Abstract
This study examined Echinococcus spp. genotypes and genetic variants isolated from humans as well as domestic and wild animals from the Qinghai-Tibetan Plateau Area using the cox1 gene. All samples except the pika isolates were identified as the Echinococcus granulosus sensu stricto. Sixteen different haplotypes with considerable intraspecific variation were detected and characterized in mitochondrial cox1 sequences. The parsimonious network of cox1 haplotypes showed star-like features, and the neutrality indexes computed via Tajima's D and Fu's Fs tests showed high negative values in E. granulosus s. s., indicating deviations from neutrality; the Fst values were low among the populations, implying that the populations were not genetically differentiated. The pika isolates were identified as E. multilocularis and E. shiquicus. Only one haplotype was recognized in the pika isolates. E. granulosus s. s. was the predominant species found in animals and humans, followed by E. multilocularis and E. shiquicus, with high genetic diversity circulating among the animals and humans in this area. Further studies are needed to cover many sample collection sites and larger numbers of pathogen isolates, which may reveal abundant strains and/or other haplotypes in the hydatid cysts infecting human and animal populations of the QTPA, China.
Key words: Cox1 gene, genetic variation, genotype, haplotypes, hydatid cyst, Qinghai-Tibetan Plateau Area
Introduction
Echinococcosis is caused by the metacestodes of Echinococcus spp. and is parasitic in the livers, lungs and/or other organs of humans and animals, resulting in serious zoonotic parasitic disease. Echinococcosis epidemic diseases not only threaten and harm the health of humans and animals but also seriously hinder the production of animal husbandry and affect the development of the national economy (Budke et al., 2006; Qian et al., 2017). This disease is prevalent in Asia, South America, North Africa, Central Europe and other regions and causes harm throughout the world (Grosso et al., 2012). China, especially the Qinghai-Tibetan Plateau Area (QTPA), has one of the highest prevalence rates in the world (Wang et al., 2014). Echinococcosis identified as a zoonotic disease with a major impact on the public health of rural populations in Qinghai-Tibetan Plateau. As our previous study showed a high seropositive rate (37.0%) of echinococcosis in Qinghai-Tibetan primary school students (Han et al., 2018). Prevalence of E. granulosus in yaks, pigs and Tibetans investigated in Qinghai-Tibetan Plateau was 6.49, 7.27 and 1.83%, and Prevalence in yaks was 3.61, 9.66 and 6.33% in 2014, 2015 and 2016, respectively (Li et al., 2017). The action plan for prevention and treatment of echinococcosis was carried out by the People's Republic of China. A series of work was being implemented, such as the management and deworming of source of infection-dog, the vaccine immunization of the livestock, the implementation of livestock slaughter standard in slaughtering house (safety disposal of diseased organs), the management of patients (patient care and rescue), health education, people training and providing safe drinking water. The seven provinces and autonomous regions in northwestern China are epidemic areas of hydatid disease, endangering nearly 50 million people and 70 million livestock, which has resulted in direct economic losses of up to 30 billion Yuan (Qian et al., 2017).
Echinococcus spp. exhibit a fixed life cycle between definitive and intermediate hosts. Carnivores, such as dogs, foxes and wolves, are the definitive hosts of the parasites. The intermediate hosts involve different species and change in different environments (Romig et al., 2017). Echinococcus spp. can be infective in metacestode hosts, such as humans, livestock and some wild animals (Thompson, 2017). With the continuous improvement of molecular and genetic knowledge of parasites, identification and classification research has surpassed morphological study (Nakao et al., 2013). Genetic analysis employing mitochondrial genes (cox1, nad1, cob and nad5 gene) and ribosomal genes might reveal the biological relationships between Echinococcus spp. and strains (Marinova et al., 2017; Kinkar et al., 2018a). Mitochondrial DNA (mtDNA) is the most suitable genetic marker for the analysis of genetic diversity, genetic differentiation and evolution of species and is widely used in species classification (Umhang et al., 2014). There are five epidemic Echinococcus spp.: E. granulosus s. l., E. multilocularis, E. oligarthrus, E. vogeli and E. shiquicus (Nakao et al., 2013).
E. granulosus s. l. is a complex of different species including E. granulosus s. s.. E. granulosus s. s. includes the G1 and G3 genotype complex groups, with the G1 genotype (sheep strain) being prevalent worldwide (animals and humans) (Kinkar et al., 2018b). The G1 genotype is also the main epidemic strain of E. granulosus s. s. in China (Zhang et al., 2014). E. granulosus s. s. G1 infections of sheep, cattle and pigs have been reported in Argentina, Brazil, Chile and Mexico in the Americas as well, with high haplotype diversity (Laurimae et al., 2016). Similar results obtained from analyses of G1 infecting intermediate host species (cattle, sheep, human, wild boar, pig and goat) in Europe (Albania, Finland, Greece, Italy, Romania, Spain and Turkey) have revealed a complex phylogeography and high genetic variation (Kinkar et al., 2016). Previously, the G2 (Tasmanian sheep strain) is regarded as a separate genotype and has been found in France, Italy and Argentina (McManus and Thompson, 2003; Casulli et al., 2008); however, the genotype G2 is currecntly treated as an invalid genotype that is belonging to the genotype G3 cluster (Kinkar et al., 2017). The G3 genotype (buffalo strain) is prevalent in India and Iran (Grosso et al., 2012; Rostami et al., 2013). However, G3 is significantly less prevalent worldwide than G1, and also exhibits high genetic diversity (Kinkar et al., 2018c). Some recently obtained E. granulosus s. l. complex genotypes can be regarded as distinct species based on the differences in the morphology, host range and life cycles of these parasites, including E. granulosus s. s. (G1 and G3), E. equinus (G4), E. ortleppi (G5) (Thompson and McManus, 2002; Kinkar et al., 2017). However, the status of genotypes G6–G10 is still under dispute (Moks et al., 2008; Thompson, 2008; Saarma et al., 2009; Knapp et al., 2011, 2015; Lymbery et al., 2015). G6/G7 and G8/G10 can be regarded as two distinct species based on recent molecular evidence based on six nuclear genes; moreover, the marked biological differences exist between G6/G7 and G8/G10 (Laurimae et al., 2018). In addition, the genotype G9 has been characterised, but the status of G9 is still uncertain (Scott et al., 1997); analysis of phylogenetic systematics showed that the dubious G9 genotype can be included in G7 (Oksanen and Lavikainen, 2015), and maybe a variant of G7. The controversial genotypes remain to be discussed in future studies.
Echinococcus spp. isolates harbour a high degree of internal variation with substantial genetic differences and inter-isolate variation in development (such as the development of eggs) in different geographical environments and different hosts. These differences cause variation in propagation dynamics, pathogenicity, antigen-antibody reactions, clinical manifestations and chemotherapy responses between hosts (McManus and Thompson, 2003; Torgerson and Heath, 2003). There are a wide range of intermediate hosts showing adaptability to E. granulosus s. s. (Onac et al., 2013). In the long evolutionary process, variations also arise in the mutual adaptation of Echinococcus spp. Therefore, studies examining polymorphism in Echinococcus spp. might be directly related to the prevention of epidemics and treatment of local hydatid disease in the QTPA (Ma et al., 2015).
