Abstract
INTRODUCTION:
Genome-wide-association studies (GWAS) discovered multiple late-onset Alzheimer’s disease (LOAD)-associated SNPs and inferred the genes based on proximity, however, the actual causal genes are yet to be identified.
METHODS:
We defined LOAD-GWAS regions by the most significantly associated SNP ±0.5Mb, and developed a bioinformatics pipeline that utilizes and integrates chromatin state segmentation track to map active enhancers and virtual 4C software to visualize interactions between active enhancers and gene-promoters. We augmented our pipeline with biomedical and functional information.
RESULTS:
We applied the bioinformatics pipeline using three ~1Mb LOAD-GWAS loci: BIN1, PICALM, CELF1. These loci contain 10–24 genes, an average of 106 active enhancers and 80 CTCF sites. Our strategy identified all genes corresponding to the promoters that interact with the active enhancer that is the closest to the LOAD-GWAS-SNP and generated a shorter list of prioritized candidate LOAD-genes (5–14/loci), feasible for post-GWAS investigations of causality.
DISCUSSION:
Interpretation of LOAD-GWAS discoveries requires the integration of brain-specific functional genomic datasets and information related to regulatory activity.
Keywords: late onset Alzheimer’s disease, GWAS, CTCF, chromatin state segmentation, virtual Circular Chromosomal Conformation Capture (4C), BIN1, PICALM, CELF1
BACKGROUND
To date, large genome-wide association studies (GWAS) have identified over 40 genomic regions associated with late-onset Alzheimer’s disease (LOAD) and many have been highly-replicated [1–7]. The large majority of LOAD-GWAS associated SNPs mapped in intergenic regions of the genome, thus, the identification of target genes is challenging due to aspects of gene density, linkage disequilibrium (LD) structure, and chromatin conformation. While the disease-associated genes have been inferred based on proximity to the most significantly associated tagging SNPs, the actual causal genes are yet to be identified and confirmed. As an example, recently Huang et al. showed that SPI1 gene rather than CELF1, the most proximal gene to the GWAS SNP, has an effect on LOAD age of onset[8].
Differential gene expression in LOAD vs. healthy controls were described in brain tissues by our team and others[9],[10, 11] and a number of evidences suggest that LOAD-risk variants may have a regulatory function. First, most LOAD-GWAS associated SNPs are located in noncoding genomic regions, possibly affecting regulatory elements including transcriptional enhancers[12, 13]. Second, expression quantitative trait loci (eQTL) studies in brain tissues from cognitively normal[14] and LOAD[15–18] samples reported overlap with LOAD-GWAS loci. Last, integration of findings from LOAD epigenome wide association (EWA) and GWA also identified a number of shared loci[19–26]. Identifying causal genes and pathways underlying LOAD-associated loci requires integrative analyses of expression and epigenetic datasets in disease-relevant brain region and cell types[27].
Herein we take LOAD-GWAS discoveries to the next level and propose an in-silico pipeline to start with GWAS discoveries, prioritize candidate functional elements, and translate them into causal genes. We modeled our strategy using three genomic regions replicated in LOAD-GWAS as highly significant loci. Two loci were identified by Lambert et. al.[1] as the most significant LOAD associated SNPs, rs6733839 (p=6.9 × 10−44) and rs10792832 (p=9.3 × 10−26), referred to by their proximate genes BIN1 and PICALM, respectively. The third locus, known as CELF1, tagged by rs10838725 (p=1.1 × 10−8) was recently investigated in depth in the context of pinpointing the target LOAD risk gene within this GWAS region [8]. The study tested for genetic association with age of onset combining with functional genomic approaches. The results suggested SPI1, rather than CELF1, as a stronger candidate causal gene within this LOAD-GWAS region. Therefore, we selected this locus to serve as a proof of concept for our bioinformatics pipeline. We found a range of 10–24 genes that mapped ±0.5Mb of the three studied GWAS-SNPs. By applying the integrated bioinformatics strategy based on potential regulatory elements we narrowed down the range to 5–14 per LOAD-GWAS region, i.e. 5, 6 and 14 genes for PICALM, BIN1 and CELF1 known regions, respectively. The candidate genes that we catalogued using the integrated computational analyses were then prioritized based on biological relevance for follow-up laboratory-based validation using in vitro and in vivo model systems.
METHODS
Selection of LOAD-associated genomic regions
The starting point for the bioinformatics analysis focused on 23 genomic regions identified by tagging SNPs in the 2013 International Genomics of Alzheimer’s Disease Project (IGAP) LOAD-GWAS[1] including the APOE-TOMM40 region (Supplementary Table S1). For each LOAD-GWAS tagging SNP, 500kb upstream and downstream defined the initial selected 1Mb region. Using a 1Mb range is a conservative boundary based on studies to predict the range of linkage disequilibrium (LD) for mapping disease genes[28, 29]. Next, we used the genes and gene prediction track, downloaded from the Table Browser[30] for the UCSC genome browser[30] (GRCh37/hg19) to identify genes that were near the boundaries of the 1Mb region. If the boundary of the 1Mb region was contained within a gene (from transcription start site to the terminator sequence in the 3’ UTR), then the coordinates for the region were extended to include the entire gene. This new set of coordinates that defined the adjusted LOAD associated regions were used for further analysis (Supplementary Table S1).
