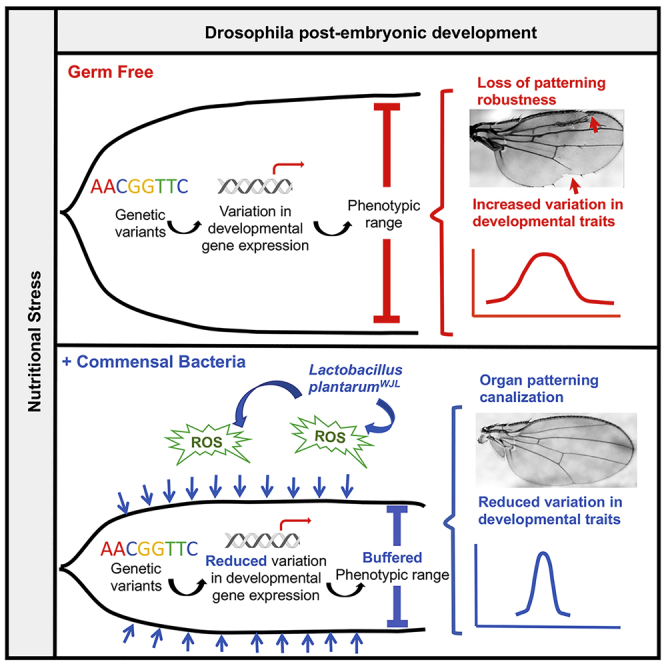
Summary
Eukaryotic genomes encode several buffering mechanisms that robustly maintain invariant phenotypic outcome despite fluctuating environmental conditions. Here we show that the Drosophila gut-associated commensals, represented by a single facultative symbiont, Lactobacillus plantarum (LpWJL), constitutes a so far unexpected buffer that masks the contribution of the host's cryptic genetic variation (CGV) to developmental traits while the host is under nutritional stress. During chronic under-nutrition, LpWJL consistently reduces variation in different host phenotypic traits and ensures robust organ patterning during development; LpWJL also decreases genotype-dependent expression variation, particularly for development-associated genes. We further provide evidence that LpWJL buffers via reactive oxygen species (ROS) signaling whose inhibition impairs microbiota-mediated phenotypic robustness. We thus identified a hitherto unappreciated contribution of the gut facultative symbionts to host fitness that, beyond supporting growth rates and maturation timing, confers developmental robustness and phenotypic homogeneity in times of nutritional stress.
Subject Areas: Biological Sciences, Microbiology, Oral Microbiology
Graphical Abstract
Highlights
-
•
Upon nutritional stress, fly commensals buffer the effects of cryptic genetic variants
-
•
Fly gut commensals buffer transcriptional variation in developmental genes
-
•
Fly commensals buffer phenotypic heterogeneity and mediate developmental canalization
-
•
Compromising ROS activities impair microbial buffering capacity
Biological Sciences; Microbiology; Oral Microbiology
Introduction
The concept of developmental robustness, or “canalization,” was first introduced by Conrad Waddington to illustrate the organisms' capacity to maintain constant and invariant phenotypic outcome in the presence of fluctuating environmental conditions and certain genetic perturbations (Huang et al., 2014, Mackay et al., 2012, Waddington, 1959). To achieve canalization, intrinsic genetic buffering programs are set in place to repress the effects of cryptic genetic variants (CGV). If these buffering mechanisms are compromised or overwhelmed by physiological or environmental stress, the CGVs can be “unlocked” to increase phenotypic variation and/or produce novel phenotypes for natural selection to act upon (Flatt, 2005, Wagner, 2007). So far, all known buffering mechanisms are encoded by the eukaryotic genome. The classic examples include the chaperone protein Hsp90 and certain microRNAs (Posadas and Carthew, 2014, Rutherford et al., 2007). Yet, the vast majority of living organisms can be viewed as the sum of the host and its associated microbial symbionts, as a result of long-term, constant, and heritable symbiosis. Whether these microbial symbionts also contribute to host developmental robustness is still poorly understood.
Symbiosis is ancient, pervasive, and diverse and in some instances is recognized as a major driving force of evolution (Brucker and Bordenstein, 2013, Gilbert, 2014). Facultative nutritional mutualism is one of the most prevalent forms of symbiosis forged by a eukaryotic host and many of its gut commensal bacteria, known collectively as the “gut microbiota.” Recent studies have established that the microbial partners contribute extensively to various aspects of host physiology, and perturbing the healthy balance of gut microbial communities often leads to undesirable developmental and fitness consequences for the host (Clemente et al., 2012, Sommer and Backhed, 2013). The nomenclature “facultative nutritional symbiosis” suggests that both partners are nonessential for each other's survival, yet neither may thrive especially under suboptimal nutritional contexts (Gilbert and Neufeld, 2014). The horizontally acquired gut commensals in Drosophila are a prototypical example of such facultative nutritional mutualists. Recent studies established that certain wild fly gut bacterial isolates can establish persistent colonization of the host's crop, a digestive organ that is unique to the adult fly but is absent in the developing larvae (Obadia et al., 2018, Pais et al., 2018), where the gut community members that comprise the microbial environment are in fact non-persistent; instead they transit rapidly through the larval gut after being ingested, are reseeded, and proliferate in the food substrate. This “farming mechanism” effectively perpetuates a mutualistic interaction with the juvenile host and confers growth advantage to the host in different nutritional context (Ma and Leulier, 2018, Storelli et al., 2018). Here for the sake of simplicity, we refer to these non-persistently commensal bacteria as “gut-associate symbionts” or “gut commensals.” Previously, we and others demonstrated that, on a standard laboratory diet, the gut commensals are dispensable for normal growth and maturation of the Drosophila host. It is only when challenged by chronic under-nutrition, germ-free (GF) larvae experienced significant growth delay (Shin et al., 2011, Storelli et al., 2011). In this context, a single gut commensal strain, Lactobacillus plantarumWJL (LpWJL) (Kim et al., 2013), can significantly accelerate the growth of the ex-germ free larvae as effectively as the entire gut-associated microbial communities (Storelli et al., 2011).
To discover host genetic variants associated to the growth benefits conferred by LpWJL during chronic under-nutrition, we first exploited the Drosophila Genetic Reference Panel (DGRP) (Huang et al., 2014, Mackay et al., 2012) by performing a genome-wide association study on the relative growth gain of LpWJL-associated compared with GF DGRP lines. During this process, we discovered that the gut-associated symbionts, represented by LpWJL, assert a previously unappreciated role that functionally resembles a broad genetic buffer. Specifically, when subjected to nutritional stress, LpWJL effectively masks the effects of the host's CGVs on developmental traits, thus conveying phenotypic homogeneity and robustness in organ patterning via reactive oxygen species (ROS) signaling. Our results qualify the fly gut community as part of the extended-host's developmental canalization program.
