Abstract
Background
It is widely believed that most xenobiotics cross biomembranes by diffusing through the phospholipid bilayer, and that the use of protein transporters is an occasional adjunct. According to an alternative view, phospholipid bilayer transport is negligible, and several different transporters may be involved in the uptake of an individual molecular type. We recognise here that the availability of gene knockout collections allows one to assess the contributions of all potential transporters, and flow cytometry based on fluorescence provides a convenient high-throughput assay for xenobiotic uptake in individual cells.
Results
We used high-throughput flow cytometry to assess the ability of individual gene knockout strains of E coli to take up two membrane-permeable, cationic fluorescent dyes, namely the carbocyanine diS-C3(5) and the DNA dye SYBR Green. Individual strains showed a large range of distributions of uptake. The range of modal steady-state uptakes for the carbocyanine between the different strains was 36-fold. Knockouts of the ATP synthase α- and β-subunits greatly inhibited uptake, implying that most uptake was ATP-driven rather than being driven by a membrane potential. Dozens of transporters changed the steady-state uptake of the dye by more than 50% with respect to that of the wild type, in either direction (increased or decreased); knockouts of known influx and efflux transporters behaved as expected, giving credence to the general strategy. Many of the knockouts with the most reduced uptake were transporter genes of unknown function (‘y-genes’). Similarly, several overexpression variants in the ‘ASKA’ collection had the anticipated, opposite effects. Similar results were obtained with SYBR Green (the range being approximately 69-fold). Although it too contains a benzothiazole motif there was negligible correlation between its uptake and that of the carbocyanine when compared across the various strains (although the membrane potential is presumably the same in each case).
Conclusions
Overall, we conclude that the uptake of these dyes may be catalysed by a great many transporters of putatively broad and presently unknown specificity, and that the very large range between the ‘lowest’ and the ‘highest’ levels of uptake, even in knockouts of just single genes, implies strongly that phospholipid bilayer transport is indeed negligible. This work also casts serious doubt upon the use of such dyes as quantitative stains for representing either bioenergetic parameters or the amount of cellular DNA in unfixed cells (in vivo). By contrast, it opens up their potential use as transporter assay substrates in high-throughput screening.
Electronic supplementary material
The online version of this article (10.1186/s12866-019-1561-0) contains supplementary material, which is available to authorized users.
Keywords: Drug transporters, Keio, Carbocyanine, SYBR Green, Membrane energisation
Background
The presence, number, nature, and physiological status of bacteria is widely assessed using fluorescent dyes [1–9]. Some of these stain particular molecules or macromolecules such as DNA [10], protein [11] or lipid [12], while others reflect physiological variables such as pH, the concentrations of other ions and small molecules, or the extent of membrane energisation [13]. In some cases (e.g. [6, 8]), cells are permeabilised before staining, such that the natural ability of stains to reach their intracellular targets is not an issue. However, studies of in vivo physiology [9, 14–16] necessarily require that native, intact cells are used. In Gram-negative cells, entry to the periplasm is mediated via porins [17], which can affect the ability of stains such as rhodamine 123 and other cations to be accumulated [18–20], while intracellular concentrations of stain may also be modified by the activities of influx and efflux transporters.
Although it is often assumed and stated that intact cells take up xenobiotics mainly according to their hydrophobicity (lipophilicity) [21–23], and generally by ‘passive diffusion’ through the limited phospholipid bilayer areas that exist, an abundance of evidence indicates that this is not in fact the case [24–26] and that ‘phospholipid bilayer transport is negligible’ [27]. Those xenobiotics that pass through the membrane, which necessarily include xenobiotic fluorescent probes, must thus be taken up by protein-based transporters, and the question arises as to which ones [28, 29]. In mammalian cells, for instance, fluorescein can be transported by an active monocarboxylate transporter [30].
Since the amount of fluorescence is often used as a quantitative indicator of the amount of the relevant determinand, it is obvious that the involvement of such transporters might significantly obscure the true values that would result if transporter-mediated membrane permeability or translocation was not occurring or was kinetically irrelevant. An early indication of this was the recognition that the apparent failure of ethidium bromide to stain intracellular DNA inside intact (live) E. coli cells was almost entirely due to the overwhelming activity of an efflux pump whose activity was much reduced at 0 °C [31]. The relevant efflux pumps were not then identified, but it is now well established that ethidium bromide is a very good substrate for ‘efflux’ transporters such as the multisubunit acrAB-tolC complex [32] and the small molecule resistance (SMR) protein emrE [33].
Concentrative transporters (whether influx or efflux) need additional sources of free energy. In E. coli these are typically the energised membrane (often seen as a membrane potential or a protonmotive force [34]) generated via electron transport (that can also be used to synthesise ATP), or ATP itself acting directly. The latter is significantly more common in prokaryotes [35]. However, it is often considered that the uptake of more or less lipophilic cations into bacteria or mitochondria is driven by a negative-inside membrane potential, whose concentration ratio reflects it according to the Nernst equation and thereby allows one to infer it (see Discussion).
Many non-fluorescent drugs, including anti-infectives, can simply be seen as xenobiotics, and systematic studies have been performed to see the extent to which the loss of effluxers (and occasionally of influxers) modulates their toxicity [36, 37]. In particular, the AcrAB-TolC complex spans inner and outer membranes, is constitutively expressed, and is considered to play a major role in multidrug resistance [38–42]. Consequently, the activities of efflux transporters are widely recognised both as major mediators of microbial resistance to antibiotics and as targets for ameliorating it [43–69]. However, such activities cannot yet be predicted reliably (e.g. [70–72]). Efflux transporters are also important in pharmacokinetics, not least by effecting the export of anticancer drugs in mammalian systems (e.g. [53, 73–76]). They can also play important roles in the biotechnological production of small molecules and/or their biotransformations [77, 78].
Since efflux transporters are required to remove such intracellular molecules that have been taken up by cells, it is reasonable that influx transporters were exploited to get them there in the first place [24–27]. However, in the case of bacteria, knowledge of the specific influx and efflux transporters for individual xenobiotics is surprisingly limited, albeit many efflux transporters can be quite promiscuous [79–81].
Although they can be interfered with by coloured compounds [82], an understanding of the transporters used in the uptake and efflux of fluorescent probes in intact microbial cells is of interest for a number of reasons: (i) as with mammalian cells [83–86], they can provide substrates for competitive or trans-stimulation-type uptake assays, (ii) they provide examples of substrates that can be used in the development of quantitative structure-activity relationships (QSARs) for the effluxers themselves, and (iii) they allow us to assess the limitations of any individual fluorescent probe assay where the expression levels of relevant transporters is not known or controlled [87]. It is already known that even lipid stains such as Nile Red [88–91] and membrane energisation stains such as bis-(1,3-dibutylbarbituric acid trimethine oxonol) (commonly known as DiBAC4(3)) are in fact efflux substrates of acrAB-tolC [92], while other widely used stains that are effluxed via acrAB-tolC include the Hoechst dye H33342 [93], berberine [94], resazurin [95], and rhodamine 6G [96]. What is much less well known is which if any other efflux transporters are involved, and which influx transporters may have been responsible for the uptake of such commonly used fluorophores in the first place.
Following the systematic genome sequencing of E. coli K12 strain MG1655 [97], and an equivalent programme in baker’s yeast [98, 99], it was soon recognised that much scientific value would accrue to the possession of a collection of single-gene knockouts (of ‘non-essential’ genes), and this was produced as the ‘Keio collection’ [100–102] http://ecoli.iab.keio.ac.jp/. Flow cytometry provides a convenient means of estimating the steady-state uptake of fluorescent probes in bacteria [9], and while efflux pumps have been analysed elegantly in this way [44] the combination of flow cytometry and the Keio collection seemed to provide an ideal opportunity to assess the contribution of individual transporter genes (i.e. their products) to the uptake and efflux of widely used fluorescent probes. We used TransportdB http://www.membranetransport.org/transporter2.php?oOID=ecol1 [103] to pick out the most pertinent subset of transporters, to which we added a few more strains whose knockouts were involved in ATP synthesis, and report the findings herein. It is concluded that every probe used can exploit a wide variety of transporters for both influx and efflux, additional to reporting on their nominal determinand of interest. This is consistent with the view [27, 29, 104] that most xenobiotics that enter cells can be taken up and/or effluxed by multiple transporters of varying specificities. A preprint has been lodged at bioRxiv (https://www.biorxiv.org/content/10.1101/603688v1).
Results
Variation of the uptake of a lipophilic carbocyanine cation
3,3′-Dipropylthiadicarbocyanine iodide (IUPAC name(3-Propyl-2-{(1E,3E,5E)-5-(3-propyl-1,3-benzothiazol-2(3H)-ylidene)-1,3-pentadien-1-yl}-1,3-benzothiazol-3-ium iodide, Fig. 1), commonly known as diS-C3(5) [105, 106], is a cationic carbocyanine dye that is accumulated by bacteria (both Gram-positive and Gram-negative) with energised membranes. Although culturability is the conventional metric for bacterial ‘viability’ [107, 108], diS-C3(5) has been exploited widely in microbiology to detect nominally ‘viable’ bacteria (at least those with intact membranes) in clinical, laboratory and environmental samples, especially using flow cytometry (e.g. [109–113]). Metabolising yeast cells can also accumulate it [114–118]. We note that the extent of fluorescence is not necessarily fully linear with concentration because of dye stacking [105, 106, 119] (and see [120]) but is at least assumed to be more or less monotonic.
Fig. 1.
Chemical structures of some molecules described in this article
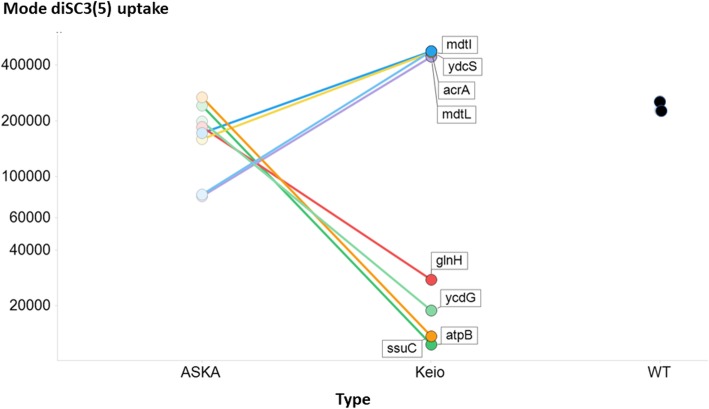
The strategy used to assess diS-C3(5) uptake, gating on light scattering in ‘forward’ and ‘side’ directions by the bacteria and assessing uptake via red fluorescence (which is not interfered with by any autofluorescence), is precisely as was described previously [121]. It is obvious (Fig. 2) that even in the wild-type strain there is considerable heterogeneity in the distribution of uptake between individual cells; this becomes even greater for the cells (ΔatpB) lacking the β subunit of the membrane ATP synthase. Figure 3 shows the mode fluorescence for the wild type strain and for knockouts of known transporter and certain other genes; the wild type and some of these are labelled in Fig. 3. The dataset (and all the other KOs described herein) is given in full as an Excel sheet in Additional file 1: Table S1. (Note that we have strong suspicions that the supposed tolC knockout was not in fact a knockout of tolC.) The very great breadth and complex shapes of the distributions is considered to be underpinned by the significant number of transporters involved, especially those driven by ATP, and their heterogeneous expression level distributions between individual cells [107].
Fig. 2.
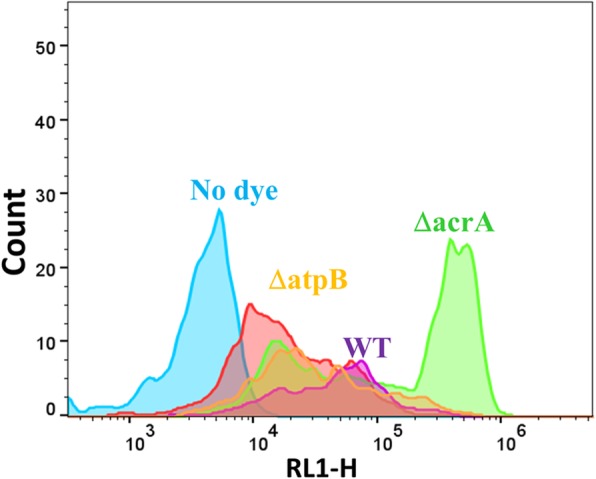
Typical cytograms of the wild-type strain (WT) stained (save for the no-dye control) with diS-C3(5) as described in Methods, along with other knockout strains. Those deleted in atpB or acrA are labelled with the relevant colours. Experiments were performed as described in the Materials and Methods section
Fig. 3.
