Most oligomer products contained from 1 to 3 amino acids and were typically enriched in hydroxy acids compared with amino acids, as would be expected from the 5:1 hydroxy acid/amino acid molar ratio used in the dry-down reactions. Similar product distributions were observed for dry-down reactions involving either glc or lac, supporting the generality of these reactions (SI Appendix, Figs. S1–S14). The presence of both ester and amide bonds within the product oligomers was confirmed with FTIR. The dry-down reaction of the glc plus Lys mixture induced shifts in the C=O band from a free acid (1,724 cm−1) to an ester (1,743 cm−1) upon drying (35), and shifts in the amide regions (amide I and amide II; SI Appendix, Fig. S15). FTIR analyses of product mixtures for the other amino acids supported the formation of ester and amide bonds in all cases, with evident shifts in the C=O band from a free acid to an ester and shifts in the amide regions (SI Appendix, Figs. S16–S26). The presence of both ester and amide bonds was further supported by degradation of the product mixture using trypsin (a protease) and porcine liver esterase. C18 HPLC analysis indicated that these enzymatic hydrolyses converted the product oligomers of glc+Lys dry-down into less hydrophobic, presumably lower molecular weight species (SI Appendix, Fig. S27). We note that, even though trypsin is known to cleave peptide chains, mainly at Lys and Arg, it has some cross-reactivity with esters (36). Control reactions involving dry-downs of amino acids in the absence of hydroxy acids failed to form oligomers (SI Appendix, Fig. S28), as expected for the mild temperatures (85 °C) used in our experiments.
Fig. 2.
Depsipeptides containing cationic amino acids are formed via dry-down reactions of mixtures of hydroxy acids and cationic amino acids. (A) Examples of possible products of dry-down reactions of glc with Lys; Lys can be amidated on either the α-amine or the ε-amine. The percentages of products shown were determined by 1H NMR analyses. (B) A mixture of glc with Lys was dried at 85 °C for 7 d, and the resulting depsipeptides were analyzed by positive-mode ESI MS. All labeled species correspond to [M+H]+ ions. (C) Examples of 3 of the many possible sequence isomers of 6glc3Lys. The molecules shown differ at the linkages in the central Lys residue.
Verification of Cationic Moieties within the Depsipeptide Product Mixtures.
The presence of free amines in the oligomeric products was supported by results of experiments with ninhydrin (SI Appendix, Fig. S29), which reacts quantitatively with primary amine groups to give a product that absorbs at 480 nm. To detect free amines in dry-down products, an aliquot from a dry-down reaction of glc and Lys was dialyzed to remove remaining monomeric Lys. Unreacted, nondialyzed Lys served as a positive control, whereas unreacted, dialyzed Lys or nondialyzed glc monomer or glc oligomers were used as negative controls. The dialyzed oligomers from the glc plus Lys dry-down reaction showed an increase in absorbance comparable to that of Lys (nondialyzed), supporting the presence of free amine groups within the product oligomers. The negative controls did not show change of absorbance at 480 nm.
The cationic properties of the product depsipeptides were further confirmed via hydrophilic-based LC (HILIC), which separates compounds based on differences in polarity (SI Appendix, Fig. S30). As indicated by the absorbance at 210 nm and LC-MS analysis, dry-down reactions of glc with Lys resulted in a variety of hydrophilic depsipeptides that were retained on the HILIC column and eluted only with polar mobile phase. In contrast to control reactions containing amino acids alone, hydrophilic depsipeptides were observed in each product mixture obtained from drying the various cationic amino acids with glc or lac (SI Appendix, Figs. S31 and S32).
Proteinaceous Amino Acids React More Efficiently and Regioselectively at the α-Amine than Nonproteinaceous Amino Acids.
In condensation reactions, a cationic amino acid such as Lys potentially can be acylated solely at the α-amine position, solely at the side-chain amine position, or at both positions (Fig. 2). A protein-like backbone topology would arise when oligomers are linked solely through the α-amino group. In contrast, linkages at side-chain amino groups would lead to nonbiological backbone topologies, and linkages at both the side chain and α-amine could yield branched oligomers (Fig. 2C). NMR experiments (described in detail below) revealed robust preferences for amidation at the α-amine groups for Lys, His, and Arg, but not for Orn, Dab, or Dpr. Acylation of the His side chain was not observed, as would be expected considering the hydrolytic instability of acyl imidazoles. As a result of the observed regioselectivity, the proteinogenic amino acids formed oligomers predominantly having an α-linked, protein-like backbone topology. These oligomers contain ester bonds instead of amide bonds at many backbone positions.