In this study, we analysed the genetic polymorphism of the mitochondrial cox1 gene of Echinococcus spp., combined with clinical data, to explore the intra-species variation of Echinococcus spp. and evaluate the population genetic structures of the three species in the QTPA. The resulting data and epidemiological information enabled us to propose evolutionary hypotheses regarding how three species of parasites have spread in the QTPA of China.
Materials and methods
Collection of samples
A total of 244 hydatid cysts were collected, including 93 human isolates (65 patients) obtained from surgical operations in the local hospitals of Qinghai Province and 38 sheep (28), 91 yak (64), and 22 wild pika (15) isolates obtained from the slaughterhouse between 2010 and 2017. The plateau pika was Ochotona curzoniae, which was identified according to the behavioural and morphological patterns of pikas by the local veterinarian. The yak species was Bos grunniens and the sheep were Tibetan sheep. Each single isolated cyst was regarded as an independent sample, and the contents of some hydatid cysts (except some lesions were calcification) were placed in tubes. In some cases, there was more than one lesion per animal/human, so two isolates were sampled from one animal/human. All the cyst samples were washed clean with phosphate-buffered saline (PBS) three times, and the samples were then preserved at −20 °C until being used for genomic DNA extraction.
DNA extraction and PCR amplification
To extract genomic DNA from cysts, including the protoscoleces and germinal layers, each individual cyst was washed at least three times with sterile distilled water and centrifuged to remove the salt ions from PBS, after which genomic DNA was extracted according to the manufacturer's instructions (TIANamp Micro DNA Kit, Code: DP316, Tiangen, Beijing, China). The genomic DNA concentration was measured using a spectrophotometer (Merck Millipore, Frankfurter, Germany), and the DNA was then stored at −20 °C until being used for PCR amplification. A fragment of the mitochondrial genes was amplified from each sample using the primers described by Liu et al. and Bowles et al. for detection and analysis (Bowles et al., 1992, 1994; Liu et al., 2015). The PCR conditions and procedures were modified slightly, and the primers were cox1F: 5′- CCTGGATTTGGTATAATTAGTCA-3′ and cox1R: 5′- ATCATGCAAAAT/CATTATCT/CAACACACA-3′ (product = 366 bp). Each PCR mixture had a total volume of 50.0 µL, containing 25.0 µL of Premix Taq™ (TaKaRa Taq™ Version 2.0, Code: R004, Takara Bio Inc, Tokyo, Japan), 2.0 µL of each primer (10.0 µm), 2.0 µL of template DNA, and 19.0 µL of deionized distilled water. Positive and negative controls were run in parallel with the PCR amplification of the DNA samples. PCR amplification was carried out in a thermocycler (Mastercycler nexus GSX1, Eppendorf, Saxony, Germany) with a 5.0 min initial denaturation step at 95.0 °C; 35 cycles of a 35 s denaturation at 94.0 °C, a 45 s of annealing at 54.5 °C and a 40.0 s of extension at 72.0 °C; and a 10.0 min final extension at 72.0 °C. In the next step, the PCR products of the cox1 gene were electrophoresed in 1.2% agarose gels and stained with nucleic acid dyes. The PCR product bands were observed under UV light and recorded as digital images with a gel documentation system (BEIJING LIUYI BIOTECHNOLOGY CO., LTD., Beijing, China).
Sequencing and alignment analysis
A total of 244 PCR products (product = 366 bp) from different hosts were sequenced using both the forward and reverse primers by the GENEWIZ Company (Suzhou, China). Sequences were identified and compared in the GenBank database through BLAST analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi?), developed by the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/). Amino acid sequences were inferred from the nucleotide sequences based on echinoderm and flatworm mitochondrial genetic codes using the ExPASy translate tool (https://web.expasy.org/translate/). Then, the sequences were subjected to multiple alignment by using the Clustal Omega alignment programme (http://www.ebi.ac.uk/Tools/msa/clustalo/) with reference sequences from different E. granulosus s. s. genotypes and Echinococcus spp..
Data analysis
DnaSP 5.0 software was used to convert the fasta format (.fas) to the Network data format (.rdf) ARLEQUIN data format (.arp), and population diversity indexes (number of haplotypes and haplotype diversity) could also be calculated with this software. Then, we used NETWORK 5.0 (http://www.fluxus-engineering.com) to construct haplotype median-joining networks (Bandelt et al., 1999). Networks were constructed from the nucleotide sequences of the mitochondrial cox1 gene of Echinococcus spp. from all samples based on parameters of weights = 10 and epsilon = 0; nuclear data showed minimal variation and were not included. We computed population diversity indexes (number of haplotypes, haplotype and nucleotide diversity, and mean number of pairwise differences) within the different host groups identified from the phylogenetic analyses with the programme ARLEQUIN 3.5 (Excoffier and Lischer, 2010), which was also employed to calculate the neutrality indexes of Tajima's D (Tajima, 1989) and Fu's Fs (Fu, 1997); finally, the degree of gene flow among the three host populations (human, yak and sheep) was estimated using a pairwise fixation index (Fst) as a relative measure of population differentiation, which was determined with the ARLEQUIN package.
Results
Sequencing analysis
PCR amplification of the cox1 gene was successfully performed for all of the isolated hydatid cysts. According to the cox1 gene nucleotide sequences obtained from the isolated samples, all the human isolates (n = 93), yak isolates (n = 91) and sheep isolates (n = 38) were identified as the E. granulosus s. s. G1 genotype. In addition, the pika isolates were identified as E. multilocularis (n = 16) and E. shiquicus (n = 6). E. granulosus s. s. G1 was clearly the most prevalent species in the animal and human isolates of hydatid cysts. None of the isolates from a given patient/animal occurred in a coinfection.
Variation in nucleotide and amino acid sequences of E. granulosus s. s.
