Abstract
Susceptibility to schizophrenia is inversely correlated with general cognitive ability at both the phenotypic and the genetic level. Paradoxically, a modest but consistent positive genetic correlation has been reported between schizophrenia and educational attainment, despite the strong positive genetic correlation between cognitive ability and educational attainment. Here we leverage published genome-wide association studies (GWASs) in cognitive ability, education, and schizophrenia to parse biological mechanisms underlying these results. Association analysis based on subsets (ASSET), a pleiotropic meta-analytic technique, allowed jointly associated loci to be identified and characterized. Specifically, we identified subsets of variants associated in the expected (“concordant”) direction across all three phenotypes (i.e., greater risk for schizophrenia, lower cognitive ability, and lower educational attainment); these were contrasted with variants that demonstrated the counterintuitive (“discordant”) relationship between education and schizophrenia (i.e., greater risk for schizophrenia and higher educational attainment). ASSET analysis revealed 235 independent loci associated with cognitive ability, education, and/or schizophrenia at p < 5 × 10−8. Pleiotropic analysis successfully identified more than 100 loci that were not significant in the input GWASs. Many of these have been validated by larger, more recent single-phenotype GWASs. Leveraging the joint genetic correlations of cognitive ability, education, and schizophrenia, we were able to dissociate two distinct biological mechanisms—early neurodevelopmental pathways that characterize concordant allelic variation and adulthood synaptic pruning pathways—that were linked to the paradoxical positive genetic association between education and schizophrenia. Furthermore, genetic correlation analyses revealed that these mechanisms contribute not only to the etiopathogenesis of schizophrenia but also to the broader biological dimensions implicated in both general health outcomes and psychiatric illness.
Keywords: cognitive ability, educational attainment, schizophrenia, GWAS, pathways, genetic correlation, pleiotropy
Introduction
It has long been observed that impaired cognitive ability is a significant aspect of the illness in schizophrenia (MIM: 181500).1, 2, 3, 4, 5 Cognitive deficits have been shown to be largely independent of clinical state and treatment status in patients with schizophrenia1, 4, 6, 7, 8, 9 and are observed (in more subtle forms) in their first-degree relatives.10, 11 Moreover, cognitive deficits precede illness onset by many years; they begin in early childhood5, 12, 13, 14 and thus result in reduced educational attainment.15, 16
Unlike phenotypic correlation, which measures covariances within the distribution of the phenotypes, genetic correlation (rg) indexes the covariance between SNP ≈phenotype genome-wide association study (GWAS) effect sizes across a pair of phenotypes measured in separate GWAS studies. Recent advances in psychiatric and cognitive genomics have reliably demonstrated that the inverse relationship between cognitive ability and risk for schizophrenia is also observed at the molecular genetic level (rg ≈ −.20).17, 18, 19, 20, 21, 22, 23 Paradoxically, genetic correlation studies have indicated a positive relationship between educational attainment and risk for schizophrenia (rg ≈ .10),20, 23, 24, 25, 26, 27 despite the fact that educational attainment and cognitive ability exhibit a very strong polygenic overlap (rg ≈ .70).18, 23, 27 Educational attainment is often considered to be a proxy for cognitive ability; however, the lack of perfect genetic overlap between the two, combined with the paradoxical genetic correlation between educational attainment and schizophrenia, suggests an opportunity to decompose distinct genetic mechanisms accounting for this pattern of results.
Whereas genetic-correlation analysis has recently become widespread because of the availability of techniques such as linkage disequilibrium (LD) score regression (LDSC),25, 28 these approaches generally result in a single, genome-wide estimate of polygenic overlap. Moreover, novel meta-analytic approaches (e.g., multi-trait analysis of GWAS [MTAG]29) for merging seemingly heterogeneous GWAS datasets tend to exploit commonalities across phenotypes rather than differences; for example, two recent studies have employed MTAG across the highly correlated cognitive and educational GWASs in order to accelerate the process of gene discovery.18, 23 In contrast, few studies have attempted to examine the counter-intuitive correlation between schizophrenia and educational attainment or to parse subsets of SNPs that might drive cross-phenotype correlations. An initial effort has successfully identified a few individual loci that act in paradoxical fashion, increasing educational attainment while simultaneously increasing risk for schizophrenia;30 two other studies have identified loci that demonstrate other pleiotropic effects.19, 31
To date, however, no studies have utilized pleiotropic meta-analytic techniques to comprehensively parse variance from cognitive, educational, and schizophrenia-focused GWASs that might pinpoint differential biological mechanisms. In order for the paradoxical pattern of genome-wide correlations to exist, there must be identifiable subsets of SNPs that are differentially involved in driving these genetic relationships. Therefore, we sought to identify differentially associated variants, which could yield crucial insights into the fine-grained genetic architecture of schizophrenia and in turn give us insights into the etiopathogenic mechanisms underlying the illness—mechanisms that standard GWAS cannot detect.
In the study reported here, we first utilized a simple subsetting approach to identify SNPs that are significantly associated either with cognitive ability or with educational attainment, but not with both (Figure S1A). We hypothesized that these SNP subsets would demonstrate stronger genetic correlations with schizophrenia than what is observed with a simple genome-wide approach. We then employed a pleiotropic meta-analytic approach, association analysis based on subsets (ASSET),32 which permits the characterization of each SNP with respect to its pattern of effects on multiple phenotypes (Figure S1B). For example, ASSET has previously been used to demonstrate that the minor allele of rs2736100 (at the TERT [MIM: 187270] locus) is positively associated with risk for pancreatic cancer (MIM: 260350), negatively associated with risk for kidney (MIM: 144700) and lung cancers (MIM: 211980), and not significantly associated with risk for cancers of the breast (MIM: 114480), bladder (MIM: 109800), or prostate (MIM: 176207); other cancer loci were demonstrated to have various other patterns of effects.32 We utilized ASSET to identify two types of loci: (1) those SNPs that are consistently associated with all three phenotypes in the expected direction (i.e., the same allele is associated with higher cognitive ability, higher educational attainment, and lower risk for schizophrenia), which we label “concordant,” and (2) SNPs that demonstrate the paradoxical association between education and schizophrenia (i.e., the same allele associated with higher educational attainment and higher risk for schizophrenia), which we labeled “discordant.” Next, we compared the statistically significant ASSET results to the output of single-trait GWASs of cognitive ability, educational attainment, or schizophrenia20, 33, 34 in order to identify novel loci suggested by ASSET. Subsequently, we conducted a series of pathway and transcriptome-wide analyses to biologically characterize differential mechanisms underlying concordant versus discordant loci. Finally, we performed a series of genetic correlation analyses to compare the overlap of concordant and discordant SNP subsets with other relevant traits. Further analytic details are covered in the Material and Methods section; the full analysis workflow is also represented in Figures S1 and S2.
Material and Methods
Stage 1: Simple Subsetting Approach Based on p Values in Cognition and Education GWAS
Note that for most purposes in this manuscript, we are using the largest GWAS for schizophrenia,35 cognitive ability,18 and educational attainment36 published prior to 2018. For each of these phenotypes, larger GWASs have been published in 2018; these were used for validation and extension as described in subsequent sections. Before we performed wany subsetting analyses, we used genome-wide genetic correlations to confirm the earlier observed genetic correlations between schizophrenia and both cognitive ability and education. In stage 1, preliminary SNP subsets were formed simply based on p value thresholds of cognitive ability and educational attainment GWAS: (1) SNPs nominally associated with cognition (p < 0.05) and not associated with education (p > 0.05) were selected, resulting in 74,470 SNPs; (2) SNPs nominally associated with cognition (p < 0.05) and not associated with education using a stricter threshold (p > 0.5) were selected, resulting in 66,657 SNPs; (3) similar procedures were carried out for SNPs nominally associated with education (p < 0.05) but not with cognition (p > 0.05), were selected, resulting in 104,807 SNPs; and (4) SNPs nominally associated with education (p < 0.05) and not cognition using a stricter threshold (p > 0.5) were selected, resulting in 44,803 SNPs.
Next, we performed a heterogeneity test between results of cognitive and educational GWAS by using METAL37 and generated sets of SNPs showing opposite effects between the two (i.e., the same allele predicts better cognitive performance but less educational attainment, and vice versa). We identified sets of SNPs of varying sizes on the basis of varying p value thresholds for the heterogeneity tests (p < 0.5; p < 0.25; p < 0.1; p < 0.05; p < 0.01; and p < 0.001).
To evaluate the degree of genetic correlation of these preliminary subsets of SNPs with respect to schizophrenia, we utilized GNOVA,38 a recently published method similar to LD score regression. GNOVA is specifically designed to examine genetic correlations using SNP subsets (rather than global genome-wide summary statistics), whereas such applications have not been explicitly tested in LD score regression and may not be robust.
Stage 2: ASSET Meta-Analysis and ASSET-Generated SNP Subsets
Schizophrenia GWAS summary statistics based on a European ancestry GWAS of schizophrenia (n = 77,096, cases = 33,640, controls = 43,456; GWAS mean χ2 = 1.677) were obtained from the Psychiatric Genomics Consortium.35 To make them compatible with effect sizes (beta weights) derived from the linear-regression-based cognition and education GWASs, we converted odds ratios from the case-control schizophrenia GWAS to beta by taking the natural logarithm of the odds ratios. Effect direction per SNP was also reversed for schizophrenia so thatit would be consistent with the interpretation of cognition and education (i.e., concordant alleles are those where the direction of effect is the same for cognitive ability, educational attainment, and schizophrenia). Summary statistics for the education GWAS were obtained from the Social Science Genomics Association Consortium (SSGAC)36 (n = 328,917, GWAS mean χ2 = 1.638). GWAS summary statistics for cognition are available via earlier inverse-variance meta-analysis of samples18 from the COGENT27 consortium (n = 107,207, GWAS mean χ2 = 1.245). We applied general quality-control parameters to the schizophrenia and cognitive GWAS summary statistics but we excluded SNPs with INFO scores < 0.6 and minor-allele frequency < 0.01; multiple quality-control parameters’ thresholds were previously reported for the education GWAS,36 and summary statistics were provided by the SSGAC. Detailed quality-control and meta-analytic procedures were reported earlier.18 Only SNPs that were present for all three phenotypes were retained as inputs to the ASSET meta-analysis, resulting in 7,306,098 SNPs for subsequent analysis.
Pooling GWAS summary statistics via conventional inverse-variance meta-analysis increases power but also poses methodological challenges when different studies are capturing heterogeneous and/or pleiotropic phenotypes. In the case of pleiotropy, individual variants are likely to be associated with only a subset of the traits analyzed, or they might even demonstrate effects in different directions for the different phenotypes under analysis. ASSET meta-analysis32 is an agnostic approach that generalizes standard fixed-effects meta-analysis by allowing a subset of the input GWASs to have no effect on a given SNP, and it exhaustively searches across all possible subsets of “non-null” GWAS inputs within a fixed-effect framework to identify the strongest association signal in both positive and negative directions. ASSET then evaluates the significance of these positive and negative associations while accounting for multiple testing. This methodology allows for a powerful pooled two-tailed Z-score test statistic that effectively combines p values for variants with strong effects in opposite directions across input GWASs. ASSET also permits the addition of a covariance term for the adjustment of overlapping samples. The genetic correlation matrix between the three input GWASs had been added to all reported ASSET analyses so that this adjustment could be performed. Recently, comparisons between cross-phenotype meta-analysis methodologies demonstrated that ASSET performed best as the number of meta-analyzed traits with null effects increased, and ASSET also did well in terms of specificity and sensitivity of the results. In addition, the ASSET approach best controlled for potential Type 1 inflation due to sample overlap and for non-uniform distribution of effect sizes.39 As such, for the purpose of the current report, we selected ASSET for its conservative effect estimates and minimal inflation.
