ABSTRACT
During Xenopus gastrulation, Wnt and FGF signaling pathways cooperate to induce posterior structures. Wnt target expression around the blastopore falls into two main categories: a horseshoe shape with a dorsal gap, as in Wnt8 expression; or a ring, as in FGF8 expression. Using ChIP-seq, we show, surprisingly, that the FGF signaling mediator Ets2 binds near all Wnt target genes. However, β-catenin preferentially binds at the promoters of genes with horseshoe patterns, but further from the promoters of genes with ring patterns. Manipulation of FGF or Wnt signaling demonstrated that ‘ring’ genes are responsive to FGF signaling at the dorsal midline, whereas ‘horseshoe’ genes are predominantly regulated by Wnt signaling. We suggest that, in the absence of active β-catenin at the dorsal midline, the DNA-binding protein TCF binds and actively represses gene activity only when close to the promoter. In contrast, genes without functional TCF sites at the promoter may be predominantly regulated by Ets at the dorsal midline and are expressed in a ring. These results suggest recruitment of only short-range repressors to potential Wnt targets in the Xenopus gastrula.
KEY WORDS: Mesodermal patterning, FGF signaling, Wnt signaling, Gastrulation, Xenopus
Highlighted Article: FGF and Wnt combinatorially regulate downstream genes, but differences in chromatin binding patterns of β-catenin and Ets may explain the differences in downstream target gene expression patterns.
INTRODUCTION
Both the Wnt and FGF signaling pathways regulate anterior-posterior pattering during gastrulation (Christen and Slack, 1997) of the vertebrate embryo. More specifically, both pathways contribute to posteriorization/caudalization of neural tissue (reviewed by Niehrs, 2004), as well as to mesodermal development (reviewed by Harland, 2004) and neural crest induction (reviewed by Pegoraro and Monsoro-Burq, 2012; Groves and LaBonne, 2014). However, whether these signaling pathways act stepwise in series or combinatorially in parallel to transcriptionally activate downstream target genes is less well studied.
The FGF ligand signals through receptor tyrosine kinases to ultimately phosphorylate and activate the Ets family of transcription factors, which comprises 30 different members (Ornitz and Itoh, 2015; Yordy and Muise-Helmericks, 2000). During gastrulation, multiple FGF ligands, but most notably the FGF8 ligand, are expressed at the posterior end of the embryo (Fletcher et al., 2006; Christen and Slack, 1997) and canonically signal through RAS-MAPK intracellular proteins (Ribisi et al., 2000; Umbhauer et al., 1995; Carballada et al., 2001). Subsequently, downstream of MAPK, Ets2 is phosphorylated and is essential for activating target genes involved in posterior patterning (Kawachi et al., 2003).
In contrast to the FGF signaling pathway, the canonical Wnt signaling pathway is activated through a low-density lipoprotein receptor-related protein 5/6 (LRP5/6) and Frizzled receptor to ultimately stabilize β-catenin, the sole transcriptional activator (Hikasa and Sokol, 2013). Subsequent to stabilization, β-catenin enters the nucleus where it competes with the repressors Groucho (also known as Tle4) or CtBP for binding to TCF/LEF transcription factors (Roose et al., 2001; Clevers, 2006; Kim et al., 2013; Brannon et al., 1999).
The repressive activity of TCF3 is essential for its role in dorsal-ventral and anterior-posterior patterning. Genes that are normally activated by β-catenin on the dorsal side of the embryo and required for organizer function must be actively repressed on the ventral side of the embryo. The ability of TCF3 to recruit Groucho keeps these genes switched off, and if TCF3 is depleted, embryos are dorsalized owing to ectopic expression of dorsal genes on the ventral side of the embryo (Houston et al., 2002). In addition, the zebrafish headless mutant has severe head defects due to a mutation in TCF3 and subsequent overactivation of Wnt target genes that suppress head formation (Kim et al., 2000).
Although these pathways signal through different proteins, the FGF and Wnt signaling pathways often crosstalk through a myriad of mechanisms to coordinate different developmental processes (Dailey et al., 2005). For example, in Ciona intestinalis pigment cell precursors, FGF signaling induces transcription of TCF, allowing Wnt signaling to activate target genes (Squarzoni et al., 2011). FGF and Wnt also act synergistically to promote proliferation, but act separately to determine cell lineage specification during limb development (ten Berge et al., 2008).
During gastrulation, crosstalk between the Wnt and FGF signaling pathways has been documented at different levels. For example, it has been shown that FGF is required downstream of Wnt signaling for induction of posterior neural markers (Domingos et al., 2001), whereas Wnt signaling is required downstream of FGF signaling for neural crest induction (Hong et al., 2008). In addition, there is evidence that both FGF and Wnt signaling directly activate expression of cdx2 (Wang and Shashikant, 2007; Keenan et al., 2006). Although targets of the Wnt/β-catenin signaling pathway at gastrulation have been identified, their expression patterns differ to an extent that may not be explained by Wnt signaling alone (Kjolby and Harland, 2017; Nakamura et al., 2016). A subset of the targets share a gapped circle, or horseshoe pattern, around the blastopore with the gap in expression at the dorsal midline, similar to expression of wnt8a. However, expression of a small subset of targets resembles that of fgf8 as a ring around the blastopore. These observations initially led us to hypothesize that genes expressed in a horseshoe pattern are mostly Wnt activated, whereas those in a ring pattern are additionally FGF activated.
We aim to discover a mechanism to explain how different expression patterns of the Wnt target genes are generated by the combinatorial activity of FGF8 and Wnt signaling. Using ChIP-seq data, we first show that all Wnt target genes during gastrulation also have Ets2 binding sites in their regulatory regions. Second, by associating β-catenin with the binding of Ets2, we found Ets2 bound to the transcription start site (TSS) of both horseshoe and ring genes, whereas β-catenin binds to the TSS of genes with a horseshoe pattern, but more distantly to genes with the ring pattern. We suggest that promoter proximal binding of TCF allows for active repression in the absence of Wnt signaling at the dorsal midline, and this cannot be overcome by Ets2, whereas more distant recruitment of repressive TCF is ineffective in repressing Ets-mediated activity. To support this, we show evidence that, at the dorsal midline, the expression of genes with a ring pattern is regulated by FGF and that Ets2 cannot induce expression of horseshoe genes in this region, whereas Wnt signaling can. We therefore hypothesize that both horseshoe and ring genes are cooperatively activated by Wnt signaling in the ventral and lateral regions, whereas negative regulation by TCF binding represses horseshoe genes in the dorsal regions. Ring genes are activated by both Wnt and FGF in parallel, but any repressive TCF binding in the dorsal domain is too distant from the promoter to inhibit gene activity.
RESULTS
Wnt target gene expression falls into two categories; a ring pattern or a horseshoe pattern
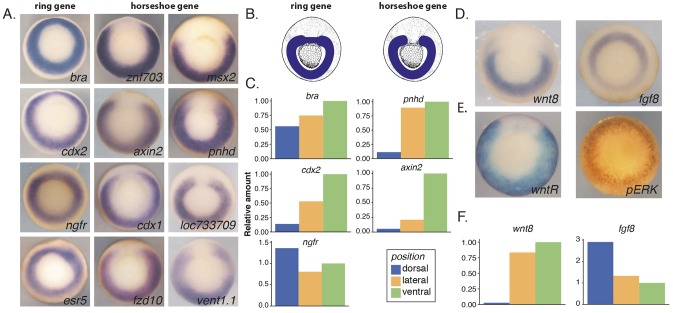
To assess the gene expression pattern of previously identified Wnt target genes during Xenopus gastrulation, in situ hybridization was carried out for 18 target genes (Fig. 1A and Fig. S1A) (Kjolby and Harland, 2017). Expression patterns fall into one of two categories: a horseshoe pattern around the blastopore with a gap in expression at the dorsal midline, or a ring shape pattern around the blastopore (Fig. 1A,B). Although the size of the gap and the thickness of the ring vary among Wnt target genes, these two categories are easily separated visually by in situ hybridization (Fig. S1B and not shown). In addition to the whole-mount in situ hybridization, we dissected mid-gastrula (stage 11.5) embryos to more accurately quantify gene expression in the different regions (Fig. S1D). We compared three regions, dorsal (roughly 60°), lateral and ventral, and measured gene expression using qPCR. Expression levels were normalized to eef1a1 (ef1a) and compared with the ventral region. As expected, chordin (chrd.1) was expressed exclusively in the dorsal region, whereas brachyury (tbxt) was slightly enriched in the ventral region and ngfr was slightly enriched in the dorsal region (Fig. 1C and Fig. S1C). Although cdx2, pnhd and axin2 appear to be weakly expressed in the dorsal region compared with the ventral region, using qPCR, we note that these values are relative to the ventral region and cannot be compared between the three different genes. Therefore, cdx2 could have more expression in the ventral region relative to the dorsal region at this stage, however still be expressed to a level that can be visualized by in situ hybridization.