A 366-nucleotide consensus cox1 sequence was used to compare and to obtain the haplotypes (Farhadi et al., 2015). Based on the comparison data on cox1 gene sequences, 16 different haplotypes (G1s) were detected, which were designated as EgQH1 to EgQH16 [GenBank accession numbers MG674403-MG674418 (Table 1)]. A total of 34 point mutations were found within the haplotypes, consisting of 27 (79.4%) synonymous and 7 (20.6%) non-synonymous substitutions. For the non-synonymous mutations, the number of transition mutations was 5, and the number of transversions was 2; interestingly, the same transition mutation was found at position 56 of EgQH4 (C to T) and EgQH11 (C to T), and the same transition mutation was found at position 111 of ten haplotypes (T to C). Based on the sequence alignment, the EgQH7 haplotype was completely identical to the G1 (AF297617) reference genotype without base mutations; EgQH1, EgQH2, EgQH4, EgQH6, EgQH8, EgQH9, EgQH11, EgQH12, EgQH13, EgQH15 and EgQH16 showed very small nucleotide variations at either one or two positions, and seven haplotypes all included the non-synonymous substitution position 111 (T to C); another five haplotypes, EgQH3, EgQH5, EgQH7, EgQH10 and EgQH14, showed a large number of nucleotide variations (3 to 6) parallel to this, resulting in 1 to 2 different amino acid substitutions. Six nucleotide variations in EgQH10 led to only 2 amino acid substitutions, but two nucleotide variations in EgQH8 also generated 2 substitutions. There was only one transition mutation in EgQH4, EgQH5 and EgQH9, leading to the substitution of an amino acid. Among all 16 haplotypes, EgQH1 was the most common variant and was observed in 53 (21.7%) isolates, comprising 19 human, 21 yak and 13 sheep isolates. EgQH2 was the second most common variant, being found in 40 (16.4%) isolates: 14 human, 15 yak and 11 sheep isolates (Table 1). The next fourteen haplotypes (EgQH3–EgQH16) were found in 129 (52.9%) isolates. In addition, while haplotype EgQH1 was the most prevalent variant (22.5%) in the animal isolates, it was also observed (20.4%) in human isolates, and the second most frequent haplotype in the animal isolates was EgQH2.
Table 1.
Echinococcus spp. haplotypes characterized by the partial mitochondrial cytochrome c oxidase subunit 1 (cox1) gene sequence (length = 366 bp) analysis
| Haplotype of isolated Qinghai strains | Host origin (number) | Accession no. | Haplotype in the Fig. 1 |
|---|---|---|---|
| EgQH1 | Human (19), yak (21), sheep (13) | MG674403 | EGH1 |
| EgQH2 | Human (14), yak (15), sheep (11) | MG674404 | EGH3 |
| EgQH3 | Human (11), yak (13), sheep (5) | MG674405 | EGH4 |
| EgQH4 | Human (9), yak (11), sheep (2) | MG674406 | EGH7 |
| EgQH5 | Human (7), yak (9), sheep (3) | MG674407 | EGH8 |
| EgQH6 | Human (6), yak (2), sheep (1) | MG674408 | EGH9 |
| EgQH7 | Human (5), yak (4), sheep (0) | MG674409 | EGH10 |
| EgQH8 | Human (3), yak (3), sheep (0) | MG674410 | EGH11 |
| EgQH9 | Human (2), yak (1), sheep (1) | MG674411 | EGH12 |
| EgQH10 | Human (3), yak (2), sheep (1) | MG674412 | EGH15 |
| EgQH11 | Human (2), yak (2), sheep (0) | MG674413 | EGH16 |
| EgQH12 | Human (2), yak (2), sheep (0) | MG674414 | EGH17 |
| EgQH13 | Human (2), yak (2), sheep (0) | MG674415 | EGH18 |
| EgQH14 | Human (3), yak (1), sheep (1) | MG674416 | EGH19 |
| EgQH15 | Human (3), yak (1), sheep (0) | MG674417 | EGH22 |
| EgQH16 | Human (2), yak (2), sheep (0) | MG674418 | EGH23 |
| EmQH1 | Pika (16) | MG674419 | EMH1 |
| EsQH1 | Pika (6) | MG674420 | ESH1 |
Haplotype networks
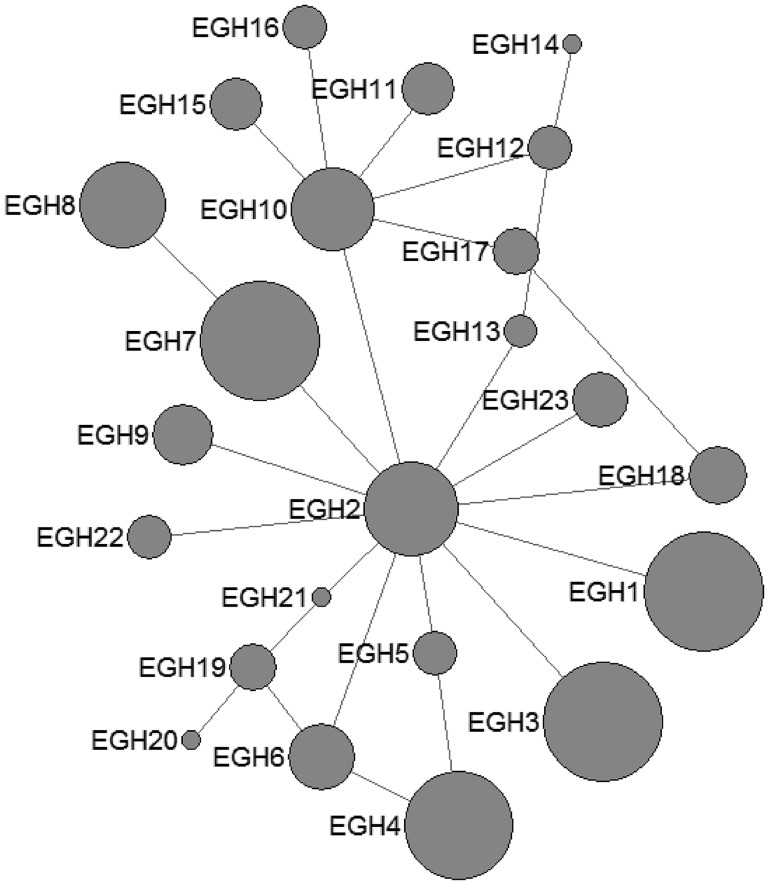
In E. granulosus s. s., 16 mtDNA haplotypes were found in 222 isolates from the QTPA of China. Nine haplotypes (EgQH1- EgQH 6, EgQH9, EgQH10 and EgQH14) were found in the populations of all three species (Fig. 1). Assuming that the ancestral haplotypes were still present in the recent populations, we constructed a statistical parsimony network to discriminate the genealogical relationships of the haplotypes among the hosts. The network showed a star-like configuration, with the common haplotype (G1) occupied the centre of the network, while G2 (EGH22) and G3 (EGH23) were linked to G1 via one mv1 (median vector) (Fig. 1). The population network presented a high-complexity structure, which included two main sub-groups (EGH4-EGH5 and EGH7-EGH12). However, the majority of haplotypes (EGH1-EGH16) and (EGH18-EGH21) all contained Chinese isolates from different hosts. Additionally, the haplotypes from different hosts were the same in the populations of other countries.
Fig. 1.
Haplotype network of the cytochrome c oxidase subunit 1 (cox1, 366 bp) gene of the Echinococcus granulosus sensu stricto complex from the QTPA of China (EGH1-EGH16) and other homologous sequences from GenBank constructed using NETWORK 5.0.
