Abstract
DNMT3B is known as a de novo DNA methyltransferase. However, its preferential target sites for DNA methylation are largely unknown. Our analysis on ChIP-seq experiment in human embryonic stem cells (hESC) revealed that DNMT3B, mCA and H3K36me3 share the same genomic distribution profile. Deletion of DNMT3B or its histone-interacting domain (PWWP) demolished mCA in hESCs, suggesting that PWWP domain of DNMT3B directs the formation of mCA landscape. In contrast to the common presumption that PWWP guides DNMT3B-mediated mCG deposition, we found that deleting PWWP does not affect the mCG landscape. Nonetheless, DNMT3B knockout led to the formation of 2985 de novo hypomethylated regions at annotated promoter sites. Upon knockout, most of these promoters gain the bivalent marks, H3K4me3 and H3K27me3. We call them spurious bivalent promoters. Gene ontology analysis associated spurious bivalent promoters with development and cell differentiation. Overall, we found the importance of DNMT3B for shaping the mCA landscape and for maintaining the fidelity of the bivalent promoters in hESCs.
INTRODUCTION
In mammals, DNA cytosines can be methylated by a specific class of enzymes known as DNA methyltransferases. Methylated cytosines in mammals are found predominantly on CG dinucleotides (1). Unlike plants, mammals lack DNA methyltransferases that specifically methylate cytosines of non-CG dinucleotides (CH) (2). Thus, CH methylations (mCH) in mammals are rare. However, recent studies show that CA methylation (mCA) can be found in mouse embryonic stem cells (mESC) (3). Moreover, whole genome bisulfite sequencing (WGBS) on the H1 human embryonic stem cell (hESC) line revealed that there is a detectable amount of mCH in the human genome, and mCA is the dominant form among all types of mCH (4). Further studies showed that pluripotent stem cells have the highest percentage of mCA in the genome (4–6). Due to the absence of CH-specific methyltransferase in mammalian cells, it has been hypothesized that de novo methyltransferases (i.e. DNMT3A and DNMT3B) could maintain mCA in mammals. Reports suggested that CA methylation levels in the genome were correlated with DNMT3B expression levels across a panel of human cell lines (5). By overexpressing DNMT3B in yeast cells, Morselli et al. reported that the CH methylation level was increased (7). Liao et al. systematically knocked out (KO) DNMT3A, DNMT3B and DNMT1 in hESC. Their result shows that both DNMT3A and DNMT3B contribute to global CA methylation levels. DNMT3B KO reduces ∼80% of global mCA levels whereas DNMT3A KO contributes to ∼20% of the global mCA level reduction (8). These studies suggested that DNMT3B could be the key enzyme for controlling CA methylation deposition. However, most of these studies only demonstrated global changes of mCA levels in the presence or absence of DNMT3B. It remains unclear whether DNMT3B deposits mCA directly or through an indirect pathway.
Unlike mCA, CG methylations (mCG) in mammalian cells have been studied intensively. mCG is deposited by the DNMT3 family and maintained by DNMT1. mCG plays important regulatory functions in gene expression (9,10). A methylated gene promoter indicates gene silencing. However, silenced genes do not necessarily have their promoters methylated. In pluripotent stem cells, there is a particular category of promoters that are defined as a bivalent promoter. Bivalent promoters are marked by both active and repressive histone marks, H3K4me3 and H3K27me3, respectively. These bivalent promoters are usually unmethylated and associated with gene silencing or low levels of gene expression. With bivalent promoters, genes are more responsive to multiple signaling pathways. This property could be crucial to pluripotent stem cells, since genes have to be activated or silenced quickly during development and cell differentiation. Nevertheless, how the bivalent promoters are established and maintained is mostly unknown. mCG is believed to be involved in the mechanism (11–19).
Evidence from previous studies indicates that DNMT3B is essential for regulating both mCA and mCG (7,8,20,21). Intriguingly, mCA and mCG exhibit distinct landscapes in the human genome. Except for active promoter loci, mCG is ubiquitous throughout the genome, whereas mCA is mainly found within active gene loci (4). It remains unclear that how DNMT3B is guided to a specific locus to regulate DNA methylation.
This study addresses gaps in our knowledge of DNMT3B-mediated DNA methylation. Several studies showed that DNMT3B interacts with histones via its PWWP domain (20,22,23), but the mechanistic function was not investigated. Also, there is lacking direct evidence to connect DNMT3B–histone interaction with de novo DNA methylation (24). Here, we established a DNMT3B-null (KO) and a DNMT3B-PWWP knock-out (ΔPWWP) H1 hESC lines and profiled their DNA methylome through WGBS and various histone marks through ChIP-seq. We also took advantage of the availability of numerous wild-type H1 hESC public datasets and integrated these data into our analysis. Investigating these data allowed us to assess the role of DNMT3B in determining the DNA methylation landscape and its crosstalk with other epigenetic marks.
MATERIALS AND METHODS
Cell culture
H1 hESCs and their derivatives were cultured on hESC-qualified Matrigel (Corning) coated plates. Cells were fed with mTeSR1 (Stemcell Technologies) daily and passaged with ReLeSR (Stemcell Technologies) every 4–6 days in the presence of 10 μM Y-27632 ROCK inhibitor (Merck). 293T were cultured in DMEM supplemented with 4500 mg/l glucose (Biowest), 1× L-glutamine (Thermo Scientific), 1× MEM non-essential amino acids (Thermo Scientific), 1× sodium pyruvate, and 10% fetal bovine serum (FBS) (Biowest). Cells were tested and free from mycoplasma contamination.
Western blot analysis
Harvested cultured cell pellets were lysed in RIPA buffer, and cleared supernatants were subjected to protein quantification by the Bradford assay (Bio-rad). About 30 μg of protein lysate was loaded into each well of a 7.5% SDS-PAGE mini gel. The gel was run in 1× Tris-glycine buffer supplemented with 0.1% SDS. Proteins were transferred onto a PVDF membrane (Bio-rad) through wet transfer. The membrane was blocked by 5% skim milk and incubated with primary antibodies followed by HRP-conjugated secondary antibody in diluted 5% BSA/TBST. Chemiluminescence was detected by exposing the X-ray film to the blot pre-incubated with either Luminata Crescendo Western HRP substrate (Millipore) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific). The list of antibodies used can be found in Supplementary Table S1.
Immunofluorescent (IF) imaging
Cells were fixed in 4% paraformaldehyde for 10 min followed by incubation with 0.1% Triton-X 100/PBS (PBST) for 15 min. The cells were blocked in 5% BSA/PBST for 1 h followed by primary antibodies, Alexa Fluor-conjugated secondary antibodies (Thermo Scientific) and DAPI (Sigma). Images were taken using an Olympus microscope. The list of antibodies used can be found in Supplementary Table S1.
Lentivirus production
pMD2.G, psPAX2 and lentivector were transfected into 10 million 293T using TransIT-LT1 reagent (Mirus). Supernatants were harvested at 48 and 72 h after transfection. Virus was concentrated 100–300 times through centrifugation after filtering through a 0.45 μm syringe filter, resuspended in DMEM-F12 medium and stored at –80°C. The viruses were titered on HeLa cell using FACS. The plasmids used in this study are listed in Supplementary Table S2.
CRISPR/Cas9
300k H1 cells were infected with the FUCas9Cherry and FgH1UTG-sgRNA viruses at 0.6 MOI with 4 μg/ml polybrene for 12 h. More than 85% double infection rates were achieved (determined by FACS-mCherry and GFP channel). The efficiency of the sgRNA was determined by the T7 endonuclease I (NEB) assay (as described in manufacturer’s protocol) on the targeted locus or western blot on targeted protein after Dox-On (1 μg/ml Dox) for 4 days. Transduced H1 hESC were broken into single cell suspensions with accutase and seeded onto Matrigel-coated 96-well plates with a density of 4 cells per well and supplemented with 10 μM Y-27632 ROCK inhibitor (Merck). After two additional rounds of clonal seeding, a pure mutant/KO clone (determined by IF) was selected for expansion and subjected to genotyping and western blot analysis. In IF screening experiments for DNMT3B PWWP mutations, the cells were treated with 0.1 μg/ml nocodazole (Sigma) for 12 h to induce mitotic metaphase arrest before fixation. The list of sgRNAs used can be found in Supplementary Table S2.
DNMT3B variants overexpression in KO H1 hESC
N-terminal flag-tagged DNMT3B variants (WT DNMT3B isoform 1, ΔPWWP DNMT3B and S270P DNMT3B) and control protein (GFP) were cloned into a modified pLX304 plasmid, in which the CMV promoter was replaced with a CAG promoter. The coding sequence (CDS) of WT DNMT3B isoform 1 was amplified from H1 total cDNA and the CDS of the DNMT3B variants was constructed by PCR mutagenesis. Lentivirus was produced and transduced into DNMT3B KO H1 with a MOI 0.6 in the presence of 4 μg/ml polybrene for 12 h. The cells were selected with 4 μg/ml of Blasticidin S (Thermo Scientific) in mTeSR1 (Stemcell Technologies) and selection medium was changed daily. The cells were expanded and harvested for western blot and ChIP assay within 2 weeks after transduction in order to avoid severe exogenous gene promoter silencing after prolonged culture (25).