Fig. 1.
Wnt target gene expression falls into two categories: a ring pattern or a horseshoe pattern. (A) In situ hybridization of Wnt target genes at mid gastrula (stage 11.5). Expression patterns fall into two main categories; ring and horseshoe shape around the blastopore. (B) Schematic of expression pattern for a ring versus horseshoe gene at mid gastrula. (C) qPCR data from eight embryos dissected into dorsal, lateral and ventral marginal zone pieces. eef1a1 was used as an internal control and values were normalized to the ventral pieces. (D) RNA in situ hybridization pattern for wnt8 and fgf8. (E) Activity of Wnt and Fgf signaling pathways at mid-gastrula. RNA in situ hybridization for GFP reporter of Wnt activity is a horseshoe pattern (left). pERK staining for activity of Fgf pathway is in a ring pattern (right). (F) qPCR on same sample as in C for fgf8 and wnt8.
The expression pattern of wnt8, thought to be the main activator of the gastrula Wnt target genes, also has a horseshoe shape, with the largest gap at the dorsal midline compared with its target genes as visualized by in situ hybridization and quantified by qPCR (Fig. 1D,F). Wnt8 is a ligand, but the extent to which it may diffuse and signal in the embryo is limited (Mii and Taira, 2009). Therefore, we used a Wnt reporter to visualize the extent of active Wnt signaling around the blastopore. The transgenic Wnt reporter frog contains 8× TCF motifs upstream of GFP (Tran et al., 2010). To visualize where Wnt signaling is active at gastrulation we fixed embryos and visualized the gfp mRNA using in situ hybridization. About 50% (13/26) of embryos had strong expression in a horseshoe pattern around the blastopore, whereas 15% (4/26) showed the same pattern but with weaker staining and ∼35% (9/26) showed no staining at all (Fig. 1E and Fig. S1E). We speculate that this is owing to the uncharacterized integration number and genotype of this Wnt reporter frog. The expression domain is restricted to the marginal zone and expression was abolished in DKK-injected embryos (Fig. S1E).
The lack of Wnt reporter expression at the dorsal midline and the extreme response to Wnt inhibition by DKK of the reporter led us to hypothesize that there may be other regulators of the Wnt target genes that are expressed circumferentially. A likely regulator is the FGF signaling pathway, because fgf8 is expressed in a ring around the blastopore and activates ERK there (Fig. 1D) (Christen and Slack, 1997; Fig. 1D,E). Supporting the previous work, we confirmed the specificity of FGF signaling, showing the signal was abolished when embryos were incubated in the FGF receptor inhibitor, SU5402 (Fig. S1F). In addition, the qPCR data show that fgf8 is expressed more highly in the dorsal versus ventral region at this stage (Fig. 1F). We therefore suspected that the expression patterns of the previously characterized Wnt target genes may be dictated by Wnt in the ventral and lateral mesodermal regions and FGF predominantly in the dorsal region. However, both activities may contribute in the lateral and ventral regions and be differently regulated in the dorsal region.
Genome-wide identification of Ets2 binding site reveals co-localization with TSS and putative enhancers
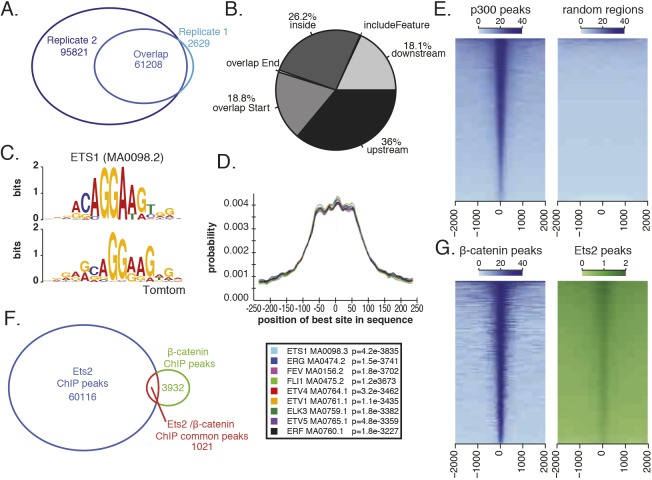
To investigate whether the FGF signaling pathway could directly regulate either all or a subset of the Wnt target genes to generate the two categories of expression, we looked at whether the downstream transcription factor of FGF signaling, Ets2, was bound near the Wnt target genes. We took a genome-wide approach and performed ChIP-seq on a triple-FLAG-tagged Ets2. As previously reported, Ets2 acts downstream of FGF signaling as the main effector during Xenopus mesoderm development (Kawachi et al., 2003). Indeed, overexpression (1 ng) of the triple-FLAG-tagged Ets2 results in a posteriorized phenotype very similar to FGF8b overexpression (Fig. S2A). For ChIP, embryos were injected with 750 pg mRNA encoding a C-terminally triple-FLAG-tagged Ets2, a dose that produced only a mild overexpression phenotype (Fig. S2A). Expression of the FLAG-tagged Ets2 was confirmed using western blotting (Fig. S2B) and mid-gastrula (stage 11.5) embryos were collected for ChIP. Success of the ChIP was validated by qPCR of previously characterized FGF responsive regions (Fig. S2C) (Haremaki et al., 2003; Degenhardt et al., 2010) and data are deposited in GEO under accession number GSE112249. A final set of 61,208 peaks was called as the intersect of replicate 1 and 2 using the R package, ChipPeakAnno (Zhu et al., 2010) (Fig. 2A). Several of these regions were further validated by ChIP-qPCR using an antibody that detects endogenous Ets2. Although this antibody was not potent enough for ChIP library construction, qPCR results demonstrate that inhibiting the FGF pathway with the small molecule inhibitor SU5402 results in reduced signal at these Ets2-bound regions (Fig. S2D).
Fig. 2.
ChIP-seq of a FLAG-tagged Ets2 at mid gastrula identifies Ets2-bound regions. (A) Number of Ets2 ChIP peaks identified by MACS2 in each replicate and the overlap. (B) Percentage of peak overlap with indicated genomic features. (C) Using MEME-ChIP, Ets1 (MA0098.2) motif is identified as enriched in Ets2 ChIP-seq peaks. Top is the Ets1 motif and bottom is the enriched motif from the ChIP-seq peaks. (D) Graph generated by CentriMo showing the probability of finding a given motif in the Ets2 peak region. Ets family transcription factors that were centrally enriched are shown (below). (E) Heatmaps of Ets2 ChIP-seq coverage on p300 peaks compared with random regions. (F) Overlap of Ets2 ChIP peaks and β-catenin ChIP peaks. Both sets of data were analyzed using MACS2 and ChIPpeakAnno R package. (G) Heatmap of Ets2 coverage on β-catenin peaks and β-catenin coverage on Ets2 peaks.
When assigning nearest genes to each Ets2 binding peak, 36% of peaks were found to be upstream of the TSS, 26% were found to overlap with gene bodies (inside, mostly in introns) and 18% overlapped with the TSS (overlap Start), suggesting that Ets2 can bind to both enhancer and promoter regions (Fig. 2B). In addition, when calculating the distance from nearest TSS to peak, there is strong enrichment at the TSS compared with random regions (Fig. S2E) and Ets2 ChIP coverage nicely centers over p300 peaks from Session et al. (2016; GSE76059); 21% of peaks overlap between Ets2 and p300 ChIP peaks, compared with 0.7% of Ets2 peaks overlapping with randomly shuffled p300 ChIP regions, suggesting that it is binding to enhancers (Fig. 2E).
To further validate our ChIP results, we performed a de novo motif search using MEME-ChIP by first extending peak regions 250 bp to either side of the summits of all Ets2 peaks (Machanick and Bailey, 2011). The most significant motif (E-value=2.2e-3832) closely matched the Ets1 motif (MA0098.2) (Fig. 2C). This is to be expected as there are no Ets2 motifs in the JASPAR database and Ets1 and Ets2 are similar and often serve redundant roles (Tymms and Kola, 1994; Hollenhorst et al., 2007). The Ets1 motif was present in 22,814 (37%) (Fig. S3D). This number may be low because closely matched motifs may be assigned to other Ets family transcription factors. Indeed, when mapping the distribution of Ets1 and Ets-related motifs, many enrich at the summits of peaks and 49% of peaks contain an Ets motif (Fig. 2D and Fig. S3D). In addition to the Ets1 motif, the MEME program found other families of transcription factor motifs centrally enriched. The TFAP2 family of transcription factor motifs are enriched at ±50 bp from the center of the peaks and the TBX family of transcription factor motifs are enriched at ±75 bp (Fig. S2F).
