Abstract
Proteins anchored to the cell surface via glycosylphosphatidylinositol (GPI) play various key roles in the human body, particularly in development and neurogenesis. As such, many developmental disorders are caused by mutations in genes involved in the GPI biosynthesis and remodeling pathway. We describe ten unrelated families with bi-allelic mutations in PIGB, a gene that encodes phosphatidylinositol glycan class B, which transfers the third mannose to the GPI. Ten different PIGB variants were found in these individuals. Flow cytometric analysis of blood cells and fibroblasts from the affected individuals showed decreased cell surface presence of GPI-anchored proteins. Most of the affected individuals have global developmental and/or intellectual delay, all had seizures, two had polymicrogyria, and four had a peripheral neuropathy. Eight children passed away before four years old. Two of them had a clinical diagnosis of DOORS syndrome (deafness, onychodystrophy, osteodystrophy, mental retardation, and seizures), a condition that includes sensorineural deafness, shortened terminal phalanges with small finger and toenails, intellectual disability, and seizures; this condition overlaps with the severe phenotypes associated with inherited GPI deficiency. Most individuals tested showed elevated alkaline phosphatase, which is a characteristic of the inherited GPI deficiency but not DOORS syndrome. It is notable that two severely affected individuals showed 2-oxoglutaric aciduria, which can be seen in DOORS syndrome, suggesting that severe cases of inherited GPI deficiency and DOORS syndrome might share some molecular pathway disruptions.
Keywords: PIGB, glycosylphosphatidylinositol, epilepsy, seizures, neuropathy, alkaline phosphatase, DOORS syndrome, intellectual disability, inherited GPI deficiency (IGD)
Introduction
Inherited glycosylphosphatidylinositol (GPI) deficiencies (IGDs) are a group of disorders found in individuals with mutations in the genes encoding proteins participating in GPI biosynthesis and modification. More than 30 proteins that are phosphatidylinositol glycan anchor biosynthesis (PIG) proteins and post-GPI-attachment-to-proteins (PGAP) proteins are involved in this process.1 GPI-anchored proteins (GPI-APs) play a variety of roles in the cell as hydrolytic enzymes, adhesion molecules, receptors, protease inhibitors, and complement regulatory proteins.2 To date, many variants in genes involved in the GPI-synthesis pathway have been linked to IGDs, and some of them are also referred to as GPIBDs (GPI biosynthesis defects [MIM: 618010]), Mabry syndrome, CHIME syndrome (coloboma, congenital heart disease, ichthyosiform dermatosis, mental retardation, and ear anomalies [MIM: 280000]), HPMRSs (hyperphosphatasia mental retardation syndromes [MIM: PS239300]), and MCAHSs (multiple congenital anomalies-hypotonia-seizures syndromes [MIM: PS614080]).3, 4 The main clinical features of IGDs include global developmental delay, intellectual disability, seizures, hypotonia, and facial dysmorphisms. Various organ anomalies such as Hirschsprung disease and renal hypoplasia can also be found in severe cases of IGDs. They also show deafness, hypoplastic nails, and brachytelephalangy, in addition to intellectual disability and seizures, all of which interestingly overlap with DOORS (deafness, onychodystrophy, osteodystrophy, mental retardation, and seizures [MIM: 220500]) syndrome, which is not considered an IGD and is most often caused by mutations in TBC1D24 [MIM: 613577] (Table S2). Here, we present a cohort of individuals from ten unrelated families with PIGB deficiency. The human PIGB [MIM: 604122], first isolated by Takahashi et al. in 1996,5 encodes an endoplasmic reticulum transmembrane protein that is made of 554 amino acids and transfers the third mannose to the GPI precursor.
Material and Methods
Samples from the Affected Individuals
The experimental procedures were approved by the institutional review boards of Osaka University, Iwate Medical University, and Yokohama City University in Japan, as well as the Centre Hospitalier Universitaire (CHU) Sainte-Justine Research Center. The samples were obtained with written informed consent by the parents.
Fluorescence-Activated Cell Sorting Analysis
The GPI-AP surface levels of granulocytes from individuals 4 and 7 or Chinese hamster ovary (CHO) cells were determined by staining cells with Alexa488-conjugated inactivated aerolysin (FLAER; Protox Biotech) and mouse anti-human CD59 (5H8), -human DAF (IA10), -human CD24 (ML5), -hamster-urokinase plasminogen activator receptor (uPAR) (5D6), and -human CD16 (3G8) monoclonal antibodies; then the cells were stained by a PE-conjugated anti-mouse IgG antibody and analyzed by flow cytometer (MACSQuant Analyzer 10, Miltenyi Biotec) with Flowjo software (v9.5.3, Tommy Digital).
Blood samples from individuals 2A and 5 were stained with FLAER-Alexa 448 (Cedarlane), PE-conjugated anti-human CD16 (BioLegend), and FITC-conjugated mouse anti-human CD55 or CD59 (BD PharMingen) for 1 h on ice in incubation buffer containing 0.5% BSA. Red blood cells were lysed in fluorescence-activated cell sorting (FACS) Lysing Solution (BD Bioscience), and the granulocytes’ GPI-AP levels were analyzed by Cytobank software.
For the fibroblasts of individuals 2A and 6B, cells were harvested at 80%–90% confluency then stained with FLAER-Alexa 448, FITC-conjugated mouse anti-human CD73, or PE-conjugated mouse anti-human CD109 (BioLegend) for 1 h on ice in the incubation buffer; then the cells were fixed in 3.7% formaldehyde. Non-specific binding was washed off before the cells were analyzed by a BD FACSCanto II system (BD Biosciences) and then analyzed by Cytobank software.
Whole-Exome Sequencing
For all the affected individuals, exome sequencing and analysis was performed, and this is described in detail in the Supplemental Material and Methods.
RNA Analysis
Total RNA was extracted with the QIAamp RNA Blood Mini Kit (QIAGEN) from the blood leukocytes of the proband (individual 6B), the mother, and a normal control. Total RNA was then reverse-transcribed with SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific) with random hexamers. Polymerase chain reaction (PCR) was performed with forward and reverse primers on exons 6 and 10 of PIGB, and the amplicon was subjected to Sanger sequencing. PCR conditions and primer sequences are available upon request.