Chromatin immunoprecipitation (ChIP)
H1 cells were resuspended into single cells after accutase treatment. About 20 million cells were counted and fixed in 1% formaldehyde/PBS for 8 min at room temperature with rotation. Excessive formaldehyde was quenched with 0.25 M glycine. Fixed cells were washed twice with PBS supplemented with 1 mM PMSF. The cell pellet was snap frozen in liquid nitrogen and stored at –80°C or processed directly. Cells were lysed in lysis buffer containing 10 mM Tris-Cl (pH 8), 100 mM NaCl, 10 mM EDTA, 0.25% Triton X-100 and EDTA-free protease inhibitor cocktail (Roche). Nuclear lysis was performed in 1% SDS lysis buffer containing 50 mM HEPES-KOH (pH 7.5), 150 mM NaCl, 1% SDS, 2 mM EDTA, 1% Triton X-100, 0.1% NaDOC and a protease inhibitor cocktail (EDTA-free). The chromatin pellet was harvested after centrifugation at 20 000 RPM using a pre-chilled JA25.50 rotor in a Beckman centrifuge. The pellet was then washed twice with 0.1% SDS lysis buffer containing 50 mM HEPES-KOH (pH 7.5), 150 mM NaCl, 0.1% SDS, 2 mM EDTA, 1% Triton X-100, 0.1% NaDOC, 1 mM PMSF and protease inhibitor cocktail (EDTA-free). Sonication with Bioruptor (15 cycles, 30-s on, 30-s off, high power) was conducted after resuspending the pellet with 0.1% SDS lysis buffer. The chromatin solution was clarified by centrifugation at 20 000 g at 4°C for 45 min and then pre-cleared with Dynabeads protein A (Thermo Scientific) for 2 h at 4°C. About 500 μl of the pre-cleared chromatin sample was incubated with 50 μl of Dynabeads protein A loaded with 3 μg antibody overnight at 4°C. The beads were washed thrice with 0.1% SDS lysis buffer, once with 0.1% SDS lysis buffer/0.35 M NaCl, once with 10 mM Tris-Cl (pH 8), 1 mM EDTA, 0.5% IGEPAL CA-630 (Sigma), 0.25 LiCl, 0.5% NaDOC and once with TE buffer (pH 8). The immunoprecipitated material was eluted from the beads by heating for 45 min at 69°C in 50 mM Tris-Cl (pH 7.5), 10 mM EDTA and 1% SDS. To reverse crosslinks, samples were incubated with 20 μg/ml proteinase K at 65°C for 8 h. The samples were then extracted with phenol:chloroform:isoamyl alcohol (25:24:1) followed by chloroform, ethanol precipitated in the presence of glycogen and re-suspended in 10 mM Tris buffer (pH 8). The ChIP-seq libraries were constructed using a ThruPLEX DNA-seq 12S kit (TakaraBio). The libraries were subjected to size selection (250–500 bp) on a 4–20% TBE PAGE gel (Thermo Scientific). The recovered libraries were sequenced on an Illumina Hiseq 2000 platform with 2 × 100 paired-end or Nextseq 500 platform with 2 × 76 paired-end. For DNMT3B, H3K4me3 and H3K36me3 ChIP-seq, two biological replicates were sequenced, and the results were validated by ChIP-qPCR with an independent biological replicate. For H3K27me3 ChIP-seq, one replicate was sequenced, and the results were validated by ChIP-qPCR with an independent biological replicate.
RNA-seq
H1 hESC and their derivatives were lysed in 1 ml of Trizol (Thermo Scientific) and extracted according to manufacturer’s protocol. The purified RNAs were subjected to DNAseI digestion and re-purified using a Qiagen RNA mini column. The RNA was quantified using Nanodrop. RIN was determined on Bioanalyzer RNA Nano 6000 Chip (Agilent). About 3 μg of RNA (RIN > 9) was used for library construction. The stranded RNA-seq libraries were constructed by Script-seq V2 RNA-seq library preparation kit (Illumina). The size selected libraries (250–500 bp) were sequenced on an Illumina Hiseq 4000 platform with 2 × 100 paired-ends or a Nextseq 500 platform with 2 × 76 paired ends.
WGBS
A 10-cm plate of cells was washed twice with cold PBS. About 2 ml of gDNA lysis buffer (50 mM Tris-HCl, pH 8, 100 mM NaCl, 25 mM EDTA and 1% SDS) was applied directly to the cells. The lysates were incubated at 65°C overnight with 2 mg of proteinase K. The lysate was diluted two times with TE buffer before adding 1 mg of RNase A and followed by a 1.5 h incubation at 37°C. The NaCl concentration was subsequently adjusted to 200 mM followed by phenol–chloroform extraction at pH 8 and isopropanol precipitation. The gDNA pellet was dissolved in 1 ml TE pH 8 buffer and incubated with RNase A with a concentration of 100 μg/ml for 1 h at 37°C. The pure gDNA was recovered by phenol–chloroform (pH 8) extraction and isopropanol precipitation and dissolved in TE pH 8 buffer. The gDNA was diluted to 100 ng/μl before sending to BGI for WGBS library construction and sequencing. Two biological replicates for each knockout sample were sequenced to approximate 30× human genome coverage (∼90 Gb) on a Hiseq X platform with 2 × 150 paired-end reads.
Quantitative PCR (qPCR)
About 1 μg of RNA was reverse transcribed using Qscript cDNA Supermix (QuantaBio). The cDNAs were diluted 10 times for expression analysis. qPCR on cDNA or ChIP-DNA was performed on 384-well plates on a QS5 system (Thermo Scientific) with GoTaq qPCR Master Mix (Promega). The fold change or percentage input of the samples was calculated based on relative standard curve method. Primers used in this study are listed in Supplementary Table S2.
Bioinformatic analysis
WGBS data and histone ChIP-seq data from H1 hESC were downloaded from the ENCODE and Roadmap Epigenomics projects. MNase-Seq was obtained from NCBI GEO (GSM1194220), published with Yazdi et al.’s study (26). The datasets used in this study are listed in Supplementary Table S3. For ChIP-seq analysis, reads were mapped by bowtie2 against human reference genome GRCh38 (27). Peak calling was performed by MACS2 (28). The resulting alignments were extended to 150 bp and then converted into signals by BEDTools in bedGraph format (29). We adjusted the average coverage of each ChIP-seq experiment to 1. Finally, the resulting files were converted to bigWig format by bedGraphToBigWig for visualization and generating heat maps. For WGBS analysis, the leading three bases and adaptor sequences were trimmed from paired-end reads by TrimGalore. The resulting FASTQ files were analyzed by BISMARK (30). PCR duplicates were removed by SAMtools rmdup (31). Then bismark_methylation_extractor continued the extraction of the DNA methylation status on every cytosine sites. DNA methylation levels were converted into bedGraph and then to bigWig format by bedGraphToBigWig. DMRs were identified by metilene (32). We only kept these that differ in absolute level of methylation by more than 50% with P-value ≤ 0.05. DMRs were associated with nearest genes by using annotatedPeak.pl in the Homer package (33). The GO analysis on DMRs was performed by GREAT (34). For RNA-seq analysis, the paired-end RNA-seq reads were trimmed of adaptor sequences by TrimGalore and then mapped by STAR to the human reference genome GRCh38 with reference gene annotation GENCODE 26 (35). PCR duplicates were removed in the paired-end alignments by samtools rmdup. Alignments with mapping quality < 20 were removed. Gene expression levels in FPKM were determined by cuffdiff in the Cufflinks package (36). Differentially expressed genes were determined by DESeq2 (37) based on the read counting result from featureCounts (38), with the fold change cutoff 2 and the q-value cutoff 0.05. Signal profiles and heat maps were based on figures generated by Deeptools (39) or ngsplot (40). Deeptools was also used for calculating genome-wise correlations among signals in bigWig files, and its bin size was set to 4000 bp. For non-genome-wise corrections, the bin size was 500 bp. FASTQ files of biological and technical replicates were merged for all analysis. The same analysis was performed on each replicate to confirm the consistency across the replicates (Supplementary Figure S13).
