Abstract
Copy-number variations (CNVs) represent a significant proportion of the genetic differences between individuals and many CNVs associate causally with syndromic disease and clinical outcomes. Here, we characterize the landscape of copy-number variation and their phenome-wide effects in a sample of 472,228 array-genotyped individuals from the UK Biobank. In addition to population-level selection effects against genic loci conferring high mortality, we describe genetic burden from potentially pathogenic and previously uncharacterized CNV loci across more than 3,000 quantitative and dichotomous traits, with separate analyses for common and rare classes of variation. Specifically, we highlight the effects of CNVs at two well-known syndromic loci 16p11.2 and 22q11.2, previously uncharacterized variation at 9p23, and several genic associations in the context of acute coronary artery disease and high body mass index. Our data constitute a deeply contextualized portrait of population-wide burden of copy-number variation, as well as a series of dosage-mediated genic associations across the medical phenome.
Keywords: genetics, genomics, copy number variation, structural variation, UK Biobank, association testing, population database, microdeletion/microduplication syndrome, selection bias
Introduction
Copy-number variants (CNVs) are a class of structural variation typically defined as large deletions or duplications of at least 50 base-pairs of genomic sequence.1, 2 CNVs exhibit substantial variability in both size and frequency in the population and have been implicated across a variety of common and rare health outcomes.3 Regional deletion and duplication syndromes have also been described at many loci, clustering near areas of segmental duplication which may potentiate non-allelic homologous recombination.4, 5, 6 For example, CNV-based architectures for neuropsychiatric (e.g., autism spectrum disorder), developmental (e.g., 16p11.2 [MIM: 611913]),7, 8 and syndromic cardiac disease (e.g., 22q11.2 [MIM: 188400]) (see GeneReviews in Web Resources) phenotypes have been well established.
Despite a growing body of research on CNV-related syndromes and disease etiologies, large-scale studies of CNV effects have been limited by their rarity in the general population. However, burden testing methods that address this rarity by pooling observed variation across gene regions have yielded reproducible associations in the context of congenital heart disease and various neurocognitive outcomes.10, 11 Moreover, as studies which include either microarray or NGS-based genotype data have grown in size and scope, it has become possible to describe the distribution of CNVs at kilobase-level resolution in the general population.12, 13 Furthermore, the aggregation of richly annotated phenotype data in biobanks has diversified the set of phenotypes available for well-powered association studies, and allows for more precise characterization of well-established pathogenic CNVs.14, 15, 16, 17
We here describe the genome-wide landscape of copy-number variation and their associations with 3,157 phenotypes in a cohort of 332,584 participants from the UK Biobank.18 We replicate well-established syndromic effects of common CNVs—namely 22q11.2 deletion (DiGeorge) syndrome and two variants of 16p11.2 deletion syndrome—and highlight associations for body mass index (BMI), acute coronary artery disease (CAD), and related adipose and cardiovascular phenotypes. Summary statistics from traditional genome-wide associations for common CNVs as well as from gene-level aggregate burden tests of rare variants across all phenotypes are available for download on the Global Biobank Engine.19
Material and Methods
CNVs were called using PennCNV v.1.0.4 on raw signal intensity data from each genotyping array. Phenotype data were derived from data-fields collected for UK Biobank corresponding to various body measurements, biomarkers, disease diagnoses, and medical procedures from medical records, as well as a questionnaire about lifestyle and medical history. Summary-level data from all statistical tests described here, as well as more thorough documentation on phenotyping, are available on the Global Biobank Engine19 and can be found in related publications.20
CNV Calling in UK Biobank
Methods for genetic data acquisition and quality control as performed by the UK Biobank have been previously described.18 In brief, two similar arrays were used for targeted genotyping within the study population: the UK BiLEVE Axiom Array (n = 49,950) by Affymetrix and the UK Biobank Axiom Array (n = 438,427), which was custom designed by Applied Biosystems. Samples and array markers were subject to threshold-based filtration and quality control prior to public release. Specifically, markers were tested for discordance across control replicates, departures from Hardy-Weinberg equilibrium, as well as effects due to batch, plate, array, and sex; affected markers were set as missing in affected batches or removed. Similarly, samples were tested for missingness (>5%) and heterozygosity across a set of high-quality markers, but samples identified as low quality (n = 968) were not excluded. We also chose to include these samples in this analysis, considering that large structural variants may have been responsible for their poor quality with respect to metrics used for filtration.
We used PennCNV v.1.0.421 to call CNVs within each of the 106 genotyping batches from UK Biobank. We first estimate genomic runs of heterozygosity (RoH) for each sample using a previously developed pipeline in PLINK22, 23 using the --homozyg option. We then select n = 100 samples with total RoH covering less than 20 Mb to train a hidden markov model (HMM) of copy state on each chromosome. HMM training was initialized with conditions optimized for Affymetrix arrays (affygw6.hmm), provided in PennCNV resources. We used the general calling mode, which performs likelihood-based testing for copy-number state (CN = 0,1,2,3,4) at each input marker using its log-normalized signal intensity and allele balance in a given sample. We also apply adjustment for GC content across sites using waviness factor correction.24 After CNV calling, we exclude 1,360 samples with more than 30 called CNVs from downstream analysis, resulting in a cohort of 472,734 individuals with 275,180 unique variants.
Gene-Level Constraint Estimation
Regional selective constraint to CNV was estimated for all autosomal protein-coding genes, with genic CNV defined as any variant overlapping within 10 kb of the HGNC gene region. We estimate a null model of structural mutation empirically as in a previous study,12 and model burden of genic CNV as a linear function of gene size, fraction of genic sequence covered by regions of segmental duplication as extracted from the UCSC Genome Browser.25, 26 We also account for biased observations due to array genotyping (as compared to exome sequence) by including the number of genic markers as a covariate. The formula for this null model can be written as:
From this model, we compute constraint z-scores for each gene using its negated standardized residual for each gene, winsorizing the negative tail at the lowest 5% of values. We also compute the probability of intolerance to CNV (akin to probability of loss of function intolerance/pLI) as the non-normalized residual over the number of expected CNV, with negative values rounded to zero.
Genetic Associations
Variant-level associations in UK Biobank were estimated with PLINK v2.00a (2 April 2019). We used the --glm firth-fallback option for computation. This option is a hybrid algorithm for logistic regression which defaults to a standard regression solver for computation, falling back to Firth’s regression in cases where one of the cells of the 2×2 contingency table is zero, or where the traditional method fails to converge in a pre-specified number of iterations. These analyses were performed in a subset of 332,584 unrelated individuals of self-reported white British ancestry with CNV genotype calls and were controlled for age, sex, and four marker-based genomic principal components from the UK Biobank PCA calculation. To ensure adequate power for estimating genetic effects, we perform these tests on 7,038 CNVs observed at a frequency of ∼0.005% (1 in ∼20,000, or 15 individuals) in the whole sample of individuals with called CNVs.
Gene-level burden tests were conducted across all gene:phenotype pairs using the same methods and cohort as the variant-level GWAS. Genic burden was encoded as a binary variable which indicates whether an individual has a CNV which contains any overlap within 10,000 base pairs of the HGNC gene region. CNVs which overlapped several gene regions were used for analysis in each gene. We treat deletions and duplications identically, with the assumption that any CNV which overlaps a gene in this fashion will disrupt its normal function. We included the following as covariates in both models: age, sex, four marker-based genomic principal components from UK Biobank’s PCA calculation, and the number and combined length of CNVs in each individual.