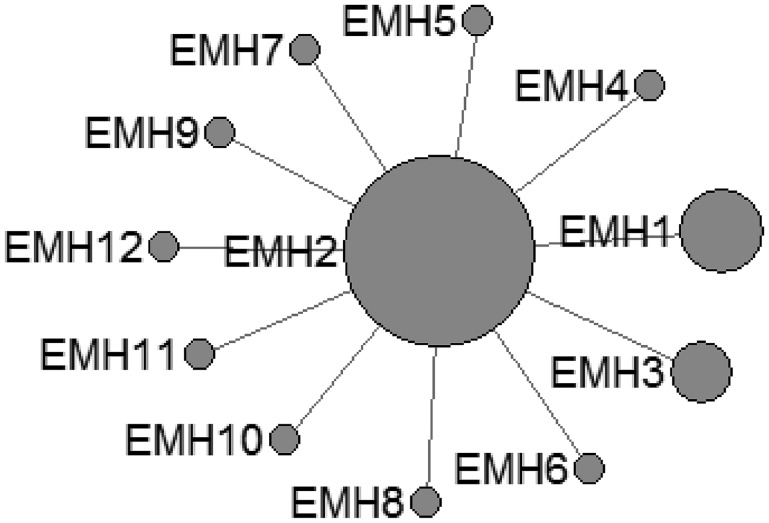
In E. multilocularis, 12 mtDNA haplotypes were found in all the isolates, which were also plotted a star-like network with one major haplotype (EMH2) (Fig. 2). The EMH1 haplotype came from three different hosts from two countries, and the EMH2 haplotype came from six different hosts from five countries, while the EMH4-EMH8 haplotypes all came from China but originated from different hosts.
Fig. 2.
Haplotype network of the cytochrome c oxidase subunit 1 (cox1, 366 bp) gene of the Echinococcus multilocularis from the QTPA of China (EMH1) and other homologous sequences from GenBank constructed using NETWORK 5.0.
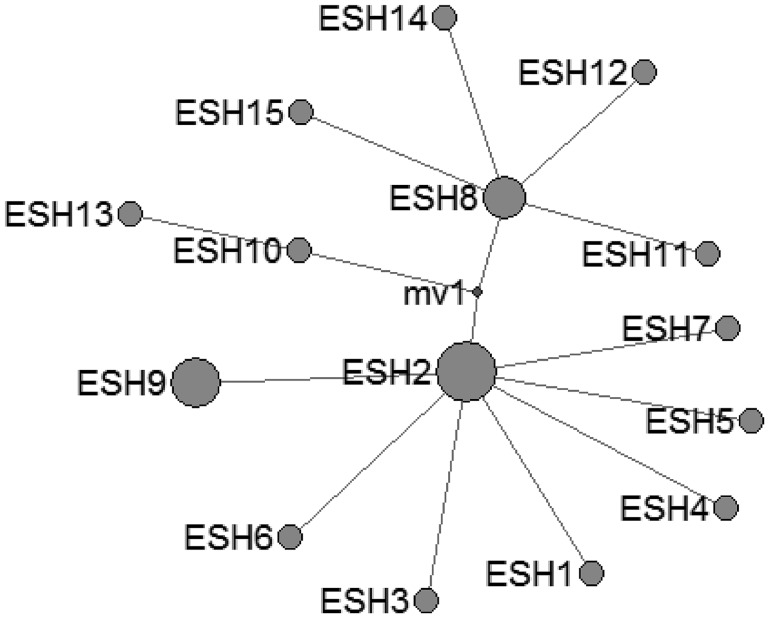
In E. shiquicus, 15 mtDNA haplotypes were found in the all isolates from China originating from four different hosts. The network was complicated with one star-like (ESH1-7 an ESH9) configuration and two main sub-groups (ESH10-ESH13 and ESH8, ESH11-ESH12, ESH14-ESH15) (Fig. 3).
Fig. 3.
Haplotype network of the cytochrome c oxidase subunit 1 (cox1, 366 bp) gene of the Echinococcus shiquicus from the QTPA of China hosts (ESH1) and other homologous sequences from GenBank constructed using NETWORK 5.0.
Diversity and neutrality indexes
The diversity indexes for the China QTPA isolates of E. granulosus s.s. from three different host species are shown in Table 2. Haplotype diversity was high for all E. granulosus s. s. populations within these three host species and was highest in E. granulosus s. s. from humans and lowest in those from sheep. In contrast, nucleotide diversity was low for all host species and ranged from 0.006 to 0.008 because of the richness of single nucleotide substitutions.
Table 2.
Diversity and neutrality indexes of the Echinococcus granulosus (sensu stricto) population calculated from the nucleotide sequence of the mitochondrial cytochrome c oxidase subunit 1 (cox1, 366 bp) gene. All P values for Tajima's D and Fu's Fs were not significant (P > 0.05)
| Host origin | n | S | K | H | Hd ± s.d. | π ± s.d. | Tajima's D | Fu's Fs |
|---|---|---|---|---|---|---|---|---|
| Humans | 93 | 21 | 2.74708 | 16 | 0.9028 ± 0.0140 | 0.007506 ± 0.004447 | −0.97737 | −3.47949 |
| Yaks | 91 | 21 | 2.60855 | 16 | 0.8781 ± 0.0161 | 0.007127 ± 0.004264 | −1.08740 | −3.89570 |
| Sheep | 38 | 15 | 2.35135 | 9 | 0.7909 ± 0.0416 | 0.006424 ± 0.003982 | −1.09838 | −1.06127 |
| Total | 222 | 21 | 2.619420 | 16 | 0.8731 ± 0.0109 | 0.007157 ± 0.004250 | −0.67886 | −1.84956 |
The neutrality indexes of E. granulosus s.s. populations from host species calculated with Tajima's D and Fu's Fs tests are also shown in Table 2; the values for these two indexes were all negative, which indicates an excess of rare polymorphic sites and a significant deviation from neutrality.
Pairwise fixation index values
The pairwise fixation indexes (Fst) for the cox1 sequences of populations of E. granulosus s.s. from different host species are shown in Supplementary Table 2. Low Fst values were observed for the majority of E. granulosus s. s. populations when compared in a pairwise manner with some populations showing negative values (humans/yaks). Since one common haplotype existed predominantly in the three host species, the Fst values between the populations were very small, ranging from 0.004 to 0.01. These low values implied that the populations were not genetically differentiated among these host species.
Discussion
In this study, the genetic diversity and population structure of Echinococcus spp. in QTPA of China was investigated. Data were obtained via sequencing of the cox1 mitochondrial gene, which had historically been demonstrated to show intraspecific variability and had been used for the study of the population structure of Echinococcus spp. from other parts of the world (Bowles et al., 1992; Nakao et al., 2010; Casulli et al., 2012; Yanagida et al., 2012). Although the new data published in October, 2018, showed that nad5 gene (680bp) was a highly suitable marker used for the differentiation of E. granulosus s. s. genotypes (Kinkar et al., 2018a). Initially, we used the partial cox1 (366 bp) to distinguish three genotypes (G1–G3) described within E. granulosus s. s (Bowles et al., 1992). The results presented in this report indicated that G1 was the most dominant distinct species of E. granulosus s. s. in the hydatid cyst samples from humans and animals in regions of the QTPA, China. The epidemiological data of this present study were in line with some previous molecular studies from China that had demonstrated G1 was the most common and dominant species of E. granulosus s. s. in humans, livestock and wild animals in China (Ma et al., 2008, 2015; Yang et al., 2009; Liu et al., 2013; Yan et al., 2013; Zhong et al., 2014; Hu et al., 2015), Turkey (Erdogan et al., 2017), Iran (Farhadi et al., 2015; Arbabi et al., 2017), and Bulgaria (Marinova et al., 2017). Likewise, many epidemiological studies conducted in the majority of regions of the world have also indicated E. granulosus s. s. G1 as the predominant species (Laurimae et al., 2016; Roinioti et al., 2016; Avila et al., 2017; Debiaggi et al., 2017; Ehsan et al., 2017; Thapa et al., 2017).
