Abstract
The transcriptome-wide association studies (TWASs) that test for association between the study trait and the imputed gene expression levels from cis-acting expression quantitative trait loci (cis-eQTL) genotypes have successfully enhanced the discovery of genetic risk loci for complex traits. By using the gene expression imputation models fitted from reference datasets that have both genetic and transcriptomic data, TWASs facilitate gene-based tests with GWAS data while accounting for the reference transcriptomic data. The existing TWAS tools like PrediXcan and FUSION use parametric imputation models that have limitations for modeling the complex genetic architecture of transcriptomic data. Therefore, to improve on this, we employ a nonparametric Bayesian method that was originally proposed for genetic prediction of complex traits, which assumes a data-driven nonparametric prior for cis-eQTL effect sizes. The nonparametric Bayesian method is flexible and general because it includes both of the parametric imputation models used by PrediXcan and FUSION as special cases. Our simulation studies showed that the nonparametric Bayesian model improved both imputation R2 for transcriptomic data and the TWAS power over PrediXcan when ≥1% cis-SNPs co-regulate gene expression and gene expression heritability ≤0.2. In real applications, the nonparametric Bayesian method fitted transcriptomic imputation models for 57.8% more genes over PrediXcan, thus improving the power of follow-up TWASs. We implement both parametric PrediXcan and nonparametric Bayesian methods in a convenient software tool “TIGAR” (Transcriptome-Integrated Genetic Association Resource), which imputes transcriptomic data and performs subsequent TWASs using individual-level or summary-level GWAS data.
Keywords: transcriptome-wide association studies, nonparametric Bayesian method, gene mapping, gene expression imputation, genetically regulated gene expression, TIGAR
Introduction
Genome-wide association studies (GWASs) have successfully identified thousands of genetic risk loci for complex traits. However, the majority of these loci are located within noncoding regions whose molecular mechanisms remain unknown.1, 2, 3 Recent studies have shown that these associated regions were enriched for regulatory elements such as enhancers (H3K27ac marks)4, 5 and expression of quantitative trait loci (eQTL),6, 7 suggesting that the genetically regulated gene expression might play a key role in explaining the etiology of complex traits. Multiple studies have recently generated rich transcriptomic datasets for diverse tissues of the human body (besides genotype data), e.g., the Genotype-Tissue Expression (GTEx) project for >44 human tissues,6 Genetic European Variation in Health and Disease (GEUVADIS) for lymphoblastoid cell lines,8 Depression Genes and Networks (DGN) for whole-blood samples,9 and the North American Brain Expression Consortium (NABEC) for cortex tissues.10 Previous studies11, 12, 13, 14, 15, 16 have also shown that integrating transcriptomic data in GWASs can help identify functional loci.
The majority of GWAS projects do not profile transcriptomic data and thus cannot enable direct integrative analysis. However, existing studies11, 12 have shown that one can impute the genetically regulated gene expression (GReX) within such GWAS projects by using reference datasets like GTEx6 and GEUVADIS8 to train gene expression imputation models, and then test for the association between imputed GReX for GWAS samples and the trait of interest—referred to as transcriptome-wide association studies (TWASs).11, 12 Specifically, the gene expression imputation models are fitted by regressing assayed gene-expression levels on cis-eQTL genotypes with reference dataset. For examples, the PrediXcan11 method uses an Elastic-Net17 variable selection model and the FUSION12 tool implements a Bayesian sparse linear mixed model (BSLMM)18 to estimate the cis-eQTL effect sizes with reference dataset. The estimated cis-eQTL effect sizes are then used to impute the GReX for GWAS samples.
In short, the Elastic-Net17 model used by PrediXcan11 assumes a combination of LASSO19 (L1) and Ridge20 (L2) penalties on the cis-eQTL effect sizes, which is equivalent to a Bayesian model with a mixture Gaussian and Laplace prior.21 In contrast, the BSLMM18 used by FUSION12 is a combination of Bayesian variable selection model (BVSR)22 and linear mixed model (LMM)23 by assuming a normal mixture prior. Since a parametric prior is assumed for the cis-eQTL effect sizes by both Elastic-Net and BSLMM, it restricts the capability of PrediXcan and FUSION for handling the underlying complex genetic architecture of transcriptomes. Existing studies11, 12 have also shown that both PrediXcan11 and FUSION12 estimated the average regression R2 (i.e., the percentage of gene expression variation that can be explained by cis-genotypes) as ∼5% for human whole-blood transcriptome, while the average genome-wide heritability of gene expression in human whole-blood transcriptome is estimated to be more than double that quantity.24, 25
Therefore, to flexibly model cis-eQTL distributions, we use a nonparametric Bayesian method that was originally proposed for genetic prediction of complex traits,26 where the prior for effect sizes is nonparametric and can be estimated from the data by assuming a Dirichlet process prior on effect-size variance. This Bayesian model is also known as latent Dirichlet process regression (DPR) model,26 which can flexibly model the underlying complex genetic architecture of transcriptomes. Thus, DPR is a more generalized model that includes Elastic-Net (implemented in PrediXcan11) and BSLMM (implemented in FUSION12) as special cases. Consequently, DPR can robustly estimate cis-eQTLs and then improve imputation R2 (the squared Pearson correlation between the observed and imputed values on test samples). Moreover, a variational Bayesian algorithm26, 27, 28 can be employed as an alternative of Monte Carlo Markov Chain (MCMC)29 to efficiently fit the Bayesian model.
