Abstract
Background
Gestational diabetes mellitus (GDM) is a common pregnancy complication which has substantial short- and long-term adverse health implications for women and their children. However, large-scale studies on genetic risk loci for GDM remain sparse.
Methods
We conducted a case-control study among 2,636 GDM cases and 6,086 non GDM controls from the Nurses’ Health Study II (NHSII) and the Danish National Birth Cohort (DNBC). One hundred and twelve susceptibility genetic variants confirmed by genome-wide association studies for type 2 diabetes (T2D) were selected and measured. A weighted genetic risk score (GRS) was created based on variants that were significantly associated with risk of GDM after correcting for false discovery rate (FDR).
Results
For the first time, we identified eight variants associated with GDM, namely rs7957197 (HNF1A), rs10814916 (GLIS3), rs3802177 (SLC30A8), rs9379084 (RREB1), rs34872471 (TCF7L2), rs7903146 (TCF7L2), rs11787792 (GPSM1), and rs7041847 (GLIS3). Additionally, we confirmed three variants, rs10830963 (MTNR1B), rs1387153 (MTNR1B), and rs4506565 (TCF7L2), that were previously significantly associated with GDM risk. Furthermore, compared to participants in the first (lowest) quartile of weighted GRS based on these 11 SNPs, the odds ratios of GDM were 1.07 (95% CI: 0.93, 1.22), 1.23 (95% CI 1.07, 1.41), and 1.53 (95% CI 1.34, 1.74) in the second, third, and fourth (highest) quartile, respectively. The significant positive associations between the weighted GRS and risk of GDM persisted across most of the strata of major risk factors for GDM including family history of type 2 diabetes, smoking status, body mass index, and age.
Conclusion
In this large-scale case-control study with women from two independent populations, eight novel GDM SNPs were identified, which offers potential to improve our understanding on GDM etiology, particularly biological mechanisms related to beta cell function.
Keywords: Gestational diabetes mellitus, genetic risk score, genetic variants
INTRODUCTION
Gestational diabetes mellitus (GDM) is one of the most common complications of pregnancy, impacting approximately 7% (ranging from 1% to 14%) of all pregnancies in the U.S. [1]. Globally, the prevalence of GDM has increased by more than 30% following worldwide trends of increasing obesity [2,3]. GDM is associated with short and long-term complications for women and their offspring. Women with GDM have an increased risk for gestational hypertension and pre-eclampsia during pregnancy and a significantly higher risk for impaired glucose tolerance and type 2 diabetes (T2D) after pregnancy [4,5]. Moreover, offspring born to women with GDM are more likely to be obese[6,7], and have impaired glucose tolerance and diabetes in childhood and early adulthood [8,9]. Several modifiable lifestyle factors of GDM have been identified for the prevention of GDM, including maintaining a normal body weight and healthful diet, being physically active, and abstaining from cigarette smoking [10–15].
Although compelling data suggest that genetic factors play a role in GDM[10,16], relatively few studies have been published on the genetic susceptibility to GDM [17,18]. Insulin resistance and defects in insulin secretion play a pivotal role in the development of GDM [19]. More than 53% of peripheral insulin sensitivity and 75% of the variation in insulin secretion can be explained by genetic components[20,21]. Previous studies have shown that genetic variants of KCNJ11[22], TCF7L2[23,24], KCNQ1[25], MTNR1B[24], and IRS1[24] were associated with GDM risk. However, these studies examined only a priori single or a small number of genetic variants. Only one genome-wide association study (GWAS) of GDM in an Asian population has been conducted and two genetic variants rs10830962 near MTNR1B and rs7754840 in CDKAL1 were identified [26]. However, this study was limited by a relatively small sample size (1399 GDM cases and 2025 controls) in a Korean population. Therefore, we conducted a comprehensive candidate gene analysis to identify genetic variants of GDM among 8,722 Whites (i.e. 2,636 GDM cases and 6,086 non GDM controls) from the Nurses’ Health Study 2 (NHSII) and the Danish National Birth Cohort (DNBC). As insulin resistance and defects in insulin secretion play a central role in the pathogenesis of both GDM and T2D, we genotyped 112 susceptibility variants identified in previous GWAS of T2D as candidate SNPs for GDM [27–32].
METHODS
Study population
The current analysis used genotyping data from two sources: 1) genomic data of 6,873 women nested within existing NHSII, and 2) candidate genotyping in a sample of 1,227 women in the DNBC [33]. All study participants gave informed consent in participating in the study.
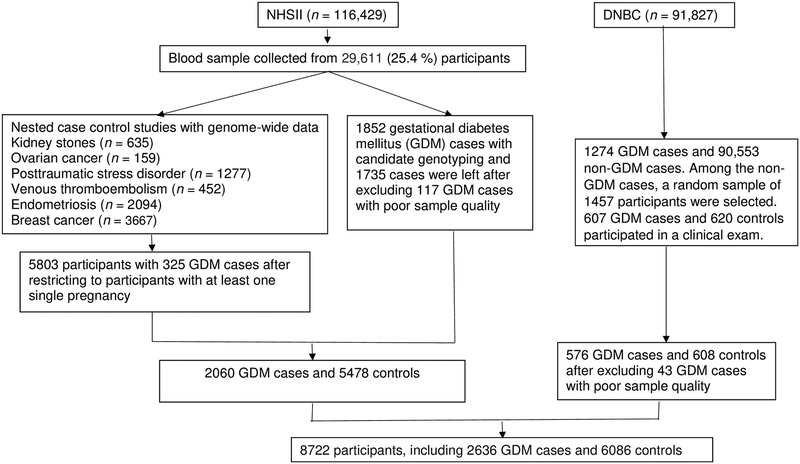
The NHSII was established in 1989 and consisted of 116,429 female registered nurses, aged 25–42 y at baseline. Detailed Questionnaire data were collected at baseline and biennially thereafter and included medical history, lifestyle, usual diet, and the occurrence of chronic diseases. In each biennial questionnaire through 2001, women were asked whether they were diagnosed as having GDM by a physician. In 2009, a questionnaire was administered to ascertain NHSII participants’ pregnancy and reproductive history. From 1996 to 2001, 29,611 NHSII participants aged 32–52 y provided blood samples. Among them, genome-wide data were available among participants of European ancestry within previous nested case-control studies of kidney stones, ovarian cancer, posttraumatic stress disorder, venous thromboembolism, endometriosis, and breast cancer [34,35]. Among all participants with genome-wide data, we restricted the current analysis to 5,803 participants with at least one pregnancy between 1989 and 2009, and 325 women reported a clinician diagnosis of GDM during pregnancy. Candidate genotyping was performed on DNA samples available from an additional 1,852 women with GDM collected as part of the Diabetes & Women’s Health (DWH) Study during 2012–2016 [33]. The flow diagram of sample selection is shown in Figure 1. The study protocols were approved by the institutional review boards of Brigham and Women’s Hospital, and the Harvard T.H. Chan School of Public Health. In a validation study among a subgroup of NHSII participants (n=120), 94% of self-reported GDM events were confirmed by medical records [10,36]. The majority of NHSII participants were screened for GDM during pregnancy. A supplemental questionnaire was sent to a random sample of parous women who did not report GDM (n=114), 83% reported undergoing a 50 g glucose screening test during pregnancy and 100% reported frequent prenatal urine glucose screening 7,32.
