Abstract
The first product of sulfate assimilation in plants, cysteine, is a proteinogenic amino acid and a source of reduced sulfur for plant metabolism. Cysteine synthesis is the convergence point of the three major pathways of primary metabolism: carbon, nitrate, and sulfate assimilation. Despite the importance of metabolic and genetic coordination of these three pathways for nutrient balance in plants, the molecular mechanisms underlying this coordination, and the sensors and signals, are far from being understood. This is even more apparent in C4 plants, where coordination of these pathways for cysteine synthesis includes the additional challenge of differential spatial localization. Here we review the coordination of sulfate, nitrate, and carbon assimilation, and show how they are altered in C4 plants. We then summarize current knowledge of the mechanisms of coordination of these pathways. Finally, we identify urgent questions to be addressed in order to understand the integration of sulfate assimilation with carbon and nitrogen metabolism particularly in C4 plants. We consider answering these questions to be a prerequisite for successful engineering of C4 photosynthesis into C3 crops to increase their efficiency.
Keywords: C4 photosynthesis, cysteine, evolution, Flaveria, primary metabolism, sulfate assimilation
This review summarizes current knowledge on the coordination of sulfur, nitrogen, and carbon metabolism and discusses sulfate assimilation in the context of evolution of C4 photosynthesis.
Introduction
Sulfur is an essential element for all living organisms, and the amino acid cysteine is perhaps the most important sulfur-containing compound in biology. Cysteine represents a remarkably versatile, highly reactive molecule involved in a number of physiological reactions, bestowing upon it a pivotal role in plant primary and secondary metabolism (Takahashi et al., 2011). The chemical reactivity of cysteine is a consequence of the thiol moiety present in its molecular structure, derived from a series of reductive enzymatic reactions in the sulfate assimilation pathway. The large atomic radius of sulfur in addition to the low dissociation energy of the S–H bond confer on cysteine the unique ability to perform both nucleophilic and redox-active functions (Pace and Weerapana, 2013).
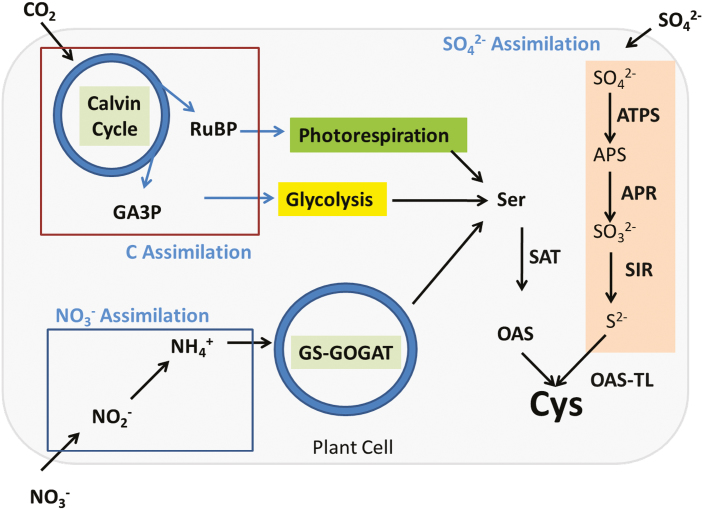
Furthermore, cysteine biosynthesis is the merging point of carbon, nitrogen, and sulfur assimilation pathways. In a simplified view, cysteine biosynthesis can be depicted in three subprocesses involving: (i) the photosynthetic assimilation of carbon dioxide (CO2), that provides the carbon backbone; (ii) the assimilation of nitrogen and its incorporation into the carbon backbone, resulting, among others, in the amino acid serine; and, finally, (iii) the reduction of inorganic sulfate into sulfide and its incorporation into the serine-derived organic compound O-acetylserine (Fig. 1; reviewed in Takahashi et al. 2011). The resulting cysteine represents the first form of reduced organic sulfur from primary metabolism in plant cells, and is therefore an important bioavailable source of sulfur for subsequent metabolic reactions (Pivato et al., 2014).
Fig. 1.
Conceptualized plant cell showing the convergence of carbon assimilation through the Calvin cycle, nitrate assimilation through reduction and the GS–GOGAT cycle, and sulfate assimilation. These three pathways converge at cysteine synthesis, making cysteine a keystone metabolite connecting primary metabolism.
The convergence of these three essential assimilatory pathways in plant metabolism raises a number of underexplored questions, starting with the coordination of mineral nutrition, namely sulfur and nitrogen, with photosynthetic capacity and performance. While the molecular components of the assimilatory pathways of carbon, nitrogen, and sulfur metabolism were uncovered in the last decades, the regulatory mechanisms integrating these pathways and their interaction are still relatively poorly understood. Scarce information exists regarding the simultaneous coordination and regulation of these pathways in the whole plant system.
In addition, evolutionary divergence of carbon assimilation strategies in higher plants imposes an extra layer of complexity on interactions with the assimilation of nitrate and sulfate, the main inorganic sources of nitrogen and sulfur in the soil. Although all plants ultimately use the enzyme Rubisco to fix CO2 into organic acid molecules, some species have evolved additional carbon-concentrating mechanisms (CCMs) to minimize the oxygenase activity of Rubisco and consequently increase carbon assimilation efficiency(Leegood, 2002).
C4 photosynthesis is the most extensively studied example of such adaptation, and can best be described as an interdependent blend of modified biochemistry, anatomy, and structural mechanisms coordinated by complex molecular entities that ultimately lead to a cell-specific spatial compartmentalization of photosynthesis-related biochemical processes (Hibberd and Covshoff, 2010). Interestingly, the cell-specific distribution of the biochemical CO2 assimilation apparatus between bundle sheath (BS) and mesophyll (M) cells observed in species employing C4 photosynthesis seems to extend to enzymatic mechanisms involved in nitrogen and sulfur assimilation, which segregate their components between these two cell types(Kopriva and Koprivova, 2005). Hence, a high degree of coordination among these processes must take place to achieve optimal growth and development under fluctuations in any of the components. While the spatial separation of sulfur assimilation machinery in C4 plants was first observed >40 years ago and has been revisited more recently with a molecular emphasis, its physiological relevance as well as its regulation and coordination with CO2 assimilation are still unknown (Weckopp and Kopriva, 2014).
Considering the number of key roles reduced sulfur compounds play in biological processes, one could argue that cysteine, while often overlooked from a nutritional perspective, is a keystone nutritional compound linking together sulfur, carbon, and nitrogen metabolism, not just in plants but in all living organisms. In this review, we will qualify this assertion by briefly presenting the biochemistry linking together carbon, nitrogen, and sulfur metabolism in plants. We will then highlight the implications this may have for C4 plants, in which these three pathways are spatially separated between two distinct cell types. Finally, we will discuss critical knowledge gaps in our understanding of plant sulfur metabolism in the context of C3 and C4 metabolism and beyond.
Cysteine: a keystone nutritional compound
Cysteine synthesis connects the assimilation of three major nutrients: carbon, nitrogen, and sulfur (Fig. 1). Carbon required for the carbon backbones of all amino acids and organic compounds is provided by the carbon reduction reactions of photosynthesis. Key among these metabolites with respect to sulfur assimilation is the amino acid serine. Serine synthesis can occur in both photosynthetic and non-photosynthetic plant cells. In plants, there are three different biochemical pathways producing serine: photorespiration, glycolysis, and the so-called ‘phosphorylated pathway’ (Fig. 1) (Ros et al., 2014; Krueger et al., 2017). For both glycolysis and the phosphorylated pathway, serine biosynthesis begins following the generation of 3-phosphoglycerate (3-PGA) derived either from the Calvin cycle or from the oxidation of sugars in glycolysis. The phosphorylated pathway is particularly important in heterotrophic tissues (Ros et al., 2014); however, in green leaves, the major pathway of serine synthesis is photorespiration, which forms one molecule of serine from two glycine molecules by the concerted action of glycine decarboxylase (GDC) and serine hydroxymethyltransferase in the mitochondria. Correspondingly, disruption of photorespiratory serine synthesis in the Arabidopsis bou-2 mutant affected leaf sulfur metabolism to a higher degree than disruption of the phosphorylated pathway (Samuilov et al., 2018a, b), which played an important role in controlling sulfur fluxes in non-photosynthetic tissues (Anoman et al., 2019).Once serine is produced, the enzyme serine acetyltransferase catalyzes the acetylation of serine to provide O-acetylserine (OAS), which is a direct precursor of cysteine (Fig. 1).
