Abstract
Purpose
Single-cell RNA-sequencing (scRNA-seq) was used to interrogate the relatively rare stem (SC) and early transit amplifying (TA) cell populations in limbal/corneal epithelia from wild-type and autophagy-compromised mice.
Methods
We conducted scRNA-seq on ocular anterior segmental tissue from wild-type and beclin 1–deficient (beclin1+/−) mice, using a 10X Gemomics pipeline. Cell populations were distinguished by t-distributed stochastic neighbor embedding. Seurat analysis was conducted to compare gene expression profiles between these two groups of mice. Differential protein expression patterns were validated by immunofluorescence staining and immunoblotting.
Results
Unbiased clustering detected 10 distinct populations: three clusters of mesenchymal and seven clusters of epithelial cells, based on their unique molecular signatures. A discrete group of mesenchymal cells expressed genes associated with corneal stromal SCs. We identified three limbal/corneal epithelial cell subpopulations designated as stem/early TA, mature TA, and differentiated corneal epithelial cells. Thioredoxin-interacting protein and PDZ-binding kinase (PBK) were identified as novel regulators of stem/early TA cell quiescence. PBK arrested corneal epithelial cells in G2/M phase of the cell cycle. Beclin1+/− mice displayed a decrease in proliferation-associated (Ki67, Lrig1) and stress-response (H2ax) genes. The most increased gene in beclin1+/− mice was transcription factor ATF3, which negatively regulates limbal epithelial cell proliferation.
Conclusions
Establishment of a comprehensive atlas of genes expressed by stromal and epithelial cells from limbus and cornea forms the foundation for unraveling regulatory networks among these distinct tissues. Similarly, scRNA-seq profiling of the anterior segmental epithelia from wild-type and autophagy-deficient mice provides new insights into how autophagy influences proliferation in these tissues.
Keywords: autophagy, cell proliferation, stem cell, transit amplifying cell
The limbal/corneal epithelia with its attending stroma is one of the most well-accepted models for studying epithelial stem cells and their progeny the transit amplifying (TA) cells. In this system, the epithelial stem cells for the corneal epithelium are preferentially located in the basal layer of the limbal epithelium.1,2 The immediate progeny of a stem cell division, the early (or young) TA cells are also located in the limbal basal layer.1,3 Many consider these early TA cells phenotypically and functionally indistinguishable from stem cells.4 Early TA cells migrate centripetally and populate the peripheral corneal epithelial basal layer.5–9 The late or more “mature” TA cells are believed to reside in the central corneal epithelial basal layer.3,10,11 It is also held that a hierarchy in proliferative capacity exists within the limbal/corneal epithelia. The stem and early TA cells have great proliferative capacity, whereas mature TA cells are more limited in their capacity to divide.1,3,11 Stem cells infrequently divide (rare cycling), whereas TA cells often divide and can be detected by pulse nucleoside incorporation (e.g., BrdU, EdU, 3H-thimidine).3 It is also believed that under resting or unperturbed conditions, young TA cells do not exercise their full proliferative potential before becoming post-mitotic differentiating cells.3 However, in situations in which the cell population needs to be expanded, such as during development or following wounding, TA cells use their full complement of divisions to restore homeostasis.3 This paradigm of corneal epithelial stem/TA cell/post-mitotic cell is more than 30 years old and many of the features of these three populations have been defined (for reviews see Refs. 12–17). In addition, the stromal microenvironment (niche) under the limbal and corneal epithelium is also becoming relatively well characterized (for reviews see Refs. 18–20). However, due to the heterogeneity of the limbal and peripheral corneal epithelial basal layer, it has been extremely difficult to isolate and characterize the stem/early TA cell population.21 Using laser capture microdissection, we isolated relatively pure populations of mouse limbal epithelial and corneal epithelial basal cells and used these populations for gene and microRNA expression profiling.22,23 Differences in gene and microRNA expression were documented between these two populations; however, this technique did not allow for the characterization of stem versus early TA versus mature TA cells.
Compared with the traditional profiling of bulk populations mentioned above, single-cell RNA-sequencing (scRNA-seq) methodologies enable the interrogation of relatively rare cell populations (e.g., stem and early TA cells). In scRNA-seq, every cell is barcoded and then grouped into different clusters or populations based on the relative similarity of gene expression.24 For example, using known genes, these groups can be defined as to epithelial versus mesenchymal. Epithelial clusters can be further subdivided as to undifferentiated versus differentiated or corneal versus conjunctival. Finally, corneal epithelial clusters can be further subclassified into early versus mature TA cells. Once classified and verified, the expression profiles of the discrete populations can be examined to uncover regulatory relationships. We have followed this paradigm to uncover new ways in which the stem/early TA cells maintain relative quiescence.
To complement this scRNA-seq approach in defining how stem/early TA cells are regulated, we conducted scRNA-seq on the cornea and limbus from beclin-1–deficient (beclin 1+/−) heterozygous mice, which have compromised autophagy.25 We have shown that autophagy is a positive regulator of proliferation in limbal/corneal epithelia.26 For example, inhibiting autophagy via treatment of primary human limbal epithelial cells with chloroquine, which disrupts the function of lysosomal enzymes,27 dramatically reduced the ability of these cells to form holoclone colonies (a marker of stem cell proliferative capacity22,28). This is consistent with findings in other systems that autophagy is a positive regulator of stem cells.29–32 In addition, we reported that beclin 1+/− mice healed corneal epithelial wounds at a slower rate compared with wild-type controls. We hypothesized that such slower healing was due to the inability of the stem/early TA population to immediately respond to wound stimuli.26 Thus, to gain insight into this problem, we compared the scRNA-seq expression profiles of the limbal/corneal stem/early TA populations of wild-type versus beclin 1+/− mice.
Herein we report that stem/early TA and mature TA cell populations were elucidated and characterized using scRNA-seq. We identified novel proteins that were preferentially localized to the human and mouse limbal basal epithelium that are involved in maintaining proliferative quiescence. Likewise, we successfully identified the stem/early TA cell populations from mice with compromised autophagy and demonstrate that this group had 50% fewer stem/TA cells and a decreased expression of genes associated with proliferation. These findings provide a mechanistic explanation for the retardation in corneal epithelial wound repair in mice deficient in autophagy.
Methods
Single-Cell Isolation of Mouse Cornea
Whole eyes were harvested from a single healthy beclin1 het (beclin1+/−)25,33 female and a healthy C57/BL6 wild-type littermate control (28 day old) female mouse. The lens, retina, iris, and trabecular network were removed leaving the entire ocular anterior segment, from which most of the conjunctiva was carefully dissected and discarded. The corneal rims were subsequently digested at 37°C for 2 hours with 2 mL of 1 mg/mL collagenase A (Sigma-Aldrich Corp., St. Louis, MO, USA) in Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS, 50 μg/mL gentamicin, and 1.25 μg/mL amphotericin B with modification of published protocols.34 After a 2-hour incubation, the corneal rims were rinsed once with PBS without Ca2+-Mg2+. Clusters were further digested with 0.25% trypsin and 1 mM EDTA at 37°C for 20 minutes with intermediate gentle pipetting to yield single cells. After trypsin digestion, cells were rinsed once with DMEM+10% fetal calf serum media to deactivate and remove trypsin. The cells were rinsed twice with sterile PBS containing 0.04% BSA. After final centrifugation, cells were resuspended in 60 μL PBS containing 0.04% BSA to achieve 700 to 1200 cells/μL, and filtered into an Eppendorf tube by using Flowmi Cell Strainer (Fisher Scientific, Lenexa, KS, USA). Cell viability was checked and accepted with at least an 80% viability score. Such cells were processed for scRNA-seq using 10X Genomics pipeline in the Northwestern University NUseq Core.
All mouse studies were approved by the institutional animal care and use committee of Northwestern University, Feinberg School of Medicine, and adhered to the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research.
Data Processing for scRNA-seq Analysis Using Cell Ranger and Loupe
Transcripts were mapped to the mm10 reference genome (GRCm38.91) using Cell Ranger Version 2.1.0. The Web summary of Cell Ranger statistics on cells, sequencing, and mapping metrics pre-quality control assessment for downstream bioinformatics analyses were as follows for wild-type and beclin, respectively: cell metrics: 2513 and 5155 estimated number of cells, 69.2% and 72.0% fraction reads in cells, 127,716 and 60,737 mean reads per cell, 3097 and 2515 median genes per cell, 18,250 and 18,585 total genes detected, 16,967 and 11,571 median UMI counts per cell; sequencing metrics: 320,951,096 and 313,103,399 total number of reads, 97.4% and 97.6% valid barcodes; mapping metrics: 89.8% and 90.2% reads mapped to genome, 86.0% and 86.5% reads mapped confidently to genome, 73.5% and 74.0% reads mapped confidently to transcriptome. The number of cells available for downstream analysis was 2513 and 5155 for the wild-type and beclin samples, respectively. Data normalization by Seurat analysis (see the next section: scRNA-Seq Clustering by Seurat) was used to account for the differences in the cell number between samples during integration and subclustering.
scRNA-Seq Clustering by Seurat
Sample clustering and comparisons were performed with the R Package Seurat,35 version 2.4. The data were first normalized and log-transformed. A list of highly variable genes was generated by calculating the average expression of each gene, placing the genes into bins, and then calculating the z-score for dispersion within each bin. Genes with an average expression between 0.0125 and 3, and with a dispersion >0.5, were selected for inclusion in the principal component analysis (PCA) calculations. The Seurat PCElbowPlot() function was used to identify the number of significant PCs (18 for wild-type, 10 for beclin) to use for clustering. Seurat's FindClusters() function was used to generate clusters for the data, with a resolution of 1. The FindClusters() function uses a procedure of embedding cells in a graph structure and then applying the Louvain algorithm (the default, used in our case) to iteratively group cells together. A classification hierarchy was constructed and clusters were merged from a random forest classifier with an Out-of-bag error (OOBE) higher than 0.05. We performed t-distributed stochastic neighbor embedding (t-SNE) in Seurat with default settings. Each of the two samples yielded eight clusters.