Figure.
A flow diagram of sample selection in the Nurses’ Health Study II (NHSII) and Danish National Birth cohort (DNBC)*.
* Characteristics of GDM cases and controls included in the final analytical population were similar to their corresponding GDM cases and controls source populations.
The DNBC (1996–2002) is a longitudinal cohort of 91,827 pregnant women in Denmark, who were recruited during their first antenatal visit to the general practitioner [37]. All women living in Denmark, who could speak Danish and were planning to carry to term, were eligible for the study. Prospective data on maternal socio-demographics, lifestyle and environmental exposures as well as clinical and perinatal conditions were collected in DNBC, through four telephone interviews at gestational weeks 12 and 30, and at 6 and 18 months postpartum.
Of the 91,827 DNBC participants, 1,274 were identified as GDM cases. Among 90,553 women who did not have GDM, a random sample of 1,457 women (controls) were selected. In the current analysis, we identified 607 GDM cases and 620 controls who participated in the DNBC clinical examination and provided bio-specimens as part of the DWH Study (2012–2014). The study was approved by the Regional Scientific Ethical Committee (VEK) of the Capital Region of Denmark (record no. H-4–2013-129). Study procedures were followed in accordance with the Declaration of Helsinki.
The methods and procedures undertaken to ascertain GDM in the DNBC have been previously described in detail [37]. Briefly, in the DNBC, questions related to GDM were asked at gestational week 30 and at 6 months postpartum. Women who either self-reported GDM in the interviews or had a GDM diagnosis recorded in the National Patient Registry (NPR) were considered as having GDM. Women who had a diabetes diagnosis recorded in the NPR prior to the index pregnancy were excluded. Medical records were retrieved for all women suspected to have GDM as well as the randomly selected control group and indicated a high sensitivity (96%). An expert panel developed criteria and guidelines for extracting relevant data and for ascertaining GDM diagnoses according to the WHO criteria [37,38].
Genotyping
Briefly, the genome-wide genotyping methods used by the NHSII have been described in detail elsewhere [39]. Genome-wide genotyping was conducted using high-density SNP marker platforms including Illumina HumanHap, Infinium OncoArray, and Infinium HumanCoreExome. Genotypes were imputed using the 1000 Genomes Project ALL Phase I Integrated Release Version 3 Haplotypes excluding monomorphic and singleton sites (2010–11 data freeze, 2012–03-14 haplotypes) as the reference panel. SNPs for which Hardy-Weinberg equilibrium testing produced a p value <1 ×10−6 were excluded. Most of the SNPs were genotyped (sample call rate = 97%) or had a high imputation quality score (r2 ≥ 0.8), as assessed with the use of MACH software. Moreover, the effective allele frequency (EAF) and imputation quality score of all SNPs genotyped in different platforms were similar (Supplementary Table 1).
Among GDM cases collected as part of the DWH Study (i.e., NHSII participants whose genome-wide data were unavailable, and DNBC participants), genotyping was performed using Taqman qPCR method. Taqman reagents and protocols for uniplex qPCR realtime-amplification and genotyping by allelic discrimination were performed per the manufacturer’s instructions (for complete details see TaqMan SNP Genotyping Assays protocol, Applied Biosystems, Foster City, CA). We excluded participants with poor sample quality (i.e., participants who had genotyping failure for more than 100 SNPs). In total, 117 participants were excluded from the NHSII (all GDM cases) and 43 participants were excluded from the DNBC. The final analytical population of the present study is composed of 6,756 women (2,014 GDM cases and 4,742 controls) from the NHSII, and 1,184 participants (576 GDM cases and 608 controls) from the DNBC
Distributions of major characteristics of these women were similar to their corresponding GDM cases and controls source populations.
Candidate SNP selection
We initially selected a total of 130 SNPs, which were significantly associated with risk of T2D based on previous GWAS [27–32]. We excluded 18 SNPs because they had minor allele frequencies <1% (rs60980157, rs2233580, rs3842770, rs7560163, and rs9552911), because they were not imputed in genome-wide genotyping in the NHSII (rs5945326 and rs12010175), or because they could not be genotyped in candidate gene genotyping (rs163182, rs10965250, rs1470579, rs312457, rs343092, rs6467136, rs7656416, rs7901695, rs34160967, rs6968865, and rs713598). In total, 112 SNPs were available for further analysis (Supplementary Table 2).
Assessment of covariates
Covariates for the NHSII and the DNBC were selected a priori. The covariates in the NHSII were ascertained from the baseline questionnaire and included age (years), smoking (never smoker vs. smoker), family history of T2D, and body mass index (BMI) calculated from self-reported height and weight. Covariates in the DNBC were ascertained from questionnaires administered during the index pregnancy and included age (years), smoking during pregnancy (yes vs. no), and pre-pregnancy BMI calculated from self-reported height and pre-pregnancy weight. In the DNBC, family history of diabetes (yes vs. no) was collected as part of the DWH Study follow-up.
Statistical analysis
We identified the risk allele of each SNP associated with risk of T2D based on previous GWAS studies of T2D (Supplementary Table 2). Logistic regression models were fitted to evaluate the association between each SNP and the risk of GDM by using an additive model in the NHSII and DNBC. Results from two cohorts were meta-analyzed using a fixed effect inverse variance model [40]. False discovery rate (FDR) was used to account for multiple testing, and the Benjamini–Yekutieli (B-Y) procedure was adopted [41]. The B-Y procedure stringently controls the proportion of false positives among rejected hypotheses, and performs well in the presence of correlation among genetic variants.