Similar to PrediXcan11 and FUSION12 methods, we employ DPR to estimate cis-eQTLs effect sizes from a reference dataset, which can then be used for downstream TWASs using either individual-level or summary-level GWAS data. In subsequent sections, we first describe the DPR26 approach for estimating cis-eQTL effect sizes from a reference dataset and how we can then use these effect sizes for a downstream TWAS. We then compare the performance of DPR with PrediXcan using both simulated data and real GWAS and transcriptomic data from the Religious Orders Study and Rush Memory Aging Project (ROS/MAP)30, 31, 32, 33 for studying Alzheimer disease (AD).
Our in-depth simulation studies demonstrated that the DPR method obtained higher imputation R2 on test samples, when ≥1% cis-SNPs are true causal and the true expression heritability is ≤0.2. Consequently, better imputation R2 resulted in improved power for follow-up association studies. Meanwhile, application of DPR to the ROS/MAP study imputed GReX for 57.8% more genes than PrediXcan. Using DPR, we also found a potentially associated gene TRAPPC6A for AD pathology indices, which was missed by PrediXcan. Further, by using the transcriptomic imputation models fitted from ROS/MAP data and summary-level GWAS data generated from the International Genomics of Alzheimer’s Project (IGAP),34 we identified three known AD loci34, 35, 36, 37, 38 that potentially affect the late-onsite AD risk through transcript abundance. We conclude with a discussion of future topics and further describe our software tool TIGAR (Transcriptome-Integrated Genetic Association Resource) implementing both parametric Elastic-Net and nonparametric Bayesian DPR methods for public use.
Material and Methods
Here, we briefly describe the underlying statistical model of gene-expression imputation. Consider the following linear regression model for estimating the cis-eQTL effect sizes from a reference study that has both genetic and transcriptomic data available,
| (Equation 1) |
where Eg denotes the gene expression levels (after corrections for confounding covariates such as age, sex, and principal components) for gene g, X denotes the genotype matrix for all cis-genotypes (encoded as the number of minor alleles or genotype dosages), w denotes the corresponding cis-eQTL effect-size vector, and denotes the error term. The intercept term is dropped in Equation 1 for assuming both Eg and X are centered at 0. Generally, SNPs within 1 Mb of the flanking 5′ and 3′ ends (cis-SNPs) are included in this regression model and non-zero will be used for follow-up analysis. The GReX will be imputed by
with cis-SNP data Xnew for GWAS samples.
Nonparametric Bayesian Method
Following the nonparametric Bayesian DPR model proposed in previous studies for genetic prediction of complex traits,26 a normal prior is assumed for the cis-eQTL effect sizes (wi, i = 1,…, p) and a Dirichlet process (DP) prior39 is assumed for the effect-size variance (as in Equation 1):
| (Equation 2) |
The prior distribution D deviates from the DP with base distribution as an inverse gamma (IG) distribution and concentration parameter . Note that can be viewed as a latent variable and integrating out will induce a nonparametric prior distribution for wi, which is equivalent to a DP normal mixture model,26, 27, 28
| (Equation 3) |
Here, the nonparametric prior distribution on wi is equivalently represented by a mixture normal prior that is a weighted sum of an infinitely number of normal distributions , corresponding weight is determined by (vl, l = 0,…, k) with a Beta prior, and in the Beta prior (the same concentration parameter as in Equation 2) determines the number of components with non-zero weights in the mixture normal prior. Conjugate hyper priors and are assumed.
Generally, the hyper parameters in the inverse gamma distributions can be set as 0.1 and in the gamma distribution can be set as (1, 0.1) to induce non-informative priors for . That is, the parameters will be adaptively estimated from the data and the nonparametric prior on wi will be data driven. The posterior estimates for w can be obtained by the MCMC29 or variational Bayesian algorithm,28, 40 from the following joint conditional posterior distribution
Particularly, the variational Bayesian algorithm28, 40 is an approximation for the MCMC29 with greatly improved computational efficiency, which is also used in our tool. Please refer to the Supplemental Material and Methods for technical details of both MCMC sampling and variational inference algorithms for obtaining the Bayesian posterior estimates for the cis-eQTL effect sizes.
Elastic-Net and BSLMM Methods
The Elastic-Net model17 (used by PrediXcan11) estimates the cis-eQTL effect sizes in Equation 1 with a combination of L1 (LASSO)19 and L2 (Ridge)20 penalties by
where denotes L2 norm, denotes L1 norm, denotes the proportion of L1 penalty, and denotes the penalty parameter. Particularly, PrediXcan11 takes and tunes the penalty parameter by a 5-fold cross validation.
As pointed out by previous studies,17, 21 the Elastic-Net model is equivalent to a Bayesian model with a mixture Gaussian and Laplace (mixture normal) prior for , that is, . In contrast, the BSLMM18 assumes a mixture of two normal as the prior for cis-eQTL effect sizes, . That is, the BSLMM18 assumes all cis-SNPs have at least a small effect, which are normally distributed with variance , and some proportion of cis-SNPs have an additional effect, normally distributed with variance . Particularly, with , BSLMM becomes BVSR,22 and with , the BSLMM becomes the LMM.23 Therefore, the DP normal mixture26, 27, 28 as assumed by the DPR method includes the parametric (mixture normal) priors used by Bayesian Elastic-Net21 and BSLMM18 as special cases, which is the main reason why DPR is a more generalized model including Elastic-Net and BSLMM as special cases. This is also why the DPR method can robustly model complex genetic architecture and improve the imputation R2.
