Abstract
Mechanosensitive channels play an important role in the adaptation of cells to hypo-osmotic shock. Among members of this channel family in Escherichia coli, the exact function and physiological role of the mechanosensitive channel homolog YbdG remain unclear. Characterization of YbdG's physiological role has been hampered by its lack of measurable transport activity. Using a nitrosoguanidine mutagenesis-aided screen in combination with next-generation sequencing, here we isolated a mutant with a point mutation in ybdG. This mutation (resulting in a I167T change) conferred sensitivity to high osmotic stress, and the mutant cells differed from WT cells in morphology during hyperosmotic stress at alkaline pH. Interestingly, unlike the cells containing the I167T variant, a null-ybdG mutant did not exhibit this sensitivity and phenotype. Although I167T was located near the putative ion-conducting pore in a transmembrane region of YbdG, no change in ion channel activities of YbdG-I167T was detected. Of note, introduction of the WT C-terminal cytosolic region of YbdG into the I167T variant complemented the osmo-sensitive phenotype. Co-precipitation of proteins interacting with the C-terminal YbdG region led to the isolation of HldD and FbaA, whose overexpression in cells containing the YbdG-I167T variant partially rescued the osmo-sensitive phenotype. This study indicates that YbdG functions as a component of a mechanosensing system that transmits signals triggered by external osmotic changes to intracellular factors. The cellular role of YbdG uncovered here goes beyond its predicted function as an ion or solute transport protein.
Keywords: bacterial signal transduction, cell biology, stress, stress response, membrane protein, mechanosensitive channel, high osmolarity, YbdG, mechanosensor, gain-of-function mutation, Escherichia coli, osmotic shock, ion channel, YbdG-I167T, signal transduction
Introduction
Bacteria are constantly exposed to various osmotic challenges in their environment; consequently, they have evolved a cellular adaptation machinery to deal with those challenges (1). The permeability of cells to ions and small carbon compounds is limited, which allows cells to control their volume. The dynamics of cell volume changes affect the cells' physiological activity and integrity. Therefore, regulatory mechanisms for controlling cell volume are key to sustaining growth and survival in extreme environments. Absorption of ions and synthesis of organic osmolytes inside cells and uptake or release of these osmolytes via solute membrane transport systems occur in response to environmental osmotic changes. Hyperosmotic shock induces a biphasic response in bacteria; the initial event involves accumulation of ions through an activated ion transport system in the membrane, triggered by osmosensing systems, and a secondary adaptation involves expression of osmolyte biosynthesis genes (2). To recover its cell volume after a hyperosmotic shock, Escherichia coli cells take up K+ through their K+-uptake transporters, Trk and Kdp. Under the same conditions, in Synechocystis sp. PCC 6803, the Trk homolog Ktr plays the main role in the supply of K+, instead of Kdp (3). After the initial response, the cells start to synthesize organic compounds, such as trehalose, glycosylglycerol, and proline, which act as substitutes for K+ in the cells (4–6). Several additional osmolytes are also taken up via transporters across the cell membrane (1). Two-component regulatory systems have been proposed to maintain cellular homeostasis in response to osmolarity changes in E. coli (7). However, osmotic sensors and/or related transporters that are directly involved in the initial osmotic response have not been fully identified.
Osmotic down-shock is also a severe stress for bacteria. Specifically, the physical forces cause an expansion of the cell volume, which increases the tension of the cell membrane. This leads to a release of solutes from the cell due to activation of mechanosensitive (MS)2 channels embedded in the cell membrane. MscS (MS channel of small conductance) and MscL (MS channel of large conductance) are the best-characterized MS channels in E. coli involved in the release of cytosolic solutes in response to hypoosmotic shock (8, 9). A study of the crystal structure of MscL and MscS and biochemical labeling experiments showed that the channels themselves undergo a large volume change during the transition between open and closed states (and also during the inactivation of MscS) (10–13). The functional unit of MscL and MscS is a pentamer and a heptamer, respectively, with each individual polypeptide containing two or three membrane-spanning domains. MscS has a long cytosolic C-terminal region that is not present in MscL. Movement of the C-terminal region of MscS participates in gating of the channel (14), and this region is in contact with other components in the cells (15). Following the identification of MscS and MscL, additional members of the MS family, MscK (potassium-dependent MS channel) and MscM (MS channel of miniconductance), were isolated (16, 17). After further genome analysis in E. coli, the expression profiles of four genes encoding MscS homologs, YjeP, YbiO, YnaI, and YbdG, were determined, and their activity was characterized by electrophysiology (17, 18). They revealed that YbiO, YjeP, and YnaI had marked ion channel conductance in patch-clamp recordings, whereas YbdG had no detectable channel current under the standard conditions. The cells expressing the YbdG V229A variant in the absence of MscS and MscK increased gating frequency (17). YbdG specifically was once described as being a component of MscM channel activity. However, the following study proposed that YjeP is the major component of MscM channel (18). Therefore, it is not yet clear whether YbdG functions as a channel. Knowledge of the physiological role of YbdG is also limited; in the case of removing mscL, mscS, and mscK simultaneously, the overexpression of ybdG could increase tolerance against osmotic down-shock (17). In contrast, the loss-of-function phenotype has not been observed in the ybdG-disruptant E. coli (17, 18).
As described above, a large number of solute transport systems have been identified in the E. coli cell membrane. However, physiologically important membrane transport systems that provide protection against osmotic up-shock have not yet been fully identified and remain to be characterized in detail. In this study, we performed a screen of E. coli mutants generated by nitrosoguanidine treatment to identify mutations that conferred sensitivity to high NaCl and KCl in the medium. Through this screen we isolated a gain-of-function mutant conferring sensitivity to high osmotic stress. This mutant was found to contain a point mutation (I167T) in the ybdG gene. The high osmotic sensitivity of this mutant (named WTD731) was not correlated with a change in ion transport activity of YbdG-I167T. Rather, our results suggest that this mutation disrupts interaction of YbdG with crucial components responsible for signal transduction processes involved in the cellular adaptation to hyperosmolarity.
Results
Isolation of an E. coli mutant with sensitivity to high-osmolarity stress
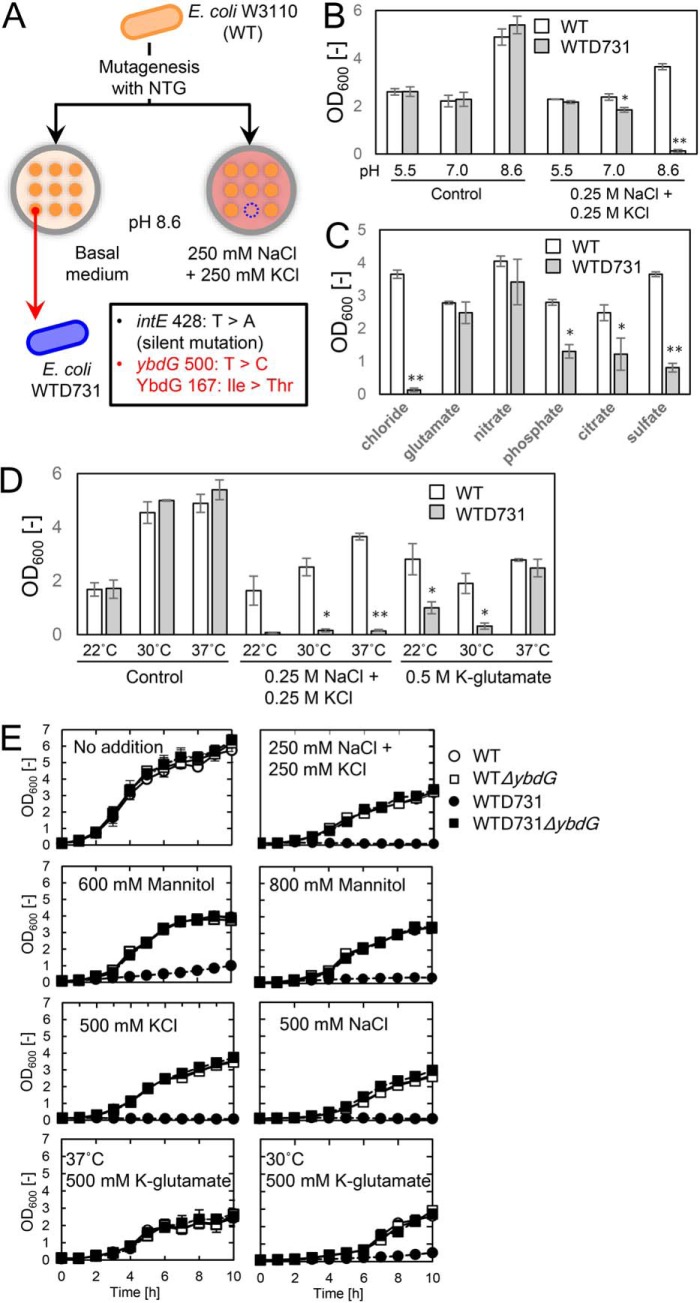
Only a few E. coli mutants sensitive to high osmotic pressure had been previously isolated with the exception of mutants that are sensitive to high sodium ion concentrations (19). We hypothesized that there are probably multiple systems operating during adaptation to high-osmolarity conditions at neutral pH in E. coli, in addition to the sodium ion extrusion systems proposed previously (20). Based on the assumption that the function of the major system declines at alkaline pH, we tried to isolate a mutant that would be hypersensitive to high osmotic pressure at alkaline pH. To reduce the possibility of isolating mutants sensitive to the cytotoxicity of high concentrations of sodium or potassium ions, medium containing both 250 mm NaCl and 250 mm KCl was used as the high osmotic condition (Fig. 1A). For mutagenesis, the WT E. coli strain W3110 was treated with 300 μg/ml N-methyl-N′-nitro-N-nitrosoguanidine (see “Experimental procedures”). A mutant, designated WTD731, grew as well as WT in the basal medium but was unable to grow in basal medium containing 250 mm NaCl and 250 mm KCl at pH 8.6 (Table S1).
