Abstract
The voltage-sensing domain (VSD) is a conserved structural module that regulates the gating of voltage-dependent ion channels in response to a change in membrane potential. Although the structures of many VSD-containing ion channels are now available, our understanding of the structural dynamics associated with gating transitions remains limited. To probe dynamics with site-specific resolution, we utilized NMR spectroscopy to characterize the VSD derived from Shaker potassium channel in 1-palmitoyl-2-hydroxy-sn-glycero-3-phospho-(1'-rac-glycerol) (LPPG) micelles. The backbone dihedral angles predicted based on secondary chemical shifts using torsion angle likeliness obtained from shift (TALOS+) showed that the Shaker-VSD shares many structural features with the homologous Kv1.2/2.1 chimera, including a transition from α-helix to 310 helix in the C-terminal portion of the fourth transmembrane helix. Nevertheless, there are clear differences between the Shaker-VSD and Kv1.2/2.1 chimera in the S2-S3 linker and S3 transmembrane region, where the organization of secondary structure elements in Shaker-VSD appears to more closely resemble the KvAP-VSD. Comparison of microsecond-long molecular dynamics simulations of Kv 1.2-VSD in LPPG micelles and a 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) bilayer showed that LPPG micelles do not induce significant structural distortion in the isolated voltage sensor. To assess the integrity of the tertiary fold, we directly probed the binding of BrMT analog 2-[2-({[3-(2-amino-ethyl)-6-bromo-1H-indol-2-yl]methoxy}k7methyl)-6-bromo-1H-indol-3-yl]ethan-1-amine (BrET), a gating modifier toxin, and identified the location of the putative binding site. Our results suggest that the Shaker-VSD in LPPG micelles is in a native-like fold and is likely to provide valuable insights into the dynamics of voltage-gating and its regulation.
Significance
The Shaker potassium channel is a model for studying voltage-gating mechanisms within ion channels, but a high-resolution structure is lacking. We have developed a robust system for overexpression, purification, and structural characterization of the isolated Shaker voltage-sensing domain (VSD), which enables further biophysical and structural analysis and deeper understanding of this important system. Our initial NMR characterization of the Shaker-VSD reveals important and interesting structural features of the Shaker-VSD that do not match the structure predicted by homology models based on the Kv1.2/2.1 chimera crystal structure. Moreover, our NMR results reveal inherent structural flexibility within the isolated Shaker-VSD, which might help explain why the isolated Shaker-VSD is able to conduct proton currents in the absence of the pore domain.
Introduction
Voltage-sensing domains (VSDs) play a fundamental role in the gating processes of voltage-gated ion channels (1). They function to sense changes in membrane voltage, control the opening and closing of these channels, and generate electrical signals. So far, all known VSDs share a conserved architecture as well as a voltage-sensing mechanism (2). The VSD is composed of four consecutive transmembrane helixes (S1-S4) and is connected to the tetrameric pore domain via a short amphipathic linker helix. Conserved positively charged arginines are found on the S4 transmembrane helix (3, 4), and their interaction with the membrane potential drives the movement of the S4 transmembrane helix in response to voltage. As the VSDs are tightly coupled to the pore domain, the resulting conformational change in the VSDs regulates the state of the pore gate (5, 6).
Numerous efforts using biochemical, electrophysiological, spectroscopic, and structural techniques have been made to characterize VSDs in an isolated form as well as in the context of full-length ion channels (5, 7, 8, 9, 10, 11, 12, 13). One of the most important model systems for functional studies of VSDs and the development of our current understanding of voltage-sensing and voltage-gating mechanisms has been the Shaker potassium channel, a voltage-gated K+-selective channel from Drosophila (14, 15, 16, 17, 18, 19, 20). Two competing models for voltage-sensor movement, the transporter model and the helical screw model, have been proposed based on structural, electrophysiological, and computational studies of the Shaker ion channel and its homologs (21, 22, 23, 24, 25, 26, 27, 28). Crystal and cryogenic electron microscopy (cryo-EM) structures of homologous ion channels (29, 30, 31) have been solved, but the structure of the Shaker potassium channel is not available. These channels differ mostly in their loop regions, which have been shown to be important for function and pharmacological specificity (32, 33, 34, 35, 36). Despite their importance, these linkers are either truncated or not visible in the available structures of homologous channels (35, 37).
In this study, we describe the use of solution NMR spectroscopy to characterize the voltage-sensing domain of the Shaker potassium channel in 1-palmitoyl-2-hydroxy-sn-glycero-3-phospho-(1'-rac-glycerol) (LPPG) micelles. NMR spectroscopy is a powerful method for studying protein structure and dynamics with residue-level resolution (38, 39, 40, 41), but the full-length tetrameric potassium channel solubilized in a membrane mimetic environment is too large for solution-phase NMR studies. The discovery of voltage-sensitive phosphatases and voltage-gated proton channels, both of which lack the canonical pore domain, shows that the VSDs of ion channels can function as independent structural modules (12, 13). We find that the backbone structure of Shaker-VSD in LPPG micelles is generally consistent with the high-resolution structure of the homologous voltage-gated K+ (Kv) channels. Our NMR structural analysis also reveals some features that were unexpectedly different from homology models based on the Kv1.2/2.1 chimera crystal structure, such as a longer S3 helix in the Shaker-VSD. Binding of the gating modifier toxin BrMT analog 2-[2-({[3-(2-amino-ethyl)-6-bromo-1H-indol-2-yl]methoxy}k7methyl)-6-bromo-1H-indol-3-yl]ethan-1-amine (BrET) to the isolated VSDs suggests that the tertiary structural fold is properly maintained in LPPG micelles and also reveals details about the toxin binding site in these channels. Overall, the results provided here show that isolated Shaker-VSD may be a suitable system for further studies of structural dynamics of voltage-sensing domains.
Materials and Methods
Cloning and overexpression of Shaker-VSD
Codon-optimized segments of Shaker (222–392, 208–380, 208–392, 208–397, or 217–397) with an N-terminal 8xHis tag followed by TEV protease cleavage site were cloned into a pET15b expression vector. To optimize the overexpression of the Shaker-VSD, pET15b-Shaker-VSD constructs were transformed into Escherichia coli BL21 (DE3) with pRARE plasmids for additional codon supply. Overexpression of Shaker-VSD was tested at three temperatures (16, 25, and 37°C) and three isopropyl β-D-1-thiogalactopyranoside concentrations (0.1, 0.5, and 1 mM) at OD 0.8–1.0 in M9 minimal media. The expression levels were evaluated using 14% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). Segment 217–397 overexpressed at 25°C and induced overnight (18–20 h) with 0.5 mM isopropyl β-D-1-thiogalactopyranoside gave the best yield and thus was used for protein production for NMR studies. For isotopic labeling with 13C, 15N, and/or 2H, the standard media components were substituted with 15NH4Cl, 13C-glucose, and/or D2O plus 0.5 g/L isotopically labeled isogro (Sigma-Aldrich, St. Louis, MO).
Reconstitution of Shaker-VSD into lysolipid micelles
To purify Shaker-VSD, cells were collected and resuspended in buffer A (50 mM Tris (pH 8.0), 100 mM KCl, 1 μg/mL pepstatin, 10 μM leupeptin, and 100 μM phenylmethylsulfonyl fluoride) with 1 mM EDTA, 5 mM β-me, 250 mM sucrose, and 1 mg/mL lysozyme and lysed by sonication. The cell debris and inclusion bodies were separated using a low speed spin (20 min at 7000 × g). The membrane fraction was isolated by spinning for 1 h at 150,000 × g. Detergent (40 mM n-decyl-β-D-maltopyranoside [DM], 40 mM n-dodecyl-β-D-maltopyranoside [DDM], 1% w/v n-octyl-β-D-glucopyranoside [OG], 1% w/v 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate [CHAPS], or 1.5% w/v foscholine-12, Anatrace, Maumee, OH) and 15 mM tris(2-carboxyethyl)phosphine (TCEP) were added to buffer A to extract Shaker-VSD from the membrane fraction. After solubilizing for 2 h with rotation at room temperature, insoluble protein was removed by centrifugation at 30,000 × g for 30 min. Only DM and foscholine-12 were able to extract Shaker-VSD. As further fast protein liquid chromatography purifications indicate that Shaker-VSD aggregates in DM micelles but not foscholine-12 micelles (data not shown), foscholine-12 is chosen as the detergent for Shaker-VSD extraction. Solubilized Shaker-VSD in foscholine-12 was applied to Ni-NTA His-bind beads (Protino, Düren, Germany) prewashed with buffer B (50 mM Tris (pH 8.0), 100 mM KCl, and 15 mM TCEP), supplemented with 0.15% w/v foscholine-12, and allowed to bind for 30 min at room temperature. The beads were washed with 10 bed volumes of buffer B, followed by 10 bed volumes of buffer C (buffer B plus 20 mM imidazole and 0.15% w/v foscholine-12).
