Abstract
Mammalian SWI/SNF chromatin remodeling complexes exist in three distinct, final-form assemblies: canonical BAF (cBAF), PBAF, and a newly-characterized non-canonical complex, ncBAF. However, their complex-specific targeting on chromatin, functions and roles in disease remain largely undefined. Here, we comprehensively mapped complex assemblies on chromatin and found that ncBAF complexes uniquely localize to CTCF sites and promoters. We identified ncBAF subunits as synthetic lethal targets specific to synovial sarcoma (SS) and malignant rhabdoid tumor (MRT), which share in common cBAF complex (SMARCB1 subunit) perturbation. Chemical and biological depletion of the BRD9 subunit of ncBAF rapidly attenuates SS and MRT cell proliferation. Notably, in cBAF-perturbed cancers, ncBAF complexes maintain gene expression at retained CTCF-promoter sites, and function in a manner distinct from fusion oncoprotein-bound complexes. Taken together, these findings unmask the unique chromatin targeting and function of ncBAF complexes and present new cancer-specific therapeutic targets.
Introduction
Mammalian SWI/SNF (mSWI/SNF) complexes are ATP-dependent chromatin remodelers that modulate genomic architecture and DNA accessibility, enabling timely and appropriate control of gene expression1–11. They are combinatorially assembled from the products of 29 total genes into three final-form complexes: canonical BAF, PBAF (polybromo-associated BAF complexes), and a newly-defined non-canonical BAF (ncBAF), with specific subunits specifying distinct complexes, such as PBRM1, ARID2, and BRD7 in PBAF complexes, ARID1A/ARID1B and DPF2 in canonical BAF (cBAF) complexes, and GLTSCR1/GLTSCR1L and BRD9 in ncBAF complexes12–15. The specific genome-wide targeting and biochemical functions of these distinct complexes to date remain poorly understood, owing in part to limitations in understanding full subunit composition and combinatorial parameters, complex assembly pathways and the paucity of robust strategies to map the relative localization of each complex on chromatin.
Mutations in the genes encoding mSWI/SNF subunits are found in over 20% of human cancers16, 17, with specific subunits mutated in specific malignancies, pointing toward subunit- and complex-specific functions. For example, >98% of cases of malignant rhabdoid tumor (MRT) exhibit biallelic loss of the SMARCB1 gene, which encodes the SMARCB1/BAF47/hSNF5 subunit of BAF and PBAF (but not ncBAF) complexes18–20. Furthermore, complex-defining subunits such as ARID1A and PBRM1 are recurrently mutated in distinct cancers, ovarian clear cell carcinoma and renal clear cell carcinoma, respectively21, 22.
While the majority of mSWI/SNF gene mutations result in loss-of-function phenotypes, the SS18-SSX fusion hallmark to synovial sarcoma (SS) results in de novo, gain-of-function targeting of BAF complexes, which activates the unique SS gene expression signature23. Incorporation of the SS18-SSX oncoprotein into BAF complexes results in protein-level destabilization of SMARCB1 (a feature shared with MRT), but this event is secondary and not required for maintenance of SS gene expression or proliferation23. Finally, genetic perturbation screens in cell lines bearing mutations in mSWI/SNF subunits that are part of paralog families (i.e. SMARCA4 and ARID1A) have unveiled synthetic lethal dependencies on residual complexes assembled with their rarely-mutated partner paralogs (i.e. SMARCA2 and ARID1B)24, 25. Collectively, these findings highlight subunit- and paralog-specific biological roles, such as those demonstrated in the context of development4, 8–10.
Defining the functional differences among the diverse array of complexes within the mSWI/SNF family represents a major goal for the field at-large. In this study, we used biochemical and bioinformatic approaches to map mSWI/SNF complexes on chromatin, which revealed an unexpected, complex-specific localization of ncBAF complexes to CTCF and promoter-proximal sites. We find that cancers driven by core cBAF subunit perturbations, such as SS and MRT, are uniquely dependent on ncBAF function. We further demonstrate that perturbation of ncBAF complexes is, unexpectedly, mechanistically distinct from perturbation of synovial sarcoma disease-driver SS18-SSX and that in both SS and MRT settings, ncBAF plays critical roles in maintaining gene expression at retained mSWI/SNF sites.
Distinct function and genome-wide localization across mSWI/SNF complex families
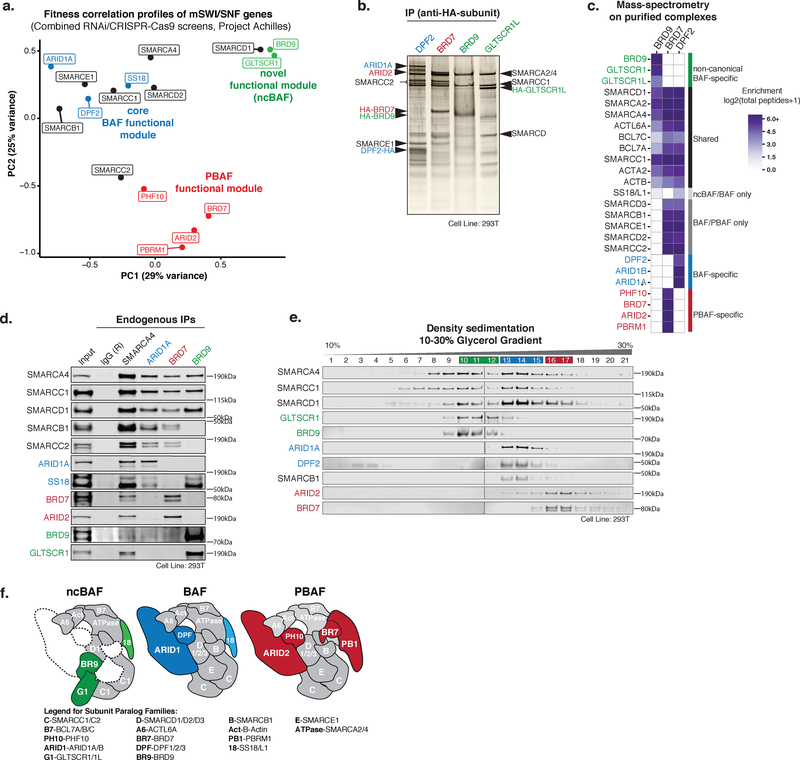
Genes involved in similar biological pathways or protein complexes often exhibit coordinated fitness variation ingenetic perturbation screens, such as those performed across hundreds of human cancer cell lines26–29. Specifically, we have previously established that mSWI/SNF complex genes cluster in to three functional modules: core BAF, PBAF, and an unexpected functional module we termed ncBAF12. Similar analyses performed on a combined shRNA- and CRISPR-Cas9-based dataset (Project Achilles, Broad Institute)26, 27, 30 as well as the Project DRIVE (Novartis) dataset29, further suggest that BAF, PBAF, and ncBAF on average represent functionally distinct entities, providing motivation to define their underlying features (Fig. 1a, Supplementary Fig. 1a). Importantly, in agreement with these functional distinctions, biochemical studies resolved complexes of distinct size and componentry, as demonstrated by complex purifications, mass-spectrometry, and density sedimentation (Fig. 1b–f, Supplementary Fig. 1b–e, Supplementary Table 1). Specifically, we found that ncBAF complexes uniquely lack core, evolutionarily conserved subunits such as SMARCB1 and SMARCE1, incorporate selective paralogs, i.e. SMARCC1 but not SMARCC2 and SMARCD1 but not SMARCD2 or SMARCD3, and contain a set of complex-specific subunits not shared by cBAF or PBAF, the GLTSCR1/1L paralogs and BRD9.
Figure 1. mSWI/SNF family complexes are biochemically and functionally distinct.
a. Principal component analysis (PCA) performed on fitness correlations between mSWI/SNF genes from combined genome-scale RNAi- and CRISPR-Cas9-based genetic perturbation screens (Project Achilles, Broad Institute).
b. SDS-PAGE and silver stain performed on purified complexes using indicated HA-tagged subunits expressed in HEK-293T cells. Purifications performed in n=3 biologically independent experiments for each bait.
c. Proteomic mass-spectrometry performed on mSWI/SNF complexes purified from HEK-293T cells expressing indicated HA-tagged mSWI/SNF subunits.
d. Immunoprecipitation for endogenous SMARCA4 (pan-mSWI/SNF complex component), ARID1A (canonical BAF-specific), BRD7 (PBAF-specific), and BRD9 (ncBAF-specific) subunits in HEK-293T nuclear extracts followed by immunoblot for select subunits. Subunits in blue, red, and green represent BAF-, PBAF-, and BRD9/GLTSCR1- specific complexes, respectively. Immunoprecipitations performed in n=2 biologically independent experiments. See also Supplementary Figure 7a.
e. Separation of HEK-293T nuclear extracts via 10–30% glycerol gradient density sedimentation followed by immunoblot for selected mSWI/SNF subunits. Glycerol gradient performed in n=3 biologically independent experiments. See also Supplementary Figure 7b.
f. Schematic depicting biochemical subunit compositions for mammalian ncBAF, canonical BAF, and PBAF complexes.
To begin to characterize these distinct assemblies and determine if differential targeting on chromatin may in part underlie their differences, we comprehensively mapped BAF, PBAF, and ncBAF complexes genome-wide by performing ChIP-seq in a mSWI/SNF-intact cell line, EoL-1, using antibodies against pan-mSWI/SNF subunits (SMARCC1 and SMARCA4) and complex-specific subunits BRD9 and GLTSCR1 for ncBAF, DPF2 for canonical BAF (cBAF), and BRD7 for PBAF (Supplementary Fig. 2a). Consistent with biochemical studies, BRD7, DPF2, and BRD9 and GLTSCR1 peaks comprise subsets of all SMARCA4 ATPase subunit peaks, and BRD9 and GLTSCR1 peaks significantly overlap one another (Fig. 2a, Supplementary Fig. 2b–d). Hierarchical clustering performed on ChIP-seq read density over the merged set of peaks across all ChIPs performed identified distinct, complex-specific enrichment on chromatin (Fig. 2b, Supplementary Fig. 2e), such as ncBAF complexes over the promoter (green shade), PBAF complex occupancy into the gene body (red shade), and cBAF complexes at distal sites (blue shade) at the VEGFA locus (Fig. 2c). ncBAF and PBAF complexes exhibited a distinct promoter-proximal distribution in comparison to cBAF complexes, which were substantially more localized to distal sites (Supplementary Fig. 2f). Additionally, at transcription start sites (TSSs), PBAF complexes were more enriched over gene bodies relative to ncBAF complexes (Fig. 2c, Supplementary Fig. 2g).