Identification of active enhancers and CCCTC-binding factor (CTCF) regions
For the identification of active enhancer elements, chromatin state segmentation data from the Roadmap Epigenomics Project[31] (http://www.roadmapepigenomics.org/) was downloaded using the UCSC Table Browser[30, 32]. Specifically, we used chromatin state segmentation data available for brain regions involved in LOAD pathology: hippocampus middle, inferior temporal lobe and mid frontal lobe. Data for peripheral blood mononuclear primary cells was included as a non-brain tissue comparator. Chromatin state segmentation was derived using ChromHMM, a multivariate Hidden Markov Model (HMM) that learns patterns of chromatin structure based on histones modification marks[33, 34]. Briefly, a common set of chromatin states (e.g. transcription start sequences, strong and weak transcription, active enhancers, genic enhancers, heterochromatin) across human tissues and cell lines were determined computationally by integrating ChIP-seq data for 6 core marks (H3K27me3, H3K36me3, H3K4me1, H3K4me3, H3K9ac, H3K9me3) and also for H3K27ac. The 18 states (Auxiliary) configuration was used to segment the defined LOAD- genomic regions and the resulting segments were annotated according to the predictions of functional elements. The set of genomic coordinates for potential enhancer elements were obtained by filtering for active enhancers (states 9, 10), including enhancers in the flanking regions of the genes. To identify CTCF binding sites, data from ENCODE/University of Washington was downloaded using the UCSC Table Browser[30, 32]. Specifically, we used the data available for the retinoic acid-induced differentiated SK-N-SH-RA neuroblastoma cell line[35].
Overlap between the active enhancers and CTCF binding sites was assessed based on the genomic coordinates of these sites, and the coordinates and number of the overlapping regions were determined. Six different types of overlaps were observed: (1) Enhancer contained within CTCF, (2) CTCF contained within enhancer, (3) Enhancer overlaps with left flanking region of CTCF, (4) Enhancer contained within CTCF but spans beyond right flanking region, (5) CTCF overlaps with left flanking region of enhancer, (6) CTCF contained within enhancer but spans beyond right flanking region.
Identification of gene promoters linked to defined enhancers using virtual 4C
Gene regulation is driven by interactions between proximal and distal regulatory elements in the genome, i.e. promoters and enhancers. Circular Chromosomal Conformation Capture (4C) is a chromatin ligation-based method to identify frequencies of chromatin interaction events between a specific “bait” locus and other proximal and distal loci[36, 37]. We used the Virtual 4C option of the 3D genome browser[38] to plot interaction frequencies between active enhancers and basal promoters of the target genes within the defined LOAD genomic regions. For the “bait” region, we used the genomic coordinates of the active enhancers that were closest to the corresponding LOAD-GWAS SNPs. Next, we utilized a feature of the 3D genome browser that displays long-range interactions between distal regulatory elements and target genes to determine the target genes linked to the active enhancers. Specifically, we applied the option to visual DNase I hypersensitive site (DHS)-linkage data to chart arcs between active enhancers and the basal promoters of the target genes. The promoter sequence of the target genes was determined as the 600 bp 5’ flanking sequence upstream of the transcription start site as shown in the UCSC Browser. For each of the studied LOAD GWAS defined regions, the results of the enhancer-promoter interaction frequency (4C reads) were plotted for the hippocampus. Each plot consists of three panels: (1) The top line graph shows the frequency of chromatin interaction events, centered on the bait enhancer element, (2) Elliptical arcs indicate the interactions between active enhancer element and promoter sites, whereas the threshold to declare an interaction and draw an arc was based on DHS-linkage data with a Pearson correlation coefficient ≥ 0.7[39]. All arcs involving the same proximal DHS are shown with the same color, (3) The arcs were overlaid on the same charts illustrating the genes’ structure and chromHMM tracks for brain hippocampus middle from the UCSC genome browser. Supplemental data includes this plot augmented with chromHMM data for brain inferior temporal lobe, brain dorsolateral prefrontal cortex and primary mononuclear cells from peripheral blood as a non-brain tissue comparator (Supplementary Figure S1).
Data and computer code availability
All computer code used for determining the overlapping genomic regions based on the various elements (enhancers, CTCF regions, promoters) is available on Github at the following link: https://github.com/NCTrailRunner/Alzheimers-bioinformatics-resource. Datasets containing the genomic regions and annotations described in the bioinformatics analysis of this paper are also available through this public resource. The virtual 4C software is available at http://promoter.bx.psu.edu/hi-c/virtual4c.php.
RESULTS AND DISCUSSION
Bioinformatics pipeline to prioritize candidate LOAD causal genes for experimental Validation
Identification of genes associated with LOAD has previously been determined by proximity to GWAS tagging SNPs[1, 4, 40]. These genes have been then mapped to several major biochemical pathways implicated in LOAD pathogenesis including, lipid metabolism, immune response, regulation of endocytosis and protein ubiquitination[41, 42]. In a recent study Amlie-Wolf et al. developed a novel software, named INFERNO, to infer the molecular mechanisms of noncoding genetic variants by integration of hundreds of functional genomics datasets spanning enhancer activity, transcription factor binding sites, and expression quantitative trait loci (eQTL) with GWAS summary statistics[12, 13]. In a subsequent work, the team applied INFERNO to IGAP GWAS data and characterized the effects of noncoding genetic variations associated with LOAD risk on gene dysregulation[12, 13].
