Abstract
Drug resistance and tumor heterogeneity are formidable challenges in cancer medicine, which is particularly relevant for KRAS-mutant cancers, the epitome of malignant tumors recalcitrant to targeted therapy efforts and first-line chemotherapy. In this study, we delineate that KRAS-mutant lung cancer cells resistant to pemetrexed (MTA) and anti-MEK drug trametinib acquire an exquisite dependency on endoplasmic reticulum (ER) stress signaling, rendering resistant cancer cells selectively susceptible to blockage of HSP90, the receptor tyrosine kinase AXL, the eukaryotic translation initiation factor 4E (eIF4E), and the unfolded protein response (UPR). Mechanistically, acquisition of drug resistance enables KRAS-mutant lung cancer cells to bypass canonical KRAS effectors but entail hyperactive AXL/eIF4E, increased protein turnover in the ER, and adaptive activation of an ER stress-relief UPR survival pathway whose integrity is maintained by HSP90. Notably, the unique dependency and sensitivity induced by drug resistance are applicable to KRAS-mutant lung cancer cells undergoing de novo intratumor heterogeneity. In line with these findings, HSP90 inhibitors synergistically enhance antitumor effects of MTA and trametinib, validating a rational combination strategy to treat KRAS-mutant lung cancer. Collectively, these results uncover collateral vulnerabilities co-occurring with drug resistance and tumor heterogeneity, informing novel therapeutic avenues for KRAS-mutant lung cancer.
Subject terms: Non-small-cell lung cancer, Oncogenes, Cancer therapeutic resistance
Introduction
Resistance to therapeutics and tumor heterogeneity inevitably limit clinical efficacy of cancer treatment, which is particularly relevant for KRAS-mutant cancers that are the most common type of human malignancies defined by genetic alterations and, ironically, the largest subset of tumors that cannot be effectively targeted by currently available therapeutics1. Oncogenic mutant KRAS is associated with striking heterogeneity2, heightened resistance to first-line chemotherapy3,4, and has proven difficult to pharmacologically target directly5, as have attempts to block synthetic lethal interactions with a mutant KRAS allele that have met with only limited success6.
Innovative efforts to inhibit KRAS-dependent tumor growth by extinguishing KRAS downstream signaling pathways have led to the important observation that suppressing the mitogen-activated protein kinase (MAPK) cascade RAF/MEK/ERK is necessary but insufficient7. Consequently, highly selective RAF, MEK, and ERK inhibitors only show marginal antitumor activity in KRAS-mutant tumors8. A major cause for the inefficiency of MAPK inhibitors, such as the clinically approved anti-MEK drug trametinib9, is the rapid and inevitable development of resistant diseases. The transient or short-lived nature of trametinib has been attributed to treatment-induced repression of negative feedback loops that reactivates the MAPK pathway via various compensatory mechanisms10–13. Nevertheless, albeit improved effects of MEK inhibitors by blocking compensatory MAPK reactivation, resistant tumors on the combination treatment still evolve14, suggesting the existence of other uncharacterized mechanisms of adaptive resistance.
In this study, we investigated the molecular mechanisms underlying adaptive resistance to currently available cancer drugs in KRAS-mutant lung cancer. We showed that HSP90/AXL/eIF4E-regulated endoplasmic reticulum (ER) stress response, or the unfolded protein response (UPR), represents an acquired dependency and confers a selective vulnerability in drug-resistant KRAS-mutant lung cancer cells that drug-naïve parental cells lack. We further demonstrated that this collateral sensitivity induced by drug resistance also applies to KRAS-mutant lung cancer cells that have undergone de novo intratumor heterogeneity. These results uncover tumor vulnerabilities that can be therapeutically exploited to overcome drug resistance and tumor heterogeneity, providing a new rationale for combination therapies to combat aggressive and difficult-to-treat KRAS-mutant lung cancer.
Results
A shared mechanism of resistance to MTA and trametinib in KRAS-mutant lung cancer
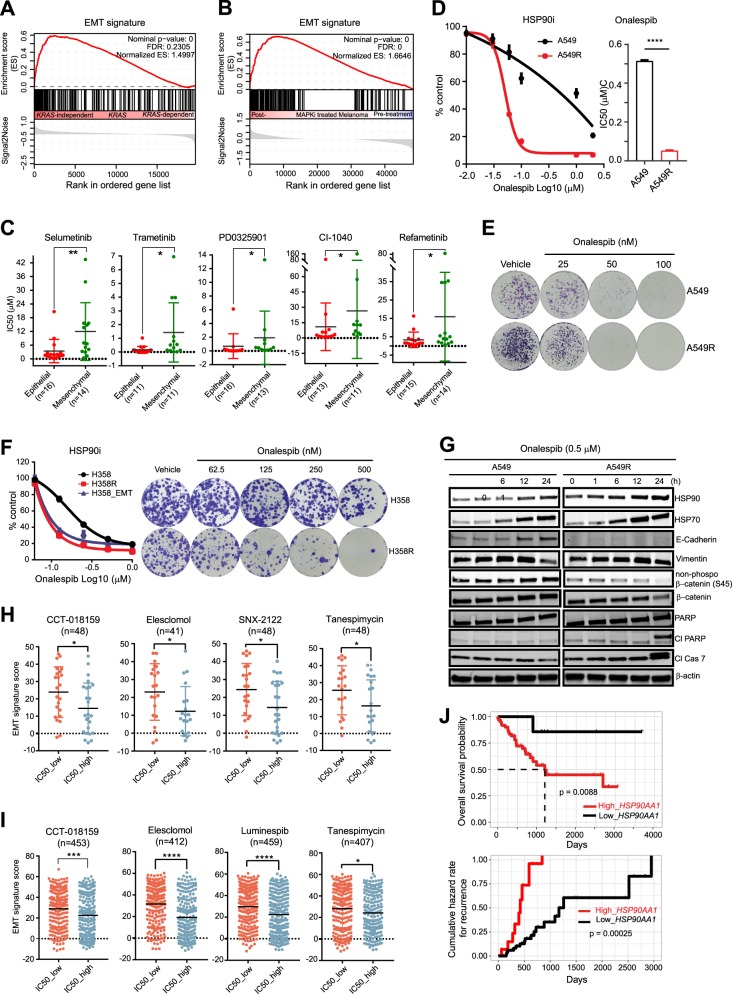
To delineate cellular and molecular mechanisms that underpin drug resistance in KRAS-mutant lung cancer, we modeled adaptive resistance by step-wisely and chronically treating A549 and H358 cells with chemotherapy drug pemetrexed (MTA) whereby resistant populations (A549R and H358R) persisted after the treatment. A549R and H358R cells showed cross-resistance to trametinib, indicating a shared mechanism of resistance to the two cancer drugs (Supplementary Fig. 1A–C). Drug-resistant A549R and H358R underwent profound changes in E-Cadherin or/and Vimentin compared to the parental cells (Supplementary Fig. 1A–C), which is characteristic of cell state switch through an epithelial-to-mesenchymal transition (EMT). Alike, de novo enforcement of mesenchymal reprogramming by treating A549 and H358 with transforming growth factor (TGF-β1) that fueled dedifferentiated subpopulations (A549_EMT, H358_EMT) co-opted acquired resistance to MTA and trametinib (Supplementary Fig. 1D, E). Supporting our results, analyzing transcriptomic data of KRAS-mutant cancer cells2 and matched tumors (pre- and post-treatment with BRAF and MEK inhibitors) from patients with BRAF-mutant melanoma15 revealed that EMT gene signatures are significantly enriched in KRAS-mutant cancer cells resistant to KRAS suppression (KRAS-independent) and in residual melanoma tumors refractory to and recurrent after MAPK (BRAF + MEK) inhibition (Fig. 1a, b). Importantly, the KRAS-mutant cancer cells independent of KRAS function, driven by mesenchymal reprogramming (high EMT gene signatures)2,16, exhibited significantly greater resistance to various MEK inhibitors (Fig. 1c). Mining the data from The Cancer Genome Atlas (TCGA) showed that EMT is positively correlated with tumor progression and predicts poor prognosis in KRAS-mutant lung cancer (Supplementary Fig. 1F, G). These results underscore the clinical relevance of our experimental models and uncover a common principle through which KRAS-mutant lung cancer cells acquire resistance to MTA and trametinib.