We combined GWAS summary statistics from schizophrenia, cognition, and education by using ASSET two-tailed meta-analysis (version 1.9.1) to obtain single cross-phenotype pleiotropic GWAS results. Default parameters were applied with the “h.traits” function. Inter-study correlations of the phenotype were first ascertained via LDSC,25, 28 which accounts for the genome-wide genetic correlation of the phenotypes and also for sample overlap. For each given SNP, ASSET generates Z scores of effect size and p values based on the strongest association from the input studies in positive and negative directions, respectively; then these p values are pooled into a single two-tailed p value for pleiotropy.32, 39 SNPs with similar relationships across the input traits (regardless of statistical significance) are then grouped into subsets identified by ASSET (see Figure 1B and Figure S1, bottom).
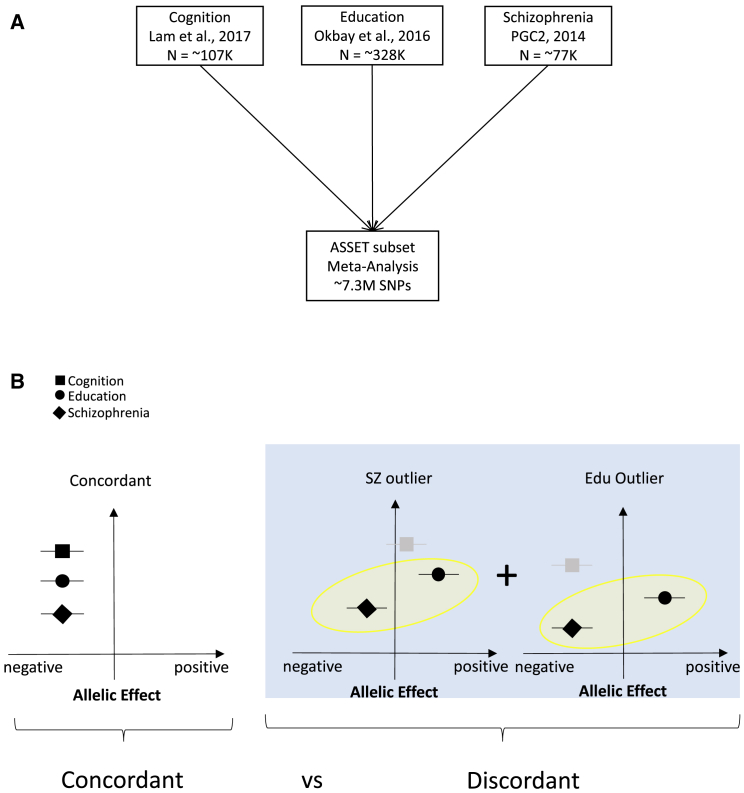
Figure 1.
Design of the Present Study
(A) Input GWAS studies used for ASSET analysis.
(B) Definition of concordant and discordant SNP subsets. Concordant SNPs have alleles that demonstrate negative effects on cognitive ability, educational attainment, and schizophrenia risk (i.e., increased schizophrenia risk, reverse-coded for consistency). Discordant SNPs have alleles that demonstrate paradoxical effects on educational attainment and schizophrenia (i.e., higher educational attainment and increased schizophrenia risk, reverse-coded). The Y axes on the forest plots represent different input summary statistics for cognition, education, and schizophrenia.
Again, as noted above, it is important to emphasize that the per-SNP direction of effect was reversed for schizophrenia so that it was consistent with the interpretation of cognition and education (i.e., higher scores are better, such that higher scores for schizophrenia are now coded as decreased risk for the disorder). Thus, in the notations to follow, ∩ represents variant subsets with the same effect directions, following the reversal of the direction of effect for the schizophrenia dataset, and | represents traits whose effect sizes are in the opposite direction of those for the other two traits (again following the reversal of the direction of effect for the schizophrenia dataset). ASSET subsets included: (1) scz ∩ edu ∩ cog (concordant, variants with an allele associated with an increase in cognitive ability and educational attainment, but a decrease in schizophrenia risk); (2) edu ∩ cog | scz (schizophrenia outliers, variants associated with an increase in cognitive ability and educational attainment but also with an increase in schizophrenia risk); (3) scz ∩ cog | edu (education outliers, variants associated with an increase in schizophrenia risk and reduced cognitive ability, but an increase in educational attainment); and (4) scz ∩ edu | cog (cognition outliers, variants associated with an increase in schizophrenia risk and reduced educational attainment, but an increase in cognitive ability) subsets. ASSET also identified SNPs where only a single trait (scz or edu or cog) was significant; these were included in a category called “single phenotype.”
Finally, to generate an appropriate contrast for the “concordant” subset, we included a combined single “discordant” subset, representing the counter-intuitive genetic correlation between education and schizophrenia, where discordant = (edu ∩ cog | scz) + (scz ∩ cog | edu) [subsets 2 and 3 above], regardless of the effect for cognition. These contrasts are also represented visually in Figure 1 and Figure S1.
Consolidation of Independent Loci
Independent genome-wide-significant loci for each ASSET meta-analysis subset were identified via SNP clumping procedures that are a part of the functional mapping and annotation (FUMA) pipeline.40 For the LD-rich MHC region, a single top SNP was retained. For all other loci, clumping procedures were carried out on the basis of the European 1000 Genomes Project phase 3 reference panel. First, “independent significant SNPs” were defined as those SNPs with a p value < 5 × 10−8 and with an LD of r2 < 0.6 and other, more significant SNPs at the same locus. Second, candidate SNPs were then identified for subsequent annotations and were defined as all SNPs that had an MAF of 0.01 and a maximum p value of 0.05 and that were in LD, with r2 ≥ 0.6, with at least one of the independent significant SNPs. To ensure that biological annotation of these loci would not be hampered by poor coverage at any locus, we included SNPs that came from the 1000 Genomes reference panel but that might not have been included in the ASSET data. Third, “lead SNPs” were defined as the independent significant SNPs that had the strongest signal at a given locus and were strictly independent from each other (r2 < 0.1). Finally, risk loci that were 250 kb or closer were merged into a single locus. The FUMA procedure was iterated across all ASSET SNP subsets, which were comprised (by definition) of non-overlapping SNPs. Additional variant annotations were conducted with ANNOVAR,41 and lookups with published GWASs were conducted with the GWAS catalog. Additional SNP lookups were performed with the input summary statistics (cognition, education, and schizophrenia GWASs18, 35, 36), recent MTAG analyses of intelligence,23 recent cognition or intelligence20, 21 GWASs, and pleiotropic analyses of cognition and schizophrenia19 as well as education and schizophrenia.31 RAggr43 was utilized for extracting SNPs within 250 kb and r2 > 0.6 from published reports to allow merging of loci generated from ASSET subsets.
MAGMA Gene-Based Analysis: Tissue Expression and Competitive Pathway Analysis
SNPs were mapped to 19,436 protein-coding genes via MAGMA as implemented in the FUMA40 pipeline. MAGMA44 gene analysis was performed with a default SNP-wide mean model that used the 1000 Genomes phase 3 reference panel and default gene annotations that are part of the FUMA pipeline. Genome-wide SNP p values and SNP-level sample sizes were included in the input files. MAGMA gene-tissue expression analysis was carried out with the Genotype-Tissue Expression (GTEx; version 745, 46, 47) resource to examine the relationship between gene expression in a specific tissue type and ASSET results. The gene-property test was performed for average expression (log2-transformed RPKM with pseudocount 1 after winsorization at 50) of 53 specific tissue types conditioning on average expression across all tissue types.
In order to identify specific biological processes linked to our sub-phenotypes of interest, we also used the results that emerged from the ASSET analysis to conduct MAGMA competitive pathway analysis. Gene sets that were tested included drug-related pathways,48, 49 as well as custom-curated neurodevelopmental and other brain-related gene sets that had gone through stringent quality control in a study originally designed to interrogate rare variants in schizophrenia.50 In the latter, pathways with more than 100 genes from Gene Ontology (release 146; June 22, 2015 release), KEGG (July 1, 2011 release), PANTHER (May 18, 2015 release), REACTOME (March 23, 2015 release), DECIPHER Developmental Disorder Genotype-Phenotype (DDG2P) database (April 13, 2015 release), and the Molecular Signatures Database (MSigDB) hallmark processes (version 4, March 26, 2015 release) were initially included. Brain-level tissue expression gene sets included the Brainspan RNA-seq dataset51 and the GTEx v7 dataset.45 MAGMA gene-based and gene-set analysis were conducted with MAGMA v1.6.44 Additional gene sets were selected on the basis of risk for schizophrenia and neurodevelopmental disorders, including those reported for schizophrenia rare variants52 (translational targets of FMRP [MIM: 309550]; components of the post-synaptic density; ion channel proteins; components of the ARC, mGluR5, and NMDAR complexes; proteins at cortical inhibitory synapses; targets of mir-137; and genes near schizophrenia common risk loci) and autism (MIM: 613436) risk (targets of CHD8 [MIM: 610528], splice targets of RBFOX [MIM: 605104], hippocampal gene expression networks, and neuronal gene lists from the Genes2Cognition database, as well as loss-of-function [LoF]-intolerant genes [pLI > 0.9 from the ExAC v0.3.1 pLI metric], ASD risk genes for FDR < 10% and 30%, and ASD or developmental disorder de novo genes hit by an LoF and/or missense de novo variant). Further details of curation of these gene sets was previously reported.50
S-PrediXcan: Brain-Tissue Expression Profiles and Hypergeometric Gene-Set Enrichment Analysis
Genetically regulated gene expression was imputed for the ASSET summary statistics with tissue models from GTEx v7 and the CommonMind Consortium via S-PrediXcan (formerly MetaXcan).53, 54, 55 GTEx v7 tissue included amygdala (n = 88), anterior cingulate cortex (n = 109), basal ganglia (n = 144), cerebellar hemisphere (n = 125), cerebellum, cortex (n = 136), frontal cortex (n = 118), hippocampus (n = 111), hypothalamus (n = 108), nucleus accumbens (n = 130), putamen (n = 111), spinal cervical-1 (n = 83), and substantia nigra (n = 80). The CommonMind consortium data consist of tissue expression data derived from the dorsolateral prefrontal cortex (DLPFC, n = 279).56 GTEX v7 Tissue expression models were trained using elastic net models that were made publicly available (see Web Resources). Elastic net models for DLPFC were contributed by collaborators from the CommonMind Consortium.56, 57, 58 Bonferroni correction was first conducted for each ASSET subset of genes. Genes that survived multiple testing corrections were entered into GENE2FUNC, which is part of the FUMA40 pipeline, to be tested for over-representation. This analysis differs from the MAGMA gene-set analysis in that the MAGMA gene-set analysis is used to examine whether gene sets, united by a known biological theme, are enriched for the phenotype under investigation. In a test of over-representation, as conducted with GENE2FUNC, the shared function of the genes of interest is unknown, and its elucidation is the goal of a test of over-representation. The over-representation test conducted with GENE2FUNC queries gene sets from (1) the Molecular Signature Database (MsigDB v 5.2), (2) WikiPathways (curated version 20161010), and (3) GWAS catalog (reported genes ver e91 20180206); to avoid spurious results, we required a minimum of three genes per pathway. For each gene set, hypergeometric tests were conducted so that the list of genes significant in each ASSET subset could be examined for overlap with gene sets within the databases stipulated above; Bonferroni correction for multiple testing was applied. To reduce the likelihood that hypergeometric pathway analysis would be influenced by the dense number of genes within the MHC region, we removed genes within the coordinates of 28,000,000–35,000,00059 on chromosome 6.