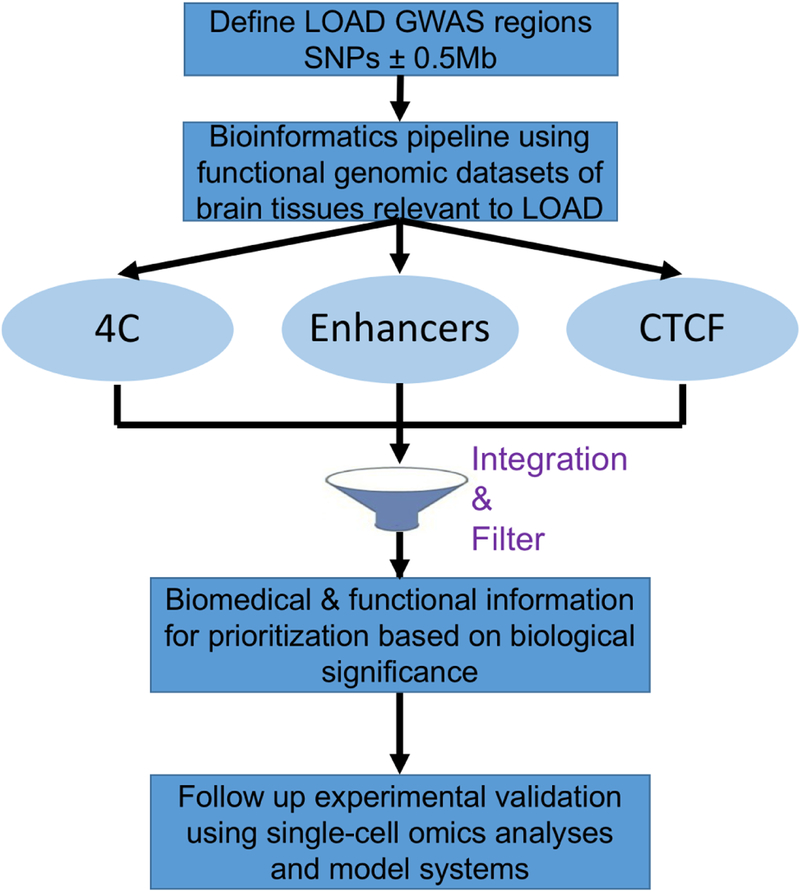
In this paper, we developed a new complementary bioinformatics strategy to prioritize candidate causal genes (vs. variants) mapped within the extended 1Mb regions tagged by LOAD GWAS[1]. The proposed bioinformatics pipeline, illustrated in Figure 1, will guide post-GWAS follow-up experimental work and validation studies. Our bioinformatics strategy is based entirely on publicly available genomic datasets including: annotation of enhancer elements and minimal promoters, definition of CTCF sites, and identification of 3D genome structure of enhancer promoter interactions. Here we integrate these genomic datasets to construct a comprehensive bioinformatics resource for the expanded (1Mb) well-replicated LOAD GWAS regions.
Figure 1.
Schematic of the bioinformatics-based strategy to prioritize LOAD-GWAS candidate risk genes for experimental follow up and validation of causality.
Defining the expanded LOAD GWAS regions and generating catalogues of genes and regulatory elements
The region tagged by each top LOAD-SNP was initially defined by anchoring the center of the region on the GWAS SNP and extending 500kb in each direction to cover a 1Mb locus. Genes on the boundary of the 1Mb region were examined and the locus extended to cover the full length of the gene if the boundary intersects within a gene. Supplementary Table S1 lists the resulting expanded 23 LOAD GWAS loci and includes the summary statistics for each tagging LOAD-associated SNP reported by Lambert et. al.[1].
The 23 expanded ~1Mb LOAD GWAS regions contain nearly 500 genes with an average of 21 genes per region (Table 1). The complete list of genes for each region appears in Supplementary Table S2. Thus, a strategy for prioritizing this extended list of target genes is needed prior proceeding with validation experiments in model systems, such as human iPSC-derived neuronal, 3D multi-culture and organoid systems, and rodent models. Towards this end, we first generated a comprehensive tissue-specific catalogue of regulatory elements, particularly, enhancer elements and CTCF binding sites, as CTCF is known to affect enhancer promoter interactions and is involved in gene regulation. The catalogue of active enhancers consists of data for brain regions affected in LOAD: hippocampus middle, inferior temporal lobe and mid frontal lobe. The catalogue of CTCF binding sites consists of data obtained from the neuroblastoma cell line, SK-N-SH-RA. The average number of active enhancers and CTCF binding sites for an expanded LOAD GWAS region was 22 and 109, respectively, with an average of 22 overlapping sites for each region (Table 1). The coordinates for all overlapping enhancer elements and CTCF binding sites and the type of overlap for each of the 23 LOAD GWAS regions are summarized as a data resource in Supplemental Table S3.
Table 1.
Summary statistics of target genes and regulatory elements positioned within LOAD associated regions
| N | average /region | SD (per region) | range (per region) | |
|---|---|---|---|---|
| Gene regions | 23 | |||
| Genes mapped to 1Mb proximal regions | 493 | 21 | 4–62 | |
| Active enhancers | 1527 | 66 | 37 | 10–117 |
| CTCF sites | 2509 | 109 | 64 | 48–281 |
| Active enhancers and CTCF site overlap | 462 | 22 | 17 | 2–70 |
Integration of the brain regulatory elements resource with visualization map of enhancerpromoter interactions: example analyses of three LOAD GWAS regions
Next, we applied our bioinformatics strategy (Figure 1) to three expanded LOAD GWAS regions denoted hereafter by the gene most proximate to the top associated SNP, i.e. BIN1, PICALM and CELF1 (Table 2). These ~1Mb loci encompass 10–24 genes each. In order to prioritize potential causal genes and focus follow-up experiments on a relatively small list of strong candidate LOAD genes, we first annotated the regulatory elements within the loci. The number of genes and brain regulatory elements for each of these three ~1Mb loci with the overlapping regions of the active enhancer and CTCF sites are summarized in Table 2. The chromHMM track for brain hippocampus middle is shown for the example loci referred to as BIN1 (Figure 2), PICALM (Figure 3) and CELF1 (Figure 4) indicating the respective location of the active enhancers (orange), transcription (green) and the transcription start site (red). Additional tissue-specific chromHMM tracks for the brain inferior temporal lobe, brain midfrontal lobe and peripheral blood mononuclear primary cells (as a non-brain comparator) are shown for these loci in Supplemental Figure S1.
Table 2.