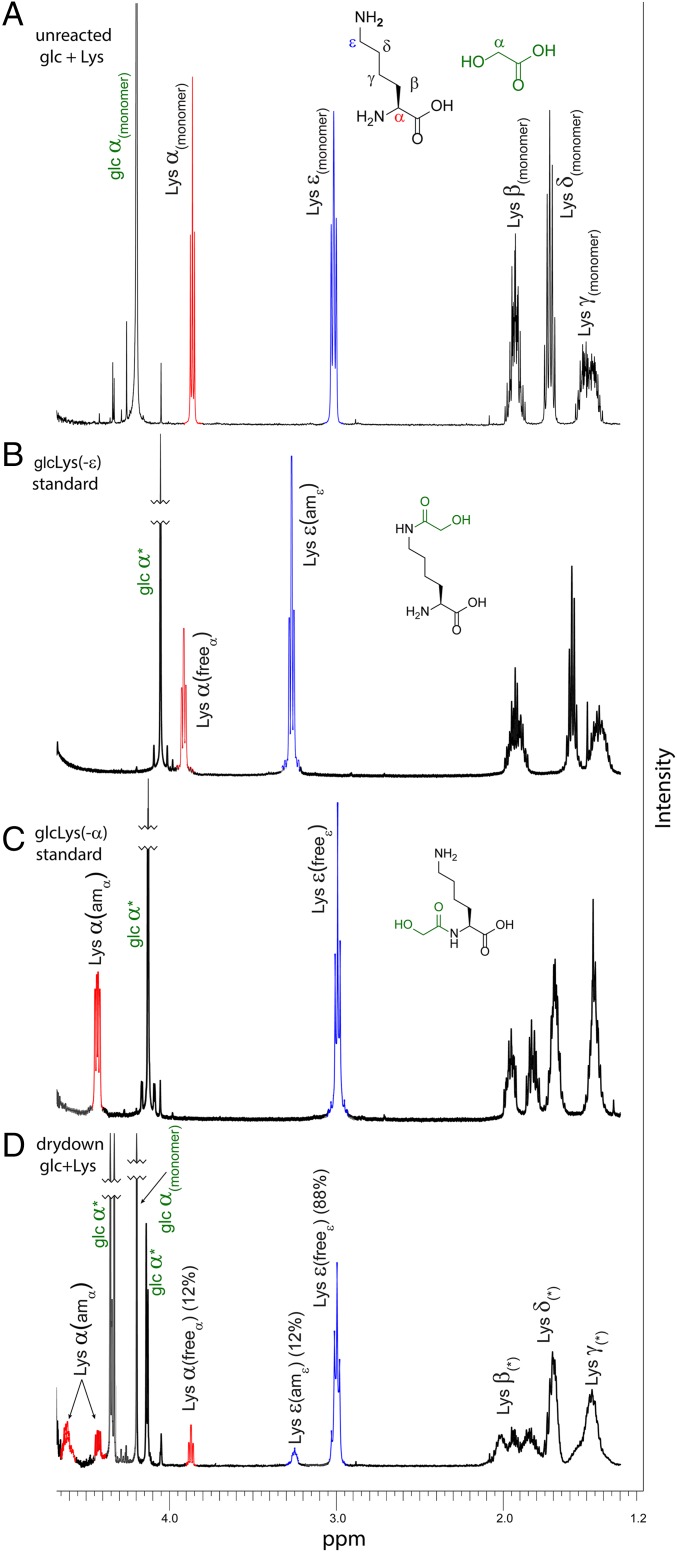
NMR characterization of the dry-down reaction for glc with Lys revealed differences in reactivities of the 2 amine positions of Lys (Fig. 3, Table 1, and SI Appendix, Fig. S33). The 1H NMR resonances were assigned using chemically synthesized standards (SI Appendix, Fig. S34). The synthesis and purification of the standards are described in SI Appendix. In the unreacted Lys starting material, the free α-proton chemical shift is centered at 3.86 parts per million (ppm), while the free ε-proton is at 3.01 ppm (Fig. 3A). Upon ε-amidation with glc, the α-proton resonance shifts slightly downfield to 3.91 ppm, and the ε-proton resonance shifts substantially downfield to 3.27 ppm (Fig. 3B). Alternatively, upon α-amidation of Lys with glc, the α-proton resonance shifts substantially downfield to 4.43 ppm, and the ε-proton resonance shifts slightly upfield to 2.99 ppm (Fig. 3C). The mixture of products produced by the dry-down reaction of glc with Lys produces a surprisingly simple set of chemical envelopes in the NMR spectra (Fig. 3D) that could readily be interpreted by comparison with spectra of the synthetic standards (Fig. 3 and SI Appendix, Figs. S35 and S36). Amidation was evident for both the α-amine and the ε-amine. However, there was significant regioselectivity between the 2 amine groups; integration of the α- and ε-proton resonances indicated that at least 88% of Lys α-amines were amidated, whereas only 12% of ε-amines were amidated (Table 1, Fig. 3D, and SI Appendix, Figs. S35–S37). An additional resonance in the vicinity of α-amidated protons (4.62 ppm) was confirmed to correspond to Lys species that are both amidated at the α-amine and esterified at the carboxylic acid by comparison with a glc-Lys(-α)-glc standard (SI Appendix, Fig. S35). The identity of this resonance as α-amidated Lys species was further supported by 1H−1H correlated spectroscopy NMR (SI Appendix, Fig. S37).
Fig. 3.
Dry-down reactions of glc and Lys produce primarily linear oligomers by amidation at the α-amine rather than the side-chain ε-amine. The 1H NMR spectra of (A) a 5:1 mixture of glc and Lys monomers, before the dry-down reaction, with blue (ε-resonance) and red (α-resonance) highlighting; (B) a synthetic standard of Lys acylated with glc at the ε-amine demonstrating the downfield shift of the ε-resonance; (C) a synthetic standard of Lys acylated with glc at the α-amine; and (D) dry-down of a 5:1 mixture of glc and Lys at 85 °C for 7 d. Upon α-amidation of Lys with glc, the Lys α-proton resonance shifted downfield to 4.43 ppm, and the ε-proton resonance shifted slightly upfield. An additional Lys resonance observed at 4.62 ppm corresponds to Lys species that are both α-amidated and esterified, as confirmed by comparison with a glc-Lys(-α)-glc standard. The colored envelopes in D indicate species that are either amidated (am) or not (free) at α or ε positions. An asterisk indicates all states, including monomer and all possible oligomeric species.
Table 1.
Product yields of dry-down reactions of 6 cationic amino acids with hydroxy acids
| Reaction | Overall conversion (%)* | α-Amidation (%)† | Side-chain amidation (%)‡ | Lactam monomer (%) | C-terminal lactam (%) | Side-chain amidation excluding lactamization (%) | α-Amidated, side-chain free amino acid in polymers (%) |
| glc+Dpr | 28 | 3–28 | 24 | ND | ND | ND | 3 |
| glc+Dab | 58 | 44 | 25 | 14 | 5 | 6 | 33 |
| glc+Orn | 94 | 62 | 60 | 32 | 28 | ND | 34 |
| glc+Lys | 88 | 88 | 12 | ND | ND | ND | 77 |
| glc+Arg | 92 | 92 | 8 | ND | ND | ND | 85 |
| glc+His | 90 | 90 | ND | ND | ND | ND | 90 |
| lac+Lys | 76 | 76 | 7 | ND | ND | ND | 71 |
Quantitated by integration of 1H NMR peaks (see Materials and Methods). ND, not detected.
Overall conversion refers to the conversion of an amino acid monomer into products.
The extent of amidation on the α-amine, independent from the extent of side-chain amidation.
The extent of amidation on the side-chain group, independent from the extent of α-amine amidation.
Based on NMR integration, at least 77% of Lys in the glc depsipeptide mixtures has retained free side-chain ε-amines (Table 1). At least 88% of monomeric Lys was converted to oligomer. Similar results were obtained for dry-downs involving lac and Lys (SI Appendix, Fig. S38), with a slightly lower conversion into oligomers (76% for lac) but with a greater regioselectivity for α- over ε-amidation (only 7% amidation at the ε-amine) (Table 1). The lower conversion for lac compared with glc likely results from steric hindrance by the methyl group on lac.
NMR analyses of reactions with various cationic amino acids indicated that Lys, Orn, Arg, and His oligomerized to a greater extent than Dpr and Dab (Table 1 and SI Appendix, Figs. S39–S56). We have compared extents of conversion of monomers into oligomers for all 6 cationic amino acids (Table 1). We confirmed the reproducibility of our results with independent replica experiments (SI Appendix, Fig. S57).