We next looked at the overlap between Ets2 peaks and our previously published β-catenin ChIP dataset (Kjolby and Harland, 2017). β-Catenin ChIP sequencing reads were re-analyzed using the same methods as the Ets2 ChIP sequencing data. The β-catenin coverage centered over p300 peaks, confirming that β-catenin binds at enhancers (Fig. S2G). A small percentage of Ets2 and β-catenin peaks (1.7% and 26%, respectively) overlap, in a total of 1021 peaks (Fig. 2F). However, when looking at the coverage of all ChIP reads, β-catenin coverage centers over Ets2 peaks as well as Ets2 coverage centering over β-catenin peaks (Fig. 2G). Therefore, we conclude that although there are unique sets of Ets2 and β-catenin peaks, there is an additional subset that is shared between the two transcriptional activators.
Differential binding of β-catenin and Ets2 at the promoters of ring genes
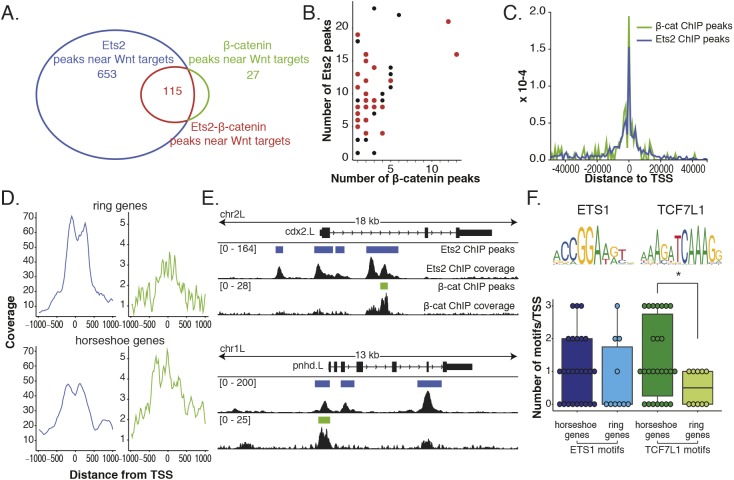
To more specifically investigate the role of differential regulation by Wnt and FGF on the Wnt target gene expression patterns, we narrowed down our list of putative regulatory regions to those that are within 50 kb of either side of the Wnt target gene set; 654 unique Ets2 peaks, 27 unique β-catenin peaks and 115 co-bound peaks remain (Fig. 3A). When compared with genome-wide peak overlap, this is a significant enrichment of co-bound regions (P-value=6e-205). Surprisingly, all Wnt target genes that have β-catenin ChIP peaks within 50 kb of either side of the coding gene also have an Ets2 peak (Fig. 3B). Next, we looked at the distribution of binding sites compared with the TSS of all Wnt target genes. Both β-catenin (as previously reported) and Ets2 are enriched near the TSS compared with random regions (Fig. 3C and Fig. S3A).
Fig. 3.
Differential overlap of Ets2 and β-catenin ChIP data at the promoters of Wnt target genes. (A) Overlap (115) of Ets2 and β-catenin subsets of peaks near previously identified Wnt target genes. (B) Number of Ets2 and β-catenin peaks near individual Wnt target genes. Genes in red are the targets that have in situ hybridization patterns described here. Genes in black are without described expression patterns. (C) Histogram of distance to TSS of both Ets2 and β-catenin ChIP peaks using subset of peaks near Wnt target genes. (D) Wnt target genes were first separated based on having either a ring or horseshoe expression pattern. Then the coverage at ±1 kb from the TSS of each gene was calculated and plotted for both Ets2 (blue) and β-catenin (green) ChIP. (E) Genome-browser view of Ets and β-catenin coverage around TSS of a ring gene (cdx2) and a horseshoe gene (pnhd). (F) Number of Ets or TCF7L1 (TCF3) motifs around TSS for horseshoe or ring genes. P-value of 0.38 between horseshoe and ring genes for Ets motif and 0.042 for TCF3 motif (*) calculated using Wilcoxon rank sum test. Data are mean±s.d. with individual data points indicated.
Although there is significant overlap of Ets and β-catenin peaks, peaks are specific to each transcription factor. Of the Ets peaks that are near Wnt target genes, 41% have TCF motifs, whereas 73% of β-catenin peaks have TCF motifs (Fig. S3D). Of the β-catenin peaks that are near Wnt target genes, only 6% have Ets1 motifs, whereas 18% of Ets peaks have Ets1 motifs (Fig. S3D).
To determine whether there are differences in the binding of Ets2 and β-catenin at promoters that could explain the expression patterns, we plotted the coverage of sequencing reads for Ets2 and β-catenin around the TSS regions of either ring or horseshoe genes (Fig. 3D). Although the coverage of Ets2 at the TSS for ring genes is greater than the coverage for horseshoe genes, the coverage for β-catenin at the TSS is greater for horseshoe genes than ring genes (Fig. 3D and Fig. S4A,B). The enrichment of β-catenin around the TSS becomes even more apparent when comparing regions 6 kb from either side of the TSS (Fig. S3B).
For example, when looking at the pileup of ChIP sequencing reads and called peaks of a ring gene, cdx2, there is a pile up of reads at the TSS for Ets2, but not for β-catenin, whereas for pnhd, a horseshoe gene, there is a pileup of reads and a peak for both Ets2 and β-catenin (Fig. 3E). To further support this finding, we counted the number of Ets1 or TCF7L1 motifs found around the TSS of either horseshoe or ring genes to see whether horseshoe genes would have more TCF motifs than ring genes (Fig. 3F). Indeed, there are about equal numbers of Ets1 motifs around the TSS for both horseshoe and ring genes (mean of 1 and 0.8 motif per TSS, respectively), but there are more TCF7L1 (TCF3) motifs discovered surrounding the TSS of horseshoe genes (Fig. 3F).
We therefore make the hypothesis that in the absence of Wnt signaling at the dorsal midline, genes with a β-catenin binding site are negatively regulated by TCF and therefore form a horseshoe pattern. Even though there is an Ets binding site at the TSS as well, perhaps it cannot overcome the negative regulation by the local recruitment of Groucho by TCF. Genes without a β-catenin binding site near the promoter are therefore not negatively regulated at the TSS at the dorsal midline and Ets2 can activate their expression, resulting in a ring pattern.
Contributions of active Wnt/β-catenin, repression in the absence of Wnt signals and FGF signaling in the marginal zone
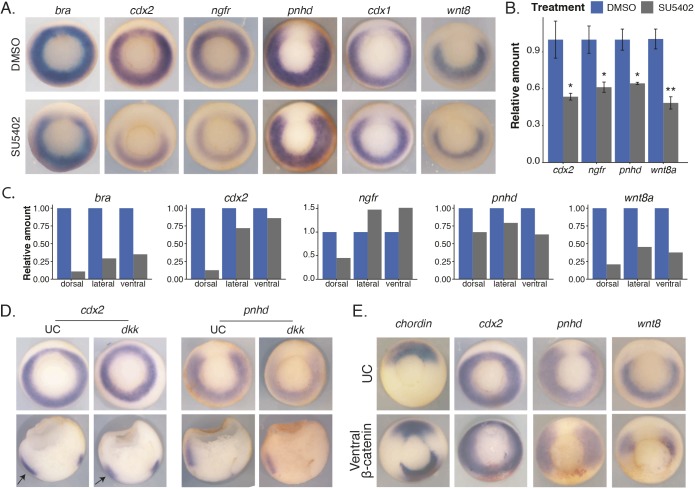
To test the contribution of FGF signaling to the expression patterns of Wnt target genes, we incubated embryos starting at stage 10 (at the first sign of blastopore lip formation) in an FGFR inhibitor, SU5402 (Mohammadi et al., 1997). Interestingly, for ring genes such as cdx2 and ngfr, although overall expression is reduced owing to FGF inhibition (visualized by in situ hybridization and quantified by qPCR), the resultant pattern of remaining expression is a horseshoe pattern (Fig. 4A,B). For genes that are normally expressed in a horseshoe pattern such as pnhd and cdx1, SU5402 appears to only slightly reduce the in situ hybridization signal, however measured using qPCR they are reduced to ∼60% of control (Fig. 4B). It is well known that FGF regulates wnt8 expression and, although the size of the dorsal gap in expression does not appear to change for wnt8, using qPCR, wnt8 expression is reduced by half at this dose of SU5402 (Fig. 4A,B) (Hong et al., 2008). These data also suggest that the gap formed in ring genes as a result of FGF inhibition is due to lack of FGF signaling, not secondarily by loss of Wnt signaling. To further confirm the result that there is greater loss of expression at the dorsal region for ring genes, we dissected gastrula (stage 11.5) control or FGF-inhibited embryos. Again, we cut the embryos into three regions: dorsal (roughly 60°), lateral, and ventral, and assessed changes in gene expression due to FGF inhibition using qPCR. Indeed, whereas horseshoe genes such as pnhd and wnt8 were reduced in expression in roughly the same amount in all three sections, ring genes such as bra, cdx2 and ngfr had the greatest change in expression in the dorsal sections (Fig. 4C).