Genetic Correlations
To examine how our ASSET concordant and discordant SNP subsets relate to other phenotypes with available GWAS data, we conducted genetic correlation tests by using GNOVA,38 an approach similar to LD score regression but capable of working with SNP subsets. Notably, GNOVA provides a corrected rg that has been demonstrated to be robust to sample overlaps, and GNOVA is able to account for LD across SNPs.38 We selected a series of neuropsychiatric, inflammatory, brain, metabolic, and cardiovascular phenotypes that have been previously demonstrated to have genetic correlations with cognitive measures and used them to interrogate the genetic overlaps of our ASSET subsets. Interpretation of GNOVA for the concordant subset was straightforward because the three input GWAS weights all follow the same direction (following the reverse coding of schizophrenia, as noted previously). In contrast, discordant SNPs have two separate potential weights (allelic β for schizophrenia versus allelic β for education); as shown in Figure 1B, a given SNP might have somewhat different effect sizes (distances from the center line) for education as compared to schizophrenia. Therefore, we weighted each SNP by the stronger value of β: variants for which the schizophrenia β was stronger than the education β were referred to as “schizophrenia type” and variants with the opposite pattern were referred to as “education type.” Nevertheless, it is important to emphasize that the discordant SNPs represent a single dimension of biology, and the net effects of all “schizophrenia type” variants were equivalent to those of the “education type” SNPs, albeit with opposite signs.
Results
Stage 1: Preliminary Evaluation of Genetic Correlations
We used GWAS summary statistics for cognition (n = 107,207)18 and education (n = 328,917)36 to evaluate preliminary genetic correlations with schizophrenia (n = 77, 096).35 Consistent with previous results, the inverse genetic correlation that GNOVA revealed between cognition and schizophrenia was significant (rg = −.21, se = 0.03, p = 1.12 × 10−12), as was the counter-intuitive positive correlation between education and schizophrenia (rg = 0.08, se = 0.02, p = 2.05 × 10−5). Note that these analyses were conducted before we reversed the direction of effect for schizophrenia.
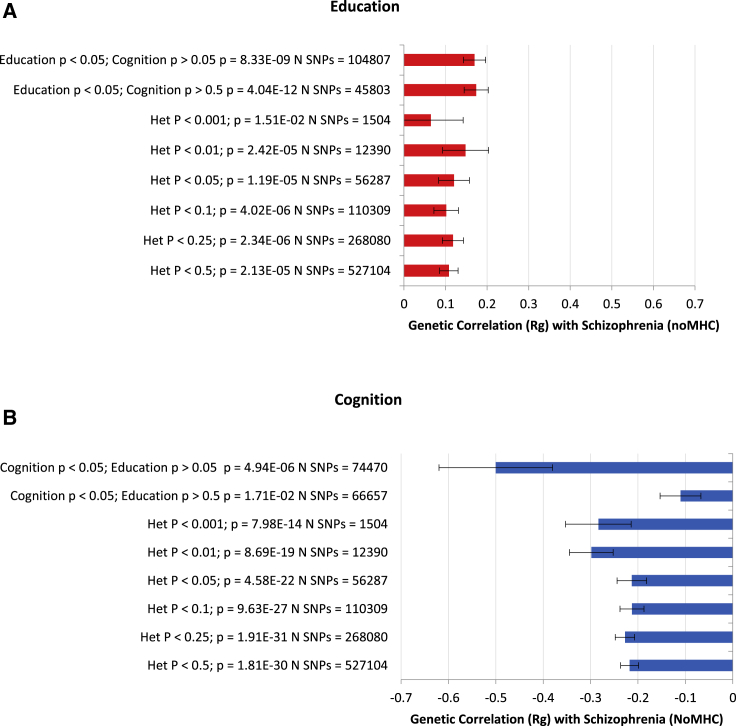
Prior to the main ASSET analysis, we used two simple approaches to examine subsets of SNPs and their association with schizophrenia (Figure S1). First, we selected SNPs that were nominally associated with education (p < 0.05) and generally not associated with cognition (p > 0.05); GNOVA revealed a slightly stronger positive correlation, rg of 0.17, between this subset of educational attainment SNPs and schizophrenia than did genome-wide summary statistics (Figure 2A). With a stricter threshold for SNPs not associated with cognition (p > 0.50), these “non-cognitive” educational attainment SNPs also attained an rg of 0.17 with schizophrenia. GNOVA analyses were repeated for SNPs nominally associated with cognition (p < 0.05), but generally not associated with education (p > 0.05), and the analyses were repeated again with the stricter threshold for education (p > 0.50). Values for rg of −.50 and −.11 were obtained between schizophrenia and these cognition subsets (Figure 2A).
Figure 2.
Genetic Correlations with Schizophrenia for SNPs Demonstrating Heterogeneity of Effects between (A) Cognitive Ability and (B) Educational Attainment
(A) Cognitive ability.
(B) Educational attainment.
Genetic correlation was carried out with GNOVA. Error bars represent standard errors. Education = Okbay PMID 27225129; cognition = Lam PMID 29186694; schizophrenia (Ripke) = PGC Schizophrenia Working Group PMID 25056061.
The second approach involved calculating the heterogeneity p values for cognition and education and identifying SNPs that have discrepant direction of effects between cognition and education. These SNPs were then binned, ranging from low probability (p < 0.5) to high probability (p < 0.001) for heterogeneous effect sizes between cognition and education (Figure 2B). GNOVA indicated that the greater the discrepancy in effect direction between SNP effects for cognition and education, the stronger the association between cognition and schizophrenia. However, this pattern was not observed for education and schizophrenia.
Stage 2: ASSET Meta-Analysis and SNP Subsets
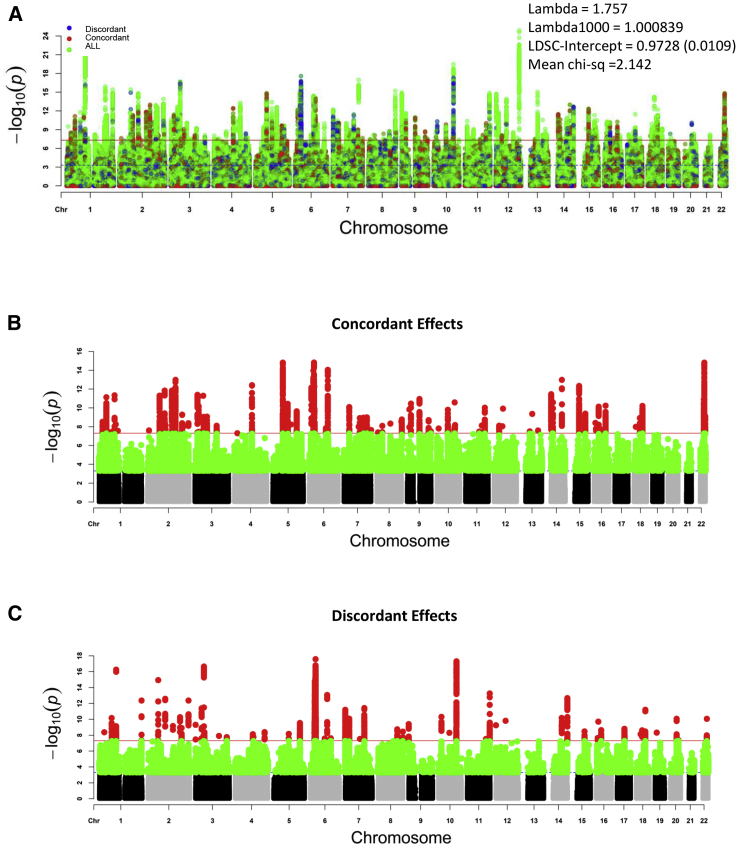
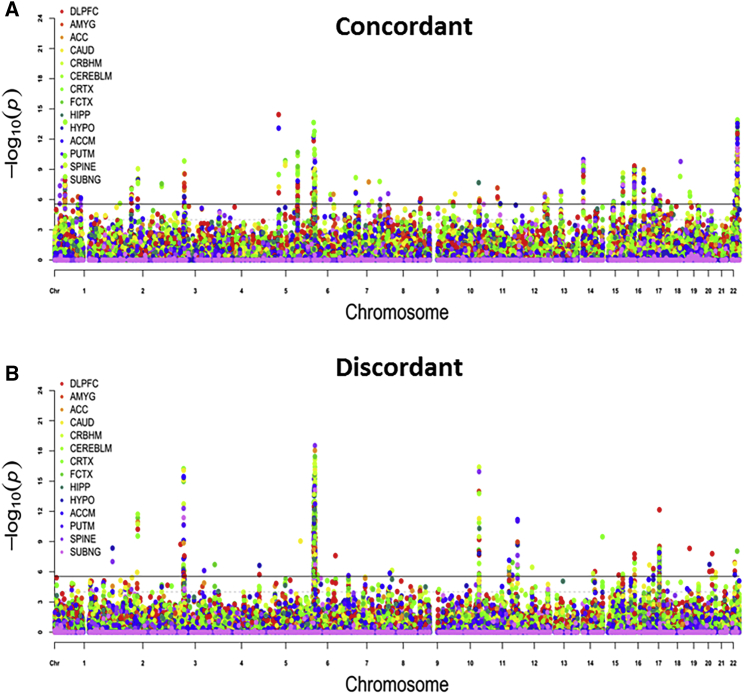
Genome-wide cross-phenotype ASSET meta-analysis across 7,306,098 SNPs revealed 300 lead SNPs (across 236 independent loci) that met the genome-wide significance threshold of p < 5 × 10−8 for the ASSET two-tailed test (see Figure 3A and Tables S1 and S2). There were 1,381,020 SNPs that demonstrated consistent direction of effects between cognition, education, and schizophrenia (i.e., lower cognitive ability, lower educational attainment, and increased risk for schizophrenia); these were assigned to the “concordant” subset, which contained 89 genome-wide-significant loci harboring 103 independent significant SNPs. By contrast, the “discordant” subset, which consisted of SNPs with counter-intuitive allelic effects for schizophrenia vis-à-vis education, encompassed 1,891,743 SNPs, with 65 genome-wide-significant loci comprising 77 independent significant SNPs (Figures 3B and 3C and Table S1). Significant loci for other ASSET subsets are also detailed in Table S2. Other supplemental tables show FUMA-derived annotations for potential functional consequences, including CADD scores (Table S3), eQTL lookups (Table S4), and prior GWAS lookups (Table S5).
Figure 3.
Manhattan Plots for ASSET Results
ASSET meta-analysis outputs:
(A) All subsets.
(B) Concordant subset.
(C) Discordant subset.
Consolidation of Independent Loci
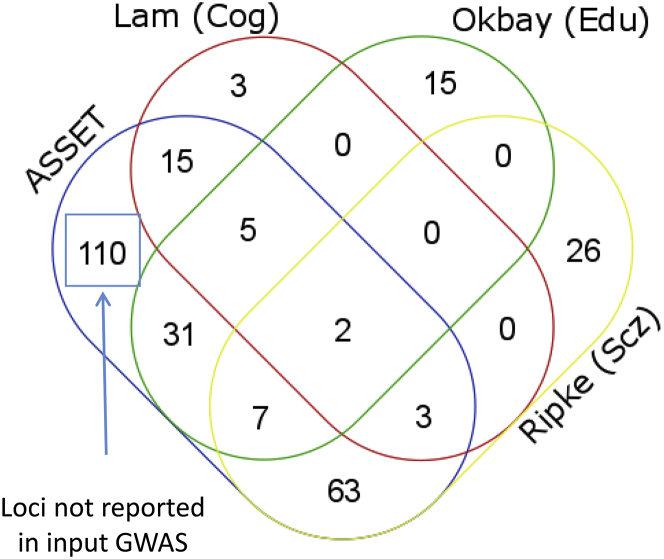
Next, we wanted to identify which loci from our ASSET results were not previously identified with respect to the three-input GWAS. Using RAggr,43 we extracted SNPs with r2 > 0.6 within a window of 250 kb of lead SNPs in reported GWASs, i.e., we extracted 101 loci from the European-ancestry cohorts of the Psychiatric Genomics Consortium GWAS of schizophrenia,35 74 loci from the SSGAC educational attainment GWAS,36 and 40 loci from the COGENT GWAS of cognitive ability.18 These were merged with the 236 loci from ASSET. As earlier described, independent loci within 250 kb were merged, resulting in 280 independent loci being identified across ASSET and the input GWAS. As shown in the resulting Venn diagram (Figure 4), 110 loci not reported earlier were identified by the ASSET meta-analysis. In contrast, 126 loci overlapped with either education or schizophrenia, whereas 44 loci were only significant in the input GWAS and not in ASSET.