We created unweighted and weighted genetic risk scores (GRS) based on SNPs that were significantly associated with risk of GDM after FDR correction (P<0.05). Specifically, unweighted GRS were created by summing up risk alleles of identified SNPs, which was the allele associated with higher risk of T2D based on a literature search (Supplementary Table 2). Weighted GRS was created by summing up risk alleles of identified SNPs multiplied by the corresponding weight estimated based on the pooled coefficient of each SNP with risk of GDM from both cohorts. Using a similar method, we created unweight and weighted GRS based on all candidate SNPs included in our study. We additionally created two sub-GRSs according to their biological functions, GRS based on 66 SNPs related to beta cell function (GRS-BC) and GRS based on 17 SNPs related to insulin resistance (GRS-IR) [42,43], and examined the potential differences in associations with risk of GDM. Participants were categorized into four groups defined by the 25th, 50th, and 75th percentile GRS scores (i.e., Group 1: ≤25%, Group 2: 25–50%, Group 3: 50–75%, and Group 4: >75%). Logistic regression models were then fitted to examine the associations of GRSs with risk of GDM using Group 1 (0–25% group) as a reference in the NHSII and DNBC, and results from both cohorts were pooled using a fixed-effects model. Given that our study did not include a replication cohort, we additionally created GRS and examined the association with risk of GDM using 10-fold cross validation.[44] We extracted a subsample with replication from the pooled sample of the NHSII and the DNBC, and the subsample was divided into 10 approximately equal bins. The association of GRS with risk of GDM was evaluated 10 times, using nine bins to estimate the weight of each SNP by obtaining coefficient of each SNP with risk of GDM and the 10th bin to examine the association of GRS with risk of GDM. We averaged the association of GRS with risk of GDM for the 10th bin across the 10 analyses. We repeated the extraction of the subsample for 1000 times to obtain the nonparametric 95% confidence interval (CI) of the association of GRS with risk of GDM.
We conducted stratified analyses by family history of T2D, and smoking status, BMI, and age at baseline. We tested for potential effect modification by these stratified variables by including interaction terms between the exposure and potential effect modifier in the multivariate adjusted model, and conducting a likelihood ratio test (LRT) comparing the models with and without interaction terms. All statistical tests were two-sided and performed using SAS (version 9.4, SAS Institute). The 10-fold cross validation was conducted using R 3.2.5.
RESULTS
Our study population included 7,538 women (2,060 GDM cases and 5,478 controls) from the NHSII, and 1,184 participants (576 GDM cases and 608 controls) from the DNBC (Table 1). Compared to controls, GDM cases were more likely to be heavier and have a family history of T2D. By pooling the results from the DNBC and NHSII, we identified 11 SNPs significantly associated with risk of GDM after FDR correction (Table 2). Of the 11 SNPs, 8 SNPs were identified for the first time as novel SNPs of GDM, namely rs7957197 (HNF1A), rs10814916 (GLIS3), rs3802177 (SLC30A8), rs9379084 (RREB1), rs34872471 (TCF7L2), rs7903146 (TCF7L2), rs11787792 (GPSM1), and rs7041847 (GLIS3). Consistent with previous literature [23,24], rs10830963 (MTNR1B), rs1387153 (MTNR1B), and rs4506565 (TCF7L2) were also associated with risk of GDM in our study. The risk allele of T2D was associated with a higher risk of GDM for all identified SNPs except rs9379084 (RREB1) and rs11787792 (GPSM1), and the results were in general consistent between cohorts. The Supplementary Table 2 shows the association of all measured individual SNPs with risk of GDM in the NHSII and DNBC.
Table 1.
Baseline characteristics of 8722 study participants including 7538 women from the Nurses’ Health Study II (NHSII) and 1184 women from the Danish National Birth Cohort (DNBC).
| Baseline characteristicsa | NHSII (1989) | |
|---|---|---|
| GDM Cases | Non-GDM Controls | |
| Number of participants | 2060 | 5478 |
| Age at baseline (years) | 33.3±4.5 | 35.4±4.3 |
| Family history of T2D, no. (%) | 559 (27.1%) | 852 (15.6%) |
| Pre-pregnancy BMI (kg/m2)b | 25.0±5.7 | 23.8±4.6 |
| Never smoker, no. (%) | 1437 (70.4 %) | 3652 (67.7%) |
| DNBC (1996) | ||
| Number of participants | 576 | 608 |
| Age at baseline (years) | 31.6±4.6 | 30.5±4.2 |
| Family history of T2D, no. (%) | 212 (37.3%) | 100 (16.6%) |
| Pre-pregnancy BMI (kg/m2)b | 27.3±5.7 | 22.8±3.9 |
| Never smoker, no. (%) | 273 (49.3%) | 320 (53.2%) |
Plus-minus values are means ±SD.
The body-mass index is the weight in kilograms divided by the square of the height in meters.
Table 2.
SNPs significantly associated with risk of gestational diabetes (GDM) after false discovery rate (FDR) correction among 8722 women from Nurses’ Health Study II (NHSII) (total N=7538, GDM cases N=2060) and the Danish National Birth Cohort (DNBC) (total N=1184, GDM cases N=576).
