Abstract
Protein tyrosine phosphatase, nonreceptor type 2 (PTPN2) is mainly expressed in hematopoietic cells, where it negatively regulates growth factor and cytokine signaling. PTPN2 is an important regulator of hematopoiesis and immune/inflammatory responses, as evidenced by loss-of-function mutations of PTPN2 in leukemia and lymphoma and knockout mice studies. Benzene is an environmental chemical that causes hematological malignancies, and its hematotoxicity arises from its bioactivation in the bone marrow to electrophilic metabolites, notably 1,4-benzoquinone, a major hematotoxic benzene metabolite. Although the molecular bases for benzene-induced leukemia are not well-understood, it has been suggested that benzene metabolites alter topoisomerases II function and thereby significantly contribute to leukemogenesis. However, several studies indicate that benzene and its hematotoxic metabolites may also promote the leukemogenic process by reacting with other targets and pathways. Interestingly, alterations of cell-signaling pathways, such as Janus kinase (JAK)/signal transducer and activator of transcription (STAT), have been proposed to contribute to benzene-induced malignant blood diseases. We show here that 1,4-benzoquinone directly impairs PTPN2 activity. Mechanistic and kinetic experiments with purified human PTPN2 indicated that this impairment results from the irreversible formation (kinact = 645 m−1·s−1) of a covalent 1,4-benzoquinone adduct at the catalytic cysteine residue of the enzyme. Accordingly, cell experiments revealed that 1,4-benzoquinone exposure irreversibly inhibits cellular PTPN2 and concomitantly increases tyrosine phosphorylation of STAT1 and expression of STAT1-regulated genes. Our results provide molecular and cellular evidence that 1,4-benzoquinone covalently modifies key signaling enzymes, implicating it in benzene-induced malignant blood diseases.
Keywords: carcinogenesis, tyrosine-protein phosphatase (tyrosine phosphatase), enzyme inactivation, leukemia, cell signaling, covalent adduct, metabolic activation, benzene, carcinogen, bioactivation, enzyme inhibition, leukemogenesis, cancer, protein tyrosine phosphatase non-receptor type 2 (PTPN2)
Introduction
Benzene is an organic compound of great industrial importance used as a solvent or starting material for the synthesis of numerous chemicals. It is also found in gasoline vapors, motor vehicle exhaust, burning coal and oil, cigarette smoke, and wood-burning fires (1, 2). Benzene has long been recognized as a group 1 carcinogen, and exposure to benzene is now established as a cause of hematological malignancies in humans (1, 3–7). Recent studies indicate that benzene is one of the major carcinogen air toxics that affect public health significantly in the United States (8). To become carcinogenic and cause leukemia, benzene must be metabolized in the bone marrow to electrophilic metabolites, such as 1,4-benzoquinone, which are considered as the ultimate hematotoxic species (1, 4, 9–11), yet the molecular mechanisms by which benzene and its metabolites exert their leukemogenic effects have not been fully elucidated (1, 12–15). Although oxidative DNA damage and chromosome alteration through alteration of topoisomerases II functions by benzene metabolites have been shown as contributors to benzene-induced leukemia, mounting evidence indicates that additional mechanisms are also involved (1, 2, 4, 12, 15, 16). Alteration of critical hematopoietic cell signaling pathways has been recently proposed as a relevant mechanism by which benzene could induce hematotoxicity and leukemia (1, 2, 4, 17). In addition, recent data indicate that chronic inflammation and aberrant immune response are likely to play a role in benzene-induced malignancies (15, 16). Interestingly, global gene expression profiling studies showed that key hematopoietic and immune signaling processes, notably the JAK6/STAT pathway, were altered in individuals occupationally exposed to benzene (4).
Maintenance of cellular homeostasis in the hematopoietic system requires the STAT transcription factors, which are activated through tyrosine phosphorylation by a variety of cytokines and growth factors (18, 19). However, aberrant activation of STAT proteins and subsequent dysregulation of STAT signaling are associated with leukemic transformation and altered immune response (19–21). Protein tyrosine phosphatases (PTPs) play a major role in the down-modulation of STAT signaling in the hematopoietic system through dephosphorylation of JAK and/or STAT proteins (18, 19, 22, 23). PTPN2 (also known as TCPTP, for T cell protein tyrosine phosphatase) is an intracellular tyrosine phosphatase highly expressed in hematopoietic cells, where it serves as a key regulator of hematopoiesis and immune and inflammatory responses (24, 25). PTPN2 deficiency in mice (PTPN2−/PTPN2−) induces severe hematopoietic defects (affecting lymphoid, myeloid, and erythroid lineages) and progressive systemic inflammation leading to death of the knockout mice within weeks (26–29). In humans, deletions or inactivating mutations of PTPN2 were identified in T-cell leukemia and non-Hodgkin's lymphoma and associated with elevated STAT signaling and changes in gene expression (25, 30–32). In addition, loss of PTPN2 may also contribute to resistance of chronic myeloid cells to imatinib through the modulation of PTPN2-dependent signals downstream of the BCR-ABL fusion protein (33). These studies highlight the important role of PTPN2 in hematopoiesis, inflammation, and immune response.
In this work, we show that 1,4-benzoquinone, the prime hematotoxic metabolite of benzene, is an irreversible inhibitor of PTPN2. Kinetic and biochemical analyses using purified human PTPN2 indicated that this irreversible inhibition of the enzyme is mainly due to rapid arylation of its catalytic cysteine by 1,4-benzoquinone. Further studies in cells expressing PTPN2 showed that exposure to 1,4-benzoquinone leads to the irreversible inhibition of the endogenous enzyme with a concomitant overactivation of the STAT1 signaling and subsequent alteration of the expression of STAT1-regulated genes. Altogether, our data provide the first evidence for the alteration of a key hematopoietic and immune signaling pathway by benzene. These findings shed a new light on our understanding of the mechanism by which benzene may induce hematological malignancies.
Results
The hematotoxic benzene metabolite 1,4-benzoquinone impairs the catalytic activity of PTPN2
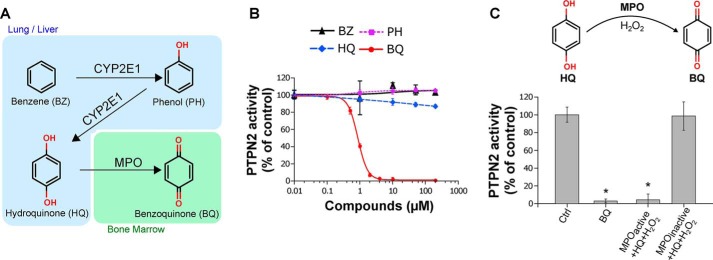
The leukemogenic properties of benzene are known to rely on its bioactivation in the bone marrow into reactive metabolites, in particular 1,4-benzoquinone (1, 4, 14). This hematotoxic quinone arises in the bone marrow mainly from myeloperoxidase-catalyzed conversions of phenolic metabolites of benzene originating from the liver, mainly phenol (PH) and hydroquinone (HQ) (Fig. 1A) (2, 34, 35). As shown in Fig. 1B, we found that, contrary to the phenolic metabolites of benzene, 1,4-benzoquinone inhibited PTPN2 phosphatase activity (IC50 = 1 μm). Whereas full inhibition of the enzyme was observed with low micromolar concentrations of 1,4-benzoquinone (2 μm), no significant effects were observed with phenol and hydroquinone even at concentrations 2 orders of magnitude higher (200 μm). To confirm that 1,4-benzoquinone inhibits PTPN2, an in vitro myeloperoxidase (MPO) activation system that mimics the bioactivation of hydroquinone into 1,4-benzoquinone in the bone marrow was used as described previously (9, 34). In the presence of a functional MPO activation system (MPO, hydroquinone, and H2O2) able to convert hydroquinone into 1,4-benzoquinone, PTPN2 activity was fully inhibited (Fig. 1C). Conversely, in reactions containing inactive MPO, no inhibition of PTPN2 was found (Fig. 1C), thus confirming that this tyrosine phosphatase is sensitive to 1,4-benzoquinone but not the phenolic metabolites of benzene.
Figure 1.
Effect of benzene and its metabolites on human PTPN2 activity. A, schematic representation of the benzene bioactivation pathway (1). B, human PTPN2 was incubated with increasing concentrations of benzene (BZ), phenol (PH), hydroquinone (HQ), or 1,4-benzoquinone (BQ) (0–200 μm) for 20 min at 37 °C, and residual PTPN2 activity was measured using a pNPP assay. Error bars, S.D. values. C, human PTPN2 was incubated with or without BQ (10 μm) or HQ in the presence of an active or inactive MPO hydroquinone-bioactivation system (including H2O2) for 20 min at 37 °C prior to residual PTPN2 activity assay. Error bars, S.D. *, p < 0.05 compared with control (Ctrl).