RESULTS
mCA levels are elevated at DNMT3B-enriched loci
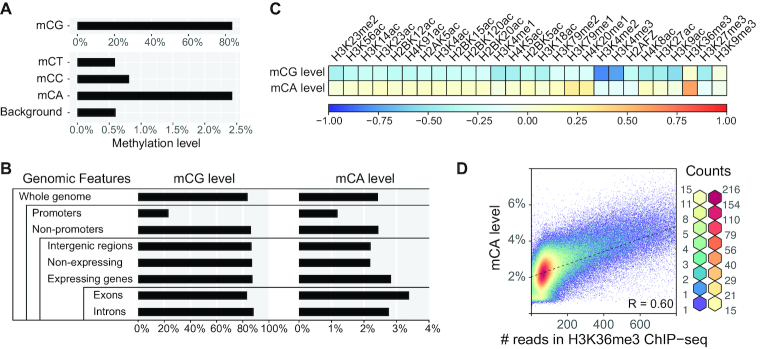
According to ENCODE data, cytosine methylation can be found on 2.43% of CpA dinucleotides in H1 hESC genome (Figure 1A and Supplementary Table S4) (5). The mCA level is relatively high in expressed gene loci, especially for regions within exons (Figure 1B). It also had high correlation with the transcription elongation mark, H3K36me3 (Figure 1C and D) (4). Previous studies had shown that DNMT3B recognizes H3K36me3 via its PWWP domain (7,20,21). We asked whether the mCA distribution observed is associated with DNMT3B–histone binding. We therefore performed DNMT3B ChIP-seq in H1 hESC.
Figure 1.
mCA distributes non-randomly and correlates with H3K36me3 histone marks in H1 hESC. (A) Global methylation levels of mCG, mCT, mCC and mCA. The background level of this WGBS experiment is estimated using spike-in unmethylated bacteriophage lambda DNA. (B) mCG and mCA levels on different genomic features. Promoters are defined by the Roadmap Project’s H1 hESC promoter annotation. (C) A heat map demonstrating Pearson correlation between mCG or mCA levels and ChIP-Seq signals of various histone marks. (D) Scatterplot illustrates the relationship between mCA levels and H3K36me3 marks. Pearson correlation is shown.
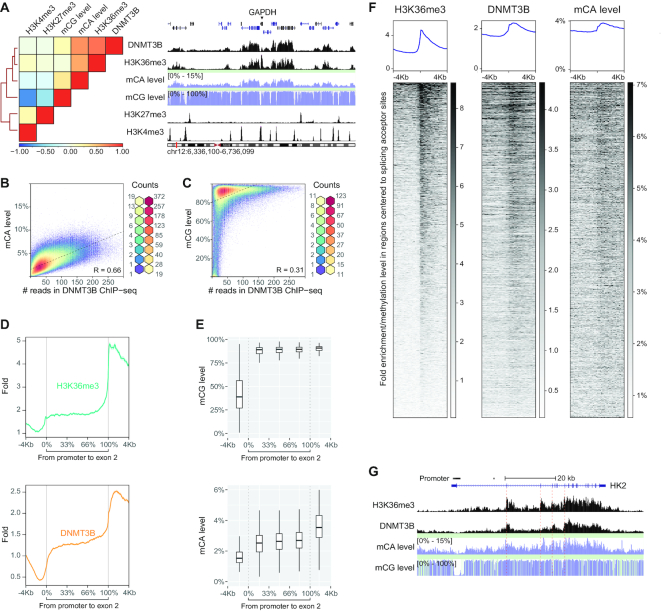
Since a ChIP-grade DNMT3B antibody was previously not available (41), we have developed a custom-made DNMT3B rabbit monoclonal antibody (Clone D3C8T, from Cell Signaling Technology) for performing endogenous DNMT3B ChIP-seq in our studies. In concordance with previous studies, we observed that the ChIP-seq signal of DNMT3B is highly correlated with H3K36me3 (Figure 2A and Supplementary Figure S1). Intriguingly, the high correlation between DNMT3B ChIP-seq signals and mCA levels was also found (Figure 2A).
Figure 2.
Profiles of DNMT3B, mCA and H3K36me3 coincide. (A) Heat map with hierarchical clustering shows the relationship among DNMT3B, mCG and mCA levels, and histone marks of H3K36me3/H3K27me3/H3K4me3. An example of their profiles is shown for the genomic interval around GAPDH. (B) Scatterplot illustrates the relationship between mCA levels and DNMT3B ChIP-seq in gene bodies (with 500 bp windows). (C) Scatterplot illustrates the relationship between mCG levels and DNMT3B ChIP-seq in gene bodies (with 500 bp windows). (D) ChIP-seq profiles demonstrating H3K36me3 and DNMT3B signals at intervals around promoters to the first splicing junction of active genes. Intervals with ambiguous promoter boundaries or with ambiguous first splice acceptor sites are excluded. (E) Box plots demonstrating mCG and mCA levels at intervals around promoters to the first splice junction of active genes. (F) H3K36me3, DNMT3B and mCA level profiles around intervals centered to splice acceptor sites that are defined by junction-spanning reads of RNA-seq. Intervals in the heat map are sorted by the strength of H3K36me3 signals in descending order. Intervals containing multiple splicing acceptor sites are excluded. (G) Profiles of H3K36me3, DNMT3B, mCA and mCG at the HK2 locus.
Since the mCA level was particularly high in transcriptionally active regions, we performed further analysis on expressed gene loci. Genes with FPKM ≥0.2 were selected. We found that the DNMT3B ChIP-seq read counts highly correlated with mCA levels at expressed gene loci (R = 0.66, Figure 2B). In contrast, correlation with mCG levels was drastically lower (R = 0.31) (Figure 2C).
DNMT3B and mCA distribution associates with RNA splicing
We observed that our DNMT3B ChIP-seq signal distribution fits into a three-level pattern along expressed gene loci: low levels at the promoter region, followed by intermediate levels before the first splicing junction and elevated levels after the first splicing junction (Figure 2D). Intriguingly, the mCA distribution also fits into this three-level pattern, whereas mCG has no noticeable change after the splicing junction and it distributed in a two-level pattern instead (Figure 2E).
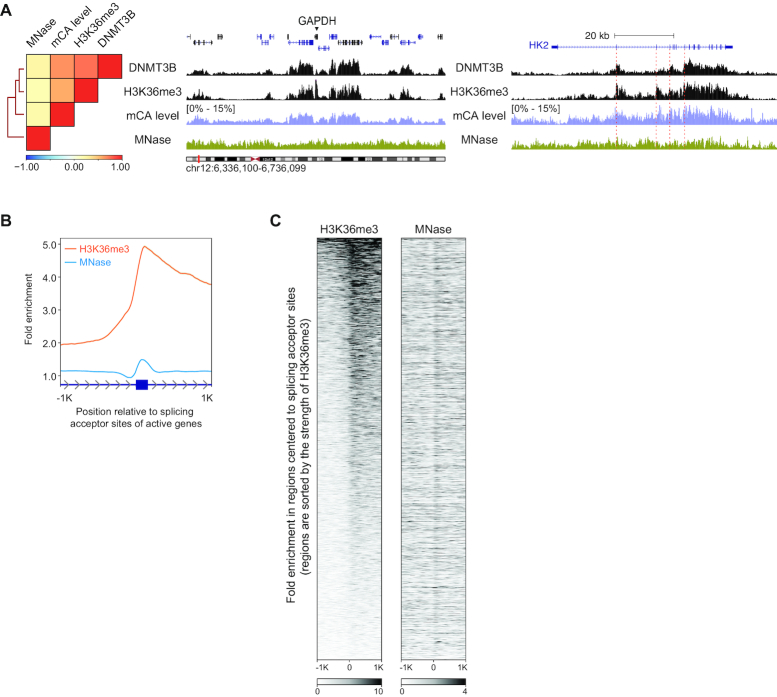
It is known that H3K36me3 can be deposited by SETD2 that interacts with the RNA POLII C-terminal domain (42,43). Studies demonstrated that SETD2 activity could be triggered by RNA splicing. Furthermore, SETD2 can be recruited by the inclusion of internal alternative exons (44). Thus, we expect that the enhancement of the DNMT3B signal and mCA level is not restricted to the first splicing junction. To test this, we investigated signals of H3K36me3/DNMT3B/mCA around all splicing junctions along the genes. Based on regions centered to splicing acceptor sites, we found that the distributions of mCA, DNMT3B and H3K36me3 follow the rhythm of RNA-splicing (Figure 2F and 2G; Supplementary Figure S13A), indicating a strong connection between DNMT3B/mCA and RNA-splicing. On the other hand, the mCG level is saturated in the gene bodies and fits poorly with H3K36me3. Its changes around splicing junctions are unnoticeable (Supplementary Figure S2). H3K36me3 is reported to be specifically enriched on exons due to the distribution of nucleosomes (45,46). To evaluate the impact of nucleosome distribution, we compared signatures between H3K36me3 ChIP-seq and MNase-Seq in H1 hESCs. On a genome-wide scale, the MNase signal did not show similarity with that of H3K36me3 (Figure 3A). Indeed, it was enriched on exons (Figure 3B). However, unlike the long-distance enrichment of H3K36me3, the influence of nucleosome occupancy was only restricted to short intervals after splice acceptor sites (Figure 3B). Moreover, the strength of the H3K36me3 signal does not follow that of MNase (Figure 3C). Hence, the nucleosome distribution is unlikely contributing to the distinct pattern of H3K36me3/DNMT3B/mCA landscape.
Figure 3.
