Abstract
Cleavage polyadenylation specificity factor (CPSF) is the core component of the 3′-end processing complex, which determines the site of 3′-end cleavage interactions of specific sequence elements within pre-mRNAs. The present study revealed that all members of the CPSF complex were overexpressed in lung cancer tissue from The Cancer Genome Atlas (TCGA) Lung Cancer Cohort compared with normal lung tissue. Analysis of overall survival and recurrence-free survival verified that only CPSF3 was associated with prognosis and recurrence of lung adenocarcinoma (LUAD), and thus could be a promising biomarker. Additionally, receiver operating characteristic curve analysis revealed that CPSF3 may function as a diagnostic biomarker to distinguish between two histological subtypes of non-small cell lung cancer. Furthermore, analysis of the association of CPSF3 expression with clinicopathological parameters indicated that CPSF3 was associated with smoking history, tumor diameter, lymph node metastasis, clinical stage and radiation therapy in LUAD. Additionally, analysis of the DNA methylation data of the TCGA-LUAD Cohort revealed that CPSF3 DNA CpG sites (cg12057242 and cg25739938) were generally hypomethylated in LUAD compared with normal lung tissue. Correlation analysis identified the CPSF3 DNA CpG site cg25739938 to be negatively correlated with CPSF3 expression, while no correlation was identified with cg12057242. In addition, correlation analysis demonstrated that the overexpression of CPSF3 was correlated with CPSF3 DNA copy number variants (CNAs). The findings indicate that abnormal expression of CPSF3 may be caused by DNA CNAs; and DNA hypermethylation and function may be a promising diagnostic and prognostic indicator for LUAD.
Keywords: cleavage and polyadenylation specificity factor, cleavage and polyadenylation specific factor 3, non-small cell lung cancer, The Cancer Genome Atlas, lung adenocarcinoma, biomarker, bioinformatics analysis
Introduction
Lung cancer is a type of malignant tumor that causes considerable mortality and morbidity worldwide (1). Out of all lung cancer cases, >85% are non-small cell lung cancer (NSCLC) (2). Surgical intervention, chemotherapy and radiation therapy are the traditional therapies for early-stage lung cancer; however, all these treatments are accompanied by undesirable adverse reactions. In recent years, molecular targeted therapies, which specifically target carcinoma cells, and reportedly have minimal adverse effects compared with conventional treatments, have become a research focus (3). Currently, a large amount of molecular therapies targeting diverse receptor tyrosine kinases (RTKs), which have been demonstrated to be frequently mutated and to promote tumorigenesis by regulating cell proliferation and survival, are widely used in patients with NSCLC (4). Numerous types of RTKs, including epidermal growth factor receptor (EGFR), hepatocyte growth factor receptor (c-Met), ALK receptor tyrosine kinase (ALK) and BRAF, which have been reported to be frequently mutated in patients with NSCLC, have been used as targets in clinical therapy (5–10). However, despite all these therapies, the majority of patients with NSCLC still exhibit a poor 5-year survival rate of 15.9% (11). Therefore, there is an urgent need to identify biomarkers with better prognostic and diagnostic potential.
The cleavage polyadenylation specificity factor (CPSF) complex is a core component of 3′-end processing and is involved in regulating mRNA maturation and alternative splicing, as well as internal introns, by modulating the cleavage and polyadenylation of mRNAs (12). The multi-subunit CPSF complex is composed of CPSF1 (also known as CPSF160), CPSF2 (also known as CPSF100), CPSF3 (the cleavage endonuclease, also known as CPSF73), CPSF4 (also known as CPSF30), factor interacting with poly(A) polymerase α and CPSF1 (FIP1L1; also known as FIP1) and WD repeat domain 33 (12). There are several studies regarding the function of the CPSF complex in cancer, particularly in lung cancer; however, CPSF1 promotes cell proliferation in human ovarian cancer (13). Decreased CPSF2 expression promotes invasion and an increased cancer stem cell population, and CPSF2 may be a novel prognostic marker for papillary thyroid carcinoma (14,15). Suppression of CPSF4 inhibits the proliferation of lung adenocarcinoma (LUAD) cells and is associated with overall survival (OS) (16–19). FIP1L1 is generated via gene fusion with platelet-derived growth factor receptor a, which is recognized as an important diagnostic marker of hematological malignancies, including chronic eosinophilic leukemia, hypereosinophilic syndrome and systemic mastocytosis, and occasionally atypical chronic myelogenous leukemia (20). Therefore, it was hypothesized that potential diagnostic and prognostic biomarkers could exist within the CPSF complex.
The Cancer Genome Atlas (TCGA; http://portal.gdc.cancer.gov) is an open-access database that is widely used globally and provides a reliable tool for the analysis of numerous types of useful clinicopathological data. In addition, TCGA data is overseen by the National Cancer Institute's Center for Cancer Genomics (21). Genetic and clinicopathological data of >1,000 patients with primary LUAD and lung squamous cell carcinoma (LUSC) are recorded in the TCGA Lung Cancer Cohort (22).
In the present study, promising biomarkers for NSCLC were identified via bioinformatics data mining. The results of the present study demonstrated that CPSF3 of the CPSF complex was overexpressed in NSCLC, and its overexpression was associated with OS and recurrence-free survival (RFS) of patients with LUAD. CPSF3 may also serve as a diagnostic biomarker for LUAD and LUSC. Additionally, CPSF3 may promote the proliferation and lymph node metastasis of LUAD. The mechanism of CPSF3 overexpression may be connected to its DNA copy number variants and DNA hypermethylation.