Targeted variant-level GWAS was performed for both BMI and CAD in the same population, methods, and covariates as in the CNV GWAS. We display summary statistics for variants imputed from the Haplotype Reference Consortium18, 27 (HRC) which overlap regions of interest as identifies in each of these analyses. Lead variants for the BMI GWAS were identified by LD-clumping these variants with PLINK’s --clump option using a p value threshold of 10−10 and r-squared cutoff of 0.2 between lead variants. The lead variant at 9p23 was selected by inspection. Correlation between lead variants and all nearby variation was computed with PLINK’s --r2 option.
Two-sample mendelian randomization was performed via the MR Base web app using GWAS summary statistics for LDLRAD3 expression QTLs from a CARDIoGRAMplusC4D meta-analysis.28 We report Wald summary statistics from inverse-variance weighted Egger regression; these are the default analysis options for the web interface.
Results
Landscape of Common and Rare CNVs in a Large Volunteer Cohort
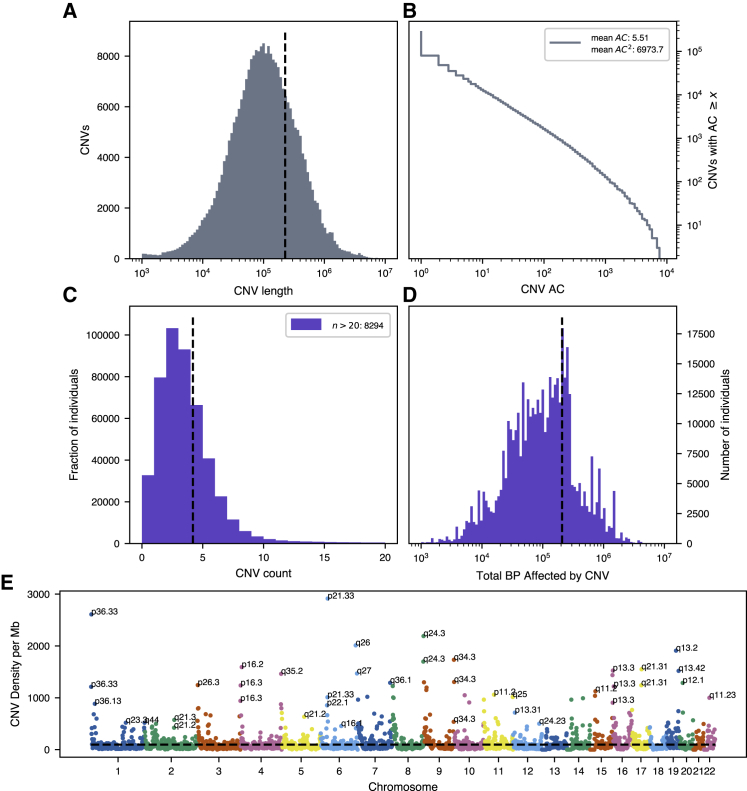
To call copy-number variants in UK Biobank, we apply PennCNV21 separately within each genotyping batch, resulting in 275,180 unique CNVs among 472,724 individuals after sample quality control. We also observe heavy-tailed distributions in size and allele count of CNVs, with average CNV length ∼226 kb and the majority of called variants singleton in the cohort (Figures 1A and 1B). This translates to notable burden of variation for nearly all individuals, with 439,464 (93.1%) of the individuals possessing at least one CNV detectable at kilobase resolution (Figures 1C and 1D). Among individuals with at least one CNV, we estimate an average burden of 5.5 variants covering >200 kb of genomic sequence (median 3 variants affecting ∼100 kb; Figures 1C and 1D). While in line with previous reports,12 these estimates of individual-level burden are likely conservative, as regions where array markers are sparse or missing limit the accuracy of variant calling. Furthermore, we are unable to call smaller (<1 kb) variants due to inconsistent marker density across all chromosomal regions on the Axiom and BiLEVE UK Biobank genotyping arrays. This limitation is visible in the histogram of called CNV lengths (Figure 1A); we call substantially fewer variants on the order of hundreds of base-pairs than on the order of thousands.
Figure 1.
Burden and Distribution of Copy-Number Variation in UK Biobank
(A) Log-scale histogram of CNV lengths. Mean length (dashed line) is 226.5 kb.
(B) Cumulative density of CNV allele count (AC), displayed in log-log axes. Average AC is 5.5, but average frequency as experienced by the population (weighted by count, hence AC2) is ∼1.6%.
(C and D) Histogram of CNV counts (C) and log-scale base-pairs affected by CNV per individual (D). Sample-level burden is heavy-tailed, with the average individual carrying 4.2 variants (dashed line), affecting mean ∼207.6 kb of genomic sequence.
(E) Genome-wide density of CNV, defined as the number of unique CNVs overlapping 10 megabase (Mb) windows tiling each chromosome. Hotspots of structural variation are labeled by cytogenic band.
We also observe a highly non-uniform burden of variation across genomic position, with breakpoints most common near the ends of chromosomes, and at known regions of segmental duplication (Figure 1E). Among them are 1p36, 8q24.3, 9q34.3, and 19q13, all of which have associated microdeletion syndromes causing developmental delay with uniquely characteristic growth patterns.29, 30, 31, 32 Other CNV hotspots like 6p21.33, which contains the major histocompatibility complex protein gene family, may be influenced by high marker density (in this case for HLA allelotyping) in addition to these biological features which underlie structural mutagenesis. However, these loci do not categorically correspond to areas where structural variation is commonly observed in the population (Figure S1). For example, 1p36 and 19q13 are also the respective sites of common CNVs overlapping RHD (MIM: 111680) and FUT2 (MIM: 182100) (Rhesus and Lewis blood groups), but there are no such common variants within the telomeric 16p13 cytoband.
Survivorship Bias due to Genetic Selection against Early-Onset Diseases
We estimate gene-level intolerance to structural variation by adapting a method for estimating regional selective constraint.12 Relative to the general population, the volunteers within the UK Biobank are described to have a “healthy-cohort” enrollment bias33 and were enrolled between the ages of 40 to 69, which informs our findings. Within the tail of positive constraint z-scores, which indicate the strongest intolerance to structural variation, we observe enrichment for genes which cause early-onset diseases, particularly cancer. Among the top 15 constrained genes (Table 1) are BRCA1 (MIM: 113705) and BRCA2 (MIM: 117305), which are associated with early-onset breast cancer;34, 35 MLH1 (MIM: 120486), MSH2 (MIM: 609309), MSH6 (MIM: 600678), which cause early-onset colorectal cancer (Lynch syndrome [MIM: 120435]);36, 37, 38 and ATM (MIM: 607585) and APC (MIM: 611731), which are involved with mismatch repair cancers.39, 40 In all we find 8,709 genes (47.9% of 18,183 protein coding autosomal genes in the analysis) to be intolerant to CNV, with probability of intolerance (see Material and Methods) above 0.9; this is a greater than 2.5-fold increase in the set of genes intolerant to loss of function variation identified in ExAC.41
Table 1.