Association Study with Univariate Phenotype
Given individual-level GWAS data (genotype data Xnew, phenotype Y, covariant matrix C) and cis-eQTL effect size estimates , the follow-up TWAS (using a burden type gene-based test41) is to test the association between and Y based on the following generalized linear regression model
| (Equation 4) |
Here, is a pre-specified link function, which can be set as identity function for quantitative phenotype or set as logit function for dichotomous phenotype. The gene-based association test is equivalent to test in Equation 4.
If only summary-level GWAS data are available, we can take the same approach as implemented by the FUSION12 method. Let Z denote the vector of Z-scores generated by single variant tests (Wald, likelihood ratio, score tests, etc.) for all cis-SNPs. The burden Z-score for gene-based association test is defined as
| (Equation 5) |
where V denotes the covariance matrix of analyzed SNPs that can be estimated from training data or reference panels such as 1000 Genomes Project42 (of the same ethnicity).
Association Study with Multivariate Phenotype
To test the association between multivariate phenotypes and imputed GReX of the focal gene, we take a similar approach as the MultiPhen method.43 For example, consider two phenotypes (Y1,Y2) and a covariate matrix C, we first adjust for the covariates by taking the residuals respectively from the linear regression models . Then we test whether the regression R2 is significantly greater than zero for the following regression model
| (Equation 6) |
That is, we test whether the multivariate phenotypes can jointly explain a non-zero percentage of variance in the imputed GReX. The p value can be calculated by using the F-statistic for the regression R2 in Equation 6.
Even when only summary-level GWAS data are available, we can first obtain a burden Z-score per phenotype from Equation 5, i.e., with two phenotypes. Then, a similar burden approach can be used to obtain a joint Z-score for multi-phenotype test,
where is the covariance matrix among multiple traits.
Simulation Study Design
We conducted in-depth simulation studies to compare the performance of both PrediXcan and DPR methods with respect to imputation R2 in the test data and the power of TWASs. Specifically, we used data from 499 ROS/MAP participants44 which contains both RNA-sequencing and genotype data as training data, and genotype data from an additional 1,200 ROS/MAP participants44 as test data. The test sample size (1,200) was chosen arbitrarily (randomly selected from the ROS/MAP study) to be comparable with the sample size (1,164) in the real association study of AD pathology indices. The genotyped and imputed genetic data for 2,799 cis-SNPs (with minor allele frequency (MAF) > 5% and Hardy-Weinberg p value > 10−5) of the arbitrarily chosen gene ABCA7 (see Figure S1 for the LD block structure) were used to simulate gene expression levels.
We performed comprehensive scenarios that varied the proportion of causal SNPs (out of 2,799 SNPs, influenced gene expression) among values in the vector pcausal = (0.001, 0.01, 0.1, 0.2). We varied the proportion of gene expression variance explained by causal SNPs (i.e., expression heritability), along with the proportion of phenotypic variance explained by simulated gene expression levels (i.e., phenotypic heritability), among values in the vector . The phenotypic heritability was selected arbitrarily with respect to expression heritability such that the follow-up association study power fell within the range of (25%, 85%). We also considered various training sample sizes (100, 300, 499) for simulation scenario with pcausal = 0.2 and .
With genotype matrix Xg of the randomly selected causal SNPs (according to pcausal), we generated effect sizes wi from N(0,1) and then re-scaled the effect sizes to ensure the targeted . Gene expression levels were generated by , with . Then the phenotype values were generated by , where was selected with respect to and .
For each scenario, we repeated simulations for 1,000 times, where we applied both PrediXcan11 and DPR methods to obtain imputation models with training samples, impute the GReX for test samples, and then conduct follow-up association studies using the imputed GReX. We did not compare with FUSION12 using BSLMM because of the computational burden of estimating cis-eQTL effect sizes by MCMC (∼2 h per gene). The association study power was calculated as the proportion of 1,000 repeated simulations with p value < 2.5 × 10−6 (genome-wide significance threshold adjusting for testing 20K independent genes).
ROS/MAP Data
Samples in the ROS/MAP data were collected from participants of the Religious Orders Study (ROS) and the Rush Memory and Aging Project (MAP), which are prospective cohort studies of studying aging and dementia.30, 31, 33 The ROS/MAP study recruited senior adults without known dementia at enrollment who underwent annual clinical evaluation. Brain autopsy was done at the time of death for each participant. All participants signed an informed consent and Anatomic Gift Act, and the studies were approved by the Institutional Review Board of Rush University Medical Center, Chicago, IL. Specifically, microarray genotype data generated for 2,093 European-decent participants44 were further imputed to the 1000 Genomes Project Phase 342 in our analysis. The post-mortem brain samples (gray matter of the dorsolateral prefrontal cortex) from ∼30% these participants were profiled for transcriptomic data by next-generation RNA seqencing.45 In this paper, we conducted TWASs for two important indices of AD pathology that were quantified with -antibody specific immunostains:30, 31, 33 neurofibrillary tangle density (tangles) with stereology and -amyloid load (amyloid) with image analysis. The neurofibrillary tangle density quantifies the average Tau tangle density within two or more 20 μm sections from eight brain regions—hippocampus, entorhinal cortex, midfrontal cortex, inferior temporal, angular gyrus, calcarine cortex, anterior cingulate cortex, and superior frontal cortex. The -amyloid load quantifies the average percent area of cortex occupied by -amyloid protein in adjacent sections from the same eight brain regions.
