Abstract
In cellular environments, proteins not only interact with their specific partners but also encounter a high concentration of bystander macromolecules, or crowders. Nonspecific interactions with macromolecular crowders modulate the activities of proteins, but our knowledge about the rules of nonspecific interactions is still very limited. In previous work, we presented experimental evidence that macromolecular crowders acted competitively in inhibiting the binding of maltose binding protein (MBP) with its ligand maltose. Competition between a ligand and an inhibitor may result from binding to either the same site or different conformations of the protein. Maltose binds to the cleft between two lobes of MBP and, in a series of mutants, the affinities increased with increasing extent of lobe closure. Here we investigated whether macromolecular crowders also have a conformational or site preference when binding to MBP. The affinities of a polymer crowder, Ficoll70, measured by monitoring tryptophan fluorescence were 3- to 6-fold higher for closure mutants than for wild-type MBP. Competition between the ligand and crowder, as indicated by fitting of titration data and directly by NMR spectroscopy, and their similar preferences for closed MBP conformations further suggest the scenario that the crowder, like maltose, preferentially binds to the inter-lobe cleft of MBP. Similar observations were obtained for bovine serum albumin as a protein crowder. Conformational and site preferences in MBP-crowder binding allude to the paradigm that nonspecific interactions can possess hallmarks of molecular recognition, which may be essential for intracellular organizations including colocalization of proteins and liquid-liquid phase separation.
Graphical Abstract
INTRODUCTION
The interactions of proteins with their specific partners are central to essentially all cellular functions. However, in addition to the specific partners, proteins also encounter a high concentration of bystander macromolecules, or crowders, in cellular environments. Whereas specific interactions are shaped by evolution such that the partners “recognize” each other, nonspecific interactions of proteins with macromolecular crowders presumably are under little evolutionary pressure. Nevertheless growing evidence indicates that nonspecific interactions modulate the activities of proteins.1 Previously we reported that macromolecular crowders acted competitively in inhibiting the binding of maltose binding protein (MBP) with its ligand maltose.2 In the present study we further investigated the mechanism behind the competitive inhibition.
Two lines of evidence in our previous study supported competitive inhibition of MBP-maltose binding by a polymer crowder, Ficoll70.1 First, data for the titration of maltose and Ficoll70 into MBP fitted well to a competitive inhibition model (Figure 1A). Second, NMR spectroscopy directly showed the displacement of Ficoll70 from MBP by a saturating amount of maltose. Specifically, nearly all of the well resolved peaks in the 1H-15N TROSY spectrum of apo MBP disappeared in the presence of 200 g/L Ficoll70. The peak disappearance could be attributed to increased viscosity or weak MBP-Ficoll70 association, as seen in other studies.3–6 However, when 1 mM maltose was then added, all the NMR peaks were recovered. The latter observation means that the most likely explanation for the peak disappearance was due to weak MBP-Ficoll70 association and, more importantly, the weak association was abrogated when MBP was bound with maltose – Ficoll70 and maltose cannot bind to MBP at the same time.
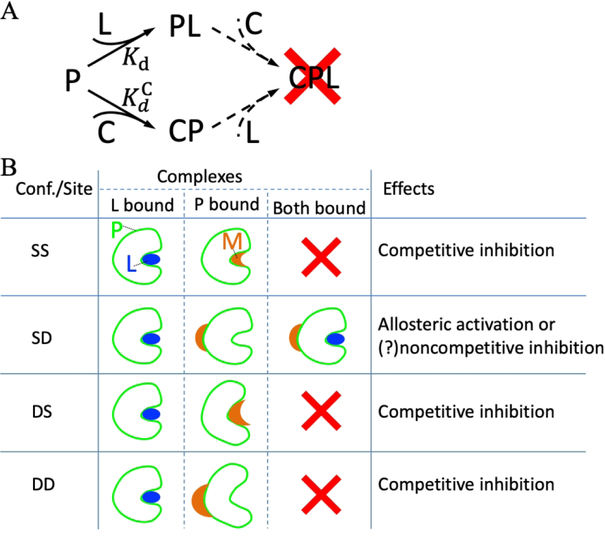
Figure 1.
(A) Competitive inhibition model, where a ligand (denoted as L) and a crowder (denoted as C) can each bind to a protein (denoted as P), but not at the same time. (B) Four scenarios for how a modulator (denoted as M) can affect protein-ligand binding, depending on whether M and L prefer the same (“S”) or distinct (“D”) conformations of and the same or distinct sites on the protein. For example, in the DS scenario, M and L prefer distinct conformations but the same site, and the effect of M is competitive inhibition.
The classical view of competitive inhibition is that a ligand and a modulator bind to the same site on a protein. This view would suggest that maltose and Ficoll70 have similar site preferences for binding to MBP. However, if the protein can sample different conformations and the ligand or modulator has preferences for certain conformations, other scenarios of competitive inhibition emerge. Depending on whether the ligand and the modulator prefer the same or distinct conformations and for the same or distinct sites of the protein, there are four possible cases (Figure 1B). The modulator appears as a competitive inhibitor in three of the four cases: same conformation and same site; and distinct conformations whether the same or distinct sites. In the remaining case, the ligand and modulator have the same conformational preference but distinct site preferences; here the modulator is an allosteric activator (or possibly a noncompetitive inhibitor). While the observed competitive inhibition of MPB-maltose binding by Ficoll70 can be explained by invoking conformational or site preference for MPB-Ficoll70 interaction, it is not clear which of the three above scenarios is at work.