Functional Analysis
HA-tagged human PIGB cDNA and its mutant, generated by site-directed mutagenesis, were subcloned into SRα (strong promoter)-driven pME vector, thymidine-kinase-promoter-driven pTK vector, or TATA-box-only pTA vector. Plasmids were transfected by electroporation into PIGB-deficient CHO cells (constitutively expressed human CD59 and DAF) with the luciferase-expressing construct to monitor the transfection efficiency. The levels of GPI-APs were analyzed by FACS, staining cells with anti-CD59 and anti-DAF, anti-uPAR antibody, and FLAER. The levels of PIGB in cells were determined by immunoblotting with anti-HA antibody (MBL) and normalized with luciferase activities for transfection efficiency and with band intensities of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) for the loading controls.
Rescue Assays of GPI-APs on the Fibroblasts from the Affected Individual 6B
Fibroblasts from the affected individual 6B were transduced with an empty-LX304 lentivirus or a wild-type (WT) PIGB-LX304 and selected by Blasticidin resistance. FACS analysis was performed as described above for fibroblasts with these transduced cells, which were compared with nontransduced cells, as well as healthy control cells.
Results
Clinical Information and Genetic Analysis of the Affected Individuals
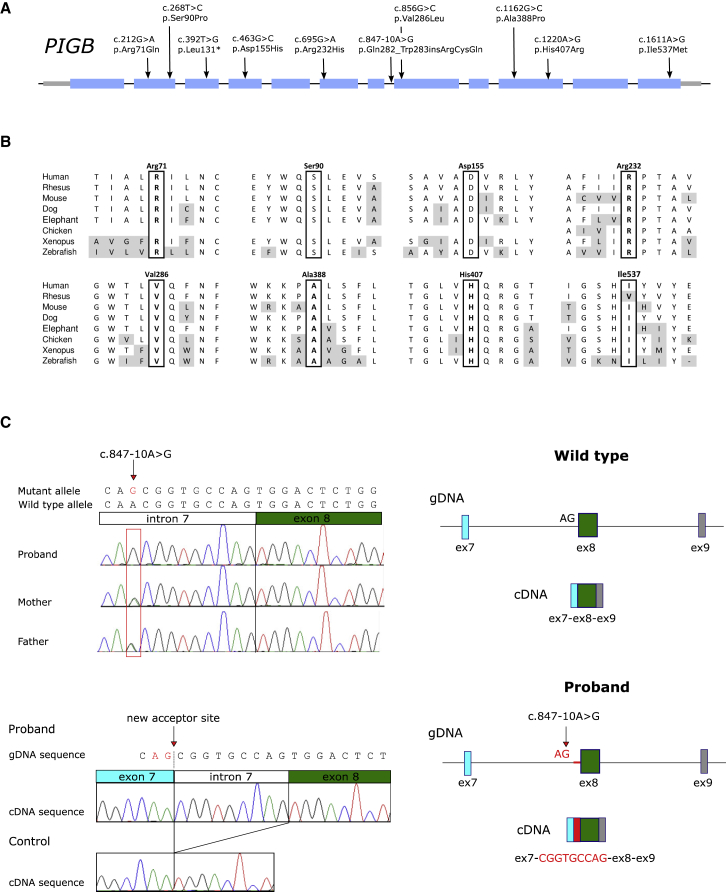
The initial family was identified as part of a pair of studies on DOORS syndrome (initially a candidate gene study looking notably at 2-oxoglutarate dehydrogenase, and then an exome study in which they were family number 12).6, 7 The probands were noted to have two mutations in PIGB (GenBank: NM_004855.4; c.212G>A [ p.Arg71Gln] and c.1162G>C [p.Ala388Pro]). The affected children had triphalangeal thumb, brachytelephalangy, and hypoplastic fingernails and toenails, in addition to seizures, developmental delay, and deafness (see Figure 1 and Table 1 for pedigrees, images, and variants for all affected families). They also showed 2-oxoglutaric aciduria (211 and 195 μmol/mmol creatinine. Reference value [RV] <9 μmol/mmol creatinine). Families 2 and 3 were identified by a search for PIGB variants in affected individuals who had clinical exomes performed at Baylor Genetics diagnostic laboratory (searching for overlapping clinical features and bi-allelic PIGB variants). Compound-heterozygous mutations c.695G>A (p.Arg232His) and c.856G>C (p.Val286Leu) were found in family 2, and the affected child in family 3 is homozygous for mutation c.1220A>G (p.His407Arg). In family 4, the affected individual, who is from Japanese non-consanguineous parents, was found through an IGD screening of CD16 cell surface presence on granulocytes by FACS analysis and then whole-exome sequencing. Subsequent families were matched to this study through Genematcher.8 The homozygous mutation c.847-10A>G was found in families 4, 6, 7, and 10. As shown in Figure 2, the replacement of A by G introduces a new acceptor site in intron 7; this results in the inclusion of 9 nucleotides of intron 7 in the mRNA and subsequently generates a splicing variant, p.Gln282_Trp283insArgCysGln (Figures 2C–E). The proband in family 5 has the compound-heterozygous mutations c.392T>G (p.Leu131∗) and c.1611A>G (p.Ile537Met), and the probands in families 8 and 9 were homozygous for the mutations c.268T>C (p.Ser90Pro) and c.463G>C (p.Asp155His), respectively.
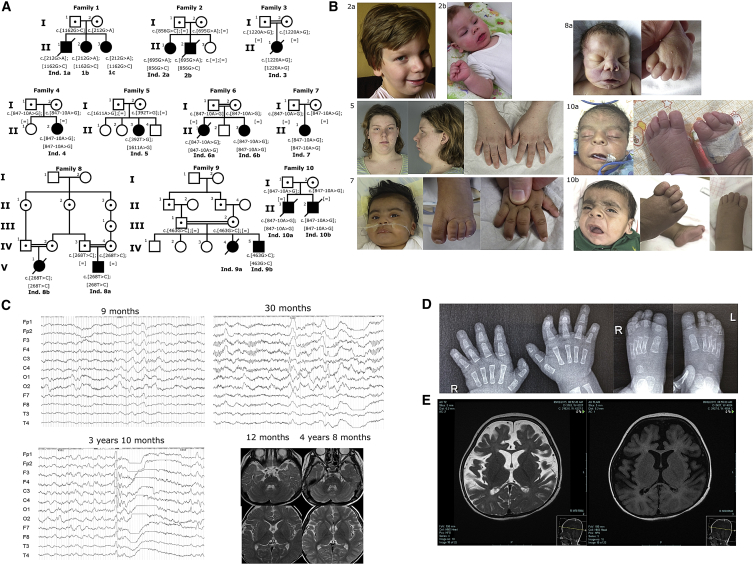
Figure 1.