Results
Mono-Association with LpWJL Reduces Size Variation of Drosophila Larvae during Chronic Under-nutrition in the DGRP Lines
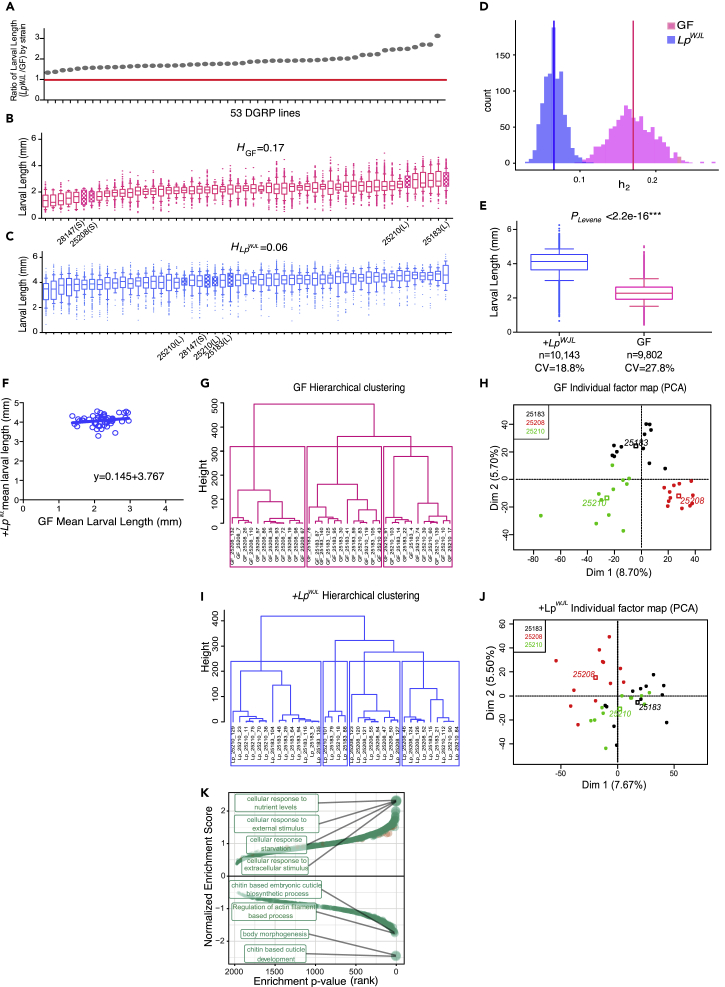
Initially, to study the host's genetic contribution to LpWJL-mediated growth during under-nutrition, we measured the body lengths of both the GF and LpWJL mono-associated larvae from 53 DGRP lines 7 days after post-embryonic development (Figures 1A–1C; Table S1) and conducted genome-wide association studies (GWAS) based on the ranking of growth gain by comparing GF and LpWJL-associated animals (Figure 1A; Table S1, column “ratio”). The GWAS yielded nine candidate variants (Table S2, Figures S1A and S1B). Through RNA interference (RNAi), we assessed the contribution of each variant-associated gene to host growth with or without LpWJL. Surprisingly, we failed to capture any obvious “loss or gain of function” of the growth benefit conferred by LpWJL. Instead, we observed that the individual RNAi-mediated knock-down of gene expression led to large phenotypic variation in GF larvae, but such variation was reduced in LpWJL, resulting in growth gain in all tested genetic crosses (Figures S1C and S1D). In parallel, we computed the respective heritability estimates (H) for the GF and LpWJL-associated DGRP populations to be 17% versus 6% (Figures 1B and 1C). Since the H values are low, we further examined the empirical distribution of the H values in the GF and LpWJL-associated populations and found that the H values of the GF samples span a significantly greater range (Figure 1D). Next, we compared the relative variability of the GF and mono-associated larval length. Since the LpWJL-associated larvae are twice the size of the GF larvae, we opted to compare the dimensionless coefficient of variation (CV) of the two populations and found that the CV is greater in the GF population despite their overall smaller average size and standard deviation (Figure 1E). These three observations indicate that genetic variants induce more pronounced size variation in GF animals, and the gut-associated symbiont such as LpWJL unexpectedly restricts growth variation despite host genetic differences. To better illustrate the buffering effect, we plotted the average GF larval length values from each DGRP line or each RNAi cross against its corresponding LpWJL-associated siblings and derived the linear regression coefficients and found that both are close to zero (0.145 and 0.06, respectively; Figures 1F and S1E). If genetic background predominantly impacts growth, then this coefficient is expected to approach one. The near-zero coefficient, the greater size variation, and the wider distribution of H values in the GF population prompts us to postulate that LpWJL presence masks the contribution of genetic variation in the DGRP lines and steers the animals to attain similar sizes despite the differences in genotype.
Figure 1.
Mono-association with LpWJL Buffers Phenotypic and Transcriptomic Variation during Growth and Development in the DGRP Lines
(A) The ranking of larval growth gain of 53 DGRP lines was used for GWAS to uncover host variants associated with growth benefits conferred by LpWJL. Each gray dot represents the quotient of average mono-associated larval length (Figure 1C) divided by the average length of GF larval length (Figure 1B) from each DGRP line on Day 7 AEL (after egg lay). The red line marks the ratio of “1,” indicating that all tested DGRP lines benefited from LpWJL presence.
(B and C) The average larval length on Day 7 AEL for each of the 53 DGRP lines. (Data are represented as mean and 10–90 percentile. Unless specified, all box plots in this manuscript present the same parameters.) Each line in the box represents the average length from pooled biological replicates containing all viable larvae from all experimental repeats. From each strain, there are between 10 and 40 viable larvae in each replicate, 3 biological replicates for each experiment, and 2 to 3 repeats of the experiments. (B): germ-free (GF, pink), (C): mono-associated (+LpWJL, blue). Note the heritability estimate (H) in the GF population is higher than in the mono-associated population (17% versus. 6%). The filled boxes denote the “small (S)” and “large (L)” DGRP lines that were selected for setting up the F2 crosses (see Figure S3A for crossing schemes).
(D) The estimation of empirical distribution of heritability indices in GF and LpWJL mono-associated larvae (p < 2.2 × 10−16, Kolmogorov-Simirnov test). The vertical lines are reported H values.
(E) Box and whiskers plots showing average larval length derived from pooled GF (pink) or LpWJL (blue) mono-associated DGRP lines. The coefficient of variation in the GF population (27.82%) is greater than that of the mono-association population (18.74%). Error bars indicate 10th to 90th percentile. Levene's test is used to evaluate homocedasticity and Mann-Whitney test for difference in the median (***p < 0.0001).
(F) Scatterplot to illustrate that LpWJL buffers size variation in ex-GF larvae in the DGRP population. Each data point represents the intercept of the average GF larvae length and its corresponding mono-associated average length at Day 7 for each DGRP line. If genetic variation was the only factor influencing growth in both GF and mono-associated flies, the slope of the scatterplot should theoretically be 1 (Null hypothesis: slope = 1. p < 0.0001: the null hypothesis is therefore rejected. A linear standard curve with an unconstrained slope was used to fit the data).