The distribution of H3K36me3 and nucleosomes. (A) A heat map with hierarchical clustering demonstrating the relationship among MNase-Seq, H3K36me3, DNMT3B and mCA levels. (B) Profiles of H3K36me3 and MNase-Seq around active splice acceptor sites. (C) Profile comparison of H3K36me3 and MNase-Seq in intervals sorted by the strength of H3K36me3 around active splice acceptor sites. The strength of MNase signals does not follow that of H3K36me3.
DNMT3B determines the mCA landscape in hESC
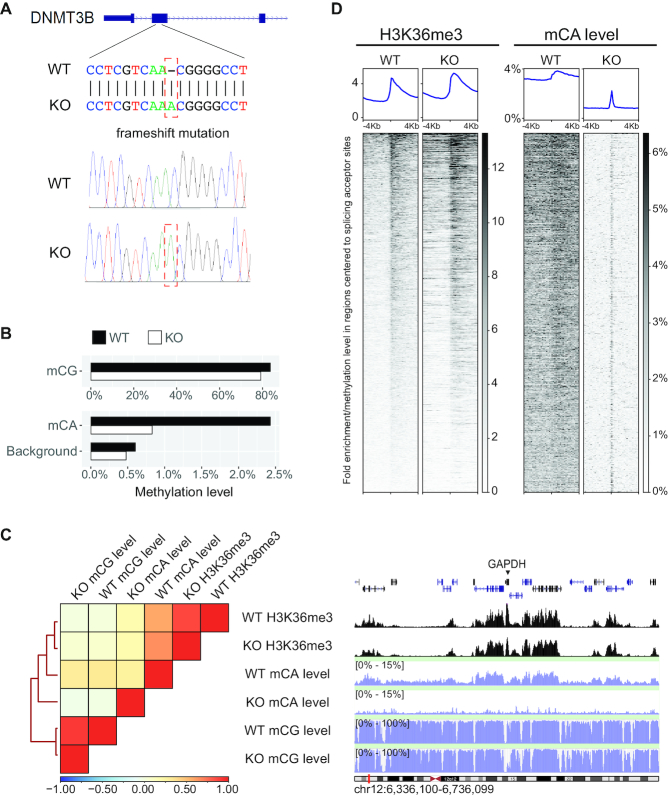
The findings presented so far strongly suggest that DNMT3B may deposit mCA. To prove this, we generated a DNMT3B null H1 cell line and studied the changes in the mCA landscape. The DNMT3B knockout (KO) was done by introducing a frameshift mutation in the second exon of DNMT3B through CRISPR-Cas9 technology (47). Homozygous mutation was confirmed by Sanger sequencing (Figure 4A) and RNA-sequencing (Supplementary Figure S3A). DNMT3B protein was undetectable by western blot analysis (Supplementary Figure S10C) or immunofluorescent staining (Supplementary Figure S3B). H3K36me3 levels were similar to those in WT cells (Supplementary Figure S10D). Pluripotent markers, such as POU5F1 and NANOG, remained expressed, and DNMT3A remained undisrupted in the KO cells (Supplementary Figure S3B and S10E).
Figure 4.
DNMT3B determines the mCA landscape in hESC. (A) A frameshift mutation was introduced into the second exon of the DNMT3B gene. Sanger sequencing results are shown at the bottom panel. (B) The change of global mCG and mCA levels after DNMT3B KO. (C) Heat map with hierarchical clustering compares H3K36me3 with mCA and mCG levels between WT and KO. An example of their profiles is shown on the genomic interval around the GAPDH gene. (D) Profile comparison of H3K36me3 and mCA levels between WT and KO on intervals around splice acceptor sites. Intervals in the heat map are sorted by the strength of WT H3K36me3 signals in descending order.
We then performed WGBS on the KO cells. We found that the global mCA level in KO cells was reduced to 0.83% from the 2.43% observed in WT. Taking the background level into account (0.48% in this dataset), global mCA was reduced by 81%, indicating that majority of mCA had been removed in KO cells (Figure 4B). Next, we performed H3K36me3 ChIP-seq to study the relationship between H3K36me3 and DNA methylation in KO cells. The data revealed that KO H3K36me3 signal distribution was similar to the signal distribution in both WT H3K36me3 and WT mCA levels (Figure 4C). They were grouped in the clustering result, whereas KO mCA level forms an out-group (Figure 4C). The clustering result suggested that KO cells have a mCA landscape distinct from WT cells. The high correlation of mCG distributions between KO and WT (R = 0.92) indicated that mCG was less affected by DNMT3B knockout (Figure 4C).
In WT cells, both the mCA level and H3K36me3 signal were elevated after the splicing acceptor sites and its level was maintained toward the 3′ end of the genes. However, in KO cells, this pattern was only observed with H3K36me3 but not with mCA (Figure 4D and Supplementary Figure S13B). This can be expected when the deposition of H3K36me3 is from an event upstream of the DNMT3B–DNA interaction, whereas the deposition of mCA is downstream. At the splicing junctions, mCA levels decreased significantly in KO cells. Around splicing acceptor sites, the mCA level was maintained. We then sorted the splicing acceptor sites according to the strength of WT H3K36me3 signal in descending order. We found that in WT cells, mCA levels appeared as descending order corresponding to the H3K36me3 signal. In contrast, splicing acceptor sites in KO cells have lower mCA levels, and the mCA level does not correspond to the strength of H3K36me3 signal. On the other hand, no noticeable change was observed in mCG levels around splicing junctions (Supplementary Figure S4). Although DNMT3B contributes to more than 80% of methylation on CA dinucleotides in hESC, the pattern of remaining mCA in KO suggested that there might be a DNMT3B-independent mCA deposition pathway, such as the DNMT3A-dependent mCA deposition pathway, in KO cells (8).
DNMT3B knockout reduces the mCG level at annotated promoter regions
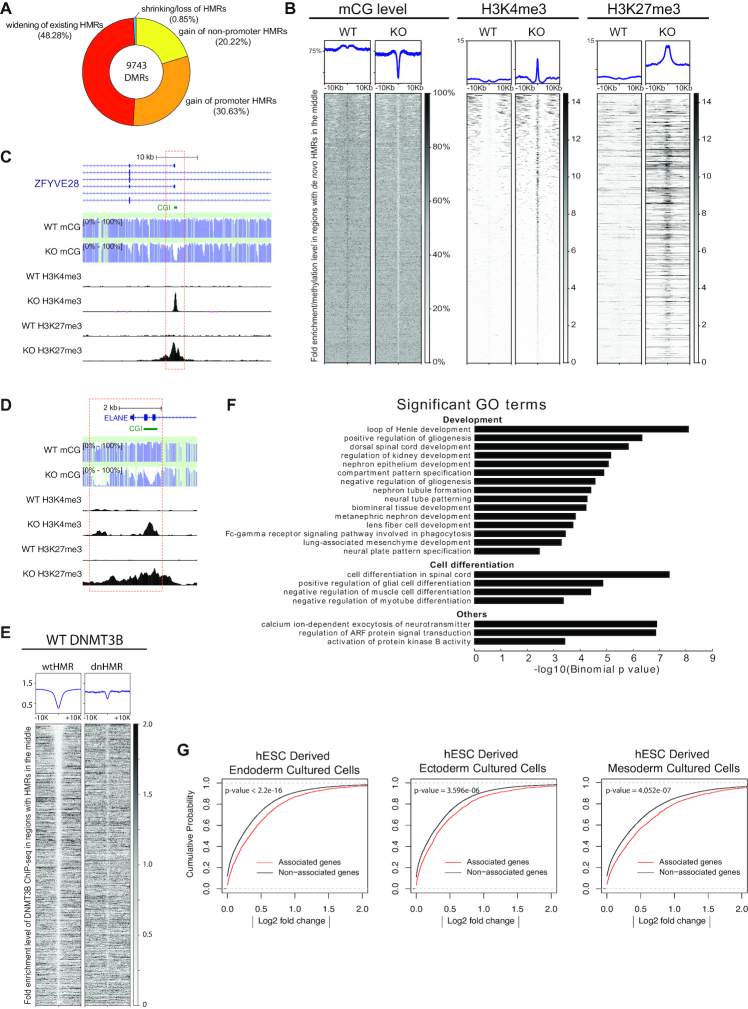
The knockout of DNMT3B decreased global mCG levels from 83.68% (WT) to 79.22% (KO). Although the change is minor, comparison of mCG methylomes between KO and the WT revealed 9743 significant differentially methylated regions (DMR) that differed in at least 50% absolute levels of methylation (Figure 5A and Supplementary Table S5). As expected, most of them (9660/9743) have lower methylation levels in the KO, in which 4704 regions widen the existing hypomethylated regions (HMR) in wild-type cells. The widening events observed at HOX gene clusters resulted in hypomethylation throughout the whole HOX loci. Intriguingly, expression of HOX genes does not change after DNMT3B KO (Supplementary Figure S5). In the other 4956 regions, the loss of mCG led to the formation of de novo hypomethylated regions (dnHMR) in KO. Since most of these dnHMRs were found in predicted promoter regions (2985/4956) from the Roadmap project collection of 129 cell types (Figure 5A and Supplementary Table S5), they might potentially have regulatory functions on gene expression. These promoter dnHMRs associate with 1736 genes nearest to them (Supplementary Table S5). However, when we compared the expression level between these dnHMR genes and the other genes, we found no tendency of dnHMR genes to be more likely up-regulated or down-regulated after DNMT3B knockout (Supplementary Figure S6). On the other hand, we found up-regulated genes do not usually be dnHMR genes. Only 8 dnHMR genes occur in the 120 significantly up-regulated genes identified by DESeq2 (Supplementary Table S6).