For reconstitution into lysolipid micelles, foscholine-12 was exchanged by washing the beads with 15 bed volumes of buffer B supplemented with 0.1% w/v 1-myristoyl-2-hydroxy-sn-glycero-3-phospho-(1'-rac-glycerol) (LMPG), LPPG, or 1-palmitoyl-2-hydroxy-sn-glycero-3-phosphocholine (LPPC). Shaker-VSD was eluted with five bed volumes of buffer B supplemented with 400 mM imidazole and 0.2% w/v LMPG, LPPG, or LPPC. For LPPG and LPPC reconstitution, eluted Shaker-VSD was dialyzed against TEV cleavage buffer (20 mM Tris (pH 8.0), 50 mM KCl, and 5 mM β-me) containing 0.0001% w/v LPPG or LPPC. For LMPG reconstitution, eluted Shaker-VSD was run through a PD-10 column equilibrated with TEV cleave buffer containing 0.2% w/v LMPG to remove imidazole. TEV protease was then added at 1:1 VSD/TEV mole ratio and incubated overnight at room temperature to remove His-tag. The next day, His-tag cleavage was verified using 14% SDS-PAGE. The solubilized and cleaved Shaker-VSD was then applied back to Ni-NTA His-bind resin to remove His-tagged TEV protease and cleaved tag. The flow through was concentrated to 0.5 mL and loaded onto a Superdex 200 column pre-equilibrated in NMR buffer (100 mM MOPS (pH 7.0), 50 mM KCl, and 4 mM TCEP) with the appropriate detergent (0.2% w/v LMPG, LPPG, or LPPC). Peak fractions containing Shaker-VSD were pooled and concentrated to 300 μL for NMR. For Shaker-VSD in LPPG/LPPC mixed micelles, samples purified with LPPG and LPPC micelles were mixed at 1:1 volume ratio before concentration. For Shaker-VSD in LPPG/Chobimalt mixed micelles, Chobimalt was added before concentration.
Reconstitution of Shaker-VSD into bicelles
Expression of Shaker-VSD was the same except the TEV protease cleavage site was replaced with a thrombin cleavage site to achieve efficient His-tag cleavage in foscholine-12. The initial extraction and Ni-NTA resin binding steps are as described above. Shaker-VSD was eluted from the Ni-NTA resin as above and then concentrated to 0.5 mL aliquots for loading directly onto a Superdex 200 column pre-equilibrated in NMR buffer (100 mM MOPS (pH 7.0), 50 mM KCl, and 4 mM TCEP) with 10 mM foscholine-12. Peak fractions were pooled, and thrombin was added to remove the His-tag.
To reconstitute Shaker-VSD into bicelles, we used our previously published protocol with slight modifications (42). Long-chain lipids (Table S1) were hydrated at 20 mg/mL in NMR buffer, bath sonicated for ∼1 min, and solubilized with 10 mM foscholine-12 for 20 min. Purified and cleaved Shaker-VSD fractions after size-exclusion purification were added to the solubilized long-chain lipids at a molar ratio of 1:100 VSD/lipids and rotated at room temperature for 3 h. Two aliquots of 45 mg Amberlite XAD-2 (Bio-Rad Laboratories, Hercules, CA) per milligram of total detergent were added to remove the detergent and incubated overnight at room temperature. Amberlite was removed by filtration, and the Shaker-VSD proteoliposomes were ultracentrifuged at 150,000 × g for 2 h at 6°C. The proteoliposome pellet was solubilized with short-chain lipids (Table S1) dissolved in NMR buffer to create q = 0.33 bicelles. Four freeze-thaw cycles produced homogeneous bicelles containing Shaker-VSD, which were flash frozen and stored at −80°C until use.
Reconstitution of Shaker-VSD into nanodiscs
MSP1D1ΔH5, a truncation mutant that produces nanodiscs with a smaller diameter, was used to assemble Shaker-VSD containing nanodiscs (43), with plasmids generously provided by Prof. Gerhard Wagner. The expression and purification of MSP1D1ΔH5 was performed according to the previously published protocol (44). The N-terminal His-tag was removed by TEV protease. Any additional His-tagged TEV protease was removed by applying back to Ni-NTA resin. His-tag cleavage and TEV removal were verified by SDS-PAGE.
Shaker-VSD was purified following the bicelle reconstitution protocol described above except that the His-tag was not cleaved. Nanodiscs were assembled according to established protocols with modifications (43, 44, 45). Briefly, MSP1D1ΔH5 and foscholine-12 solubilized lipids (Table S1) were incubated with Shaker-VSD at a molar ratio of VSD/MSP1D1ΔH5/lipid of 1:20:200 overnight at room temperature in 50 mM Tris (pH 8.0), 100 mM KCl, 4 mM TCEP, and 100 mM foscholine-12. The next day, three aliquots of 10 mg Amberlite XAD-2 (Bio-Rad Laboratories) per milligram of total detergent were added to remove the detergent, with incubation 4–12 h at room temperature after each addition. Amberlite was removed by filtration, and the nanodisc-containing solution was incubated with Ni-NTA beads at room temperature for 30 min to separate Shaker-VSD containing nanodiscs. The Ni-NTA beads were then washed with 10 bed volumes of buffer B followed by 10 bed volumes of buffer D (buffer B plus 20 mM imidazole). VSD-containing nanodiscs were eluted with five bed volumes of elution buffer (buffer B plus 400 mM imidazole). Elutions were concentrated to 0.5 mL and loaded onto a Superdex 200 column pre-equilibrated in NMR buffer (100 mM MOPS (pH 7.0), 50 mM KCl, and 4 mM TCEP). Peak fractions were pooled and concentrated to 300 μL for NMR sample.
Reconstitution of Shaker-VSD into Amphipol A8-35
Shaker-VSD was reconstituted into Amphipol A8-35 according to the published protocol (46). Shaker-VSD was purified the same way as for bicelle reconstitution and mixed with A8-35 polymer at a mass ratio VSD/A8-35 of 1:5 and incubated at room temperature for 1 h. Two aliquots of 10 mg Amberlite XAD-2 (Bio-Rad Laboratories) per milligram of total detergent were added to remove the detergent with an incubation for 2 h at room temperature after each addition. Amberlite was removed by filtration, and the Shaker-VSD in A8-35 was concentrated to 0.5 mL and loaded onto a Superdex 200 column pre-equilibrated in NMR buffer (100 mM MOPS (pH 7.0), 50 mM KCl, and 4 mM TCEP). Fractions containing Shaker-VSD were pooled and concentrated to 300 μL for an NMR sample.
NMR experiments for the screening of membrane mimetics and assignment of backbone resonance
NMR samples contained 0.2–0.5 mM Shaker-VSD in various membrane mimetics with 10% D2O and 5% NaN3. Spectra were acquired at the National Magnetic Resonance Facility at Madison (University of Wisconsin-Madison, Madison, WI). Transverse relaxation optimized spectroscopy (TROSY)-HNCO, TROSY-HN(ca)CO, TROSY-HNCA, TROSY-HN(co)CA, and TROSY-HNCACB were analyzed on a 750-MHz Bruker spectrometer with cryoprobe and processed using NMRPipe (47) and analyzed with CCPNMR Analysis (48). Backbone walk experiments were supplemented with 1-13C′-amino acid-specific labeling (49). The backbone assignments have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu) as BMRB: 27886.