We explored the possibility of chirality-based differences in reactivity by comparing the dry-down of glc with l-Lys to that of glc with d-Lys or to a dry-down reaction of glc with racemic Lys. No differences in the overall conversion of Lys into oligomers or the ratio of α-amidation to ε-amidation were observed (Fig. 3 and SI Appendix, Figs. S58 and S59). These results are in agreement with Forsythe et al. (5), which reported that no differences were observed in the product distribution upon dry-down of l-lac with l-Ala or with d-Ala. We also tested dry-down reactions of glc with analogs of Lys and Dpr having methylated side chains. We found that dry-down of glc with either Lys(Me)2-OH or with H-Lys(Me)3-OH resulted in oligomerization of these amino acids through their α-amine to an extent similar to that of free Lys. As would be expected, no acylation at the methylated ε-amine was observed. Also consistent with the oligomerization involving Lys, we observed a slight upfield shift of the nonamidated ε-protons, indicative of oligomerization through the α-amine in both cases (SI Appendix, Figs. S60 and S61). Dry-down of glc with H-Dpr(Me)2-OH resulted in the formation of new peaks in the ∼1.5-ppm region in the 1H NMR spectra (which are not present in the amino acid), so it appears that this amino acid largely degraded during the dry-down reaction. There was no evidence consistent with incorporation of H-Dpr(Me)2-OH into oligomers (SI Appendix, Fig. S62).
Nonproteinaceous Amino Acids Orn and Dab Cyclize during Dry-Downs.
NMR analyses indicated that both Orn and Dab underwent cyclization into lactams, composed of a 6-membered ring for Orn and a 5-membered ring for Dab (Table 1 and SI Appendix, Figs. S42, S45, S54, and S55). The identified lactams were either monomeric or C-terminal lactams in polymers, amidated both at their α-amine and side-chain amine. Cyclization products were identified via comparison with lactam standards of Orn and Dab (SI Appendix, Figs. S54 and S55) and MS analysis, which indicated that dry-down reactions with Orn or Dab produced depsipeptides with additional water losses (SI Appendix, Figs. S3 and S4). As supported by essentially insignificant extent of lactam formation in control dry-down reactions of Orn and Dab without any hydroxy acid (SI Appendix, Fig. S28), these intramolecular cyclizations result from ester−amide exchange, which is the same mechanism that leads to the formation of depsipeptides in dry-down reactions.
Dry-Down Reactions Chemically Select for Incorporation of Proteinaceous Amino Acids.
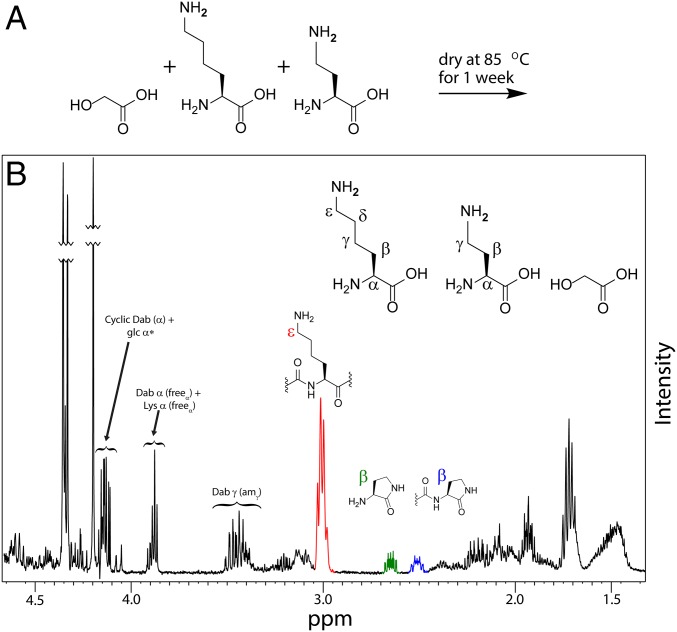
The extensive cyclization of Dab and Orn into lactams and lower conversion for Dpr in dry-down conditions suggested that proteinaceous amino acids might outcompete these nonproteinaceous amino acids for incorporation into oligomers when more than one amino acid were simultaneously present. To test this prediction, we carried out dry-down reactions using a mixture of Lys and Dab with glc (10:1:1 ratio of glc:Dab:Lys) or Lys and Orn with glc (10:1:1 ratio of glc:Orn:Lys). Although the complexity of the resulting product mixtures prevented complete characterization by 1H NMR, we can make some relevant observations. In the mixture containing Dab and Lys, the reaction produced depsipeptides that incorporated most of the Lys that was present, whereas at least 40% of Dab was excluded completely from the oligomers as a lactam monomer (18.5%) or incorporated only as a C-terminal lactam (21.5%) (Fig. 4). Similar to what was observed in the reaction containing only Lys and glc, in this competition reaction, Lys oligomerized regioselectively via the α-amino group and maintained free ε-amines. However, it is notable that Lys and Dab reacted differently in the ternary mixture of glc+Lys+Dab than in the binary mixtures of either glc+Lys or glc+Dab. For instance, we observed more side-chain amidation for Dab in the presence than in the absence of Lys, which contributed to an overall higher conversion of Dab (86%) in this reaction compared with the glc+Dab reaction (58%). Moreover, there was less side-chain amidation for Lys in the presence (<5%) than in the absence (12%) of Dab. This higher regioselectivity contributed to a somewhat lower overall conversion of Lys into oligomers in the ternary (63%) than in the binary (88%) system. Similar results were obtained for a ternary mixture of glc+Lys+Orn. Robust cyclization of Orn was evident, while Lys oligomerized regioselectively via the α-amino group and maintained free ε-amines (SI Appendix, Fig. S63). Overlap in 1H NMR spectra prevented quantitation of the extent of conversion of Orn and Lys. Overall, the findings here support a mechanism by which proteinaceous cationic amino acids could have been chemically selected over nonproteinaceous competitors for incorporation into cationic depsipeptides.
Fig. 4.
Dry-down reactions chemically select for incorporation of proteinaceous amino acids into depsipeptide products. (A) A schematic diagram for the dry-down reaction used for the competition experiment between Lys and Dab in the presence of glc. (B) The 1H NMR spectrum of the dry-down reaction indicated that Lys maintains free ε-amines in depsipeptide products (red) whereas Dab with free side-chain amines is extensively excluded from the oligomers. Rather, Dab is observed mainly as lactams (green, lactam monomer; blue, α-amidated lactam).