Integrating Wild-Type and Beclin Samples, and Subclustering
To compare two separate sets of samples, the data were integrated and analyzed using Seurat software. The samples from wild-type and Beclin-1+/− mice were “integrated” via canonical correlation analysis (CCA) for four goals: (1) identifying cell types present in both datasets; (2) obtaining cell-type markers that are conserved in both control and experimental cells; (3) comparing the datasets to find cell-type specific responses to treatment; and (4) controlling for the batch effects of having the disparity in the numbers of cells/number of reads per cell.
Starting with the Seurat objects generated above, the union of the top 2000 genes with the highest dispersion (var/mean) from both datasets was processed. A CCA was performed to combine the two datasets into a single object and store the canonical correlation (CC) vectors. To choose CCs for downstream analysis, Seurat's MetageneBicorPlot() function was used with inclusion of the top 25 CCs. The CCA subspaces were aligned (using Seurat's AlignSubspace() function) to prepare the data for clustering. The unsupervised clustering procedure described above was repeated by merging clusters with an OOBE higher than 0.05, and the clusters were visualized with a t-SNE plot.
The Seurat FindConservedMarkers() function was used to obtain a list of marker genes conserved across conditions, and to identify cluster four as being probable epithelial cells. The epithelial cluster designated was isolated and further processed for subclustering as described above (i.e., generating a list of highly variable genes, selecting genes with average expression between 0.0125 and 3 and with a dispersion >0.5 for inclusion in the PCA calculations, using the PCElbowPlot() function to select an appropriate number of significant PCs, choosing the 36 most significant principal components (PCs), and using them in FindClusters() to generate subclusters). The integrated epithelial cluster samples yielded nine subclusters.
Immunohistochemistry
Three separate human corneas, ages 27, 42, and 62, were used with postmortem delay of no more than 72 hours (Eversight Eye Bank, Chicago, IL, USA). Frozen sections (5 μm) of optimal cutting temperature compound-embedded human and mouse corneas were fixed in 4% paraformaldehyde, blocked in 10% goat serum in PBS and incubated overnight with the following primary antibodies: rabbit polyclonal antibodies against Kl67 (SP6- 1:500 dil; Sigma-Aldrich Corp., St. Louis, MO, USA), PBK (16110–1-AP-1:100dil; Proteintech, Rosemont, IL, USA), TXNIP (40–3700- 1:100 dil; Thermo Scientific, Waltham, MA, USA), H2AX (10856–1-AP- 1:50 dil; Proteintech), ATF3 (HPA001562–1:50 dil; Sigma-Aldrich Corp.) or a mouse monoclonal antibody against K15 (MA5–11244–1:100 dil; Thermo Scientific). Appropriate secondary antibodies conjugated to Alexa-555 or Alexa-488 (Invitrogen, Carlsbad, CA, USA) at 1:300 dilution were used to detect primary antibodies. DAPI (4′,6-diamidino-2-phenylindole) was used to label nuclei, and mounted using Gelvatol. Images were acquired using a 20X 0.5 EC Plan-Neofluar objective on an epiflourescence microscope system (AxioVision Z1; Carl Zeiss, Thornwood, NY, USA) fitted with an Apotome slide module and a digital camera (AxioCam MRm).
Cell Culture
hTCEpi cells were cultured in keratinocyte–serum-free media (K-SFM) with supplements (Invitrogen). Primary HCEKs and HLEKs were isolated from cadaver donor corneas provided by Midwest Eye Bank (Ann Arbor, MI, USA) and cultured in CnT-20 media with supplements (CellnTech, Bern, Switzerland) on collagen IV-coated plates (Corning, Corning, NY, USA). To silence gene expression, small interfering RNAs (siRNAs) (20 nM) targeting ATF3 (Catalog #AM16708; Invitrogen), PBK (Catalog #4390824; siPBK#1: Assay ID s31708, siPBK#2: Assay ID s31707, siPBK#3: Assay ID s31706; Invitrogen) or a scrambled negative control (Catalog #465372; Invitrogen) were transiently transfected into cells using RNAiMAX (Invitrogen). Although we have tested three siRNA sequences for PBK1, only two of them (#1 and #3) reduced the expression efficiently.
Cell Proliferation and Cell Cycle Analysis
To examine cell proliferation, cells were subjected to WST-1 assays.22 To analyze the phases of the cell cycle, cells were trypsinized, harvested, and fixed in 1 mL 80% cold ethanol in test tubes and stained with propidium iodine (50 μg/mL) containing 0.2 mg/mL RNase A (Sigma; St. Louis, MO). Cell cycle distribution was calculated from 30,000 cells using a FACSCalibur Flow Cytometer (BD Biosciences, San Jose, CA, USA).
Immunoblotting
Western blots were performed as described previously.36 The following antibodies were used: PBK (Catalog #16110–1-AP-1; 1:1000 dil; Proteintech), ATF3 (Catalog #33593 - 1:1000 dil; Cell Signaling Technologies, Danvers, MA, USA), GAPDH (sc-32233–1:1000 dil; Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Real-Time Quantitative PCR Analysis
Whole eyes were harvested from beclin1+/− and C57/BL6 wild-type littermate control (28 day old) mice (n = 4). Corneal epithelial sheets were isolated as described previously.37 Total RNA from epithelial sheets was purified using a miRNeasy kit (Qiagen, Valencia, CA, USA), and cDNA was prepared using a Superscript III reverse transcription kit (Invitrogen). Real-time qPCR was performed on a Lightcycler 96 real-time PCR system (Roche, Indianapolis, IN, USA) using a quantitative SYBR green PCR kit (Roche). Mouse Aft3 primers were as follows: FWD 5′-GGC AGG AAG AGC CAA AGA TAA; REV 5′-GTG CCA TTA ACA TCC CAC AAT G. Mouse 18S RNA was used as the internal control. Values are fold change over wild-type littermate controls.
Statistical Analysis
In column plots, all values are expressed as mean ± SD. The significance of the differences between two groups was evaluated by an unpaired Student's t-test. For violin plots, the Wilcoxon rank sum test was conducted. Parameters with values P < 0.05 were considered significant.
Results and Discussion
scRNA-seq From the Limbus and Cornea of Wild-Type and Beclin1+/− Mice
The limbus and cornea with underlying stroma was dissociated with collagenase, partitioned into single cells, and processed for scRNA-seq using the 10X Genomics platform. In total, we sequenced 2513 cells from the wild-type limbus and cornea and 5155 cells from the beclin 1+/− limbus and cornea: To ensure that an adequate number of mRNA transcripts were sequenced, we generated more than 127,000 reads per wild-type cell and 60,000 reads per beclin 1+/− cell. It has been shown that 50,000 reads per cell is sufficient for accurate cell-type classification and biomarker identification.38 The median number of genes profiled per wild-type cell was 3100 vs. 2500 per beclin 1+/− cell.
Presently, there is no established approach to handle biological and/or technical replicates of scRNA-seq data, and an established set of standards regarding replicates in scRNA-seq is being explored. scRNA-seq differential analyses are only confined within the sample and each cell is considered as an independent measurement. However, a minimum of three replicates was used for downstream analysis of the data (i.e., immunostaining, proliferation, cell cycle) to answer specific biological questions and define patterns.
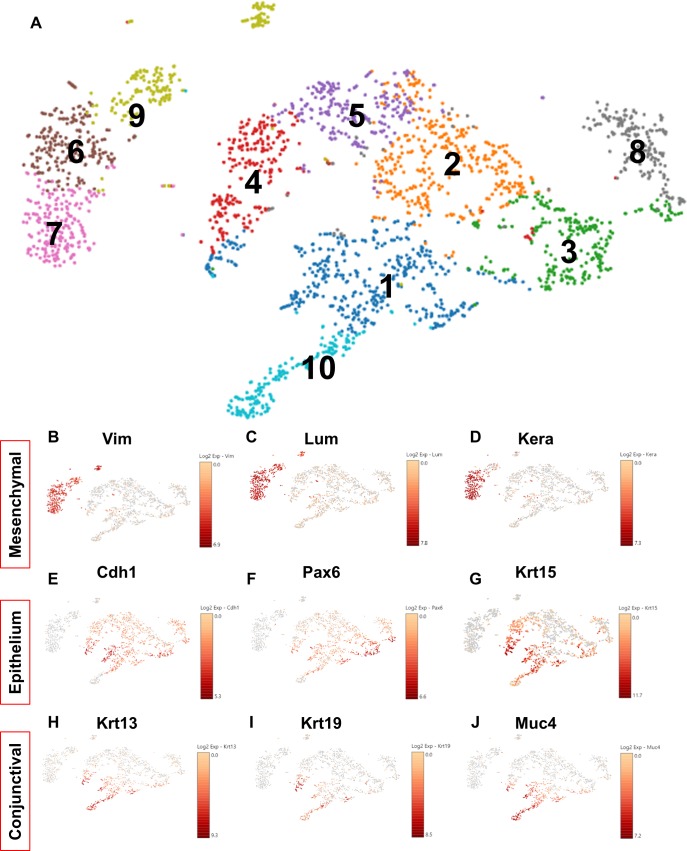
A general approach in analyzing scRNA-seq data is to determine clusters and subclusters, based on prior published and proven markers. This is a calculated approach, and leads to identification of novel genes that are residing within the already determined clusters.39,40 Therefore, to evaluate the heterogeneity among the single cells from the wild-type limbus and cornea, data generated from the scRNA-seq were subjected to unsupervised clustering using the 10X Genomics Loupe analysis program (Fig. 1). The t-SNE analysis revealed 10 distinct clusters and the top genes/cluster were used to manually identify each of the clusters (Fig. 1A). Three clusters expressed high levels of vimentin (Vim), lumican (Lum), and keratocan (Kera) (Figs. 1 B–D), which are genes commonly associated with corneal stromal keratocytes41 and thus we assigned these clusters as mesenchymal cells (Figs. 1A–D). Interestingly, within the mesenchymal cluster was a small subcluster of cells that (1) had low levels of keratocan, (2) expressed Six2 and Scf, and (3) had high levels of Fhl1 and Cd90 (Thy1) (Supplementary Fig. S1). Fhl1 and Thy1 are markers associated with corneal stromal stem cells (CSSCs).42 Furthermore, the absence of keratocan in this cluster suggests a less differentiated cell-type and thus we are postulating that these cells may represent CSSCs.