Figure 4.
Venn Diagram Comparing Significant ASSET Loci to Significant Loci from Input GWASs
Education = Okbay PMID 27225129; Cognition = Lam PMID 29186694; Schizophrenia (Ripke) = PGC Schizophrenia Working Group PMID 25056061; ASSET = results from ASSET meta-analysis.
Very recently, new GWASs have been published for schizophrenia, cognitive ability, and educational attainment, and these studies are larger than the input GWAS used for our ASSET analysis.20, 33, 34 This permitted us to perform a lookup of our 110 “novel” ASSET SNPs, thus providing an opportunity to validate ASSET as a tool for locus discovery (Table S6). We also performed lookup in a paper that utilized MTAG to examine intelligence20 and several recent papers that applied pleiotropic approaches to these phenotypes.19, 23, 31 We found that 75% of the loci were in fact reported as significant in the later GWASs with larger sample sizes, and 28 of the 110 loci were independent from other single-phenotype GWAS reports. These loci are reported in Table 1. Notably, three of these loci (indexed by rs207338, rs708212, and rs11617058) were identified in secondary analyses (using MTAG) performed in the study of educational attainment,34 and one locus (rs67652508) has been reported as being genome-wide significant for association to putamen volumes as measured on magnetic resonance imaging (MRI) scans of the brain.60 Further ANNOVAR41 annotations are available for novel loci (Table S7).
Table 1.
Loci Identified by ASSET
| Lead SNPs | Chr | Base Position Start (bp) | Base Position End (bp) | GWAS p Value | Beta (β) | SE | Nearest Gene | SNP Function |
|---|---|---|---|---|---|---|---|---|
| rs13010104 | 2 | 208331258 | 208369715 | 9.30 × 10-9 | −0.0148 | 0.0026 | ENSG00000223725 | ncRNA_intronic |
| rs207338 | 4 | 19053350 | 19070123 | 4.99 × 10-8 | −0.0108 | 0.0020 | ENSG00000248238 | intergenic |
| rs6844280 | 4 | 31371970 | 31387144 | 3.53 × 10-8 | 0.0113 | 0.0021 | ENSG00000251434 | intergenic |
| rs71615297 | 4 | 179013479 | 179210420 | 2.17 × 10-8 | −0.0115 | 0.0021 | RNU1-45P | intergenic |
| rs73260443 | 5 | 113404038 | 113464825 | 6.30 × 10-9 | −0.0147 | 0.0025 | ENSG00000251628 | intergenic |
| rs9372208 | 6 | 109507533 | 109648130 | 2.96 × 10-9 | −0.0120 | 0.0020 | ENSG00000233908 | intergenic |
| rs9371912 | 6 | 155907483 | 156009420 | 1.06 × 10-10 | −0.0168 | 0.0026 | RNU7-152P | intergenic |
| rs1870571 | 8 | 80129136 | 80296429 | 4.69 × 10-9 | 0.0124 | 0.0021 | ENSG00000253659 | intergenic |
| rs11786685 | 8 | 81220185 | 81316928 | 2.79 × 10-8 | −0.0116 | 0.0021 | ENSG00000253237 | intergenic |
| rs3890699 | 8 | 110164392 | 110344596 | 7.31 × 10-9 | 0.0122 | 0.0021 | NUDCD1 | intergenic |
| rs11166628 | 8 | 137022220 | 137091947 | 3.27 × 10-8 | 0.0113 | 0.0020 | ENSG00000253248 | ncRNA_intronic |
| rs10993909 | 9 | 136924744 | 136942560 | 3.13 × 10-8 | 0.0116 | 0.0021 | BRD3 | intronic |
| rs10994707 | 10 | 62919886 | 63096664 | 1.14 × 10-9 | 0.0202 | 0.0033 | TMEM26 | intergenic |
| rs72945305 | 11 | 81164016 | 81210528 | 3.34 × 10-8 | −0.0128 | 0.0023 | ENSG00000254747 | intergenic |
| rs556587 | 11 | 92317365 | 92554716 | 1.57 × 10-8 | −0.0163 | 0.0029 | FAT3 | intronic |
| rs75261250 | 11 | 124276497 | 124303201 | 2.16 × 10-8 | −0.0188 | 0.0034 | OR8X1P | intergenic |
| rs708212 | 12 | 31517960 | 31769501 | 7.79 × 10-9 | 0.0133 | 0.0023 | DENND5B | intronic |
| rs3741434 | 12 | 53605344 | 53605344 | 1.20 ×10-10 | 0.0191 | 0.0030 | RARG | UTR3 |
| rs7321596 | 13 | 44390857 | 44514022 | 3.16 × 10-8 | −0.0109 | 0.0020 | LACC1 | intergenic |
| rs11617058 | 13 | 85176690 | 85305386 | 2.95 × 10-11 | 0.0183 | 0.0028 | LINC00333 | intergenic |
| rs67652508 | 14 | 55487496 | 55567991 | 2.88 × 10-8 | −0.0140 | 0.0025 | MAPK1IP1L | intergenic |
| rs11130 | 16 | 15687755 | 15837246 | 4.24 × 10-8 | 0.0110 | 0.0020 | NDE1 | UTR3 |
| rs7214058 | 17 | 9968014 | 9995284 | 6.32 × 10-9 | 0.0118 | 0.0020 | GAS7 | intronic |
| rs28584904 | 17 | 68984046 | 69007006 | 2.36 × 10-8 | 0.0144 | 0.0026 | ENSG00000271101 | intergenic |
| rs56791590 | 18 | 26259012 | 26496051 | 7.59 × 10-9 | 0.0117 | 0.0020 | ENSG00000265994 | intergenic |
| rs12462428 | 19 | 16665215 | 16738369 | 2.20 × 10-8 | 0.0143 | 0.0026 | ENSG00000141979:MED26:CTC-429P9.4 | intronic:intronic:intronic |
| rs5767976 | 22 | 48133458 | 48183889 | 8.14 × 10-10 | −0.0125 | 0.0020 | RP11-191L9.4 | ncRNA_intronic |
| rs68178377 | 22 | 50742346 | 50771464 | 4.28 × 10-8 | 0.0122 | 0.0022 | DENND6B | exonic |
Abbreviations are as follows: Chr = chromosome; SE = standard error
MAGMA Gene-Based Analysis: Tissue-Expression and Competitive-Pathway Analysis
MAGMA gene-based analysis was conducted on all ASSET subsets. 772 genes survived Bonferroni correction in the overall ASSET analysis, with 306 genes in the concordant subset and 304 genes within the discordant subset (Table S8). MAGMA gene property analysis revealed significant association (p < 0.000926, Bonferroni-corrected) of gene expression of ASSET SNP subsets across GTExv7 brain tissues (Figure S3 and Table S9). There were no significant differences between concordant and discordant result subsets; both subsets were significantly enriched (positive beta weights) across all brain compartments.
Because of the significant enrichment in brain tissues, we next performed MAGMA competitive-pathway analyses by using neurodevelopmental and other brain-related gene sets as curated in a recent publication;50 full results are reported in Table S10. Although there was considerable overlap of pathway enrichment across ASSET categories, several gene sets were uniquely associated with either the concordant or the discordant result subsets (Table 2). Specifically, the CHD8 pathway, reflecting genes involved in early neurodevelopment, was uniquely associated with the concordant subset (p = 7.11 × 10−6). In contrast, a number of synaptic pathways (e.g., ion-channel and synaptic-density pathways) and constrained gene sets appeared to be uniquely associated with the discordant subset. Only two gene sets were enriched in both the concordant and the discordant results, and these were rather generic: brain-enriched genes and schizophrenia GWAS results (Table S10). It is notable that when the MHC region was removed from the pathway analysis, the overall pattern of results remained (see Table S10).
Table 2.
MAGMA Significant Gene Sets for Concordant and Discordant SNP Subsets
|
Concordant | ||||||
|---|---|---|---|---|---|---|
| MAGMA Gene Sets | NGENES | p | Pbon | BETA | BETA_STD | SE |
| Cotney2015:hNSC_Chd8_prom∗ | 8,186 | 4.05 × 10-9 | 7.11×10-6 | 0.115 | 0.0571 | 0.0199 |
| Cotney2015:hNSC+human+mouse_Chd8_prom | 1,902 | 2.81 × 10-5 | 0.049458 | 0.123 | 0.0378 | 0.0304 |
| Sugathan2014:Chd8_binding | 5,314 | 1.69 × 10-5 | 0.029744 | 0.0874 | 0.04 | 0.0211 |
| Discordant | ||||||
| Trapnell2012: constrained_genes_0_10∗ | 1,705 | 3.41 × 10-7 | 0.0006 | 0.131 | 0.0383 | 0.0263 |
| Lek2016: constrained_genes_pLI_90∗ | 2,972 | 3.33 × 10-7 | 0.000585 | 0.104 | 0.0387 | 0.021 |
| Darnell2011:fmrp_targets∗ | 765 | 1.41 × 10-5 | 0.024693 | 0.162 | 0.0327 | 0.0387 |
| ddg2p:dominant_mis_all_brain | 137 | 4.42 × 10-6 | 0.007768 | 0.411 | 0.0357 | 0.0925 |
| g2cdb:bayes_collins_mouse_psd_consensus∗ | 918 | 8.89 × 10-6 | 0.015615 | 0.152 | 0.0333 | 0.0353 |
| g2cdb:bayes_collins_mouse_psd_full∗ | 1,442 | 1.26 × 10-6 | 0.002214 | 0.134 | 0.0363 | 0.0284 |
| g2cdb:human_psd∗ | 1,001 | 1.40 × 10-6 | 0.002461 | 0.159 | 0.0363 | 0.0339 |
| g2cdb:human_psp∗ | 1,039 | 2.04 × 10-6 | 0.003579 | 0.154 | 0.0358 | 0.0334 |
| GOBP:membrane_depolarization | 105 | 2.17 × 10-5 | 0.038129 | 0.439 | 0.0334 | 0.107 |
| GOBP:synaptic_transmission | 549 | 6.58 × 10-6 | 0.011554 | 0.205 | 0.0353 | 0.0471 |
| GOMF:voltage-gated_cation_channel_activity∗ | 144 | 6.43 × 10-7 | 0.00113 | 0.45 | 0.0401 | 0.093 |
| Sanders2015:asd_fdr10∗ | 64 | 4.67 × 10-6 | 0.008207 | 0.622 | 0.037 | 0.14 |
| Sanders2015:asd_lof_genes∗ | 559 | 3.52 × 10-6 | 0.006178 | 0.203 | 0.0352 | 0.0451 |
Abbreviations are as follows: NGENES = number of genes in pathway; Pbon = Bonferroni-corrected p value; BETA_STD = standardized beta; GOBP = gene ontology biological process; GOMF = gene ontology biological molecular function; Cotney = PMID 25752243; hNSC = human neural stem cells; prom = promotor; Sugathan = PMID 25294932; Trapnell = PMID 22383036; Lek = PMID 27535533; Darnell = PMID 21784246; ddg2p = DECIPHER developmental disorder genotype-phenotype; dominant_mis_all_brain = dominant mode of effect/loss-of-function missense/brain affected; g2cdb = Genes2cognition database; psd = post-synaptic density; psp = post-synaptic proteomes; Sanders = 26402605; asd_fdr_10 = autism risk genes, FDR < 0.10; and asd_lof_genes = ASD loss-of-function genes.
