Abstract
Background
Acrylic acid (AA) is a widely used commodity chemical derived from non-renewable fossil fuel sources. Alternative microbial-based production methodologies are being developed with the aim of providing “green” acrylic acid. These initiatives will benefit from component sensing tools that facilitate rapid and easy detection of in vivo AA production.
Results
We developed a novel transcriptional sensor facilitating in vivo detection of acrylic acid (AA). RNAseq analysis of Escherichia coli exposed to sub-lethal doses of acrylic acid identified a selectively responsive promoter (PyhcN) that was cloned upstream of the eGFP gene. In the presence of AA, eGFP expression in E. coli cells harbouring the sensing construct was readily observable by fluorescence read-out. Low concentrations of AA (500 μM) could be detected whilst the closely related lactic and 3-hydroxy propionic acids failed to activate the sensor. We further used the developed AA-biosensor for in vivo FACS-based screening and identification of amidase mutants with improved catalytic properties for deamination of acrylamide to acrylic acid.
Conclusions
The transcriptional AA sensor developed in this study will benefit strain, enzyme and pathway engineering initiatives targeting the efficient formation of bio-acrylic acid.
Electronic supplementary material
The online version of this article (10.1186/s12934-019-1185-y) contains supplementary material, which is available to authorized users.
Background
Acrylic acid (AA; also known as 2-propenoic acid) is a low molecular weight commodity chemical synthesized from petroleum-derived products. It is a feedstock for acrylate esters that are extensively used in the manufacture of paint-additives, adhesives, textiles and super absorbent materials such as diapers. Acrylic acid is conventionally generated by oxidation of propylene or propane, and the main challenge to this process is dependence on non-renewable fossil-fuel sources subject to unpredictable price fluctuations. Alternative “green” bio-based routes to produce acrylic acid from renewable sources such as sugars have therefore been proposed and investigated [1–4].
Early research focused on endogenous acrylic acid producers, such as the obligate anaerobes Clostridium propionicum and Megasphaera elsdenii that are capable of reducing lactic acid to propionic acid via an acrylyl-CoA intermediate [5]. In the presence of an electron acceptor (e.g. oxygen or methylene blue) acrylate can accumulate in Clostridium propionicum via oxidation of propionate [2, 6]. More recently, it was demonstrated that acrylate pathway enzymes from Clostridium propionicum can be expressed in a heterologous host (E. coli), enabling propionic acid biosynthesis from d-lactic acid via an acrylate intermediate [7]. Total biosynthesis of AA has further been described in engineered E. coli via enzymatic dehydration of 3-hydroxy propionic acid (3-HP) [8, 9]. However, given the low AA yields reported, significant pathway and enzyme engineering will be required to develop a commercially viable process. This endeavor will be expedited by tools facilitating rapid analysis of AA, routinely detected by low throughput, time consuming chromatographic techniques requiring sample preparation. High throughput screening campaigns interrogating sizeable mutant enzyme diversity and metabolic pathway iterations would particularly benefit from such tools. To this end, genetically encoded transcriptional sensors have proven useful for the clonal, real-time, and quantitative assessment of intracellular analyte concentrations [10]. These typically comprise cognate promoter elements of an analyte-responsive transcription factor that drive expression of a coupled read-out module, often a fluorescent protein [11]. Using this approach, sensors have been developed for a diverse array of analytes including amino acids, alcohols, flavonoids, organic acids, sugars and antibiotics [10–16]. When read-outs are compatible with high-throughput analytical platforms such as FACS, these sensors have facilitated significant improvements in product yields.
Here, we employ transcriptome analysis to identify E. coli genes selectively up-regulated in the presence of acrylic acid. Candidate promoters regulating these genes were coupled to an eGFP reporter module and sensitivity to acrylic acid confirmed. We further validate the lead sensor by in vivo selection of amidase variants showing improved catalytic conversion of acrylamide to acrylic acid.
Results and discussion
Identification of AA-responsive genes by transcriptome analysis
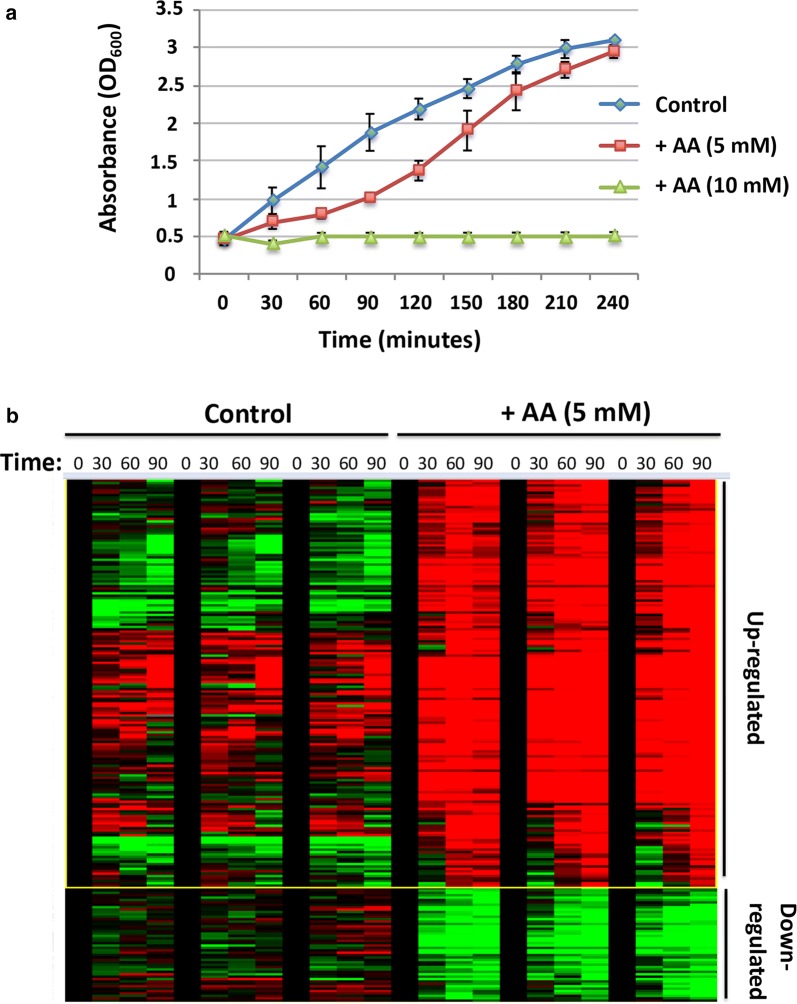
Escherichia coli growth curves in the presence of acrylic acid indicated that concentrations > 5 mM were lethal. At 5 mM AA, cells recover after an initial delay (~ 90 min), and follow the growth characteristics of untreated cells (Fig. 1a). In order to study gene expression changes upon exposure to acrylic acid, we carried out RNAseq analysis of E. coli cells treated with 5 mM AA at early time points (0, 30, 60, 90 min) on the growth curve. Notable variations were observed in global expression profiles between treated and untreated samples (Fig. 1b). Greater than twofold transcriptional up-regulation occurred in 34, 58 and 83 genes at the respective 30, 60 and 90 min time points (Additional file 1).
