Abstract
A total of 24 Colletotrichum isolates were isolated from diseased Japanese plum (Prunus salicina) fruits showing chlorotic regions with whitish-brown sunken necrotic lesions and phylogenetic relationships among the collected Colletotrichum isolates were determined. A subset of 11 isolates was chosen for further taxonomic study based on morphology and molecular characteristics identified using the internal transcribed spacer (ITS) and beta-tubulin (TUB2) genes. Isolates in the C. acutatum complex were analyzed using partial sequencing of five gene regions (ITS, GAPDH, ACT, TUB2, and CHS), and C. gloeosporioides sensu lato (s.l.) isolates were analyzed using seven gene regions (ITS, TUB2, GAPDH, ACT, CAL, CHS-1, and ApMat). Morphological assessments in combination with phylogenetic analysis delineated four species of Colletotrichum including C. gloeosporioides sensu stricto (s.s.), C. nymphaeae, C. foriniae, and C. siamense; these data identify Colletotrichum fioriniae and C. siamense two new species associated with plum anthracnose in South Korea. Finally, the pathogenicity of these four species in the development of plum anthracnose in South Korea was confirmed by inoculations of plum fruit.
Subject terms: Fungal pathogenesis, Pathogens
Introduction
Japanese plums (Prunus salicina Lindl.) are delicious stone fruits, which have a wide variety of uses. Consumers typically prefer to eat fresh Japanese plums for their characteristic taste, though a small percentage prefer them dry. They can also be used in jams or jellies. The fruits are rich in carbohydrates (sucrose, glucose, and fructose), malic acid, phenolic compounds (chlorogenic acid, neochlorogenic acid), anthocyanins (cyanidin-3-glucoside, cyaniding-3-rutinoside), vitamin C, β-carotene, and minerals (potassium, phosphorus)1. Though native to China the name Japanese plum derives from the fruit tree first being imported into the USA from Japan2. Japanese plums are cultivated along with apples, peaches, oranges, and Asian pears in South Korea. Both the cultivation area and production of Japanese plums in South Korea increased from 2007 (5,803 ha, 64,816 tons) to 2015 (5,920 ha, 67,810 tons)3. The production of Japanese plum fruits in Korea can be negatively impacted by various factors including different diseases including fungal diseases (brown rot, gray mold, leaf spot, plum pocket, and powdery mildew) and bacterial diseases (bacterial black spot, shot hole, etc.)4–8. Recently anthracnose of Japanese plum caused by Colletotrichum species has been reported in Korea3,9.
Most Colletotrichum species are plurivorous anthracnose pathogens that cause disease in a wide range of hosts, including fruit trees and vegetables10,11. The most characteristic symptom enabling the recognition of anthracnose is the presence of sunken necrotic lesions on leaves, stems, flowers, and fruit, which limits the quality of agricultural products (fruits, flowers). Colletotrichum species have also been reported to caused anthracnose in common fruits in Korea, such as apples, grapes, peaches, and persimmons12–15. Multiple Colletotrichum species can infect a single fruit cultivar. In Korea, Colletotrichum acutatum and C. gloeosporioides sensu stricto (s.s.) are responsible for bitter rot of apples and anthracnose of peaches; C. acutatum, C. gloeosporioides s.s., and C. viniferum for ripe rot of grapes; and C. acutatum, C. gloeosporioides s.s., C. horii, and C. siamense for anthracnose of persimmons12–14,16,17. To date, C. acutatum, C. gloeosporioides s.s., and C. nymphaeae have been reported as the causal agents of plum anthracnose in Korea3,9. Lee et al. identified C. acutatum and C. gloeosporioides s.s. (causal agents of plum anthracnose) based on morphology and internal transcribed spacers (ITS) sequence data9. Methods for identifying Colletotrichum species based on morphology and ITS sequences are not reliable for species discrimination within Colletotrichum, although they can be helpful in the resolution of species complexes or clades18–20. A recently developed multi-locus sequence analysis approach combined with morphological evaluation revealed that C. gloeosporioides sensu lato (s.l.) and C. acutatum s.l., each comprise a species complex20,21. C. gloeosporioides s.s. is a strictly defined species ((Colletotrichum gloeosporioides (Penz.) Penz. & Sacc)) excluding other species within the C. gloeosporioides species complex, while C. gloeosporioides s.l. includes other Colletotrichum species of this complex20. In addition, the ApMat marker gene has been used to resolve and improve the systematic classification of Colletotrichum species complexes, such as C. gloeosporioides and C. siamense complexes22–24. The use of a five genes phylogenetic analyses along with morphological characters to identify C. nymphaeae as the causative agent of plum anthracnose revealed that Colletotrichum species associated with plum anthracnose in Korea may have a remarkable species diversity9.
Therefore, this study sought to investigate species diversity within Colletotrichum isolates related to plum anthracnose in Sangju, Korea based on combined morphological and multigene phylogenetic strategies, followed by a pathogenesis analysis of the different identified Colletotrichum species on plum fruit.