LOAD GWAS loci used to demonstrate the bioinformatics pipeline for prioritizing candidate LOAD genes
| Gene | Variant | Chr | Location | #CTCF | # enhancers | # genes | Interacting genes | Location of nearest Enhancer | CTCF/enhancer overlap | # overlapping regions |
|---|---|---|---|---|---|---|---|---|---|---|
| CELF1 | rs10838725 | 11 | 47,557,871 | 99 | 90 | 24 | CELF1, SPI1, ACP2, AGBL2, ARFGAP2, DDB2, FNBP4, MADD, MTCH2, NRIH3, NUP160, PSMC3, PTPRJ, SLC39A13 | 47,557,800–47,558,200 | CTCF contained within enhancer | 6 |
| Enhancer contained within CTCF but spans beyond right flanking region | 2 | |||||||||
| Enhancer overlaps with left flanking region of CTCF | 6 | |||||||||
| PICALM | rs10792832 | 11 | 85,867,875 | 75 | 116 | 10 | PICALM, SYTL2, EED, CCDC83, CCDC89 | 85,867,400–85,867,800 | CTCF contained within enhancer | 6 |
| Enhancer contained within CTCF but spans beyond right flanking region | 1 | |||||||||
| Enhancer overlaps with left flanking region of CTCF | 2 | |||||||||
| BIN1 | rs6733839 | 2 | 127,892,810 | 65 | 112 | 10 | BIN1, ERCC3, GYPC, IWS1, MYO7B, PROC | 127,893,000–127,893,400 | CTCF contained within enhancer | 4 |
| Enhancer contained within CTCF but spans beyond right flanking region | 1 | |||||||||
| Enhancer overlaps with left flanking region of CTCF | 1 |
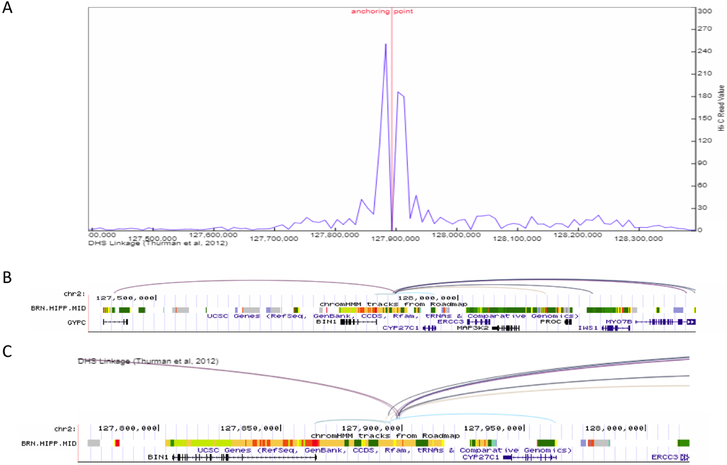
Figure 2.
Enhancer-promoter interaction plot for the >1Mb defined BIN1 LOAD-GWAS region, tagged by SNP rs6733839. (A) Line plot of the interaction frequency for Hi-C reads in the hippocampus, anchoring point at chr2:127,893,000 (5’ end of enhancer). (B) gene structure and chromHMM track for brain hippocampus middle to show the location of genomic structures including active enhancers (orange), transcription (green) and the transcription start site (red). Proximate active enhancer (type 9) was defined at chr2:127,893,000–127,893,400 (400 bp), 190bp from the tagging SNP rs6733839 (chr2:127,892,810). Arcs show interactions between the defined enhancer and promoters (Pearson correlation coefficient ≥ 0.7) in the target genes based on DNase I hypersensitive site (DHS)-linkage data. All arcs involving the same proximal DHS are drawn with the same color. (C) Inset shows a magnified version of the arcs and ChromHMM for an approximately 200Kb subset of the full genomic region. The inset shows the origin of the arcs in the enhancer.
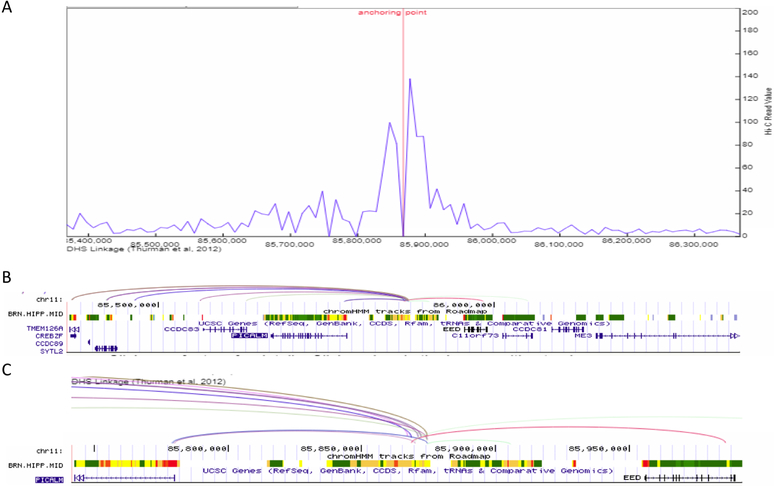
Figure 3.
Enhancer-promoter interaction plot for the >1Mb defined PICALM LOAD-GWAS region, tagged by SNP rs10792832. (A) Line plot of the interaction frequency for Hi-C reads in the hippocampus, anchoring point at chr11:85,867,400 (5’ end of enhancer). (B) gene structure and chromHMM track for brain hippocampus middle to show the location of genomic structures including active enhancers (orange), transcription (green) and the transcription start site (red). Proximate active enhancer (type 9) was defined at chr11:85,867,400–85,867,800 (400 bp), 75bp from the tagging SNP rs10792832 (chr11:85,867,875). Arcs show interactions between the defined enhancer and promoters (Pearson correlation coefficient ≥ 0.7 between enhancer and promoter) in the target genes based on DNase I hypersensitive site (DHS)-linkage data. All arcs involving the same proximal DHS are drawn with the same color. (C) Inset shows a magnified version of the arcs and ChromHMM for an approximately 200Kb subset of the full genomic region. The inset shows the origin of the arcs in the enhancer.