Figure 1.
scRNA-seq identifies 10 clusters of cells from wild-type mouse limbal/corneal tissues. (A) t-SNE plot of 2513 cells is visualized. Cells are colored by the clusters. (B–J) t-SNEs colored by the normalized log-transformed expression of the mesenchymal markers Vim (B), Lum (C), Kera (D), epithelial markers Cdh1 (E), Pax6 (F), Krt15 (G), conjunctival epithelial markers Krt13 (H), Krt19 (I), and Muc4 (J).
The remaining large cluster of cells expressed high levels of Pax6, adherens, and tight junctional as well as keratin genes (Figs. 1E–G), all markers of epithelial cells.43–45 Within the epithelial cluster, a subcluster expressed high levels of keratin 13 and 19 genes (Figs. 1H, 1I), as well as mucin 4 (Fig. 1J), mucin 1, and mucin 20 (Supplementary Fig. S2), markers for conjunctival epithelial cells.46,47 We suggest that this cluster represents a small amount of bulbar conjunctival tissue immediately next to the limbus.
The Limbal/Corneal Epithelial Cluster From Wild-Type Mice Can Be Classified Into Stem/Early TA, Mature TA, and Relatively Differentiated Cells
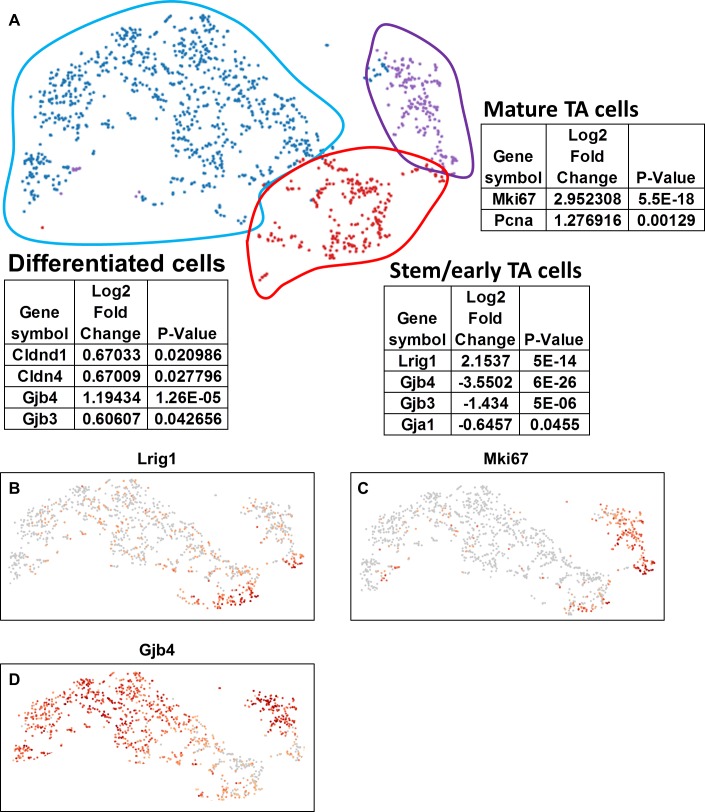
After removal of the cluster representing the conjunctival epithelial cells, unbiased clustering identified clusters of cells that were assigned to three primary cell populations (Fig. 2A) based on the expression of genes that were related to stem cells, proliferation, and differentiation.
Figure 2.
scRNA-seq identifies stem/early TA, mature TA and differentiated corneal epithelial cell populations. (A) t-SNE plot of limbal/corneal epithelial cells are visualized. Cells are colored by the cell types as follows: stem/early TA = red, mature TA = purple, and differentiated corneal epithelial cells = blue. Several top signature genes in each cluster are listed. (B–D) t-SNEs colored by the normalized log-transformed expression of Lrig1 (B), MKi67 (C), and Gjb4 (D). Color represents log2 (expression level).
Stem Cells
Leucine-rich repeats and immunoglobulin-like domains 1 (LRIG1) was initially proposed as a marker of quiescent stem cells in the epidermis.48 In the intestine, lineage tracing demonstrated that LRIG1 marks intestinal epithelial stem cells.49 More recently, LRIG1 was detected in all of the limbal label-retaining cells (LRCs),50 which identify stem cells as rarely (slow) cycling cells.1,3 scRNA-seq data indicated preferential Lrig1 expression in a cluster of cells that we designated stem/early TA cells (Fig. 2B), as well as a small group of cells at the boundary between the stem/early TA cells and the mature TA cell cluster (Fig. 2B).
Proliferation and Differentiation
We use Kl67 expression to aid in defining the proliferative status of the cell populations, as this marker visualizes cells that are cycling.51 Immunohistochemical detection of Kl67, is low in the limbal basal epithelial cells, which infrequently cycle1 (Supplementary Fig. S3). Conversely, Kl67 expression is high in the central corneal epithelial basal cells, which proliferate more frequently3 (Supplementary Fig. S3). Using Ki67 expression as a marker for TA populations, the upper right cluster of cells (outlined in purple; Figs. 2A, 2C) had high expression, the stem/early TA cluster (outlined in red; Figs. 2A, 2C) had relatively low expression and the large cluster on the left (outlined in blue; Figs. 2A, 2C) also demonstrated low expression. Besides having relatively low levels of Ki67, the stem/early TA cell cluster had low levels of connexins (Gjb4; Figs. 2A, 2D), which is a characteristic of stem cell–enriched tissues.22,52–54 The large cluster of cells with the lowest Ki67 expression (outlined in blue; Figs. 2A, 2C), exhibited high expression of genes associated with tight and gap junctions (Figs. 2A, 2D), which are found on more differentiated cells,55–57 hence we believe that this population of cells are primarily post-mitotic differentiated cells (Fig. 2). To interrogate the clusters further, we used gene-annotation enrichment analysis (GO by DAVID58,59). Not surprisingly GO analysis of the stem/early TA cell cluster (Supplementary Fig. S4B) indicated enrichment for cell fate commitment and regulation of pluripotency of stem cells. Conversely, GO analysis of the mature TA cell cluster (Supplementary Fig. S4G) indicated enrichment of cell division and cell cycle.
scRNA-seq Elucidates Novel Regulatory Genes in the Stem/Early TA and Mature TA Cell Clusters
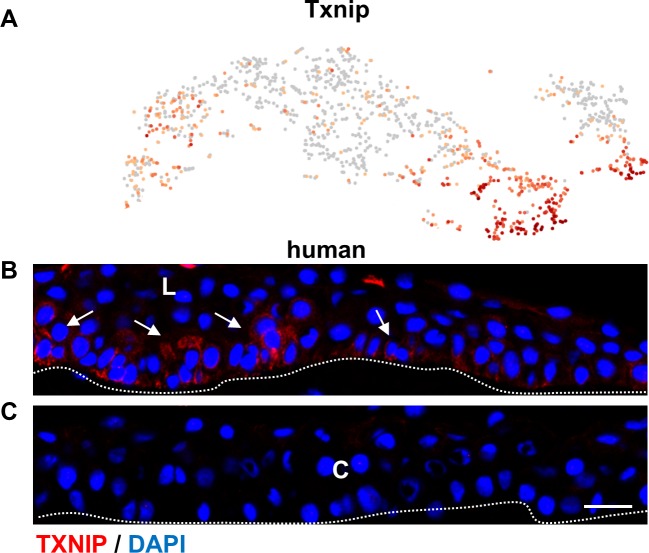
Having identified a discrete population of stem/early TA cells, we investigated highly expressed genes with regulatory properties. One of the highest expressed genes was thioredoxin-interacting protein (Txnip; Fig. 3A, Supplementary Table S1). Txnip is a metabolic gene involved in redox regulation that can also function as a tumor suppressor.60 TXNIP functions to maintain quiescence through G0/G1 cell cycle arrest via p27kip1.61 Immunofluorescence studies revealed that TXNIP was expressed on human limbal epithelial basal cells (arrows) as well as a few suprabasal cells, which are likely to be the immediate progeny of stem/early TA cells due to asymmetric division (Figs. 3B, 3C). This makes excellent biological sense, as quiescence is one of the features of limbal epithelial stem cells22 and Txnip may be a novel limbal epithelial-preferred gene that contributes, in part, to stem cell maintenance. In addition, preferential immunofluorescent localization of TXNIP to the limbal basal cell population (Fig. 3B) further validates the designation of stem/early TA cell to this cluster (Fig. 2A). Although we tested two different TXNIP antibodies under different fixation conditions, the staining pattern with these antibodies in mouse eyes was inconsistent.
Figure 3.
scRNA-seq identifies Txnip as preferentially expressed in stem/early TA cells in the limbal epithelium. (A) t-SNEs colored by the normalized log-transformed expression of Txnip. (B, C) Immunofluorescence staining with TXNIP antibody in a frozen section of human limbal and corneal epithelium. TXNIP was primarily restricted to the limbal epithelial basal layer (B) and virtually absent in the corneal epithelium (C). C, cornea; L, limbus. Arrows mark the positively stained cells. Scale bar: 20 μm. Dotted lines mark the basement membrane. n = 3.