Phenotypes of Affected Individuals
(A) The pedigrees of families with PIGB mutations. In family 5, the unaffected siblings were genotyped, and none were compound heterozygous. However, the family does not wish their carrier status to be published.
(B) Photographs of affected individuals. Individual 2A at 9 years old (note a wide nasal bridge, a long and smooth philtrum, a thin upper lip, a horizontal chin crease, and upturned earlobes), 2B at 11 months old (note similar features), individual 5 at 20 years old (note a horizontal chin crease, a prominent philtrum, and slightly upturned earlobes), individual 7 at 8 months old (note a wide nasal bridge, brachytelephalangy, and nail hypoplasia), individual 8A at birth (note a wide nasal bridge, a long and smooth philtrum, and a thin upper lip), individual 10A at birth, and individual 10B at 2.5 months old (note for both hypertrichosis, a wide nasal bridge, coarse facial features, a long and smooth philtrum, a pointed chin with a horizontal crease, uplifted earlobes, and toe and nail hypoplasia).
(C) EEG findings for individual 4. The top left panel depicts a sleep EEG at 9 months old, before any treatment; diffuse, low-voltage fast waves seem to be characteristic in this individual. Spindle waves are observed. High-amplitude slow waves are sometimes seen at the frontal regions predominantly. The top right panel depicts a sleep EEG at 30 months old; there are 0.5 s 12 Hz rhythmic waves at the frontal region and very small spike and high-voltage slow wave complexes. There is also a paucity of sleep markers and bilateral central spikes. The bottom left panel depicts a sleep EEG at 3 years and 10 months old. Generalized spike and wave discharge followed by voltage attenuation were seen. In the bottom right pictures, brain MRI did not demonstrate any brain anomalies such as dysplasia, atrophy, or delayed myelination.
(D) Radiographs of the hands and feet of individual 10B at 2.5 months of age showing aplasia of the terminal phalange of the fifth finger bilaterally and hypoplasia or aplasia of the terminal phalanges of the toes.
(E) MRI of individual 9A at 2 years and 3 months old. The left image depicts an axial T-2 sequence showing diffuse cerebral volume loss characterized by widening of the ventricles and extra CSF spaces with a prominent interhemispheric fissure anteriorly. There is suspicion of polymicrogyria along the bilateral occipital lobes, more pronounced on the right. The right image depicts an axial FLAIR sequence showing diffuse cerebral volume loss. There were signs of hypomyelination with a hyperintense signal in peri-ventricular and subcortical white matter, more pronounced at the occipital and frontal lobes bilaterally.
Table 1.
Mutations Identified
| Family | Genomic Variant (hg19) | mRNA Variant (GenBank:NM_004855.4) | Protein Variant | ExAC Minor Allele Frequency | gnomAD Minor Allele Frequency |
|---|---|---|---|---|---|
| 1 | chr15:g.55612521G>A | c.212G>A | p.Arg71Gln | 1.682 × 10−5 | 1.07 × 10−5 |
| chr15:g.55642935G>C | c.1162G>C | p.Ala388Pro | Not found | Not found | |
| 2 | chr15:g.55626106G>A | c.695G>A | p.Arg232His | 5.174 × 10−5 | 4.52 × 10−5 |
| chr15:g.55632819G>C | c.856G>C | p.Val286Leu | Not found | Not found | |
| 3 | chr15:g.55642993A>G | c.1220A>G | p.His407Arg | Not found | Not found |
| 4 | chr15:g.55632800A>G | c.847-10A>G | p.Gln282_Trp283insArgCysGln | 0.0001592 | 6.51 × 10−5 |
| 5 | chr15:g.55613563T>G | c.392T>G | p.Leu131∗ | Not found | 4.29 × 10−6 |
| chr15:g.55647576A>G | c.1611A>G | p.Ile537Met | Not found | Not found | |
| 6 | chr15:g.55632800A>G | c.847-10A>G | p.Gln282_Trp283insArgCysGln | 0.0001592 | 6.51 × 10−5 |
| 7 | chr15:g.55632800A>G | c.847-10A>G | p.Gln282_Trp283insArgCysGln | 0.0001592 | 6.51 × 10−5 |
| 8 | chr15: 55612577T>C | c.268T>C | p.Ser90Pro | Not found | Not found |
| 9 | chr15:55619774G>C | c.463G>C | p.Asp155His | 1.684 × 10−5 | 1.43 × 10−5 |
| 10 | chr15:55632800A>G | c.847-10A>G | p.Gln282_Trp283insArgCysGln | 0.0001592 | 6.51 × 10−5 |
Figure 2.
PIGB Variants
(A) PIGB variants found in affected individuals.
(B) Alignment of the PIGB sequence where missense mutations were found.
(C) Analysis of the PIGB splicing mutation. The upper left depicts Sanger sequencing of proband 6B and the parents that used their genomic DNA showed that the c.847-10A>G mutation was homozygous in the proband and heterozygous in both parents. The lower left depicts the cDNA analysis performed with leukocytes from the proband; the analysis showed the insertion of the last nine bases of intron 7 before the canonical exon 8 as a result of the activation of an aberrant splice acceptor site. The proband did not express wild-type mRNA, which was observed in a control. At right, a graphic description of the activation of the aberrant splice acceptor site at exon 8 in PIGB occurred in the proband. Abbreviations are as follows: gDNA = genomic DNA and Ex = exon.
All affected individuals have global development delay and/or intellectual disability (Tables 2 and S1). The individuals from families 1–3, 6, and 8–10 were severely affected, and one in each family died at early ages of complications of their epileptic encephalopathy. Neurological abnormalities including axonal degenerative polyneuropathy and demyelinating sensorimotor polyneuropathy, hearing loss, and visual impairment were observed in the severely affected individuals. All individuals suffered from seizures that started in infancy. One individual has hypertonia, spasticity, and joint contractures. On MRI, two had polymicrogyria, two had hypomyelination, three had a thin corpus collosum, and three had enlarged ventricles (Figure 1E).
Table 2.