For the nitrogen found in amino acids, including serine, and other metabolites, plants take up nitrate from the soil and reduce it to ammonium, which is incorporated into amino acids. The first enzyme involved in nitrate assimilation is nitrate reductase, which catalyzes the reduction of nitrate to nitrite. Nitrite is toxic for plant cells, and is immediately transported from the cytosol into plastids, where it is reduced by nitrite reductase into ammonium. The ammonium generated is then used by the glutamate synthetase–glutamine oxoglutarate aminotransferase (GS–GOGAT) cycle to produce glutamate and glutamine (Krapp, 2015).
The last constituent of cysteine is sulfur. Plant sulfate assimilation requires a complex series of biochemical reactions (reviewed by Koprivova and Kopriva, 2014). Sulfate, which is taken up at the plant root by sulfate transporters, is very stable and requires activation by ATP to form adenosine 5'-phosphosulfate (APS). The enzyme involved in this step of the pathway is ATP sulfurylase. There are two divergent pathways utilizing APS, one for the creation of reduced sulfur compounds and one for the synthesis of sulfated compounds; however, the pathway leading to reduced sulfur compounds is the dominant pathway in most circumstances and occurs exclusively in the plastids. To produce sulfite, two electrons are transferred from glutathione (GSH) to APS by APS reductase (APR). Reduction of sulfite is catalyzed by sulfite reductase (SIR), and the resultant sulfide reacts with OAS for cysteine synthesis. The cysteine-producing reaction is catalyzed by O-acetylserine (thiol)lyase (OAS-TL) (Fig. 1).
Cysteine as an indicator of evolution
Cysteine residues usually account for one of the less abundant amino acids in proteins, although evolutionary studies show an increased frequency of cysteine incorporation into proteins in different taxa over time. Evidence suggests that cysteine enrichment in proteomes may reflect the evolution of the genetic code itself, and consequently the amino acid composition relative to ancestral proteins, implying that the usage of cysteine may further expand in descendants (Brooks et al., 2002; Jordan et al., 2005; Liu et al., 2010; Yampolsky et al., 2017). One plausible explanation for this evolutionary increase in cysteine abundance in the amino acid composition of proteins might be related to the physicochemical properties of this molecule. Despite the low frequency, cysteine residues are more frequent and prevalent at functionally important sites within protein scaffolds, such as the CxxC motifs characteristic of zinc fingers and oxidoreductase active sites, or in regions that are not functionally characterized yet, representing a vast area to explore regarding the possible numerous functional aspects involving cysteine (Pe’er et al., 2004; Poznański et al., 2018).
Examples can be found already in the sulfate assimilation pathway. APR in plants possesses an iron–sulfur cluster as cofactor, which is bound to the protein by two cysteine pairs (Kopriva et al., 2001). These cysteine pairs are not present in related enzymes from yeast and most cyanobacteria, which instead need a second activation step for sulfate reduction from APS to 3'-phosphoadenosine 5'-phosphosulfate (PAPS) (Kopriva et al., 2002a). In addition, several sulfate assimilation enzymes are known to be redox regulated in plants but not in other organisms (Hell and Bergmann, 1990; Bick et al., 2001; Jez et al., 2004, 2016; Hothorn et al., 2006). Redox regulation seems to control the partitioning of sulfur between primary sulfate reduction and secondary sulfation pathways (Koprivova and Kopriva, 2016). While APR is activated by oxidation, the enzyme catalyzing entry of sulfate into secondary metabolism, APS kinase, is activated by reduction (Hothorn et al., 2006). However, while APS kinase is a ubiquitous enzyme, its redox regulation has been described only for plants and, correspondingly, only plant APS kinases possess the redox-active cysteine pair (Ravilious et al., 2012). Another enzyme connected to secondary sulfur metabolism present in all kingdoms of life is the phosphoadenosine phosphate phosphatase SAL1. SAL1 is inactivated by oxidation in a retrograde stress signaling process only in plants and not in other organisms, again relying on additional cysteine residues conserved only in the plant proteins (Chan et al., 2016). The glutamate-cysteine ligase (γ-ECS), which is the first and rate-limiting enzyme in GSH biosynthesis, also exhibits several layers or redox regulation that correlate well with the evolutionary history of the enzyme. While one redox-active cysteine pair is present in γ-ECS proteins from all sources, plant enzymes possess another level of redox regulation by a second cysteine pair. Under oxidizing conditions, the disulfide bridges bring the dimeric complex together and dramatically increase the enzymatic activity, while under reducing conditions the disulfide bridges are reduced, causing the dissociation of the dimer into monomers with substantially decreased activity (Hothorn et al., 2006). Thus, cysteine residues can be indicators of increasing regulatory complexity and evolutionary advancement.
Coordination of carbon, nitrogen, and sulfur metabolism
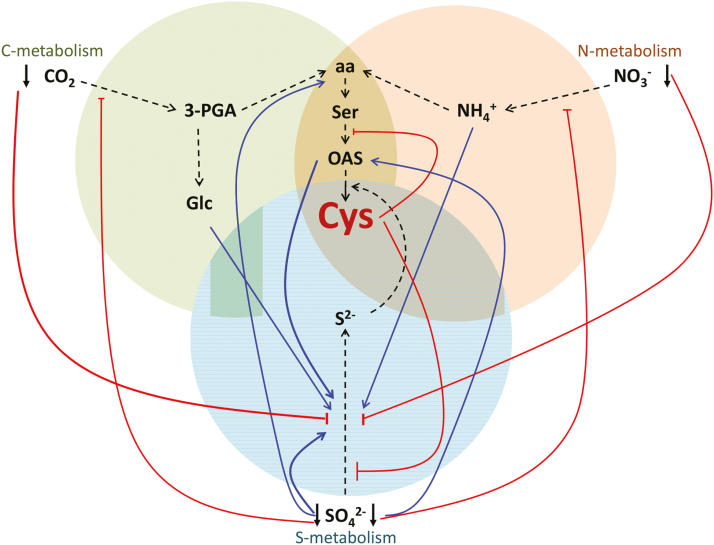
The need for regulatory interconnections between the metabolism of carbon, nitrogen, and sulfur is obvious, but the mechanisms, sensors, and signals are far from understood (Koprivova and Kopriva, 2014). Sulfur nutrition is strongly coordinated with nitrogen due to the need for both in protein synthesis (Fig. 2). Thus, sulfate deficiency reduces nitrate uptake and reduction, and, vice versa, nitrogen deficiency leads to diminished sulfate uptake and reduction rate (Fig. 2). On the other hand, the key assimilatory enzymes APR and nitrate reductase are induced by reduced nitrogen (ammonium and amino acids) and sulfur (cysteine) compounds, respectively (Kopriva and Rennenberg, 2004). In the very few available reports, the coordination of sulfate assimilation with carbon metabolism seems to be similar to nitrogen—the pathway was down-regulated by low CO2 availability and induced by glucose and sucrose (Fig. 2) (Kopriva et al., 2002b; Hesse et al., 2003). Sulfate deficiency results in a general slowdown of metabolism, leading to a decrease in carbon assimilation and photosynthesis, and to a reprogramming of metabolism and developmental programs towards economizing resources for seed production (Nikiforova et al., 2005).
Fig. 2.
Carbon and nitrogen metabolism impact sulfur signaling. Red lines signify repression or down-regulation, while blue lines represent activation or up-regulation. Black lines represent biochemical pathways
While the interconnection of these pathways is well described on physiological as well as on systems biology levels, the sensors and signals triggering the metabolic adaptations are unknown. Among the signals discussed, the cysteine precursor OAS was previously and controversially considered a mediator of plant sulfur status (Hirai et al., 2003; Hopkins et al., 2005) or a signal connecting nitrogen and sulfur metabolism (Koprivova et al., 2000). Using a systems biology approach, Hubberten et al. (2012) identified six genes whose expression was highly correlated with accumulation of OAS, the ‘OAS cluster’, which included genes shown previously to be strongly up-regulated by sulfate deficiency. Thus, OAS displays a signaling function leading to changes in transcript levels of a specific gene set irrespective of the sulfur status of the plant, and seems to play a specific part in the sulfate response. Given the increase in OAS accumulation and OAS cluster transcript levels in conditions not connected to sulfur deficiency (Espinoza et al., 2010; Caldana et al., 2011), the true function of OAS as a signal and the OAS cluster genes might be less linked to sulfur deficiency response but might instead function in more general coordination of the assimilatory pathways.