The cox1 haplotypes of E. granulosus s. s. found in this study did not reveal apparent systematic phylogeographic structuring in the QTPA of China. The parsimony network analysis revealed that the haplotypes exhibited a star-like expansion from a main global founder haplotype, suggesting that the populations of the QTPA of China were not fully differentiated from each other. It was noteworthy that the founder was predominant in the world population, which suggested that one particular lineage of E. granulosus s. s. was globally widespread in geographically unrelated populations. Nakao suggested that the mutation of the founder haplotypes were not advantageous, because the amino acid sequences of the founder were the same as those of certain other haplotypes (Nakao et al., 2010). Recently, Kinkar et al. (2018b) had shown that there was no such a founder haplotype for geographically unrelated populations of E. granulosus s.s. G1 based on a significantly longer mtDNA sequences and using a much larger number of samples, Moreover, such a founder also cannot be identified in case of E. granulosus s.s. G3 (Kinkar et al., 2018c). The high haplotype diversity observed in E. granulosus s.s. coupled with the low nucleotide diversity observed in the isolates of the QTPA of China was similar to that reported from Tunisia (Boufana et al., 2014), Jordan, Iran (Yanagida et al., 2012) and Europe (Casulli et al., 2012). On the other hand, the population genetic structures of E. granulosus s. s. comparing among various endemic areas clarified its worldwide dispersal. The neutrality indexes of Tajima's D obtained in the current study were negative, which suggested a bias towards the presence of nucleotide variants and was a feature of recent population expansion. Additionally, the neutrality index values Fu's Fs values was also negative for all populations, which indicated that the incidence of rare haplotypes was lower than expected under neutrality, and the values pointed to bottleneck events and/or purifying selection events that might have occurred in the past (Nakao et al., 2010; Boufana et al., 2014). Furthermore, the extremely low values of the fixation index Fst also supported genetic non-differentiation between the local populations, indicating limited gene flow.
The occurrence of most haplotypes (EgQH1, 3, 4, 7, 12, 13, 16) of E. granulosus s.s. in the QTPA of China was consistent with previously reported results (Ma et al., 2008, 2015). This distribution of the haplotypes indicates the importance of sheep and yak in maintaining potential reservoir infections for humans and definitive hosts, which further suggested it might cycle among these host species. The high frequency of the dominant E. granulosus s. s. haplotypes in the QTPA suggested that it may be the ancestral haplotype in the QTPA of China. In another surveillance report of Echinococcus isolates from the QTPA of China, a total of 105 haplotypes (H1-H105) were detected, and 177 variable sites were recorded in 521 samples using the cox1 mtDNA marker gene (Ma et al., 2015). Our results were different from these, our animal isolates collected from the slaughterhouse and the transported animals usually came from the same places, so the results were also different.
In the current study, E. multilocularis (n = 16) and E. shiquicus (n = 6) were present in pika isolates, and no co-infections were observed in individual isolated samples from the Golog Tibetan Autonomous Prefecture. Unfortunately, we were only allowed to conduct sampling in the Golog prefecture at this time, which provided an explanation for why only the same genotype was found here. In our future work, we will conduct sampling and analyses elsewhere, and more samples will be collected and analysed. The identified sequences were highly similar to the referenced E. multilocularis (AB033406) and E. shiquicus sequences (AB208064). E. shiquicus was first identified in the Tibetan fox Vulpes ferrilata (adult stage) and the plateau pika Ochotona curzoniae (larval stage) and was characterized based on its morphological, genetic and ecological features (Xiao et al., 2005, 2006). Thereafter, E. shiquicus infections in dogs were also reported in the eastern QTPA; E. shiquicus was being transferred from its natural host, the Tibetan fox to the domestic dog (Boufana et al., 2013), which will threaten human health, although no cases of E. shiquicus human infection have yet been reported. Fan et al., conducted a genetic diversity analysis of E. shiquicus isolates from the plateau pika in Darlag County of Qinghai Province, and the genetic diversity of the nad1 and cox1 genes was shown to vary by 0.1–1.2% and 0.1–1.0%, respectively, with 6 haplotypes ranging from 4.2 to 29.6% (Fan et al., 2016); however, only one E. shiquicus haplotype was found in this study. Fan et al. trapped 71 plateau pika samples; we obtained 22 samples and conducted one gene (cox1) sequence genetic diversity analysis. The number of samples and sample collection sites for analysis may need to be expanded in future studies. E. multilocularis represents the second greatest echinococcosis threat to the local people following E. granulosus s. l., and some E. multilocularis haplotypes are also shared by domestic animals (sheep, yaks, and dogs) and humans (Ma et al., 2015); however, only one E. multilocularis haplotype was found in the plateau pika and not in the yaks, sheep and humans in this study, but the threat of E. multilocularis to humans and livestock should not be neglected. E. multilocularis has been detected in domestic animals (dogs and cats) and wild hosts (deer mice, meadow voles and southern red backed voles), not only in China but also in other countries (Liccioli et al., 2014; Poulle et al., 2017). Echinococcosis can also be a foodborne disease, and host factors and environmental risk factors can also serve as important transmission routes that pose a threat to humans. Further studies are needed to cover many sample collection sites and a larger number of pathogen isolates, which may identify abundant strains and haplotypes in the hydatid cysts infecting human and animal populations of the QTPA, China.
Acknowledgements
The authors would like to express their gratitude and appreciation to the staff working in the slaughterhouse and the operating rooms of the hospitals in Qinghai Provincial People's Hospital for their assistance in collecting the samples required for the study.
Author ORCIDs
Zhang Xueyong, 0000-0001-5076-9211.