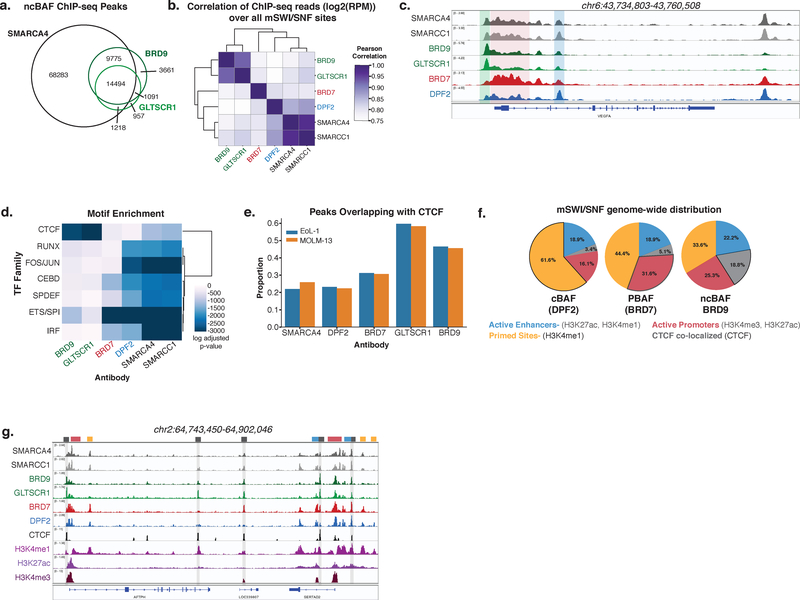
Figure 2. Differential localization of mSWI/SNF complexes, ncBAF, cBAF, and PBAF, on chromatin.
a. Venn diagram of peaks from BRD9, GLTSCR1, and SMARCA4 ChIP-seq experiments.
b. Heatmap representing correlations between normalized ChIP-seq reads (Log2(RPM)) over a merged set of all mSWI/SNF subunit peaks. ChIP performed in n=2 independent samples for each.
c. Localization of ncBAF, BAF, and PBAF complexes at the VEGFA locus. ChIP performed in n=2 independent samples for each.
d. Heatmap of CentriMo log adjusted p-values for top motifs returned by MEME-ChIP analysis for each ChIP-seq experiment. ChIP performed in n=2 independent samples for each, p-values were calculated using binomial test.
e. Proportion of peaks from ChIP-seq experiments using indicated antibodies overlapping CTCF ChIP-seq peaks in MOLM-13 and EoL-1 cell lines.
f. Pie graphs reflecting proportion of ncBAF-, BAF-, and PBAF- specific peaks overlapping with specified chromatin features (see also Supplementary Fig. 2i).
g. Example tracks demonstrating differential mSWI/SNF complex family enrichment across the AFTPH locus.
ChIP performed in n=2 independent samples for each.
Motif analyses revealed central enrichment of cBAF complexes over known transcription factor (TF) motifs, including FOS/JUN, AP-1, SPDEF, and ETS (with PBAF complexes enriched at a subset of these), however, ncBAF complexes specifically localized to CTCF sequence motifs, sites well characterized to play roles in the maintenance of DNA architecture31–34 (Fig. 2d). We performed ChIP-seq for CTCF and indeed found that ncBAF complexes strongly and selectively co-localized with CTCF across cell lines (Fig. 2e, Supplementary Fig. 2h). cBAF complexes were most enriched at active enhancers (H3K27ac and H3K4me1) and at primed sites (H3K4me1), suggesting roles for cBAF in enhancer regulation (Fig. 2f,g, Supplementary Fig. 2i–k). In contrast, a greater proportion of PBAF complexes were localized to active promoters (H3K27ac and H3K4me3), at which PBAF complexes were also the most enriched among the three complexes. Finally, ncBAF complexes were most enriched at CTCF sites, particularly CTCF sites co-localized with H3K4me1 (Supplementary Fig. 2l). These CTCF co-localized sites comprised a greater portion of all ncBAF peaks relative to cBAF and PBAF peaks. Thus, while the localization and biological roles for mSWI/SNF complexes have been most extensively explored at enhancers35–38, these results implicate specialized roles for ncBAF and PBAF complexes at CTCF sites and promoters, respectively, and demonstrate mSWI/SNF complex-specific chromatin localization.
Genome-scale fitness screening reveals cancer-specific dependencies on ncBAF complexes
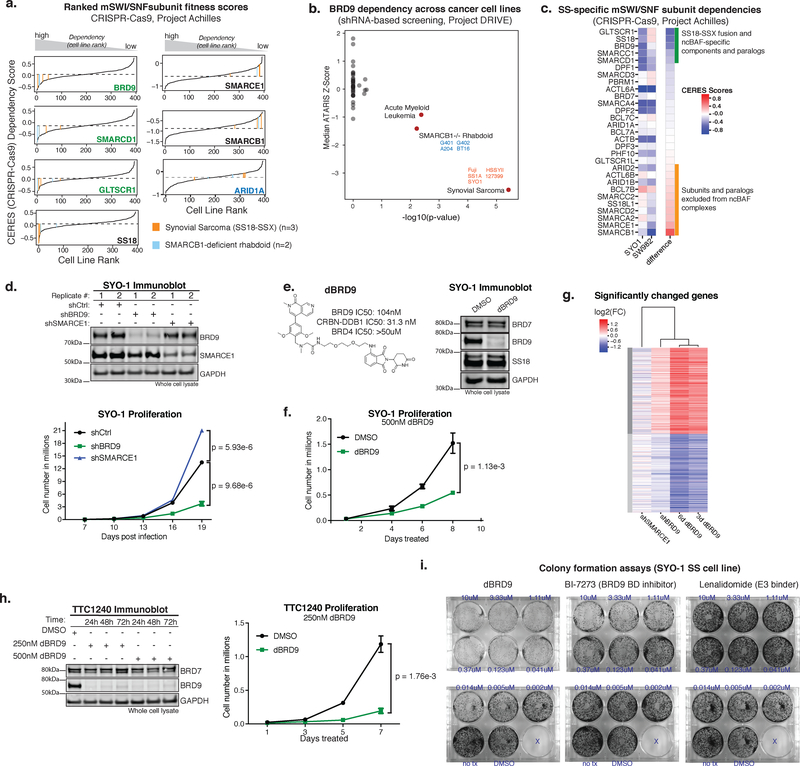
To determine whether ncBAF subunits were uniquely required for proliferative maintenance of any cancer types, analyzed CRISPR-Cas9-based screens performed across 387 cancer cell lines26 and performed screens in 3 new synovial sarcoma (SS) cell lines (Supplementary Fig. 3a; Achilles 18Q3 dataset, https://cds.team/depmap/#). These screens identified significant, selective sensitivity of both SS and MRT cell lines to perturbation of ncBAF complex subunits BRD9, GLTSCR1, and SMARCD1 (Fig. 3a). These dependency profiles were specific to SS and MRT, both of which are sarcomas, and not to other soft-tissue malignancies (Supplementary Fig. 3b). To corroborate these results, we analyzed shRNA-based fitness screens from Project DRIVE29 and again found that SS (n=5) and MRT (n=4) cell lines were selectively sensitive to BRD9 suppression (Fig. 3b, Supplementary Fig. 3c). We further confirmed that ncBAF dependency was specific to SS18-SSX fusion oncoprotein-positive SS, as an SS histological mimic cell line, SW982, which lacks the SS18-SSX fusion, was insensitive to ncBAF component perturbation (Fig. 3c).
Figure 3. ncBAF components are selective synthetic lethal dependencies in synovial sarcoma and rhabdoid tumor cell lines.
a. Waterfall plots for CERES scores across n=393 cell lines screened using CRISPR-Cas9 (Project Achilles). SS (orange) and MRT (blue) cell lines are indicated. Dashed line represents median dependency.
b. BRD9 sensitivity across n=387 cell lines in Project DRIVE. Two-sided Fisher’s exact test -log10(pvalue) for BRD9 sensitivity (ATARIS score<-.75) against median z-score in each cancer type. Annotations with FDR<0.1 are colored in red.
c. Heatmap of CERES scores in SYO-1 (SS18-SSX-driven SS) and SW982 (histological SS mimic without SS18-SSX translocation) ranked by difference in dependency.
d. (Top) Immunoblot on total cell lysates from SYO-1 cells in each knockdown condition (n=2 biologically independent experiments); (Bottom) Proliferation experiments in SYO-1 representative of n=3 biologically-independent experiments (with separate lentiviral infection and cell number calculations) performed. Each data point represents mean +/- SD, p-value calculated by two-sided t-test on day 19. See also Supplementary Figure 7c, Supplementary Table 2.
e. (Left) Chemical structure of dBRD9 degron compound; (Right) Immunoblot on total cell lysates from SYO-1 cells treated with DMSO vehicle control or dBRD9 (500nM) for 3 days (n=2 biologically independent experiments). See also Supplementary Figure 7d.
f. Proliferation experiments in SYO-1 representative of n=3 biologically-independent experiments (separate treatments and cell number calculations). Each data point represents mean +/- SD, p-value calculated by two-sided t-test on day 8. See also Supplementary Table 2.
g. Heatmap of gene expression changes of significantly changed genes (p-adjusted<.001 and log2(fc)>.59; Benjamini-Hochberg) in any one of the four treatments. Genes were k-means clustered into 2 groups, samples were clustered hierarchically. n=2 biological replicates for each RNA-seq experiment.
h. (Top) Immunoblot on total cell lysates from TTC1240 malignant rhabdoid tumor cells treated with either DMSO vehicle control or dBRD9 (250nM) (n=2 biologically independent experiments); (Bottom) Proliferation experiments in TTC1240 representative of n=3 biologically-independent experiments (separate treatments and cell number calculations). Each data point represents mean +/- SD, p-value calculated by two-sided t-test on day 7. See also Supplementary Figure 7e, Supplementary Table 2.
i. Representative colony formation assay performed on SYO-1 cells treated with dBRD9, BI-7273, or lenalidomide as a control (n=3 biologically independent experiments).