Figure 1.
Isolation of E. coli mutants impaired in adaptation to high osmolarity. A, E. coli strain W3110 (WT) treated with N-methyl-N′-nitro-N-nitrosoguanidine (NTG) was grown on basal agar medium. Colonies were transferred by replica-plating to basal agar medium containing 250 mm NaCl and 250 mm KCl. Colonies unable to grow on the high-osmolarity plates were recovered from the basal medium. One of these colonies was designated WTD731 and further analyzed. B, growth of E. coli strains WT and WTD731 was determined in basal medium without or with 250 mm KCl and 250 mm NaCl at pH 5.5, pH 7.0, or pH 8.6, for 10 h. C, growth of E. coli strains WT and WTD731 determined in basal medium containing different anions, 250 mm KCl and 250 mm NaCl (chloride), 500 mm potassium glutamate (glutamate), 125 mm KNO3 and 125 mm NaNO3 (nitrate), 125 mm KH2PO4 and 125 mm NaH2PO4 (phosphate), 250 mm Na3C6H5O7 (citrate), or 250 mm Na2SO4 (sulfate) at 37 °C (pH 8.6) for 10 h. D, growth of E. coli strains WT and WTD731 determined in basal medium (pH 8.6) without or with 250 mm KCl and 250 mm NaCl or 500 mm potassium glutamate at 22, 30, and 37 °C for 10 h. E, time course of the growth of E. coli strains WT, WTD731, WTΔybdG, and WTD731ΔybdG were grown at 30 or 37 °C in basal medium (pH 8.6) containing NaCl, KCl, potassium glutamate, or mannitol at the concentrations indicated. Error bars, S.D. (n = 3). *, p < 0.05; **, p < 0.001, Student's t test.
To identify the mutations in the genome of WTD731 responsible for the lack of growth in the high osmotic medium, we performed bacteriophage P1 transduction using E. coli strains in the Keio Collection library as donor strains (21). The strain containing the kanamycin resistance gene inserted within ybdG exhibited the highest recovery value (95–100%) from the hypersensitive phenotype of WTD731 (data not shown). These data indicated that genes located in the vicinity of ybdG (12.99 min in the E. coli chromosome) had mutations in WTD731. Next, we determined the DNA sequence around ybdG and identified only one point mutation in the ybdG (a T-to-C mutation at nucleotide 500 of ybdG, converting Ile at position 167 to Thr (I167T)). To confirm the cause of the hypersensitivity of WTD731, we introduced the plasmid expressing ybdG in mobile plasmid collection into WTD731 (22). Unexpectedly, the hyperosmotic sensitivity was not rescued. We used next-generation sequencing of the chromosomal DNA of WTD731 to identify other mutations in the chromosome. Comparison of chromosomal DNA sequences of the mutant strain WTD731 and its parental laboratory strain W3110 revealed only two nucleotide replacements in WTD731: the same mutation of ybdG (I167T) and a T-to-A mutation at nucleotide 428 of intE, which was silent (no change of the corresponding amino acid) (Table S1). In consequence, we concluded that WTD731 had a gain-of-function mutation in ybdG (YbdG-I167T) but not loss of function mutation.
Phenotype of the mutant containing YbdG-I167T under high osmotic conditions
We characterized WTD731 as shown in Fig. 1, B–E. Reduced growth of WTD731 in high osmotic medium (250 mm NaCl and 250 mm KCl) was observed only at pH 8.6 and not at pH 7.0 or pH 5.5 (Fig. 1B). At pH 8.6, chloride had the strongest effect on growth compared with five different anions (glutamate, nitrate, phosphate, citrate, and sulfate), all supplied as sodium salts (Fig. 1C). The same growth inhibition was also seen when cultures were grown at different temperatures, 22, 30, and 37 °C (Fig. 1D) in medium containing 250 mm NaCl and 250 mm KCl. In contrast, potassium glutamate (500 mm) inhibited growth of WTD731 only at lower temperatures (22 and 30 °C).
To corroborate that the sensitivity of WTD731 to high osmotic medium was caused by a specific alteration of YbdG function but not by complete loss of YbdG activity, we eliminated ybdG from WT and WTD731 by replacing it with the chloramphenicol resistance gene (Fig. S1). The resulting mutants were designated as WTΔybdG and WTD731ΔybdG, respectively. The addition of 250 mm NaCl and 250 mm KCl, 500 mm NaCl, 500 mm KCl, and 600 or 800 mm mannitol to the media did not reduce growth of either WTΔybdG or WTD731ΔybdG when compared with the WT (Fig. 1E). Unlike WTD731, the ybdG null mutant was not sensitive to hyperosmotic shock. In addition, neither of the two null mutants showed the cold-sensitive phenotype in the presence of 500 mm potassium glutamate at 30 °C (Fig. 1E). The above results confirmed a gain-of-function mutation, but not loss of function mutation, in WTD731, conferring the sensitivity to high osmolarity.
Morphological differences between WT and WTD731 under hyperosomotic conditions
There are some reports that mutations in mechanosensitive channel genes in microorganisms and in plastids of Arabidopsis thaliana lead to changes in cell shape (15, 23, 24). We compared the shape and dimension of intact WT and WTD731 cells (Fig. 2). Under hyperosmotic stress (250 mm NaCl and 250 mm KCl), the cells of the WT took on a bent shape, which was not observed for WTD731 cells (Fig. 2A). WT cultures in high-osmolarity medium contained a higher number of bent cells with a higher degree of curvature, compared with nontreated WT or with both treated and nontreated WTD731 cells (Fig. 2B). These results indicate that the I167T change in YbdG caused morphological changes under high osmotic conditions.
Figure 2.
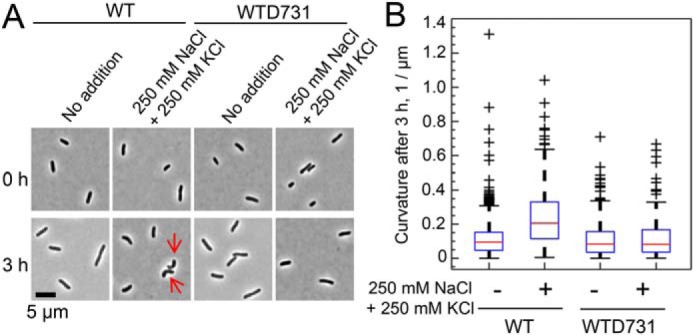
Differences in cell morphology between WT and WTD731 induced by high osmotic treatments. A, high salinity induced bending of WT cells (red arrows). WT and WTD731 cells were precultured overnight in basal medium at 37 °C and then transferred into basal medium without or with 250 mm NaCl and 250 mm KCl at an OD600 = 0.1. After incubation for 3 h, cell shape was observed by phase-contrast microscopy. B, box plots represent distribution of curvature of the cells. The median, lower and upper quartiles, and extreme values are indicated by red lines, blue boxes, and whiskers, respectively. The number of cells analyzed for each condition was as follows: WT (no addition), 931; WT (addition of 250 mm NaCl and KCl), 233; WTD731 (no addition), 510; WTD731 (addition of 250 mm NaCl and KCl), 130.
Location of Ile-167 and ion transport activity of YbdG-I167T
Based on a sequence alignment of YbdG with MscS, Ile at position 167 in the YbdG was predicted to be located in the vicinity of the pore-forming region on the N-terminal side of the last transmembrane helix of YbdG (17) (Fig. S2). YbdG has an additional region inserted in the middle of the C-terminal domain, which is not present in MscS. Ile-167 in YbdG corresponds to Ser-95 in MscS. To confirm this location, we constructed four types of fusions with alkaline phosphatase (PhoA), which is active when located in the periplasm but not when in the cytoplasm (25). Ile-167 was predicted to be located in the external loop between the presumed transmembrane domains (Fig. 3A and Fig. S2). Accordingly, the PhoA fusion sites were selected to be at the N-terminal end of the transmembrane domain preceding Ile-167 (K138-PhoA), at the position immediately after the loop containing Ile-167 (G173-PhoA), at the position after the transmembrane domain following Ile-167 (D206-PhoA), or at the C-terminal end of YbdG-I167T (Q415-PhoA). Cells containing G173-PhoA displayed high alkaline phosphatase activity, whereas cells expressing the three other constructs showed only low activity, similar to the negative control (empty vector (EV)) (Fig. 3B). Immunoblot analysis of the membrane protein fractions detected the fusion proteins at sizes corresponding to their predicted molecular mass (Fig. S3). The lower amount of K138-PhoA protein was probably due to the instability of PhoA in the cytoplasm (26). These biochemical assays indicated that Ile-167 was located in the external loop or at the N-terminal side of the last transmembrane helix.