Fig. 1. HSP90 inhibitors selectively target KRAS-mutant lung cancer cells resistant to chemo and anti-MEK drugs.
a, b Gene set enrichment analysis (GSEA) of KRAS-mutant cancer cell lines (a) and matched tumor samples (pre- and post-treatment with MAPK (BRAF + MEK) inhibitors of patients with BRAF-mutant melanoma (b) revealed significant enrichment of EMT gene signatures in KRAS-independent cell lines (a) and in residual melanoma resistant to and recurrent after MAPK inhibition (b). The Gene Expression Omnibus (GEO) dataset GSE15126 (a) and GSE65185 (b) were used for GSEA. c Mesenchymal phenotype is inversely associated with sensitivity to MEK inhibitors in KRAS-mutant cancer cell lines. Dichotomization of epithelial and mesenchymal subgroups is based on an EMT gene signature defined by the GSE15126 dataset. *p < 0.05, **p < 0.01. d, e Dose–response curves (d) and clonogenic assay (e) of A549R (mesenchymal) and A549 (epithelial) cells using the HSP90 inhibitor onalespib. The half-maximal inhibitory concentration (IC50) is shown to the right (d). ****p < 0.0001, unpaired two-sided t-test. f Dose–response curves (left) and clonogenic assay (right) of mesenchymal (H358R, H358_EMT) and parental H358 cells using the HSP90 inhibitor onalespib. g Immunoblots of A549 and A549R cells treated with onalespib (0.5 µM) for the indicated time. h, i Association of the sensitivity to HSP90 inhibitors with mesenchymal cell states in lung adenocarcinoma (h) and solid cancer cell lines (i). Gene expression data and IC50 values are downloaded from the Cancer Cell Line Encyclopedia and the Genomics of Drug Sensitivity in Cancer, respectively. Dichotomization of IC50_low and IC50_high is based on the median IC50 value. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. j Kaplan–Meier analysis of KRAS-mutant lung cancer based on HSP90AA1 expression. Patients with high- (in red) or low-HSP90AA1 (in black) were stratified by optimal cutoff value of the HSP90AA1 across all patients using the surv_cutpoint function in the R “maxstat” package. Overall survival and cumulative hazard rates are analyzed and plotted using the R “survival” and “survminer” package. The p value is calculated by the log-rank test
HSP90 dictates adaptive resistance to MTA and trametinib in KRAS-mutant lung cancer cells
To identify cellular processes that represent selective dependencies of drug-resistant cells, we performed drug screening using small molecules targeting various pathways (Supplementary Table 1). Among the compounds tested, only onalespib and R428, inhibitors of the molecular chaperone HSP90 and the receptor tyrosine kinase (RTK) AXL, differentially impaired the resistant versus parental cells, causing over 10- and 3-fold reduction of the half-maximal inhibitory concentration (IC50) in A549R compared to A549, respectively (Fig. 1d, e; Supplementary Fig. 2A, B). In contrast, other compounds, including inhibitors of the RTKs (FGFR, EGFR) and KRAS effectors (PI3K, STAT3) previously reported to mediate resistance to MEK inhibitors12,17–20 showed no differential effect on A549R and A549. Similarly, PU-WS13, a selective inhibitor of the ER-resident HSP90 isoform GRP94 (ref. 21), was equal for A549R and A549 (Supplementary Fig. 2A), indicating that the selective effect of onalespib on A549R is due to the inhibition of the cytosolic HSP90. Further analyses using other HSP90 inhibitors (luminespib, ganetespib) and drug-resistant cells (A549_EMT, H358R, and H358_EMT) led to similar findings (Fig. 1f; Supplementary Fig. 2B–G).
HSP90 is moderately overexpressed in drug-resistant relative to parental cells, but HSP90 inhibition with onalespib (0.5 µM) selectively induced apoptosis in resistant cells only, marked by dramatic increase of cleaved poly(ADP-ribose) polymerase (Cl PARP) and caspase 7 (Cl Cas7) at 24 h (Fig. 1g; Supplementary Figs. 2E and 3A). Notably, onalespib blocked EMT in A549 and H358 cells, evidenced by upregulated E-Cad and non-phospho-β-catenin, but failed to do so in drug-resistant cells (Fig. 1g; Supplementary Figs. 2E and 3A), indicating that an intact HSP90 is essential for drug-resistant mesenchymal cell state. In agreement, the EMT gene signature is a biomarker that positively determines the sensitivity of KRAS-mutant and pan-solid cancer cells to various HSP90 inhibitors (Fig. 1h, i) and high levels of HSP90AA1 (HSP90) are associated with tumor progression, poor survival, and high recurrence rates in KRAS-mutant and pan-cancers (Fig. 1j; Supplementary Fig. 3B–E). These results identify HSP90 as a particular dependency of KRAS-mutant lung cancer cells acquiring resistance to MTA and trametinib.
