The overuse of antibiotics has led to the evolution of drug-resistant bacteria that are becoming increasingly dangerous to human health. According to the Centers for Disease Control and Prevention, antibiotic-resistant bacteria cause at least 2 million illnesses and 23,000 deaths in the United States annually. Traditionally, antibiotics are bactericidal or bacteriostatic agents that place selective pressure on bacteria, leading to the expansion of antibiotic-resistant strains.
KEYWORDS: Clostridium difficile, Escherichia coli, Salmonella, enteropathogens, microbiota
ABSTRACT
The overuse of antibiotics has led to the evolution of drug-resistant bacteria that are becoming increasingly dangerous to human health. According to the Centers for Disease Control and Prevention, antibiotic-resistant bacteria cause at least 2 million illnesses and 23,000 deaths in the United States annually. Traditionally, antibiotics are bactericidal or bacteriostatic agents that place selective pressure on bacteria, leading to the expansion of antibiotic-resistant strains. In addition, antibiotics that are effective against some pathogens can also exacerbate their pathogenesis and may lead to severe progression of the disease. Therefore, alternative strategies are needed to treat antibiotic-resistant bacterial infections. One novel approach is to target bacterial virulence to prevent or limit pathogen colonization, while also minimizing tissue damage and disease comorbidities in the host. This review focuses on the interactions between enteric pathogens and naturally occurring small molecules in the human gut as potential therapeutic targets for antivirulence strategies. Individual small molecules in the intestines modulate enteric pathogen virulence and subsequent intestinal fitness and colonization. Targeted interruption of pathogen sensing of these small molecules could therefore attenuate their virulence. This review highlights the paths of discovery for new classes of antimicrobials that could potentially mitigate the urgent problem of antibiotic resistance.
INTRODUCTION
The first antibiotic, penicillin, was discovered by Sir Alexander Fleming in 1928 and was introduced as a drug in the early 1940s to treat bacterial infections. Within a few years, penicillin-resistant strains of Staphylococcus aureus were identified as a severe problem in hospitals and in the community (1). The next decades witnessed a surge in the development of many antibiotics. Their initial successes led to the premature belief that infectious diseases were on the road to eradication, which dampened further interest in developing new classes of antibacterial drugs. Bacteria have the means to become resistant to antibiotics with diverse mechanisms of action (2, 3). According to the Global Antimicrobial Resistance Surveillance System (GLASS) report for 2014 by the World Health Organization (WHO), antibiotic-resistant strains were ubiquitously found worldwide (4). The report, including data from 22 countries, revealed that Escherichia coli, Klebsiella pneumoniae, Staphylococcus aureus, Streptococcus pneumoniae, and Salmonella spp. were the most commonly reported antibiotic-resistant strains (4). In the United States alone, more than 2 million people are affected by antibiotic-resistant bacteria each year, according to the CDC. Thus, infections by antibiotic-resistant bacteria are a growing threat to the health care system worldwide.
Antibiotics impart their antibacterial effects through two general mechanisms of action, killing bacterial cells (bactericidal) and inhibition of bacterial growth (bacteriostatic) (5). Broadly, bactericidal agents include antibiotics that target bacterial cell wall synthesis (e.g., β-lactams), DNA synthesis (e.g., quinolones), or RNA synthesis (e.g., rifamycins). Protein synthesis inhibitors (e.g., macrolides, amphenicols such as chloramphenicol, and streptogramins) that target bacterial ribosomal subunits are usually bacteriostatic because they interfere with the translational machinery, which stalls cellular growth (6). In contrast, protein synthesis inhibitors such as aminoglycosides bind to ribosomal subunits and promote protein mistranslation by incorporating inappropriate amino acids, which imparts their bactericidal activities (6). Antibiotic resistance arises from the overuse of bactericidal and bacteriostatic drugs, which impart selective pressures to bacterial populations. The development of antimicrobial resistance is a multifactorial process that includes, but is not limited to, decreased antibiotic uptake, structural alterations to the antibiotic target, increased activity of efflux pumps, posttranslational modifications, and enzymatic modification of antibiotics (2). Therefore, there is an urgent need to develop novel strategies that counteract and/or bypass these mechanisms of acquiring antibiotic resistance to treat bacterial infections.
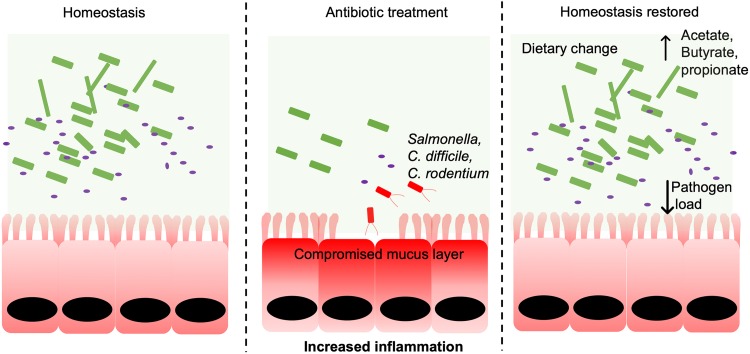
In addition to having effects on the microevolution of bacterial populations, antibiotics can enhance the virulence potential of pathogens by directly altering pathogen function and through their effects on the host microbiome. For example, antibiotics exacerbate the progression of disease induced by the foodborne human pathogen enterohemorrhagic E. coli (EHEC) O157:H7 by triggering the production of a phage-encoded virulence factor, Shiga toxin (Stx) (7). Antibiotics act as a stress signal that triggers the lysogenic phage (integrated into the EHEC genome) to enter into its lytic cycle. The stress signals may also induce the prophages that contain Stx genes, leading to an increase in the transcription and production of Stx (7, 8). During the lytic cycle, the phage expresses lytic-phase genes that enable rapid viral replication and Stx release. Together, this results in EHEC cell lysis and extracellular release of Stx, which exacerbates EHEC-induced human disease that includes the development of sometimes fatal comorbidities (e.g., hemolytic-uremic syndrome [HUS], seizures, cerebral edema, and/or coma) (9–11). The arsenal of virulence factors encoded in pathogen genomes is often essential for establishing successful infection by enabling rapid niche formation and antagonistic competition with the microbiota. The activation of these virulence programs also results in pathological damage to host tissues and disruption of homeostatic physiological processes. Antibiotic treatment can further accelerate pathogen colonization of the intestines by profoundly impacting the composition of the microbiota. Antibiotics can also target endogenous bacteria, which opens otherwise unavailable niches and nutrient sources and enables the overgrowth of invading pathogens and endogenous pathobionts, including Salmonella enterica serovar Typhimurium, Escherichia coli, and Clostridium difficile (12) (Fig. 1).