Site preferences in nonspecific interactions have been reported. Similar to the NMR observations on Ficoll70 and maltose competition for MBP binding,2 Luh et al.6 acquired 1H-15N SOFAST-HMQC spectra of a WW domain, showing peak disappearance in living cells or cell extracts and peak recovery upon binding of a substrate peptide. Importantly, this study further showed that nonspecific binding with intracellular components was abrogated not only by substrate binding but also by a mutation in the substrate recognition pocket, lending support to the contention that the latter is the preferred site for both substrate binding and nonspecific interactions. Furthermore, weak binding leading to NMR peak disappearance of the WW domain could be recreated by ovalbumin but not by Ficoll70, indicating, as seen in a prior NMR study of chymotrypsin inhibitor 2,4 a difference between a protein crowder and a polymer crowder in whether forming weak attractive interactions with some test proteins. In a recent molecular dynamics simulation study of concentrated villin solutions, weak association between villin molecules was found to significantly slow down their rotational diffusion.7 The weak association preferentially occurred on one face of villin that was enriched in charged residues, indicating the electrostatic nature of the weak association. In another simulation study of the Influenza M2 tetrameric proton channel in lipid bilayers, weak association between tetramers leading to rotational slowdown was hydrophobic in nature and driven by shape complementarity.8
While the present work focuses on protein-crowder weak attraction, which, as already alluded to, varies case by case, it should be noted that protein-crowder interactions always have an excluded-volume component.9 If only the excluded-volume component were present, it would drive test proteins from open to closed conformations,10–12 and thereby enhancing the MBP-maltose binding affinity (reminiscent of the SD scenario in Figure 1B, which leads to allosteric activation). This prediction contradicts the observed inhibitory effect of Ficoll70, indicating once again the presence of MBP-Ficoll70 weak attraction.2 Little is known about any conformational preference possibly exhibited by protein-crowder weak attraction, although this is a prominent feature in the binding between many specific partners. Several reports suggested that weak attraction by protein crowders or polymer crowders counteracts the conformational contraction of intrinsically disordered proteins by the excluded volume component.13–15 The counteraction arises from preferentially binding of the crowders to more open conformations of the disordered proteins, which presumably are more accessible for crowder attraction than the more closed conformations. In the case of globular proteins, a recent molecular dynamics simulation study found competition of fatty acids and amyloid-β peptides for binding to human serum albumin (HSA).16 Fatty acids, by binding to high-affinity buried sites, quenched the conformational flexibility and preferred a relatively small region of the HSA conformational space. In contrast, with amyloid-β peptides weakly bound to surface sites, HSA remained flexible and sampled multiple regions in conformational space. This situation bears resemblance to the DD scenario of competitive inhibition in Figure 1B, where ligands (here fatty acids) and modulators (here amyloid-β peptides) have both distinct conformational preferences and distinct site preferences.
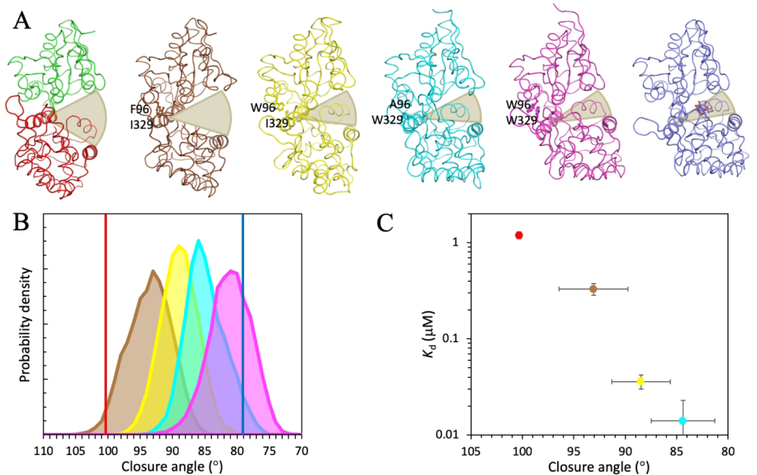
MBP has a bi-lobed structure, with the ligand-binding site located within the inter-lobe cleft. Crystal structures of apo and ligand-bound MBP show a significant closure of the lobes upon maltose binding.17, 18 In the apo form, MBP samples mostly open conformations, but can make excursions to semi-closed conformations, which may be important for the ligand binding process.19–21 The extent of lobe closure can be increased by changing small residues (Ala96 and Ile329) in the hinge region into bulky ones (Phe or Trp; Figure 2A), and the maltose binding affinities increase progressively as the lobes become more and more closed in the apo structures.22, 23 We refer to MBP variants as closure mutants.
Figure 2.
(A) Crystal structures of apo (green and red, from PDB 1OMP) and maltose-bound (blue; from PDB 1ANF) MBP and representative conformations of four closure mutants from molecular dynamics simulations (brown, yellow, cyan, and magenta). The two colors of apo MBP represent the N- and C-terminal lobes. The two mutated residues in the hinge region of the closure mutants and the maltose ligand in the holo structure are shown as ball-and-stick. Two helices, spanning residues 314–327 in the hinge and residues 209–219 in the C-lobe, used for defining the lobe closure angle are highlighted with thicker ribbon. Shaded arcs illustrate increasing lobe closure from left to right structures. (B) Values of lobe closure angle in the apo (red vertical line) and maltose-bound (blue vertical line) structures and histograms of lobe closure angle in the molecular dynamics simulations of the four closure mutants (brown, yellow, cyan, and magenta shaded curves for the A96F, A96W, I329W, and A96W/I329W mutants, respectively). (C) Correlation between maltose binding affinities and average closure angles of wild-type and three closure mutants. Kd values were from fitting of maltose titration data to a two-state binding model. Error bars in closure angle represent standard deviations among snapshots sampled from molecular dynamics simulations; error bars in Kd represent uncertainties in fitting crowder-free titration data to a two-state model.
Here we used the closure mutants to investigate whether crowder binding to MBP has conformational preference. Relative to wild-type MBP, the closure mutants showing increased affinities for maltose also bind Ficoll70 with 3- to 6-fold higher affinities. The competition between maltose and Ficoll70 for MBP binding was verified by NMR titration. Taken together, these results suggest that both maltose and Ficoll70 preferentially bind to closed conformations of MBP, and their competition further indicates that the crowder too binds to the inter-lobe cleft, following the SS scenario in Figure 1 (with maltose and Ficoll70 having the same conformational preference and same site preference). Similar results were obtained for bovine serum albumin (BSA) as a protein crowder. Computation based on atomistic modeling and exhaustive sampling confirmed stronger attractive interactions between BSA and MBP with a closed cleft than with an open cleft.
MATERIALS AND METHODS
Protein Expression and Purification.