Results
Isolation and preliminary identification of Colletotrichum species
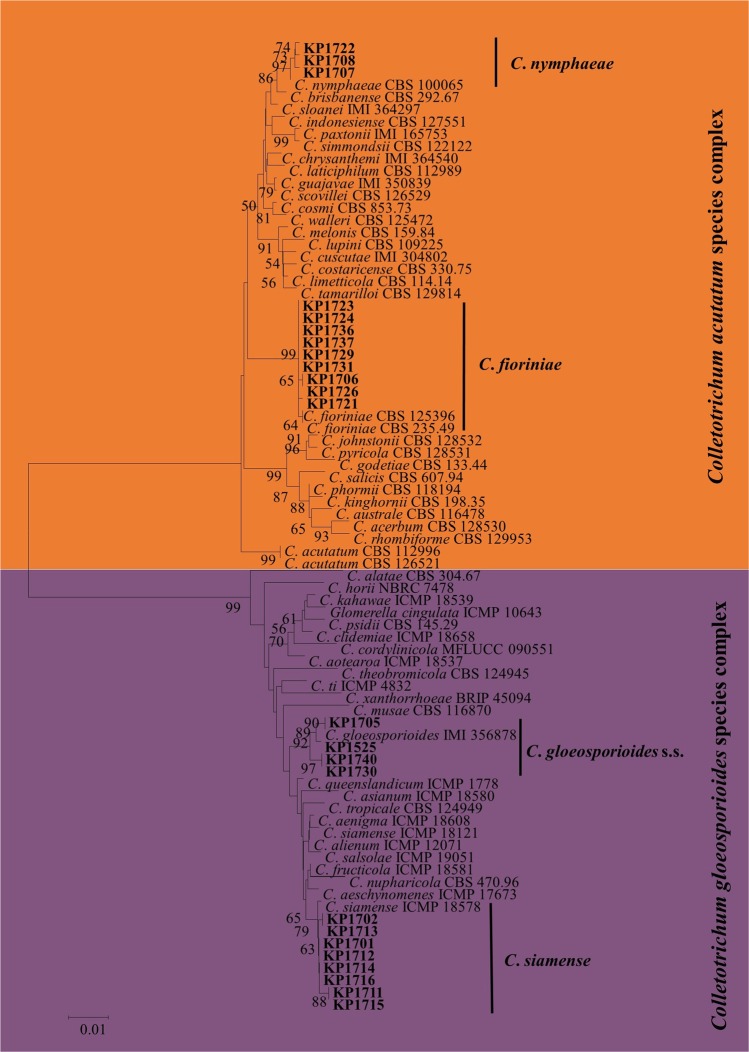
A total of 24 Colletotrichum isolates were isolated from Japanese plum fruits collected from different commercial orchards exhibiting anthracnose in Sangju, South Korea (Gyeongbuk Province). The Colletotrichum spp. were isolated and preliminarily identified based on colony and conidial morphology. Among the 24 isolates, 15 colonies were gray to white and produced subcylindrical to cylindrical conidia similar to that of C. gloeosporioides s.l.20. Colonies of the remaining nine isolates were pinkish in color and produced fusiform conidia, which is common of fungi in the C. acutatum species complex21. Colletotrichum isolates belonging to C. gloeosporioides s.l., (12 isolates) and C. acutatum s.l., (12 isolate) were first delineated using the combined ITS and TUB2 align sequence data set for phylogenetic analysis (Fig. 1). Six isolates of C. gloeosporioides s.l., (four from C. siamense clade and two from C. gloeosporioides s.s clade) and five isolates of C. acutatum s.l., (three from C. fioriniae clade and two from C. nymphaeae clade) identified based on ITS and TUB2 sequences data were selected for further phylogenetic analysis (Fig. 1).
Figure 1.
Neighbor-joining (NJ) tree derived from concatenated sequence alignment of ITS and TUB2 showing the separation of Colletotrichum isolates into the C. acutatum species complex and C. gloeosporioides s.l. (indicated by colored blocks). Bootstrap support values (ML > 50) are given at the nodes.
Phylogenetic analyses of the combined datasets
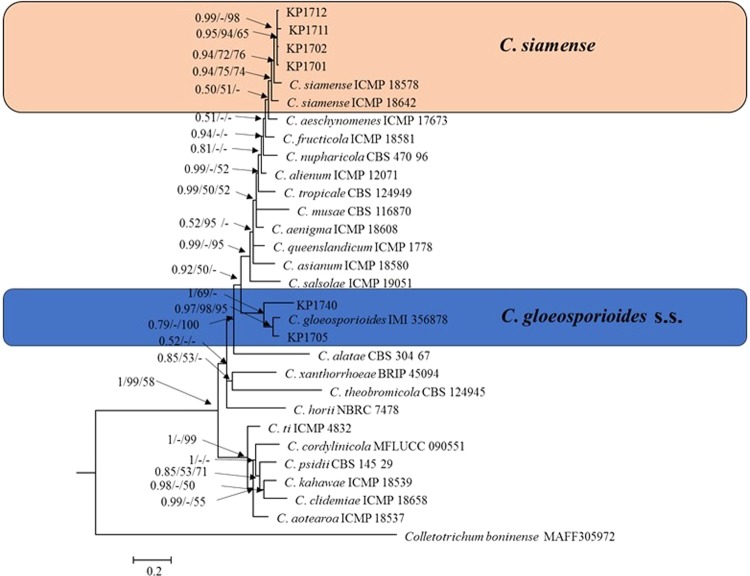
Colletotrichum gloeosporioides s.l. isolates were identified at the species level using a six-gene phylogenetic analysis (Fig. 2). Thirty sequences were present in the combined aligned data matrix (ITS, TUB2, GAPDH, ACT, CAL, CHS-1), which included C. boninense (CBS 123755) as the outgroup and 1,566 characters, as well as gaps in the alignment. The C. gloeosporioides species complex phylogram showed that isolates of the plum clustered in two clades (Fig. 2). Two isolates (KP1705 and KP1740) clustered with C. gloeosporioides s.s., ex-type isolate (IMI356878) with a high bootstrap support/posterior probability value (69%/1.00) and could be identified with confidence as C. gloeosporioides s.s. The remaining four isolates (KP1701, KP1702, KP1711 and KP1712) formed a sister clade with C. siamense ex-type isolates (ICMP I8578and ICMP I8642). The isolates KP1701, KP1702, KP1711 and KP1712 were further confirmed as C. siamense by phylogenetic analysis using ApMat sequences data (Appendix 1).
Figure 2.
Bayesian phylogeny (BI) based on a 50% majority rule consensus tree using combined sequence alignment of ITS, TUB2, GAPDH, ACT, CAL, and CHS-1. Colored blocks indicate the two clades containing plum isolates. Bayesian posterior probability values ≥ 0.5 and bootstrap support values ≥ 50% of maximum parsimony analysis and maximum likelihood analysis are given at the nodes. The scale bar shows the number of substitutions expected per site. Colletotrichum boninense MAFF305972 was used as the out-group.
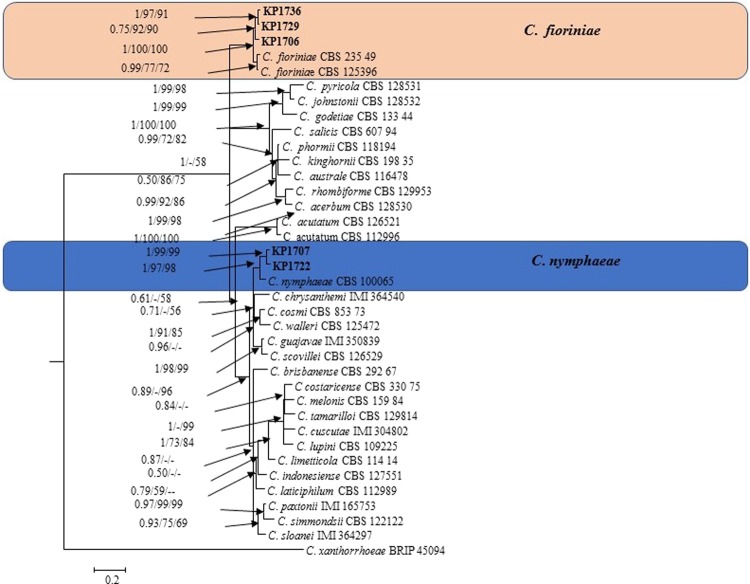
The phylogram in Fig. 3 shows the isolates identified in the C. acutatum species complex. The five-gene (ITS, TUB2, GAPDH, ACT, and CHS-1) phylogenetic analysis of C. acutatum s.l. contained 38 sequences, including the outgroup C. xanthorrhoeae (BRIP 45094). Three isolates (KP1706, KP1729, and KP1736) could be identified as C. fioriniae as it was in the same clade as C. fioriniae isolates CBS 23549 and CBS 125396 and showed robust posterior probability and bootstrap support values (1.00 and 100%) (Fig. 3). Two isolates (KP1707, and KP1722) clustered with the C. nymphaeae ex-type isolate CBS 100065 (bootstrap support/posterior probability value 97%/1.00) and were identified as C. nymphaeae.
