Abstract
Large-scale expression profiling of micro-RNAs (miRNAs) in bovine granulosa cells from dominant and subordinate follicles on Day 19 of the estrous cycle revealed enriched micro-RNA-183-96-182 cluster miRNAs in preovulatory dominant follicles that coordinately regulate the forkhead box protein O1 (FOXO1) gene. However, little is known about the role of this cluster in bovine granulosa cell function. We used an in vitro granulosa cell culture model to investigate this role. Granulosa cells aspirated from small growing follicles (3–5 mm in diameter) were cultured in Dulbecco modified Eagle medium/F-12 medium supplemented with fetal bovine serum and transfected with locked nucleic acid-based miRNA mimics, inhibitors, and corresponding negative controls. Overexpression of the miRNA cluster resulted in suppression of FOXO1 mRNA and protein, whereas inhibition of the cluster increased expression of FOXO1 mRNA. Overexpression also increased the relative rate of cell proliferation, whereas inhibition slowed it down. Similarly, the proportion of cells under G0/G1 arrest declined, whereas the ratio of cells in S phase increased in response to miR-183-96-182 overexpression. Selective knockdown of FOXO1 mRNA using anti-FOXO1 small interfering RNA increased the rate of granulosa cell proliferation, decreased the proportion of cells under G0/G1 arrest, and increased the proportion of cells in the S phase of cell cycle. Our data suggest that miR-183-96-182 cluster miRNAs promote proliferation and G1/S transition of bovine granulosa cells by coordinately targeting FOXO1, suggesting a critical role in granulosa cell function. MicroRNA-183-96-182 cluster regulates bovine granulosa cell function by targeting FOXO1 gene.
Keywords: cell cycle, cell proliferation, FOXO1, granulosa cell, miR-183-96-182 cluster
Introduction
Bovine ovarian follicular growth is a complex process in which cohort follicles of various sizes undergo a series of maturational waves accompanied by dramatic changes in the shape and function of granulosa cells [1]. Among the factors involved in regulating follicular development, the pituitary gonadotropic hormones (mainly follicle-stimulating hormone [FSH] and luteinizing hormone [LH]) are known to play critical roles in controlling ovarian functions by triggering several downstream pathways [2]. This triggering action in turn leads to various morphological and functional changes in follicular cells [3]. These sequential changes in morphology and function of granulosa cells during various stages of follicular development are tightly regulated by expression of a multitude of genes [4, 5] and intraovarian growth factors and steroids [3].
The wingless type MMTV integration site family member 4 (WNT4) and members of the transforming growth factor-β (TGF-β) superfamily are among the major genes involved in follicular growth and granulosa cell functions [6, 7]. Transcription factors like the forkhead box (FOXO) transcription factor have emerged as important regulators of mammalian ovarian functions [8]. The FOXO genes work coordinately with the phosphatidylinositide 3-kinase (PI3K)/AKT signaling pathway for their activation [9]. Translational modifications regulate the activity of FOXO with nonphosphorylated [10] FOXO binding to FOXO-recognized elements (FRE) within the promoter regions of target genes to transactivate or repress the expression of downstream transcripts [11]. Phosphorylation of FOXO proteins causes their nuclear exclusion and subsequent degradation in cytoplasm, which leads to inhibition of FOXO-mediated transcription [12]. The FOXO genes are involved in primordial follicle activation, oocyte maintenance, and granulosa cell proliferation and differentiation [2]. Based on these facts, it is predicted that the large set of genes that play a significant role in mammalian follicular development and granulosa cell proliferation and differentiation, in one way or another will be regulated transcriptionally or post-transcriptionally.
MicroRNAs (miRNAs) are a class of small noncoding RNAs that have emerged as regulatory molecules involved in modulating the expression of genes at the post-transcriptional level. This occurs either by degrading the messenger RNA (mRNA) or by suppressing protein translation [13]. In addition to the expression profiling miRNAs in follicular cells [14, 15], there is growing evidence for involvement of miRNAs in various important cellular physiological processes, including granulosa cell proliferation [10, 16, 17], cell cycle transition [18], apoptosis [19–22], hormone biosynthesis [23], oocyte developmental competence and cell-to-cell communication [24, 25], and ovarian disorders like polycystic ovary syndrome [26].
We have demonstrated differential expression of miRNAs (miRs) between granulosa cells of subordinate and dominant follicles obtained on Days 3, 7 (early luteal phase) [14], and 19 (late follicular phase) [15] of the bovine estrous cycle. Among the various clusters, miR-183-96-182 cluster miRNAs were found to be the top three most significantly upregulated in granulosa cells of preovulatory dominant follicles compared to their subordinate counterparts [15]. In silico analysis of the miR-183-96-182 cluster miRNAs revealed an array of predicted target genes that are known to be involved in various physiological processes, including apoptosis, the cell cycle, and metabolism.
A wet-laboratory dual-luciferase reporter assay technique confirmed that all miRNAs of the miR-183-96-182 cluster coordinately target the FOXO1 gene [15]. In that same study, as opposed to showing expression of the miRNA cluster, we showed that FOXO1 mRNA is highly enriched in granulosa cells of bovine subordinate follicles compared to their preovulatory dominant counterparts. Thus, further understanding of the molecular crosstalk between the miRNA cluster and the FOXO1 transcription factor and the phenotypic changes in bovine granulosa cell function are needed. This information would widen our understanding of the role of miRNAs in regulating important transcription factors in bovine folliculogenesis. We aimed to decipher the functional role of miR-183-96-182 cluster miRNAs in granulosa cells by using an in vitro loss-and-gain functional analysis. Results provide evidence of the regulatory role of these miRNAs in bovine granulosa cell proliferation and cell cycle transition by targeting the FOXO1 gene as a transcriptional factor, which subsequently regulates the expression of other downstream transcripts.
Materials and Methods
Ovarian Sample Collection and Granulosa Cell Isolation
Bovine ovaries were collected from a local abattoir and transported in vacuum flasks containing warm physiologic saline solution (0.9% NaCl) to the laboratory. Afterward, ovarian samples were processed as previously described by Gebremedhn et al. [15]. Briefly, upon arrival, ovaries were washed twice with warm (37°C) phosphate buffered saline without Ca2+/Mg2+ (PBS−). They were then rinsed in 70% warm ethanol for 30 sec, followed by washing three times with PBS−. Granulosa cells were aspirated from small healthy, growing follicles (3- to 5-mm diameter) using 20-gauge sterile needles (B-Braun, Melsungen, Germany) and transferred into a 15-ml sterilized tube (Falcon; Thermo Fisher Scientific, Dreieich, Germany) containing warm PBS−.
Cumulus-oocyte complexes (COCs) were left to settle at the bottom of the tube. The upper suspension of follicular fluid with floating granulosa cells was carefully transferred into 15-ml tubes and centrifuged at 750 × g for 7 min. The supernatant follicular fluid was removed, and granulosa cell pellets were resuspended in red blood cell lysis buffer for 1 min. Next, the pellets were washed with Dulbecco modified Eagle medium/F-12 Ham culture medium (DMEM/F-12 Ham; Sigma-Aldrich, Darmstadt, Germany) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich). After granulosa cells were centrifuged briefly and washed with PBS−, samples were resuspended in DMEM/F-12 Ham culture medium supplemented with 10% FBS. Cell viability was assessed using trypan blue (Sigma-Aldrich) exclusion.