(G–J) Hierarchical clustering (G: GF and I: mono-associated) and principal-component analyses (PCA) (H: GF and J: mono-associated) based on individual larvae transcriptome analyses show that the samples cluster more based on genotypes when germ-free (G and H, G: Pgenotype = 1.048 × 10−8, H: R2 Dim1 = 0.73, Pgenotype = 7.81 × 10−10, R2Dim2 = 0.72, Pgenotype = 1.12 × 10−9) than mono-associated (I and J, I: Pgenotype = 0.000263, J: R2Dim1 = 0.42, Pgenotype = 0.00017, R2Dim2 = 0.31, Pgenotype = 0.00269). A scaled PCA using the genotype as categorical supplementary variable was performed. A hierarchical clustering on principle components (HCPC) was applied on the PCA results, and the trees were automatically cut based on inertia drop (Figure S2F). Both PCA and HCPC were performed with the R package FactoMineR on the voom corrected read counts. Correlations between the genotype variable and PCA dimensions or HCPC clusters were assessed by χ2 tests. The dots represent the different samples according to genotype, and the empty squares are the calculated centers for each genotype.
(K) Gene set enrichment analysis based on the change in standard deviation of gene expression. Positive enrichment indicates gene sets that are enriched in the genes whose expression level variation increases in response to LpWJL mono-association. Negative gene sets are those that are enriched in the genes whose expression level variation decreases in response to LpWJL mono-association. The top four positively and negatively enriched sets are labeled. The genes whose expression levels are reduced by LpWJL mono-association predominantly act in chitin biosynthesis and morphogenesis (see also Figure S2).
Mono-Association with LpWJL Decreases Variability in Gene Expression of Developmentally Related Genes
Since LpWJL reduces host growth variation phenotypically, and phenotypic variation is often the manifestation of transcriptomic variation due to genetic differences (Lehner, 2013), we explored if LpWJL also decreases gene expression variation during larval development. To do so, we conducted BRB-seq (Alpern et al., 2019) on 36 mono-associated and 36 GF individual larvae from 3 DGRP lines and specifically compared transcriptional variation in individual LpWJL mono-associated larvae with that of age-matched GF samples (Figures S2A). To minimize the size difference between the GF and mono-associated larvae to ensure sample homogeneity, we provided the GF larvae with 33% more yeast in the diet (more detailed description later) and analyzed the individual transcriptomes at an earlier time point (Day 4 post-embryonic development) where the size difference between the GF and mono-associated larvae is subtle. First, we observed that the transcriptomes moderately cluster by genotype and LpWJL status after batch effect correction (Figures S2B and S2C, Table S3). Specifically, samples from line 25,208, a “weak GF grower,” showed the greatest transcriptomic changes and growth response to LpWJL association, whereas samples from line 25,210, a “strong GF grower,” tend to cluster more based on genotype. Second, the overall transcriptomic changes associated with LpWJL presence corroborate several previous studies, including our own (Dobson et al., 2016, Erkosar et al., 2014). For example, genes involved in immune response and proteolysis, such as LysB, PGRP-SC1a&b are significantly up-regulated (Figure S2D). In addition, GO terms such as “immune response,” “defense response,” and “cellular component assembly involved in morphogenesis” are among the most up-regulated gene sets by mono-association (Figure S2E, top panel), and genes associated to “response to nutrient levels,” “cellular response to starvation,” and “tRNA modification” were down-regulated by LpWJL (Figure S2E, bottom panel). Therefore, both microbe sensing and nutrient adaptation drive the most significantly detected transcriptomic changes in mono-associated larvae.
Interestingly, we found that genotype was a stronger clustering driver for GF transcriptomes than for LpWJL mono-associated ones. When we added “genotype” as an illustrative variable in the principal component analysis based on bacterial presence, we observed that genotype has higher coefficients of correlation in the two first axes of variation in GF samples (compare Figures 1G versus 1I., and 1H. versus 1J. and S2F). These observations suggest that LpWJL can mask host genetic differences also at the transcriptomic level. Next, we compared the standard deviation (SD) of each expressed gene in both conditions and found that mono-association can either elevate or reduce expression variation in different gene sets (Figures S2G and S2H). Among the genes whose expression variation decreased the most upon LpWJL association are Ssrp, a member of the FACT chromatin complex (Saunders et al., 2003, Shimojima et al., 2003), and many cuticle-related proteins (Figure S2G, left panel), whereas for genes induced by LpWJL, such as Larval serum proteins (Lsp1s), more expression variation is detected (Figure S2G, right panel). This result suggests that mono-association does not indiscriminately reduce variation in the entire transcriptome, even though the GF transcriptomes tended to show an overall increase in expression variation (Figure S2H, red line), and this trend was more apparent in genes that were non-differentially expressed between the GF and mono-associated conditions (Figure S2I, middle panel, gray lines). Finally, we found that genes whose expression variation was most decreased by LpWJL are enriched in developmental processes such as “body morphogenesis” and “cuticle development” (Figure 1K). These data reveal that LpWJL mono-association dampens genotype-dependent expression variation, especially of genes linked to developmental processes, which in turn may account for the ability of LpWJL to reduce larval size variation.
LpWJL Broadly Buffers Variation in Different Physical Fitness Traits in Genetically Diverse Populations
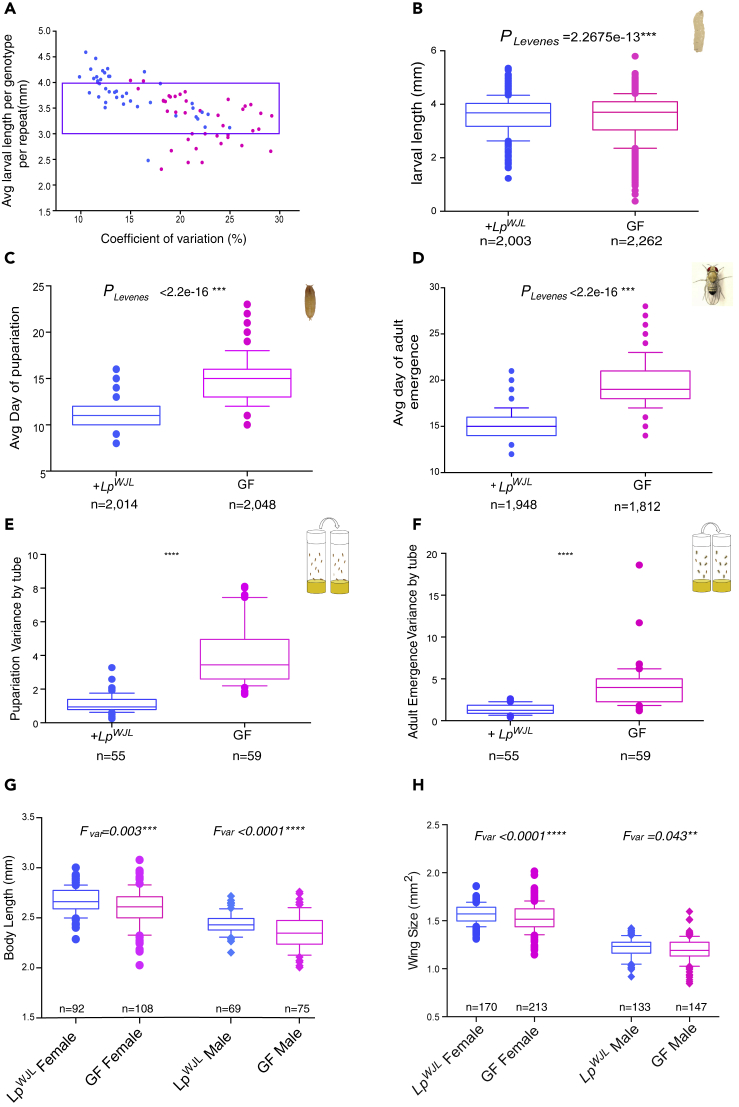