Phenotypic Summary of the Affected Individuals
| Family | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | Total | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Individual | 1A | 1B | 2A | 2B | 3 | 4 | 5 | 6A | 6B | 7 | 8A | 9B | 10A | 10B | |
| DD/ID | + | + | + | + | + | + | + | + | + | + | NA | + | + | + | 13/13 |
| Seizures | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 14/14 |
| Dysmorphic features | + | + | − | + | + | − | + | + | + | + | + | − | + | + | 11/14 |
| Deafness | + | + | + | − | + | − | − | − | − | + | − | − | + | + | 7/14 |
| Ophthalmological anomalies | + | + | + | + | + | − | − | + | + | + | NA | NA | NA | NA | 8/10 |
| Hand and feet anomalies | + | + | − | + | + | − | + | − | − | + | + | − | + | + | 9/14 |
| Brain MRI or CT anomalies | + | + | − | + | + | − | − | − | − | + | + | NA | + | + | 8/13 |
| Elevated serum alkaline phosphatase | NA | NA | − | NA | + | + | + | + | + | + | NA | NA | + | + | 8/9 |
| 2-oxoglutaric aciduria | + | + | NA | − | − | − | NA | − | NA | − | NAa | NAa | NA | − | 2/8 |
Thorough clinical details are not available for individuals 1C, 8B, and 9A. NA = details not available.
Urine organic acids were normal in the other affected relative shown in Figure 1.
Coarse facial features were noted in five individuals, ear anomalies in ten (notably upturned earlobes in seven), micrognathia in two, a tented mouth in two, full cheeks in four, a broad nasal bridge in four, shortening of distal phalanges in six, and hypoplastic nails in seven (Figures 1B and 1D).
Elevated plasma alkaline phosphatase (ALP) was noted in most tested individuals (Table S1). The fetal phenotype of the second child of family 10 included increased nuchal thickness at 19 weeks gestation and polyhydramnios with a protuberant abdomen and upper lip noted at the 30 weeks antenatal scan.
Flow Cytometric Analysis of Blood Cells
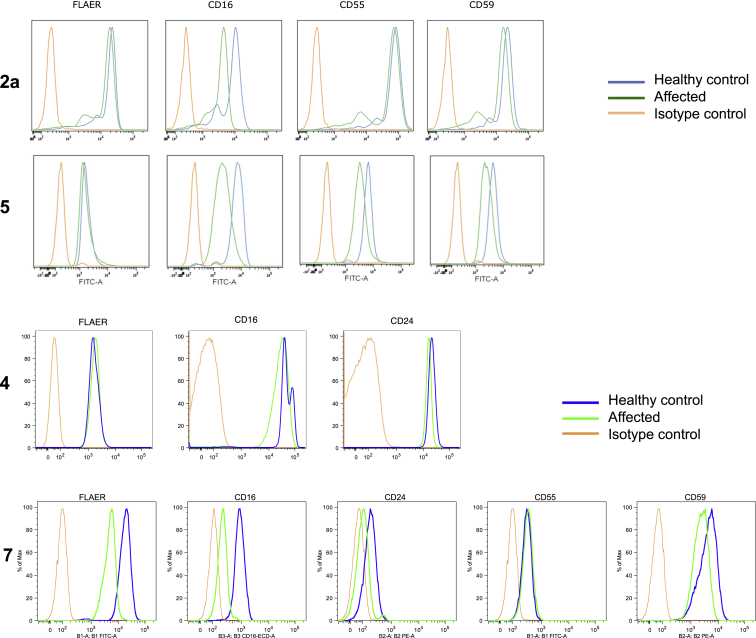
The levels of cell surface GPI-AP abundance in available blood samples from the affected individuals of families 2, 4, 5, and 7 were examined by flow cytometric analysis that used various GPI-AP markers, including fluorescence-labeled aerolysin (FLAER), CD16, CD24, CD55, CD56, and CD59. The CD16 presence on the cell surface of granulocytes from individuals 2A, 4, 5, and 7 was decreased to 53%, 56%, 35%, and 25% of that from the healthy control, respectively (Figure 3). The presences of CD55 and CD59 on the cell surface of granulocytes from individual 5 were both decreased to 60%, and in individual 7, CD59 was reduced to 50% (Figure 3). Lymphoblastoid cell lines established by immortalization of the B lymphocytes showed a decrease to 55% for the FLAER signal in individual 2A and a decrease to 55% for CD55 in individual 5 compared to that in the healthy control (Figure S1).
Figure 3.
Flow Cytometry Analysis of Cell Surface GPI-APs of Granulocytes
Blood samples collected from affected individuals and controls were stained with FLAER and antibodies against GPI-APs (CD16, CD24, CD55, and CD59) and were analyzed by the BD Scanto II system or the MACSQuant system. The granulocyte population was gated on the basis of cellular granularity, cell size, and CD45 level.
Flow Cytometric Analysis of Fibroblasts
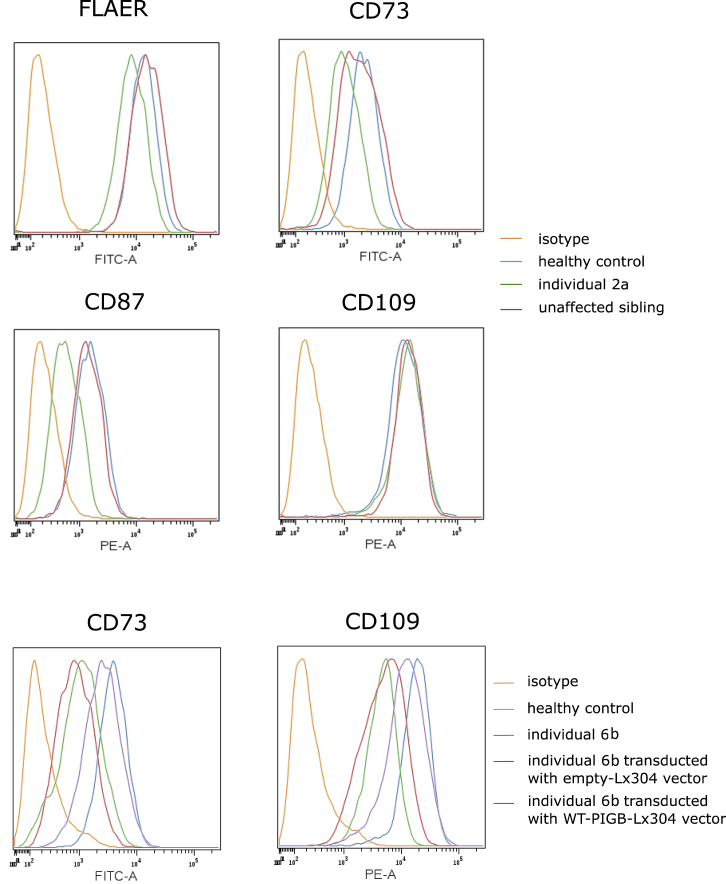
In fibroblasts established from skin biopsies of individuals 2A and 6B, we also found low GPI-AP levels. The fibroblasts from individual 2A showed decreased FLAER (35%), CD73 (54%), and CD87 (58%) compared to those from the healthy control, whereas the unaffected sibling showed normal levels of GPI-APs (Figure 4). In individual 6B, CD73 and CD109 levels were decreased to 30% and 26%, respectively, but were rescued up to 70% of the healthy control by transduction with a PIGB-encoding-Lx304 lentiviral vector. This suggested that the mutation in PIGB is responsible for the reduced detection of GPI-APs on the fibroblasts from individual 6B (Figure 4).