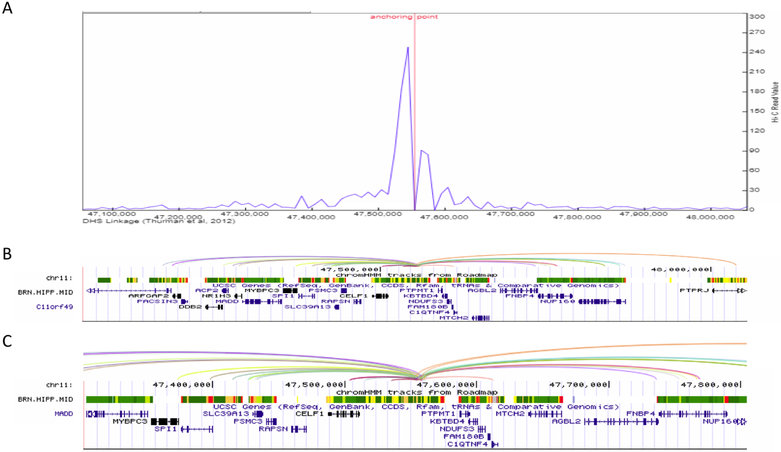
Figure 4.
Enhancer-promoter interaction plot for the >1Mb defined CELF1 LOAD-GWAS region, tagged by SNP rs10838725. (A) Line plot of the interaction frequency for Hi-C reads in the hippocampus, anchoring point at chr11:47,557,800 (5’ end of enhancer). (B) gene structure and chromHMM track for brain hippocampus middle to show the location of genomic structures including active enhancers (orange), transcription (green) and the transcription start site (red). Proximate active enhancer (type 11) was defined at chr11: 47,557,800–47,558,200 (400 bp), 71bp from the tagging SNP rs10838725 (chr11:47,557,871). Arcs show interactions between the defined enhancer and promoters (Pearson correlation coefficient ≥ 0.7 between enhancer and promoter) in the target genes based on DNase I hypersensitive site (DHS)-linkage data. All arcs involving the same proximal DHS are drawn with the same color. (C) Inset shows a magnified version of the arcs and ChromHMM for an approximately 200Kb subset of the full genomic region. The inset shows the origin of the arcs in the enhancer.
Next, we identified and visualized the interactions between distal and proximal regulatory elements, i.e. the annotated enhancer elements and the minimal promoter of the genes within the BIN1, PICALM and CELF1 LOAD GWAS regions. We used the data resource described above for the hippocampus to define the ‘bait’ for the visualization of the interactions map as the active enhancer site closest to the top LOAD GWAS SNPs. Specifically, Table 2 indicates the SNP rs number and chromosomal location, and the coordinates of the closest active enhancer element used as an anchor for plotting the promoter interactions. The closest active enhancers were determined 190bp, 75bp and 71bp from the anchor point set as the LOAD GWAS SNPs, for BIN1, PICALM and CELF1 loci, respectively (Figures 2–4, Table 2). The coordinates for the closest active enhancers for each of the 23 LOAD GWAS SNPs are provided in Supplemental Table S2. Subsequently, the 3D genome browser was used to construct the visualization maps of the genome organization[38] in the hippocampus for the example loci, known as: BIN1 (Figure 2), PICALM (Figure 3) and CELF1 (Figure 4). The arcs show interactions between the defined closest active enhancer for each region and the promoters of the target genes based on DNase I hypersensitive site (DHS)-linkage data (Pearson correlation coefficient ≥ 0.7 between enhancer and promoter) and were verified by checking the location of the promoter as described in the Methods. The genes linked to the promoters are listed as interacting genes in Table 2.
The filtering stage that used the 3D genome organization maps to identify enhancer-promoter interactions reduced the overall number of LOAD candidate genes by 40%−50% (Table 2). Of note, the virtual 4C analysis of the CELF1 loci (Figure 3) identified the interaction between the proximal enhancer and the promoter of the SPI1 gene. SPI1 encodes PU.1, a transcription factor that is critical for myeloid cell development and function. Our results provide, retrospectively, a bioinformatics validation to a recently published study that reported an association between a SNP in the SPI1 gene, positioned in the CELF1 LOAD risk locus that delayed LOAD onset[8]. In addition, the study found SPI1 eQTL association in monocytes and macrophages for this same SNP, and provided evidence suggesting that PU.1 may regulate the expression of multiple LOAD associated genes in myeloid cells[8]. Last, overexpression and down regulation of PU.1 levels in mouse microglial cells affected the expression of mouse orthologs of several LOAD risk genes and the phagocytic activity. While CELF1 is the most proximate gene to the LOAD-SNP at this locus, SPI1, separated by 87,357bp, was suggested as a stronger candidate causal gene for LOAD risk based on functional lines of evidence. Collectively, the results for SPI1 demonstrated the concept that a gene different from the most proximal gene to the GWAS SNP contributes to LOAD pathogenesis. This concept was also applied for the interpretation of novel loci identified through the largest LOAD GWAS meta-analysis[43]. As a prioritization strategy for ranking the genes located in LOAD-GWAS significant loci the authors used several criteria including, gene expression and eQTL effect on the gene in tissue relevant to LOAD, expression correlation with Braak stage and LOAD-associated differential expression, as well as involvement in biological pathways enriched in LOAD. Our study and the two studies discussed above [8][43] integrated functional genomic information to interpret LOAD-GWAS discoveries; while the others prioritize primarily based on expression traits our strategy uniquely utilize the genome features/states and chromatin organization and focus on the interplay between noncoding regulatory element and 3D chromatin structure.
Biological insights to further narrow down candidate for experimental follow-up
Subsequent to the bioinformatics analysis, we used biological knowledge and outcomes from previous studies related to the interacting genes (Table 2 and Supplementary Table S4) to refine the list of target genes for post-GWAS follow up studies that will validate and characterize their putative contribution to LOAD pathogenesis. We annotated each of the interacting genes based on functional and biomedical information including: encoded protein function, expression profiles, related pathways and associated diseases (Supplementary Table S4). Herein we discuss pathways that presumably have a biological relevance to LOAD and were implicated for a subset of the interacting genes (Table 3). Overall this approach narrowed down the highest priority genes for experimental exploration using model systems to 2–7 genes per locus from the interacting genes identified by the bioinformatic pipeline.