Another mechanism coordinating sulfur assimilation with other metabolic processes and plant growth has recently been uncovered (Dong et al., 2017). It has long been known that growth of eukaryotic cells is regulated by amino acid availability through a gene called target of rapamycin (TOR). However, the well-established transducing molecules (TOR-interacting proteins: RAG GTPase, TSC1/2, and RHEB) for TOR signaling are absent in plants. Thus, how plants sense amino acids and regulate TOR signaling has been an open question in the field. Interestingly, a recent publication has shown that, unlike in mammalian systems that sense amino acids directly, plants may sense amino acid precursors (Dong et al. 2017). More specifically, in the case of cysteine, the precursors OAS and sulfide (S2−) appear to be selectively sensed and can activate TOR signaling via two distinct pathways. Under conditions of low carbon or nitrogen status, decreased levels of OAS can activate TOR signaling in a GCN2- (general control nonderepressible 2) mediated manner, while under low sulfur status decreased sulfide levels can activate TOR signaling via glucose–TOR signaling (Dong et al., 2017). While uncovering TOR signaling mechanisms represents a major breakthrough in understanding the regulatory integration of sulfur metabolism in global plant metabolism, the downstream mechanisms and transcription factors remain to be identified.
Cysteine synthesis: C4 plants are different
The group of plants with probably the highest divergence in primary metabolism compared with the model plant Arabidopsis is C4 plants. CO2 fixation in C3 photosynthesis is inherently inefficient due to Rubisco’s poor ability to discriminate between CO2 and O2. The oxygenation results in generation of a two-carbon product, 2-phosphoglycolate. Since this compound is toxic, it is rapidly metabolized and a portion of the carbon is regenerated through photorespiration, which incurs significant energy costs (Hagemann and Bauwe, 2016). To overcome this inefficiency, plants using C4 photosynthesis fix CO2 initially into a four-carbon compound (oxaloacetate) via the enzyme phosphoenolpyruvate (PEP) carboxylase, which is not affected by oxygen (Gilbert and Wilhelm, 2019). CO2 is then released from the C4 compounds for re-fixation by Rubisco.
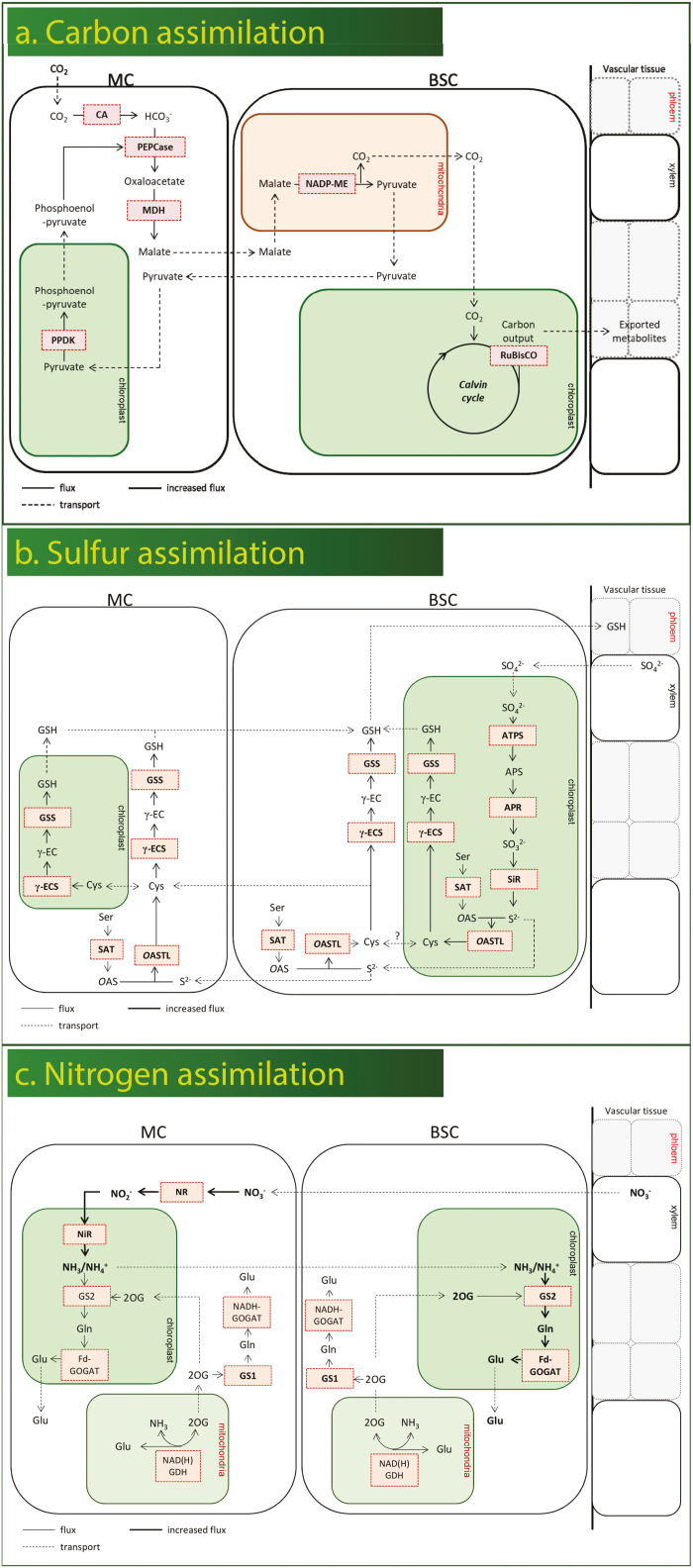
However, for C4 photosynthesis to be more efficient than C3 photosynthesis, these steps must be separated into two compartments—one where PEP carboxylase can capture CO2 from a high O2 environment, and one where Rubisco can fix CO2 in a low O2 environment (Fig. 3a). In most C4 plants, these two processes have been spatially separated into two distinct photosynthetic cell types. C4 M cells perform the C4 carbon capture reactions, while BS cells perform carbon assimilation using Rubisco and the Calvin cycle. Importantly, this compartmentalization requires major alterations in typical C3 leaf structure giving rise to a repeating pattern of cells within C4 leaves. While C3 leaves are structurally characterized by having many M cells between the BS cells and the veins (V), C4 plants tend to form a repeating pattern of V–BS–M–M–BS–V. This cellular arrangement results in fewer M cells, decreased M area, and higher V density in C4 plants compared with C3 plants. Finally, in addition to this cellular rearrangement, C4 plants have more abundant chloroplasts in the BS cells compared with C3 plants. Collectively, these changes in tissue arrangement and BS chloroplast number are referred to as Kranz anatomy (Lundgren et al., 2014). Thus, C4 metabolism requires highly coordinated changes in tissue structure to achieve Kranz anatomy as well as the development of cell type-specific photosynthetic biochemistry to achieve spatial separation of carbon fixation and carbon assimilation (Junqueira et al., 2018). A detailed analysis of monocot species revealed that C4 photosynthesis evolved in lineages with higher BS:M ratios; that is, the anatomical pre-adaptation was the prerequisite for evolution of a full C4 cycle (Christin et al., 2013).
Fig. 3.
Model showing separation of enzymes between the M and BS in carbon assimilation (a), nitrogen assimilation (b), and sulfate assimilation (c) in C4 plants.
C4 photosynthesis conveys improved water use efficiency, making it more efficient than C3 carbon assimilation, particularly in hot, dry, and/or saline environmental conditions where photorespiration is high (Sage et al., 2018). Despite its complexity, this extraordinarily successful metabolic trait has evolved independently >60 times and is present in many of the most successful arid and semi-arid grasses, eudicot herbs and shrubs in low to mid latitudes, and major crops, such as maize, sorghum, and sugar cane (Sage et al., 1999; Still et al., 2003; Edwards et al., 2010). Since its discovery >50 years ago, there has been substantial interest in C4 metabolism, much of which transcends plant metabolism (e.g. see Beerling and Osborne, 2006 for C4 effects on climate; Bobe and Behrensmeyer, 2004; Sage and Zhu, 2011 for information on C4 influence on meat and sugar production).