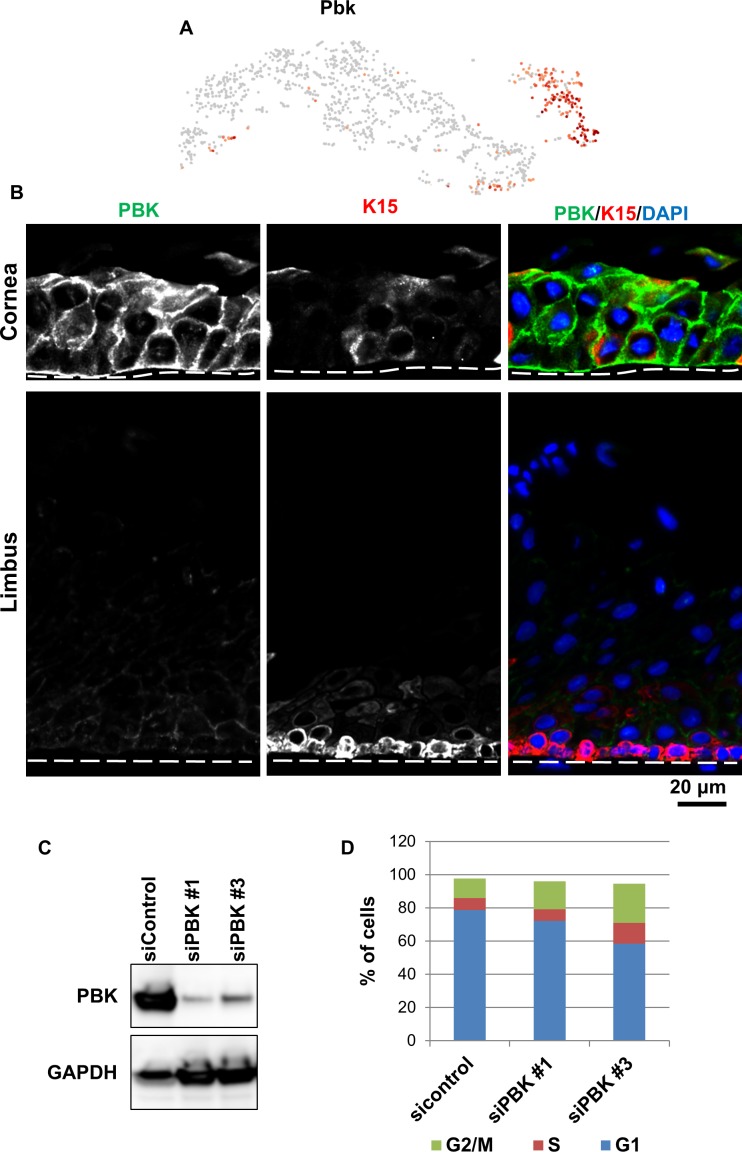
Conversely, PDZ-binding kinase (Pbk) was highly expressed in the mature TA cell cluster (Fig. 4A, Supplementary Table S2) and is one of the lowest expressed genes in the stem/early TA cluster (Fig. 4A). PBK is a mitogen-activated protein kinase (MAPKK)62 that is associated with mitosis, and downregulation leads to quiescence.63 Immunofluorescence experiments indicated high expression of PBK in the corneal epithelium (Fig. 4B). PBK in mouse and human limbal epithelial basal cells was markedly lower than central corneal epithelial cells (Fig. 4B). Low levels of PBK in the limbal epithelial basal cells may help maintain the aforementioned quiescence. PBK activity and expression has been shown to contribute to the release of proliferating cells from mid-mitotic checkpoint control and maintain a timely exit from mitosis.64 Therefore, we investigated how genetic manipulation of PBK in a limbal-derived corneal epithelial cell line (hTCEpi65) affected the cell cycle. We knocked down PBK levels in hTCEpi cells with two different siRNA constructs (Fig. 4C) and analyzed the cell cycle distribution of these cells (Fig. 4D). Following transduction, cells were FACS sorted. Analysis indicated 78% of the control si-transduced cells were in G0/G1, 7% were in S, and 12% were in G2/M (Fig. 4D). In contrast, 60% to 70% of si-PBK transduced cells were in G0/G1, 6% to 12% were in S, and 17% to 24% were in G2/M (Fig. 4D). Thus, low levels of PBK in hTCEpi cells arrested these cells in the G2/M phase of the cell cycle. This may explain, in part, why autophagy-compromised mice have delayed corneal epithelial wound healing26 (see below). A reduced PBK expression pattern in the limbal epithelium validates the stem/early TA and mature TA cell cluster designations.
Figure 4.
scRNA-seq identifies Pbk as preferentially expressed in mature TA cells in the corneal epithelium. (A) t-SNEs colored by the normalized log-transformed expression of Pbk. (B) Immunofluorescence staining with PBK antibody in a frozen section of human limbal and corneal epithelium. PBK expression was dramatically lower in limbal epithelium than in the corneal epithelium. Cor, cornea; K15, Keratin 15; Lim, limbus. Scale bar: 20 μm. Dotted lines mark the basement membrane. n = 3. (C, D) hTCEpi cells were transfected with two different siRNAs recognizing PBK transcripts (siPBK no. 1 and no. 3) or sicontrol. Immunoblotting showed that siPBK transfection dramatically reduced PBK protein expression (C). Cell cycle distribution was analyzed by flow cytometry, showing that a loss of PBK induced a G2/M phase arrest (D). n = 4.
scRNA-seq of the Limbus and Cornea From Beclin 1+/− Mice Reveal Diminished Proliferative Capacity
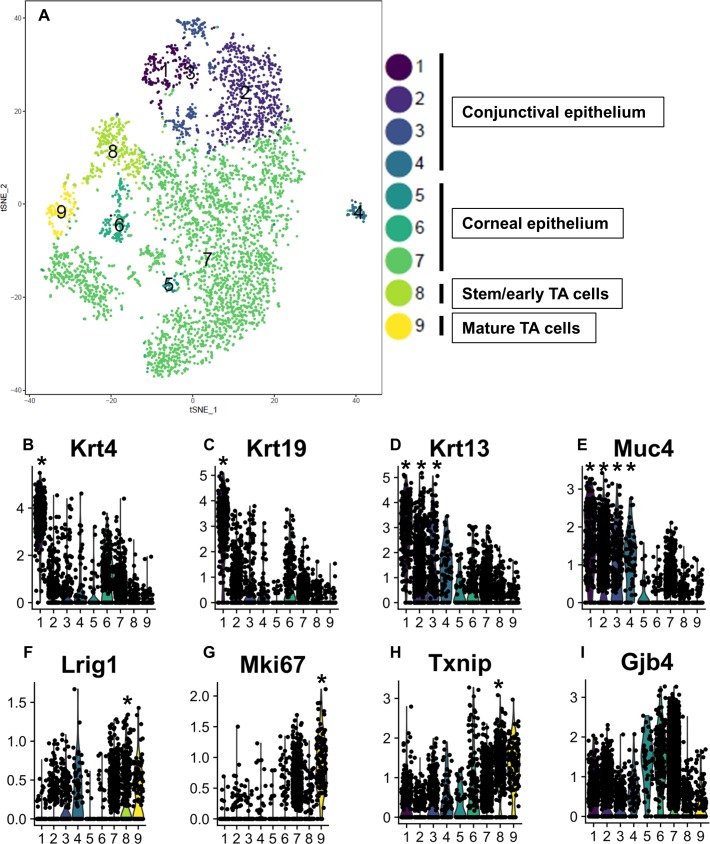
To interrogate the regulatory mechanisms underlying the delay in corneal epithelial wound healing in mice with compromised autophagy,26 data from scRNA-seq of wild-type and beclin 1+/− heterozygous mouse epithelia were normalized and merged using the Seurat package to obtain an integrated data set (Fig. 5A). Such analysis revealed nine distinct clusters (Fig. 5A), which we grouped into conjunctival (clusters 1–4), and corneal (clusters 5–9) based on the expression of known genes associated with each tissue (Fig. 5). Within the corneal clusters, we identified the stem/early TA (cluster 8) and the mature TA (cluster 9) populations. Violin plots, which show the distribution of all the data points for a given set, indicated that the highest frequency and distribution of the conjunctival-associated keratins 4, 19, and 13 were in clusters 1 to 4 (Fig. 5B–D). Muc-4, a conjunctival mucin, was also highly expressed in clusters 1 to 4 (Fig. 5E). In contrast, (1) Lrig1, a stem/early TA cell marker was high in cluster 8 (Fig. 5F); (2) Ki67, a maker of rapidly proliferating cells was low in cluster 8 (Fig. 5G); (3) Txnip, a novel marker for stem/early TA cell was high in cluster 8 (Fig. 5H); and (4) the negative stem cell marker, connexin (Gjb4), was low in cluster 8 (Fig. 5I). Collectively, this suggests that cluster 8 represents the stem/early TA cells. Comparison of the number of cells in cluster 8 from wild-type versus beclin1+/− mice revealed approximately 50% fewer stem/early TA cells in autophagy-compromised mice (Table). Such a reduction in stem cells and their immediate progeny is consistent with the marked reduction in holoclone colonies from human limbal epithelial cells with compromised autophagy.26 Cluster 9 had the highest expression of Ki67 (Fig. 5G), a marker of rapidly proliferating cells, and we are designating this cluster as representing the mature TA cells (Fig. 5A).
Figure 5.
scRNA-seq identified nine subclusters of cells from integrated samples of wild-type and Beclin 1+/− mouse limbal/corneal epithelial cells. (A) Cell-type clusters. We used t-SNE to visualize the re-clustering (color coding) of the original epithelial cell clusters from the integrated samples of wild-type and beclin 1+/− epithelial cells, based on the expression of known marker genes. The percentage of stem/early TA cells (cluster 8) in total corneal/limbal epithelial cells from beclin+/− mice is 50% less than wild-type mice. For the table, we used the “prop.table” command from the R base library. (B–I) Violin plots show the expression of gene markers from the nine clusters. Clusters are color coded. P values are tested by the Wilcoxon rank sum test. *P < 0.05.