Figure 5.
Spurious bivalent promoters emerge after DNMT3B KO. (A) The pie chart demonstrates different types of hypomethylated regions (HMRs) induced following DNMT3B KO. (B) Profiles of mCG, H3K4me3 and H3K27me3 comparing WT and KO on intervals around dnHMRs. (C and D) Genome browser tracks of mCG, H3K4me3 and H3K27me3 comparing WT and KO at the ZFYVE28 and ELANE loci. (E) The occupancy of DNMT3B in dnHMR regions and in wild-type HMR regions. (F) GO analysis of genes proximate to dnHMRs. (G) Kolmogorov–Smirnov test for comparing dnHMR-associated genes and non-associated genes in hESC-derived endoderm, ectoderm and mesoderm cultured cells. Processed RNA-seq data in the Roadmap Epigenomics Project were used.
Spurious bivalent promoters emerge after DNMT3B knockout
Since we observed almost no increase in RNA level in genes with promoter dnHMRs, we expected that these promoter dnHMR might be repressed by PRC2 complex as reported by others in mouse ESCs (48). Intriguingly, our ChIP-seq revealed that most of the promoter dnHMRs were coated not only with H3K27me3, but also with H3K4me3 marks (Figure 5B–D; Supplementary Figure S7 and S13C). We use ‘spurious bivalent promoters’ to describe these promoter dnHMRs that possess both H3K4me3 and H3K27me3 marks. Bivalent promoters are known to associate with low gene expression, possibly explaining our observation that no increase in RNA was observed in spite of decreased methylation of their promoter. Furthermore, we detected an elevated level of H2AK119ub at the selected spurious bivalent promoters through ChIP-qPCR (Supplementary Figure S7D, I and N), suggesting that the PRC1 complex may be involved in spurious bivalent promoter formation after DNMT3B KO.
In wild-type cells, these spurious bivalent promoter loci were hypermethylated. We also observed that the WT DNMT3B ChIP-seq signals at dnHMR loci were 2.89 times stronger than at wild-type HMR loci, suggesting that the spurious bivalent promoter loci were occupied with DNMT3B before the knockout (Figure 5E and Supplementary Figure S13D). Knockout of DNMT3B led to the formation of bivalency. Furthermore, DNMT3B knockout had no observable effect on canonical bivalent promoters or active promoters in WT cells (Supplementary Figure S8). Our results suggested that DNMT3B might be acting as a safeguard to prevent the formation of these spurious bivalent promoters.
Spurious bivalent promoters are involved in development and cell differentiation
It has been proposed that bivalent promoters regulate the differentiation of pluripotent stem cells (11,18). We asked whether dnHMRs have any relevant functions. Promoter dnHMRs with significant H3K4me3 peaks (2117/2985) were selected and interpreted by GREAT, a tool designed for predicting functions of cis-regulatory regions (34). The result showed that these promoter dnHMRs were proximate to genes that are involved in development and cell differentiation (Figure 5F; Supplementary Tables S7 and S8). In contrast, those differential methylated regions (DMRs) that widen the existing HMRs did not lead to any significant GO terms. Moreover, with hESC-derived cells from the Roadmap Epigenomics Project, the Kolmogorov–Smirnov test revealed that dnHMR-associated genes exhibited greater gene expression changes upon hESC differentiation (P < 0.001 in all cell types examined, Figure 5G and Supplementary Figure S9). Furthermore, 69% of dnHMR-associated genes were up-regulated more than 2-fold in at least one of the examined cell types. In contrast, 59% of these genes could be down-regulated in at least one cell type (Supplementary Table S9). These analyses suggest that DNMT3B regulates specific functions via dnHMRs. It may play a role in maintaining the repository of bivalent promoters that control genes for development and cell differentiation. Interestingly, only 26% of dnHMRs possess CGI, suggesting that they tend to be dynamically regulated. It is worth to note that angiogenesis pathway is significantly enriched in PANTHER pathway analysis (P = 3.31e-9) (Supplementary Table S7).
DNMT3B–H3K36me3 interaction is crucial for mCA landscape formation
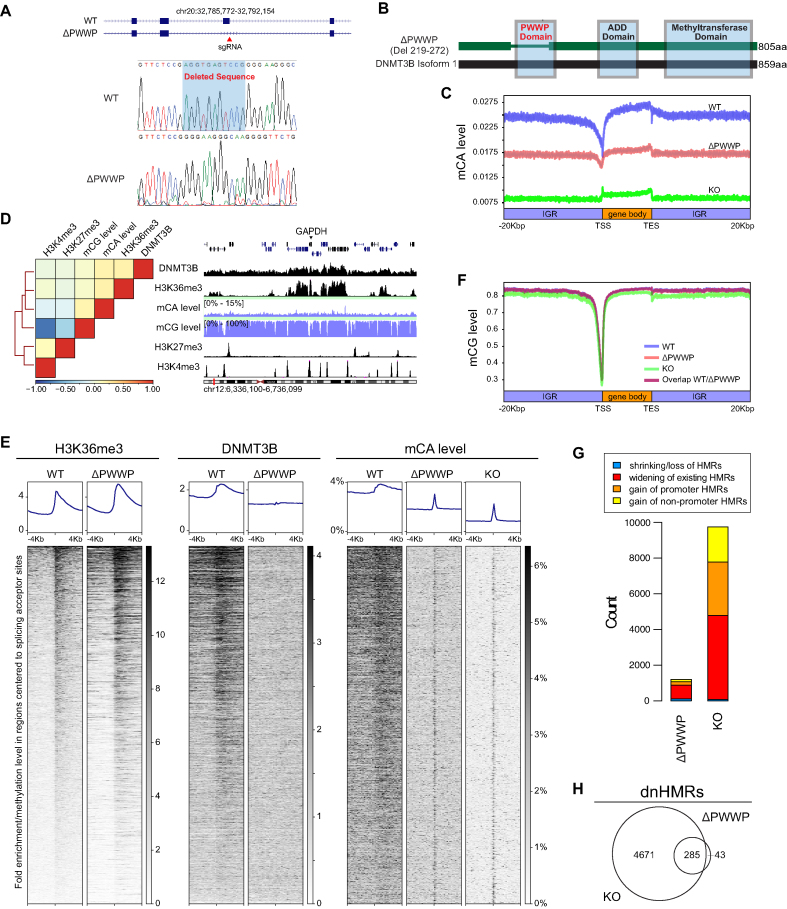
To study how the DNA methylome is regulated by the DNMT3B–H3K36me3 interaction, we deleted the DNMT3B PWWP domain. We introduced a small homologous deletion of the splicing donor of Exon 7 in the DNMT3B gene (Figure 6A). Exon 7 encodes over 90% of the PWWP domain. This small deletion of a splicing junction induces exon skipping of Exon 7 during transcription without frameshift mutations (Figure 6B; Supplementary Figure S10A and C). Thus, we generated an H1 hESC line that expresses PWWP-null DNMT3B (ΔPWWP). As is the case for the DNMT3B PWWP mutants reported before (22), PWWP-null DNMT3B disperses all over the nucleus, rather than showing colocalization with metaphase chromatin (Supplementary Figure S10B). Also, ΔPWWP cells express pluripotent markers such as POU5F1 and NANOG (Supplementary Figure S10E). It is worth to note that ΔPWWP DNMT3B retains the ability to interact with DNMT3A in the cell (Supplementary Figure S10F).
Figure 6.
The DNMT3B–H3K36me3 interaction is crucial for shaping the mCA but not the mCG landscape. (A) Homologous deletion of the splice donor of exon 7 in the DNMT3B gene. (B) The deleted region largely overlaps with the PWWP domain in the DNMT3B protein. (C) Profiles of mCA in WT, KO and ΔPWWP H1 hESC are shown across genes; IGR, intergenic region. Intervals that overlap with other intervals are excluded. (D) A heat map of ΔPWWP cells with hierarchical clustering demonstrating the relationship among DNMT3B, mCG and mCA levels, and H3K36me3/H3K27me3/H3K4me3 histone marks. An example of their profiles is shown on the genomic interval around the GAPDH gene. (E) Profile comparison of H3K36me3 and mCA levels between WT and ΔPWWP at intervals around splice acceptor sites. Intervals in the heat map are sorted by the strength of WT H3K36me3 signals in descending order. (F) Profiles of mCG in WT, KO and ΔPWWP H1 hESC are shown across genes. (G) Comparison of the amount of DMR types between ΔPWWP and KO. (H) Venn diagram shows that dnHMRs in ΔPWWP are a subset of those in KO cells.