PTPN2 is irreversibly inhibited by 1,4-benzoquinone through rapid covalent adduction with its catalytic cysteine
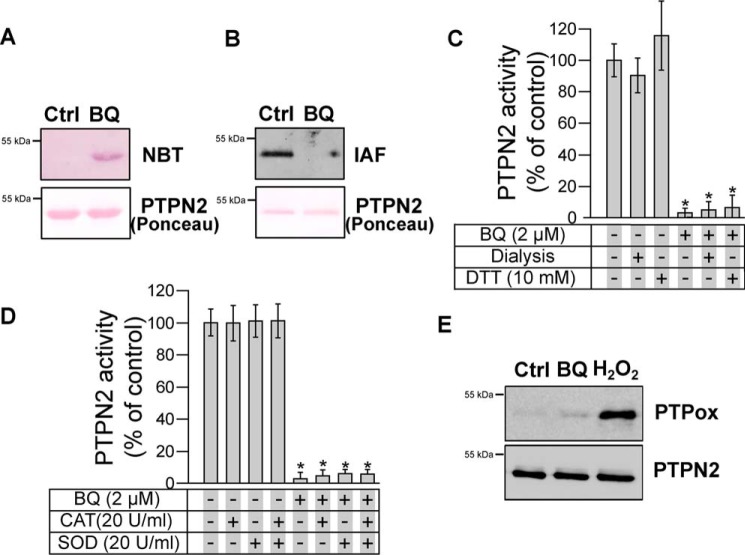
1,4-Benzoquinone is known to arylate directly protein thiol groups through Michael addition (35–37). This arylating ability of 1,4-benzoquinone is considered as a major mechanism responsible for the toxicological effects of this quinone, in particular in the bone marrow (1, 2, 38, 39). Exposure of purified PTPN2 to 1,4-benzoquinone led to the covalent arylation of the enzyme as evidenced by the visualization of protein–quinone adducts on nitrocellulose membranes using the quinone-specific nitro blue tetrazolium (NBT) staining (40) (Fig. 2A). Moreover, the covalent binding of 1,4-benzoquinone to PTPN2 was accompanied by the concomitant loss of cysteine labeling by fluorescein-iodoacetamide (IAF), thus supporting that 1,4-benzoquinone arylation occurs on cysteine residues of PTPN2 (Fig. 2B). As shown in Fig. 2C, the activity of 1,4-benzoquinone–inhibited PTPN2 could not be recovered by dialysis nor by DTT treatment, further supporting the irreversible covalent nature of the reaction between 1,4-benzoquinone and PTPN2 cysteine residues. As shown in Fig. S1, we found that inhibition of PTPN2 by 1,4-benzoquinone was inhibited by GSH, a major physiological reducing agent known to react with 1,4-benzoquinone (35). To further ascertain that inhibition of PTPN2 relies on the arylating ability of 1,4-benzoquinone but not on redox cycling mechanisms (that generate reactive oxygen species), the inhibition assays were carried out in the presence of catalase and/or superoxide dismutase as described previously (41–43). As shown in Fig. 2D, no protection from 1,4-benzoquinone–dependent inhibition was afforded by superoxide dismutase or catalase enzymes, thus confirming that inhibition of PTPN2 by 1,4-benzoquinone is due to its arylating properties (35, 37, 44). Western blot analyses with an antibody that detects specifically oxidized forms of the catalytic cysteine of PTPN2 (45, 46) further support that 1,4-benzoquinone does not inhibit the enzyme through redox cycling and oxidation of the catalytic cysteine residue (Fig. 2E).
Figure 2.
Arylation of PTPN2 cysteine residues by BQ. A, human PTPN2 was incubated with BQ (2 μm) for 20 min. Samples were separated by SDS-PAGE and transferred onto nitrocellulose membrane. Quinone adducts were detected by NBT staining. Ponceau red staining is shown as a loading control. B, human PTPN2 was incubated with BQ (2 μm) for 20 min at 37 °C and then incubated with 10 μm IAF for 10 min. Samples were separated by SDS-PAGE and transferred onto nitrocellulose membrane. IAF adducts were detected by fluorescence. Ponceau red staining is shown as a loading control. C, human PTPN2 was incubated with BQ (2 μm) for 20 min at 37 °C. Samples were either dialyzed overnight at 4 °C or incubated with DTT for 30 min at 37 °C. Residual PTPN2 activity was measured using a pNPP assay. Error bars, S.D. values. *, p < 0.05 compared with controls. D, human PTPN2 was incubated with BQ (2 μm) for 20 min at 37 °C in the presence or absence of catalase (20 units/ml) or superoxide dismutase (20 units/ml), and residual PTPN2 activity was measured. Error bars, S.D. values. *, p < 0.05 compared with controls. E, human PTPN2 was incubated with BQ (2 μm) or H2O2 (100 μm) for 20 min. Samples were subjected to immunoblotting analysis using anti-oxidized PTP active site (PTPox) and anti-PTPN2 antibody.
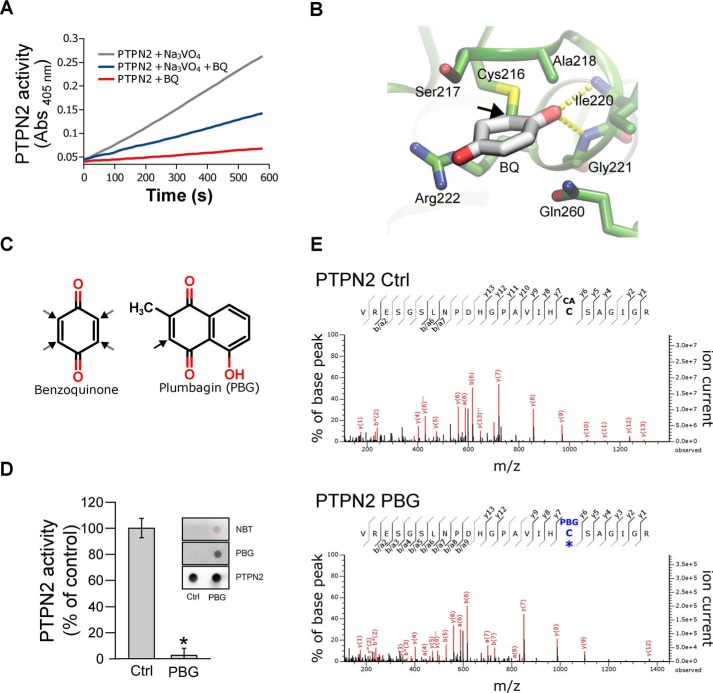
Taking into account the data reported above, we next carried out experiments to test whether 1,4-benzoquinone–dependent inhibition of PTPN2 was due to covalent binding of the quinone to the catalytic cysteine residue of the enzyme. To this end, we conducted active-site protection assays using orthovanadate, a reversible inhibitor of protein tyrosine phosphatases, as described previously (43). As shown in Fig. 3A, the inhibition of PTPN2 by 1,4-benzoquinone was slowed down by the addition of the competitive PTPN2 inhibitor orthovanadate, thereby indicating that 1,4-benzoquinone reacts with the active site of the enzyme. Computational covalent docking approaches further indicated that 1,4-benzoquinone could react at the active site of PTPN2 and form a covalent adduct with the catalytic cysteine of the enzyme (Cys-216) (Fig. 3B).
Figure 3.
Irreversible inhibition of PTPN2 by BQ occurs through modification of active-site cysteine. A, recombinant PTPN2 was incubated with BQ (2 μm) for 20 min in the presence or absence of 1 mm orthovanadate (Na3VO4). Progress curves for residual PTPN2 activity were obtained by monitoring pNPP hydrolysis over time (absorbance at 405 nm). B, molecular docking model of BQ bound to human PTPN2 protein structure. BQ adduct atom coordinates were placed inside the PTPN2 active-site pocket as described under “Materials and methods.” BQ is displayed covalently bonded to the Cys-216 residue. C, chemical structures of BQ and PBG. Black arrows, potential Michael adduction sites. D, recombinant PTPN2 was incubated with PBG (50 μm) for 20 min at 37 °C. Residual PTPN2 activity was measured using a pNPP assay. Error bars, S.D. values. *, p < 0.05 compared with control (Ctrl). NBT staining and immunoblotting analysis (using anti-PBG and anti-PTPN2 antibodies) of treated and nontreated samples are shown. E, recombinant PTPN2 was incubated with PBG (100 μm) for 20 min and then incubated with iodoacetamide for 60 min. Samples were separated by SDS-PAGE and analyzed by MS. Mass spectra derived from iodoacetamide or PBG-modified PTPN2 active-site cysteine (Cys-216) peptides are depicted.
To ascertain that the active-site cysteine of PTPN2 reacts covalently with 1,4-benzoquinone, MS analysis was carried out. However, no individual amino acid residues were identified in these experiments. Similar technical difficulty was reported by Bender et al. (47) for the identification of 1,4-benzoquinone adducts on topoisomerase IIα. This is likely to be due to the ability of 1,4-benzoquinone to cross-link multiple residues in the protein (47). To overcome this technical difficulty, we followed the approach used by Bender et al. (47) to identify covalent adducts of quinones in topoisomerase IIa cysteine residues. These authors used plumbagin (PBG; a naphthoquinone that has only a single reactive site) as a surrogate of 1,4-benzoquinone (Fig. 3C) (47). Interestingly, certain naphthoquinones have been described as protein tyrosine phosphatase inhibitors (48). In addition, as shown in Fig. 3D, PBG was found to be able to covalently bind to PTPN2 and to inhibit the enzyme. Mass spectrometry experiments were thus carried out using PTPN2 treated or not with PBG. Free cysteine residues were alkylated with iodoacetamide prior to trypsin digestion and MS. As shown in Fig. 3E, for the PBG-inhibited PTPN2, LC-MS/MS analysis of tryptic digests showed an increase in mass of 130.02 Da of ion peptides containing the catalytic cysteine of PTPN2 (y7, y9, and y12 peptide ions). These data confirm the formation of a PBG adduct at the catalytic cysteine (note that 130.02 Da corresponds to the difference between the monoisotopic masses of the PBG adduct and the iodoacetamide-driven carboxyamidomethyl modification, which are 187.04 and 57.02 Da, respectively).