| SNP | Chr | Gene | Functional annotation of the SNP | Effect allele of T2Da | EAFb | Pooled OR of GDM | p value | p value after FDR correction | OR of GDM in the NHSII | OR of GDM in the DNBC |
|---|---|---|---|---|---|---|---|---|---|---|
| Newly identified SNPs | ||||||||||
| rs7957197 | 12 | HNF1A | Intron region, beta cell function | T | 0.76 | 1.22 (1.12, 1.33) | 1×10−5 | 0.003 | 1.20 (1.09, 1.32) | 1.30 (1.05, 1.60) |
| rs10814916 | 9 | GLIS3 | Intron region, beta cell function | C | 0.58 | 1.16 (1.08, 1.24) | 4×10−5 | 0.005 | 1.18 (1.09, 1.27) | 1.05 (0.89, 1.23) |
| rs3802177 | 8 | SLC30A8 | Intron region, beta cell function | G | 0.74 | 1.17 (1.08, 1.26) | 8×10−5 | 0.008 | 1.14 (1.05, 1.24) | 1.32 (1.09, 1.60) |
| rs9379084 | 6 | RREB1 | NA | A | 0.09 | 0.80 (0.71, 0.90) | 1×10−4 | 0.009 | 0.78 (0.69, 0.88) | 0.93 (0.69, 1.25) |
| rs34872471 | 10 | TCF7L2 | Intron region, beta cell function | G | 0.09 | 1.14 (1.06, 1.23) | 4×10−4 | 0.02 | 1.12 (1.03, 1.21) | 1.26 (1.06, 1.50) |
| rs7903146 | 10 | TCF7L2 | Intron region, beta cell function | T | 0.54 | 1.15 (1.06, 1.24) | 4×10−4 | 0.02 | 1.13 (1.04, 1.23) | 1.24 (1.03, 1.49) |
| rs11787792 | 9 | GPSM1 | NA | A | 0.75 | 0.87 (0.80, 0.94) | 5×10−4 | 0.03 | 0.83 (0.76, 0.91) | 1.02 (0.86, 1.21) |
| rs7041847 | 9 | GLIS3 | Intron region, beta cell function | A | 0.66 | 1.13 (1.05, 1.20) | 5×10−4 | 0.03 | 1.15 (1.07, 1.24) | 1.02 (0.87, 1.20) |
| Previously identified SNPs | ||||||||||
| rs10830963 | 11 | MTNR1B | Intron region, beta cell function | G | 0.26 | 1.27 (1.18, 1.37) | 2×10−9 | <0.001 | 1.26 (1.16, 1.37) | 1.31 (1.10, 1.56) |
| rs4506565 | 10 | TCF7L2 | Intron region, beta cell function | T | 0.28 | 1.16 (1.08, 1.24) | 7×10−5 | 0.008 | 1.14 (1.05, 1.23) | 1.24 (1.04, 1.48) |
| rs1387153 | 11 | MTNR1B | Intron region, beta cell function | T | 0.35 | 1.17 (1.09, 1.26) | 2×10−5 | 0.005 | 1.16 (1.07, 1.26) | 1.24 (1.04, 1.48) |
T2D: type 2 diabetes.
Effect allele frequency.
Logistic regression model adjusted for baseline age.
The weighted GRS based on the 11 SNPs was significantly associated with a higher risk of GDM in both the NHSII and DNBC, and the results were consistent between cohorts (P for heterogeneity >0.05) (Table 3). Compared to participants in the lowest quartile of the weighted GRS, the OR of GDM across increasing quartile of GRS was 1.07 (95% CI 0.93, 1.22), 1.23 (95% CI 1.07, 1.41), and 1.53 (95% CI 1.34, 1.74) (p for trend <0.001). Specifically, per allele increase in GRS was associated with higher risk of GDM (OR=1.04, 95% CI 1.03, 1.05). The associations for the risk of GDM persisted and remained significant when using two other types of GRS as the main exposure: unweighted GRS and weighted GRS created using 10-fold cross validation. Furthermore, we created GRS by additionally including four SNPs that were found to be associated with the risk of GDM in the previous studies[24,26] (rs7756992 [CDKAL1], rs7754840 [CDKAL1], rs9939609 [FTO], and rs1801278 [IRS]), by including all 112 candidate SNPs, and by creating GRS-BC and GRS-IR. As expected, these GRS were associated with higher risk of GDM (Table 3). However, the association of per allele increase in GRS with risk of GDM was strongest based on the 11 SNPs identified in our study. The magnitude of association of GRS-BC with the risk of GDM was stronger than that of GRS-IR. Additionally, we examined associations of weighted GRS with risk of GDM by pooling both cohorts rather than meta-analysis, and results were unchanged.
Table 3.
Associations of genetic risk score (GRS) with risk of gestational diabetes (GDM) among 8722 women from Nurses’ Health Study II (NHSII) (total N=7538, GDM cases N=2060) and the Danish National Birth Cohort (DNBC) (total N=1184, GDM cases N=576).
| Quartile 1 | Quartile 2 | Quartile 3 | Quartile 4 | Per allele increase | |
|---|---|---|---|---|---|
| GRS based on significant SNPs | |||||
| GRS based on 11 SNPs with p value <0.05 after FDR correction | |||||
| Weighted GRS | |||||
| NHSII | 1.00 | 1.03 (0.88, 1.19) | 1.21 (1.04, 1.40) | 1.47 (1.27, 1.70) | 1.03 (1.02, 1.04) |
| DNBC | 1.00 | 1.27 (0.92, 1.76) | 1.34 (0.97, 1.86) | 1.84 (1.33, 2.55) | 1.06 (1.03, 1.08) |
| Pooled | 1.00 | 1.07 (0.93, 1.22) | 1.23 (1.07, 1.41) | 1.53 (1.34, 1.74) | 1.04 (1.03, 1.05) |
| Weighted GRS using 10-fold cross validation | 1.00 | 1.08 (0.91, 1.30) | 1.29 (1.10, 1.53) | 1.77 (1.53, 2.03) | 1.05 (1.03, 1.06) |
| Unweighted GRS | |||||
| NHSII | 1.00 | 1.05 (0.91, 1.22) | 1.27 (1.09, 1.47) | 1.53 (1.32, 1.76) | 1.05 (1.03, 1.06) |
| DNBC | 1.00 | 1.16 (0.84, 1.61) | 1.58 (1.14, 2.18) | 1.77 (1.29, 2.44) | 1.08 (1.04, 1.12) |
| Pooled | 1.00 | 1.07 (0.94, 1.23) | 1.32 (1.15, 1.51) | 1.57 (1.37, 1.79) | 1.05 (1.04, 1.07) |
| GRS based on 15 SNPs additionally including rs7756992, rs7754840, rs9939609, and rs1801278 according to previous studies | |||||
| Weighted GRS | |||||
| NHSII | 1.00 | 1.10 (0.95, 1.28) | 1.27 (1.09, 1.47) | 1.56 (1.35, 1.80) | 1.03 (1.02, 1.04) |
| DNBC | 1.00 | 1.09 (0.79, 1.51) | 1.49 (1.08, 2.06) | 1.95 (1.41, 2.71) | 1.05 (1.03, 1.07) |
| Pooled | 1.00 | 1.10 (0.96, 1.26) | 1.30 (1.14, 1.49) | 1.62 (1.42, 1.85) | 1.03 (1.02, 1.04) |
| GRS based on all SNPs | |||||