Figure 3.
Bayesian phylogeny (BI) according to a 50% majority rule consensus tree using combined sequence alignment of ITS, TUB2, GAPDH, ACT, and CHS-1. Colored blocks indicate the two clades containing plum isolates. Bayesian posterior probability values ≥ 0.5 and bootstrap support values ≥ 50% of maximum parsimony analysis and maximum likelihood analysis are given at the nodes. The scale bar shows the number of substitutions expected per site. C. xanthorrhoeae BRIP 45094 was used as the out-group.
Pairwise homoplasy index (PHI) test
The concept of Genealogical Concordance Phylogenetic Species Recognition (GCPSR) was used to analyze phylogenetically related but ambiguous species. The pairwise homoplasy index (PHI) test found significant recombination between C. siamense and four closely related strains (KP1701, KP1702, KP1711 and KP1712) (Φw = <0.001), C. gloeosporioides s.s., and two closely related strains (KP1705and KP1740) (Φw = <0.003), C. nymphaeae and two closely related strains (KP1707 and KP1722) (Φw = <0.002), and C. fioriniae and three closely related strains (KP1706, KP1729, and KP1736) (Φw = <0.01) (Appendix 2).
Taxonomy
Colletotrichum siamense Prihastuti, L. Cai and K.D. Hyde, Fungal Diversity 39: 158. 2009 Fig. 4.
Figure 4.
C. siamense (KP1702), C. gloeosporioides s.s., (KP1740), C. fioriniae (KP1706) and C. nymphaeae (KP1707) (from left to right). (a,b) Colonies on PDA of different Colletotrichum species isolates (KP1702, KP1740, KP1706 and KP1707 from left to right). (c) Conidia of different isolates of Colletotrichum species isolates (KP1702, KP1740, KP1706 and KP1707 from left to right). (d) Appressoria of different isolates of Colletotrichum species isolates (KP1702, KP1740, KP1706 and KP1707 from left to right). (e) Symptoms of anthracnose on artificially inoculated plum fruits after 12 days of inoculation by the wounding method (KP1702, KP1740, KP1706 and KP1707 from left to right). (f) Symptoms of anthracnose on naturally infected plum fruits. Scale: (c,d) = 10 μm.
Description
Colonies on Difco potato dextrose agar (PDA) grew to 70–76 mm in diameter at a rate of 10.9 mm/day after seven days at 28 °C in the dark. Colonies were creamy white with aerial mycelium, and there were yellowish white masses of conidial ooze. The colonies were pale yellow in reverse. Conidia were hyaline, aseptate, smooth, cylindrical, straight or slightly curved, slightly tapered toward the end, 17.8–24.2 × 5.0–7.3 μm, av ± SD = 19.8 ± 1.7 × 6.3 ± 0.60, L/W ratio = 3.0, n = 50. Appressoria globose to ellipsoid, without lobes, dark brown, unbranched, 6.8–12.20 × 6.7–10.3 μm, av ± SD 8.70 ± 1.20 × 8.21 ± 0.80 μm, L/W ratio = 1.1, n = 50 (Table 1; Fig. 4).
Table 1.
Morphological data of Colletotrichum isolates.
| Taxon | Isolates | Colony morphology | Conidia morphology | Appressoria | morphology | ||
|---|---|---|---|---|---|---|---|
| Length (average ± SD) | Width (average ± SD) | Shape | Length (average ± SD) | Width (average ± SD) | |||
| C. siamense | KP1701 | Creamy white, formed a thin layer over the PDA with white aerial mycelium and conidia produced across the PDA plate | 19.40 ± 1.10 μm | 5.90 ± 0.44 μm | Cylindrical, straight to slight curve, not fusiform but rather slightly tapered toward the end | 9.54 ± 1.12 μm | 7.20 ± 0.54 μm |
| KP1702 | Creamy white, formed a thin layer over the PDA with white aerial mycelium and conidia produced across the PDA plate | 19.81 ± 1.71 μm | 6.32 ± 60 μm | Cylindrical, straight to slight curve, not fusiform but rather slightly tapered toward the end | 8.70 ± 1.20 μm | 8.21 ± 0.80 μm | |
| KP1711 | Creamy white, formed a thin layer over the PDA with white aerial mycelium and conidia produced across the PDA plate | 19.32 ± 1.23 μm | 5.60 ± 0.60 μm | Cylindrical, straight to slight curve, not fusiform but rather slightly tapered toward the end | 9.17 ± 1.41 μm | 7.20 ± 0.74 μm | |
| KP1712 | Creamy white, formed a thin layer over the PDA with white aerial mycelium and conidia produced across the PDA plate | 19.41 ± 1.10 μm | 7.03 ± 0.62 μm | Cylindrical, straight, obtuse end | 9.81 ± 1.00 μm | 7.31 ± 0.80 μm | |
| C. gloeosporioides s.s. | KP1705 | Dense cottony, pale orange | 16.63 ± 1.74 μm | 7.27 ± 0.52 μm | Cylindrical, round at both ends | 9.60 ± 1.08 μm | 8.08 ± 1.42 μm |
| KP1740 | Dense cottony, gray | 17.90 ± 1.60 μm | 5.93 ± 0.80 μm | Cylindrical, round at both ends | 12.95 ± 1.60 μm | 8.66 ± 1.00 μm | |
| C. fioriniae | KP1706 | Pink with white aerial mycelium | 14.52 ± 1.01 μm | 5.90 ± 0.71 μm | Fusiform | 9.25 ± 1.02 μm | 8.12 ± 0.52 μm |
| KP1729 | Pink with white aerial mycelium | 14.62 ± 1.41 μm | 5.61 ± 0.71 μm | Fusiform | 11.08 ± 1.35 μm | 7.26 ± 1.02 μm | |
| KP1736 | Pink with white aerial mycelium | 12.89 ± 1.35 μm | 4.90 ± 0.84 μm | Fusiform | 11.86 ± 1.78 μm | 8.20 ± 1.04 μm | |
| C. nymphaeae | KP1707 | Gray with light aerial mycelium | 10.37 ± 1.92 μm | 4.32 ± 0.65 μm | Subcylindrical, round at both ends or slightly tapered at one end | 9.83 ± 1.24 μm | 7.71 ± 1.54 μm |
| KP1722 | Gray with light aerial mycelium | 9.73 ± 1.51 μm | 4.23 ± 0.64 μm | Subcylindrical, round at both ends or slightly tapered at one end | 10.80 ± 1.78 μm | 7.15 ± 1.05 μm | |
Materials examined
SOUTH KOREA, Gyeongbuk Province, Sangju City, from diseased fruit of Prunus salicina, 22 Jul. 2017, O. Hassan, culture KP1701, KP1702, KP1711 and KP1712.