Fig. 1.
a Growth curves of E. coli cells treated with indicated concentrations of acrylic acid. Values represent average ± SD (n = 2). b RNAseq cluster heatmap indicating significant changes in up-regulated (red) and down-regulated (green) genes in E. coli exposed to acrylic acid (5 mM). Experiments were carried out in triplicate over the four indicated time points
Validation of the acrylic acid—responsive genes
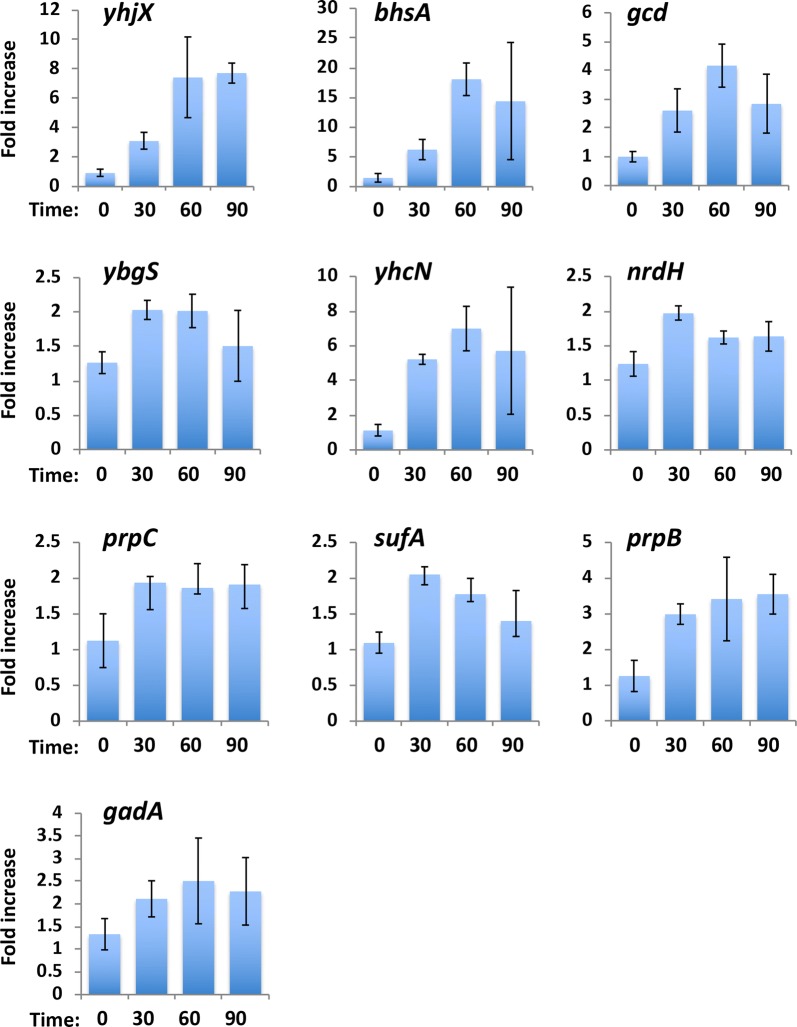
Transcript levels of the top 10 genes up-regulated by AA at the 30 min sampling point (Table 1) were further investigated by quantitative real time PCR. In all cases, transcriptional up-regulation induced by AA was observed, with maximum increments over controls ranging between 2- and 18-fold (Fig. 2). Signals plateaued at 30–60 min for all genes, correlating with the 90 min time interval post AA-treatment when E. coli growth reverts to normal. A subset of these genes (yhjX, bhsA, yhcN and prpB) displayed improved signal to noise ratios due to persistent low level activity in the absence of AA (Additional file 2: Fig. S1), and were further evaluated.
Table 1.
Top 10 genes upregulated after 30 min in E. coli exposed to acrylic acid
| Gene | Fold up-regulation | Function |
|---|---|---|
| yhjX | 6.5 | Putative carboxylic acid antiporter |
| bhsA | 5.7 | Multiple stress protein |
| gcd | 5.4 | Glucose dehydrogenase |
| ybgS | 5.4 | Unknown |
| yhcN | 4.5 | Cellular response to acidity/peroxide |
| nrdH | 4.3 | Glutaredoxin-like protein |
| prpC | 4.3 | Citrate synthase |
| sufA | 4.0 | Iron-sulphur cluster assembly |
| prpB | 4.0 | Methyl isocitrate lyase |
| gadA | 3.8 | Glutamate decarboxylase |
Fig. 2.
Transcription levels of candidate AA-responsive genes in E. coli measured by qPCR over indicated times (minutes) after AA treatment (5 mM). Values represent fold increase over levels of the adk housekeeping gene transcript (n = 3 ± SD)
Development of transcriptional sensor detecting acrylic acid
Promoter sequences of the 4 candidate genes (PyhjX, PbhsA, PyhcN and PprpB) (Additional file 2: Fig. S2) were cloned upstream of eGFP and E. coli cells transformed with plasmids encoding the putative sensors. Cells were treated with 5 mM acrylic acid and fluorescence monitored over 24 h (Additional file 2: Fig. S3). The PyhcN, PprpB and PyhjX sensors displayed varying increases in fluorescence over no treatment and lactic acid controls. This was most pronounced for PyhcN, exhibiting ~ 50% increased signal over controls 2 h post-treatment that persisted over the 24 h time course. Relatively lower signal gains for the PprpB and PyhjX sensors (15–20%) were observed from the 6 h time point onwards. Efforts to improve the signal to noise ratio for the PyhcN-eGFP sensor by truncation of the yhcN promoter sequence did not boost sensor performance (Additional file 2: Fig. S4).