Financial support
This research was supported by the Project for Medical Science and Technology of the Qinghai National Health and Family Planning Commission (2017-wjzdx-22), the International Cooperation Project of the Qinghai Science and Technology Department (2019-HZ-803) and the National Natural Science Foundation of China (No. 31860700) and the Qinghai University Young and Middle Research Foundation Project (2018-QNY-1).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
This study was reviewed strictly by the ethics committee of Qinghai Provincial People's Hospital in accordance with relevant medical ethics regulations, because of the study of hydatid cysts isolated from patients, which was related to the category of medical ethics. Therefore, the study was performed under the supervision of ethics committee, to ensure that the study met the relevant requirements of the ethics committee and maintained the rights of patient. This study was only focus on the hydatid cysts isolated from patients, it was not involved any personal privacy information (individual details, images or videos), and before hydatid cysts removal surgery, the patients were all informed that the hydatid cysts would be further identified or used for scientific research, they all knew and agreed to publish the hydatid cysts biological information data.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0031182019000544.
click here to view supplementary material
References
- Arbabi M, Pirestani M, Delavari M, Hooshyar H, Abdoli A and Sarvi S (2017) Molecular and morphological characterizations of Echinococcus granulosus from human and animal isolates in Kashan, Markazi Province, Iran. Iranian Journal of Parasitology 12, 177–187. [PMC free article] [PubMed] [Google Scholar]
- Avila HG, Santos GB, Cucher MA, Macchiaroli N, Perez MG, Baldi G, Jensen O, Perez V, Lopez R, Negro P, Scialfa E, Zaha A, Ferreira HB, Rosenzvit M and Kamenetzky L (2017) Implementation of new tools in molecular epidemiology studies of Echinococcus granulosus sensu lato in South America. Parasitology International 66, 250–257. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P and Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16, 37–48. [DOI] [PubMed] [Google Scholar]
- Boufana B, Qiu J, Chen X, Budke CM, Campos-Ponce M and Craig PS (2013) First report of Echinococcus shiquicus in dogs from eastern Qinghai-Tibet plateau region, China. Acta Tropica 127, 21–24. [DOI] [PubMed] [Google Scholar]
- Boufana B, Lahmar S, Rebai W, Ben Safta Z, Jebabli L, Ammar A, Kachti M, Aouadi S and Craig PS (2014) Genetic variability and haplotypes of Echinococcus isolates from Tunisia. Transactions of the Royal Society of Tropical Medicine and Hygiene 108, 706–714. [DOI] [PubMed] [Google Scholar]
- Bowles J, Blair D and McManus DP (1992) Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Molecular and Biochemical Parasitology 54, 165–173. [DOI] [PubMed] [Google Scholar]
- Bowles J, Blair D and McManus DP (1994) Molecular genetic characterization of the cervid strain (‘northern form’) of Echinococcus granulosus. Parasitology 109(Pt2), 215–221. [DOI] [PubMed] [Google Scholar]
- Budke CM, Deplazes P and Torgerson PR (2006) Global socioeconomic impact of cystic echinococcosis. Emerging Infectious Diseases 12, 296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casulli A, Manfredi MT, La Rosa G, Cerbo AR, Genchi C and Pozio E (2008) Echinococcus ortleppi and E. granulosus g1, G2 and G3 genotypes in Italian bovines. Veterinary Parasitology 155, 168–172. [DOI] [PubMed] [Google Scholar]
- Casulli A, Interisano M, Sreter T, Chitimia L, Kirkova Z, La Rosa G and Pozio E (2012) Genetic variability of Echinococcus granulosus sensu stricto in Europe inferred by mitochondrial DNA sequences. Infection, Genetics and Evolution 12, 377–383. [DOI] [PubMed] [Google Scholar]
- Debiaggi MF, Soriano SV, Pierangeli NB, Lazzarini LE, Pianciola LA, Mazzeo ML, Moguillansky S and Farjat JAB (2017) Genetic characterization of human hydatid cysts shows coinfection by Echinococcus canadensis G7 and Echinococcus granulosus sensu stricto G1 in Argentina. Parasitology Research 116, 2599–2604. [DOI] [PubMed] [Google Scholar]
- Ehsan M, Akhter N, Bhutto B, Arijo A and Ali Gadahi J (2017) Prevalence and genotypic characterization of bovine Echinococcus granulosus isolates by using cytochrome oxidase 1 (Co1) gene in Hyderabad, Pakistan. Veterinary Parasitology 239, 80–85. [DOI] [PubMed] [Google Scholar]
- Erdogan E, Ozkan B, Mutlu F, Karaca S and Sahin I (2017) Molecular characterization of Echinococcus granulosus isolates obtained from different hosts. Mikrobiyoloji Bulteni 51, 79–86. [DOI] [PubMed] [Google Scholar]
- Excoffier L and Lischer HE (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10, 564–567. [DOI] [PubMed] [Google Scholar]
- Fan YL, Lou ZZ, Li L, Yan HB, Liu QY, Zhan F, Li JQ, Liu CN, Cai JZ, Lei MT, Shi WG, Yang YR, McManus DP and Jia WZ (2016) Genetic diversity in Echinococcus shiquicus from the plateau pika (Ochotona curzoniae) in Darlag County, Qinghai, China. Infection, Genetics and Evolution 45, 408–414. [DOI] [PubMed] [Google Scholar]
- Farhadi M, Fazaeli A and Haniloo A (2015) Genetic characterization of livestock and human hydatid cyst isolates from northwest Iran, using the mitochondrial cox1 gene sequence. Parasitology Research 114, 4363–4370. [DOI] [PubMed] [Google Scholar]
- Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosso G, Gruttadauria S, Biondi A, Marventano S and Mistretta A (2012) Worldwide epidemiology of liver hydatidosis including the Mediterranean area. World Journal of Gastroenterology 18, 1425–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han XM, Cai QG, Wang W, Wang H, Zhang Q and Wang YS (2018) Childhood suffering: hyper endemic echinococcosis in Qinghai-Tibetan primary school students, China. Infectious Diseases of Poverty 7, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Song X, Wang N, Zhong X, Wang J, Liu T, Jiang Z, Dawa T, Gu X, Peng X and Yang G (2015) Molecular identification of Echinococcus granulosus on the Tibetan Plateau using mitochondrial DNA markers. Genetics and Molecular Research 14, 13915–13923. [DOI] [PubMed] [Google Scholar]