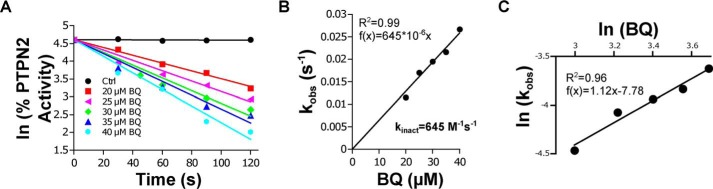
Kinetic analyses of PTPN2 inhibition by 1,4-benzoquinone were carried out under pseudo-first-order conditions. As shown in Fig. 4A, the enzyme was inhibited in a time- and dose-dependent manner by 1,4-benzoquinone, which is in agreement with the irreversible nature of the inhibition of PTPN2 by 1,4-benzoquinone. Replotting the observed first-order constants (kobs) against 1,4-benzoquinone concentration gave a straight line passing through the origin consistent with a single-step bimolecular reaction with a second-order rate constant (kinact) equal to 645 m−1·s−1 (Fig. 4B). The stoichiometry of PTPN2 inhibition by 1,4-benzoquinone was determined by replotting ln(kobs) against ln(1,4-benzoquinone). The slope was found to be very close to 1 (1.12), which further supports that the inhibition of PTPN2 by 1,4-benzoquinone is due to the reaction of one molecule of 1,4-benzoquinone with one molecule of PTPN2 (1:1 stoichiometry). Altogether, the data reported above support the notion that 1,4-benzoquinone irreversibly impairs PTPN2 phosphatase activity through covalent adduction of its catalytic cysteine residue.
Figure 4.
kinetic analysis of PTPN2 inhibition by BQ. A, recombinant PTPN2 was incubated with increasing concentrations of BQ. Aliquots were taken every 30 s, and residual PTPN2 activity was measured using a pNPP assay. For each BQ concentration, the natural logarithm (ln) of the percentage of residual PTPN2 activity was plotted as a function of time. The apparent first-order inhibition constant (kobs) was calculated from the linear regression (slope). B, kobs values were plotted against BQ concentrations, and the second-order inhibition constant (kinact) was extracted from the slope. C, ln(kobs) were plotted as a function of ln[BQ], and the stoichiometry of the reaction was extracted from the slope.
PTPN2 is inhibited in HEK293T cells exposed to 1,4-benzoquinone
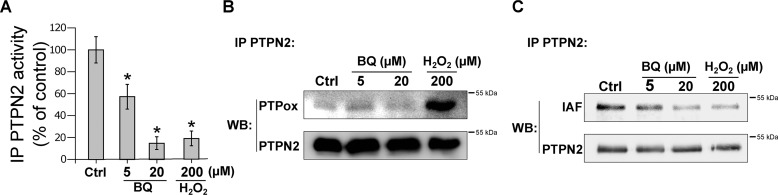
To test whether the inhibition of PTPN2 by 1,4-benzoquinone occurs in a cellular context, the FLAG-tagged enzyme was transiently transfected in HEK293T cells. Cell monolayers were exposed to 1,4-benzoquinone or H2O2, and the FLAG-tagged enzyme was immunoprecipitated. As shown in Fig. 5A, exposure of HEK293T cells to 1,4-benzoquinone led to the inhibition of transfected PTPN2. Exposure to H2O2 (which is known to inhibit PTPN2 through oxidation of the catalytic cysteine) also led to enzyme inhibition as reported previously (49, 50). In addition, whereas H2O2 induced the oxidative modification of PTPN2 catalytic cysteine residue, 1,4-benzoquinone did not (Fig. 5B). Labeling of the immunoprecipitates with IAF indicated that inhibition of transfected PTPN2 by 1,4-benzoquinone or H2O2 is concomitant with the modification of cysteine residues as shown with the purified enzyme (Fig. 5C). Overall, these experiments are consistent with our data obtained using the recombinant enzyme, supporting the notion that 1,4-benzoquinone mainly impairs PTPN2 activity through covalent modification of the active-site cysteine of the enzyme.
Figure 5.
Exposure of HEK293T cells to BQ leads to PTPN2 inhibition through an arylation mechanism. A, HEK293T cells were transfected with a FLAG-PTPN2 plasmid and exposed to BQ or H2O2 for 30 min. Cells were lysed, and PTPN2 was isolated by immunoprecipitation (IP) using an anti-PTPN2 antibody. PTPN2 activity of immunoprecipitates was measured by RP-UFLC. Error bars, S.D. *, p < 0.05 compared with control (Ctrl). B, immunoprecipitated PTPN2 was incubated with IAF for 20 min at room temperature. Immunobeads were separated and analyzed by Western blotting (WB) using anti-oxidized PTP (PTPox). C, detection of IAF labeling was done by fluorescence. Anti-PTPN2 antibody was used as loading control.
1,4-Benzoquinone impairs endogenous PTPN2 and increases STAT1 signaling in Jurkat cells
Jurkat cells were exposed to different concentrations of 1,4-benzoquinone. As shown in Fig. S2, endogenous PTPN2 was inhibited by 1,4-benzoquinone in a dose-dependent manner.
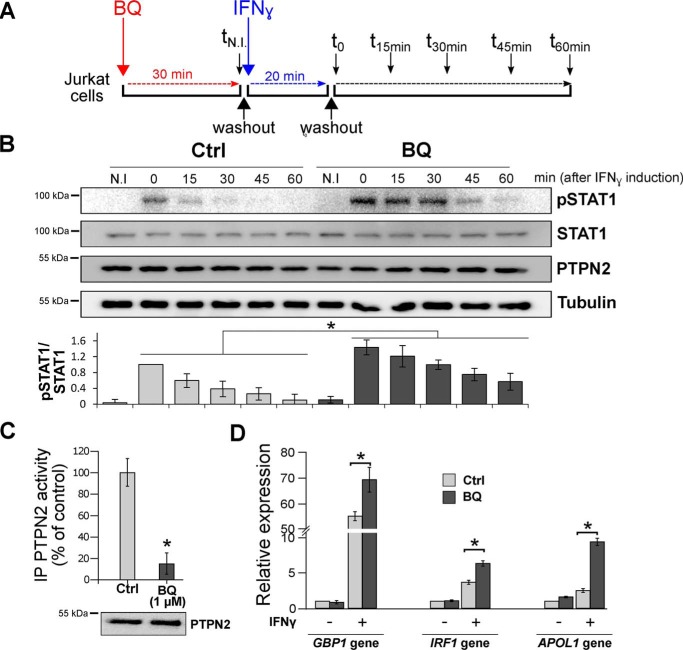
To further analyze the effects of 1,4-benzoquinone on endogenous PTPN2 and STAT1 signaling, we exposed Jurkat cells to 1,4-benzoquinone prior to stimulation of STAT1 signaling by IFNγ (Fig. 6A). In contrast to Jurkat cells not exposed to 1,4-benzoquinone, we observed a significant increase in STAT1 tyrosine phosphorylation in cells pretreated with 1,4-benzoquinone (Fig. 6B). In line with these results, we found that cellular PTPN2 was impaired in Jurkat cells exposed to 1,4-benzoquinone (Fig. 6C). To further confirm that 1,4-benzoquinone increases STAT1 signaling, we measured the expression of three genes (GBP1, IRF1, and APOL1) known to be regulated by STAT1 (51, 52). As shown in Fig. 6D, qPCR analyses indicated that the expression of GBP1, IRF1, and APOL1 is significantly increased in Jurkat cells exposed to 1,4-benzoquinone. Altogether, these data show that 1,4-benzoquinone inhibits PTPN2 in cells with subsequent increase in STAT1 tyrosine phosphorylation and higher expression of STAT1-regulated genes.
Figure 6.