15 Genes Most Intolerant to Copy-Number Variation
| Gene | Constraint z | Probability of CNV Intolerance |
|---|---|---|
| BRCA2 | 3.402 | 0.9911 |
| BRCA1 | 2.570 | 0.9840 |
| APC | 2.086 | 0.9456 |
| ATM | 1.242 | 0.9892 |
| MSH2 | 1.241 | 0.9883 |
| MLH1 | 1.224 | 0.9962 |
| MSH6 | 0.957 | 0.9933 |
| RB1 | 0.905 | 0.9577 |
| SBDS | 0.861 | 0.9741 |
| SPATA31D1 | 0.853 | 0.9979 |
| CYP3A4 | 0.846 | 0.9979 |
| PABPC3 | 0.831 | 0.9923 |
| OTOP1 | 0.830 | 0.9930 |
| KRT16 | 0.828 | 0.9979 |
| ZNF302 | 0.827 | 0.9979 |
Columns are gene label, constraint z-score, and probability of CNV intolerance (see Material and Methods for definitions).
Selections from the most highly constrained pathways from Gene Ontology Consortium42 resources (Table 2) also suggest strong intolerance to CNV among genes involved with core biological processes like protein binding, cellular structural integrity (keratinization), development (growth hormone receptor binding), and immune regulation (natural killer cell activation). Similar results at the gene and pathway level are observed for deletion-specific constraint (Tables S1 and S2), whereas duplication-specific analysis suggests autoimmune-related genes and pathways are most strongly intolerant to dosage effects (Tables S1 and S2). Analysis of medical terms from the Human Phenotype Ontology project43 further validates the observation that genes with carcinogenic variation are enriched among those most intolerant to CNV (Table 3). The most constrained HPO terms include carcinomas, neoplasms, and other conditions like chronic fatigue. Deletion-specific analysis of HPO terms (Table S3) also follows this trend, while duplication-specific constraint suggests strong intolerance to variation altering normal developmental pathways. HPO terms most strongly intolerant to putatively dosage-altering variation include an array of nervous and musculoskeletal abnormalities. These results indicate strong selective effects occurring prior to enrollment in the UK Biobank during childhood and early adulthood against loss of function variation in core developmental, metabolic, and tumor-suppressing genes, and against dosage-altering variation in immune-related genes.
Table 2.
15 Pathways from Gene Ontology Consortium Most Enriched for Constrained Genes
| GO ID | CNV-Intolerant Pathway Name | Delta z | p |
|---|---|---|---|
| GO:0000137 | Golgi cis cisterna | 0.4086 | 7.14E−30 |
| GO:0045095 | keratin filament | 0.2594 | 8.20E−30 |
| GO:0031436 | BRCA1-BARD1 complex | 1.4562 | 2.18E−28 |
| GO:0005515 | protein binding | 0.0707 | 5.52E−23 |
| GO:0000800 | lateral element | 0.4642 | 1.48E−21 |
| GO:0031424 | keratinization | 0.1816 | 1.50E−20 |
| GO:0032301 | MutSalpha complex | 1.0987 | 7.98E−20 |
| GO:0008194 | UDP-glycosyltransferase activity | 0.3525 | 1.06E−18 |
| GO:0052697 | xenobiotic glucuronidation | 0.4715 | 4.53E−18 |
| GO:0070200 | establishment of protein localization to telomere | 0.9767 | 1.58E−17 |
| GO:0032300 | mismatch repair complex | 0.5164 | 4.29E−17 |
| GO:0008274 | gamma-tubulin ring complex | 0.3902 | 2.72E−16 |
| GO:0008202 | steroid metabolic process | 0.2776 | 6.76E−16 |
| GO:0042954 | lipoprotein transporter activity | 0.4233 | 9.53E−15 |
| GO:0015020 | glucuronosyltransferase activity | 0.3365 | 1.24E−14 |
Columns are GO pathway ID/name, change in z-score between set and non-set members, indicating mean strength of selective effect in the pathway, and p value (t test, gene set members versus all others).
Table 3.
15 Pathways from Human Phenotype Ontology Consortium Most Enriched for Constrained Genes
| HPO ID | CNV-Intolerant HPO Term | Delta z | P |
|---|---|---|---|
| HP:0006725 | Pancreatic adenocarcinoma | 0.5545 | 2.50E−46 |
| HP:0012432 | Chronic fatigue | 0.7301 | 3.00E−39 |
| HP:0025318 | Ovarian carcinoma | 0.6659 | 1.94E−38 |
| HP:0003003 | Colon cancer | 0.4031 | 2.95E−37 |
| HP:0004389 | Intestinal pseudo-obstruction | 0.6735 | 3.16E−36 |
| HP:0100787 | Prostate neoplasm | 0.5044 | 2.72E−34 |
| HP:0012125 | Prostate cancer | 0.5044 | 2.72E−34 |
| HP:0100273 | Neoplasm of the colon | 0.3444 | 9.78E−34 |
| HP:0030406 | Primary peritoneal carcinoma | 0.5883 | 8.40E−32 |
| HP:0012334 | Extrahepatic cholestasis | 0.5551 | 4.59E−28 |
| HP:0003002 | Breast carcinoma | 0.3147 | 3.65E−27 |
| HP:0002885 | Medulloblastoma | 0.5220 | 2.38E−26 |
| HP:0002254 | Intermittent diarrhea | 0.5046 | 4.04E−26 |
| HP:0100834 | Neoplasm of the large intestine | 0.2734 | 7.87E−26 |
| HP:0009592 | Astrocytoma | 0.5033 | 2.38E−24 |
Columns are HPO ID/term, change in z-score between set and non- set members, indicating mean strength of selective effect in the pathway, and p value (t test, gene set members versus all others).
Association Testing Identifies CNVs at Several Genomic Loci
We compute genome-wide associations across 3,157 phenotypes for 7,038 common CNVs observed at 0.005% allele frequency (1 in 20,000) in our GWAS cohort, using regression as implemented in the analysis software PLINK.44 We also perform aggregate rare-variant burden tests, pooled by gene. For these tests, we measure the net effect of rare CNVs (AF < 0.1%) overlapping within 10 kb of the gene region as defined by HGNC45 for 16,250 autosomal protein coding genes with at least five individuals with overlapping CNV. In sum, we find 14,182 CNV-level associations (about 4 per phenotype) and 102,606 gene-level associations (about 32 per phenotype) with Bonferroni-corrected p < 0.05/7,038 (7.1 × 10−6, for GWAS) or 0.05/16,250 (3.1 × 10−6, burden test). It is noteworthy that many of our phenotype observations are correlated (e.g., right/left hand grip strength), and aggregate gene-level tests are also correlated (e.g., cases where a single rare variant overlaps several genes, as in DiGeorge syndrome). A complete list of phenotypes analyzed is available on the Global Biobank Engine (Web Resources). Here, we describe representative results for one common disease and one quantitative measure with established genetic risk factors and large sample sizes in UK Biobank: acute coronary artery disease (CAD) and body mass index (BMI).