Table 3.
Highest priority candidate LOAD causal genes: Biomedical and Functional information
| Gene name | Gene coordinate | GWAS-SNP (coordinate) | Biomedical/Functional category | Brain expression |
|---|---|---|---|---|
| CELF1 | Chr11: 47,487,489–47,587,121 | rs10838725 (Chr11: 47,557,871) | LOAD as a systematic disease, RNA metabolism related | ++ |
| SPI1 | Chr11: 53,773,960–53,810,230 | LOAD as a systematic disease, Neurological and Alzheimer’s associated, HIV related, DNA damage and repair related | ++ | |
| DDB2 | Chr11: 47,236,493–47,260,769 | DNA damage and repair related | + | |
| MADD | Chr11: 47,290,712–47,351,582 | Neurological and Alzheimer’s associated | ++ | |
| MTCH2 | Chr11: 47,639,858–47,664,206 | LOAD as a systematic disease | ++ | |
| NR1H3 | Chr11: 47,269,851–47,290,401 | LOAD as a systematic disease | ++ | |
| PSMC3 | Chr11: 47,440,320–47,448,024 | DNA damage and repair related | +++ | |
| PICALM | Chr11: 85,668,214–85,780,924 | rs10792832 (Chr11:85,867,875) | Neurological and Alzheimer’s associated | +++ |
| EED | Chr11: 85,955,586–85,989,855 | HIV related | + | |
| BIN1 | Chr2: 127,048,023–127,107,400 | rs6733839 (Chr2:127,892,810) | Neurological and Alzheimer’s associated | +++ |
| ERCC3 | Chr2: 128,014,866–128,051,752 | HIV related, RNA metabolism related, DNA damage and repair related | ++ | |
| IWS1 | Chr2: 128,193,783–128,284,462 | HIV related, RNA metabolism related | + |
Four of the interacting genes identified in this study positioned across all studied loci, PICALM, BIN1, SPI1 and MADD, have been well-studied in relation to neurological pathways and LOAD. This category includes PICALM and BIN1 genes that were previously associated with LOAD based on their proximity to the GWAS-SNPs. PICALM was shown to affect LOAD risk by modulating processes affecting beta-amyloid accumulation[44, 45], and BIN1 was suggested to be involved in LOAD related pathways including, synaptic vesicle endocytosis and BACE1 recycling that in-turn impacts beta-amyloid endocytic production[45]. The CELF1 locus harbors SPI1 with potential contribution to LOAD as described above[8]. In addition, the SPI1 gene was implicated in neuronal cell death in Huntington’s disease, and its inhibition in mouse models led to memory deficits and increased levels of amyloid-beta plaques, suggesting a possible role in LOAD pathogenesis[46]. Last, down regulation of MADD was correlated with neuronal cell death in LOAD[47].
LOAD has been investigated as a brain disease, however, it has been suggested that LOAD is a systemic disease involving other organs such as the liver and gut, and affecting various biological processes including lipid metabolism and immune response[48]. Furthermore, APOE, the most reproducible LOAD genetic risk factor with the strongest effect size, plays a role in cholesterol and lipid metabolism. Interestingly the CELF1 LOAD-GWAS expanded locus is enriched in genes involved in lipid and cholesterol metabolism: CELF1, MTCH2, NR1H3, generating the hypothesis that co-regulation of genes in this cluster may lead to an additive or synergistic effect on disease risk; the forth gene mapped to this pathway from our focused analysis is SPI1.
Four interacting genes have been related to aspects of HIV including EED, ERCC3, IWS1, and SPI1 positioned across all three LOAD-GWAS loci used as examples in this paper. The relationship between HIV and cognitive impairment has been studied for decades[49] and HIV-associated neurocognitive disorder (HAND) presents similar symptoms to Alzheimer’s disease[50]. The genetic intersection between HIV and LOAD is intriguing and warrants further investigations using experimental models to better understand shared mechanisms of disease susceptibility.
CELF1, IWS1, and ERCC3 all play roles in the regulation of RNA transcription, splicing, and metabolism. CELF1 and IWS1 both exert a role on regulating mRNA translation and splicing, while ERCC3 plays a role in RNA transcriptional initiation and promotion. Dysregulation of gene expression and changes in transcriptional programs have been described in aging and neurodegenerative diseases including LOAD[51]. Noteworthy IWS1 and ERCC3 located in the BIN1 LOAD-GWAS locus are also HIV-related genes, implying a possible connection between deficiency in RNA metabolisms and cognition in the context shared mechanisms in LOAD and HAND.
Four interacting genes are known to be associated with DNA repair and damage response including SPI1, DDB2, PSMC3, ERCC3, mapped to the BIN1 and CELF1 LOAD-GWAS loci. High rates of DNA damage have been reported in aging and in the progression of neurodegenerative diseases such as LOAD, mild cognitive impairment[52], and Parkinson’s[53].
CONCLUDING REMARKS
LOAD is a genetically heterogenous disease involving multiple genomic loci that mediate their effect on pathogenesis via genetic and epigenetic mechanisms. GWAS and whole genome/exome sequencing implicated about 40 loci in LOAD navigating LOAD genetic research to specific genomic regions of interest. Post-GWAS research that applies a multifaceted strategy combining in silico, in vitro and in vivo approaches is needed for the identification and validation of the precise causal gene/s within the associated loci. Our overall strategy is based on the hypothesis that dysregulation of gene expression contributes, at least in part, to LOAD pathogenesis. This paper focuses on in silico analysis approach and proposes a new bioinformatics pipeline (Figure 1) that utilizes publicly available functional genomic and epigenomic datasets specifically, chromatin state segmentation based on hippocampus-specific histone modification marks from the Epigenome Roadmap, and enhancer-promoter interactions based on hippocampus specific 4C data. Using three model LOAD regions, we demonstrated that our computational strategy is feasible to identify the highest priority LOAD candidate causal genes and to guide follow up laboratory experiments.