Figure 3.
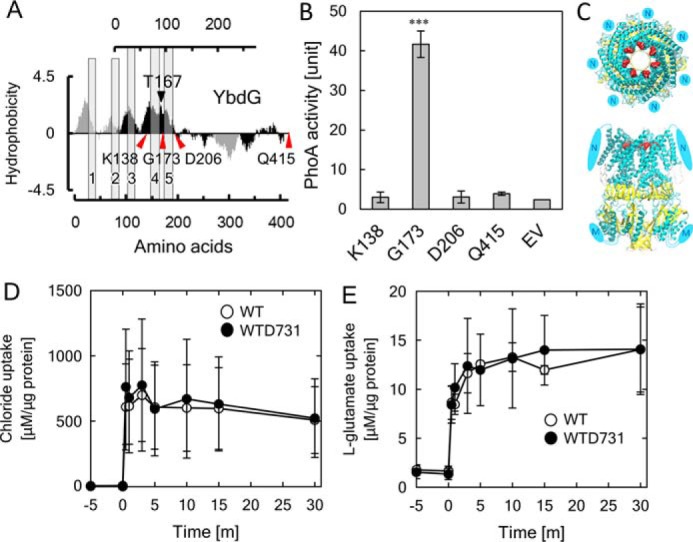
Characterization of YbdG-I167T. A, localization of the Ile at position 167 in YbdG. The hydropathy plots of MscS and YbdG were created using the method of Kyte and Doolittle with a window of 19 amino acids (17). The black arrowhead points to the position of Thr-167, and red arrowheads indicate PhoA fusion sites. Detailed sequences are given in Fig. S2. B, quantification of PhoA activity of the fusion proteins. Error bars, S.D. (n = 3). ***, p < 0.001, one-way analysis of variance followed by Dunnett's multiple-comparison test. C, protein structure models of YbdG and predicted interactions of residues. Top and front views of the structure of YbdG (open states), shown as ribbon models. N and M, the additional N-terminal region (N) and additional sequence inserted into the middle of the C-terminal region (M) of YbdG; both are not present in MscS. Ile-167 in each of the seven subunits of YbdG is represented by red spheres. D and E, measurements of solute transport in WT, WTD731, and WTD731ΔybdG. Transport activity for chloride (D) and l-glutamate (E) in WT and WTD731 is shown. Error bars, S.D. (n = 3).
Based on the published crystal structures of MscS in the open and closed state, homology modeling of YbdG in both states was performed (Fig. 3C and Fig. S4). The resulting structure placed the side chain of Ile-167 in YbdG facing in the opposite direction of the ion-conductive pore and participating in a hydrophobic cluster with the surrounding residues in the closed state. The hydrophobic interactions in the closed state seem more stable than those in the open state. Given that in the mutant, Ile-167 was replaced with Thr, the hydrophobic cluster would be disturbed in the closed state, and a hydrogen bond between the hydroxyl group (-OH) of Thr-167 and the ϵ-oxygen (or main-chain oxygen) of Gln-163 could form in the open state (Fig. 3C). Therefore, this substitution might result in increased stability of the open state of YbdG.
We examined whether the amino acid replacement in YbdG-I167T increased its ion channel conductance. Cellular contents of Cl− and glutamate concentrations in WT and WTD731 were determined after the addition of 250 mm NaCl and 250 mm KCl or 500 mm potassium glutamate (Fig. 3, D and E). No significant differences between the two strains were observed. As an alternative approach, we tested the K+-uptake activity of WT YbdG and YbdG-I167T in a complementation growth assay (26). Neither of the two constructs were able to complement the K+-uptake deficiency of the LB2003 strain (Fig. S5A). Next, we performed stopped-flow measurements to compare the cell volume changes of the WT and WTD731 strains in response to hyperosmotic shock with 250 mm NaCl and 250 mm KCl or 500 mm potassium glutamate (Fig. S5B). Based on these results, there were no apparent changes in the channel activity of YbdG-I167T.
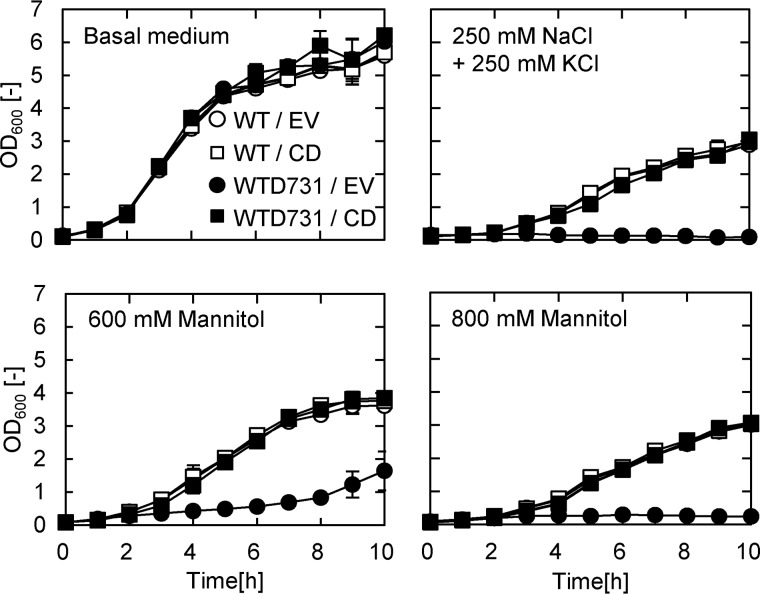
Rescue of hyperosmotic sensitivity of WTD731 by expression of the C-terminal cytosolic region of YbdG
Several reports indicate that subtle dynamic alterations of the transmembrane domains in MscS affect its cytosolic function (27, 28). Because no significant differences in the ion transport activity of YbdG and YbdG-I167T could be detected (Fig. 3D and Fig. S5), we predicted that the I167T substitution led to a modification of the function of the extended cytosolic C-terminal region (∼200 residues). It has been reported that in its inactive state, the C-terminal region of MscS is bound to FtsZ, a protein that functions in cell division (15). To investigate the physiological importance of the cytosolic C-terminal region of YbdG in E. coli, the region immediately following the last transmembrane helix was expressed in WTD731 (Fig. 4). Expression of the C-terminal region in WTD731 restored the growth rate under high osmotic conditions (supplemented with 250 mm NaCl and 250 mm KCl or 600/800 mm mannitol) to the same level as that of the corresponding WT control. Two truncated forms of the C-terminal domain containing either the first or the second half of the C-terminal region did not complement the phenotype of the WTD731 mutant (data not shown). Only the complete C-terminal region rescued the hypersensitive mutation in WTD731.
Figure 4.
Overexpression of the C-terminal region of YbdG in WTD731 complemented the high osmolarity–sensitive phenotype in WTD731. WT and WTD731 containing EV or CD were grown in basal medium containing the indicated concentrations of NaCl/KCl or mannitol at 37 °C (pH 8.6). Error bars, S.D. (n = 3).
Protein–protein interactions with the C-terminal cytosolic region of YbdG
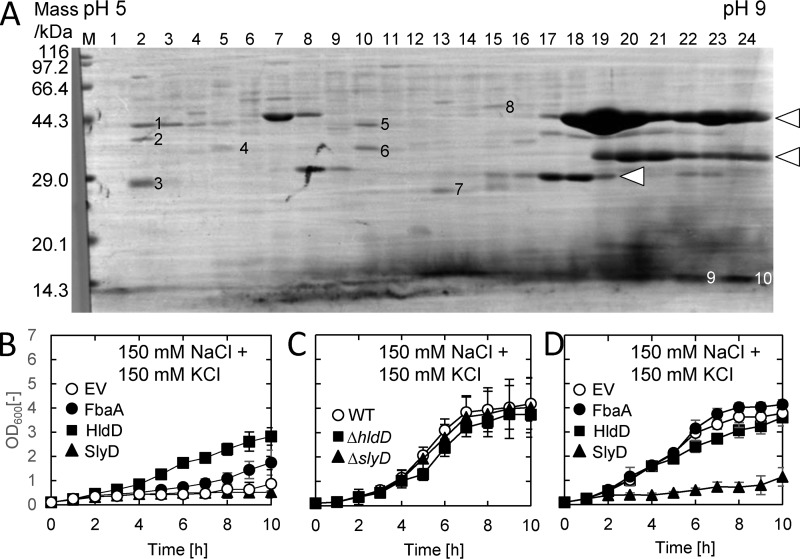
To identify potential protein interactors of the C-terminal cytosolic domain of YbdG, this region was fused to GST and expressed in E. coli. GST pulldown experiments were performed with crude extracts of the transformed E. coli. Pulled-down proteins were separated by two-dimensional electrophoresis, and sequencing of the N-terminal amino acids of 10 different protein bands, numbered 1–10 in Fig. 5A, was performed. The sequences of five of them could not be identified as E. coli proteins (Table 1). The remaining five proteins were identified as FbaA (fructose-bisphosphate aldolase), HldD (ADP-l-glycero-d-mannoheptose 6-epimerase), SlyD (peptidyl-prolyl cis-trans-isomerase), Pseudomonas GST derived from the bait fusion proteins, and E. coli GST, most likely isolated due to its affinity with the GSH beads. To examine whether expression of the corresponding genes would rescue the hyperosmotic stress sensitivity phenotype of WTD731, the fbaA, hldD, and slyD genes were independently introduced into WTD731. Medium containing 250 mm NaCl and KCl did not allow growth of WTD731 expressing FbaA and HldD. However, when we reduced the concentration of NaCl and KCl to 150 mm (Fig. 5, B–D), we observed complementation of the transformed mutants. Expression of hldD partially rescued the growth defect of WTD731 under high osmotic stress, and fbaA also slightly increased growth under these conditions (Fig. 5B). In contrast, expression of slyD did not allow growth of WTD731 in the same medium. To eliminate the possibility of unexpected “side effects” due to the expression of fbaA, slyD, and hldD, the growth of the corresponding deletion mutants as well as the effect of overexpression of fbaA, slyD, and hldD in the WT strain were examined (Fig. 5, C and D). A ΔfbaA strain could not be tested because fbaA is an essential gene (21). Both the ΔhldD mutant and the ΔslyD mutant showed a similar growth rate to the WT under high osmotic conditions (Fig. 5C). Overexpression of either fbaA or hldD had no effect on growth in the WT (Fig. 5D). However, overexpression of slyD led to reduced growth in the WT under high osmotic conditions, which was seen in WTD731 (Fig. 5B). In addition, cell morphology of WTD731 expressing FbaA or HldD was different from the WT containing the empty vector (Fig. S6A), with cells remaining attached to each other. This phenotype was also different from that of WTD731 expressing YbdG, which increased the number of bent cells (Fig. S6B).