Expression and purification of MBP constructs were carried out as described previously.2 For the two mutants I329W and A96W/I329W with the highest maltose affinities, a denaturation/desalting/refolding procedure was followed to remove any residual bound oligosaccharides. All MBP constructs were checked to be in the apo form by the peak wavelength (around 346 nm) of tryptophan fluorescence.
Fluorescence Titration.
Maltose and Ficoll70 binding affinities for MBP constructs were measured as done previously.2 In particular, binding was monitored by the peak wavelength of tryptophan fluorescence (excitation wavelength at 280 nm) on a Cary 300 fluorometer, while maintaining an MBP concentration of 200 nM in 100 mM sodium phosphate buffer (pH 7.2) at 25 °C. Maltose titration was achieved by sequentially adding small volumes of maltose stocks (with concentrations ranging from 1 μM to 1 M), to pure apo MBP or apo MBP premixed with a fixed concentration of Ficoll70. Ficoll70 titration was carried out in separate samples where apo MBP was mixed with increasing concentrations of Ficoll70.
To monitor maltose titration into apo MBP premixed with fixed concentrations of BSA, a cysteine mutation at residue Ser337 was introduced to attach a fluorophore, nitrobenzoxadiazole (NBD). The fluorescence intensity at 550 nm (excitation wavelength at 500 nm) was used as the probe of binding. As a check, maltose binding to NBD-labelled MBP premixed with Ficoll70 was monitored by both NBD fluorescence intensity at 550 nm and peak wavelength of tryptophan fluorescence. Data for each titration curve were collected in triplicates.
Data Analysis.
Following our previous work,2 maltose titration data in the absence and presence of a fixed concentration of Ficoll70 were first fitted to a normal two-state binding model. The model predicts the following dependence of the peak wavelength, λ, of tryptophan fluorescence on the total maltose concentration [L]T and MBP concentration [P]T:
| (1) |
where λf and λb, denoting peak wavelengths for maltose-free and maltose-bound forms of MBP, respectively, and Kd;app, denoting apparent dissociation constant, are fitting parameters. Maltose titration data monitored by NBD fluorescence intensity were fitted to a formula analogous to eq 1, with λ, λf, and λb replaced by the fluorescence intensities of the corresponding mixture or species. The mean values of replicated data points were used for fitting; errors reported for fitting parameters (e.g., Kd;app) represented uncertainties of the fit.
Data for titrating Ficoll70 into apo MBP were also fitted to a two-state model, with quantities in eq 1 replaced by their counterparts for MBP-Ficoll70 binding. Given the weak MBP Ficoll70 binding (with dissociation constant ), the peak wavelength becomes independent of [P]T and its dependence on Ficoll70 concentration [C]T is simplified to
| (2) |
where λp and λCP denote peak wavelengths for unbound and Ficoll70-bound forms of MBP. All the maltose and Ficoll70 titration data were also globally fitted to a competitive inhibition model (Figure 1A). Under the condition , the peak wavelength can again be written in the form of eq 1, but with λf and Kd;app dependent on [C]T. Specifically,
| (3) |
| (4) |
where Kd is the maltose dissociation constant in the absence of Ficoll70. The predicted peak wavelength was obtained by substituting eqs 3 and 4 into eq 1. The sum of squared deviations between predicted and measured peak wavelengths for maltose titration (at zero and three or four nonzero Ficoll70 concentrations) and for Ficoll70 titration (in the absence of maltose) was minimized by varying five global parameters: λP, λCP, λb, Kd and .
NMR Titration.
1H-15N HSQC-TROSY experiments were performed on a Bruker 700 MHz spectrometer for MBP samples titrated with Ficoll70 and a Varian 600 MHz spectrometer for the ones titrated with BSA. MBP concentration was maintained at 100 μM in 10 mM sodium phosphate buffer (pH 7.2) with 12% D2O and 0.2% sodium azide added. All NMR experiments were performed at 37 °C. Bruker-based data were analyzed using TopSpin 3.5 and Varian-based data by NMRPipe.
Molecular Dynamics Simulations.
The equilibrium conformations of MBP closure mutants were investigated by molecular dynamics simulations. Mutations were introduced to the maltose-bound closed structure [Protein Data Bank (PDB) entry 1ANF]. Then the maltose molecule was stripped and the mutant structures were allowed to relax by running simulations in GROMACS24 with force field version 54a7.25
Each mutant protein was solvated in a cubic box filled with simple point charge (SPC) water molecules,26 and the simulation system was electrically neutralized by adding counter ions. After 5000 steps of energy minimization, the system was gradually heated up to 300 K in 60 ps at constant volume. The simulation then continued for 150 ns at constant temperature (300 K) and constant pressure (1 bar). Nonbonded interactions were truncated with a 10 Å cutoff, and long-range electrostatic interactions were treated by the particle mesh Ewald method.27 The last 100 ns, saved at 10 ps intervals, of each simulation was used for analysis. Here we measured the lobe closure angle as the angle defined by three points, located at the Cα centers of geometry of the following three groups of residues: the N-lobe (residues 7 to 111 and 260 to 312), the hinge helix spanning residues 314 to 327, and the C-lobe helix spanning residues 209 to 219 (Figure 2A).
Calculations of MBP-BSA Interaction Energies.
The free energy of transferring MBP with either an open cleft or with a closed cleft (structure from PDB entry 1OMP or 1ANF) from a dilute solution to a BSA solution at 110 g/L concentration was calculated by a method called fast Fourier transform-based modeling of atomistic protein-crowder interactions, or FMAP.28 The interaction energy function is a sum of terms over all pairs of atoms between protein and crowder molecules; each atom pair contributes three terms: a steric repulsion term, a term in the form of a Lennard-Jones potential modeling nonpolar interactions, and a term in the form of a Debye-Hückel potential modeling electrostatic interactions. Eight BSA molecules in a cubic box with side length of 200 Å were used to represent the crowder solution. FMAP yielded the interaction energies of an MBP molecule fictitiously placed at 3.73 × 107 points on a cubic grid covering the crowder box with a 0.6 Å spacing. This calculation was repeated 54,000 times with MBP in different orientations. The Boltzmann average of these interaction energies gave the transfer free energy.