Specificity and sensitivity of the acrylic acid sensor
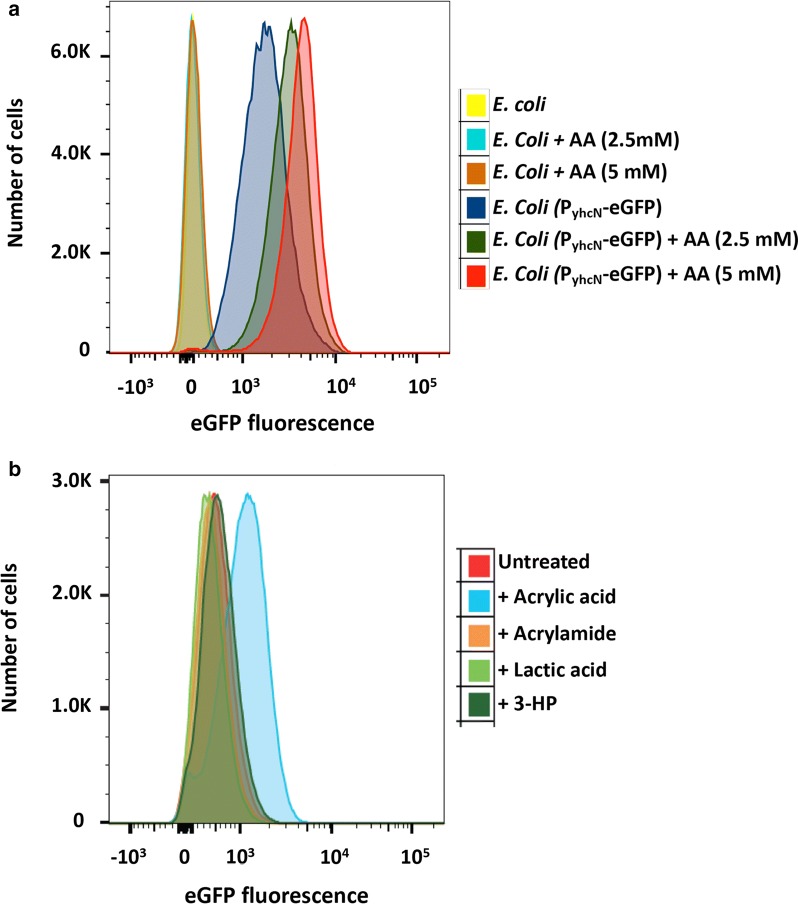
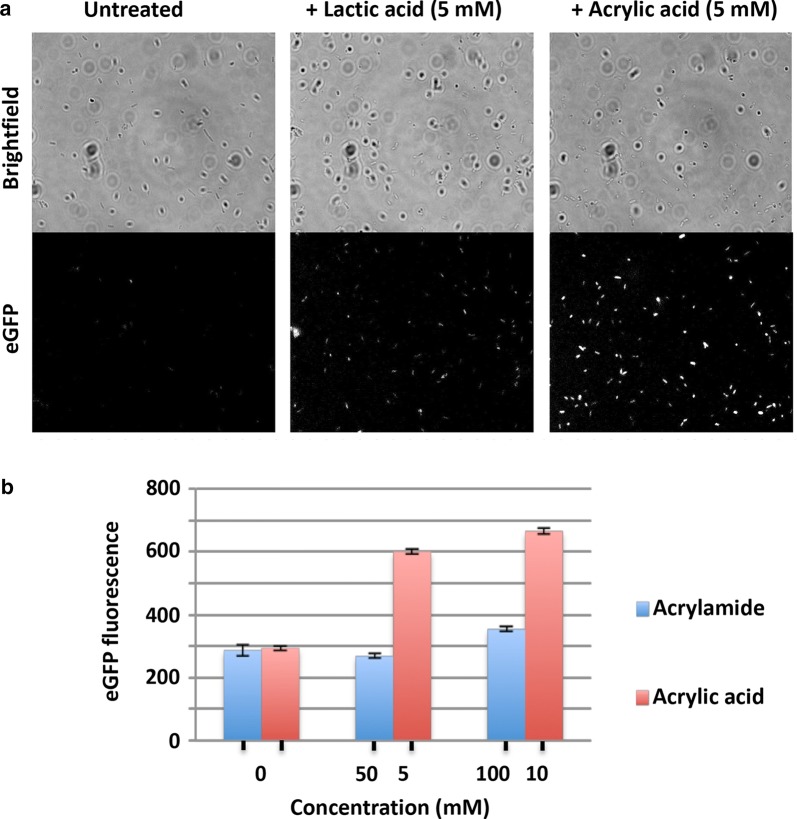
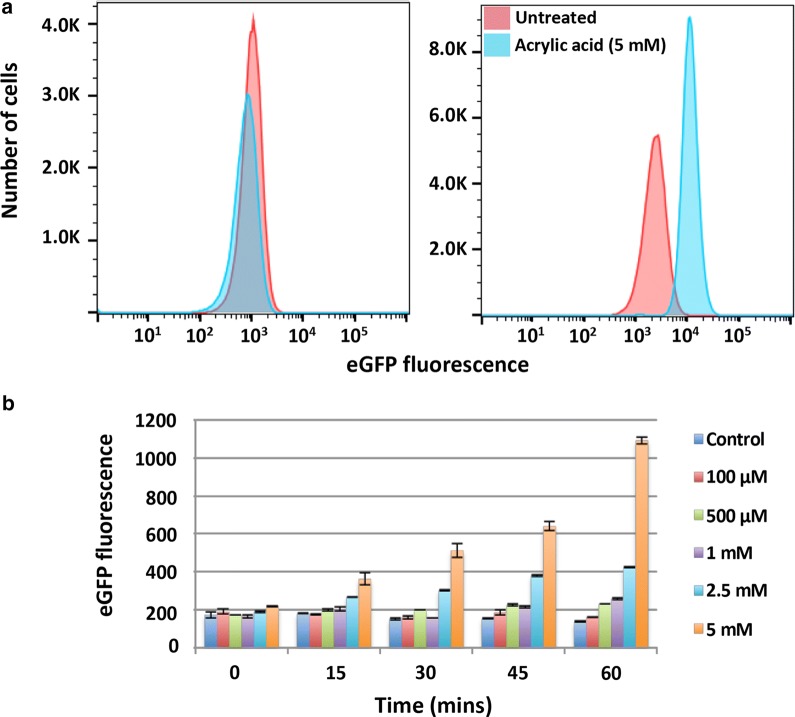
The PyhcN-eGFP sensor strain was next evaluated by FACS analysis. Cells treated with acrylic acid (0, 2.5, 5 mM) showed a dose responsive increase in eGFP fluorescence indicated by right shifting of the histogram (Fig. 3a). Treatment with the potential acrylic acid precursors acrylamide, lactic acid and 3-hydroxy propionic acid (5 mM) did not result in appreciable signal gain, confirming specificity of the PyhcN-eGFP sensor (Fig. 3b). This was further confirmed by fluorescence imaging of the PyhcN-eGFP sensor strain 6 h post treatment with AA (5 mM) which showed clear enhancement of signal over no-treatment and lactic acid (5 mM) controls (Fig. 4a). Additionally, high concentrations of acrylamide (100 mM) did not activate the sensor (Fig. 4b).