Notes
Colletotrichum siamense has been identified as the causative agent of anthracnose of Malus pumila and Diospyros kaki in South Korea17,25. Colletotrichum siamense is believed to have first infect coffee berries in Thailand; it has been reported as a pathogen on various hosts and is now considered a biologically and geographically diverse species17,20,26. Based on multi-locus (ITS, TUB2, GAPDH, ACT, CAL, and CHS-1) phylogenetic analysis, KP1701, KP1702, KP1711 and KP1712 isolates were identified as C. siamense (Fig. 2). A phylogenetic tree based on ApMat sequences also revealed that C. siamense species formed different clades, and KP1701, KP1702, KP1711 and KP1712 clustered together with one C. siamense species clade (Appendix 1). This result is consistent with recent publications from Sharma et al.23,24. Although C. siamense species isolates clustered in different clades, they are considered a single species rather than a species complex23,24,27. The closest matches (99% identity) in a BLAST search using ApMat sequence of previously identified strains were YT02, SQ01, and LQ22 from China28.
Colletotrichum gloeosporioides (Penz.) Penz. & Sacc., Atti Reale Ist. Veneto Sci. Lett. Arti., Serie 6, 2: 670. 1884.
For detailed description of C. gloeosporioides s.s., see Cannon et al.29 and Weir et al.20.
Materials examined
SOUTH KOREA, Gyeongbuk Province, Sangju City, from diseased fruit of Prunus salicina, 20 Jul. 2017, O. Hassan, Culture KP1705 and KP1740 (Fig. 4).
Notes: Colletotrichum gloeosporioides s.s., was reported to be the causative agent of anthracnose on various host plants, including Malus prunifolia, Ficus carica, Liriodendron chinense, Prunus avium, and Diospyros kaki in South Korea6,30–33. Previously, C. gloeosporioides s.s. was isolated from Prunus salicina from Daegu area3. This species was isolated from Prunus salicina from the Sangju area in the present study. This study identified the isolates KP1705 and KP1740 as C. gloeosporioides s.s. based on morphology and multi-locus (ITS, TUB2, GAPDH, ACT, CAL, and CHS-1) phylogenetic analysis. In the phylogram, these isolates clustered in the same clade with C. gloeosporioides s.s. (IMI 356878) with 70% bootstrap support and posterior probability value of 1.00 (Fig. 2).
Colletotrichum fioriniae (Marcelino & Gouli) R.G. Shivas & Y.P. Tan, Fungal Diversity 39: 117. 2009. Description and illustrations: see Damm et al.21.
Materials examined: SOUTH KOREA, Gyeongbuk Province, Sangju City, from diseased fruit of Prunus salicina, 21 Jul. 2017, O. Hassan, Culture KP1706, KP1729 and KP1736 (Fig. 4).
Notes: Colletotrichum fioriniae has been reported as the causative agent of anthracnose on various host plants, including Lycium chinense and Solanum melongena in Korea6,14,34. In this study, C. fioriniae was isolated from Prunus salicina in the Sangju area, Korea. The isolates KP1706, KP1729 and KP1736 were identified based on multi-locus (ITS, TUB2, GAPDH, ACT, and CHS-1) phylogenetic analysis and morphological characteristics.
Colletotrichum nymphaeae (Pass.) Aa, Netherlands Journal of Plant Pathology, Supplement 1 84: 110. 1978.
For detailed illustrations of C. nymphaeae see Damm et al.21.
Materials examined
SOUTH KOREA, Gyeongbuk Province, Sangju City, from diseased fruit of Prunus salicina, 21 Jul. 2017, O. Hassan, Culture KP1707 and KP1722 (Fig. 4).
Notes: KP1707 and KP1722 in our study were confidently identified as C. nymphaeae based on multi-locus (ITS, TUB2, GAPDH, ACT, and CHS-1) phylogenetic analysis. Colony color, conidia (shape), and appressoria (shape) is comparable with some Colletotrichum species with in the C. gloeosporioides and C. acutatum species complex20,21. Colletotrichum nymphaeae separated clearly from other species based on multi - locus (ITS, TUB2, GAPDH, ACT, and CHS-1) molecular analysis, rather than morphological characteristics. C. nymphaeae was reported in our recent publication as the causative agent of plum anthracnose9.
Pathogenicity assay
The pathogenicity of the Colletotrichum isolates was evaluated on detached plum fruits for confirmation of Koch’s postulates. All isolates of Colletotrichum showed anthracnose symptoms on plum fruit inoculated using the wounding approach, while only C. siamense, C. nymphaeae and C. fioriniae were capable of infecting non-wounded fruits as shown in Table 2. The C. siamense isolates produced the largest lesions on wounded fruits. Colletotrichum siamense, C. nymphaeae and C. fioriniae showed less virulence on non-wounded fruits in term of both disease incidence and lesion size.
Table 2.
Pathogenicity testing of Colletotrichum species from Japanese Plum (Prunus salicina).
| Species and isolates | Mean infection incidence (%) | Lesion diameter on fruits (mm) | |||
|---|---|---|---|---|---|
| Wounding | Non-wounding | Wounding | Non-wounding | ||
| C. siamense | KP1701 | 100 | 10 | 50.60 ± 3.30 | 9.3 ± 1.2 |
| KP1702 | 100 | 10 | 50.72 ± 3.50 | 8.0 ± 2.0 | |
| KP1711 | 100 | 5 | 49.20 ± 3.92 | 6.3 ± 1.5 | |
| KP1712 | 100 | 10 | 42.62 ± 1.50 | 7.3 ± 1.5 | |
| C. gloeosporioides s.s. | KP1705 | 100 | 0 | 20.60 ± 0.63 | 0 |
| KP1740 | 100 | 0 | 22.31 ± 0.35 | 0 | |
| C. fioriniae | KP1706 | 100 | 5 | 19.14 ± 2.33 | 4.7 ± 0.42 |
| KP1729 | 100 | 10 | 18.50 ± 1.01 | 4.6 ± 0.72 | |
| KP1736 | 100 | 25 | 19.82 ± 1.23 | 5.12 ± 1.02 | |
| C. nymphaeae | KP1707 | 100 | 5 | 18.75 ± 1.70 | 3.25 ± 1.0 |
| KP1722 | 100 | 0 | 19.45 ± 1.36 | 0 | |
Discussion
Anthracnose and other diseases caused by Colletotrichum spp. on the leaves, stems and fruits of numerous important crops have become increasingly common in South Korea. The disease of anthracnose has severely limited commercial production of various important fruit crops, such as apples, peaches, persimmons, grapes, and others across South Korea12–14,16,17. Anthracnose on fruits causes severe losses because of both pre and post-harvest fruit decay, which makes the fruits completely unmarketable. Very recently, Colletotrichum anthracnose has been reported in Japanese plums in Korea3,9. In previous research, morphological and ITS sequence approaches has been used to identify Colletotrichum spp. responsible for anthracnose on Japanese plums3. Morphological characteristics along with ITS sequence analysis may be more beneficial for identifying isolates to species complex rather than specific species. In the present study, Colletotrichum species associated with plum anthracnose from Sangju, Korea were identified using a multilocus phylogenetic analysis approach followed by an evaluation of their pathogenicity. Four isolates were identified as C. siamense, two isolates as C. gloeosporioides s.s., three isolates as C. fioriniae and two isolates as C. nymphaeae.