We also further investigated possible site preferences that favorable MBP-BSA pairwise interactions might exhibit. For each of 2 × 106 MBP placements with the most favorable interaction energies with the eight BSA molecules, we switched to a more expensive but accurate atom-based method to obtain all the MBPBSA pairwise interaction energies. [A fraction (approximately 24%) of the 2 × 106 placements were eliminated due to MBP-BSA clashes newly identified by the atom-based method.] These pairwise interaction energies were used to identify “hot” regions around MBP, where a crowder such as BSA binds preferentially.
RESULTS
MBP Closure Mutants Exhibit Correlation Between Closure Angle and Maltose Affinity.
Four closure mutants, A96F, A96W, I329W, and A96W/I329W, were engineered by changing small residues in the hinge region into bulky ones (Figure 2A). In the crystal structures of the apo and maltose-bound forms of MBP,17, 18 the closure angles are 100.3° and 79.1°, respectively. In molecular dynamics simulations of the four closure mutants, the closure angles averaged at 93.1°, 88.5°, 84.4°, and 80.6°, respectively, with standard deviations all around 3° (Figure 2B). As the closure angles progressively reduced, the maltose binding affinities increased by 100-fold, from a Kd of 1.2 ± 0.1 μM for the wild-type protein to a Kd of 14 ± 9 nM for the I329W mutant, in line with previous studies22, 23 (Figure 2C; the maltose affinity for the A96W/I329W mutant was too high for accurate determination on the fluorometer). Values of Kd and other parameters from fitting of titration data are listed in Table S1.
Ficoll70 Inhibits Maltose Binding to Closure Mutants.
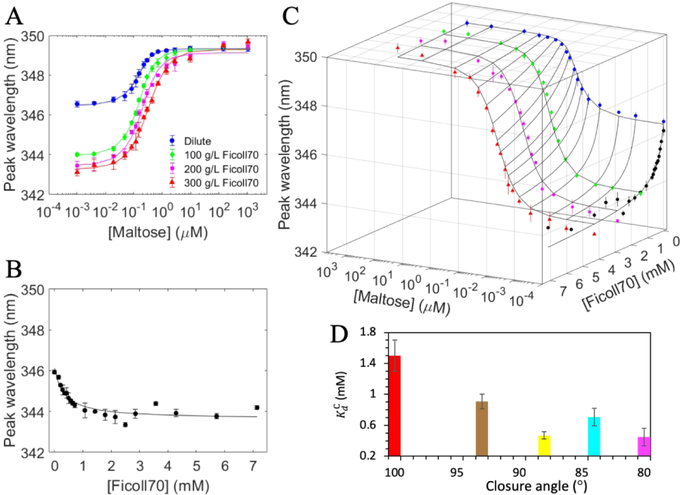
Our previous study demonstrated that Ficoll70 competitively inhibits maltose binding to MBP.2 In order to probe the underlying mechanism, here we studied the effects of Ficoll70 on the maltose binding affinities of the MBP closure mutants. To that end, we carried out maltose titrations into the A96F, A96W, and I329W mutants not only in the absence but also in the presence of 100, 200, and 300 g/L Ficoll70, by monitoring the peak wavelength of tryptophan fluorescence (Figures 3A, S1A, and S2A). As found for the wild-type protein,2 the apparent maltose dissociation constant, Kd;app, for each of the three closure mutants increased with increasing Ficoll70 concentration (Table S1), demonstrating once again the inhibitory effects of Ficoll70. The increase in Kd;app was approximately linear with respect to Ficoll70 concentration (Figure S3), as is expected of a three-state competitive inhibition model (eq 4).
Figure 3.
(A) Two-state fit of binding isotherms from titrating maltose into the A96W mutant in the absence or presence of fixed concentrations of Ficoll70. (B) Two-state fit of the binding isotherm from titrating Ficoll70. (C) Three-state fit of all the binding isotherms. (D) (MBP-Ficoll70 dissociation constant) plotted against average closure angle; red, brown, yellow, cyan, and magenta bars are for wild-type MBP and the A96F, A96W, I329W, and A96W/I329W mutants, respectively. Results were from three-state fitting for all but the last mutant and from two-state fitting for the last mutant. Error bars in (A)-(C) represent variance among triplicate measurements at a given titrant concentration; error bars of in (D) represent uncertainties of the fit; data used in the fit were mean values of triplicate measurements.
Ficoll70 Titration Indicates Higher Affinities for Closure Mutants Than for Wild-Type MBP.
The three-state model calls for direct binding between apo MBP and Ficoll70, and indeed the Ficoll70 titration data, including those for the A96W/I329W mutant, could be fitted to a two-state binding model (Figures 3B, S1B, S2B and S4). Importantly, all the four closure mutants had higher Ficoll70 affinities than wild-type MBP, by 3- to 6-fold ( listed in Table S1). This observation suggests that Ficoll70, like the maltose ligand, preferentially binds to closed conformations of MBP.
We fitted all the maltose and Ficoll70 titration data for each mutant to the three-state competitive inhibition model (Figures 3C, S1C, and S2C), in order to explain the data with a minimum set of fitting parameters. Values of from fitting to the two-state and three-state models were largely consistent (Table S1), although mild differences were possible (e.g., for A96W, is 0.35 ± 0.04 mM from two-state fitting but 0.47 ± 0.05 mM from three-state fitting). We deemed the three-state fitting values for more reliable, and display them in Figure 3D for wild-type MBP2 and the A96F, A96W, and I329W mutants. Relative to wild-type MBP, the mutant, A96F, with the smallest reduction in closure angle also had the smallest increase in Ficoll70 ( changing from 1.5 ± 0.2 mM to 0.91 ± 0.09 mM). However, the next two mutants, A96W and I329W, had comparable values and exhibited no clear correlation with the closure angle. A value similar to those of the latter two mutants was also obtained, though only from two-state fitting, for A96F/I329W, the mutant with the largest reduction in closure angle. Overall, the data show that Ficoll70 preferentially binds to closed conformations; moderate lobe closure leads to a moderate increase in Ficoll70 affinity but, with further reduction in closure angle, Ficoll70 affinity appears to reach a plateau.