Hyperactive AXL/eIF4E defines drug-resistant KRAS-mutant lung cancer cells
Next, we investigated signaling events that underlie HSP90 requirement in drug-resistant KRAS-mutant lung cancer cells. A549R and A549 cells displayed similar basal activities in RAF/MEK/ERK and PI3K/AKT/mTOR pathways, as were the cells treated with onalespib (Supplementary Fig. 3F), matching the results of drug screening using inhibitors of these pathways (Supplementary Fig. 2A). In contrast, the drug-resistant cells (A549R, H358R, and H358_EMT) overexpressed MYC (also known as c-MYC), MNK1 (MAP kinase-interacting serine/threonine-protein kinase 1), and eIF4E, and showed overwhelming phosphorylation (activation) in AXL (p-AXL), MNK1 (p-MNK1), and eIF4E (p-eIF4E). However, onalespib (0.5 µM) acutely reduced AXL, MYC, and p-eIF4E whereas the total eIF4E was not affected, as did R428 (Fig. 2a, b; Supplementary Fig. 3A). Notably, like onalespib, R428 induced apoptosis (Cl Cas7) in A549R only, whereas it blocked EMT by inducing a mesenchymal-to-epithelial transition (MET) in A549 (Fig. 2b). This effect was faithfully translated to an increased toxicity of R428 on drug-resistant cells (Fig. 2c). In agreement with other studies22,23, we demonstrated that AXL complexed with HSP90 (Supplementary Fig. 4A) and that AXL gene and protein levels were positively correlated with EMT (Supplementary Fig. 4B–D). Importantly, the AXL mRNA level is a predictive marker for poor prognosis in lung cancer patients (Supplementary Fig. 4E, F).
Fig. 2. Hyperactive AXL/MYC/eIF4E defines drug-resistant KRAS-mutant lung cancer cells.
a, b Immunoblots of A549 and A549R cells treated with onalespib (a) or R428 (b) for the indicated time. c–g Dose–response curves to R428 (c), JQ-1 (d), 4EGI-1 (e, g), and CGP57380 (f) assayed on resistant (A549R, A549_EMT, H358R, H358_EMT) and parental (A549, H358) cells. Representative results of clonogenic assays are shown to the right. h Immunoblots of A549R cells transfected with EIF4E-specific and control siRNAs. Growth curves of the transfected cells are shown to the right. i Dose–response curves to pemetrexed (MTA) and trametinib determined in A549R cells transfected with MYC- or EIF4E-specific and control shRNAs or siRNAs. j Clonogenic assay of A549R cells expressing EIF4E-specific or control siRNAs after treatment with the indicated doses of onalespib. k Kaplan–Meier analysis of a TCGA cohort of patients with KRAS-mutant lung cancer. Stratification of patients into high_ EIF4E (in red) and low_ EIF4E (in black) is based on the optimal cutoff value of EIF4E transcripts across all patients by using the surv_cutpoint function in R “maxstat” package. Overall survival curves and cumulative hazard rates were analyzed and plotted by using R “survival” and “survminer” packages. The p value is calculated using the log-rank test
Small-molecule inhibitors of MYC (JQ-1), MNK1 (CGP57380), and eIF4E/eIF4G complex (4EGI-1) more strongly suppressed the resistant than parental cells (Fig. 2d–g), as did MYC and EIF4E knockdown that barely affected cell proliferation but markedly reduced resistance to MTA and trametinib and augmented the sensitivity to onalespib (Fig. 2h–j; Supplementary Fig. 4G). Further, the MYC expression is positively associated with AXL, EIF4E, and EMT signatures (Supplementary Fig. 4H) and both mRNA and protein levels of the EIF4E are of prognostic significance in KRAS-mutant lung cancer, lung adenocarcinoma, and lung cancer (Fig. 2k; Supplementary Fig. 5A–C). These data, in line with a positive role for eIF4E in EMT24, establish AXL/MYC/eIF4E as central signaling nodes required for adaptive resistance in KRAS-mutant lung cancer cells.
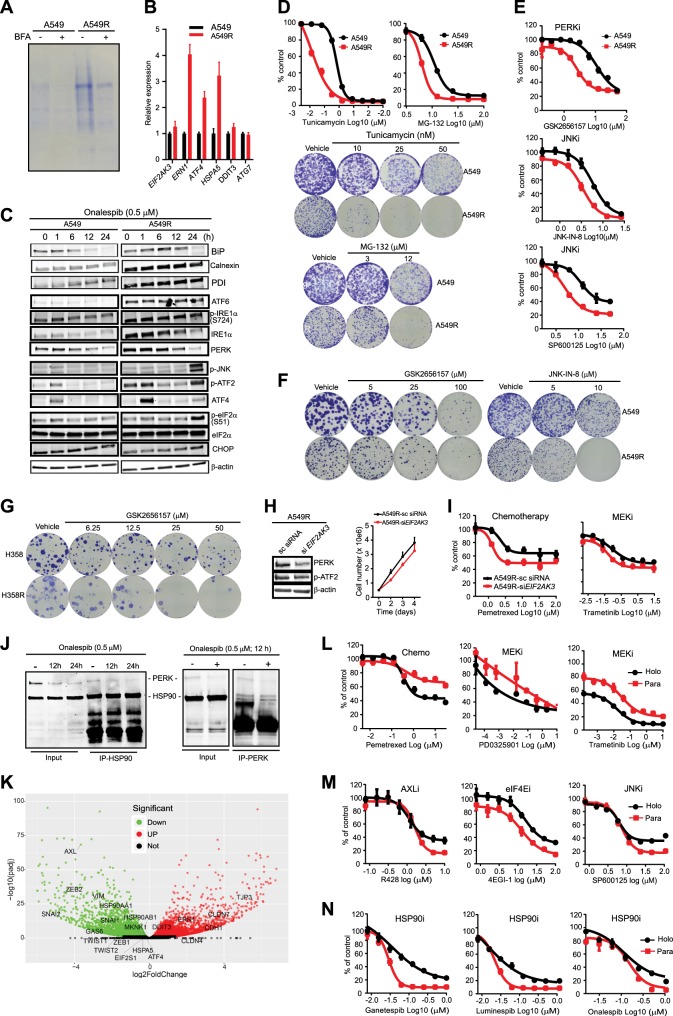
Deregulation of AXL/eIF4E activates an ER stress-relief UPR survival mechanism
To dissect the effector pathways downstream of AXL/MYC/eIF4E signaling, we conducted an integrative analysis of TCGA data, which significantly linked eIF4E to ER-related functions (Supplementary Fig. 5D–F). We therefore reasoned that deregulation of AXL/eIF4E induces ER stress and activates an ER stress-responsive mechanism. Indeed, The EMT status and AXL expression were significantly correlated with the UPR gene signature (Supplementary Fig. 5G, H). Importantly, compared to parental cells, A549R showed a higher magnitude of protein secretion (Fig. 3a), upregulated expression of UPR genes [HSPA5 (GRP78/BiP), EIF2AK3 (PERK), ERN1 (IRE1α), and ATF4 (ATF4)] (Fig. 3b), increased levels of ER chaperons (BiP, Calnexin, and PDI), ER stress sensors (ATF6, IRE1α, and PERK), and UPR effectors (p-JNK and p-ATF2) (Fig. 3c). In sharp contrast, eIF2α and CHOP that catalyze a malfunctional UPR and induce apoptosis25,26 were repressed in A549R (Fig. 3c). Consequently, further perturbations of ER homeostasis by treating with tunicamycin, MG-132, and thapsigargin that induce persistent ER stress preferentially impaired A549R (Fig. 3d; Supplementary Fig. 6A). Importantly, A549R and H358R were particularly susceptible to inhibitors of PERK (GSK2656157), JNK (JNK-IN-8, SP600125), but not of IREα (4µ8c), p38 (LY2228820), or eIF2α (Salubrinal), compared to the parental cells (Fig. 3e–g; Supplementary Fig. 6B). Genetic depletion of PERK reduced p-ATF2 and re-sensitized A549R cells to MTA and trametinib (Fig. 3h, i), whereas it induced an MET in A549 cells (Supplementary Fig. 6C). In line with these results, analyzing gene ontology terms and TCGA data significantly associated ATF2 with EMT, PERK with ATF2 and JNK but not with MAPK14 (p38) in KRAS-mutant lung cancer (Supplementary Fig. 6D–F).