NMR-detected BrET binding studies
BrET-observed one-dimensional 1H spectra with titrated Shaker-VSD were recorded on a 600-MHz Bruker spectrometer. Samples contained 0.1 mM BrET and 0.2% w/v (∼3.9 mM) LPPG in NMR buffer in a 1.7-mm NMR tube at 25°C to track Shaker-VSD concentration-dependent chemical shift perturbations of the HN in the indole ring of BrET. Because the Shaker-VSD and BrET have extremely limited solubility in water and are only soluble within the detergent micelles, traditional concentration units using the total sample volume are not appropriate. Instead, the mole fraction of Shaker-VSD and BrET in each sample was calculated relative to the moles of LPPG in each sample. The NMR chemical shift perturbation data were fit to determine the binding affinity using the equation below (50):
| (1) |
where Δωobs is the observed chemical shift perturbation of HN, Δωmax is the maximal chemical shift perturbation, [P]t is the total concentration of Shaker-VSD in each sample in mole percentage relative to LPPG, [L]t is the BrET concentration in mole percentage relative to LPPG, and Kd is the molar percentage binding affinity.
TROSY-HNCO spectra of Shaker-VSD without BrET were recorded with ∼0.2 mM Shaker-VSD under the same conditions on a 750-MHz Bruker spectrometer. After the recording of TROSY-HNCO spectrum without BrET, 0.5 mg lyophilized BrET was incubated with the NMR sample overnight at room temperature (final molar percentage concentration relative to LPPG is ∼9%), and the TROSY-HNCO spectrum was re-recorded. Chemical shift perturbation and peak volume were analyzed using CCPNMR Analysis (48).
Molecular dynamics simulations
Crystal structure of Kv1.2-VSD (Protein Data Bank [PDB]: 3LUT) (51) served as the starting structure. The initial Kv1.2-VSD LPPG micelle model and Kv.2-VSD 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) bilayer model were built with the web-based program CHARMM-GUI (52, 53, 54). For the micelle model, 150 LPPG molecules were added around the Kv1.2-VSD to form a torus-shaped micelle. The protein-micelle complex was solvated in a cube box of water with a side length of 115 Å. For the bilayer model, Kv1.2-VSD was embedded in a POPC lipid bilayer with a side length of 80 Å. A water box with a thickness of 15 Å was added to the top and the bottom of the lipid bilayer, and 150 mM KCl ions were randomly placed in the water.
Both simulations used the OpenMM package (55) with GPU-acceleration. The CHARMM36 force field (56, 57) with TIP3P water model (58) was applied in constant particle number, pressure, and temperature ensemble. The system was first energy minimized for 500 steps. Then, a six-step equilibration was carried out before the production simulation with decreasing constraint forces on the backbone carbons, side chain heavy atoms, and micelle/lipid bilayer as recommended by CHARMM-GUI (59, 60). Production simulations were carried out for 1000 ns at 298 K. Langevin Integrator was used with 1-fs time step and a collision frequency of 1 ps−1. MonteCarloBarostat was applied at 1 atm with a frequency of 100 fs−1. Nonbonded method used in the simulation was particle-mesh Ewald (PME) with a nonbonded cutoff of 12 Å and a switch distance of 10 Å. Bonds to hydrogens were constrained, and rigidWater option was implemented. Finally, periodic boundary condition was implemented at the edge of the water box.
Results
Isolated Shaker voltage-sensing domain is properly folded in LPPG micelles and is suitable for NMR study
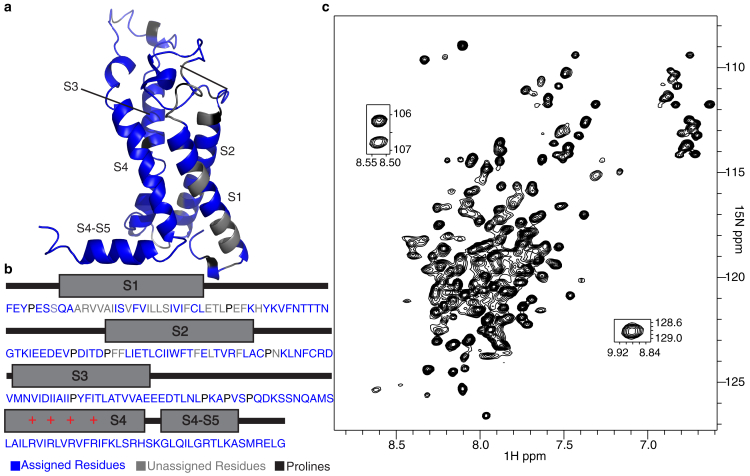
To determine the conditions for studying isolated Shaker-VSD using NMR, we screened various truncation segments of the Shaker channel, testing five different constructs spanning the voltage-sensing domain for expression in E. coli (BL21) (Fig. S1). Only segment 217–397 gave a high-level expression (Fig. 1, a and b); the other tested constructs did not express sufficiently for detection on SDS-PAGE with coomassie blue stain, indicating yields well below the amounts needed for NMR. Thus, this construct, hereafter referred to as Shaker-VSD, was used for all further studies. We then screened 31 different membrane mimetics to determine the optimal conditions for NMR (Table S1), as assessed by the number of resolved signals, peak dispersion, resolution, and uniformity of peak intensity across the 1H-15N TROSY-heteronuclear single quantum coherence (HSQC) spectrum. The best 1H-15N TROSY-HSQC spectra were obtained in LPPG and LMPG detergent micelles at 45°C, which is consistent with NMR studies of isolated VSDs from other ion channels (Figs. S2 and S3; (45, 61, 62, 63, 64, 65)). The 1H-15N TROSY-HSQC spectra obtained in other membrane mimetics had fewer peaks or worse resolution, indicating a lack of stable, well-folded tertiary structure, and thus were excluded from further NMR study (Fig. S3).
Figure 1.
Isolated Shaker-VSD construct optimized for NMR study. (a) Shown is a homology model and (b) amino acid sequence of the isolated Shaker-VSD (217–397), which spans from the S1 transmembrane helix to the S4-S5 linker. Residues with assigned backbone resonances are colored blue. Prolines are colored black. Unassigned residues are colored gray. (c) 1H-15N TROSY-HSQC NMR spectrum of the isolated Shaker-VSD construct in LPPG micelle at pH 7.0 and 45°C is shown. Sequence assignments are shown in Fig. S4.