Granulosa Cell Culture
Isolated granulosa cells were seeded at 2 × 105 cells per well in a tissue culture-treated 24-well plate (Starlab, Hamburg, Germany) in DMEM/F-12 Ham supplemented with 10% FBS, 1% penicillin-streptomycin (Sigma-Aldrich), and 1% fungizone (Sigma-Aldrich). Cells were incubated at 37°C in a humidified chamber with 5% CO2 atmosphere. Culture medium was replaced with fresh medium every 48 h. To determine the effect of plating on the expression of the miR-183-96-182 cluster miRNAs and gene markers, cultured granulosa cells were harvested using 0.25% trypsin-EDTA (Sigma-Aldrich) at 24, 48, 96, and 144 h after being plated. Freshly isolated granulosa cells were snap frozen immediately and used as controls for timed expression patterns of miRNAs and genes.
Locked Nucleic Acid-Oligonucleotide Transfection
To determine the role of miR-183-96-182 cluster miRNAs in bovine granulosa cells, an in vitro gain-and-loss of function experiment was performed. The experiment was done using Locked Nucleic Acid (LNA)-mediated oligonucleotide miRNA mimics, mimic negative control, inhibitors, and inhibitor negative control (Exiqon, Vedbæk, Denmark). Oligonucleotide transfection was performed in subconfluent (75–80%) plated granulosa cells, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) transfection reagent in Opti-MEM I reduced-serum medium (Invitrogen).
According to the manufacturers' instructions, 75 nM miRNA mimic, inhibitor, or corresponding negative control was added to each well of the 24-well plate. After 24 h of incubation, culture medium was replaced with fresh medium. The efficiency of the LNA-oligonucleotide transfection in either decreasing or increasing the expression of target miRNAs was assessed 48 h later by using quantitative real-time PCR (qPCR). Similarly, the sponge effect of miRNA inhibition was assessed using qPCR. In this phenomenon, inhibition of one miRNA in a cluster results in a decrease in the expression of another miRNA within the cluster.
RNA Interference
Two antisense LNA GapmeRs (Exiqon) targeting bovine FOXO1 mRNA (FOXO1-short interfering RNAs [siRNAs]) and negative control siRNA (NC-siRNA) (Supplemental Table S1; supplemental data are available online at www.biolreprod.org) were used for targeted knockdown of bovine FOXO1 mRNA. For this, 2 × 105 cells were seeded onto a 24-well plate. Subconfluent cells were transiently transfected with pooled 75 nM FOXO1-siRNA or NC-siRNA, using Lipofectamine 2000 (Invitrogen) in Opti-MEM I reduced-serum medium. Twenty-four h post transfection, culture medium was replaced with fresh medium. Cells were trypsinized 48 h post transfection and used for subsequent molecular and cellular phenotype analyses.
Cell Proliferation Assay
We evaluated the effect of overexpression or inhibition of miR-183-96-182 cluster miRNAs and selective degradation of FOXO1 mRNA on the proliferation rate of granulosa cells. A 96-well plate was seeded with 2 × 104 cells, and subconfluent cells were transfected with 75 nM miR-183-96-182 cluster miRNA mimics or mimic negative control, miRNA inhibitors, or inhibitor negative control, and FOXO1-siRNA or NC-siRNA. Cell viability was measured using Cell Counting kit-8 (CCK-8; Dojindo Molecular Technology, Kumamoto, Japan) at 48, 72, 96, and 120 h post transfection according to the manufacturer's instructions. Briefly, 10 μl of CCK-8 was added to each well, and plates were incubated for 3 h at 37°C in 5% CO2 atmosphere. The optical density (OD) of released formazan dye was measured as an indicator of the number of living cells at a wave length of 450 nm, using a microplate reader (BioTek Instruments Inc, Friedrichshall, Germany). Blank OD measurements were obtained from wells containing only culture medium and used for normalization. The increase in viable cell number relative to the first measurement at 48 h post transfection (given an arbitrary value of 1) was used to calculate the relative proliferation of cells.
Cell Cycle Assay
We next determined the role of miR-183-96-182 cluster miRNAs and the FOXO1 gene in granulosa cell cycle regulation. Cultured granulosa cells were transfected with 75 nM miR-183-96-182 cluster miRNA mimics or mimic negative control, miRNA inhibitors or inhibitor negative control, and FOXO1-siRNA or NC-siRNA. Cells were trypsinized 48 h later and collected in a 15-ml Falcon tube, followed by centrifugation at 750 × g for 5 min and being washed twice with 1× PBS−. A minimum of 1 × 106 cells were fixed in ice-cold 70% ethanol at 4°C overnight. Cells were then centrifuged briefly, and the cell pellets were washed twice with 500 μl of 1× PBS−. Cells were stained with 50 μg/ml propidium iodide and 50 μg/ml RNase. Cells were then incubated at 37°C for 30 min and processed using an LSRFortessa flow cytometer (BD Biosciences, San Jose, CA). Cell cycle distribution was analyzed using ModFit LT software (http://www.vsh.com/products/mflt/index.asp).
Western Blot Analysis
Whole-cell protein lysate was prepared from granulosa cell samples collected 48 h after transfection, using 1× passive lysis buffer (Promega, Madison, WI). Protein concentration was determined using the Bradford method [27]. Equal amounts (30 μg) of protein lysate from each sample were electrophoresed in 8–10% gradient SDS-PAGE and transferred onto nitrocellulose membrane (Whatman-protran; Sigma-Aldrich). Membranes were blocked in 1× Roti-Block blocking solution (Carl Roth, Karlsruhe, Germany) at room temperature for 1 h. They were then incubated overnight at 4°C with diluted primary antibodies of polyclonal rabbit anti-FOXO1 (1:150 dilution; code sc-11350; Santa Cruz Biotechnology, Dallas, TX) or with diluted mouse monoclonal anti-β-actin (1:1000 dilution; Santa Cruz Biotechnology) as an internal control.
After being washed three times with diluted Tris-buffered saline with Tween 20 (1× TBST), the membranes were incubated with horseradish peroxidase-conjugated goat-anti-rabbit (1:5000 dilution) or goat-anti-mouse (1:5000 dilution) secondary antibodies for 2 h at room temperature. Subsequently, membranes were washed three times with 1× TBST, and specific signals were visualized with enhanced chemiluminescence, using Clarity Western ECL substrate (Bio-Rad, Munich, Germany). Images were acquired using Quantity One 1-D software (Bio-Rad) and a Gel Doc XRS+ imaging system (Bio-Rad).
RNA Isolation and qPCR
Total RNA enriched with miRNA was isolated from all granulosa cell samples by using the miRNeasy mini kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. On-column DNA digestion was performed to remove genomic DNA contamination by using RNase-free DNase (Qiagen). Total RNA samples were quantified using a NanoDrop 8000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). A 1-μg portion of total RNA sample from each treatment group was reverse-transcribed to cDNA by using First-Strand cDNA synthesis kit (Thermo Fisher Scientific) with oligo(dT)18 primers.
Gene-specific primers were designed using Primer3web version 4.0.0 (http://bioinfo.ut.ee/primer3/). For semiquantitative PCR, thermocycling consisted of preincubation at 95°C for 5 min, then 40 cycles of 30-sec denaturation at 95°C, 30-sec of annealing at 55°C (GAPDH), and 57°C (FSHR and LHR) for 30 sec, extension at 72°C for 1 min, and final extension at 72°C for 10 min, and maintenance at 4°C. The specificity of each primer was confirmed by sequencing the PCR products and BLAST analysis of the sequence results. Finally, 10 μl of PCR product was mixed with 5 μl of loading buffer, loaded into 2% agarose gel stained with ethidium bromide, and visualized under UV light, using Gel Doc XRS+ imaging system (Bio-Rad).