Variation in the mode uptake of diS-C3(5) into different knockout strains of the Keio collection as judged by flow cytometry. Experiments were performed as described in the legend to Fig. 3. Experiments were performed as described in the Materials and Methods section
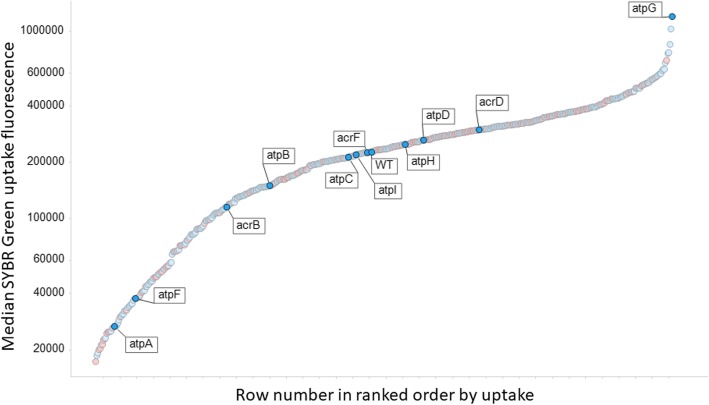
The shape of the curve of Fig. 3 is of interest, with two major break points, one after the lowest 75 and one before the highest 15 in terms of their effects on the steady-state uptake of DiS-C3(5). Several other important features emerge from this plot (Fig. 3). (i) the range of steady-state uptake over the family of single-gene knockouts is very substantial, i.e. some 36-fold between the lowest and highest mode uptake, (ii) this range is somewhat asymmetric, in that the highest values are just two-fold greater than is that of the wild type (although we cannot exclude truncation due to fluorescence quenching [105]), while the lowest value is ~ 18 times lower; (iii) the number of (y-)genes of unknown function that are uptake transporters (or whose removal most decreases the uptake of diS-C3(5)), encoded in red, is far greater than is the number of unknown effluxers (or those whose removal most increases its uptake; the three highest such y-genes are ygfQ, ydcS and ybbW, Table 1), (iv) the five highest values include four known drug effluxers (mdtJ, acrA, mdtI, mdtA), giving confidence in the strategy; (v) 297 knockouts are higher and 244 lower. If a cutoff of 50% above or below the WT is taken (fluorescence levels < 128,000 or > 384,000), we still have 115 knockouts below and 33 knockouts above these thresholds. While we do not know which ones are operating under each condition, this does imply the potential contribution of a very considerable number of transporters to the steady-state uptake of the dye, which is, of course, consistent with the very broad range of uptakes of an individual dye in a given cytogram (Fig. 2); (vi) even if all of the uptake in the most ‘potent’ knockout was through the phospholipid bilayer, and none through ion-coupled transporters, for instance, this uptake (one thirty-sixth of that of the highest strain) would be less than 3% of it, which we regard as negligible. We note that ybbW has been annotated as a putative uptake transporter for allantoin [136] (http://www.tcdb.org), but in this case it must at least be an antiporter, since its knockout causes a large increase in dye uptake. Such findings also illustrate that there remain many mis-annotations or incomplete annotations of the E. coli transporterome.
Table 1.
A selected subset of the most effective knockouts in terms of their ability to affect the uptake of diS-C3(5)
| Gene | Comments | Uniprot ID | Representative reference(s) |
|---|---|---|---|
| atpB | F1-ATPase subunit | P0ABB0 | [122] |
| atpA | F1-ATPase subunit | P0AB98 | [122] |
| ycdG (rutG) | Broad-specificity pyrimidine permease | P75892 | [123–125] |
| rbsB | Ribose ABC transporter periplasmic binding protein | P02925 | [126] |
| ybiO | mechanosensitive ion channel | P75783 | [127] |
| yifK | probable transport protein (possibly amino acid) | P27837 | [128] |
| yliA/gsiA | ATP-driven Glutathione importer | P75796 | [129] |
| ybiR | Inner membrane protein | P75788 | (none) |
| yccS | Inner membrane protein | P75870 | (none) |
| phoR | Phosphate sensor regulon | P08400 | [130] |
| yejA | Putative oligopeptide ABC transporter periplasmic binding protein | P33913 | [131] |
| mdtI | Spermidine export protein | P69210 | [132, 133] |
| mdtJ | Spermidine export protein | P69212 | [133] |
| mdtL | Multidrug resistance protein (e.g. vs chloramphenicol) | P31462 | [133] |
| mdtA | Multidrug resistance protein (e.g. vs novobiocin) | P76397 | [134] |
| acrA | Multidrug efflux pump subunit | P0AE06 | [135] |
| ybbW | Putative allantoin permease | P75712 | [136] |
| ydcS | Part of ABC transporter complex, possibly involved in poly-β-hydroxybutyrate synthesis/ polyamine efflux | P76108 | [137] |
| ygfQ (ghxQ) | Hypoxanthine-guanine permease | Q46817 | [138] |
Those whose knockout increased uptake are given in bold face
A referee has helpfully pointed out that some of the y-genes (as designated in the Keio collection terminology we have used (and retain) throughout), now have more secure functions, and these alternative non-y-gene designations appear in Table 1 and Additional file 1. Other updates, including those emerging after this paper, may be found in Ecocyc.
Although it was not financially reasonable to determine any compensating or pleiotropic changes [139] at the level of the genome-wide transcriptome for these many hundreds of experiments, the simplest explanation for such data is that an increase in uptake following a knockout denotes the removal of an efflux transporter, while a decrease denotes that an influx transporter (or its source of free energy) has been knocked out. Either there are a massive number of pleiotropic effects on a smaller number of transporters or there are least 115 influx and 34 efflux transporters for diS-C3(5) (or both). Similarly, this vast (36-fold) range of uptake levels is not obviously consistent with the fact that uptake might reflect a membrane potential, although the activity of at least acrAB/tolC is in fact considered to be enhanced by an energised membrane. Indeed, the lowering of uptake in the ΔatpA/B knockouts, that would be expected to have a higher membrane potential [140], makes it absolutely clear that the uptake cannot mainly be driven by such a potential, but instead by ATP directly (generated by substrate-level phosphorylation). We note that a tolC knockout did not result in major changes in uptake (although it might be expected to inhibit many RND-type efflux pump activities), but it seems that this strain may have had a mutation elsewhere as tolC was still present as judged by colony PCR and our own attempts to remove it were unsuccessful. Similar suspicion applies to acrB (Fig. 3, which would be anticipated to lie near acrA). But the general principle remains clear: individual gene knockouts lead to a huge range in the steady-state uptake of the dye that cannot be ascribed to changes in a membrane potential with which its distribution might equilibrate.
To confirm these general findings, we took a few of the gene designations whose removal in the Keio collection led to the greatest or lowest uptake, and assessed their behaviours in the corresponding strains in the ASKA collection of overexpressed genes. As anticipated, overexpression of these effluxers lowered the uptake levels to less than that of the wildtype, while overexpression of putative uptake transporters raised the steady-state level of their uptake considerably (Fig. 4), albeit not to the levels of the highest effluxer KOs, implying some spare kinetic capacity in the latter.
Fig. 4.
Effect of overexpression and knockout of a series of genes whose knockout causes major changes (both up and down) in the modal uptake of diSC3(5) relative to that of the wild type. Experiments were performed as described in the Materials and Methods section
To analyse more closely the main conclusion of this section – that the steady-state uptake level of the diS-C3(5) molecule is determined significantly by multiple proteins including transporters – we highlight some of the most significant knockouts in Table 1. A number are unsurprising, which gives confidence in the idea that our basic method is sound. Among the most potent of the non-transporters at inhibiting uptake were knockouts of the genes encoding the two main (α and β) subunits of the membrane ATP synthase, implying strongly that the uptake transporters were powered directly by ATP, rather than (e.g.) by an electron-transport-derived energised membrane (often referred to as a protonmotive force). This is consistent with the relative prevalence of ATP-driven transporters in prokaryotes [35].
The complement of y-genes of unknown or poorly annotated function stands at around 35% of the total, with transporters being over-represented among them [141]. It is of particular interest that so many y-genes are represented among the KOs showing the biggest effects; clearly we have much to learn, as with mammalian cell transporters of pharmaceutical drugs [142, 143], about their ‘natural’ substrates.
It is of course entirely arbitrary to pick the top few only (all are given in the Additional file 1), and we would add that one set of interest is represented by potC and potE (as well as other products of pot genes) that are components of a (cationic) sperm(id) ine import/efflux system [144–147]. Obviously it is entirely reasonable that this might serve, when active, to remove a cation such as diS-C3(5), and the fact that the potent effluxers mdtI and mdtJ are also spermi(di)ne effluxers (Table 1) lends weight to this view. In a similar vein, the KOs of mdtH and mdtK [132] have a significantly reduced uptake; although they were originally tagged as effluxers (in the sense of multidrug transporters), it seems more likely that they are in fact antiporters, normally contributing to the uptake (here) of the cyanine dye.
SYBR Green
SYBR Green is another cationic dye, and increases its fluorescence massively on binding to (especially) double-stranded DNA [10, 148]. It is considered to be ‘membrane-permeable’ (by whatever means) and it is widely used both in environmental and general microbiology (e.g. [10, 148–152]) and in mammalian cell biology (e.g. [153]). SYBR Green also contains a benzothiazole motif (Fig. 1), and using our standard methods of cheminformatics [143, 154, 155], we noted that SYBR Green and DiS-C3(5) have a Tanimoto similarity to each other of 0.731 when encoded using the RDKit (www.rdkit.org) “patterned” fingerprint.
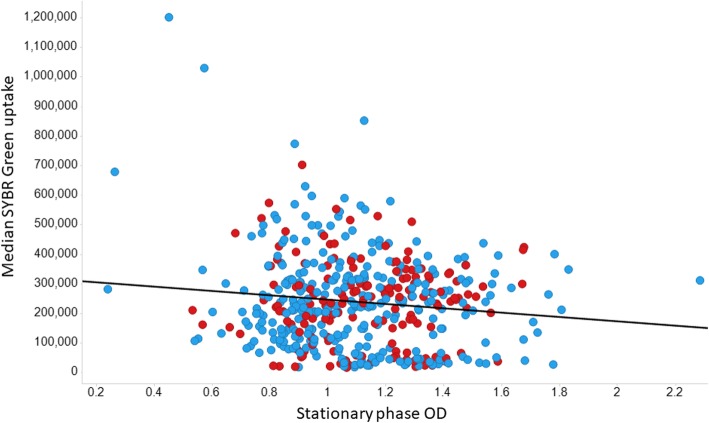
A similar experiment to that performed with diS-C3(5) was performed with SYBR Green. Typical cytograms of SYBR Green uptake are shown in Fig. 5, and the full set of results (slightly fewer KOs than for diSC3(5)) shown in Fig. 6. Again there is a huge range of uptakes (69-fold), the lowest being ~five-fold lower and the largest approximately thirteen-fold greater than that of the wild type. Because of the greater range, and the occasionally bimodal peaks such as that for ΔatpG in Fig. 5, the median fluorescence values are given on a logarithmic scale. All data are again given in Additional file 1: Table S1. Some of this variation can of course reflect differences in the DNA content of the cells, since this is a function of the earlier growth rate [156, 157], though this typically does not exceed about 8 chromosome contents even at the fastest growth rates [121, 158], so variations in DNA content alone could not conceivably explain this range. We therefore also performed growth rate experiments for almost all the strains tested; there was no correlation between the uptake of SYBR green and either the growth rate (Fig. 7, r2 ~ 0.02) or the stationary phase OD (Fig. 8, r2 ~ 0.015), indicating clearly that variations in DNA content were not a significant contributor to these findings in intact cells.
Fig. 5.