Fig. 3. AXL/eIF4E-regulated ER stress-relief UPR underpins drug resistance and intratumor heterogeneity in KRAS-mutant lung cancer.
a A549 and A549R cells without or with brefeldin A (5 µg/ml) treatment (6 h) are profiled for secreted proteins. Shown are representative data of three independent experiments (n = 3). b Transcriptional quantification of UPR genes in A549R and A549 cells by qRT-PCR. c Immunoblots of A549 and A549R cells treated onalespib (0.5 µM) for the indicated time. d–g Dose–response curves to tunicamycin (d), MG-132 (d), and inhibitors to PERK and JNK (e–g) assayed on resistant (A549R, H358R) versus parental (A549, H358) cells. Representative results of clonogenic assays are shown to the right. h, i A549R cells transfected with PERK-specific and control siRNAs were subjected to immunoblotting (h) and dose–responsive curves to pemetrexed and trametinib (i). Growth curves of the transfected cells are also shown (h). j Protein lysates (200 μg) prepared from A549R cells treated with onalespib or vehicle were subjected to coimmunoprecipitation with HSP90 and PERK antibodies. Proteins in the precipitates (IP) and the starting material (input) were visualized by western blotting. k Volcano plots of transcriptomic profiling (RNAseq) of the A549_holo and A549_para clones. The X-axis is the fold-change (log2) of individual genes in the holoclone versus paraclone. Genes significantly downregulated (adjusted p value <0.05) in the holo compared to paraclones are shown in green and genes significantly upregulated in the A549_holo versus A549_para are marked in red. Note that HSP90AA1 and HSP90AB1 (encoding HSP90) and the genes involved in EMT, AXL signaling, and the UPR are enriched in the A549_para clone. l–n Dose–response curves to MTA and the MEK inhibitors PD0325901 and trametinib (l), inhibitors of AXL, eIF4E, and JNK (m) and HSP90 (n) as determined on A549_holo_clone (black) and A549_para_clone (red) subpopulations. Data are mean ± s.d. of three biological replicates (n = 3)
R428 strikingly increased p-eIF2α and CHOP on top of the induction of apoptosis (cleaved caspase 7), indicating that hyperactive AXL/eIF4E signaling is functionally important for PERK/JNK/ATF2-dependent UPR (Fig. 2b). Similarly, EIF4E knockdown perturbed the UPR by substantially inducing p-ATF2 and p-eIF2α (Fig. 2h). Thus, deregulation of AXL/eIF4E in drug-resistant cancer cells invokes ER stress, which in turn activates a stress-relief UPR mediated by the PERK/JNK/ATF2 cascade.
HSP90 controls PERK/JNK/ATF2 integrity and protects from a malfunctional UPR
Next, we sought evidence that connects PERK/JNK/ATF2-dependent UPR with HSP90. Blocking HSP90 function with onalespib profoundly interfered with ER stress responses, measured by dramatic decrease of the PERK and IREα protein levels27 (Fig. 3c), and marked inhibition of the HSP90/PERK complex formation (Fig. 3j) in A549R cells. Notably, onalespib treatment enormously upregulated ER stress-induced UPR genes (Supplementary Fig. 7A) and pro-apoptotic UPR effectors (p-eIF2α and CHOP) at 24 h (Fig. 3c), a time coincident with the induction of apoptotic markers (Cl PARP, Cl Cas7) (Fig. 1g). These results indicate that an intact HSP90 is critical for PERK/JNK/ATF2-dependent UPR that is pro-survival and that HSP90 blockage tips the balance from survival towards apoptosis by turning on a malfunctional UPR25,26 executed by the eIF2α/CHOP axis. In agreement, the UPR gene signature is significantly correlated with unfavorable clinical outcomes in patients with lung and pan-cancers (Supplementary Fig. 7B–E).
HSP90/AXL/eIF4E-regulated UPR regulates de novo intratumor heterogeneity of KRAS-mutant lung cancer cells
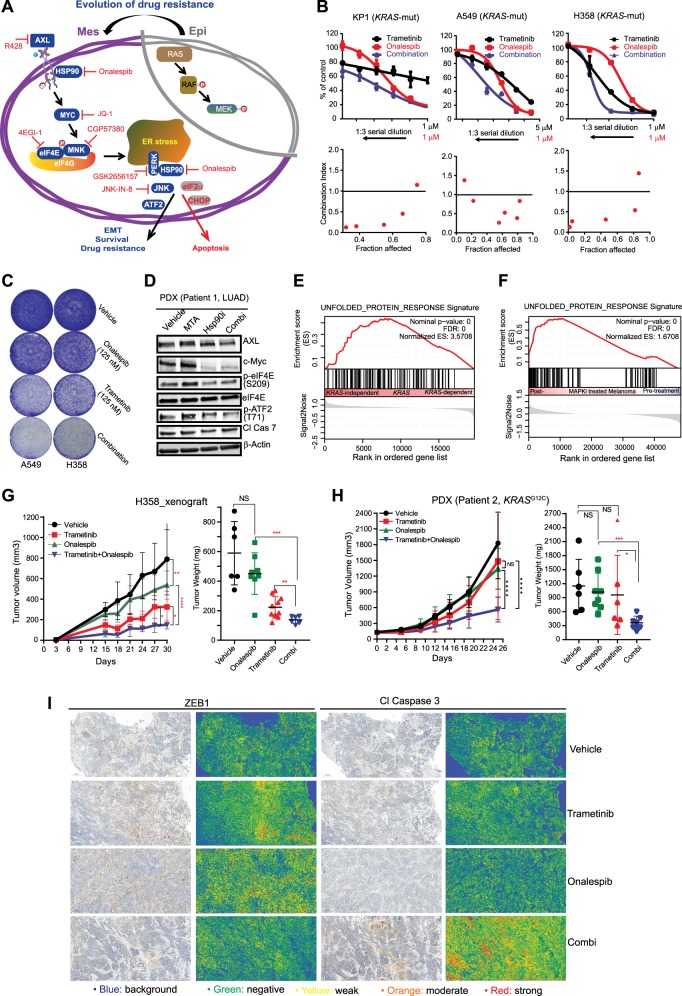
Our models recapitulate tumor progression upon cancer drugs, which might differ from that occurring under pathological circumstances such as de novo intratumor heterogeneity. A549 cells displayed marked heterogeneity28–30, evidenced by coexistence of epithelial (holoclone) and mesenchymal (paraclone) subpopulations under standard culture conditions (Supplementary Fig. 8A). Transcriptomic profiling of the A549 holoclone and paraclone30 revealed overexpression of epithelial [CDH1 (E-Cadherin), TJP3 (tight junction protein ZO-3), CLDN4/7 (epithelial tight junction proteins Claudin 4/7)], and mesenchymal (SNAI2, ZEB2 and VIM) markers, respectively. Notably, numerous genes encoding key components of the resistance pathway, most prominently GAS6 [encoding growth arrest-specific protein 6 (GAS-6), a cognate ligand of AXL], AXL (AXL), MKNK1 (MNK1), HSP90AA1, and HSP90AB1 that encode the cytosolic isoforms HSP90α and HSP90β, respectively, were expressed at significantly higher levels in A549 paraclones than the holoclone (Fig. 3k). Importantly, the A549 paraclone that displayed heightened resistance to MTA and MEK inhibitors (trametinib, PD0325901) was highly susceptible to inhibitors of AXL, eIF4E, JNK, and HSP90 compared to the A549 holoclone (Fig. 3l–n). Thus, HSP90/AXL/eIF4E-regulated UPR that underpins adaptive resistance to MTA and trametinib also regulates intratumor heterogeneity of KRAS-mutant lung tumor cells that occurs under pathological conditions, extrapolating our findings to a generalized principle that governs tumor progression (Fig. 4a).