Next, we performed WGBS and ChIP-seq of DNMT3B, H3K36me3, H3K27me3 and H3K4me3 in ΔPWWP cells. The WGBS results revealed that the global mCA level of ΔPWWP is in between the mCA levels observed in WT and KO (Supplementary Figure S11). Only the mCA in the WT corresponded to genomic features such as the TSS and gene bodies (Figure 6C). Interestingly, gene body mCA levels in ΔPWWP and KO are slightly higher compared to those in intergenic regions (Figure 6C).
We then compared the clustering analysis between ΔPWWP (Figure 6D) and WT (Figure 2A). We found that the relationship between mCG levels, H3K27me3 and H3K4me3 in ΔPWWP cells (Figure 6D) remained similar to those in WT cells (Figure 2A). However, in ΔPWWP cells, the Pearson correlations between DNMT3B, H3K36me3 and mCA levels dropped dramatically to <0.20 (Figure 6D) from more than 0.64 observed in WT cells (Figure 2A).
The expression level of ΔPWWP DNMT3B in our CRISPR-line was lower than the WT DNMT3B in WT H1 (Supplementary Figure S10C). We asked whether the loss of ΔPWWP DNMT3B–H3K36me3 correlation was due to the loss of PWWP domain or the lower expression level of ΔPWWP DNMT3B. We overexpressed WT DNMT3B and ΔPWWP DNMT3B in DNMT3B KO H1 cells and performed DNMT3B ChIP-qPCR. The mutant S270P DNMT3B, which has been reported to lose the ability to bind H3K36me3, was included as a control as well (49). The results demonstrated that only WT DNMT3B, but neither ΔPWWP DNMT3B nor S270P DNMT3B, binds to the gene-body and follows the H3K36me3 distribution (Supplementary Figure S12). The results suggested that the PWWP domain is crucial for DNMT3B–H3K36me3 interaction. The deletion of the PWWP domain in ΔPWWP cells led to the loss of the DNMT3B–H3K36me3 correlation.
We then asked whether the loss of DNMT3B-mCA correlations in ΔPWWP cells was due to the loss of DNMT3B–H3K36me3 interaction. We found that the Pearson correlation between H3K36me3 and mCA levels in WT, ΔPWWP and KO were 0.58, 0.19 and 0.20, respectively. ΔPWWP and KO have a similar H3K36me3–mCA level correlation, suggesting that ΔPWWP DNMT3B was unable to maintain the mCA landscape at H3K36me3 positive loci. To validate the correlation results, we plotted signals of H3K36me3, DNMT3B and mCA around splice acceptor sites. The genomic regions were sorted according to the strength of the WT H3K36me3 signals. H3K36me3 signals of ΔPWWP rose at splice acceptor sites as in WT cells (Figure 6E and Supplementary Figure S13E). Deleting the PWWP domain did not affect H3K36me3 deposition as an upstream event. The results demonstrate that the DNMT3B signal in ΔPWWP no longer followed the trend of the H3K36me3 signal, and no elevation was observed at the junction site. The mCA pattern of ΔPWWP also did not follow the H3K36me3 signal. Although the reduced DNMT3B protein level in the ΔPWWP cells could possibly compromise the interpretation, we expected that the mCA level in ΔPWWP would correspond to the H3K36me3 signal if the ΔPWWP DNMT3B could maintain the mCA landscape at H3K36me3 positive loci. However, the strength of junction mCA in ΔPWWP was independent of H3K36me3 as observed in the KO, indicating that ΔPWWP DNMT3B cannot maintain mCA at these junctions (Figure 6E and Supplementary Figure S13E). Overall, these observations, alongside the well-described interaction of the PWWP domain with H3K36me3 (49), suggest that the DNMT3B–H3K36me3 interaction is crucial to determine the mCA landscape in hESC, even though some effect of the differences in expression levels between the WT and ΔPWWP DNMT3B cannot be formally excluded in our experimental settings.
The PWWP domain is dispensable for DNMT3B-mediated mCG deposition
We have shown that loss of promoter mCG is common in KO and leads to the formation of dnHMRs and spurious bivalent promoters. Surprisingly, although the PWWP domain determines the localization of DNMT3B, ΔPWWP hESCs do not lose promoter mCG as observed in KO cells (Figure 6F). dnHMRs are rare in ΔPWWP, as we only identified 195 promoter dnHMRs in ΔPWWP, a relatively low in amount compared to 2985 promoter dnHMRs found in KO cells (Figure 6G). The widening event of the existing hypomethylated regions is common in ΔPWWP. However, it is far less than the event observed in KO (Figure 6G). It is worth noting that all dnHMRs found in ΔPWWP are almost a subset of those in KO (Figure 6H). Overall, our results suggest that DNMT3B is involved in the regulation of promoter mCG and its bivalency, but the DNMT3B–H3K36me3 interaction is unnecessary.
DISCUSSION
To date, most studies of DNA methylation have focused on mCG, while relatively little was known about the non-CG methylation (mCH) landscape in mammalian cells. While the enzymes responsible for mCG deposition and maintenance have been identified, it is uncertain which account for mCA in mammalian cells. Biochemical assays suggested that both DNMT3A and DNMT3B could methylate CH dinucleotides in vitro (50), and that total mCA levels correlated with DNMT3B expression (5,51). Knockout of DNMT3B diminished mCA levels in hESC (8). Also, two independent studies from Morselli et al. and Baubec et al. demonstrated that DNMT3B localizes with H3K36me3 (7,20). These observations led us to study the connection among mCA deposition, DNMT3B and its chromatin interaction in H1 hESC.
One of the challenges in this study was to identify DNMT3B-binding loci in hESC. High quality endogenous DNMT3B ChIP is known to be difficult to perform, partially due to weak binding of DNMT3B to chromatin. More problematic was the lack of ChIP-grade DNMT3B antibodies (41) (Supplementary Figure S14). We screened a panel of custom-made DNMT3B mouse and rabbit monoclonal antibodies and selected a specific antibody for ChIP experiments. Our endogenous DNMT3B ChIP-seq result is consistent with a previous flag-tagged Dnmt3b ChIP-seq performed in mESC (20). Both results suggest that DNMT3B colocalizes with H3K36me3. Furthermore, our analysis revealed that the mCA distribution mirrors the DNMT3B distribution in hESC.
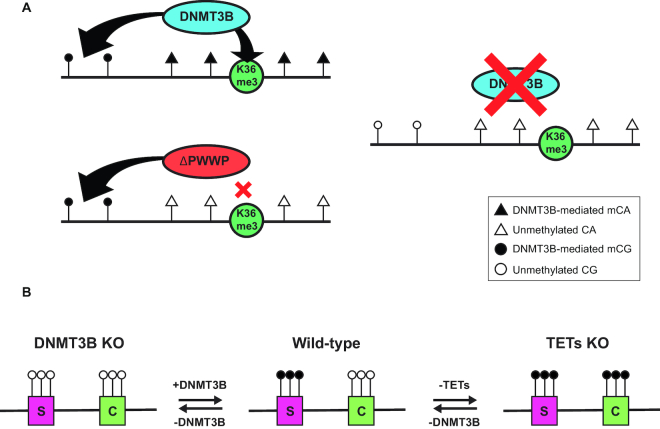
One test whether DNMT3B deposits mCA is to profile the mCA landscape in DNMT3B-null H1 hESC. There are more than 20 DNMT3B isoforms reported, and most do not contain the C5 methyltransferase domain (52,53). It has been reported that DNMT3B isoforms that lack the methyltransferase domain could contribute to DNA methylation through forming complexes with DNMT3A harboring an intact catalytic domain (54,55). Instead of deleting the C5 methyltransferase domain of DNMT3B as described by Liao et al. (8), we decided to introduce a frameshift mutation in DNMT3B exon 2, which is shared by all annotated DNMT3B isoforms, to disrupt all functional DNMT3B enzymes. Although we employed a different KO strategy, our observation is similar to Liao et al.’s report. We observed that knockout of DNMT3B largely reduced the deposition of mCA, with only minimal changes in mCG. This may be due to active maintenance of mCG in the cell by DNMT1 (8). However, DNMT1 cannot maintain asymmetric methylation such as mCA (3,56). Thus, we strengthen the evidence that DNMT3B is a de novo methyltransferase as well as a maintenance methyltransferase for mCA (Figure 7A).
Figure 7.