Similarly, qPCR experiments were performed using a reaction volume of 20 μl with 7.4 μl of ddH2O, 0.3 μl of forward primer, 0.3 μl of reverse primer, 10 μl of 1× SYBR Green I Master Mix (Bio-Rad), and 2 μl of cDNA template. Thermocycling conditions consisted of preheating at 95°C for 3 min, followed by 40 cycles of amplification at 95°C for 15 sec, and 1 min at 60°C. The specificity of PCR amplification was determined by melting curve analysis generated at the end of each qPCR run. Due to the stable expression of GAPDH in granulosa cells of various stages (before and after LH surge) [4], the expression of GAPDH was used as an internal control to normalize the expression of genes of interest.
Candidate miRNAs were quantified as described previously [14, 15]. Briefly, cDNA was synthesized from 80 ng of miRNA-enriched total RNA, using a miRCURY LNA Universal cDNA synthesis kit (Exiqon) according to the manufacturer's instructions. The resulting cDNA was dilution 40× and used for qPCR analysis of candidate miRNAs by using ExiLENT SYBR Green Master Mix (Exiqon). Thermal cycling conditions were preheating at 95°C for 10 min, followed by 40 cycles of amplification at 95°C for 10 sec and 60°C for 1 min. The specificity of each miRNA amplification was evaluated by melting curve analysis. The geometric mean of the expression of U6 small noncoding small nuclear RNA (snRNA) and 5S ribosomal RNA was used to normalize the expression values of candidate miRNAs. qPCR data were analyzed using the comparative cycle threshold (Ct) method [28]. The list of primers used in the experiment is available in Supplemental Table S2.
Statistical Analysis
Statistical analysis was performed using Prism version 5 software (GraphPad, LaJolla, CA). Data are mean ± SEM biological replicates. Statistical differences in mean expression values of two treatment groups were compared using a two-tailed Student t-test. For time course expression analysis, the normality of expression data was first assessed using a D'Agostino and Pearson omnibus normality test for the residual values of the normalized expression values. Equality of variance was evaluated using the Bartlett test. Statistical differences among means were then analyzed using one-way ANOVA followed by a Dunnett post-hoc test. Statistical significance was defined as a P value of ≤0.05.
Results
Expression of MiR-183-96-182 Cluster miRNAs in Bovine Granulosa Cells Increased While FOXO1 mRNA Decreased Upon Plating
We determined whether in vitro granulosa cell plating affected the identity of granulosa cells and transformed them into luteal cells. To achieve this, the presence of FSHR and LHR was determined at different time points during plating as marker genes of granulosa and luteal cells, respectively. We also examined the expression of ovulatory genes across plating time that are known to be induced by LH surge, namely PTX3 and PTGS2. FSHR was detected in granulosa cells before and after plating. LHR was not detected in any of the times before and after plating of granulosa cells (Supplemental Fig. S1A). Similarly, expression levels of PTX3 and PTGS2 were not induced during the culture period, as shown in Supplemental Figure S1B.
The relative abundance of miR-183-96-182 cluster miRNAs in granulosa cells was analyzed in cultures collected at 24, 48, 96, and 144 h post plating. Results revealed that the relative expression of all mature miRNAs of the cluster increased with time, reaching significant levels starting at 48 h after plating (Fig. 1, A–C). Interestingly, FOXO1 expression was significantly reduced during the culture period (Fig. 1D), indicating a reciprocal expression pattern with the miRNA cluster.
Fig. 1.
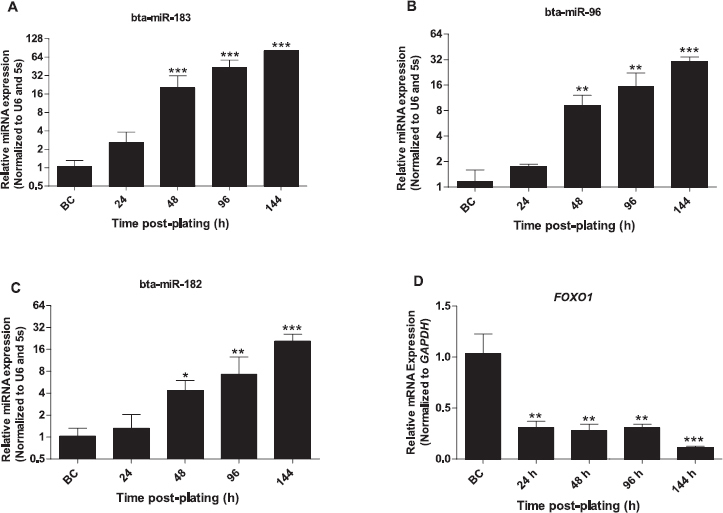
Expression of miR-183-96-182 cluster miRNAs increased in response to in vitro culture. Effect of granulosa cells culture on the expression of miR-183 (A), miR-96 (B), miR-182 (C). Expression pattern of FOXO1 mRNA in response to in vitro culture (D). The Y-axis indicates relative expression of miRNAs or mRNA, and the X-axis indicates the time post plating (before culture [BC], 24, 48, 96, and 144 h post plating). Expression values of target miRNAs and mRNA were normalized to those of the geometric mean of U6 small noncoding small nuclear RNA (U6 snRNA) and 5s ribosomal RNA (5s rRNA) and GAPDH, respectively. One-way ANOVA followed by a Dunnett post-hoc test was used to detect statistical differences. Data are mean ± SEM of n = 3 (*P < 0.05, **P < 0.01, ***P < 0.001).
LNA-Mediated Inhibition of Individual Members of miR-183-96-182 Cluster Resulted in Sponge Effect in Cultured Granulosa Cells
To modulate expression of cluster miR-183-96-182 in vitro, we used LNA-mediated knockdown of components of the miRNAs, both individually and pooled. Interestingly, inhibition of individual miRNAs resulted in a sponge effect (Fig. 2, A–C), in which transfection of cells with miR-183 inhibitor caused significant reduction in the expression of all three miRNAs of the miRNA cluster. Similarly, inhibition of miR-96 led to a measurable reduction in the level of all three miRNAs. However, inhibition of miR-182 reduced the expression of miR-183 and caused no significant reduction in miR-96 expression level. Transfection using pooled inhibitors resulted in coalescent reduction in the expression of all three miRNAs.
Fig. 2.