Fig. 4. HSP90 antagonism enhances antitumor effects of MTA and trametinib in preclinical cancer models.
a Schematic overview of acquired vulnerabilities resulting from drug resistance. Epi epithelial cell state, Mes mesenchymal cell state. b Growth inhibition of KRAS-mutant murine (KP1) and human (A549, H358) lung adenocarcinoma cell lines after treated with trametinib, MTA, and onalespib, alone or in combinations. Each drug was dosed at the indicated concentrations for single treatment or mixed following threefold serial dilutions for combination treatment. Data are mean ± s.d. of three biological replicates (n = 3). Plots of fraction affected (Fa) versus combination index (CI) was determined by the CompuSyn software. CI < 1 indicates synergism. c HSP90 inhibition enhances trametinib effectiveness by preventing adaptive drug resistance. H358 and A549 cells treated (24 h) with onalespib and trametinib, alone or in combination, were washed with PBS and subjected to further culture for additional 10 days in the absence of the drug. d Immunoblots of a lung adenocarcinoma patient (patient 1)-derived xenograft tumors cultured ex vivo and treated with vehicle, pemetrexed (1 µM), and onalespib (0.5 µM), alone or in combination for 24 h. e, f GSEA of KRAS-mutant cancer cell lines (e) and matched tumor samples (pre- and post-treatment with MAPK inhibitors) of patients with BRAF-mutant melanoma (f) show significant enrichment of UPR gene signatures in KRAS-independent cell lines (e) and in residual melanoma that survived BRAF inhibitors (f). The GEO dataset GSE15126 and GSE65185 were used for the analysis. g, h Volumes of H358 xenografts (g) and a KRAS-mutant lung cancer patient (patient 2)-derived xenograft model (h) treated with vehicle or the indicated drugs. *p < 0.05, **p < 0.01, ****p < 0.0001 by two-way ANOVA. Tumor weights at the end of the treatment are shown to the right. *p < 0.05, **p < 0.01, ****p < 0.0001 by unpaired two-sided t-test. i Immunohistochemistry (IHC) analysis for EMT (ZEB1) and apoptosis (Cl caspase 3) in H358 xenograft tumors after treated with the indicated drugs. Original overall magnification ×100 (scale bar: 200 μM). Images were taken and processed using CaseViewer software. Both the original IHC slides and the corresponding slides with gradient map visualization were shown. Blue: insignificant (background); green: moderate significant; yellow: significant; orange: more significant; red: most significant
HSP90 blockage enhances antitumor effects of MTA and trametinib in lung cancer models
Our finding that HSP90/AXL/eIF4E-regulated UPR fosters tumor evolution and heterogeneity provides a conceptual framework for developing rational therapeutic strategies to treat KRAS-mutant lung cancer. As proof of concept, we tested if antagonizing HSP90 enhances antitumor effects of MTA and trametinib. Onalespib strongly synergized with trametinib and MTA in inhibiting the proliferation of KRAS-mutant A549, H358, and murine KP cells but not of KRAS wild-type H3122 and PC9 cells (Fig. 4b; Supplementary Fig. 8B, C). Consistently, short-term exposure (24 h) to the combined treatment with trametinib and onalespib dramatically suppressed A549 and H358 cell growth to a much greater extent than single drug alone (Fig. 4c). Ex vivo organotypic culture and treatment (24 h) of patient-derived xenograft (PDX) tumors revealed that MTA activated the adaptive resistance mechanism, gauged by marked increase of MYC, p-eIF4E, and p-ATF2, which, however, was effectively counteracted by addition of onalespib (Fig. 4d). In agreement, analyzing transcriptomic data from previous studies2,15 revealed that the UPR gene signature was significantly enriched in KRAS-mutant cancer cells that underwent an EMT and acquired KRAS independence (Fig. 4e) and in residual BRAF-mutant melanoma resistant to and relapsed after the concurrent treatment with BRAF and MEK inhibitors (Fig. 4f). These results underscore the ability of MAPK inhibitors to activate the UPR in preclinical models and cancer patients.
Finally, we assessed in vivo efficacy of the combined treatment with trametinib and onalespib, both of which were administrated at doses significantly below clinically achievable levels9,31. The combination showed potent antitumor effects in H358 xenografts and a PDX model of KRAS-mutant lung cancer without apparent toxicities (Fig. 4g, h; Supplementary Fig. 8D). Importantly, residual tumors after the combination therapy showed augmented apoptosis (Cl Cas3) and reduced proliferation (Ki-67), accompanied by marked attenuation of adaptive properties, e.g., mesenchymal phenotypes (ZEB1), compared to those treated with trametinib alone (Fig. 4i; Supplementary Fig. 8E). These results interrogate our in vitro and ex vivo results and the clinical data of patients, emphasizing the importance to investigate and harness acquired dependency (collateral sensitivity) resulting from tumor evolution and heterogeneity to treat KRAS-mutant lung cancer and perhaps other malignancies as well.
Discussion
In this study, we uncovered that AXL/eIF4E-regulated UPR signaling network, with HSP90 at the helm of the regulatory hierarchy, is an acquired dependency of and confers a selective vulnerability in KRAS-mutant lung cancer cells resistant to chemo and anti-MEK cancer drugs. This non-genetic mechanism, unprecedented for MTA resistance31 and different from previously reported ones that contribute to adaptive resistance to MEK inhibitors10–12,17–20, is clinically relevant in that it underpinned KRAS-independent survival of KRAS-mutant cancer cells and was activated in patients’ tumors that had acquired resistance to MEK inhibitors. Notably, this collateral sensitivity induced by drug resistance is also applicable to KRAS-mutant lung cancer cells that have undergone intratumor heterogeneity under pathological circumstances. Therefore, pharmacological and/or genetic blockage of key nodes of the HSP90/AXL/eIF4E/UPR signaling cascade selectively induces apoptotic cell death of drug-resistant cancer cells, limits tumor heterogeneity in vitro, and precludes the emergence of adaptive phenotypes in vivo.