Table.
Percentage of Cells Observed From Each Cluster in Total Limbal/Corneal Epithelial Cell Numbers of Wild-Type and Beclin 1+/− Mouse scRNA-seq Clusters From Integrated Samples
The higher percentage of cells in beclin 1+/− clusters compared with wild-type are highlighted in green and the lower percentage of beclin 1+/− clusters compared with wild-type are highlighted in orange.
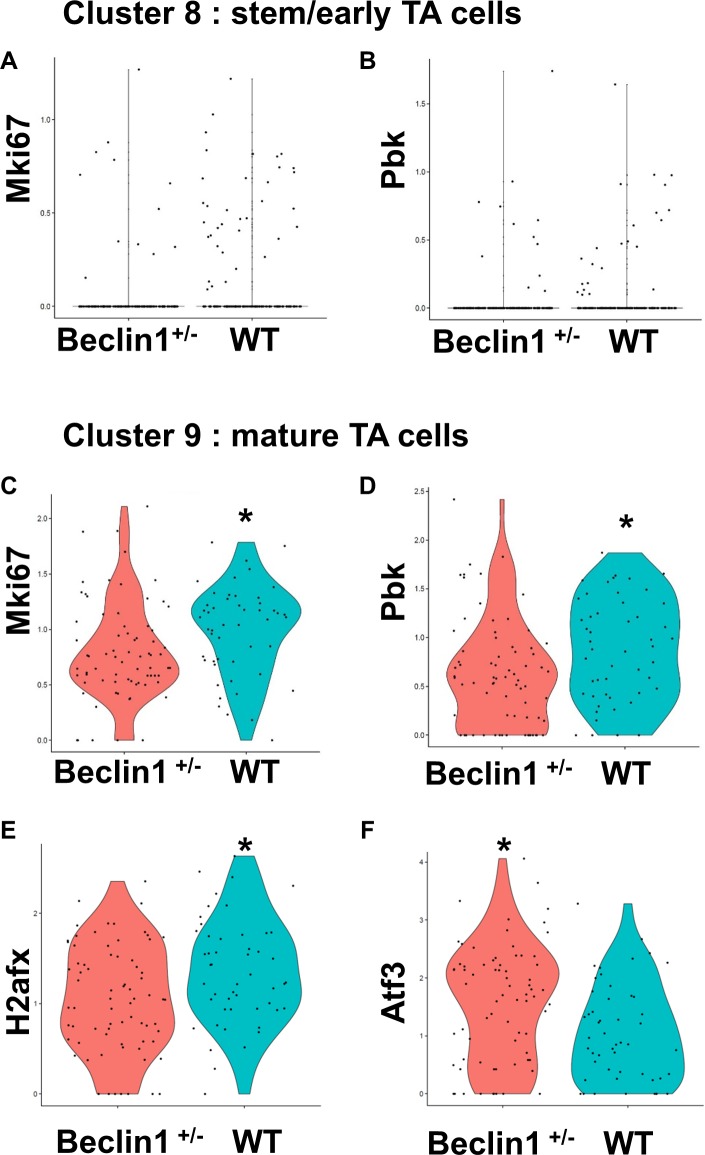
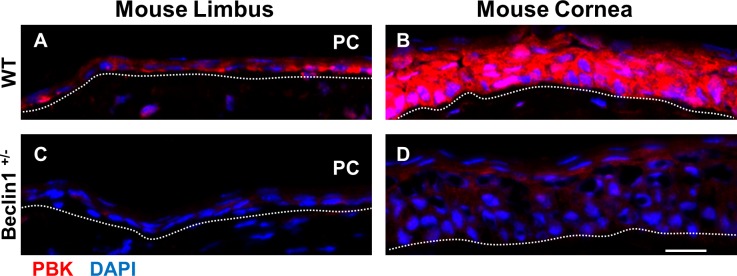
A decrease in expression of Ki67 was seen in the beclin 1+/− mice versus wild-type in both the stem/early TA (cluster 8) and mature TA cell (cluster 9) populations (Figs. 6A, 6C). Similarly, a decrease was seen in PBK expression in beclin 1+/− mice compared with wild-type mice (Figs. 6B, 6D). This observation was confirmed immunohistochemically, as PBK was barely detected in the corneal epithelium from the autophagy-deficient beclin 1+/− mice and virtually absent in the peripheral corneal epithelium (Figs. 7C, 7D). Wild-type mice had high expression of PBK in the corneal epithelium (Fig. 7B). We suggest that such a reduction in PBK might help explain the compromised wound-healing response in the beclin 1+/− mice.66 In wild-type mice, 24 hours following a central corneal epithelial wound, peripheral corneal epithelial basal cells are induced to divide, going from a labeling index of 7% to 24% after wounding. In addition, the cells have a markedly shortened cell cycle time (from 72 to 24 hours).3 This accounts for the efficiency in corneal epithelial wound healing. Low levels of PBK in hTCEpi cells results in an arrest of these cells in G2/M (Fig. 4D) and the low levels of PBK seen in the peripheral corneal epithelium of the beclin 1+/− mice (Figs. 7C, 7D) could impede cells from routinely entering the cell cycle and be a mechanism for the observed delay in wound response seen in these mice.26
Figure 6.
scRNA-seq identifies differentially expressed genes between wild-type and beclin 1+/− mouse limbal/corneal epithelial cells. Violin plots show the differential expression of the genes between wild-type and beclin 1+/− from clusters 8 (A, B) and 9 (C–F). *P < 0.05. P values are tested by the Wilcoxon rank sum test.
Figure 7.
scRNA-seq identifies a reduction of PBK expression in Beclin 1+/− mouse corneal epithelial cells. Immunofluorescence staining with PBK antibody in a frozen section of mouse limbal and corneal epithelium. In wild-type mice, PBK is preferentially expressed in the corneal epithelium (B) compared with the limbal epithelium (A). PBK expression is significantly decreased in the corneal and limbal epithelium from Beclin 1+/− mice (C, D). PC, peripheral cornea. Scale bar: 50 μm. Dotted lines mark the basement membrane. n = 4.
Beclin 1+/− Mice Reveal Reciprocal Expression Patterns for Stress-Related Response Genes
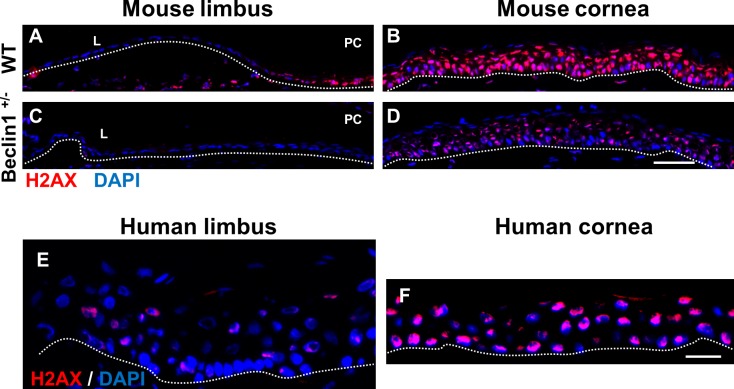
Autophagy is the means by which cells adapt and respond to differing intrinsic and extrinsic cellular-related stress situations.67–70 Thus, it was not surprising that one of the most downregulated genes in the beclin 1+/− scRNA-seq data set was histone H2ax (Fig. 6E; Supplementary Table S3). Phosphorylation of histone H2AX is a well-known modification that regulates the DNA damage signaling pathway and maintains genome integrity.71–73 H2AX was minimally expressed in the limbal epithelium of wild-type mice and humans (Figs. 8A, 8E); however, H2AX was prominently detected in the basal, wing, and superficial cells of the corneal epithelium (Figs. 8B, 8F). A marked reduction in H2AX expression was observed throughout the corneal epithelium of the beclin 1+/− mice (Fig. 8D), suggesting that these mice may have an attenuated ability to repair damaged DNA.
Figure 8.
scRNA-seq identifies a reduction in H2ax expression in beclin 1+/− mouse corneal epithelial cells. (A–D) Immunofluorescence staining with H2AX antibody in a frozen section of mouse limbal and corneal epithelium. In wild-type mice, H2AX is preferentially expressed in the corneal epithelium (B) compared with limbal epithelium (A). H2AX expression is significantly decreased in the corneal epithelium from beclin 1+/− mice (D). n = 4. Scale bar: 50 μm. (E, F) Immunofluorescence staining with H2AX antibody in a frozen section of human limbal (F) and corneal (E) epithelium. H2AX was primarily restricted to the corneal epithelium. Scale bar: 20 μm. Dotted lines mark the basement membrane. n = 3.