Both SS and MRT exhibit perturbations to the cBAF core functional module12. SS is uniformly characterized by the t(X;18) chromosomal translocation which produces the SS18-SSX fusion oncoprotein, a stable and dedicated mSWI/SNF complex subunit that destabilizes SMARCB139, 40, and MRT and atypical teratoid/rhabdoid tumor (AT/RT) cell lines are driven by biallelic loss of the SMARCB1 gene (encoding the SMARCB1/BAF47/SNF5/INI1 subunit)18–20 (Supplementary Fig. 3d). In SS, loss of proliferative fitness resulting from ncBAF subunit perturbation was comparable to SS18 perturbation, the driver of disease (Fig. 3a). Both SS and MRT cell lines exhibited higher sensitivity to BRD9 loss than AML cell lines, which have been previously been reported to be sensitive to BRD9 knockdown41, 42 (Fig. 3a, Supplementary Fig. 3c). We found that AML cell lines were near uniformly sensitive to depletion of a wide range of mSWI/SNF complex subunits rather than ncBAF components selectively (Supplementary Fig. 3e). In SS and MRT, SMARCB1 (destabilized and deleted in these cancers) as well as other cBAF and PBAF subunits such as SMARCE1, ARID1A, and BRD7, did not score as dependencies (Fig. 3a, Supplementary Fig 3c), highlighting the selective sensitivity to ncBAF subunit disruption in these cancer types.
To validate these findings, we biologically and chemically depleted BRD9 using shRNA and dBRD9 chemical degradation43 strategies. Suppression of BRD9 in SYO-1 synovial sarcoma cells significantly attenuated proliferation, as compared to either control shRNA or shRNA directed against SMARCE1, a structurally essential component of cBAF and PBAF complexes which is not a part of ncBAF, further confirming screening results (Fig. 3d, Supplementary Table 2). dBRD9-treated SS cells exhibited near complete depletion of BRD9 from whole cell lysates and proliferative attenuation, approaching that which results from SS18-SSX oncoprotein knockdown (Fig. 3e–f, Supplementary Fig. 3f,g, Supplementary Table 2). Knockdown of GLTSCR1 in SYO-1 cells also attenuated proliferation, supporting the role of ncBAF complexes in SS cell maintenance (Supplementary Fig. 3h). Cell cycle analysis performed on cells treated with dBRD9 revealed a decrease in cells in S phase and an increase in cells in sub G1 relative to DMSO vehicle control, and Annexin V staining demonstrated an increase in apoptotic cells (Supplementary Fig. 3i,j). Further, global transcriptional profiling revealed similar effects on gene expression between dBRD9 and shBRD9 treatments, while shSMARCE1 resulted in discordant changes and minimal transcriptional effect (Fig. 3g). Finally, dBRD9 treatment of SMARCB1-deficient MRT cell lines TTC1240 and G401 resulted in reduced proliferation (Fig. 3h, Supplementary Fig. 3k, Supplementary Table 2), while treatment in a SMARCB1-intact epithelioid sarcoma (EpS) cell line, ESX, did not (Supplementary Fig. 3l). As mSWI/SNF complexes in SS and MRT/ATRT/EpS disease settings exhibit the shared feature of cBAF perturbation and SMARCB1 (BAF47) loss or destabilization, these results unmask a selective ncBAF dependency in two aggressive and intractable BAF-mutant cancer types.
Finally, we compared total degradation of BRD9 (dBRD9 degrader) versus BRD9 bromodomain inhibition using BI-7273. Colony formation assays in SYO-1, HSSYII, and Aska SS cell lines demonstrated that dBRD9 treatment was substantially and reproducibly more potent than BI-7273 (or treatment with lenalidomide control) (Fig. 3i, Supplementary Fig. 3m,n). Moreover, these treatments in cell lines lacking BAF complex perturbations did not result in colony formation inhibition (Supplementary Fig. 3o–q). Knockout of BRD9 in HEK-293T cells resulted in destabilized ncBAF complexes (Supplementary Fig. 3r), perhaps underlying the increased potency of dBRD9 relative to BI-7273. Taken together, these data suggest that total loss of BRD9 is more potent than bromodomain inhibition, supporting the possibility that other domains of BRD9 may be important for BRD9 subunit function.
CRISPR guide RNA tiling experiments define required domains on GLTSCR1 and BRD9 ncBAF subunits
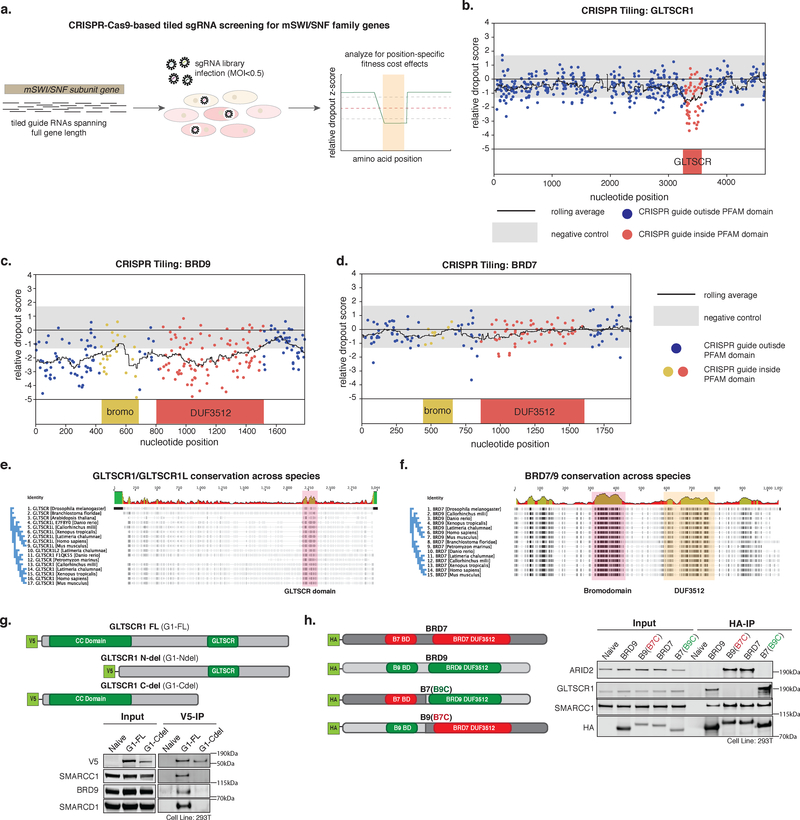
We next sought to identify specific regions on BRD9 as well as other ncBAF components that uniquely underlie the synthetic lethality in SS. We performed CRISPR tiling screens with an array of guide RNAs against the exonic sequences of several subunits and their paralogs in the SYO-1 synovial sarcoma cell line (Fig. 4a, Supplementary Table 2,3). Guides targeting regions of the disease driver, SS18, part of the SS18-SSX fusion, drop out as expected (Supplementary Fig. 4a,b). Consistent with CRISPR and RNAi screening results above (Fig. 3), cBAF/PBAF-specific subunits SMARCB1 and SMARCE1 did not drop out (Supplementary Fig. 4c,d), and ncBAF-specific paralogs SMARCD1 and SMARCC1 exhibited more drop out relative to SMARCD2/3 and SMARCC2, respectively, paralogs which do not assemble in to ncBAF (Supplementary Fig. 4e–h). Interestingly, guides targeting the GLTSCR domain of GLTSCR1 and most of the coding region of BRD9, including the bromodomain and the DUF3512, exhibited significant fitness dropout (Fig. 4b,c). In contrast, guides targeting the bromodomain and DUF3512 of PBAF-specific BRD7, the paralog of BRD9, did not result in reduced fitness (Fig. 4d). These data highlight the synthetic lethal specificity for ncBAF components, and demonstrate the importance of the GLTSCR and DUF3512 domains of GLTSCR1/1L and BRD9, respectively, in ncBAF function in SS.
Figure 4. ncBAF subunit domains underlie complex-specific synthetic lethalities.
a. Schematic for tiled gRNA CRISPR-Cas9 screening performed across mSWI/SNF subunit genes in SYO-1 SS cells.
b. Tiled CRISPR screening for the GLTSCR1 gene in SYO-1 cells.
c,d. Tiled CRISPR screening for the BRD9 gene (c) and BRD7 gene (d) in SYO-1 cells.
e. Alignment of GLTSCR1 amino acid sequences across species. GLTSCR domain is highlighted.
f. Alignment of amino acid sequences for BRD9 and BRD7 across species. Bromodomain and DUF3512 are highlighted.
g. (Top) Construct design for GLTSCR domain experiments in HEK-293T cells. (Bottom) Immunoprecipitation of V5-tagged constructs followed by immunoblot (n=2 biologically independent experiments). See also Supplementary Figure 7f.
h. (Left) Construct design for C-terminal swap experiments for BRD9 and BRD7 in HEK-293T cells (BD = bromodomain). (Right) Immunoprecipitation of BRD9, BRD7, and BRD7(B9C) and BRD9(B7C) C-terminal swap variants followed by immunoblot in HEK-293T cells (n=2 biologically independent experiments). See also Supplementary Figure 7g.