Colletotrichum siamense is a member of C. gloeosporioides s.l. and is described here for the first time as responsible for plum anthracnose in Sangju, Korea. C. gloeosporioides s.s., and C. nymphaeae are species from the C. gloeosporioides species complex and the C. acutatum species complex respectively, that have been previously reported to cause anthracnose in plums in Korea3.9. Colletotrichum fioriniae was first reported as a species of C. acutatum s.l., to have cause plum anthracnose in Korea. Phylogenetic analysis and morphological data including colony characters and conidial measurements were previously used to distinguish four Colletotrichum species20,21. Morphological characteristics of C. siamense and C. gloeosporioides s.s., including colony characters, conidial measurements, and appressoria measurements overlapped with those of other species in C. gloeosporioides s.l. Identifying Colletotrichum species within C. gloeosporioides s.l., based on morphological characteristics is uncertain because of: (1) overlapping morphological characteristics among the species20 and (2) slight morphological differences that can be due to different growing conditions, temperature, light regime, and geographic isolates20. Multilocus (ITS, TUB2, GAPDH, ACT, CAL, and CHS) phylogeny analysis clearly showed that the present isolates, C. siamense and C. gloeosporioides s.s., clustered in a distinct phylogenetic clade with in C. gloeosporioides s.l., with a high posterior probability value (0.92) (Fig. 2). C. siamense isolates were further confirmed via both phylogenetic analysis and BLAST search using ApMat sequence data. ApMat is a potentially powerful gene disentangling both C. gloeosporioides s.l., and C. siamense complexes22,24. Colletotrichum siamense was previously considered as a species complex, but recent studies have shown C. siamense to be a single species based on molecular analyses using GCPSR as well as coalescent methods General Mixed Yule Coalescent and Poisson Tree Processes23,24,28. The PHI test result in the present study also found significant recombination among C. siamense species of different geographic origins. Colletotrichum siamense has been associated with anthracnose in various commercial crops17,20,24. To the best of our knowledge, this is the first report of anthracnose of plums caused C. siamense in Korea.
C. gloeosporioides s.s., is the most frequently reported plant pathogen causing anthracnose in a variety of hosts in Korea6,30–33. However, this is only the second report on plum anthracnose caused by C. gloeosporioides s.s. in Korea. It was previously identified based on morphological characteristics and the ITS sequence analysis, whereas here we identified it using multilogue phylogenetic analysis, which was supported by morphological characteristics evaluations. Colletotrichum fioriniae can be easily identified by the colony pigment on PDA, which is pink cottony with gray aerial mycelium in compact tufts from above and pink with flecking in reverse35. Colletotrichum fioriniae was previously reported as the causative agent of anthracnose on a variety of hosts6,14,34. To our knowledge, this is the first report of C. fioriniae causing anthracnose of plums in South Korea. Colletotrichum nymphaeae is reported as the causal agent of plum anthracnose for the second time here9.
The pathogenicity tests showed that the four species of Colletotrichum evaluated in this study are pathogenic to plum fruits and could be differentiated by the degree of virulence and lesion size in inoculated fruits. All Colletotrichum isolates tested caused anthracnose on wounded fruit, whereas only C. siamense, C. nymphaeae and C. fioriniae isolates were able to infect unwounded fruits. C. siamense isolates produced larger lesions on plum fruits followed by C. gloeosporioides s.s., C. nymphaeae and C. fioriniae. Koch’s postulates were fulfilled by re-isolating the fungus from the lesions of inoculated fruits and reidentifying them at the species level using morphological and multi-locus sequences approaches.
In conclusion, this study identified 2 species within C. gloeosporioides s.l., and 2 species within the C. acutatum complex. This investigation included only one area (Sangju) of Korea, which highlights the importance of further research on Colletotrichum strains isolated from different Korean regions to mitigate the risk to the plum fruit industry in Korea.
Material and Methods
Sample collection and isolation
Japanese plum fruits with visible anthracnose were collected in 2017 from different commercial orchards in Sangju Korea. The fruits were characterized by sunken, round, and brown necrotic lesions. Three diseased fruits were selected from each orchard for the isolation causal agents, and fruits were washed with distilled water. Causal agents were isolated from necrotic tissue of diseased fruits as follows. Small pieces (2 mm2) of necrotic tissue were removed aseptically with a scalpel, disinfected with a 1% NaOCl solution (w/v) for 1 min followed by three washes in sterile distilled water. After drying by blotting, the disinfected tissues were placed on water agar (WA) petri plates supplemented with streptomycin (0.05 g/L) and incubated at 25 °C in the dark. Newly emerging hyphae from the tissue were transferred onto fresh potato dextrose agar (PDA) petri plates and incubated at 25 °C in the dark. Pure fungal cultures were obtained using the single spore isolation technique from 7-day PDA cultures18. Conidial suspensions were prepared in sterile distilled water. The concentration of each conidial suspension was determined by using a hemocytometer. Then, the conidial suspensions (~104 conidia/ml) were made from the concentrated suspensions. Conidial suspensions were then sprayed on to PDA plates and incubated in the dark at 25 °C. Single germinating spores were collected with a sterilized needle after overnight incubation and placed on fresh PDA plates and incubated in the dark at 25 °C. Seven-day-old cultures were grouped based on culture morphology and conidial shape.