Figure 4.
Decreased Level of GPI-AP in the Fibroblasts and Rescue by PIGB-Expressing Lentivector
Skin fibroblasts derived from individuals 2A and 6B were stained with FLAER, CD73, CD87, and CD109 and were analyzed by the BD FACScanto II system and then by Cytobank software. Fibroblasts from individual 6B were transduced with PIGB-expressing-Lx304 lentivirus or empty-vector lentivirus, and these fibroblasts were stained with CD73 and CD109 as described above. The figure shows representative results from experiments done in triplicate.
Functional Studies with PIGB-Deficient CHO Cells
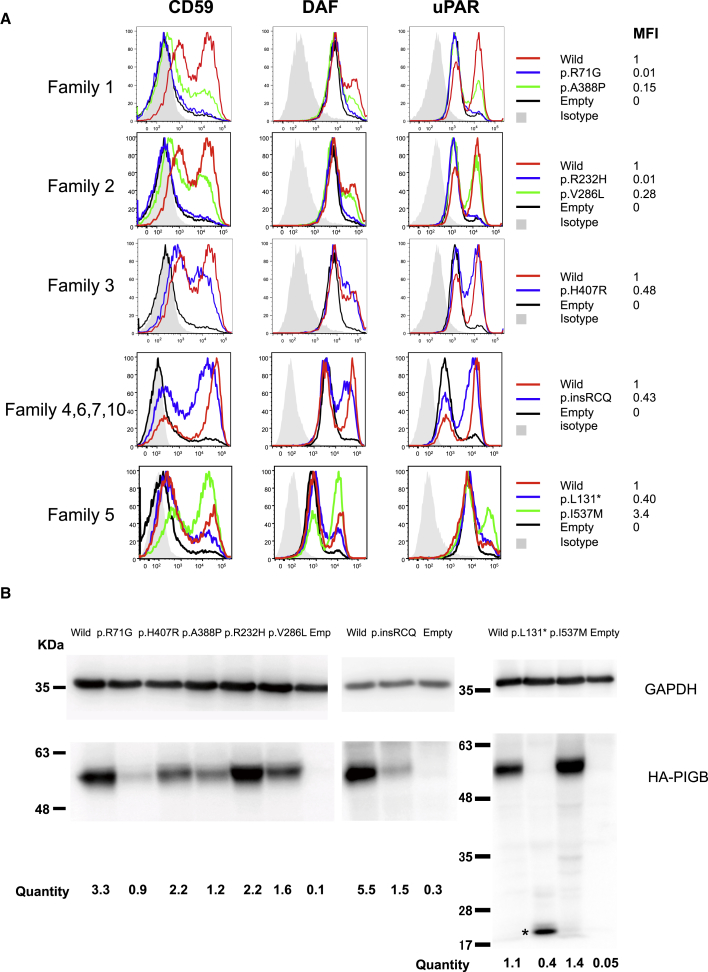
PIGB-deficient CHO cells9 were transfected with wild-type or the various mutant PIGB cDNAs driven by the strong (pME) promoter. FACS analysis for the presence of GPI-APs, such as CD59, CD55, and CD87 (uPAR), at the cell surface and immunoblotting were performed three days later (Figure 5). PIGB cDNA bearing the p.Ala388Pro variant only slightly restored the surface presence of GPI-APs compared to wild-type cDNA (15% of wild type), and PIGB cDNA with the p.Arg71Gln variant showed null activity even with the strong-promoter-driven constructs. These data suggested that both variants found in Family 1 greatly reduced PIGB activity, leading to clinical severity. The levels of the p.Arg71Gln variant were severely decreased; the p.Ala388Pro variant also decreased to 50% of the wild-type PIGB (Figure 5B). Of the variants found in family 2, p.Val286Leu only partially restored GPI-APs (28% of wild type), and p.Arg232His showed almost null activity, suggesting again greatly reduced PIGB activity. The variant protein level was slightly reduced for p.Val286Leu. The variant found in family 3, p.His407Arg, partially restored the surface levels of GPI-APs (48% of wild type), and its protein level was reduced to 70%. The splicing variant p.Gln282_Trp283insArgCysGln, found in families 4, 6, 7, and 10, partially restored the surface levels of GPI-APs (43% of wild type), and the protein level was severely reduced, suggesting that the variant protein is unstable and the actual PIGB activity in the affected individuals might be more severely decreased. The p.Ile537Met and p.Leu131∗ variants found in family 5 showed divergent results: p.Leu131∗ showed partial restoration (40% of wildtype), whereas the p.Ile537Met variant restored GPI-APs more efficiently than the wild type, presumably because of elevated protein levels. Note that the transcript encoding p.Leu131∗ most likely undergoes nonsense-mediated decay (NMD) in vivo; in these experiments, however, the cDNA does not have introns, and thus there is no NMD. When the p.Ile537Met mutant was produced by constructs driven by weaker promoters (pTK and pTA), slightly decreased activities were detected, suggesting that mild reduction of PIGB activity leads to a mild phenotype (as seen in individual 5; Figure S2).
Figure 5.
Functional Analysis of the Mutant PIGB cDNAs Found in the Families
(A) PIGB-deficient CHO cells were transiently transfected with wild-type and mutant HA-tagged PIGB cDNAs subcloned in pME (strong SRα promoter-driven vector). Restoration of the surface expression of CD59, CD55 (DAF), and uPAR was assessed two days later by flow cytometry. Black lines indicate the empty vector; green and blue lines indicate various types of mutant PIGB; the red line indicates wild-type PIGB; and light gray shadows indicate isotype controls. Various missense variants, except for I537M, driven by a strong promoter only partially rescued the expression of GPI-APs compared to wild-type PIGB.