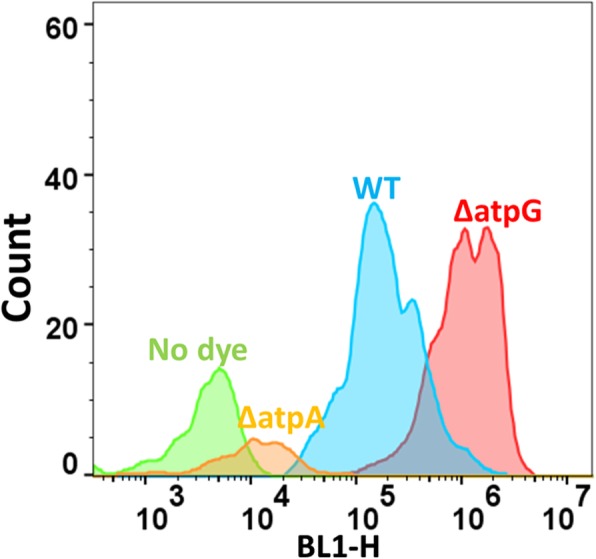
Typical cytograms of the uptake of SYBR Green in the wild-type strain (WT) and some other, knockout strains stained with SYBR Green as described in Methods (save for the no-dye control), along with other knockout strains
Fig. 6.
Median uptake in ranked order of SYBR Green uptake into E. coli single-gne knockout strains, with a small subset of gene names marked. Y-genes are encoded in red. Experiments were performed as described in the Materials and Methods section
Fig. 7.
Lack of relationship between the median extent of uptake of SYBR Green and growth rate. Y-genes are encoded in red. Experiments were performed as described in the Materials and Methods section
Fig. 8.
Lack of relationship between the median extent of uptake of SYBR Green and stationary phase OD. As before, y-genes are encoded in red. Experiments were performed as described in the Materials and Methods section
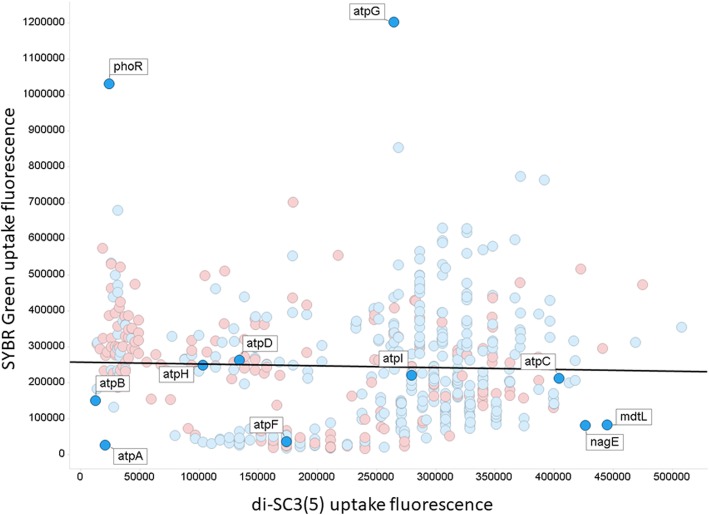
By contrast with the diS-C3(5) data (Fig. 3), there are considerable differences (Fig. 9), in that the phoR knockout now shows the second greatest uptake (rather than the eighth lowest). Again there is a considerable preponderance of y-genes in the part of the figure where their removal lowers uptake relative to the wild type. A subset of genes with the ‘highest’ and ‘lowest’ effects is given in Table 2.
Fig. 9.
Relationship between the uptake of SYBR Green (ordinate) and of diS-C3(5) (abscissa). The line is a line of best fit (r2~0.002). Also shown are some of the knockout labels as discussed in the text. Symbols for y-genes are in red, others in blue. Experiments were performed as described in the Materials and Methods section
Table 2.
A selected subset of the most effective knockouts in terms of their ability to affect the uptake of Sybr Green
| Gene | Comments | Uniprot ID | Representative reference(s) |
|---|---|---|---|
| yiaM | 2,3-diketo-L-gulonate transporter | P37674 | (none) |
| setC | Putative sugar efflux system | B7L779 | [159] |
| tsgA | Uncharacterised transporter | P60778 | (none) |
| yhjV | Uncharacterised transporter | P37660 | [160] |
| yggT | Unknown transporter involved in osmotolerance | P64564 | [161] |
| actP | Cation/acetate symporter | P32705 | [162] |
| yrbG | Possible inorganic cation transporter | P45394 | [163, 164] |
| yidK | Uncharacterised symporter | P31448 | (none) |
| aaeB | p-hydroxybenzoate efflux pump | P46481 | [165–167] |
| atpG | ATP synthase gamma chain | P0ABA6 | (none) |
| phoR | Phosphate sensor regulon | P08400 | [130] |
| cysU (cysT) | Sulphate transport permease | P16701 | (none) |
| mdtD | Putative multidrug resistance transporter | P36554 | [159] |
| dctA | Dicarboxylate transport protein | P0A830 | [168] |
| yehW | Non-osmoprotecting glycine betaine uptake system (presumably antiporter) | P33359 | [169] |
Those whose knockout increased uptake are given in bold face
Despite the fact that both dyes contain a benzothiazole motif, there is absolutely no correlation (r2 ~ 0.002) between the uptake of the two dyes in individual knockouts (Fig. 6). Thus, mdtL and nagE are among the highest for diS-C3(5) but among the lowest for SYBR Green, while the converse is true for phoR, zntA (zinc/lead-transportng ATPase) and yifK (‘probable amino acid transporter protein’). This is despite the presumption that the putative membrane potential with which their uptake might equilibrate is the same in the same strains. Such findings imply that some transporters have a fairly unusual specificity, even if they are labelled (as is mdtL) as multidrug resistance proteins. In the case of SYBR Green, the heterogeneity in uptake can clearly contribute to the heterogeneity in staining observed [10] when cells are not permeabilised.
It was also of interest to assess the effect of chemical inhibitors on the ability of transporter knockouts to take up SYBR Green. In contrast to earlier work [170] where we studied effects on gene transcription, the interest here was in direct, acute effects. Table 3 shows the effects of a few such molecules, that of chlorpromazine, a known inhibitor of acrAB activity [171–173], and clozapine (a second-generation anti-psychotic) [174] being particularly striking. Note (see Methods) that different conditions were used from those of Figs. 2, 3, 4, 5 and 6. The very large increase upon chlorpromazine addition (more than 25-fold) might be taken to imply that it can inhibit multiple effluxers, and suggests that it might be a useful adjunct therapy in cases of antimicrobial resistance.
Table 3.
Effects of various modifiers on steady-state uptake of diSC3(5) and SYBR Green by wild-type E. coli. Experiments were performed as described in Materials and Methods
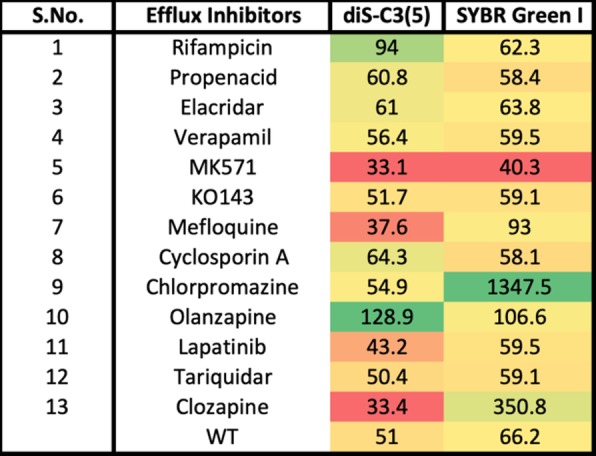
WT means no inhibitors added. Red and green highlight the most significant decreases or increases, respectively
Discussion
There is present lively debate as to whether a majority of xenobiotic uptake through cellular membranes occurs via whatever phospholipid bilayer may be present, or whether Phospholipid Bilayer diffusion Is Negligible (a view referred to as “PBIN”; [27, 175]). In this latter view, it is recognised that potentially a great many transporters can and do interact with a given molecule [27, 175] (see also [29]). This would be unsurprising given that the typical known numbers of binding targets for pharmaceutical drugs is ~six [26, 176]. The present data are entirely consistent with (indeed lend strong support to) this view.
If a closed biological cell or organelle maintains a transmembrane electrical potential difference Δψ relative to that of its adjacent, external phase, it is in principle possible to estimate that potential by allowing a freely membrane-permeable (usually lipophilic) ion to come into a Nernstian equilibrium with it (e.g. [177–186]). For a negative-inside potential, and concentrative uptake by a lipophilic cation such as methyltriphenylphosphonium (TPMP+), Δψ may be related to the ratio of internal ain and external aout activities of the ions by:
| 1 |
where T is the absolute temperature, R the universal gas constant, F Faraday’s constant, and z the charge on the lipophilic cation. RT/F is about 60 mV at room temperature, such that a Δψ of 60 mV equates to a concentration ratio for a monovalent cation of 10:1, a Δψ of 120 mV a concentration ratio of 100, and so on. It is often assumed that the uptake of such lipophilic cationic dyes by bacteria reflects the existence and magnitude of such a transmembrane potential. However, import and export transporters invalidate such estimations of Δψ, their activities respectively causing an over- and under-estimate, and this has been shown clearly to occur for the uptake of dibenzyldimethylammonium in baker’s yeast [70] and of Tl+ in bacteria [187–189]. Confidence in any such estimations of Δψ is bolstered if the equilibrium uptake ratio is independent not only of the concentration but the nature of the lipophilic cations employed, and also of any anions that may be present. In practice, and although such difficulties are commonly ignored, these requirements are rarely if ever met [177, 179, 190, 191].
However, in the present work we have shown clearly that a variety of cations enter cells via a large number of transporters, especially those driven by ATP directly, so whatever their uptake is reflecting it cannot simply be a bulk transmembrane potential. Indeed, the loss of ATP synthesis by electron transport-linked phosphorylation in the ΔatpA and ΔatpB strains might be expected to increase the level of any such membrane potential [140] but instead the extent of uptake is substantially reduced. This is very strong evidence against any such equilibration of uptake with a membrane potential. Similarly, the almost complete lack of correlation between the two cations studies make it inconceivable that the steady-state uptake reflects the value of any membrane potential as it is presumably the same for each strain when the two dyes are compared. In the case of SYBR Green, the data also call into question the use of that dye for estimating quantitatively the amount of DNA in live, non-permeabilised cells, and indeed (using growth rate as a surrogate [156, 192] for DNA content) no such relationship was found.
By contrast, a possible benefit of our findings, for those interested in estimating transporter activities, is that if overexpression of a particular transporter causes most of the uptake (or efflux) flux to occur via it (as in Fig. 4), competition or trans-stimulation assays with a fluorophore provide a powerful and potentially high-throughput [44, 193] method for measuring Quantitative structure-activity relationshipss. The fact that the substrate specificities of individual transporters are typically rather different from each other (Fig. 6) implies that there could indeed be much value in pursuing this more widely.
Conclusions
Despite a widespread assumption that xenobiotics, (including pharmaceutical drugs such as antibiotics) cross biomembranes by diffusion through whatever phospholipid bilayer exists, there does not seem to have been a systematic attempt to assess the contribution of such a flux to cellular uptake. The availability of systematic gene knockout collections such as the Keio collection allows an assessment of every candidate transporter (and other genes) to the uptake of a substance of interest, and fluorescence flow cytometry provides a convenient means of assessing the extent of that uptake in individual cells. We have done this for what seems to be the first time. The very large number of gene knockouts with an extent of uptake of one or more dyes that is reduced substantially (i.e. less than half) relative to that of the wild type, as well as the large ranges between ‘lowest’ and ‘highest’ uptake, even in only single-gene knockouts, really does attest to the fact that phospholipid bilayer transport is indeed negligible. It also casts considerable doubt on the utility of these dyes in vivo in assessing quantitatively any membrane potential that might exist, or the DNA content of the cells.
Materials and methods
Bacterial strains
E. coli (K-12, MG1655) was taken from the laboratory collection of Prof R. Goodacre [194, 195]). The Keio collection of E. coli (K-12, MG1655) was obtained from the SYNBIOChem group (University of Manchester, UK) from which a collection of 530 knockouts (188 y-genes) was selected for the study, mainly those with a transporter protein gene knocked out. The full list is given in Additional file 1. We recognise that one or two transport proteins may have been missed (e.g. nupC, as pointed out by a referee), but the conclusions drawn are not affected by this. A few strains were selected from the ASKA collection of E. coli (BW38029) also taken from SYNBIOChem group. No permissions were required to use these strains.
Culture
E. coli strains were grown from single colonies on agar plates in conical flasks using Lysogeny broth (LB) to an optical density (600 nm) of 1.5–2.0, representing stationary phase in this medium. They were held in stationary phase for 2–4 h before being inoculated at a concentration of 105 cells.mL− 1 into LB.