FIG 1.
Dysbiosis is associated with enteric pathogen expansion in the gut. Antibiotic treatment causes the depletion of the gut microbiota and a reduction in microbial metabolites. Opportunistic pathogens like Salmonella and C. difficile thrive in such an environment and cause disease. Acute dietary interventions change the metabolic landscape and help to reduce the pathogen load as well as overall pathogenesis of the disease, leading to restored homeostasis. Commensal bacteria are indicated in green, gut pathogens are indicated in red, and purple dots represent gut small molecules like acetate, butyrate, and propionate.
With the advent of metabolomics, we now appreciate that the intestines are home to a complex milieu of small molecules (13). These small molecules are derived from the intestinal microbiota, the host, or dietary sources and can act as chemical signals that functionally modulate bacteria, including invading pathogens. Microbiota-derived small molecules include oligopeptides synthesized by bacteria such as antibiotics (<40 amino acids) and bacteriocins (>40 amino acids) and products generated through bacterial metabolism of diet and host nutrient sources, including indole and short-chain fatty acids (SCFAs). Host-derived small molecules also include metabolic by-products as well as hormones such as epinephrine that modulate bacterial function and pathogenesis. Intestinal pathogens directly sense many of these small molecules as a means to regulate their virulence programs, which can be either conducive or detrimental to the overall virulence programs (14, 15). The majority of these small molecules do not directly inhibit pathogen growth and are naturally present in the gut. Thus, developing therapeutics that target these interspecies and interkingdom signaling pathways represent an alternative approach to combat bacterial infections. These antivirulence strategies aim to limit the activation of pathogen virulence repertoires (Fig. 2) and thus modify pathogens into nonpathogenic versions of themselves, which are more likely to be outcompeted by the resident microbiota. In addition to augmenting colonization resistance to limit infection, these antivirulence strategies may also restore the symbiosis between the host and the microbiota and accelerate the reestablishment of gut homeostasis, including gut immune tone, through their effects on the microbiota and its associated metabolome. In this review, we discuss four different strategies of antivirulence approaches that target gut pathogen interactions with small molecules. We focus primarily on the small molecules present in the human gut and the following enteric pathogens: the attaching and effacing (A/E) pathogens EHEC and Citrobacter rodentium, the invasive enteric pathogen Salmonella Typhimurium, and the opportunistic pathobiont Clostridium difficile.
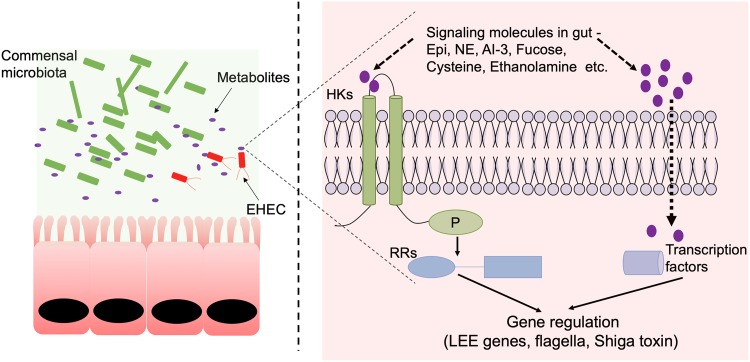
FIG 2.
EHEC senses gut metabolites to regulate gene expression. In the healthy gut, small molecules/metabolites accumulate through microbial/host metabolic processes. These small molecules are present in abundance in the gut and are important for maintaining homeostasis. For successful infection, an intestinal pathogen like EHEC needs to sense these signals to coordinately regulate its gene expression, especially those involved in virulence and colonization. Small molecules in the gut can affect the virulence of enteric pathogens either by directly sensing a cytoplasmic transcription factor (e.g., CutR and EutR for cysteine and ethanolamine, respectively) or by sensing through bacterial two-component systems. Small molecules like epinephrine, norepinephrine, autoinducer-3, and fucose are sensed by histidine kinases like QseC. QseE, and FusK, respectively. These histidine kinases then transmit the message to their cognate response regulators that directly mediate different aspects of bacterial gene expression, including regulation of the virulence gene program.
EHEC is a foodborne pathogen found throughout the world. It is responsible for major outbreaks of bloody diarrhea and HUS, a type of kidney failure. Annually in the United States, EHEC is responsible for an estimated 73,000 illnesses, 1,800 to 3,600 hospitalizations, and 61 to 541 deaths, with combined annual economic costs exceeding $400 million (https://www.cdc.gov) (16). EHEC colonizes the large intestine and forms A/E lesions on intestinal epithelial cells. Most of the genes required to form these lesions are contained within a chromosomal pathogenicity island termed the locus of enterocyte effacement (LEE). EHEC also expresses Stx in the intestine, and this potent inhibitor of protein synthesis can be absorbed systemically. Stx binds to receptors found in the kidneys and central nervous system (CNS), causing HUS, seizures, cerebral edema, and/or coma. There are few, if any, good treatment options for HUS. Antibiotics and antimotility agents are contraindicated for EHEC infections. In fact, antibiotics promote the expression and release of Stx, thereby increasing the occurrence and severity of HUS and CNS involvement (10, 11). Consequently, innovative, cost-effective EHEC treatments are urgently needed to address a significant unmet health care need.
EHEC does not form A/E lesions on the intestines of mice, and study of the EHEC-host relationship at the level of intestinal disease must rely on expensive and genetically intractable animal models. An alternative contemplated by many investigators in the field is to study the surrogate natural murine pathogen Citrobacter rodentium, which harbors the LEE and forms A/E lesions on the intestines of mice, leading to colonic hyperplasia (17, 18). All of the known virulence genes of EHEC have been validated in vivo using C. rodentium murine infections (18–22). The utilization of the C. rodentium model capitalizes on merging the powerful genetic tractability of host and pathogen to unravel the mechanisms involved in host recognition and infection.
Salmonella enterica includes significant human pathogens responsible for food poisoning and enteric fever (23). Nontyphoidal Salmonella (NTS) infections (i.e., “food poisoning” or salmonellosis) result in mild to moderate diarrhea, fever, nausea, and cramps, which normally resolve without treatment in 4 to 7 days. Nevertheless, nontyphoidal infections have a large health impact, with an estimated ∼1.4 million infections, 16,000 hospitalizations, and 400 to 600 deaths annually in the United States. In economic terms, the impact has been estimated at $2.4 billion for 2005 (24). Studies from around the world show that Salmonella has become increasingly resistant to many antibiotics (25–30). First-line antibiotics against NTS, including ampicillin, chloramphenicol, sulfonamides, streptomycin, and tetracycline, have all been observed to be ineffective. Multidrug-resistant strains of NTS are associated with a higher risk of invasive disease and increased deaths than susceptible strains (31, 32). Clearly, new treatments for Salmonella infections are needed now. Ideally, any new drugs would target new mechanisms and avoid resistance.