(B) Levels of the mutant PIGB. Lysates of the transfectants were subjected to SDS-PAGE and immunoblotting. For quantification, levels of PIGB were determined by dividing PIGB band intensities by GAPDH band intensities and luciferase activity to normalize for both loading and transfection efficiencies. ∗ = truncated PIGB. Expression levels of mutant PIGB, except for the p.I5le37Met variant, were decreased to various degrees.
Discussion
IGDs show clinical heterogeneity and overlap with various other syndromes and diseases. For example, Mabry syndrome, also known as hyperphosphatasia with mental retardation syndrome, is now known to be an IGD. Some individuals with Fryns syndrome (MIM: 229850), characterized by diaphragmatic hernia, gastrointestinal and genitourinary malformations, coarse facies, and distal digital hypoplasia, also have pathogenic PIGN (MIM: 606097) or PIGV (MIM: 610274) mutations. CHIME syndrome, characterized by colobomas, heart defects, ichthyosiform dermatosis, intellectual disability, and either ear defects or epilepsy is also an IGD. Toriello-Carey syndrome is also caused by PGAP3 mutations10, 11 (MIM: 611801). These syndromes were initially defined by a combination of characteristic symptoms, and GPI pathway genes were subsequently implicated by genetic analyses.
FACS analysis is useful for the screening of IGDs. There are more than 150 different GPI-APs, and each precursor protein has a GPI attachment signal (20∼30 hydrophobic amino acids) at the C terminus. The GPI attachment signals in different proteins have different signal strengths. When the production of GPI is limited by the mutations, those precursor proteins that have strong GPI attachment signals are preferentially modified by GPI. Thus, the degree of decrease differs among the various GPI-APs. Taking into consideration that different kinds of cells express different kinds of GPI-APs, we applied the most sensitive markers known to examine the pathogenicity of the mutations.
In this report, we describe an IGD: PIGB deficiency. Levels of various GPI-APs on granulocytes and fibroblasts were decreased in the affected individuals; however, the degree of decrease was not correlated with the clinical severity. On the basis of functional analysis that used PIGB-deficient CHO cells, the residual activities of the mutant PIGB from the families are in a rank order from high to low: p.Ile537Met, p.Leu131∗ (family 5), > p.His407Arg (family 3), ≥ p.Gln282_Trp283insArgCysGln (families 4, 6, 7, and 10), > p.Arg232His, p.Val28 6Leu (family 2), > p.Arg71Gln, and p.Ala388Pro (family 1). Note that the c.847-10A>G mutation has a minor allele frequency of 0.046% in South Asians and 0.002% in Europeans. It is thus not so rare in South Asians, but no homozygotes were found in gnomAD. Nevertheless, it still does not have such a high frequency as the pathogenic PGAP3 GenBank: NM_033419.4; c.∗559C>T mutation that is found in Europeans (0.6%) and was implicated in another IGD (HPMRS4; MIM: 615716).
These results correlate with the clinical severities: the individuals from families 1 and 2 showed severe development delay, axonal degenerative polyneuropathy, dysmorphic facies and ears, hearing loss, visual impairment, and finger abnormalities, whereas the one from family 5 showed no dysmorphism and experienced seizures later in infancy. Notably, some affected individuals (family 1) with a severe phenotype were diagnosed with DOORS syndrome. Because the defining symptoms of DOORS syndrome (deafness, onycho-osteodystrophy, mental retardation, and seizures) completely fit with severe forms of IGDs, this clinical description also fits reported cases of IGDs caused by mutation in other genes.12, 13 DOORS syndrome is mainly caused by bi-allelic mutations in TBC1D24. It is important to clarify whether clinical overlap between IGDs and TBC1D24 deficiency are related to a chance similarity of symptoms or whether they have some link in pathophysiology, especially when considering the management of these individuals (Table S2). TBC1D24 was reported to negatively regulate both RAB35 and ARF6 by working as a GTPase-activating protein (GAP) for these small G proteins.14, 15, 16 Effector proteins modulate the crosstalk between RAB35 and ARF6 and thereby regulate endocytosis and the recycling of surface proteins, including GPI-APs.16, 17 Thus, TBC1D24 deficiency might affect the surface levels of GPI-APs on neurons, suggesting some pathological relationship between IGDs and DOORS syndrome. The evidence that two of the individuals affected with severe PIGB deficiency showed 2-oxoglutaric aciduria, an intermediate in the TCA cycle, sharing the characteristic metabolic abnormality of DOORS syndrome, supports the suggestion above. Individuals who have DOORS syndrome with TBC1D24 deficiency show 2-oxoglutaric aciduria in a high proportion of cases; however, the mechanism of this is still unknown. 2-oxoglutaric aciduria can be caused by the decreased activity of the 2-oxoglutaric dehydrogenase in the TCA cycle, which requires vitamin B1 as a coenzyme. Vitamin B1 is absorbed from the intestine as a form of thiamine, either after de-phosphorylation of thiamine monophosphate or thiamine diphosphate. Intestinal alkaline phosphatase (ALP) and tissue non-specific ALP, which are both GPI-APs, play a role in this de-phosphorylation. We speculate that the vitamin B1 concentration in the neurons from IGD is decreased because the surface level of ALP would be decreased,18 and this leads to accumulation of 2-oxoglutaric acid. 2-oxoglutaric aciduria in TBC1D24 deficiency might be similarly caused by a decreased level of ALP. These hypotheses should be further investigated. It is interesting to note that abnormally high signals within the brainstem on the diffusion-weighted images of MRI are often observed in severe cases of IGDs, suggesting mitochondrial dysfunction or other metabolic abnormalities,19, 20 and affected individuals with PIGA mutations (MIM: 300868) have evidence of mitochondrial dysfunction on biopsied tissues.20 However, it should be considered that there are many other causes of 2-oxoglutric aciduria (e.g., multiple mitochondrial diseases, Amish type microcephaly, physiologic immaturity in newborns, high carbohydrate diets, and citrate-based food additives). Yet, in these situations, there usually are not multiple malformations as is seen in IGDs and TBC1D24 mutations. Because 2-oxoglutaric acid in the urine (as part of a urinary organic acid profile) is not routinely examined in general hospitals, clinicians request it only when they suspect an inborn error of intermediary metabolism. In our cohort, this was measured in eight individuals, and only the severely affected siblings of family 1 showed 2-oxoglutaric aciduria, thus there might be a correlation with the severity of the disease. Note that the levels were not always elevated in both siblings.7 Such a correlation between 2-oxoglutaric aciduria and disease severity has been proposed for DOORS syndrome,21 but instead it was demonstrated that the levels can fluctuate with time between normal and elevated and did not correlate with disease severity.22, 23, 24 In addition, because thiamine pyrophosphate is abundantly contained in animal food products, 2-oxoglutaric aciduria can be affected by the diet. It is necessary to measure 2-oxoglutaric acid in the urine from more IGD-affected individuals to clarify the mechanism for this sign.