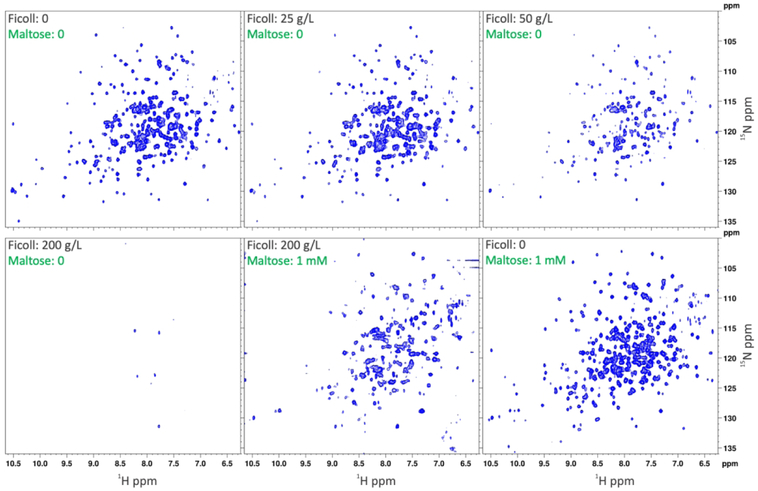
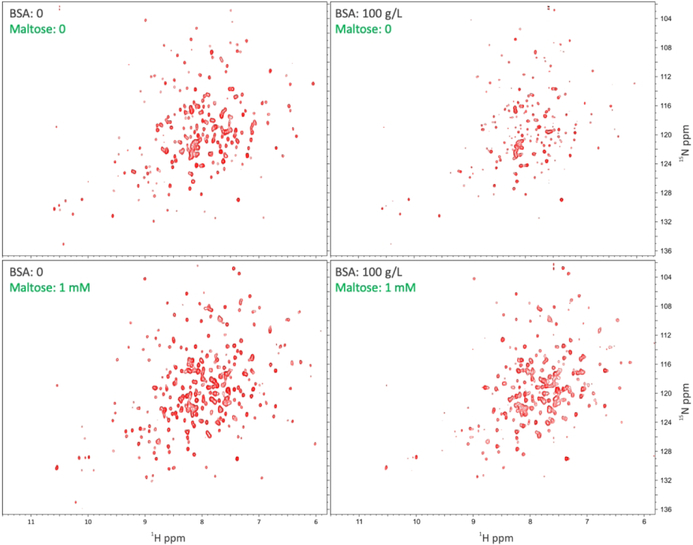
NMR Spectra of A96W mutant Provide Direct Evidence for Ficoll70 and Maltose Competition.
As demonstrated previously for wild-type MBP and for other proteins,2–6 NMR spectroscopy presents a powerful technique for ascertaining weak association with crowders. Here we performed similar 1H-15N HSQC-TROSY experiments on the A96W mutant (Figure 4), which had the highest Ficoll70 affinity. In the absence of Ficoll70, the protein showed well resolved resonances. At 25 g/L Ficoll70, the intensities of most resonances were noticeably reduced, and a few resonances disappeared. At 50 g/L Ficoll70, more resonances disappeared, and finally at 200 g/L Ficoll70, most of the resonances disappeared. Importantly, when 1 mM maltose was added to the latter sample, nearly all the lost resonances were recovered, and the spectrum resembles that of maltose-bound A96W in buffer.
Figure 4.
1H-15N HSQC-TROSY spectra of the A96W mutant in buffer, in increasing concentrations of Ficoll70, in both 200 g/L Ficoll70 and 1 mM maltose, and in 1 mM maltose only, at 700 MHz.
Four significant conclusions can be drawn. First, the recovery of the resonances strongly indicates that the loss of resonances in the presence of Ficoll70 is due to weak association, not due to increased viscosity. Second, the observation that a saturating amount of maltose could chase out Ficoll70 from MBP directly demonstrates competition. Third, considerable resonance disappearance occurred between 25 and 50 g/L Ficoll70, suggesting that these concentrations bracket the value for this mutant. These concentrations correspond to 0.36 and 0.71 mM, while = 0.47 mM according to tryptophan fluorescence. The latter value is thus corroborated by Ficoll70 titration in the NMR tube. Last, the extent of resonance disappearance at 200 g/L Ficoll70 was more complete for the A96W mutant than for wild-type MBP,2 thus providing additional evidence that Ficoll70 has a higher affinity for the closure mutant than for wild-type MBP.
MBP Also Weakly Associates with BSA.
To demonstrate that weak association was not unique to Ficoll70, we also studied the interactions of MBP with a protein crowder, BSA. Again, we titrated maltose into MBP in the presence of a fixed concentration of BSA. To be able to monitor the maltose binding, we labeled the protein with the NBD dye at residue 337, which lines the entrance to the inter-lobe cleft. The presence of the dye weakened the maltose affinity, by 5-fold as monitored by the peak wavelength of tryptophan fluorescence and 24-fold by NBD fluorescence intensity (Table S1). We contend the former probe is less susceptible to artifacts, but the latter probe was a necessity for studying protein crowders. In the presence of Ficoll70, these two probes continued to differ in the reported absolute values of Kd;app, with the latter probe yielding Kd;app approximately 2-fold higher than the former probe. Still, the effects of 100 and 200 g/L Ficoll70 on Kd;app, with 2.1- and 5.0-fold increases, respectively, reported by NBD fluorescence intensity were comparable to the counterparts, with 1.6- and 3.3-fold increases, reported by tryptophan fluorescence for the wild-type MBP (free of any mutations or labeling).
Based on NBD fluorescence, 100 g/L BSA increased Kd;app by 16-fold, compared with the 2.1-fold increase by 100 g/L Ficoll70. Even with the caveat just noted for this probe, we can still safely conclude that MBP shows weak association with BSA.
To further support the conclusion of MBP-BSA weak association, we acquired 1H-15N HSQC-TROSY spectra of the A96W mutant in buffer, in the presence of 100 g/L BSA, in the presence of both 100 g/L BSA and S1 mM maltose, and in the presence of only 1 mM maltose (Figure 5). 100 g/L BSA clearly led to resonance disappearance and intensity decrease, while further addition of saturating maltose led to resonance recovery. Therefore the NMR spectra show that BSA also competes with maltose in binding to MBP.