Clostridium difficile is a pathobiont. This is a spore-forming bacterium that is a member of the gastrointestinal (GI) microbiota. Upon germination, it colonizes the large intestine and causes colitis through the action of two toxins: toxin A (TcdA) and toxin B (TcdB). The majority of C. difficile infections (CDI) are nosocomial; however, there is also an increase in community-acquired infections. C. difficile colonizes the mammalian GI tract without causing disease. However, antibiotic use can lead to C. difficile-mediated colitis (33).
INHIBITING PATHOGEN SENSING OF PROVIRULENCE SIGNALS
It is well established that bacteria sense a diverse array of environmental signals to gather information about their local environment, which in turn alters bacterial growth, function, and virulence. For example, the host neurotransmitters epinephrine (Epi) and norepinephrine (NE) can be sensed by the bacterial histidine kinase (HK) QseC, which is encoded in the core genomes of numerous proteobacterial families, including Enterobacteriaceae (34, 35). Epi and NE are abundantly found throughout the gut, with concentrations highest near the intestinal epithelial interface and lowest in luminal contents (36). Extensive research has demonstrated that Epi and NE directly impact host susceptibility to infectious diseases, including infections by A/E pathogens such as EHEC, NTS, Vibrio parahaemolyticus, and E. coli pathobionts (37).
The effects of QseC-mediated sensing of Epi and NE on pathogen virulence have been best studied in A/E pathogens. This functional class of enteric pathogens, which includes EHEC and its murine surrogate Citrobacter rodentium, harbor the LEE pathogenicity island that encodes a type III secretion system (T3SS) essential for establishing colonic infection (38–40). A/E pathogens use this T3SS to intimately attach to colonic epithelial cells, which results in the formation of A/E lesions characterized by the effacement of the microvilli and the rearrangement of the epithelial cytoskeleton (38–40). Indeed, mutants lacking a functional LEE-encoded T3SS fail to effectively colonize the intestines and cause disease. In the presence of Epi or NE, QseC becomes autophosphorylated and transfers this phosphate to downstream response regulators in the bacterial cytoplasm, resulting in the transcriptional regulation of downstream genes and altered bacterial responses (41). QseC sensing of Epi and NE in EHEC and C. rodentium transcriptionally activates the LEE, which results in enhanced LEE-encoded T3SS activity, exacerbated A/E lesion formation, and more-severe enteric disease (38–40). QseC activation by NE and Epi also enhances the flagellar motility of EHEC by modulating the transcription of the master flagellar transcriptional regulators, FlhDC (34, 42). Thus, inhibition of QseC sensing of Epi/NE represents a promising antivirulence strategy to limit initial pathogen colonization and to attenuate disease in the host.
To identify putative QseC inhibitors, a high-throughput screen was performed with a library of 150,000 small molecules, which revealed LED209 {N-phenyl-4-[(phenylamino) thioxomethyl]amino-benzenesulfonamide} as a potent inhibitor of QseC (43). Extensive structure-activity relationships revealed that LED209 is a potent prodrug and is highly specific for QseC (35). Mechanistically, LED209 covalently modifies specific lysine residues in QseC, which prevents the activation of virulence gene repertoires by allosterically preventing activating signals (e.g., Epi, NE, and autoinducer-3) from binding the QseC receptor. In vitro studies demonstrated that LED209 attenuates EHEC virulence by decreasing the expression of LEE genes and reducing A/E lesion formation on epithelial cells. In preclinical models of inflammatory bowel diseases, LED209 confers protection against adhesive-invasive E. coli (AIEC), a pathobiont associated with Crohn’s disease (44). LED209 was also effective in combating murine infections by S. Typhimurium and Francisella tularensis (43). Importantly, LED209 did not cause any toxicity in treated mice. Moreover, pharmacokinetic studies demonstrated that the administration of LED209 via the oral route appeared to be more desirable. Bacteriostatic effects have not been reported for LED209 on the pathogens tested so far, which reduces the likelihood that resistant strains will emerge (35).
Efficacious antivirulence strategies have also been demonstrated in infections by the endogenous opportunistic pathogen C. difficile, a spore-forming member of the intestinal microbiota. Upon spore germination, C. difficile colonizes the large intestines, usually without causing disease. However, under certain contexts, such as antibiotic ingestion, C. difficile can cause a recurring pseudomembranous colitis through the actions of its two exotoxins, toxin A (TcdA) and toxin B (TcdB). Upon endocytosis by host cells, glucosylating toxins TcdA and TcdB are activated through the proteolytic activities of their cysteine protease domain (CPD) (45). The autoprocessing of the toxins releases the glucosyltransferase domain in the cytoplasm, which leads to the glucosylation of Rho GTPases, resulting in inhibition of its function and eventually cell death (45, 46). Inhibition of CPD activity in TcdA and TcdB could therefore diminish the progression of C. difficile-mediated disease. Indeed, a high-throughput screen identified ebselen as a potent inhibitor of CPD activity, which was consequently shown to decrease C. difficile-associated pathology and mortality in mice without affecting C. difficile growth (47). Taken together, preclinical studies of A/E pathogen and opportunistic infections clearly demonstrate that the virulence potential of enteric pathogens can be attenuated by (i) identifying the signaling mechanisms utilized by these pathogens to modulate their virulence strategies and (ii) precisely inhibiting this pathogen-holobiont cross talk to disrupt pathogen growth and disease causation.
PREVENTING INITIAL COLONIZATION
The composition of the intestinal microbiota is dynamic and changes with shifts in diet and other environmental factors, such as antibiotic use (48). Our consumption of diet and xenobiotics influences the availability of carbon and nitrogen sources in the gut that can be used for energy by the commensal flora. Members of the microbiota degrade complex polysaccharides into simpler sugars, which can then be utilized for their growth. Upon infection, enteric pathogens also utilize these carbon and nitrogen sources for growth and as environmental cues that regulate the expression of their virulence genes.
Reversing antibiotic-mediated dysbiosis.