To understand the roles of the GLTSCR and DUF3512 domains in ncBAF complexes, we assessed evolutionary conservation of these regions (Fig. 4e,f). The most evolutionarily conserved region of the GLTSCR1/1L paralogs is the GLTSCR domain, suggesting it could possibly serve an important structural role. Indeed, immunoprecipitation followed by immunoblot of N-terminal and C-terminal truncation mutants of mammalian GLTSCR1 demonstrated that this domain is required for interaction with ncBAF complexes and thus serves as an ncBAF-specific binding region (Fig. 4g, Supplementary Fig. 4i). In contrast, although the bromodomain and DUF3512 regions are evolutionarily conserved between BRD9 and BRD7 homologs across species (Fig. 4f), mammalian BRD9 and BRD7 paralogs incorporate into ncBAF and PBAF complexes, respectively. To determine if the DUF3512 is involved in complex-specific binding of the BRD9 and BRD7 subunits, we performed domain swapping experiments in which we fused the C-terminal DUF-containing region of BRD9 to the N-terminus of BRD7 and vice versa (Fig. 4h). Swapping of BRD9 and BRD7 DUF3512 regions resulted in switched complex specification, with BRD9-(BRD7 DUF) binding PBAF complexes and BRD7-(BRD9 DUF) binding ncBAF complexes (Fig. 4h). Taken together, these results implicate the BRD9 DUF3512 and the GLTSCR1 GLTSCR domains as ncBAF complex binding domains that underlie critical dependencies in SS cell contexts.
ncBAF is not required for SS18-SSX fusion-mediated gene expression and primarily regulates retained fusion-independent sites
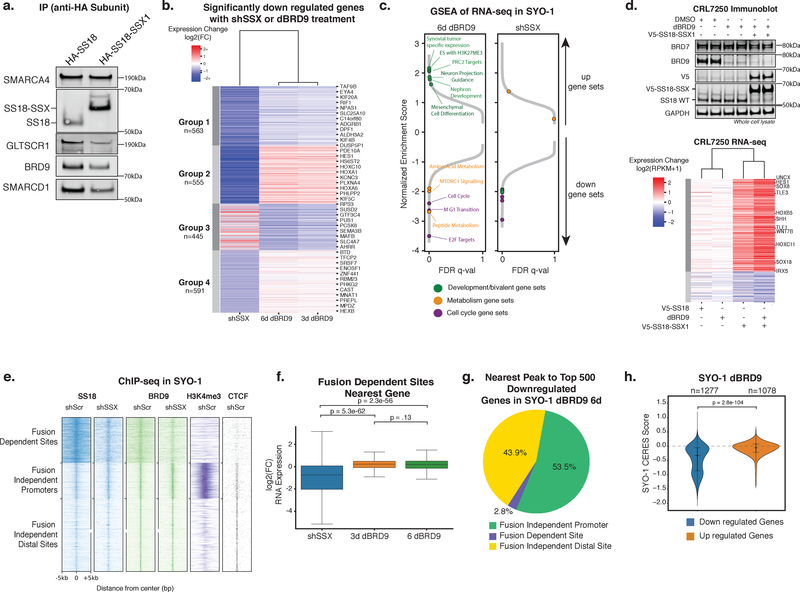
SS18 is a subunit of both cBAF and ncBAF complexes (Fig. 1), and the SS18-SSX fusion protein is a dedicated and stable subunit in cBAF complexes in SS39. To understand the sensitivity of SS cells to ncBAF complex depletion, we examined if SS18-SSX incorporates into ncBAF complexes. Complex purifications via HA-tagged wild-type SS18 and SS18-SSX1 revealed that while ncBAF subunits co-purify with SS18-SSX, they are less robustly captured in the SS18-SSX1 purification than the SS18 purification, relative to SMARCA4 (Fig. 5a).
Figure 5. ncBAF is not required for SS18-SSX1-mediated gene expression and primarily regulates fusion-independent sites.
a. Immunoblot for ncBAF components in HA-SS18 and HA-SS18-SSX complex purifications (n=1 biologically independent experiment). See also Supplementary Figure 7h.
b. Heatmap of significantly downregulated genes (p-adjusted<1e-3 and log2(fold change)< -0.59) in shSS18-SSX (7 days post infection) and dBRD9 (3 and 6 days) conditions in SYO-1 cells k-means clustered into 4 groups. n=2 biological replicates for each RNA-seq experiment, p-adjusted values are Benjamini-Hochberg adjusted Wald p-values.
c. GSEA of RNA-seq data for shSS18-SSX and dBRD9 (day 6) conditions in (b). Specific pathways and gene sets are indicated.
d. (Top) Immunoblot on CRL7250 whole cell lysates described in Figure S5B (n=1 biologically independent experiment); (Bottom) Heatmap of log2(fold change) of gene expression in CRL7250 fibroblast cells treated with DMSO, dBRD9, or dBRD9 followed by lentiviral introduction of V5-SS18 or V5-SS18-SSX (n=2 biological replicates for each RNA-seq experiment). Genes included were expressed (>1 RPKM) and had a log2(fold change) of at least +/-.59 in at least one of the conditions. Genes were k-means clustered into 2 groups and samples were clustered hierarchically. See also Supplementary Figure 7i.
e. Heatmap of ChIP-seq read density of SS18, BRD9, and H3K4me3 over SS18 sites in SYO-1 cells (shScr (control hairpin) and shSSX (targeting SS18-SSX) conditions), clustered into 3 groups.
f. Box plot of log2(fold change) in gene expression of genes closest to fusion-dependent sites (n=595) in shSS18-SSX and dBRD9 conditions. n=2 independent samples for each ChIP performed, significance was calculated by two tailed t-test. Box represents interquartile range (IQR), bar in center shows data median. Minima and maxima shown extend to from the box +/- 1.5*IQR.
g. Pie chart representing chromatin landscape (fusion-dependent, fusion-independent promoter, fusion-independent distal) of the nearest BRD9 peak to the top 500 most downregulated genes.
h. Violin plot of CERES scores for genes that changed with a significance of p-adjusted<1e-3 after 6 days of dBRD9 treatment in SYO-1 cells. P-value between conditions calculated by two tailed t-test. Violin plot represents kernel density estimation with data quartiles represented as lines, the data median is shown as a dot.
Since the SS18-SSX1 fusion protein destabilizes SMARCB1, a core subunit in cBAF complexes but not present in ncBAF complexes, we sought to determine whether fusion-containing ncBAF complexes may drive oncogenesis and the hallmark gene expression phenotypes of SS tumors. Thus, we performed RNA-seq on SYO-1 synovial sarcoma cells treated with either an shRNA targeting SS18-SSX (shSSX) or dBRD9. Intriguingly, while treatment with dBRD9 resulted in proliferative attenuation similar to knockdown of disease-driver SS18-SSX (Fig. 3), few genes were concordantly affected by both treatments (Fig. 5b, Supplementary Fig. 5a). Specifically, although both BRD9 and SS18-SSX perturbations similarly affected cell cycle pathways consistent with proliferative attenuation, we found discordant effects on mesenchymal stem cell genes, neural differentiation and bivalent polycomb target genes, gene sets hallmark to the SS-specific oncogenic signature, suggesting different underlying mechanisms (Fig. 5c)23, 39.
To determine if BRD9 and hence ncBAF complexes were required for de novo SS18-SSX-mediated gene activation, we performed RNA-seq in CRL7250 human fibroblasts in which we expressed either wild-type V5-SS18 or V5-SS18-SSX1 fusion with or without 24-hour pre-treatment with dBRD9 followed by sustained dBRD9 treatment (Supplementary Fig. 5b). Despite full degradation of BRD9 protein, dBRD9 treatment did not attenuate SS18-SSX-mediated gene activation and polycomb target genes associated with H3K27me3-mediated repression were equally activated irrespective of dBRD9 treatment (Fig. 5d, Supplementary Fig. 5c). These data in the SYO-1 and CRL7250 settings suggest that the function of ncBAF complexes is distinct from that of SS18-SSX-bound canonical BAF complexes known to oppose polycomb at cancer-specific sites on the genome23. Additionally, ncBAF is not required for the de novo activation of SS-specific gene signatures driven by the SS18-SSX fusion protein, pointing toward a distinct mechanism underlying ncBAF dependency in synovial sarcoma.
We next sought to define the divergent gene regulatory effects between SS18-SSX1 and BRD9 perturbation. In SS, the SS18-SSX fusion directs targeting of BAF complexes to a cancer-specific set of sites on chromatin which are crucial for oncogenesis23 (Fig. 5e). To assess whether the SS18-SSX fusion hijacks BRD9 to such cancer-specific sites, we performed ChIP-seq for BRD9 before and after SS18-SSX knockdown and found that BRD9 is minimally retargeted by the SS18-SSX fusion to broad-peak fusion-dependent sites (Fig. 5e). However, fusion-independent sites (sites retained irrespective of SS18-SSX knockdown) are largely marked by H3K4me3 and CTCF (Fig. 5e), two hallmarks of ncBAF complex targeting (Fig. 2), whereas fusion-dependent sites are not.
We found that genes closest to fusion-dependent sites were strongly downregulated by SS18-SSX knockdown23, but expression of these genes did not change with BRD9 degradation (Fig. 5f, Supplementary Fig. 5d). Instead, the most downregulated genes upon dBRD9 treatment were closest to fusion-independent sites (Fig. 5g, Supplementary Fig. 5e). This result is consistent with the lack of requirement for BRD9 in mediating de novo activation of fusion-dependent genes in CRL7250 fibroblasts and the divergent transcriptional effects between shSS18-SSX and dBRD9 treatments in SYO-1 SS cells. Finally, we compared changes in gene expression upon dBRD9 treatment with gene dependency scores derived from CRISPR screening, and found that genes downregulated by dBRD9 treatment significantly enriched for sensitivities (Fig. 5h). However, dBRD9 treatment in a BAF-intact cancer cell line, such as MOLM-13, did not result in preferential downregulation of genes that are enriched for dependencies (Supplementary Fig. 5f). Taken together, these results support a model in which BRD9/ncBAF complexes are important for regulation of gene expression at fusion-independent sites. We propose that ncBAF complexes, which preferentially associate with wild-type SS18 and are, in contrast to canonical BAF complexes, less perturbed by the incorporation of the fusion protein, are critical for maintenance of essential genes at fusion-independent sites in a setting in which SS18-SSX has further targeted canonical BAF complexes away from these sites.