Fig. 4.
Contributions of active Wnt/β-catenin, repression in the absence of Wnt signals, and FGF signaling in the marginal zone. (A) RNA in situ hybridization on mid-gastrula-stage embryos incubated in either DMSO or 50 µM SU5402 at stage 10.5 until collection. (B) Embryos were treated as in A, but collected in Trizol for RNA extraction and subsequent qPCR. Data shown is from three replicates of single embryos. Data are mean±s.d. *P<0.05; **P<0.01; Student's t-test. (C) Embryos were either incubated in DMSO or 50 µM SU5402 starting at stage 10.5 until dissection at stage 11.5. qPCR data from dorsal, lateral or ventral equatorial dissections. Results are from a single experiment using 10 embryos for each condition. (D) In situ hybridization on gastrula-stage embryos injected with 100 pg dkk1 in all four blastomeres of a four-cell-stage embryo. Arrows are pointing to the ventral domain. Sections were made by bisecting embryos. (E) In situ hybridization on gastrula-stage embryos injected in both ventral blastomeres at the four-cell stage with β-catenin and β-gal tracer. UC, uninjected control embryos.
We next tested the contribution of Wnt signaling to the expression patterns of the Wnt target genes. We injected 100 pg of dkk1 into all blastomeres of a four-cell-stage embryo and performed in situ hybridization. Although the activity of the Wnt reporter was abolished, expression of both ring and horseshoe genes persisted to some extent, even though the same dose of dkk was used (Fig. 4D and Figs S1E, S5A). In addition, for ring genes, expression was reduced mainly in the ventral and lateral regions, whereas expression remained in the dorsal midline (Fig. 4D). This further suggests that expression in the dorsal midline domain is not controlled by Wnt signaling, but rather by FGF.
One prediction from these results is that, if we induce a second organizer, at which zygotic Wnt expression is inactivated, then we should be able to induce a second gap in horseshoe genes but not ring genes. To mimic the early Nieuwkoop center signal, we injected β-catenin mRNA at the four-cell stage into the two prospective ventral blastomeres and traced our injection with β-galactosidase (β-gal) (Fig. 4). Injection of β-catenin indeed produced a second organizer as evidenced by ectopic chordin and gsc expression at the site of injection (Fig. 4E and Fig. S5A). This second organizer did not generate a gap in expression for ring genes such as cdx2, but for horseshoe genes such as axin2 and pnhd, an ectopic gap in expression was formed at the site of injection (Fig. 4E and Fig. S5B).
These data show that when we inhibit FGF signaling, a gap in expression is formed in ring genes, whereas if we remove Wnt signaling to a level that abolishes reporter expression, there is still residual expression of both horseshoe and ring genes; notably at the dorsal region for the ring genes. In addition, if we generate a second organizer and thus a secondary gap in wnt8 expression, we form an ectopic gap in in horseshoe gene expression. This suggests that ring genes require FGF signaling at the dorsal midline and both FGF and Wnt in the lateral and ventral domains to form the correct pattern, whereas horseshoe genes simply require proper Wnt signaling for the correct induction of ventrolateral expression and repression of dorsal expression.
FGF/Ets is not sufficient to induce ectopic expression of the horseshoe Wnt target genes at the dorsal midline, whereas Wnt signaling is
To test whether Ets2 is sufficient for expression of the horseshoe Wnt target genes at the dorsal midline, we injected ets2 mRNA together with β-gal tracer in the two prospective dorsal blastomeres at the four-cell stage. Although overexpression of ets2 appears to increase expression at the dorsal midline for ring genes such as cdx2, Ets2 overexpression cannot ectopically induce expression of horseshoe genes at the dorsal midline (Fig. 5A). In addition, dorsal injection of ets2 was able to rescue the gap formed at the dorsal midline as a result of FGF inhibition by SU5402 for ring genes (Fig. S6A). Occasionally, we saw ectopic expression in more anterior regions at the dorsal midline, suggesting that this region has other competence factors that more easily allow Ets2 to activate expression (Fig. S6B). However, in contrast to the inability of Ets2 to activate dorsal expression of horseshoe genes, the gap was easily filled in when Wnt signaling was overactivated by incubation in the Wnt agonist, BIO (Fig. 5B).
Fig. 5.
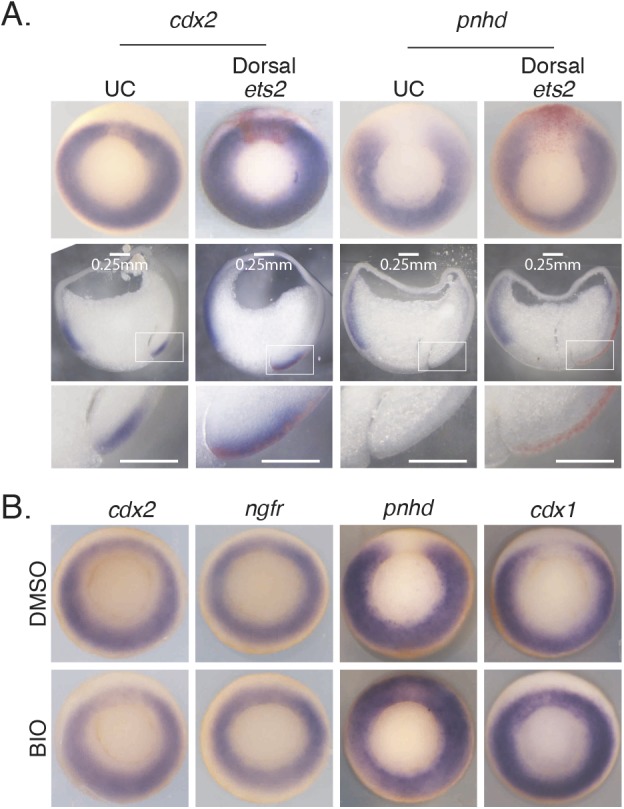
FGF/Ets is not sufficient for expression of the horseshoe Wnt target genes at the dorsal midline, whereas Wnt signaling is. (A) In situ hybridization on gastrula-stage embryos injected with ets2 mRNA and β-gal tracer (red) in the two dorsal blastomeres at the four-cell stage. Sections are of whole embryos pictured. Bottom row shows higher magnification of boxed areas from row above. (B) In situ hybridization on gastrula-stage embryos incubated in the Wnt agonist BIO starting at stage 8-9 until collection.
Ets2 is sufficient for expression of ring genes in dorsal mesoderm explants but not for horseshoe genes
Animal cap experiments are a powerful tool to study gene regulation during Xenopus development, as they can be removed and manipulated to form different tissue types. To determine the ability of Ets2 to drive expression of ring pattern genes but not horseshoe pattern genes, we induced animal caps to form mesoderm with either dorsal or ventral character to serve as substrates for the assay (Fig. 6A,B). To form dorsal mesoderm, embryos were first dorsalized by incubation in 0.3M LiCl between the eight- to 16-cell stages for 6 min. At this dose and duration of incubation, embryos had an average dorsoanterior index (DAI) of 8.5 compared with 5.1 for control, 5.8 for 3 min treatment and 9.3 for 10 min treatment (Fig. S7A). For ventral mesoderm, embryos were treated with UV light 1 h after fertilization (incubated at 16°C) before cortical rotation was initiated. The dose of UV produced a DAI of 1 compared with a DAI of 5 for the control (Fig. S7B).
Fig. 6.