C. difficile expansion and colitis induction are associated with antibiotic therapy (49). Almost all common classes of antibiotics have been associated with the development of C. difficile infections (CDI), including metronidazole and vancomycin, which are often prescribed to treat these infections (33, 50). The normal intestinal microflora, which serves as the main protective barrier against CDI, is compositionally disrupted upon antibiotic treatment. Additionally, antibiotic treatment increases the concentrations of free sialic acids in the intestines, which are subsequently used by endogenous C. difficile for its own expansion (12). Taken together, antibiotic treatment disrupts the intestinal microbiota and local metabolome to enable the pathogenic expansion of toxigenic C. difficile, thus leading to the development of CDI (33, 51).
An antivirulence strategy that is now used in the clinic to treat CDI is known as fecal microbiota transplantation (FMT). In conjunction with the complete withdrawal of antibiotic treatment, oral or rectal transplantation of fecal contents from healthy donors has successfully treated recurrent CDI in 90% of reported cases (52, 53). FMT reverses the dysbiotic effects of antibiotics on the microbiota by reintroducing bacteria that were unintentionally vanquished, thus restoring the ability of the microbiota to resist expansion of pathobionts such as C. difficile (54). Moreover, unlike with antibiotics, this therapeutic strategy may not impart selection pressures on the microbiota, which can enable the expansion of resistant endogenous bacteria. However, although FMT is effective in treating CDI, the long-term effects of FMT on the transplant recipient have not been extensively studied. Therefore, in addition to the administration of compounds that neutralize C. difficile toxins, these studies introduce an alternative approach that utilizes FMT to minimize C. difficile infection in at-risk populations (i.e., recipients of antibiotics), which could be implemented together with the targeted inhibition of key virulence strategies employed by C. difficile.
Dietary fiber.
The precise compositional features of the intestinal microbiome that maximizes the prevention of infectious diseases have not been fully characterized. However, 16S rRNA profiling of gut-associated microbial communities in humans and in mouse models of infectious colitis have established key correlations between diet, microbiome composition, and resistance against infectious disease. For example, the presence of certain members of the Bacteroidetes and Firmicutes phyla in FMT is associated with effectiveness in treating CDI (55, 56). Moreover, disease in CDI patients is associated with a marked depletion of butyrogenic bacteria (56), which are typically members of the Bacteroidetes and Firmicutes. Butyrogenic bacteria anaerobically ferment complex polysaccharides such as dietary fiber and release short-chain fatty acids (SCFAs) such as butyrate as metabolic by-products. Indeed, long-term dietary fiber consumption in mice increases intestinal SCFA levels. Interestingly, butyrate counteracts C. difficile growth in vitro and could therefore also reduce burdens of this opportunistic pathogen in the gut (57). The protective effects of dietary fiber may not be limited to CDI. A second study utilizing the C. rodentium colitis model demonstrated that mice deprived of dietary fiber exhibited increased inflammation with C. rodentium infection. The authors proposed that in the absence of dietary fiber, an important source of nutrition for endogenous Bacteroides spp., these commensals instead “eat” the host mucus layer, which increases C. rodentium access to its preferred replicative niche, the apical epithelial surface (58). Thus, dietary fiber indirectly inhibits pathogen outgrowth in the gut through its effects on the pathogen itself and on the microbiota.
Amino acid availability.
Enteric infection significantly changes the metabolic landscape of the gut, which in turn can alter the milieu of available nutrients and impact enteric pathogen function (59, 60). For example, infection with enteric pathogens alters amino acid availability in the gut and consequently affects the development and progression of infectious disease. Using RNA sequencing and metabolomics in a mouse model of CDI, infection was associated with decreased availability of amino acids, especially proline and branched-chain amino acids. These metabolomic changes corresponded with increased bacterial transcription of amino acid utilization genes (61), therefore corroborating the limited availability of amino acids with CDI. Interestingly, administration of a low-protein or low-proline diet attenuated the expansion of C. difficile in ex-germfree mice transplanted with a human dysbiotic microbiota. Moreover, inactivation of proline utilization in C. difficile significantly reduced gut colonization (62). Together, these studies suggest that C. difficile preferentially utilizes proline for its overgrowth during CDI.
In contrast, infection with C. rodentium increases the availability of the amino acid l-cysteine in the murine colon (59). A high-throughput screen identified significant regulators involved in LEE regulation, including CutR (63). The presence of cysteine is sensed by CutR, which serves as an activator of the LEE in the presence of cysteine. Indeed, a cutR mutant is attenuated for pathogenesis in mice. Similarly, mice fed a high-cysteine diet succumb to infection with C. rodentium earlier than mice fed a low-cysteine diet (63). CutR is prevalent among members of the Enterobacteriaceae, including EHEC and Salmonella, making it an important target for antivirulence strategies. These findings demonstrate that acute dietary interventions alter the colonization potential of enteric pathogens.
Fucose as a free carbon source.
Enteric pathogens utilize free carbon sources for metabolism and regulation of virulence genes. The effects of fucose in regulating the virulence and metabolism of EHEC have been uncoupled (64). Bacteroides thetaiotamicron, an endogenous member of the human microbiota, harbors multiple fucosidases that release fucose from host glycans in the lumen (65). Commensal E. coli and enteric pathogens like EHEC can then utilize fucose as a carbon source to grow in the lumen. EHEC also senses fucose, resulting in decreased expression of virulence genes and increased expression of metabolic genes to maintain its niche. This sensing mechanism is signaled through the two-component system FusK/FusR. A mutant of FusK has a colonization defect because of its inability to regulate virulence gene expression and fucose metabolism and is therefore outcompeted by the microbiota (64). Thus, an inhibitor that specifically targets the FusK histidine kinase (HK) may potentially act as a novel antivirulence therapeutic.
EA.
Ethanolamine (EA) in the gut is continuously derived from the diet or by the breakdown of bacterial and host cell membranes (66). Bacteria in the gut can break down EA into ammonia and acetaldehyde by an EA ammonia lyase. The proteins required for transport and catabolism of EA are encoded in an EA utilization (eut) operon. The eut operon is present across species, although there are considerable differences in gene content and organization (66). Bacterial pathogens, including Salmonella Typhimurium and EHEC, utilize EA as a nitrogen source for growth and as a chemical signal to promote disease by increasing virulence gene expression (67, 68). The bacterial transcription factor EutR, encoded within the eut operon, directly binds to the LEE in EHEC to increase virulence gene expression. EutR also increases the expression of Salmonella pathogenicity island 2 to promote the survival of Salmonella within macrophages (68, 69). Additionally, EA is considered an important cue for bacterial adherence to epithelial cells. High-throughput analysis of EHEC in the presence or absence of EA indicated that EA increased the expression of fimbrial genes (70). However, EA can also be used by particular members of the microbiota to outcompete invading pathogens as a mechanism of colonization resistance (71). A human commensal E. coli strain senses and utilizes EA to outwit EHEC through currently unknown mechanisms (71). These observations warrant further investigation to elucidate the effects of EA in animal models of infection. The wide prevalence of EA in the gastrointestinal environment makes it an ideal target to study novel antivirulence strategies.