Fig. 3.
a Parental E. coli and cells expressing plasmid-encoded acrylic acid sensor were treated with indicated acrylic acid concentrations (3 h) and eGFP fluorescence measured by FACS. Traces for both control and treated parental E. coli cells (yellow, cyan and brown) overlap. b Response of PyHCN-eGFP sensor to indicated compounds (5 mM)
Fig. 4.
a Cells expressing PyHCN-eGFP sensor were treated as indicated for 6 h and imaged by fluorescence microscopy. b eGFP fluorescence of cells treated with AA (0, 5, 10 mM) or acrylamide (0, 50, 100 mM)
Stable integration of the transcriptional acrylic acid sensor into E. coli cells
The PyhcN-eGFP reporter cassette was next stably integrated into E. coli using bacteriophage lambda mediated recombination [17]. The resultant strain showed improved sensor turn-on in the presence of AA when measured by FACS (Fig. 5a). Dose-dependent signal read-out was also readily detected by fluorescence measurements using a plate reader, with relatively low concentrations (500 μM) of AA measurable after 45 min using the whole-cell PyhcN-eGFP biosensor (Fig. 5b). Imaging of the stably transformed strain exposed to AA showed clear analyte-dependent eGFP fluorescence (Additional file 2: Fig. S5), as observed with the plasmid-based sensor (Fig. 4a).
Fig. 5.
a Control E. coli (left) or stably transduced E. coli (PyHCN-eGFP)(right) were treated with acrylic acid (5 mM) overnight and analysed by FACS. b Stably transduced AA reporter cells treated with indicated concentrations of AA and fluorescence measured over time
Selection of improved G. pallidus RAPc8 amidase variants
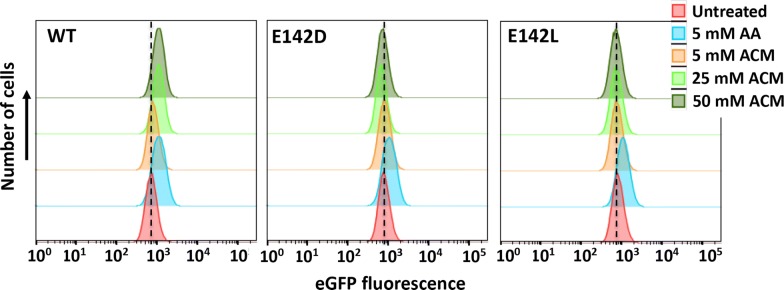
Acrylic acid can be made via enzymatic deamination of acrylamide as demonstrated using aliphatic amidases [18, 19]. We coupled this relatively simple pathway to AA sensor output by expressing the G. pallidus RAPc8 amidase [19] in E. coli and measuring sensor fluorescence by FACS after incubation with acrylamide. A clear upwards shift in the population of eGFP-expressing cells is observed compared to the no-substrate control (Fig. 6). This shift is not observed when the catalytically inactive E142D and E142L amidase mutants [20] were expressed. We next created a library of randomly mutated RAPc8 amidase variants (n = ~ 5 × 105) and screened for improved in vivo acrylamide deamination by FACS. After 2 rounds of selection, secondary screening was carried out on 62 individually sorted clones using plate-based measurement of reporter fluorescence. The top 5 clones from this analysis were further analysed by FACS, and this confirmed 4 of these clones to show improved activity over WT (Additional file 2: Fig. S6).
Fig. 6.
FACS analysis indicating eGFP fluorescence of stably transduced E. coli (PyHCN-eGFP) cells expressing RAPc8 amidase (WT) or the inactive E142D and E142L mutants. Cells were treated with indicated concentrations of acrylic acid (AA, positive control) or acrylamide (ACM) for 2 h prior to analysis
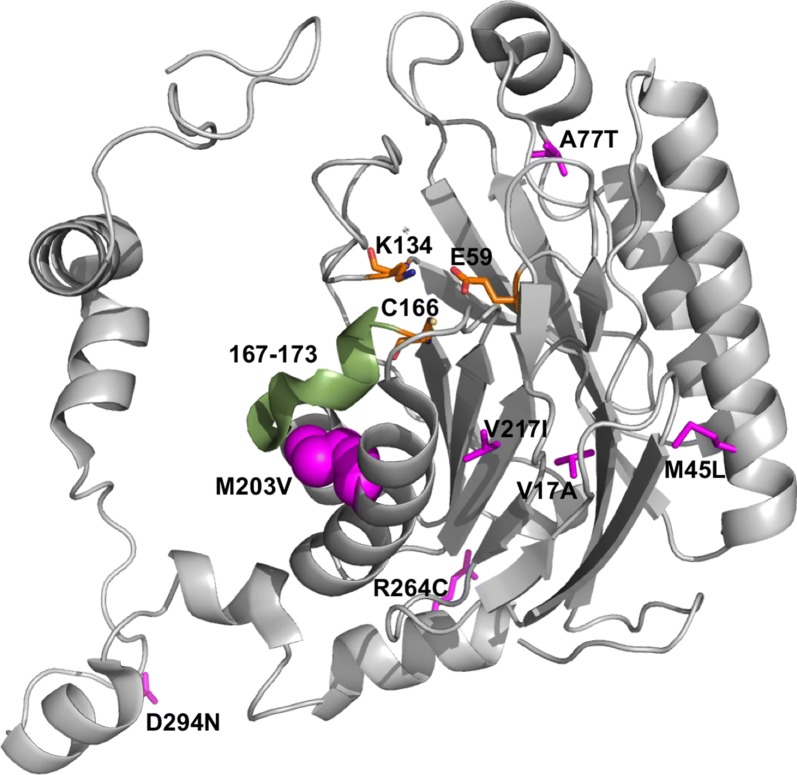
Sequence analysis of the amidase gene in 3 of these selectants (C26, C60, C65) highlighted 5 mutations in C60 (M45L, A77T, M203V, D294N, K342E), and 3 mutations in C65 (V17A, V217I, R264C). Interestingly, whilst C26 encoded WT enzyme, two silent mutations were present in codons 208 (GCG to GCA) and 326 (ACT to ACC) that likely contributed to increased amidase expression and acrylamide turnover via codon-optimisation effects [21, 22]. Mutations in C60 and C65 were distributed throughout the enzyme’s tertiary structure (Fig. 7). Comparative sequence analysis indicated common phylogenetic variation at positions 77 (alternatively T or S) and 203 (alternatively V or I), highlighting natural selection for the A77T and M203V mutations in C60 identified here by laboratory evolution. Notably, positions 77 and 203 were correspondingly S and V in the Hydrogenovibrio kuenenii aliphatic amidase (84% sequence identity to RAPc8 amidase). Kinetic parameters of purified RAPc8-C60 for the acyl transfer reaction indicated ~ 1.6-fold improvement in kcat/Km over wild-type enzyme (Table 2) that was driven by an elevated kcat. The M203V mutation contributed towards this improved activity of C60, with RAPc8-M203V showing ~ 1.3-fold kcat/Km increase over WT. This is likely due to repositioning of an adjacent distorted helix (residues 167–173) that precedes C166 of the catalytic triad (Fig. 7). Whilst RAPc8-A77T displayed ~ twofold reduced Km compared to WT, this was accompanied by reduced kcat. Kinetic paramters of the RAPc8-A77T/M203V double mutant were essentially the same as RAPc8-A77T. Other mutations present in C60 therefore likely contributed to the overall improved phenotype.