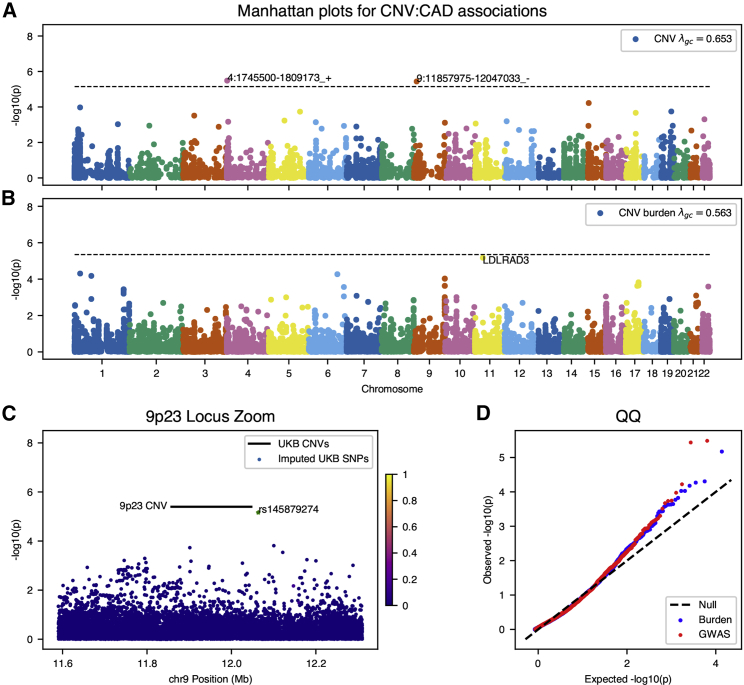
For acute CAD, we identify two statistically significant (p < 9 × 10−6) associations after Bonferroni correction for the common CNV GWAS: an intergenic deletion at chromosome 9p23 and a putative gene fusion event on chromosome 4 (Figure 2A). The association of the duplicated FGFR3-TACC3 fusion (MIM: 134934) is unclear; only two individuals with this variant appear in gnomAD SV resource and no previous experimental or genetic data link this locus to cardiovascular disease. However, intergenic variants at the 9p21 locus have been implicated in previous association studies of blood-based biomarkers relevant to cardiac outcomes, specifically, decreases in hematocrit and hemoglobin concentration,46 as well as carotid plaque burden.47 A recent meta-analysis48 using data from UK Biobank and CARDIoGRAMplusC4D identified a lead variant in the vicinity of this locus (rs2891168) associated with 6% unit increase in risk for similarly defined coronary artery disease. However, the CNV we here identify confers an estimated 12.4-fold increased risk (95%CI: 7.2–21.3, p = 3.7 × 10−6) and is at least 2 Mb distant from the nearest SNPs (rs10961206) at genome-wide significance near the 9p21/9p23 locus in the meta-analysis. This and the absence of linkage between the 9p23 CNVs and rs10961206 (r2 = 0.013) are suggestive of independent effects. However, translocation of flanking regulatory elements has been suggested as a mechanism for CNV-derived phenotypic effect;49 given the proximity of this variant to a well-established susceptibility region (9p21) for CAD, we cannot rule out the possibility that trans-regulatory effect on known regions drives this association.
Figure 2.
Genome-wide CNV Associations for Acute Coronary Artery Disease (CAD)
(A and B) Manhattan plots for (A) genome-wide association of common copy-number variants and (B) genome-wide burden test of rare variants for genes with at least ten individuals observed with CNVs.
(C) Locus inset of 9p23 CNV and summary statistics from GWAS of coronary artery disease using variants imputed on the same study population used in the CNV analysis. Variants are colored by marker LD with lead regional GWAS SNPs (rs145879274) from the analysis. This marker is highly stratified by continental ancestry and does not show significant correlation with any other variant in the region.
(D) Quantile-quantile plots for genome-wide summary statistics from CNV associations.
Gene-level burden testing of rare CNVs in individuals with CAD implicates LDLRAD3 (MIM: 617986), a member of the low-density lipoprotein (LDL) receptor family that did not meet pre-specified significance criteria in revised analyses but remains strongly associated with disease, and is a clear outlying genome-wide signal (Figure 2B). The CNVs called in this gene are predominantly deletions affecting the coding sequence—in aggregate (n = 27), these variants confer an estimated 11-fold increase in risk of acute CAD (95% CI: 6.5–19.0, p = 6.7 × 10−6). Though the role of lipoprotein receptors in cholesterol metabolism is a well-established mechanism of risk for cardiovascular disease, LDLRAD3 is not known to participate in cholesterol metabolism. It is, however, a receptor widely expressed throughout adult tissues which may participate in proteolysis in the central nervous system.50, 51 We therefore sought to replicate these findings using two-sample mendelian randomization52 on expression quantitative trait loci (eQTLs) from CAD summary statistics from a CARDIoGRAMplusC4D meta-analysis.28 We identify a nominally significant protective effect between an eQTL increasing expression of LDLRAD3 and CAD (OR = 0.85 [95%CI: 0.62–0.97], p = 0.012), the direction of which is consistent with a dosage model of LDLRAD3-mediated risk for CAD.
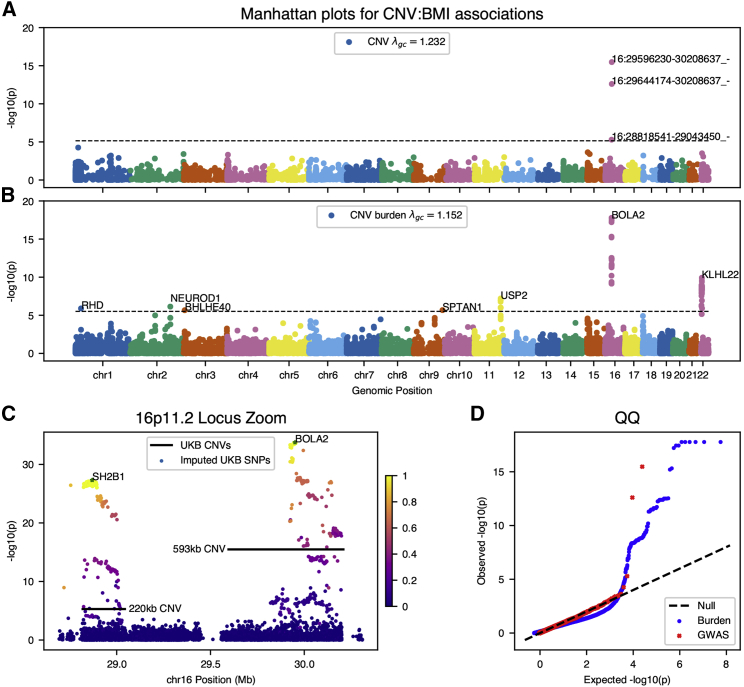
We find multiple significant associations for BMI; three deletions at chromosome 16p11.2, a locus implicated in syndromic early-onset obesity and developmental delay (Figure 3A). Each of these CNVs appears to correspond to a distinct form of 16p11.2 deletion syndrome. The smaller ∼220kb deletion (β = 0.92 SD [95%CI: 0.72–1.12], p = 5.1 × 10−6, AC = 24 [MIM: 613444]) has been associated with early-onset obesity and spans nine distinct genes, with SH2B1 [MIM: 608937] the suspected causal obesity gene.7 Obesity is also a phenotypic consequence of a larger ∼593kb deletion (β = 1.35 SD [95%CI: 1.18–1.51], p = 3.3 × 10−16, AC = 37 [MIM: 611913]), which is further associated with neurodevelopmental delay and related conditions.8 However, this deletion spans a wholly distinct set of genes which are suspected to play complex dosage-dependent roles in the phenotypic consequences of the syndrome.53 As both subtypes of 16p11.2 deletion syndrome may present in early childhood, it is noteworthy that the effect we measure on BMI is in a cohort comprised entirely of older individuals, indicating burden of adult disease associated with the CNV locus. Moreover, these effects are consistent, though slightly higher, than previous meta-analysis or targeted study of this locus in UK Biobank.14, 54
Figure 3.