Figure 5.
1H-15N HSQC-TROSY spectra of the A96W mutant in buffer, in 100 g/L BSA, in both 100 g/L BSA and 1 mM maltose, and in 1 mM maltose only, at 600 MHz.
Atomistic Modeling Reveals Stronger Attraction between BSA and Closed MBP.
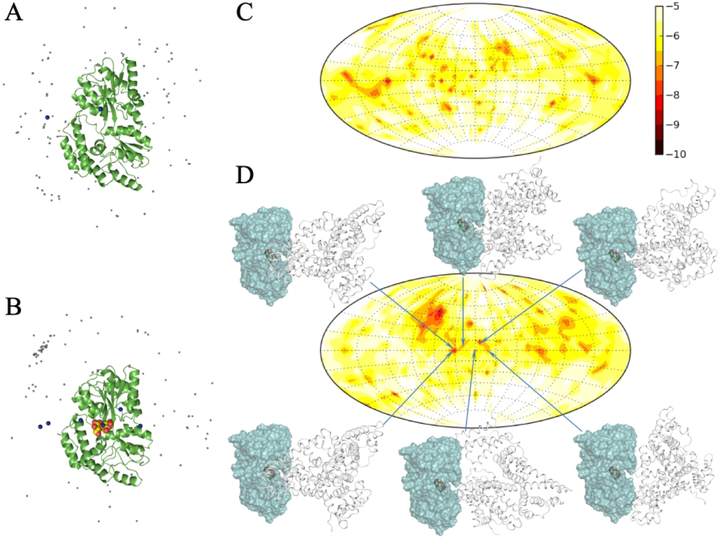
Our FMAP method was developed specifically to model protein-crowder interactions.28 In this method, a test protein (in the present case, MBP) is placed at many locations inside a crowder solution to calculate the protein-crowder interaction energies (Figure S5). This calculation shows that the closed form of MBP, as modeled by the crystal structure of the maltose-bound protein but with the ligand stripped, interacts with BSA more favorably than the apo, or open form of MBP (Figure S6). The Boltzmann averages of the interaction energies at the 500,000 most favorable locations were −13.0 kcal/mol for the closed form and −11.4 kcal/mol for the open form.
To gain further insight into possible site preferences in the interactions of BSA with MBP, we collected the interaction energies of MBP-BSA pairs and the corresponding poses, when MBP was placed at the most favorable locations inside the crowder solution. The top 100 poses are displayed in Figure 6A,B for the open and closed forms of MBP, with BSA represented by a dot located at its center of geometry. Two differences emerged. First, whereas the BSA poses around open MBP were largely uniform over the MBP surface, the BSA poses around closed MBP were more clustered. Second, fewer BSA molecules were found over the open inter-lobe cleft than over the closed inter-lobe cleft. The histograms of the MBP-BSA interaction energies collected from 1.22 × 107 pairs, presented on a Hammer projection of a spherical surface around MBP (Figure 6C,D), confirmed that closed MBP, relative to the open form, had stronger attraction for BSA. Part of this difference came from stronger binding to the top of the N-lobe of the closed form of MBP, but the closed cleft also presented a hotter region for BSA binding. Structures of poses where BSA is directly over the closed cleft (Figure 6D) show that, with the cleft closed, the “front” face of MBP is flat and thus presents an extensive flat surface for a large molecule like BSA to dock.
Figure 6.
(A) Poses of 100 BSA molecules (represented by gray dots located at the centers of geometry) with the lowest interaction energies with open MBP. Two blue dots highlight poses over the inter-lobe cleft. (B) Poses of 100 BSA molecules with the lowest interaction energies with closed MBP. Maltose is shown as spheres to indicate the binding site, but was not present in the calculation of interaction energies. Six blue dots highlight poses over the inter-lobe cleft. (C) Map of interaction energies between BSA and open MBP, presented on a Hammer projection of a spherical surface around the MBP molecule. The inter-lobe cleft is parallel to the equator. Interaction energies are displayed as colors according to a scale (in kcal/mol) shown on the right. (D) Map of interaction energies between BSA and closed MBP. Six poses where BSA molecules are directly over the inter-lobe cleft are shown in side view; maltose is included only to indicate the biding site.
DISCUSSION
Using both tryptophan fluorescence and NMR spectroscopy, we have demonstrated that maltose and Ficoll70 compete in binding to MBP and, most importantly, both prefer closed conformations of MBP for binding. The particular mechanism behind the competitive inhibition of MBP-maltose binding by Ficoll70 is thus the SS scenario of Figure 1B, where the ligand and the crowder have the same conformational and site preferences. While site preferences have been reported in experimental and computational studies,6–8 this is the first time, to the best of our knowledge, that conformational preferences have been shown for nonspecific interactions. While it is well established that nonspecific interactions with crowders can modulate proteins folding stability, meaning that the strengths of nonspecific interactions are different for the folding and unfolded protein, what we observed here is measurable response of crowders to relatively subtle conformational changes of a folded protein. This finding has enormous implications for the functions of proteins in cellular environments, where nonspecific interactions are ever present.
Knowing that Ficoll70 prefers closed conformations of MBP puts us one step closer toward elucidating the mechanism behind its competitive inhibition of maltose binding, which was first observed on the wild-type protein2 and now shown also for the closure mutants. With various combinations of conformational and site preferences possessed by the ligand and the crowder, three scenarios of competitive inhibition exist (Figure 1). If the ligand and crowder have the same conformational preference, as our data seem to indicate, then competitive inhibition is achieved only if they both also have the same site preference. Since maltose is known to bind to the inter-lobe cleft, that would suggest that the crowder also preferentially binds to this site. Our FMAP calculations have indeed shown that another crowder, BSA, preferentially binds around the cleft when MBP is in closed conformations, because the two lobes then move closer to form a nearly contiguous flat surface well suited for the docking of a large crowder molecule. The FMAP calculations indicated that, at this site, protein-crowder attraction was dominated by nonpolar (van der Waals and hydrophobic) interactions, but electrostatic interactions also contributed. One may anticipate that the crowder molecule can further protrude into the narrowed cleft and thereby block the ligand from binding. While this scenario seems very plausible, we cannot rule out the possibility that large crowders like Ficoll70 most prefer semi-closed MBP conformations, whereas maltose prefers the fully closed conformations. In that case, maltose binding quenches MBP to its preferred conformations (fully closed ones in the present circumstance) and thereby abrogates Ficoll70 binding (regardless where the latter prefers to bind), and vice versa.