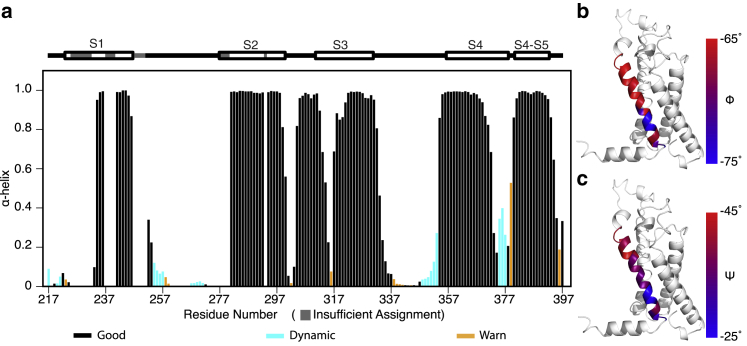
NMR chemical shifts are highly sensitive to local structure, which affects the distribution of electrons around a nucleus and thus the shielding of the nucleus from the applied magnetic field of the spectrometer. Backbone chemical shifts are particularly sensitive to backbone torsion angles, hydrogen bonding, and primary amino acid sequence (66, 67). The backbone torsion angle and secondary structure propensity of a protein can thus be predicted based on the backbone chemical shift data and the known amino acid sequence. The torsion angle likeliness obtained from shift (TALOS)+ program uses the empirical relationship between backbone 13C, 15N, and 1H chemical shifts and backbone φ and ψ angles to reliably predict the backbone dihedral angles and secondary structure elements in proteins. Comparison of backbone torsion angles predicted from the NMR chemical shift data with the corresponding crystal structure across a 200-protein data set revealed significant deviations for only 2.5% of residues, some of which likely reflect actual structural differences between the solution and the crystal (68). This demonstrates the accuracy of chemical shift-based torsion angle prediction, and this approach is widely used in NMR structure calculation (69, 70). Therefore, to check that Shaker-VSD was properly folded in LPPG detergent micelles, we first performed backbone assignment for secondary structure evaluation. 86% of the backbone amide resonances were assigned for Shaker-VSD with TROSY-HNCO, HN(ca)CO, HNCA, HN(co)CA, and HNCACB NMR experiments plus 1-13C′ Gly, Phe, Ala, Tyr, Leu, Ile, and Val specifically labeled samples (Figs. 1 and S4; (49)). The secondary structure of Shaker-VSD was calculated by TALOS+ using the chemical shifts of backbone HN, 15N, 13CO, 13Cα, and 13Cβ (68), and the dihedral angles were found to be most consistent with the expected secondary structural features observed in the Kv 1.2/2.1 chimera and full-length KvAP (Figs. 2 a and S5; (7, 29, 30)). As noted previously, the S4 transmembrane helix switches from α-helix to a 310-helix toward the C-terminus. Given the high homology between the Kv 1.2/2.1 chimera and Shaker channel in this region, we might expect that the Shaker-VSD will also exhibit this feature. Remarkably, we find that the NMR-derived backbone dihedral angles predicted by TALOS+ are consistent with classic 310-helix throughout the C-terminal end of the S4 transmembrane helix (Fig. 2, b and c).
Figure 2.
Secondary structure and backbone dihedral angles of the isolated Shaker-VSD. TALOS+ analysis of the backbone NMR chemical shifts (HN, N, Co, Cα, and Cβ) can reliably predict the backbone dihedral angles and secondary structure elements in proteins. The α-helical propensity of the isolated Shaker-VSD under the conditions used for NMR studies is shown in (a), with numbering according to the full-length Shaker sequence. The expected secondary structure is shown above the data with lines and helices designating loops and rectangles, respectively, and regions with insufficient chemical shift assignments for TALOS+ analysis are drawn in gray. For the predicted α-helical propensity, bars are colored according to the quality of prediction indicated by TALOS+, with black indicating good prediction, cyan indicating dynamic conformations, and orange indicating ambiguous prediction. The data correspond quite well with the expected helical segments. The backbone dihedral angles, φ (b) and ψ (c), of the S4 transmembrane helix are plotted on the homology model of isolated Shaker-VSD with a blue to red color scale to illustrate that the reduced α-helical propensity in the C-terminal half of S4 is due to a transition to 310-helix. Backbone dihedral angles characteristic of α-helix are colored red, and torsion angles characteristic of 310-helix are colored blue in both (b) and (c).
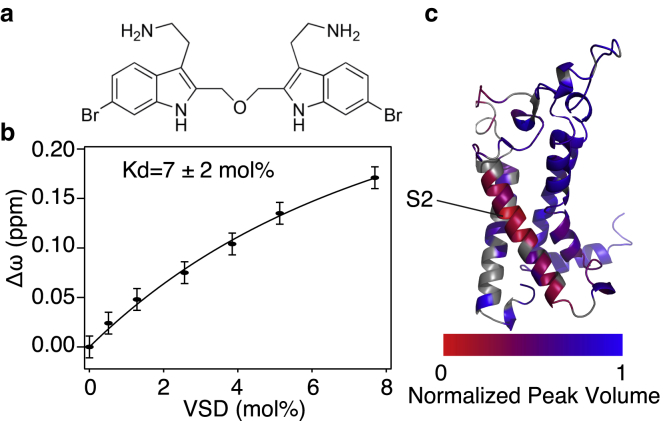
Next, we probed the integrity of the tertiary structure of isolated VSD by assaying its ability to bind the toxin BrET (Fig. 3 a). BrET is a synthetic derivative of a naturally occurring gating modifier gastropod toxin BrMT (6-bromo-2-mercaptotryptamine) (71). BrMT induces cooperativity in the Shaker potassium, presumably by interacting with the VSD (72). The BrMT binding site is proposed to lie within the S1, S2, and/or S3 transmembrane helices, but its binding has not been measured directly (71, 72, 73, 74). We performed separate NMR experiments monitoring either BrET- or Shaker-VSD to confirm direct and specific binding of BrET to Shaker-VSD at 25°C. Specific chemical shift perturbation of a well-resolved BrET peak corresponding to the −NH group in the indole ring was observed upon titration with unlabeled Shaker-VSD. Fitting this data yields an apparent binding affinity of 7 ± 2 mol% (Figs. 3 b and S6). Results are reported as mol% because the highly hydrophobic BrET has minimal solubility in water and is only soluble in the LPPG micelles. Given that most of the VSD is likely to be in an activated conformation in the absence of membrane potential, it is not surprising that the BrET, which has a preference for the resting conformation (72, 74), binds weakly to the isolated VSD.
Figure 3.
BrET interacts directly with Shaker-VSD. (a) Chemical structure of BrET is shown. (b) Binding curve was generated from NMR titration of BrET with the Shaker-VSD. The BrET 1H NMR spectra used to generate this curve are shown in Fig. S8. (c) The addition of BrET to the isolated Shaker-VSD causes a localized perturbation of the three-dimensional TROSY-HNCO NMR spectrum, indicating a specific binding interaction. The normalized change in peak volume in the Shaker-VSD upon BrET binding is plotted on a homology model of the isolated Shaker-VSD with a blue to red color scale as shown. Significant peak volume reduction is observed for residues from S2 transmembrane helix. Small chemical shift perturbations are also observed, as shown in Fig. S7.
To further identify the binding site of BrET within Shaker-VSD, we recorded three-dimensional TROSY-HNCO spectra of Shaker-VSD with and without BrET at 45°C. We observed a significant volume reduction of peaks from residues on the S2 transmembrane helix (Fig. 3 c). BrET was previously hypothesized to bind to the same region based on electrophysiology studies (71, 72, 74). The significant line broadening and loss of peak intensity observed upon BrET addition is consistent with the rapid on and off of a weak binding ligand coupled with a structural transition upon ligand binding (75). Moreover, we also observed a small but significant chemical shift perturbation throughout the entire protein (Fig. S7), suggesting that the protein undergoes a global conformational change upon binding. These results confirm direct binding of BrET to Shaker-VSD with the binding site centered on the S2 helix, indicating that the architecture of the toxin binding site in isolated Shaker-VSD in LPPG micelles is conserved.
Multiple structural states of the isolated Shaker-VSD
Upon assigning the backbone resonances of Shaker-VSD, we observed peak doubling for residues in the S0 helix, S1-S2 linker, S4 transmembrane helix, and S4-S5 linker in the TROSY-HNCO and HNCA spectra (Fig. S8). This indicates that these regions of Shaker-VSD exist in two distinct conformations under our NMR experimental condition. One possibility is that these reflect the resting and activated states of the isolated VSDs, but charge-voltage measurements of Shaker-VSD shows that these channels are more than 90% activated at 0 mV (76). Alternatively, the isolated voltage sensors are known to undergo a transition into a relaxed state upon prolonged activation (77). Because these isolated domains do not experience any voltage gradient in the micelles, it is quite likely that the observed peak doubling particularly in the S4 and S4-S5 linker reflects the conformational transitions between the activated and relaxed states. Nevertheless, additional experiments would be necessary to determine whether these conformational transitions correspond to the functional states identified by electrophysiologists. In any case, the observation of peak doubling highlights the inherent flexibility of this region of the VSD and exactly where the functionally relevant structural transitions are expected to occur.