Results
Simulation Studies
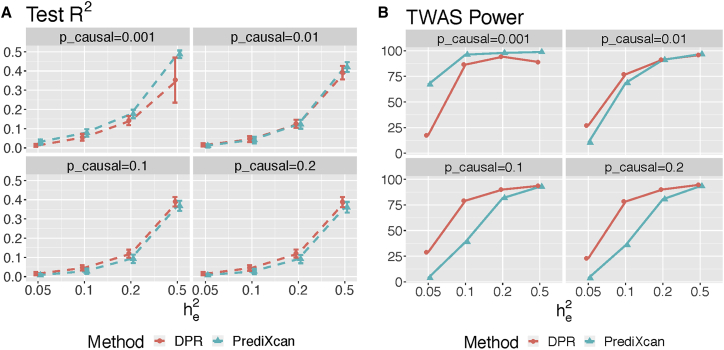
In the simulation studies, we observed that the DPR method performed robustly with respect to different causal proportions and gene expression heritability. Specifically, when pcausal > 0.01 DPR outperformed PrediXcan across all expression heritability values, giving higher imputation R2 in test data (Figure 1A). For example, when pcausal = 0.2, the average imputation R2 of 1,000 simulations was estimated as 4.55% by using DPR versus 2.64% by using PrediXcan with , while the average imputation R2 was estimated as 12.02% by using DPR versus 9.13% by using PrediXcan with (Table 1). When pcausal = 0.01, DPR performed slightly out-performed PrediXcan with and PrediXcan outperformed DPR with 0.5 (Table 1, Figure 1). On the other hand, under a sparse cis-eQTL causality model with pcausal = 0.001 (i.e., with 3 true causal cis-eQTL), the Elastic-Net method resulted in higher imputation R2 and TWAS power on test data (Figure 1).
Figure 1.
Performance Comparison of DPR versus PrediXcan
Plots of average imputation R2 (A) and TWAS power (B) in test samples by DPR and PrediXcan, with various proportions of true causal SNPs pcausal = (0.001, 0.01, 0.1, 0.2) and true expression heritability . TWAS power was evaluated with paired expression and phenotype heritability .
Table 1.
Simulation Prediction R2 Comparison
|
Causal Proportion 0.01 |
Causal Proportion 0.2 |
|||
|---|---|---|---|---|
| DPR | PrediXcan | DPR | PrediXcan | |
| 0.05 | 1.60%∗ | 1.12% | 1.54%∗ | 0.76% |
| 0.1 | 4.54%∗ | 4.13% | 4.55%∗ | 2.64% |
| 0.2 | 12.54%∗ | 12.29% | 12.02%∗ | 9.13% |
| 0.5 | 39.31% | 42.05%∗ | 38.78%∗ | 36.04% |
Various simulation scenarios were considered, with the proportion of true causal SNPs pcausal = (0.01, 0.2) and expression heritability . The best prediction R2 per scenario is indicated with asterisk (∗).
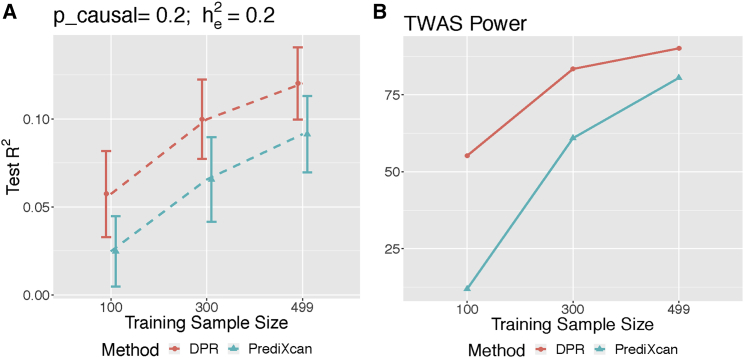
Consequently, when pcausal ≥ 0.01 and , the power of association studies was higher by using DPR than using PrediXcan imputation models (Figure 1B). When , using both imputation models led to comparable power for association studies (Figure 1B). Even though both methods had similar over-estimated training R2 (Figure S2), the DPR method resulted in higher imputation R2 for test data (Table 1; Figures 1A) and higher power for association studies under cis-eQTL causality models with pcausal ≥ 0.01 and (Figure 1B). In addition, from the simulation studies with various training sample sizes (100, 300, 499), pcausal = 0.2, and , the imputation R2 and TWAS power increases as sample size increases while the DPR method consistently outperforms PrediXcan (Figure 2). Overall, these results demonstrated the advantages of the DPR method for modeling the complex genetic architecture of transcriptomes, especially when the causal proportions ≥0.01 and the expression heritability ≤0.2.
Figure 2.
Performance of DPR and PrediXcan with Respect to Various Training Sample Sizes
Test R2 (A) and TWAS power (B) from simulation studies with causal proportion pcausal = 0.2, expression heritability and phenotype heritability and various training sample sizes (100, 300, 499).
Real Applications to ROS/MAP Data
To illustrate the performance of the DPR method in real studies, we applied both DPR and PrediXcan on the ROS/MAP data (see Material and Methods). We trained the gene expression imputation models using 499 samples that have both transcriptomic data for prefrontal cortex tissues and genotype data (imputed to 1000 Genomes Phase 3, with MAF > 5%, Hardy-Weinberg p value > 10−5, and genotype imputation R2 > 0.3). A total of 15,583 genes had gene expression levels after standard RNA-sequencing quality control. The gene expression levels were first adjusted for age at death, sex, postmortem interval, study (ROS or MAP), batch effects, RNA integrity number scores, and cell type proportions (with respect to oligodendrocytes, astrocytes, microglia, neurons) by linear regression models. For each gene, cis-SNPs within the 1 Mb of the flanking 5′ and 3′ ends were used in the imputation models as predictors.