Results remaining significant after removal of MHC region variants.
To see whether the distinction between the concordant and discordant subsets harbors potential implications for drug targeting (for schizophrenia and/or cognitive enhancement), we performed drug-based and drug family competitive gene set analyses on our MAGMA results. These analyses revealed that the class of drugs associated with voltage-gated calcium channel genes was over-represented among in the results from the discordant subset (Bonferroni-corrected p = 0.02), as was Abacavir (nucleoside reverse transcriptase inhibitor; Bonferroni-corrected p = 0.00018). Although both of these sets showed similar direction of effects with respect to the concordant subset, no drug-related gene sets attained Bonferroni-corrected significance in the results from the concordant subset (Table S11).
S-Predixcan: Brain Tissue Expression Profiles and Gene-Set Enrichment Analysis
Transcriptome-wide association analysis (TWAS) was carried out via S-Predixcan to identify top expressed genes within GTEXv746 and CommonMind Consortium54, 56, 57, 58 brain tissue models (Figure 5 and Table S12). The top brain-expressed genes unique to the discordant subsets were CYP21A1P (MIM: 613815), CFB (MIM: 138470), and C4A (MIM: 120810), along with 177 additional genes that were significantly expressed in the discordant, but not the concordant, subsets. On the other hand, ELOVL7 (MIM: 614451), NAGA (MIM: 609241), and 201 other genes were uniquely associated with the concordant subset. Significant genes identified by S-Predixcan were subjected to GENE2FUNC hypergeometric gene-set analysis (excluding MHC genes, which were over-represented due to significant LD; see Material and Methods for more details). The goal of this analysis was to examine whether the genes identified in the TWAS overlapped with those found in known biological systems. As shown in Table 3, the results of the TWAS consistently identified genes found in cell adhesion and membrane protein gene sets for the concordant subset. In contrast, synaptic (specifically dendritic) pathways, as well as chromosomal repair pathways, were consistently identified by the TWAS during examination of the discordant subset.
Figure 5.
S-PrediXcan Transcriptome-Wide Association Results
(A) Concordant and (B) discordant subsets.
Table 3.
GENE2FUNC Pathway Analysis of GO Genesets with MHC Filtered
| Category | GeneSet | N_genes | N_overlap | p | Pbon |
|---|---|---|---|---|---|
| Concordant | |||||
| GO_bp | GO_CELL_CELL_ADHESION_VIA_PLASMA_MEMBRANE_ADHESION_MOLECULES | 202 | 10 | 6.89 × 10-9 | 3.1 × 10-5 |
| GO_bp | GO_RESPONSE_TO_XENOBIOTIC_STIMULUS | 105 | 7 | 6.22 × 10-8 | 2.8 × 10-4 |
| GO_bp | GO_HOMOPHILIC_CELL_ADHESION_VIA_PLASMA_MEMBRANE_ADHESION_MOLECULES | 151 | 8 | 7.74 × 10-8 | 3.4 × 10-4 |
| GO_bp | GO_CELL_CELL_ADHESION | 604 | 14 | 4.46 × 10-7 | 2.0 × 10-3 |
| GO_mf | GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H_QUINONE_OR_SIMILAR_COMPOUND_AS_ACCEPTOR | 52 | 4 | 6.39 × 10-6 | 5.8 × 10-3 |
| GO_cc | GO_MEMBRANE_MICRODOMAIN | 286 | 8 | 1.53 × 10-5 | 8.9 × 10-3 |
| GO_cc | GO_MEMBRANE_PROTEIN_COMPLEX | 1018 | 16 | 1.58 × 10-5 | 9.2 × 10-3 |
| GO_bp | GO_PURINE_RIBONUCLEOSIDE_BISPHOSPHATE_METABOLIC_PROCESS | 20 | 3 | 2.77 × 10-6 | 1.2 × 10-2 |
| GO_cc | GO_MITOCHONDRION | 1633 | 21 | 2.47 × 10-5 | 1.4 × 10-2 |
| GO_bp | GO_BIOLOGICAL_ADHESION | 1027 | 17 | 4.56 × 10-6 | 2.0 × 10-2 |
| GO_bp | GO_MACROMOLECULAR_COMPLEX_ASSEMBLY | 1388 | 20 | 6.87 × 10-6 | 3.0 × 10-2 |
| GO_bp | GO_MITOCHONDRIAL_RESPIRATORY_CHAIN_COMPLEX_I_BIOGENESIS | 56 | 4 | 9.25 × 10-6 | 4.1 × 10-2 |
| Discordant | |||||
| GO_cc | GO_DENDRITIC_SHAFT | 37 | 3 | 1.28 × 10-5 | 7.4 × 10-3 |
| GO_bp | GO_DOUBLE_STRAND_BREAK_REPAIR | 165 | 6 | 3.89 × 10-6 | 1.7 × 10-2 |
| GO_cc | GO_NUCLEAR_CHROMOSOME | 522 | 9 | 3.84 × 10-5 | 2.2 × 10-2 |
| GO_mf | GO_PROTEIN_DOMAIN_SPECIFIC_BINDING | 620 | 10 | 3.14 × 10-5 | 2.8 × 10-2 |
| GO_bp | GO_NON_RECOMBINATIONAL_REPAIR | 70 | 4 | 7.97 × 10-6 | 3.5 × 10-2 |
| GO_cc | GO_DENDRITE | 451 | 8 | 6.94 × 10-5 | 4.0 × 10-2 |
Note: N_gene = Genes identified in pathway; N_overlap = input genes overlapping with pathway; Pbon = Bonferoni-adjusted p value
Genetic Correlations
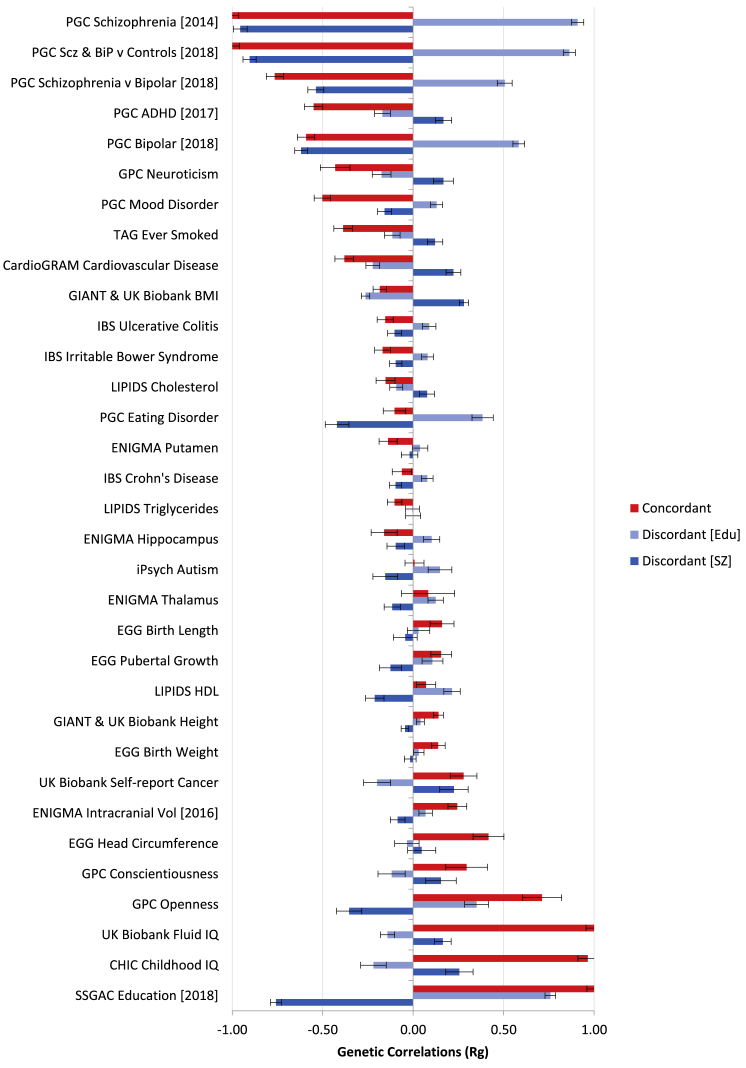
A series of psychiatric, personality, structural-brain-imaging, metabolic, cardiovascular, and anthropometric traits were selected for GNOVA modeling with the ASSET subset results (see Figure 6 and Table S13); multiple testing was adjusted on the basis of the false discovery rate (FDR).The concordant subset demonstrated significant (FDR < .05) genetic correlations, in the expected direction, with many forms of psychopathology in addition to schizophrenia (such forms included ADHD [MIM: 143465], bipolar disorder [MIM: 125480], and major depressive disorder [MDD, MIM: 608516], as well as neuroticism and smoking). This subset also demonstrated a significant (FDR < .05) positive genetic correlation (i.e., better cognition/higher education/lower risk for schizophrenia) with larger volumes of several brain regions (including total intracranial volume) as measured by structural MRI, as well as several measures of infant size and adult height. Significant positive associations were also seen with the personality dimensions of openness and conscientiousness, and (surprisingly) with self-reported cancer; significant negative associations were seen for total cholesterol and triglycerides, as well as the presence of ulcerative colitis and inflammatory bowel disease. Additionally, a negative genetic correlation was observed for the concordant subset with BMI and measures of cardiovascular disease (i.e., lower cognition/lower education/greater risk for schizophrenia associated with greater BMI and risk for cardiovascular disease).
Figure 6.
Genetic Correlations for Concordant and Discordant Subsets with Other Relevant Phenotypes
Genetic correlation analysis was carried out with GNOVA. Error bars represent standard errors. Summary statistics of selected phenotypes were downloaded from the LD Hub and Psychiatric Genomics Consortium websites. See Web Resources for further detail.
The discordant subset was strongly associated with schizophrenia and education, by definition, in a manner demonstrating the paradoxical relationship (higher education with greater risk for schizophrenia, Figure 6). (It is important to note that the light blue bars and dark blue bars in Figure 6 are essentially mirror images of each other and are therefore providing somewhat redundant information; both sets of bars are included to indicate the both sides of this dimension). Interestingly, a similar pattern was observed for bipolar disorder (higher education/greater risk for schizophrenia—greater risk for bipolar disorder). Similar relationships were also observed, at a nominally significant level, for autism spectrum disorder and eating disorders (MIM: 606788), which were not associated with the concordant subset, as well as for MDD. The reverse relationship, however, was observed with ADHD (i.e., higher education/greater risk for schizophrenia—lower risk for ADHD). This pattern was also observed for the smoking, BMI, and cardiovascular disease phenotypes. A counter-intuitive pattern was observed for the relationship between the discordant subset and neuroticism, which was the opposite of that observed for MDD (despite the fact that MDD and neuroticism are themselves highly correlated).