Materials and methods
TCGA Lung Cancer Cohort
TCGA Lung Cancer Cohort data, (http://tcga.cancer.gov/dataportal; accessed June 2016), which include the LUAD cohort, comprising 706 primary LUAD tissues and normal lung tissues, and the LUSC cohort, comprising 554 primary LUSC tissues and normal lung tissues, were downloaded from the University of California Santa Cruz Xena Browser (https://xenabrowser.net). Excel 2016 (Microsoft Corporation) was used for further data processing. From the cohorts, 520 primary LUAD cases and 506 primary LUSC cases with existing RNA-seq and intact clinical parameters were selected for further analysis. In addition, DNA methylation data (‘Illumina 450k methylation’) and CNA data (‘gene-level GISTIC2-processed’) were downloaded from this database to detect the possible mechanisms of abnormal expression of CPSF3 in LUAD.
Cancer cell line encyclopedia (CCLE)
CCLE (https://portals.broadinstitute.org/ccle/about) is a database containing genomic data, including copy number, mRNA expression, reverse-phase protein microarray and reduced representation bisulfite sequencing, from >1,100 cell lines. LUAD cell line data (based on The Global Bioresource Center; http://www.atcc.org) were downloaded to examine the association between CPSF3 expression and its DNA methylation.
Statistical analysis
GraphPad Prism version 5.0 (GraphPad Software, Inc.) or SPSS version 16.0 (SPSS, Inc.) were used for data analysis. Unless specifically mentioned, all values are presented as the mean ± SD. Two-group independent sample comparisons were performed using a Student's t-test (two-tailed) when the two groups had equal variances, while Welch's t-test was used for unequal variances. Multi-group samples statistics were analyzed via one-way ANOVA if the variances were equal; if not, Welch's ANOVA was performed. Bonferroni post hoc tests were performed for all ANOVAs. Samples from TCGA Lung Cancer Cohort were divided into two groups, according to median values. OS and RFS curves were plotted using the Kaplan-Meier method, and OS and RFS differences were assessed using the log-rank test. Receiver operating characteristic (ROC) curves were constructed using CPSF3 expression data, and the area under the curve (AUC) was estimated to investigate the feasibility of distinguishing LUAD from LUSC. Pearson correlation analysis was used to evaluate the correlations among CPSF3 mRNA expression and CPSF3 DNA methylation and DNA copy number variants (CNAs). P<0.05 was considered to indicate a statistically significant difference.
Results
CPSF complex is significantly elevated in NSCLC tissues compared with normal lung tissues (NTL)
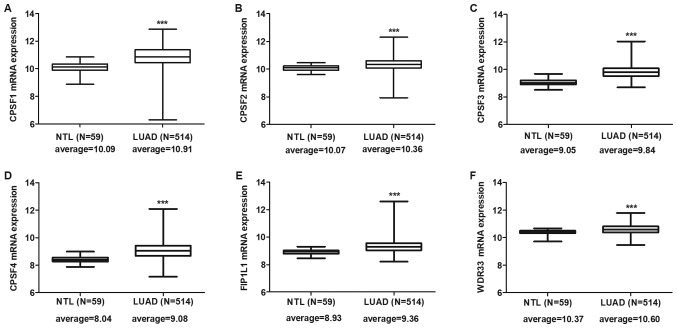
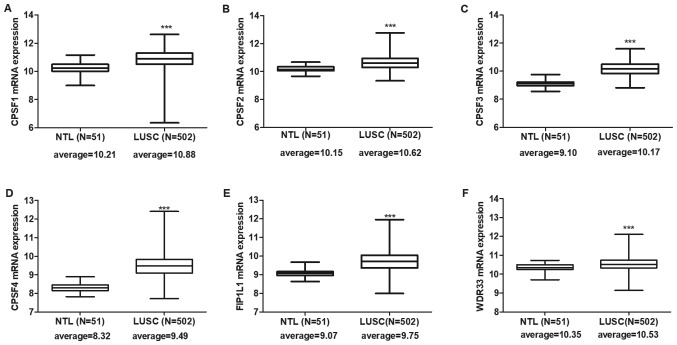
Expression levels of CPSF complex components were individually assessed in the TCGA LUAD and LUSC cohorts. The results indicated that all CPSF complex components were significantly overexpressed in LUAD (Fig. 1) and LUSC (Fig. 2) tissues compared with NTL.
Figure 1.
CPSF complex component mRNA expression in The Cancer Genome Atlas-LUAD tissues. CPSF complex components, including (A) CPSF1, (B) CPSF2, (C) CPSF3, (D) CPSF4, (E) FIP1L1 and (F) WDR33, were significantly overexpressed in LUAD tissues compared with in NTL. ***P<0.0001. CPSF, cleavage polyadenylation specificity factor; NTL, normal lung tissue; LUAD, lung adenocarcinoma; FIP1L1, factor interacting with poly(A) polymerase α and CPSF1; WDR33, WD repeat domain 33.
Figure 2.
CPSF complex component mRNA expression in The Cancer Genome Atlas-LUSC tissues. All CPSF complex components, including (A) CPSF1, (B) CPSF2, (C) CPSF3, (D) CPSF4, (E) FIP1L1 and (F) WDR33, were significantly overexpressed in LUSC tissues compared with in NTL. ***P<0.0001. CPSF, cleavage polyadenylation specificity factor; NTL, normal lung tissue; LUSC, lung squamous cell carcinoma; FIP1L1, factor interacting with poly(A) polymerase α and CPSF1; WDR33, WD repeat domain 33.