Our results provide a strong rationale for combining current chemo and anti-MEK cancer drugs with inhibitors of the HSP90/AXL/eIF4E/UPR pathway to treat KRAS-mutant lung cancer and perhaps other malignancies for which no effective therapy available for advanced diseases. As preclinical proof of concept, drug combinations of clinically advanced HSP90 inhibitors32 with trametinib or MTA potently suppressed cancer cell viability and tumor growth in various tumor models, including a PDX model of KRAS-mutant lung adenocarcinoma, validating a long-sought and readily implemented therapy for the most devastating subset of lung cancer. It is also conceivable that HSP90/AXL/eIF4E signaling strength, ER stress magnitudes, and UPR status may stratify subsets of patients with KRAS-mutant lung cancer who likely benefit from the combination therapy.
Materials and methods
Cell culture and compounds
KRAS-mutant (A549, H358) and KRAS wild-type (H3122 and PC9) lung cancer cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Murine KRASG12D; p53−/− cells (KP) were derived from a murine lung adenocarcinoma generated as previously described33. Cells were cultured in RPMI-1640 (Sigma-Aldrich, St. Louis, MO, USA), supplemented with 10% fetal bovine seurm (FBS) (Life Technologies, Grand Island, NY, USA) and 1% penicillin/streptomycin (Sigma-Aldrich) at 37 °C in a humid incubator with 5% CO2. All cell lines were authenticated by DNA fingerprinting and negative for mycoplasma. Chemical inhibitors used in this study, including those targeting AXL34, MYC35, eIF4E36,37, and PERK38, are listed in Supplementary Table 1.
In vitro modeling of drug resistance
Drug-resistant populations (A549R and H358R) were generated by chronical treatment (6 months) of A549 and H358 cells with stepwise incremental concentrations of MTA (0.1–1.0 µM for A549; 0.5–10 µM for H358) as previously reported31,39. Mesenchymal populations (A549_EMT, H358_EMT), the cellular products emanating from mesenchymal reprogramming of A549 and H358 cells, were obtained after 3-month treatment (5 ng/ml for A549; 2.5 ng/ml for H358) with TGF-β1 (AF-100-21C; Peprotech, London, UK).
Cell viability assay and quantitative analysis of drug synergy
Cells seeded in triplicate at 96-well plates (1000–1500 cells/well) were drugged 24 h later with various compounds as indicated. Cell viability was determined 72 h post-treatment by the Acid Phosphatase Assay Kit (ab83367; Abcam, Cambridge, UK) according to the manufacturer’s protocol. Drug synergism was determined by CompuSyn software, which is based on the median-effect principle and the combination index–isobologram theorem40. CompuSyn software generates fraction affected (Fa) and combination index (CI) values. CI < 1.0, synergism; CI = 1.0, additive effects; CI > 1.0, antagonism.
Clonogenic assay
Cells seeded in triplicate at 103–104 cells/well (six-well plates) were allowed to adhere overnight before treated with various inhibitors or vehicle control for 24 h. The treated cells were subsequently cultured in drug-free medium for 7–14 days depending on growth rates, with culture media replaced every 3–4 days. Surviving cells were then fixed with methanol (1%) and formaldehyde (1%), visualized by staining with crystal violet (0.5% dissolved in 25% methanol). All experiments were performed in three biological replicates and shown were representative experiments.
Immunoblotting, immunoprecipitation, and immunohistochemistry
Cell lysates were prepared using RIPA buffer (Cell Signaling Technology, Hercules, CA, USA) containing protease inhibitor cocktail and phosphatase inhibitors (Santa Cruz, Dallas, TX, USA). Equal amounts of total proteins were separated by SDS-PAGE (4561033; Bio-Rad) and transferred to nitrocellulose membranes (170-4156; Bio-Rad). After brief incubation with blocking buffer (927-4000; Li-COR Biosciences, Bad Homburg, Germany) at room temperature, the membranes were blotted with primary antibodies (Supplementary Table 2) and anti-rabbit (926–32211) or anti-mouse (926-68020) secondary antibodies (Promega, Madison, WI, USA). Membrane-bound secondary antibodies were visualized by the Odyssey Infrared Imaging System (Li-COR Biosciences).
For immunoprecipitation, precleared cell lysates (200 µg) and monoclonal anti-mouse HSP90 or anti-rabbit PERK antibodies conjugated with Protein G (37478 P) or Protein A agarose beads (9863 P) were mixed by gentle rocking overnight at 4 °C. The beads were washed five times with lysis buffer and bound proteins were eluted by SDS sample buffer. Proteins in the precipitates were analyzed by immunoblotting.
For immunohistochemistry, tissues were fixed in formalin, embedded in paraffin and cut into 4 μm sections. Sections were stained with hematoxylin and eosin and subjected to immunohistochemical staining following standard protocols. Visualization was performed using the Bond Polymer Refine Detection kit (Leica Biosystems, Newcastle, UK) as instructed by the manufacturer. Images were acquired using PANNORAMIC® whole slide scanners and processed using CaseViewer (3DHISTECH Ltd, Budapest, Hungary). In parallel, the staining intensity was shown as blue (background), green (negative), yellow (weak), orange (moderate), and red (strong) using the gradient map visualization plugin within CaseViewer. The following primary antibodies were used: Ki-67 (M7240; Dako, Santa Clara, CA, USA), ZEB1 (HPA027524; Sigma), and cleaved caspase 3 (9664; Cell Signaling Technology).
qRT-PCR
Total RNA was isolated and purified using RNeasy Mini Kit (74106; Qiagen, Hilden, Germany). Complementary DNA (cDNA) was synthesized by the High capacity cDNA reverse transcription kit (4368814; Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. Real-time PCR was performed in triplicate on a 7500 Fast Real-Time PCR System (Applied Biosystems) using TaqMan primer/probes (Applied Biosystems): HSPA5, Hs00607129_gH; ERN1, Hs00980095_m1; EIF2AK3, Hs00984003_m1; ATF4, Hs00909569_g1; DDIT3, Hs00358796_g1; BBC3, Hs00248075_m1, and ATG7, Hs00893766_m1. GAPDH (Hs02786624_g1) and ACTB (Hs01060665_g1) were used as endogenous normalization controls.
Gene silencing by small interfering (siRNA) and short-hairpin RNAs (shRNA)
Cells cultured in triplicate at 50–70% confluency were transfected using SiTran1.0 (TT300001; Origene Technologies, Rockville, MD, USA) according to the manufacturer’s protocol. EIF4E and EIF2AK3 were knocked down by specific pooled siRNA duplexes (SR320018 and SR306267; OriGene Technologies), with control siRNA Duplex as a negative control.