| GRS based on all 112 SNPs | |||||
| Weighted GRS | |||||
| NHSII | 1.00 | 1.09 (0.94, 1.27) | 1.25 (1.07, 1.44) | 1.64 (1.42, 1.90) | 1.01 (1.01, 1.01) |
| DNBC | 1.00 | 1.36 (0.98, 1.88) | 1.61 (1.16, 2.23) | 2.91 (2.08, 4.06) | 1.02 (1.01, 1.02) |
| Pooled | 1.00 | 1.13 (0.99, 1.30) | 1.30 (1.14, 1.49) | 1.80 (1.57, 2.05) | 1.01 (1.01, 1.01) |
| Weighted GRS using 10-fold cross validation | 1.00 | 1.27 (1.04, 1.55) | 1.50 (1.25, 1.83) | 2.11 (1.80, 2.54) | 1.01 (1.01, 1.02) |
| Unweighted GRS | |||||
| NHSII | 1.00 | 0.93 (0.81, 1.08) | 1.08 (0.93, 1.25) | 1.31 (1.14, 1.52) | 1.01 (1.01, 1.02) |
| DNBC | 1.00 | 1.52 (1.09, 2.11) | 1.81 (1.31, 2.51) | 2.35 (1.69, 3.26) | 1.04 (1.03, 1.05) |
| Pooled | 1.00 | 1.01 (0.89, 1.16) | 1.17 (1.03, 1.34) | 1.44 (1.26, 1.64) | 1.02 (1.01, 1.02) |
| GRS based on 66 SNPs related to beta cell function (GRS-BC) | |||||
| Weighted GRS | |||||
| NHSII | 1.00 | 1.07 (0.93, 1.25) | 1.06 (0.91, 1.23) | 1.35 (1.17, 1.57) | 1.01 (1.00, 1.01) |
| DNBC | 1.00 | 1.60 (1.14, 2.23) | 1.94 (1.39, 2.71) | 2.78 (1.98, 3.89) | 1.03 (1.02, 1.04) |
| Pooled | 1.00 | 1.15 (1.00, 1.31) | 1.17 (1.03, 1.34) | 1.52 (1.33, 1.73) | 1.01 (1.01, 1.02) |
| GRS based on 17 SNPs related to insulin resistance (GRS-IR) | |||||
| Weighted GRS | |||||
| NHSII | 1.00 | 1.10 (0.95, 1.27) | 1.06 (0.91, 1.22) | 1.26 (1.09, 1.46) | 1.01 (1.00, 1.01) |
| DNBC | 1.00 | 1.03 (0.74, 1.43) | 1.09 (0.78, 1.51) | 1.33 (0.96, 1.84) | 1.00 (1.00, 1.01) |
| Pooled | 1.00 | 1.08 (0.95, 1.24) | 1.06 (0.93, 1.21) | 1.27 (1.11, 1.45) | 1.01 (1.00, 1.01) |
Logistic regression model adjusted for baseline age was used.
The positive associations between weighted GRS and risk of GDM did not materially change across different strata of stratification variables (i.e., family history of T2D, smoking, BMI and age) although the magnitude of the associations were stronger among participants without family history of T2D (ORs across increasing quartiles of GRS were 1.00, 1.10, 1.33, 1.64 for participants with family history of T2D vs. 1.00, 1.01, 0.98, and 1.25 for participants without family history of T2D) (all p value for interaction were >0.10) (Table 4).
Table 4.
Associations of weighted genetic risk score (GRS) based on the 11 SNPs with risk of gestational diabetes (GDM) stratified by family history of type 2 diabetes, age, smoking status, and BMI at baseline among 8722 women from Nurses’ Health Study II (NHSII) (total N=7538, GDM cases N=2060) and the Danish National Birth Cohort (DNBC) (total N=1184, GDM cases N=576).
| Quartile 1 | Quartile 2 | Quartile 3 | Quartile 4 | Per allele increase | p for interaction | |
|---|---|---|---|---|---|---|
| Family history of T2D | ||||||
| Yes | 1.00 | 1.01 (0.76, 1.34) | 0.98 (0.74, 1.30) | 1.25 (0.95, 1.64) | 1.01 (0.99, 1.03) | |
| No | 1.00 | 1.10 (0.94, 1.29) | 1.33 (1.14, 1.56) | 1.64 (1.41, 1.92) | 1.04 (1.03, 1.06) | 0.15 |
| Age at baseline | ||||||
| <35 years | 1.00 | 1.07 (0.89, 1.28) | 1.17 (0.98, 1.39) | 1.46 (1.23, 1.74) | 1.03 (1.02, 1.04) | |
| ≥35 years | 1.00 | 1.06 (0.86, 1.31) | 1.32 (1.07, 1.62) | 1.62 (1.32, 1.98) | 1.04 (1.03, 1.06) | 0.93 |
| Smoking | ||||||
| No | 1.00 | 1.03 (0.87, 1.22) | 1.29 (1.09, 1.52) | 1.54 (1.31, 1.81) | 1.04 (1.03, 1.06) | |
| Yes | 1.00 | 1.12 (0.89, 1.41) | 1.11 (0.88, 1.40) | 1.48 (1.18, 1.87) | 1.02 (1.00, 1.04) | 0.56 |
| BMI | ||||||
| BMI ≥25 kg/m2 | 1.00 | 0.95 (0.76, 1.21) | 1.20 (0.95, 1.51) | 1.56 (1.24, 1.97) | 1.03 (1.02, 1.05) | |
| BMI <25 kg/m2 | 1.00 | 1.09 (0.92, 1.30) | 1.26 (1.06, 1.49) | 1.49 (1.26, 1.76) | 1.04 (1.02, 1.05) | 0.81 |
DISCUSSION
In this study with 8,722 participants including 2,636 GDM cases, we identified 11 SNPs that were significantly associated with the risk of GDM after FDR correction, of which 8 SNPs were identified for the first time. The GRS based on the 11 SNPs was significantly associated with risk of GDM, and the positive associations remained significant within most of the subgroups stratified by family history of T2D, smoking, BMI, and age at baseline. Additionally, we found that most of the 11 identified SNPs were related to beta-cell function, and the association of GRS-BC with risk of GDM was stronger than that of GRS-IR, indicating that T2D SNPs related to insulin biosynthesis and secretion play an important role in the development of GDM.
Among the previously identified T2D SNPs, TCF7L2 has been the strongest genetic predictor of T2D to date [45]. For the first time, we identified two other TCF7L2-related SNPs (rs34872471 and rs7903146) whose T2D-associated risk alleles were also associated with a higher risk of GDM. Rs34872471 and rs7903146 are located in the intron region of TCF7L2, and are in linkage disequilibrium with rs4506565, whose risk allele increased the risk of GDM by 44–49% in previous studies [23,46] and was confirmed to be associated with a higher risk of GDM in our study. These three SNPs might impair the expression of glucagon-like peptide 1 (GLP-1) in enteroendocrine cells by interfering with β-catenin-mediated transcriptional activation of its glucagon gene (GCG) [47]. This could in turn result in a defective or poorly expressed GCG protein and lead to decreased insulin secretion and consequently hyperglycemia [47].