Discussion
A consistent finding in recent schizophrenia, cognition, and education GWASs has been the involvement of both neurodevelopmental pathways and synaptic processes;19, 20, 39, 61, 62, 92 the present study aimed to at least partially disentangle these mechanisms. In this study, we leveraged the genetic pleiotropy underlying three partially overlapping, complex phenotypes in order to identify homogeneous subsets of SNPs with distinct characteristics. Specifically, we were able to parse a subset of SNPs with alleles that were associated in the expected fashion across our three phenotypes of interest: lower cognitive ability, lower educational attainment, and greater risk for schizophrenia. These “concordant” SNPs were characterized by their association with genes and pathways relevant to early neurodevelopmental processes. By contrast, SNPs that demonstrated a counterintuitive, discordant pattern of association (higher educational attainment yet greater risk for schizophrenia) were primarily associated with genes and pathways involved in synaptic function of mature neurons.
This distinction was robustly observed across several methods of functional annotation. First, MAGMA competitive gene-set analysis revealed a significant enrichment of CHD8-related genes in the concordant subset (Table 2). CHD8, encoding a chromatin remodeling protein, is a gene that has been robustly associated with autism62, 63, 64, 65 but that to date has only limited or anecdotal evidence for association to schizophrenia.66, 67 Disruption of the homologous gene (Chd8) in animal models has demonstrated that the resulting protein plays a key role in very early neurodevelopmental processes, including neuronal proliferation and differentiation68, 69 as well as cell adhesion and axon guidance.70 On the other hand, MAGMA competitive gene-set analysis revealed a significant enrichment of discordant genes for functions including synaptic transmission and postsynaptic density, as well as membrane depolarization and voltage-gated cation channel activity. Although these processes have been commonly associated with both schizophrenia33, 35 and cognitive phenotypes,20, 21, 24, 71, 72, 73, 74 our study is the first to demonstrate that these synaptic mechanisms operate in a surprising manner: the same synaptic functions that increase risk for schizophrenia also serve to enhance educational attainment.
The linkage of early neurodevelopmental processes to SNPs associated with impaired cognition and increased risk for schizophrenia is consistent with a large body of literature demonstrating that cognitive deficits are often observed early on in the lifespans of these individuals.5, 13, 14, 75 At the same time, the discordant variant subset implicates more mature neuronal regulation, and synaptic pruning mechanisms that are most prominent later in childhood, adolescence, and into adulthood, ostensibly as part of a neuroplasticity mechanism for making more “efficient” connections within the brain.77 However, the dysregulation of such mechanisms has been shown to be intricately linked to schizophrenia psychopathology.78 It is important to note that these results are obtained from separate GWASs of two different phenotypes and do not represent a subset of highly educated individuals with schizophrenia. Rather, it is plausible to posit an inverted U relationship such that efficient synaptic pruning processes are essential mechanisms underlying academic performance but might be carried too far in disorders such as schizophrenia.
Additionally, transcriptome-wide analysis using S-Predixcan pointed toward the same distinction between concordant and discordant genes and pathways. Two of the strongest genes with differential expression in the concordant subset were NAGA (an enzyme cleaving specific moieties from glycoconjugates) and NDUFAF2 (part of the mitochondrial complex [MIM: 609653]); rare mutations in each of these genes are associated with early and severe neurodevelopmental disorders.79, 80 TWAS of the discordant subset revealed synaptic genes including C4A, which plays a key role in synaptic pruning,78 as well as other genes essential to synapse structure and function; such genes include ARL3 (MIM: 604695), FXR1 (MIM: 600819), and CNNM2 (MIM: 607803). Moreover, pathway analysis of S-Predixcan results (Table 3) demonstrated that the strongest gene set associated with the concordant subset was cell-to-cell adhesion via plasma-membrane adhesion molecules (GO: 0098742); this gene set encompasses processes such as those necessary for neural tube closure, cerebral cortex migration, and neuronal-glial interactions. In contrast, the discordant subset transcriptome was significantly enriched for genes located at dendrites, as well as for genes associated with DNA repair. Recently, the role of DNA repair in modulating neuronal activity-induced gene expression has been shown to be crucial for synaptic plasticity and related processes of learning and memory;81 impairments in DNA repair have been linked to neurodegeneration82, 83
ASSET also permitted the identification of SNPs for cognition-related phenotypes. Lookups of the full ASSET results revealed that ∼75% of the additional 110 loci, which were not identified in the input GWAS studies,18, 35, 36 were in fact replicated in an MTAG study examining intelligence20 and in more recent follow-up GWASs20, 33, 34 that used larger samples and were better powered for variant discovery. This result strongly supports the validity of the ASSET methodology and demonstrates that the approach indeed improves power for cross-phenotype discovery of new loci, as previously discussed by the developers of the method.32 Notably, several of our loci were associated with eQTLs, suggesting new potential biological mechanisms for individual variation in cognitive and psychiatric phenotypes. For example, one of the loci strongly implicates variation in PLXNB2 (MIM: 604293), a gene associated with GABA and glutamate synapses in the hippocampus.84 Another locus shows strong eQTL signal with NDE1 (MIM: 609449), a neurodevelopmental gene at the 16p13.11 locus, where copy-number variants have been associated with neurodevelopmental disorders.85 Our work supports and extends a recent study by Bansal and colleagues,30 whose paper is the only published report (to our knowledge) that has deeply examined the paradoxical relationship between educational attainment and schizophrenia. Using a proxy-phenotype approach, these investigators identified two loci, implicating the FOXO6 (MIM: 611457) and SLITRK1 (MIM: 609678) genes, with pleiotropic (i.e., “discordant”) effects across the two phenotypes. Using ASSET, we also uncovered those genes among our 110 loci (one of which was also not identified in any of the updated single-phenotype GWAS, see Table 1). Several other studies18, 19, 23, 31 have employed other statistical approaches to identify pleiotropy and/or overlap across cognitive/educational and schizophrenia GWAS and have uncovered a subset of the loci identified by ASSET. By utilizing ASSET, we were able to systematically and powerfully identify concordant and discordant pleiotropic loci across the genome and to then characterize underlying biological mechanisms. Future studies could also apply ASSET and related techniques to further understand other reported polygenic overlaps such as that between schizophrenia and creativity86 or that between cognitive ability and smoking status.26
In addition to functional characterization via pathway analyses, we were able to characterize the concordant and discordant SNP sets with respect to genetic overlap with other relevant phenotypes. To our knowledge, this is the first study to examine genetic correlations with dimensional sub-sets rather than global correlations with full GWASs. Although the concordant subset followed the expected patterns of genetic correlation with several forms of psychopathology, as well as brain and head size, results for the discordant subset were somewhat surprising. For example, we had anticipated that the discordant subset might be significantly related to personality as a non-cognitive trait that could promote greater educational attainment. However, correlations with conscientiousness, openness, and neuroticism were stronger for the concordant than for the discordant subset.
On the other hand, significant correlations for the discordant subset were observed with risk for autism, which has previously been shown to demonstrate a counter-intuitive positive genetic correlation with cognition.87 Given that variants within the discordant subset tend to index regulation of synaptic function and pruning processes, our results suggest that these mechanisms should be investigated with respect to their impact on autism, eating disorders, and bipolar disorder. Moreover, it is noteworthy that autism, despite being a neurodevelopmental disorder, did not demonstrate a significant genetic correlation with the concordant subset, indicating that it does not share the specific neurodevelopmental pathways implicated in the common variant genetic overlap between schizophrenia risk and impaired cognition. It is also intriguing that bipolar disorder demonstrated a very similar pattern of GNOVA results to schizophrenia, despite prior reports that bipolar disorder is not significantly correlated at the genetic level with general cognitive ability.87, 88 Thus, our approach was able to refine components of neurodevelopment and synaptic function that are shared across cognitive phenotypes, schizophrenia, and bipolar disorder. Further research is needed to identify components of cognition that differentiate schizophrenia and bipolar disorder.
One limitation of this study is that only common SNPs (MAF > 0.01) were examined. The genetic architecture of cognitive ability and educational attainment is composed of causal variants in LD with common SNPs (cognitive ability h2 = 22.7%, education h2 = 15.6%) as well as with causal variants in LD with rare and less-common SNPs (cognitive ability h2 = 31.3%, education h2 = 28.1%); rarer variants make greater contributions to cognitive differences than more common variants do.89 Rare variants are also known to explain some of the differences in schizophrenia prevalence.50 However, GNOVA, used in the identification of genetic correlations across data-independent datasets using summary GWAS data, can only capture the contributions made by common genetic effects. Future work aiming to investigate the concordant and discordant effect of rare variants across cognitive ability, schizophrenia, and education is needed.90 Additionally, the input GWASs for ASSET were of somewhat different sample sizes and power, and the cognitive GWAS demonstrated smaller mean effect sizes than those for schizophrenia and educational attainment; the effects of such differences on ASSET results are not fully understood, although ASSET has been benchmarked as the best available approach to handling non-uniform distribution of effect sizes.39
Now that the utility and validity of the ASSET approach has been demonstrated, future studies are planned that can further exploit this method using larger, and more varied, input GWASs. Recent studies have demonstrated that genetic correlations exist across seemingly disparate brain-related phenotypes.91 However, such genetic correlations only describe the average genetic effect between pairs of traits. As such, they are not informative as to which variants are associated across traits, nor if a minority of these variants have effects across traits that are the opposite of what would be expected on the basis of the direction of the genetic correlation. The application of the ASSET approach to these datasets would help researchers to move beyond the analysis of shared genetic variance and begin to identify shared genetic variants that, as shown in the current study, might be composed of variants with different combinations of protective and deleterious effects. Future studies, employing additional statistical techniques and incorporating rare variants and novel annotation resources, are needed to further decompose the early neurodevelopmental and adult synaptic pathways highlighted in the present report.
Declaration of Interests
The authors declare no competing interests.
Published: August 1, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.06.012.