Exposure of Jurkat cells to BQ leads to inhibition of endogenous PTPN2 and subsequent overactivation of the STAT1 pathway. A, after a 30-min treatment with BQ (1 μm) or vehicle (Ctrl) (corresponding to time tN.I.), Jurkat cells were incubated with IFNγ for 20 min (corresponding to time t0). Cells were then washed and cultured in fresh medium to remove IFNγ and harvested every 15 min up to 60 min (corresponding to times t15, t30, t45, and t60). B, Jurkat cells treated as described in A were taken at different times (tN.I. (30 min after BQ treatment) and t0, t15, t30, t45, and t60 (minutes after IFN treatment)) and were analyzed by Western blotting using anti-pSTAT1, anti-STAT1, anti-PTPN2, and anti-tubulin. Quantification of STAT1 phosphorylation in Western blots was carried out using ImageJ software. Error bars, S.D. *, p < 0.05 compared with control (Ctrl). C, Jurkat cells treated with 1 μm BQ for 30 min (corresponding to time tN.I.) were washed and lysed, and endogenous PTPN2 was isolated by immunoprecipitation (IP) using an anti-PTPN2 antibody. PTPN2 activity of immunoprecipitates was measured by RP-UFLC. Error bars, S.D. values. *, p < 0.05 compared with control (Ctrl). D, Jurkat cells were treated with BQ (1 μm) for 30 min and then incubated or not with IFNγ for 180 min. Relative gene expression of GBP1, IRF1, and APOL1 was analyzed by RT-qPCR. Error bars, S.D. values. *, p < 0.05 compared with control (Ctrl).
Discussion
PTPN2 is a protein tyrosine phosphatase that plays a key role in hematopoiesis notably through the negative regulation of the JAK/STAT pathway (25). In connection with this, inactivating mutations or deletions of PTPN2 were found in leukemia and lymphoma and are associated with aberrant STAT signaling (30–32). Knockout mouse models further support the important role of PTPN2 in the homeostasis of the hematopoietic system (26, 27, 29). Interestingly, alteration of critical hematopoietic signaling pathways, notably JAK/STAT, has been proposed as a mechanism contributing to benzene-induced leukemia (1, 4). In this study, we report that 1,4-benzoquinone, a major leukemogenic metabolite of benzene, impairs the tyrosine phosphatase activity of PTPN2. We provide mechanistic and kinetic evidence that the inhibition of PTPN2 by 1,4-benzoquinone occurs through the irreversible formation of a covalent adduct of 1,4-benzoquinone with the catalytic cysteine residue (Cys-216) of the enzyme. As for other PTP enzymes, PTPN2 activity depends on a conserved cysteine residue (22). It has been shown that oxidative or covalent modification of this cysteine residue in PTPN2 and other PTPs can lead to irreversible impairment of the enzyme functions (22, 53). Interestingly, PTPs structurally related to PTPN2 have been shown to be inhibited by quinones such as naphthoquinones through covalent adduction of their catalytic cysteine (48, 54, 55). It is known that the toxic effects of quinones, such as 1,4-benzoquinone, rely at least in part on their reaction with cysteine residues in target proteins, resulting in their loss of function (35, 36). Protection assays using orthovanadate, NBT staining, and all MS analysis indicate that 1,4-benzoquinone covalently binds to the catalytic cysteine of PTPN2. This is also in agreement with the fact that 1,4-benzoquinone acts mainly as an arylating quinone that covalently reacts with cysteine rather than a redox-cycling quinone that generates reactive species that oxidize cysteine residues (35–37). Western blotting studies using an antibody that detects specifically oxidized catalytic cysteines of PTPs further support that PTPN2 is inhibited by 1,4-benzoquinone through direct covalent binding to its active-site cysteine (Figs. 2 and 5). The leukemogenic properties of benzene are known to rely on its bioactivation into reactive metabolites in the bone marrow, such as 1,4-benzoquinone (1, 14). Irreversible inhibition of topoisomerase II enzymes by 1,4-benzoquinone has been put forward as an important mechanism involved in benzene-dependent leukemogenesis through DNA damage (3, 9, 56, 57). As observed for PTPN2, irreversible inhibition of topoisomerases II by 1,4-benzoquinone is due to covalent adduct formations with cysteine residues of the enzyme (47). In addition, as also reported for topoisomerase II enzymes, 1,4-benzoquinone was much more efficient at inhibiting PTPN2 than benzene or phenol and hydroquinone metabolites, which were found to have poor or no inhibitory action (9, 56). These results are in agreement with the strong reactive electrophilic nature of 1,4-benzoquinone and its propensity to readily react with reactive thiols to form covalent adducts (35, 36). In line with this, 1,4-benzoquinone has recently been shown to inhibit urease activity through rapid adduction of its catalytic cysteine with a kinact value of 1240 m−1·s−1, close to the value found for PTPN2 (645 m−1·s−1) (58). In accordance with the data obtained with purified enzyme, the irreversible inhibition of PTPN2 was also observed in transfected HEK293T cells exposed to 1,4-benzoquinone. As found with recombinant human PTPN2, exposure to 1,4-benzoquinone led to the modification of cysteine residues of the cellular enzyme. In addition, no significant oxidation of the catalytic cysteine residue of transfected enzyme upon exposure of cells to 1,4-benzoquinone could be evidenced. This is in agreement with the properties of 1,4-benzoquinone, which is known to readily arylate cysteine residues through Michael addition (36, 37). The arylation of proteins by 1,4-benzoquinone is considered as a major mechanism involved in the toxicity of 1,4-benzoquinone, in particular in the bone marrow (1, 38, 39). Further experiments carried out with Jurkat cells indicated that 1,4-benzoquinone inhibited endogenous PTPN2, led to increased tyrosine phosphorylation of STAT1, and increased expression of STAT1-regulated genes. These data support the notion that 1,4-benzoquinone may perturb STAT1 signaling pathway by impairing the activity of PTPN2. Interestingly similar effects were observed in Jurkat cells, where PTPN2 was knocked down by RNAi (30). In addition, 1,4-benzoquinone has also been shown to increase the extracellular signal–regulated kinase/mitogen-activated protein kinase signaling pathway (59). In addition, certain PTPs structurally related to PTPN2, such as PTP1B, have been shown to be inhibited by other arylating quinones (54). For instance, 1,2-naphthoquinone was found to increase EGFR tyrosine phosphorylation through irreversible inhibition of PTP1B due to covalent adduction of active-site cysteine residues of the enzyme (48). Interestingly, alteration of signaling pathways critical for the hematopoietic system, such as JAK/STAT, by benzene and/or its hematotoxic metabolites has been shown as a mechanism contributing to benzene-dependent leukemia (1, 2, 17). Our data showing irreversible inhibition and altered STAT1 signaling by 1,4-benzoquinone through inhibition of PTPN2 support this notion. Recent studies also suggest that chronic inflammation and aberrant immune response are involved in benzene-related leukemogenesis (15, 16). Interestingly, impaired PTPN2 expression/activity is known to lead to altered immune response and systemic inflammation (24, 25, 27).
Materials and methods
Chemicals
Benzene, hydroquinone, phenol, 1,4-benzoquinone, hydrogen peroxide, N-ethylmaleimide (NEM), fluorescein 5-iodoacetamide (IAF), plumbagin, p-nitrophenyl phosphate (pNPP), sodium orthovanadate, NBT, DMSO, protein A–agarose, bovine liver catalase, bovine erythrocyte superoxide dismutase, His-select nickel resin, protease inhibitors, and polyclonal anti-PTPN2 antibody were from Sigma-Aldrich (Saint-Quentin-Fallavier, France). Benzene, phenol, hydroquinone, and 1,4-benzoquinone were diluted in DMSO at a stock concentration of 100 mm. The anti-plumbagin antibody was a kind gift of Dr. Hiroyuki Tanaka (Kyushu University).
Expression and purification of recombinant human PTPN2 enzyme
The complementary DNA (cDNA) coding for full-length human PTPN2 (TC45 variant) was cloned into pET28a plasmid and used to produce His6-tagged PTPN2. pET28a-PTPN2 plasmid was transformed into Escherichia coli BL21 (DE3) cells for production and purification of the PTPN2 enzyme. Briefly, transformed bacterial cells were grown overnight at 16 °C in the presence of 0.5 mm isopropyl-1-thio-β-d-galactopyranoside. Cells were harvested by centrifugation and resuspended in lysis buffer (20 mm Tris-HCl, pH 8, 300 mm NaCl, 0.2% Triton X-100, 1 mg/ml lysozyme, and protease inhibitors). The lysate was subjected to sonication on ice and pelleted (12,000 × g, 30 min, 4 °C). The supernatant was incubated with His-select nickel resin (Sigma) in the presence of 20 mm imidazole for 2 h at 4 °C. Resin was poured into an empty column and washed with washing buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl and 35 mm imidazole). Proteins were eluted in Elution Buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 300 mm imidazole), reduced with 10 mm DTT (20 min on ice), and then applied to a PD-10 column (GE Healthcare, Paris, France). Proteins were eluted with Tris-HCl buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl) and quantified using Bradford reagent. The purity of the purified PTPN2 was assessed by SDS-PAGE. Proteins were kept at −80 °C until use.
Determination of PTPN2 activity using p-nitrophenyl phosphate (pNPP assay)
The phosphatase activity of recombinant PTPN2 was assayed using the chromogenic phosphatase substrate pNPP as described previously (60). Samples containing PTPN2 were diluted 10 times with 100 mm sodium acetate, pH 6, containing 10 mm pNPP, and the formation of the product (p-nitrophenol) was measured by reading the absorbance at 405 nm at 37 °C using a thermostated microplate reader (BioTek, France). The final concentration of PTPN2 during the assay was 50 nm in a total volume of 250 μl/microplate well. Recombinant PTPN2 (0.5 μm) was incubated with different concentrations of benzene, phenol, hydroquinone, or 1,4-benzoquinone in 100 mm sodium acetate, pH 6, at 37 °C (in a total volume of 50 μl). After 20 min of incubation, samples were diluted 10 times in 100 mm sodium acetate, pH 6, in the presence of 10 mm pNPP. The formation of p-nitrophenol was monitored by reading the absorbance at 405 nm at 37 °C using a thermostated microplate reader (BioTek, Colmar, France).