So far we have found that LpWJL reduces both phenotypic and transcriptional fluctuations during chronic under-nutrition, thus conferring a biological function that resembles various canonical buffering mechanisms that maintain phenotypic homogeneity by masking the effects of cryptic genetic variation (Mestek Boukhibar and Barkoulas, 2016, Posadas and Carthew, 2014, Rohner et al., 2013, Rutherford et al., 2007), despite the presence of a persistent nutritional stress signal. Since our studies insofar were conducted only in homozygous inbred DGRP lines, we sought to test if the observed buffering also operates in a population of genetically heterogeneous individuals. Therefore, based on their GF growth profile, we selected two DGRP strains from each end of the phenotypic extremes (Figures 1B and 1C, patterned pink and blue bars), established seven different inter-strains crosses, and compared the growth variation in the GF and mono-associated F2 progenies (Figure S3A, Transparent Methods). As in the RNA-seq experiment, we also supplemented the GF larvae with 33% more yeast (8g.L−1 versus 6g.L−1) to address two possible caveats: first, we wished to exclude that LpWJL might simply act as an additional inert source of nutrients. Several recent studies have demonstrated that live fly gut-associated symbionts can provide different micronutrients to the host, thus boosting growth and lifespan (Keebaugh et al., 2018, Wong et al., 2014, Yamada et al., 2015). Our previous findings demonstrate that the Lp symbionts need to be alive to assert their full beneficial impact in growth. For example, inoculating 1X living bacteria works far more efficiently to promote larval growth than adding three doses of 100X heat-inactivated bacteria within the first 7 days of development (Storelli et al., 2018). However, it is important to assess if increasing dietary yeast content can also reduce the variability in GF growth to the same extent as the gut bacteria, so that the observed buffering effect by the bacteria may be generally attributed to augmented “inert food effect,” a somewhat trivial conclusion. Second, greater yeast content accelerates GF growth, thus allowing us to compare variation in size-matched GF and mono-associated larvae en masse, while minimizing the size and stage differences between the GF and mono-associated larvae. As GF larvae are growth-delayed and take longer to reach the same physical size compared with their mono-associated siblings, they can accumulate more developmental noise as a consequence of aberrantly protracted physiological responses, which in turn may contribute to higher phenotypic variation. In the same line of reasoning, at the same age, the faster developing mono-associated larvae have had less time to accumulate developmental noise and are closer to maturation than the germ-free larvae; thus, they can appear more uniform phenotypically, which may also account for less variability. To limit such bias imposed by the potential difference in developmental stage, we chose to augment the yeast content of the food for the GF larvae, and with the “boost” in GF growth, we compared the variances of growth when both GF and mono-associated larvae reach similar physical size during a comparable growth period.
In our initial testing trials, we found that the additional yeast invariably accelerated GF growth in different genetic backgrounds, sometimes to the same extent of the LpWJL presence (Figure S3C). However, when pooled based on dietary yeast content, the GF larvae that have received more yeast were longer but showed greater variation in lengths (Figure S3D). Moreover, in the genetically heterogeneous F2 larvae, the CV and SD values tend to separate into two distinct groups, as driven by LpWJL presence (Figures 2A and S3B). Overall, the F2 LpWJL mono-associated larvae were slightly longer, but their GF siblings varied more in length, regardless of yeast content or larval age (Figure S3E). In the size-matched pools (Figure 2A, purple bracket), GF size still fluctuated more than that of the LpWJL mono-associated siblings (Figure 2B), despite the fact that they were raised on a richer diet. Therefore, we first confirm that augmenting yeast content fails to recapitulate the same buffering effect mediated by living commensals. This is consistent with our previous observation that Lp-mediated transcriptomic buffering is readily visible, even if the GF transcriptomes are derived from larvae that have been raised on greater yeast quantities (Figures 1 and S2). More importantly, we conclude that phenotypic buffering by the gut microbe LpWJL indeed operates in a genetically diverse host population facing a nutritional challenge, hence qualifying the gut microbiota as a previously unappreciated buffering agent of cryptic genetic variation.
Figure 2.
In the Genetically Diverse DGRP F2 Population, LpWJL Reduces Variation in Different Physical Fitness Traits
(A) A scatterplot showing how coefficient of variation (CV) changes as a function of larval length and how such change differs in the DGRP F2 GF (pink) and LpWJL mono-associated (blue) populations (see Figure S3A and Methods for detailed schemes). Each data point represents the intercept of a CV value and its corresponding average larval length in a particular cross. Each CV, SD, and average value was derived from larvae measurements gathered from at least three biological replicates from either GF or LpWJL mono-associated conditions. Each replicate contains 10–40 larvae. Based on multivariate ANOVA analysis, the factors affecting variants in this plot are: larval age∗ (p = 0.053), bacterial presence∗∗∗(p = 3.02 × 10−6), and larval length (p = 8.27 × 10−15∗∗∗). The purple bracket indicates the arbitrarily selected experiments where the average larval length for each cross falls between 3 and 4 mm for size-matching purpose.
(B) The average larval length of the F2 progeny pooled from experiments demarcated by the purple bracket in Figure 2A. The average size is perfectly matched (GF Avg Length = 3.522mm, LpWJL Avg Length = 3.582mm, p = 0.857ns, Mann-Whitney test), whereas the GF population exhibits greater variation than the LpWJL mono-associated population (VarGF = 0.642, CVGF = 22.8%, VarLp = 0.427, CVLp = 18.3%).
(C) Variance and mean comparisons for the average day of pupariation for individual larva in the F2 GF and mono-associated populations. (Difference in mean p < 0.0001∗∗∗, Mann-Whitney test, Var GF = 2.42, VarLp = 1.22).
(D) Variance comparison for average day of adult emergence in the F2 GF and mono-associated populations (Difference in mean p < 0.0001∗∗∗, VarLp = 1.84, VarGF = 5.27).
(E) Box plots comparing the variances of pupariation derived from each tube containing approximately 40 larvae. The average variance per tube for the GF population = 3.99; the average variance per tube for the LpWJL-associated population = 1.12. VarLp = 0.54, VarGF = 1.76. Note that these values are the “variance of variances.”
(F) Box plots comparing the variances for adult emergence from each tube containing approximately 40 larvae (difference in mean p < 0.0001∗∗∗). The average variance per tube for the GF population = 4.06; the average variance per tube for the LpWJL associated population = 1.34. For “variance of the variances,” VarLp = 1.33, VarGF = 4.2.