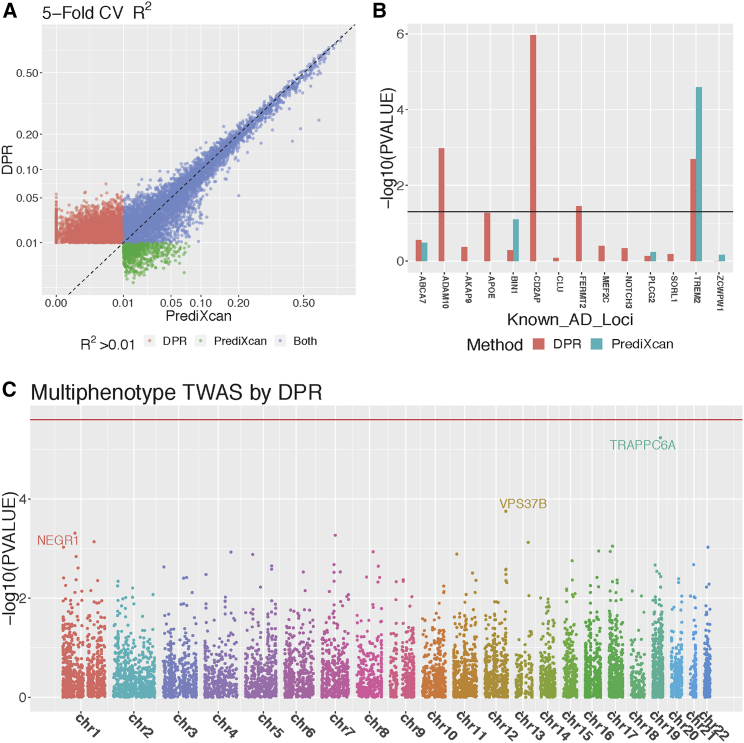
First, we compared transcriptome-wide 5-fold cross validation (CV) regression R2 estimated by using both DPR and PrediXcan methods. Specifically, we randomly split 499 training samples into 5 folds, where the imputation R2 of each fold was calculated using the model trained with the other 4-fold samples. If the training model is null, we take the imputation R2 as 0 and take the average imputation R2 across all 5-fold test samples as 5-fold CV R2. The transcriptome-wide median of 5-fold CV R2 is 0.013 by DPR versus 0.005 by PrediXcan. The 5-fold CV R2 was used as the criterion for selecting significant imputation models (R2 > 0.01 as used by previous studies11, 46). From Figure 3A, we can see that the DPR method obtained more imputation models and higher imputation R2 when 5-fold CV R2 is in the range of (0.01, 0.05), which is also consistent with our simulation studies. Overall, the DPR method obtained significant imputation models for 8,752 genes versus 5,547 genes by PrediXcan (with 57.8% increases). Thus, the DPR method featuring data-driven nonparametric prior for the cis-eQTL is preferred in real studies for identifying more genes with imputable expression levels.
Figure 3.
TWAS Results of Studying Alzheimer's Disease
Transcriptome-wide 5-fold cross validation R2 (A) by PrediXcan and DPR with 499 ROS/MAP training samples, with different colors denoting whether the imputation R2 > 0.01 by DPR, PrediXcan, or both methods (genes with R2 > 0.01 by both DPR and PrediXcan were excluded from the plot). TWAS results (B) at known AD loci using GWAS summary-level statistics from IGAP and imputation models fitted from ROS/MAP data, where missing values are due to NULL imputation models by PrediXcan. Manhattan plot (C) for the multiphenotype TWAS (with neurofibrillary tangle density and -amyloid load), using individual-level ROS/MAP data.
Second, to investigate how both DPR and PrediXcan methods perform in real studies with independent prediction cohort, we used the ROS cohort (256 samples) to train gene expression imputation models and then used the MAP cohort (243 samples) as a test dataset. Specifically, we compared the median prediction R2 by both DPR and PrediXcan with MAP test cohort. As shown in Table 2, the DPR method obtained higher median prediction R2 than PrediXcan among 8,752 genes that have 5-fold CV R2 > 0.01 by DPR (0.011 versus 0.003), performed similarly as PrediXcan among 5,547 genes that have 5-fold CV R2 > 0.01 by PrediXcan (0.026 versus 0.026), obtained slightly lower median predication R2 among 4,819 genes that have 5-fold CV R2 > 0.01 by both DPR and PrediXcan (0.033 versus 0.036). These results are also consistent with our simulation results and 5-fold cross validation results with ROS/MAP data. That is, PrediXcan method is preferred for genes with sparse causal eQTL that have relatively large effect sizes, whereas DPR is preferred for genes with less sparse causal eQTL that have minor effect sizes due to low expression heritability.
Table 2.
Real Study Prediction R2 Comparison
Median prediction R2 in MAP test cohort by using imputation models trained with ROS cohort with both DPR and PrediXcan methods.
Genes that have 5-fold CV R2 > 0.01 by DPR.
Genes that have 5-fold CV R2 > 0.01 by PrediXcan.
Genes that have 5-fold CV R2 > 0.01 by both DPR and PrediXcan.