Additional approaches.
Initial adherence to the host surface facilitated by type I and P pili and expression of T3SSs are important arsenals utilized by several pathogens to initiate successful colonization. Toward this end, several inhibitors targeting the T3SS (e.g., Yersinia spp., EHEC, Salmonella, etc.), pilicides aiming at type I and P pili (e.g., uropathogenic E. coli [UPEC]), and curlicides targeting curli subunits (e.g., enteroaggregative E. coli [EAEC]) have been developed and are extensively reviewed elsewhere (72–74). Bacteria have the means to communicate with each other in a process known as quorum sensing (QS) by producing chemical signaling molecules. These chemical signals, called autoinducers (AI), alter bacterial gene expression and regulate multiple physiological processes, including virulence and biofilm formation, in several pathogens (75, 76). Disruption of QS signaling may act as an antivirulence approach. For example, the addition of the probiotic Lactobacillus acidophilus to the feed of weaning pigs harboring EHEC in their gut inhibited the activity of AI-2 and prevented biofilm formation, without affecting the growth of EHEC (77). AI may also dictate the presence of a certain microbial species, thereby changing the overall metabolic landscape of the gut. In an antibiotic-induced dysbiotic gut (microbiota with a diminished Firmicutes population), the addition of AI-2 in the murine gut using engineered E. coli favored Firmicutes expansion and reshaped the composition of the microbiota (78). Collectively, engineered probiotics, live microorganisms that confer health benefits, can be administered to manipulate the microbiota as well as the concentration and activity of small molecules present in the human gut as an effective antivirulence strategy.
Humans are frequently accidental hosts of many enteric pathogens. Therefore, inhibiting pathogen expansion may limit the selection pressures placed on pathogens that enable the rapid evolution of resistant strains.
COUNTERACTING INFLAMMATION-INDUCED PATHOGEN OVERGROWTH
Inflammation alters the composition of the microbiota and promotes the overgrowth of Enterobacteriaceae (79, 80). Metagenomic sequencing of intestinal microbial communities revealed that formate dehydrogenases and molybdenum cofactor-dependent metabolic pathways are metabolic signatures of inflammation-associated dysbiosis (81, 82). Respiratory reductases and formate dehydrogenases contain molybdenum within a cofactor. Enterobacteriaceae such as E. coli utilize molybdenum cofactor-dependent anaerobic respiratory enzymes to bloom in the inflamed gut (81). Tungsten, with chemical properties similar to those of molybdenum, competitively displaces molybdenum, thus inactivating molybdenum cofactor-dependent respiratory enzymes. Indeed, administration of tungsten during inflammation inhibits anaerobic respiration in Enterobacteriaceae, which leads to decreased Enterobacteriaceae growth, reduced inflammation, and restoration of homoeostasis. Importantly, tungsten treatment has minimal compositional effects on the inflammation-associated microbiota (82). Enterobacteriaceae populations within the microbiota are comprised of facultative anaerobes that in healthy individuals are at lower abundances and are helpful to the host in maintaining homeostasis. However, the overgrowth of some Enterobacteriaceae species can exacerbate inflammation in susceptible hosts (80). Therefore, tungsten treatment provides a unique way to limit specific bacterial populations in the gut to restore and maintain homeostasis.
ALTERING METABOLIC ACTIVITIES OF THE MICROBIOTA
Direct effects.
Fermentation of dietary fibers by commensal bacteria results in the production of short-chain fatty acids (SCFAs), including acetate, propionate, and butyrate (83). Intake of fiber-rich diets further increases the abundance of SCFAs in the gut (84). SCFAs alter the local pH in the gut, which impacts growth and confers colonization resistance to some enteric pathogens, such as Salmonella and EHEC (85, 86). For example, propionate disrupts intracellular pH homeostasis in S. Typhimurium and confers colonization resistance to S. Typhimurium infection (87). SCFAs inhibit bacterial growth in vitro, and this correlates with changes in SCFA concentrations in the intestines (57).
Enteric pathogens sense chemical alterations in the gut to counteract mechanisms of colonization resistance. For example, EHEC senses alterations in pH and membrane integrity through the HK CpxA (88). In response to these signals, CpxA phosphorylates its cognate response regulator, CpxR, resulting in the transcriptional regulation of virulence genes. Tryptophan metabolites such as indole are small molecules that are abundantly present in the human gut (14). Indole is a microbiota-derived small molecule produced as a by-product of tryptophan metabolism by bacteria. The highest concentrations of indole are present in the lumen, which decreases in concentration toward the mucus layer and epithelium (89). Enteric pathogens may sense this indole gradient and regulate the expression of virulence genes. A recent study has shown that indole signaling also occurs through CpxA, and increased indole concentrations decrease the autophosphorylation activity of CpxA, leading to decreased LEE expression (89). Synthetically engineering the microbiota in susceptible mice to alter indole production affects disease outcome. Mice with an indole-producing microbiota are less susceptible to C. rodentium infection (89). When this pathogen is in a zone of higher indole concentration, it represses virulence gene expression. Meanwhile, at the epithelial lining, which is a region of lower indole concentration, it expresses the T3SS to promote infection. This spatial regulation of the virulence repertoire minimizes energy expenditure.
Indirect effects via the host.
SCFAs, including propionate and butyrate, are also important in providing colonization resistance to Salmonella infection. Antibiotic treatment decreases the abundance of SCFA-producing members of the microbiota. Antibiotic-mediated depletion of endogenous Clostridium spp. decreases butyrate levels in the gut and consequently increases epithelial oxygenation (90). Increased availability of oxygen fuels the expansion of aerobically respiring enteric pathogens, including S. Typhimurium and C. rodentium (90, 91).
Together, these studies demonstrate the impact of altering the metabolic activity of microbiota that may potentially help to limit pathogen colonization and infection.