Model of how DNMT3B maintains the DNA methylation landscape in hESC. (A) DNMT3B deposits mCA through interactions with H3K36me3 (K36me3). DNMT3B without the PWWP domain (ΔPWWP) cannot interact with H3K36me3 and results in loss of mCA but not mCG. (B) DNMT3B maintains the fidelity of the bivalent promoter landscape. Unmethylated spurious bivalent promoters (labeled as S) emerge after DNMT3B KO. From a previous study (Verma et al., 2017), DNMT3B can methylate canonical bivalent promoters (labeled as C) in the absence of TET proteins. In the model, black circles represent methylated CA (A) or methylated CG (B) and white circles represent unmethylated CA (A) or unmethylated CG (B).
The function of mCA is mainly unknown. Some studies have shown that mCA may regulate gene silencing (57). Our observations of the mCA landscape indicate that mCA may be involved in the regulation of splicing. We observed that DNMT3B deposition and mCA levels always increase at splice junctions, presumably as the result of H3K36me3 distribution, which is affected by splicing. Interestingly, recent studies found that mCA could be recognized by MeCP2 (58,59). It is reported that MeCP2 could regulate alternative splicing through DNA methylation (60). Thus, it is possible that mCA could affect alternative splicing.
Although there are minor changes on mCG, we observed some dnHMRs after DNMT3B KO. The observation suggested that DNMT3B regulates mCG at specific loci. Intriguingly, most of these dnHMRs are found at annotated promoters. The previous report in mESC found that dnHMRs after deletion of DNA methyltransferase are coated with H3K27me3 but not H3K4me3 (48). However, we found that these dnHMRs are marked by both H3K27me3 and H3K4me3 in hESC. We defined these dnHMRs with both H3K27me3 and H3K4me3 as spurious bivalent promoters. The discrepancy between our observations with other studies may be explained by the different stage of pluripotency between murine and human ESC. We found that these spurious bivalent promoters are clustered in differentiation genes, and genes involve in the angiogenesis pathway. This finding may shed light on explaining why DNMT3B knockout mouse embryos die at an embryonic stage (61), as deficiency of blood vessel formation was reported in Dnmt3b-/- mouse embryos (62).
The PWWP domain in DNMT3B is known to guide the enzyme to active gene bodies coated with H3K36me3. Disrupting the PWWP domain interrupts DNMT3B–histone interactions, and this leads to the loss of gene-body binding preference in mutant DNMT3B (Supplementary Figures S1 and S12). Intriguingly, similar to its ΔPWWP DNMT3B distribution profile, the mCA in ΔPWWP cells was distributed across the genome without obvious preference for any genomic loci. Furthermore, both the ΔPWWP DNMT3B and mCA distributions are distinct from that observed in WT cells. These findings suggest that loss of the PWWP domain could disrupt the mCA landscape observed in WT cells. Moreover, ΔPWWP cells have a higher level of global mCA than KO cells, suggesting that DNMT3B without the PWWP domain may retain the methyltransferase activity and could randomly deposit mCA across the genome. ΔPWWP cells have lower global mCA levels than that in WT cells. This may be due to the loss of interaction between ΔPWWP DNMT3B and H3K36me3, which led to reduced deposition of mCA across the genome. Nevertheless, we cannot rule out the possibility that the lower expression level of ΔPWWP DNMT3B in the CRISPR-line also contributed to the observation of lower mCA level in ΔPWWP cells.
PWWP domain is thought to be important for mCG de novo methylation (20,21,49). However, our data suggested that DNMT3B could maintain mCG without PWWP domain. DNMT3B does not require on H3K36me3 interaction for gene-body mCG deposition or maintenance. Intriguingly, despite of the lower expression of ΔPWWP DNMT3B, the mCG landscape was well-maintained in ΔPWWP cells. Nonetheless, we cannot rule out the possibility that ΔPWWP DNMT3B could complex with DNMT3A (Supplementary Figure S10F) harboring an intact PWWP domain to maintained mCG at those loci.
In Figures 4D and 6E, we observed the H3K36me3 ChIP-seq signal demonstrated a slight decrease in the WT cells. Since the WT H3K36me3 ChIP-seq data were downloaded from ENCODE, it is likely that the observed difference in the WT H3K36me3 ChIP-seq was caused by using different antibodies or ChIP-seq protocols in the studies. We have shown that the H3K36me3 levels were the same in WT, ΔPWWP and KO cells (Supplementary Figure S10D). The discrepancy observed in the WT H3K36me3 ChIP-seq is unlikely to affect the interpretation of our results or our conclusions.
Recently, Verma et al. reported that TET proteins could act antagonistically with DNMT3B in terms of regulating DNA methylation of bivalent promoters (41). Our study complements their finding on the role of DNMT3B on bivalent promoter methylation. They found that TET proteins could safeguard bivalent promoters from de novo methylation deposited by DNMT3B, whereas we found that DNMT3B protects lineage-specific promoters from becoming unmethylated spurious bivalent promoters. Altogether, the results strongly suggest that mCG on bivalent promoters is mainly deposited by DNMT3B in hESCs.
We propose that bivalent promoters consist of two types: canonical bivalent promoter and spurious bivalent promoters. The native landscape of bivalent promoters is pre-defined by unknown factors in hESC. DNMT3B could methylate both types of bivalent promoters, and TET proteins could demethylate canonical bivalent promoters. In WT hESC, in which DNMT3B and TET proteins co-exist, the canonical bivalent promoters are unmethylated and spurious bivalent promoters are methylated. When TET proteins are depleted, canonical bivalent promoters methylated by DNMT3B could not be reversed, leading to the loss of bivalent marks and subsequently a defect in the ability to differentiate. When DNMT3B is depleted instead, both canonical and spurious bivalent promoters could not be methylated, the spurious bivalent promoter regains bivalent marks, and spurious bivalent promoters may lead to a differentiation defect during development (Figure 7B).
Our study highlights the role of DNMT3B in determining the mCA landscape and bivalent promoter landscape in human pluripotent stem cells. We provide evidence to demonstrate that DNMT3B shapes the mCA landscape by following the guide of its PWWP domain to H3K36me3 coated loci. Surprisingly, DNMT3B–H3K36me3 interaction is not essential for mCG deposition. In addition, these studies revealed the connection between DNMT3B and bivalent promoters. By integrating recent findings on TET-null hESCs, we propose a model to describe the methylation status of the bivalent promoters, which is controlled by both DNMT3B and TET proteins. Hypermethylation on bivalent promoters demolishes bivalent marks, whereas demethylation restores bivalent marks. An improper bivalent promoter landscape may lead to a defect in the differentiation potential of hESCs.
AVAILABILITY OF DATA AND MATERIALS
The NGS data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO) under accession number GSE113881.
Supplementary Material
ACKNOWLEDGEMENTS
Authors’ contributions: A.D.R. and D.G.T. proposed the project. H.K.T., C.S.W., A.D.R. and D.G.T. designed the experiments. H.K.T. and Z.H.T. performed experiments. J.R.H. and C.J.F. produced anti-human DNMT3B rabbit monoclonal antibodies. C.S.W., J.L. and H.Y. performed bioinformatics analyses. H.K.T., C.S.W., A.D.R. and D.G.T. wrote the manuscript. All authors participated in discussion of the manuscript. All authors read and approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the Singapore Ministry of Health’s National Medical Research Council under its Singapore Translational Research (STaR) Investigator Award, the National Research Foundation Singapore and the Singapore Ministry of Education under its Research Centres of Excellence initiative; the Singapore Ministry of Education Academic Research Fund Tier 3 [MOE2014-T3-1-006 to D.G.T.]; the National Cancer Institute of the National Institutes of Health [R35CA197697 and CA66996 to D.G.T., R00CA188595 to A.D.R.]; the National Heart, Lung, and Blood Institute of the National Institutes of Health [HL131477 to D.G.T.]; the Italian Association for Cancer Research (AIRC) [Start-up 15347 to A.D.R.]; the Giovanni Armenise-Harvard Foundation (to A.D.R.). Funding for open access charge: [MOE2014-T3-1-006].
Conflict of interest statement. None declared.