DNA extraction, PCR amplification, and sequencing
Fungal mycelia were acquired with a sterile scalpel from 4-day-old cultures of isolates grown on PDA, and total genomic DNA was extracted using a HiGeneTM Genomic DNA Prep Kit (Yuseong-Gu, Daejeon, Korea), following the manufacturer’s instructions. For C. gloeosporioides s.l. isolates, seven targeted genes were selected for PCR amplification and sequencing: internal transcribed spacer regions and intervening 5.8S nrRNA gene (ITS), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), actin (ACT), beta-tubulin (TUB2), calmodulin (CAL), chitin synthase (CHS-1) and the Apn2–Mat1–2 intergenic spacer and partial mating type (Mat1–2) gene (ApMat). For C. acutatum s.l. isolates, five targeted genes were selected for PCR amplification and sequencing: ITS, GAPDH, ACT, TUB2, and CHS-1. The primer sets used in this study are listed in Table 3. The PCR amplifications were carried out in a simpliAmp™ thermal cycler (Thermo Fisher Scientific Inc). Each 25 μL PCR mixture consisted of 18.8 μL UV-sterilized ultra-filtered water, 2.5 µL 10x F-star Taq buffer, 0.5 µl dNTP Mix (each 10 mM), 1 µL forward primer (10 pmol), 1 µL reverse primer (10 pmol), 1 µL genomic DNA, and 0.2 µL F-star Taq DNA polymerase (BIOFACT, Korea). The PCR conditions were the same as the conditions applied for amplification of ITS using the universal primers ITS1F/ITS4, except for the annealing temperatures20. Locus-specific annealing temperatures are shown in Table 1. Purification and sequencing of the PCR product were performed commercially at Macrogen, Inc. (Seoul, Korea).
Table 3.
Primers used in this study, including sequences and sources.
| Gene | Primer Name | Direction | Sequence (5′-3′) | Annealing temperature (°C) | References |
|---|---|---|---|---|---|
| GAPDH | GDF | Forward | GCC GTC AAC GAC CCC TTC ATT GA | 60 | Templeton et al.44 |
| GDR | Reverse | GGG TGG AGT CGT ACT TGA GCA TGT | 60 | Templeton et al.44 | |
| ITS | ITS-1F | Forward | CTT GGT CAT TTA GAG GAA GTA A | 55 | Gardes & Bruns45 |
| ITS-4 | Reverse | TCC TCC GCT TAT TGA TAT GC | 55 | White et al.46 | |
| CAL | CL1C | Forward | GAA TTC AAG GAG GCC TTC TC | 59 | Weir et al.20 |
| CL2C | Reverse | CTT CTG CAT CAT GAG CTG GAC | 59 | Weir et al.20 | |
| Actin | ACT-512F | Forward | ATG TGC AAG GCC GGT TTC GC | 58 | Carbone & Kohn47 |
| ACT-783R | Reverse | TAC GAG TCC TTC TGG CCC AT | 58 | Carbone & Kohn47 | |
| ApMat | AM-F | Forward | TCA TTC TAC GTA TGT GCC CG | 62 | Silva et al.22 |
| AM-R | Reverse | CCA GAA ATA CAC CGA ACT TGC | 62 | Silva et al.22 | |
| TUB2 | Bt2a | Forward | GGT AAC CAA ATC GGT GCT GCT TTC | 55 | Glass & Donaldson48 |
| Bt2b | Reverse |
ACC CTC AGT GTA GTG ACC CTT GGC |
55 | Glass & Donaldson48 | |
| CHS-1 | CHS-79F | Forward | TGG GGC AAG GAT GCT TGG AAG AAG | 58 | Carbone & Kohn47 |
| CHS-345R | Reverse | TGG AAG AAC CAT CTG TGA GAG TTG | 58 | Carbone & Kohn47 |
Phylogenetic analysis
The accession numbers for all sequences were acquired after depositing the resulting consensus sequences in GenBank (accession numbers are listed in Table 4). The generated sequences from the present isolates and those retrieved from GenBank (Table 4) for each gene were aligned using the MUSCLE multiple sequence alignment programs of MEGA v. 6.036. Manually edited (if necessary) multiple sequence alignments were constructed for each gene, all gaps were treated as missing data and concatenated with Mesquite v. 2.7537. The phylogenetic analyses were performed using concatenated aligned sequences of different gene combinations. Neighbor-joining (NJ), maximum likelihood (ML), and maximum parsimony (MP) phylogenetic analyses were performed using MEGA v. 6.036. Bayesian inference (BI) phylogenetic analyses were performed with MrBayes v. 3.2.238. GTR + I + gamma mode determined using MrModeltest v. 2.3 was utilized to construct the Bayesian phylogenetic tree39. MCMC analysis of four chains based on the full dataset was run in parallel from a random tree topology, the heat parameter was set at 0.15, and trees were sampled every 100 generations. The MCMC analysis was stopped when the average standard deviation of split frequencies reached 0.01 (stop value). The first 25% of the generations were set as burn-in after which the likelihood values remained stationary. Consensus BI phylogenetic trees were viewed in FigTree v 1.3.140. For the preliminarily identification of Colletotrichum isolates belonging to C. gloeosporioides s.l. and C. acutatum s.l., both the ITS and TUB2 alignment sequences were used for phylogenetic analysis. The sequences of five genes (ITS, TUB2, GAPDH, CHS-1, and ACT) were used for the phylogenetic analysis of isolates belonging to the C. acutatum species complex. The sequences of six genes (ITS, TUB2, GAPDH, ACT, CAL, and CHS-1) were used to analyze isolates belonging to C. gloeosporioides s.l. ApMat sequences were used for proper identification of C. siamense isolates.
Table 4.
GenBank accession numbers of the Colletotrichum isolates used in this study for molecular data analyses.
*Ex-holotype or ex-epitype cultures.
Genealogical concordance phylogenetic species recognition analysis
The Genealogical Concordance Phylogenetic Species Recognition (GCPSR) model was used to analyze the phylogenetically related, but ambiguous species as described by Quaedvlieg et al. by performing a pairwise homoplasy index (Φw, PHI) test41. The PHI test was performed in Splits Tree 442,43. A six-locus concatenated dataset (ITS, TUB2, GAPDH, ACT, CAL, and CHS-1) of closely related species (Fig. 2) and a five-locus concatenated dataset (ITS, TUB2, GAPDH, ACT, and CHS-1) of closely related species (Fig. 3) were used to determine the recombination level and both the LogDet transformation and splits decomposition options were selected41. The PHI test value below a 0.05 threshold (Φw < 0.05) indicated significant recombination in the dataset.
Morphological characterization
All selected isolates were described based on culture morphology and growth rate, and conidia as well as appressoria shape and size. Cultures were grown on PDA using mycelial discs (5 mm diameter) from 5-day-old cultures at 25 °C under 16 h light/8 h dark conditions. Culture diameter was measured each day, and the appearance was evaluated after 7 days of growth. The daily growth rate was calculated based on measurement from six replicates. Conidial characteristics (size and shape) were determined using conidia taken from the conidial mass on the culture and mounted on glass slides in clear lactic acid; the length and wide of 50 conidia were measured for each isolate. For appressoria production, conidia mounted on glass slides in distilled water were placed in Petri dishes containing a moistened tissue and incubated at 25 °C under 16 h light/8 h dark conditions. After two days of incubation, appressoria that formed across the underside of the coverslip were measured; the size of 50 appressoria was measured for each isolate. Conidia and appressoria sizes were measured with a stage micrometer under an Olympus BX43 microscope (Olympus Corporation, Japan) at 400× magnification.