Inherent structural features of Shaker-VSD
The crystal structure of Kv1.2/2.1 chimera has been widely used as a model for functional studies of Shaker potassium channel (6, 20, 78, 79, 80). Because of their highly conserved sequences, the structure of Shaker-VSD is expected to be almost identical to that of Kv1.2/2.1 chimera, except for S1-S2 and S3-S4 linker whose length and sequence are very different. As previously noted, the NMR-derived backbone torsion angles are consistent with a transition from α-helix to 310-helix in the C-terminal portion of S4 as observed in Kv1.2/2.1. The NMR chemical shift-based secondary structure prediction also shows that the Shaker S3 transmembrane helix is composed of two individual helices (Fig. S3, a and b) like Kv 1.2/2.1(Figs. 2 a and S5). However, S3b is ∼1–2 helical turns longer and is separated from S3a by a short S3 loop in Shaker, and the loop between S2 and S3 helices is about seven residues shorter in Shaker-VSD than in Kv1.2/2.1-VSD. At this point, it is not clear whether these observed structural differences between the Shaker and Kv1.2/2.1 chimera are due to differences in detergents, primary structure, or both.
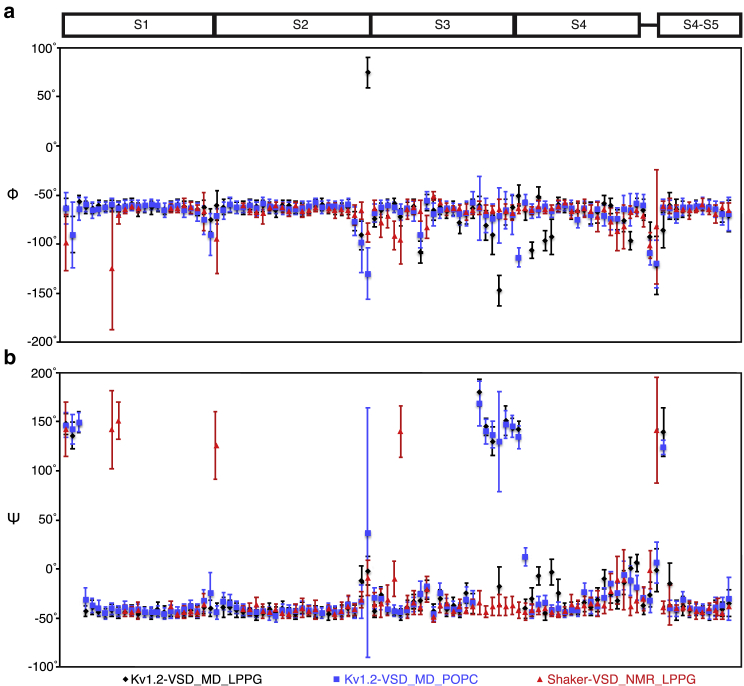
To test whether the LPPG detergent micelles can alter the VSD conformation, we performed molecular dynamics (MD) simulations of the VSD of Kv1.2 in both LPPG micelles and POPC bilayer for comparison. These simulations were performed with the Kv1.2 because of the availability of the high-resolution structural model (51). We extracted the average backbone dihedral angles and their variances from 1 μs MD trajectories for comparison with the NMR data. We focused our comparison on the transmembrane helices because the sequences of the transmembrane helices are highly conserved between Shaker-VSD and Kv1.2-VSD. The length and sequence in the loop regions of Shaker-VSD vary significantly from that of Kv1.2-VSD (Figs. 4 and S9), so we excluded these regions from the analysis. Except for a few residues at the beginning and end of each transmembrane helix, no major secondary structure differences were observed between Kv1.2-VSD in LPPG micelles and POPC bilayer. Therefore, these observations support the notion that the LPPG micelles are unlikely to distort the secondary structure differences of the isolated voltage sensors.
Figure 4.
Comparison Shaker-VSD with simulated backbone torsion angles of Kv1.2-VSD. MD simulations of Kv1.2-VSD were performed in both LPPG micelle and POPC bilayers to evaluate the effect of the micelle environment on the VSD structure. Kv1.2-VSD was chosen because of its homology to Shaker and the availability of a crystal structure. Backbone (a) φ and (b) ψ torsion angles are shown for helical segments in which the homology between Shaker-VSD and Kv1.2-VSD is high (sequence alignment shown in Fig. S9). Comparison of backbone torsion angles of Kv1.2-VSD after 1 μs MD simulations in LPPG micelles (black) and POPC bilayers (blue) shows that they are nearly identical, indicating that the lysolipid micelle does not significantly distort the VSD structure. The experimentally determined backbone torsion angles for Shaker-VSD in LPPG micelles (red, data from Fig. 2) also match the Kv1.2-VSD simulation data, except where there are sequence differences between Shaker and Kv1.2.
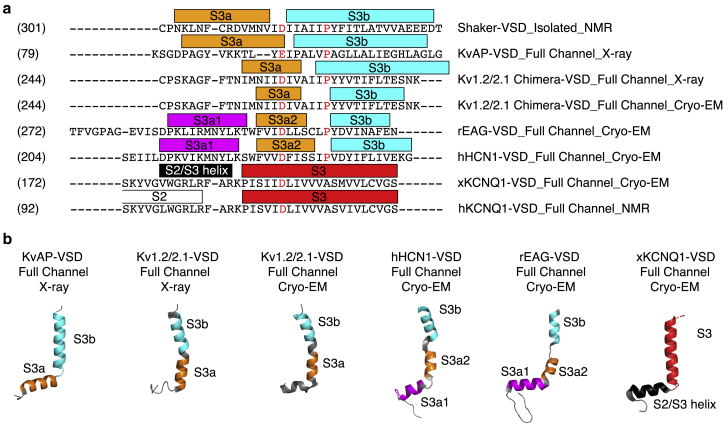
We further compared the secondary structure of isolated Shaker-VSD with high-resolution x-ray and cryo-EM structures or NMR studies of other Kv-VSDs (7, 29, 30, 63, 81, 82, 83). Despite an overall conserved four-helix bundle architecture, we observed significant secondary structural diversity especially in the S3 transmembrane helix (Figs. 5 and S10). KCNQ1 has a straight S3 helix ∼5 helical turns long, both in humans and Xenopus (63, 83). In the cryo-EM structure of X. laevis KCNQ1-VSD, the linker connecting the S2 and S3 segments, is observed to form a short helix that is denoted as the S2/S3 helix. However, this is not present in isolated hKCNQ1-VSD, according to torsion angles derived from NMR secondary chemical shift analysis using TALOS+. rEAG-VSD and hHCN1-VSD share similar secondary structural configurations in this region (81, 82). The S3b helices are ∼3–4 helical turns long, with an unstructured region connecting the S3a and the S3b helices following a proline residue. A Thr or Ser in the middle of the S3a helix breaks it into two subhelices, each of which are ∼3 helical turns long and is denoted as S3a1 and S3a2, respectively. The crystal and cryo-EM structures of the Kv1.2/2.1 chimera-VSD obtained in different membrane mimetics (detergent/lipid mixed micelles and nanodiscs, respectively) are almost identical to each other but show differences compared to the rEAG and hHCN channel structures (29, 30). The S3 helix in Kv1.2/2.1 chimera is also broken into S3a and S3b following a proline residue, but the S3a is not further subdivided into smaller helices. This architecture is similar to the KvAP-VSD in which the bend between the S3a and S3b helix is more pronounced. Overall, the backbone torsion angles and secondary structure assignments based on NMR secondary chemical shift analysis are in agreement with the high-resolution structures of related channels, which strongly indicate that the isolated Shaker-VSD in LPPG micelles is in a native-like fold. The backbone structure of the Shaker-VSD is similar to the Kv1.2/2.1 chimera as expected, particularly in the charged residue containing S4 helix. In contrast, comparison of the variable S3 helix (Figs. 5, S5, and S11) suggests that the length of S3a and S3b and the size of the intervening loop in Shaker-VSD might be more similar to KvAP. However, the current data are insufficient to evaluate whether the angle between the S3a and S3b helices matches full-length KvAP.
Figure 5.