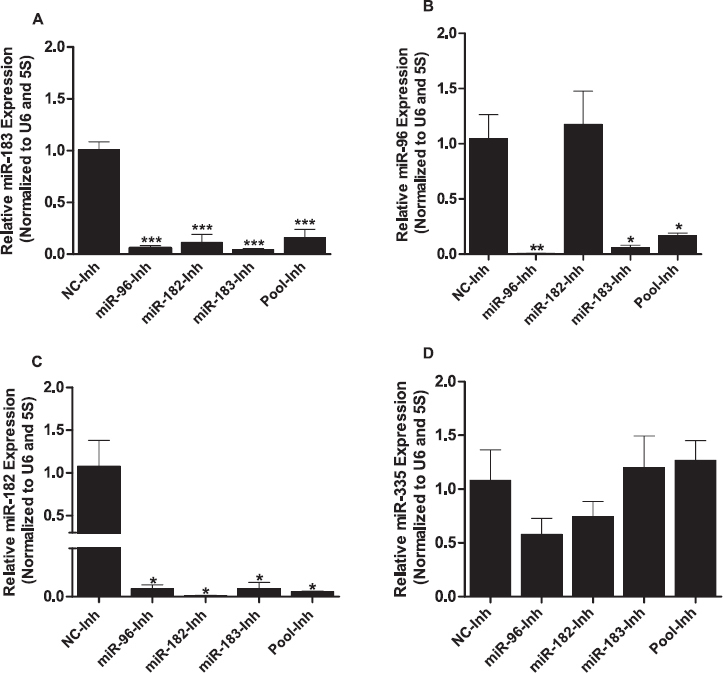
Sponge effect was observed upon inhibition of individual miRNAs of the miR-183-96-182 cluster. LNA-based miRNA inhibitors effectively inhibited individual miRNAs of the miR-183-96-182 cluster and resulted in a sponge effect (A, B, C). Inhibitions of either individual or pooled miRNAs of the miR-183-96-182 cluster did not produce the sponge effect on nonmember miRNA of the cluster (D). Expression values of target miRNAs were normalized to those of the geometric mean of U6 small noncoding small nuclear RNA (U6 snRNA) and 5s ribosomal RNA (5s rRNA). Two-tailed Student t-test was used to detect statistical differences. Data are mean ± SEM of n = 3 (*P < 0.05, **P < 0.01, ***P < 0.001).
To determine the specificity of the LNA-oligonucleotide miRNA inhibitors, we checked for the expression of miR-335, which does not belong to the miR-183-96-182 cluster and has no sequence homology. Inhibition of targeted miRNAs using either individual or pooled miRNA inhibitors of the miR-183-96-182 cluster had no significant impact on the expression of miR-335 (Fig. 2D). In contrast to the sponge effect observed upon transfection of granulosa cells with miRNA inhibitors, transfection of granulosa cells with miRNA mimics showed a selective increment in expression of the specific miRNAs on the cluster. Subsequently, cells transfected with pooled miRNA mimics showed increased expression of all miRNAs in the cluster (Fig. 3, A–C).
Fig. 3.
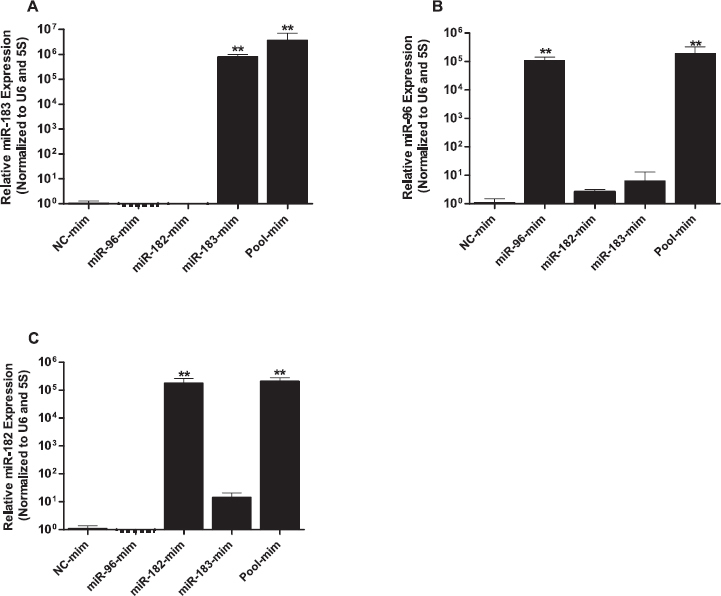
Transfection of granulosa cells with individual miRNA mimics of the miR-183-96-182 cluster selectively increased the expression of corresponding miRNAs (A–C). Expression values of target miRNAs were normalized to those of the geometric mean of U6 snRNA and 5s rRNA. Two-tailed Student t-test was used to detect statistical differences. Data are mean ± SEM of n = 3 (**P < 0.01).
miR-183-96-182 Cluster Regulates Expression of FOXO1 and Subsequently Its Pro-Apoptotic Downstream Transcript in Bovine Granulosa Cells In Vitro
We previously demonstrated that FOXO1 is a target gene of all miR-183-96-182 cluster miRNAs [15]. To further determine the role of miR-183-96-182 cluster miRNAs in modulating the expression of FOXO1 in bovine granulosa cells in vitro, FOXO1 mRNA and protein levels were measured in granulosa cells transfected with individual or pooled miRNA mimics, inhibitors, mimic negative control, and inhibitor negative control. qPCR analysis revealed that FOXO1 mRNA was significantly decreased in both the individual and the pooled miRNA mimic-transfected granulosa cells compared to granulosa cells transfected with mimic-negative control (Fig. 4A). In agreement with the mRNA expression pattern, FOXO1 protein expression level was markedly decreased in granulosa cells treated with both individual and pooled miRNA mimics (Fig. 4B).
Fig. 4.
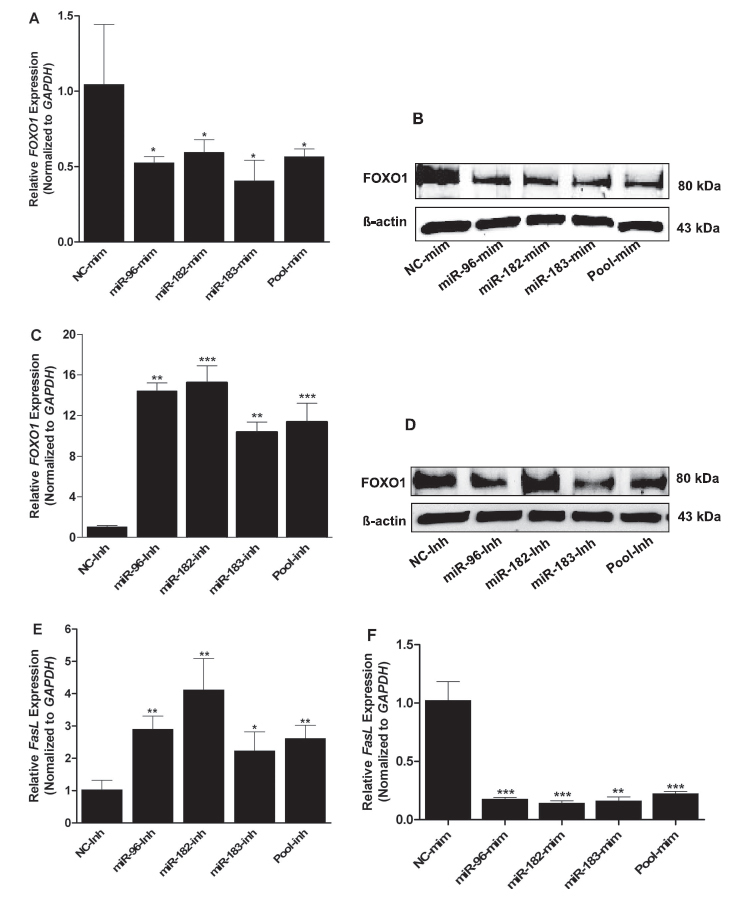
Modulation of miR-183-96-182 cluster miRNAs in granulosa cells led to a change in mRNA and protein of FOXO1 gene. Expression of FOXO1 mRNA (A) and protein (B) decreased significantly upon overexpression of miR-183-96-182 cluster miRNAs. Inhibition of miR-183-96-182 cluster miRNAs significantly increased the expression of FOXO1 mRNA (C) and FOXO1 protein expression (D). Expression of FASL mRNAs was decreased upon overexpression (E) and increased upon inhibition (F) of miR-183-96-182 cluster miRNAs. GAPDH was used as internal control, and two-tailed Student t-test was used to detect statistical differences. Data are mean ± SEM of n = 3 (*P < 0.05, **P < 0.01, ***P < 0.001).