Knockdown of MYC was achieved via lentiviral delivery of MYC Human shRNA Plasmid Kit (TL311323; OriGene Technologies). A scrambled shRNA was used as a control. Lentiviral particles were generated and cells infected according to the protocol from Broad Institute. The supernatant containing lentiviruses was collected, filtered through 0.45 µm filters, and stored in aliquots at −80 °C, or immediately used to infect recipient cells. After infection, cells were selected in puromycin (1.5 µg/ml) and further passaged in culture for functional assays.
Secretion assay
5 × 106 cells seeded in 10-cm dishes were cultured in FBS-free media for 6 h, with or without treated with 5 µg/ml Brefeldin A (B6542; Sigma-Aldrich). Proteins in the supernatant were precipitated by trichloroacetic acid, separated by SDS-PAGE and visualized by Coomassie blue staining.
Mining public databases (TCGA, CCLE, GDSC, and GEO), GSEA, and gene ontology terms analysis
Transcriptomic and reverse phase protein array data of cancer patients were obtained from the Cancer Genome Atlas (TCGA). The gene expression and corresponding survival data were extracted for correlation and prognostic analysis using the corresponding packages in R (“corrplot” and “Hmisc” packages for correlation analysis; “maxstat”, “survival”, and “survminer” packages for prognostic analysis). Analysis of the enriched biological pathway, process, and molecular function from Gene Ontology, as well as gene interaction map, were performed using Funrich software41. The expression data and drug sensitivity for cancer cell lines were obtained from the Cancer Cell Line Encyclopedia (CCLE)42 and Genomics of Drug Sensitivity in Cancer (GDSC)43, respectively. Transcriptomic data of KRAS-dependent and -independent cancer cell lines (GSE15126)2, matched melanoma pre- and post-treated with MAPK inhibitors (GSE65185)15 were downloaded from the Gene Expression Omnibus (GEO) database44. Gene set enrichment analysis (GSEA) was performed using GSEA software45.
EMT and UPR gene signatures
The EMT status of tumors and cell lines was determined by the EMT gene signature18, scored as the sum of a mesenchymal gene set (FN1 + VIM + ZEB1 + ZEB2 + TWIST1 + TWIST2 + SNAI1 + SNAI2 + CDH2) minus that of epithelial genes (CLDN4 + CLDN7 + TJP3 + MUC1 + CDH1). The UPR gene signature was calculated using the sum of gene expression of MMP11, FN1, METRN, BGN, SLC7A5, SPP1, CREB3L1, HIST1H1C, COL5A2, ARNT2, COL1A2, TMED3, MTHFD2, SLC2A6, IER3 + CDH11, CTSD, SLC20A1, PDIA4, COL12A1, ECM1, MAGED2 and PTRH1 as previously described46.
Patient samples
Experiments with surgically resected tumor specimens from lung cancer patients (patient #1, 57-year-old female, lung adenocarcinoma; patient #2, 67-year-old male, KRAS-mutant lung adenocarcinoma) were approved by the institutional review board, with informed consent obtained from all patients as per protocol.
Animal studies
All mouse experiments approved by ethics committee were performed in accordance with institutional regulations governing animal care. Age- and gender-matched NSG (NOD-SCID IL2Rγnull) mice were used for animal experiments with human cell lines and PDXs. For H358 xenografts, tumor cells mixed with Matrigel (356231; Corning) were subcutaneously inoculated in left and right flanks. For the PDX model, tumor tissues were cut into small pieces (5 µm × 5 µm) and inserted into a subcutaneous pocket. Treatment was initiated when tumors were palpable and drugs were prepared in the following solvent: onalespib (2% DMSO + 30% PEG 300 in H2O) and trametinib (4% DMSO in Corn Oil). Trametinib (0.3 mg/kg for H358 xenografts, 0.5 mg/kg for PDX) was administered 5 days/week (p.o.), and onalespib (15 mg/kg for H358 xenografts and 30 mg/kg for PDX) twice/week (i.p.). Tumor size/volume was calculated by the formula: (D × d2)/2, where “D” refers to the long tumor diameter and “d” the short one.
Ex vivo organotypic culture
Ex vivo organotypic culture and treatment were processed as previously described39,47. In brief, freshly explanted PDX tumors were mounted on agarose, soaked in ice-cold sterile phosphate-buffered saline containing antibiotic/antimycotic, and cut into slices (300–500 μM) by Vibratome VT1200 (Leica Biosystems). Tissue slices were placed in ultra-low attachment plates (3471; Corning Incorporated, Corning, NY, USA) and covered by Dulbecco's modified Eagle's medium medium supplemented with 20% FBS and 1% penicillin/streptomycin solution. After overnight culturing at 37 °C in a humid atmosphere with 5% CO2 and 95% air, drug treatment was initiated and lasted for up to 72 h.
Statistical analysis
Data were presented as mean ± s.d., with the indicated sample size (n) representing biological replicates. Data analysis was performed by GraphPad Prism 7 (GraphPad Software, Inc., San Diego, CA, USA). Group size was determined based on preliminary experiments but no statistical method was used to predetermine sample size. Group allocation was performed in a blinded manner. All samples that met proper experimental conditions were included in the analysis. Gene expression and survival data derived from the public database, as well as correlation coefficient (Pearson and Spearman), were analyzed using R (version 3.4.3). For survival analysis, patients were grouped by gene expression, where “high” and “low” expression groups were stratified by the optimal cutoff value. Statistical significance was determined by one-way/two-way analysis of variance (ANOVA), Bonferroni’s multiple comparison test, and Student’s t-test using GraphPad Prism 7, unless otherwise indicated. P < 0.05 was considered statistically significant.
Supplementary information
Acknowledgements
We thank Lung Cancer Center, Bern University Hospital and Tissue Bank Bern, Institute of Pathology, University of Bern, in acquiring patient tissues, Ursina Lüthi (Department of Tumor Immunology, Bern University Hospital) for assistance of PDXs, Erik Vassella (Institute of Pathology, University of Bern) for mutational profiling of patients’ tumors. This work was supported by Swiss Cancer League (#KFS-3772-08 2015; to R-W.P.), Cancer League of the Canton of Bern (to R-W. P.), Ph.D. fellowships from China Scholarship Council (H.Y., Z.Y., and Y.G.).
Author contributions
H.Y. and S-Q.L. performed experiments, analyzed data, and wrote the paper. D.X., Z.Y., T.M., and Y.G. performed experiments and analyzed data. G.J.K. and H.Z. provided clinical samples and experimental materials. R.A.S. provided clinical samples, financial resources, and supervised the study; R.-W.P. conceived the project, analyzed data, supervised the study, and wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ralph A. Schmid, Phone: +41(0)31 632 23 64, Email: Ralph.Schmid@insel.ch
Ren-Wang Peng, Phone: +41(0)31 632 40 81, Email: Renwang.Peng@insel.ch.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41389-019-0158-7).