Discussion
Experimental evaluation of models of early protein evolution is critical for understanding the origins of life. Models of chemical evolution in which unactivated amino acids directly condense to form polypeptide face 3 main obstacles which have been partially resolved:
-
1)
Formation of peptide bonds is thermodynamically unfavorable in aqueous solution (37, 38). The resolution is as follows: The free energy of peptide bond formation is modulated by water activity. In dilute aqueous solution, peptide bond cleavage by hydrolysis is thermodynamically favorable. Under “dry-down” conditions, peptide bond formation by condensation dehydration is thermodynamically favorable. Depsipeptides, which contain mixtures of ester and amide linkages (Fig. 1A), form spontaneously under mild conditions during dry-down of mixtures of hydroxy acids and amino acids (5–9).
-
2)
Formation of peptide bonds is kinetically frustrated by high-energy barriers. The resolution is as follows: Pathways with low activation energies for peptide bond formation are provided by esterification preceding ester−amide exchange (Fig. 1D) (5–9, 39).
-
3)
Formation of peptide oligomers or polymers is hindered by cyclization of dipeptides into DKP side products (11, 12, 40–43). The resolution is as follows: Facile ring opening of cyclic hydroxy acid dimers (dilactones, i.e., lactide and glycolide) and cyclic amino acid−hydroxy acid heterodimers (morpholinediones) promotes oligomerization.
Selective Reactivity.
The dry-down reactions here with mixtures of various cationic amino acids and hydroxy acids produced depsipeptides. The proteinaceous amino acids Lys, Arg, and His oligomerized more readily and preferentially at the α-amine than the nonproteinaceous amino acids Orn, Dab, and Dpr. Before our work here, there was no experimental demonstration of selective incorporation of Lys, Arg, and His over Orn, Dab, and Dpr into proto-proteins. The exclusion of Orn is especially significant considering that it is abundant in biological systems and has been proposed as a component of primitive oligomers (44). The results here are consistent with a model in which Lys, Arg, and His yielded longer, potentially more functional oligomers in prebiotic systems, contributing to their ultimate selection in coded translation. The combined data support the idea that intrinsic amino acid reactivity, especially in regard to oligomerization potential, may have played a role in selection of proteinogenic amino acids. Along these lines, a previous study has demonstrated that Pro incorporates much more rapidly than linear N-alkyl amino acids in the ribosome (45), in accordance with expectations based on inherent sterics and basicity.
In addition to a higher extent of incorporation into products, we also observed preferential acylation of the α-amine over the side-chain amine for Lys, Arg, and His in plausible prebiotic oligomerization reactions. Side-chain−acylated His would not be expected considering the hydrolytic instability of acyl imidazoles. However, we were initially surprised by the substantially lower degrees of side-chain acylation for Lys and Arg compared with Orn, Dab, and Dpr. The striking preference for linkage at the α-amine over the side-chain amine for certain amino acids is of special interest because this regioselectivity generates a “proteinaceous”-like backbone topology rather than polymers that are cross-linked and that have nonbiological topology. Our results provide an empirical chemical basis for the selection of proteinaceous cationic amino acids over these nonproteinaceous amino acids on early Earth. It was previously suggested that the selection of Lys over shorter analogs (Orn, Dab, and Dpr) is related to the ability of Lys to form superior ion-pairing interactions with negatively charged amino acids in helical structures (46); however, that model was undermined by the observation that replacement of Lys with Dab can increase the stability of protein−protein interactions in other contexts (47). An alternative mechanism that achieves regioselective α-amidation of Lys involving oxidation of amino thioacids has recently been put forth (48). The robust cyclization of Orn and Dab under dry-down conditions may have served as a selective force to exclude them from the repertoire of coded amino acids. Peptides containing Orn are labile because of internal lactamization by the side-chain amino group that cleaves the peptide backbone (49, 50). Further, the putative aminoacyl ester of transfer RNA (tRNA) containing an Orn or Dab ester would suffer from intramolecular cyclization to the lactam form, akin to what was observed in our dry-down reactions, which would hinder incorporation of these amino acids into proteins during translation (49).
Differences in pKa (negative logarithm of the acid dissociation constant) between the α-amines and side-chain amines may account, in part, for the observed selectivity (51). The pKas of the α-amines in Arg, Lys, Orn, Dab, and Dpr are ∼2 to 3 units lower the pKas of the corresponding side-chain amines (Table 2), resulting in a higher fraction of deprotonated, nucleophilic amine at the α-position under the acidic conditions of the reaction (52–54). Furthermore, esterification of the amino acids during the dry-down would cause the pKas of their α-amines to be lowered, whereas the pKas of the side-chain amines would be largely unchanged, likely affording higher regioselectivity (55, 56). Preferential α-amine reactivity based on differences in amine pKa values would only be expected at lower pH values where relative differences in protonated and unprotonated amine forms could influence the reaction; indeed, under more basic aqueous conditions, preferences for side-chain amidation over α-amidation have been reported for reactions of Lys, Orn, Dab, and Dpr (57, 58).
Table 2.
pKa values of the cationic amino acids used in this study
| Amino acid | pKa α-ammonium | pKa protonated side chain | pKa differences (side chain minus α-ammonium) |
| Dpr | 6.4 | 9.4 | 3.0 |
| Dab | 8.2 | 10.2 | 2.0 |
| Orn | 8.7 | 10.8 | 2.1 |
| Lys | 8.9 | 10.8 | 1.9 |
| Arg | 9.0 | 12.5 | 3.5 |
| His | 9.2 | 6.0 | −3.2 |
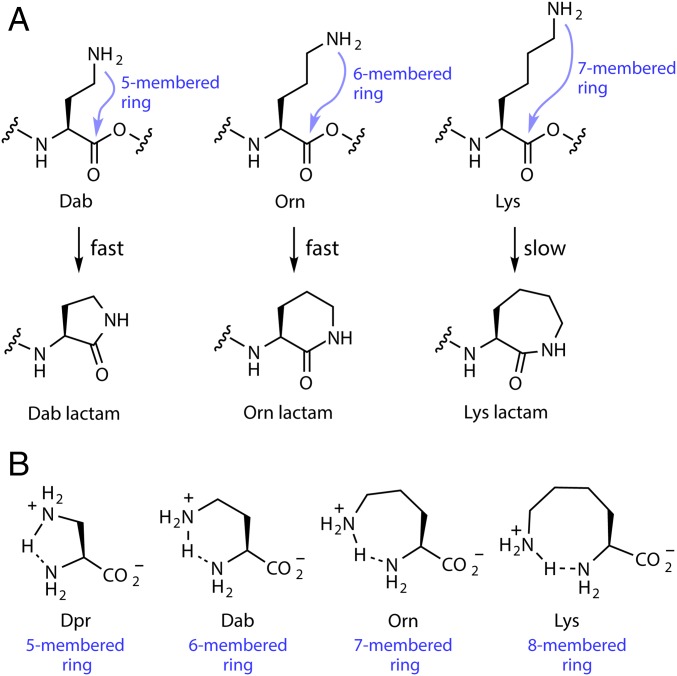
Differences in pKa values alone do not fully explain our observations, because these differences incorrectly predict that Arg, Lys, Orn, Dab, and Dpr would exhibit similar degrees of regioselectivity (since their α vs. side-chain pKa differences are similar). Instead, we observed superior regioselectivity for Arg and Lys compared with these nonproteinaceous amino acids. It seems likely that the relatively high levels of side-chain amidation for Orn and Dab are due mostly (if not exclusively) to intramolecular amidation. When Orn or Dab are esterified, the side-chain amine can attack the ester in an intramolecular reaction to form a 6-membered (Orn) or 5-membered ring (Dab). On the other hand, a similar intramolecular process with esterified Lys would require the formation of a 7-membered ring, which is enthalpically and entropically more difficult than the smaller ring analogs (Fig. 5A). The lower reactivities of Dpr and Dab might be explained by intramolecular hydrogen bonding between their 2 amino groups, forming more thermodynamically stable 5- or 6-membered rings compared with amino acids bearing more methylene groups in the side chain, or similarly, due to internal “salt bridge” formation (Fig. 5B) (59).