Fig. 7.
Position of mutations present in selected RAPc8 C60 and C65 variants (magenta). Residues depicted in orange (E59, K134, C166) comprise the catalytic triad. M203 is shown in spheres to highlight packing against the distorted helix (residues 167–173 denoted in green) that likely undergoes structural changes to adapt to the loss of interactions when M203 is mutated to V with a smaller sidechain
(adapted from the crystal structure 2PLQ [28] and drawn using PyMOL [29])
Table 2.
Kinetic parameters for RAPc8 and indicated mutants
| RAPc8 enzyme variant | Vmax (uM min−1) |
Km (mM) |
kcat (min−1) |
kcat/Km (mM−1 min−1) |
|---|---|---|---|---|
| WT | 106.5 ± 6.1 | 7.3 ± 0.1 | 7096.3 ± 404.7 | 978.9 ± 63.3 |
| A77T | 66.7 ± 2.1 | 4.5 ± 0.2 | 4447.7 ± 139.2 | 996 ± 21.4 |
| M203V | 146.8 ± 16.7 | 7.2 ± 1.1 | 9784.7 ± 1113.3 | 1374.5 ± 70 |
| A77T + M203V | 64.7 ± 4.6 | 4.6 ± 0.4 | 4315 ± 308.4 | 935.7 ± 35.9 |
| C60 | 176.3 ± 16.1 | 7.4 ± 1.3 | 11,754.3 ± 1073.3 | 1607.1 ± 143.5 |
Values represent average ± SD (n = 3)
In the selection protocol we describe there is the possibility that diffusion of AA from within cells expressing active amidase genes would co-activate the sensor in other cells expressing inactive mutants, resulting in increased false postives and higher background. Future selections could mitigate this issue by segregating cells utilising compartmentalisation [23] or performing colony-based selection on agar plates [24].
Up-regulation of yhcN in response to stress
The AA-responsive yhcN gene identified in this study encodes an 87 amino acid polypeptide with a predicted periplasmic location. It has previously been ascribed to a network of E. coli stress-induced proteins, with its expression up-regulated by hydrogen peroxide and cadmium exposure. It is also implicated in regulating biofilm formation in response to stress [25]. Furthermore, in addition to 4 other markedly AA-responsive genes identified in this study (yhjX, nrdH, gcd and gadA)(Table 1), yhcN is up-regulated in low-pH conditions in the E. coli strain K-12 W3110 [26, 27]. In our experiments, the PyhcN-eGFP reporter was selectively responsive to acrylic acid, showing no activation by lactic acid or 3-HP. Given the similar pKa values of these acids, the differential response may arise from the different yhcN promoter contexts (endogenous versus synthetic construct) and/or differences in effective intracellular concentrations of these acids. As lactic acid and 3-HP are both direct precursors of AA in synthetic pathways grafted into E. coli [7–9], the developed AA sensor should expedite efforts to increase yields. This could be achieved through rational pathway engineering and/or mutagenesis of key enzymes linked to selection, as shown in this study by identification of improved amidase variants.
Conclusions
In summary, we have developed and applied a transcriptional biosensor facilitating detection of acrylic acid in vivo. This was achieved through transcriptome analysis of E. coli exposed to acrylic acid and validation of candidate promoters upstream of highly up-regulated genes. We further applied the biosensor to report on E. coli acrylic acid production linked to the simple enzymatic deamination of exogenous acrylamide. This tool should expedite both development and optimisation of E. coli strains capable of producing sustainable and economically viable bio-acrylic acid.
Materials and methods
Chemicals and reagents
All chemicals including acrylic acid, acrylamide, lactic acid, 3-hydroxy propionic acid and DMSO used in this study were purchased from Sigma-Aldrich (USA).
Oligonucleotide primers
| Real-time PCR primers | |
| yhjX-F | 5′-TGCTGACGCTCTCTAACTGC-3′ |
| yhjX-R | 5′-GCAATCGCTCCCCAAATCAC-3′ |
| bhsA-F | 5′-TGTCATTTGCCAGCTTTGCG-3′ |
| bhsA-R | 5′-TACGGAAAGATTTTGCGCCC-3′ |
| gcd-F | 5′-TGGTCGCAATCAGGAAGGTC-3′ |
| gcd-R | 5′-ATCGGCGTCACTTCATTGGT-3′ |
| ybgS-F | 5′-ATGTCGCGCCAAATAACGTC-3′ |
| ybgS-R | 5′-TATCCGGACAGCGACCATCT-3′ |
| yhcN-F | 5′-TCTCTTTCGGTGCATTCGCT-3′ |
| yhcN-R | 5′-TAATCTGGTAGGCCGTTGCG-3′ |
| nrdH-F | 5′-GTAACGATTGCGTTCAGTGCC-3′ |
| nrdH-R | 5′-CAGACCAGCTAAGATCGCCA-3′ |
| prpC-F | 5′-CGAGTTTAACGCCTCCACCT-3′ |
| prpC-R | 5′-TCGTAGCGTTGCTGGATCTC-3′ |
| sufA-F | 5′-CTTAGGCGTGAAGCAAACGG-3′ |
| sufA-R | 5′-TCGACTTCCGTGCCATCAAT-3′ |
| prpB-F | 5′-GCTGCCCGATCTCGGTATTT-3′ |
| prpB-R | 5′-CGCACCGGCTTTAATCATCG-3′ |
| gadA-F | 5′-CTGCTGGCATAAATTCGCCC-3′ |
| gadA-R | 5′-GTGTAGGTCACGCCGAAAGT-3′ |
| adk-F | 5′-ATCCGCCGAAAGTAGAAGGC-3′ |
| adk-R | 5′-TTACCCGCTTCCGCTTCTTT-3′ |
| gyrB-F | 5′-TGGTTGTGGTATCGGTCGTG-3′ |
| gyrB-R | 5′-GCTGAGCGATGTAGACGTGA-3′ |
| Primers to clone promoters and GFP into pUC19 vector | |
| GFP BamHI | cgactctagaggatccATGGTGAGCAAGGGCGAGG |
| GFP NdeI | tgagagtgcaccatatgTTATCTAGACTTGTACAGCTCGTCCATGCCG |
| PyhjX for | ccttttgctcacatgtCGTAACAGTCACAATTGAAACCATTAAATAAC |
| PyhjX rev | GCCCTTGCTCACCATGGCAGTATTCCTGCAGTAATAAAAAGG |
| PbhsA for | ccttttgctcacatgtGATGCCGTTGTACCTGGTGAC |
| PbhsA rev | GCCCTTGCTCACCATAATAGTGGCCTTATGCAGATGAATGAC |
| PyhcN for | ccttttgctcacatgtTCTCTGCCCCGTCGTTTC |
| PyhcN rev | GCCCTTGCTCACCATGATTTTTACCTCGACATAATCTTTTAGCTGG |
| PprpB for | ccttttgctcacatgtAGCGCACCGCAAAGTTAAGAAAC |
| PprpB rev | GCCCTTGCTCACCATAGCCCATCCTTTGTTATCAACTTGTTATTTG |
| Sequencing primers | |
| eGFP mid rev | 5′-AGGGTCAGCTTGCCGTAGG-3′ |
| pET Upstream | 5′-ATGCGTCCGGCGTAGA-3′ |