Keio collection sample preparation
Singer ROTOR HDA (Singer Instruments, UK), a high throughput robotic replicator and colony picker, was used to pick single strains from stock 96-well plates stored at -80 °C and stamped on nutrient agar plates in 96-well format. The agar plates were grown overnight and strains were then transferred in a 96-well plate with 200μL of LB in each well. The resulting set of six 96-well plates was incubated overnight at 37 °C and 200 rpm shaking and used later for analysis.
DiSC3(5) and Sybr Green I uptake measurement
Singer RePads 96 long (Singer Instruments, UK), 96-pin plastic replica-plating pads were used to replicate and culture the new subset of the Keio collection grown overnight. 150μL of LB was added in each well with 3 μM (final concentration) of DiSC3(5) This was chosen because it was an amount that gave a signal roughly an order of magnitude greater than the undyed control in the wild type. (Thermo Fisher Scientific, UK) in six U-bottom 96-well plates. The Singer RePads were used to transfer individual strains from overnight cultured plates to the new plates. The plates were sealed with Breathable Film (Starlab UK, Ltd.) and incubated at 37 °C for 2 min in the dark. The plates were vortexed at 200 rpm for 2 min and screened on a Sartorius Intellicyt iQue Screener Plus™ flow cytometer.
Our variant of this flow cytometer has three fixed excitation lasers (405, 488, 640 nm), forward and side scatter (from the 488 nm excitation) and 13 fluorescence channels based on filters. The channels we report here are mainly BL1 (Ex 488, Em 530 ± 15 nm) and RL1 (Ex 640 nm, Em 675 ± 15 nm). To detect bacteria we gated via forward scatter and side scatter. This instrument does not have user-adjustable photomultiplier tubes so the numbers simply reflect the extent of fluorescence it registers. The following settings [121] were used for the flow cytometry: automatic prime – 60 s (in Qsol buffer); pre-plate shake − 15 s and 900 r.p.m.; sip time − 2 s (actual sample uptake); additional sip time − 0.5 s (the gap between sips); pump speed − 29 r.p.m. (1.5 μl.s− 1 sample uptake); plate model – U- bottom well plate (for 96-well plates); mid-plate clean-up – after every well (4 washes; 0.5 s each in Qsol buffer); inter-well shake − 900 rpm; after 11 wells, 4 s in Qsol buffer; flush and clean – 30 s with Decon and Clean buffers followed by 60 s with de-ionized water. Fluorescence intensity was measured with RL1 channel and the Forecyt™ software supplied with the instrument was used to perform the analysis.
For Sybr Green I (Thermo Fisher Scientific, UK) uptake measurements, the samples were prepared similarly to those for DiSC3(5) except that Sybr Green I was added at 10,000x diluted concentration from the original stock as provided by the supplier (catalogue number S7585). The sample plates were incubated for 15 min and fluorescence was measured using the BL1 channel.
The fluorescence kinetics experiment was performed on a Sony SH800 FACS machine. An overnight-grown culture of E. coli was diluted in LB to a concentration of 105 CFU.mL− 1. diSC3(5) dye was added to 10 mL of the bacterial solution at a final concentration of 3 μM in a 50 mL amber falcon tube (to prevent photobleaching) and incubated in the dark at 37 °C for 2 min. Post-vortexing, the falcon tube was placed in the flow cytometer and the fluorescence intensity of the sample was measured for 1 h continuously. Same was repeated with SYBR Green I.
SYBR Green I uptake measurement on fixed Keio strains
A further subset of 20 strains from the previously analysed Keio collection subset was taken and fixed by injection in 70% ice-cold ethanol. The cells were washed twice by centrifugation in 0.1 M-Tris/HCl buffer, pH 7.4, before being resuspended in PBS at the concentration of 106 CFU.mL− 1. Sybr Green I was added to the sample at 10,000x diluted concentration from the stock and the samples were kept at 37 °C in the dark for 15 min. Samples were added in a U-bottom 96-well plate and fluorescence intensity was measured using the BL1 channel.
Effect of inhibitors on the fluorescence of dye uptake in E. coli
A few efflux inhibitors were tested for their effects on the fluorescence intensity of the bacterial cells. The stock solutions of the inhibitors were made at 1 mM in DMSO and 5uL was added in two 96-well plates in triplicates. A vacuum centrifuge was used to evaporate all the DMSO in the plates. 200μL of overnight-cultured E. coli was added in each well of the plates at 105 CFU.mL− 1 final concentrations. The plates were sealed and kept at 30 °C and 900 rpm shaking for 30 min. 3uM of DiSC3(5) and 10,000x diluted Sybr Green I was then added to the different plates. The plate with DiSC3(5) was incubated for 2 min while one with Sybr Green I was incubated for 15 min at 37 °C before being analysed using the flow cytometer.
Screening ASKA collection strains for dye uptake
Selected strains from the ASKA collection were taken from -80 °C stock and resuscitated in LB with 34 μg.mL− 1 Chloramphenicol. The strains were cultured overnight in M9 media (M9 Minimal Media: 1x M9 Salts, 2 mM MgSO4, 0.1 mM CaCl2, 0.2% glucose, 10 μg.mL− 1 Thiamine HCl and 0.2% Casamino Acids) and 0.5 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added in the morning and grown for another hour. The strains were diluted and their fluorescence upon uptake of DiSC3(5) was measured using the same steps as performed above with the Keio strains.
Growth assessment of Keio collection strains by bulk OD measurement
The Growth Profiler 960 (Enzyscreen, NL) is a commercial instrument (http://www.enzyscreen.com/growth_profiler.htm) that estimates growth rates using camera-based measurements in up to 10 96-well plates. E. coli was taken from an overnight culture and diluted to a concentration of 106 CFU.mL− 1. The Keio collection strains were sub-cultured and samples were prepared using stamping method as described above. The strains were stamped in CR1496c Polystyrene transparent square 96-half-deepwell microtiter plates used with CR1396b Sandwich covers in the Growth Profiler. The samples were prepared in 10 96-well plates and incubated in the instrument at 37 °C with 225 rpm shaking (recommended settings). The pictures of the plates were recorded at 15-min interval. The G values obtained from these pictures were converted to their respective OD600 values using the manufacturer’s software, Eq. 1, and pre-calculated values of the constants (a = 0.0234, b = 1, c = 1.09E-6, d = 3.41, e = 1.56E-13 and f = 6.58).
| 2 |
In some cases the estimates were clearly far too high due to outliers; growth rates were truncated at 3 doubling times.h− 1.
Additional file
Modal and mean uptake data for diS-C3(5) and Sybr green as described in the body of the paper. (XLSX 55 kb)
Acknowledgments
None beyond the funders.
Abbreviations
- DiBAC4(3)
bis-(1,3-dibutylbarbituric acid trimethine oxonol)
- diS-C3(5)
3-Propyl-2-{(1E,3E,5E)-5-(3-propyl-1,3-benzothiazol-2(3H)-ylidene)-1,3-pentadien-1-yl}-1,3-benzothiazol-3-ium iodide
- SYBR Green
N,N-dimethyl-N′-[(4E)-4-[(3-methyl-1,3-benzothiazol-3-ium-2-yl)methylidene]-1-phenyl-2,3-dihydroquinolin-2-yl]-N′-propylpropane-1,3-diamine
Authors’ contributions
SJ performed most of the experiments, including the flow cytometry. LY created some of the knockout and overexpression strains. PD and DBK designed the study and obtained funding for it. All authors contributed to the writing of the manuscript and approved it. All authors read and approved the final manuscript.
Funding
We thank the BBSRC (grants BB/R000093/1 and BB/P009042/1) and the Novo Nordisk Foundation (grant NNF10CC1016517) for financial support. The funding bodies had no role nor involvement in the design of the study and collection, analysis, and interpretation of data nor in writing the manuscript.
Availability of data and materials
The raw flow cytometric datasets generated and/or analysed during the current study are not publicly available due to the fact that they were simply massive, but are available from the corresponding author on reasonable request. Relevant derived data are given in supplementary information as a spreadsheet.
Ethics approval and consent to participate
No ethics approval was required for this work.
Consent for publication
Consent to publish beyond that of the authors was not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Srijan Jindal, Email: srijan.jindal@postgrad.manchester.ac.uk.
Lei Yang, Email: leiya@biosustain.dtu.dk.
Philip J. Day, Email: Philip.j.day@manchester.ac.uk
Douglas B. Kell, Email: dbk@liv.ac.uk, Email: doukel@biosustain.dtu.dk
References
- 1.Guilini C, Baehr C, Schaeffer E, Gizzi P, Rufi F, Haiech J, Weiss E, Bonnet D, Galzi JL. New fluorescein precursors for live Bacteria detection. Anal Chem. 2015;87(17):8858–8866. doi: 10.1021/acs.analchem.5b02100. [DOI] [PubMed] [Google Scholar]
- 2.Müller S, Losche A, Bley T. Staining procedures for flow cytometric monitoring of bacterial populations. Acta Biotechnol. 1993;13(3):289–297. doi: 10.1002/abio.370130311. [DOI] [Google Scholar]
- 3.Wang Y, Hammes F, De Roy K, Verstraete W, Boon N. Past, present and future applications of flow cytometry in aquatic microbiology. Trends Biotechnol. 2010;28(8):416–424. doi: 10.1016/j.tibtech.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Müller S, Nebe-von-Caron G. Functional single-cell analyses: flow cytometry and cell sorting of microbial populations and communities. FEMS Microbiol Rev. 2010;34(4):554–587. doi: 10.1111/j.1574-6976.2010.00214.x. [DOI] [PubMed] [Google Scholar]
- 5.Prest EI, Hammes F, Kotzsch S, van Loosdrecht MCM, Vrouwenvelder JS. Monitoring microbiological changes in drinking water systems using a fast and reproducible flow cytometric method. Water Res. 2013;47(19):7131–7142. doi: 10.1016/j.watres.2013.07.051. [DOI] [PubMed] [Google Scholar]
- 6.Van Nevel S, Koetzsch S, Proctor CR, Besmer MD, Prest EI, Vrouwenvelder JS, Knezev A, Boon N, Hammes F. Flow cytometric bacterial cell counts challenge conventional heterotrophic plate counts for routine microbiological drinking water monitoring. Water Res. 2017;113:191–206. doi: 10.1016/j.watres.2017.01.065. [DOI] [PubMed] [Google Scholar]
- 7.Shapiro HM. Practical flow cytometry, 3rd edition. 3. New York: John Wiley; 1995. [Google Scholar]
- 8.Kepner RL, Pratt JR. Use of fluorochromes for direct enumeration of total bacteria in environmental samples - past and present. Microbiol Rev. 1994;58(4):603–615. doi: 10.1128/mr.58.4.603-615.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davey HM, Kell DB. Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analysis. Microbiol Rev. 1996;60:641–696. doi: 10.1128/mr.60.4.641-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nescerecka A, Hammes F, Juhna T. A pipeline for developing and testing staining protocols for flow cytometry, demonstrated with SYBR Green I and propidium iodide viability staining. J Microbiol Methods. 2016;131:172–180. doi: 10.1016/j.mimet.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 11.Hawe A, Sutter M, Jiskoot W. Extrinsic fluorescent dyes as tools for protein characterization. Pharm Res. 2008;25(7):1487–1499. doi: 10.1007/s11095-007-9516-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez V, Henary M. Nile red and Nile blue: applications and syntheses of structural analogues. Chemistry. 2016;22(39):13764–13782. doi: 10.1002/chem.201601570. [DOI] [PubMed] [Google Scholar]
- 13.Mason D, Gant VA. The application of flow cytometry to the estimation of bacterial antibiotic susceptibility. J Antibiotic Chemother. 1995;36:441–443. doi: 10.1093/jac/36.2.441. [DOI] [PubMed] [Google Scholar]