CONCLUSIONS
With the advances made in the field of synthetic biology, we are preparing for a future of an engineered microbiota that can precisely sense and respond to the chemical environment of the gut as a novel means to restore and maintain homeostasis (92). Manipulation of the gut microbiota is routinely performed by employing germfree mice or microbiota-depleted mice through the administration of antibiotics. These mice can be reconstituted with a single bacterial species, a consortium of cultivatable defined microbiota, or a more complex undefined or xenografted microbiota to further manipulate the concentration of small molecules present in the gut (93). Working with defined and/or complex microbiota, it is also important to investigate the microbiota-microbiota and microbiota-host interactions. The gut microbiome is extremely dynamic owing to the presence of differences in diet, antibiotic treatment, and genetic and environmental factors. The murine intestinal microbiota has been computationally modeled using mathematical time series approaches to better understand the complex dynamics of these microbial communities (94). Therefore, mathematical modeling can be used as a tool to predict favorable interactions between the host and the microbiome network when employing antivirulence approaches such as FMT, and subsequently the hypothesis can be tested using in vivo models.
Bacteria gauge gut biogeography by sensing concentration gradients of different signals. Manipulating the concentrations and localization of these small molecules to modulate the virulence of enteric pathogens represents a futuristic approach to prevent and/or treat enteric infections. However, it is important to keep in mind that the regulation of virulence programs is sensitive to the activities of multiple signaling pathways. Thus, it will be essential to define the precise mechanisms by which these signaling pathways maximize the virulence potentials of bacterial pathogens in order to develop successful and safe antivirulence therapies for infectious diseases.
Much future research is needed to advance these strategies into therapies. A more profound understanding of the mechanisms engaged in antivirulence approaches is needed. One also needs to keep in mind how antivirulence approaches may impact resident microbial communities and what this would mean in terms of potential dysbiosis. Although it is believed that virulence approaches engender less evolutionary pressure toward drug resistance, it is likely that resistance will eventually develop, and different strategies must be constantly developed and evaluated in the background. Treatment of immunocompromised hosts with antivirulence approaches may also pose a challenge, and combinatorial strategies with current antimicrobials should also be an avenue of research. Finally, the development of antivirulence drugs is a brand-new world in drug development. It requires more refined and costly testing of drugs, far beyond the MIC test currently employed for antibiotics. The rapid evolution of microbes will always be a challenge for drug development; the key is for investigators to always think two steps ahead and to continue to engage in creative and collaborative research to combat infectious diseases.
ACKNOWLEDGMENTS
This study was supported by NIH grants AI053067, AI05135, AI077613, and AI114511.
REFERENCES
- 1.Ventola CL. 2015. The antibiotic resistance crisis: part 1: causes and threats. P T 40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 2.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spellberg B, Gilbert DN. 2014. The future of antibiotics and resistance: a tribute to a career of leadership by John Bartlett. Clin Infect Dis 59(Suppl 2):S71–S75. doi: 10.1093/cid/ciu392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 5.Sengupta S, Chattopadhyay MK, Grossart HP. 2013. The multifaceted roles of antibiotics and antibiotic resistance in nature. Front Microbiol 4:47. doi: 10.3389/fmicb.2013.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bielaszewska M, Idelevich EA, Zhang W, Bauwens A, Schaumburg F, Mellmann A, Peters G, Karch H. 2012. Effects of antibiotics on Shiga toxin 2 production and bacteriophage induction by epidemic Escherichia coli O104:H4 strain. Antimicrob Agents Chemother 56:3277–3282. doi: 10.1128/AAC.06315-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mühldorfer I, Hacker J, Keusch GT, Acheson DW, Tschäpe H, Kane AV, Ritter A, Olschläger T, Donohue-Rolfe A. 1996. Regulation of the Shiga-like toxin II operon in Escherichia coli. Infect Immun 64:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGannon CM, Fuller CA, Weiss AA. 2010. Different classes of antibiotics differentially influence Shiga toxin production. Antimicrob Agents Chemother 54:3790–3798. doi: 10.1128/AAC.01783-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimmitt PT, Harwood CR, Barer MR. 1999. Induction of type 2 Shiga toxin synthesis in Escherichia coli O157 by 4-quinolones. Lancet 353:1588–1589. doi: 10.1016/s0140-6736(99)00621-2. [DOI] [PubMed] [Google Scholar]
- 11.Kimmitt PT, Harwood CR, Barer MR. 2000. Toxin gene expression by Shiga toxin-producing Escherichia coli: the role of antibiotics and the bacterial SOS response. Emerg Infect Dis 6:458–465. doi: 10.3201/eid0605.000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. 2013. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. 2009. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A 106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donia MS, Fischbach MA. 2015. Small molecules from the human microbiota. Science 349:1254766. doi: 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee WJ, Hase K. 2014. Gut microbiota-generated metabolites in animal health and disease. Nat Chem Biol 10:416–424. doi: 10.1038/nchembio.1535. [DOI] [PubMed] [Google Scholar]
- 16.Kaper JB, O'Brien AD. 1998. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains, 1st ed ASM Press, Washington, DC. [Google Scholar]
- 17.Mundy R, Pickard D, Wilson RK, Simmons CP, Dougan G, Frankel G. 2003. Identification of a novel type IV pilus gene cluster required for gastrointestinal colonization of Citrobacter rodentium. Mol Microbiol 48:795–809. doi: 10.1046/j.1365-2958.2003.03470.x. [DOI] [PubMed] [Google Scholar]
- 18.Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, Vazquez A, Barba J, Ibarra JA, O'Donnell P, Metalnikov P, Ashman K, Lee S, Goode D, Pawson T, Finlay BB. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci U S A 101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finlay BB. 1999. Bacterial disease in diverse hosts. Cell 96:315–318. doi: 10.1016/S0092-8674(00)80544-9. [DOI] [PubMed] [Google Scholar]
- 20.Gruenheid S, Sekirov I, Thomas NA, Deng W, O'Donnell P, Goode D, Li Y, Frey EA, Brown NF, Metalnikov P, Pawson T, Ashman K, Finlay BB. 2004. Identification and characterization of NleA, a non-LEE-encoded type III translocated virulence factor of enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol 51:1233–1249. doi: 10.1046/j.1365-2958.2003.03911.x. [DOI] [PubMed] [Google Scholar]
- 21.Dougan G, Ghaem-Maghami M, Pickard D, Frankel G, Douce G, Clare S, Dunstan S, Simmons C. 2000. The immune responses to bacterial antigens encountered in vivo at mucosal surfaces. Philos Trans R Soc Lond B Biol Sci 355:705–712. doi: 10.1098/rstb.2000.0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemrajani C, Marches O, Wiles S, Girard F, Dennis A, Dziva F, Best A, Phillips AD, Berger C, Mousnier A, Crepin VF, Kruidenier L, Woodward MJ, Stevens MP, La Ragione RM, Macdonald TT, Frankel G. 2008. Role of NleH, a type III secreted effector from attaching and effacing pathogens, in colonization of the bovine, ovine and murine gut. Infect Immun 76:4804–4813. doi: 10.1128/IAI.00742-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyle EC, Bishop JL, Grassl GA, Finlay BB. 2007. Salmonella: from pathogenesis to therapeutics. J Bacteriol 189:1489–1495. doi: 10.1128/JB.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frenzen P. 2006. Data feature: a Web-based tool for calculating the cost of foodborne illness. https://www.ers.usda.gov/amber-waves/2006/june/data-feature/. Accessed 2 April 2007.