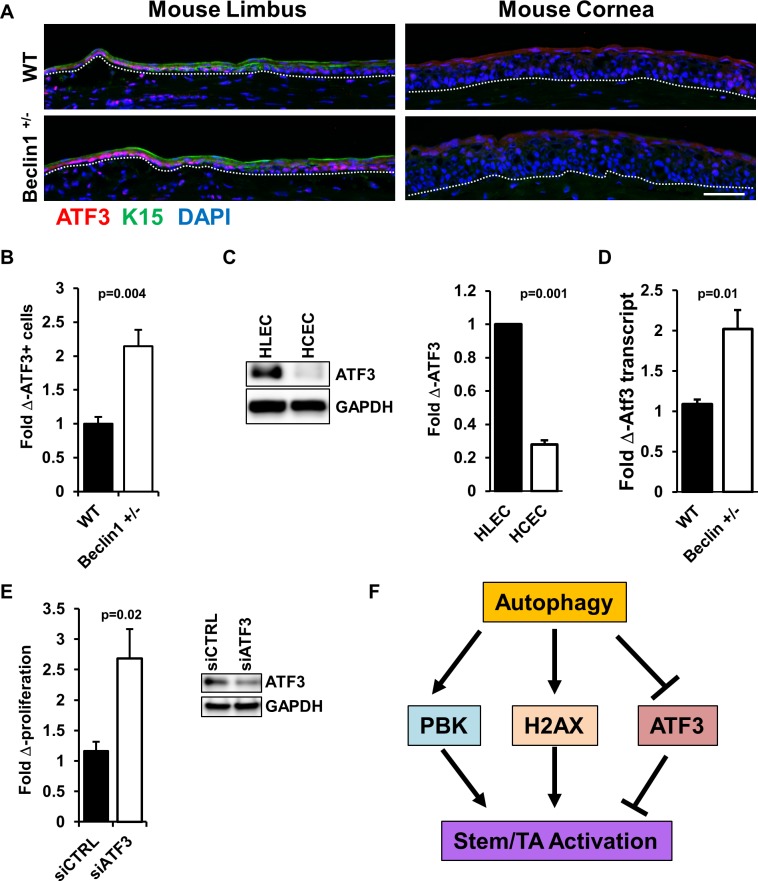
Activating transcription factor 3 (ATF3) responds to diverse cellular stresses and can be activated by several important signal transduction pathways.74,75 It is well established that following stress, downregulation of mammalian target of rapamycin (mTOR) initiates autophagy.76,77 Recently, Sirolimus, an mTOR inhibitor, was demonstrated to decrease ATF3 expression in 3D organotypic skin culture models.78 Based on this finding, the prediction is that compromised autophagy will result in an increase in ATF3 expression. Indeed, Atf3 was one of the highest upregulated genes in the beclin 1+/− scRNA-seq data (Fig. 6F, Supplementary Table S3). ATF3 expression in wild-type mice was most prominent in the nuclei of limbal epithelial basal cells (Fig. 9A). Such expression was markedly enhanced in the limbal epithelial basal cells from the beclin 1+/− mice (Fig. 9A), with a significant increase in ATF3+ cell number in limbal epithelia of beclin 1+/− mice (Fig. 9B), and is consistent with the scRNA-seq data. Although ATF3 antibodies tested under different fixation conditions failed give a consistent nuclear staining pattern with human cornea sections, immunoblots from lysates of human limbal and corneal epithelial cells revealed strong expression of ATF3 in the limbal epithelial cells (Fig. 9C). Moreover, real-time quantitative PCR of the total RNA isolated from the cornea showed increased mRNA transcript level for ATF3 in beclin 1+/− mice compared with the wild-type littermate controls (Fig. 9D). In a mouse model of heart failure, ATF3 reduced autophagy via suppression of the beclin-1–dependent pathway.79 This negative regulation of autophagy via ATF3 is in line with the present findings of an increase in ATF3 in the limbal epithelium of mice with compromised autophagy (Fig. 9A). In addition to stress response, ATF3 has been demonstrated to both positively and negatively affect proliferation in a variety of oncogenic settings.80–83 We have shown that autophagy plays a positive role in corneal epithelial proliferation,26 and thus wanted to examine how modulation of ATF3 affects proliferation in these cells. Using a genetic approach, we treated hTCEpi cells with siATF3 and noted a significant increase in cell growth compared with the control siRNA (Fig. 9E). This suggests that in the context of normal, resting corneal epithelial cells, ATF3 dampens proliferation and may, in part, contribute to the maintenance of quiescence in the stem cell–enriched limbal basal cells. Furthermore, the marked increase in ATF3 expression in limbal epithelium of beclin1+/− mice may contribute to the delayed corneal epithelial wound-healing response observed in these autophagy-compromised mice.26
Figure 9.
scRNA-seq identifies an increase in Atf3 expression in beclin 1+/− mouse corneal epithelial cells. (A) Immunofluorescence staining with ATF3 antibody in frozen sections of mouse limbal and corneal epithelium. In wild-type mice, ATF3 is preferentially expressed in the limbal epithelium compared with the corneal epithelium. (B) Bar graph shows a significant increase in ATF3-expressing cells in the limbal epithelium from beclin 1+/− mice as calculated from immunofluorescence images using ImageJ (http://imagej.nih.gov/ij/; provided in the public domain by the National Institutes of Health, Bethesda, MD, USA) (ATF3+ cells, n = 4). Scale bar: 50 μm. Dotted lines mark the basement membrane. (C) Immunoblotting showed that ATF3 expression was significantly higher in human limbal epithelial cells (HLECs) than in human corneal epithelial cells (HCECs). n = 3. (D) Atf3 mRNA expression in beclin 1+/− mice corneal epithelium as compared with its littermate controls (wild-type) shows an increase in Atf3 expression. Values are fold change over wild-type. n = 4. (E) hTCEpi cells were transfected with siATF3 or sicontrol (siCTRL). Cell proliferation was analyzed by WST-1 assay, showing that loss of ATF3 increased proliferation. Immunoblotting showed that siATF3 transfection dramatically reduced ATF3 protein expression. n = 3. (F) Model proposing how autophagy regulates epithelial proliferation via increased PBK and H2AX expression and decreased ATF3 expression, resulting in stem/TA cell activation.
Conclusions
The experiments reported here allow, for the first time, an in-depth survey of the transcriptional profile of mesenchymal and limbal/corneal epithelial cells. More importantly, use of scRNA-seq allowed direct assessment of specific rare cell types, such as the stem/early TA cells and mature TA cells that contribute to limbal/corneal homeostasis. During the evaluation and validation of the scRNA-seq data, novel genes were identified that may contribute to the preservation of cell cycle quiescence, which is a hallmark of limbal epithelial stem cells. The establishment of this comprehensive atlas of genes expressed by stromal and epithelial cells from limbus and cornea forms the foundation for future studies directed at unraveling regulatory networks between and among these distinct tissues.
We used scRNA-seq of limbal/corneal tissue from wild-type mice and autophagy-deficient mice to identify autophagy-associated transcriptomic changes in specific cell types that has not been feasible until recently. We found an overall decrease in proliferation-associated genes. Furthermore, dysregulation of ATF3 and PBK, genes associated with the cell cycle and stress response, may provide valuable insight into how autophagy functions as a positive regulator of proliferation in the limbal/corneal epithelium (Fig. 9F). In this model, we propose that in the context of re-epithelialization following wounding,26 one of the functions of autophagy is to ensure a timely upregulation of epithelial proliferation. To accomplish this, PBK and H2AX expression is promoted, whereas ATF3 expression is attenuated, resulting in stem/TA cell activation (Fig. 9F). This study was primarily aimed to distinguish and characterize the stem/TA cell populations in limbal/corneal epithelia and how autophagy affects this population. Therefore, most of the emphasis was given to analysis and characterization of the subpopulation of epithelial cells that had the stem/TA signature. Nonetheless, we identified and partially characterized certain nonepithelial clusters (Supplementary Fig. S1). Although it is beyond the scope of this article to conduct an in-depth analysis of these clusters, the mesenchymal cell population could be subclustered into other cell types. It is interesting, but not surprising, that we did not detect a subcluster of cells with an immune signature within the mesenchymal cell population. Possible explanations are (1) the limbus and corneas were acquired from healthy resting mice and such animals as well as humans do not have a preponderance of immune cells (i.e., dendritic cells)84; and (2) the numbers of immune cells normally present in the limbus were below scRNA-seq detection levels.
The ability of scRNA-seq to provide information with translational relevance is clearly demonstrated in the present comparative study of limbal/corneal epithelial-derived stem/early TA and mature TA cells from wild-type and autophagy-deficient mice. The scRNA-seq approach appears to be particularly well suited to investigating the corneal abnormalities associated with diabetes (see Ref. 85 and references therein). For example, corneal epithelial stem cells have decreased expression of putative stem cell markers, leading to the idea that this population of cells is compromised.86 Using approaches outlined in the present study, the stem/early TA cell and mature TA cell populations from limbal/corneas of wild-type and mouse models of diabetes (see Ref. 85 and references therein) could be compared. In this way, corneal keratopathies, such as delayed wound healing, persistent corneal epithelial erosions, barrier defects, and stem cell dysfunction,85 could be more deeply interrogated. Dry eye is a multifactorial disease of the ocular surface (see Refs. 87 and 88 and references therein). In a mouse model of experimental dry eye, desiccation stress resulted in abnormalities in corneal epithelial proliferation.89 Changes in the stem/early TA and mature TA populations from limbal/corneas of wild-type and experimental dry-eye mouse models90 should be relatively easily probed using scRNA-seq techniques. Information from such studies will give new understanding, not only on how dry eye affects the proliferative and differentiative status of limbal/corneal epithelium, but on how desiccation stress modulates autophagy in these epithelia.
Supplementary Material
Acknowledgments
The authors thank Tung-Tien (Henry) Sun, Alex Schermer, and George Cotsarelis, of our original team who established the concept that the limbus is the location of corneal epithelial stem cells.
Supported by National Institutes of Health Grants EY028560, EY019463 (RML); Dermatology Foundation Research Grant and Career Development Award (HP); Eversight Award (HP). The Northwestern University Skin Disease Research Center (NU-SDRC) Morphology and Phenotyping Core facility assisted in the morphologic analyses. The NU-SDRC is supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant AR057216. Single-cell RNA-sequencing services were performed by the Northwestern NUseq Core.