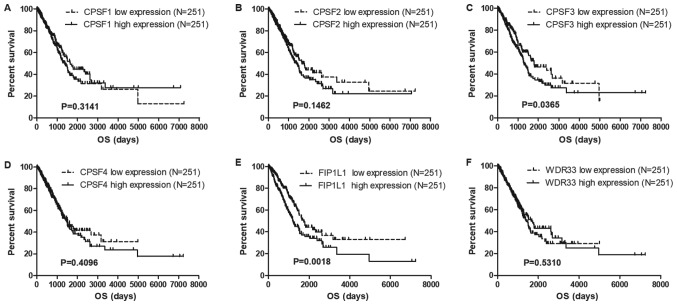
CPSF3 is the only component associated with lung cancer OS and RFS in the CPSF complex
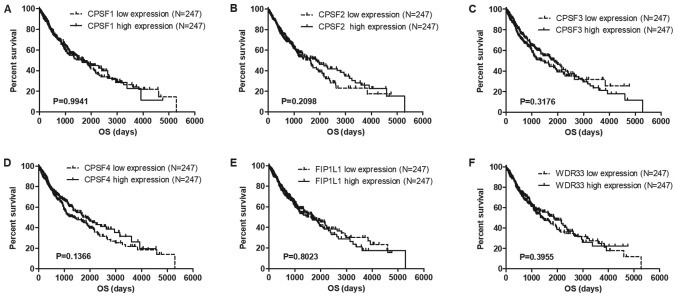
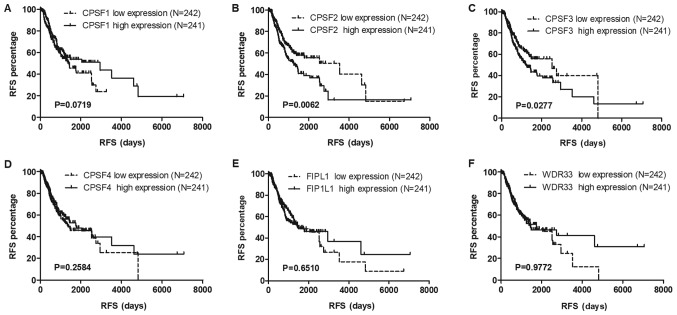
To identify potential prognostic biomarkers in the CPSF complex, the associations among the CPSF complex components and OS and RFS in the TCGA Lung Cancer Cohort were assessed. The results of the present study demonstrated that overexpression of CPSF3 and FIP1L1 in patients with LUAD was associated with decreased survival times compared with patients with low expression levels of CPSF3 and FIP1L1 (Fig. 3). In patients with LUSC, no CPSF complex components were significantly associated with OS (Fig. 4). Additionally, analysis of the association between the CPSF complex component proteins and RFS was investigated. Patients with LUAD with high CPSF2 and CPSF3 had shorter RFS than those in the low expression groups (Fig. 5), whereas expression of the components had no effect on RFS of patients with LUSC (Fig. 6). Taken together, CPSF3 was the only CSPF complex component that affected OS and RFS, and it may thus function as a promising prognostic biomarker for NSCLC. Therefore, CPSF3 was considered as a potential biomarker and focused on in further research.
Figure 3.
Association between CPSF complex components and OS in TCGA-LUAD patients. Kaplan-Meier analysis of OS time of TCGA-LUAD Cohort patients. The patients were divided into high and low expression groups depending on the average expression level of CPSF complex (A) CPSF1 exhibited no association with OS in LUAD, (B) CPSF2 showed no association with OS in LUAD, (C) CPSF3 high expression showed reduced survival time compared with the patient with CPSF3 low expression in LUAD (D) CPSF4 exhibited no association with OS in LUAD, (E) FIP1L1 high expression showed reduced survival time compared with the patient with CPSF3 low expression in LUAD and (F) WDR33 showed no association with OS in LUAD.
Figure 4.
Association between CPSF complex components and OS in TCGA-LUSC patients. Kaplan-Meier analysis of OS time of TCGA-LUSC Cohort patients. The patients were divided into high and low expression groups depending on the average expression level of each gene. CPSF complex components, including (A) CPSF1, (B) CPSF2, (C) CPSF3, (D) CPSF4, (E) FIP1L1 and (F) WDR33, exhibited no connection with OS in LUSC. TCGA, The Cancer Genome Atlas; LUSC, lung squamous cell carcinoma; CPSF, cleavage polyadenylation specificity factor; OS, overall survival; FIP1L1, factor interacting with poly(A) polymerase α and CPSF1; WDR33, WD repeat domain 33.
Figure 5.
Association of CPSF complex components and RFS in TCGA-LUAD patients. Kaplan-Meier analysis of the RFS of TCGA-LUAD patients. The patients were stratified depending on the average expression level of (A) CPSF1, (B) CPSF2, (C) CPSF3, (D) CPSF4, (E) FIP1L1 and (F) WDR33. High expression levels of CPSF2 and CPSF3 were associated with the RFS of patients with LUAD. Overexpression of CPSF1, CPSF4, FIP1L1 and WDR33 had no influence on RFS. TCGA, The Cancer Genome Atlas; LUAD, lung adenocarcinoma; CPSF, cleavage polyadenylation specificity factor; RFS, recurrence-free survival; FIP1L1, factor interacting with poly(A) polymerase α and CPSF1; WDR33, WD repeat domain 33.