(G and H) In both male (lozenge) and female (circle) adults, the variances in body size (G the difference in mean body length: for females, p = 0.003∗∗∗, for males, p < 0.0001∗∗∗∗) and wing size (H, the difference in mean wing area for females, p < 0.0001∗∗∗∗ for males, p = 0.043∗∗) are greater in the GF population than in the mono-associated population. The adult datasets presented in Figures 2G and 2H and in S3G and S3H take on normal distribution by D'Agostino and Pearson omnibus normality test, F variances are therefore calculated and compared. Data are represented as mean and 10–90 percentile in all panels.
During chronic under-nutrition, LpWJL sustains growth rate as effectively as an entire gut associated commensal community (Storelli et al., 2011). We thus wondered if a natural and more complex gut-associated community can also buffer growth variation like LpWJL. To address this question, we rendered a population of wild flies collected in a nearby garden germ-free, and re-associated them with their own fecal microbial community (Tefit et al., 2018). In three of four experimental repeats, growth variation is significantly reduced in the larval population fed on food inoculated with their parents' fecal microbes (Figure S3F and data not shown), and the cumulative CV and variances derived from each food cap were significantly higher in the GF population (Figures S3G and S3H). This suggests that the gut-associated microbial community of wild flies indeed decreases growth variation of a natural Drosophila population. However, since the wild-derived microbes did not consistently buffer larval growth, probably due to the difficulty to precisely control the quantity and composition of the inoculated fecal microbiota, we returned to the mono-association model for subsequent studies.
If the observed growth variation in GF larvae indeed reflects the “unleashing” of the host's genetic potential due to the loss of a buffering mechanism provided by gut microbes, then we posit that other physical fitness traits in a fertile surviving GF population should in principle also exhibit greater phenotypic variation. We therefore examined the variances in pupariation timing and adult emergence in the F2 progeny of the inter-DGRP strain crosses (Figure S3A). First, individual GF larvae pupariated and eclosed later, but the variances in the pooled data were greater than that of mono-associated counterparts (Figures 2C and 2D); from each vial containing an equal number of larvae, the variances of pupariation and eclosion were also greater in the GF samples (Figures 2E and 2F). Therefore, both inter-individual and among-population variances in developmental timing and adult emergence are reduced upon mono-association. Lastly, GF adults were slightly shorter (Figure 2F); the sizes of representative organs, expressed as area of the eye and the wing, were also smaller, yet the variances in these traits were greater (Figures 2H and S3I). Furthermore, the wing/body-length allometric slopes remained unaltered, but the individual GF values were more dispersed along the slope (Figures S3J and S3K); when taken as a ratio (wing length/body-length), the variance was greater in the GF flies (Figure S3L). These observations indicate that gut microbes, represented by LpWJL, confer phenotypic homogeneity in various physical fitness traits in a genetically diverse host population under nutritional stress.
LpWJL Conveys Robustness in Organ-Patterning under Nutritional Stress
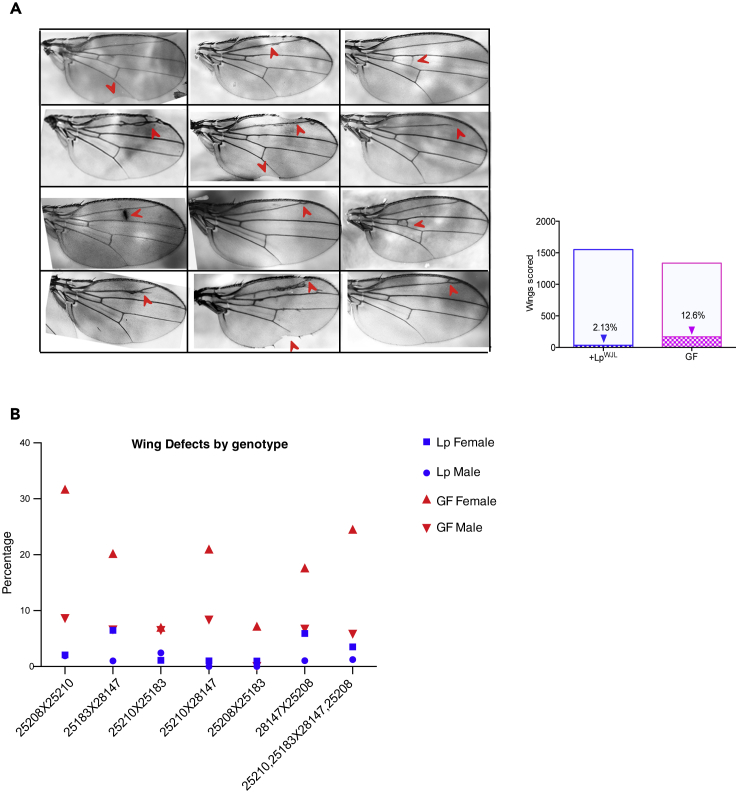
We have thus far shown that LpWJL association confers transcriptomic stability and phenotypic constancy to the developing host facing nutritional stress, in a fashion that is reminiscent of the host's own genetic buffering mechanism. For example, reducing Hsp90 activity has been shown to increase organ size variation in both plants and animals (Queitsch et al., 2002, Rohner et al., 2013, Rutherford and Lindquist, 1998). Moreover, compromising Hsp90 can lead to morphological aberrations that are otherwise “hidden” (Rutherford and Lindquist, 1998). Similarly, we also found that a significant fraction of the GF F2 flies bore aberrant wing patterns such as missing margins, incomplete vein formations, and ectopic vein tissue (Figure 3A). The incidence of wing anomalies differed according to the genotype, and females were more affected than males (Figure 3B). In contrast, the most visible “defect” in their LpWJL associated siblings, if any, were rare and hardly discernable (Figures 3A and S4A). Furthermore, gross patterning anomalies were absent in the viable adults from the GF parental homozygous strains or in F2 adults reared on a standard diet (data not shown), supporting the notion that the fly gut-associated commensals likely act as a canalization mechanism by suppressing the contribution of cryptic genetic variation to developmental phenotypes in the presence of nutritional stress. Organ patterning is a robust process; changes in nutrition, humidity, temperature, and crowding can alter the final adult body and wing size; yet wing patterning is usually invariant and reproducible (Mirth and Shingleton, 2012). Surprisingly, we found that, in GF flies, constant nutritional stress can in fact unveil the effects of preexisting “silent” mutations that manifest themselves as visible wing patterning anomalies. Furthermore, as the patterning defects appear only in nutritionally challenged F2 flies devoid of their gut-associated commensals, we conclude that these defects reflect a breach of the canalization process during developmental patterning when the hidden effects of genetic variants are unlocked (Waddington, 1959) and the gut-associated symbionts buffer the effects of these otherwise seemingly “neutral” variants to confer robustness to the canalized process of organ patterning.
Figure 3.
In the DGRP F2 Progeny, LpWJL Association Provides Robustness in Wing Developmental Patterning
(A) A compilation of representative images illustrating wing patterning anomalies in the GF DGRP F2 progeny, indicated by red arrows. The number of such patterning anomalies are compiled together for GF and LpWJL mono-associated flies (χ2 test, p < 0.0001***, NLp = 1,551 NGF = 1,335), and the percentage of defects are indicated inside each bar.