Fig. 5.
Possible rationale for the observed lower chemical reactivity and incorporation of nonproteinaceous amino acids Dpr, Dab, and Orn. (A) A schematic diagram showing lactam formation for Dab, Orn, and Lys. Closure of the Lys lactam requires the formation of a 7-membered ring, which is enthalpically and entropically less favorable compared with the smaller ring lactams of Dab and Orn. (B) Intramolecular hydrogen bonding between amine moieties may explain the relatively lower observed conversions for Dpr and Dab compared with Orn and Lys.
Abiotic Amino Acids.
The plausibility of prebiotic production of cationic amino acids is complex and unresolved. Amino acids with shorter cationic side chains, such as Orn and Dab, have been found in meteorites and produced in model prebiotic reactions. Orn and Dab are thought to have been abundant on prebiotic Earth (26–30). Prebiotic routes have been proposed for production of both Arg and His (33, 34), and Lys has been found in meteorites (31, 32). It has been proposed that His might have prebiotic roots from imidazole-4-acetaldehyde through the Strecker synthesis (34). A prebiotic route for production of Arg has been proposed that relies upon a cyanide-rich reducing environment containing hydrogen sulfide (33). It will be interesting, in the future, to explore the nature of the chemical reactivity of other nonproteinogenic amino acids. For instance, we performed a dry-down reaction of glc with l-2-amino-3-guanidinopropionic acid (Agp), which is an analog of Arg that is shorter by 2 methylene groups in its side chain. Oligomers of glc and Agp were observed by MS, and a complete conversion of the amino acid into products was observed by 1H NMR. MS indicated that the oligomers of glc and Agp corresponded mainly to species with additional water loss. This water loss is likely due to cyclization between the Agp side chain and carboxyl group to form a dihydropyrimidinone (SI Appendix, Figs. S64 and S65), in analogy with the tendency of Orn and Dab to form lactams. An additional/complementary mechanism for generating cationic depsipeptides would be the oligomerization of cationic hydroxy acids, such as isoserine, serine, 2-hydroxy-4-aminobutyric acid (hydroxy acid analog of Dab), or 2-hydroxy-6-aminohexanoic acid (hydroxy acid analog of Lys).
Peptide Evolution—Working Model.
The studies reported here represent our first step in exploring the following working model for ribosomal origins and evolution (60–62). Interactions between short, unstructured proto-peptide oligomers and proto-ribosomal RNA drove an evolutionary process of molecular refinement in which the unstructured proto-peptides converted over time into simple antiparallel β−β secondary structures, which then collapsed into β-strand protein domains, which eventually gave rise to complex folds composed of α-helices and β-strands. In this model, protein synthesis and protein folding coevolved along with the translation system. At the earliest stages, proto-peptide synthesis cooperated with differential rates of hydrolytic degradation, mediated by folding and assembly, to drive chemical evolution (1, 63–66). This model is consistent with incremental conversion of noncoded depsipeptides (which are unstructured, in part, due to their ester bonds, below) to coded polypeptide (which can fold to well-defined functional domains). This model is consistent with the following: 1) facile synthesis of cationic depsipeptides in dry-down reactions, as demonstrated here; 2) the association of cationic depsipeptides with RNA; 3) limited folding, self-assembly, and stability of depsipeptide because of unbalanced numbers of backbone amide hydrogen bond donors and acceptors and weakened ester backbone hydrogen bonding acceptors, as well as higher flexibility of the ester bonds compared with amide bonds; 4) greater lifetimes of peptide than depsipeptide due to higher rates of hydrolysis and intramolecular cyclization in esters than in amides; 5) conversion of esters into amides via ester−amide exchange; and 6) close analogy of ester−amide exchange in dry-down reactions and in the ribosome (67). Peptide bonds are formed in the ribosome when the nascent peptide, preesterified at its C terminus to the 3′ end of the P-site tRNA, undergoes ester−amide exchange with a free amine of an amino acid linked to the A-site aminoacyl tRNA. Similarly, depsipeptides in dry-down reactions are formed in condensation reactions with an ester precursor that undergoes ester−amide exchange with an amino acid.
Molecules in Mutualism.
As a next step in evaluating the above model of peptide evolution, we are actively exploring whether incorporation of cationic amino acids in depsipeptides confers functional properties such as interactions with RNA or catalytic functions. Since the side chains are substantially unlinked and ionizable in the depsipeptides produced by the proteinaceous amino acids, the resulting depsipeptides are cationic around neutral pH. In analogy with peptides, depsipeptides decorated with diverse functional groups should exhibit a variety of functions and interactions with other polymers. Szostak and coworkers (68) reported that short cationic peptide amphiphiles can drive RNA binding to protocell membranes and thus increase the local concentration of the RNA; cationic depsipeptides might be expected to show similar properties. Likewise, cationic depsipeptides might engage in mutualistic interactions with fatty acids, as has recently been described for cationic peptides by Greenwald and coworkers (69). Cationic side chains in proto-peptides could have facilitated important interactions with other proto-polymers, similar to their crucial role in today’s biology (70–72), that could have shaped the early stages of evolution (73). Moreover, cationic depsipeptides could have exhibited ligand binding and/or catalytic activities such as aldol condensation, Michael addition, and ester aminolysis (8, 74–79). For example, Adamala et al. (76) showed that, when encapsulated within fatty acid vesicles, the dipeptide Ser−His catalyzed ester aminolysis to form a second, hydrophobic dipeptide. The newly formed dipeptide localized to the vesicle membranes, which imparted enhanced affinity for fatty acids and thus promoted vesicle growth.