We have demonstrated both Ficoll70 and BSA weakly associate with MBP, and both lead to competitive inhibition of the latter’s binding with its ligand. This observation highlights the prevalence of weak association between proteins and bystander macromolecules (crowders) in their cellular environments as well as the anticipated effects on the functions of the proteins. Nearly 40 years ago McConkey29 proposed the term “quinary structure” to denote transient interactions of cellular proteins. Nowadays quinary interactions have sometimes been used to denote nonspecific interactions between proteins and crowders.7, 30–35 We should recall that McConkey’s motivation was to explain apparently slower than expected (based on sequence comparison of small soluble proteins) evolution of cellular proteins. From the examples he listed (e.g., interactions of the ribosome with translational factors), it is clear that McConkey meant quinary for denoting specific interactions, between proteins (or protein complexes) and their partners that must work together to accomplish a cellular function (e.g., translation). This is distinct from the type of nonspecific interactions that we suggest to be prevalent between proteins and other macromolecules, which may not even co-exist in vivo (and hence evolved together). By contrast, McConkey assumed that quinary interactions were conserved during evolution (or, were tuned by evolutionary pressure). In essence, we propose that nonspecific interactions are intrinsic to all proteins and are prevalent without the need for evolutionary pressure. In fact, as suggested by a recent study,36 nonspecific interactions may be so prevalent as to present the risk that a few mutations may turn them into detrimental, specific interactions. So there may be evolutionary pressure to keep nonspecific interactions in check. Underscoring this point are studies linking gene dosage toxicity to over-population, driven by mass action, of promiscuously or weakly interacting complexes of disordered or structured proteins.37, 38 Nonspecific interactions have also been found to modulate the conformational ensemble and stability of the unfolded state of a foreign protein overexpressed in Escherichia coli.30
Site preference of nonspecific interactions already blurs their divide from specific interactions.1 With conformational preference, the divide is even more blurred. These types of preferences are hallmarks of molecular recognition; possessing them affords bystander macromolecules higher ability to fine-tune protein activities. For example, a common mechanism for regulating a receptor’s activity is by controlling its conformation; bystander macromolecules with conformational preference in their interactions with the receptor can thus contribute to this regulatory mechanism. In particular, they may help determine the constitutive activity of the receptor, and promote or suppress its activation, i.e., transition from the inactive form to the active form. Lastly, we note that, in addition to directly modulating protein activities, nonspecific interactions may be essential for transient intracellular organizations of macromolecules, including colocalization of proteins (such as metabolic enzymes) and liquid-liquid phase separation of proteins (often along with RNA). A recent study has shown that site preference of nonspecific interactions, as modeled by attractive patches on spherical particles, is a very important determinant of liquid-liquid phase separation.39
Supplementary Material
Funding Sources
This work was supported by National Institutes of Health Grant GM118091 (to H.-X.Z.) and the Pukyong National University Research Abroad Fund (C-D-2017–0984, to M.Y.).
ABBREVIATIONS
- BSA
bovine serum albumin
- FMAP
fast Fourier transform-based modeling of atomistic protein-crowder interactions
- HSA
human serum albumin
- MBP
maltose binding protein
- NBD
nitrobenzoxadiazole
- PDB
Protein Data Bank
- SPC
simple point charge
Footnotes
Publisher's Disclaimer: This document is confidential and is proprietary to the American Chemical Society and its authors. Do not copy or disclose without written permission. If you have received this item in error, notify the sender and delete all copies.
Accession Code
Maltose binding protein, UniProtKB P0AEX9 (MALE_ECOLI)
Supporting Information. Addition experimental data and MBP-BSA interaction energy results are given in Table S1 and Figures S1 – S6.
REFERENCES
- [1].Qin S, and Zhou HX (2017) Protein folding, binding, and droplet formation in cell-like conditions, Curr Opin Struct Biol 43, 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Miklos AC, Sumpter M, and Zhou HX (2013) Competitive interactions of ligands and macromolecular crowders with maltose binding protein, PLoS One 8, e74969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Li C, Charlton LM, Lakkavaram A, Seagle C, Wang G, Young GB, Macdonald JM, and Pielak GJ (2008) Differential dynamical effects of macromolecular crowding on an intrinsically disordered protein and a globular protein: implications for in-cell NMR spectroscopy, J Am Chem Soc 130, 6310–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wang Y, Li C, and Pielak GJ (2010) Effects of proteins on protein diffusion, J Am Chem Soc 132, 9392–9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Barnes CO, Monteith WB, and Pielak GJ (2011) Internal and global protein motion assessed with a fusion construct and in-cell NMR spectroscopy, ChemBioChem 12, 390–391. [DOI] [PubMed] [Google Scholar]
- [6].Luh LM, Hansel R, Lohr F, Kirchner DK, Krauskopf K, Pitzius S, Schafer B, Tufar P, Corbeski I, Guntert P, and Dotsch V (2013) Molecular crowding drives active Pin1 into nonspecific complexes with endogenous proteins prior to substrate recognition, J Am Chem Soc 135, 13796–13803. [DOI] [PubMed] [Google Scholar]