- 25.Davis MA, Hancock DD, Besser TE, Rice DH, Gay JM, Gay C, Gearhart L, DiGiacomo R. 1999. Changes in antimicrobial resistance among Salmonella enterica Serovar typhimurium isolates from humans and cattle in the Northwestern United States, 1982–1997. Emerg Infect Dis 5:802–806. doi: 10.3201/eid0506.990610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plant JE, Blackwell JM, O'Brien AD, Bradley DJ, Glynn AA. 1982. Are the Lsh and Ity disease resistance genes at one locus on mouse chromosome 1? Nature 297:510–511. doi: 10.1038/297510a0. [DOI] [PubMed] [Google Scholar]
- 27.Crump JA, Barrett TJ, Nelson JT, Angulo FJ. 2003. Reevaluating fluoroquinolone breakpoints for Salmonella enterica serotype Typhi and for non-Typhi salmonellae. Clin Infect Dis 37:75–81. doi: 10.1086/375602. [DOI] [PubMed] [Google Scholar]
- 28.Nakaya H, Yasuhara A, Yoshimura K, Oshihoi Y, Izumiya H, Watanabe H. 2003. Life-threatening infantile diarrhea from fluoroquinolone-resistant Salmonella enterica typhimurium with mutations in both gyrA and parC. Emerg Infect Dis 9:255–257. doi: 10.3201/eid0902.020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weill FX, Bertrand S, Guesnier F, Baucheron S, Cloeckaert A, Grimont PA. 2006. Ciprofloxacin-resistant Salmonella Kentucky in travelers. Emerg Infect Dis 12:1611–1612. doi: 10.3201/eid1210.060589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samrakandi MM, Zhang C, Zhang M, Nietfeldt J, Kim J, Iwen PC, Olson ME, Fey PD, Duhamel GE, Hinrichs SH, Cirillo JD, Benson AK. 2004. Genome diversity among regional populations of Francisella tularensis subspecies tularensis and Francisella tularensis subspecies holarctica isolated from the US. FEMS Microbiol Lett 237:9–17. doi: 10.1016/j.femsle.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 31.Prestinaci F, Pezzotti P, Pantosti A. 2015. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health 109:309–318. doi: 10.1179/2047773215Y.0000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akullian A, Montgomery JM, John-Stewart G, Miller SI, Hayden HS, Radey MC, Hager KR, Verani JR, Ochieng JB, Juma J, Katieno J, Fields B, Bigogo G, Audi A, Walson J. 2018. Multi-drug resistant non-typhoidal Salmonella associated with invasive disease in western Kenya. PLoS Negl Trop Dis 12:e0006156. doi: 10.1371/journal.pntd.0006156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leffler DA, Lamont JT. 2015. Clostridium difficile infection. N Engl J Med 372:1539–1548. doi: 10.1056/NEJMra1403772. [DOI] [PubMed] [Google Scholar]
- 34.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. 2006. The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci U S A 103:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curtis MM, Russell R, Moreira CG, Adebesin AM, Wang C, Williams NS, Taussig R, Stewart D, Zimmern P, Lu B, Prasad RN, Zhu C, Rasko DA, Huntley JF, Falck JR, Sperandio V. 2014. QseC inhibitors as an antivirulence approach for Gram-negative pathogens. mBio 5:e02165. doi: 10.1128/mBio.02165-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asano Y, Hiramoto T, Nishino R, Aiba Y, Kimura T, Yoshihara K, Koga Y, Sudo N. 2012. Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. Am J Physiol Gastrointest Liver Physiol 303:G1288–G1295. doi: 10.1152/ajpgi.00341.2012. [DOI] [PubMed] [Google Scholar]
- 37.Freestone PP, Sandrini SM, Haigh RD, Lyte M. 2008. Microbial endocrinology: how stress influences susceptibility to infection. Trends Microbiol 16:55–64. doi: 10.1016/j.tim.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 38.Stevens MP, Frankel GM. 2014. The locus of enterocyte effacement and associated virulence factors of enterohemorrhagic Escherichia coli. Microbiol Spectr 2:EHEC-0007-2013. doi: 10.1128/microbiolspec.EHEC-0007-2013. [DOI] [PubMed] [Google Scholar]
- 39.Deng W, Li Y, Vallance BA, Finlay BB. 2001. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect Immun 69:6323–6335. doi: 10.1128/IAI.69.10.6323-6335.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A 92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jung K, Fried L, Behr S, Heermann R. 2012. Histidine kinases and response regulators in networks. Curr Opin Microbiol 15:118–124. doi: 10.1016/j.mib.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 42.Clarke MB, Sperandio V. 2005. Transcriptional regulation of flhDC by QseBC and sigma (FliA) in enterohaemorrhagic Escherichia coli. Mol Microbiol 57:1734–1749. doi: 10.1111/j.1365-2958.2005.04792.x. [DOI] [PubMed] [Google Scholar]
- 43.Rasko DA, Moreira CG, Li DR, Reading NC, Ritchie JM, Waldor MK, Williams N, Taussig R, Wei S, Roth M, Hughes DT, Huntley JF, Fina MW, Falck JR, Sperandio V. 2008. Targeting QseC signaling and virulence for antibiotic development. Science 321:1078. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rooks MG, Veiga P, Reeves AZ, Lavoie S, Yasuda K, Asano Y, Yoshihara K, Michaud M, Wardwell-Scott L, Gallini CA, Glickman JN, Sudo N, Huttenhower C, Lesser CF, Garrett WS. 2017. QseC inhibition as an antivirulence approach for colitis-associated bacteria. Proc Natl Acad Sci U S A 114:142–147. doi: 10.1073/pnas.1612836114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen A, Lupardus PJ, Gersch MM, Puri AW, Albrow VE, Garcia KC, Bogyo M. 2011. Defining an allosteric circuit in the cysteine protease domain of Clostridium difficile toxins. Nat Struct Mol Biol 18:364–371. doi: 10.1038/nsmb.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Genth H, Dreger SC, Huelsenbeck J, Just I. 2008. Clostridium difficile toxins: more than mere inhibitors of Rho proteins. Int J Biochem Cell Biol 40:592–597. doi: 10.1016/j.biocel.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 47.Bender KO, Garland M, Ferreyra JA, Hryckowian AJ, Child MA, Puri AW, Solow-Cordero DE, Higginbottom SK, Segal E, Banaei N, Shen A, Sonnenburg JL, Bogyo M. 2015. A small-molecule antivirulence agent for treating Clostridium difficile infection. Sci Transl Med 7:306ra148. doi: 10.1126/scitranslmed.aac9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pereira FC, Berry D. 2017. Microbial nutrient niches in the gut. Environ Microbiol 19:1366–1378. doi: 10.1111/1462-2920.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelly CP, Pothoulakis C, Lamont JT. 1994. Clostridium difficile colitis. N Engl J Med 330:257. doi: 10.1056/NEJM199401273300406. [DOI] [PubMed] [Google Scholar]