| Duet Down1 | 5′-GATTATGCGGCCGTGTACAA-3′ |
| Duet Up2 | 5′-TTGTACACGGCCGCATAATC-3′ |
| T7 terminator | 5′-GCTAGTTATTGCTCAGCGG-3′ |
| ACYCDuetUP1 | 5′-GGATCTCGACGCTCTCCCT-3′ |
| pETF2 | 5′-CATCGGTGATGTCGGCGAT-3′ |
| petR | 5′-CGGATATAGTTCCTCCTTTCAGCA-3′ |
| Stable integration primers | |
| attP-F | cacagaattcCGTCTGTTACAGGTCACTAATACCATCT |
| attPSOE-R | ACATTTCCCCGAAAAGTGCCACCTGAACATCACCGG GAAATCAAATAATGAT |
| pSR158-F | 5′-GATCCGGCTGCTAACAAAGCC-3′ |
| pSR158-R | 5′-GATTTTTACCTCGACATAATCTTTTAGCTGGG-3′ |
| EcoliAttB-F | CTG AAA ATG TGT TCA CAG GTT GCT |
| EcoliattB-R | GCA ATG CCA TCT GGT ATC ACT |
| TEM1prom-F | TTC AGG TGG CAC TTT TCG GGG AAA TGT |
| TEM1prom-R | TGT GGA ATT CCT ACA CTA GAA GGA CAG TAT TTG GTA TCT GC |
| Cloning RApc8 amidase WT and mutants | |
| GpAmidase-F | gtataagaaggagatataCATATGCGTCATGGAGATATTAGCTCCTC |
| GpAmidase-R | cagcggtttctttaccagaCTCGAGTTAGTGGTGGTGGTGG |
| E142D-F | 5′-CTTGGTGCCCCATCGAtGGGTGGTACCCTGGCG-3′ |
| E142D-R | 5′-CGCCAGGGTACCACCCaTCGATGGGGCACCAAG-3′ |
| E142L-F | 5′-CCTTGGTGCCCCATCctgGGGTGGTACCCTGGC-3′ |
| E142L-R | 5′-GCCAGGGTACCACCCcagGATGGGGCACCAAGG-3′ |
| A77T-F | 5′-GAAATGTTCGCGACAGCCaCCAGCATTCCAGGGG-3′ |
| A77T-R | 5′-CCCCTGGAATGCTGGtGGCTGTCGCGAACATTTC-3′ |
| M203V-F | 5′-GAACAGCAAATAATGgTGGCTAAAGCAATGG-3′ |
| M203V-R | 5′-CCATTGCTTTAGCCAcCATTATTTGCTGTTC-3′ |
Identification of acrylic acid up-regulated genes by RNAseq analysis
Acrylic acid treatment and RNA isolation
Escherichia coli BL21 cells cultured in LB medium (37 °C) were exposed to increasing concentration of acrylic acid at the mid-log phase (OD600 = 0.5), and the sublethal dose determined from the growth curves. For RNAseq experiments, E. coli BL21 culture was treated with either 0 or 5 mM acrylic acid at the mid-log phase and the cells were harvested at 0, 30, 60 and 90 min time points. Total RNA was extracted using RNeasy Mini kit (Qiagen) following the manufacturer’s protocol, and rRNA was removed using ribo-zero rRNA removal kit (Epicentre). RNA was quantified and quality confirmed by Bioanalyzer quality analysis (RIN values > 9.0). Experiments were carried out in triplicate. Illumina sequencing libraries were generated from the RNA using the NEBNext RNA-seq library preparation kit following manufacturer’s guidelines (New England Biolabs). Each library was barcoded during PCR, libraries were then quantified by qPCR and equimolar aliquots of each library were pooled together. Sequencing was done on an Illumina HiSeq 2500 instrument.
Gene-based expression matrix
Cuffnorm v2.2.0, a program that is part of Cufflinks [30], was used to generate a gene-based expression matrix from the BAM files. Cuffnorm was run with the options of “–library-type fr-unstranded” and “–library-norm-method classic-fpkm”. The resulting expression matrix was normalized for library size and values were represented as FPKM (fragments per kilobase of exon per million fragments mapped).
Hierarchical clustering
Hierarchical clustering was performed on the FPKM expression matrix using R v3.1.0. The expression matrix was first transformed to the log-2 space before computing the distance matrix based on the Euclidean distance measure using the dist function of R. The Spearman’s rank correlation was next calculated using the cor function before being plotted using the heatmap.2 function from the gplots package of CRAN.
Differential expression analysis
Cuffdiff v2.1.1 (part of Cufflinks program) was used to identify differentially expressed genes at each time point. Default parameters were used except for the option of “–multi-read-correct” and “–max-bundle-frags 100000000”. A threshold of FDR < 0.05 and absolute log2 fold change > 2.0 was employed for significance. SAM (Significance Analysis of Microarrays) [31] was used to identify genes differentially expressed across time points. Analysis was performed in R v3.1.0 using the samr package from CRAN with the following options: resp.type = “Two class unpaired timecourse”, nperms = 100 and time.summary.type = “slope”. Genes having a log-2 fold change > 2.0 were assigned as up-regulated, while those having a log-2 fold change smaller than − 2 were assigned as down-regulated.
Analysis of endogenous transcript levels by qPCR
qPCR was carried out using the iScript Reverse Transcription Supermix (Bio-Rad). 1 µg RNA was used for a 20 µl reaction following manufacturer’s protocol. For RT-qPCR, SsoAdvanced SYBR Green Supermix (Bio-Rad) kit was used and 10 µl reactions were set up following manufacturer’s protocol using real time PCR primers for each gene analysed.
Plasmid construction and live cell biosensor development
eGFP was amplified using GFP-BamHI and GFP-NdeI primers and cloned into pUC19 vector. Candidate promoter regions of the top four up-regulated genes (arbitrarily predicted to be located within the first 300 bp of the non-coding region preceding the ORF of candidate up-regulated genes) (Additional file 2: Fig. S2) were amplified by PCR using Pprom-F and Pprom-R primers respectively and cloned upstream of the eGFP gene in the plasmid pUC19-eGFP cut using PciI and BamHI restriction by infusion cloning method. Reporter plasmid constructs contain an ampicillin resistance gene, E. coli origin of replication and an acrylic acid inducible putative promoter region followed by the eGFP coding sequence. To make the truncated yhcN promoter Δ108, pUC19-PyhcN-eGFP was cut with PciI and re-ligated.