- 14.Novo DJ, Perlmutter NG, Hunt RH, Shapiro HM. Multiparameter flow cytometric analysis of antibiotic effects on membrane potential, membrane permeability, and bacterial counts of Staphylococcus aureus and Micrococcus luteus. Antimicrob Agents Chemother. 2000;44(4):827–834. doi: 10.1128/AAC.44.4.827-834.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nebe-von-Caron G, Stephens PJ, Hewitt CJ, Powell JR, Badley RA. Analysis of bacterial function by multi-colour fluorescence flow cytometry and single cell sorting. J Microbiol Methods. 2000;42(1):97–114. doi: 10.1016/S0167-7012(00)00181-0. [DOI] [PubMed] [Google Scholar]
- 16.Lewis G, Taylor IW, Nienow AW, Hewitt CJ. The application of multi-parameter flow cytometry to the study of recombinant Escherichia coli batch fermentation processes. J Ind Microbiol Biotechnol. 2004;31(7):311–322. doi: 10.1007/s10295-004-0151-8. [DOI] [PubMed] [Google Scholar]
- 17.Westfall DA, Krishnamoorthy G, Wolloscheck D, Sarkar R, Zgurskaya HI, Rybenkov VV. Bifurcation kinetics of drug uptake by gram-negative bacteria. PLoS One. 2017;12(9):e0184671. doi: 10.1371/journal.pone.0184671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirota N, Matsuura S, Mochizuki N, Mutoh N, Imae Y. Use of lipophilic cation-permeable mutants for measurement of transmembrane electrical potential in metabolizing cells of Escherichia coli. J Bacteriol. 1981;148(2):399–405. doi: 10.1128/jb.148.2.399-405.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuyama T. Staining of living bacteria with rhodamine 123. FEMS Microbiol Lett. 1984;21:153–157. doi: 10.1111/j.1574-6968.1984.tb00202.x. [DOI] [Google Scholar]
- 20.Kaprelyants AS, Kell DB. Rapid assessment of bacterial viability and vitality using rhodamine 123 and flow cytometry. J Appl Bacteriol. 1992;72:410–422. doi: 10.1111/j.1365-2672.1992.tb01854.x. [DOI] [Google Scholar]
- 21.Pliška V, Testa B, van de Waterbeemd H. Lipophilicity in drug action and toxicology. Weinheim: VCH; 1996. [Google Scholar]
- 22.Edwards MP, Price DA. Role of physicochemical properties and ligand lipophilicity efficiency in addressing drug safety risks. Ann Rep Med Chem. 2010;45:381–391. [Google Scholar]
- 23.Waring MJ. Lipophilicity in drug discovery. Expert Opin Drug Discovery. 2010;5(3):235–248. doi: 10.1517/17460441003605098. [DOI] [PubMed] [Google Scholar]
- 24.Dobson PD, Kell DB. Carrier-mediated cellular uptake of pharmaceutical drugs: an exception or the rule? Nat Rev Drug Disc. 2008;7:205–220. doi: 10.1038/nrd2438. [DOI] [PubMed] [Google Scholar]
- 25.Kell DB, Dobson PD, Oliver SG. Pharmaceutical drug transport: the issues and the implications that it is essentially carrier-mediated only. Drug Disc Today. 2011;16(15/16):704–714. doi: 10.1016/j.drudis.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 26.Kell DB, Dobson PD, Bilsland E, Oliver SG. The promiscuous binding of pharmaceutical drugs and their transporter-mediated uptake into cells: what we (need to) know and how we can do so. Drug Disc Today. 2013;18(5/6):218–239. doi: 10.1016/j.drudis.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Kell DB, Oliver SG. How drugs get into cells: tested and testable predictions to help discriminate between transporter-mediated uptake and lipoidal bilayer diffusion. Front Pharmacol. 2014;5:231. doi: 10.3389/fphar.2014.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winter GE, Radic B, Mayor-Ruiz C, Blomen VA, Trefzer C, Kandasamy RK, Huber KVM, Gridling M, Chen D, Klampfl T, et al. The solute carrier SLC35F2 enables YM155-mediated DNA damage toxicity. Nat Chem Biol. 2014;10:768–773. doi: 10.1038/nchembio.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanthaler K, Bilsland E, Dobson P, Moss HJ, Pir P, Kell DB, Oliver SG. Genome-wide assessment of the carriers involved in the cellular uptake of drugs: a model system in yeast. BMC Biol. 2011;9:70. doi: 10.1186/1741-7007-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuwayama K, Miyauchi S, Tateoka R, Abe H, Kamo N. Fluorescein uptake by a monocarboxylic acid transporter in human intestinal Caco-2 cells. Biochem Pharmacol. 2002;63(1):81–88. doi: 10.1016/S0006-2952(01)00853-X. [DOI] [PubMed] [Google Scholar]
- 31.Jernaes MW, Steen HB. Staining of Escherichia coli for flow cytometry: influx and efflux of ethidium bromide. Cytometry. 1994;17:302–309. doi: 10.1002/cyto.990170405. [DOI] [PubMed] [Google Scholar]
- 32.Paixão L, Rodrigues L, Couto I, Martins M, Fernandes P, de Carvalho CCCR, Monteiro GA, Sansonetty F, Amaral L, Viveiros M. Fluorometric determination of ethidium bromide efflux kinetics in Escherichia coli. J Biol Eng. 2009;3:18. doi: 10.1186/1754-1611-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tal N, Schuldiner S. A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc Natl Acad Sci U S A. 2009;106(22):9051–9056. doi: 10.1073/pnas.0902400106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholls DG, Ferguson SJ. Bioenergetics 2. London: Academic Press; 1992. [Google Scholar]
- 35.Darbani B, Kell DB. Borodina I: energetic evolution of cellular transportomes. BMC Genomics. 2018;19:418. doi: 10.1186/s12864-018-4816-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishino K, Yamaguchi A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J Bacteriol. 2001;183(20):5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sulavik MC, Houseweart C, Cramer C, Jiwani N, Murgolo N, Greene J, DiDomenico B, Shaw KJ, Miller GH, Hare R, et al. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob Agents Chemother. 2001;45(4):1126–1136. doi: 10.1128/AAC.45.4.1126-1136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weston N, Sharma P, Ricci V, Piddock LJV. Regulation of the AcrAB-TolC efflux pump in Enterobacteriaceae. Res Microbiol. 2017. [DOI] [PubMed]
- 39.Nikaido H. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin Infect Dis. 1998;27(Suppl 1):S32–S41. doi: 10.1086/514920. [DOI] [PubMed] [Google Scholar]
- 40.Zgurskaya HI, Nikaido H. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc Natl Acad Sci U S A. 1999;96(13):7190–7195. doi: 10.1073/pnas.96.13.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (mar) mutants. J Bacteriol. 1996;178(1):306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto K, Tamai R, Yamazaki M, Inaba T, Sowa Y, Kawagishi I. Substrate-dependent dynamics of the multidrug efflux transporter AcrB of Escherichia coli. Sci Rep. 2016;6:21909. doi: 10.1038/srep21909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Du D, Wang-Kan X, Neuberger A, van Veen HW, Pos KM, Piddock LJV, Luisi BF. Multidrug efflux pumps: structure, function and regulation. Nat Rev Microbiol. 2018;16:523–539. doi: 10.1038/s41579-018-0048-6. [DOI] [PubMed] [Google Scholar]
- 44.Haynes MK, Garcia M, Peters R, Waller A, Tedesco P, Ursu O, Bologa CG, Santos RG, Pinilla C, Wu TH, et al. High-throughput flow cytometry screening of multidrug efflux systems. Methods Mol Biol. 1700;2018:293–318. doi: 10.1007/978-1-4939-7454-2_16. [DOI] [PubMed] [Google Scholar]
- 45.Piddock LJ. Multidrug-resistance efflux pumps - not just for resistance. Nat Rev Microbiol. 2006;4(8):629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 46.Piddock LJV. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev. 2006;19(2):382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piddock L, Garneau-Tsodikova S, Garner C. Ask the experts: how to curb antibiotic resistance and plug the antibiotics gap? Future Med Chem. 2016;8(10):1027–1032. doi: 10.4155/fmc-2014-0032. [DOI] [PubMed] [Google Scholar]
- 48.Holmes AR, Cardno TS, Strouse JJ, Ivnitski-Steele I, Keniya MV, Lackovic K, Monk BC, Sklar LA, Cannon RD. Targeting efflux pumps to overcome antifungal drug resistance. Future Med Chem. 2016;8(12):1485–1501. doi: 10.4155/fmc-2016-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cannon RD, Lamping E, Holmes AR, Niimi K, Baret PV, Keniya MV, Tanabe K, Niimi M, Goffeau A, Monk BC. Efflux-mediated antifungal drug resistance. Clin Microbiol Rev. 2009;22(2):291–321. doi: 10.1128/CMR.00051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swick MC, Morgan-Linnell SK, Carlson KM, Zechiedrich L. Expression of multidrug efflux pump genes acrAB-tolC, mdfA, and norE in Escherichia coli clinical isolates as a function of fluoroquinolone and multidrug resistance. Antimicrob Agents Chemother. 2011;55(2):921–924. doi: 10.1128/AAC.00996-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willers C, Wentzel JF, du Plessis LH, Gouws C, Hamman JH. Efflux as a mechanism of antimicrobial drug resistance in clinical relevant microorganisms: the role of efflux inhibitors. Expert Opin Ther Targets. 2017;21(1):23–36. doi: 10.1080/14728222.2017.1265105. [DOI] [PubMed] [Google Scholar]
- 52.Kourtesi C, Ball AR, Huang YY, Jachak SM, Vera DM, Khondkar P, Gibbons S, Hamblin MR, Tegos GP. Microbial efflux systems and inhibitors: approaches to drug discovery and the challenge of clinical implementation. Open Microbiol J. 2013;7:34–52. doi: 10.2174/1874285801307010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tegos GP, Haynes M, Strouse JJ, Khan MM, Bologa CG, Oprea TI, Sklar LA. Microbial efflux pump inhibition: tactics and strategies. Curr Pharm Des. 2011;17(13):1291–1302. doi: 10.2174/138161211795703726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar S, Mukherjee MM, Varela MF. Modulation of bacterial multidrug resistance efflux pumps of the major fFacilitator sSuperfamily. Int J Bacteriol. 2013;2013:204141. doi: 10.1155/2013/204141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li XZ, Plésiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in gram-negative bacteria. Clin Microbiol Rev. 2015;28(2):337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakashima R, Sakurai K, Yamasaki S, Hayashi K, Nagata C, Hoshino K, Onodera Y, Nishino K, Yamaguchi A. Structural basis for the inhibition of bacterial multidrug exporters. Nature. 2013;500(7460):102–106. doi: 10.1038/nature12300. [DOI] [PubMed] [Google Scholar]
- 57.Sjuts H, Vargiu AV, Kwasny SM, Nguyen ST, Kim HS, Ding X, Ornik AR, Ruggerone P, Bowlin TL, Nikaido H, et al. Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc Natl Acad Sci U S A. 2016;113(13):3509–3514. doi: 10.1073/pnas.1602472113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Venter H, Mowla R, Ohene-Agyei T, Ma ST. RND-type drug efflux pumps from gram-negative bacteria: molecular mechanism and inhibition. Front Microbiol. 2015;6:377. doi: 10.3389/fmicb.2015.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abdali N, Parks JM, Haynes KM, Chaney JL, Green AT, Wolloscheck D, Walker JK, Rybenkov VV, Baudry J, Smith JC, et al. Reviving antibiotics: efflux pump inhibitors that interact with AcrA, a membrane fusion protein of the AcrAB-TolC multidrug efflux pump. ACS Infect Dis. 2017;3(1):89–98. doi: 10.1021/acsinfecdis.6b00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alibert S, N'gompaza Diarra J, Hernandez J, Stutzmann A, Fouad M, Boyer G, Pagès JM. Multidrug efflux pumps and their role in antibiotic and antiseptic resistance: a pharmacodynamic perspective. Expert Opin Drug Met Toxicol. 2017;13(3):301–309. doi: 10.1080/17425255.2017.1251581. [DOI] [PubMed] [Google Scholar]
- 61.Amaral L, Martins A, Spengler G, Molnar J. Efflux pumps of gram-negative bacteria: what they do, how they do it, with what and how to deal with them. Front Pharmacol. 2014;4:168. doi: 10.3389/fphar.2013.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun J, Deng Z, Yan A. Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochem Biophys Res Commun. 2014;453(2):254–267. doi: 10.1016/j.bbrc.2014.05.090. [DOI] [PubMed] [Google Scholar]