Third, we used all 499 training samples to fit imputation models for genes with respective 5-fold CV R2 > 0.01 by both DPR and PrediXcan, and then used these models to impute the GReX for all GWAS samples. We conducted univariate phenotype association studies (Material and Methods) using all GWAS samples (n = 1,164) that have the AD pathology indices (neurofibrillary tangle density and -amyloid load, with Pearson correlation 0.48) quantified. Possible confounding covariates including age at death, sex, study (ROS or MAP), smoking, education, and first three genotype principle components were adjusted in the association studies. Interestingly, the association studies for both AD pathology indices using the DPR imputation models identified the same top significant gene TRAPPC6A (within the 2 Mb region from the major risk gene APOE, encoding apolipoprotein E, but independent of APOE) with p values 1.64 × 10−5 and 5.35 × 10−5 (Figures S3A and S4A). Moreover, the multivariate phenotype association studies (Material and Methods) for both AD pathology indices identified TRAPPC6A as the most significant gene with p value 5.81 × 10−6 and FDR 0.08 (Figure 3C). On the other hand, the PrediXcan failed to obtain a transcriptomic imputation model for TRAPPC6A (Figures S3B, S4B, and S6). Quantile-quantile plots for these TWAS p values were presented in Figure S5.
In addition, for 14 known common and rare loci of late-onset AD34, 35, 36, 37, 38 with significant imputation models, we conducted association studies using transcriptomic imputation models (DPR and PrediXcan) fitted from ROS/MAP data and summary-level GWAS data from IGAP.34 Using the imputation models fit by DPR, we identified three significant loci with FDR < 0.05 (Figure 3B)—ADAM10, CD2AP, and TREM2—that potentially affect late-onset AD risk through transcriptomic changes. Here, TREM2 was also identified by using the PrediXcan imputation model (Figure 3B). Particularly, the PrediXcan method imputed GReX for only 5 out of these 14 loci. In summary, these results show that the DPR method has superior power for follow-up TWASs.
Discussion
In this paper, by both in-depth simulations and real applications using individual-level ROS/MAP30, 31, 32, 33 and summary-level IGAP34 GWAS data, we demonstrated that the nonparametric Bayesian DPR method is preferred for imputing gene expression when the proportion of causal cis-eQTL ≥ 0.01 and the true gene expression heritability ≤ 0.2. The advantage of DPR model is due to the flexible nonparametric modeling of cis-eQTL effect sizes that results in improved imputation R2 for gene expression levels and higher power for TWASs. Here, we provide an integrated tool (freely available on GITHUB), referred as Transcriptome-Integrated Genetic Association Resource (TIGAR), which integrates both parametric Elastic-Net and nonparametric Bayesian DPR models as two options for transcriptomic data imputation, along with TWAS options using individual-level and summary-level GWAS data for univariate and multi-variate phenotypes. TIGAR also conducts 5-fold cross validation by default and output significant imputation models with CV R2 > 0.01.
With respect to user-friendly interface and computational efficiency, TIGAR can (1) take standard input files such as genotype files in VCF and dosage formats, phenotype files in PED format, and a combined text file for gene annotations and expression levels; (2) load input data per gene by TABIX for memory efficiency; (3) filter SNPs based on input thresholds of MAF and Hardy-Weinberg p value; (4) provide options of training both Elastic-Net (use Python3 scripts) and DPR (generate input files and call the executable tool developed with C++26) imputation models with unified output format; and (5) implement multi-threaded computation to take full advantage of multi-core clusters. These features make TIGAR a preferred tool for saving tedious data preparation and computation time for users. For example, TIGAR can complete training imputation models for ∼20K genes and ∼1K samples within ∼20 h and TWAS within ∼1 h with a 2.4 GHz 16-core CPU.
It is important to notice that imputing GReX with cis-eQTL effect sizes estimated from a training dataset is analogous to the idea of estimating polygenic risk scores (PRSs).47 Even though studies of population heterogeneity are lacked for imputing GReX, the same philosophy of estimating PRSs still applies because of the same underlying statistical models. That is, given both genetic and transcriptomic heterogeneities across different populations, one needs to be cautious not using training dataset of a different ethnicity for a TWAS.47
As observed in the real ROS/MAP studies, there remains a large gap between the 5-fold CV R2 using cis-eQTL predictors (∼5%) and the average genome-wide heritability of gene expression levels (21.8% estimated by GCTA48 based on a LMM). This is likely due to the large trans-acting contribution to transcript abundance documented for most genes. Thus, we hypothesize that it is promising to further improve the imputation R2 by fitting transcriptomic imputation models with genome-wide variants as predictors. Scalable Bayesian inference techniques such as the Expectation Maximization MCMC (EM-MCMC) algorithm49 are required for incorporating genome-wide variants.
Another limitation of existing TWAS methods is that the uncertainty of cis-eQTL effect-size estimates has not been taken into accounted in the association studies. A Bayesian framework can also be derived by taking the standard errors of these cis-eQTL effect-size estimates as prior standard deviations, which is part of our continuing research.
Besides the follow-up gene-based association studies (i.e., TWASs) described in this paper, the transcriptomic imputation models can be further extended by incorporating environmental contributions. The imputed transcript abundance levels can then be used for gene network analysis, differential gene expression analysis, and transcriptome mediation analysis with GWAS data. Validation of transcriptomic prediction accuracy in independent datasets will be critical in this regard, but unfortunately multiple large and similar datasets are not yet generally available for tissues other than peripheral blood.
In conclusion, we expect our work will provide a convenient and improved tool for transcriptomic imputation using the currently available rich reference datasets, as well as enhanced gene mapping for better understanding the genetic etiology of complex traits.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
J.Y. was supported by the startup funding from Department of Human Genetics at Emory University School of Medicine. A.P.W. and T.S.W. were supported by National Institutes of Health (NIH) R01AG056533. M.P.E. was supported by NIH R01GM11796. L.C.T. was supported by the Dermatology Foundation, the Arthritis National Research Foundation, the National Psoriasis Foundation, and NIH K01AR072129. ROS/MAP study data were provided by the Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, IL. Data collection was supported through funding by NIA grants P30AG10161, R01AG15819, R01AG17917, R01AG30146, R01AG36836, U01AG32984, and U01AG46152, the Illinois Department of Public Health, and the Translational Genomics Research Institute. In addition, we thank Thanneer Perumal and Benjamin Logsdon for performing quality control of the ROS/MAP RNA-sequencing data and for creating the brain cell type proportions.