- Kinkar L, Laurimae T, Simsek S, Balkaya I, Casulli A, Manfredi MT, Ponce-Gordo F, Varcasia A, Lavikainen A, Gonzalez LM, Rehbein S, VDG J, Sprong H and Saarma U (2016) High-resolution phylogeography of zoonotic tapeworm Echinococcus granulosus sensu stricto genotype G1 with an emphasis on its distribution in Turkey, Italy and Spain. Parasitology 143, 1790–1801. [DOI] [PubMed] [Google Scholar]
- Kinkar L, Laurimae T, Sharbatkhori M, Mirhendi H, Kia EB, Ponce-Gordo F, Andresiuk V, Simsek S, Lavikainen A, Irshadullah M, Umhang G, Oudni-M'rad M, Acosta-Jamett G, Rehbein S and Saarma U (2017) New mitogenome and nuclear evidence on the phylogeny and taxonomy of the highly zoonotic tapeworm Echinococcus granulosus sensu stricto. Infection, Genetics and Evolution 52, 52–58. [DOI] [PubMed] [Google Scholar]
- Kinkar L, Laurimae T, Acosta-Jamett G, Andresiuk V, Balkaya I, Casulli A, Gasser RB, Gonzalez LM, Haag KL, Zait H, Irshadullah M, Jabbar A, Jenkins DJ, Manfredi MT, Mirhendi H, M'Rad S, Rostami-Nejad M, Oudni-M'rad M, Pierangeli NB, Ponce-Gordo F, Rehbein S, Sharbatkhori M, Kia EB, Simsek S, Soriano SV, Sprong H, Snabel V, Umhang G, Varcasia A and Saarma U (2018a) Distinguishing Echinococcus granulosus sensu stricto genotypes G1 and G3 with confidence: a practical guide. Infection, Genetics and Evolution 64, 178–184. [DOI] [PubMed] [Google Scholar]
- Kinkar L, Laurimae T, Acosta-Jamett G, Andresiuk V, Balkaya I, Casulli A, Gasser RB, van der Giessen J, Gonzalez LM, Haag KL, Zait H, Irshadullah M, Jabbar A, Jenkins DJ, Kia EB, Manfredi MT, Mirhendi H, M'Rad S, Rostami-Nejad M, Oudni-M'rad M, Pierangeli NB, Ponce-Gordo F, Rehbein S, Sharbatkhori M, Simsek S, Soriano SV, Sprong H, Snabel V, Umhang G, Varcasia A and Saarma U (2018b) Global phylogeography and genetic diversity of the zoonotic tapeworm Echinococcus granulosus sensu stricto genotype G1. International Journal for Parasitology 48, 729–742. [DOI] [PubMed] [Google Scholar]
- Kinkar L, Laurimae T, Balkaya I, Casulli A, Zait H, Irshadullah M, Sharbatkhori M, Mirhendi H, Rostami-Nejad M, Ponce-Gordo F, Rehbein S, Kia EB, Simsek S, Snabel V, Umhang G, Varcasia A and Saarma U (2018c) Genetic diversity and phylogeography of the elusive, but epidemiologically important Echinococcus granulosus sensu stricto genotype G3. Parasitology 145, 1613–1622. [DOI] [PubMed] [Google Scholar]
- Knapp J, Nakao M, Yanagida T, Okamoto M, Saarma U, Lavikainen A and Ito A (2011) Phylogenetic relationships within Echinococcus and Taenia tapeworms (Cestoda: Taeniidae): an inference from nuclear protein-coding genes. Molecular Phylogenetics and Evolution 61, 628–638. [DOI] [PubMed] [Google Scholar]
- Knapp J, Gottstein B, Saarma U and Millon L (2015) Taxonomy, phylogeny and molecular epidemiology of Echinococcus multilocularis: from fundamental knowledge to health ecology. Veterinary Parasitology 213, 85–91. [DOI] [PubMed] [Google Scholar]
- Laurimae T, Kinkar L, Andresiuk V, Haag KL, Ponce-Gordo F, Acosta-Jamett G, Garate T, Gonzalez LM and Saarma U (2016) Genetic diversity and phylogeography of highly zoonotic Echinococcus granulosus genotype G1 in the americas (Argentina, Brazil, Chile and Mexico) based on 8279 bp of mtDNA. Infection, Genetics and Evolution 45, 290–296. [DOI] [PubMed] [Google Scholar]
- Laurimae T, Kinkar L, Moks E, Romig T, Omer RA, Casulli A, Umhang G, Bagrade G, Irshadullah M, Sharbatkhori M, Mirhendi H, Ponce-Gordo F, Soriano SV, Varcasia A, Rostami-Nejad M, Andresiuk V and Saarma U (2018) Molecular phylogeny based on six nuclear genes suggests that Echinococcus granulosus sensu lato genotypes G6/G7 and G8/G10 can be regarded as two distinct species. Parasitology 145, 1929–1937. [DOI] [PubMed] [Google Scholar]
- Li K, Zhang L, Zhang H, Lei Z, Luo H, Mehmood K, Shahzad M, Lan Y, Wang M and Li J (2017) Epidemiological investigation and risk factors of Echinococcus granulosus in yaks (Bos grunniens), Tibetan pigs and Tibetans on Qinghai Tibetan plateau. Acta Tropica 173, 147–152. [DOI] [PubMed] [Google Scholar]
- Liccioli S, Kutz SJ, Ruckstuhl KE and Massolo A (2014) Spatial heterogeneity and temporal variations in Echinococcus multilocularis infections in wild hosts in a North American urban setting. International Journal for Parasitology 44, 457–465. [DOI] [PubMed] [Google Scholar]
- Liu Q, Cao L, Zhang Y, Xu D, Shang L, Wang X, Wei F, Xiao L, Ma R, Cai J and Zhao Q (2013) Genotypes of Echinococcus granulosus in animals from Yushu, Northeastern China. Vector-Borne and Zoonotic Diseases 13, 134–137. [DOI] [PubMed] [Google Scholar]
- Liu CN, Lou ZZ, Li L, Yan HB, Blair D, Lei MT, Cai JZ, Fan YL, Li JQ, Fu BQ, Yang YR, McManus DP and Jia WZ (2015) Discrimination between E. granulosus sensu stricto, E. multilocularis and E. shiquicus using a Multiplex PCR Assay. PLoS Neglected Tropical Diseases 9, e0004084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lymbery AJ, Jenkins EJ, Schurer JM and Thompson RC (2015) Echinococcus canadensis, E. borealis, and E. intermedius. What's in a name? Trends in Parasitology 31, 23–29. [DOI] [PubMed] [Google Scholar]
- Ma SM, Maillard S, Zhao HL, Huang X, Wang H, Geng PL, Bart JM and Piarroux R (2008) Assessment of Echinococcus granulosus polymorphism in Qinghai province, People's Republic of China. Parasitology Research 102, 1201–1206. [DOI] [PubMed] [Google Scholar]
- Ma J, Wang H, Lin G, Zhao F, Li C, Zhang T, Ma X, Zhang Y, Hou Z, Cai H, Liu P and Wang Y (2015) Surveillance of Echinococcus isolates from Qinghai, China. Veterinary Parasitology 207, 44–48. [DOI] [PubMed] [Google Scholar]
- Marinova I, Spiliotis M, Wang J, Muhtarov M, Chaligiannis I, Sotiraki S, Rainova I, Gottstein B and Boubaker G (2017) Molecular characterization of Echinococcus granulosus isolates from Bulgarian human cystic echinococcosis patients. Parasitology Research 116, 1043–1054. [DOI] [PubMed] [Google Scholar]
- McManus DP and Thompson RC (2003) Molecular epidemiology of cystic echinococcosis. Parasitology 127(Suppl), S37–S51. [DOI] [PubMed] [Google Scholar]
- Moks E, Jogisalu I, Valdmann H and Saarma U (2008) First report of Echinococcus granulosus G8 in Eurasia and a reappraisal of the phylogenetic relationships of ‘genotypes’ G5-G10. Parasitology 135, 647–654. [DOI] [PubMed] [Google Scholar]