- 63.Spengler Gabriella, Kincses Annamária, Gajdács Márió, Amaral Leonard. New Roads Leading to Old Destinations: Efflux Pumps as Targets to Reverse Multidrug Resistance in Bacteria. Molecules. 2017;22(3):468. doi: 10.3390/molecules22030468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schuster S, Vavra M, Schweigger TM, Rossen JWA, Matsumura Y, Kern WV. Contribution of AcrAB-TolC to multidrug resistance in an Escherichia coli sequence type 131 isolate. Int J Antimicrob Agents. 2017;50(3):477–481. doi: 10.1016/j.ijantimicag.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 65.Li X-Z, Elkins CA. Zgurskaya HI (eds.): efflux-mediated antimicrobial resistance in bacteria: mechanisms, regulation and clinical implications. Berlin: Springer; 2016. [Google Scholar]
- 66.Blair JM, Richmond GE, Piddock LJV. Multidrug efflux pumps in gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. 2014;9(10):1165–1177. doi: 10.2217/fmb.14.66. [DOI] [PubMed] [Google Scholar]
- 67.Poole K. Efflux pumps as antimicrobial resistance mechanisms. Ann Med. 2007;39(3):162–176. doi: 10.1080/07853890701195262. [DOI] [PubMed] [Google Scholar]
- 68.Rahman T, Yarnall B, Doyle DA. Efflux drug transporters at the forefront of antimicrobial resistance. Eur Biophys J. 2017;46(7):647–653. doi: 10.1007/s00249-017-1238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Phillips-Jones MK, Harding SE. Antimicrobial resistance (AMR) nanomachines-mechanisms for fluoroquinolone and glycopeptide recognition, efflux and/or deactivation. Biophys Rev. 2018;10(2):347–362. doi: 10.1007/s12551-018-0404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barts PWJA, Hoeberichts JA, Klaassen A, Borst-Pauwels GWFH. Uptake of the lipophilic cation dibenzyldimethylammonium into Saccharomyces cerevisiae. Interaction with the thiamine transport system. Biochim Biophys Acta. 1980;597(1):125–136. doi: 10.1016/0005-2736(80)90156-X. [DOI] [PubMed] [Google Scholar]
- 71.Brown DG, May-Dracka TL, Gagnon MM, Tommasi R. Trends and exceptions of physical properties on antibacterial activity for gram-positive and gram-negative pathogens. J Med Chem. 2014;57(23):10144–10161. doi: 10.1021/jm501552x. [DOI] [PubMed] [Google Scholar]
- 72.Prabhala BK, Aduri NG, Sharma N, Shaheen A, Sharma A, Iqbal M, Hansen PR, Brasen C, Gajhede M, Rahman M, et al. The prototypical proton-coupled oligopeptide transporter YdgR from Escherichia coli facilitates chloramphenicol uptake into bacterial cells. J Biol Chem. 2018;293(3):1007–1017. doi: 10.1074/jbc.M117.805960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bugde P, Biswas R, Merien F, Lu J, Liu DX, Chen M, Zhou S, Li Y. The therapeutic potential of targeting ABC transporters to combat multi-drug resistance. Expert Opin Ther Targets. 2017;21(5):511–530. doi: 10.1080/14728222.2017.1310841. [DOI] [PubMed] [Google Scholar]
- 74.Gameiro M, Silva R, Rocha-Pereira C, Carmo H, Carvalho F, de Lourdes BM, Remião F. Cellular models and in vitro assays for the screening of modulators of P-gp, MRP1 and BCRP. Molecules. 2017;22:600. doi: 10.3390/molecules22040600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen Z, Shi T, Zhang L, Zhu P, Deng M, Huang C, Hu T, Jiang L, Li J. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family in multidrug resistance: a review of the past decade. Cancer Lett. 2016;370(1):153–164. doi: 10.1016/j.canlet.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 76.Szabó E, Türk D, Telbisz Á, Kucsma N, Horváth T, Szakács G, Homolya L, Sarkadi B, Várady G. A new fluorescent dye accumulation assay for parallel measurements of the ABCG2, ABCB1 and ABCC1 multidrug transporter functions. PLoS One. 2018;13(1):e0190629. doi: 10.1371/journal.pone.0190629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kell DB, Swainston N, Pir P, Oliver SG. Membrane transporter engineering in industrial biotechnology and whole-cell biocatalysis. Trends Biotechnol. 2015;33:237–246. doi: 10.1016/j.tibtech.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 78.Kell DB: Control of metabolite efflux in microbial cell factories: current advances and future prospects. OSF preprints 2018:xg9jh.
- 79.Yamaguchi A, Nakashima R, Sakurai K. Structural basis of RND-type multidrug exporters. Front Microbiol. 2015;6:327. doi: 10.3389/fmicb.2015.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nikaido H. Structure and mechanism of RND-type multidrug efflux pumps. Adv Enzymol Relat Areas Mol Biol. 2011;77:1–60. doi: 10.1002/9780470920541.ch1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nikaido H, Pagès JM. Broad-specificity efflux pumps and their role in multidrug resistance of gram-negative bacteria. FEMS Microbiol Rev. 2012;36(2):340–363. doi: 10.1111/j.1574-6976.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brown AR, Ettefagh KA, Todd D, Cole PS, Egan JM, Foil DH, Graf TN, Schindler BD, Kaatz GW, Cech NB. A mass spectrometry-based assay for improved quantitative measurements of efflux pump inhibition. PLoS One. 2015;10(5):e0124814. doi: 10.1371/journal.pone.0124814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tegos GP, Evangelisti AM, Strouse JJ, Ursu O, Bologa C, Sklar LA. A high throughput flow cytometric assay platform targeting transporter inhibition. Drug Disc Today Technol. 2014;12:e95–e103. doi: 10.1016/j.ddtec.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Strouse JJ, Ivnitski-Steele I, Waller A, Young SM, Perez D, Evangelisti AM, Ursu O, Bologa CG, Carter MB, Salas VM, et al. Fluorescent substrates for flow cytometric evaluation of efflux inhibition in ABCB1, ABCC1, and ABCG2 transporters. Anal Biochem. 2013;437(1):77–87. doi: 10.1016/j.ab.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fardel O, Le Vee M, Jouan E, Denizot C, Parmentier Y. Nature and uses of fluorescent dyes for drug transporter studies. Expert Opin Drug Metab Toxicol. 2015;11(8):1233–1251. doi: 10.1517/17425255.2015.1053462. [DOI] [PubMed] [Google Scholar]
- 86.Nerada Z, Hegyi Z, Szepesi A, Tóth S, Hegedüs C, Várady G, Matula Z, Homolya L, Sarkadi B, Telbisz Á. Application of fluorescent dye substrates for functional characterization of ABC multidrug transporters at a single cell level. Cytometry A. 2016;89(9):826–834. doi: 10.1002/cyto.a.22931. [DOI] [PubMed] [Google Scholar]
- 87.Blair JMA, Piddock LJV: How to measure export via bacterial multidrug resistance efflux pumps. MBio 2016, 7(4). [DOI] [PMC free article] [PubMed]
- 88.Bohnert JA, Schuster S, Kern WV, Karcz T, Olejarz A, Kaczor A, Handzlik J, Kieć-Kononowicz K. Novel Piperazine Arylideneimidazolones inhibit the AcrAB-TolC pump in Escherichia coli and simultaneously act as fluorescent membrane probes in a combined real-time influx and efflux assay. Antimicrob Agents Chemother. 2016;60(4):1974–1983. doi: 10.1128/AAC.01995-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Greenspan P, Fowler SD. Spectrofluorometric studies of the lipid probe, nile red. J Lipid Res. 1985;26(7):781–789. [PubMed] [Google Scholar]
- 90.Bohnert JA, Schuster S, Szymaniak-Vits M, Kern WV. Determination of real-time efflux phenotypes in Escherichia coli AcrB binding pocket phenylalanine mutants using a 1,2′-dinaphthylamine efflux assay. PLoS One. 2011;6(6):e21196. doi: 10.1371/journal.pone.0021196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bohnert JA, Karamian B, Nikaido H. Optimized Nile red efflux assay of AcrAB-TolC multidrug efflux system shows competition between substrates. Antimicrob Agents Chemother. 2010;54(9):3770–3775. doi: 10.1128/AAC.00620-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Iyer R, Ferrari A, Rijnbrand R, Erwin AL. A fluorescent microplate assay quantifies bacterial efflux and demonstrates two distinct compound binding sites in AcrB. Antimicrob Agents Chemother. 2015;59(4):2388–2397. doi: 10.1128/AAC.05112-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Coldham NG, Webber M, Woodward MJ, Piddock LJV. A 96-well plate fluorescence assay for assessment of cellular permeability and active efflux in Salmonella enterica serovar Typhimurium and Escherichia coli. J Antimicrob Chemother. 2010;65(8):1655–1663. doi: 10.1093/jac/dkq169. [DOI] [PubMed] [Google Scholar]
- 94.Zhou XY, Ye XG, He LT, Zhang SR, Wang RL, Zhou J, He ZS. In vitro characterization and inhibition of the interaction between ciprofloxacin and berberine against multidrug-resistant Klebsiella pneumoniae. J Antibiot (Tokyo) 2016;69(10):741–746. doi: 10.1038/ja.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vidal-Aroca F, Meng A, Minz T, Page MGP, Dreier J. Use of resazurin to detect mefloquine as an efflux-pump inhibitor in Pseudomonas aeruginosa and Escherichia coli. J Microbiol Methods. 2009;79(2):232–237. doi: 10.1016/j.mimet.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 96.Horiyama T, Yamaguchi A, Nishino K. TolC dependency of multidrug efflux systems in Salmonella enterica serovar Typhimurium. J Antimicrob Chemother. 2010;65(7):1372–1376. doi: 10.1093/jac/dkq160. [DOI] [PubMed] [Google Scholar]
- 97.Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, ColladoVides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277(5331):1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 98.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. Life with 6000 genes. Science. 1996;274(5287):546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 99.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418(6896):387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 100.Long CP, Antoniewicz MR. Metabolic flux analysis of Escherichia coli knockouts: lessons from the Keio collection and future outlook. Curr Opin Biotechnol. 2014;28:127–133. doi: 10.1016/j.copbio.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yamamoto N, Nakahigashi K, Nakamichi T, Yoshino M, Takai Y, Touda Y, Furubayashi A, Kinjyo S, Dose H, Hasegawa M, et al. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol Syst Biol. 2009;5:335. doi: 10.1038/msb.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006–0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Elbourne LDH, Tetu SG, Hassan KA, Paulsen IT. TransportDB 2.0: a database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic Acids Res. 2017;45(D1):D320–D324. doi: 10.1093/nar/gkw1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.O'Hagan S, Kell DB. The apparent permeabilities of Caco-2 cells to marketed drugs: magnitude, and independence from both biophysical properties and endogenite similarities. PeerJ. 2015;3:e1405. doi: 10.7717/peerj.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sims PJ, Waggoner AS, Wang CH, Hoffman JF. Studies on the mechanism by which cyanine dyes measure membrane potential in red blood cells and phosphatidylcholine vesicles. Biochemistry. 1974;13(16):3315–3330. doi: 10.1021/bi00713a022. [DOI] [PubMed] [Google Scholar]
- 106.Waggoner AS. Dye indicators of membrane potential. Annu Rev Biophys Bioeng. 1979;8:47–68. doi: 10.1146/annurev.bb.08.060179.000403. [DOI] [PubMed] [Google Scholar]
- 107.Kell DB, Potgieter M, Pretorius E. Individuality, phenotypic differentiation, dormancy and ‘persistence’ in culturable bacterial systems: commonalities shared by environmental, laboratory, and clinical microbiology. F1000Research. 2015;4:179. doi: 10.12688/f1000research.6709.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kell DB, Kaprelyants AS, Weichart DH, Harwood CL, Barer MR. Viability and activity in readily culturable bacteria: a review and discussion of the practical issues. Antonie Van Leeuwenhoek. 1998;73:169–187. doi: 10.1023/A:1000664013047. [DOI] [PubMed] [Google Scholar]
- 109.Sträuber H, Müller S. Viability states of bacteria: specific mechanisms of selected probes. Cytometry A. 2010;77(7):623–634. doi: 10.1002/cyto.a.20920. [DOI] [PubMed] [Google Scholar]