More recently, interest has grown in engineering C4 metabolism into agronomically important C3 crops, such as rice and wheat, to improve stress tolerance and carbon fixation efficiency (Hibberd et al., 2008; Jones, 2010; von Caemmerer et al., 2012; von Caemmerer and Furbank, 2016; Wang et al., 2016). However, for these approaches to be successful, we must first recognize and understand the metabolic, biochemical, and structural consequences of spatially separating carbon capture from carbon assimilation, which is the hallmark of C4 metabolism. To this end, extensive scientific literature examining these consequences exists for carbon assimilation and, to a much lesser extent, for nitrogen assimilation.
In addition to spatial separation of carbon assimilation, C4 plants have also been shown to spatially separate nitrate assimilation (Fig. 3b). The reduction of nitrate to nitrite occurs exclusively in M cells, while the incorporation of reduced nitrogen into glutamate and glutamine (GS–GOGAT) occurs either in the BS or in the M and BS (Rathnam and Edwards, 1976; Moore and Black, 1979; Becker et al., 2000). C4 species have also been shown to exhibit higher nitrogen use efficiency than C3 species, reportedly due to the strict localization of Rubisco in BS leading to a decreased Rubisco quantity per leaf area (Brown, 1999; Ghannoum et al., 2010). It is unclear if spatial separations of nitrate reduction or the strong decrease in photorespiratory ammonium recycling contribute to improved nitrogen use efficiency in C4 plants. Also, cysteine synthesis might be affected: the restriction of photorespiration to BS of C4 plants could limit the ability of M cells to synthesize serine—an essential precursor of cysteine. Indeed, very little is known regarding how the massive biochemical and structural rearrangements that accompany the evolution of C4 metabolism impact sulfate assimilation and cysteine synthesis.
Sulfate assimilation in C4 plants
As early as the 1970s, there was evidence suggesting that sulfate assimilation in C4 grasses is somewhat different from that in C3 plants. Gerwick and Black (1979) demonstrated, using enzymatic assays, that 90% of the ATPS activity in crabgrass leaves was found specifically in the BS, suggesting that the bulk of sulfate assimilation was occurring in these cells. An extension of this work showed BS localization of ATPS in a broader assessment of 18 different C4 plant species (Gerwick et al., 1980). Shortly afterwards, Schmutz and Brunold (1984) confirmed the ATPS observations of Gerwick et al. in maize and wheat, and further showed that APR and SIR were also preferentially expressed in the BS, while OAS-TL was expressed in both BS and M cells. Burgener et al. (1998) then showed that isolated BS strands export cysteine into the medium, suggesting that in C4 plants, sulfate assimilation and cysteine synthesis occur exclusively in the BS (Fig. 3c).
In an attempt to find out whether the BS localization of sulfate assimilation is a consequence or prerequisite of C4 photosynthesis, Koprivova et al. (2001) analyzed the pathway in plants of the genus Flaveria, which contains species with C3 and C4 photosynthesis as well as a number of C3–C4 intermediate plants that show different degrees of C4 characteristics (Ku et al., 1991). Surprisingly, in both C3 and C4 species of Flaveria, APR transcript and protein localized to both the M and the BS. Because Flaveria is a eudicot species, these results could not be easily reconciled with previous observations in monocot systems and were interpreted as a difference between eudicot and monocot C4 species. Further questions regarding the importance of spatial compartmentalization of sulfate assimilation in C4 plants later came from the C3 model organism Arabidopsis thaliana. Using a systems biology approach, Aubry et al. (2014) obtained a translatome of Arabidopsis BS cells to identify transcripts enriched in this cell type. Surprisingly, they found preferential expression of ATPS and APR, as well as transcripts associated with secondary sulfur metabolism and transport of sulfur-containing compounds in Arabidopsis BS (Aubry et al., 2014). While it has been previously proposed that BS spatial separation of key metabolic enzymes is a pre-condition for evolution of C2 photosynthesis and has been observed for GDC in several Brassicaceae species (Adwy et al., 2015), the significance of BS-specific expression of sulfur enzymes is unclear. Indeed, a recent review proposes that anatomical modifications are the rate-limiting step in the C4 trajectory, not metabolic changes (Edwards, 2019). When viewed in context with the results from Flaveria, two major questions arising from these studies remain: (i) what is the ancestral state/characteristics of plant sulfur metabolism, especially regarding preferential expression in the BS; and (ii) why do we not observe compartmentalization of sulfur assimilation genes in C4Flaveria, the only C4 eudicot to be evaluated to date?
A partial answer to the second question was recently given by detailed analyses of sulfate assimilation in Flaveria species with a gradient of C4 photosynthetic characteristics. Overall, across a range of Flaveria species, cysteine and GSH levels roughly followed the C3–C4 gradient, with C4 lines having the highest foliar thiol levels, C3 species having the lowest, and C2 intermediates showing increasing foliar cysteine and GSH levels with increasing C4 characteristics (Gerlich et al., 2018). However, Gerlich et al. (2018) not only measured sulfur metabolites in these species, but also performed reciprocal grafting experiments to evaluate the contribution of roots to sulfur nutrition in Flaveria. Surprisingly, they found that in C4 but not C3Flaveria, both sulfate assimilation and GSH biosynthesis occur predominantly in the roots (Gerlich et al., 2018). This suggests that compartmentalization of sulfur assimilation also occurs in eudicot C4 plants; however, the separation does not take place between M and BS cells as observed in C4 monocots, but rather between roots and shoots. Interestingly, the study also suggested that the localization of sulfate assimilation and GSH synthesis may be driven by serine synthesis, proposing a new avenue for exploration of the control of the spatial localization (Gerlich et al., 2018). This important discovery shows that during the transition from C3 to C4 metabolism, multiple evolutionary solutions leading to alteration in C4 sulfur metabolism may exist (Williams et al., 2013). Furthermore, this study highlights the importance of evaluating the contributions of root nitrogen and sulfur metabolism in the evolution of C4 metabolism, which in the case of sulfur metabolism has been poorly appreciated in monocots to date. From an engineering perspective, characterizing these natural evolutionary solutions may help us better understand how to overcome genetic obstacles and transfer C4 characteristics into C3 crops. Regarding the ancestral state or ancestral characteristics of sulfate assimilation, this question is more difficult to address at this time as cell-specific characterization of sulfur metabolism has been performed in very few C3 species.
The differential spatial localization of sulfate and nitrate assimilation in C4 plants indicates that cysteine may play an additional role in metabolic coordination. Indeed, experiments performed in maize suggest that, unlike in C3 plants, cysteine but not GSH triggers the demand-driven control of sulfate assimilation (Bolchi et al., 1999). In this experiment, sulfur-starved maize seedlings were treated with buthionine sulfoximine (BSO), a potent inhibitor of glutathione biosynthesis, and subsequently fed either l-cysteine or d-cysteine. Only seedlings fed with l-cysteine showed a decreased sulfur deficiency response, suggesting that the effect was not due to thiol feeding alone, but was specific for the biologically relevant form of cysteine and not dependent on GSH biosynthesis, as in C3 plants (Bolchi et al., 1999; Lappartient et al., 1999). This experiment has suggested that cysteine is the major regulator of sulfate assimilation in maize—a role typically associated with GSH.
Knowledge gaps and new directions
Perhaps the most pressing and difficult to answer questions surrounding sulfate (and nitrate) assimilation in C4 plants are centered on the consequences of confining sulfate assimilation to the BS (and nitrate assimilation to the M). Given the number of independent evolutionary origins of C4 photosynthesis, why is this spatial configuration so often recruited by C4 plants and, importantly, why are there exceptions, as seems to be the case for Flaveria? Additionally, why is nitrate reduction confined to M and sulfate reduction confined to BS in some species? Both of these processes are metabolically expensive in terms of energy and redox equivalents. Thus, if energy or redox status were providing the selective force to drive this metabolic rewiring, we might expect that sulfate and nitrate reduction would be confined to the same cell type. Despite the surprising metabolic differences recently identified in sulfur metabolism of C3 and C4Flaveria roots, it remains unknown if these metabolic alterations were accompanied by changes in root nitrogen metabolism. Considering the well-documented alterations in Flaveria leaf nitrogen metabolism between C3 and C4 species and the potential role these metabolic changes played in driving the transition toward C4 metabolism, additional studies to unravel root nitrogen metabolism in Flaveria are warranted (Mallmann et al., 2014). It also needs to be tested whether M-specific localization of nitrate reduction might be a consequence of the C2 cycle. The confinement of GDC to the BS results in a net transfer of ammonium from the M to BS (Mallmann et al., 2014), which might be counteracted by an increase in nitrate reduction rate in M and a decrease in BS. A more thorough investigation of nitrogen metabolism in C2 and C3–C4 plants will help to test this hypothesis. Finally, it is unclear if partitioning of sulfate assimilation is a consequence of C4 metabolism, or if it is a prerequisite for C4 metabolism. Answers to these questions are likely to be connected to one another and may be revealed through focused genetic and biochemical studies aided by systems biology and evolutionary insights, particularly as such analyses are now feasible in non-model organisms. Thus, we would like to highlight a few promising directions that might help identify key regulatory nodes of sulfur metabolism in C4 plants and pave the way to deeper mechanistic understanding.