Published: June 20, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.05.018.
Web Resources
IGAP data, http://web.pasteur-lille.fr/en/recherche/u744/igap/igap_download.php
PrediXcan, https://github.com/hakyim/PrediXcan
RADC Research Resource Sharing Hub, http://www.radc.rush.edu/
ROS/MAP data, https://www.synapse.org/#!Synapse:syn3219045
Supplemental Data
References
- 1.Visscher P.M., Brown M.A., McCarthy M.I., Yang J. Five years of GWAS discovery. Am. J. Hum. Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarthy M.I., Abecasis G.R., Cardon L.R., Goldstein D.B., Little J., Ioannidis J.P., Hirschhorn J.N. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 3.Huang Q. Genetic study of complex diseases in the post-GWAS era. J. Genet. Genomics. 2015;42:87–98. doi: 10.1016/j.jgg.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Farh K.K., Marson A., Zhu J., Kleinewietfeld M., Housley W.J., Beik S., Shoresh N., Whitton H., Ryan R.J., Shishkin A.A. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsoi L.C., Stuart P.E., Tian C., Gudjonsson J.E., Das S., Zawistowski M., Ellinghaus E., Barker J.N., Chandran V., Dand N. Large scale meta-analysis characterizes genetic architecture for common psoriasis associated variants. Nat. Commun. 2017;8:15382. doi: 10.1038/ncomms15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration &Visualization—EBI. Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz. Lead analysts. Laboratory, Data Analysis &Coordinating Center (LDACC) NIH program management. Biospecimen collection. Pathology. eQTL manuscript working group Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [Google Scholar]
- 7.Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lappalainen T., Sammeth M., Friedländer M.R., ’t Hoen P.A., Monlong J., Rivas M.A., Gonzàlez-Porta M., Kurbatova N., Griebel T., Ferreira P.G., Geuvadis Consortium Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Battle A., Mostafavi S., Zhu X., Potash J.B., Weissman M.M., McCormick C., Haudenschild C.D., Beckman K.B., Shi J., Mei R. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibbs J.R., van der Brug M.P., Hernandez D.G., Traynor B.J., Nalls M.A., Lai S.L., Arepalli S., Dillman A., Rafferty I.P., Troncoso J. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamazon E.R., Wheeler H.E., Shah K.P., Mozaffari S.V., Aquino-Michaels K., Carroll R.J., Eyler A.E., Denny J.C., Nicolae D.L., Cox N.J., Im H.K., GTEx Consortium A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W., Jansen R., de Geus E.J., Boomsma D.I., Wright F.A. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Z., Zhang F., Hu H., Bakshi A., Robinson M.R., Powell J.E., Montgomery G.W., Goddard M.E., Wray N.R., Visscher P.M., Yang J. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 14.Mancuso N., Shi H., Goddard P., Kichaev G., Gusev A., Pasaniuc B. Integrating Gene Expression with Summary Association Statistics to Identify Genes Associated with 30 Complex Traits. Am. J. Hum. Genet. 2017;100:473–487. doi: 10.1016/j.ajhg.2017.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su Y.R., Di C., Bien S., Huang L., Dong X., Abecasis G., Berndt S., Bezieau S., Brenner H., Caan B. A Mixed-Effects Model for Powerful Association Tests in Integrative Functional Genomics. Am. J. Hum. Genet. 2018;102:904–919. doi: 10.1016/j.ajhg.2018.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Y., Li M., Lu Q., Weng H., Wang J., Zekavat S.M., Yu Z., Li B., Gu J., Muchnik S., Alzheimer’s Disease Genetics Consortium A statistical framework for cross-tissue transcriptome-wide association analysis. Nat. Genet. 2019;51:568–576. doi: 10.1038/s41588-019-0345-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou H., Hastie T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Series B Stat. Methodol. 2005;67:301–320. [Google Scholar]
- 18.Zhou X., Carbonetto P., Stephens M. Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet. 2013;9:e1003264. doi: 10.1371/journal.pgen.1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tibshirani R. Regression Shrinkage and Selection via the Lasso. J. R. Stat. Soc. B. 1996;58:267–288. [Google Scholar]
- 20.Hoerl A.E., Kennard R.W. Ridge Regression: Biased Estimation for Nonorthogonal Problems. Technometrics. 2000;42:80–86. [Google Scholar]
- 21.Li Q., Lin N. The Bayesian elastic net. Bayesian Anal. 2010;5:151–170. [Google Scholar]
- 22.Guan Y.T., Stephens M. Bayesian Variable Selection Regression for Genome-Wide Association Studies and Other Large-Scale Problems. Ann. Appl. Stat. 2011;5:1780–1815. [Google Scholar]
- 23.Yu J., Pressoir G., Briggs W.H., Vroh Bi I., Yamasaki M., Doebley J.F., McMullen M.D., Gaut B.S., Nielsen D.M., Holland J.B. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 24.Huan T., Liu C., Joehanes R., Zhang X., Chen B.H., Johnson A.D., Yao C., Courchesne P., O’Donnell C.J., Munson P.J., Levy D. A systematic heritability analysis of the human whole blood transcriptome. Hum. Genet. 2015;134:343–358. doi: 10.1007/s00439-014-1524-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lloyd-Jones L.R., Holloway A., McRae A., Yang J., Small K., Zhao J., Zeng B., Bakshi A., Metspalu A., Dermitzakis M. The Genetic Architecture of Gene Expression in Peripheral Blood. Am. J. Hum. Genet. 2017;100:371. doi: 10.1016/j.ajhg.2017.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng P., Zhou X. Non-parametric genetic prediction of complex traits with latent Dirichlet process regression models. Nat. Commun. 2017;8:456. doi: 10.1038/s41467-017-00470-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blei D.M., Jordan M.I. Variational inference for Dirichlet process mixtures. Bayesian Anal. 2006;1:121–143. [Google Scholar]