- 110.Mason DJ, López-Amorós R, Allman R, Stark JM, Lloyd D. The ability of membrane potential dyes and calcofluor white to distinguish between viable and non-viable bacteria. J Appl Bacteriol. 1995;78(3):309–315. doi: 10.1111/j.1365-2672.1995.tb05031.x. [DOI] [PubMed] [Google Scholar]
- 111.Boi P, Manti A, Pianetti A, Sabatini L, Sisti D, Rocchi MB, Bruscolini F, Galluzzi L, Papa S. Evaluation of Escherichia coli viability by flow cytometry: a method for determining bacterial responses to antibiotic exposure. Cytometry B Clin Cytom. 2015;88(3):149–153. doi: 10.1002/cytob.21214. [DOI] [PubMed] [Google Scholar]
- 112.Shapiro HM. Multiparameter flow cytometry of bacteria: implications for diagnostics and therapeutics. Cytometry. 2001;43(3):223–226. doi: 10.1002/1097-0320(20010301)43:3<223::AID-CYTO1054>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 113.Shapiro HM. Flow cytometry of bacterial membrane potential and permeability. Methods Mol Med. 2008;142:175–186. doi: 10.1007/978-1-59745-246-5_14. [DOI] [PubMed] [Google Scholar]
- 114.Gášková D, Čadek R, Chaloupka R, Vacata V, Gebel J, Sigler K. Monitoring the kinetics and performance of yeast membrane ABC transporters by diS-C3(3) fluorescence. Int J Biochem Cell Biol. 2002;34(8):931–937. doi: 10.1016/S1357-2725(02)00013-4. [DOI] [PubMed] [Google Scholar]
- 115.Szczepaniak J, Łukaszewicz M, Krasowska A. Estimation of Candida albicans ABC transporter behavior in real-time via fluorescence. Front Microbiol. 2015;6:1382. doi: 10.3389/fmicb.2015.01382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bartosiewicz D, Krasowska A. Inhibitors of ABC transporters and biophysical methods to study their activity. Z Naturforsch C. 2009;64(5–6):454–458. doi: 10.1515/znc-2009-5-625. [DOI] [PubMed] [Google Scholar]
- 117.Peña A, Sánchez NS, Calahorra M. Estimation of the electric plasma membrane potential difference in yeast with fluorescent dyes: comparative study of methods. J Bioenerg Biomembr. 2010;42(5):419–432. doi: 10.1007/s10863-010-9311-x. [DOI] [PubMed] [Google Scholar]
- 118.Čadek R, Chládková K, Sigler K, Gášková D. Impact of the growth phase on the activity of multidrug resistance pumps and membrane potential of S. cerevisiae: effect of pump overproduction and carbon source. Biochim Biophys Acta. 2004;1665(1–2):111–117. doi: 10.1016/j.bbamem.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 119.Smith JC. Potential-sensitive molecular probes in membranes of bioenergetic relevance. Biochim Biophys Acta. 1990;1016(1):1–28. doi: 10.1016/0005-2728(90)90002-L. [DOI] [PubMed] [Google Scholar]
- 120.Smiley ST, Reers M, Mottolo-Hartshorn C, Lin M, Chen A, Smith TW, Steele GD, Chen LB. Intracellular heterogeneity in mitochondrial membrane potentials revealed by a J-aggregate-forming lipophilic cation JC-1. Proc Natl Acad Sci. 1991;88:3671–3675. doi: 10.1073/pnas.88.9.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jindal S, Thampy H, J DP, Kell DB. Very rapid flow cytometric assessment of antimicrobial susceptibility during the apparent lag phase of bacterial (re)growth. Microbiology. 2019;165:439–454. doi: 10.1099/mic.0.000777. [DOI] [PubMed] [Google Scholar]
- 122.Nakanishi-Matsui M, Sekiya M, Futai M. ATP synthase from Escherichia coli: mechanism of rotational catalysis, and inhibition with the epsilon subunit and phytopolyphenols. Biochim Biophys Acta. 2016;1857(2):129–140. doi: 10.1016/j.bbabio.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 123.Loh KD, Gyaneshwar P, Markenscoff Papadimitriou E, Fong R, Kim KS, Parales R, Zhou Z, Inwood W, Kustu S. A previously undescribed pathway for pyrimidine catabolism. Proc Natl Acad Sci U S A. 2006;103(13):5114–5119. doi: 10.1073/pnas.0600521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen CS, Sullivan S, Anderson T, Tan AC, Alex PJ, Brant SR, Cuffari C, Bayless TM, Talor MV, Burek CL, et al. Identification of novel serological biomarkers for inflammatory bowel disease using Escherichia coli proteome chip. Mol Cell Proteomics. 2009;8(8):1765–1776. doi: 10.1074/mcp.M800593-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Botou M, Lazou P, Papakostas K, Lambrinidis G, Evangelidis T, Mikros E, Frillingos S. Insight on specificity of uracil permeases of the NAT/NCS2 family from analysis of the transporter encoded in the pyrimidine utilization operon of Escherichia coli. Mol Microbiol. 2018;108(2):204–219. doi: 10.1111/mmi.13931. [DOI] [PubMed] [Google Scholar]
- 126.Clifton LA, Holt SA, Hughes AV, Daulton EL, Arunmanee W, Heinrich F, Khalid S, Jefferies D, Charlton TR, Webster JRP, et al. An accurate in vitro model of the E. coli envelope. Angew Chem Int Ed Engl. 2015;54:1–5. doi: 10.1002/anie.201504287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Edwards MD, Black S, Rasmussen T, Rasmussen A, Stokes NR, Stephen TL, Miller S, Booth IR. Characterization of three novel mechanosensitive channel activities in Escherichia coli. Channels (Austin) 2012;6(4):272–281. doi: 10.4161/chan.20998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yang Q, Figueroa-Bossi N, Bossi L. Translation enhancing ACA motifs and their silencing by a bacterial small regulatory RNA. PLoS Genet. 2014;10(1):e1004026. doi: 10.1371/journal.pgen.1004026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Suzuki H, Koyanagi T, Izuka S, Onishi A, Kumagai H. The yliA, −B, −C, and -D genes of Escherichia coli K-12 encode a novel glutathione importer with an ATP-binding cassette. J Bacteriol. 2005;187(17):5861–5867. doi: 10.1128/JB.187.17.5861-5867.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Marzan LW, Shimizu K. Metabolic regulation of Escherichia coli and its phoB and phoR genes knockout mutants under phosphate and nitrogen limitations as well as at acidic condition. Microb Cell Factories. 2011;10:39. doi: 10.1186/1475-2859-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Novikova M, Metlitskaya A, Datsenko K, Kazakov T, Kazakov A, Wanner B, Severinov K. The Escherichia coli Yej transporter is required for the uptake of translation inhibitor microcin C. J Bacteriol. 2007;189(22):8361–8365. doi: 10.1128/JB.01028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bay DC, Stremick CA, Slipski CJ, Turner RJ. Secondary multidrug efflux pump mutants alter Escherichia coli biofilm growth in the presence of cationic antimicrobial compounds. Res Microbiol. 2017;168(3):208–221. doi: 10.1016/j.resmic.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 133.Higashi K, Ishigure H, Demizu R, Uemura T, Nishino K, Yamaguchi A, Kashiwagi K, Igarashi K. Identification of a spermidine excretion protein complex (MdtJI) in Escherichia coli. J Bacteriol. 2008;190(3):872–878. doi: 10.1128/JB.01505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Baranova N, Nikaido H. The baeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J Bacteriol. 2002;184(15):4168–4176. doi: 10.1128/JB.184.15.4168-4176.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Aono R. Improvement of organic solvent tolerance level of Escherichia coli by overexpression of stress-responsive genes. Extremophiles. 1998;2(3):239–248. doi: 10.1007/s007920050066. [DOI] [PubMed] [Google Scholar]
- 136.Cusa E, Obradors N, Baldoma L, Badía J, Aguilar J. Genetic analysis of a chromosomal region containing genes required for assimilation of allantoin nitrogen and linked glyoxylate metabolism in Escherichia coli. J Bacteriol. 1999;181(24):7479–7484. doi: 10.1128/jb.181.24.7479-7484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dai D, Reusch RN. Poly-3-hydroxybutyrate synthase from the periplasm of Escherichia coli. Biochem Biophys Res Commun. 2008;374(3):485–489. doi: 10.1016/j.bbrc.2008.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Papakostas K, Botou M, Frillingos S. Functional identification of the hypoxanthine/guanine transporters YjcD and YgfQ and the adenine transporters PurP and YicO of Escherichia coli K-12. J Biol Chem. 2013;288(52):36827–36840. doi: 10.1074/jbc.M113.523340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Boyarskiy S, Davis López S, Kong N, Tullman-Ercek D. Transcriptional feedback regulation of efflux protein expression for increased tolerance to and production of n-butanol. Metab Eng. 2016;33:130–137. doi: 10.1016/j.ymben.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 140.Kell DB, John P, Ferguson SJ. The protonmotive force in phosphorylating membrane vesicles from Paracoccus denitrificans: magnitude, sites of generation and comparison with the phosphorylation potential. Biochem J. 1978;174:257–266. doi: 10.1042/bj1740257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ghatak S, King ZA, Sastry A, Palsson BO. The y-ome defines the 35% of Escherichia coli genes that lack experimental evidence of function. Nucleic Acids Res. 2019;47(5):2446–2454. doi: 10.1093/nar/gkz030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.César-Razquin A, Snijder B, Frappier-Brinton T, Isserlin R, Gyimesi G, Bai X, Reithmeier RA, Hepworth D, Hediger MA, Edwards AM, et al. A call for systematic research on solute carriers. Cell. 2015;162(3):478–487. doi: 10.1016/j.cell.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 143.O'Hagan S, Kell DB. Consensus rank orderings of molecular fingerprints illustrate the ‘most genuine’ similarities between marketed drugs and small endogenous human metabolites, but highlight exogenous natural products as the most important ‘natural’ drug transporter substrates. ADMET & DMPK. 2017;5(2):85–125. doi: 10.5599/admet.5.2.376. [DOI] [Google Scholar]
- 144.Kashiwagi K, Shibuya S, Tomitori H, Kuraishi A, Igarashi K. Excretion and uptake of putrescine by the PotE protein in Escherichia coli. J Biol Chem. 1997;272(10):6318–6323. doi: 10.1074/jbc.272.10.6318. [DOI] [PubMed] [Google Scholar]
- 145.Tomitori H, Kashiwagi K, Igarashi K. Structure and function of polyamine-amino acid antiporters CadB and PotE in Escherichia coli. Amino Acids. 2012;42(2–3):733–740. doi: 10.1007/s00726-011-0989-9. [DOI] [PubMed] [Google Scholar]
- 146.Guerra PR, Herrero-Fresno A, Pors SE, Ahmed S, Wang D, Thofner I, Antenucci F, Olsen JE. The membrane transporter PotE is required for virulence in avian pathogenic Escherichia coli (APEC) Vet Microbiol. 2018;216:38–44. doi: 10.1016/j.vetmic.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 147.Higashi K, Sakamaki Y, Herai E, Demizu R, Uemura T, Saroj SD, Zenda R, Terui Y, Nishimura K, Toida T, et al. Identification and functions of amino acid residues in PotB and PotC involved in spermidine uptake activity. J Biol Chem. 2010;285(50):39061–39069. doi: 10.1074/jbc.M110.186536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Feng J, Wang T, Zhang S, Shi W, Zhang Y. An optimized SYBR Green I/PI assay for rapid viability assessment and antibiotic susceptibility testing for Borrelia burgdorferi. PLoS One. 2014;9(11):e111809. doi: 10.1371/journal.pone.0111809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Barbesti S, Citterio S, Labra M, Baroni MD, Neri MG, Sgorbati S. Two and three-color fluorescence flow cytometric analysis of immunoidentified viable bacteria. Cytometry. 2000;40(3):214–218. doi: 10.1002/1097-0320(20000701)40:3<214::AID-CYTO6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 150.Arnoldini M, Heck T, Blanco-Fernandez A, Hammes F. Monitoring of dynamic microbiological processes using real-time flow cytometry. PLoS One. 2013;8(11):e80117. doi: 10.1371/journal.pone.0080117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Grégori G, Citterio S, Ghiani A, Labra M, Sgorbati S, Brown S, Denis M. Resolution of viable and membrane-compromised bacteria in freshwater and marine waters based on analytical flow cytometry and nucleic acid double staining. Appl Environ Microbiol. 2001;67(10):4662–4670. doi: 10.1128/AEM.67.10.4662-4670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Berney M, Vital M, Hülshoff I, Weilenmann HU, Egli T, Hammes F. Rapid, cultivation-independent assessment of microbial viability in drinking water. Water Res. 2008;42(14):4010–4018. doi: 10.1016/j.watres.2008.07.017. [DOI] [PubMed] [Google Scholar]