For the first time, the current study identified several T2D-associated risk alleles were associated with a higher risk of GDM, namely rs7957197 (HNF1A), rs10814916 (GLIS3), rs3802177 (SLC30A8), and rs7041847 (GLIS3). These SNPs were not previously identified significantly related to GDM. There is distinct biological plausibility for the associations of these SNPs with GDM. Rs7957197 is located in the intronic region of HNF1A, which encodes a transcription factor required for the expression of GLUT1 and GLUT2 transporter in pancreatic beta cells [48]. Furthermore, defects in HNF1A are a cause of maturity onset diabetes of the young type 3 (MODY3) [49]. Rs3802177 is located in the intronic region of SLC30A8, which encodes a zinc transporter expressed solely in the secretory vesicles of beta cells and involved in the final stages of insulin biosynthesis and secretion [45]. Previous studies have shown that reduced zinc transport activity increases T2D risk [50], while overexpression of SLC30A8 in pancreatic cells increases glucose-stimulated insulin secretion [51]. Rs10814916 is located in the intronic region of GLIS3, a member of the GLI-similar zinc finger protein family and encodes a nuclear protein with five C2H2-type zinc finger domains. This protein is highly expressed in pancreatic β cells, and variants in this gene have been associated with neonatal diabetes [52]. Of note, we also observed that the T2D-associated risk alleles of rs9379084 (RREB1) and rs11787792 (GPSM1) were associated with lower risk of GDM. The reason for the inverse association between the two SNPs and GDM risk needs further investigation.
Our findings that SNPs rs10830963 and rs1387153 were significantly associated with GDM risk is consistent with several other studies on GDM [24,26,53]. A meta-analysis involving 8,204 GDM cases and 15,221 controls demonstrated that out of six T2D risk variants, rs10830963 near MTNR1B was most strongly associated with GDM risk [24]. In addition, rs10830963 was one of the two GDM-associated variants identified in a prior GWAS conducted among Korean women [26]. Furthermore, rs10830963 and rs1387153 were associated with higher levels of glucose during pregnancy in Greek and Chinese women [54,55]. MTNR1A and MTNR1B are receptors of melatonin, which is best known as a regulator of seasonal and circadian rhythms [56]. Rs10830963 is located within the intron region of MTNR1, and carriers of the risk allele of rs10830963 exhibit increased expression of MTNR1B in pancreatic beta cells, which leads to impaired insulin secretion [56].
Our study has several unique strengths. First, our study is, thus far, the largest study of genetic variants of GDM which allowed greater statistical power to detect potential associations after correcting for false discoveries. Second, for the identified SNPs, we observed consistent association of weighted GRS with risk of GDM in two independent cohorts of White women, the NHSII in the US and the DNBC in Denmark, further demonstrating the validity of the identified SNPs and the GRS. In addition, we were able to examine whether the SNPs-GDM associations were modified by other major risk factors of GDM.
Our study also has several potential limitations. First, we included only candidate SNPs that were known to be associated with risk of T2D, which limits our capacity of discovering novel variants of GDM beyond these candidate SNPs of T2D. However, our current effort represented the initial step of our endeavor in investigating genetic variants of GDM, and further demonstrated at least partially shared etiology of GDM with T2D. Second, given that only candidate SNPs were genotyped in the DNBC and the majority of GDM cases in the NHSII, our study did not adjust for population stratification resulting from a systematic difference in allele frequencies between populations. However, all of the participants included in the DNBC and the NHSII were self-reported Whites, and the population in Denmark has been shown to have high population homogeneity [57]. Third, potential misclassification of GDM cases might exist in both cohorts, attenuating associations between SNPs and risk of GDM and limiting our study power. However, the validation study conducted in the NHSII showed that the majority of NHSII participants were screened for GDM during pregnancy and most of the self-reported GDM events could be confirmed by medical records [10,36]. Fourth, given that there is not sufficient a priori data on genetic studies of GDM for deriving weights of GRS, we used our own data to calculate GRS. However, we created a weighted GRS using a cross-weight method, and the associations between cross-weighted GRS and risk of GDM were significant and consistent in both cohorts. Finally, the generalizability of our findings to other populations with differing genetic and other characteristics needs further investigation. However, the homogeneity of our population minimizes bias related to population stratification.
In summary, among two independent populations of women, we identified 8 novel SNPs for GDM, and confirmed 3 previously known GDM SNPs. In addition, the GRS based on identified SNPs was significantly and positively associated with GDM risk. These findings potentially provide novel information to improve our understanding of GDM etiology, particularly biological mechanisms related to GDM and insulin biosynthesis and secretion.
Supplementary Material
Research in context.
What is already known about this subject? (maximum of 3 bullet points)
Gestational diabetes mellitus (GDM) has a genetic preposition.
Only several variants associated with GDM have been identified.
The identified genetic variants explained limited heritability of GDM.
What is the key question? (one bullet point only)
Given that GDM has shared genetic basis with type 2 diabetes (T2D), can we identify novel genetic variants associated with risk of GDM using a T2D-associated candidate gene approach?
What are the new findings? (maximum of 3 bullet points)
Our studyidentified eight variants significantly associated with GDM among 2,636 GDM cases and 6,086 non GDM controls from the Nurses’ Health Study II (NHSII) and the Danish National Birth Cohort (DNBC).
Additionally, we confirmed three variants that were previously significantly associated with GDM risk.
Higher genetic risk score based on the 11 variants was significantly associated with higher risk of GDM.
How might this impact on clinical practice in the foreseeable future? (one bullet point only)
Our research offers potential to improve our understanding on GDM etiology, particularly biological mechanisms related to GDM and insulin biosynthesis and secretion.
Funding
This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development at the National Institutes of Health [contract numbers HHSN275201000020C, HHSN275201500003C, HHSN275201300026I, HSN275201100002I]. The Nurses’ Health Study II cohort is supported by the National Institutes of Health [grant number R01 CA67262, UM1 CA176726, R01 CA50385, and NICHD contract HHSN275201000020C]. Financial support for the Danish component was received from: March of Dimes Birth Defects Foundation [6-FY-96–0240, 6-FY97–0553, 6-FY97–0521, 6-FY00–407], Innovation Fund Denmark [grant number 09–067124 and 11–115923, ‘Centre for Fetal Programming’], the Health Foundation [11/263–96], the Heart Foundation [96–2-4–83-22450] and EU (FP7–289346-EarlyNutrition).”