REFERENCES
- 1. Smith Z.D., Meissner A.. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013; 14:204–220. [DOI] [PubMed] [Google Scholar]
- 2. Chan S.W., Henderson I.R., Jacobsen S.E.. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 2005; 6:351–360. [DOI] [PubMed] [Google Scholar]
- 3. Ramsahoye B.H., Biniszkiewicz D., Lyko F., Clark V., Bird A.P., Jaenisch R.. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:5237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.-M et al.. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009; 462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ziller M.J., Müller F., Liao J., Zhang Y., Gu H., Bock C., Boyle P., Epstein C.B., Bernstein B.E., Lengauer T. et al.. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLos Genet. 2011; 7:e1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Laurent L., Wong E., Li G., Huynh T., Tsirigos A., Ong C.T., Low H.M., Kin Sung K.W., Rigoutsos I., Loring J. et al.. Dynamic changes in the human methylome during differentiation. Genome Res. 2010; 20:320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morselli M., Pastor W.A., Montanini B., Nee K., Ferrari R., Fu K., Bonora G., Rubbi L., Clark A.T., Ottonello S. et al.. In vivo targeting of de novo DNA methylation by histone modifications in yeast and mouse. eLife. 2015; 4:e06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liao J., Karnik R., Gu H., Ziller M.J., Clement K., Tsankov A.M., Akopian V., Gifford C.A., Donaghey J., Galonska C. et al.. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat. Genet. 2015; 47:469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhu H., Wang G., Qian J.. Transcription factors as readers and effectors of DNA methylation. Nat. Rev. Genet. 2016; 17:551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012; 13:484–492. [DOI] [PubMed] [Google Scholar]
- 11. Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. et al.. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006; 125:315–326. [DOI] [PubMed] [Google Scholar]
- 12. Azuara V., Perry P., Sauer S., Spivakov M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M. et al.. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006; 8:532–538. [DOI] [PubMed] [Google Scholar]
- 13. Sharov A.A., Nishiyama A., Piao Y., Correa-Cerro L.S., Amano T., Thomas M., Mehta S., Ko M.S.. Responsiveness of genes to manipulation of transcription factors in ES cells is associated with histone modifications and tissue specificity. BMC Genomics. 2011; 12:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Badeaux A.I., Shi Y.. Emerging roles for chromatin as a signal integration and storage platform. Nat. Rev. Mol. Cell Biol. 2013; 14:211–224. [PubMed] [Google Scholar]
- 15. Voigt P., Tee W.W., Reinberg D.. A double take on bivalent promoters. Genes Dev. 2013; 27:1318–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao X.D., Han X., Chew J.L., Liu J., Chiu K.P., Choo A., Orlov Y.L., Sung W.K., Shahab A., Kuznetsov V.A. et al.. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007; 1:286–298. [DOI] [PubMed] [Google Scholar]
- 17. Pan G., Tian S., Nie J., Yang C., Ruotti V., Wei H., Jonsdottir G.A., Stewart R., Thomson J.A.. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007; 1:299–312. [DOI] [PubMed] [Google Scholar]
- 18. Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P. et al.. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007; 448:553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gu T., Lin X., Cullen S.M., Luo M., Jeong M., Estecio M., Shen J., Hardikar S., Sun D., Su J. et al.. DNMT3A and TET1 cooperate to regulate promoter epigenetic landscapes in mouse embryonic stem cells. Genome Biol. 2018; 19:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baubec T., Colombo D.F., Wirbelauer C., Schmidt J., Burger L., Krebs A.R., Akalin A., Schübeler D.. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015; 520:243–247. [DOI] [PubMed] [Google Scholar]
- 21. Neri F., Rapelli S., Krepelova A., Incarnato D., Parlato C., Basile G., Maldotti M., Anselmi F., Oliviero S.. Intragenic DNA methylation prevents spurious transcription initiation. Nature. 2017; 543:72–77. [DOI] [PubMed] [Google Scholar]
- 22. Ge Y.-Z., Pu M.-T., Gowher H., Wu H.-P., Ding J.-P., Jeltsch A., Xu G.-L.. Chromatin targeting ofde NovoDNA methyltransferases by the PWWP Domain. J. Biol. Chem. 2004; 279:25447–25454. [DOI] [PubMed] [Google Scholar]
- 23. Chen T., Tsujimoto N., Li E.. The PWWP domain of Dnmt3a and Dnmt3b Is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol. Cell Biol. 2004; 24:9048–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. He Y., Ecker J.R.. Non-CG methylation in the human genome. Annu. Rev. Genomics Hum. Genet. 2015; 16:55–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xia X., Zhang Y., Zieth C.R., Zhang S.-C.. Transgenes delivered by lentiviral vector are suppressed in human embryonic stem cells in a promoter-dependent manner. Stem Cells Dev. 2007; 16:167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yazdi P.G., Pedersen B.A., Taylor J.F., Khattab O.S., Chen Y.H., Chen Y., Jacobsen S.E., Wang P.H.. Nucleosome organization in human embryonic stem cells. PLoS One. 2015; 10:e0136314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krueger F., Andrews S.R.. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011; 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jühling F., Kretzmer H., Bernhart S.H., Otto C., Stadler P.F., Hoffmann S.. metilene: fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome Res. 2016; 26:256–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G.. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010; 28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L.. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 39. Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shen L., Shao N., Liu X., Nestler E.. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014; 15:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Verma N., Pan H., Doré L.C., Shukla A., Li Q.V., Pelham-Webb B., Teijeiro V., González F., Krivtsov A., Chang C.-J. et al.. TET proteins safeguard bivalent promoters from de novo methylation in human embryonic stem cells. Nat. Genet. 2017; 50:83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li B., Howe L., Anderson S., Yates J.R., Workman J.L.. The set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 2003; 278:8897–8903. [DOI] [PubMed] [Google Scholar]
- 43. Vojnic E., Simon B., Strahl B.D., Sattler M., Cramer P.. Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36Methylation to transcription. J. Biol. Chem. 2006; 281:13–15. [DOI] [PubMed] [Google Scholar]
- 44. de Almeida S.F., Grosso A.R., Koch F., Fenouil R., Carvalho S., Andrade J., Levezinho H., Gut M., Eick D., Gut I. et al.. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat. Struct. Mol. Biol. 2011; 18:977–983. [DOI] [PubMed] [Google Scholar]
- 45. Spies N., Nielsen C.B., Padgett R.A., Burge C.B.. Biased chromatin signatures around polyadenylation sites and exons. Mol. Cell. 2009; 36:245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schwartz S., Meshorer E., Ast G.. Chromatin organization marks exon-intron structure. Nat. Struct. Mol. Biol. 2009; 16:990–995. [DOI] [PubMed] [Google Scholar]
- 47. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. King Andrew D., Huang K., Rubbi L., Liu S., Wang C.-Y., Wang Y., Pellegrini M., Fan G.. Reversible regulation of promoter and enhancer histone landscape by DNA methylation in mouse embryonic stem cells. Cell Rep. 2016; 17:289–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rondelet G., Dal Maso T., Willems L., Wouters J.. Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B. J. Struct. Biol. 2016; 194:357–367. [DOI] [PubMed] [Google Scholar]
- 50. Aoki A., Suetake I., Miyagawa J., Fujio T., Chijiwa T., Sasaki H., Tajima S.. Enzymatic properties of de novo-type mouse DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 2001; 29:3506–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee J.-H., Park S.-J., Nakai K.. Differential landscape of non-CpG methylation in embryonic stem cells and neurons caused by DNMT3s. Sci. Rep. 2017; 7:11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gopalakrishnan S., Van Emburgh B.O., Shan J., Su Z., Fields C.R., Vieweg J., Hamazaki T., Schwartz P.H., Terada N., Robertson K.D.. A novel DNMT3B splice variant expressed in tumor and pluripotent cells modulates genomic dna methylation patterns and displays altered DNA binding. Mol. Cancer Res. 2009; 7:1622–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ostler K.R., Davis E.M., Payne S.L., Gosalia B.B., Expósito-Céspedes J., Beau M.M.L., Godley L.A.. Cancer cells express aberrant DNMT3B transcripts encoding truncated proteins. Oncogene. 2007; 26:5553–5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Duymich C.E., Charlet J., Yang X., Jones P.A., Liang G.. DNMT3B isoforms without catalytic activity stimulate gene body methylation as accessory proteins in somatic cells. Nat. Commun. 2016; 7:11453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gordon C.A., Hartono S.R., Chedin F.. Inactive DNMT3B splice variants modulate de novo DNA methylation. PLoS One. 2013; 8:e69486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lin I.G., Han L., Taghva A., O’Brien L.E., Hsieh C.L.. Murine de novo methyltransferase Dnmt3a demonstrates strand asymmetry and site preference in the methylation of DNA in vitro. Mol. Cell Biol. 2002; 22:704–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Barrès R., Osler M.E., Yan J., Rune A., Fritz T., Caidahl K., Krook A., Zierath J.R.. Non-CpG methylation of the PGC-1α promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009; 10:189–198. [DOI] [PubMed] [Google Scholar]
- 58. Greally J.M., Lagger S., Connelly J.C., Schweikert G., Webb S., Selfridge J., Ramsahoye B.H., Yu M., He C., Sanguinetti G. et al.. MeCP2 recognizes cytosine methylated tri-nucleotide and di-nucleotide sequences to tune transcription in the mammalian brain. PLos Genet. 2017; 13:e1006793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen L., Chen K., Lavery L.A., Baker S.A., Shaw C.A., Li W., Zoghbi H.Y.. MeCP2 binds to non-CG methylated DNA as neurons mature, influencing transcription and the timing of onset for Rett syndrome. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:5509–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Maunakea A.K., Chepelev I., Cui K., Zhao K.. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013; 23:1256–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Okano M., Bell D.W., Haber D.A., Li E.. DNA methyltransferases Dnmt3a and Dnmt3b Are Essential for De novo methylation and mammalian development. Cell. 1999; 99:247–257. [DOI] [PubMed] [Google Scholar]
- 62. Ueda Y. Roles for Dnmt3b in mammalian development: a mouse model for the ICF syndrome. Development. 2006; 133:1183–1192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The NGS data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO) under accession number GSE113881.