Genome-wide CNV Associations for Body Mass Index (BMI)
(A and B) Manhattan plots for (A) genome-wide association of common copy-number variants and (B) genome-wide burden test of rare variants for genes with at least five individuals observed with CNVs.
(C) Locus inset of 16p11.2 CNVs and summary statistics from GWAS of BMI using variants imputed on the same study population used in the CNV analysis. Variants are colored by marker LD with lead regional GWAS SNPs overlapping each CNV (rs62037365 in SH2B1; rs12716975 in non-coding BOLA2).
(D) Quantile-quantile plots for genome-wide summary statistics from CNV associations.
After controlling for multiple comparisons, burden testing for BMI identifies KLHL22 at chromosome 22q11.2, recapitulates the 16p11.2 deletions at the gene level (SH2B1, BOLA2 [MIM: 613182]), and associations at five additional loci each with strong evidence for causality (Figure 3B). Mechanisms for these loci are related to risk of diabetes; mouse knockouts of USP2 (MIM: 604725) reduce insulin resistance,55 NEUROD1 (MIM: 601724) variation is a known cause of diabetes,56 and BHLHE40 (MIM: 604256) may modify diabetes in mice via disturbances in circadian rhythm.57 While SPTAN1 (MIM: 182810) has been previously associated with severe neurological disease,58 neither SPTAN1 nor RHD have a previous association with BMI. Variation at the 22q11.2 locus, also known as DiGeorge syndrome, has variable phenotypic consequences including craniofacial dysmorphisms and conotruncal congenital heart disease, along with increased risk for an adverse cardiovascular outcomes and neuropsychiatric disease later in life. Among individuals affected with 22q11.2 deletion syndrome, obesity is a recognized manifestation of disease, and we estimate a 1.5–2.0 point increase in BMI for genic CNVs near 22q11.2 in KLHL22. as well as 3.0–3.8 for genic CNV at 16p11.2—these effects are attenuated relative to clinically estimated effect of DiGeorge syndrome on obesity,59 our own estimates of the effect of 16p11.2 deletion, and previous studies,14, 54 likely due to the inclusion of non-causal variants which overlap non-candidate genes in these regions. The presence of these associations in a large volunteer cohort offers further evidence that these potentially pathogenic CNV contribute to population-scale risk for common diseases.
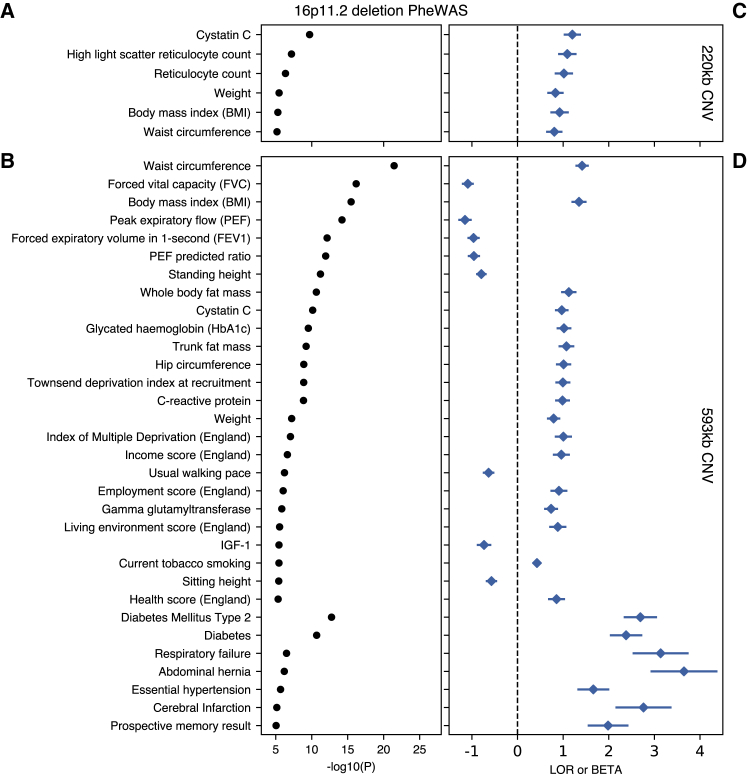
Phenome-wide associations for each of the CNVs at 16p11.2 further highlight changes in biomarkers, biomeasures, and increased risk of common disease, consistent with high BMI over the course of a lifetime (Figure 4). Genome-wide significant phenotypes for the 220 kb CNV recapitulate the established syndromic effects from early-onset obesity. We observe significant increases, on the order of one standard deviation, in weight, BMI, hip and waist circumference, reticulocyte count, and Cystatin C measures for these individuals. The larger 593 kb CNV associates with similar measures of body size and fat, as well as hypertension, diabetes/HbA1c, and abdominal hernia. These results are also indicative of effects due to developmental delay; namely, decreased measures of memory, higher Townsend deprivation (an index of material deprivation which considers employment, home/auto ownership, and household overcrowding in a person’s neighborhood), and lower lung capacity (FEV, FVC), with higher associated risk of respiratory failure. Taken together, these results highlight the variable expressivity of CNV-related disease, as well as its long-term effects across the medical phenome.
Figure 4.
PheWAS of 16p11.2 CNVs
Selected genome-wide significant (p < 9 × 10−6) associations for 220 kb (top) and 593 kb (bottom) 16p11.2 CNVs, with n > 500 binary cases or 15,000 quantitative values. Traits are grouped by type (binary/quantitative) then sorted by p value (left). Log-odds ratio and standardized betas (right) align with trait names on the y axis, with the horizontal dashed line separating positive and negative direction of association.
Discussion
In calling copy-number variants and performing genetic association studies at scale from a large cohort of array-genotyped individuals with richly annotated phenotype data, we provide a portrait of the phenome-wide burden of genomic copy-number variation. Our estimates of the individual-level burden of CNV and population-wide allele frequencies are consistent with previous reports, and the deep phenotypic information available in the UK Biobank permits more finely tuned measures of the genic intolerance to CNV which include estimates of variation absent from our cohort of predominantly healthy, middle-aged individuals.
Our study has significant limitations that inform our analysis. While arrays are an inexpensive way to genotype large cohorts, permitting straightforward algorithms to infer the presence of structural variation, the resulting CNV calls are limited by the density and placement of markers across chromosomes. For UK Biobank genotyping arrays in particular, there are large portions of genomic sequence with low marker density (in particular near centromeric regions) which bias our resulting genotype calls away from such regions. Array-derived CNV likewise cannot differentiate structural events like inversions or translocations, or determine breakpoint position at base-pair resolution.60 Complicating these barriers is the fact that our sample was genotyped on two distinct arrays, which may cause identical CNVs to present with different breakpoints across individuals in the call set—as evident in the two calls of the 593 kb form of 16p11.2 deletion syndrome (Figure 3A). Our choice to present gene-level burden tests which include the vast majority of variants included in our CNV GWAS was informed by this realization. Given the release of exome-sequence data for 50,000 UK Biobank participants,61 it is worth noting that NGS-based analysis of structural variants is a natural extension of this work which would complement the limitations of our genotyping.
Our associations are also heavily impacted by a known “healthy-cohort” bias, which may influence null results for phenotypes with known genetic contributions; notably, there are no genome-wide significant hits in our burden test for breast cancer. With this in mind, our constraint scores constitute a sobering observation of genetic survivorship bias. We take this opportunity to honor these non-participating individuals and their implicit contribution to our understanding of genetic disease. Though consideration of genic intolerance to variation may complement association studies, we find no novel candidate genes for early-onset disease among our results. However, the observation of selection bias in UK Biobank suggests that findings from biobank studies around the world will be influenced by implicit and explicit barriers to participation.