ncBAF is required for the maintenance of gene expression via retained co-localization with CTCF in SMARCB1-deficient cancers
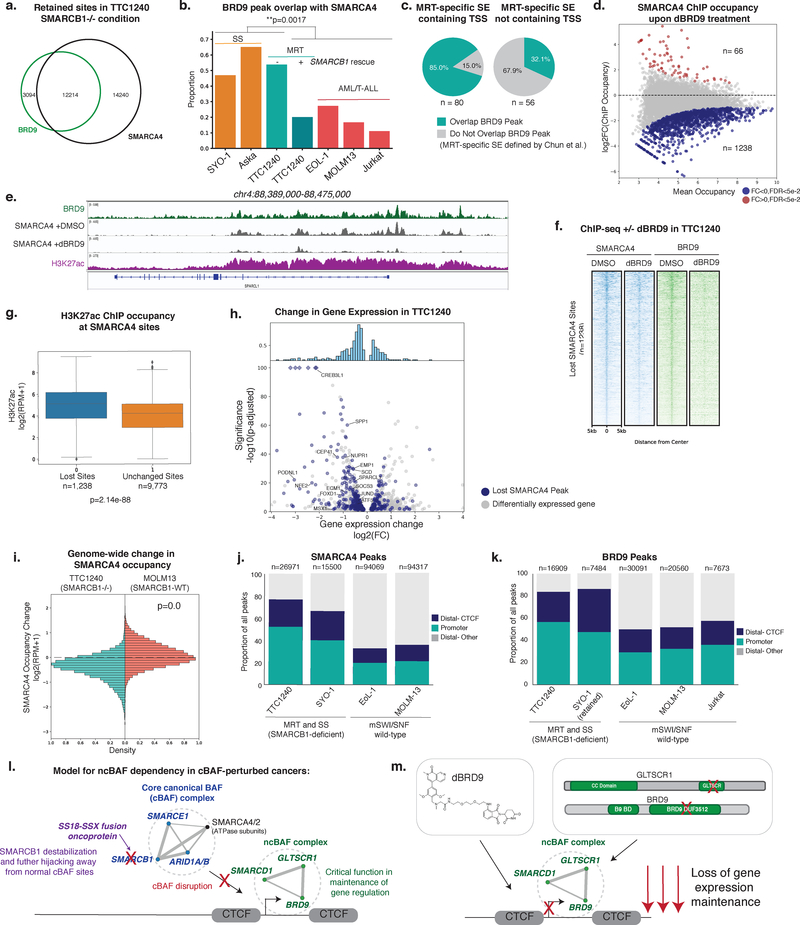
In the absence of SMARCB1 in MRT cell lines, residual SMARCA4-marked mSWI/SNF complexes are substantially more localized to promoter-proximal sites and are deficient in enhancer targeting37, 38. Previous studies using Brg1 (Smarca4) conditional KO in mouse models identified that Smarcb1-deficient cells are still dependent on Smarca4 for survival44, and these data have been more recently corroborated in large-scale dependency screens in cancer cell lines. Thus, we asked whether these residual mSWI/SNF complexes in MRT would primarily represent intact ncBAF complexes. We performed ChIP-seq for BRD9 in MRT cell line TTC1240, finding that BRD9 localizes to a large proportion of SMARCA4 sites (Fig. 6a). In contrast, ncBAF complexes co-localize with approximately 1/3 or fewer of all SMARCA4 sites in mSWI/SNF- intact settings, such as MOLM13 and Jurkat cells, and in MRT TTC1240 cells in which SMARCB1 has been rescued (Fig. 6b). Thus, a large percentage of residual mSWI/SNF complexes required for proliferative maintenance in SMARCB1-deficient cell lines represent ncBAF complexes.
Figure 6. ncBAF is required for maintenance of gene expression and retains co-localization with promoters and CTCF in SMARCB1-deficient cancers.
a. Venn diagram of BRD9 and SMARCA4 ChIP-seq peaks in TTC1240.
b. Bar plot of proportion of SMARCA4 peaks that overlap with a BRD9 peak in SS, MRT and mSWI/SNF-intact hematopoietic cancer cell lines. n=2 independent ChIPs, p-value calculated with two tailed t-test.
c. Proportion of MRT-specific super-enhancers (SE) (Chun et al.) overlapping a BRD9 peak in TTC1240.
d. log2(fold change) in SMARCA4 occupancy against mean occupancy between DMSO and dBRD9 treated TTC1240 cells. Highlighted, occupancy change of FDR<5e-2. n=2 biological replicates per ChIP, FDR values are multiple test corrected Wilcox test p-values.
e. Tracks showing BRD9, H3K27ac, and SMARCA4 +DMSO and +dBRD9 occupancy in TTC1240 at SPARCL1 (n=2 independent samples for each ChIP performed).
f. Spike-in normalized heatmap of SMARCA4 and BRD9 ChIP occupancy across lost SMARCA4 sites in TTC1240 upon dBRD9 treatment ranked by SMARCA4 occupancy in DMSO condition.
g. Boxplot of H3K27ac ChIP occupancy in TTC1240 cells at SMARCA4 sites lost or retained upon dBRD9 treatment. Box represents interquartile range (IQR), bar in center shows data median. Minima and maxima shown extend to from the box +/- 1.5*IQR with data falling outside of that range shown as points.
h. Volcano plot of gene expression changes in TTC1240 with dBRD9 treatment (7 days). Blue indicates genes with TSS within 100kb of a lost site. MRT disease-associated genes (Chun et al.) with TSS<100kb from peak are labeled. n=2 biological replicates for each RNA-seq experiment, p-adjusted values are Benjamini-Hochberg adjusted Wald p-values.
i. Histograms of log2(fold change) in SMARCA4 ChIP occupancy across SMARCA4 peaks in TTC1240 and MOLM-13 after dBRD9 treatment.
j,k. SMARCA4 (j) and BRD9 (k) peak distribution in BAF-perturbed (TTC1240 and SYO-1) and BAF-wild-type (EoL-1, MOLM-13, Jurkat) settings.
l, m. Model for ncBAF dependency in cancers driven by cBAF perturbations. Perturbations in the core BAF functional module (SMARCB1, SMARCE1, ARID1A/B, with the exception of the ATPase subunits) result in loss of cBAF gene regulatory function and reliance on ncBAF for gene expression maintenance at hallmark ncBAF promoters and CTCF sites (l). Chemical or biologic ncBAF disruption results in loss of gene expression maintenance (m).
BAF complex localization and function at enhancers and superenhancers (SEs) have been shown to be aberrant in MRT37, 38. We examined BRD9 targeting to MRT-specific SEs, defined by Chun et al. in primary tumors and cell lines compared to hESC lines and fetal brain tissue45. The TTC1240 cell line exhibits strong overlap with these primary tumor-associated MRT-specific enhancers and SEs (Supplementary Fig. 6a) and BRD9-marked ncBAF complexes localized to a large number of these MRT-specific SEs, particularly those which encompass a TSS (Fig. 6c). To investigate the role for BRD9 at these genes in MRT, we treated TTC1240 cells with dBRD9 and performed ChIP-seq and RNA-seq. Treatment with dBRD9 resulted in a significant decrease in SMARCA4 occupancy, particularly, at BRD9-marked sites genome-wide (Fig. 6d–f, Supplementary Fig. 6b). Consistent with overlap at SEs, lost SMARCA4 peaks were highly enriched in H3K27Ac relative to peaks that did not change (Fig. 6g). Additionally, many downregulated genes had BRD9 occupancy at their promoters and significantly changing genes had higher H3K27Ac and BRD9 occupancy than non-significantly changing active genes (Supplementary Fig. 6c). Genes that were downregulated by dBRD9 and lost SMARCA4 occupancy were enriched for those genes overexpressed in MRT compared to wild-type tissue or defined as regulated by MRT-specific super enhancers (i.e. JUND, VGF, ID3, HOXC9, and CREB3L1) (Fig. 6h)45 and involved in development and differentiation (Supplementary Fig. 6d). Finally, loss of SMARCA4 occupancy was specific to the MRT cell setting, as dBRD9 treatment in MOLM13, a BAF-intact cell line, did not result in similar loss of SMARCA4 (Fig. 6i). Similar to SS, these data support a model in which ncBAF complexes, the only mSWI/SNF family complexes not perturbed by SMARCB1 loss, are critical for gene expression maintenance and subsequently the proliferative capacity, of MRT cells.
Since SS and MRT, which share in common core cBAF (particularly SMARCB1) perturbation, are dependent on ncBAF complexes that regulate gene expression at retained mSWI/SNF sites, we investigated if there were any convergent features between these sites in these two distinct disease settings. Importantly, we found that SMARCA4-marked mSWI/SNF complexes in MRT and SS both converge on a largely promoter-proximal and CTCF co-localized distribution, two hallmarks of ncBAF complex localization (Fig. 2), relative to SMARCA4 in mSWI/SNF-intact cell types (Fig. 6j, Supplementary Fig. 6e,f). In both SS and MRT cell lines, enrichment of BRD9 at CTCF sites remained unchanged upon SS18-SSX knockdown or SMARCB1 re-introduction, respectively, further highlighting that default, hallmark ncBAF complex-specific targeting to promoters and CTCF sites occurs irrespective of BAF complex perturbations (Fig. 6k, Supplementary Fig. 6g,h ).
These data support in a model in which ncBAF complexes maintain gene expression at retained, promoter-proximal and CTCF sites when regulatory functions of the core cBAF functional module containing SMARCB1, SMARCE1, and ARID1A/B are perturbed (Fig. 6l). ncBAF complex disruption (chemical or biological)leads to a loss of gene expression maintenance, defining the cancer-specific sensitivities of cBAF-deficient cancers such as SS and MRT (Fig. 6m).
Discussion
In this study, we generated comprehensive chromatin binding profiles for all three mSWI/SNF family complexes relative to defined genomic features. Occupancy patterns of ncBAF, BAF, and PBAF complexes to CTCF sites, active enhancers, and active promoters, may in part underlie complex-specific functional roles implicated in cell fitness screening efforts. These mSWI/SNF localization studies provide an important platform upon which to identify specific complex-chromatin interaction mechanisms governing preferential genome-wide targeting profiles. Differences in histone binding affinities, interactions with general or tissue-specific transcription factors, or complex-specific steric constraints for binding chromatin landscape features are likely at play, alone and in combination, to direct targeting.
Importantly, we identify a synthetic lethal relationship in specific cancers with perturbations to the core cBAF functional module: synovial sarcoma (driven by the SS18-SSX fusion) and SMARCB1-deficient malignant rhabdoid tumor. These cancers are uniquely and specifically dependent on ncBAF complexes for proliferative maintenance, unveiling ncBAF as a potential target for therapeutic intervention in these cancers (relative to other cancer types spanning hundreds of other lineages). Our findings are particularly exciting given recent development of selective small molecule inhibitors and chemical degraders targeting BRD9 41–43, 46. Further studies will be needed to carefully define in vivo properties of these compounds. Our identification of domains within ncBAF-specific subunits that underlie these dependencies, such as the GLTSCR domain of GLTSCR1/1L and the DUF3512 of BRD9, further expands therapeutic opportunities.
In characterizing the convergent mechanism of ncBAF dependency in synovial sarcoma and malignant rhabdoid tumor, unexpectedly, we found that although ncBAF complexes do incorporate the SS18-SSX fusion that drives synovial sarcoma, perturbation of BRD9 and disruption of SS18-SSX are mechanistically distinct. ncBAF complexes primarily regulate retained fusion-independent sites, linking to SMARCB1-deficient MRT in which ncBAF complexes comprise a large share of essential residual complexes that likewise maintain gene expression at retained mSWI/SNF sites. The retained sites in both of these disease settings share in common CTCF co-localization and promoter proximity, the two hallmarks of ncBAF complex localization. Thus, this work now provides a complex-specific basis for previously identified dependencies on SMARCA4 (BRG1) in SMARCB1-deficient cancers44, and highlights the importance of understanding subunit-specific contributions to complex assembly and function when designing strategies to target mSWI/SNF-perturbed cancers. Excitingly, our data suggest the possibility that other cBAF-perturbed cancers, such as those with loss of ARID1A/B or SMARCE147, 48, may exhibit similar ncBAF dependencies.