Figure 6.
Association between CPSF complex expression and RFS in TCGA-LUSC patients. Kaplan-Meier analysis of the RFS of TCGA-LUSC patients. The patients were stratified depending on the average expression level of each gene. CPSF complex members, including (A) CPSF1, (B) CPSF2, (C) CPSF3, (D) CPSF4, (E) FIP1L1 and (F) WDR33, exhibited no connection with RFS in LUSC. TCGA, The Cancer Genome Atlas; LUSC, lung squamous cell carcinoma; CPSF, cleavage polyadenylation specificity factor; RFS, recurrence-free survival; FIP1L1, factor interacting with poly(A) polymerase α and CPSF1; WDR33, WD repeat domain 33.
CPSF3 serves as a biomarker to distinguish between histological subtypes of NSCLC
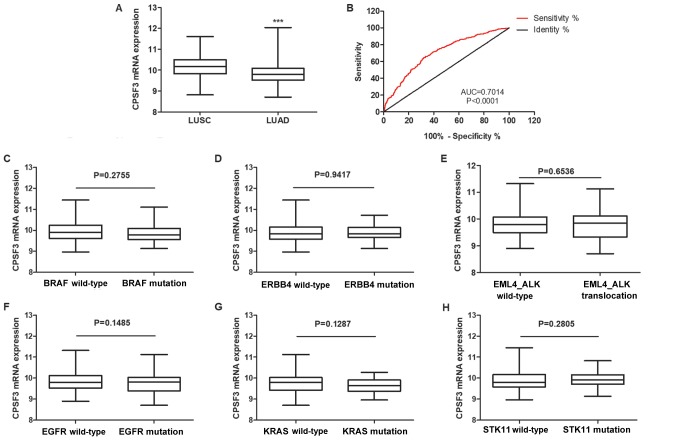
To further test whether CPSF3 could serve as a biomarker to distinguish between LUAD and LUSC, CPSF3 expression status of patients with LUAD and LUSC in the TCGA Lung Cancer Cohort was compared. As shown in Fig. 7A, the average expression level of CPSF3 among 502 patients with LUSC was higher than that of 512 patients with LUAD. ROC analysis further demonstrated that CPSF3 expression could be a single significant parameter to discriminate between LUAD and LUSC, with an AUC of 0.7014 (Fig. 7B). To further explore the clinical implications of CPSF3, associations among CPSF3 expression and different molecular subtypes were investigated within different types of adenocarcinoma classified according to the genotypes of BRAF, erb-b2 receptor tyrosine kinase 4 (ERBB4), EGFR, echinoderm microtubule associated protein like 4, KRAS and serine/threonine kinase 11, and ALK translocation, in the TCGA-LUAD cohort. There was no difference in CPSF3 between the wild type and mutant samples categorized by any of the aforementioned genes of interest (Fig. 7C-H). Overall, CPSF3 was shown to be a novel and efficient diagnostic biomarker for distinguishing between LUAD and LUSC in NSCLC.
Figure 7.
CPSF3 functions as a biomarker to distinguish between the histological subtypes of non-small cell lung cancer. (A) Expression levels of CPSF3 were markedly higher in patients with LUSC than in patients with LUAD. (B) Receiver operating characteristic curve analysis revealed the potential use of CPSF3 expression to discriminate between patients with LUAD and LUSC in the TCGA Cohort. Comparison of the relative expression of CPSF3 between two groups of patients with LUAD in the TCGA Cohort, classified by molecular subtype based on (C) BRAF, (D) ERBB4, (E) ALK, (F) EGFR, (G) KRAS and (H) STK11 mutations, respectively. None of the mutations were identified to be associated with CPSF3 expression. ***P<0.0001. TCGA, The Cancer Genome Atlas; CPSF3, cleavage polyadenylation specific factor 3; LUSC, lung squamous cell carcinoma; LUAD, lung adenocarcinoma; AUC, area under the curve; ERBB4, erb-b2 receptor tyrosine kinase 4; EML4, echinoderm microtubule associated protein like 4; ALK, ALK receptor tyrosine kinase; EGFR, epidermal growth factor receptor; STK11, serine/threonine kinase 11.
Association of CPSF3 expression with clinicopathological features in LUAD
To explore the role of high CPSF3 expression in LUAD progression, the association of clinicopathological features with CPSF3 expression was evaluated. The present study revealed that CPSF3 expression was associated with smoking history, tumor diameter, lymph node metastasis, TNM stage and radiation therapy, whereas there was no association with age, gender, distant metastasis and targeted molecular therapy in the TCGA-LUAD Cohort (Table I). Therefore, it was hypothesized that CPSF3 may promote proliferation and metastasis, and may be associated with radiation therapy.
Table I.