Regarding the neuropathy, there are interesting correlates to make when considering other known cause of neuropathies. Considering the mitochondrial dysfunction hypothesis mentioned above, demyelinating neuropathy is a known feature of such mitochondrial disorders as neuropathy, ataxia, and retinitis pigmentosa (NARP [MIM: 551500]) and myoneurogastrointestinal encephalopathy (MNGIE [MIM: 603041]). Another condition is inherited CD59 (a GPI-anchored complement regulator) deficiency (MIM: 612300), which is associated with immune-mediated polyneuropathy.25 Also, a polyneuropathy is seen in vitamin B1 deficiency (see above) and in folate deficiency (the folate receptor is another GPI-AP); in both cases the neuropathy is axonal. Finally, a PLP (pyridoxal 5′-phosphate)-responsive primary axonal peripheral neuropathy and optic atrophy have recently been associated with recessive mutations in PDXK (MIM: 179020), encoding a gene involved in converting pyridoxal to the active form of B6, PLP.26 Given the fact that some children with IGDs have pyridoxine-responsive seizures, this is interesting because alkaline phosphatase also converts PLP to pyridoxal, and this process is essential for B6 to reach the central nervous system (CNS). The mechanism at play in the neuropathy seen in the individuals described here would benefit from model organism studies.
In summary, we characterize an IGD due to PIGB deficiency associated with global developmental and/or intellectual delay, early onset epilepsy, and an axonal neuropathy in severe cases. There was clinical overlap with DOORS syndrome, not only with regard to clinical signs, but also at the level of metabolic anomalies. 2-oxoglutaric aciduria in IGDs might be unnoticed, therefore urine organic acids should be measured in individuals with IGDs. This suggests a possible converging molecular mechanism that will need to be assessed in future studies.
Declaration of Interests
The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing conducted at Baylor Genetics. The authors declare no competing interests.
Acknowledgments
This work is supported in part by a grant for Research on Measures for Intractable Diseases, a grant for Comprehensive Research on Disability Health and Welfare, the Strategic Research Program for Brain Science (SRPBS) (JP19dm0107090) (N.M.), and the Practical Research Project for Rare/Intractable Diseases from the Japan Agency for Medical Research and Development (AMED) (JP19ek0109280, JP19ek0109301, JP18kk0205001, and JP19ek0109348 for N.M.); Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (KAKENHI) under grant numbers JP17K15639 (N.M.) and 17K10080 (S.M.); grants from the Ministry of Health, Labor and Welfare (for N.M. and Y.M.); and the Takeda Science Foundation (N.M.). P.M.C. and E.R. are supported by the Canadian Institutes of Health Research (CIHR) (grant number RN 324373) and T.T.M.N. is supported by the Savoy Foundation. T.B.H. was supported by the German Bundesministerium für Bildung und Forschung (BMBF) through the Juniorverbund in der Systemmedizin “mitOmics” (FKZ 01ZX1405C). S.E. is a Wellcome Senior Investigator. We thank Hessa Alsaif, Mohammed Alamoudi, Saori Umeshita, and Kana Miyanagi for technical assistance.
Published: June 27, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.05.019.
Contributor Information
Taroh Kinoshita, Email: tkinoshi@biken.osaka-u.ac.jp.
Philippe M. Campeau, Email: p.campeau@umontreal.ca.
Web Resources
ExAC Browser, http://exac.broadinstitute.org/
OMIM, https://omim.org/
UCSC Genome Browser, https://genome.ucsc.edu/
UniProt, https://www.uniprot.org/uniprot/
GeneMatcher (GM), https://www.genematcher.org/
Supplemental Data
References
- 1.Kinoshita T., Fujita M. Biosynthesis of GPI-anchored proteins: Special emphasis on GPI lipid remodeling. J. Lipid Res. 2016;57:6–24. doi: 10.1194/jlr.R063313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.UniProt Consortium UniProt: A hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knaus A., Pantel J.T., Pendziwiat M., Hajjir N., Zhao M., Hsieh T.C., Schubach M., Gurovich Y., Fleischer N., Jäger M. Characterization of glycosylphosphatidylinositol biosynthesis defects by clinical features, flow cytometry, and automated image analysis. Genome Med. 2018;10:3. doi: 10.1186/s13073-017-0510-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellai-Dussault K., Nguyen T.T.M., Baratang N.V., Jimenez-Cruz D.A., Campeau P.M. Clinical variability in inherited glycosylphosphatidylinositol deficiency disorders. Clin. Genet. 2019;95:112–121. doi: 10.1111/cge.13425. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi M., Inoue N., Ohishi K., Maeda Y., Nakamura N., Endo Y., Fujita T., Takeda J., Kinoshita T. PIG-B, a membrane protein of the endoplasmic reticulum with a large lumenal domain, is involved in transferring the third mannose of the GPI anchor. EMBO J. 1996;15:4254–4261. [PMC free article] [PubMed] [Google Scholar]
- 6.Campeau P.M., Kasperaviciute D., Lu J.T., Burrage L.C., Kim C., Hori M., Powell B.R., Stewart F., Félix T.M., van den Ende J. The genetic basis of DOORS syndrome: An exome-sequencing study. Lancet Neurol. 2014;13:44–58. doi: 10.1016/S1474-4422(13)70265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Bever Y., Balemans W., Duval E.L., Jespers A., Eyskens F., van Hul W., Courtens W. Exclusion of OGDH and BMP4 as candidate genes in two siblings with autosomal recessive DOOR syndrome. Am. J. Med. Genet. A. 2007;143A:763–767. doi: 10.1002/ajmg.a.31641. [DOI] [PubMed] [Google Scholar]