Conversely, inhibition of either the individual or the cluster miRNAs using LNA-miRNA inhibitors resulted in sharp increase in FOXO1 mRNA (Fig. 4C). However, the increment in FOXO1 protein in granulosa cells transfected with miR-183-96-182 cluster inhibitors was not in full agreement with the increase in FOXO1 mRNA (Fig. 4D). FOXO1 as a transcription factor regulates the expression of apoptosis-related genes, including the Fas ligand (FASL) [29]. To further confirm whether the alteration in expression of FOXO1 by the miR-183-96-182 cluster miRNAs could affect the expression of this FOXO1 downstream gene, we checked the expression of FASL, a pro-apoptotic downstream gene of FOXO1. Interestingly, similar to the trend of FOXO1, the expression of FASL mRNA increased in granulosa cells transfected with miR-183-96-182 cluster inhibitors (Fig. 4E). However, the expression decreased in granulosa cells transfected with miR-183-96-182 cluster mimics (Fig. 4F).
Overexpression of miR-183-96-182 Cluster Promotes Granulosa Cell Proliferation
To assess the role of the miR-183-96-182 cluster in granulosa cell proliferation, cells were transfected with miRNA mimics, inhibitors, and corresponding negative controls. The relative rate of cell proliferation treated with the mimics was higher than that in the control group (Fig. 5A). In contrast, the relative rate of granulosa cell proliferation transfected with the inhibitors was lower than that with the inhibitor control (Fig. 5B).
Fig. 5.
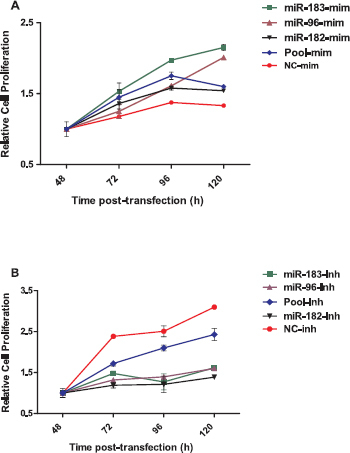
Inhibition of miR-183-96-182 cluster miRNAs reduced the rate of granulosa cell proliferation. Cell proliferation assay of granulosa cells transfected with miR-183-96-182 cluster mimics and mimic negative control (A), cells transfected with miR-183-96-182 cluster inhibitors and inhibitor negative control (B). The relative rate of cell proliferation is indicated on the Y-axis, and time post transfection is indicated on the X-axis. Data are mean ± SEM of n = 3.
Overexpression of miR-183-96-182 Cluster Promotes G1/S Cell Cycle Transition in Bovine Granulosa Cells
We further examined the effect of miR-183-96-182 cluster miRNAs in modulating cell cycle transition in granulosa cells. Granulosa cells transfected with the mimics showed reduction in the proportion of cells in G0/G1 arrest, as well as an increment in the percentage of cells in S phase compared to granulosa cells transfected with mimic-negative control (Fig. 6). MiR-183 mimic caused a higher reduction in the proportion of cells in G0/G1 phase (∼12%) and an increment of cells in S phase (10%) (Fig. 6B), followed by pooled mimics (Fig. 6E), miR-96 mimic (Fig. 6C), and miR-182 mimic (Fig. 6D). This increased level of miRNAs in granulosa cells could enhance cell proliferation by promoting G1/S cell cycle transition. In contrast, inhibition of miR-183-96-182 cluster miRNAs did not result in measurable changes in the cell cycle profiles of granulosa cells (Supplemental Table S3).
Fig. 6.
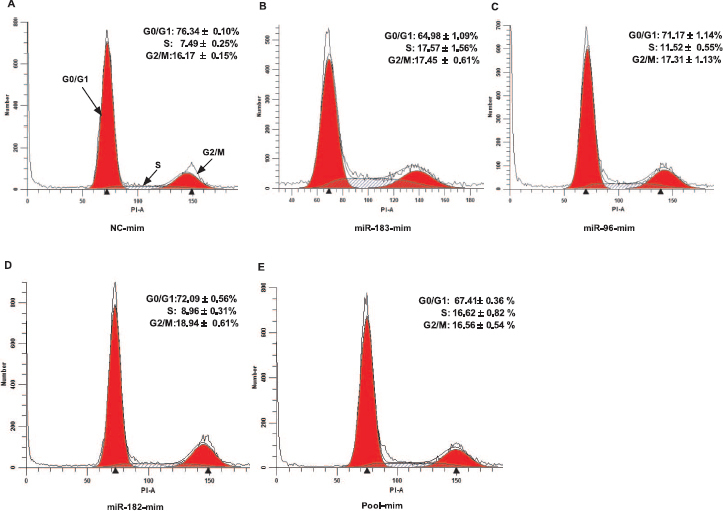
Overexpression of miR-183-96-182 cluster changes the cell cycle profile of granulosa cells. Representative histograms of flow cytometric analysis of granulosa cells transfected with miRNA mimic negative control (A), miR-183 mimics (B), miR-96 mimic (C), miR-182 mimic (D), and pooled mimics (E). The analyzed cell counts are indicated on the Y-axis, and the DNA content of cells detected by PI staining is indicated on the X-axis. Data are mean ± SEM of n = 3.
Selective Knockdown of FOXO1 by siRNA Promotes Granulosa Cell Proliferation and Induces G1/S Cell Cycle Transition
To determine the involvement of FOXO1 in granulosa cell proliferation and cell cycle transition and to confirm the regulatory role of miRNAs in FOXO1 expression, the siRNA technique was used to selectively knockdown the FOXO1 mRNA in granulosa cells. Granulosa cells transfected with FOXO1 siRNA showed a significant reduction in the level of FOXO1 mRNA (∼72%) compared to that in cells treated with NC-siRNA (Fig. 7A). In agreement with the mRNA expression, the FOXO1 protein markedly declined in granulosa cells transfected with FOXO1 siRNA compared to that in the negative control (Fig. 7B). Degradation of FOXO1 also resulted in reduced expression of downstream pro-apoptotic FASL mRNA (Fig 7C).
Fig. 7.