Transcriptional regulators
Despite the realization that sulfate assimilation undergoes spatial reconfiguration in C4 plants and to some extent also in C3 eudicots, little work in understanding the molecular basis for this restructuring has been performed. Generally, very little is known about transcriptional regulators of sulfate assimilation beyond Arabidopsis. For example, homologs of the main transcriptional regulator of sulfur deficiency response in Arabidopsis, SLIM1, have been identified in maize and other monocots; however, to date, none of these has been linked with sulfur nutrition in these organisms (Gallie and Young, 2004; Mao et al., 2006). However, two rice homologs of SLIM1 were able to complement the Arabidopsis mutant, suggesting that they are functionally conserved in C3 monocots. It remains an open question as to whether SLIM1 homologs from C4 plants are also involved in sulfur responses and if they have been recruited to orchestrate some of the changes observed in C4 sulfur metabolism. However, a function for SLIM1 in the spatial distribution of the pathway is unlikely as SLIM1 transcript does not show such a localization in Arabidopsis (Aubry et al., 2014). Answers to these questions will probably require cell-specific investigations.
Transporters
The differential spatial distribution of the metabolic pathways can function only if the substrates, intermediates, and products can be easily transported between the cells. However, very little is known regarding the cell to cell transport of sulfur metabolites, including cysteine and GSH. Our current understanding of C4 sulfate assimilation suggests that primary cysteine synthesis occurs exclusively in the BS. Thus, cysteine transport to the M and other cell types is necessary (Burgener et al., 1998; Kopriva and Koprivova, 2005). Plants are known to possess amino acid transporters, and many of them are capable of transporting cysteine (Miranda et al., 2001; Tegeder, 2012); however, specific genes involved in establishing or maintaining this spatial gradient have not been identified to date. It is currently unclear if specific cysteine transporters exist in plants or if cysteine transport occurs via permeases or some unidentified mechanism. It is also unknown whether sulfate is transported into M cells to contribute to anion homeostasis. Similarly, nothing is known about the oxidized pathway of sulfate assimilation. For example, is APS kinase also differently expressed between M and BS? Are sulfotransferases catalyzing biological sulfations present in the M of C4 plants? Is there a specific PAPS transporter between BS and M similar to those found in plastid membranes in Arabidopsis (Ashykhmina et al., 2018)? Judging from Arabidopsis, which can be considered to have a C4-like localization of sulfate assimilation, this might be the case. APS kinase, several sulfotransferases, and the PAPST1 transporter are highly enriched in the BS, together with genes for sulfate assimilation and glucosinolate synthesis (Aubry et al., 2014). However, the significance of sulfated metabolites other than glucosinolates is poorly understood, and this aspect of sulfur metabolism in C4 plants awaits initial studies.
In contrast, GSH transporters in plants have been identified. Transport of both the reduced and oxidized forms of GSH is very important for C4 monocots as oxidized glutathione (GSSG) cannot be reduced to GSH in the BS. This is because glutathione reductase (GR), a key enzyme in the glutathione–ascorbate cycle, is not expressed in these cells. Thus, GSSG must be transported to M cells for recycling (Kingston‐Smith and Foyer, 2000). Furthermore, recent work in Flaveria suggests that GSH biosynthesis largely occurs in the roots of this C4 eudicot. Thus, GSH must be transported to the shoots and unloaded from the vasculature into the BS and from there to the M, suggesting that a number of GSH transporters are required in Flaveria (Gerlich et al., 2018).
Thus far, all of the confirmed plant GSH transporters belong to the oligopeptide family of transporters and are homologs of the high-affinity yeast GSH transporter ScOPT1. The first plant GSH transporter, BjGT1, was cloned from Brassica juncea; however, functional homologs have been identified in several other species, including maize (Bogs et al., 2003; Pang et al., 2010). Most recently, mutants in two previously characterized GSH transporters from Arabidopsis, AtOPT4 and AtOPT6, were identified as having GSH-related phenotypes, including decreased GSH in floral organs and disrupted ionomic profiles when exposed to Cd (Zhang et al., 2016; Wongkaew et al., 2018). These studies suggest that OPTs do indeed play a role in GSH or sulfur metabolism. Unfortunately, the affinity and specificity of these transporters are much lower than observed for ScOPT1 and they are probably only minor contributors to the high GSH flux observed within most plant cells, suggesting that additional high-affinity transporters may exist. Clearly, the molecular nature of cysteine and GSH transport between cells and tissues remains an intriguing question for the future not only in connection with C4 plants.
Sensors and signaling molecules
The sequestration of sulfate assimilation into the BS of C4 plants sets up an intriguing metabolic scenario where sulfate is imported into the BS and a reduced sulfur compound (cysteine or possibly GSH) is exported to the rest of the plant to satisfy its sulfur needs. Thus, sulfur assimilation within the BS could be regulated by the supply (availability of sulfate through import) or by the demand (rate of utilization of the exported reduced sulfur). At a whole-plant level, it is thought that plants sense and respond to lowered internal sulfate content (i.e. lack of sulfate availability); however, the exact sensor(s) have not been identified (Lee et al., 2012). In the case of C4 BS, the regulatory mechanisms and signaling molecules are likely to be even more complex, as fine-tuning of sulfate assimilation requires tight control over sulfate import as well as cysteine and/or GSH export to maintain appropriate levels of reduced sulfur throughout the plant. One intriguing possibility is that cysteine itself acts as a signaling molecule in C4 plants. As discussed previously, cysteine is thought to undergo long-distance transport in maize, and supplying exogenous cysteine to maize ameliorated sulfur deficiency responses while exogenous GSH did not. However, this response has not been demonstrated in other C4 plants.
Sulfated peptides have been identified in C3 plants as signaling molecules, including phytosulfokines, root meristem growth factors (RGFs), or Casparian strip integrity factors (CIF1 and CIF2) (Matsuzaki et al., 2010; Sauter, 2015; Doblas et al., 2017). These peptides are sulfated by a tyrosylprotein sulfotransferase in the trans-Golgi, and then secreted into the apoplast, where they are proteolyticaly cleaved. Once in the apoplast, the secreted peptides can be perceived by membrane-bound receptor kinases belonging to the leucine-rich repeat family. These important signaling molecules have been implicated in many processes. Phytosulfokines have been shown to promote plant growth, act in funicular pollen tube guidance, affect ethylene production, and alter immune responses (Sauter, 2015). RGFs are important for maintenance of the root meristem and control cell proliferation (Matsuzaki et al., 2010), while CIFs control formation of the root Casparian strip barrier (Doblas et al., 2017). These target activities, cell proliferation and diffusion barriers, might also be important for establishment of C4 photosynthesis, making sulfated peptides attractive candidates for new signals. Because their activity is regulated by sulfation, they are ultimately regulated by sulfur metabolism, potentially linking these processes with sulfur. Thus, it is possible that multiple sulfur sensors and signaling molecules exist in C4 plants, and these molecules may or may not be broadly conserved across C4 species due to multiple evolutionary origins of C4 metabolism. However, as is the case in C3 plants, the identity of sulfur sensors remains elusive.
Temporal regulation
One final and speculative area for future studies is the open question of temporal regulation of sulfur metabolism. While C4 metabolism seeks to increase carbon assimilation efficiency by spatially separating the site of carbon capture and carbon assimilation, some plants living in extremely arid regions instead separate carbon fixation and carbon assimilation in a temporal manner to conserve water. In crassulacean acid metabolism (CAM), carbon is only captured at night while temperatures are low and the CO2 is stored in the vacuole as the four-carbon acid malic acid (Borland and Taybi, 2004). This minimizes water loss through stomata. During the daytime, the stomata close and the malic acid is metabolized back into CO2 and pyruvate to support the Calvin cycle. Similar to C4 metabolism, this metabolic strategy has multiple evolutionary origins having arisen independently at least 35 times (Heyduk et al., 2016). While it is unclear if these plants have also developed novel temporal regulation of sulfur assimilation, it is easy to speculate that the demands for reduced sulfur compounds in these plants are dramatically different from those of C3 and C4 plants. The temporal differences in carbon metabolism can be expected to pose a challenge for the coordination with nitrogen and sulfur assimilation for cysteine synthesis. Thus, the potential for novel temporal regulation of sulfur (and nitrogen) assimilation in CAM plants remains a tantalizing possibility for future research.