Despite selection against high-penetrance alleles causing early-onset disease, we detect a strong association for coronary artery disease at LDLRAD3. While this locus has prior putative association with bone mineral density,62 existing large-scale GWASs do not detect a strong association with coronary artery disease or established cardiometabolic risk-factors. In our study, CNVs at this locus are associated with some established cardiometabolic risk factors, such as diabetes onset, smoking status, and arterial stiffness, but not obesity or other fat-related phenotypes (Figure S6). Consistent with our findings that a decrease in LDLRAD3 dosage increases the risk of disease, a strong eQTL increasing LDLRAD3 expression decreases the risk of disease when used as an instrument in a two-sample mendelian randomization in a large-scale study of coronary artery disease. These results highlight the utility of analyzing genic CNV which, when directly impacting mRNA dosage, offer an interpretable mechanism distinct from alterations of protein structure or small changes in transcriptional regulation.
The observation of variation at the 16p11.2 and 22q11.2 loci sheds further light on the penetrance of potentially pathogenic CNVs in the general population. The 16p11.2 recurrent microdeletion syndrome has been previously described in individuals with autism and neuropsychiatric disease and may include seizures, and brain and other anatomic abnormalities. People carrying variation at the 22q11.2 locus within the general population are known to be at increased risk of neuropsychiatric diseases63 for which variable phenotypic penetrance is well recognized.64 To wit, individuals with genetic variation at both loci were by and large sufficiently healthy and capable of volunteering to participate in UK Biobank. Our findings support a growing recognition that the penetrance and effect sizes of syndromic alleles may require revision in the context of broad population-based surveys of rare genetic variation.65, 66
These described associations suggest a role of structural variation in population-wide burden of common disease and suggest loci where CNV-derived syndromic disease may exist. As such, these resources may be of immediate use by genetic clinicians in classification of CNV detected in clinical testing and for follow-up functional study. Of particular interest would be an analysis of “human knockout” individuals with both gene copies altered by CNV or other loss-of-function variation, as determined by SNP genotype data from UK Biobank. As with single-nucleotide variation, the functional consequence of and pathogenicity of genic structural variation is difficult to classify. One consequence is a dosage effect upon mRNA transcription; alternatively, dosage compensation to modulate mRNA levels67 or fusion of flanking regions may drive phenotypic alteration.68, 69 Summary statistics from association studies described here, as well as for all phenotypes present on the Global Biobank Engine, are freely available for download on the site. We hope that these data will be leveraged to empower future analyses of the phenome-wide effects of structural variation and gene-level dosage effects.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
This research has been conducted using the UK Biobank Resource under application numbers 24983, 16698, 13721, and 15860. We thank all the participants in the study. The primary and processed data used to generate the analyses presented here are available in the UK Biobank access management system for application 24983, “Generating effective therapeutic hypotheses from genomic and hospital linkage data,” and the results are displayed in the Global Biobank Engine.
M.A.R. is supported by Stanford University and a National Institute of Health center for Multi- and Trans-ethnic Mapping of Mendelian and Complex Diseases grant (5U01 HG009080). This work was supported by National Human Genome Research Institute (NHGRI) of the National Institutes of Health (NIH) under awards R01HG010140. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. J.P. is supported by the Stanford University Department of Pediatrics, National Heart Lung and Blood Institute (R00 HL130523), and Chan-Zuckerberg Biohub.
Published: July 25, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.07.001.
Web Resources
GeneReviews, McDonald-McGinn, D.M., Emanuel, B.S., and Zackai, E.H. (1999). 22q11.2 deletion syndrome, https://www.ncbi.nlm.nih.gov/books/NBK1523/
Global Biobank Engine (summary statistics), https://biobankengine.stanford.edu/downloads
OMIM, https://www.omim.org/
UK Biobank Application 24983, http://www.ukbiobank.ac.uk/wp-content/uploads/2017/06/24983-Dr-Manuel-Rivas.pdf
UK Biobank CNV Analysis, https://github.com/priestlab/cnv-ukb
Supplemental Data
References
- 1.Sudmant P.H., Mallick S., Nelson B.J., Hormozdiari F., Krumm N., Huddleston J., Coe B.P., Baker C., Nordenfelt S., Bamshad M. Global diversity, population stratification, and selection of human copy-number variation. Science. 2015;349:aab3761. doi: 10.1126/science.aab3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mills R.E., Walter K., Stewart C., Handsaker R.E., Chen K., Alkan C., Abyzov A., Yoon S.C., Ye K., Cheetham R.K., 1000 Genomes Project Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mikhail F.M. Copy number variations and human genetic disease. Curr. Opin. Pediatr. 2014;26:646–652. doi: 10.1097/MOP.0000000000000142. [DOI] [PubMed] [Google Scholar]
- 4.Carvill G.L., Mefford H.C. Microdeletion syndromes. Curr. Opin. Genet. Dev. 2013;23:232–239. doi: 10.1016/j.gde.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 6.Stankiewicz P., Lupski J.R. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 7.Bachmann-Gagescu R., Mefford H.C., Cowan C., Glew G.M., Hing A.V., Wallace S., Bader P.I., Hamati A., Reitnauer P.J., Smith R. Recurrent 200-kb deletions of 16p11.2 that include the SH2B1 gene are associated with developmental delay and obesity. Genet. Med. 2010;12:641–647. doi: 10.1097/GIM.0b013e3181ef4286. [DOI] [PubMed] [Google Scholar]
- 8.Zufferey F., Sherr E.H., Beckmann N.D., Hanson E., Maillard A.M., Hippolyte L., Macé A., Ferrari C., Kutalik Z., Andrieux J., Simons VIP Consortium. 16p11.2 European Consortium A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. J. Med. Genet. 2012;49:660–668. doi: 10.1136/jmedgenet-2012-101203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raychaudhuri S., Korn J.M., McCarroll S.A., Altshuler D., Sklar P., Purcell S., Daly M.J., International Schizophrenia Consortium Accurately assessing the risk of schizophrenia conferred by rare copy-number variation affecting genes with brain function. PLoS Genet. 2010;6:e1001097. doi: 10.1371/journal.pgen.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Priest J.R., Osoegawa K., Mohammed N., Nanda V., Kundu R., Schultz K., Lammer E.J., Girirajan S., Scheetz T., Waggott D. De Novo and Rare Variants at Multiple Loci Support the Oligogenic Origins of Atrioventricular Septal Heart Defects. PLoS Genet. 2016;12:e1005963. doi: 10.1371/journal.pgen.1005963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruderfer D.M., Hamamsy T., Lek M., Karczewski K.J., Kavanagh D., Samocha K.E., Daly M.J., MacArthur D.G., Fromer M., Purcell S.M., Exome Aggregation Consortium Patterns of genic intolerance of rare copy number variation in 59,898 human exomes. Nat. Genet. 2016;48:1107–1111. doi: 10.1038/ng.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirov G., Kendall K., Rees E., Escott-Price V., Hewitt J., Thomas R., O’Donovan M., Owen M., Walters J. The Uk Biobank: A Resource For Cnv Analysis. Eur. Neuropsychopharmacol. 2017;27:S491. [Google Scholar]