(B) The incidence of wing patterning defects separated by F2 genotypes. The y axis denotes the percentage of wings with aberrant patterning as represented in Figure 3A.
Compromising ROS Activity Impairs the Buffering Capacity of LpWJL without Affecting Bacterial Growth
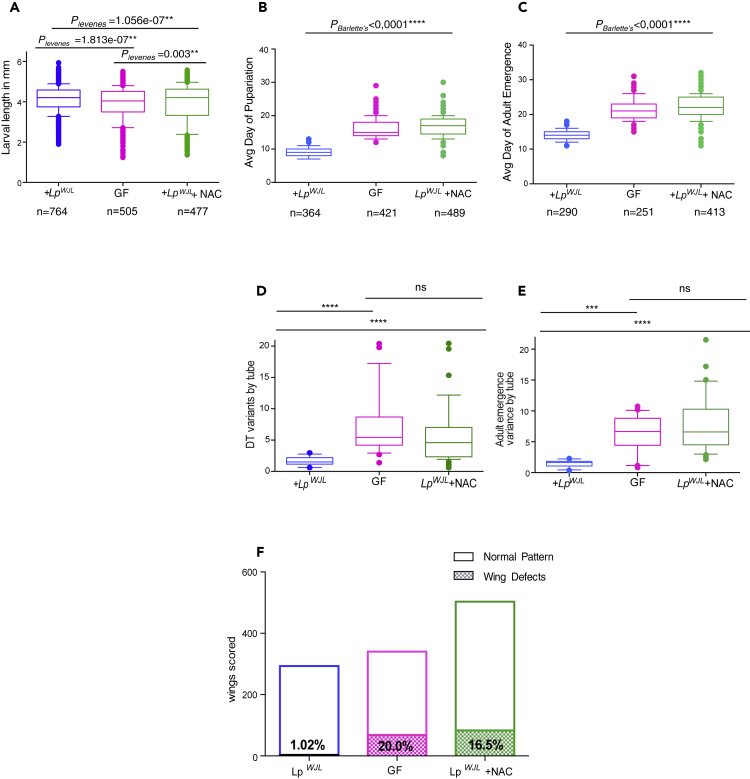
The wing anomalies in the GF F2 progeny highly resemble the phenotypes reported by Santabarbara-Ruiz et al., who blocked ROS by feeding the larvae with antioxidants, such as N-acetylcysteine (NAC), and induced regeneration defects in the wing (Santabarbara-Ruiz et al., 2015). NAC is a widely used and safe antioxidant that has been repeatedly used to block ROS without adversely affecting adult feeding behavior (Atkuri et al., 2007, Bailey et al., 2015, Santabarbara-Ruiz et al., 2015, Sun et al., 2012). We therefore repeated the DGRP F2 cross experiment with an additional condition by mixing the antioxidant molecule NAC in the diet of mono-associated flies. First, germ-free larvae fed on NAC tended to be small, yet the variance in their size is comparable with that of mono-associated flies fed with NAC and of germ-free flies never exposed to NAC (Figure S4B). However, over 95% of NAC-fed germ-free larvae failed to pupariate, making it impossible to assess variances in developmental timing and adult traits. Furthermore, NAC feeding did not compromise bacterial growth (Figure S4C) but significantly diminished the buffering capacity of LpWJL (Figure 4). Specifically, variation in larval size (Figure 4A), developmental timing (Figures 4B and 4D), and adult emergence (Figures 4C and 4E) were significantly increased in NAC-fed larvae mono-associated with LpWJL, to a level similar to or even higher than that in GF larvae. Wing patterning anomalies were also unmasked (Figure 4F). Therefore, blocking ROS activity through NAC feeding suppresses the genetic buffering effect mediated by the gut bacteria.
Figure 4.
Blocking ROS Activity by N-acetylcysteine (NAC) Compromises the LpWJL Buffering Capacity
(A) In the DGRP F2 progeny, feeding LpWJL mono-associated animals with food supplemented with NAC increases the variances in size-matched larvae. Average Lp larval size: 4.08mm; average GF larval size: 3.83mm; average LpWJL + NAC larval size: 3.94mm. There is no size difference between GF and NAC-treated flies associated with LpWJL, p = 0.064. CVLp = 15.8%, CVGF = 20.8%; CVLp+NAC = 24.0%.
(B and C) NAC treatment to the Lp-associated animals also increases the variances of pupariation (B) and adult emergence (C). The average day to become a pupa for LpWJL mono-associated larva: Day 8.9 (Var = 2.13); for a GF larva: Day 16.1 (Var = 8.27); for a NAC-treated, mono-associated larva: Day 16.8 (Var = 8.36). The average day for an LpWJL mono-associated adult to emerge is: Day14.1 (Var = 2.08), for a GF adult: Day 21 (Var = 8.3), and for an NAC-treated, mono-associated adult: Day 21.7 (Var = 11.3).
(D and E) NAC treatment to the LpWJL mono-associated animals also increases the among-population variances of pupariation and adult emergence. Each data point represents the variance calculated based on the average day of pupariation (D) or adult emergence (E) from each tube housing approximately 40 animals.
(F) Morphological defects in the wings are also significantly increased in NAC-treated mono-associated adults (χ2 test, p < 0.0001***) pink: GF (N = 340); Blue: +LpWJL (N = 293), Green: + LpWJL + NAC (N = 503). Data are represented as mean and 10–90 percentile in all panels.
Discussion
Here we show that a single Drosophila gut commensal strain LpWJL functionally resembles a general buffer mechanism that safeguards the host's genetic potential and confers developmental robustness in times of nutritional stress. This conclusion emerged from our analyses in different genetic contexts, such as the DGRP lines, the RNAi knock-down crosses, and the heterozygous F2 crosses, in which we observed that LpWJL mono-associated flies not only grow better than their germ-free counter parts but also show less variation in transcriptional and phenotypic traits related to growth and maturation. Microbial buffering also operates in wild-derived flies associated with their endogenous gut communities, which implies that such buffering may be a universal feature of many beneficial microbes. In Drosophila, nutritional mutualism with commensals is inconstant and volatile by nature (Broderick et al., 2014, Storelli et al., 2018, Wong et al., 2013), in that the gut community composition highly varies among individuals and along the life stages of each individual. Consequently, the rapid acquisition or loss of particular gut community members can alter the functionality and capacity of the gut-associated symbionts, which in turn affects how the developing host population adjusts its phenotypic range to adapt to the changing environment during their life. Furthermore, under nutritional stress, genetically diverse fly populations devoid of their gut microbes manifest wing patterning defects that are masked by the presence of a gut microbe. The action of genetic buffering by the gut commensal bacteria therefore maintains organ patterning robustness for the developing population while facing nutritional stress.