Figure 5.
Complementation of the WTD731 phenotype by FbaA and HldD. A, two-dimensional electrophoresis of proteins co-precipitated with the YbdG cytoplasmic domain fused to GST. Proteins were separated by pI-based electrophoresis using linear gradient gel (horizontal axis) followed by SDS-PAGE (vertical axis). Proteins were transferred to PVDF membrane and stained with Coomassie Brilliant Blue. The numbered protein bands were subjected to amino acid sequencing analysis. Protein bands indicated by arrowheads cross-reacted with the anti-GST antibodies (data not shown), and they most likely correspond to GST-fused YbdG itself. B, WTD731 cells expressing fbaA, hldD, or slyD were grown in basal media containing 150 mm NaCl plus 150 mm KCl at 37 °C (pH 8.6). C, the deletion strains ΔhldD and ΔslyD were grown under the same conditions as the strains in B. ΔfbaA was not tested because fbaA is an essential gene. D, WT strains expressing fbaA, hldD, or slyD were grown under the same conditions as cells in B. Error bars, S.D. (n = 3).
Table 1.
List of proteins identified by pulldown assay with the C-terminal region of YbdG
| Mass | Amino acid sequence | Protein | Gene | |
|---|---|---|---|---|
| kDa | ||||
| 1 | IVQ | MPILGYWKIKGLVQPSW | GST | Fusionsa |
| 2 | 34.1 | MIIVTGGAGFIGSNI | ADP-l-glycero-d-manno-heptose-6-epimerase | hldD |
| 3 | 21.7 | MKVADLVVSLAYYVQb | FKBP-type peptidyl-prolyl cis-trans-isomerase | slyD |
| 4 | AEITAVL | NDc | ||
| 5 | 39.6 | SKIFDFVKPGVITGD | Fructose-bisphosphate aldolase class 2 | fbaA |
| 6 | APVDNTK | ND | ||
| 7 | MPLFYKPGAVSLASP | GST | gstAd | |
| 8 | MLG | ND | ||
| 9 | XXXXFLVM | ND | ||
| 10 | XXXXLXDVIAEFA | ND |
a Pseudomonas GST in bait fusion proteins.
b One lysine residue is missing in the sequence of SlyD: MKVAKDLVVSLAYYVQ.
c ND, not determined.
d E. coli endogenous GST (number 7) was isolated due to its affinity to glutathione beads.
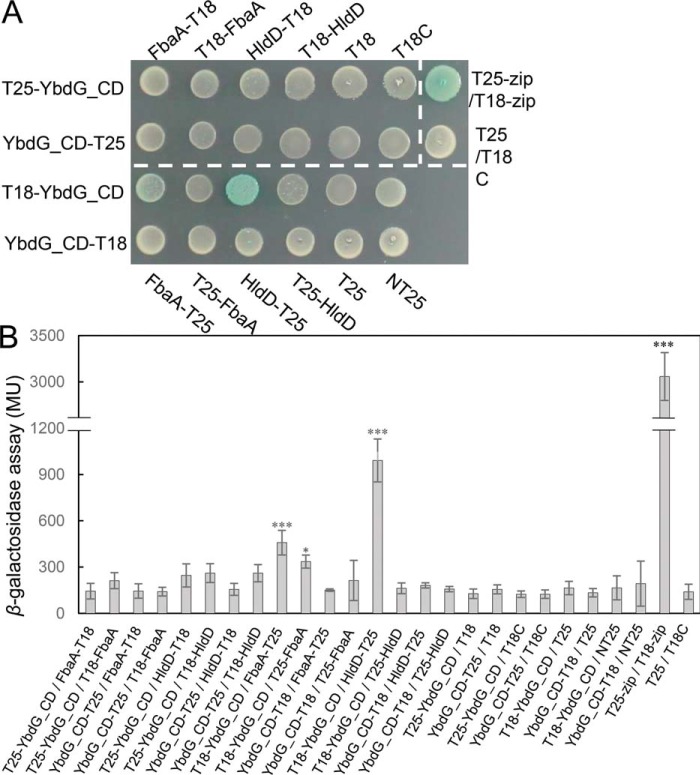
Next, we evaluated in vivo protein–protein interactions between YbdG and FbaA or HldD using a bacterial two-hybrid (BACTH) assay (29) (Fig. 6). Due to the toxicity of expression of slyD in the cells, SlyD was not used in further experiments. An E. coli strain co-expressing T18-YbdG_CD and FbaA-T25 or HldD-T25 yielded light blue colonies, indicating direct interaction between these proteins (Fig. 6A). Quantification of the β-gal activity confirmed protein–protein interactions between FbaA or HldD and YbdG (Fig. 6B). These results implied that the long C-terminal cytosolic region of YbdG interacted with intracellular proteins like FbaA and HldD and that YbdG might participate in the process of osmotic regulation in concert with these molecules.
Figure 6.
In vivo protein–protein interactions between the C-terminal cytoplasmic domain of YbdG and HldD or FbaA. A, blue colonies indicate protein–protein interactions between YbdG and FbaA or HldD. Cells containing YbdG together with pKT25-zip or pUT18-zip were used as a positive control. White colonies indicate that no interaction was detected, and cotransformation of YbdG with pKT25 and pUT18 was used a negative control. Transformed E. coli was incubated at 30 °C for 48 h. The experiments were repeated three times, and a representative plate is shown. B, quantification of β-gal activity of the same strains tested in A. Error bars, S.D. (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001, one-way analysis of variance followed by Dunnett's multiple-comparison test. MU, Miller units.
Discussion
Mechanosensitive channels mediate the efflux of intracellular solutes to prevent cell rupture during rapid decreases in external osmolarity (8, 9). E. coli contains as least seven Ms channel homologous channels, MscL, MscS, MscK, YjeP, YbiO, YnaI, and YbdG. Among them, YbdG alone has not shown its ion channel activity in electrophysiological measurement (17, 18). In particular, the channel property of YbdG remains elusive after the reports on the identification of YjeP as a major component of MscM (18). In accordance with this, no flux of ions or solutes mediated by WT YbdG or YbdG-I167T was detected (Fig. 3 (D and E) and Fig. S5) (17, 18, 30). Absence of channel activity of YbdG has hampered earlier studies of YbdG function. In this study, performing a negative selection screen led to the identification of a gain-of-function mutation in ybdG that decreased tolerance to hyperosmotic shock. Characterization of this mutant shed light on the physiological role of YbdG in osmoadaptation because the null mutants, WTΔybdG and WTD731ΔybdG, did not show the same hypersensitive phenotype as WTD731 (Fig. 1E). The hypersensitive phenotype in high osmotic medium and the slow growth of WTD731 in the presence of 500 mm potassium glutamate at lower temperatures (Fig. 1, D and E) confirmed that YbdG function was required for cell growth after osmotic up-shock. An osmotic rather than an ionic effect was likely the main cause of the slow growth of WTD731. These results strongly suggested that YbdG plays a crucial role in cellular protection against high osmotic pressure in E. coli. This property of YbdG was further supported by the previous finding that expression of ybdG increased in response to the addition of 500 mm NaCl to the media (17).