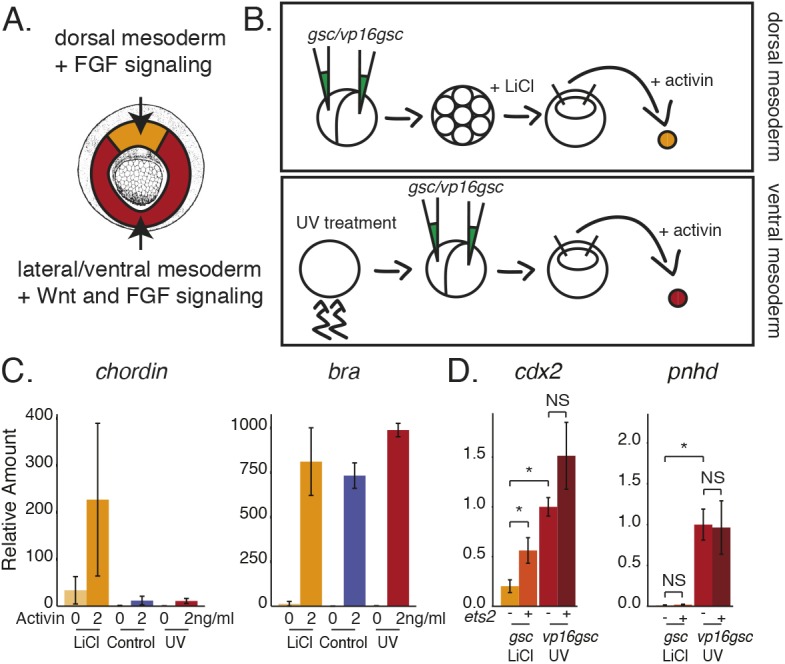
Ets is sufficient for expression of ring genes in dorsal mesoderm explants but not for expression of horseshoe genes. (A) Schematic of location of dorsal versus lateral/ventral mesoderm in mid-gastrula-stage embryos. (B) Schematic of experiment to make either dorsal or ventral mesoderm from animal cap explants. (C) qPCR from triplicate experiments showing expression of either chordin or brachyury in Activin-treated caps derived from either control, dorsalized (LiCl) or ventralized (UV) embryos. (D) qPCR from triplicate experiments showing expression of either cdx2 or pnhd from dorsal and ventral mesodermal caps with or without the addition of ets2 mRNA. Data are mean±s.d. *P<0.05, Student's t-test. NS, not significant.
Early blastula animal caps were then cut from either LiCl-treated or UV-treated embryos and cultured in 2 ng/ml Activin. This dose of Activin was sufficient to elongate animal caps and to turn on the skeletal muscle marker immunostained for the 12/101 antibody in animal caps derived from either control or UV-treated embryos (Fig. S7D). When animal caps derived from dorsalized embryos were incubated in 2 ng/ml Activin they also turned on markers of dorsal mesoderm such as Tor70 (Fig. S7D) (Bolce et al., 1992). In addition, dorsal animal caps expressed the dorsal mesodermal marker, chordin, and all animal caps treated with Activin expressed the mesodermal marker, bra (Fig. 6C). To further ensure that ventral mesoderm caps had Wnt activity, ventralized embryos were also injected with an activator form of gsc, vp16gsc, which promotes expression of wnt8, and to ensure that there was no Wnt activity in the dorsal mesodermal caps, dorsalized embryos were injected with gsc, the normal role of which is to inhibit expression of wnt8 (Yao and Kessler, 2001). Indeed, in both ventralized and dorsalized mesodermal animal caps, vp16gsc induced expression of wnt8 compared with caps derived from embryos injected with gsc (Fig. S7E).
Having established the substrate for the assay, we then tested the expression levels of cdx2 and pnhd in our dorsal versus ventral mesodermal caps. Based on whole-mount in situ hybridization expression patterns and our dissections, we expected that both cdx2 and pnhd would be expressed to a greater level in the ventral mesodermal caps compared with the dorsal mesodermal caps. Indeed, both were significantly more highly expressed, as assayed by qPCR, in the ventral mesodermal caps as there is Wnt activity regulating this expression (Fig. 6D).
To test whether ets2 could further increase expression we co-injected ets2 with either gsc in our dorsal mesodermal caps or with vp16gsc in our ventral mesodermal caps. Based on our previous results, we expected that cdx2 should be responsive to ets2 induction whereas pnhd should not be responsive in the absence of Wnt signaling. Indeed, in the dorsal mesodermal caps, the addition of ets2 significantly increased expression of cdx2, but not of pnhd (Fig. 6D). We hypothesize that this is because of active repression by TCF at the promoters of horseshoe genes such as pnhd. In the ventral mesodermal caps, we expected that ets2 might increase expression in an already high Wnt environment. Although not statistically significant, ets2 does modestly increase expression of cdx2 in ventral mesodermal caps, however it does not increase expression of pnhd (Fig. 6D). We speculate that Wnt signaling is high in these caps and expression is already saturated such that addition of ets2 has minimal effect. Thus, overall, these experiments confirm the different responsiveness of horseshoe and ring genes in dorsal and ventrolateral tissues.
DISCUSSION
Both the Wnt and FGF signaling pathways induce and are required for posterior fates in the embryo. They are both expressed around the blastopore of gastrula-stage embryos. However, whether these pathways act directly on a set of common target genes to achieve this was not clear, and some studies had suggested a sequential pathway. In addition, although overexpression or inhibition of either pathway results in similar phenotypes, there are obvious differences that could be a result of differential regulation on a set of common target genes. Our results suggest that there is a set of common targets that are regulated in at least one of two ways: The common target is expressed in a ring pattern around the blastopore, being activated by FGF and Wnt in the lateral and ventral regions and by FGF alone in the dorsal region, or the common target is expressed in a horseshoe pattern, again being activated by Wnt and FGF in the lateral and ventral regions, but is repressed in the dorsal region by lack of Wnt signaling. The difference in patterns can be accounted for by differential binding of TCF3 to the promoters of horseshoe genes and ring genes.
Activity of signaling pathways mimic two categories of expression patterns
By comparing the expression patterns of previously identified Wnt target genes, we found that the patterns fall into two main categories that mimic the expression patterns for wnt8 and fgf8 (Fig. 1A,D). The patterns more closely mimic the patterns of the active signaling pathways, with Wnt active in the lateral and ventral regions and FGF active around the entire blastopore, the highest activity being seen in the dorsal region (Fig. 1E). Removal of FGF signaling by incubation in an FGF inhibitor caused ring pattern genes to appear as horseshoe pattern genes, suggesting that this part of the pattern is driven by Wnt (Fig. 4A). In contrast, when we inhibit Wnt signaling, ring pattern genes lose expression predominantly in the lateral and ventral region, although they maintain expression in the dorsal region, suggesting that a large fraction of this lateral and ventral expression is regulated by Wnt signaling (Fig. 4D).
Combinatorial regulation by Wnt and FGF
Our data suggest that Wnt and FGF both act directly on a common set of target genes during Xenopus gastrulation. Although FGF may also contribute to activating Wnt gene expression and vice-versa, our ChIP data show that both β-catenin and Ets2 bind regulatory regions of a common set of genes (Fig. 3). In fact, we show evidence that FGF does regulate wnt8 expression as embryos incubated in the FGF inhibitor, SU5402, showed reduced expression of wnt8 using in situ hybridization and qPCR (Fig. 4A-C). However, this does not preclude the possibility of the two pathways acting directly at the transcriptional level.
FGF may not be the only other signaling pathway involved in forming the expression patterns of the Wnt target genes. The Wnt/β-catenin signaling pathway has also been shown to interact with the BMP signaling pathway to directly activate target genes such as msx2 during gastrulation (Hussein et al., 2003). It will be interesting to follow up with ChIP experiments to ask what subset of Wnt/β-catenin targets are also directly regulated by BMP signaling, as BMP activity is greatest in the ventral region of the gastrula (Faure et al., 2002).
Ets2 cannot overcome active repression by TCF3 at promoters
The repressive activity of TCF in the absence of β-catenin was first shown by Brannon and Kimelman, with TCF-binding sites required for repression of siamois (sia1) on the ventral side of the early gastrula (Brannon et al., 1997). This conclusion was reinforced by experiments knocking down TCF3, which resulted in activation of dorsal gene expression (Houston et al., 2002). In Drosophila, mutation of dTcf (Pan) binding sites in the dpp enhancer also results in ectopic reporter expression (Yang et al., 2000). In another study, mutating dTCF sites in an eve reporter suggested that, in the absence of Wg/Wnt signaling, dTCF serves to abrogate synergistic activities of Smads and Tin in eve activation (Knirr and Frasch, 2001).