Gut commensals stimulate host ROS production, which consequently elicits diverse physiological consequences. Jones et al. previously reported that acute exposure to Lactobacillus plantarum stimulates the dNox-dependent production of ROS in larval enterocytes and subsequently increases the expression of genes involved in the Nrf2-mediated cyto-protection program (Jones et al., 2013, Jones et al., 2015). In adult flies, Lp-derived lactic acid stimulates ROS production and leads to shortened lifespan, which is rescued by blocking ROS with NAC feeding (Iatsenko et al., 2018). Therefore, the microbial regulation of ROS is highly complex and seems to mediate antagonistic outcomes depending on host life-stages. Our results further add to such complexity. We identified an unexpected role of ROS in mediating microbial buffering of host phenotypic variance. Blocking ROS in germ-free flies leads to maturation failure, but without further increasing variation in larval growth. This result suggests that ROS-mediated microbial buffering of growth is separable from its involvement in metamorphosis. Future explorations are required to reconcile how ROS activity can be integrated into the molecular dialogue between the host and its gut microbiome to maintain robustness during development.
A recent study by Elgart et al. showed that, when raised on standard food, the wild-type, axenic embryonic transcriptome showed accelerated maternal-zygotic-transition (MZT) and a shortened period of embryogenesis. Moreover, four Drosophila strains, each bearing a single, heterozygous genetic mutation manifested greater variance in pupariation timing in the germ-free progeny than in their axenic parents (Elgart et al., 2016). Whether the accelerated MZT in germ-free, wild-type embryos can account for a mechanism inducing greater larval maturation variability in the mutant progeny remains to be elucidated. However, the fact that in axenic flies, hidden phenotypic variation is revealed in the next generation is consistent with our finding that microbial buffering acts through unmasking CGVs. As greater phenotypic, transcriptomic variation and organ patterning anomaly are only observed in germ-free flies under nutritional stress, our results indicate that microbial buffering may be a natural outcome of long-term co-evolution with the host under strong selection pressure. Therefore, we propose that the facultative gut commensals not only increase the host’s fitness in a stressful environment during its lifetime but also enable its evolutionary adaptation by preserving the host's CGVs in the long run.
Limitations of the Study
We have observed the effects of microbial buffering in a genetically diverse population, making the search for “buffering genes” a challenge, as classic genetic screens in Drosophila do not apply. By the same token, to alter ROS activity in the genetically diverse F2s, we had to resort to NAC treatment, which is a conventional and widely accepted approach to block ROS in the broad literature. We obtained the expected results, namely, the buffering capacity of Lactobacillus plantarum is compromised, but future QTL studies are required confirm the role of the ROS pathway and identify genes and variants affected by such microbial buffering activities.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We would like to thank Benjamin Prud'homme and colleagues at ENS de Lyon and EPFL for their critical reading of the manuscript and valuable suggestions, the ArthroTools platform of the SFR Biosciences (UMS3444/US8) for the fly equipment and facility, and the Bloomington Stock Centre and VDRC for fly lines. This work was supported by an international collaborative grant through the French "Agence Nationale de la Recherche" and the "Fond National Suisse pour la Recherche" (ANR-15-CE14-0028-01) awarded to F.L. and B.D., an ERC starting grant (FP7/2007-2013-N_309704) awarded to F.L., a SystemsX.ch (AgingX) Grant awarded to B.D., and Institutional Support by the EPFL (B.D.). C.-E.I. was funded by a Ph.D. fellowship from the Rhone-Alps region. F.L. is supported by the Finovi, FRM, and FSER foundations and the EMBO Young Investigator Program. M.B.-S., M.F., M.L., and B.D. were supported by AgingX (SystemsX.ch) and/or Institutional Support by the EPFL.
Author Contributions
D.M., M.B.-S., B.D., and F.L. conceived the project and designed the experiments; D.M. and C.-E.I. conducted all fly-related experiments; M.B.-S. and M.L. conducted the GWAS analysis; M.B.-S, M.F., and V.B. prepared the libraries and conducted single-larvae transcriptome analyses. P.J. conducted the multivariate statistical analyses; G.S., has identified the effect of NAC on Lp-mediated larval phenotypes. D.M., M.B.-S., B.D., and F.L. analyzed the data. D.M. drafted the manuscript, D.M., M.B.-S, B.D., and F.L revised the paper and wrote the final draft together.
Declaration of Interests
The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to francois.leulier@ens-lyon.fr or bart.deplancke@epfl.ch.
Published: September 27, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.07.048.
Contributor Information
Bart Deplancke, Email: bart.deplancke@epfl.ch.
François Leulier, Email: francois.leulier@ens-lyon.fr.
Supplemental Information
References
- Alpern D., Gardeux V., Russeil J., Mangeat B., Meireles-Filho A., Breysse R., Hacker D., Deplancke B. BRB-seq: ultra-affordable high-throughput transcriptomics enabled by bulk RNA barcoding and sequencing. Genome Biol. 2019;20 doi: 10.1186/s13059-019-1671-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkuri K.R., Mantovani J.J., Herzenberg L.A., Herzenberg L.A. N-Acetylcysteine–a safe antidote for cysteine/glutathione deficiency. Curr. Opin. Pharmacol. 2007;7:355–359. doi: 10.1016/j.coph.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A.P., Koster G., Guillermier C., Hirst E.M., MacRae J.I., Lechene C.P., Postle A.D., Gould A.P. Antioxidant role for lipid droplets in a stem cell Niche of Drosophila. Cell. 2015;163:340–353. doi: 10.1016/j.cell.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick N.A., Buchon N., Lemaitre B. Microbiota-induced changes in drosophila melanogaster host gene expression and gut morphology. MBio. 2014;5 doi: 10.1128/mBio.01117-14. e01117–01114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brucker R.M., Bordenstein S.R. The hologenomic basis of speciation: gut bacteria cause hybrid lethality in the genus Nasonia. Science. 2013;341:667–669. doi: 10.1126/science.1240659. [DOI] [PubMed] [Google Scholar]
- Clemente J.C., Ursell L.K., Parfrey L.W., Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A.J., Chaston J.M., Douglas A.E. The Drosophila transcriptional network is structured by microbiota. BMC Genomics. 2016;17:975. doi: 10.1186/s12864-016-3307-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgart M., Stern S., Salton O., Gnainsky Y., Heifetz Y., Soen Y. Impact of gut microbiota on the fly's germ line. Nat. Commun. 2016;7:11280. doi: 10.1038/ncomms11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkosar B., Defaye A., Bozonnet N., Puthier D., Royet J., Leulier F. Drosophila microbiota modulates host metabolic gene expression via IMD/NF-kappaB signaling. PLoS One. 2014;9:e94729. doi: 10.1371/journal.pone.0094729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T. The evolutionary genetics of canalization. Q. Rev. Biol. 2005;80:287–316. doi: 10.1086/432265. [DOI] [PubMed] [Google Scholar]