- Nakao M, Li T, Han X, Ma X, Xiao N, Qiu J, Wang H, Yanagida T, Mamuti W, Wen H, Moro PL, Giraudoux P, Craig PS and Ito A (2010) Genetic polymorphisms of Echinococcus tapeworms in China as determined by mitochondrial and nuclear DNA sequences. International Journal for Parasitology 40, 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao M, Lavikainen A, Yanagida T and Ito A (2013) Phylogenetic systematics of the genus Echinococcus (cestoda: Taeniidae). International Journal for Parasitology 43, 1017–1029. [DOI] [PubMed] [Google Scholar]
- Oksanen A and Lavikainen A (2015) Echinococcus canadensis transmission in the North. Veterinary Parasitology 213, 182–186. [DOI] [PubMed] [Google Scholar]
- Onac D, Gyorke A, Oltean M, Gavrea R and Cozma V (2013) First detection of Echinococcus granulosus G1 and G7 in wild boars (Sus scrofa) and red deer (Cervus elaphus) in Romania using PCR and PCR-RFLP techniques. Veterinary Parasitology 193, 289–291. [DOI] [PubMed] [Google Scholar]
- Poulle ML, Bastien M, Richard Y, Josse-Dupuis E, Aubert D, Villena I and Knapp J (2017) Detection of Echinococcus multilocularis and other foodborne parasites in fox, cat and dog faeces collected in kitchen gardens in a highly endemic area for alveolar echinococcosis. Parasite 24, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian MB, Abela-Ridder B, Wu WP and Zhou XN (2017) Combating echinococcosis in China: strengthening the research and development. Infectious Diseases of Poverty 6, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roinioti E, Papathanassopoulou A, Theodoropoulou I, Simsek S and Theodoropoulos G (2016) Molecular identification of Echinococcus granulosus isolates from ruminants in Greece. Veterinary Parasitology 226, 138–144. [DOI] [PubMed] [Google Scholar]
- Romig T, Deplazes P, Jenkins D, Giraudoux P, Massolo A, Craig PS, Wassermann M, Takahashi K and de la Rue M (2017) Ecology and Life Cycle Patterns of Echinococcus Species. Advances in Parasitology 95, 213–314. [DOI] [PubMed] [Google Scholar]
- Rostami S, Talebi S, Babaei Z, Sharbatkhori M, Ziaali N, Rostami H and Harandi MF (2013) High resolution melting technique for molecular epidemiological studies of cystic echinococcosis: differentiating G1, G3, and G6 genotypes of Echinococcus granulosus sensu lato. Parasitology Research 112, 3441–3447. [DOI] [PubMed] [Google Scholar]
- Saarma U, Jogisalu I, Moks E, Varcasia A, Lavikainen A, Oksanen A, Simsek S, Andresiuk V, Denegri G, Gonzalez LM, Ferrer E, Garate T, Rinaldi L and Maravilla P (2009) A novel phylogeny for the genus Echinococcus, based on nuclear data, challenges relationships based on mitochondrial evidence. Parasitology 136, 317–328. [DOI] [PubMed] [Google Scholar]
- Scott JC, Stefaniak J, Pawlowski ZS and McManus DP (1997) Molecular genetic analysis of human cystic hydatid cases from Poland: identification of a new genotypic group (G9) of Echinococcus granulosus. Parasitology 114(Pt1), 37–43. [DOI] [PubMed] [Google Scholar]
- Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa NK, Armua-Fernandez MT, Kinzang D, Gurung RB, Wangdi P and Deplazes P (2017) Detection of Echinococcus granulosus and Echinococcus ortleppi in Bhutan. Parasitology International 66, 139–141. [DOI] [PubMed] [Google Scholar]
- Thompson RC (2008) The taxonomy, phylogeny and transmission of Echinococcus. Experimental Parasitology 119, 439–446. [DOI] [PubMed] [Google Scholar]
- Thompson RC (2017) Biology and systematics of Echinococcus. Advances in Parasitology 95, 65–109. [DOI] [PubMed] [Google Scholar]
- Thompson RC and McManus DP (2002) Towards a taxonomic revision of the genus Echinococcus. Trends in Parasitology 18, 452–457. [DOI] [PubMed] [Google Scholar]
- Torgerson PR and Heath DD (2003) Transmission dynamics and control options for Echinococcus granulosus. Parasitology 127(Suppl), S143–S158. [DOI] [PubMed] [Google Scholar]
- Umhang G, Chihai O and Boue F (2014) Molecular characterization of Echinococcus granulosus in a hyperendemic European focus, the Republic of Moldova. Parasitology Research 113, 4371–4376. [DOI] [PubMed] [Google Scholar]
- Wang Q, Huang Y, Huang L, Yu W, He W, Zhong B, Li W, Zeng X, Vuitton DA, Giraudoux P, Craig PS and Wu W (2014) Review of risk factors for human echinococcosis prevalence on the Qinghai-Tibet Plateau, China: a prospective for control options. Infectious Diseases of Poverty 3, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao N, Qiu J, Nakao M, Li T, Yang W, Chen X, Schantz PM, Craig PS and Ito A (2005) Echinococcus shiquicus n. sp., a taeniid cestode from Tibetan fox and plateau pika in China. International Journal for Parasitology 35, 693–701. [DOI] [PubMed] [Google Scholar]
- Xiao N, Qiu J, Nakao M, Li T, Yang W, Chen X, Schantz PM, Craig PS and Ito A (2006) Echinococcus shiquicus, a new species from the Qinghai-Tibet plateau region of China: discovery and epidemiological implications. Parasitology International 55(Suppl), S233–S236. [DOI] [PubMed] [Google Scholar]
- Yan N, Nie HM, Jiang ZR, Yang AG, Deng SJ, Guo L, Yu H, Yan YB, Tsering D, Kong WS, Wang N, Wang JH, Xie Y, Fu Y, Yang DY, Wang SX, Gu XB, Peng XR and Yang GY (2013) Genetic variability of Echinococcus granulosus from the Tibetan plateau inferred by mitochondrial DNA sequences. Veterinary Parasitology 196, 179–183. [DOI] [PubMed] [Google Scholar]
- Yanagida T, Mohammadzadeh T, Kamhawi S, Nakao M, Sadjjadi SM, Hijjawi N, Abdel-Hafez SK, Sako Y, Okamoto M and Ito A (2012) Genetic polymorphisms of Echinococcus granulosus sensu stricto in the Middle East. Parasitology International 61, 599–603. [DOI] [PubMed] [Google Scholar]
- Yang YR, Liu T, Bai X, Boufana B, Craig PS, Nakao M, Ito A, Zhang JZ, Giraudoux P and McManus DP (2009) Natural infection of the ground squirrel (Spermophilus spp.) with Echinococcus granulosus in China. PLoS Neglected Tropical Diseases 3, e518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Yang D, Zeng Z, Zhao W, Liu A, Piao D, Jiang T, Cao J, Shen Y, Liu H and Zhang W (2014) Genetic characterization of human-derived hydatid cysts of Echinococcus granulosus sensu lato in Heilongjiang Province and the first report of G7 genotype of E. canadensis in humans in China. PLoS One 9, e109059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Wang N, Hu D, Wang J, Liu T, Gu X, Wang S, Peng X and Yang G (2014) Sequence analysis of cytb gene in Echinococcus granulosus from Western China. The Korean Journal of Parasitology 52, 205–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0031182019000544.
click here to view supplementary material