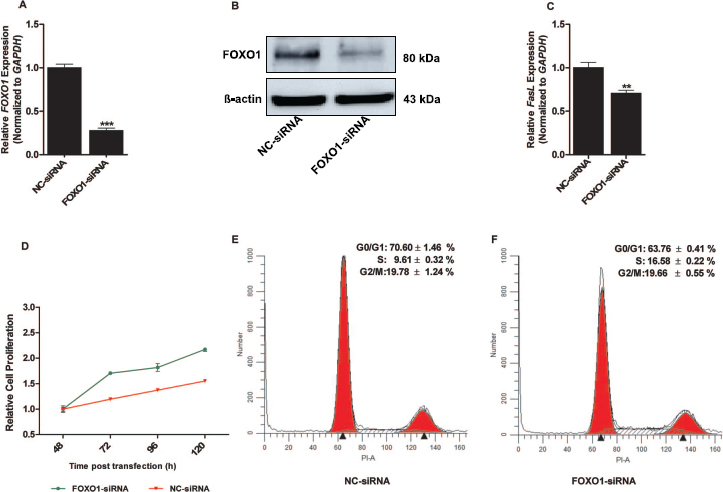
Targeted suppression of FOXO1 expression promotes cell proliferation and altered the cell cycle profile of granulosa cells. QPCR analysis of FOXO1 mRNA levels in granulosa cells transfected with FOXO-siRNA or NC-siRNA are shown (A). Western blot analyses of FOXO1 protein in granulosa cells transfected with FOXO-siRNA or NC-siRNA (B). FASL mRNA levels in granulosa cells transfected with FOXO-siRNA or NC-siRNA, as determined by qPCR (C). The relative rate of granulosa cell proliferation transfected with FOXO-siRNA or NC-siRNA as determined by CCK-8 (D). Representative histograms of flow cytometric cell cycle analysis of cells transfected with NC-siRNA (E) and FOXO-siRNA (F). GAPDH was used as an internal control for gene expression analysis, and two-tailed Student t-test was used to detect statistical differences in gene expression analysis. Data are mean ± SEM of n = 3 experiments (**P < 0.01, ***P < 0.001).
Similarly, results of cell proliferation assays showed a relative rate of cell proliferation in granulosa cells transfected with FOXO1 siRNA that was increased compared to that in the negative control (Fig. 7D), indicating an antiproliferative role for FOXO1 in bovine granulosa cells. Interestingly, G0/G1 cell cycle arrest was markedly decreased in granulosa cells transfected with FOXO1-siRNA. These cells showed a proportion of cells in the S phase of the cell cycle that was increased (Fig. 7E) compared to NC-siRNA transfected cells (Fig. 7F).
Discussion
Bovine folliculogenesis is a well-orchestrated biological process in which granulosa cells produce steroid hormones, growth factors, and cytokines [30, 31]. Large-scale transcriptome profiling of granulosa cells showed the expression and interaction of a multitude of genes and transcription factors in regulating granulosa cell function in various stages of follicular development [4, 5, 32]. Recently, miRNAs have been shown to play regulatory roles in ovarian functions, including activation of primordial follicles, follicular recruitment, and granulosa cell functions [33]. The aim of the present study was to decipher the functional role of miR-183-96-182 cluster miRNAs in bovine granulosa cells. We demonstrated that modulation of these miRNAs leads to changes in the proliferative potential of granulosa cells and transition in the cell cycle profile through coordinated targeting of the FOXO1 transcription factor. The regulatory role of the miRNA cluster in granulosa cell function through FOXO1 could be confirmed by selective degradation of FOXO1 mRNAs in bovine granulosa cells.
The transcription of the cluster miRNAs increased whereas the expression of their target gene FOXO1 decreased in response to plating. However, this trend of expression was not accompanied by luteinization of the granulosa cells during in vitro culture, which was confirmed by the absence of LHR throughout the culture period. This absence of LHR could be associated with the follicular size from which the granulosa cells were aspirated, as the acquisition of LHR in bovine granulosa cells is correlated with follicular diameter [34, 35]. Maintenance of the identity of granulosa cells during plating was validated by detection of FSHR throughout the culture period.
Even though the mechanisms that govern transcription of miRNAs are not fully understood, miRNAs are known to be transcribed either as individuals or in clusters [36]. MiRNA clusters are groups of miRNAs adjacently located on the same chromosome that are transcribed as one pre-miRNA transcript and further processed into individual mature miRNAs [37]. The chromosomal span of miRNA clusters ranges from hundreds to several kilobase (kb) pairs and often shares a common promoter [38]. The miR-183-96-182 cluster is a class of conserved polycistronic miRNAs transcribed from a 4.6-kb–long intergenic region of bovine chromosome 4 of the bovine genome (Supplemental Fig. S2A). This cluster is evolutionarily conserved across mammalian species (Supplemental Fig. S2B). Members of this miRNA cluster are transcribed in the same direction from telomere to centromere and function synergistically [39]. Studies have shown that the miR-183-96-182 cluster miRNAs are enriched in most breast cancers [40], regulate oxidative stress-induced apoptosis [41], and inhibit invasion and metastasis in lung cancers [42] by regulating the expression of several key genes.
The LNA-mediated inhibition of individual members of the miR-183-96-182 cluster resulted in a sponge effect (Fig. 2, A–C), which could be attributed to the partial sequence homology between miRNAs in the same cluster (Supplemental Fig. S2A). In addition to the conserved seed sequences, the identical directional transcription of individual miRNAs in the same cluster suggests their identical origin of a primary miRNA transcript that contributes to the sponge effect in miRNA knockdown experiments [39, 41]. In addition to the sponge effect, high sequence homology between miRNAs in the cluster permits them to target similar genes and play a vital role by coordinately regulating key genes in cellular processes [43, 44].
In bovine granulosa cells, overexpression of the miR-183-96-182 cluster miRNAs resulted in modulation of both mRNA and protein expression of FOXO1 (Fig. 4, A and B). This is attributed to the conserved seed sequence and coexpression of the cluster miRNAs, which resulted in cotargeting of FOXO1 and subsequently affected molecular pathways in the signaling axis [45]. The abundance of FOXO1 protein after inhibition of the cluster was not in full agreement with FOXO1 mRNA expression (Fig. 4, C and D). Discrepancies could be due to the fact that binding sites of several other miRNAs reside within the 3′-untranslated region (UTR) of FOXO1, and inhibiting only the miR-183-96-182 cluster might not produce the desired increment in FOXO1 protein level.
The FOXO transcription factor family consists of FOXO1, FOXO3, FOXO4, and FOXO6. It is known to regulate genes involved in follicular growth [46] and numerous cellular processes, including proliferation, apoptosis, cell cycle arrest, cell differentiation, and metabolism [12, 47]. FOXO1 is abundantly expressed in granulosa cells of growing follicles [48], anovulatory subordinate follicles [15], and small atretic bovine follicles [32], signifying its potential involvement in ovarian follicular development. Its involvement in the apoptotic process during follicular atresia is evidenced by the depletion and enrichment of Foxo1 in rat granulosa cells of healthy antral follicles and small atretic follicles, respectively [49].
Despite the fact that FASL is not among the conserved target genes of the miR-183-96-182 cluster miRNAs in bovine (Supplemental Fig. S3), its expression was affected by modulation of the cluster miRNAs (Fig. 4, E and F) and by modulation of FOXO1 mRNA (Fig. 7C). This was in agreement with the observations by Shen et al. [50]. They showed that induction of Foxo1 through oxidative stress in mouse granulosa cells induced the expression of its downstream transcripts Fasl, Bim, and Trail, which were associated with cell cycle arrest and apoptosis.
Proliferation of granulosa cells is ultimately important in follicular growth and the creation of a unique microenvironment for oocyte maturation [51]. Accumulated evidence is available from the involvement of miRNAs in regulating granulosa cell proliferation [10, 16, 17]. In the present study, the rate of granulosa cell proliferation increased upon miR-183-96-182 cluster miRNAs overexpression, which was accompanied by downregulation of the antiproliferative FOXO1 gene. This is further supported by the shift in proportion of cells from G0/G1 arrest to the S phase of the cell cycle.