- 8.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashida H., Hong Y., Murakami Y., Shishioh N., Sugimoto N., Kim Y.U., Maeda Y., Kinoshita T. Mammalian PIG-X and yeast Pbn1p are the essential components of glycosylphosphatidylinositol-mannosyltransferase I. Mol. Biol. Cell. 2005;16:1439–1448. doi: 10.1091/mbc.E04-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maddirevula S., Alsahli S., Alhabeeb L., Patel N., Alzahrani F., Shamseldin H.E., Anazi S., Ewida N., Alsaif H.S., Mohamed J.Y. Expanding the phenome and variome of skeletal dysplasia. Genet. Med. 2018;20:1609–1616. doi: 10.1038/gim.2018.50. [DOI] [PubMed] [Google Scholar]
- 11.Maddirevula S., Alzahrani F., Al-Owain M., Al Muhaizea M.A., Kayyali H.R., AlHashem A., Rahbeeni Z., Al-Otaibi M., Alzaidan H.I., Balobaid A. Autozygome and high throughput confirmation of disease genes candidacy. Genet. Med. 2019;21:736–742. doi: 10.1038/s41436-018-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alessandri J.L., Gordon C.T., Jacquemont M.L., Gruchy N., Ajeawung N.F., Benoist G., Oufadem M., Chebil A., Duffourd Y., Dumont C. Recessive loss of function PIGN alleles, including an intragenic deletion with founder effect in La Réunion Island, in patients with Fryns syndrome. Eur. J. Hum. Genet. 2018;26:340–349. doi: 10.1038/s41431-017-0087-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleming L., Lemmon M., Beck N., Johnson M., Mu W., Murdock D., Bodurtha J., Hoover-Fong J., Cohn R., Bosemani T. Genotype-phenotype correlation of congenital anomalies in multiple congenital anomalies hypotonia seizures syndrome (MCAHS1)/PIGN-related epilepsy. Am. J. Med. Genet. A. 2016;170A:77–86. doi: 10.1002/ajmg.a.37369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uytterhoeven V., Kuenen S., Kasprowicz J., Miskiewicz K., Verstreken P. Loss of skywalker reveals synaptic endosomes as sorting stations for synaptic vesicle proteins. Cell. 2011;145:117–132. doi: 10.1016/j.cell.2011.02.039. [DOI] [PubMed] [Google Scholar]
- 15.Falace A., Buhler E., Fadda M., Watrin F., Lippiello P., Pallesi-Pocachard E., Baldelli P., Benfenati F., Zara F., Represa A. TBC1D24 regulates neuronal migration and maturation through modulation of the ARF6-dependent pathway. Proc. Natl. Acad. Sci. USA. 2014;111:2337–2342. doi: 10.1073/pnas.1316294111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheehan P., Waites C.L. Coordination of synaptic vesicle trafficking and turnover by the Rab35 signaling network. Small GTPases. 2019;10:54–63. doi: 10.1080/21541248.2016.1270392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai B., Katafiasz D., Horejsi V., Naslavsky N. Pre-sorting endosomal transport of the GPI-anchored protein, CD59, is regulated by EHD1. Traffic. 2011;12:102–120. doi: 10.1111/j.1600-0854.2010.01135.x. [DOI] [PubMed] [Google Scholar]
- 18.Murakami Y., Kanzawa N., Saito K., Krawitz P.M., Mundlos S., Robinson P.N., Karadimitris A., Maeda Y., Kinoshita T. Mechanism for release of alkaline phosphatase caused by glycosylphosphatidylinositol deficiency in patients with hyperphosphatasia mental retardation syndrome. J. Biol. Chem. 2012;287:6318–6325. doi: 10.1074/jbc.M111.331090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato M., Saitsu H., Murakami Y., Kikuchi K., Watanabe S., Iai M., Miya K., Matsuura R., Takayama R., Ohba C. PIGA mutations cause early-onset epileptic encephalopathies and distinctive features. Neurology. 2014;82:1587–1596. doi: 10.1212/WNL.0000000000000389. [DOI] [PubMed] [Google Scholar]
- 20.Tarailo-Graovac M., Sinclair G., Stockler-Ipsiroglu S., Van Allen M., Rozmus J., Shyr C., Biancheri R., Oh T., Sayson B., Lafek M. The genotypic and phenotypic spectrum of PIGA deficiency. Orphanet J. Rare Dis. 2015;10:23. doi: 10.1186/s13023-015-0243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajab A., Riaz A., Paul G., Al-Khusaibi S., Chalmers R., Patton M.A. Further delineation of the DOOR syndrome. Clin. Dysmorphol. 2000;9:247–251. doi: 10.1097/00019605-200009040-00003. [DOI] [PubMed] [Google Scholar]
- 22.Patton M.A., Krywawych S., Winter R.M., Brenton D.P., Baraitser M. DOOR syndrome (deafness, onycho-osteodystrophy, and mental retardation): Elevated plasma and urinary 2-oxoglutarate in three unrelated patients. Am. J. Med. Genet. 1987;26:207–215. doi: 10.1002/ajmg.1320260131. [DOI] [PubMed] [Google Scholar]
- 23.Félix T.M., de Menezes Karam S., Della Rosa V.A., Moraes A.M. DOOR syndrome: Report of three additional cases. Clin. Dysmorphol. 2002;11:133–138. doi: 10.1097/00019605-200204000-00012. [DOI] [PubMed] [Google Scholar]
- 24.James A.W., Miranda S.G., Culver K., Hall B.D., Golabi M. DOOR syndrome: Clinical report, literature review and discussion of natural history. Am. J. Med. Genet. A. 2007;143A:2821–2831. doi: 10.1002/ajmg.a.32054. [DOI] [PubMed] [Google Scholar]
- 25.Nevo Y., Ben-Zeev B., Tabib A., Straussberg R., Anikster Y., Shorer Z., Fattal-Valevski A., Ta-Shma A., Aharoni S., Rabie M. CD59 deficiency is associated with chronic hemolysis and childhood relapsing immune-mediated polyneuropathy. Blood. 2013;121:129–135. doi: 10.1182/blood-2012-07-441857. [DOI] [PubMed] [Google Scholar]
- 26.Chelban V., Wilson M.P., Warman Chardon J., Vandrovcova J., Zanetti M.N., Zamba-Papanicolaou E., Efthymiou S., Pope S., Conte M.R., Abis G. PDXK mutations cause polyneuropathy responsive to PLP supplementation. Annals of Neurology. 2019 doi: 10.1002/ana.25524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.