A particularly pertinent example in Xenopus is the study of the cdx4 intronic regulatory region in which the authors showed that injection of TCF3 reduces the activation of a cdx4 intronic reporter by bFGF (Haremaki et al., 2003). Although cdx4 is not on our list of Wnt targets because it did not pass a significance threshold for differential expression in our original RNA-seq analysis (Kjolby and Harland, 2017), cdx4 is known to be regulated by both Wnt and FGF signaling (Haremaki et al., 2003; Keenan et al., 2006; Shimizu et al., 2005), is expressed in a ring around the blastopore (Northrop and Kimelman, 1994) and lacks β-catenin ChIP coverage at the TSS. At first sight, this conflicts with our finding that cdx4 would not be repressed by TCF3 in the dorsal region (Fig. 3D), but in the reporter experiments, Haremaki et al. (2003) fused the intronic enhancer to the reporter, such that the TCF binding site is now close to the TSS, making it subject to TCF repression in the absence of Wnt signaling. This enhancer is also found to be a TCF/β-catenin binding region in our experiments, but in the normal cdx4 gene is located 4 kb downstream of the TSS, making it refractory to TCF-mediated repression. In retrospect, these experiments provide a good test and verification of our model for the repressive activity of TCF only in proximity to the promoter.
Our data suggest that this type of negative regulation is occurring for horseshoe genes where TCF3, and Groucho or CtBP at the promoters of these Wnt/FGF target genes, represses their expression at the dorsal midline. Even though these genes are also targets of FGF signaling, it appears that even overexpression of Ets2 is not sufficient to overcome the repression by the TCF3 complex (Figs 5A and 6D). Although Groucho was initially thought to act as a long-range repressor (Barolo and Levine, 1997), it has also been described as acting locally over short ranges rather than in a dominant long-range fashion (Nibu et al., 2001). In addition, with the increase in resolution of ChIP data in recent years, others have found that Groucho does not appear to oligomerize along DNA, but rather appears in discrete peaks that span less than a kilobase (Kaul et al., 2015). Alternatively, the short-range repressor CtBP may be the dominant corepressor bound in this context and subject the promoter to short-range repression (Brannon et al., 1997; Nibu et al., 2001).
Our initial goal with these experiments was to identify all the Wnt and FGF target genes in posterior development of the gastrula. All of the targets that we found are expressed in a ring of posterior mesoderm or ectoderm during gastrulation, though several of them are not expressed in the organizer region. The induction of posterior development is a property of the entire marginal zone (Kolm and Sive, 1997), whereas the organizer region can promote anterior development in part by the exclusion of signal transcripts such as wnt8 and wnt3 from the organizer, and protect the anterior domain from posterior signals through the expression of Wnt antagonists. Although the organizer does progressively become differentiated into anterior and posterior domains of gene expression and signaling activity (Zoltewicz and Gerhart, 1997), the exclusion of a subset of Wnt and FGF target transcripts from the organizer is likely to be important to continue to protect anterior regions from posterior determination. As development proceeds, the Wnt and FGF targets further alter their expression (Kjolby and Harland, 2017) to mediate events such as neural crest induction or further posterior development of the tailbud. The network of signaling and enhancer activity that leads to neural crest formation is now being dissected (Simões-Costa and Bronner, 2015), but the determinants of differences between neural crest and posterior germ layer differentiation remains not fully understood.
MATERIALS AND METHODS
Embryo culture and manipulation
X. laevis embryos were obtained (Sive et al., 2010) and staged (Nieuwkoop and Faber, 1967) as previously described. All controls are uninjected controls (UC) unless otherwise indicated. All animal work was conducted under the auspices of the Institutional Animal Care and Use Committee; NIH assurance AS107-01.
Animal caps
For ventralized mesodermal explants, embryos were incubated at 16°C after fertilization and de-jellied 45 min postfertilization with 3% cysteine, then placed on quartz glass (Ted Pella, 26012) and exposed to UV radiation in a UV Stratalinker 1800 (Stratagene) turned upside down using the Auto Crosslink setting. For dorsalized mesodermal explants, again embryos were de-jellied 45 min postfertilization, then placed in 0.3 M LiCl solution for 6 min between the eight- to 16-cell stages. Explants were cut at stage 9 from the animal region and cultured in 2 ng/ml Activin (VWR, 10007062). For stage 11.5 equatorial explants, the depth of the archenteron was used to recognize the dorsal side during dissection.
Wnt reporter
The Wnt reporter frog has been described previously (Tran and Vleminckx, 2013). An in situ hybridization probe was generated for GFP after digesting the pcs2 GFP plasmid with BamHI.
FGF inhibitor treatments
Embryos were cultured in 75 µM SU5402 (Millipore, CAS215543-92-3) starting at stage 10 until stage 11.5 to inhibit FGF signaling. The dose used was empirically determined as a dose that does not inhibit mesoderm induction but still blocks FGF signaling. Given variation between lots, the dose used was empirically determined as a dose that inhibits the phosphorylation of ERK (Fig S1F) and inhibits FGF-dependent expression of mesodermal genes, but does not block Wnt-dependent gene expression in the mesoderm (as shown in Fig. 4).
DNA constructs and RNA synthesis
The triple-FLAG-epitope-tagged X. laevis Ets2 (Ets2-3×FLAG) was generated using primers for Ets2 including restriction enzyme recognition sites for ClaI and AvrII (underlined): F, TGCAATCGATATGACAGAGTTTGGAATTCG; R, AATCTCCTAGGGCTTCGTCTGTGTCGGGC. The ets2 fragment was amplified from cDNA and then ligated into the pcs108 plasmid containing a triple-FLAG-epitope tag as previously described (Kjolby and Harland, 2017). Plasmids for synthesis of vp16-gsc and gsc mRNA were provided graciously by Dan Kessler (Perelman School of Medicine at the University of Pennsylvania, Philadelphia, USA).
Capped RNAs were synthesized using an mMessage mMachine (Ambion). dkk1 pCS108, β-catenin-3×FLAG pCS108 and ets2-3×FLAG pCS108 were digested using AscI and transcribed with SP6 RNA polymerase. All RNAs were injected in 5nl or 10nl bursts along with mCherry RNA to serve as a tracer.
Whole-mount in situ hybridization and β-gal staining
Embryos were stained using RNA in situ hybridization as previously described (Harland, 1991). For β-gal staining, embryos were collected in MEMFA 100 mM MOPS (pH 7.4), 2 mM EGTA, 1 mM MgSO4, 3.7% formaldehyde) for 30 min at room temperature and then rinsed twice with Ptween (PBS+0.1% Tween-20). Embryos were incubated for 20 min at 37°C in β-gal stain (250ul Ferricyanide, 250 µl Ferrocyanide, 125 µl Red-Gal, 10 µl 1 M MgCl2, 4.365 µl Ptween). Embryos for sectioning were mounted in a PBS solution containing 20% sucrose, 30% SNA and 4.9% gelatin, and fixed with 1.5% glutaraldehyde (Walentek et al., 2012). Embedded embryos were sectioned at 50 μm using a Pelco 101 vibratome.
pERK staining
Whole-mount pERK staining was performed using a primary anti-pERK antibody (Cell Signaling Technology, #9101) at 1:400 followed by biotin-labeled secondary anti-rabbit antibody (Invitrogen, #31820) at 1:3000. HRP-labeled Streptavidin at 1:300 was then used to detect this secondary antibody followed by visualization with diaminobenzidine and hydrogen peroxide (Sive et al., 2010).
qPCR
For quantitative PCR (qPCR) analysis cDNA was made using 1ug of total RNA using iScript (Bio-Rad) and reactions were amplified using a CFX96 light cycler (Bio-Rad) with SsoAdvanced Universal SYBR Green supermix (Bio-Rad) (see Kjolby and Harland, 2017). In all samples eef1a1 was used as the internal control. Data shown is from RNA extracted from single embryos from three independent experiments (n=3).
Chromatin immunoprecipitation (ChIP)
We injected 750 pg of triple-FLAG C-terminus-tagged Ets2 into both blastomeres of X. laevis embryos at the two-cell stage. Embryos were collected at stage 11.5 and processed for ChIP as previously described (Wills et al., 2014; Blythe et al., 2009) using an anti-FLAG antibody (Sigma-Aldrich, F3165, xxdilutionxx). Sequencing libraries were made from immunoprecipitates of between 200-500 embryos and input chromatin (chromatin before immunoprecipitation) using the Illumina TruSeq ChIP prep kit according to the manufacturer's instructions. Libraries were made by the UC Berkeley Functional Genomics Laboratory and sequenced by the UC Berkeley Vincent J. Coates Genomics Sequencing Laboratory. The antibodies used in Fig. S3A were anti-FLAG (Sigma-Aldrich, F3165, 1:20) and anti-beta Actin (GeneTex, GT5512, 1:1000). For endogenous Ets2 ChIP, embryos were incubated in SU5402 from stage 10 to 11.5 and processed as described above using anti-ETS2 antibody (Cell Signaling Technology, 66476S, 1:500).