Materials and Methods
Standards.
Synthesis of standards is presented in SI Appendix.
Dry-Down Reactions.
For formation of cationic depsipeptides, aqueous solutions of hydroxy acids and amino acids at a 5:1 molar ratio (in favor of the hydroxy acid) were allowed to dry at 85 °C under unbuffered, mildly acidic conditions (initial pH of ∼3) for 1 wk. The amino acids were all used in their HCl form, and no additional salt was added to the reactions. Control reactions contained either a hydroxy acid alone or an amino acid alone. For testing possible chirality-based differences between l-Lys and d-Lys, glc was dried with a racemic mixture of Lys at a 10:1:1 molar ratio (glc:l-Lys:d-Lys). For the competition experiments, glc was dried with Lys and Dab at a 10:1:1 molar ratio (glc:Lys:Dab) or with Lys and Orn at a 10:1:1 molar ratio (glc:Lys:Orn). Before analysis, dry-down samples were resuspended in ultrapure water to 100 mM concentration based on original amino acid concentration, vortexed, sonicated in ice, and centrifuged at 15,294 × g for 5 min. The supernatant, which contained nearly all of the amino acid present in the reaction based on NMR analysis, was collected and diluted to the specified concentration.
NMR Spectroscopy.
NMR spectra were recorded on a Bruker Avance II-500. A long relaxation delay time of 15 s was used for dry-down mixtures to ensure quantitative integration of the resonances. Data were processed and spectra were plotted with MestReNova and TopSpin software packages. NMR spectra for standards and dry-down reactions not shown in the manuscript are contained in SI Appendix.
Overall conversion (Table 1) is the lowest percent estimate of conversion of amino acid monomer into products. With the exception of His, the overall conversion was estimated from integration of the free, nonamidated α-proton 1H NMR resonance. The overall conversion for His was estimated from integration of nonreacted vs. reacted imidazole proton resonance.
The extent of amidation on the α-amine (Table 1) was estimated by integration of 1H NMR resonances. A range is given for Dpr due to noise in the 1H NMR spectra; the upper limit is the conversion extent of Dpr (based on the free α-proton region) with the assumption that there are no species that are esterified but not amidated. This assumption is supported by the absence of resonances for species that are esterified but not amidated in the 1H NMR spectra of glc and Lys dry-down reactions. The lower Dpr limit is estimated from the integration of the β-proton resonance, which is shifted upfield upon α-amidation in the absence of side-chain amidation. The extent of amidation of the α-amine for Dab and Orn was estimated from downfield shifted α-proton resonances, and excluding the free lactam, which was shifted downfield as well. For Orn, the upper limit for α-amidation is 62% (94% conversion minus 32% lactam monomer). For Dab, the upper limit for α-amidation is 44% (58% conversion minus 14% lactam monomer). For Arg and His, estimates of the extent of α-amidation assumed Arg and His gave α-resonance shifts that are similar to those of Lys.
The extent of amidation at side-chain amines was quantified by integration of the resonance corresponding to methylene protons adjacent to the side-chain amine. These resonances shift downfield upon side-chain amidation. The extent of formation of lactam monomer of Dab or Orn was quantified by integration of the most downfield β-proton of the lactam monomer. The extent of C-terminal lactam formation for Dab or Orn was quantified by integration of the second most downfield β-resonance, corresponding to the C-terminal lactam (confirmed via standards). Side-chain amidation excluding lactamization was calculated by subtraction of the lactam fraction (as either monomers or as C-terminal lactams) from the fraction that represents side-chain amidated amino acid for the corresponding amino acid (Dab or Orn). For Arg, the downfield chemical shifts of the δ-protons are consistent with side-chain amidation, as noted for the other amino acids.
For Dpr, the lower limit of α-amidated, side-chain free amino acid in oligomers was quantified by integration of upfield β-protons that had shifted upfield. For Dab and Orn, the estimate was based on subtraction of the C-terminal lactam and side-chain amidated amino acid (which excludes lactamization products) from the fraction of α-amidated amino acid. For Lys, Arg, and His, the lower limit was calculated by (xα-amidated) times (1 − xside-chain amidated). For instance, for the glc+Lys mixture, given that the conversion of amino acid monomer into oligomers was at least 88%, and that a maximum of only 12% of linkages involved the ε-amino group, we can conclude that at least 77% [88 × (1 − 0.12)] of Lys in these depsipeptide mixtures has retained free side-chain ε-amines.
MS.
LC-MS data were collected on an Agilent 1260 HPLC coupled to an Agilent 6130 single quadrupole mass spectrometer (Agilent Technologies) and an inline Agilent UV absorbance detector (210 nm) using a 3.0-kV electrospray ionization (ESI) capillary voltage. Samples were directly infused into the mass spectrometer by ESI.
Processing of MS data were conducted using a suite of macros (Igor Pro-8.0). The macros, upon user input of reactants, search the spectrum for m/z values corresponding to condensation−dehydration reactions and automatically label peaks with this information. This processing package, along with a how-to-guide, is freely available at https://github.com/GATech-HudLab/Mass-Spec-Processor. LC-MS data are compiled in SI Appendix.
Supplementary Material
Acknowledgments
We thank the L.D.W. and N.V.H. group members for fruitful discussions and Amy Mu for technical assistance. We thank the Scripps Research Center for Mass Spectrometry and Automated Synthesis Facility for high-resolution MS analyses. This research was supported by the NSF and the NASA Astrobiology Program under the NSF Center for Chemical Evolution (CHE-1504217). M.F.P. acknowledges the NASA postdoctoral fellowship program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904849116/-/DCSupplemental.