Diversified secondary structure configuration of S2-S3 linker and S3 helix in different Kv-VSDs. (a) Sequence alignment of the S2-S3 linker and S3 helix region of different Kv-VSDs is shown. Colored rectangles above the sequences indicate the segments corresponding to S3a1 (magenta), S3a2 (orange), S3b (cyan), straight S3 helix (blue), S2-S3 helix (gray), or S2 helix (white). Backbone φ and ψ angles extracted from the Protein Data Bank structures were extracted and analyzed (Ramachandran plots in Fig. S10) to define the helical regions: isolated Shaker-VSD (our NMR results); KvAP-VSD in full channel, x-ray (1ORQ); Kv1.2/2.1-VSD in full channel, x-ray (2R9R); Kv1.2/2.1-VSD in full channel, cryo-EM (6EBK); human KCNQ1-VSD, NMR; X. laevis KCNQ1-VSD in full channel, cryo-EM (5VMS); EAG-VSD in full channel, cryo-EM (5K7L); and HCN1-VSD in full channel, cryo-EM (5U6O). (b) The defined helical regions are shown in the same color scheme plotted on the segment of the structure corresponding to the sequence alignment in (a) for each of the different Kv-VSDs, highlighting the structural diversity in this region. Comparison of the variable S3 helix (also Figs. S5 and S11) suggests that the length and position of helices within this region in Shaker-VSD might be more similar to KvAP.
Discussion
The Shaker potassium channel is a model system for studying the mechanisms of voltage gating and regulation. It is well characterized mainly in terms of its electrophysiological behavior, but very little is known about the structure of these channels. The related Kv 1.2/2.1 chimera high-resolution structure is widely used as a model for interpreting the functional effects of structural perturbations. Nevertheless, it is clear that there are significant differences between the Shaker and Kv 1.2 channels. For instance, Islas et al. have shown that the Kv 1.2 channels move ∼25% less charge than the Shaker potassium channel in response to a depolarizing voltage pulse (84). Biophysical and structural characterization of Shaker-VSD has been hindered, in part, by the difficulty in obtaining milligram quantities of properly folded and functional samples (85). In this study, we have generated a robust protocol for heterogeneous overexpression and purification of isolated Shaker-VSD. We assessed the structural integrity of the Shaker-VSD in 31 different membrane mimetics (86, 87, 88) and found conditions suitable for NMR studies. Among all the membrane mimetics that were screened, the best 1H-15N TROSY-HSQC spectra were obtained in lysophosphatidylglycerol (LMPG and LPPG) detergent micelles. The secondary structure predicted from our backbone resonance assignments, as well as the BrET binding studies, support a native-like fold for Shaker-VSD in LPPG micelles. Interestingly, LPPG micelles have also been used for NMR studies of the VSDs of KCNQ1, Nav 1.4, and hERG as well as the sensing domain of TRPM8 (45, 61, 62, 63, 64), suggesting that Lyso-PG detergent micelles are best suited for studying isolated VSDs.
Although the overall secondary structure of the Shaker potassium channel is consistent with the structures of other closely related potassium channels such as Kv 1.2/2.1 chimera, there were a few notable differences. First, the increased length of the Shaker S3a and the S3b helices and their separation by a short unstructured loop region as well as the break within the S3a helix make this region more closely resemble the structure of KvAP than Kv1.2/2.1. Second, our studies reveal an inherent flexibility of the isolated Shaker-VSD. The 1H-15N TROSY-HSQC spectrum of isolated Shaker-VSD is crowded with significant variation in peak intensity across the spectrum (Fig. 1 c). In contrast, the NMR peaks from isolated KvAP-VSD and hKCNQ1-VSD are well dispersed and more uniform (45, 63, 65, 89). A single set of well-resolved resonances is the hallmark of a tightly packed, stable tertiary structure in which each residue is locked in a single unique environment. Protein motion leads to increased line broadening due to enhanced relaxation or the averaging effect of sampling multiple different structural states, depending on the timescale. Conformational heterogeneity and dynamic sampling of multiple states on microsecond-millisecond timescales also average the chemical shift for a single resonance across different environments and reduce chemical dispersion. For several regions of the Shaker-VSD, we also observed peak doubling, indicating that those regions in the channel are sampling at least two different conformations on a millisecond or slower timescale. This peak doubling further contributes to the crowding in the NMR spectra of Shaker-VSD and emphasizes the inherent dynamics of the Shaker-VSD. Recent electrophysiological studies of the isolated Shaker-VSD show that the voltage-sensing domain is able to conduct proton currents in the absence of pore domain, whereas these proton currents are not observed in the full-length channel (76). One interpretation is that in the absence of the stabilizing influence of the pore domain, the voltage-sensing domain of the Shaker potassium channel is more dynamic and allows protons to pass through. Note that inside the gating pore of the Shaker-VSD, the extracellular and the intracellular solutions are separated by a single aromatic charge transfer center. Therefore, it is not difficult to envision that this separation is not as tight in the isolated VSD. However, specific conclusions about whether the observation of proton current through isolated Shaker-VSD is related to the flexibility here by NMR is not possible without additional experiments. Comparison with equivalent data for KCNQ and KvAP-VSDs may be helpful in testing this hypothesis.
In conclusion, our study for the first time to our knowledge establishes a robust system for biochemical, biophysical, and structural study of isolated Shaker-VSD. We have characterized secondary structural features of the isolated Shaker-VSD using NMR spectroscopy and provided insight into the channel dynamics. Our studies also shed light on the binding site of a small-molecule gating modifier of the Shaker potassium channel, BrET. Follow-up studies will allow us to gain a deeper structural understanding of the Shaker-VSD for comparison with functional studies to gain more insight into the voltage-sensing mechanism of voltage-gated ion channels.
Author Contributions
H.C., B.C., and K.A.H.-W. designed the research. H.C. executed the experiments. H.C. and K.A.H.-W. analyzed the data. J.P. performed MD simulations and analyzed the data under the supervision of Q.C. and B.C. D.M.G. and C.D. synthesized BrET for BrET binding experiments. H.C., B.C., and K.A.H.-W. wrote the manuscript.
Acknowledgments
We thank Scott Wildman for generating the homology model of Shaker-VSD, Marco Tonelli and Claudia Cornilescu for technical assistance with NMR experiments, Prof. Jon Sack for helpful communications and suggestions regarding BrET-related experiments, Prof. Gerhard Wagner for kindly proving the plasmids for MSP1D1 expression, Vilius Kurauskas for help generating Ramachandran plots, and Vadim Klenchin for help with protein expression and purification at the initial stage.
We thank the National Institutes of Health (NIH) for grant support NS081293 (B.C.). This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH grant P41GM103399 (National Institute of General Medical Sciences) (old number: P41RR002301). Equipment was purchased with funds from the University of Wisconsin-Madison, the NIH P41GM103399, S10RR02781, S10RR08438, S10RR023438, S10RR025062, S10RR029220), the National Science Foundation (DMB-8415048, OIA-9977486, BIR-9214394), and the U.S. Department of Agriculture.
Disha M. Gandhi’s present address is University of North Carolina-Chapel Hill, Chapel Hill, North Carolina 27599.
Footnotes
Editor: Sudha Chakrapani.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.06.020.
Contributor Information
Baron Chanda, Email: chanda@wisc.edu.
Katherine A. Henzler-Wildman, Email: henzlerwildm@wisc.edu.