- [7].Nawrocki G, Wang PH, Yu I, Sugita Y, and Feig M (2017) Slow-down in diffusion in crowded protein solutions correlates with transient cluster formation, J Phys Chem B 121, 11072–11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Paulino J, Pang X, Hung I, Zhou HX, and Cross TA (2019) Influenza a M2 channel clustering at high protein to lipid ratios: Viral budding implications, Biophys J, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhou HX, Rivas G, and Minton AP (2008) Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences, Annu Rev Biophys 37, 375–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Qin S, Minh DDL, McCammon JA, and Zhou H-X (2010) Method to predict crowding effects by postprocessing molecular dynamics trajectories: application to the flap dynamics of HIV-1 protease, J Phys Chem Lett 1, 107–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dong H, Qin S, and Zhou HX (2010) Effects of macromolecular crowding on protein conformational changes, PLoS Comput Biol 6, e1000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dhar A, Samiotakis A, Ebbinghaus S, Nienhaus L, Homouz D, Gruebele M, and Cheung MS (2010) Structure, function, and folding of phosphoglycerate kinase are strongly perturbed by macromolecular crowding, Proc Natl Acad Sci U S A 107, 17586–17591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Goldenberg DP, and Argyle B (2014) Minimal effects of macromolecular crowding on an intrinsically disordered protein: a small-angle neutron scattering study, Biophys J 106, 905–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Banks A, Qin S, Weiss KL, Stanley CB, and Zhou H-X (2018) Intrinsically disordered protein exhibits both compaction and expansion under macromolecular crowding, Biophys. J 114, 1067–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nguemaha V, Qin S, and Zhou HX (2018) Atomistic modeling of intrinsically disordered proteins under polyethylene glycol crowding: Quantitative comparison with experimental data and implication of protein-crowder attraction, J Phys Chem B 122, 11262–11270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Guo C, and Zhou HX (2018) Fatty acids compete with abeta in binding to serum albumin by quenching its conformational flexibility, Biophys J 116, 248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sharff AJ, Rodseth LE, Spurlino JC, and Quiocho FA (1992) Crystallographic evidence of a large ligand-induced hinge-twist motion between the two domains of the maltodextrin binding protein involved in active transport and chemotaxis, Biochemistry 31, 10657–10663. [DOI] [PubMed] [Google Scholar]
- [18].Quiocho FA, Spurlino JC, and Rodseth LE (1997) Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor, Structure 5, 997–1015. [DOI] [PubMed] [Google Scholar]
- [19].Stockner T, Vogel HJ, and Tieleman DP (2005) A salt-bridge motif involved in ligand binding and large-scale domain motions of the maltose-binding protein, Biophys J 89, 3362–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tang C, Schwieters CD, and Clore GM (2007) Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR, Nature 449, 1078–1082. [DOI] [PubMed] [Google Scholar]
- [21].Bucher D, Grant BJ, and McCammon JA (2011) Induced fit or conformational selection? The role of the semi-closed state in the maltose binding protein, Biochemistry 50, 10530–10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Millet O, Hudson RP, and Kay LE (2003) The energetic cost of domain reorientation in maltose-binding protein as studied by NMR and fluorescence spectroscopy, Proc Natl Acad Sci U S A 100, 12700–12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Seo MH, Park J, Kim E, Hohng S, and Kim HS (2014) Protein conformational dynamics dictate the binding affinity for a ligand, Nat Commun 5, 3724. [DOI] [PubMed] [Google Scholar]
- [24].Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, and Berendsen HJ (2005) GROMACS: fast, flexible, and free, J Comput Chem 26, 1701–1718. [DOI] [PubMed] [Google Scholar]
- [25].Schmid N, Eichenberger AP, Choutko A, Riniker S, Winger M, Mark AE, and van Gunsteren WF (2011) Definition and testing of the GROMOS force-field versions 54A7 and 54B7, Eur Biophys J 40, 843–856. [DOI] [PubMed] [Google Scholar]
- [26].Berweger CD, van Gunsteren WF, and Müller-Plathe F (1995) Force field parametrization by weak coupling. Re-engineering SPC water, Chem Phys Lett 232, 429–436. [Google Scholar]
- [27].Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, and Pedersen LG (1995) A smooth particle mesh Ewald method, J Chem Phys 103, 8577–8593. [Google Scholar]
- [28].Qin S, and Zhou HX (2014) Further development of the FFT-based method for atomistic modeling of protein folding and binding under crowding: Optimization of accuracy and speed, J Chem Theory Comput 10, 2824–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].McConkey EH (1982) Molecular evolution, intracellular organization, and the quinary structure of proteins, Proc Natl Acad Sci U S A 79, 3236–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cohen RD, and Pielak GJ (2017) Quinary interactions with an unfolded state ensemble, Protein Sci 26, 1698–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gao MM, Held C, Patra S, Arns L, Sadowski G, and Winter R (2017) Crowders and cosolvents—major contributors to the cellular milieu and efficient means to counteract environmental stresses, ChemPhysChem 18, 2951–2972. [DOI] [PubMed] [Google Scholar]
- [32].Cheung MS, and Gasic AC (2018) Towards developing principles of protein folding and dynamics in the cell, Phys Biol 15, 063001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Davis CM, Gruebele M, and Sukenik S (2018) How does solvation in the cell affect protein folding and binding?, Curr Opin Struct Biol 48, 23–29. [DOI] [PubMed] [Google Scholar]
- [34].Guseman AJ, Speer SL, Goncalves GMP, and Pielak GJ (2018) Surface charge modulates protein-protein interactions in physiologically relevant environments, Biochemistry 57, 1681–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ribeiro S, Ebbinghaus S, and Marcos JC (2018) Protein folding and quinary interactions: creating cellular organisation through functional disorder, FEBS Lett 592, 3040–3053. [DOI] [PubMed] [Google Scholar]
- [36].Cohen-Khait R, Dym O, Hamer-Rogotner S, and Schreiber G (2017) Promiscuous protein binding as a function of protein stability, Structure 25, 1867–1874. [DOI] [PubMed] [Google Scholar]
- [37].Vavouri T, Semple JI, Garcia-Verdugo R, and Lehner B (2009) Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity, Cell 138, 198–208. [DOI] [PubMed] [Google Scholar]
- [38].Bhattacharyya S, Bershtein S, Yan J, Argun T, Gilson AI, Trauger SA, and Shakhnovich EI (2016) Transient protein-protein interactions perturb E. coli metabolome and cause gene dosage toxicity, Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Nguemaha V, and Zhou HX (2018) Liquid-liquid phase separation of patchy particles illuminates diverse effects of regulatory components on protein droplet formation, Sci Rep 8, 6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.