Biosensor development and validation
Live cell biosensors were developed by transforming chemically competent E. coli BL21 cells with the four plasmid constructs containing putative AA-responsive promoter sequences. Cells were grown in LB medium at 37 °C to mid-log phase followed by treatment with AA (or other chemicals) for various time points. The cells were collected by centrifugation, washed with phosphate-buffered saline (PBS) and resuspended in the same buffer to normalise cell concentrations to OD600 = 1. Expression of eGFP was measured by either FACS using the BD FACSAria cell sorter (BD Biosciences) or plate reader (PerkinElmer 2104), measuring green fluorescence (Ex/Em = 488/509 nm) of 100 µl resuspended cells (OD600 = 1.0). Cells were also visualised by fluorescence microscopy (AxioImager Z1 upright fluorescent microscope, Zeiss) to detect eGFP fluourescence. Treated or control cells were washed with PBS and diluted to OD600 = 1.0. One drop of the cell suspension was placed on a microscope slide, air dried and covered with a coverslip. The 63 × oil immersion lens was used to visualise cells that were imaged with 500 ms exposure time.
Generation of the E. coli BL21 PyhcN-eGFP sensor strain
pET22b-Amp-attP-PyhcN-eGFP was amplified using primers pETF2 and TEM1prom-R to create minicircles of attP-PyhcN-eGFP. The C3INT-HIS-PET22b(+) plasmid [17] was amplified with petF2 and petR and the PCR products were intramolecularly ligated to produce attP-PyhcN-eGFP and C3INT-HIS minicircles. A total of 100 ng of C3INT-HIS minicircle and 100 ng attP-PyhcN-eGFP minicircle were combined and electroporated into 25 µl electrocompetent BL21 cells. The cells were allowed to recover for 1 h at 37 °C before being plated on varying concentrations of ampicillin-LB agar plates (0.07 and 0.1 mg/ml). Incubation was carried out at 37 °C for 12–14 h to allow for expression of C3 integrase and chromosomal integration of the ampicillin-resistance cassette. Positive clones were confirmed by treating cells with 5 mM AA to turn on GFP expression. Genomic integration into the attB site was confirmed by sequencing of PCR products generated using primers EcoliAttB-F and TEM1prom-R along with EcoliattB-R and TEM1prom-R.
Selection of improved RAPc8 amidase variants
The RAPc8 amidase gene was randomly mutated (using the Agilent Gene Morph II random mutagenesis kit) and cloned into the plasmid pDuet-Amp-PyhcN-eGFP cut with NdeI/XhoI. The pDuet-Amp-RapC8 amidase + PyhcN-eGFP plasmid was transformed into BL21 cells to create a library (~ 5 × 105 transformants). The library was grown to OD600 ~ 0.5, induced with 0.1 mM IPTG and acrylamide added thereafter (5 mM). After 60 min, cells were diluted in 1X PBS and analysed by FACS. The top 5% and 1% brightest cells were respectively sorted in rounds 1 and 2. The amidase gene in R1 selectants was amplified by PCR prior to digestion and ligation into cut plasmid pDuet-Amp-PyhcN-eGFP and transformation into BL21 cells. Individual clones post R2 (n = 62) were assayed using the plate-based fluorescence assay with a subset of these showing improved activity over WT enzyme further validated by FACS.
Determination of kinetic parameters
Amidase activity was determined by quantifying the production of ammonia via the modified Berthelot reaction [20]. 15 nM amidase enzyme was incubated in 100 μl volume of reaction buffer (20 mM potassium phosphate pH 7.2, 150 mM NaCl, 2 mM DTT) with acrylamide titrated up to a final concentration of 40 mM. The mixture was incubated at 37 °C for 20 min and ammonia production detected colourimetrically at 670 nm using the ammonia assay kit (Abcam). Standards were prepared using ammonium chloride. Measurements were performed in triplicate and kinetic parameters obtained using the nonlinear regression curve fitting analysis in GraphPad Prism 7 (GraphPad Software, Inc., CA).
Expression and purification of RAPc8 amidase variants
The RAPc8 amidase constructs were cloned with a C-terminal 6xHIS tag and transformed into Escherichia coli BL21(DE3) (Invitrogen) competent cells. These were grown in LB medium at 37 °C and induced at OD600 ~ 0.5 at 30 °C with 0.1 mM IPTG for 6 h for RAPc8, RAPc8-A77T and RAPc8-M203V. Cells expressing RAPc8-A77T/M203V and RAPC60 were induced at 25 °C with 0.5 mM IPTG and incubated overnight. Cells were then harvested by centrifugation, sonicated and clarified by centrifugation. The clarified cell lysates were applied to a His-GraviTrap column (GE Healthcare) and purified according to the manufacturer’s protocol. Protein purity as assessed by SDS-PAGE were ~ 95%, and fractions were pooled and buffer exchanged into PBS buffer with 0.5 mM DTT. All proteins were concentrated using Amicon-Ultra (3 kDa MWCO) concentrator (Millipore). Protein concentration were determined using the Bradford protein assay kit (BioRad) with bovine serum albumin as standard.
Additional files
Acknowledgements
None.
Authors’ contributions
SSR, SC, NHQA, JP and SJW carried out experimental work. JL carried out transcriptomic analysis with guidance from NN. SSR, FJG and CSV conceived experiments. FJG and SSR wrote manuscript. All authors interpreted data and reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Agency for Science, Technology and Research (Singapore).