References
- 1.Runnels C. M., et al. , Folding, assembly, and persistence: The essential nature and origins of biopolymers. J. Mol. Evol. 86, 598–610 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rose G. D., Fleming P. J., Banavar J. R., Maritan A., A backbone-based theory of protein folding. Proc. Natl. Acad. Sci. U.S.A. 103, 16623–16633 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xia Y., Levitt M., Simulating protein evolution in sequence and structure space. Curr. Opin. Struct. Biol. 14, 202–207 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Fahnestock S., Rich A., Ribosome-catalyzed polyester formation. Science 173, 340–343 (1971). [DOI] [PubMed] [Google Scholar]
- 5.Forsythe J. G., et al. , Ester-mediated amide bond formation driven by wet-dry cycles: A possible path to polypeptides on the prebiotic earth. Angew. Chem. Int. Ed. Engl. 54, 9871–9875 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu S., et al. , Elongation of model prebiotic proto-peptides by continuous monomer feeding. Macromolecules 50, 9286–9294 (2017). [Google Scholar]
- 7.Forsythe J. G., et al. , Surveying the sequence diversity of model prebiotic peptides by mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 114, E7652–E7659 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doran D., Abul-Haija Y. M., Cronin L., Emergence of function and selection from recursively programmed polymerisation reactions in mineral environments. Angew. Chem. Int. Ed. Engl. 10.1002/anie.201902287 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouza M., et al. , Compositional characterization of complex protopeptide libraries via triboelectric nanogenerator orbitrap mass spectrometry. Rapid Commun. Mass Spectrom. (2019). [DOI] [PubMed] [Google Scholar]
- 10.Ross D. S., Deamer D., Dry/wet cycling and the thermodynamics and kinetics of prebiotic polymer synthesis. Life (Basel) 6, E28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ura Y., et al. , Dynamic polythioesters via ring-opening polymerization of 1,4-thiazine-2,5-diones. Org. Biomol. Chem. 7, 2878–2884 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Olsén P., Odelius K., Albertsson A. C., Thermodynamic presynthetic considerations for ring-opening polymerization. Biomacromolecules 17, 699–709 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller S. L., A production of amino acids under possible primitive Earth conditions. Science 117, 528–529 (1953). [DOI] [PubMed] [Google Scholar]
- 14.Bernstein M. P., Dworkin J. P., Sandford S. A., Cooper G. W., Allamandola L. J., Racemic amino acids from the ultraviolet photolysis of interstellar ice analogues. Nature 416, 401–403 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Peltzer E. T., Bada J. L., α-Hydroxycarboxylic acids in the Murchison meteorite. Nature 272, 443–444 (1978). [Google Scholar]
- 16.Neveu M., Kim H. J., Benner S. A., The “strong” RNA world hypothesis: Fifty years old. Astrobiology 13, 391–403 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Higgs P. G., Lehman N., The RNA world: Molecular cooperation at the origins of life. Nat. Rev. Genet. 16, 7–17 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Joyce G. F., RNA evolution and the origins of life. Nature 338, 217–224 (1989). [DOI] [PubMed] [Google Scholar]
- 19.Orgel L. E., Prebiotic chemistry and the origin of the RNA world. Crit. Rev. Biochem. Mol. Biol. 39, 99–123 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Hager A. J., Pollard J. D., Szostak J. W., Ribozymes: Aiming at RNA replication and protein synthesis. Chem. Biol. 3, 717–725 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Wieczorek R., Dörr M., Chotera A., Luisi P. L., Monnard P. A., Formation of RNA phosphodiester bond by histidine-containing dipeptides. ChemBioChem 14, 217–223 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Dworkin J. P., Lazcano A., Miller S. L., The roads to and from the RNA world. J. Theor. Biol. 222, 127–134 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Griesser H., Bechthold M., Tremmel P., Kervio E., Richert C., Amino acid-specific, ribonucleotide-promoted peptide formation in the absence of enzymes. Angew. Chem. Int. Ed. Engl. 56, 1224–1228 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Vázquez-Salazar A., Lazcano A., Early life: Embracing the RNA world. Curr. Biol. 28, 1166–1167 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Levy M., Ellington A. D., Peptide-templated nucleic acid ligation. J. Mol. Evol. 56, 607–615 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Meierhenrich U. J., Muñoz Caro G. M., Bredehöft J. H., Jessberger E. K., Thiemann W. H., Identification of diamino acids in the Murchison meteorite. Proc. Natl. Acad. Sci. U.S.A. 101, 9182–9186 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson A. P., et al. , The Miller volcanic spark discharge experiment. Science 322, 404 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Ruiz-Bermejo M., Menor-Salván C., Osuna-Esteban S., Veintemillas-Verdaguer S., The effects of ferrous and other ions on the abiotic formation of biomolecules using aqueous aerosols and spark discharges. Orig. Life Evol. Biosph. 37, 507–521 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Zaia D. A., Zaia C. T., De Santana H., Which amino acids should be used in prebiotic chemistry studies? Orig. Life Evol. Biosph. 38, 469–488 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Hattori Y., Kinjo M., Ishigami M., Nagano K., Formation of amino acids from CH4-rich or CO2-rich model atmosphere. Orig. Life 14, 145–150 (1984). [DOI] [PubMed] [Google Scholar]
- 31.Georgiou C. D., Functional properties of amino acid side chains as biomarkers of extraterrestrial life. Astrobiology 18, 1479–1496 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cobb A. K., Pudritz R. E., Nature’s starships. I. Observed abundances and relative frequencies of amino acids in meteorites. Astrophys. J. 783, 140 (2014). [Google Scholar]
- 33.Patel B. H., Percivalle C., Ritson D. J., Duffy C. D., Sutherland J. D., Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nat. Chem. 7, 301–307 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen C., Yang L., Miller S. L., Oró J., Prebiotic synthesis of imidazole-4-acetaldehyde and histidine. Orig. Life Evol. Biosph. 17, 295–305 (1987). [DOI] [PubMed] [Google Scholar]
- 35.Pandey A., Pandey G. C., Aswath P. B., Synthesis of polylactic acid-polyglycolic acid blends using microwave radiation. J. Mech. Behav. Biomed. Mater. 1, 227–233 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Walsh K. A., Trypsinogens and trypsins of various species. Methods Enzymol. 19, 41–63 (1970). [Google Scholar]
- 37.Rodriguez-Garcia M., et al. , Formation of oligopeptides in high yield under simple programmable conditions. Nat. Commun. 6, 8385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imai E., Honda H., Hatori K., Brack A., Matsuno K., Elongation of oligopeptides in a simulated submarine hydrothermal system. Science 283, 831–833 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Goodman M., Stueben K. C., Peptide syntheses via amino acid active Esters1. J. Am. Chem. Soc. 81, 3980–3983 (1959). [Google Scholar]
- 40.Lahav N., White D., Chang S., Peptide formation in the prebiotic era: Thermal condensation of glycine in fluctuating clay environments. Science 201, 67–69 (1978). [DOI] [PubMed] [Google Scholar]