Conclusions
The key role cysteine synthesis plays in linking together sulfur, carbon, and nitrogen assimilation is obvious, and the importance of understanding how these pathways are coordinated cannot be understated. While this is true for both C3 and C4 organisms, C4 plants pose an additional challenge due to the spatial organization of carbon, nitrogen, and sulfur metabolisms. We presume that the spatial rewiring of sulfur assimilation confers some advantage to C4 plants or, alternatively, failure to do so results in some penalty. Yet, because it is more difficult to assess the cost–benefit relationships associated with sulfur compounds than for simple carbon compounds, correlative relationships have thus far been the only measure available. However, these correlative measures fail to assess the order in which these adaptations occur, namely did changes in sulfur metabolism occur before the development of C4 metabolism, after the development of C4 metabolism, or did they co-evolve alongside C4 metabolism. Answers to these questions could have major impacts on efforts to engineer C4 metabolism into C3 crops.
While differences in sulfur assimilation between C3 and C4 species are known to exist, it is still unclear if these differences apply broadly to all C4 species or if major differences exist between monocot and eudicot C4 species. Furthermore, because C4 metabolism has evolved independently >60 times, even if many of the mechanisms underlying sulfur (and nitrogen) metabolism are conserved between monocot and eudicot C4 species, it is likely that there are numerous subtle differences in regulation. While we are beginning to appreciate the spatial redistribution of sulfate assimilation in C4 plants, the underlying regulatory networks, including transcriptional regulators, transporters, and sensors, have yet to be explored. Thus, despite our growing knowledge of plant sulfur assimilation, it is clear that many major discoveries have yet to be made.
Acknowledgements
IZ is funded by a DAAD Stipend. PRK obtained funding from the Iran Ministry of Science, Research and Technology. Research in SK’s laboratory is funded by the Deutsche Forschungsgemeinschaft (DFG) under Germany´s Excellence Strategy – EXC 2048/1 – project 390686111.
References
- Adwy W, Laxa M, Peterhansel C. 2015. A simple mechanism for the establishment of C2-specific gene expression in Brassicaceae. The Plant Journal 84, 1231–1238. [DOI] [PubMed] [Google Scholar]
- Anoman AD, Flores-Tornero M, Benstein RM, Blau S, Rosa-Téllez S, Bräutigam A, Fernie AR, Muņoz-Bertomeu J, Schilasky S, Meyer AJ. 2019. Deficiency in the phosphorylated pathway of serine biosynthesis perturbs sulfur assimilation. Plant Physiology 180, 153–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashykhmina N, Lorenz M, Frerigmann H, Koprivova A, Hofsetz E, Stührwohldt N, Flügge UI, Haferkamp I, Kopriva S, Gigolashvili T. 2018. PAPST2 plays a critical role for PAP removal from the cytosol and subsequent degradation in plastids and mitochondria. The Plant Cell 31, 231–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry S, Smith-Unna RD, Boursnell CM, Kopriva S, Hibberd JM. 2014. Transcript residency on ribosomes reveals a key role for the Arabidopsis thaliana bundle sheath in sulfur and glucosinolate metabolism. The Plant Journal 78, 659–673. [DOI] [PubMed] [Google Scholar]
- Becker TW, Carrayol E, Hirel B. 2000. Glutamine synthetase and glutamate dehydrogenase isoforms in maize leaves: localization, relative proportion and their role in ammonium assimilation or nitrogen transport. Planta 211, 800–806. [DOI] [PubMed] [Google Scholar]
- Beerling DJ, Osborne CP. 2006. The origin of the savanna biome. Global Change Biology 12, 2023–2031. [Google Scholar]
- Bick JA, Setterdahl AT, Knaff DB, Chen Y, Pitcher LH, Zilinskas BA, Leustek T. 2001. Regulation of the plant-type 5'-adenylyl sulfate reductase by oxidative stress. Biochemistry 40, 9040–9048. [DOI] [PubMed] [Google Scholar]
- Bobe R, Behrensmeyer AK. 2004. The expansion of grassland ecosystems in Africa in relation to mammalian evolution and the origin of the genus Homo. Palaeogeography, Palaeoclimatology, Palaeoecology 207, 399–420. [Google Scholar]
- Bogs J, Bourbouloux A, Cagnac O, Wachter A, Rausch T, Delrot S. 2003. Functional characterization and expression analysis of a glutathione transporter, BjGT1, from Brassica juncea: evidence for regulation by heavy metal exposure. Plant, Cell & Environment 26, 1703–1711. [Google Scholar]
- Bolchi A, Petrucco S, Tenca PL, Foroni C, Ottonello S. 1999. Coordinate modulation of maize sulfate permease and ATP sulfurylase mRNAs in response to variations in sulfur nutritional status: stereospecific down-regulation by l-cysteine. Plant Molecular Biology 39, 527–537. [DOI] [PubMed] [Google Scholar]
- Borland AM, Taybi T. 2004. Synchronization of metabolic processes in plants with Crassulacean acid metabolism. Journal of Experimental Botany 55, 1255–1265. [DOI] [PubMed] [Google Scholar]
- Brooks DJ, Fresco JR, Lesk AM, Singh M. 2002. Evolution of amino acid frequencies in proteins over deep time: inferred order of introduction of amino acids into the genetic code. Molecular Biology and Evolution 19, 1645–1655. [DOI] [PubMed] [Google Scholar]
- Brown RH. 1999. Agronomic implications of C4 photosynthesis. In: Sage RF, Monsoon RK, eds. C4 plant biology. San Diego: Academic Press, 473–507. [Google Scholar]
- Burgener M, Suter M, Jones S, Brunold C. 1998. Cyst(e)ine is the transport metabolite of assimilated sulfur from bundle-sheath to mesophyll cells in maize leaves. Plant Physiology 116, 1315–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C, Degenkolbe T, Cuadros-Inostroza A, Klie S, Sulpice R, Leisse A, Steinhauser D, Fernie AR, Willmitzer L, Hannah MA. 2011. High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. The Plant Journal 67, 869–884. [DOI] [PubMed] [Google Scholar]
- Chan KX, Mabbitt PD, Phua SY, Mueller JW, Nisar N, Gigolashvili T, Stroeher E, Grassl J, Arlt W, Estavillo GM. 2016. Sensing and signaling of oxidative stress in chloroplasts by inactivation of the SAL1 phosphoadenosine phosphatase. Proceedings of the National Academy of Sciences, USA 113, E4567–E4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin P-A, Osborne CP, Chatelet DS, Columbus JT, Besnard G, Hodkinson TR, Garrison LM, Vorontsova MS, Edwards EJ. 2013. Anatomical enablers and the evolution of C4 photosynthesis in grasses. Proceedings of the National Academy of Sciences 110, 1381–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblas VG, Smakowska-Luzan E, Fujita S, Alassimone J, Barberon M, Madalinski M, Belkhadir Y, Geldner N. 2017. Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science 355, 280–284. [DOI] [PubMed] [Google Scholar]
- Dong Y, Silbermann M, Speiser A, Forieri I, Linster E, Poschet G, Samami AA, Wanatabe M, Sticht C, Teleman AA. 2017. Sulfur availability regulates plant growth via glucose–TOR signaling. Nature Communications 8, 1174. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Edwards EJ. 2019. Evolutionary trajectories, accessibility, and other metaphors: the case of C4 and CAM photosynthesis. New Phytologist https://doi.org/ 10.1111/nph.15851 [DOI] [PubMed] [Google Scholar]
- Edwards EJ, Osborne CP, Strömberg CAE, Smith SA, Consortium CG. 2010. The origins of C4 grasslands: integrating evolutionary and ecosystem science. Science 328, 587–591. [DOI] [PubMed] [Google Scholar]
- Espinoza C, Degenkolbe T, Caldana C, Zuther E, Leisse A, Willmitzer L, Hincha DK, Hannah MA. 2010. Interaction with diurnal and circadian regulation results in dynamic metabolic and transcriptional changes during cold acclimation in Arabidopsis. PLoS One 5, e14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Young TE. 2004. The ethylene biosynthetic and perception machinery is differentially expressed during endosperm and embryo development in maize. Molecular Genetics and Genomics 271, 267–281. [DOI] [PubMed] [Google Scholar]