- 28.Blei D.M., Kucukelbir A., McAuliffe J.D. Variational Inference: A Review for Statisticians. J. Am. Stat. Assoc. 2017;112:859–877. [Google Scholar]
- 29.Casella G. Empirical Bayes Gibbs sampling. Biostatistics. 2001;2:485–500. doi: 10.1093/biostatistics/2.4.485. [DOI] [PubMed] [Google Scholar]
- 30.Bennett D.A., Schneider J.A., Arvanitakis Z., Wilson R.S. Overview and findings from the religious orders study. Curr. Alzheimer Res. 2012;9:628–645. doi: 10.2174/156720512801322573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennett D.A., Schneider J.A., Buchman A.S., Barnes L.L., Boyle P.A., Wilson R.S. Overview and findings from the rush Memory and Aging Project. Curr. Alzheimer Res. 2012;9:646–663. doi: 10.2174/156720512801322663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng B., White C.C., Klein H.U., Sieberts S.K., McCabe C., Patrick E., Xu J., Yu L., Gaiteri C., Bennett D.A. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat. Neurosci. 2017;20:1418–1426. doi: 10.1038/nn.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennett D.A., Buchman A.S., Boyle P.A., Barnes L.L., Wilson R.S., Schneider J.A. Religious Orders Study and Rush Memory and Aging Project. J. Alzheimers Dis. 2018;64(s1):S161–S189. doi: 10.3233/JAD-179939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., European Alzheimer’s Disease Initiative (EADI) Genetic and Environmental Risk in Alzheimer’s Disease. Alzheimer’s Disease Genetic Consortium. Cohorts for Heart and Aging Research in Genomic Epidemiology Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reitz C. Genetic loci associated with Alzheimer’s disease. Future Neurol. 2014;9:119–122. doi: 10.2217/fnl.14.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reitz C. Novel susceptibility loci for Alzheimer’s disease. Future Neurol. 2015;10:547–558. doi: 10.2217/fnl.15.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sims R., van der Lee S.J., Naj A.C., Bellenguez C., Badarinarayan N., Jakobsdottir J., Kunkle B.W., Boland A., Raybould R., Bis J.C., ARUK Consortium. GERAD/PERADES, CHARGE, ADGC, EADI Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet. 2017;49:1373–1384. doi: 10.1038/ng.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan X.Z., Sun S., Tan C.C., Yu J.T., Tan L. The Role of ADAM10 in Alzheimer’s Disease. J. Alzheimers Dis. 2017;58:303–322. doi: 10.3233/JAD-170061. [DOI] [PubMed] [Google Scholar]
- 39.Müller P., Mitra R. Bayesian Nonparametric Inference - Why and How. Bayesian Anal. 2013;8:8. doi: 10.1214/13-BA811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carbonetto P., Stephens M. Scalable Variational Inference for Bayesian Variable Selection in Regression, and Its Accuracy in Genetic Association Studies. Bayesian Anal. 2012;7:73–107. [Google Scholar]
- 41.Li B., Leal S.M. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Reilly P.F., Hoggart C.J., Pomyen Y., Calboli F.C., Elliott P., Jarvelin M.R., Coin L.J. MultiPhen: joint model of multiple phenotypes can increase discovery in GWAS. PLoS ONE. 2012;7:e34861. doi: 10.1371/journal.pone.0034861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Jager P.L., Shulman J.M., Chibnik L.B., Keenan B.T., Raj T., Wilson R.S., Yu L., Leurgans S.E., Tran D., Aubin C., Alzheimer’s Disease Neuroimaging Initiative A genome-wide scan for common variants affecting the rate of age-related cognitive decline. Neurobiol. Aging. 2012;33:1017.e1–1017.e15. doi: 10.1016/j.neurobiolaging.2011.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Jager P.L., Srivastava G., Lunnon K., Burgess J., Schalkwyk L.C., Yu L., Eaton M.L., Keenan B.T., Ernst J., McCabe C. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat. Neurosci. 2014;17:1156–1163. doi: 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu L., Shi W., Long J., Guo X., Michailidou K., Beesley J., Bolla M.K., Shu X.O., Lu Y., Cai Q., NBCS Collaborators. kConFab/AOCS Investigators A transcriptome-wide association study of 229,000 women identifies new candidate susceptibility genes for breast cancer. Nat. Genet. 2018;50:968–978. doi: 10.1038/s41588-018-0132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin A.R., Gignoux C.R., Walters R.K., Wojcik G.L., Neale B.M., Gravel S., Daly M.J., Bustamante C.D., Kenny E.E. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am. J. Hum. Genet. 2017;100:635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang J., Fritsche L.G., Zhou X., Abecasis G., International Age-Related Macular Degeneration Genomics Consortium A Scalable Bayesian Method for Integrating Functional Information in Genome-wide Association Studies. Am. J. Hum. Genet. 2017;101:404–416. doi: 10.1016/j.ajhg.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.