The new bioinformatics pipeline presented here utilized datasets of brain regulatory elements obtained from normal brains. While gene regulation in health is beneficial to interpret gene dysregulation in disease, it lacks the context of disease. Thus, we suggest augmenting our basic pipeline by integration of comprehensive epi/genomic datasets, generated through ongoing and prospective projects, from LOAD brains. Moreover, the publicly available omics databases were generated using bulk brain tissues. Brain tissue homogenates consist of heterogenous cell-types, i.e. different neurons and various types of glia cells, and therefore introduce bias and sample-to-sample variations related to cell-type composition. In addition, molecular phenotypes determined using bulk brain tissues are not informative regarding the brain cell-type responsible for the differential molecular/omics profile. These limitations underscore the need for single brain cell-type specific omics data from healthy and LOAD individuals to further enhance the interpretation of LOAD-GWAS discoveries. Finally, the top LOAD candidate genes and variants, identified through genetic association studies and bioinformatics analyses, will be subject for validation and in-depth characterization of their pathogenic effects using in vitro and in vivo model systems.
Supplementary Material
RESEARCH IN CONTEXT.
Systematic review: The authors reviewed the Literature using Pubmed, meeting abstracts and presentations and downloaded publicly available genome and epigenome datasets. While LOAD-GWAS genes have been inferred based on proximity to the top significantly associated tagging SNPs, the actual causal genes are yet to be identified.
Interpretation: Our findings support the concept that LOAD causal genes may not be simply inferred as the most proximate gene but that the interpretation of GWAS discoveries requires the integration of functional genomic datasets and information related to regulatory activity in the context of LOAD. This will facilitate cataloguing the highest priority LOAD candidate genes for post-GWAS follow-up experiments.
Future directions: We propose a framework for in-depth investigation of causality in the following directions: (a) Exploring the effect of the top priority target genes on LOAD related phenotypes using in vitro and in vivo model systems; (b) Applying the bioinformatics pipeline using the extended list of LOAD-associated SNPs and depositing the data as a resource for the community of researchers in the field of LOAD genetics; (c) Identifying regulatory genetic variants within the active enhancer elements and characterizing their effects on the expression of the strongest candidate LOAD genes.
Highlights.
LOAD-GWAS regions defined by the top associated SNP ±0.5 Mb encompass an average of 21 genes.
Gene/s within LOAD-associated loci, not necessarily the most proximal gene to the GWAS SNP, may be the LOAD causal genes and play a role in disease pathogenesis.
We propose a bioinformatics pipeline that integrates brain active enhancers information and promoter-enhancer interaction maps to prioritize candidate LOAD causal genes for experimental validation and further exploration of their pathogenic effects.
Applying our strategy using three ~1Mb LOAD-GWAS regions resulted in a list of 40–50% fewer candidate LOAD causal genes vs the physical map in the genome browser. Biomedical information facilitated further sharpened the focus on 2–7 top priority target genes per region.
Acknowledgments
We thank Dr. Feng Yue at Pennsylvania State University College of Medicine for providing support and advice in using the Virtual 4C genome browser software developed by his team.
FUNDING SOURCES
This work was funded in part by the National Institutes of Health/National Institute of Aging (NIH/NIA) [R01 AG057522 to O.C-F.].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts
The authors declare no competing interests.
REFRENCES
- [1].Lambert J-C, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet.43:436–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet.43:429–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jansen I, Savage J, Watanabe K, Bryois J, Williams D, Steinberg S, et al. Genetic meta-analysis identifies 9 novel loci and functional pathways for Alzheimers disease risk. bioRxiv. 2018. [Google Scholar]
- [6].Marioni R, Harris SE, McRae AF, Zhang Q, Hagenaars SP, Hill WD, et al. GWAS on family history of Alzheimer’s disease. bioRxiv. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Naj AC, Boland A, et al. Meta-analysis of genetic association with diagnosed Alzheimer’s disease identifies novel risk loci and implicates Abeta, Tau, immunity and lipid processing. bioRxiv. 2018. [Google Scholar]
- [8].Huang KL, Marcora E, Pimenova AA, Di Narzo AF, Kapoor M, Jin SC, et al. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat Neurosci. 2017;20:1052–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Linnertz C, Anderson L, Gottschalk W, Crenshaw D, Lutz MW, Allen J, et al. The cis-regulatory effect of an Alzheimer’s disease-associated poly-T locus on expression of TOMM40 and apolipoprotein E genes. Alzheimers Dement. 2014;10:541–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Matsui T, Ingelsson M, Fukumoto H, Ramasamy K, Kowa H, Frosch MP, et al. Expression of APP pathway mRNAs and proteins in Alzheimer’s disease. Brain Res. 2007;1161:116–23. [DOI] [PubMed] [Google Scholar]
- [11].Zarow C, Victoroff J. Increased apolipoprotein E mRNA in the hippocampus in Alzheimer disease and in rats after entorhinal cortex lesioning. Experimental neurology. 1998;149:79–86. [DOI] [PubMed] [Google Scholar]