Materials and Methods
Cell lines and tissue culture
HEK-293T, G401, TTC1240, ESX, IMR-90, BJ Fibroblast, CRL7250, and NCIH-1437 cells were grown in DMEM (Gibco) supplemented with 10% FBS, 1% GlutaMAX (Gibco), and 1% penicillin-streptomycin (Gibco). ES-2 cells were grown in McCoy’s 5A (Gibco) supplemented with 10% FBS, 1% GlutaMAX (Gibco), and 1% penicillin-streptomycin (Gibco). EoL-1 and MOLM-13 were grown in RPMI (Gibco) supplemented with 10% FBS, 1% GlutaMAX (Gibco), and 1% penicillin-streptomycin (Gibco). RD were cultured in DMEM (Gibco) supplemented with 10% FBS. HCT116 were grown in McCoy’s 5A (Gibco) supplemented with 10% FBS. Calu-6 were grown in EMEM (ATCC 30–2003) supplemented with 10% FBS. SYO-1 was grown in DMEM without sodium pyruvate (Gibco) supplemented with 10% FBS, 1% GlutaMAX (Gibco), and 1% penicillin-streptomycin (Gibco). The SYO-1 cell line was a gift from Akira Kawai (National Cancer Center Hospital, Japan. ASKA and HS-SY-II cell lines were obtained from RIKEN49. The CRL7250 human fibroblast cell line was a kind gift from Drs. Berkeley Gryder and Javed Khan (National Cancer Institute, Bethesda, MD).
Constructs and cloning
Lentiviral shRNA hairpins targeting BRD9 (RHS4430–200302441), SMARCE1 (RHS4430–200219172), and a non-silencing control (RHS4346) were constitutively expressed from the pGIPZ vector and obtained from GE Dharmacon; hairpins targeting GLTSCR1 were inducibly expressed from the pTRIPZ vector (#RHS4696) from GE Dharmacon.
shRNA hairpins targeting SS18-SSX (5’-CAGTCACTGACAGTTAATAAA-3’) or a non-targeting scramble control (5’-CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG-3’) were constitutively expressed from the pLKO.1 vector with puromycin selection.
All expression constructs were cloned using In-Fusion HD (cat. 639650) per manufacturer’s recommendations. V5-GLTSCR1 and corresponding N-Del and C-Del mutants were synthesized and cloned into a modified pTight vector by GenScript Biotech Corporation. GLTSCR1L (clone 40146333) was cloned with a V5 tag into a modified pTight vector using In-Fusion HD. HA tag sequence was included in the primers for human BRD9, BRD7, SMARCD1, DPF2, and GLTSCR1L and cloned into a modified pTight vector under constitutive EF1alpha-driven expression with a blasticidin resistance gene. HA-BRD9(B7C) contains amino acids 1–265 of BRD9 and amino acids 266–651 of BRD7. HA-BRD7(B9C) contains amino acids 1–265 of BRD7 and amino acids 266–597 of BRD9. HA-BRD9(B7C) and HA-BRD7(B9C) were cloned in steps, with the N- and C-terminal fragments amplified independently, followed by mixing N- and C-terminal PCR products in equal quantities in a subsequent PCR reaction to fuse the two fragments together, into the same modified pTight vector with a puromycin resistance gene. All primers used for cloning are listed in Supplementary Table 6.
Lentiviral Production and Transduction
shRNA or gene delivery vectors, psPAX2, and pMD2.g were transfected into HEK-293T cells at a ratio of 4:3:1 using PEI (Polysciences, Inc.). Media was filtered through 0.4micron filters 72h post transfection and lentiviral particles were concentrated at 20,000rpm for 2.5h at 4°C. Lentiviral particles were resuspended in 200μl PBS and cells were transduced using 1:1000 polybrene (Santa Cruz Biotechnology, cat. sc-134220). Two days post-infection, cells were selected with 2μg/mL puromycin or 10μg/mL blasticidin.
Proliferation Curves
25,000–40,000 cells were plated per well of 12 well plates, or 50,000–60,000 per 10cm plate, depending on cell line. Cell counts were performed in biological triplicate using a Vi-CELL XR Cell Counter (Beckman Coulter) on days indicated.
Colony Formation Assays
Colony formation assays were performed in 6-well or 12-well format. For each cell line, appropriate cell seeding conditions were first identified in a 14-day assay format. A titration of cell number of each of the cell lines was performed to identify the best cell seeding condition. Compounds were treated continuously for 14 days at the indicated concentration, with change of medium every 5 days. By day 14, medium was removed, and cells were washed with PBS and stained using 500uL of 0.005% (w/v) crystal violet solution in 25% (v/v) methanol for at least 1 hour at room temperature.
Cell Cycle Analysis
Cell cycle analysis was performed using the Click-iT™Plus EdU Flow Cytometry Assay (Invitrogen). Apoptosis assay was performed using the Annexin V-FITC Apoptosis Detection Kit (Sigma A9210). Assays were performed according to the manufacturer’s protocol. SYO-1 cells were treated for 8 days and compound was refreshed every 5 days.
CRISPR Domain Scanning
To perform high density sgRNA tiling screen, sgRNA library against BAF complex subunits were custom synthesized at Cellecta (Mountain View, CA) following Cellecta’s recommendation. Negative and positive control sgRNA were included in the library for quality control check with the screening results. Negative controls consist of 200sgRNAs that do not target human genome. The positive controls are sgRNAs targeting essential genes (CDC16, GTF2B, HSPA5, HSPA9, PAFAH1B1, PCNA, POLR2L, RPL9 and SF3A3). All positive and negative control sgRNA are listed in Supplementary Table 3. Procedure for virus production, cell infection and performing sgRNA screen were previously described 26, 50. For each sgRNA, 50 counts were added to the sequencing counts and the resulting counts were normalized to the total number of counts. The log2 of the ratio between the counts (defined as dropout ratio) at day 24 and day 1 post-infection was calculated (Supplementary Table 4). For negative control sgRNAs the 2.5 and 97.5 percentile of the log2 dropout ratio of all non-targeting sgRNAs was calculated and considered as background (grey box in the graph). Protein domains were obtained from PFAM regions defined for the following UNIPROT identifiers: BRD9 : Q9H8M2, BICRA : Q9NZM4, BRD7 : Q9NPI1, SMARCA2 : P51531, SMARCA4 : Q9HBD4, SMARCB1 : G5E975, SMARCC1 : Q92922, SMARCC2 : Q8TAQ2, SMARCD1 : Q96GM5, SMARCD2 : Q92925, SMARCE1 : Q969G3, SS18 : Q15532, SSX2 : Q16385. Rolling averages were calculated for each nucleotide in the corresponding transcript by selecting a 150 nucleotide window centered at the corresponding position and calculating the average of log2 dropout ratio of the sgRNAs in the window.
mSWI/SNF Complex Purification
Mammalian SWI/SNF complexes were purified as previously described 51. In this study, complexes were purified from HEK-293T cells stably expressing HA-tagged constructs (as indicated). Cells were scraped from plates and washed with cold PBS. Suspension was centrifuged at 3000 rpm for 5 min at 4°C and pellets were resuspended in hypotonic buffer (HB) containing 10mM Tris HCl pH 7.5, 10mM KCl, 1.5 mM MgCl2, 1mM DTT, 1mM PMSF and incubated on ice for 5min. Suspension was centrifuged at 5000 rpm for 5 min at 4°C, and pellets were resuspended in 5 volumes of fresh HB containing protease inhibitor cocktail and homogenized using glass Dounce homogenizer. Suspension was layered onto HB sucrose cushion containing 30% sucrose w/v, centrifuged at 5000 rpm for 1 hour at 4°C and cytosol-containing layer was discarded. Nuclear pellets were resuspended in high salt buffer (HSB) containing 50mM Tris HCl pH 7.5, 300mM KCl, 1mM MgCl2, 1mM EDTA, 1mM, 1% NP40, 1mM DTT, 1mM PMSF and protease inhibitor cocktail. Homogenate was incubated on rotator for 1H. Homogenates then were centrifuged at 20,000 rpm (30,000 x g) for 1 hour at 4°C using an SW32Ti rotor. Chromatin pellets were discarded and high salt nuclear extract was filtered through a 0.45μm filter and incubated overnight with HA magnetic resin. HA beads were washed in HSB and eluted with HSB containing 1mg/ml of HA peptide for 4 times 1.5 hour each. Eluted proteins were then subjected to density gradient centrifugation or dialysis.
Protein Extraction Methods
Ammonium sulfate nuclear extraction was performed as described previously38. Pellets were resuspended in IP buffer (300 mM NaCl, 50 mM Tris-HCl pH 7.5, 1 mM EDTA and 1% Triton-X100 with protease inhibitor, 1 mM DTT and 1 mM PMSF) for subsequent experiments.
For whole cell lysates, cells were washed in PBS and resuspended in ~5 volumes of extraction buffer (20mM Tris and 1.5% SDS). Chromatin was solubilized via sonication, and proteins were quantified using BCA.
Immunoprecipitation
Nuclear extracts were quantified using BCA, and 1mg of protein at 1mg/mL in IP buffer supplemented with protease inhibitors was used per IP with 2–5ug of antibody or with 25uL of Pierce Anti-HA Magnetic Beads (cat. 88837) overnight with rotation at 4°C. Nuclear extract + antibody solution was incubated with 30uL of Protein G Dynabeads (Thermo Fisher) for 2h at 4°C with rotation and washed 5 times with IP buffer. Immunoprecipitated proteins were eluted with sample buffer (2X NuPAGE LDS buffer with 100mM DTT) and loaded onto 4–12% Bis-Tris NuPAGE Gels (Life Technologies). See Reporting Summary for antibodies used in this study.