Web Resources
FUMA, https://fuma.ctglab.nl/
Genes2Cognition database, http://www.genes2cognition.org
NHGRI-EBI catalog of published genome-wide association studies, https://www.genome.gov/gwastudies/
OMIM, https://www.omim.org/
PredictDB, http://predictdb.org/
Psychiatric Genomics Consortium, https://www.med.unc.edu/pgc/results-and-downloads/
S-Predixcan, https://github.com/hakyimlab/MetaXcan
Social Science Genetic Association Consortium, https://www.thessgac.org/data
Supplemental Data
References
- 1.Keefe R.S.E. Should cognitive impairment be included in the diagnostic criteria for schizophrenia? World Psychiatry. 2008;7:22–28. doi: 10.1002/j.2051-5545.2008.tb00142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mesholam-Gately R.I., Giuliano A.J., Goff K.P., Faraone S.V., Seidman L.J. Neurocognition in first-episode schizophrenia: a meta-analytic review. Neuropsychology. 2009;23:315–336. doi: 10.1037/a0014708. [DOI] [PubMed] [Google Scholar]
- 3.Heinrichs R.W., Zakzanis K.K. Neurocognitive deficit in schizophrenia: a quantitative review of the evidence. Neuropsychology. 1998;12:426–445. doi: 10.1037//0894-4105.12.3.426. [DOI] [PubMed] [Google Scholar]
- 4.Bilder R.M., Goldman R.S., Robinson D., Reiter G., Bell L., Bates J.A., Pappadopulos E., Willson D.F., Alvir J.M.J., Woerner M.G. Neuropsychology of first-episode schizophrenia: initial characterization and clinical correlates. Am. J. Psychiatry. 2000;157:549–559. doi: 10.1176/appi.ajp.157.4.549. [DOI] [PubMed] [Google Scholar]
- 5.Reichenberg A., Caspi A., Harrington H., Houts R., Keefe R.S.E., Murray R.M., Poulton R., Moffitt T.E. Static and dynamic cognitive deficits in childhood preceding adult schizophrenia: a 30-year study. Am. J. Psychiatry. 2010;167:160–169. doi: 10.1176/appi.ajp.2009.09040574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottesman I.I., Gould T.D. The endophenotype concept in psychiatry: etymology and strategic intentions. Am. J. Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 7.Glahn D.C., Almasy L., Blangero J., Burk G.M., Estrada J., Peralta J.M., Meyenberg N., Castro M.P., Barrett J., Nicolini H. Adjudicating neurocognitive endophenotypes for schizophrenia. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2007;144B:242–249. doi: 10.1002/ajmg.b.30446. [DOI] [PubMed] [Google Scholar]
- 8.Lencz T., Smith C.W., McLaughlin D., Auther A., Nakayama E., Hovey L., Cornblatt B.A. Generalized and specific neurocognitive deficits in prodromal schizophrenia. Biol. Psychiatry. 2006;59:863–871. doi: 10.1016/j.biopsych.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Goldberg T.E., Burdick K.E., McCormack J., Napolitano B., Patel R.C., Sevy S.M., Goldman R., Lencz T., Malhotra A.K., Kane J.M., Robinson D.G. Lack of an inverse relationship between duration of untreated psychosis and cognitive function in first episode schizophrenia. Schizophr. Res. 2009;107:262–266. doi: 10.1016/j.schres.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornblatt B., Obuchowski M., Roberts S., Pollack S., Erlenmeyer-Kimling L. Cognitive and behavioral precursors of schizophrenia. Dev. Psychopathol. 1999;11:487–508. doi: 10.1017/s0954579499002175. [DOI] [PubMed] [Google Scholar]
- 11.Snitz B.E., Macdonald A.W., 3rd, Carter C.S. Cognitive deficits in unaffected first-degree relatives of schizophrenia patients: a meta-analytic review of putative endophenotypes. Schizophr. Bull. 2006;32:179–194. doi: 10.1093/schbul/sbi048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reichenberg A., Weiser M., Rabinowitz J., Caspi A., Schmeidler J., Mark M., Kaplan Z., Davidson M. A population-based cohort study of premorbid intellectual, language, and behavioral functioning in patients with schizophrenia, schizoaffective disorder, and nonpsychotic bipolar disorder. Am. J. Psychiatry. 2002;159:2027–2035. doi: 10.1176/appi.ajp.159.12.2027. [DOI] [PubMed] [Google Scholar]
- 13.Bilder R.M., Reiter G., Bates J., Lencz T., Szeszko P., Goldman R.S., Robinson D., Lieberman J.A., Kane J.M. Cognitive development in schizophrenia: follow-back from the first episode. J. Clin. Exp. Neuropsychol. 2006;28:270–282. doi: 10.1080/13803390500360554. [DOI] [PubMed] [Google Scholar]
- 14.Lam M., Lee J., Rapisarda A., See Y.M., Yang Z., Lee S.-A., Abdul-Rashid N.A., Kraus M., Subramaniam M., Chong S.-A., Keefe R.S.E. longitudinal cognitive changes in young individuals at ultrahigh risk for psychosis. JAMA Psychiatry. 2018;75:929–939. doi: 10.1001/jamapsychiatry.2018.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacCabe J.H., Lambe M.P., Cnattingius S., Torrång A., Björk C., Sham P.C., David A.S., Murray R.M., Hultman C.M. Scholastic achievement at age 16 and risk of schizophrenia and other psychoses: a national cohort study. Psychol. Med. 2008;38:1133–1140. doi: 10.1017/S0033291707002048. [DOI] [PubMed] [Google Scholar]
- 16.Chong S.A., Subramaniam M., Lee I.-M., Pek E., Cheok C., Verma S., Wong J. Academic attainment: a predictor of psychiatric disorders? Soc. Psychiatry Psychiatr. Epidemiol. 2009;44:999–1004. doi: 10.1007/s00127-009-0027-3. [DOI] [PubMed] [Google Scholar]
- 17.Lencz T., Knowles E., Davies G., Guha S., Liewald D.C., Starr J.M., Djurovic S., Melle I., Sundet K., Christoforou A. Molecular genetic evidence for overlap between general cognitive ability and risk for schizophrenia: a report from the Cognitive Genomics Consortium (COGENT) Mol. Psychiatry. 2014;19:168–174. doi: 10.1038/mp.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lam M., Trampush J.W., Yu J., Knowles E., Davies G., Liewald D.C., Starr J.M., Djurovic S., Melle I., Sundet K. Large-scale cognitive GWAS meta-analysis reveals tissue-specific neural expression and potential nootropic drug targets. Cell Rep. 2017;21:2597–2613. doi: 10.1016/j.celrep.2017.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smeland O.B., Frei O., Kauppi K., Hill W.D., Li W., Wang Y., Krull F., Bettella F., Eriksen J.A., Witoelar A., NeuroCHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology) Cognitive Working Group Identification of genetic loci jointly influencing schizophrenia risk and the cognitive traits of verbal-numerical reasoning, reaction time, and general cognitive function. JAMA Psychiatry. 2017;74:1065–1075. doi: 10.1001/jamapsychiatry.2017.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savage J.E., Jansen P.R., Stringer S., Watanabe K., Bryois J., de Leeuw C.A., Nagel M., Awasthi S., Barr P.B., Coleman J.R.I. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat. Genet. 2018;50:912–919. doi: 10.1038/s41588-018-0152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies G., Lam M., Harris S.E., Trampush J.W., Luciano M., Hill W.D., Hagenaars S.P., Ritchie S.J., Marioni R.E., Fawns-Ritchie C. Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat. Commun. 2018;9:2098. doi: 10.1038/s41467-018-04362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill W.D., Davies G., Liewald D.C., McIntosh A.M., Deary I.J., CHARGE Cognitive Working Group Age-dependent pleiotropy between general cognitive function and major psychiatric disorders. Biol. Psychiatry. 2016;80:266–273. doi: 10.1016/j.biopsych.2015.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill W.D., Marioni R.E., Maghzian O., Ritchie S.J., Hagenaars S.P., McIntosh A.M., Gale C.R., Davies G., Deary I.J. A combined analysis of genetically correlated traits identifies 187 loci and a role for neurogenesis and myelination in intelligence. Mol. Psychiatry. 2019;24:169–181. doi: 10.1038/s41380-017-0001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies G., Marioni R.E., Liewald D.C., Hill W.D., Hagenaars S.P., Harris S.E., Ritchie S.J., Luciano M., Fawns-Ritchie C., Lyall D. Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N=112 151) Mol. Psychiatry. 2016;21:758–767. doi: 10.1038/mp.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bulik-Sullivan B.K., Loh P.-R., Finucane H.K., Ripke S., Yang J., Patterson N., Daly M.J., Price A.L., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagenaars S.P., Harris S.E., Davies G., Hill W.D., Liewald D.C.M., Ritchie S.J., Marioni R.E., Fawns-Ritchie C., Cullen B., Malik R., METASTROKE Consortium, International Consortium for Blood Pressure GWAS. SpiroMeta Consortium. CHARGE Consortium Pulmonary Group, CHARGE Consortium Aging and Longevity Group Shared genetic aetiology between cognitive functions and physical and mental health in UK Biobank (N=112 151) and 24 GWAS consortia. Mol. Psychiatry. 2016;21:1624–1632. doi: 10.1038/mp.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trampush J.W., Yang M.L.Z., Yu J., Knowles E., Davies G., Liewald D.C., Starr J.M., Djurovic S., Melle I., Sundet K. GWAS meta-analysis reveals novel loci and genetic correlates for general cognitive function: a report from the COGENT consortium. Mol. Psychiatry. 2017;22:336–345. doi: 10.1038/mp.2016.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R., Duncan L., Perry J.R., Patterson N., Robinson E.B., ReproGen Consortium. Psychiatric Genomics Consortium. Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3 An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. Bulik-Sullivan, B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turley P., Walters R.K., Maghzian O., Okbay A., Lee J.J., Fontana M.A., Nguyen-Viet T.A., Wedow R., Zacher M., Furlotte N.A., 23andMe Research Team. Social Science Genetic Association Consortium Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet. 2018;50:229–237. doi: 10.1038/s41588-017-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bansal V., Mitjans M., Burik C.A.P., Linnér R.K., Okbay A., Rietveld C.A., Begemann M., Bonn S., Ripke S., de Vlaming R. Genome-wide association study results for educational attainment aid in identifying genetic heterogeneity of schizophrenia. Nat. Commun. 2018;9:3078. doi: 10.1038/s41467-018-05510-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Hellard S., Wang Y., Witoelar A., Zuber V., Bettella F., Hugdahl K., Espeseth T., Steen V.M., Melle I., Desikan R., Schizophrenia Working Group of the Psychiatric Genomics Consortium Identification of gene loci that overlap between schizophrenia and educational attainment. Schizophr. Bull. 2017;43:654–664. doi: 10.1093/schbul/sbw085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhattacharjee S., Rajaraman P., Jacobs K.B., Wheeler W.A., Melin B.S., Hartge P., Yeager M., Chung C.C., Chanock S.J., Chatterjee N., GliomaScan Consortium A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am. J. Hum. Genet. 2012;90:821–835. doi: 10.1016/j.ajhg.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pardiñas A.F., Holmans P., Pocklington A.J., Escott-Price V., Ripke S., Carrera N., Legge S.E., Bishop S., Cameron D., Hamshere M.L., GERAD1 Consortium. CRESTAR Consortium Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 2018;50:381–389. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J.J., Wedow R., Okbay A., Kong E., Maghzian O., Zacher M., Nguyen-Viet T.A., Bowers P., Sidorenko J., Karlsson Linnér R., 23andMe Research Team. COGENT (Cognitive Genomics Consortium) Social Science Genetic Association Consortium Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 2018;50:1112–1121. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okbay A., Beauchamp J.P., Fontana M.A., Lee J.J., Pers T.H., Rietveld C.A., Turley P., Chen G.-B., Emilsson V., Meddens S.F.W., LifeLines Cohort Study Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–542. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu Q., Li B., Ou D., Erlendsdottir M., Powles R.L., Jiang T., Hu Y., Chang D., Jin C., Dai W. A powerful approach to estimating annotation-stratified genetic covariance via GWAS summary statistics. Am. J. Hum. Genet. 2017;101:939–964. doi: 10.1016/j.ajhg.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Z., Anttila V., Smoller J.W., Lee P.H. Statistical power and utility of meta-analysis methods for cross-phenotype genome-wide association studies. PLoS ONE. 2018;13:e0193256. doi: 10.1371/journal.pone.0193256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe K., Taskesen E., van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 44.de Leeuw C.A., Mooij J.M., Heskes T., Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gamazon E.R., Segrè A.V., van de Bunt M., Wen X., Xi H.S., Hormozdiari F., Ongen H., Konkashbaev A., Derks E.M., Aguet F., GTEx Consortium Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat. Genet. 2018;50:956–967. doi: 10.1038/s41588-018-0154-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gaspar H.A., Breen G. Pathways analyses of schizophrenia GWAS focusing on known and novel drug targets. bioRxiv. 2016 [Google Scholar]