- 41.Parker E. T., et al. , A plausible simultaneous synthesis of amino acids and simple peptides on the primordial Earth. Angew. Chem. Int. Ed. Engl. 53, 8132–8136 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Fuchida S., Masuda H., Shinoda K., Peptide formation mechanism on montmorillonite under thermal conditions. Orig. Life Evol. Biosph. 44, 13–28 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Brack A., Ehler K. W., Orgel L. E., N, N′-carbonyldiimidazole-induced diketopiperazine formation in aqueous solution in the presence of adenosine-5′-monophosphate. J. Mol. Evol. 8, 307–310 (1976). [DOI] [PubMed] [Google Scholar]
- 44.Hartman H., Smith T. F., The evolution of the ribosome and the genetic code. Life (Basel) 4, 227–249 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavlov M. Y., et al. , Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc. Natl. Acad. Sci. U.S.A. 106, 50–54 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng R. P., Girinath P., Ahmad R., Effect of lysine side chain length on intra-helical glutamate–lysine ion pairing interactions. Biochemistry 46, 10528–10537 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Virdee S., Macmillan D., Waksman G., Semisynthetic Src SH2 domains demonstrate altered phosphopeptide specificity induced by incorporation of unnatural lysine derivatives. Chem. Biol. 17, 274–284 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Okamoto R., et al. , Regioselective α-peptide bond formation through the oxidation of amino thioacids. Biochemistry 58, 1672–1678 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Weber A. L., Miller S. L., Reasons for the occurrence of the twenty coded protein amino acids. J. Mol. Evol. 17, 273–284 (1981). [DOI] [PubMed] [Google Scholar]
- 50.Gracia Lux C., Olejniczak J., Fomina N., Viger M. L., Almutairi A., Intramolecular cyclization assistance for fast degradation of ornithine-based poly(ester amide)s. J. Polym. Sci. A Polym. Chem. 51, 3783–3790 (2013). [Google Scholar]
- 51.King J. F., Rathore R., Lam J. Y. L., Guo Z. R., Klassen D. F., pH optimization of nucleophilic reactions in water. J. Am. Chem. Soc. 114, 3028–3033 (1992). [Google Scholar]
- 52.Dawson R. M. C., Elliott D. C., Elliott W. H., Jones K. M., Data for Biochemical Research (Clarendon Press, Oxford, UK, 1959). [Google Scholar]
- 53.Lan Y., et al. , Incorporation of 2,3-diaminopropionic acid into linear cationic amphipathic peptides produces pH-sensitive vectors. ChemBioChem 11, 1266–1272 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sigel A., Sigel H., Sigel R. K. O., Nickel and Its Surprising Impact in Nature (Metal Ions in Life Sciences, Wiley, Chichester, UK, 2008), vol. 2. [Google Scholar]
- 55.Aso K., Kodaka H., Trypsin-catalyzed oligomerization of L-lysine esters. Biosci. Biotechnol. Biochem. 56, 755–758 (1992). [DOI] [PubMed] [Google Scholar]
- 56.Bruice T. C., Schmir G. L., Imidazole catalysis. II. The reaction of substituted imidazoles with phenyl acetates in aqueous solution. J. Am. Chem. Soc. 80, 148–156 (1958). [Google Scholar]
- 57.Leclerc J., Benoiton L., On the selectivity of acylation of unprotected diamino acid. Can. J. Biochem. 46, 1047–1051 (1968). [DOI] [PubMed] [Google Scholar]
- 58.Schallenberg E. E., Calvin M., Ethyl thioltrifluoroacetate as an acetylating agent with particular reference to peptide synthesis. J. Am. Chem. Soc. 77, 2779–2783 (1955). [Google Scholar]
- 59.Hay R. W., Morris P. J., Proton ionisation constants and kinetics of base hydrolysis of some α-amino-acid esters in aqueous solution. Part III. Hydrolysis and intramolecular aminolysis of αω-diamino-acid methyl esters. J. Chem. Soc., Perkin Trans. 2, 1021–1029 (1972). [Google Scholar]
- 60.Kovacs N. A., Petrov A. S., Lanier K. A., Williams L. D., Frozen in time: The history of proteins. Mol. Biol. Evol. 34, 1252–1260 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petrov A. S., et al. , Evolution of the ribosome at atomic resolution. Proc. Natl. Acad. Sci. U.S.A. 111, 10251–10256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petrov A. S., et al. , History of the ribosome and the origin of translation. Proc. Natl. Acad. Sci. U.S.A. 112, 15396–15401 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brack A., Selective emergence and survival of early polypeptides in water. Orig. Life Evol. Biosph. 17, 367–379 (1987). [DOI] [PubMed] [Google Scholar]
- 64.Abkevich V. I., Gutin A. M., Shakhnovich E. I., How the first biopolymers could have evolved. Proc. Natl. Acad. Sci. U.S.A. 93, 839–844 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hud N. V., Anet F. A., Intercalation-mediated synthesis and replication: A new approach to the origin of life. J. Theor. Biol. 205, 543–562 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Peters J. W., Williams L. D., The origin of life: Look up and look down. Astrobiology 12, 1087–1092 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Krishnamurthy R., Life’s biological chemistry: A destiny or destination starting from prebiotic chemistry? Chemistry 24, 16708–16715 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Kamat N. P., Tobé S., Hill I. T., Szostak J. W., Electrostatic localization of RNA to protocell membranes by cationic hydrophobic peptides. Angew. Chem. Int. Ed. Engl. 54, 11735–11739 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bomba R., Kwiatkowski W., Sánchez-Ferrer A., Riek R., Greenwald J., Cooperative induction of ordered peptide and fatty acid aggregates. Biophys. J. 115, 2336–2347 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bashan A., Yonath A., Ribosome crystallography: Catalysis and evolution of peptide-bond formation, nascent chain elongation and its co-translational folding. Biochem. Soc. Trans. 33, 488–492 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Harms J., et al. , High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107, 679–688 (2001). [DOI] [PubMed] [Google Scholar]
- 72.van der Gulik P. T., Speijer D., How amino acids and peptides shaped the RNA world. Life (Basel) 5, 230–246 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Blanco C., Bayas M., Yan F., Chen I. A., Analysis of evolutionarily independent protein-RNA complexes yields a criterion to evaluate the relevance of prebiotic scenarios. Curr. Biol. 28, 526–537.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 74.Zozulia O., Dolan M. A., Korendovych I. V., Catalytic peptide assemblies. Chem. Soc. Rev. 47, 3621–3639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rufo C. M., et al. , Short peptides self-assemble to produce catalytic amyloids. Nat. Chem. 6, 303–309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Adamala K., Szostak J. W., Competition between model protocells driven by an encapsulated catalyst. Nat. Chem. 5, 495–501 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shu W., et al. , Selective formation of Ser-His dipeptide via phosphorus activation. Orig. Life Evol. Biosph. 48, 213–222 (2018). [DOI] [PubMed] [Google Scholar]
- 78.Poláčková V., Cˇmelová P., Górová R., Sˇebesta R., Peptide-catalyzed stereoselective Michael addition of aldehydes and ketones to heterocyclic nitroalkenes. Monatshefte für Chemie Chemical Monthly 149, 729–736 (2018). [Google Scholar]
- 79.De Duve C., Singularities: Landmarks on the Pathways of Life (Cambridge University Press, Cambridge, UK, 2005). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.