- 153.Briggs C, Jones M. SYBR Green I-induced fluorescence in cultured immune cells: a comparison with Acridine Orange. Acta Histochem. 2005;107(4):301–312. doi: 10.1016/j.acthis.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 154.O'Hagan S, Swainston N, Handl J, Kell DB. A ‘rule of 0.5′ for the metabolite-likeness of approved pharmaceutical drugs. Metabolomics. 2015;11(2):323–339. doi: 10.1007/s11306-014-0733-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.O'Hagan S, Kell DB. MetMaxStruct: a Tversky-similarity-based strategy for analysing the (sub)structural similarities of drugs and endogenous metabolites. Front Pharmacol. 2016;7:266. doi: 10.3389/fphar.2016.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cooper S, Helmstetter CE. Chromosome replication and the division cycle of Escherichia coli B/r. J Mol Biol. 1968;31:519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- 157.Skarstad K, Katayama T. Regulating DNA replication in bacteria. Cold Spring Harb Perspect Biol. 2013;5(4):a012922. doi: 10.1101/cshperspect.a012922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Skarstad K, Steen HB, Boye E. Cell cycle parameters of slowly growing Escherichia coli B/r studied by flow cytometry. J Bacteriol. 1983;154:656–662. doi: 10.1128/jb.154.2.656-662.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Koita K, Rao CV. Identification and analysis of the putative pentose sugar efflux transporters in Escherichia coli. PLoS One. 2012;7(8):e43700. doi: 10.1371/journal.pone.0043700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sargentini NJ, Gularte NP, Hudman DA. Screen for genes involved in radiation survival of Escherichia coli and construction of a reference database. Mutat Res. 2016;793-794:1–14. doi: 10.1016/j.mrfmmm.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 161.Ito T, Uozumi N, Nakamura T, Takayama S, Matsuda N, Aiba H, Hemmi H, Yoshimura T. The implication of YggT of Escherichia coli in osmotic regulation. Biosci Biotechnol Biochem. 2009;73(12):2698–2704. doi: 10.1271/bbb.90558. [DOI] [PubMed] [Google Scholar]
- 162.Borghese R, Canducci L, Musiani F, Cappelletti M, Ciurli S, Turner RJ, Zannoni D. On the role of a specific insert in acetate permeases (ActP) for tellurite uptake in bacteria: functional and structural studies. J Inorg Biochem. 2016;163:103–109. doi: 10.1016/j.jinorgbio.2016.06.029. [DOI] [PubMed] [Google Scholar]
- 163.Sääf A, Baars L, von Heijne G. The internal repeats in the Na+/Ca2+ exchanger-related Escherichia coli protein YrbG have opposite membrane topologies. J Biol Chem. 2001;276(22):18905–18907. doi: 10.1074/jbc.M101716200. [DOI] [PubMed] [Google Scholar]
- 164.Sperandeo P, Pozzi C, Dehò G, Polissi A. Non-essential KDO biosynthesis and new essential cell envelope biogenesis genes in the Escherichia coli yrbG-yhbG locus. Res Microbiol. 2006;157(6):547–558. doi: 10.1016/j.resmic.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 165.Van Dyk TK, Templeton LJ, Cantera KA, Sharpe PL, Sariaslani FS. Characterization of the Escherichia coli AaeAB efflux pump: a metabolic relief valve? J Bacteriol. 2004;186(21):7196–7204. doi: 10.1128/JB.186.21.7196-7204.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lee S, Nam D, Jung JY, Oh MK, Sang BI, Mitchell RJ. Identification of Escherichia coli biomarkers responsive to various lignin-hydrolysate compounds. Bioresour Technol. 2012;114:450–456. doi: 10.1016/j.biortech.2012.02.085. [DOI] [PubMed] [Google Scholar]
- 167.Licandro-Seraut H, Roussel C, Perpetuini G, Gervais P, Cavin JF. Sensitivity to vinyl phenol derivatives produced by phenolic acid decarboxylase activity in Escherichia coli and several food-borne gram-negative species. Appl Microbiol Biotechnol. 2013;97(17):7853–7864. doi: 10.1007/s00253-013-5072-x. [DOI] [PubMed] [Google Scholar]
- 168.Karinou E, Hoskisson PA, Strecker A, Unden G, Javelle A. The E. coli dicarboxylic acid transporters DauA act as a signal transducer by interacting with the DctA uptake system. Sci Rep. 2017;7(1):16331. doi: 10.1038/s41598-017-16578-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lang S, Cressatti M, Mendoza KE, Coumoundouros CN, Plater SM, Culham DE, Kimber MS, Wood JM. YehZYXW of Escherichia coli is a low-affinity, non-Osmoregulatory betaine-specific ABC transporter. Biochemistry. 2015;54(37):5735–5747. doi: 10.1021/acs.biochem.5b00274. [DOI] [PubMed] [Google Scholar]
- 170.Grixti J, O'Hagan S, Day PJ, Kell DB. Enhancing drug efficacy and therapeutic index through cheminformatics-based selection of small molecule binary weapons that improve transporter-mediated targeting: a cytotoxicity system based on gemcitabine. Front Pharmacol. 2017;8:155. doi: 10.3389/fphar.2017.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bailey AM, Paulsen IT, Piddock LJ. RamA confers multidrug resistance in Salmonella enterica via increased expression of acrB, which is inhibited by chlorpromazine. Antimicrob Agents Chemother. 2008;52(10):3604–3611. doi: 10.1128/AAC.00661-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Baugh S, Phillips CR, Ekanayaka AS, Piddock LJV, Webber MA. Inhibition of multidrug efflux as a strategy to prevent biofilm formation. J Antimicrob Chemother. 2014;69(3):673–681. doi: 10.1093/jac/dkt420. [DOI] [PubMed] [Google Scholar]
- 173.Lawler AJ, Ricci V, Busby SJW, Piddock LJV. Genetic inactivation of acrAB or inhibition of efflux induces expression of ramA. J Antimicrob Chemother. 2013;68(7):1551–1557. doi: 10.1093/jac/dkt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dickens D, Rädisch S, Chiduza GN, Giannoudis A, Cross MJ, Malik H, Schaeffeler E, Sison-Young RL, Wilkinson EL, Goldring CE, et al. Cellular uptake of the atypical antipsychotic clozapine is a carrier-mediated process. Mol Pharm. 2018;15(8):3557–3572. doi: 10.1021/acs.molpharmaceut.8b00547. [DOI] [PubMed] [Google Scholar]
- 175.Kell DB. What would be the observable consequences if phospholipid bilayer diffusion of drugs into cells is negligible? Trends Pharmacol Sci. 2015;36(1):15–21. doi: 10.1016/j.tips.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 176.Mestres J, Gregori-Puigjané E, Valverde S, Solé RV. The topology of drug-target interaction networks: implicit dependence on drug properties and target families. Mol BioSyst. 2009;5(9):1051–1057. doi: 10.1039/b905821b. [DOI] [PubMed] [Google Scholar]
- 177.Tomov TC. Pyronin G as a fluorescent probe for quantitative determination of the membrane potential of mitochondria. J Biochem Biophys Methods. 1986;13(1):29–38. doi: 10.1016/0165-022X(86)90005-9. [DOI] [PubMed] [Google Scholar]
- 178.Bunting JR, Phan TV, Kamali E, Dowben RM. Fluorescent cationic probes of mitochondria. Metrics and mechanism of interaction. Biophys J. 1989;56(5):979–993. doi: 10.1016/S0006-3495(89)82743-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bunting JR. Influx and efflux kinetics of cationic dye binding to respiring mitochondria. Biophys Chem. 1992;42(2):163–175. doi: 10.1016/0301-4622(92)85006-P. [DOI] [PubMed] [Google Scholar]
- 180.Ross MF, Kelso GF, Blaikie FH, James AM, Cocheme HM, Filipovska A, Da Ros T, Hurd TR, Smith RA, Murphy MP. Lipophilic triphenylphosphonium cations as tools in mitochondrial bioenergetics and free radical biology. Biochemistry (Mosc) 2005;70(2):222–230. doi: 10.1007/s10541-005-0104-5. [DOI] [PubMed] [Google Scholar]
- 181.Davidson SM, Yellon D, Duchen MR. Assessing mitochondrial potential, calcium, and redox state in isolated mammalian cells using confocal microscopy. Methods Mol Biol. 2007;372:421–430. doi: 10.1007/978-1-59745-365-3_30. [DOI] [PubMed] [Google Scholar]
- 182.Solaini G, Sgarbi G, Lenaz G, Baracca A. Evaluating mitochondrial membrane potential in cells. Biosci Rep. 2007;27(1–3):11–21. doi: 10.1007/s10540-007-9033-4. [DOI] [PubMed] [Google Scholar]
- 183.Monfort P, Baleux B. Cell cycle characteristics and changes in membrane potential during growth of Escherichia coli as determined by a cyanine fluorescent dye and flow cytometry. J Microbiol Meth. 1996;25(1):79–86. doi: 10.1016/0167-7012(95)00089-5. [DOI] [Google Scholar]
- 184.Waggoner A. Optical probes of membrane potential. J Membr Biol. 1976;27(4):317–334. doi: 10.1007/BF01869143. [DOI] [PubMed] [Google Scholar]
- 185.Azzone GF, Pietrobon D, Zoratti M. Determination of the proton electrochemical gradient across biological membranes. Curr Top Bioenerg. 1984;13:1–77. doi: 10.1016/B978-0-12-152513-2.50008-8. [DOI] [Google Scholar]
- 186.Rottenberg H. Proton electrochemical potential gradient in vesicles, organelles, and prokaryotic cells. Methods Enzymol. 1989;172:63–84. doi: 10.1016/S0076-6879(89)72008-5. [DOI] [PubMed] [Google Scholar]
- 187.Damper PD, Epstein W, Rosen BP, Sorensen EN. Thallous ion is accumulated by potassium transport systems in Escherichia coli. Biochemistry. 1979;18(19):4165–4169. doi: 10.1021/bi00586a018. [DOI] [PubMed] [Google Scholar]
- 188.Bakker EP, Harold FM. Energy coupling to potassium transport in Streptococcus faecalis. Interplay of ATP and the protonmotive force. J Biol Chem. 1980;255(2):433–440. [PubMed] [Google Scholar]
- 189.McCarthy JE, Ferguson SJ, Kell DB. Estimation with an ion-selective electrode of the membrane potential in cells of Paracoccus denitrificans from the uptake of the butyltriphenylphosphonium cation during aerobic and anaerobic respiration. Biochem J. 1981;196(1):311–321. doi: 10.1042/bj1960311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tedeschi H. The transport of cations in mitochondria. Biochim Biophys Acta. 1981;639(3–4):157–196. doi: 10.1016/0304-4173(81)90009-4. [DOI] [PubMed] [Google Scholar]
- 191.Ritchie RJ. A critical assessment of the use of lipophilic cations as membrane potential probes. Prog Biophys Mol Biol. 1984;43(1):1–32. doi: 10.1016/0079-6107(84)90002-6. [DOI] [PubMed] [Google Scholar]
- 192.Skarstad K, Steen HB, Boye E. Escherichia coli DNA distributions measured by flow cytometry and compared with theoretical computer simulations. J Bacteriol. 1985;163:661–668. doi: 10.1128/jb.163.2.661-668.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ivnitski-Steele I, Larson RS, Lovato DM, Khawaja HM, Winter SS, Oprea TI, Sklar LA, Edwards BS. High-throughput flow cytometry to detect selective inhibitors of ABCB1, ABCC1, and ABCG2 transporters. Assay Drug Dev Technol. 2008;6(2):263–276. doi: 10.1089/adt.2007.107. [DOI] [PubMed] [Google Scholar]
- 194.AlRabiah H, Xu Y, Rattray NJ, Vaughan AA, Gibreel T, Sayqal A, Upton M, Allwood JW, Goodacre R. Multiple metabolomics of uropathogenic E. coli reveal different information content in terms of metabolic potential compared to virulence factors. Analyst. 2014;139(17):4193–4199. doi: 10.1039/C4AN00176A. [DOI] [PubMed] [Google Scholar]
- 195.Dawson SE, Gibreel T, Nicolaou N, AlRabiah H, Xu Y, Goodacre R, Upton M. Implementation of Fourier transform infrared spectroscopy for the rapid typing of uropathogenic Escherichia coli. Eur J Clin Microbiol Infect Dis. 2014;33(6):983–988. doi: 10.1007/s10096-013-2036-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Modal and mean uptake data for diS-C3(5) and Sybr green as described in the body of the paper. (XLSX 55 kb)
Data Availability Statement
The raw flow cytometric datasets generated and/or analysed during the current study are not publicly available due to the fact that they were simply massive, but are available from the corresponding author on reasonable request. Relevant derived data are given in supplementary information as a spreadsheet.