Association of CPSF3 with clinicopathological features in The Cancer Genome Atlas lung adenocarcinoma patients.
| Variable | N | CPSF3 (mean ± SE) | t-value | P-value |
|---|---|---|---|---|
| Sex | ||||
| Female | 277 | 9.805±0.4380 | 1.804 | 0.0717 |
| Male | 237 | 9.876±0.4555 | ||
| Age, years | ||||
| <65 | 220 | 9.871±0.4691 | 1.693 | 0.0911 |
| ≥65 | 275 | 9.876±0.4252 | ||
| Smoking history | ||||
| Smoked | 75 | 9.700±0.4516 | 2.937 | 0.0035a |
| Never smoked | 425 | 9.766±0.3928 | ||
| Tumor diameter, cm | ||||
| ≤3 | 169 | 9.876±0.4677 | 2.638 | 0.0086a |
| >3 | 342 | 9.766±0.3928 | ||
| Lymph node metastasis | ||||
| No | 330 | 9.805±0.4598 | 2.763 | 0.0059a |
| Yes | 172 | 9.766±0.4188 | ||
| Distant metastasis | ||||
| M0 | 346 | 9.853±0.4440 | 1.611 | 0.1079 |
| M1 | 25 | 9.921±0.4796 | ||
| TNM stage | ||||
| I | 425 | 9.792±0.4115 | 4.178 | 0.0062a |
| II | 122 | 9.820±0.4917 | ||
| III | 84 | 9.961±0.4288b | ||
| IV | 25 | 9.979±0.4837b | ||
| Radiation therapy | ||||
| Yes | 60 | 9.812±0.4398 | 2.742 | 0.0063a |
| No | 397 | 9.981±0.4847 | ||
| Molecular therapy | ||||
| No | 303 | 9.804±0.4477 | 1.946 | 0.0523 |
| Yes | 152 | 9.891±0.4482 |
P<0.01
P<0.01 vs. stage I. Mean age, 65 years. TNM stage statistics were analyzed by one-way ANOVA and Bonferroni post hoc test. Other statistics were performed using a two-tailed Student's t-test. CPSF3, cleavage polyadenylation specific factor 3; TNM, tumor-node-metastasis.
DNA methylation and DNA CNAs are correlated with CPSF3 expression in LUAD
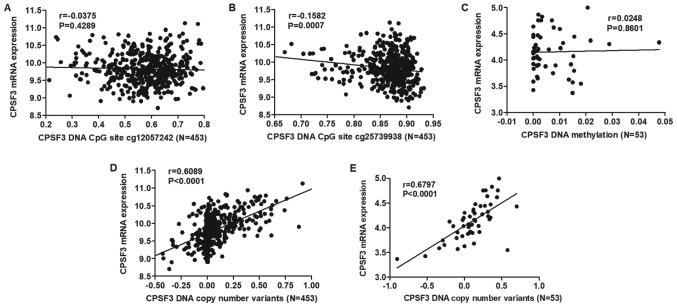
DNA methylation is one of the most common factors associated with abnormal gene expression. By analyzing CPSF3 DNA methylation and RNA-seq data in the TCGA-LUAD Cohort, two CPSF3 DNA methylation CpG sites (cg12057242 and cg25739938) were identified to be differentially methylated in TCGA-LUAD tissues compared with normal lung tissues (Table II). The correlation between the two differentially expressed DNA CpG sites and RNA expression was assessed, demonstrating that cg25739938 exhibited a negative correlation with CPSF3 expression (Fig. 8A and B). However, the same analysis conducted in 53 LUAD cell lines from CCLE indicated there was no correlation between CPSF3 expression and methylation (Fig. 8C). DNA CNAs are another mechanism that leads to aberrant RNA expression. Therefore, the association between CPSF3 DNA CNAs and RNA expression was further analyzed. The results demonstrated that CPSF3 CNAs were positively correlated with CPSF3 mRNA expression in TCGA-LUAD samples (Fig. 8D) and CCLE-LUAD cell lines (Fig. 8E).
Table II.
CPSF3 DNA methylation CpG sites analysis in The Cancer Genome Atlas-LUAD.
| CpG site | Tissue | CpG methylation (mean ± SE) | t-value | P-value |
|---|---|---|---|---|
| cg23889771 | NTL | 0.0700±0.0196 | 0.0107 | 0.9915 |
| LUAD | 0.0700±0.0110 | |||
| cg24873957 | NTL | 0.0837±0.0137 | 0.5146 | 0.6071 |
| LUAD | 0.0825±0.0124 | |||
| cg20361001 | NTL | 0.0388±0.0083 | 0.3778 | 0.7058 |
| LUAD | 0.0040±0.0108 | |||
| cg20093808 | NTL | 0.0276±0.0067 | 0.0731 | 0.9417 |
| LUAD | 0.0274±0.0092 | |||
| cg07814910 | NTL | 0.0801±0.0187 | 0.3878 | 0.6983 |
| LUAD | 0.0792±0.0128 | |||
| cg20549545 | NTL | 0.0864±0.0178 | 1.826 | 0.0685 |
| LUAD | 0.0916±0.0139 | |||
| cg08937729 | NTL | 0.0948±0.0213 | 0.1190 | 0.9054 |
| LUAD | 0.0944±0.0173 | |||
| cg07179925 | NTL | 0.0288±0.0134 | 1.171 | 0.2421 |
| LUAD | 0.0268±0.0090 | |||
| cg00024812 | NTL | 0.0601±0.0152 | 1.133 | 0.2578 |
| LUAD | 0.0573±0.0133 | |||
| cg26306976 | NTL | 0.0795±0.0482 | 0.6744 | 0.5004 |
| LUAD | 0.7806±0.1175 | |||
| cg07974891 | NTL | 0.6744±0.0430 | 0.2672 | 0.7894 |
| LUAD | 0.6685±0.1237 | |||
| cg18666330 | NTL | 0.8918±0.0274 | 1.260 | 0.2083 |
| LUAD | 0.8856±0.0261 | |||
| cg12057242 | NTL | 0.5333±0.0448 | 2.207 | 0.0278a |
| LUAD | 0.5772±0.1117 | |||
| cg25739938 | NTL | 0.8485±0.8658 | 2.137 | 0.0331a |
| LUAD | 0.8658±0.0445 | |||
| cg18794882 | NTL | 0.8378±0.0475 | 1.774 | 0.0767 |
| LUAD | 0.8187±0.0595 |
P<0.05. CPSF3, cleavage polyadenylation specific factor 3; LUAD, lung adenocarcinoma; NTL, normal lung tissue.