Effects of benzene and benzene-phenolic metabolites on PTPN2 activity
Recombinant PTPN2 (0.5 μm) was incubated with different concentrations of benzene, phenol, hydroquinone, or 1,4-benzoquinone in 100 mm sodium acetate, pH 6, at 37 °C (total volume of 50 μl). After a 20-min incubation, samples were assayed for residual PTPN2 activity using a pNPP assay. Effects of PBG on PTPN2 activity were analyzed as described above for benzene and its metabolites.
MPO bioactivation of hydroquinone into 1,4-benzoquinone
Bioactivation of HQ into 1,4-benzoquinone by MPO was carried out as described (39). Briefly, MPO (0.1 units/ml) was incubated with hydroquinone (50 μm) and H2O2 (50 μm) in 100 mm sodium acetate, pH 6.5, for 30 min at room temperature (in a total volume of 500 μl). Conversion of HQ to 1,4-benzoquinone was monitored by spectrophotometry (UV-1650PC, Shimadzu, France) as described previously (39). Heat-inactivated MPO (boiled for 30 min) was used as a negative control. 3 μl of reaction mix was then added to recombinant PTPN2 (0.5 μm final) in 100 mm sodium acetate, pH 6, and incubated for 20 min at 37 °C (total volume of 50 μl). After incubation, the residual activity of PTPN2 was measured using the pNPP assay.
Effects of dialysis or DTT on 1,4-benzoquinone–inhibited PTPN2
PTPN2 (0.5 μm) was first incubated with 1,4-benzoquinone (2 μm) in 100 mm sodium acetate, pH 6, for 20 min at 37 °C (total volume of 50 μl). Samples were then either dialyzed overnight at 4 °C against 100 mm sodium acetate, pH 6, or incubated for 30 min with 10 mm DTT at 37 °C prior to measurement of residual PTPN2 activity using a pNPP assay.
Effects of superoxide dismutase or catalase on 1,4-benzoquinone–dependent inhibition of PTPN2
PTPN2 (0.5 μm) was incubated with 1,4-benzoquinone (2 μm) in 100 mm sodium acetate, pH 6, containing catalase (20 units/ml) or superoxide dismutase (20 units/ml) for 20 min at 37 °C. Samples were then diluted 10 times in 100 mm sodium acetate, 10 mm pNPP, pH 6, and samples were assayed for residual PTPN2 activity using a pNPP assay.
Fluorescein-conjugated iodoacetamide labeling, nitro blue tetrazolium staining, and detection of oxidized PTPN2 catalytic cysteine
PTPN2 (0.5 μm) was first incubated with or without 1,4-benzoquinone (2 μm) in 100 mm sodium acetate, pH 6, at 37 °C for 20 min followed by the addition of 10 μm IAF (final concentration) for 10 min at 37 °C. For the detection of IAF labeling and 1,4-benzoquinone adducts, samples containing 200 and 400 ng of PTPN2 were separated by SDS-PAGE and transferred onto nitrocellulose membrane, respectively. IAF labeling of PTPN2 cysteine residues was detected by fluorescence analysis of the membranes (excitation, 494 nm; emission, 512 nm) using a LAS-4000 apparatus (Fujifilm, Montigny le Bretonneux, France). 1,4-Benzoquinone adducts were detected by incubating the membrane for 30 min in the dark with 0.6 mg/ml NBT diluted in 2 m potassium glycinate, pH 10, as described previously (40). The total amount of PTPN2 in membranes was detected using Ponceau red staining.
For the detection of oxidized PTPN2 catalytic cysteine, samples (50 ng of PTPN2) treated or not with 1,4-benzoquinone (2 μm) or H2O2 (100 μm) were separated by SDS-PAGE and analyzed by Western blotting with an anti-oxidized PTP active-site cysteine antibody (R&D Systems, Lille, France) as described previously (45). An anti-PTPN2 mouse mAb was used to detect total PTPN2 enzyme (R&D Systems) after stripping of the nitrocellulose membranes.
Detection of PBG covalent adducts on PTPN2 treated or not with (50 μm PBG) was carried out using NBT staining (see above) of dot-blotted nitrocellulose membranes. PBG adducts were also assessed on the dot-blotted membranes using a specific antibody against PBG adducts (61) as well as the anti-PTPN2 mAb.
Kinetic analysis of PTPN2 inhibition by 1,4-benzoquinone
The kinetic data were analyzed as reported by Copeland (62) for irreversible inhibitors. The data were fitted and plotted using Qtiplot software. Briefly, PTPN2 (0.5 μm) was incubated with various concentrations of 1,4-benzoquinone under pseudo-first-order conditions (excess of 1,4-benzoquinone) in 100 mm sodium acetate pH 6 at 37 °C. At different time points, aliquots were removed and assayed for residual PTPN2 activity using a pNPP assay. The equation rate of inhibition of PTPN2 by 1,4-benzoquinone can be represented as follows: ln(% residual activity) = kobs × t (where t is time and kobs is the apparent first-order inhibition rate constant). Provided that 1,4-benzoquinone is present in substantial excess compared with enzyme, the apparent first-order inhibition rate constants (kobs = kinact × [1,4-benzoquinone]) can be obtained for each 1,4-benzoquinone concentration from the slope of the natural log (ln) of the percentage of residual activity plotted against time. The second-order rate constant (kinact) was extracted from the slope of kobs plotted against 1,4-benzoquinone concentrations.
Determination of the reaction order of the reaction of PTPN2 enzyme with 1,4-benzoquinone was carried out by replotting the ln(kobs) values as a function of ln[1,4-benzoquinone].
Protection of PTPN2 from 1,4-benzoquinone–dependent inhibition by orthovanadate (Na3VO4)
PTPN2 (0.5 μm) was first preincubated with 1 mm Na3VO4 for 5 min on ice and then incubated with 1,4-benzoquinone (2 μm) in 100 mm sodium acetate, pH 6, at 37 °C for 20 min. Samples were diluted 10 times in 100 mm sodium acetate, pH 6, containing 1 mm EDTA, and residual PTPN2 activity was measured using a pNPP assay.
Docking of 1,4-benzoquinone in PTPN2 active site
1,4-Benzoquinone structure was built with Maestro11 (Schrodinger, LLC) and minimized by Merck molecular force field. The PTPN2 structure (PDB code 1L8K) was prepared by the Protein Preparation Wizard. Waters were removed, and hydrogens were added. Finally, the hydrogens' orientations were optimized with OPLS3 force field (63). The covalent docking was performed by the CovDock module. Cys-216 was set as the reactive residue and as the docking site center. The reaction type was set to be Michael addition. After docking, the conformations were minimized by Prime, and the poses with the top MMGBSA score were selected for further analysis. The structural representation was made by VMD1.9.3 (64).
A similar approach was used to dock PBG in PTPN2 active site.
Mass spectrometry analysis
PTPN2 (0.5 μm) was incubated with 100 μm PBG for 20 min at 37 °C in 100 mm sodium acetate, pH 6. Unmodified thiol moieties of cysteines were blocked by the addition of 10 mm NEM for 10 min, and reactions were stopped by the addition of Laemmli sample buffer containing 100 mm DTT. Samples were separated on a 12% SDS-polyacrylamide gel and stained with Coomassie Blue R250. PTPN2 bands (∼45 kDa) were extracted from the gel and incubated overnight at 37 °C with trypsin (Roche Applied Science) at 12.5 ng/μl in 25 mm ammonium bicarbonate, 0.05% CaCl2. After the addition of 100 μl of 5% aqueous formic acid (FA), the supernatant containing extracted peptides was concentrated to 10 μl using a SpeedVac, desalted on C18 stage tips (ZipTipTM, Millipore), and eluted in 4 μl of 60% ACN, 0.1% FA. Desalted samples were then diluted 10 times in buffer A (buffer A: 98% water, 2% ACN, 0.1% FA), and 2 μl were injected on a nano-LC HPLC system (RSLC, Thermo Fisher Scientific) coupled to a hybrid quadrupole-Orbitrap mass spectrometer (Q Exactive, Thermo Fisher Scientific). Peptides were loaded on a reverse-phase C18 μ-precolumn (C18 PepMap100, 5 μm, 100A, 300-μm inner diameter × 5 mm) at 15 μl/min and separated on a C18 column (Acclaim® PepMap100, 75-μm inner diameter × 50 cm) at a constant flow rate of 220 nl/min, with a 45-min gradient of 2–40% buffer B (buffer B: 10% water, 90% ACN, 0.1% FA). MS/MS analysis consisted of a Top10 DDA experiment (one survey MS scan 400–2,000 m/z; resolution 70,000 followed by 10 MS/MS of the 10 most intense precursors, with a dynamic exclusion time of 30 s).