- 14.Crawford K., Bracher-Smith M., Owen D., Kendall K.M., Rees E., Pardiñas A.F., Einon M., Escott-Price V., Walters J.T.R., O’Donovan M.C. Medical consequences of pathogenic CNVs in adults: analysis of the UK Biobank. J. Med. Genet. 2019;56:131–138. doi: 10.1136/jmedgenet-2018-105477. [DOI] [PubMed] [Google Scholar]
- 15.Owen D., Bracher-Smith M., Kendall K.M., Rees E., Einon M., Escott-Price V., Owen M.J., O’Donovan M.C., Kirov G. Effects of pathogenic CNVs on physical traits in participants of the UK Biobank. BMC Genomics. 2018;19:867. doi: 10.1186/s12864-018-5292-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kendall K.M., Rees E., Escott-Price V., Einon M., Thomas R., Hewitt J., O’Donovan M.C., Owen M.J., Walters J.T.R., Kirov G. Cognitive Performance Among Carriers of Pathogenic Copy Number Variants: Analysis of 152,000 UK Biobank Subjects. Biol. Psychiatry. 2017;82:103–110. doi: 10.1016/j.biopsych.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 17.Warland A., Kendall K.M., Rees E., Kirov G., Caseras X. Schizophrenia-associated genomic copy number variants and subcortical brain volumes in the UK Biobank. Mol. Psychiatry. 2019 doi: 10.1038/s41380-019-0355-y. Published online January 24, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McInnes G., Tanigawa Y., DeBoever C., Lavertu A., Olivieri J.E., Aguirre M., Rivas M.A. Global Biobank Engine: enabling genotype-phenotype browsing for biobank summary statistics. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty999. Published online December 8, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeBoever C., Tanigawa Y., Lindholm M.E., McInnes G., Lavertu A., Ingelsson E., Chang C., Ashley E.A., Bustamante C.D., Daly M.J., Rivas M.A. Medical relevance of protein-truncating variants across 337,205 individuals in the UK Biobank study. Nat. Commun. 2018;9:1612. doi: 10.1038/s41467-018-03910-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K., Li M., Hadley D., Liu R., Glessner J., Grant S.F.A., Hakonarson H., Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howrigan D.P., Simonson M.A., Davies G., Harris S.E., Tenesa A., Starr J.M., Liewald D.C., Deary I.J., McRae A., Wright M.J. Genome-wide autozygosity is associated with lower general cognitive ability. Mol. Psychiatry. 2016;21:837–843. doi: 10.1038/mp.2015.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diskin S.J., Li M., Hou C., Yang S., Glessner J., Hakonarson H., Bucan M., Maris J.M., Wang K. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res. 2008;36:e126. doi: 10.1093/nar/gkn556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey J.A., Yavor A.M., Massa H.F., Trask B.J., Eichler E.E. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haeussler M., Zweig A.S., Tyner C., Speir M.L., Rosenbloom K.R., Raney B.J., Lee C.M., Lee B.T., Hinrichs A.S., Gonzalez J.N. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2019;47(D1):D853–D858. doi: 10.1093/nar/gky1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K., Haplotype Reference Consortium A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikpay M., Goel A., Won H.-H., Hall L.M., Willenborg C., Kanoni S., Saleheen D., Kyriakou T., Nelson C.P., Hopewell J.C. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordan V.K., Zaveri H.P., Scott D.A. 1p36 deletion syndrome: an update. Appl. Clin. Genet. 2015;8:189–200. doi: 10.2147/TACG.S65698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akbaroghli S., Tonekaboni S.H., Kariminejad R., Liehr T., Coci E.G. De-novo interstitial 2.33cMb deletion in 8q24.3: new insights on a very rare partial monosomy syndrome. Clin. Dysmorphol. 2018;27:97–100. doi: 10.1097/MCD.0000000000000224. [DOI] [PubMed] [Google Scholar]
- 31.Iwakoshi M., Okamoto N., Harada N., Nakamura T., Yamamori S., Fujita H., Niikawa N., Matsumoto N. 9q34.3 deletion syndrome in three unrelated children. Am. J. Med. Genet. A. 2004;126A:278–283. doi: 10.1002/ajmg.a.20602. [DOI] [PubMed] [Google Scholar]
- 32.Cario H., Bode H., Gustavsson P., Dahl N., Kohne E. A microdeletion syndrome due to a 3-Mb deletion on 19q13.2--Diamond-Blackfan anemia associated with macrocephaly, hypotonia, and psychomotor retardation. Clin. Genet. 1999;55:487–492. doi: 10.1034/j.1399-0004.1999.550616.x. [DOI] [PubMed] [Google Scholar]
- 33.Fry A., Littlejohns T.J., Sudlow C., Doherty N., Adamska L., Sprosen T., Collins R., Allen N.E. Comparison of Sociodemographic and Health-Related Characteristics of UK Biobank Participants With Those of the General Population. Am. J. Epidemiol. 2017;186:1026–1034. doi: 10.1093/aje/kwx246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall J.M., Friedman L., Guenther C., Lee M.K., Weber J.L., Black D.M., King M.C. Closing in on a breast cancer gene on chromosome 17q. Am. J. Hum. Genet. 1992;50:1235–1242. [PMC free article] [PubMed] [Google Scholar]
- 35.Wooster R., Bignell G., Lancaster J., Swift S., Seal S., Mangion J., Collins N., Gregory S., Gumbs C., Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 36.Papadopoulos N., Nicolaides N.C., Wei Y.F., Ruben S.M., Carter K.C., Rosen C.A., Haseltine W.A., Fleischmann R.D., Fraser C.M., Adams M.D. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 37.Papadopoulos N., Nicolaides N.C., Liu B., Parsons R., Lengauer C., Palombo F., D’Arrigo A., Markowitz S., Willson J.K., Kinzler K.W. Mutations of GTBP in genetically unstable cells. Science. 1995;268:1915–1917. doi: 10.1126/science.7604266. [DOI] [PubMed] [Google Scholar]
- 38.Fishel R., Lescoe M.K., Rao M.R., Copeland N.G., Jenkins N.A., Garber J., Kane M., Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 39.Savitsky K., Bar-Shira A., Gilad S., Rotman G., Ziv Y., Vanagaite L., Tagle D.A., Smith S., Uziel T., Sfez S. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 40.Horii A., Nakatsuru S., Miyoshi Y., Ichii S., Nagase H., Kato Y., Yanagisawa A., Nakamura Y. The APC gene, responsible for familial adenomatous polyposis, is mutated in human gastric cancer. Cancer Res. 1992;52:3231–3233. [PubMed] [Google Scholar]
- 41.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Gene Ontology Consortium The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Köhler S., Carmody L., Vasilevsky N., Jacobsen J.O.B., Danis D., Gourdine J.-P., Gargano M., Harris N.L., Matentzoglu N., McMurry J.A. Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources. Nucleic Acids Res. 2019;47(D1):D1018–D1027. doi: 10.1093/nar/gky1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Povey S., Lovering R., Bruford E., Wright M., Lush M., Wain H. The HUGO Gene Nomenclature Committee (HGNC) Hum. Genet. 2001;109:678–680. doi: 10.1007/s00439-001-0615-0. [DOI] [PubMed] [Google Scholar]