Supporting Material
References
- 1.Swartz K.J. Sensing voltage across lipid membranes. Nature. 2008;456:891–897. doi: 10.1038/nature07620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chanda B., Bezanilla F. A common pathway for charge transport through voltage-sensing domains. Neuron. 2008;57:345–351. doi: 10.1016/j.neuron.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 3.Lee S.Y., Lee A., MacKinnon R. Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane. Proc. Natl. Acad. Sci. USA. 2005;102:15441–15446. doi: 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Payandeh J., Scheuer T., Catterall W.A. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roux B. Dissecting the coupling between the voltage sensor and pore domains. Neuron. 2006;52:568–569. doi: 10.1016/j.neuron.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Fernández-Mariño A.I., Harpole T.J., Chanda B. Gating interaction maps reveal a noncanonical electromechanical coupling mode in the Shaker K+ channel. Nat. Struct. Mol. Biol. 2018;25:320–326. doi: 10.1038/s41594-018-0047-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang Y., Lee A., MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 8.Ruta V., MacKinnon R. Localization of the voltage-sensor toxin receptor on KvAP. Biochemistry. 2004;43:10071–10079. doi: 10.1021/bi049463y. [DOI] [PubMed] [Google Scholar]
- 9.Chakrapani S., Cuello L.G., Perozo E. Structural dynamics of an isolated voltage-sensor domain in a lipid bilayer. Structure. 2008;16:398–409. doi: 10.1016/j.str.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Q., Wanderling S., Perozo E. Structural basis of lipid-driven conformational transitions in the KvAP voltage-sensing domain. Nat. Struct. Mol. Biol. 2014;21:160–166. doi: 10.1038/nsmb.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cowgill J., Klenchin V.A., Chanda B. Bipolar switching by HCN voltage sensor underlies hyperpolarization activation. Proc. Natl. Acad. Sci. USA. 2019;116:670–678. doi: 10.1073/pnas.1816724116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramsey I.S., Moran M.M., Clapham D.E. A voltage-gated proton-selective channel lacking the pore domain. Nature. 2006;440:1213–1216. doi: 10.1038/nature04700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q., Wanderling S., Perozo E. Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat. Struct. Mol. Biol. 2014;21:244–252. doi: 10.1038/nsmb.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liman E.R., Hess P., Koren G. Voltage-sensing residues in the S4 region of a mammalian K+ channel. Nature. 1991;353:752–756. doi: 10.1038/353752a0. [DOI] [PubMed] [Google Scholar]
- 15.Aggarwal S.K., MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 16.Papazian D.M., Schwarz T.L., Jan L.Y. Cloning of genomic and complementary DNA from Shaker, a putative potassium channel gene from Drosophila. Science. 1987;237:749–753. doi: 10.1126/science.2441470. [DOI] [PubMed] [Google Scholar]
- 17.Hoshi T., Zagotta W.N., Aldrich R.W. Shaker potassium channel gating. I: transitions near the open state. J. Gen. Physiol. 1994;103:249–278. doi: 10.1085/jgp.103.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zagotta W.N., Hoshi T., Aldrich R.W. Shaker potassium channel gating. II: transitions in the activation pathway. J. Gen. Physiol. 1994;103:279–319. doi: 10.1085/jgp.103.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zagotta W.N., Hoshi T., Aldrich R.W. Shaker potassium channel gating. III: evaluation of kinetic models for activation. J. Gen. Physiol. 1994;103:321–362. doi: 10.1085/jgp.103.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tao X., Lee A., MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328:67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y., Ruta V., MacKinnon R. The principle of gating charge movement in a voltage-dependent K+ channel. Nature. 2003;423:42–48. doi: 10.1038/nature01581. [DOI] [PubMed] [Google Scholar]
- 22.Long S.B., Campbell E.B., Mackinnon R. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 2005;309:903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 23.Jensen M.O., Borhani D.W., Shaw D.E. Principles of conduction and hydrophobic gating in K+ channels. Proc. Natl. Acad. Sci. USA. 2010;107:5833–5838. doi: 10.1073/pnas.0911691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broomand A., Elinder F. Large-scale movement within the voltage-sensor paddle of a potassium channel-support for a helical-screw motion. Neuron. 2008;59:770–777. doi: 10.1016/j.neuron.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 25.Starace D.M., Stefani E., Bezanilla F. Voltage-dependent proton transport by the voltage sensor of the Shaker K+ channel. Neuron. 1997;19:1319–1327. doi: 10.1016/s0896-6273(00)80422-5. [DOI] [PubMed] [Google Scholar]
- 26.Bezanilla F. Voltage sensor movements. J. Gen. Physiol. 2002;120:465–473. doi: 10.1085/jgp.20028660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starace D.M., Bezanilla F. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 2004;427:548–553. doi: 10.1038/nature02270. [DOI] [PubMed] [Google Scholar]
- 28.Treptow W., Tarek M. Environment of the gating charges in the Kv1.2 Shaker potassium channel. Biophys. J. 2006;90:L64–L66. doi: 10.1529/biophysj.106.080754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthies D., Bae C., Swartz K.J. Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs. eLife. 2018;7:e37558. doi: 10.7554/eLife.37558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long S.B., Tao X., MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 31.Long S.B., Campbell E.B., Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 32.Mathur R., Zheng J., Sigworth F.J. Role of the S3-S4 linker in Shaker potassium channel activation. J. Gen. Physiol. 1997;109:191–199. doi: 10.1085/jgp.109.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez C., Rosenman E., Latorre R. Modulation of the Shaker K(+) channel gating kinetics by the S3-S4 linker. J. Gen. Physiol. 2000;115:193–208. doi: 10.1085/jgp.115.2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez C., Rosenman E., Latorre R. Periodic perturbations in Shaker K+ channel gating kinetics by deletions in the S3-S4 linker. Proc. Natl. Acad. Sci. USA. 2001;98:9617–9623. doi: 10.1073/pnas.171306298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Priest M.F., Lacroix J.J., Bezanilla F. S3-S4 linker length modulates the relaxed state of a voltage-gated potassium channel. Biophys. J. 2013;105:2312–2322. doi: 10.1016/j.bpj.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sand R., Sharmin N., Gallin W.J. Fine-tuning of voltage sensitivity of the Kv1.2 potassium channel by interhelix loop dynamics. J. Biol. Chem. 2013;288:9686–9695. doi: 10.1074/jbc.M112.437483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sørensen J.B., Cha A., Bezanilla F. Deletion of the S3-S4 linker in the Shaker potassium channel reveals two quenching groups near the outside of S4. J. Gen. Physiol. 2000;115:209–222. doi: 10.1085/jgp.115.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imai S., Osawa M., Shimada I. Structural basis underlying the dual gate properties of KcsA. Proc. Natl. Acad. Sci. USA. 2010;107:6216–6221. doi: 10.1073/pnas.0911270107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brettmann J.B., Urusova D., Henzler-Wildman K.A. Role of protein dynamics in ion selectivity and allosteric coupling in the NaK channel. Proc. Natl. Acad. Sci. USA. 2015;112:15366–15371. doi: 10.1073/pnas.1515965112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang H., Kuenze G., Sanders C.R. Mechanisms of KCNQ1 channel dysfunction in long QT syndrome involving voltage sensor domain mutations. Sci. Adv. 2018;4:eaar2631. doi: 10.1126/sciadv.aar2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maruyama T., Imai S., Osawa M. Functional roles of Mg2+ binding sites in ion-dependent gating of a Mg2+ channel, MgtE, revealed by solution NMR. eLife. 2018;7:e31596. doi: 10.7554/eLife.31596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrison E.A., Henzler-Wildman K.A. Reconstitution of integral membrane proteins into isotropic bicelles with improved sample stability and expanded lipid composition profile. Biochim. Biophys. Acta. 2012;1818:814–820. doi: 10.1016/j.bbamem.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hagn F., Etzkorn M., Wagner G. Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J. Am. Chem. Soc. 2013;135:1919–1925. doi: 10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ritchie T.K., Grinkova Y.V., Sligar S.G. Chapter 11 - reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shenkarev Z.O., Lyukmanova E.N., Arseniev A.S. Lipid-protein nanodiscs as reference medium in detergent screening for high-resolution NMR studies of integral membrane proteins. J. Am. Chem. Soc. 2010;132:5628–5629. doi: 10.1021/ja9097498. [DOI] [PubMed] [Google Scholar]