Glycerol Gradient
Linear 10–30% glycerol gradients were prepared in 14 x 89mm polyallomer centrifuge tubes (Beckman Coulter, cat. 331327) by overlaying 10% glycerol solution in HEMG buffer on 30% glycerol solution with mixing by a Gradient Master. 500–1000ug of nuclear extracts were resuspended in 200μL 0% glycerol HEMG and overlaid on the gradient. Purified protein complexes were loaded in their elution buffers. Gradients were centrifuged in an SW41 rotor at 40,000rpm for 16h at 4°C, and 0.55mL fractions were collected for analysis.
Mass Spectrometry Sample Preparation and Analysis
Purified complex elutions (BRD9, BRD7, Mock) or glycerol gradient fractions (DPF2, fractions 13–14) were concentrated using StrataClean beads, loaded onto 4–12% SDS PAGE gels, migrated 2cm into the gel, and stained with colloidal blue (Invitrogen). Stained samples were excised and sent to Taplin Biological Mass Spectrometry Facility at Harvard Medical School for analysis. Heatmap displaying log2(number of total peptides+1) was created using Seaborn.
Protein SDS PAGE
Proteins were run on 4–12% Bis-Tris NuPAGE gels (Life Technologies). For Western blot, proteins were wet transferred onto PVDF membranes at 300mA for 2.5h, blocked for 1h with 10% milk PBS-T, and visualized using LI-COR Odyssey CLx. For silver stain, gels were stained using SilverQuest Silver Staining Kit (Thermo Fisher) according to manufacturer’s protocol.
Chromatin Immunoprecipitation (ChIP)
Cells were fixed in 1% formaldehyde (Sigma Aldrich, F8775) for 10min at 37°C and quenched with 125mM glycine for 5min at 37°C. Cells were subsequently washed with cold PBS and stored at -80°C until use. 10M cells per ChIP were used for EoL-1, MOLM-13, and Jurkat cell lines, and 5M cells per ChIP were used for SYO-1 and TTC1240 cell lines. Nuclei were extracted and chromatin was sonicated using the adaptive focused acoustics technology with a Covaris sonicator. Sonicated chromatin was used in immunoprecipitation reactions with indicated antibodies (see Reporting Summary) overnight followed by capture using Protein G Dynabeads (Thermo Fisher). For ChIP-seq using spike-in chromatin, 15ng of spike-in Drosophila chromatin (Active Motif, cat#530830) was added to each sample with 2ug of spike-in antibody (Active Motif, cat#61686). Captured antibody-chromatin complexes were washed, eluted, and treated with RNAse A (Roche 11 119 915 011) for 30min at 37°C and Proteinase K (Life Technologies 100005393) for 3 hours at 65°C. ChIP DNA was extracted using SPRI beads (Beckman Coulter Agencourt AMP Xpure), washed, and eluted.
RNA-seq Sample Preparation
RNA was collected from 2 million cells per condition, in biological duplicate, using the RNeasy Mini Kit (QIAGEN) according to manufacturer’s protocol.
Library Preparation and Sequencing
Library preparation and sequencing of ChIP DNA and RNA was performed by the Molecular Biology Core Facilities at the Dana-Farber Cancer Institute (75bp single end on Illuminia Nextseq 500).
ChIP-Seq Data Alignment
For alignment of ChIP-seq data, Bowtie2, version 2.1.052 was used to map reads to the hg19 human reference genome, using the parameter –k 1.
For spike-in normalization, Drosophila DNA was aligned to the dm3 genome using Bowtie2 version 2.1.0 with the parameter –k 1. Duplicated reads were removed using samtools rmdup with the –b option. (SAMtools v1.3.1) As per manufacturer instructions, normalization ratios were calculated using the ratio of the total number of non-redundant mapped reads in each sample in comparison to the sample with the fewest non-redunant mapped reads.
ChIP-Seq Data Analysis
Data processing
MACS253 version 2.1.0 was used to call peaks against input with a cutoff of q=.001. In EoL-1, MOLM-13 and TTC1240 narrow peaks were called for all SWI/SNF antibodies and CTCF while broad peaks were used for all histone marks. In SYO-1, broad peaks were called for all antibodies. Peaks that fell in ENCODE blacklisted regions or were mapped to unmappable chromosomes (not chr1–22, X or Y) were removed. Quality control metrics are available in Supplementary Table 5. All down stream analysis was performed on bam files with duplicates removed using the samtools rmdup command with the –b option. ChIP-seq tracks were generated using the bedGraphToBigWig script downloaded from UCSC. Bedgraph files were generated with MACS2 using the –B –SPMR options. For TTC1240 SMARCA4 tracks shown, the bedGraph file values were multiplied by the spike-in normalization ratios calculated as described above.
Overlaps for ChIP venn diagrams were created using the ChIPPeakAnno54 v3.10.1 bioconductor package, peak files were read in using the toGRanges() command, values were determined using the getVennCounts() function with maxgap=0. Data was visualized using matplotlib. The number of overlapping peaks displayed in pie charts, bar charts and heatmaps was determined using the pybedtools55 intersect function. Proportions were calculated by dividing the number of overlapping peaks by the number of total peaks.
Read count across peak sets of interest were calculated by calling the Rsubread56 v1.26.1 bioconductor package function featureCounts() on duplicate removed bam files. These values were divided by the total number of mapped reads divided by one million, giving a normalized value of reads per million mapped reads for each interval in the input bed.
Peak distance from TSS elements was determined using BEDtools v2.26.0 closest function with the hg19 ref Flat TSS annotation.
Determination of super enhancers was performed using ROSE57, 58 with all default settings using the TTC1240 H3K27ac ChIP-seq file and TTC1240 H3K27ac peak file as input. MRT-specific super enhancers were downloaded from Chun et al45 and merged using bedtools merge, as many of their published enhancers abutted one another.
Data analysis and visualization
Metagene plots and heatmaps were generated created using HTSeq59 v0.9.1. To account for the 200bp average fragment length selected for in sonication, fragment length was extended 200 bp from the edge of each genomic interval. Total read counts for each interval were normalized to reads per million mapped reads (RPM). For each antibody the resulting matrix has a width of the number of bp in the window (in this study primarily 5000) and a height of the number of peaks in the indicated set. Strandedness of the interval was not considered, except for the TSS metagene plot in Supplementary Figure 2g. Metagene plots show the average RPM at each position. Heatmaps were visualized using python. Heatmaps were ordered by the maximum value in each matrix row of the indicated antibody. Heatmaps were colored such that the midpoint of the color spectrum is equivalent to the median of the set of maximum values in each row. For heatmaps where multiple peak sets are shown, these color values were calculated for each antibody across both sets together. For the spike-in normalized heatmap, all heatmap data was calculated as described but then multiplied by the normalization factor, described above, before plotting.
The EoL-1 histone mark and CTCF heatmap in Supplementary Fig. 2i the HTSEQ procedure described above was carried over peaks that had been split into 100 bins and 2500 bp on either side of the peak. The resulting matrix was k-means clustered to 4 clusters. This was carried out over the merged set of all EoL-1 mSWI/SNF peaks.
The SYO-1 differential heatmap (Fig. 5e) was ordered by the ratio of the row means for BRD9 in the +/- shSSX conditions. Any interval that had more than or equal to a 25% increase in mean BRD9 ChIP occupancy upon shSSX treatment was considered gained. Any interval that had more than or equal to 25% decrease in mean BRD9 ChIP occupancy upon shSSX treatment was considered lost. Intervals that did not change more than 25% in either direction were considered retained.
Differential occupancy of SMARCA4 in TTC1240 upon dBRD9 treatment was determined using the DiffBind v2.4.8 bioconductor package (https://bioconductor.org/packages/release/bioc/html/DiffBind.html ), with all default settings. Peak files and duplicate removed bam files were provided for each SMARCA4 sample in each condition, along with the bam files corresponding to the input in each condition. The package functions count(), contrast(), analyze(), and report() were used in sequence.
Gene ontology of genes near lost SMARCA4 sites in TTC1240 (Fig. S6d) was performed using Genomic Regions Enrichment of Annotations Tool (GREAT)60.
Motif Analysis
A fasta sequence for a region of 250 bp on either side of the center of each peak was generated using the bedtools getfasta function. Motif analysis on these sequences was done was done using the MEME-ChIP suite61.
In Fig. 2d, for each antibody the motif with the highest CentriMo log-adjusted p-value in the indicated TF Family was selected. These log-adjusted p-values were used to make a heatmap using the Seaborns clustermap function, clustered by correlation.
Enrichment plots for the motifs are the average number of the CentriMo site counts for each antibody in the window around the indicated motif split into bins of 10 bp.
RNA-Seq Data Analysis
Data processing
RNA-seq data reads were mapped using default parameters to hg19 using STAR62 version 2.5.2a.
RPKM values were calculated using GFOLD version 1.1.463. Unless otherwise noted, log2 fold change and Bonferri-corrected p values were generated using DESEQ2 v1.16.1, with reads mapped using RSUBREAD56 v1.26.1. Genes were considered significantly changing if they had an adjusted p-value <.001 and a log2 fold change of at least .59 (approximately 50% change). All RNA-seq experiments were performed in biological replicate. Quality control metrics are available in Supplementary Table 5.
RNA BigWig files were generated using the bamCoverage command from deepTools release 2.464 with all default settings.
Data analysis/visualization
The input for Gene Set Enrichment Analysis (GSEA)65 was created by calculating the log2 fold change between the mean RPKM of the replicates in each condition +1. Noncoding genes (SNO and MIR RNA’s) were excluded, as were genes that did not have an expression level of at least 1 RPKM in any condition of the comparison. GSEA Preranked was run over these files with default settings.
Gene ontology and pathway terms of gene clusters in Synovial Sarcoma (Fig. S5a) were determined using Metascape66.
Genes associated with MRT superenhancers were downloaded from Chun et al.45. Differential expression files of genes between MRT and normal tissues were downloaded from Chun et al., genes that were overexpressed in MRT with Bonferroni-adjusted p-value < .01 were considered overexpressed in MRT.
CRISPR-Cas9 and shRNA synthetic lethal screening data analysis
DRIVE data is publicly available and downloaded from the Novartis DRIVE Data Portal28. Statistical analysis was performed using the scipy.stats package.
Significance values for shBRD9 in tissue types were calculated using a Fisher’s Exact Test, and FDR corrected using the Benjamini-Hochberg procedure. An ATARIS score of -.75 was used as the cutoff for sensitivity.