The database search was performed against the PTPN2 sequence with Mascot version 2.5 software with the following parameters: tryptic peptides only with up to 4 missed cleavages; variable modifications: cysteine carbamidomethylation, cysteine plumbagin or cysteine NEM, methionine oxidation, and N-terminal acetylation. MS and MS/MS error tolerances were set to 10 ppm and 0.02 Da, respectively. Protein identification was validated using myProms software (65), and the candidate sequences modified by plumbagin were manually inspected for de novo sequencing.
PTPN2 transfection in HEK293T cells and immunoprecipitation
HEK293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum, 1 mm l-glutamine, and 50 mg/ml penicillin/streptomycin at 37 °C, 5% CO2. Cells (4 × 106 cells) were seeded in a 10-cm dish and transfected with 5 μg of FLAG-tagged human PTPN2 plasmid (Origene) using Metafectene (Biontex, Munich, Germany). One day after transfection, cells were exposed to 1,4-benzoquinone (5 and 20 μm) or H2O2 (200 μm) in PBS for 30 min.
Cells were washed twice with PBS and resuspended in lysis buffer (PBS, 1% Triton X-100, 1 mm sodium orthovanadate, and protease inhibitor mixture (Sigma-Aldrich)) for 20 min. Cell lysates were centrifuged at 15,000 × g for 10 min at 4 °C, and supernatants (whole-cell extracts) were collected. PTPN2 was immunoprecipitated by incubating 500 μg of whole-cell extracts with 2 μg of polyclonal PTPN2 antibody (Sigma-Aldrich) overnight at 4 °C. Samples were then rocked for 2 h at 4 °C in the presence of protein A-agarose (Sigma-Aldrich). The immunobeads were collected by centrifugation, washed three times with lysis buffer, and split into two pools. The immunobeads from the first pool were incubated with 50 μm FAM-pSTAT1 peptide to measure residual PTPN2 activity by RP-UFLC as described previously (50). Immunobeads from the second pool were incubated with 10 μm IAF for 20 min (room temperature) washed with PBS and eluted with Laemmli sample buffer containing 100 mm β-mercaptoethanol. Samples were separated by SDS-PAGE and analyzed by Western blotting using the specific anti-oxidized PTP active-site antibody (see above) followed by stripping and reprobing using an anti-PTPN2 antibody. IAF labeling of proteins on membranes was detected by fluorescence (excitation, 494 nm; emission, 512 nm) using an LAS-4000 apparatus (Fujifilm, France).
Jurkat cell treatment and STAT1 phosphorylation kinetics in cells
Jurkat cells were maintained in RPMI medium supplemented with 10% heat-inactivated fetal bovine serum, 1 mm l-glutamine, and 50 mg/ml penicillin/streptomycin at 37 °C, 5% CO2. Cells were treated with DMSO (control) or 1 μm 1,4-benzoquinone for 30 min in PBS. Endogenous PTPN2 activity was assessed by immunoprecipitation of PTPN2 followed by RP-UFLC analysis as described above.
For the STAT1 phosphorylation kinetic, cells were treated with DMSO (control) or 1 μm 1,4-benzoquinone for 30 min in PBS, washed with fresh medium, and incubated for 20 min in medium supplemented with 25 ng/ml human IFNγ. Cells were then washed and resuspended in fresh medium. Aliquots (2 × 106 cells) were collected at different time points and washed in PBS. Cell extracts were done, and STAT1 activation was assayed by Western blotting using anti-phospho-STAT1 (phosphorylated tyrosine 701). Membranes were stripped and successively reprobed with anti-STAT1 (Cell Signaling, St-Cyr l'Ecole, France), anti-PTPN2, and anti-tubulin antibodies. Quantification of STAT1 phosphorylation in Western blots was carried out using ImageJ software (National Institutes of Health).
RNA extraction, cDNA synthesis, and RT-qPCR
Jurkat cells were treated with DMSO (control) or 1 μm 1,4-benzoquinone for 30 min in PBS, washed with fresh medium, and incubated for 180 min in fresh medium supplemented with 25 ng/ml human IFNγ. Cells were then collected by centrifugation, and total RNA was extracted using TRIzol (Invitrogen, Paris, France). cDNA synthesis was performed using the M-MLV RT kit (Invitrogen) according to the manufacturer's instructions. LightCycler 480 SYBR Green I Master (Roche, Boulogne Billancourt, France) was then used to amplify specific cDNA fragments in the 7300 real-time PCR system (Applied Biosystems, Illkirch, France), with oligonucleotides for different STAT1-regulated genes: GBP1, IRF, and APOL1 (51, 52). RPL19 was used as reference as reported earlier (66). Primer sequences are given in Table S1.
Statistical analysis
The results are presented as means ± S.D. of three experiments. Statistical analysis was performed using one-way analysis of variance followed by Bonferroni's post hoc test using Qtiplot software. If only two groups were compared, a mean comparison t test was used.
Author contributions
R. D., L.-C. B., F. G., C. C., and F. R.-L. conceptualization; R. D., L.-C. B., F. G., C. C., and F. R.-L. data curation; R. D., L.-C. B., C. M., Q. N., J. B., X. X., I. H., J. V., J.-M. D., F. B., F. G., C. C., and F. R.-L. formal analysis; R. D. and F. R.-L. supervision; R. D., J.-M. D., F. B., C. C., and F. R.-L. funding acquisition; R. D., L.-C. B., C. M., Q. N., J. B., X. X., I. H., J. V., J.-M. D., F. G., C. C., and F. R.-L. validation; R. D., L.-C. B., C. M., Q. N., J. B., X. X., I. H., J. V., J.-M. D., and F. R.-L. investigation; R. D., L.-C. B., C. M., Q. N., J. B., X. X., I. H., J. V., J.-M. D., F. B., F. G., C. C., and F. R.-L. methodology; R. D., L.-C. B., and F. R.-L. writing-original draft; R. D., C. C., and F. R.-L. project administration; R. D., L.-C. B., C. M., Q. N., J. B., X. X., I. H., J. V., J.-M. D., F. B., F. G., C. C., and F. R.-L. writing-review and editing; L.-C. B. and C. M. visualization; X. X. and I. H. software.
Supplementary Material
Acknowledgments
We thank the staff of the Bioprofiler platform for HPLC analysis.
This work was supported by University Paris Diderot, CNRS, INSERM, and Plan Cancer AAP Environnement et Cancer 2013 (to C. C. and F. R. -L.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1 and S2.
- JAK
- Janus kinase
- STAT
- signal transducers and activators of transcription
- PTP
- protein tyrosine phosphatase
- PTPN2
- protein tyrosine phosphatase, non-receptor type 2
- BQ
- 1,4-benzoquinone
- HQ
- hydroquinone
- MPO
- myeloperoxidase
- NBT
- nitro blue tetrazolium
- IAF
- fluorescein-iodoacetamide
- PBG
- plumbagin
- pNPP
- para-nitrophenyl phosphate
- NEM
- N-ethylmaleimide
- FA
- formic acid
- ACN
- acetonitrile
- IFN
- interferon
- qPCR
- quantitative PCR.