ChIP-seq analysis
Single-end sequencing reads of 50 bp were aligned to the X. laevis genome (assembly version 9.1) using bowtie2. Alignment rates were as follows: ChIP replicate 1, 95.4%; ChIP replicate 2, 95.56%; input replicate 1, 95.39%; input replicate 2, 95.71%. Peaks were called using MACS2 (Zhang et al., 2008) (https://github.com/taoliu/MACS/) and a final set of peaks was called using the R package, ChIPpeakAnno (Zhu et al., 2010). Both Ets2 ChIP as described in this manuscript as well as previously published β-catenin ChIP sequencing data (Kjolby and Harland, 2017) were analyzed using the same tools and parameters. ChIPpeakAnno was used to generate Venn diagrams and coverage heat maps. Bedtools (Quinlan and Hall, 2010) and custom scripts were used to calculated distance to TSS.
Peaks and read pile ups were visualized with Integrative Genomics Viewer (IGV) (Robinson et al., 2011). De novo motif analysis was performed using MEMEchip (Machanick and Bailey, 2011).
Supplementary Material
Acknowledgements
We thank Dan Kessler for donating vp16GSC and GSC expression plasmids, and Ryan Morrie for careful reading and editing.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Formal analysis: R.A.S.K.; Investigation: R.A.S.K., M.T.-G., S.I., R.M.H.; Writing - original draft: R.A.S.K., R.M.H.; Writing - review & editing: R.A.S.K., R.M.H.; Supervision: R.M.H.; Funding acquisition: R.M.H.
Funding
This work was supported by the National Institutes of Health (GM42341, GM086321, MH112158) and the Siebel Stem Cell Institute. Deposited in PMC for release after 12 months.
Data availability
ChIP-seq data are deposited in GEO under accession number GSE112249
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.179580.supplemental
References
- Barolo S. and Levine M. (1997). hairy mediates dominant repression in the Drosophila embryo. EMBO J. 16, 2883-2891. 10.1093/emboj/16.10.2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe S. A., Reid C. D., Kessler D. S. and Klein P. S. (2009). Chromatin immunoprecipitation in early Xenopus laevis embryos. Dev. Dyn. 238, 1422-1432. 10.1002/dvdy.21931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolce M. E., Hemmati-Brivanlou A., Kushner P. D. and Harland R. M. (1992). Ventral ectoderm of Xenopus forms neural tissue, including hindbrain, in response to activin. Development 115, 681-688. [DOI] [PubMed] [Google Scholar]
- Brannon M., Gomperts M., Sumoy L., Moon R. T. and Kimelman D. (1997). A beta-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes. Dev. 11, 2359-2370. 10.1101/gad.11.18.2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannon M., Brown J. D., Bates R., Kimelman D. and Moon R. T. (1999). XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development 126, 3159-3170. [DOI] [PubMed] [Google Scholar]
- Carballada R., Yasuo H. and Lemaire P. (2001). Phosphatidylinositol-3 kinase acts in parallel to the ERK MAP kinase in the FGF pathway during Xenopus mesoderm induction. Development 128, 35-44. [DOI] [PubMed] [Google Scholar]
- Christen B. and Slack J. M. W. (1997). FGF-8 is associated with anteroposterior patterning and limb regeneration in Xenopus. Dev. Biol. 192, 455-466. 10.1006/dbio.1997.8732 [DOI] [PubMed] [Google Scholar]
- Clevers H. (2006). Wnt/beta-catenin signaling in development and disease. Cell 127, 469-480. 10.1016/j.cell.2006.10.018 [DOI] [PubMed] [Google Scholar]
- Dailey L., Ambrosetti D., Mansukhani A. and Basilico C. (2005). Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor. Rev. 16, 233-247. 10.1016/j.cytogfr.2005.01.007 [DOI] [PubMed] [Google Scholar]
- Degenhardt K. R., Milewski R. C., Padmanabhan A., Miller M., Singh M. K., Lang D., Engleka K. A., Wu M., Li J., Zhou D. et al. (2010). Distinct enhancers at the Pax3 locus can function redundantly to regulate neural tube and neural crest expressions. Dev. Biol. 339, 519-527. 10.1016/j.ydbio.2009.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingos P. M., Itasaki N., Jones C. M., Mercurio S., Sargent M. G., Smith J. C. and Krumlauf R. (2001). The Wnt/beta-catenin pathway posteriorizes neural tissue in Xenopus by an indirect mechanism requiring FGF signalling. Dev. Biol. 239, 148-160. 10.1006/dbio.2001.0431 [DOI] [PubMed] [Google Scholar]
- Faure S., De Santa Barbara P., Roberts D. J. and Whitman M. (2002). Endogenous patterns of BMP signaling during early chick development. Dev. Biol. 244, 44-65. 10.1006/dbio.2002.0579 [DOI] [PubMed] [Google Scholar]
- Fletcher R. B., Baker J. C. and Harland R. M. (2006). FGF8 spliceforms mediate early mesoderm and posterior neural tissue formation in Xenopus. Development 133, 1703-1714. 10.1242/dev.02342 [DOI] [PubMed] [Google Scholar]
- Groves A. K. and LaBonne C. (2014). Setting appropriate boundaries: fate, patterning and competence at the neural plate border. Dev. Biol. 389, 2-12. 10.1016/j.ydbio.2013.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haremaki T., Tanaka Y., Hongo I., Yuge M. and Okamoto H. (2003). Integration of multiple signal transducing pathways on Fgf response elements of the Xenopus caudal homologue Xcad3. Development 130, 4907-4917. 10.1242/dev.00718 [DOI] [PubMed] [Google Scholar]
- Harland R. M. (1991). Appendix G: in situ hybridization: an improved whole-mount method for Xenopus embryos. In Xenopus laevis: Practical Uses in Cell and Molecular Biology: Methods in Cell Biology (ed. Kay B. and Peng H.), pp. 685-695. Elsevier. [DOI] [PubMed] [Google Scholar]
- Harland R. M. (2004). Dorsoventral patterning of the mesoderm. In Gastrulation: from Cells to Embryo (C. D. Stern, ed.), pp. 373-388 Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Hikasa H. and Sokol S. Y. (2013). Wnt signaling in vertebrate axis specification. Cold Spring Harbor Perspect. Biol. 5, a007955 10.1101/cshperspect.a007955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst P. C., Shah A. A., Hopkins C. and Graves B. J. (2007). Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 21, 1882-1894. 10.1101/gad.1561707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C. S., Park B. Y. and Saint-Jeannet J. P., (2008). Fgf8a induces neural crest indirectly through the activation of Wnt8 in the paraxial mesoderm. Development 135, 3903-3910. 10.1242/dev.026229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston D. W., Kofron M., Resnik E., Langland R., Destree O., Wylie C. and Heasman J. et al. (2002). Repression of organizer genes in dorsal and ventral Xenopus cells mediated by maternal XTcf3. Development 129, 4015-4025. [DOI] [PubMed] [Google Scholar]
- Hussein S. M., Duff E. K. and Sirard C. (2003). Smad4 and beta-catenin co-activators functionally interact with lymphoid-enhancing factor to regulate graded expression of Msx2. J. Biol. Chem. 278, 48805-48814. 10.1074/jbc.M305472200 [DOI] [PubMed] [Google Scholar]
- Kaul A. K., Schuster E. F. and Jennings B. H., (2015). Recent insights into Groucho co-repressor recruitment and function. Transcription 6, 7-11. 10.1080/21541264.2014.1000709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawachi K., Masuyama N. and Nishida E. (2003). Essential role of the transcription factor Ets-2 in Xenopus early development. J. Biol. Chem. 278, 5473-5477. 10.1074/jbc.M211054200 [DOI] [PubMed] [Google Scholar]
- Keenan I. D., Sharrard R. M. and Isaacs H. V. (2006). FGF signal transduction and the regulation of Cdx gene expression. Dev. Biol. 299, 478-488. 10.1016/j.ydbio.2006.08.040 [DOI] [PubMed] [Google Scholar]