- [12].Amlie-Wolf A, Tang M, Mlynarski EE, Kuksa PP, Valladares O, Katanic Z, et al. INFERNO: inferring the molecular mechanisms of noncoding genetic variants. Nucleic Acids Res. 2018;46:8740–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Amlie-Wolf A, Tang M, Way J, Dombroski B, Jiang M, Vrettos N, et al. Inferring the molecular mechanisms of noncoding Alzheimer’s disease-associated genetic variants. bioRxiv. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet.6:e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Allen M, Zou F, Chai HS, Younkin CS, Crook J, Pankratz VS, et al. Novel late-onset Alzheimer disease loci variants associate with brain gene expression. Neurology. 2012;79:221–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zou F, Chai HS, Younkin CS, Allen M, Crook J, Pankratz VS, et al. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS genetics. 2012;8:e1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Karch CM, Jeng AT, Nowotny P, Cady J, Cruchaga C, Goate AM. Expression of novel Alzheimer’s disease risk genes in control and Alzheimer’s disease brains. PLoS One. 2012;7:e50976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Karch CM, Ezerskiy LA, Bertelsen S, Alzheimer’s Disease Genetics C, Goate AM. Alzheimer’s Disease Risk Polymorphisms Regulate Gene Expression in the ZCWPW1 and the CELF1 Loci. PloS one. 2016;11:e0148717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Smith AR, Smith RG, Condliffe D, Hannon E, Schalkwyk L, Mill J, et al. Increased DNA methylation near TREM2 is consistently seen in the superior temporal gyrus in Alzheimer’s disease brain. Neurobiol Aging. 2016;47:35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhao J, Zhu Y, Yang J, Li L, Wu H, De Jager PL, et al. A genome-wide profiling of brain DNA hydroxymethylation in Alzheimer’s disease. Alzheimer’s & dementia : the journal of the Alzheimer’s Association. 2017;13:674–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lunnon K, Smith R, Hannon E, De Jager PL, Srivastava G, Volta M, et al. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nat Neurosci. 2014;17:1164–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L, et al. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci. 2014;17:1156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chibnik LB, Yu L, Eaton ML, Srivastava G, Schneider JA, Kellis M, et al. Alzheimer’s loci: epigenetic associations and interaction with genetic factors. Ann Clin Transl Neurol. 2015;2:636–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yu L, Chibnik LB, Srivastava GP, Pochet N, Yang J, Xu J, et al. Association of Brain DNA methylation in SORL1, ABCA7, HLA-DRB5, SLC24A4, and BIN1 with pathological diagnosis of Alzheimer disease. JAMA Neurol. 2015;72:15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Watson CT, Roussos P, Garg P, Ho DJ, Azam N, Katsel PL, et al. Genome-wide DNA methylation profiling in the superior temporal gyrus reveals epigenetic signatures associated with Alzheimer’s disease. Genome Med. 2016;8:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nativio R, Donahue G, Berson A, Lan Y, Amlie-Wolf A, Tuzer F, et al. Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease. Nat Neurosci. 2018;21:497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, et al. Comprehensive functional genomic resource and integrative model for the human brain. Science (New York, NY. 2018;362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ott J Predicting the range of linkage disequilibrium. Proc Natl Acad Sci U S A. 2000;97:2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ernst J, Kellis M. Chromatin-state discovery and genome annotation with ChromHMM. Nat Protoc. 2017;12:2478–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Biedler JL, Helson L, Spengler BA. Morphology and growth, tumorigenicity, and cytogenetics of human neuroblastoma cells in continuous culture. Cancer Res. 1973;33:2643–52. [PubMed] [Google Scholar]
- [36].Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet. 2006;38:1348–54. [DOI] [PubMed] [Google Scholar]
- [37].Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–7. [DOI] [PubMed] [Google Scholar]
- [38].Wang Y, Song F, Zhang B, Zhang L, Xu J, Kuang D, et al. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018;19:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nature genetics. 2009;41:1094–9. [DOI] [PubMed] [Google Scholar]
- [41].Pimenova AA, Raj T, Goate AM. Untangling Genetic Risk for Alzheimer’s Disease. Biol Psychiatry. 2018;83:300–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Karch CM, Cruchaga C, Goate AM. Alzheimer’s disease genetics: from the bench to the clinic. Neuron. 2014;83:11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nature genetics. 2019;51:414–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Xu W, Tan L Fau - Yu J-T, Yu JT. The Role of PICALM in Alzheimer’s Disease. [DOI] [PubMed]
- [45].Guimas Almeida C, Sadat Mirfakhar F, Perdigao C, Burrinha T. Impact of late-onset Alzheimer’s genetic risk factors on beta-amyloid endocytic production. Cell Mol Life Sci. 2018;75:2577–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Citron BA, Saykally JN, Cao C, Dennis JS, Runfeldt M, Arendash GW. Transcription factor Sp1 inhibition, memory, and cytokines in a mouse model of Alzheimer’s disease. American journal of neurodegenerative disease. 2015;4:40–8. [PMC free article] [PubMed] [Google Scholar]
- [47].Del Villar K, Miller CA. Down-regulation of DENN/MADD, a TNF receptor binding protein, correlates with neuronal cell death in Alzheimer’s disease brain and hippocampal neurons. [DOI] [PMC free article] [PubMed]
- [48].Morris JK, Honea RA, Vidoni ED, Swerdlow RH, Burns JM. Is Alzheimer’s disease a systemic disease? [DOI] [PMC free article] [PubMed]
- [49].Grant I, Atkinson JH, Hesselink JR, Kennedy CJ, Richman DD, Spector SA, et al. Evidence for early central nervous system involvement in the acquired immunodeficiency syndrome (AIDS) and other human immunodeficiency virus (HIV) infections: studies with neuropsychologic testing and magnetic resonance imaging. Annals of Internal Medicine. 1987;107:828–36. [DOI] [PubMed] [Google Scholar]
- [50].Chakradhar S A tale of two diseases: Aging HIV patients inspire a closer look at Alzheimer’s disease. Nature Medicine. 2018;24:376. [DOI] [PubMed] [Google Scholar]
- [51].Chen X-F, Zhang Y-w, Xu H, Bu G. Transcriptional regulation and its misregulation in Alzheimer’s disease. Molecular brain. 2013;6:44-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Fabio C, Lucia M. DNA Damage and Repair in Alzheimers Disease. Current Alzheimer Research. 2009;6:36–47. [DOI] [PubMed] [Google Scholar]
- [53].Tagliafierro L, Zamora ME, Chiba-Falek O. Multiplication of the SNCA locus exacerbates neuronal nuclear aging. Human Molecular Genetics. 2018:ddy355–ddy. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.