Pathogenicity tests
All eleven isolates were subjected to pathogenicity tests on Japanese plum. Mature detached Japanese plum fruits were collected from Sangju Emart and used for the pathogenicity assay. The collected plum fruits were washed with tap water and then disinfected for 3 minutes in 1% sodium hypochlorite, followed by washing with sterile distilled water three times. Disinfected fruits were placed in a plastic container and inoculated with conidial suspension of the respective isolates using both nonwounding and wounding methods. A 106 conidia/mL conidial suspension was made from 7-day-old cultures of each isolate, as described above. For the wounding method, fruits were wounded by pricking with a sterile needle and a 10 µL droplet of the conidial suspension was placed at the wounded point. For the non-wounding method, the conidial suspension was sprayed over the fruits surface until surface runoff was observed. Control fruits for both methods received distilled water. Ten fruits were used for each treatment. After inoculation the plastic containers were sealed and incubated at 25 °C in the dark under high humidity conditions in an incubator. After 5 days of incubation, anthracnose lesions were observed on fruits inoculated with fungal conidia. Control fruits remained symptom-free. The disease incidence (DI) was expressed as the percentage of infected fruits compared to the total number of inoculated fruits. A ruler was used to measure lesion diameters (LDs). Causal agents were isolated from infected fruits, cultured on a new PDA plate, and then identified according to the methods described above to confirm Koch’s postulates.
Statistical analysis
MS Excel was used to calculate the average and standard deviation of each data sets. Values for daily growth rate, conidia and appressor sizes, and lesion diameters expressed as the average ± standard deviation (av ± SD).
Comment
The photograph(s) in figure 4 were obtained from Sangju, Korea, and the images were taken by Oliul Hassan (O.H) and Taehyun Chang (T.C).
Supplementary information
Acknowledgements
We would like to thank all the members of the Plant Pathology Lab, School of Ecology & Environmental System, Kyungpook National University, Sangju, Gyeongbuk 37224, Korea (Republic of) for their help conducting the experiments.
Author Contributions
Conceived and designed the experiments: O.H. Performed the experiments: O.H. Analyzed the data: O.H. and Y.S.L. Wrote the paper: O.H. and T.C. Revised and approved the final version of the paper: Y.S.L. and T.C. This is the first submission of the manuscript, and we confirm that it is not being considered for publication elsewhere in whole or in part. All authors have approved the submission of this manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-48108-1.
References
- 1.Roussos, P. A., Efstathios, N., Intidhar, B., Denaxa, N. K. & Tsafouros, A. Plum (Prunus domestica L. and P. salicina Lindl.) in Nutritional Composition of Fruit Cultivars (ed. Simmonds, M. & Preedy, V.) 639–666 (Elsevier, 2016).
- 2.Fanning KJ, Topp B, Russell D, Stanley R, Netzel M. Japanese plums (Prunus salicina Lindl.) and phytochemicals – breeding, horticultural practice, postharvest storage, processing and bioactivity. J. Sci. Food Agric. 2014;94:2137–2147. doi: 10.1002/jsfa.6591. [DOI] [PubMed] [Google Scholar]
- 3.Lee, Y.-S. et al. Isolation and characterization of Colletotrichum isolates causing anthracnose of Japanese plum fruit. Korean J. Environ. Agric. 36, 299–305 (In Korean, abstract in English) (2017)
- 4.Choi, J. E., Lee, E. J. & Park, Y. S. Shot hole of peach and Japanese plum caused by Xanthomonas campestris pv. pruni and Erwinia nigrifluens in Korea. Res. Plant Dis. 6, 10–14 (In Korean, abstract in English) (2000).
- 5.Kim Y, Lee HB, Yu SH. First report of leaf spot on Japanese plum caused by an Alternaria sp. in Korea. Plant Dis. 2005;89:343. doi: 10.1094/PD-89-0343A. [DOI] [PubMed] [Google Scholar]
- 6.The Korean Society of Plant Pathology. List of plant diseases in Korea. 5th ed. pp. 779 (in Korean) (2009).
- 7.Lee, S. C., Han, K. S., Cho, S. E., Park, J. H. & Shin, H. D. Occurrence of powdery mildew of Japanese plum caused by Podosphaera tridactyla in Korea. Res. Plant Dis. 18, 49–53 (In Korean, abstract in English) (2012).
- 8.Ryu, Y. H., Lee, J. H., Kwon, T. Y., Kim, S. H. & Kim, D. G. Occurrence of bacterial black spot on plum by Xanthomonas aboricola pv. pruni and its pathogenicity on varieties of some stone fruits. Res. Plant Dis. 18, 40–44 (In Korean, abstract in English) (2012).