- 46.Zoonens M., Zito F., Popot J.-L. Membrane Proteins Production for Structural Analysis. Springer; 2014. Amphipols: a general introduction and some protocols; pp. 173–204. [Google Scholar]
- 47.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 48.Vranken W.F., Boucher W., Laue E.D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 49.Takeuchi K., Ng E., Wagner G. 1-13C amino acid selective labeling in a 2H15N background for NMR studies of large proteins. J. Biomol. NMR. 2007;38:89–98. doi: 10.1007/s10858-007-9152-z. [DOI] [PubMed] [Google Scholar]
- 50.Williamson M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 51.Chen X., Wang Q., Ma J. Structure of the full-length Shaker potassium channel Kv1.2 by normal-mode-based X-ray crystallographic refinement. Proc. Natl. Acad. Sci. USA. 2010;107:11352–11357. doi: 10.1073/pnas.1000142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jo S., Kim T., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 53.Cheng X., Jo S., Im W. CHARMM-GUI micelle builder for pure/mixed micelle and protein/micelle complex systems. J. Chem. Inf. Model. 2013;53:2171–2180. doi: 10.1021/ci4002684. [DOI] [PubMed] [Google Scholar]
- 54.Wu E.L., Cheng X., Im W. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eastman P., Friedrichs M.S., Pande V.S. Openmm 4: a reusable, extensible, hardware independent library for high performance molecular simulation. J. Chem. Theory Comput. 2013;9:461–469. doi: 10.1021/ct300857j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacKerell A.D., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 57.Best R.B., Zhu X., Mackerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 59.Brooks B.R., Brooks C.L., III, Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee J., Cheng X., Im W. Charmm-gui input generator for namd, gromacs, amber, openmm, and charmm/openmm simulations using the charmm36 additive force field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ng H.Q., Kim Y.M., Kang C. Purification and structural characterization of the voltage-sensor domain of the hERG potassium channel. Protein Expr. Purif. 2012;86:98–104. doi: 10.1016/j.pep.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 62.Rath P., Hilton J.K., Van Horn W.D. Implications of human transient receptor potential melastatin 8 (trpm8) channel gating from menthol binding studies of the sensing domain. Biochemistry. 2016;55:114–124. doi: 10.1021/acs.biochem.5b00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peng D., Kim J.H., Sanders C.R. Purification and structural study of the voltage-sensor domain of the human KCNQ1 potassium ion channel. Biochemistry. 2014;53:2032–2042. doi: 10.1021/bi500102w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paramonov A.S., Lyukmanova E.N., Shenkarev Z.O. NMR investigation of the isolated second voltage-sensing domain of human Nav1.4 channel. Biochim. Biophys. Acta Biomembr. 2017;1859:493–506. doi: 10.1016/j.bbamem.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 65.Shenkarev Z.O., Paramonov A.S., Arseniev A.S. NMR structural and dynamical investigation of the isolated voltage-sensing domain of the potassium channel KvAP: implications for voltage gating. J. Am. Chem. Soc. 2010;132:5630–5637. doi: 10.1021/ja909752r. [DOI] [PubMed] [Google Scholar]
- 66.de Dios A.C., Pearson J.G., Oldfield E. Secondary and tertiary structural effects on protein NMR chemical shifts: an ab initio approach. Science. 1993;260:1491–1496. doi: 10.1126/science.8502992. [DOI] [PubMed] [Google Scholar]
- 67.Spera S., Bax A. Empirical correlation between protein backbone conformation and c.Alpha. And c.Beta. 13c nuclear magnetic resonance chemical shifts. J. Am. Chem. Soc. 1991;113:5490–5492. [Google Scholar]
- 68.Shen Y., Delaglio F., Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Linge J.P., Habeck M., Nilges M. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics. 2003;19:315–316. doi: 10.1093/bioinformatics/19.2.315. [DOI] [PubMed] [Google Scholar]
- 70.Lemak A., Gutmanas A., Arrowsmith C.H. A novel strategy for NMR resonance assignment and protein structure determination. J. Biomol. NMR. 2011;49:27–38. doi: 10.1007/s10858-010-9458-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dockendorff C., Gandhi D.M., Sack J.T. Synthetic analogues of the snail toxin 6-bromo-2-mercaptotryptamine dimer (brmt) reveal that lipid bilayer perturbation does not underlie its modulation of voltage-gated potassium channels. Biochemistry. 2018;57:2733–2743. doi: 10.1021/acs.biochem.8b00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sack J.T., Aldrich R.W. Binding of a gating modifier toxin induces intersubunit cooperativity early in the Shaker K channel’s activation pathway. J. Gen. Physiol. 2006;128:119–132. doi: 10.1085/jgp.200609492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kelley W.P., Wolters A.M., Gilly W.F. Characterization of a novel gastropod toxin (6-bromo-2-mercaptotryptamine) that inhibits shaker K channel activity. J. Biol. Chem. 2003;278:34934–34942. doi: 10.1074/jbc.M301271200. [DOI] [PubMed] [Google Scholar]
- 74.Sack J.T., Aldrich R.W., Gilly W.F. A gastropod toxin selectively slows early transitions in the Shaker K channel’s activation pathway. J. Gen. Physiol. 2004;123:685–696. doi: 10.1085/jgp.200409047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Palmer A.G., III, Kroenke C.D., Loria J.P. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 76.Zhao J., Blunck R. The isolated voltage sensing domain of the Shaker potassium channel forms a voltage-gated cation channel. eLife. 2016;5:e18130. doi: 10.7554/eLife.18130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Villalba-Galea C.A., Sandtner W., Bezanilla F. S4-based voltage sensors have three major conformations. Proc. Natl. Acad. Sci. USA. 2008;105:17600–17607. doi: 10.1073/pnas.0807387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Infield D.T., Lee E.E.L., Ahern C.A. Replacing voltage sensor arginines with citrulline provides mechanistic insight into charge versus shape. J. Gen. Physiol. 2018;150:1017–1024. doi: 10.1085/jgp.201812075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ottosson N.E., Silverå Ejneby M., Elinder F. A drug pocket at the lipid bilayer-potassium channel interface. Sci. Adv. 2017;3:e1701099. doi: 10.1126/sciadv.1701099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wood M.L., Freites J.A., Tobias D.J. Atomistic modeling of ion conduction through the voltage-sensing domain of the shaker k+ ion channel. J. Phys. Chem. B. 2017;121:3804–3812. doi: 10.1021/acs.jpcb.6b12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Whicher J.R., MacKinnon R. Structure of the voltage-gated K+ channel Eag1 reveals an alternative voltage sensing mechanism. Science. 2016;353:664–669. doi: 10.1126/science.aaf8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee C.H., MacKinnon R. Structures of the human HCN1 hyperpolarization-activated channel. Cell. 2017;168:111–120.e11. doi: 10.1016/j.cell.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun J., MacKinnon R. Cryo-EM structure of a KCNQ1/CaM complex reveals insights into congenital long QT syndrome. Cell. 2017;169:1042–1050.e9. doi: 10.1016/j.cell.2017.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ishida I.G., Rangel-Yescas G.E., Islas L.D. Voltage-dependent gating and gating charge measurements in the Kv1.2 potassium channel. J. Gen. Physiol. 2015;145:345–358. doi: 10.1085/jgp.201411300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sokolova O., Kolmakova-Partensky L., Grigorieff N. Three-dimensional structure of a voltage-gated potassium channel at 2.5 nm resolution. Structure. 2001;9:215–220. doi: 10.1016/s0969-2126(01)00578-0. [DOI] [PubMed] [Google Scholar]
- 86.Columbus L., Lipfert J., Lesley S.A. Mixing and matching detergents for membrane protein NMR structure determination. J. Am. Chem. Soc. 2009;131:7320–7326. doi: 10.1021/ja808776j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Poget S.F., Girvin M.E. Solution NMR of membrane proteins in bilayer mimics: small is beautiful, but sometimes bigger is better. Biochim. Biophys. Acta. 2007;1768:3098–3106. doi: 10.1016/j.bbamem.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Warschawski D.E., Arnold A.A., Marcotte I. Choosing membrane mimetics for NMR structural studies of transmembrane proteins. Biochim. Biophys. Acta. 2011;1808:1957–1974. doi: 10.1016/j.bbamem.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 89.Butterwick J.A., MacKinnon R. Solution structure and phospholipid interactions of the isolated voltage-sensor domain from KvAP. J. Mol. Biol. 2010;403:591–606. doi: 10.1016/j.jmb.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.