- 46.Astle W.J., Elding H., Jiang T., Allen D., Ruklisa D., Mann A.L., Mead D., Bouman H., Riveros-Mckay F., Kostadima M.A. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell. 2016;167:1415–1429.e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pott J., Burkhardt R., Beutner F., Horn K., Teren A., Kirsten H., Holdt L.M., Schuler G., Teupser D., Loeffler M. Genome-wide meta-analysis identifies novel loci of plaque burden in carotid artery. Atherosclerosis. 2017;259:32–40. doi: 10.1016/j.atherosclerosis.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 48.van der Harst P., Verweij N. Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ. Res. 2018;122:433–443. doi: 10.1161/CIRCRESAHA.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fudenberg G., Pollard K.S. Chromatin features constrain structural variation across evolutionary timescales. Proc. Natl. Acad. Sci. USA. 2019;116:2175–2180. doi: 10.1073/pnas.1808631116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ranganathan S., Noyes N.C., Migliorini M., Winkles J.A., Battey F.D., Hyman B.T., Smith E., Yepes M., Mikhailenko I., Strickland D.K. LRAD3, a novel low-density lipoprotein receptor family member that modulates amyloid precursor protein trafficking. J. Neurosci. 2011;31:10836–10846. doi: 10.1523/JNEUROSCI.5065-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noyes N.C., Hampton B., Migliorini M., Strickland D.K. Regulation of Itch and Nedd4 E3 Ligase Activity and Degradation by LRAD3. Biochemistry. 2016;55:1204–1213. doi: 10.1021/acs.biochem.5b01218. [DOI] [PubMed] [Google Scholar]
- 52.Hemani G., Zheng J., Elsworth B., Wade K.H., Haberland V., Baird D., Laurin C., Burgess S., Bowden J., Langdon R. The MR-Base platform supports systematic causal inference across the human phenome. eLife. 2018;7:7. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiu Y., Arbogast T., Lorenzo S.M., Li H., Tang S.C., Richardson E., Hong O., Cho S., Shanta O., Pang T. Oligogenic Effects of 16p11.2 Copy Number Variation on Craniofacial Development. bioRxiv. 2019 doi: 10.1016/j.celrep.2019.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Macé A., Tuke M.A., Deelen P., Kristiansson K., Mattsson H., Nõukas M., Sapkota Y., Schick U., Porcu E., Rüeger S. CNV-association meta-analysis in 191,161 European adults reveals new loci associated with anthropometric traits. Nat. Commun. 2017;8:744. doi: 10.1038/s41467-017-00556-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saito N., Kimura S., Miyamoto T., Fukushima S., Amagasa M., Shimamoto Y., Nishioka C., Okamoto S., Toda C., Washio K. Macrophage ubiquitin-specific protease 2 modifies insulin sensitivity in obese mice. Biochem. Biophys. Rep. 2017;9:322–329. doi: 10.1016/j.bbrep.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malecki M.T., Jhala U.S., Antonellis A., Fields L., Doria A., Orban T., Saad M., Warram J.H., Montminy M., Krolewski A.S. Mutations in NEUROD1 are associated with the development of type 2 diabetes mellitus. Nat. Genet. 1999;23:323–328. doi: 10.1038/15500. [DOI] [PubMed] [Google Scholar]
- 57.Takeshita S., Suzuki T., Kitayama S., Moritani M., Inoue H., Itakura M. Bhlhe40, a potential diabetic modifier gene on Dbm1 locus, negatively controls myocyte fatty acid oxidation. Genes Genet. Syst. 2012;87:253–264. doi: 10.1266/ggs.87.253. [DOI] [PubMed] [Google Scholar]
- 58.Syrbe S., Harms F.L., Parrini E., Montomoli M., Mütze U., Helbig K.L., Polster T., Albrecht B., Bernbeck U., van Binsbergen E. Delineating SPTAN1 associated phenotypes: from isolated epilepsy to encephalopathy with progressive brain atrophy. Brain. 2017;140:2322–2336. doi: 10.1093/brain/awx195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Voll S.L., Boot E., Butcher N.J., Cooper S., Heung T., Chow E.W.C., Silversides C.K., Bassett A.S. Obesity in adults with 22q11.2 deletion syndrome. Genet. Med. 2017;19:204–208. doi: 10.1038/gim.2016.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Collins R.L., Brand H., Karczewski K.J., Zhao X., Alföldi J., Khera A.V., Francioli L.C., Gauthier L.D., Wang H., Watts N.A. An open resource of structural variation for medical and population genetics. bioRxiv. 2019 [Google Scholar]
- 61.Van Hout C.V., Tachmazidou I., Backman J.D., Hoffman J.X., Ye B., Pandey A.K., Gonzaga-Jauregui C., Khalid S., Liu D., Banerjee N. Whole exome sequencing and characterization of coding variation in 49,960 individuals in the UK Biobank. bioRxiv. 2019 doi: 10.1038/s41586-020-2853-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Medina-Gomez C., Kemp J.P., Trajanoska K., Luan J., Chesi A., Ahluwalia T.S., Mook-Kanamori D.O., Ham A., Hartwig F.P., Evans D.S. Life-Course Genome-wide Association Study Meta-analysis of Total Body BMD and Assessment of Age-Specific Effects. Am. J. Hum. Genet. 2018;102:88–102. doi: 10.1016/j.ajhg.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Olsen L., Sparsø T., Weinsheimer S.M., Dos Santos M.B.Q., Mazin W., Rosengren A., Sanchez X.C., Hoeffding L.K., Schmock H., Baekvad-Hansen M. Prevalence of rearrangements in the 22q11.2 region and population-based risk of neuropsychiatric and developmental disorders in a Danish population: a case-cohort study. Lancet Psychiatry. 2018;5:573–580. doi: 10.1016/S2215-0366(18)30168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klaassen P., Duijff S., Swanenburg de Veye H., Beemer F., Sinnema G., Breetvelt E., Schappin R., Vorstman J. Explaining the variable penetrance of CNVs: Parental intelligence modulates expression of intellectual impairment caused by the 22q11.2 deletion. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2016;171:790–796. doi: 10.1002/ajmg.b.32441. [DOI] [PubMed] [Google Scholar]
- 65.Castel S.E., Cervera A., Mohammadi P., Aguet F., Reverter F., Wolman A., Guigo R., Iossifov I., Vasileva A., Lappalainen T. Modified penetrance of coding variants by cis-regulatory variation contributes to disease risk. Nat. Genet. 2018;50:1327–1334. doi: 10.1038/s41588-018-0192-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang N.K., Chiang J.P.W. Increasing evidence of combinatory variant effects calls for revised classification of low-penetrance alleles. Genet. Med. 2019;21:1280–1282. doi: 10.1038/s41436-018-0347-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maynard T.M., Haskell G.T., Peters A.Z., Sikich L., Lieberman J.A., LaMantia A.-S. A comprehensive analysis of 22q11 gene expression in the developing and adult brain. Proc. Natl. Acad. Sci. USA. 2003;100:14433–14438. doi: 10.1073/pnas.2235651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rippey C., Walsh T., Gulsuner S., Brodsky M., Nord A.S., Gasperini M., Pierce S., Spurrell C., Coe B.P., Krumm N. Formation of chimeric genes by copy-number variation as a mutational mechanism in schizophrenia. Am. J. Hum. Genet. 2013;93:697–710. doi: 10.1016/j.ajhg.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walsh T., McClellan J.M., McCarthy S.E., Addington A.M., Pierce S.B., Cooper G.M., Nord A.S., Kusenda M., Malhotra D., Bhandari A. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.