Disclosure: N. Kaplan, None; J. Wang, None; B. Wray, None; P. Patel, None; W. Yang, None; H. Peng, None; R.M. Lavker, None
References
- 1.Cotsarelis G, Cheng SZ, Dong G, Sun T-T, Lavker RM. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: implications on epithelial stem cells. Cell. 1989;57:201–209. doi: 10.1016/0092-8674(89)90958-6. [DOI] [PubMed] [Google Scholar]
- 2.Schermer A, Galvin S, Sun T-T. Differentiation-related expression of a major 64K corneal keratin in vivo and in culture suggests limbal location of corneal epithelial stem cells. J Cell Biol. 1986;103:49–62. doi: 10.1083/jcb.103.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehrer MS, Sun TT, Lavker RM. Strategies of epithelial repair: modulation of stem cell and transit amplifying cell proliferation. J Cell Sci. 1998;111(Pt. 19):2867–2875. doi: 10.1242/jcs.111.19.2867. [DOI] [PubMed] [Google Scholar]
- 4.Potten CS, Loeffler M. Stem cells: attributes, cycles, spirals, pitfalls and uncertainties. Lessons for and from the crypt. Development. 1990;110:1001–1020. doi: 10.1242/dev.110.4.1001. [DOI] [PubMed] [Google Scholar]
- 5.Amitai-Lange A, Altshuler A, Bubley J, Dbayat N, Tiosano B, Shalom-Feuerstein R. Lineage tracing of stem and progenitor cells of the murine corneal epithelium. Stem Cells. 2015;33:230–239. doi: 10.1002/stem.1840. [DOI] [PubMed] [Google Scholar]
- 6.Buck RC. Measurement of centripetal migration of normal corneal epithelial cells in the mouse. Invest Ophthalmol Vis Sci. 1985;26:1296–1299. [PubMed] [Google Scholar]
- 7.Collinson JM, Chanas SA, Hill RE, West JD. Corneal development, limbal stem cell function, and corneal epithelial cell migration in the Pax6(+/-) mouse. Invest Ophthalmol Vis Sci. 2004;45:1101–1108. doi: 10.1167/iovs.03-1118. [DOI] [PubMed] [Google Scholar]
- 8.Collinson JM, Morris L, Reid AI, et al. Clonal analysis of patterns of growth, stem cell activity, and cell movement during the development and maintenance of the murine corneal epithelium. Dev Dyn. 2002;224:432–440. doi: 10.1002/dvdy.10124. [DOI] [PubMed] [Google Scholar]
- 9.Di Girolamo N, Bobba S, Raviraj V, et al. Tracing the fate of limbal epithelial progenitor cells in the murine cornea. Stem Cells. 2015;33:157–169. doi: 10.1002/stem.1769. [DOI] [PubMed] [Google Scholar]
- 10.Beebe DC, Masters BR. Cell lineage and the differentiation of corneal epithelial cells. Invest Ophthalmol Vis Sci. 1996;37:1815–1825. [PubMed] [Google Scholar]
- 11.Lavker RM, Tseng SC, Sun TT. Corneal epithelial stem cells at the limbus: looking at some old problems from a new angle. Exp Eye Res. 2004;78:433–446. doi: 10.1016/j.exer.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 12.Dong Y, Peng H, Lavker RM. Emerging therapeutic strategies for limbal stem cell deficiency. J Ophthalmol. 2018;2018:7894647. doi: 10.1155/2018/7894647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez G, Sasamoto Y, Ksander BR, Frank MH, Frank NY. Limbal stem cells: identity, developmental origin, and therapeutic potential. Wiley Interdiscip Rev Dev Biol. 2018;7:e303. doi: 10.1002/wdev.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li D-Q, Pflugfelder SC, Huang AJW. Ocular surface epithelial stem cells. In: Low WC, Verfaillie CM, editors. Stem Cells and Regenerative Medicine. London: Wold Scientific Publishing Company;; 2008. pp. 111–141. [Google Scholar]
- 15.Li J, Xiao Y, Coursey TG, et al. Identification for differential localization of putative corneal epithelial stem cells in mouse and human. Sci Rep. 2017;7:5169. doi: 10.1038/s41598-017-04569-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, Hayashida Y, Chen YT, Tseng SC. Niche regulation of corneal epithelial stem cells at the limbus. Cell Res. 2007;17:26–36. doi: 10.1038/sj.cr.7310137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saghizadeh M, Kramerov AA, Svendsen CN, Ljubimov AV. Concise review: stem cells for corneal wound healing. Stem Cells. 2017;35:2105–2114. doi: 10.1002/stem.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dziasko MA, Daniels JT. Anatomical features and cell-cell interactions in the human limbal epithelial stem cell niche. Ocul Surf. 2016;14:322–330. doi: 10.1016/j.jtos.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Stepp MA, Zieske JD. The corneal epithelial stem cell niche. Ocul Surf. 2005;3:15–26. doi: 10.1016/s1542-0124(12)70119-2. [DOI] [PubMed] [Google Scholar]
- 20.Yazdanpanah G, Haq Z, Kang K, Jabbehdari S, Rosenblatt ML, Djalilian AR. Strategies for reconstructing the limbal stem cell niche. Ocul Surf. 2019;17:230–240. doi: 10.1016/j.jtos.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo ZH, Zhang W, Jia YYS, Liu QX, Li ZF, Lin JS. An insight into the difficulties in the discovery of specific biomarkers of limbal stem cells. Int J Mol Sci. 2018;19:1982–2003. doi: 10.3390/ijms19071982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng H, Park JK, Katsnelson J, et al. microRNA-103/107 family regulates multiple epithelial stem cell characteristics. Stem Cells. 2015;33:1642–1656. doi: 10.1002/stem.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou M, Li XM, Lavker RM. Transcriptional profiling of enriched populations of stem cells versus transient amplifying cells. A comparison of limbal and corneal epithelial basal cells. J Biol Chem. 2006;281:19600–19609. doi: 10.1074/jbc.M600777200. [DOI] [PubMed] [Google Scholar]
- 24.Tang F, Barbacioru C, Wang Y, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 25.He C, Bassik MC, Moresi V, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park JK, Peng H, Katsnelson J, et al. MicroRNAs-103/107 coordinately regulate macropinocytosis and autophagy. J Cell Biol. 2016;215:667–685. doi: 10.1083/jcb.201604032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boya P, Gonzalez-Polo RA, Poncet D, et al. Mitochondrial membrane permeabilization is a critical step of lysosome-initiated apoptosis induced by hydroxychloroquine. Oncogene. 2003;22:3927–3936. doi: 10.1038/sj.onc.1206622. [DOI] [PubMed] [Google Scholar]
- 28.Barrandon Y, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci U S A. 1987;84:2302–2306. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aymard E, Barruche V, Naves T, et al. Autophagy in human keratinocytes: an early step of the differentiation? Exp Dermatol. 2011;20:263–268. doi: 10.1111/j.1600-0625.2010.01157.x. [DOI] [PubMed] [Google Scholar]
- 30.Guan JL, Simon AK, Prescott M, et al. Autophagy in stem cells. Autophagy. 2013;9:830–849. doi: 10.4161/auto.24132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phadwal K, Watson AS, Simon AK. Tightrope act: autophagy in stem cell renewal, differentiation, proliferation, and aging. Cell Mol Life Sci. 2013;70:89–103. doi: 10.1007/s00018-012-1032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vessoni AT, Muotri AR, Okamoto OK. Autophagy in stem cell maintenance and differentiation. Stem Cells Dev. 2012;21:513–520. doi: 10.1089/scd.2011.0526. [DOI] [PubMed] [Google Scholar]
- 33.Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie HT, Chen SY, Li GG, Tseng SC. Isolation and expansion of human limbal stromal niche cells. Invest Ophthalmol Vis Sci. 2012;53:279–286. doi: 10.1167/iovs.11-8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng H, Kaplan N, Hamanaka RB, et al. microRNA-31/factor-inhibiting hypoxia-inducible factor 1 nexus regulates keratinocyte differentiation. PNAS. 2012;109:14030–14034. doi: 10.1073/pnas.1111292109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gipson IK, Grill SM. A technique for obtaining sheets of intact rabbit corneal epithelium. Invest Ophthalmol Vis Sci. 1982;23:269–273. [PubMed] [Google Scholar]
- 38.Pollen AA, Nowakowski TJ, Shuga J, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol. 2014;32:1053–1058. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borcherding N, Voigt AP, Liu V, Link BK, Zhang W, Jabbari A. Single-cell profiling of cutaneous T-cell lymphoma reveals underlying heterogeneity associated with disease progression. Clin Cancer Res. 2019;25:2996–3005. doi: 10.1158/1078-0432.CCR-18-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Usoskin D, Furlan A, Islam S, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18:145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 41.Syed-Picard FN, Du Y, Lathrop KL, Mann MM, Funderburgh ML, Funderburgh JL. Dental pulp stem cells: a new cellular resource for corneal stromal regeneration. Stem Cells Transl Med. 2015;4:276–285. doi: 10.5966/sctm.2014-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Funderburgh ML, Du Y, Mann MM, SundarRaj N, Funderburgh JL. PAX6 expression identifies progenitor cells for corneal keratocytes. FASEB J. 2005;19:1371–1373. doi: 10.1096/fj.04-2770fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rubsam M, Broussard JA, Wickstrom SA, Nekrasova O, Green KJ, Niessen CM. Adherens junctions and desmosomes coordinate mechanics and signaling to orchestrate tissue morphogenesis and function: an evolutionary perspective. Cold Spring Harb Perspect Biol. 2018;10:1–21. doi: 10.1101/cshperspect.a029207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun TT, Eichner R, Nelson WG, et al. Keratin classes: molecular markers for different types of epithelial differentiation. J Invest Dermatol. 1983;81:109s–115s. doi: 10.1111/1523-1747.ep12540831. [DOI] [PubMed] [Google Scholar]
- 45.Tsukita S, Tanaka H, Tamura A. The claudins: from tight junctions to biological systems. Trends Biochem Sci. 2019;44:141–152. doi: 10.1016/j.tibs.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 46.Gipson IK. Distribution of mucins at the ocular surface. Exp Eye Res. 2004;78:379–388. doi: 10.1016/s0014-4835(03)00204-5. [DOI] [PubMed] [Google Scholar]