- Gerlich SC, Walker BJ, Krueger S, Kopriva S. 2018. Sulfate metabolism in C4 Flaveria species is controlled by the root and connected to serine biosynthesis. Plant Physiology 178, 565–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwick BC, Black CC. 1979. Sulfur assimilation in C4 plants: intercellular compartmentation of adenosine 5'-triphosphate sulfurylase in crabgrass leaves. Plant Physiology 64, 590–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwick BC, Ku SB, Black CC. 1980. Initiation of sulfate activation: a variation in C4 photosynthesis plants. Science 209, 513–515. [DOI] [PubMed] [Google Scholar]
- Ghannoum O, Evans JR, von Caemmerer S. 2010. Nitrogen and water use efficiency of C4 plants . In: Raghavendra A, Sage R, eds. C4 photosynthesis and related CO2 concentrating mechanisms. Dordrecht: Springer, 129–146. [Google Scholar]
- Gilbert M, Wilhelm C. 2019. Biomass production: biological basics. In: Kaltschmitt M, ed. Energy from organic materials (biomass). A volume in the encyclopedia of sustainability science and technology, 2nd edn, New York: Springer New York, 17–52. [Google Scholar]
- Hagemann M, Bauwe H. 2016. Photorespiration and the potential to improve photosynthesis. Current Opinion in Chemical Biology 35, 109–116. [DOI] [PubMed] [Google Scholar]
- Hell R, Bergmann L. 1990. λ-Glutamylcysteine synthetase in higher plants: catalytic properties and subcellular localization. Planta 180, 603–612. [DOI] [PubMed] [Google Scholar]
- Hesse H, Trachsel N, Suter M, Kopriva S, von Ballmoos P, Rennenberg H, Brunold C. 2003. Effect of glucose on assimilatory sulphate reduction in Arabidopsis thaliana roots. Journal of Experimental Botany 54, 1701–1709. [DOI] [PubMed] [Google Scholar]
- Heyduk K, McKain MR, Lalani F, Leebens-Mack J. 2016. Evolution of a CAM anatomy predates the origins of Crassulacean acid metabolism in the Agavoideae (Asparagaceae). Molecular Phylogenetics and Evolution 105, 102–113. [DOI] [PubMed] [Google Scholar]
- Hibberd JM, Covshoff S. 2010. The regulation of gene expression required for C4 photosynthesis. Annual Review of Plant Biology 61, 181–207. [DOI] [PubMed] [Google Scholar]
- Hibberd JM, Sheehy JE, Langdale JA. 2008. Using C4 photosynthesis to increase the yield of rice—rationale and feasibility. Current Opinion in Plant Biology 11, 228–231. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K. 2003. Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. The Plant Journal 33, 651–663. [DOI] [PubMed] [Google Scholar]
- Hopkins L, Parmar S, Błaszczyk A, Hesse H, Hoefgen R, Hawkesford MJ. 2005. O-Acetylserine and the regulation of expression of genes encoding components for sulfate uptake and assimilation in potato. Plant Physiology 138, 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M, Wachter A, Gromes R, Stuwe T, Rausch T, Scheffzek K. 2006. Structural basis for the redox control of plant glutamate cysteine ligase. Journal of Biological Chemistry 281, 27557–27565. [DOI] [PubMed] [Google Scholar]
- Hubberten HM, Klie S, Caldana C, Degenkolbe T, Willmitzer L, Hoefgen R. 2012. Additional role of O-acetylserine as a sulfur status-independent regulator during plant growth. The Plant Journal 70, 666–677. [DOI] [PubMed] [Google Scholar]
- Jez JM, Cahoon RE, Chen S. 2004. Arabidopsis thaliana glutamate-cysteine ligase: functional properties, kinetic mechanism, and regulation of activity. Journal of Biological Chemistry 279, 33463–33470. [DOI] [PubMed] [Google Scholar]
- Jez JM, Ravilious GE, Herrmann J. 2016. Structural biology and regulation of the plant sulfation pathway. Chemico-Biological Interactions 259, 31–38. [DOI] [PubMed] [Google Scholar]
- Jones MB. 2010. C4 species as energy crops. In: Raghavendra A, Sage R, eds. C4 photosynthesis and related CO2 concentrating mechanisms. Dordrecht: Springer, 379–397. [Google Scholar]
- Jordan IK, Kondrashov FA, Adzhubei IA, Wolf YI, Koonin EV, Kondrashov AS, Sunyaev S. 2005. A universal trend of amino acid gain and loss in protein evolution. Nature 433, 633–638. [DOI] [PubMed] [Google Scholar]
- Junqueira NEG, Ortiz-Silva B, Leal-Costa MV, Alves-Ferreira M, Dickinson HG, Langdale JA, Reinert F. 2018. Anatomy and ultrastructure of embryonic leaves of the C4 species Setaria viridis. Annals of Botany 121, 1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston‐Smith AH, Foyer CH. 2000. Bundle sheath proteins are more sensitive to oxidative damage than those of the mesophyll in maize leaves exposed to paraquat or low temperatures. Journal of Experimental Botany 51, 123–130. [PubMed] [Google Scholar]
- Kopriva S, Büchert T, Fritz G, Suter M, Benda R, Schünemann V, Koprivova A, Schürmann P, Trautwein AX, Kroneck PMH. 2002. a The presence of an iron–sulfur cluster in adenosine 5'-phosphosulfate reductase separates organisms utilizing adenosine 5'-phosphosulfate and phosphoadenosine 5'-phosphosulfate for sulfate assimilation. Journal of Biological Chemistry 277, 21786–21791. [DOI] [PubMed] [Google Scholar]
- Kopriva S, Büchert T, Fritz G, Suter M, Weber M, Benda R, Schaller J, Feller U, Schürmann P, Schünemann V. 2001. Plant adenosine 5'-phosphosulfate reductase is a novel iron–sulfur protein. Journal of Biological Chemistry 276, 42881–42886. [DOI] [PubMed] [Google Scholar]
- Kopriva S, Koprivova A. 2005. Sulfate assimilation and glutathione synthesis in C4 plants. Photosynthesis Research 86, 363–372. [DOI] [PubMed] [Google Scholar]
- Kopriva S, Rennenberg H. 2004. Control of sulphate assimilation and glutathione synthesis: interaction with N and C metabolism. Journal of Experimental Botany 55, 1831–1842. [DOI] [PubMed] [Google Scholar]
- Kopriva S, Suter M, von Ballmoos P, Hesse H, Krähenbühl U, Rennenberg H, Brunold C. 2002b. Interaction of sulfate assimilation with carbon and nitrogen metabolism in Lemna minor. Plant Physiology 130, 1406–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A, Kopriva S. 2014. Molecular mechanisms of regulation of sulfate assimilation: first steps on a long road. Frontiers in Plant Science 5, 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A, Kopriva S. 2016. Sulfation pathways in plants. Chemico-Biological Interactions 259, 23–30. [DOI] [PubMed] [Google Scholar]
- Koprivova A, Melzer M, von Ballmoos P, Mandel T, Brunold C, Kopriva S. 2001. Assimilatory sulfate reduction in C3, C3–C4, and C4 species of Flaveria. Plant Physiology 127, 543–550. [PMC free article] [PubMed] [Google Scholar]
- Koprivova A, Suter M, den Camp RO, Brunold C, Kopriva S. 2000. Regulation of sulfate assimilation by nitrogen in Arabidopsis. Plant Physiology 122, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A. 2015. Plant nitrogen assimilation and its regulation: a complex puzzle with missing pieces. Current Opinion in Plant Biology 25, 115–122. [DOI] [PubMed] [Google Scholar]
- Krueger S, Benstein RM, Wulfert S, Anoman AD, Flores-Tornero M, Ros R. 2017. Studying the function of the phosphorylated pathway of serine biosynthesis in Arabidopsis thaliana. Methods in Molecular Biology 1653, 227–242. [DOI] [PubMed] [Google Scholar]
- Ku MS, Wu J, Dai Z, Scott RA, Chu C, Edwards GE. 1991. Photosynthetic and photorespiratory characteristics of Flaveria species. Plant Physiology 96, 518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappartient AG, Vidmar JJ, Leustek T, Glass AD, Touraine B. 1999. Inter-organ signaling in plants: regulation of ATP sulfurylase and sulfate transporter genes expression in roots mediated by phloem-translocated compound. The Plant Journal 18, 89–95. [DOI] [PubMed] [Google Scholar]