References
- 1. Smith M. T., Zhang L., McHale C. M., Skibola C. F., and Rappaport S. M. (2011) Benzene, the exposome and future investigations of leukemia etiology. Chem. Biol. Interact. 192, 155–159 10.1016/j.cbi.2011.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Snyder R. (2012) Leukemia and benzene. Int. J. Environ. Res. Public Health. 9, 2875–2893 10.3390/ijerph9082875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang L., He X., Bi Y., and Ma Q. (2012) Stem cell and benzene-induced malignancy and hematotoxicity. Chem. Res. Toxicol. 25, 1303–1315 10.1021/tx3001169 [DOI] [PubMed] [Google Scholar]
- 4. McHale C. M., Zhang L., Lan Q., Vermeulen R., Li G., Hubbard A. E., Porter K. E., Thomas R., Portier C. J., Shen M., Rappaport S. M., Yin S., Smith M. T., and Rothman N. (2011) Global gene expression profiling of a population exposed to a range of benzene levels. Environ. Health Perspect. 119, 628–634 10.1289/ehp.1002546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Greim H., Kaden D. A., Larson R. A., Palermo C. M., Rice J. M., Ross D., and Snyder R. (2014) The bone marrow niche, stem cells, and leukemia: impact of drugs, chemicals, and the environment. Ann. N.Y. Acad. Sci. 1310, 7–31 10.1111/nyas.12362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Loomis D., Guyton K. Z., Grosse Y., El Ghissassi F., Bouvard V., Benbrahim-Tallaa L., Guha N., Vilahur N., Mattock H., Straif K., and International Agency for Research on Cancer Monograph Working Group (2017) Carcinogenicity of benzene. Lancet Oncol. 18, 1574–1575 10.1016/S1470-2045(17)30832-X [DOI] [PubMed] [Google Scholar]
- 7. Goldstein B. D. (2011) Hematological and toxicological evaluation of formaldehyde as a potential cause of human leukemia. Hum. Exp. Toxicol. 30, 725–735 10.1177/0960327110381682 [DOI] [PubMed] [Google Scholar]
- 8. Zhou Y., Li C., Huijbregts M. A., and Mumtaz M. M. (2015) Carcinogenic air toxics exposure and their cancer-related health impacts in the United States. PLoS One 10, e0140013 10.1371/journal.pone.0140013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Frantz C. E., Chen H., and Eastmond D. A. (1996) Inhibition of human topoisomerase II in vitro by bioactive benzene metabolites. Environ. Health Perspect. 104, Suppl. 6, 1319–1323 10.1289/ehp.961041319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Whysner J., Reddy M. V., Ross P. M., Mohan M., and Lax E. A. (2004) Genotoxicity of benzene and its metabolites. Mutat. Res. 566, 99–130 10.1016/S1383-5742(03)00053-X [DOI] [PubMed] [Google Scholar]
- 11. Kolachana P., Subrahmanyam V. V., Meyer K. B., Zhang L., and Smith M. T. (1993) Benzene and its phenolic metabolites produce oxidative DNA damage in HL60 cells in vitro and in the bone marrow in vivo. Cancer Res. 53, 1023–1026 [PubMed] [Google Scholar]
- 12. Meek B., Cloosen S., Borsotti C., Van Elssen C. H., Vanderlocht J., Schnijderberg M. C., van der Poel M. W., Leewis B., Hesselink R., Manz M. G., Katsura Y., Kawamoto H., Germeraad W. T., and Bos G. M. (2010) In vitro-differentiated T/natural killer-cell progenitors derived from human CD34+ cells mature in the thymus. Blood 115, 261–264 10.1182/blood-2009-05-223990 [DOI] [PubMed] [Google Scholar]
- 13. Zhang L., McHale C. M., Rothman N., Li G., Ji Z., Vermeulen R., Hubbard A. E., Ren X., Shen M., Rappaport S. M., North M., Skibola C. F., Yin S., Vulpe C., Chanock S. J., Smith M. T., and Lan Q. (2010) Systems biology of human benzene exposure. Chem. Biol. Interact. 184, 86–93 10.1016/j.cbi.2009.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eastmond D. A., Keshava N., and Sonawane B. (2014) Lymphohematopoietic cancers induced by chemicals and other agents and their implications for risk evaluation: an overview. Mutat. Res. Rev. Mutat. Res. 761, 40–64 10.1016/j.mrrev.2014.04.001 [DOI] [PubMed] [Google Scholar]
- 15. Sauer E., Gauer B., Nascimento S., Nardi J., Göethel G., Costa B., Correia D., Matte U., Charão M., Arbo M., Duschl A., Moro A., and Garcia S. C. (2018) The role of B7 costimulation in benzene immunotoxicity and its potential association with cancer risk. Environ. Res. 166, 91–99 10.1016/j.envres.2018.05.029 [DOI] [PubMed] [Google Scholar]
- 16. Gross S. A., and Paustenbach D. J. (2018) Shanghai Health Study (2001–2009): What was learned about benzene health effects? Crit. Rev. Toxicol. 48, 217–251 10.1080/10408444.2017.1401581 [DOI] [PubMed] [Google Scholar]
- 17. Minciullo P. L., Navarra M., Calapai G., and Gangemi S. (2014) Cytokine network involvement in subjects exposed to benzene. J. Immunol. Res. 2014, 937987 10.1155/2014/937987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baker S. J., Rane S. G., and Reddy E. P. (2007) Hematopoietic cytokine receptor signaling. Oncogene 26, 6724–6737 10.1038/sj.onc.1210757 [DOI] [PubMed] [Google Scholar]
- 19. Dorritie K. A., Redner R. L., and Johnson D. E. (2014) STAT transcription factors in normal and cancer stem cells. Adv. Biol. Regul. 56, 30–44 10.1016/j.jbior.2014.05.004 [DOI] [PubMed] [Google Scholar]
- 20. Benekli M., Baumann H., and Wetzler M. (2009) Targeting signal transducer and activator of transcription signaling pathway in leukemias. J. Clin. Oncol. 27, 4422–4432 10.1200/JCO.2008.21.3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O'Shea K. M., Hwang S. A., and Actor J. K. (2015) Immune activity of BCG infected mouse macrophages treated with a novel recombinant mouse lactoferrin. Ann. Clin. Lab. Sci. 45, 487–494 [PubMed] [Google Scholar]
- 22. Tonks N. K. (2006) Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 7, 833–846 10.1038/nrm2039 [DOI] [PubMed] [Google Scholar]
- 23. Böhmer F. D., and Friedrich K. (2014) Protein tyrosine phosphatases as wardens of STAT signaling. JAKSTAT 3, e28087 10.4161/jkst.28087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tiganis T., and Bennett A. M. (2007) Protein tyrosine phosphatase function: the substrate perspective. Biochem. J. 402, 1–15 10.1042/BJ20061548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pike K. A., and Tremblay M. L. (2016) TC-PTP and PTP1B: regulating JAK-STAT signaling, controlling lymphoid malignancies. Cytokine 82, 52–57 10.1016/j.cyto.2015.12.025 [DOI] [PubMed] [Google Scholar]
- 26. You-Ten K. E., Muise E. S., Itié A., Michaliszyn E., Wagner J., Jothy S., Lapp W. S., and Tremblay M. L. (1997) Impaired bone marrow microenvironment and immune function in T cell protein tyrosine phosphatase-deficient mice. J. Exp. Med. 186, 683–693 10.1084/jem.186.5.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bourdeau A., Dubé N., Heinonen K. M., Théberge J. F., Doody K. M., and Tremblay M. L. (2007) TC-PTP-deficient bone marrow stromal cells fail to support normal B lymphopoiesis due to abnormal secretion of interferon-γ. Blood 109, 4220–4228 10.1182/blood-2006-08-044370 [DOI] [PubMed] [Google Scholar]
- 28. Heinonen K. M., Bourdeau A., Doody K. M., and Tremblay M. L. (2009) Protein tyrosine phosphatases PTP-1B and TC-PTP play nonredundant roles in macrophage development and IFN-γ signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 9368–9372 10.1073/pnas.0812109106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wiede F., Chew S. H., van Vliet C., Poulton I. J., Kyparissoudis K., Sasmono T., Loh K., Tremblay M. L., Godfrey D. I., Sims N. A., and Tiganis T. (2012) Strain-dependent differences in bone development, myeloid hyperplasia, morbidity and mortality in ptpn2-deficient mice. PLoS One 7, e36703 10.1371/journal.pone.0036703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kleppe M., Lahortiga I., El Chaar T., De Keersmaecker K., Mentens N., Graux C., Van Roosbroeck K., Ferrando A. A., Langerak A. W., Meijerink J. P., Sigaux F., Haferlach T., Wlodarska I., Vandenberghe P., Soulier J., and Cools J. (2010) Deletion of the protein tyrosine phosphatase gene PTPN2 in T-cell acute lymphoblastic leukemia. Nat. Genet. 42, 530–535 10.1038/ng.587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kleppe M., Tousseyn T., Geissinger E., Kalender Atak Z., Aerts S., Rosenwald A., Wlodarska I., and Cools J. (2011) Mutation analysis of the tyrosine phosphatase PTPN2 in Hodgkin's lymphoma and T-cell non-Hodgkin's lymphoma. Haematologica 96, 1723–1727 10.3324/haematol.2011.041921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kleppe M., Soulier J., Asnafi V., Mentens N., Hornakova T., Knoops L., Constantinescu S., Sigaux F., Meijerink J. P., Vandenberghe P., Tartaglia M., Foa R., Macintyre E., Haferlach T., and Cools J. (2011) PTPN2 negatively regulates oncogenic JAK1 in T-cell acute lymphoblastic leukemia. Blood 117, 7090–7098 10.1182/blood-2010-10-314286 [DOI] [PubMed] [Google Scholar]
- 33. Nishiyama-Fujita Y., Shimizu T., Sagawa M., Uchida H., and Kizaki M. (2013) The role of TC-PTP (PTPN2) in modulating sensitivity to imatinib and interferon-α in CML cell line, KT-1 cells. Leuk. Res. 37, 1150–1155 10.1016/j.leukres.2013.05.008 [DOI] [PubMed] [Google Scholar]