Figure 8.
Correlation of CPSF3 mRNA expression, and its DNA methylation and DNA copy number variants in TCGA-LUAD and in Cancer Cell Line Encyclopedia-LUAD cell lines. Correlation analysis revealed that (A) CPSF3 mRNA expression was not significantly correlated with DNA methylation at CpG site cg12057242, while (B) it was negatively correlated with its DNA methylation at CpG site cg25739938 in TCGA-LUAD. (C) Correlation analysis indicated that CPSF3 mRNA expression had no association with its DNA methylation in 53 LUAD cell lines. (D) Correlation analysis indicated that CPSF3 mRNA expression was positively correlated with its DNA copy number variants in TCGA-LUAD. (E) CPSF3 was positively correlated with its DNA copy number variants in LUAD cell lines. TCGA, The Cancer Genome Atlas; LUAD, lung adenocarcinoma; CPSF3, cleavage polyadenylation specific factor 3.
Discussion
Previously, a number of promising potential biomarkers have been identified in a series of secondary analyses using TCGA data. For example, DNA methylation of SOX30 is correlated with myelodysplastic syndrome progression, and has been reported to act as a potential predictive and prognostic biomarker in acute myeloid leukemia (23). Higher FOS expression is associated with a better outcome in breast cancer datasets (24). Using public data from TCGA, a 22-gene signature that demonstrated the best predictive value for assessing the clinical benefit of postoperative chemoradiotherapy was established (25).
Compound gene analysis is a more effective way of elucidating novel biomarkers and assessing the interactions among individual genes. Targeting one of these genes may lead to a large feedback effect. CPSF complex components have been demonstrated to regulate the cleavage and polyadenylation of mRNAs during the mRNA maturation and alternative splicing processes (12). However, there are few reports regarding the role of CPSF4 in lung cancer (16–19), and, to the best of our knowledge, no reports on the other components. Therefore, TCGA data was utilized to elucidate potential prognostic and diagnostic biomarkers within the complex in the present study.
In the present study, CPSF complex components were first evaluated in terms of their expression levels in NSCLC using TCGA data, as biomarkers must be differentially expressed between cancer tissue and normal tissue. The results of the present study indicated that expression of all CPSF complex components was increased in NSCLC. A promising biomarker should affect prognosis and recurrence, and may therefore serve as a predictor. The associations of CPSF complex expression with OS and RFS were assessed, which demonstrated that CPSF3 was the only component that affected OS and RFS in LUAD. Additionally, there is only one previous report regarding the role of CPSF3 in cancer (26), and, to the best of our knowledge, no reports regarding the role of CPSF3 in lung cancer; therefore, the present study focused on CPSF3 for further research, as it appeared to be a potentially promising biomarker. Further ROC curve analysis demonstrated that CPSF3 may serve as a diagnostic biomarker for NSCLC to distinguish between LUAD and LUSC.
A previous study in prostate cancer demonstrated that knockdown of CPSF3 by a specific siRNA induces apoptosis (26), which verified that CPSF3 is associated with the proliferation of malignant carcinoma. To further assess the role of CPSF3 in LUAD, the association between clinicopathological features and CPSF3 was assessed. The present study revealed that CPSF3 expression was associated with smoking history, tumor diameter, lymph node metastasis, TNM stage and radiation therapy; thus, CPSF3 may affect LUAD proliferation and metastasis, consistent with the results of the aforementioned study in prostate cancer (26).
The mechanism of gene dysregulation in NSCLC is complex. Among all the mechanisms, genetic and epigenetic alterations, including DNA amplification, DNA methylation and somatic mutations, commonly lead to abnormal gene expression accompanied by anomalous cancer cell behavior. For instance, heteroclite sulfatase 2 methylation acts as a prognostic marker for lung cancer survival (27). ERBB2 amplification results in erlotinib resistance in EGFR-L858R mutated tyrosine kinase inhibitor-naïve LUAD (28). c-Met overexpression, HER-2 gene amplification and spectrin β non-erythrocytic 1-ALK gene fusion have been reported to coexist in LUAD, and this may become a novel biomarker for cancer that is refractory to crizotinib, chemotherapy and radiotherapy, and poor prognosis (29). Promoter methylation of cadherin 13 is strongly associated with LUAD and may function as a promising diagnostic biomarker for LUAD (30). Thus, the present study assessed whether DNA CNA and abnormal DNA methylation were the mechanism underlying the aberrant mRNA expression of CPSF3, similar to the aforementioned gene. In the present study, DNA methylation levels in TCGA LUAD tissues were compared with levels in normal lung tissues, which revealed two differentially expressed CpG sites. Further correlation analysis confirmed that cg25739938 was negatively correlated with CPSF3 expression, while cg12057242 in TCGA-LUAD Cohort patients and in LUAD cell lines has no correlation with CPSF3 expression. The aforementioned results suggested that the hypermethylation of cg25739938 may be the potential mechanism affecting CPSF3 mRNA expression in LUAD. Furthermore, the DNA amplification status of CPSF3 was detected. The results demonstrated that DNA CNAs were closely associated with increased CPSF3 expression, in TCGA LUAD Cohort patients and 53 LUAD cell lines. Overall, the dysregulation of CPSF3 may be caused by DNA methylation and DNA CNAs.