- Kim C.-H., Oda T., Itoh M., Jiang D., Artinger K. B., Chandrasekharappa S. C., Driever W. and Chitnis A. B. (2000). Repressor activity of Headless/Tcf3 is essential for vertebrate head formation. Nature 407, 913-916. 10.1038/35038097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-E., Huang H., Zhao M., Zhang X., Zhang A., Semonov M. V., Macdonald B. T., Zhang X., Abreu J. G., Peng L. et al. (2013). Wnt stabilization of β-catenin reveals principles for morphogen receptor-scaffold assemblies. Science 340, 867-870. 10.1126/science.1232389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjolby R. A. S. and Harland R. M. (2017). Genome-wide identification of Wnt/b-catenin transcriptional targets during Xenopus gastrulation. Dev. Biol. 426, 165-175. 10.1016/j.ydbio.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knirr S. and Frasch M. (2001). Molecular integration of inductive and mesoderm-intrinsic inputs governs even-skipped enhancer activity in a subset of pericardial and dorsal muscle progenitors. Dev. Biol. 238, 13-26. 10.1006/dbio.2001.0397 [DOI] [PubMed] [Google Scholar]
- Kolm P. J. and Sive H. L., (1997). Retinoids and posterior neural induction: a reevaluation of Nieuwkoop's two-step hypothesis. Cold Spring Harbor Symp. Quant. Biol. 62, 511-521. 10.1101/SQB.1997.062.01.058 [DOI] [PubMed] [Google Scholar]
- Machanick P. and Bailey T. L. (2011). MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696-1697. 10.1093/bioinformatics/btr189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mii Y. and Taira M. (2009). Secreted Frizzled-related proteins enhance the diffusion of Wnt ligands and expand their signalling range. Development 136, 4083-4088. 10.1242/dev.032524 [DOI] [PubMed] [Google Scholar]
- Mohammadi M., Mcmahon G., Sun L., Tang C., Hirth P., Yeh B. K., Hubbard S. R. and Schlessinger J. (1997). Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science 276, 955-960. 10.1126/science.276.5314.955 [DOI] [PubMed] [Google Scholar]
- Nakamura Y., de Paiva Alves E., Veenstra G. J. and Hoppler S. (2016). Tissue- and stage-specific Wnt target gene expression is controlled subsequent to β-catenin recruitment. Development 143, 1914-1925. 10.1242/dev.131664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibu Y., Zhang H. and Levine M. (2001). Local action of long-range repressors in the Drosophila embryo. EMBO J. 20, 2246-2253. 10.1093/emboj/20.9.2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C. (2004). Regionally specific induction by the Spemann–Mangold organizer. Nat. Rev. Genet. 5, 425-434. 10.1038/nrg1347 [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P. D. and Faber J. (1967). Normal Table of Xenopus laevis (Daudin). Amsterdam, The Netherlands: North-Holland Publishing Company. [Google Scholar]
- Northrop J. L. and Kimelman D. (1994). Dorsal-ventral differences in Xcad-3 expression in response to FGF-mediated induction in Xenopus. Dev. Biol. 161, 490-503. 10.1006/dbio.1994.1047 [DOI] [PubMed] [Google Scholar]
- Ornitz D. M. and Itoh N. (2015). The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 4, 215-266. 10.1002/wdev.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro C. and Monsoro-Burq A. H. (2012). Signaling and transcriptional regulation in neural crest specification and migration: lessons from Xenopus embryos. Wiley Interdiscip. Rev. Dev. Biol. 2, 247-259. 10.1002/wdev.76 [DOI] [PubMed] [Google Scholar]
- Quinlan A. R. and Hall I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841-842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribisi S., Mariani F. V., Aamar E., Lamb T. M., Frank D. and Harland R. M. (2000). Ras-mediated FGF signaling is required for the formation of posterior but not anterior neural tissue in Xenopus laevis. Dev. Biol. 227, 183-196. 10.1006/dbio.2000.9889 [DOI] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G. and Mesirov J. P. (2011). Integrative genomics viewer. Nat. Publishing Group 29, 24-26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose J., Molenaar M., Peterson J., Hurenkamp J., Brantjes H., Moerer P., van de Wetering M., Destrée O. and Clevers H. (2001). The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nucleic Acids Res. 395, 1410-1419. 10.1038/26989 [DOI] [PubMed] [Google Scholar]
- Session A. M., Uno Y., Kwon T., Chapman J. A., Toyoda A., Takahashi S., Fukui A., Hikosaka A., Suzuki A., Kondo M. et al. (2016). Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336-343. 10.1038/nature19840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Bae Y.-K., Muraoka O. and Hibi M. (2005). Interaction of Wnt and caudal-related genes in zebrafish posterior body formation. Dev. Biol. 279, 125-141. 10.1016/j.ydbio.2004.12.007 [DOI] [PubMed] [Google Scholar]
- Simões-Costa M. and Bronner M. E. (2015). Establishing neural crest identity: a gene regulatory recipe. Development 142, 242-257. 10.1242/dev.105445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive H. L., Grainger R. M. and Harland R. M. (2010). Isolation of Xenopus oocytes. Cold Spring Harb. Protoc. 12, pdb.prot5534 10.1101/pdb.prot5534 [DOI] [PubMed] [Google Scholar]
- Squarzoni P., Parveen F., Zanetti L., Ristoratore F. and Spagnuolo A. (2011). FGF/MAPK/Ets signaling renders pigment cell precursors competent to respond to Wnt signal by directly controlling Ci-Tcf transcription. Development 138, 1421-1432. 10.1242/dev.057323 [DOI] [PubMed] [Google Scholar]
- ten Berge D., Brugmann S. A., Helms J. A. and Nusse R. (2008). Wnt and FGF signals interact to coordinate growth with cell fate specification during limb development. Development 135, 3247-3257. 10.1242/dev.023176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H. T. and Vleminckx K. (2013). Design and use of transgenic reporter strains for detecting activity of signaling pathways in Xenopus. Methods 66, 422-432. 10.1016/j.ymeth.2013.06.028 [DOI] [PubMed] [Google Scholar]
- Tran H. T., Sekkali B., Van Imschoot G., Janssens S. and Vleminckx K. (2010). Wnt/beta-catenin signaling is involved in the induction and maintenance of primitive hematopoiesis in the vertebrate embryo. Proc. Natl Acad. Sci. USA 107, 16160-16165. 10.1073/pnas.1007725107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymms M. J. and Kola I. (1994). Regulation of gene expression by transcription factors Ets-1 and Ets-2. Mol. Reprod. Dev. 39, 208-214. 10.1002/mrd.1080390214 [DOI] [PubMed] [Google Scholar]
- Umbhauer M., Marshall C. J., Mason C. S., Old R. W. and Smith J. C. (1995). Mesoderm induction in Xenopus caused by activation of MAP kinase. Nature 376, 58-62. 10.1038/376058a0 [DOI] [PubMed] [Google Scholar]
- Walentek P., Beyer T., Thumberger T., Schweickert A. and Blum M. (2012). ATP4a is required for Wnt- dependent Foxj1 expression and leftward flow in Xenopus left-right development. Cell Rep. 1, 516-527. 10.1016/j.celrep.2012.03.005 [DOI] [PubMed] [Google Scholar]
- Wang W. C. H. and Shashikant C. S., (2007). Evidence for positive and negative regulation of the mouse Cdx2 gene. J. Exp. Zool. 308B, 308-321. 10.1002/jez.b.21154 [DOI] [PubMed] [Google Scholar]
- Wills A. E., Gupta R., Chuong E. and Baker J. C. (2014). Chromatin immunoprecipitation and deep sequencing in Xenopus tropicalis and Xenopus laevis. Methods 66, 410-421. 10.1016/j.ymeth.2013.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., van Beest M., Clevers H., Jones T., Hursh D. A. and Mortin M. A. (2000). decapentaplegic is a direct target of dTcf repression in the Drosophila visceral mesoderm. Development 127, 3695-3702. [DOI] [PubMed] [Google Scholar]
- Yao J. and Kessler D. S. (2001). Goosecoid promotes head organizer activity by direct repression of Xwnt8 in Spemann's organizer. Development 128, 2975-2987. [DOI] [PubMed] [Google Scholar]
- Yordy J. S. and Muise-Helmericks R. C., (2000). Signal transduction and the Ets family of transcription factors. Oncogene 19, 6503-6513. 10.1038/sj.onc.1204036 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nussbaum C., Myers R. M., Brown M., Li W. et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L. J., Gazin C., Lawson N. D., Pagès H., Lin S. M., Lapointe D. S. and Green M. R. (2010). ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11, 237 10.1186/1471-2105-11-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltewicz J. S. and Gerhart J. C. (1997). The spemann organizer of Xenopus is patterned along its anteroposterior axis at the earliest gastrula stage. Dev. Biol. 192, 482-491. 10.1006/dbio.1997.8774 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.