Inhibition of miR-183-96-182 cluster miRNAs decreased the relative rate of granulosa cell proliferation. Similar results have been reported in breast cancer cells, where the enrichment of miR-183-96-182 cluster miRNAs promotes cell proliferation, cell migration, and change in the cell cycle profile [40]. The rate at which granulosa cells proliferate was higher in cells where FOXO1 mRNA was suppressed than in the control group in the RNA interference experiment, which indicates an antiproliferative role for FOXO1. Consistent with the present findings, another study of cultured granulosa cells indicated that FOXO1 can regulate genes associated with cell proliferation and apoptosis [52].
Selective degradation of FOXO1 using siRNA in granulosa cells resulted in a decline in the proportion of cells under G0/G1 arrest and an increase of cells in the S phase of the cell cycle. This suggests that overexpression of FOXO1 in bovine granulosa cells maintains the cells in G0/G1 arrest and prevents cells from entering S phase and progressing in the cell cycle. Mammalian FOXO proteins are involved in regulating the G1/S phase cell cycle transition and commit the cells to cell cycle progression [53, 54]. Evidence for this was further shown in mouse granulosa cells, where Foxo1 was depleted and showed promoted cell cycle transition [52]. Taken together, enrichment of FOXO1 in granulosa cells has an inhibitory role in cell cycle transition by preventing the induction of positive regulators of cell cycle.
In conclusion, the present study provides new insights into the role of miR-183-96-182 cluster miRNAs in coordinated regulation of the FOXO1 transcription factor in bovine granulosa cells. These miRNAs exert subsequent influence on bovine granulosa cell proliferation and cell cycle transition.
Supplementary Material
Acknowledgment
We acknowledge assistance of the Flow Cytometry Core Facility at the Institute of Molecular Medicine, University of Bonn, supported in part by SFB 704.
References
- 1. Robker RL, Richards JS.. Hormonal control of the cell cycle in ovarian cells: proliferation versus differentiation. Biol Reprod 1998; 59:476–482. [DOI] [PubMed] [Google Scholar]
- 2. Makker A, Goel MM, Mahdi AA.. PI3K/PTEN/Akt and TSC/mTOR signaling pathways, ovarian dysfunction, and infertility: an update. J Mol Endocrinol 2014; 53:14–0220. [DOI] [PubMed] [Google Scholar]
- 3. Hirshfield AN. Development of follicles in the mammalian ovary. Int Rev Cytol 1991; 124:43–101. [DOI] [PubMed] [Google Scholar]
- 4. Douville G, Sirard MA.. Changes in granulosa cells gene expression associated with growth, plateau and atretic phases in medium bovine follicles. J Ovarian Res 2014; 7:1757–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hatzirodos N, Irving-Rodgers HF, Hummitzsch K, Harland ML, Morris SE, Rodgers RJ.. Transcriptome profiling of granulosa cells of bovine ovarian follicles during growth from small to large antral sizes. BMC Genomics 2014; 15:1471–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boyer A, Lapointe E, Zheng X, Cowan RG, Li H, Quirk SM, DeMayo FJ, Richards JS, Boerboom D.. WNT4 is required for normal ovarian follicle development and female fertility. Faseb J 2010; 24:3010–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fenwick MA, Mora JM, Mansour YT, Baithun C, Franks S, Hardy K.. Investigations of TGF-beta signaling in preantral follicles of female mice reveal differential roles for bone morphogenetic protein 15. Endocrinology 2013; 154:3423–3436. [DOI] [PubMed] [Google Scholar]
- 8. Uhlenhaut NH, Treier M.. Forkhead transcription factors in ovarian function. Reproduction 2011; 142:489–495. [DOI] [PubMed] [Google Scholar]
- 9. Hannenhalli S, Kaestner KH.. The evolution of Fox genes and their role in development and disease. Nat Rev Genet 2009; 10:233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang Q, Sun H, Jiang Y, Ding L, Wu S, Fang T, Yan G, Hu Y.. MicroRNA-181a suppresses mouse granulosa cell proliferation by targeting activin receptor IIA. PLoS One 2013; 8:e59667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shen M, Liu Z, Li B, Teng Y, Zhang J, Tang Y, Sun SC, Liu H.. Involvement of FoxO1 in the effects of follicle-stimulating hormone on inhibition of apoptosis in mouse granulosa cells. Cell Death Dis 2014; 5:e1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang H, Tindall DJ.. Dynamic FoxO transcription factors. J Cell Sci 2007; 120:2479–2487. [DOI] [PubMed] [Google Scholar]
- 13. Ying SY, Chang DC, Miller JD, Lin SL.. The microRNA: overview of the RNA gene that modulates gene functions. Methods Mol Biol 2006; 342:1–18. [DOI] [PubMed] [Google Scholar]
- 14. Salilew-Wondim D, Ahmad I, Gebremedhn S, Sahadevan S, Hossain MD, Rings F, Hoelker M, Tholen E, Neuhoff C, Looft C, Schellander K, Tesfaye D.. The expression pattern of microRNAs in granulosa cells of subordinate and dominant follicles during the early luteal phase of the bovine estrous cycle. PLoS One 2014; 9:e106795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gebremedhn S, Salilew-Wondim D, Ahmad I, Sahadevan S, Hossain MM, Hoelker M, Rings F, Neuhoff C, Tholen E, Looft C, Schellander K, Tesfaye D.. MicroRNA expression profile in bovine granulosa cells of preovulatory dominant and subordinate follicles during the late follicular phase of the estrous cycle. PLoS One 2015; 10:e0125912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yan G, Zhang L, Fang T, Zhang Q, Wu S, Jiang Y, Sun H, Hu Y.. MicroRNA-145 suppresses mouse granulosa cell proliferation by targeting activin receptor IB. FEBS Lett 2012; 586:3263–3270. [DOI] [PubMed] [Google Scholar]
- 17. Yao G, Yin M, Lian J, Tian H, Liu L, Li X, Sun F.. MicroRNA-224 is involved in transforming growth factor-beta-mediated mouse granulosa cell proliferation and granulosa cell function by targeting Smad4. Mol Endocrinol 2010; 24:540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiang L, Huang J, Li L, Chen Y, Chen X, Zhao X, Yang D.. MicroRNA-93 promotes ovarian granulosa cells proliferation through targeting CDKN1A in polycystic ovarian syndrome. J Clin Endocrinol Metab 2015; 100:E729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou J, Liu J, Pan Z, Du X, Li X, Ma B, Yao W, Li Q, Liu H.. The let-7g microRNA promotes follicular granulosa cell apoptosis by targeting transforming growth factor-beta type 1 receptor. Mol Cell Endocrinol 2015; 409:103–112. [DOI] [PubMed] [Google Scholar]
- 20. Rui C, Wangjun W, Xiaolong Z, Kaiqing L, Bojiang L, Xianju H, Zhang Y, Honglin L.. Let-7g induces granulosa cell apoptosis by targeting MAP3K1 in the porcine ovary. Int J Biochem Cell Biol 2015; 20:30006–30006. [DOI] [PubMed] [Google Scholar]
- 21. Liu J, Du X, Zhou J, Pan Z, Liu H, Li Q.. MicroRNA-26b functions as a proapoptotic factor in porcine follicular granulosa cells by targeting Sma-and Mad-related protein 4. Biol Reprod 2014; 91:13. [DOI] [PubMed] [Google Scholar]