In summary, the present study is the first to elucidate the potential role of the CPSF complex, particularly CPSF3, in proliferation and migration in lung cancer. As the present study was conducted via bioinformatics analysis, experimental studies regarding the role of CPSF3 in LUAD will be performed in order to verify the results of the present study. In conclusion, aberrant CPSF3 expression may be regulated by DNA CNAs, and it may function as a promising prognostic and diagnostic biomarker for LUAD.
Acknowledgements
Not applicable.
Funding
This study was funded by the National Nature Science Foundation of China (grant no. 81401391) and the National Nature Science Foundation of Guangdong Province (grant no. 2015A030313452).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request. Data were obtained from The Cancer Genome Atlas (TCGA; http://portal.gdc.cancer.gov), CCLE (https://portals.broadinstitute.org/ccle/about); The Global Bioresource Center (http://www.atcc.org) and the University of California Santa Cruz Xena Browser (https://xenabrowser.net).
Authors' contributions
YN conceived and designed the study. WL performed the bioinformatics analysis. XG helped design the project and wrote the paper. XX analyzed the data, reviewed and checked the manuscript. YZ proposed the concept of the study, analyzed and interpreted the data, supervised the research throughout, managed the project and funding and revised the article for important intellectual content. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Thomas A, Liu SV, Subramaniam DS, Giaccone G. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat Rev Clin Oncol. 2015;12:511–526. doi: 10.1038/nrclinonc.2015.90. [DOI] [PubMed] [Google Scholar]
- 3.Gadgeel SM, Ramalingam SS, Kalemkerian GP. Treatment of lung cancer. Radiol Clin North Am. 2012;50:961–974. doi: 10.1016/j.rcl.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Daga A, Ansari A, Patel S, Mirza S, Rawal R, Umrania V. Current drugs and drug targets in Non-small cell lung cancer: Limitations and opportunities. Asian Pac J Cancer Prev. 2015;16:4147–4156. doi: 10.7314/APJCP.2015.16.10.4147. [DOI] [PubMed] [Google Scholar]
- 5.Godin-Heymann N, Ulkus L, Brannigan BW, McDermott U, Lamb J, Maheswaran S, Settleman J, Haber DA. The T790M ‘gatekeeper’ mutation in EGFR mediates resistance to low concentrations of an irreversible EGFR inhibitor. Mol Cancer Ther. 2008;7:874–879. doi: 10.1158/1535-7163.MCT-07-2387. [DOI] [PubMed] [Google Scholar]
- 6.Coate LE, John T, Tsao MS, Shepherd FA. Molecular predictive and prognostic markers in non-small-cell lung cancer. Lancet Oncol. 2009;10:1001–1010. doi: 10.1016/S1470-2045(09)70155-X. [DOI] [PubMed] [Google Scholar]
- 7.Yu HA, Sima CS, Huang J, Solomon SB, Rimner A, Paik P, Pietanza MC, Azzoli CG, Rizvi NA, Krug LM, et al. Local therapy with continued EGFR tyrosine kinase inhibitor therapy as a treatment strategy in EGFR-mutant advanced lung cancers that have developed acquired resistance to EGFR tyrosine kinase inhibitors. J Thorac Oncol. 2013;8:346–351. doi: 10.1097/JTO.0b013e31827e1f83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soucheray M, Capelletti M, Pulido I, Kuang Y, Paweletz CP, Becker JH, Kikuchi E, Xu C, Patel TB, Al-Shahrour F, et al. Intratumoral heterogeneity in EGFR-Mutant NSCLC results in divergent resistance mechanisms in response to EGFR tyrosine kinase inhibition. Cancer Res. 2015;75:4372–4383. doi: 10.1158/0008-5472.CAN-15-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciuffreda L, Incani UC, Steelman LS, Abrams SL, Falcone I, Curatolo AD, Chappell WH, Franklin RA, Vari S, Cognetti F, et al. Signaling intermediates (MAPK and PI3K) as therapeutic targets in NSCLC. Curr Pharm Des. 2014;20:3944–3957. doi: 10.2174/13816128113196660763. [DOI] [PubMed] [Google Scholar]
- 10.Schrank Z, Chhabra G, Lin L, Iderzorig T, Osude C, Khan N, Kuckovic A, Singh S, Miller RJ, Puri N. Current molecular-targeted therapies in NSCLC and their mechanism of resistance. Cancers (Basel) 2018;10(pii):E224. doi: 10.3390/cancers10070224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldstraw P, Ball D, Jett JR, Le Chevalier T, Lim E, Nicholson AG, Shepherd FA. Non-small-cell lung cancer. Lancet. 2011;378:1727–1740. doi: 10.1016/S0140-6736(10)62101-0. [DOI] [PubMed] [Google Scholar]
- 12.Misra A, Green MR. From polyadenylation to splicing: Dual role for mRNA 3′end formation factors. RNA Biol. 2016;13:259–264. doi: 10.1080/15476286.2015.1112490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B, Liu Y, Liu D, Yang L. Targeting cleavage and polyadenylation specific factor 1 via shRNA inhibits cell proliferation in human ovarian cancer. J Biosci. 2017;42:417–425. doi: 10.1007/s12038-017-9701-x. [DOI] [PubMed] [Google Scholar]