- Lee B-R, Huseby S, Koprivova A, Chetelat A, Wirtz M, Mugford ST, Navid E, Brearley C, Saha S, Mithen R. 2012. Effects of fou8/fry1 mutation on sulfur metabolism: is decreased internal sulfate the trigger of sulfate starvation response? PLoS One 7, e39425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegood RC. 2002. C4 photosynthesis: principles of CO2 concentration and prospects for its introduction into C3 plants. Journal of Experimental Botany 53, 581–590. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang J, Ni F, Dong X, Han B, Han D, Ji Z, Zhao Y. 2010. Genome wide exploration of the origin and evolution of amino acids. BMC Evolutionary Biology 10, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren MR, Osborne CP, Christin PA. 2014. Deconstructing Kranz anatomy to understand C4 evolution. Journal of Experimental Botany 65, 3357–3369. [DOI] [PubMed] [Google Scholar]
- Mallmann J, Heckmann D, Bräutigam A, Lercher MJ, Weber AP, Westhoff P, Gowik U. 2014. The role of photorespiration during the evolution of C4 photosynthesis in the genus Flaveria. eLife 3, e02478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Wang S, Jia Q, Wu P. 2006. OsEIL1, a rice homolog of the Arabidopsis EIN3 regulates the ethylene response as a positive component. Plant Molecular Biology 61, 141–152. [DOI] [PubMed] [Google Scholar]
- Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. 2010. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329, 1065–1067. [DOI] [PubMed] [Google Scholar]
- Miranda M, Borisjuk L, Tewes A, Heim U, Sauer N, Wobus U, Weber H. 2001. Amino acid permeases in developing seeds of Vicia faba L.: expression precedes storage protein synthesis and is regulated by amino acid supply. The Plant Journal 28, 61–71. [DOI] [PubMed] [Google Scholar]
- Moore R, Black CC. 1979. Nitrogen assimilation pathways in leaf mesophyll and bundle sheath cells of C4 photosynthesis plants formulated from comparative studies with Digitaria sanguinalis (L.) Scop. Plant Physiology 64, 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, Hesse H, Hoefgen R. 2005. Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiology 138, 304–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace NJ, Weerapana E. 2013. Diverse functional roles of reactive cysteines. ACS Chemical Biology 8, 283–296. [DOI] [PubMed] [Google Scholar]
- Pang S, Li X-F, Liu ZQ, Wang C-J. 2010. ZmGT1 transports glutathione conjugates and its expression is induced by herbicide atrazine. Progress in Biochemistry and Biophysics 37, 1120–1127. [Google Scholar]
- Pe’er I, Felder CE, Man O, Silman I, Sussman JL, Beckmann JS. 2004. Proteomic signatures: amino acid and oligopeptide compositions differentiate among phyla. Proteins 54, 20–40. [DOI] [PubMed] [Google Scholar]
- Pivato M, Fabrega-Prats M, Masi A. 2014. Low-molecular-weight thiols in plants: functional and analytical implications. Archives of Biochemistry and Biophysics 560, 83–99. [DOI] [PubMed] [Google Scholar]
- Poznański J, Topiński J, Muszewska A, Dębski KJ, Hoffman-Sommer M, Pawłowski K, Grynberg M. 2018. Global pentapeptide statistics are far away from expected distributions. Scientific Reports 8, 15178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathnam CK, Edwards GE. 1976. Distribution of nitrate-assimilating enzymes between mesophyll protoplasts and bundle sheath cells in leaves of three groups of C4 plants. Plant Physiology 57, 881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravilious GE, Nguyen A, Francois JA, Jez JM. 2012. Structural basis and evolution of redox regulation in plant adenosine-5′-phosphosulfate kinase. Proceedings of the National Academy of Sciences, USA 109, 309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros R, Muñoz-Bertomeu J, Krueger S. 2014. Serine in plants: biosynthesis, metabolism, and functions. Trends in Plant Science 19, 564–569. [DOI] [PubMed] [Google Scholar]
- Sage RF, Monson RK, Ehleringer JR, Adachi S, Pearcy RW. 2018. Some like it hot: the physiological ecology of C4 plant evolution. Oecologia 187, 941–966. [DOI] [PubMed] [Google Scholar]
- Sage RF, Wedin DA, Li M. 1999. The biogeography of C4 photosynthesis: patterns and controlling factors. In: Sage RF, Monsoon RK, eds. C4 plant biology. San Diego: Academic Press, 313–373. [Google Scholar]
- Sage RF, Zhu XG. 2011. Exploiting the engine of C4 photosynthesis. Journal of Experimental Botany 62, 2989–3000. [DOI] [PubMed] [Google Scholar]
- Samuilov S, Brilhaus D, Rademacher N, Flachbart S, Arab L, Alfarraj S, Kuhnert F, Kopriva S, Weber APM, Mettler-Altmann T. 2018a. The photorespiratory BOU gene mutation alters sulfur assimilation and its crosstalk with carbon and nitrogen metabolism in Arabidopsis thaliana. Frontiers in Plant Science 9, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuilov S, Rademacher N, Brilhaus D, Flachbart S, Arab L, Kopriva S, Weber APM, Mettler-Altmann T, Rennenberg H. 2018b. Knock-down of the phosphoserine phosphatase gene effects rather N- than S-metabolism in Arabidopsis thaliana. Frontiers in Plant Science 9, 1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter M. 2015. Phytosulfokine peptide signalling. Journal of Experimental Botany 66, 5161–5169. [DOI] [PubMed] [Google Scholar]
- Schmutz D, Brunold C. 1984. Intercellular localization of assimilatory sulfate reduction in leaves of Zea mays and Triticum aestivum. Plant Physiology 74, 866–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Still CJ, Berry JA, Collatz GJ, DeFries RS. 2003. Global distribution of C3 and C4 vegetation: carbon cycle implications. Global Biogeochemical Cycles 17, 1–6. [Google Scholar]
- Takahashi H, Kopriva S, Giordano M, Saito K, Hell R. 2011. Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annual Review of Plant Biology 62, 157–184. [DOI] [PubMed] [Google Scholar]
- Tegeder M. 2012. Transporters for amino acids in plant cells: some functions and many unknowns. Current Opinion in Plant Biology 15, 315–321. [DOI] [PubMed] [Google Scholar]
- von Caemmerer S, Furbank RT. 2016. Strategies for improving C4 photosynthesis. Current Opinion in Plant Biology 31, 125–134. [DOI] [PubMed] [Google Scholar]
- von Caemmerer S, Quick WP, Furbank RT. 2012. The development of C4 rice: current progress and future challenges. science 336, 1671–1672. [DOI] [PubMed] [Google Scholar]
- Wang P, Vlad D, Langdale JA. 2016. Finding the genes to build C4 rice. Current Opinion in Plant Biology 31, 44–50. [DOI] [PubMed] [Google Scholar]
- Weckopp SC, Kopriva S. 2014. Are changes in sulfate assimilation pathway needed for evolution of C4 photosynthesis? Frontiers in Plant Science 5, 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BP, Johnston IG, Covshoff S, Hibberd JM. 2013. Phenotypic landscape inference reveals multiple evolutionary paths to C4 photosynthesis. eLife 2, e00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongkaew A, Asayama K, Kitaiwa T, Nakamura SI, Kojima K, Stacey G, Sekimoto H, Yokoyama T, Ohkama-Ohtsu N. 2018. AtOPT6 protein functions in long-distance transport of glutathione in Arabidopsis thaliana. Plant & Cell Physiology 59, 1443–1451. [DOI] [PubMed] [Google Scholar]
- Yampolsky LY, Wolf YI, Bouzinier MA. 2017. Net evolutionary loss of residue polarity in drosophilid protein cores indicates ongoing optimization of amino acid composition. Genome Biology and Evolution 9, 2879–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xie Q, Jobe TO, Kau AR, Wang C, Li Y, Qiu B, Wang Q, Mendoza-Cózatl DG, Schroeder JI. 2016. Identification of AtOPT4 as a plant glutathione transporter. Molecular Plant 9, 481–484. [DOI] [PMC free article] [PubMed] [Google Scholar]