Abbreviations:
- GDM
Gestational diabetes mellitus
- GRS
genetic risk score
- NHSII
Nurses’ Health Study 2
- DNBC
Danish National Birth Cohort
Footnotes
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Data availability
We have enforced a restriction on data used in our analysis to protect the identity of our participants involved in the analysis. Please contact with Dr. Cuilin Zhang (zhangcu@mail.nih.gov) to access the data.
Reference
- 1.(2004) Gestational diabetes mellitus. Diabetes Care 27 Suppl 1: S88–90. [DOI] [PubMed] [Google Scholar]
- 2.Zhu Y, Zhang C (2016) Prevalence of Gestational Diabetes and Risk of Progression to Type 2 Diabetes: a Global Perspective. Curr Diab Rep 16: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng M, Fleming T, Robinson M, Thomson B, Graetz N, et al. (2014) Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 384: 766–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryson CL, Ioannou GN, Rulyak SJ, Critchlow C (2003) Association between gestational diabetes and pregnancy-induced hypertension. Am J Epidemiol 158: 1148–1153. [DOI] [PubMed] [Google Scholar]
- 5.Bellamy L, Casas JP, Hingorani AD, Williams D (2009) Type 2 diabetes mellitus after gestational diabetes: a systematic review and meta-analysis. Lancet 373: 1773–1779. [DOI] [PubMed] [Google Scholar]
- 6.Li S, Zhu Y, Yeung E, Chavarro JE, Yuan C, et al. (2017) Offspring risk of obesity in childhood, adolescence and adulthood in relation to gestational diabetes mellitus: a sex-specific association. Int J Epidemiol 46: 1533–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Y, Olsen SF, Mendola P, Yeung EH, Vaag A, et al. (2016) Growth and obesity through the first 7 y of life in association with levels of maternal glycemia during pregnancy: a prospective cohort study. Am J Clin Nutr 103: 794–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hillier TA, Pedula KL, Schmidt MM, Mullen JA, Charles MA, et al. (2007) Childhood obesity and metabolic imprinting: the ongoing effects of maternal hyperglycemia. Diabetes Care 30: 2287–2292. [DOI] [PubMed] [Google Scholar]
- 9.Clausen TD, Mathiesen ER, Hansen T, Pedersen O, Jensen DM, et al. (2008) High prevalence of type 2 diabetes and pre-diabetes in adult offspring of women with gestational diabetes mellitus or type 1 diabetes: the role of intrauterine hyperglycemia. Diabetes Care 31: 340–346. [DOI] [PubMed] [Google Scholar]
- 10.Solomon CG, Willett WC, Carey VJ, Rich-Edwards J, Hunter DJ, et al. (1997) A prospective study of pregravid determinants of gestational diabetes mellitus. JAMA 278: 1078–1083. [PubMed] [Google Scholar]
- 11.Tobias DK, Zhang C, Chavarro J, Bowers K, Rich-Edwards J, et al. (2012) Prepregnancy adherence to dietary patterns and lower risk of gestational diabetes mellitus. Am J Clin Nutr 96: 289–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C, Solomon CG, Manson JE, Hu FB (2006) A prospective study of pregravid physical activity and sedentary behaviors in relation to the risk for gestational diabetes mellitus. Arch Intern Med 166: 543–548. [DOI] [PubMed] [Google Scholar]
- 13.Zhang C, Tobias DK, Chavarro JE, Bao W, Wang D, et al. (2014) Adherence to healthy lifestyle and risk of gestational diabetes mellitus: prospective cohort study. BMJ 349: g5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C, Rawal S, Chong YS (2016) Risk factors for gestational diabetes: is prevention possible? Diabetologia 59: 1385–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C, Ning Y (2011) Effect of dietary and lifestyle factors on the risk of gestational diabetes: review of epidemiologic evidence. Am J Clin Nutr 94: 1975S–1979S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin AO, Simpson JL, Ober C, Freinkel N (1985) Frequency of diabetes mellitus in mothers of probands with gestational diabetes: possible maternal influence on the predisposition to gestational diabetes. Am J Obstet Gynecol 151: 471–475. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe RM (2011) Inherited destiny? Genetics and gestational diabetes mellitus. Genome Med 3: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarnowski M, Malinowski D, Pawlak K, Dziedziejko V, Safranow K, et al. (2017) GCK, GCKR, FADS1, DGKB/TMEM195 and CDKAL1 Gene Polymorphisms in Women with Gestational Diabetes. Can J Diabetes 41: 372–379. [DOI] [PubMed] [Google Scholar]
- 19.Buchanan TA, Xiang AH (2005) Gestational diabetes mellitus. J Clin Invest 115: 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poulsen P, Levin K, Petersen I, Christensen K, Beck-Nielsen H, et al. (2005) Heritability of insulin secretion, peripheral and hepatic insulin action, and intracellular glucose partitioning in young and old Danish twins. Diabetes 54: 275–283. [DOI] [PubMed] [Google Scholar]
- 21.Zhang C, Bao W, Rong Y, Yang H, Bowers K, et al. (2013) Genetic variants and the risk of gestational diabetes mellitus: a systematic review. Hum Reprod Update 19: 376–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaat N, Ekelund M, Lernmark A, Ivarsson S, Almgren P, et al. (2005) Association of the E23K polymorphism in the KCNJ11 gene with gestational diabetes mellitus. Diabetologia 48: 2544–2551. [DOI] [PubMed] [Google Scholar]
- 23.Shaat N, Lernmark A, Karlsson E, Ivarsson S, Parikh H, et al. (2007) A variant in the transcription factor 7-like 2 (TCF7L2) gene is associated with an increased risk of gestational diabetes mellitus. Diabetologia 50: 972–979. [DOI] [PubMed] [Google Scholar]
- 24.Wu L, Cui L, Tam WH, Ma RC, Wang CC (2016) Genetic variants associated with gestational diabetes mellitus: a meta-analysis and subgroup analysis. Sci Rep 6: 30539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Q, Zhang K, Li W, Liu JT, Hong J, et al. (2009) Association of KCNQ1 gene polymorphism with gestational diabetes mellitus in a Chinese population. Diabetologia 52: 2466–2468. [DOI] [PubMed] [Google Scholar]