- 14.Nilubol N, Boufraqech M, Zhang L, Kebebew E. Loss of CPSF2 expression is associated with increased thyroid cancer cellular invasion and cancer stem cell population, and more aggressive disease. J Clin Endocrinol Metab. 2014;99:E1173–E1182. doi: 10.1210/jc.2013-4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sung TY, Kim M, Kim TY, Kim WG, Park Y, Song DE, Park SY, Kwon H, Choi YM, Jang EK, et al. Negative expression of CPSF2 predicts a poorer clinical outcome in patients with papillary thyroid carcinoma. Thyroid. 2015;25:1020–1025. doi: 10.1089/thy.2015.0079. [DOI] [PubMed] [Google Scholar]
- 16.Yi C, Wang Y, Zhang C, Xuan Y, Zhao S, Liu T, Li W, Liao Y, Feng X, Hao J, et al. Cleavage and polyadenylation specific factor 4 targets NF-κB/cyclooxygenase-2 signaling to promote lung cancer growth and progression. Cancer Lett. 2016;381:1–13. doi: 10.1016/j.canlet.2016.07.016. [DOI] [PubMed] [Google Scholar]
- 17.Chen W, Qin L, Wang S, Li M, Shi D, Tian Y, Wang J, Fu L, Li Z, Guo W, et al. CPSF4 activates telomerase reverse transcriptase and predicts poor prognosis in human lung adenocarcinomas. Mol Oncol. 2014;8:704–716. doi: 10.1016/j.molonc.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Z, Yu W, Zhang C, Zhao S, Yu Z, Xiao X, Tang R, Xuan Y, Yang W, Hao J, et al. CREB-binding protein regulates lung cancer growth by targeting MAPK and CPSF4 signaling pathway. Mol Oncol. 2016;10:317–329. doi: 10.1016/j.molonc.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen W, Guo W, Li M, Shi D, Tian Y, Li Z, Wang J, Fu L, Xiao X, Liu QQ, et al. Upregulation of cleavage and polyadenylation specific factor 4 in lung adenocarcinoma and its critical role for cancer cell survival and proliferation. PLoS One. 2013;8:e82728. doi: 10.1371/journal.pone.0082728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Appiah-Kubi K, Lan T, Wang Y, Qian H, Wu M, Yao X, Wu Y, Chen Y. Platelet-derived growth factor receptors (PDGFRs) fusion genes involvement in hematological malignancies. Crit Rev Oncol Hematol. 2017;109:20–34. doi: 10.1016/j.critrevonc.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The cancer genome atlas Pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang JT, Lee YM, Huang RS. The impact of the Cancer Genome Atlas on lung cancer. Transl Res. 2015;166:568–585. doi: 10.1016/j.trsl.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou JD, Wang YX, Zhang TJ, Li XX, Gu Y, Zhang W, Ma JC, Lin J, Qian J. Identification and validation of SRY-box containing gene family member SOX30 methylation as a prognostic and predictive biomarker in myeloid malignancies. Clin Epigenetics. 2018;10:92. doi: 10.1186/s13148-018-0523-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fisler DA, Sikaria D, Yavorski JM, Tu YN, Blanck G. Elucidating feed-forward apoptosis signatures in breast cancer datasets: Higher FOS expression associated with a better outcome. Oncol Lett. 2018;16:2757–2763. doi: 10.3892/ol.2018.8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Fu G, Chen Y, Zhu G, Wang Z. Gene-expression signature predicts survival benefit from postoperative chemoradiotherapy in head and neck squamous cell carcinoma. Oncol Lett. 2018;16:2565–2578. doi: 10.3892/ol.2018.8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Z, Yu YP, Shi Y, Nelson JB, Luo J. CSR1 induces cell death through inactivation of CPSF3. Oncogene. 2009;28:41–51. doi: 10.1038/onc.2008.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tessema M, Yingling CM, Thomas CL, Klinge DM, Bernauer AM, Liu Y, Dacic S, Siegfried JM, Dahlberg SE, Schiller JH, Belinsky SA. SULF2 methylation is prognostic for lung cancer survival and increases sensitivity to topoisomerase-I inhibitors via induction of ISG15. Oncogene. 2012;31:4107–4116. doi: 10.1038/onc.2011.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carney BJ, Rangachari D, VanderLaan PA, Gowen K, Schrock AB, Ali SM, Costa DB. De novo ERBB2 amplification causing intrinsic resistance to erlotinib in EGFR-L858R mutated TKI-naive lung adenocarcinoma. Lung Cancer. 2017;114:108–110. doi: 10.1016/j.lungcan.2017.08.018. [DOI] [PubMed] [Google Scholar]
- 29.Gu FF, Zhang Y, Liu YY, Hong XH, Liang JY, Tong F, Yang JS, Liu L. Lung adenocarcinoma harboring concomitant SPTBN1-ALK fusion, c-Met overexpression, and HER-2 amplification with inherent resistance to crizotinib, chemotherapy, and radiotherapy. J Hematol Oncol. 2016;9:66. doi: 10.1186/s13045-016-0296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pu W, Geng X, Chen S, Tan L, Tan Y, Wang A, Lu Z, Guo S, Chen X, Wang J. Aberrant methylation of CDH13 can be a diagnostic biomarker for lung adenocarcinoma. J Cancer. 2016;7:2280–2289. doi: 10.7150/jca.15758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request. Data were obtained from The Cancer Genome Atlas (TCGA; http://portal.gdc.cancer.gov), CCLE (https://portals.broadinstitute.org/ccle/about); The Global Bioresource Center (http://www.atcc.org) and the University of California Santa Cruz Xena Browser (https://xenabrowser.net).