Microscopic observations revealed that WT E. coli that underwent osmotic stress took on a bent shape and then continued to grow (Fig. 2). In contrast, these morphological changes were not seen in the WTD731 mutants. Similarly, in Arabidopsis thaliana, two plastid envelope membrane-localized MscS-like proteins, MSL2 and MSL3, influence the size and shape of the plastids (23). They also regulate Z-ring formation in chloroplasts as components of the chloroplast division machinery (31). A bent cell shape is the consequence of increased tension caused by the osmotic pressure (32). Cell shape is determined by the cytoskeleton and the cell wall consisting of peptidoglycan. It has been reported that the eukaryotic actin homolog MreB, which is specifically localized at curved areas of the cell, is a primary determinant for cell shape and proliferation (33). MreB communicates with peptidoglycan synthetic enzymes via the inner-membrane intrinsic proteins, MreC/D and RodZ in E. coli. An abnormal interaction of YbdG with proteins that are part of the cytoskeleton or with peptidoglycan-forming proteins might be the reason for the observed growth defect of WTD731 in high osmotic medium (Fig. 2, A and B). Increased cell curvature may be advantageous for cell proliferation under the mechanical stress conditions (33–35). The drastic change in cell shape observed in the WT may activate cytoskeleton rearrangements and initiate recovery of the normal cell shape by promoting cell wall synthesis for cell growth (35, 36).
Substantial molecular dynamics are involved in the transition from open to closed states in MscL and MscS (11, 13, 37). The C-terminal regions are pulled up toward the membrane in the open state (38). Our model of YbdG-I167T (Fig. S4) predicted that the I167T change increased the stability of the open state of YbdG due to an increase in the hydrophobicity of the space surrounding position 167. However, contrary to the prediction, YbdG-I67T did not show higher transport activity (Fig. 3 (D and E) and Fig. S6). Moreover, hypersensitivity to osmotic up-shock was observed only in WTD731 and not the ybdG knockout strains, WTΔybdG and WTD731ΔybdG (Fig. 1). This strongly suggested that the channel activity of YbdG and YbdG-I167T itself was not responsible for the WTD731 phenotype.
In contrast to the large conducting MscL, MscS and YbdG have relatively long cytosolic C-terminal regions following the transmembrane region (17). YbdG has a relatively long cytosolic C-terminal region compared with that of MscS due to an inserted sequence in the region (Fig. S2). The homology between the C-terminal regions of YbdG and MscS is low. The presence of different features in the C-terminal regions of Msc homologs may indicate that they also have different physiological functions. Ion channels interact with a variety of signaling and scaffolding proteins (39). Given that mechanosensitive channels are part of the osmoadaptation signaling network, YbdG is likely to be associated with other proteins. The C-terminal region of the MscS homolog from the cyanobacterium Synechocystis sp. PCC 6803, PamA, interacts with the nitrogen regulatory protein PII (40). Similarly, several proteins that co-precipitate with the cytosolic C-terminal region of MscS were identified in E. coli (15). One of these interacting proteins was FtsZ, a key component of the Z-ring, an important structure in cell division in bacteria. FtsZ is the bacterial homolog of tubulin, a major component of the eukaryotic cytoskeleton, and it has been proposed that binding of FtsZ to the C-terminal domain of inactive MscS may provide a connection to the cell wall–remodeling complex. MscS therefore clearly has multiple tasks not only in ion conduction but also in the modulation of FtsZ-dependent cell division and in the process of cell wall synthesis.
Biochemical and electrophysiological investigation of MscS also showed that the flexible large C-terminal region is essential for the regulation of channel activity and its stability (14, 41, 42). The sensitivity of WTD731 to high osmolarity could be complemented by introduction of the C-terminal region of YbdG into WTD731. This supported the interpretation that the point mutation in YbdG-I167T interrupted or adversely affected a physical connection with components of the osmotic signaling pathway and that the proper connection could be restored by reintroduction of the WT C-terminal region (Fig. 4). The complementation of the mutation of WTD731 by overexpression of hldD and fbaA indicated that recovery of a direct interaction between YbdG and HldD or FbaA was required for osmoadaptation (Figs. 5 and 6). HldD is a key enzyme in the lipopolysaccharide core biosynthesis pathway, which is closely connected to outer-membrane biogenesis (43). The association of YbdG with HldD might be involved in maintaining cell wall integrity under various conditions. FbaA is also an essential component for cell growth in E. coli. Therefore, an altered interaction with YbdG-I167T might impair the function of FbaA in WTD731. All evidence suggests that YbdG belongs to a mechanosensing system responsible for signal transmission to intracellular factors to cope with rapid changes in external osmotic pressure. However, YbdG was unlikely a main component for hyperosmotic stress response because the inactivation of ybdG (WTΔybdG and WTD731ΔybdG) did not affect the cell growth in the medium containing 250 mm NaCl and 250 mm KCl at pH 8.6 (Fig. 1). This is similar to the results for the minor contribution of YbdG to hypo-osmotic shock (17). At alkaline pH, the function of a major transport system relevant to osmoregulation like Na+/H+ antiporters declines (20). Therefore, aberrant behavior of YbdG-I167T might cause an enhanced perturbation of components involved in the osmoadaptation pathway during osmotic up-shock at pH 8.6 (Fig. 1).
Lack of significant channel property of YbdG is reminiscent of the silent potassium channel AtKC1 in Arabidopsis thaliana (Fig. 3 (D and E) and Fig. S6). Tetrameric assembly of AtKC1 with other active K+ channels leads to modulate K+ flux across the membrane (44). The overall structure of YbdG resembles that of MscS except for an additional stretch of sequence in the N-terminal region of YbdG and an insertion in the middle of the C-terminal region (Figs. S2 and S4), and YbdG, YbiO, YnaI, YjeP, and MscS form heptamers (12, 17, 18, 42). YbdG might function as a modulator for other dominant MS channels via heteromerization to suppress MS channel activity. In this case, if YbdG were able to form heteromers with other ion channels like MscS, incorporation of the aberrant form YbdG-I167T might disturb the proper function of MS channels in E. coli, which would consequently adversely affect the growth of WTD731 under high-osmolarity conditions (Fig. 1, B–E).
In this study, due to this weak nature, it was necessary to investigate in limited experimental conditions to elucidate the function and role of YbdG. Given that there is no loss of function phenotype due to ybdG mutation, the WTD731 conferring a gain-of-function phenotype at alkaline pH in this study provided the only clue for investigation of the role of YbdG in E. coli. The isolation of WTD731 demonstrated that YbdG, a member of the Ms channel family, participates in the physiological protection against not only hypoosmotic shock but also hyperosmotic stress, functioning as a mediator and/or modulator of hyperosmotic stress signaling beyond its possible function as an ion or solute transport system.
Experimental procedures
Bacterial strains, plasmids, and medium
E. coli strains constructed in this study are listed in Table S1. E. coli was grown in LB medium (1% polypeptone, 0.5% yeast extract, 1% NaCl) at 37 °C with aeration at 150 rpm. When necessary, medium were supplemented with 25 μg/ml chloramphenicol or 50 μg/ml ampicillin. For growth tests, E. coli strains were cultured in basal medium containing 100 mm Tricine, 10 mm K2HPO4, 0.5% tryptone, 0.25% yeast extract, 0.5% glucose (pH 8.6 with KOH), and 250 mm NaCl/250 mm KCl, 600 or 800 mm mannitol, or 500 mm potassium glutamate were added as indicated. Disruption of ybdG (ΔybdG) in the genome of E. coli W3110 (WT) and WTD731 was generated by a PCR-based method using a set of primers, YbdG-H1-P1 and YbdG-H1-P2 (Table S2), as described (45). E. coli mutants, ΔhldD and ΔslyD (Keio collection), and the plasmids containing fbaA, hldD, and slyD (ASKA clone) were purchased from the National BioResource Project, Japan (21, 46).
Isolation of mutants sensitive to hyperosmotic stress
E. coli strain W3110 was treated with N-methyl-N′-nitro-N-nitrosoguanidine (300 μg/ml) in LB medium for 1 h at 37 °C. After washing with LB medium, the treated cells were allowed to segregate in LB medium for 2 h at 37 °C. The mutant cells were then enriched by culturing in basal medium containing 250 mm NaCl, 250 mm KCl, and penicillin G (200 units/ml) overnight at 37 °C. After washing with LB medium, the cells were cultured in basal medium overnight at 37 °C. This enrichment procedure was repeated twice. Then colonies that grew on plates of basal medium but not on plates of basal medium containing 250 mm NaCl and 250 mm KCl were selected, and one of these selected colonies was designated WTD731.
Expression of YbdG and YbdG-I167T in E. coli
P1 bacteriophage transduction was performed using the standard protocol (47). To construct the plasmid for expression of YbdG or YbdG-I167T (with a T to C conversion at position 500 from the A of the start codon), DNA fragments were amplified by PCR with a set of primers, YbdG_BamHI_FP and YbdG_SalI_RP (Table S2) using chromosomal DNA of E. coli W3110 or WTD731 as template. The amplified PCR fragments were digested with BamHI/SalI and ligated into corresponding sites of pPAB404. The resulting plasmids were introduced into E. coli LB2003, a strain lacking the K+-uptake activity of Trk, Kup, and Kdp (Table S1). Growth tests at different K+ concentrations were carried out as described previously (26).
Expression of the cytoplasmic domain (CD) of YbdG
For the construction of the expression plasmid of GSH S-transferase (GST) fused to the CD of YbdG (corresponding to the amino acid residues at positions 254–415), the domain was amplified by PCR using E. coli chromosomal DNA as a template with primers YbdG_CD_F and YbdG_CD_R (Table S2). The PCR fragment was digested with BamHI/EcoRI and ligated into corresponding sites of pGEX-2T.