- 34. Mondrala S., and Eastmond D. A. (2010) Topoisomerase II inhibition by the bioactivated benzene metabolite hydroquinone involves multiple mechanisms. Chem. Biol. Interact. 184, 259–268 10.1016/j.cbi.2009.12.023 [DOI] [PubMed] [Google Scholar]
- 35. Bolton J. L., and Dunlap T. (2017) Formation and biological targets of quinones: cytotoxic versus cytoprotective effects. Chem. Res. Toxicol. 30, 13–37 10.1021/acs.chemrestox.6b00256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mbiya W., Chipinda I., Siegel P. D., Mhike M., and Simoyi R. H. (2013) Substituent effects on the reactivity of benzoquinone derivatives with thiols. Chem. Res. Toxicol. 26, 112–123 10.1021/tx300417z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Y., Jongberg S., Andersen M. L., Davies M. J., and Lund M. N. (2016) Quinone-induced protein modifications: Kinetic preference for reaction of 1,2-benzoquinones with thiol groups in proteins. Free Radic. Biol. Med. 97, 148–157 10.1016/j.freeradbiomed.2016.05.019 [DOI] [PubMed] [Google Scholar]
- 38. Rappaport S. M., Waidyanatha S., Yeowell-O'Connell K., Rothman N., Smith M. T., Zhang L., Qu Q., Shore R., Li G., and Yin S. (2005) Protein adducts as biomarkers of human benzene metabolism. Chem. Biol. Interact. 153, 103–109 10.1016/j.cbi.2005.03.014 [DOI] [PubMed] [Google Scholar]
- 39. Eastmond D. A., Mondrala S. T., and Hasegawa L. (2005) Topoisomerase II inhibition by myeloperoxidase-activated hydroquinone: a potential mechanism underlying the genotoxic and carcinogenic effects of benzene. Chem. Biol. Interact. 153, 207–216 10.1016/j.cbi.2005.03.024 [DOI] [PubMed] [Google Scholar]
- 40. Paz M. A., Flückiger R., Boak A., Kagan H. M., and Gallop P. M. (1991) Specific detection of quinoproteins by redox-cycling staining. J. Biol. Chem. 266, 689–692 [PubMed] [Google Scholar]
- 41. Verrax J., Delvaux M., Beghein N., Taper H., Gallez B., and Buc Calderon P. (2005) Enhancement of quinone redox cycling by ascorbate induces a caspase-3 independent cell death in human leukaemia cells. An in vitro comparative study. Free Radic. Res. 39, 649–657 10.1080/10715760500097906 [DOI] [PubMed] [Google Scholar]
- 42. Rodríguez F., Allende C. C., and Allende J. E. (2005) Protein kinase casein kinase 2 holoenzyme produced ectopically in human cells can be exported to the external side of the cellular membrane. Proc. Natl. Acad. Sci. U.S.A. 102, 4718–4723 10.1073/pnas.0501074102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Seiner D. R., LaButti J. N., and Gates K. S. (2007) Kinetics and mechanism of protein tyrosine phosphatase 1B inactivation by acrolein. Chem. Res. Toxicol. 20, 1315–1320 10.1021/tx700213s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tapper M. A., Sheedy B. R., Hammermeister D. E., and Schmieder P. K. (2000) Depletion of cellular protein thiols as an indicator of arylation in isolated trout hepatocytes exposed to 1,4-benzoquinone. Toxicol. Sci. 55, 327–334 10.1093/toxsci/55.2.327 [DOI] [PubMed] [Google Scholar]
- 45. Karisch R., Fernandez M., Taylor P., Virtanen C., St-Germain J. R., Jin L. L., Harris I. S., Mori J., Mak T. W., Senis Y. A., Östman A., Moran M. F., and Neel B. G. (2011) Global proteomic assessment of the classical protein-tyrosine phosphatome and “Redoxome”. Cell 146, 826–840 10.1016/j.cell.2011.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ostman A., Frijhoff J., Sandin A., and Böhmer F. D. (2011) Regulation of protein tyrosine phosphatases by reversible oxidation. J. Biochem. 150, 345–356 10.1093/jb/mvr104 [DOI] [PubMed] [Google Scholar]
- 47. Bender R. P., Ham A. J., and Osheroff N. (2007) Quinone-induced enhancement of DNA cleavage by human topoisomerase IIα: adduction of cysteine residues 392 and 405. Biochemistry 46, 2856–2864 10.1021/bi062017l [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Iwamoto N., Sumi D., Ishii T., Uchida K., Cho A. K., Froines J. R., and Kumagai Y. (2007) Chemical knockdown of protein-tyrosine phosphatase 1B by 1,2-naphthoquinone through covalent modification causes persistent transactivation of epidermal growth factor receptor. J. Biol. Chem. 282, 33396–33404 10.1074/jbc.M705224200 [DOI] [PubMed] [Google Scholar]
- 49. Gurzov E. N., Tran M., Fernandez-Rojo M. A., Merry T. L., Zhang X., Xu Y., Fukushima A., Waters M. J., Watt M. J., Andrikopoulos S., Neel B. G., and Tiganis T. (2014) Hepatic oxidative stress promotes insulin-STAT-5 signaling and obesity by inactivating protein tyrosine phosphatase N2. Cell Metab. 20, 85–102 10.1016/j.cmet.2014.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Duval R., Bui L. C., Berthelet J., Dairou J., Mathieu C., Guidez F., Dupret J. M., Cools J., Chomienne C., and Rodrigues-Lima F. (2015) A RP-UFLC assay for protein tyrosine phosphatases: focus on protein tyrosine phosphatase non-receptor type 2 (PTPN2). Sci. Rep. 5, 10750 10.1038/srep10750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Reardon C., and McKay D. M. (2007) TGF-β suppresses IFN-γ-STAT1-dependent gene transcription by enhancing STAT1-PIAS1 interactions in epithelia but not monocytes/macrophages. J. Immunol. 178, 4284–4295 10.4049/jimmunol.178.7.4284 [DOI] [PubMed] [Google Scholar]
- 52. Satoh J., and Tabunoki H. (2013) A comprehensive profile of ChIP-Seq-based STAT1 target genes suggests the complexity of STAT1-mediated gene regulatory mechanisms. Gene Regul. Syst. Bio. 7, 41–56 10.4137/GRSB.S11433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Böhmer F., Szedlacsek S., Tabernero L., Ostman A., and den Hertog J. (2013) Protein tyrosine phosphatase structure-function relationships in regulation and pathogenesis. FEBS J. 280, 413–431 10.1111/j.1742-4658.2012.08655.x [DOI] [PubMed] [Google Scholar]
- 54. Wang Q., Dubé D., Friesen R. W., LeRiche T. G., Bateman K. P., Trimble L., Sanghara J., Pollex R., Ramachandran C., Gresser M. J., and Huang Z. (2004) Catalytic inactivation of protein tyrosine phosphatase CD45 and protein tyrosine phosphatase 1B by polyaromatic quinones. Biochemistry 43, 4294–4303 10.1021/bi035986e [DOI] [PubMed] [Google Scholar]
- 55. Klotz L. O., Hou X., and Jacob C. (2014) 1,4-naphthoquinones: from oxidative damage to cellular and inter-cellular signaling. Molecules 19, 14902–14918 10.3390/molecules190914902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lindsey R. H. Jr, Bromberg K. D., Felix C. A., and Osheroff N. (2004) 1,4-Benzoquinone is a topoisomerase II poison. Biochemistry 43, 7563–7574 10.1021/bi049756r [DOI] [PubMed] [Google Scholar]
- 57. Lindsey R. H. Jr, Bender R. P., and Osheroff N. (2005) Effects of benzene metabolites on DNA cleavage mediated by human topoisomerase II α: 1,4-hydroquinone is a topoisomerase II poison. Chem. Res. Toxicol. 18, 761–770 10.1021/tx049659z [DOI] [PubMed] [Google Scholar]
- 58. Mazzei L., Cianci M., Musiani F., and Ciurli S. (2016) Inactivation of urease by 1,4-benzoquinone: chemistry at the protein surface. Dalton Trans. 45, 5455–5459 10.1039/C6DT00652C [DOI] [PubMed] [Google Scholar]
- 59. Ruiz-Ramos R., Cebrian M. E., and Garrido E. (2005) Benzoquinone activates the ERK/MAPK signaling pathway via ROS production in HL-60 cells. Toxicology 209, 279–287 10.1016/j.tox.2004.12.035 [DOI] [PubMed] [Google Scholar]
- 60. Montalibet J., Skorey K. I., and Kennedy B. P. (2005) Protein tyrosine phosphatase: enzymatic assays. Methods 35, 2–8 10.1016/j.ymeth.2004.07.002 [DOI] [PubMed] [Google Scholar]
- 61. Sakamoto S., Taura F., Tsuchihashi R., Putalun W., Kinjo J., Tanaka H., and Morimoto S. (2010) Expression, purification, and characterization of anti-plumbagin single-chain variable fragment antibody in Sf9 insect cell. Hybridoma 29, 481–488 10.1089/hyb.2010.0052 [DOI] [PubMed] [Google Scholar]
- 62. Copeland R. A. (2005) Evaluation of Enzyme Inhibitors in Drug Discovery, pp. 214–248, Wiley-Interscience, Hoboken, NJ [Google Scholar]
- 63. Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J. Y., Wang L., Lupyan D., Dahlgren M. K., Knight J. L., Kaus J. W., Cerutti D. S., Krilov G., Jorgensen W. L., Abel R., and Friesner R. A. (2016) OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 12, 281–296 10.1021/acs.jctc.5b00864 [DOI] [PubMed] [Google Scholar]
- 64. Humphrey W., Dalke A., and Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- 65. Poullet P., Carpentier S., and Barillot E. (2007) myProMS, a web server for management and validation of mass spectrometry-based proteomic data. Proteomics 7, 2553–2556 10.1002/pmic.200600784 [DOI] [PubMed] [Google Scholar]
- 66. Li Z., Levast B., and Madrenas J. (2017) Staphylococcus aureus downregulates IP-10 production and prevents Th1 cell recruitment. J. Immunol. 198, 1865–1874 10.4049/jimmunol.1601336 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.