- 9.Chang T, Hassan O, Lee Y-S. First report of anthracnose of Japanese plum (Prunus salicina) caused by Colletotrichum nymphaeae in Korea. Plant Dis. 2018;102:1461. doi: 10.1094/PDIS-01-18-0018-PDN. [DOI] [Google Scholar]
- 10.Hyde KD, et al. Colletotrichum: a catalogue of confusion. Fungal Divers. 2009;39:1–17. [Google Scholar]
- 11.Cannon PF, Damm U, Johnston PR, Weir BS. Colletotrichum - current status and future directions. Stud. Mycol. 2012;73:181–213. doi: 10.3114/sim0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee DH, Kim DH, Jeon YU, Uhm JY, Hong SB. Molecular and cultural characterization of Colletotrichum spp. causing bitter rot of apples in Korea. Plant Pathol. J. 2007;23:37–44. doi: 10.5423/PPJ.2007.23.2.037. [DOI] [Google Scholar]
- 13.Kim WG, Hong SK. Occurrence of anthracnose on peach tree caused by Colletotrichum species. Plant Pathol. J. 2008;24:80–83. doi: 10.5423/PPJ.2008.24.1.080. [DOI] [Google Scholar]
- 14.Oo MM, Oh S-K. Identification and characterization of new record of grape ripe rot disease caused by Colletotrichum viniferum in Korea. Mycobiol. 2017;45:421–425. doi: 10.5941/MYCO.2017.45.4.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeon JY, et al. Anthracnose of persimmon (Diospyros kaki) caused by Colletotrichum horii in Sangju, Korea. Plant Dis. 2017;101:1035. doi: 10.1094/PDIS-01-17-0085-PDN. [DOI] [Google Scholar]
- 16.Hong SK, Kim WG, Yun HK, Choi KJ. Morphological variations, genetic diversity and pathogenicity of Colletotrichum species causing grape ripe rot in Korea. Plant Pathol. J. 2008;24:269–278. doi: 10.5423/PPJ.2008.24.3.269. [DOI] [Google Scholar]
- 17.Hassan O, Jeon JY, Chang T, Shin JS, Oh NK. Molecular and morphological characterization of Colletotrichum species in the Colletotrichum gloeosporioides complex associated with persimmon anthracnose in South Korea. Plant Dis. 2018;102:1015–1024. doi: 10.1094/PDIS-10-17-1564-RE. [DOI] [PubMed] [Google Scholar]
- 18.Cai L, et al. A polyphasic approach for studying. Colletotrichum. Fungal Divers. 2009;39:183–204. [Google Scholar]
- 19.Crouch JA, Clarke BB, Hillman BI. What is the value of ITS sequence data in Colletotrichum systematics and species diagnosis? A case study using the falcate-spored graminicolous Colletotrichum group. Mycologia. 2009;101:648–656. doi: 10.3852/08-231. [DOI] [PubMed] [Google Scholar]
- 20.Weir BS, Johnston PR, Damm U. The Colletotrichum gloeosporioides species complex. Stud. Mycol. 2012;73:115–180. doi: 10.3114/sim0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Damm U, Cannon PF, Woudenberg JH, Crous PW. The Colletotrichum acutatum species complex. Stud. Mycol. 2012;73:37–113. doi: 10.3114/sim0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva DN, et al. Application of the Apn2/MAT locus to improve the systematics of the Colletotrichum gloeosporioides complex: an example from coffee (Coffea spp.) hosts. Mycologia. 2012;104:396–409. doi: 10.3852/11-145. [DOI] [PubMed] [Google Scholar]
- 23.Sharma G, Kumar N, Weir BS, Hyde KD, Shenoy BD. The ApMat marker can resolve Colletotrichum species: a case study with Mangifera indica. Fungal Divers. 2013;61:117–138. doi: 10.1007/s13225-013-0247-4. [DOI] [Google Scholar]
- 24.Sharma G, Pinnaka AK, Shenoy BD. Resolving the Colletotrichum siamense species complex using ApMat marker. Fungal Divers. 2015;71:247–264. doi: 10.1007/s13225-014-0312-7. [DOI] [Google Scholar]
- 25.Park MS, Kim B-R, Park I-H, Hahm S-S. First Report of two Colletotrichum species associated with bitter rot on apple fruit in Korea – C. fructicola and C. siamense. Mycobiol. 2018;46:1–5. doi: 10.1080/12298093.2018.1454012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prihastuti H, Cai L, Chen H, McKenzie E, Hyde K. Characterization of Colletotrichum species associated with coffee berries in northern Tailand. Fungal Divers. 2009;39:89. [Google Scholar]
- 27.Liu F, Wang M, Damm U, Crous PW, Cai L. Species boundaries in plant pathogenic fungi: a Colletotrichum case study. BMC Evol. Biol. 2016;16:81. doi: 10.1186/s12862-016-0649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang X. Comparative analysis of the mitochondrial genomes of Colletotrichum gloeosporioides sensu lato: insights into the evolution of a fungal species complex interacting with diverse plants. BMC Genom. 2017;18:171. doi: 10.1186/s12864-016-3480-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cannon PF, Buddie AG, Bridge PD. The typification of Colletotrichum gloeosporioides. Mycotaxon. 2008;104:189–204. [Google Scholar]
- 30.Cheon W, Kim YS, Jeon YH. First report of anthracnose caused by Colletotrichum gloeosporioides on Malus prunifolia in Korea. Plant Dis. 2012;96:766. doi: 10.1094/PDIS-01-12-0063-PDN. [DOI] [PubMed] [Google Scholar]
- 31.Choi O, Choi O, Kwak Y-S, Kim J, Kwon J-H. Spot anthracnose disease caused by Colletotrichum gloeosporioides on tulip tree in Korea. Mycobiol. 2012;40:82–84. doi: 10.5941/MYCO.2012.40.1.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi IY, Park JH, Cho SE, Shin HD. First confirmed report of anthracnose fruit rot caused by Colletotrichum gloeosporioides on common fig in Korea. Plant Dis. 2013;97:1119. doi: 10.1094/PDIS-01-13-0109-PDN. [DOI] [PubMed] [Google Scholar]
- 33.Lee JH, Kwak Y-S. First report of anthracnose of Ledebouriella seseloides caused by Colletotrichum gloeosporioides in Korea. J. Phytopathol. 2014;162:342–344. doi: 10.1111/jph.12192. [DOI] [Google Scholar]
- 34.Xu SJ, Aktaruzzaman M, Kim BS, Kim JY, Shin HD. First report of anthracnose caused by Colletotrichum fioriniae on eggplant fruits in Korea. Plant Dis. 2018;102:2642. doi: 10.1094/PDIS-04-18-0711-PDN. [DOI] [Google Scholar]
- 35.Shivas RG, Tan YP. A taxonomic re-assessment of Colletotrichum acutatum, introducing C. fioriniae comb. et stat. nov. and C. simmondsii sp. nov. Fungal Divers. 2009;39:111–122. [Google Scholar]
- 36.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maddison, W. P. & Maddison, D. R. Mesquite: a modular system for evolutionary analysis. Version 2.75, http://mesquiteproject.org (2011).
- 38.Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nylander JA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY: A system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- 40.Rambaut, A. & Drummond, A. FigTree v1. 3.1: Tree figure drawing tool. Institute of Evolutionary Biology, Edinburgh, UK. http://tree.bio.ed.uk/sofware/fgtree (2009).
- 41.Quaedvlieg W, et al. Introducing the consolidated species concept to resolve species in the Teratosphaeriaceae. Persoonia. 2014;33:1–40. doi: 10.3767/003158514X681981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huson DH. SplitsTree: Analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 43.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 44.Templeton MD, Rikkerink EHA, Solon SL, Crowhurst RN. Cloning and molecular characterization of the glyceraldehyde-3-phosphate dehydrogenase-encoding gene and cDNA from the plant pathogenic fungus Glomerella cingulata. Gene. 1992;122:225–230. doi: 10.1016/0378-1119(92)90055-T. [DOI] [PubMed] [Google Scholar]
- 45.Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 46.White, T. J., Bruns, T., Lee, S. & Taylor, J. W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics in PCR Protocols: A Guide to Methods and Applications. (ed. Innis, M. A., Gelfand, D. H., Sninsky, J. J. & White, T. J.) 315–322 (Academic Press: New York, 1990).
- 47.Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. doi: 10.1080/00275514.1999.12061051. [DOI] [Google Scholar]
- 48.Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.