SOLiD DNA sequencing
Total genomic DNA was isolated from E. coli W3110 or WTD731 during exponential growth, as described (31). Sequencing runs were performed using cycled ligation sequencing on a SOLiD analyzer (Life Technologies, Inc.). 10–20 μg of purified genomic DNA of strain WTD731 and its parental strain W3110 were sheared into ∼125-bp fragments with the Covaris S2 system (Covaris, Woburn, MA). The sheared DNA fragments were blunted using NEB Next NR enzyme (New England Biolabs, Beverly, MA) and then purified using the Pure Link PCR purification kit (Invitrogen, Life Technologies). The purified DNA fragments were ligated to short SOLiD P1 and P2 adapters by T4 ligase (Invitrogen, Life Technologies). Adapter-ligated DNA was purified using the Pure Link PCR purification kit. These DNA fragments were nick-translated, amplified by PCR, and then purified using the Pure Link PCR purification kit. The purified PCR products were separated by SOLiD Size Selection gel (2% agarose gel; Invitrogen, Life Technologies), and fragments corresponding to 150–200 bp of DNA were collected. These DNA fragment libraries were quantified by quantitative PCR using a SOLiD TaqMan probe. In preparation for sequencing, the DNA fragments were clonally amplified by emulsion PCR using beads with P1 primer covalently attached to the surface. Emulsions were broken with butanol, and emulsion PCR beads were enriched by hybridization with P2-coated capture beads (Life Technologies). dTs were added at the 3′-end of DNA molecules on the beads in the presence of terminal transferase to connect to the slide glass. About 60 million beads were deposited onto one-fourth of a glass surface of a 25 × 75-mm SOLiD slide. The slide was loaded onto a SOLiD instrument, and 50-base sequences were obtained according to the manufacturer's protocol.
SNV analysis
DNA sequence data (csfasta) were analyzed using CLC Genomics workbench (Qiagen, Venlo, Netherlands). Sequence reads of two strains were mapped to the genome sequence of E. coli strain W3110 obtained from KEGG (http://www.genome.jp/kegg/),3 and SNVs were called using the default parameters.
Cell curvature analysis
Cells were grown to stationary phase in basal medium at 37 °C with aeration at 200 rpm. Then cells were diluted 50 times into basal medium without or with 250 mm NaCl and 250 mm KCl. After a 3-h cultivation at 37 °C, cells were sampled and placed on 1% agarose pads containing the growth medium (basal medium without or with 250 mm NaCl and 250 mm KCl). Phase-contrast images were taken on an Eclipse E600 microscope (Nikon, Tokyo) equipped with a phase-contrast objective CFI PlanApo DM ×100/1.40 numerical aperture (Nikon, Tokyo) and an ORCA-Flash4.0 version 3 camera (Hamamatsu Co., Hamamatsu, Japan). To obtain cell outlines, phase-contrast images were analyzed using Oufti software (48). Cell curvature was analyzed based on the cell outlines using the curvhist function in MicrobeTracker software (49). Box plots in Fig. 2B and Fig. S6A were generated with the boxplot function in MATLAB (MathWorks).
For the complementation assays with WTD731, cells were grown to stationary phase in basal medium containing 25 μg/ml chloramphenicol in test tubes at 37 °C with aeration at 200 rpm. Then cells were diluted 50 times into basal medium containing 0.1 mm isopropyl-β-thiogalactopyranoside (IPTG) and chloramphenicol without or with 150 mm NaCl and 150 mm KCl. Cells were transferred to a 96-well plate and cultured at 37 °C with continuous shaking using a microplate reader (Multiskan FC, Thermo Fisher Scientific). To monitor cell growth, optical density was measured at a wavelength of 620 nm. Early exponential cells (OD < 0.4) were sampled to perform microscopic observations and curvature analyses as described above, except that a Ti2-E microscope (Nikon, Tokyo) was used to take images.
Alkaline phosphatase assay
The experiment was performed essentially as described previously (26). To construct plasmids encoding YbdG-PhoA fusion proteins, PCR-amplified ybdG DNA fragments generated using primers listed in Table S2 were digested with BamHI/SalI and then ligated into the corresponding sites of pPAB404. PhoA fusion constructs were introduced into E. coli strain UT5600 (F− ara-14 leuB6 secA6 lacY1 proC14 tsx-67 Δ(ompT-fepC)266 entA403 trpE38 rfbD1 rpsL109 xyl-5 mtl-1 thi-1), a common host for alkaline phosphatase assays. Cells were grown at 30 °C in LB medium supplemented with 50 μg/ml ampicillin, 0.1 mm IPTG, 1.5% agar, and 40 μg/ml 5-bromo-4-chloro-3-indoylphosphate as a chromogenic indicator. For the measurement of PhoA activity, cells were cultured in LB liquid medium until mid-logarithmic phase, and then 1 mm IPTG was added into the medium. After 3 h, cells were collected by centrifugation, and PhoA activity was measured as described (51).
Immunoblot detection
E. coli UT5600 containing PhoA fusion plasmids was grown in LB medium containing 50 μg/ml ampicillin, 0.1 mm IPTG, and 1.5% agar at 30 °C, harvested, washed in 10 mm Tris-HCl phosphate buffer (pH 8.0), and resuspended in the same buffer. The suspension containing 0.1 mg/ml cells was frozen and thawed, and cells were disrupted by sonication. Cell debris was removed by centrifugation (17,000 × g for 1 min). The supernatant was subjected to ultracentrifugation (100,000 × g) for 30 min, and the resulting pellets were resuspended in 10 mm Tris-HCl buffer (pH 8.0). Protein concentration was determined by the Bradford protein assay (52). Aliquots were subjected to SDS-PAGE, and then proteins were transferred electrophoretically onto polyvinylidene difluoride membrane (PVDF, Immobilon-P, Millipore). Immunodetection was performed according to the protocol for ECL prime Western blotting detection reagent (GE Healthcare) with an anti-PhoA antibody (Rockland) and anti-rabbit IgG horseradish peroxidase–linked antibody (Merck Millipore) and analyzed with a luminescent image analyzer, LAS-3000 (Fujifilm).
Measurement of chloride and glutamate content
E. coli strains were precultured overnight in basal medium at 30 °C. Cells were then inoculated into fresh basal medium at an OD600 = 0.1. When the culture reached OD600 = 1.0, basal medium containing concentrated NaCl, KCl, or potassium glutamate was added to a final concentration of 50 mm NaCl plus 50 mm KCl or 100 mm potassium glutamate (time = 0). Aliquots of cells were collected at the indicated times. After harvest, cells were treated at 100 °C for 10 min for cell lysis and then centrifuged at 17,000 × g for 5 min. Glutamate content in the supernatant was determined with the l-Glutamate Assay Kit II (Yamasa Co., Tokyo Japan). Cl− content was measured using an ion chromatograph (Metrohm, Herisau, Switzerland). Protein concentrations were determined by the Bradford protein assay (52).
Stopped-flow measurements
E. coli cells grown at 30 °C or 37 °C overnight were diluted 1:100 with basal medium and cultured at the same temperature until mid-logarithmic phase. The cultures were then rapidly mixed with an equal volume of hyperosmotic medium containing 500 mm NaCl plus 500 mm KCl or 1 m glutamate in basal medium. Kinetic reactions after hyperosmotic shock were measured by a temperature-controlled stopped-flow apparatus (Unisoku, Co., Ltd., Osaka, Japan) as described previously (53) with some modifications. A 532-nm diode pumped solid-state laser was used to illuminate the flow cell, and the time course of cell volume change was monitored by detecting 90° angle light scattering for 1 s at 1-ms intervals.
Homology modeling of YbdG
The model structures of YbdG in the open and closed state were constructed as follows. First, a BLAST search for YbdG (UniProtKB P0AAT4, 415 aa) against the UniProtKB database was conducted because no template sequence for YbdG was found in the Protein Data Bank (PDB). In this BLAST search, MscMJ (UniProtKB Q57634, 350 aa) was retrieved with an E value of 8 × 10−5 (query cover of 31%). A BLAST search for MscMJ against the PDB database was also conducted, and MscS (PDB code 2OAU, 306 aa) was found with an E value of 2 × 10−23 (query cover of 64%). Then, using these three sequences (YbdG, MscMJ, and MscS), a multiple-sequence alignment was created using ClustalW. In addition, based on the known structural features of open MscS (PDB code 2VV5) (13) and closed MscS (PDB code 2OAU) (54), the alignment of YbdG and MscS was modified manually (Fig. S2) within the helix-loop-helix structure (green) and by removing the ∼50-residue insertion in the C-terminal half of YbdG that is not present in MscS (blue, not modeled) as well as the N- and C-terminal regions (blue) that are not conserved. Finally, homology modeling of the open and closed structures of YbdG (residues 91–271 and 323–409) based on the modified alignment was performed using Discovery Studio version 4.0 (Accelrys). The resultant model structures are shown in Fig. 4. For the above processes, the UniProt, PDB, GenomeNet, and NCBI databases were used for sequences, structures, and ClustalW and BLAST analyses, respectively.