Availability of data and materials
The data and materials used/analysed in this study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
CSV is the founder/scientific consultant of Sinopsee Therapeutics, a biotechnology company developing molecules for therapeutic purposes; the current work has no conflict with the company. All other authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gao C, Ma C, Xu P. Biotechnological routes based on lactic acid production from biomass. Biotechnol Adv. 2011;29:930–939. doi: 10.1016/j.biotechadv.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 2.Danner H, Urmos M, Gartner M, Braun R. Biotechnological production of acrylic acid from biomass. Appl Biochem Biotechnol. 1998;70–72:887–894. doi: 10.1007/BF02920199. [DOI] [PubMed] [Google Scholar]
- 3.Choi S, Song CW, Shin JH, Lee SY. Biorefineries for the production of top building block chemicals and their derivatives. Metab Eng. 2015;28:223–239. doi: 10.1016/j.ymben.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Straathof AJ, Sie S, Franco TT, van der Wielen LA. Feasibility of acrylic acid production by fermentation. Appl Microbiol Biotechnol. 2005;67:727–734. doi: 10.1007/s00253-005-1942-1. [DOI] [PubMed] [Google Scholar]
- 5.Akedo M, Cooney CL, Sinskey AJ. Direct demonstration of lactate-acrylate interconversion in Clostridium propioinicum. Bio/Technology. 1983;1:791–794. doi: 10.1038/nbt1183-791. [DOI] [Google Scholar]
- 6.O’’Brien DJ, Panzer CC, Eisele WP. Biological production of acrylic acid from cheese whey by resting cells of Clostridium propionicum. Biotechnol Progr. 1990;6:237–242. doi: 10.1021/bp00004a001. [DOI] [Google Scholar]
- 7.Kandasamy V, Vaidyanathan H, Djurdjevic I, Jayamani E, Ramachandran KB, Buckel W, Jayaraman G, Ramalingam S. Engineering Escherichia coli with acrylate pathway genes for propionic acid synthesis and its impact on mixed-acid fermentation. Appl Microbiol Biotechnol. 2013;97:1191–1200. doi: 10.1007/s00253-012-4274-y. [DOI] [PubMed] [Google Scholar]
- 8.Chu HS, Ahn JH, Yun J, Choi IS, Nam TW, Cho KM. Direct fermentation route for the production of acrylic acid. Metab Eng. 2015;32:23–29. doi: 10.1016/j.ymben.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Liu Z, Liu T. Production of acrylic acid and propionic acid by constructing a portion of the 3-hydroxypropionate/4-hydroxybutyrate cycle from Metallosphaera sedula in Escherichia coli. J Ind Microbiol Biotechnol. 2016;43:1659–1670. doi: 10.1007/s10295-016-1843-6. [DOI] [PubMed] [Google Scholar]
- 10.Cheng F, Tang XL, Kardashliev T. Transcription factor-based biosensors in high-throughput screening: advances and applications. Biotechnol J. 2018;13:e1700648. doi: 10.1002/biot.201700648. [DOI] [PubMed] [Google Scholar]
- 11.Binder S, Schendzielorz G, Stabler N, Krumbach K, Hoffmann K, Bott M, Eggeling L. A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biol. 2012;13:R40. doi: 10.1186/gb-2012-13-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dietrich JA, Shis DL, Alikhani A, Keasling JD. Transcription factor-based screens and synthetic selections for microbial small-molecule biosynthesis. ACS Synth Biol. 2013;2:47–58. doi: 10.1021/sb300091d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sana B, Chia KHB, Raghavan SS, Ramalingam B, Nagarajan N, Seayad J, Ghadessy FJ. Development of a genetically programed vanillin-sensing bacterium for high-throughput screening of lignin-degrading enzyme libraries. Biotechnol Biofuels. 2017;10:32. doi: 10.1186/s13068-017-0720-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers JK, Guzman CD, Taylor ND, Raman S, Anderson K, Church GM. Synthetic biosensors for precise gene control and real-time monitoring of metabolites. Nucleic Acids Res. 2015;43:7648–7660. doi: 10.1093/nar/gkv616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strachan CR, Singh R, VanInsberghe D, Ievdokymenko K, Budwill K, Mohn WW, Eltis LD, Hallam SJ. Metagenomic scaffolds enable combinatorial lignin transformation. Proc Natl Acad Sci U S A. 2014;111:10143–10148. doi: 10.1073/pnas.1401631111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu H, Wang N, Huo W, Zhang Y, Zhang W, Yang Y, Chen Z, Huo YX. Establishment of BmoR-based biosensor to screen isobutanol overproducer. Microb Cell Fact. 2019;18:30. doi: 10.1186/s12934-019-1084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siau JW, Chee S, Makhija H, Wai CM, Chandra SH, Peter S, Droge P, Ghadessy FJ. Directed evolution of lambda integrase activity and specificity by genetic derepression. Protein Eng Des Sel. 2015;28:211–220. doi: 10.1093/protein/gzv015. [DOI] [PubMed] [Google Scholar]
- 18.Thakur N, Kumar V, Sharma NK, Thakur S, Bhalla TC. Aliphatic amidase of Rhodococcus rhodochrous PA-34: purification, characterization and application in synthesis of acrylic acid. Protein Pept Lett. 2016;23:152–158. doi: 10.2174/0929866523666151215105442. [DOI] [PubMed] [Google Scholar]
- 19.Makhongela HS, Glowacka AE, Agarkar VB, Sewell BT, Weber B, Cameron RA, Cowan DA, Burton SG. A novel thermostable nitrilase superfamily amidase from Geobacillus pallidus showing acyl transfer activity. Appl Microbiol Biotechnol. 2007;75:801–811. doi: 10.1007/s00253-007-0883-2. [DOI] [PubMed] [Google Scholar]
- 20.Weber BW, Kimani SW, Varsani A, Cowan DA, Hunter R, Venter GA, Gumbart JC, Sewell BT. The mechanism of the amidases: mutating the glutamate adjacent to the catalytic triad inactivates the enzyme due to substrate mispositioning. J Biol Chem. 2013;288:28514–28523. doi: 10.1074/jbc.M113.503284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spadiut O, Olsson L, Brumer H., 3rd A comparative summary of expression systems for the recombinant production of galactose oxidase. Microb Cell Fact. 2010;9:68. doi: 10.1186/1475-2859-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Athey J, Alexaki A, Osipova E, Rostovtsev A, Santana-Quintero LV, Katneni U, Simonyan V, Kimchi-Sarfaty C. A new and updated resource for codon usage tables. BMC Bioinform. 2017;18:391. doi: 10.1186/s12859-017-1793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghadessy FJ, Ong JL, Holliger P. Directed evolution of polymerase function by compartmentalized self-replication. Proc Natl Acad Sci USA. 2001;98:4552–4557. doi: 10.1073/pnas.071052198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porter JL, Rusli RA, Ollis DL. Directed evolution of enzymes for industrial biocatalysis. ChemBioChem. 2016;17:197–203. doi: 10.1002/cbic.201500280. [DOI] [PubMed] [Google Scholar]
- 25.Lee J, Hiibel SR, Reardon KF, Wood TK. Identification of stress-related proteins in Escherichia coli using the pollutant cis-dichloroethylene. J Appl Microbiol. 2010;108:2088–2102. doi: 10.1111/j.1365-2672.2009.04543.x. [DOI] [PubMed] [Google Scholar]
- 26.Maurer LM, Yohannes E, Bondurant SS, Radmacher M, Slonczewski JL. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J Bacteriol. 2005;187:304–319. doi: 10.1128/JB.187.1.304-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kannan G, Wilks JC, Fitzgerald DM, Jones BD, Bondurant SS, Slonczewski JL. Rapid acid treatment of Escherichia coli: transcriptomic response and recovery. BMC Microbiol. 2008;8:37. doi: 10.1186/1471-2180-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agarkar VB, Kimani SW, Cowan DA, Sayed MF, Sewell BT. The quaternary structure of the amidase from Geobacillus pallidus RAPc8 is revealed by its crystal packing. Acta Crystallogr, Sect F: Struct Biol Cryst Commun. 2006;62:1174–1178. doi: 10.1107/S1744309106043855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeLano WL. The PyMOL molecular graphics system. San Carlos: DeLano Scientific; 2002. [Google Scholar]
- 30.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and materials used/analysed in this study are available from the corresponding author on reasonable request.