- 22. Carletti MZ, Fiedler SD, Christenson LK.. MicroRNA 21 blocks apoptosis in mouse periovulatory granulosa cells. Biol Reprod 2010; 83:286–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu S, Sun H, Zhang Q, Jiang Y, Fang T, Cui I, Yan G, Hu Y.. MicroRNA-132 promotes estradiol synthesis in ovarian granulosa cells via translational repression of Nurr1. Reprod Biol Endocrinol 2015; 13:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sohel MM, Hoelker M, Noferesti SS, Salilew-Wondim D, Tholen E, Looft C, Rings F, Uddin MJ, Spencer TE, Schellander K, Tesfaye D.. Exosomal and non-exosomal transport of extra-cellular micrornas in follicular fluid: implications for bovine oocyte developmental competence. PLoS One 2013; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. da Silveira JC, Veeramachaneni DN, Winger QA, Carnevale EM, Bouma GJ.. Cell-secreted vesicles in equine ovarian follicular fluid contain miRNAs and proteins: a possible new form of cell communication within the ovarian follicle. Biol Reprod 2012; 86. [DOI] [PubMed] [Google Scholar]
- 26. Hossain MM, Cao M, Wang Q, Kim JY, Schellander K, Tesfaye D, Tsang BK.. Altered expression of miRNAs in a dihydrotestosterone-induced rat PCOS model. J Ovarian Res 2013; 6:1757–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976; 72:248–254. [DOI] [PubMed] [Google Scholar]
- 28. Livak KJ, Schmittgen TD.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 29. Cui M, Huang Y, Zhao Y, Zheng J.. New insights for FOXO and cell-fate decision in HIV infection and HIV associated neurocognitive disorder. Adv Exp Med Biol 2009; 665:143–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Richards JS. Hormonal control of gene expression in the ovary. Endocr Rev 1994; 15:725–751. [DOI] [PubMed] [Google Scholar]
- 31. Toda K, Hayashi Y, Ono M, Saibara T.. Impact of ovarian sex steroids on ovulation and ovulatory gene induction in aromatase-null mice. Endocrinology 2012; 153:386–394. [DOI] [PubMed] [Google Scholar]
- 32. Hatzirodos N, Hummitzsch K, Irving-Rodgers HF, Harland ML, Morris SE, Rodgers RJ.. Transcriptome profiling of granulosa cells from bovine ovarian follicles during atresia. BMC Genomics 2014; 15:1471–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maalouf SW, Liu WS, Pate JL.. MicroRNA in ovarian function. Cell Tissue Res 2016; 363:7–18. [DOI] [PubMed] [Google Scholar]
- 34. Evans AC, Ireland JL, Winn ME, Lonergan P, Smith GW, Coussens PM, Ireland JJ.. Identification of genes involved in apoptosis and dominant follicle development during follicular waves in cattle. Biol Reprod 2004; 70:1475–1484. [DOI] [PubMed] [Google Scholar]
- 35. Beg MA, Bergfelt DR, Kot K, Wiltbank MC, Ginther OJ.. Follicular-fluid factors and granulosa-cell gene expression associated with follicle deviation in cattle. Biol Reprod 2001; 64:432–441. [DOI] [PubMed] [Google Scholar]
- 36. Lai EC, Tomancak P, Williams RW, Rubin GM.. Computational identification of Drosophila microRNA genes. Genome Biol 2003; 4:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mathelier A, Carbone A.. Large scale chromosomal mapping of human microRNA structural clusters. Nucleic Acids Res 2013; 41:4392–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ryazansky SS, Gvozdev VA, Berezikov E.. Evidence for post-transcriptional regulation of clustered microRNAs in Drosophila. BMC Genomics 2011; 12:1471–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weeraratne SD, Amani V, Teider N, Pierre-Francois J, Winter D, Kye MJ, Sengupta S, Archer T, Remke M, Bai AH, Warren P, Pfister SM et al. . Pleiotropic effects of miR-183-96-182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol 2012; 123:539–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li P, Sheng C, Huang L, Zhang H, Cheng Z, Zhu Q.. MiR-183/-96/-182 cluster is up-regulated in most breast cancers and increases cell proliferation and migration. Breast Cancer Res 2014; 16:473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tang H, Bian Y, Tu C, Wang Z, Yu Z, Liu Q, Xu G, Wu M, Li G.. The miR-183/96/182 cluster regulates oxidative apoptosis and sensitizes cells to chemotherapy in gliomas. Curr Cancer Drug Targets 2013; 13:221–231. [DOI] [PubMed] [Google Scholar]
- 42. Kundu ST, Byers LA, Peng DH, Roybal JD, Diao L, Wang J, Tong P, Creighton CJ, Gibbons DL.. The miR-200 family and the miR-183-96-182 cluster target Foxf2 to inhibit invasion and metastasis in lung cancers. Oncogene 2016; 35:173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dambal S, Shah M, Mihelich B, Nonn L.. The microRNA-183 cluster: the family that plays together stays together. Nucleic Acids Res 2015; 43:7173–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ebert MS, Neilson JR, Sharp PA.. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 2007; 4:721–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hausser J, Zavolan M.. Identification and consequences of miRNA-target interactions–beyond repression of gene expression. Nat Rev Genet 2014; 15:599–612. [DOI] [PubMed] [Google Scholar]
- 46. Liu Z, Castrillon DH, Zhou W, Richards JS.. FOXO1/3 depletion in granulosa cells alters follicle growth, death and regulation of pituitary FSH. Mol Endocrinol 2013; 27:238–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barthel A, Schmoll D, Unterman TG.. FoxO proteins in insulin action and metabolism. Trends Endocrinol Metab 2005; 16:183–189. [DOI] [PubMed] [Google Scholar]
- 48. Liu Z, Rudd MD, Hernandez-Gonzalez I, Gonzalez-Robayna I, Fan HY, Zeleznik AJ, Richards JSFSH.. and FOXO1 regulate genes in the sterol/steroid and lipid biosynthetic pathways in granulosa cells. Mol Endocrinol 2009; 23:649–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shi F, LaPolt PS.. Relationship between FoxO1 protein levels and follicular development, atresia, and luteinization in the rat ovary. J Endocrinol 2003; 179:195–203. [DOI] [PubMed] [Google Scholar]
- 50. Shen M, Lin F, Zhang J, Tang Y, Chen WK, Liu H.. Involvement of the up-regulated FoxO1 expression in follicular granulosa cell apoptosis induced by oxidative stress. J Biol Chem 2012; 287:25727–25740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Maruo T. [Expression of oncogenes, growth factors and their receptors in follicular growth, regression and atresia: their roles in granulosa cell proliferation and differentiation]. Nihon Sanka Fujinka Gakkai Zasshi 1995; 47:738–750. [PubMed] [Google Scholar]
- 52. Park Y, Maizels ET, Feiger ZJ, Alam H, Peters CA, Woodruff TK, Unterman TG, Lee EJ, Jameson JL, Hunzicker-Dunn M.. Induction of cyclin D2 in rat granulosa cells requires FSH-dependent relief from FOXO1 repression coupled with positive signals from Smad. J Biol Chem 2005; 280:9135–9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Medema RH, Kops GJ, Bos JL, Burgering BM.. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 2000; 404:782–787. [DOI] [PubMed] [Google Scholar]
- 54. Dijkers PF, Medema RH, Pals C, Banerji L, Thomas NS, Lam EW, Burgering BM, Raaijmakers JA, Lammers JW, Koenderman L, Coffer PJ.. Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27(KIP1). Mol Cell Biol 2000; 20:9138–9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lewis BP, Burge CB, Bartel DP.. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;January14;120(1):15–20. [DOI] [PubMed] [Google Scholar]
- 56. Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP.. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 2007; 27:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Friedman RC, Farh KK, Burge CB, Bartel DP.. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009; 19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