GST pulldown assay
WTΔybdG transformed with the pGEX-2T plasmid encoding the C-terminal cytoplasmic domain of YbdG fused to GST was cultured in LB medium. The overnight culture was diluted 1:100 into fresh basal medium supplemented with 250 mm NaCl/250 mm KCl and incubated until the OD600 = 0.6–0.8. Then 1 mm IPTG was added, and the cells were cultured for an additional 4 h. The cells were harvested by centrifugation, suspended in ice-cold PBS containing a protease inhibitor mixture (Roche), and lysed by sonication. Triton X-100 was added to the suspension to a final concentration of 1%, and the mix was incubated on ice for 10 min. The lysate was subjected to centrifugation at 15,000 rpm for 10 min at 4 °C. The supernatant was incubated with GSH Sepharose 4B beads (GE Healthcare) for 1 h at room temperature. The beads were collected by centrifugation at 6,000 rpm for 1 min. The precipitated beads were washed with ice-cold PBS four times. GST-fused proteins were dissociated from the beads by the addition of buffer comprising 20 mm reduced GSH, 100 mm Tris-HCl (pH 8.0), and 120 mm NaCl.
Two-dimensional electrophoresis and protein sequence analysis
For pI-based protein separation, a 3100 OFFGEL Fractionator (Agilent) and ReadyStrip IPG strip pH 5–8 (Bio-Rad) was used according to the manufacturer's protocol. A 20-μl aliquot of each electrophoresis fraction was subjected to SDS-PAGE on a 10% acrylamide gel. Then proteins were transferred to PVDF membrane (Immobilon-P, Millipore). The PVDF membrane was stained with Coomassie Brilliant Blue G-250 (Wako Co., Tokyo, Japan). Protein extraction and amino acid sequencing by a gas-phase Edman sequencer, PPSQ-53A gradient system (Shimadzu, Kyoto, Japan) was performed as described previously (55). Peptide Match (http://research.bioinformatics.udel.edu/peptidematch/index.jsp)4 (56) was used for peptide sequence identification.
Bacterial two-hybrid assays
fbaA and hldD were amplified by PCR using specific primers listed in Table S2 and cloned into the BamHI sites of the pKT25, pKNT25, pUT18, and pUT18C vectors (29). Fusions of the cytoplasmic C-terminal domain of ybdG (CD) and of fbaA and hldD to the T25 and T18 fragments of adenylate cyclase were expressed in E. coli BTH101. Cells were spotted onto LB plates supplemented with 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), 0.1 mm IPTG, 100 μg/ml ampicillin, and 25 μg/ml kanamycin and grown for 48 h at 30 °C. Cells were cultured in liquid medium for quantitative analysis of β-gal activity.
Author contributions
S. A., H. T., and S. H. performed growth tests and biochemical experiments. H. S. and H. K. isolated the mutants. K. I. determined the sequence of the E. coli genome. M. K., R. K., and S. K. performed microscopic observations. K. K. conducted the stopped-flow measurements. S. A. and T. F. performed the structure modeling. S. H. and N. U. designed the research and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Naomi Hoshi and Kasumi Sakamoto for technical assistance and Anke Reinders for critical reading of the manuscript. Setsu Kato is supported through the HIRAKU consortium under MEXT.
This work was supported by Grants-in-Aid for Scientific Research 16H06558, 19K22264, and 19H02880 (to N. U.) and 15K186780 (to S. H.) from Japan Society for the Promotion of Science (JSPS). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1 and S2 and Figs. S1–S6.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- MS
- mechanosensitive
- MscS
- MS channel of small conductance
- MscL
- MS channel of large conductance
- MscK
- potassium-dependent MS channel
- MscM
- MS channel of miniconductance
- GST
- glutathione S-transferase
- CD
- cytoplasmic domain
- IPTG
- isopropyl-β-thiogalactopyranoside
- PVDF
- polyvinylidene difluoride membrane
- EV
- empty vector
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- LB
- lysogeny broth
- OD
- optical density
- aa
- amino acids
- PDB
- Protein Data Bank.
References
- 1. Wood J. M. (2015) Bacterial responses to osmotic challenges. J. Gen. Physiol. 145, 381–388 10.1085/jgp.201411296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Reed R. H., Warr S. R. C., Richardson D. L., Moore D. J., and Stewart W. D. P. (1985) Multiphasic osmotic adjustment in a euryhaline cyanobacterium. FEMS Microbiol. Lett. 28, 225–229 10.1111/j.1574-6968.1985.tb00796.x [DOI] [Google Scholar]
- 3. Nanatani K., Shijuku T., Takano Y., Zulkifli L., Yamazaki T., Tominaga A., Souma S., Onai K., Morishita M., Ishiura M., Hagemann M., Suzuki I., Maruyama H., Arai F., and Uozumi N. (2015) Comparative analysis of kdp and ktr mutants reveals distinct roles of the potassium transporters in the model cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 197, 676–687 10.1128/JB.02276-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dinnbier U., Limpinsel E., Schmid R., and Bakker E. P. (1988) Transient accumulation of potassium glutamate and its replacement by trehalose during adaptation of growing cells of Escherichia coli K-12 to elevated sodium chloride concentrations. Arch. Microbiol. 150, 348–357 10.1007/BF00408306 [DOI] [PubMed] [Google Scholar]
- 5. Marin K., Zuther E., Kerstan T., Kunert A., and Hagemann M. (1998) The ggpS gene from Synechocystis sp. strain PCC 6803 encoding glucosyl-glycerol-phosphate synthase is involved in osmolyte synthesis. J. Bacteriol. 180, 4843–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Whatmore A. M., Chudek J. A., and Reed R. H. (1990) The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J. Gen. Microbiol. 136, 2527–2535 10.1099/00221287-136-12-2527 [DOI] [PubMed] [Google Scholar]
- 7. Mizuno T., and Mizushima S. (1987) Isolation regulatory proteins and characterization genes OmpC for expression and OmpF of deletion mutants of ompR and envZ, of the outer membrane in Escherichia coli. J. Biochem. 101, 387–396 10.1093/oxfordjournals.jbchem.a121923 [DOI] [PubMed] [Google Scholar]
- 8. Martinac B., Buechner M., Delcour A. H., Adler J., and Kung C. (1987) Pressure-sensitive ion channel in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 84, 2297–2301 10.1073/pnas.84.8.2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sukharev S. I., Martinac B., Arshavsky V. Y., and Kung C. (1993) Two types of mechanosensitive channels in the Escherichia coli cell envelope: solubilization and functional reconstitution. Biophys. J. 65, 177–183 10.1016/S0006-3495(93)81044-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Record M. T. Jr., Courtenay E. S., Cayley D. S., and Guttman H. J. (1998) Responses of E. coli to osmotic stress: large changes in amounts of cytoplasmic solutes and water. Trends Biochem. Sci. 23, 143–148 10.1016/S0968-0004(98)01196-7 [DOI] [PubMed] [Google Scholar]
- 11. Sukharev S., Betanzos M., Chiang C. S., and Guy H. R. (2001) The gating mechanism of the large mechanosensitive channel MscL. Nature 409, 720–724 10.1038/35055559 [DOI] [PubMed] [Google Scholar]
- 12. Bass R. B., Strop P., Barclay M., and Rees D. C. (2002) Crystal structure of Escherichia coli MscS, a voltage-modulated and mechanosensitive channel. Science 298, 1582–1587 10.1126/science.1077945 [DOI] [PubMed] [Google Scholar]
- 13. Wang W., Black S. S., Edwards M. D., Miller S., Morrison E. L., Bartlett W., Dong C., Naismith J. H., and Booth I. R. (2008) The structure of an open form of an E. coli mechanosensitive channel at 3.45 Å resolution. Science 321, 1179–1183 10.1126/science.1159262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koprowski P., and Kubalski A. (2003) C termini of the Escherichia coli mechanosensitive ion channel (MscS) move apart upon the channel opening. J. Biol. Chem. 278, 11237–11245 10.1074/jbc.M212073200 [DOI] [PubMed] [Google Scholar]
- 15. Koprowski P., Grajkowski W., Balcerzak M., Filipiuk I., Fabczak H., and Kubalski A. (2015) Cytoplasmic domain of MscS interacts with cell division protein FtsZ: a possible non-channel function of the mechanosensitive channel in Escherichia coli. PLoS One 10, e0127029 10.1371/journal.pone.0127029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Y., Moe P. C., Chandrasekaran S., Booth I. R., and Blount P. (2002) Ionic regulation of MscK, a mechanosensitive channel from Escherichia coli. EMBO J. 21, 5323–5330 10.1093/emboj/cdf537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schumann U., Edwards M. D., Rasmussen T., Bartlett W., van West P., and Booth I. R. (2010) YbdG in Escherichia coli is a threshold-setting mechanosensitive channel with MscM activity. Proc. Natl. Acad. Sci. U.S.A. 107, 12664–12669 10.1073/pnas.1001405107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edwards M. D., Black S., Rasmussen T., Rasmussen A., Neil R., Stephen T., Miller S., Booth I. R., Edwards M. D., Black S., Rasmussen T., Rasmussen A., Stokes N. R., Stephen T. L., Miller S., et al. (2012) Characterization of three novel mechanosensitive channel activities in Escherichia coli. Channels (Austin) 6, 272–281 10.4161/chan.20998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ohyama T., Igarashi K., and Kobayashi H. (1994) Physiological role of the chaA gene in sodium and calcium circulations at a high pH in Escherichia coli. J. Bacteriol. 176, 4311–4315 10.1128/jb.176.14.4311-4315.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kobayashi H., Saito H., and Kakegawa T. (2000) Bacterial strategies to inhabit acidic environments. J. Gen. Appl. Microbiol. 46, 235–243 10.2323/jgam.46.235 [DOI] [PubMed] [Google Scholar]