- 49.Gaspar H.A., Breen G. Drug enrichment and discovery from schizophrenia genome-wide association results: an analysis and visualisation approach. Sci. Rep. 2017;7:12460. doi: 10.1038/s41598-017-12325-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh T., Walters J.T.R., Johnstone M., Curtis D., Suvisaari J., Torniainen M., Rees E., Iyegbe C., Blackwood D., McIntosh A.M., INTERVAL Study. UK10K Consortium The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat. Genet. 2017;49:1167–1173. doi: 10.1038/ng.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang H.J., Kawasawa Y.I., Cheng F., Zhu Y., Xu X., Li M., Sousa A.M., Pletikos M., Meyer K.A., Sedmak G. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purcell S.M., Moran J.L., Fromer M., Ruderfer D., Solovieff N., Roussos P., O’Dushlaine C., Chambert K., Bergen S.E., Kähler A. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barbeira A.N., Dickinson S.P., Bonazzola R., Zheng J., Wheeler H.E., Torres J.M., Torstenson E.S., Shah K.P., Garcia T., Edwards T.L., GTEx Consortium Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 2018;9:1825. doi: 10.1038/s41467-018-03621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dobbyn A., Huckins L.M., Boocock J., Sloofman L.G., Glicksberg B.S., Giambartolomei C., Hoffman G.E., Perumal T.M., Girdhar K., Jiang Y., CommonMind Consortium Landscape of conditional eQTL in dorsolateral prefrontal cortex and co-localization with schizophrenia GWAS. Am. J. Hum. Genet. 2018;102:1169–1184. doi: 10.1016/j.ajhg.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gamazon E.R., Wheeler H.E., Shah K.P., Mozaffari S.V., Aquino-Michaels K., Carroll R.J., Eyler A.E., Denny J.C., Nicolae D.L., Cox N.J., Im H.K., GTEx Consortium A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fromer M., Roussos P., Sieberts S.K., Johnson J.S., Kavanagh D.H., Perumal T.M., Ruderfer D.M., Oh E.C., Topol A., Shah H.R. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Senthil G., Dutka T., Bingaman L., Lehner T. Genomic resources for the study of neuropsychiatric disorders. Mol. Psychiatry. 2017;22:1659–1663. doi: 10.1038/mp.2017.29. [DOI] [PubMed] [Google Scholar]
- 58.Hauberg M.E., Zhang W., Giambartolomei C., Franzén O., Morris D.L., Vyse T.J., Ruusalepp A., Sklar P., Schadt E.E., Björkegren J.L.M., Roussos P., CommonMind Consortium Large-scale identification of common trait and disease variants affecting gene expression. Am. J. Hum. Genet. 2017;100:885–894. doi: 10.1016/j.ajhg.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trowsdale J., Knight J.C. Major histocompatibility complex genomics and human disease. Annu. Rev. Genomics Hum. Genet. 2013;14:301–323. doi: 10.1146/annurev-genom-091212-153455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen C.-H., Wang Y., Lo M.-T., Schork A., Fan C.-C., Holland D., Kauppi K., Smeland O.B., Djurovic S., Sanyal N. Leveraging genome characteristics to improve gene discovery for putamen subcortical brain structure. Sci. Rep. 2017;7:15736. doi: 10.1038/s41598-017-15705-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Dushlaine C., Kenny E., Heron E., Donohoe G., Gill M., Morris D., Corvin A., International Schizophrenia Consortium Molecular pathways involved in neuronal cell adhesion and membrane scaffolding contribute to schizophrenia and bipolar disorder susceptibility. Mol. Psychiatry. 2011;16:286–292. doi: 10.1038/mp.2010.7. [DOI] [PubMed] [Google Scholar]
- 62.Neale B.M., Kou Y., Liu L., Ma’ayan A., Samocha K.E., Sabo A., Lin C.-F., Stevens C., Wang L.-S., Makarov V. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bernier R., Golzio C., Xiong B., Stessman H.A., Coe B.P., Penn O., Witherspoon K., Gerdts J., Baker C., Vulto-van Silfhout A.T. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krumm N., Turner T.N., Baker C., Vives L., Mohajeri K., Witherspoon K., Raja A., Coe B.P., Stessman H.A., He Z.-X. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang T., Guo H., Xiong B., Stessman H.A.F., Wu H., Coe B.P., Turner T.N., Liu Y., Zhao W., Hoekzema K. De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat. Commun. 2016;7:13316. doi: 10.1038/ncomms13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCarthy S.E., Gillis J., Kramer M., Lihm J., Yoon S., Berstein Y., Mistry M., Pavlidis P., Solomon R., Ghiban E. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol. Psychiatry. 2014;19:652–658. doi: 10.1038/mp.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kimura H., Wang C., Ishizuka K., Xing J., Takasaki Y., Kushima I., Aleksic B., Uno Y., Okada T., Ikeda M. Identification of a rare variant in CHD8 that contributes to schizophrenia and autism spectrum disorder susceptibility. Schizophr. Res. 2016;178:104–106. doi: 10.1016/j.schres.2016.08.023. [DOI] [PubMed] [Google Scholar]
- 68.Durak O., Gao F., Kaeser-Woo Y.J., Rueda R., Martorell A.J., Nott A., Liu C.Y., Watson L.A., Tsai L.-H. Chd8 mediates cortical neurogenesis via transcriptional regulation of cell cycle and Wnt signaling. Nat. Neurosci. 2016;19:1477–1488. doi: 10.1038/nn.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gompers A.L., Su-Feher L., Ellegood J., Copping N.A., Riyadh M.A., Stradleigh T.W., Pride M.C., Schaffler M.D., Wade A.A., Catta-Preta R. Germline Chd8 haploinsufficiency alters brain development in mouse. Nat. Neurosci. 2017;20:1062–1073. doi: 10.1038/nn.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suetterlin P., Hurley S., Mohan C., Riegman K.L.H., Pagani M., Caruso A., Ellegood J., Galbusera A., Crespo-Enriquez I., Michetti C. Altered neocortical gene expression, brain overgrowth and functional over-connectivity in chd8 haploinsufficient mice. Cereb. Cortex. 2018;28:2192–2206. doi: 10.1093/cercor/bhy058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sniekers S., Stringer S., Watanabe K., Jansen P.R., Coleman J.R.I., Krapohl E., Taskesen E., Hammerschlag A.R., Okbay A., Zabaneh D. Genome-wide association meta-analysis of 78,308 individuals identifies new loci and genes influencing human intelligence. Nat. Genet. 2017;49:1107–1112. doi: 10.1038/ng.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davies G., Armstrong N., Bis J.C., Bressler J., Chouraki V., Giddaluru S., Hofer E., Ibrahim-Verbaas C.A., Kirin M., Lahti J., Generation Scotland Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N=53949) Mol. Psychiatry. 2015;20:183–192. doi: 10.1038/mp.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hill W.D., Davies G., van de Lagemaat L.N., Christoforou A., Marioni R.E., Fernandes C.P.D., Liewald D.C., Croning M.D.R., Payton A., Craig L.C.A. Human cognitive ability is influenced by genetic variation in components of postsynaptic signalling complexes assembled by NMDA receptors and MAGUK proteins. Transl. Psychiatry. 2014;4:e341. doi: 10.1038/tp.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fernández E., Collins M.O., Frank R.A.W., Zhu F., Kopanitsa M.V., Nithianantharajah J., Lemprière S.A., Fricker D., Elsegood K.A., McLaughlin C.L. Arc requires PSD95 for assembly into postsynaptic complexes involved with neural dysfunction and intelligence. Cell Rep. 2017;21:679–691. doi: 10.1016/j.celrep.2017.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reichenberg A., Weiser M., Rapp M.A., Rabinowitz J., Caspi A., Schmeidler J., Knobler H.Y., Lubin G., Nahon D., Harvey P.D., Davidson M. Elaboration on premorbid intellectual performance in schizophrenia: premorbid intellectual decline and risk for schizophrenia. Arch. Gen. Psychiatry. 2005;62:1297–1304. doi: 10.1001/archpsyc.62.12.1297. [DOI] [PubMed] [Google Scholar]
- 77.Denève S., Alemi A., Bourdoukan R. The Brain as an efficient and robust adaptive learner. Neuron. 2017;94:969–977. doi: 10.1016/j.neuron.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 78.Sekar A., Bialas A.R., de Rivera H., Davis A., Hammond T.R., Kamitaki N., Tooley K., Presumey J., Baum M., Van Doren V., Schizophrenia Working Group of the Psychiatric Genomics Consortium Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Keulemans J.L., Reuser A.J., Kroos M.A., Willemsen R., Hermans M.M., van den Ouweland A.M., de Jong J.G., Wevers R.A., Renier W.O., Schindler D. Human alpha-N-acetylgalactosaminidase (alpha-NAGA) deficiency: new mutations and the paradox between genotype and phenotype. J. Med. Genet. 1996;33:458–464. doi: 10.1136/jmg.33.6.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ghaloul-Gonzalez L., Goldstein A., Walsh Vockley C., Dobrowolski S.F., Biery A., Irani A., Ibarra J., Morton D.H., Mohsen A.-W., Vockley J. Mitochondrial respiratory chain disorders in the Old Order Amish population. Mol. Genet. Metab. 2016;118:296–303. doi: 10.1016/j.ymgme.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 81.Shiwaku H., Okazawa H. Impaired DNA damage repair as a common feature of neurodegenerative diseases and psychiatric disorders. Curr. Mol. Med. 2015;15:119–128. doi: 10.2174/1566524015666150303002556. [DOI] [PubMed] [Google Scholar]
- 82.Madabhushi R., Pan L., Tsai L.-H. DNA damage and its links to neurodegeneration. Neuron. 2014;83:266–282. doi: 10.1016/j.neuron.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Su Y., Ming G.L., Song H. DNA damage and repair regulate neuronal gene expression. Cell Res. 2015;25:993–994. doi: 10.1038/cr.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McDermott J.E., Goldblatt D., Paradis S. Class 4 Semaphorins and Plexin-B receptors regulate GABAergic and glutamatergic synapse development in the mammalian hippocampus. Mol. Cell. Neurosci. 2018;92:50–66. doi: 10.1016/j.mcn.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grayton H.M., Fernandes C., Rujescu D., Collier D.A. Copy number variations in neurodevelopmental disorders. Prog. Neurobiol. 2012;99:81–91. doi: 10.1016/j.pneurobio.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 86.Power R.A., Steinberg S., Bjornsdottir G., Rietveld C.A., Abdellaoui A., Nivard M.M., Johannesson M., Galesloot T.E., Hottenga J.J., Willemsen G. Polygenic risk scores for schizophrenia and bipolar disorder predict creativity. Nat. Neurosci. 2015;18:953–955. doi: 10.1038/nn.4040. [DOI] [PubMed] [Google Scholar]
- 87.Hill W.D., Harris S.E., Deary I.J. What genome-wide association studies reveal about the association between intelligence and mental health. Curr. Opin. Psychol. 2018;27:25–30. doi: 10.1016/j.copsyc.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 88.Gale C.R., Batty G.D., McIntosh A.M., Porteous D.J., Deary I.J., Rasmussen F. Is bipolar disorder more common in highly intelligent people? A cohort study of a million men. Mol. Psychiatry. 2013;18:190–194. doi: 10.1038/mp.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hill W.D., Arslan R.C., Xia C., Luciano M., Amador C., Navarro P., Hayward C., Nagy R., Porteous D.J., McIntosh A.M. Genomic analysis of family data reveals additional genetic effects on intelligence and personality. Mol. Psychiatry. 2018;23:2347–2362. doi: 10.1038/s41380-017-0005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Evans L.M., Tahmasbi R., Vrieze S.I., Abecasis G.R., Das S., Gazal S., Bjelland D.W., de Candia T.R., Goddard M.E., Neale B.M., Haplotype Reference Consortium Comparison of methods that use whole genome data to estimate the heritability and genetic architecture of complex traits. Nat. Genet. 2018;50:737–745. doi: 10.1038/s41588-018-0108-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brainstorm Consortium. Anttila V., Bulik-Sullivan B., Finucane H.K., Walters R.K., Bras J., Duncan L., Escott-Price V., Falcone G.J., Gormley P. Analysis of shared heritability in common disorders of the brain. Science. 2018;360:eaap8757. doi: 10.1126/science.aap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat. Neurosci. 2015;18:199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.