- 47.Tseng SC, Jarvinen MJ, Nelson WG, Huang JW, Woodcock-Mitchell J, Sun TT. Correlation of specific keratins with different types of epithelial differentiation: monoclonal antibody studies. Cell. 1982;30:361–372. doi: 10.1016/0092-8674(82)90234-3. [DOI] [PubMed] [Google Scholar]
- 48.Jensen KB, Collins CA, Nascimento E, et al. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell. 2009;4:427–439. doi: 10.1016/j.stem.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Powell AE, Wang Y, Li Y, et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parfitt GJ, Kavianpour B, Wu KL, Xie Y, Brown DJ, Jester JV. Immunofluorescence tomography of mouse ocular surface epithelial stem cells and their niche microenvironment. Invest Ophthalmol Vis Sci. 2015;56:7338–7344. doi: 10.1167/iovs.15-18038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ross W, Hall PA. Ki67: from antibody to molecule to understanding? Clin Mol Pathol. 1995;48:M113–117. doi: 10.1136/mp.48.3.m113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Z, Evans WH, Pflugfelder SC, Li DQ. Gap junction protein connexin 43 serves as a negative marker for a stem cell-containing population of human limbal epithelial cells. Stem Cells. 2006;24:1265–1273. doi: 10.1634/stemcells.2005-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matic M, Evans WH, Brink PR, Simon M. Epidermal stem cells do not communicate through gap junctions. J Invest Dermatol. 2002;118:110–116. doi: 10.1046/j.0022-202x.2001.01623.x. [DOI] [PubMed] [Google Scholar]
- 54.Matic M, Petrov IN, Chen S, Wang C, Dimitrijevich SD, Wolosin JM. Stem cells of the corneal epithelium lack connexins and metabolite transfer capacity. Differentiation. 1997;61:251–260. doi: 10.1046/j.1432-0436.1997.6140251.x. [DOI] [PubMed] [Google Scholar]
- 55.Kinoshita S, Adachi W, Sotozono C, et al. Characteristics of the human ocular surface epithelium. Prog Retin Eye Res. 2001;20:639–673. doi: 10.1016/s1350-9462(01)00007-6. [DOI] [PubMed] [Google Scholar]
- 56.Miyashita H, Shimmura S, Kobayashi H, et al. Collagen-immobilized poly(vinyl alcohol) as an artificial cornea scaffold that supports a stratified corneal epithelium. J Biomed Mater Res B Appl Biomater. 2006;76:56–63. doi: 10.1002/jbm.b.30332. [DOI] [PubMed] [Google Scholar]
- 57.Morita K, Itoh M, Saitou M, et al. Subcellular distribution of tight junction-associated proteins (occludin, ZO-1, ZO-2) in rodent skin. J Invest Dermatol. 1998;110:862–866. doi: 10.1046/j.1523-1747.1998.00209.x. [DOI] [PubMed] [Google Scholar]
- 58.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 60.Nagaraj K, Lapkina-Gendler L, Sarfstein R, et al. Identification of thioredoxin-interacting protein (TXNIP) as a downstream target for IGF1 action. Proc Natl Acad Sci U S A. 2018;115:1045–1050. doi: 10.1073/pnas.1715930115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamaguchi F, Takata M, Kamitori K, et al. Rare sugar D-allose induces specific up-regulation of TXNIP and subsequent G1 cell cycle arrest in hepatocellular carcinoma cells by stabilization of p27kip1. Int J Oncol. 2008;32:377–385. [PubMed] [Google Scholar]
- 62.Gaudet S, Branton D, Lue RA. Characterization of PDZ-binding kinase, a mitotic kinase. Proc Natl Acad Sci U S A. 2000;97:5167–5172. doi: 10.1073/pnas.090102397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park JH, Nishidate T, Nakamura Y, Katagiri T. Critical roles of T-LAK cell-originated protein kinase in cytokinesis. Cancer Sci. 2010;101:403–411. doi: 10.1111/j.1349-7006.2009.01400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Herbert KJ, Ashton TM, Prevo R, Pirovano G, Higgins GS. T-LAK cell-originated protein kinase (TOPK): an emerging target for cancer-specific therapeutics. Cell Death Dis. 2018;9:1089. doi: 10.1038/s41419-018-1131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robertson DM, Li L, Fisher S, et al. Characterization of growth and differentiation in a telomerase-immortalized human corneal epithelial cell line. Invest Ophthalmol Vis Sci. 2005;46:470–478. doi: 10.1167/iovs.04-0528. [DOI] [PubMed] [Google Scholar]
- 66.Park J, Peng H, Katsnelson J, et al. microRNAs-103/107 coordinately regulate macropinocytosis and autophagy. J Cell Biol. 2016;215:667–685. doi: 10.1083/jcb.201604032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eskelinen EL, Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim Biophys Acta. 2009;1793:664–673. doi: 10.1016/j.bbamcr.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 68.Martin LM, Jeyabalan N, Tripathi R, et al. Autophagy in corneal health and disease: a concise review. Ocul Surf. 2019;17:186–197. doi: 10.1016/j.jtos.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peng H, Park JK, Lavker RM. Autophagy and macropinocytosis: keeping an eye on the corneal/limbal epithelia. Invest Ophthalmol Vis Sci. 2017;58:416–423. doi: 10.1167/iovs.16-21111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peng H, Park JK, Lavker RM. Eyeing autophagy and macropinocytosis in the corneal/limbal epithelia. Autophagy. 2017;13:975–977. doi: 10.1080/15548627.2017.1287658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ji JH, Min S, Chae S, et al. De novo phosphorylation of H2AX by WSTF regulates transcription-coupled homologous recombination repair. Nucleic Acids Res. 2019;47:6299–6314. doi: 10.1093/nar/gkz309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Turinetto V, Giachino C. Multiple facets of histone variant H2AX: a DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 2015;43:2489–2498. doi: 10.1093/nar/gkv061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yuan J, Adamski R, Chen J. Focus on histone variant H2AX: to be or not to be. FEBS Lett. 2010;584:3717–3724. doi: 10.1016/j.febslet.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hai T, Wolford CC, Chang YS. ATF3, a hub of the cellular adaptive-response network, in the pathogenesis of diseases: is modulation of inflammation a unifying component? Gene Expr. 2010;15:1–11. doi: 10.3727/105221610x12819686555015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao J, Li X, Guo M, Yu J, Yan C. The common stress responsive transcription factor ATF3 binds genomic sites enriched with p300 and H3K27ac for transcriptional regulation. BMC Genomics. 2016;17:335. doi: 10.1186/s12864-016-2664-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schaper-Gerhardt K, Walter A, Schmitz-Rode C, Satzger I, Gutzmer R. The mTOR-inhibitor Sirolimus decreases the cyclosporine-induced expression of the oncogene ATF3 in human keratinocytes. J Dermatol Sci. 2018;92:172–180. doi: 10.1016/j.jdermsci.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 79.Lin H, Li HF, Chen HH, et al. Activating transcription factor 3 protects against pressure-overload heart failure via the autophagy molecule Beclin-1 pathway. Mol Pharmacol. 2014;85:682–691. doi: 10.1124/mol.113.090092. [DOI] [PubMed] [Google Scholar]
- 80.Li X, Zang S, Cheng H, Li J, Huang A. Overexpression of activating transcription factor 3 exerts suppressive effects in HepG2 cells. Mol Med Rep. 2019;19:869–876. doi: 10.3892/mmr.2018.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wan H, Dopping-Hepenstal PJ, Gratian MJ, et al. Striate palmoplantar keratoderma arising from desmoplakin and desmoglein 1 mutations is associated with contrasting perturbations of desmosomes and the keratin filament network. Br J Dermatol. 2004;150:878–891. doi: 10.1111/j.1365-2133.2004.05996.x. [DOI] [PubMed] [Google Scholar]
- 82.Wang Z, Yan C. Emerging roles of ATF3 in the suppression of prostate cancer. Mol Cell Oncol. 2016;3:e1010948. doi: 10.1080/23723556.2015.1010948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wolford CC, McConoughey SJ, Jalgaonkar SP, et al. Transcription factor ATF3 links host adaptive response to breast cancer metastasis. J Clin Invest. 2013;123:2893–2906. doi: 10.1172/JCI64410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mastropasqua L, Nubile M, Lanzini M, et al. Epithelial dendritic cell distribution in normal and inflamed human cornea: in vivo confocal microscopy study. Am J Ophthalmol. 2006;142:736–744. doi: 10.1016/j.ajo.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 85.Ljubimov AV. Diabetic complications in the cornea. Vision Res. 2017;139:138–152. doi: 10.1016/j.visres.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saghizadeh M, Soleymani S, Harounian A, et al. Alterations of epithelial stem cell marker patterns in human diabetic corneas and effects of c-met gene therapy. Mol Vis. 2011;17:2177–2190. [PMC free article] [PubMed] [Google Scholar]
- 87.Stevenson W, Chauhan SK, Dana R. Dry eye disease: an immune-mediated ocular surface disorder. Arch Ophthalmol. 2012;130:90–100. doi: 10.1001/archophthalmol.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pflugfelder SC, de Paiva CS. The pathophysiology of dry eye disease: what we know and future directions for research. Ophthalmology. 2017;124:S4–S13. doi: 10.1016/j.ophtha.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fabiani C, Barabino S, Rashid S, Dana MR. Corneal epithelial proliferation and thickness in a mouse model of dry eye. Exp Eye Res. 2009;89:166–171. doi: 10.1016/j.exer.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stern ME, Pflugfelder SC. What we have learned from animal models of dry eye. Int Ophthalmol Clin. 2017;57:109–118. doi: 10.1097/IIO.0000000000000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.