- Gilbert J.A., Neufeld J.D. Life in a world without microbes. PLoS Biol. 2014;12:e1002020. doi: 10.1371/journal.pbio.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert S.F. Symbiosis as the way of eukaryotic life: the dependent co-origination of the body. J. Biosci. 2014;39:201–209. doi: 10.1007/s12038-013-9343-6. [DOI] [PubMed] [Google Scholar]
- Huang W., Massouras A., Inoue Y., Peiffer J., Ramia M., Tarone A.M., Turlapati L., Zichner T., Zhu D., Lyman R.F. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 2014;24:1193–1208. doi: 10.1101/gr.171546.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iatsenko I., Boquete J.P., Lemaitre B. Microbiota-derived lactate activates production of reactive oxygen species by the intestinal NADPH oxidase Nox and shortens Drosophila lifespan. Immunity. 2018;49:929–942.e5. doi: 10.1016/j.immuni.2018.09.017. [DOI] [PubMed] [Google Scholar]
- Jones R.M., Desai C., Darby T.M., Luo L., Wolfarth A.A., Scharer C.D., Ardita C.S., Reedy A.R., Keebaugh E.S., Neish A.S. Lactobacilli modulate epithelial cytoprotection through the Nrf2 pathway. Cell Rep. 2015;12:1217–1225. doi: 10.1016/j.celrep.2015.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R.M., Luo L., Ardita C.S., Richardson A.N., Kwon Y.M., Mercante J.W., Alam A., Gates C.L., Wu H., Swanson P.A. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 2013;32:3017–3028. doi: 10.1038/emboj.2013.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keebaugh E.S., Yamada R., Obadia B., Ludington W.B., Ja W.W. Microbial quantity impacts Drosophila nutrition, development, and lifespan. iScience. 2018;4:247–259. doi: 10.1016/j.isci.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.K., Park Y.M., Lee O.Y., Lee W.J. Draft genome sequence of Lactobacillus plantarum strain WJL, a Drosophila gut symbiont. Genome Announc. 2013;1 doi: 10.1128/genomeA.00937-13. e00937–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B. Genotype to phenotype: lessons from model organisms for human genetics. Nat. Rev. Genet. 2013;14:168–178. doi: 10.1038/nrg3404. [DOI] [PubMed] [Google Scholar]
- Ma D., Leulier F. The importance of being persistent: the first true resident gut symbiont in Drosophila. PLoS Biol. 2018;16:e2006945. doi: 10.1371/journal.pbio.2006945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T.F., Richards S., Stone E.A., Barbadilla A., Ayroles J.F., Zhu D., Casillas S., Han Y., Magwire M.M., Cridland J.M. The Drosophila melanogaster genetic reference panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestek Boukhibar L., Barkoulas M. The developmental genetics of biological robustness. Ann. Bot. 2016;117:699–707. doi: 10.1093/aob/mcv128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirth C.K., Shingleton A.W. Integrating body and organ size in Drosophila: recent advances and outstanding problems. Front. Endocrinol. (Lausanne) 2012;3:49. doi: 10.3389/fendo.2012.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obadia B., Keebaugh E.S., Yamada R., Ludington W.B., Ja W.W. Diet influences host-microbiota associations in Drosophila. Proc. Natl. Acad. Sci. U S A. 2018;115:E4547–E4548. doi: 10.1073/pnas.1804948115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pais I.S., Valente R.S., Sporniak M., Teixeira L. Drosophila melanogaster establishes a species-specific mutualistic interaction with stable gut-colonizing bacteria. PLoS Biol. 2018;16:e2005710. doi: 10.1371/journal.pbio.2005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posadas D.M., Carthew R.W. MicroRNAs and their roles in developmental canalization. Curr. Opin. Genet. Dev. 2014;27:1–6. doi: 10.1016/j.gde.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C., Sangster T.A., Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- Rohner N., Jarosz D.F., Kowalko J.E., Yoshizawa M., Jeffery W.R., Borowsky R.L., Lindquist S., Tabin C.J. Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in cavefish. Science. 2013;342:1372–1375. doi: 10.1126/science.1240276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S., Hirate Y., Swalla B.J. The Hsp90 capacitor, developmental remodeling, and evolution: the robustness of gene networks and the curious evolvability of metamorphosis. Crit. Rev. Biochem. Mol. Biol. 2007;42:355–372. doi: 10.1080/10409230701597782. [DOI] [PubMed] [Google Scholar]
- Rutherford S.L., Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- Santabarbara-Ruiz P., Lopez-Santillan M., Martinez-Rodriguez I., Binagui-Casas A., Perez L., Milan M., Corominas M., Serras F. ROS-induced JNK and p38 signaling is required for unpaired cytokine activation during Drosophila regeneration. PLoS Genet. 2015;11:e1005595. doi: 10.1371/journal.pgen.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A., Werner J., Andrulis E.D., Nakayama T., Hirose S., Reinberg D., Lis J.T. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003;301:1094–1096. doi: 10.1126/science.1085712. [DOI] [PubMed] [Google Scholar]
- Shimojima T., Okada M., Nakayama T., Ueda H., Okawa K., Iwamatsu A., Handa H., Hirose S. Drosophila FACT contributes to Hox gene expression through physical and functional interactions with GAGA factor. Genes Dev. 2003;17:1605–1616. doi: 10.1101/gad.1086803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S.C., Kim S.H., You H., Kim B., Kim A.C., Lee K.A., Yoon J.H., Ryu J.H., Lee W.J. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science. 2011;334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- Sommer F., Backhed F. The gut microbiota–masters of host development and physiology. Nat. Rev. Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- Storelli G., Defaye A., Erkosar B., Hols P., Royet J., Leulier F. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 2011;14:403–414. doi: 10.1016/j.cmet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- Storelli G., Strigini M., Grenier T., Bozonnet L., Schwarzer M., Daniel C., Matos R., Leulier F. Drosophila perpetuates nutritional mutualism by promoting the fitness of its intestinal Symbiont Lactobacillus plantarum. Cell Metab. 2018;6:362–377. doi: 10.1016/j.cmet.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Komatsu T., Lim J., Laslo M., Yolitz J., Wang C., Poirier L., Alberico T., Zou S. Nutrient-dependent requirement for SOD1 in lifespan extension by protein restriction in Drosophila melanogaster. Aging Cell. 2012;11:783–793. doi: 10.1111/j.1474-9726.2012.00842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefit M.A., Gillet B., Joncour P., Hughes S., Leulier F. Stable association of a Drosophila-derived microbiota with its animal partner and the nutritional environment throughout a fly population’s life cycle. J. Insect Physiol. 2018;106(Pt 1):2–12. doi: 10.1016/j.jinsphys.2017.09.003. [DOI] [PubMed] [Google Scholar]
- Waddington C.H. Canalization of development and genetic assimilation of acquired characters. Nature. 1959;183:1654–1655. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- Wagner A. Princeton University Press; 2007. Robustness and Evolvability in Living Systems. Paperback edn. [Google Scholar]
- Wong A.C., Chaston J.M., Douglas A.E. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J. 2013;7:1922–1932. doi: 10.1038/ismej.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.C., Dobson A.J., Douglas A.E. Gut microbiota dictates the metabolic response of Drosophila to diet. J. Exp. Biol. 2014;217:1894–1901. doi: 10.1242/jeb.101725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada R., Deshpande S.A., Bruce K.D., Mak E.M., Ja W.W. Microbes promote amino acid harvest to rescue undernutrition in Drosophila. Cell Rep. 2015;10:865–872. doi: 10.1016/j.celrep.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.