- 26.Kwak SH, Kim SH, Cho YM, Go MJ, Cho YS, et al. (2012) A genome-wide association study of gestational diabetes mellitus in Korean women. Diabetes 61: 531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, et al. (2014) Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet 46: 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, et al. (2012) Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 44: 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, et al. (2010) Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 42: 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manning AK, Hivert MF, Scott RA, Grimsby JL, Bouatia-Naji N, et al. (2012) A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet 44: 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, et al. (2012) Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet 44: 991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, et al. (2010) New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 42: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C, Hu FB, Olsen SF, Vaag A, Gore-Langton R, et al. (2014) Rationale, design, and method of the Diabetes & Women’s Health study--a study of long-term health implications of glucose intolerance in pregnancy and their determinants. Acta Obstet Gynecol Scand 93: 1123–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindstrom S, Loomis S, Turman C, Huang H, Huang J, et al. (2017) A comprehensive survey of genetic variation in 20,691 subjects from four large cohorts. PLoS One 12: e0173997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pardo LM, Li WQ, Hwang SJ, Verkouteren JA, Hofman A, et al. (2017) Genome-Wide Association Studies of Multiple Keratinocyte Cancers. PLoS One 12: e0169873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solomon CG, Willett WC, Rich-Edwards J, Hunter DJ, Stampfer MJ, et al. (1996) Variability in diagnostic evaluation and criteria for gestational diabetes. Diabetes Care 19: 12–16. [DOI] [PubMed] [Google Scholar]
- 37.Olsen SF, Houshmand-Oeregaard A, Granstrom C, Langhoff-Roos J, Damm P, et al. (2017) Diagnosing gestational diabetes mellitus in the Danish National Birth Cohort. Acta Obstet Gynecol Scand 96: 563–569. [DOI] [PubMed] [Google Scholar]
- 38.(2014) Diagnostic criteria and classification of hyperglycaemia first detected in pregnancy: a World Health Organization Guideline. Diabetes Res Clin Pract 103: 341–363. [DOI] [PubMed] [Google Scholar]
- 39.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, et al. (2007) A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet 39: 870–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borenstein M, Hedges LV, Higgins JP, Rothstein HR (2010) A basic introduction to fixed-effect and random-effects models for meta-analysis. Res Synth Methods 1: 97–111. [DOI] [PubMed] [Google Scholar]
- 41.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I (2001) Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125: 279–284. [DOI] [PubMed] [Google Scholar]
- 42.Gan W, Walters RG, Holmes MV, Bragg F, Millwood IY, et al. (2016) Evaluation of type 2 diabetes genetic risk variants in Chinese adults: findings from 93,000 individuals from the China Kadoorie Biobank. Diabetologia 59: 1446–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qi Q, Stilp AM, Sofer T, Moon JY, Hidalgo B, et al. (2017) Genetics of Type 2 Diabetes in U.S. Hispanic/Latino Individuals: Results From the Hispanic Community Health Study/Study of Latinos (HCHS/SOL). Diabetes 66: 1419–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Machiela MJ, Chen CY, Chen C, Chanock SJ, Hunter DJ, et al. (2011) Evaluation of polygenic risk scores for predicting breast and prostate cancer risk. Genet Epidemiol 35: 506–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, et al. (2007) A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445: 881–885. [DOI] [PubMed] [Google Scholar]
- 46.Lauenborg J, Grarup N, Damm P, Borch-Johnsen K, Jorgensen T, et al. (2009) Common type 2 diabetes risk gene variants associate with gestational diabetes. J Clin Endocrinol Metab 94: 145–150. [DOI] [PubMed] [Google Scholar]
- 47.Florez JC, Jablonski KA, Bayley N, Pollin TI, de Bakker PI, et al. (2006) TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med 355: 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luni C, Marth JD, Doyle FJ 3rd (2012) Computational modeling of glucose transport in pancreatic beta-cells identifies metabolic thresholds and therapeutic targets in diabetes. PLoS One 7: e53130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ellard S (2000) Hepatocyte nuclear factor 1 alpha (HNF-1 alpha) mutations in maturity-onset diabetes of the young. Hum Mutat 16: 377–385. [DOI] [PubMed] [Google Scholar]
- 50.Nicolson TJ, Bellomo EA, Wijesekara N, Loder MK, Baldwin JM, et al. (2009) Insulin storage and glucose homeostasis in mice null for the granule zinc transporter ZnT8 and studies of the type 2 diabetes-associated variants. Diabetes 58: 2070–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chimienti F, Devergnas S, Pattou F, Schuit F, Garcia-Cuenca R, et al. (2006) In vivo expression and functional characterization of the zinc transporter ZnT8 in glucose-induced insulin secretion. J Cell Sci 119: 4199–4206. [DOI] [PubMed] [Google Scholar]
- 52.Yang Y, Chang BH, Samson SL, Li MV, Chan L (2009) The Kruppel-like zinc finger protein Glis3 directly and indirectly activates insulin gene transcription. Nucleic Acids Res 37: 2529–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huopio H, Cederberg H, Vangipurapu J, Hakkarainen H, Paakkonen M, et al. (2013) Association of risk variants for type 2 diabetes and hyperglycemia with gestational diabetes. Eur J Endocrinol 169: 291–297. [DOI] [PubMed] [Google Scholar]
- 54.Vlassi M, Gazouli M, Paltoglou G, Christopoulos P, Florentin L, et al. (2012) The rs10830963 variant of melatonin receptor MTNR1B is associated with increased risk for gestational diabetes mellitus in a Greek population. Hormones (Athens) 11: 70–76. [DOI] [PubMed] [Google Scholar]
- 55.Liao S, Liu Y, Tan Y, Gan L, Mei J, et al. (2012) Association of genetic variants of melatonin receptor 1B with gestational plasma glucose level and risk of glucose intolerance in pregnant Chinese women. PLoS One 7: e40113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mulder H, Nagorny CL, Lyssenko V, Groop L (2009) Melatonin receptors in pancreatic islets: good morning to a novel type 2 diabetes gene. Diabetologia 52: 1240–1249. [DOI] [PubMed] [Google Scholar]
- 57.Athanasiadis G, Cheng JY, Vilhjalmsson BJ, Jorgensen FG, Als TD, et al. (2016) Nationwide Genomic Study in Denmark Reveals Remarkable Population Homogeneity. Genetics 204: 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.