References
- 1.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh A, et al. A gene expression signature associated with “K-Ras Addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janne PA, et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 2013;14:38–47. doi: 10.1016/S1470-2045(12)70489-8. [DOI] [PubMed] [Google Scholar]
- 4.Liang SQ, et al. mTOR mediates a mechanism of resistance to chemotherapy and defines a rational combination strategy to treat KRAS-mutant lung cancer. Oncogene. 2019;38:622–636. doi: 10.1038/s41388-018-0479-6. [DOI] [PubMed] [Google Scholar]
- 5.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: mission possible? Nat. Rev. Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan DA, Giaccia AJ. Harnessing synthetic lethal interactions in anticancer drug discovery. Nat. Rev. Drug Discov. 2011;10:351–364. doi: 10.1038/nrd3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samatar AA, Poulikakos PI. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat. Rev. Drug Discov. 2014;13:928–942. doi: 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- 9.Blumenschein GR, et al. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC) Ann. Oncol. 2015;26:894–901. doi: 10.1093/annonc/mdv072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lito P, et al. Disruption of CRAF-mediated mek activation is required for effective MEK inhibition in kras mutant tumors. Cancer Cell. 2014;25:697–710. doi: 10.1016/j.ccr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamba S, et al. RAF suppression synergizes with MEK inhibition in KRAS mutant cancer cells. Cell Rep. 2014;8:1475–1483. doi: 10.1016/j.celrep.2014.07.033. [DOI] [PubMed] [Google Scholar]
- 12.Manchado E, et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature. 2016;534:647–651. doi: 10.1038/nature18600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith MP, Wellbrock C. Molecular pathways: maintaining MAPK inhibitor sensitivity by targeting nonmutational tolerance. Clin. Cancer Res. 2016;22:5966–5970. doi: 10.1158/1078-0432.CCR-16-0954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mandal R, Becker S, Strebhardt K. Stamping out RAF and MEK1/2 to inhibit the ERK1/2 pathway: an emerging threat to anticancer therapy. Oncogene. 2016;35:2547–2561. doi: 10.1038/onc.2015.329. [DOI] [PubMed] [Google Scholar]
- 15.Hugo W, et al. Non-genomic and immune evolution of melanoma acquiring MAPKi resistance. Cell. 2015;162:1271–1285. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao DD, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun C, et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep. 2014;7:86–93. doi: 10.1016/j.celrep.2014.02.045. [DOI] [PubMed] [Google Scholar]
- 18.Kitai H, et al. Epithelial-to-mesenchymal transition defines feedback activation of receptor tyrosine kinase signaling induced by MEK inhibition in KRAS-mutant lung cancer. Cancer Discov. 2016;6:754–769. doi: 10.1158/2159-8290.CD-15-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee HJ, et al. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell. 2014;26:207–221. doi: 10.1016/j.ccr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 20.Salt MB, Bandyopadhyay S, McCormick F. Epithelial-to-mesenchymal transition rewires the molecular path to PI3K-dependent proliferation. Cancer Discov. 2014;4:186–199. doi: 10.1158/2159-8290.CD-13-0520. [DOI] [PubMed] [Google Scholar]
- 21.Patel PD, et al. Paralog-selective Hsp90 inhibitors define tumor-specific regulation of HER2. Nat. Chem. Biol. 2013;9:677–684. doi: 10.1038/nchembio.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boshuizen J, Koopman LA, Krijgsman O, Shahrabi A, Gresnigt-van den Heuvel E, Ligtenberg MA, et al. Cooperative targeting of melanoma heterogeneity with an AXL antibody-drug conjugate and BRAF/MEK inhibitors. Nat. Med. 2018;24:203–212. doi: 10.1038/nm.4472. [DOI] [PubMed] [Google Scholar]
- 23.Krishnamoorthy GP, et al. Molecular mechanism of 17-allylamino-17-demethoxygeldanamycin (17-AAG)-induced AXL receptor tyrosine kinase degradation. J. Biol. Chem. 2013;288:17481–17494. doi: 10.1074/jbc.M112.439422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robichaud N, et al. Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene. 2015;34:2032–2042. doi: 10.1038/onc.2014.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hetz C, Papa FR. The unfolded protein response and cell fate control. Mol. Cell. 2018;69:169–181. doi: 10.1016/j.molcel.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 26.Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006;7:880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcu MG, et al. Heat shock protein 90 modulates the unfolded protein response by stabilizing IRE1 alpha. Mol. Cell Biol. 2002;22:8506–8513. doi: 10.1128/MCB.22.24.8506-8513.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye XQ, et al. Mitochondrial and energy metabolism-related properties as novel indicators of lung cancer stem cells. Int. J. Cancer. 2011;129:820–831. doi: 10.1002/ijc.25944. [DOI] [PubMed] [Google Scholar]
- 29.Andriani F, et al. Conversion to stem-cell state in response to microenvironmental cues is regulated by balance between epithelial and mesenchymal features in lung cancer cells. Mol. Oncol. 2016;10:253–271. doi: 10.1016/j.molonc.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tieche CC, et al. Tumor initiation capacity and therapy resistance are differential features of EMT-related subpopulations in the NSCLC cell line A549. Neoplasia. 2019;21:185–196. doi: 10.1016/j.neo.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang SQ, et al. Blocking the epithelial-to-mesenchymal transition pathway abrogates resistance to anti-folate chemotherapy in lung cancer. Cell Death Dis. 2015;6:e1284. doi: 10.1038/cddis.2015.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Canella A, et al. Efficacy of Onalespib, a long-acting second-generation HSP90 inhibitor, as a single agent and in combination with Temozolomide against malignant gliomas. Clin. Cancer Res. 2017;23:6215–6226. doi: 10.1158/1078-0432.CCR-16-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scaltriti M, Elkabets M, Baselga J. Molecular pathways: AXL, a membrane receptor mediator of resistance to therapy. Clin. Cancer Res. 2016;22:1313–1317. doi: 10.1158/1078-0432.CCR-15-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:903–916. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graff JR, et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J. Clin. Invest. 2007;117:2638–2648. doi: 10.1172/JCI32044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moerke NJ, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 38.Wang M, Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat. Rev. Cancer. 2014;14:581–597. doi: 10.1038/nrc3800. [DOI] [PubMed] [Google Scholar]
- 39.Xu D, et al. Increased sensitivity to apoptosis upon endoplasmic reticulum stress-induced activation of the unfolded protein response in chemotherapy-resistant malignant pleural mesothelioma. Br. J. Cancer. 2018;119:65–75. doi: 10.1038/s41416-018-0145-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 41.Pathan M, et al. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 42.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang WJ, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barrett T, et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng YX, et al. Cancer-specific PERK signaling drives invasion and metastasis through CREB3L1. Nat. Commun. 2017;8:1079. doi: 10.1038/s41467-017-01052-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaira V, et al. Preclinical model of organotypic culture for pharmacodynamic profiling of human tumors. Proc. Natl Acad. Sci. USA. 2010;107:8352–8356. doi: 10.1073/pnas.0907676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.