Principal components analysis of fitness data from Project Achilles
Datasets were obtained from the Project Achilles Data Portal (https://portals.broadinstitute.org/achilles/about ). The CRISPR data (Avana-18Q1) and the RNAi data (2.20.2) for BAF subunits were scaled across cell lines. In the RNAi dataset, cell lines were omitted if fitness scores were not available for all BAF genes. The fitness scores from both datasets were concatenated and correlated across genes, and principal components analysis was performed on the resulting correlation matrix (R prcomp, default settings). The first two principle components were plotted.
All heatmaps and plots were generated using matplotlib and/or seaborns. Unless otherwise noted, all default parameters were used for the seaborn clustermap function.
Code Availability Statement
Code used in this paper is available from the corresponding author upon request.
Data Availability Statement
The ChIP-seq and RNA-seq data sets generated and/or analyzed during the current study have been deposited in the Gene Expression Omnibus (GEO) repository under accession number GSE113042 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113042).
Other data sets that were previously published and used in this study have been deposited in the Gene Expression Omnibus (GEO) repository under accession numbers GSE90634 and GSE108025 available at (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE90634 ) and (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108025 ) respectively.The fitness data were derived from Project Achilles through the Project Achilles Data Portal (https://portals.broadinstitute.org/achilles/about ). The data-set derived from this resource that supports the findings of this study is available at https://portals.broadinstitute.org/achilles/datasets/all. The fitness data were also derived from Project DRIVE. The data-set derived from this resource that supports the findings of this study is available at https://oncologynibr.shinyapps.io/drive/.
All proteomics/ mass-spectrometry data is deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD011103.
Statistics and Reproducibility
All statistics performed on data in this manuscript are detailed above, and statistical tests and their parameters used are indicated in the legends. Representative data are shown from independently repeated experiments with similar results.
Supplementary Material
Acknowledgments
We are grateful to members of the Kadoch Laboratory for thoughtful discussions and insights relating to this study. We thank Fred Winston, Robert Kingston, and Yang Shi for advice and critical review of the results. We thank A. Kuo and G.R. Crabtree for the BAF155 (SMARCC1) homemade polyclonal antibody used in these studies. We thank Zach Herbert, Maura Berkeley and members of the Molecular Biology Core Facility (MBCF) for library preparation and sequencing. We also thank Wenqian Wang, Ze Zhang, Beibei Li for the CRISPR screening and cellular proliferation experiments. This work was supported in part by awards from the NIH DP2 New Innovator Award 1DP2CA195762–01(C.K.), the American Cancer Society Research Scholar Award RSG-14–051-01-DMC (C.K.), and the Pew- Stewart Scholars in Cancer Research Grant (C.K.). B.C.M. holds the Albert J. Ryan Fellowship granted by the Division of Medical Sciences (Harvard Medical School). Additionally, this work was supported in part by NIH Grant Number 5 T32 GM095450–04 (M.J.M.), and by the Harvard University Graduate School of Arts and Sciences (GSAS) Fellowship (M.J.M.), the Ford Foundation Fellowship (A.M.V), and the National Science Foundation Graduate Research Fellowship Program (J.P.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Competing Financial Interest Statement
C.K. is a scientific founder, fiduciary Board of Directors member, Scientific Advisory Board member, shareholder, and consultant for Foghorn Therapeutics, Inc. (Cambridge, MA, USA). H.M.C., Q.Z, M.B., L.M.M.S are employees and shareholders of Foghorn Therapeutics. Nathanael Gray is a scientific founder, SAB member and equity holder in Gatekeeper, Syros, Petra, Soltego and C4 Therapeutics. J.E.B. is an employee and shareholder of the Novartis Institutes for Biomedical Research (Cambridge, MA, USA). The other authors declare no competing interests.
References
- 1.Narlikar Geeta J., Sundaramoorthy R & Owen-Hughes T Mechanisms and Functions of ATP-Dependent Chromatin-Remodeling Enzymes. Cell 154, 490–503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clapier CR & Cairns BR The biology of chromatin remodeling complexes. Annu Rev Biochem 78, 273–304 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Ho L et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proceedings of the National Academy of Sciences of the United States of America 106, 5181–5186 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lessard J et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55, 201–215 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lickert H et al. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature 432, 107–112 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Priam P et al. SMARCD2 subunit of SWI/SNF chromatin-remodeling complexes mediates granulopoiesis through a CEBPvarepsilon dependent mechanism. Nat Genet 49, 753–764 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Witzel M et al. Chromatin-remodeling factor SMARCD2 regulates transcriptional networks controlling differentiation of neutrophil granulocytes. Nat Genet 49, 742–752 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staahl BT et al. Kinetic analysis of npBAF to nBAF switching reveals exchange of SS18 with CREST and integration with neural developmental pathways. J Neurosci 33, 10348–10361 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoo AS, Staahl BT, Chen L & Crabtree GR MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460, 642–646 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoo AS et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pedersen TA, Kowenz-Leutz E, Leutz A & Nerlov C Cooperation between C/EBPalpha TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev 15, 3208–3216 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan J et al. Interrogation of Mammalian Protein Complex Structure, Function, and Membership Using Genome-Scale Fitness Screens. Cell Syst 6, 555–568 e557 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alpsoy A & Dykhuizen EC Glioma tumor suppressor candidate region gene 1 (GLTSCR1) and its paralog GLTSCR1-like form SWI/SNF chromatin remodeling subcomplexes. J Biol Chem (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang W et al. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev 10, 2117–2130 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Kaeser MD, Aslanian A, Dong MQ, Yates JR 3rd & Emerson BM BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J Biol Chem 283, 32254–32263 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadoch C et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 45, 592–601 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shain AH & Pollack JR The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One 8, e55119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biegel JA et al. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer research 59, 74–79 (1999). [PubMed] [Google Scholar]
- 19.Eaton KW, Tooke LS, Wainwright LM, Judkins AR & Biegel JA Spectrum of SMARCB1/INI1 mutations in familial and sporadic rhabdoid tumors. Pediatr Blood Cancer 56, 7–15 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Versteege I et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394, 203–206 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Jones S et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330, 228–231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varela I et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McBride MJ et al. The SS18-SSX Fusion Oncoprotein Hijacks BAF Complex Targeting and Function to Drive Synovial Sarcoma. Cancer Cell 33, 1128–1141 e1127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helming KC et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat Med 20, 251–254 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffman GR et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proceedings of the National Academy of Sciences of the United States of America 111, 3128–3133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyers RM et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet 49, 1779–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsherniak A et al. Defining a Cancer Dependency Map. Cell 170, 564–576 e516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang T et al. Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell 168, 890–903 e815 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonald ER 3rd et al. Project DRIVE: A Compendium of Cancer Dependencies and Synthetic Lethal Relationships Uncovered by Large-Scale, Deep RNAi Screening. Cell 170, 577–592 e510 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Cowley GS et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data 1, 140035 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bell AC & Felsenfeld G Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405, 482–485 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Bell AC, West AG & Felsenfeld G The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98, 387–396 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Hark AT et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405, 486–489 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Kanduri C et al. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol 10, 853–856 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Alver BH et al. The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat Commun 8, 14648 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathur R et al. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat Genet 49, 296–302 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat Genet 49, 289–295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakayama RT et al. SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat Genet 49, 1613–1623 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kadoch C & Crabtree GR Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell 153, 71–85 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clark J et al. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet 7, 502–508 (1994). [DOI] [PubMed] [Google Scholar]
- 41.Hohmann AF et al. Sensitivity and engineered resistance of myeloid leukemia cells to BRD9 inhibition. Nature chemical biology 12, 672–679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin LJ et al. Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor. Journal of medicinal chemistry 59, 4462–4475 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Remillard D et al. Degradation of the BAF Complex Factor BRD9 by Heterobifunctional Ligands. Angewandte Chemie (International ed. in English) 56, 5738–5743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X et al. Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex. Cancer research 69, 8094–8101 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chun HJ et al. Genome-Wide Profiles of Extra-cranial Malignant Rhabdoid Tumors Reveal Heterogeneity and Dysregulated Developmental Pathways. Cancer Cell 29, 394–406 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Theodoulou NH et al. Discovery of I-BRD9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition. Journal of medicinal chemistry 59, 1425–1439 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coatham M et al. Concurrent ARID1A and ARID1B inactivation in endometrial and ovarian dedifferentiated carcinomas. Mod Pathol 29, 1586–1593 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Tauziede-Espariat A et al. Loss of SMARCE1 expression is a specific diagnostic marker of clear cell meningioma: a comprehensive immunophenotypical and molecular analysis. Brain Pathol (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naka N et al. Synovial sarcoma is a stem cell malignancy. Stem Cells 28, 1119–1131 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Munoz DM et al. CRISPR Screens Provide a Comprehensive Assessment of Cancer Vulnerabilities but Generate False-Positive Hits for Highly Amplified Genomic Regions. Cancer Discov 6, 900–913 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Mashtalir N et al. Autodeubiquitination protects the tumor suppressor BAP1 from cytoplasmic sequestration mediated by the atypical ubiquitin ligase UBE2O. Mol Cell 54, 392–406 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu LJ et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11, 237 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dale RK, Pedersen BS & Quinlan AR Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics 27, 3423–3424 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao Y, Smyth GK & Shi W The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41, e108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loven J et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whyte WA et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anders S, Pyl PT & Huber W HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McLean CY et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Machanick P & Bailey TL MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feng J et al. GFOLD: a generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics 28, 2782–2788 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Ramirez F, Dundar F, Diehl S, Gruning BA & Manke T deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42, W187–191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tripathi S et al. Meta- and Orthogonal Integration of Influenza "OMICs" Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe 18, 723–735 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ChIP-seq and RNA-seq data sets generated and/or analyzed during the current study have been deposited in the Gene Expression Omnibus (GEO) repository under accession number GSE113042 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113042).
Other data sets that were previously published and used in this study have been deposited in the Gene Expression Omnibus (GEO) repository under accession numbers GSE90634 and GSE108025 available at (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE90634 ) and (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108025 ) respectively.The fitness data were derived from Project Achilles through the Project Achilles Data Portal (https://portals.broadinstitute.org/achilles/about ). The data-set derived from this resource that supports the findings of this study is available at https://portals.broadinstitute.org/achilles/datasets/all. The fitness data were also derived from Project DRIVE. The data-set derived from this resource that supports the findings of this study is available at https://oncologynibr.shinyapps.io/drive/.
All proteomics/ mass-spectrometry data is deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD011103.