- 21. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., and Mori H. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saka K., Tadenuma M., Nakade S., Tanaka N., Sugawara H., Nishikawa K., Ichiyoshi N., Kitagawa M., Mori H., Ogasawara N., and Nishimura A. (2005) A complete set of Escherichia coli open reading frames in mobile plasmids facilitating genetic studies. DNA Res. 12, 63–68 10.1093/dnares/12.1.63 [DOI] [PubMed] [Google Scholar]
- 23. Haswell E. S., and Meyerowitz E. M. (2006) MscS-like proteins control plastid size and shape in Arabidopsis thaliana. Curr. Biol. 16, 1–11 10.1016/j.cub.2005.11.044 [DOI] [PubMed] [Google Scholar]
- 24. Nanatani K., Shijuku T., Akai M., Yukutake Y., Yasui M., Hamamoto S., Onai K., Morishita M., Ishiura M., and Uozumi N. (2013) Characterization of the role of a mechanosensitive channel in osmotic down shock adaptation in Synechocystis sp PCC 6803. Channels 7, 238–242 10.4161/chan.25350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Manoil C., and Beckwith J. (1986) A genetic approach to analyzing membrane topology. Science 233, 1403–1408 10.1126/science.3529391 [DOI] [PubMed] [Google Scholar]
- 26. Uozumi N., Nakamura T., Schroeder J. I., and Muto S. (1998) Determination of transmembrane topology of an inward-rectifying potassium channel from Arabidopsis thaliana based on functional expression in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 95, 9773–9778 10.1073/pnas.95.17.9773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grajkowski W., Kubalski A., and Koprowski P. (2005) Surface changes of the mechanosensitive channel MscS upon its activation, inactivation, and closing. Biophys. J. 88, 3050–3059 10.1529/biophysj.104.053546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nomura T., Sokabe M., and Yoshimura K. (2008) Interaction between the cytoplasmic and transmembrane domains of the mechanosensitive channel MscS. Biophys. J. 94, 1638–1645 10.1529/biophysj.107.114785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karimova G., Ullmann A., and Ladant D. (2001) Protein-protein interaction between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J. Mol. Microbiol. Biotechnol. 3, 73–82 [PubMed] [Google Scholar]
- 30. Berrier C., Besnard M., Ajouz B., Coulombe A., and Ghazi A. (1996) Multiple mechanosensitive ion channels from Escherichia coli, activated at different thresholds of applied pressure. J. Membr. Biol. 151, 175–187 10.1007/s002329900068 [DOI] [PubMed] [Google Scholar]
- 31. Wilson M. E., Jensen G. S., and Haswell E. S. (2011) Two mechanosensitive channel homologs influence division ring placement in Arabidopsis chloroplasts. Plant Cell. 23, 2939–2949 10.1105/tpc.111.088112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang K. C., Mukhopadhyay R., Wen B., Gitai Z., and Wingreen N. S. (2008) Cell shape and cell-wall organization in Gram-negative bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 19282–19287 10.1073/pnas.0805309105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shi H., Bratton B. P., Gitai Z., and Huang K. C. (2018) How to build a bacterial cell: MreB as the foreman of E. coli construction. Cell 172, 1294–1305 10.1016/j.cell.2018.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bartlett T. M., Bratton B. P., Duvshani A., Miguel A., Sheng Y., Martin N. R., Nguyen J. P., Persat A., Desmarais S. M., VanNieuwenhze M. S., Huang K. C., Zhu J., Shaevitz J. W., and Gitai Z. (2017) A periplasmic polymer curves Vibrio cholerae and promotes pathogenesis. Cell 168, 172–185.e15 10.1016/j.cell.2016.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cabeen M. T., Charbon G., Vollmer W., Born P., Ausmees N., Weibel D. B., and Jacobs-Wagner C. (2009) Bacterial cell curvature through mechanical control of cell growth. EMBO J. 28, 1208–1219 10.1038/emboj.2009.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Amir A., Babaeipour F., McIntosh D. B., Nelson D. R., and Jun S. (2014) Bending forces plastically deform growing bacterial cell walls. Proc. Natl. Acad. Sci. U.S.A. 111, 5778–5783 10.1073/pnas.1317497111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yoshimura K., and Sokabe M. (2010) Mechanosensitivity of ion channels based on protein-lipid interactions. J. R. Soc. Interface 7, S307–S320 10.1098/rsif.2010.0095.focus [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rowe I., Anishkin A., Kamaraju K., Yoshimura K., and Sukharev S. (2014) The cytoplasmic cage domain of the mechanosensitive channel MscS is a sensor of macromolecular crowding. J. Gen. Physiol. 143, 543–557 10.1085/jgp.201311114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee A., Fakler B., Kaczmarek L. K., and Isom L. L. (2014) More than a pore: ion channel signaling complexes. J. Neurosci. 34, 15159–15169 10.1523/JNEUROSCI.3275-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Osanai T., Sato S., Tabata S., and Tanaka K. (2005) Identification of PamA as a PII-binding membrane protein important in nitrogen-related and sugar-catabolic gene expression in Synechocystis sp. PCC 6803. J. Biol. Chem. 280, 34684–34690 10.1074/jbc.M507489200 [DOI] [PubMed] [Google Scholar]
- 41. Miller S., Bartlett W., Chandrasekaran S., Simpson S., Edwards M., and Booth I. R. (2003) Domain organization of the MscS mechanosensitive channel of Escherichia coli. EMBO J. 22, 36–46 10.1093/emboj/cdg011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miller S., Edwards M. D., Ozdemir C., and Booth I. R. (2003) The closed structure of the MscS mechanosensitive channel: cross-linking of single cysteine mutants. J. Biol. Chem. 278, 32246–32250 10.1074/jbc.M303188200 [DOI] [PubMed] [Google Scholar]
- 43. Joloba M. L., Clemmer K. M., Sledjeski D. D., and Rather P. N. (2004) Activation of the gab operon in an RpoS-dependent manner by mutations that truncate the inner core of lipopolysaccharide in Escherichia coli. J. Bacteriol. 186, 8542–8546 10.1128/JB.186.24.8542-8546.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jeanguenin L., Alcon C., Duby G., Boeglin M., Chérel I., Gaillard I., Zimmermann S., Sentenac H., and Véry A. A. (2011) AtKC1 is a general modulator of Arabidopsis inward Shaker channel activity. Plant J. 67, 570–582 10.1111/j.1365-313X.2011.04617.x [DOI] [PubMed] [Google Scholar]
- 45. Datsenko K. A., and Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kitagawa M., Ara T., Arifuzzaman M., Ioka-Nakamichi T., Inamoto E., Toyonaga H., and Mori H. (2005) Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12, 291–299 10.1093/dnares/dsi012 [DOI] [PubMed] [Google Scholar]
- 47. Thomason L. C., Costantino N., and Court D. L. (2007) E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. Chapter 1, Unit 1.17 10.1002/0471142727.mb0117s79 [DOI] [PubMed] [Google Scholar]
- 48. Paintdakhi A., Parry B., Campos M., Irnov I., Elf J., Surovtsev I., and Jacobs-Wagner C. (2016) Oufti: an integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol. Microbiol. 99, 767–777 10.1111/mmi.13264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sliusarenko O., Heinritz J., Emonet T., and Jacobs-Wagner C. (2011) High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 80, 612–627 10.1111/j.1365-2958.2011.07579.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Deleted in proof.
- 51. Brickman E., and Beckwith J. (1975) Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and phi80 transducing phages. J. Mol. Biol. 96, 307–316 10.1016/0022-2836(75)90350-2 [DOI] [PubMed] [Google Scholar]
- 52. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 53. Murata A., Ito Y., Kashima R., Kanbayashi S., Nanatani K., Igarashi C., Okumura M., Inaba K., Tokino T., Takahashi S., and Kamagata K. (2015) One-dimensional sliding of p53 along DNA is accelerated in the presence of Ca2+ or Mg2+ at millimolar concentrations. J. Mol. Biol. 427, 2663–2678 10.1016/j.jmb.2015.06.016 [DOI] [PubMed] [Google Scholar]
- 54. Steinbacher S., Bass R., Strop P., and Rees D. C. (2007) Structures of the prokaryotic mechanosensitive channels MscL and MscS. Curr. Top. Membr. 58, 1–24 10.1016/S1063-5823(06)58001-9 [DOI] [Google Scholar]
- 55. Kera K., Nagayama T., Nanatani K., Saeki-Yamoto C., Tominaga A., Souma S., Miura N., Takeda K., Kayamori S., Ando E., Higashi K., Igarashi K., and Uozumi N. (2018) Reduction of spermidine content resulting from inactivation of two arginine decarboxylases increases biofilm formation in Synechocystis sp. strain PCC 6803. J. Bacteriol. 200, e00664–17 10.1128/JB.00664-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen C., Li Z., Huang H., Suzek B. E., Wu C. H., and UniProt Consortium (2013) A fast peptide match service for UniProt Knowledgebase. Bioinformatics 29, 2808–2809 10.1093/bioinformatics/btt484 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.