- 50.Bingley PJ, Harding GM. 1987. Clostridium difficile colitis following treatment with metronidazole and vancomycin. Postgrad Med J 63:993–994. doi: 10.1136/pgmj.63.745.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Czepiel J, Dróżdż M, Pituch H, Kuijper EJ, Perucki W, Mielimonka A, Goldman S, Wultańska D, Garlicki A, Biesiada G. 2019. Clostridium difficile infection: review. Eur J Clin Microbiol Infect Dis 38:1211. doi: 10.1007/s10096-019-03539-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kassam Z, Lee CH, Yuan Y, Hunt RH. 2013. Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am J Gastroenterol 108:500–508. doi: 10.1038/ajg.2013.59. [DOI] [PubMed] [Google Scholar]
- 53.Borody TJ, Brandt LJ, Paramsothy S. 2014. Therapeutic faecal microbiota transplantation: current status and future developments. Curr Opin Gastroenterol 30:97–105. doi: 10.1097/MOG.0000000000000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le Bastard Q, Ward T, Sidiropoulos D, Hillmann BM, Chun CL, Sadowsky MJ, Knights D, Montassier E. 2018. Fecal microbiota transplantation reverses antibiotic and chemotherapy-induced gut dysbiosis in mice. Sci Rep 8:6219. doi: 10.1038/s41598-018-24342-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song Y, Garg S, Girotra M, Maddox C, von Rosenvinge EC, Dutta A, Dutta S, Fricke WF. 2013. Microbiota dynamics in patients treated with fecal microbiota transplantation for recurrent Clostridium difficile infection. PLoS One 8:e81330. doi: 10.1371/journal.pone.0081330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Antharam VC, Li EC, Ishmael A, Sharma A, Mai V, Rand KH, Wang GP. 2013. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J Clin Microbiol 51:2884–2892. doi: 10.1128/JCM.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hryckowian AJ, Van Treuren W, Smits SA, Davis NM, Gardner JO, Bouley DM, Sonnenburg JL. 2018. Microbiota-accessible carbohydrates suppress Clostridium difficile infection in a murine model. Nat Microbiol 3:662–669. doi: 10.1038/s41564-018-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, Young VB, Henrissat B, Wilmes P, Stappenbeck TS, Nunez G, Martens EC. 2016. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167:1339–1353.e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Curtis MM, Hu Z, Klimko C, Narayanan S, Deberardinis R, Sperandio V. 2014. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe 16:759–769. doi: 10.1016/j.chom.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jenior ML, Leslie JL, Young VB, Schloss PD. 2018. Clostridium difficile alters the structure and metabolism of distinct cecal microbiomes during initial infection to promote sustained colonization. mSphere 3:e00261-18. doi: 10.1128/mSphere.00261-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fletcher JR, Erwin S, Lanzas C, Theriot CM. 2018. Shifts in the gut metabolome and Clostridium difficile transcriptome throughout colonization and infection in a mouse model. mSphere 3:e00089-18. doi: 10.1128/mSphere.00089-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Battaglioli E, Hale V, Chen J, Jeraldo P, Ruiz-Mojica C, Schmidt B, Rekdal V, Till L, Huq L, Smits S, Moor W, Jones-Hall Y, Smyrk T, Khanna S, Pardi D, Grover M, Patel R, Chia N, Nelson H, Sonnenburg J, Farrugia G, Kashyap P. 2018. Clostridioides difficile uses amino acids associated with gut microbial dysbiosis in a subset of patients with diarrhea. Sci Transl Med 10:eaam7019. doi: 10.1126/scitranslmed.aam7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pifer R, Russell RM, Kumar A, Curtis MM, Sperandio V. 2018. Redox, amino acid, and fatty acid metabolism intersect with bacterial virulence in the gut. Proc Natl Acad Sci U S A 115:E10712–E10719. doi: 10.1073/pnas.1813451115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. 2012. Fucose sensing regulates bacterial intestinal colonization. Nature 492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fischbach MA, Sonnenburg JL. 2011. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe 10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garsin DA. 2010. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat Rev Microbiol 8:290–295. doi: 10.1038/nrmicro2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kendall MM, Gruber CC, Parker CT, Sperandio V. 2012. Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. mBio 3:e00050-12. doi: 10.1128/mBio.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anderson CJ, Clark DE, Adli M, Kendall MM. 2015. Ethanolamine signaling promotes Salmonella niche recognition and adaptation during infection. PLoS Pathog 11:e1005278. doi: 10.1371/journal.ppat.1005278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luzader DH, Clark DE, Gonyar LA, Kendall MM. 2013. EutR is a direct regulator of genes that contribute to metabolism and virulence in enterohemorrhagic Escherichia coli O157:H7. J Bacteriol 195:4947–4953. doi: 10.1128/JB.00937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gonyar LA, Kendall MM. 2014. Ethanolamine and choline promote expression of putative and characterized fimbriae in enterohemorrhagic Escherichia coli O157:H7. Infect Immun 82:193–201. doi: 10.1128/IAI.00980-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rowley CA, Anderson CJ, Kendall MM. 2018. Ethanolamine influences human commensal Escherichia coli growth, gene expression, and competition with enterohemorrhagic E. coli O157:H7. mBio 9:e01429-18. doi: 10.1128/mBio.01429-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johnson BK, Abramovitch RB. 2017. Small molecules that sabotage bacterial virulence. Trends Pharmacol Sci 38:339–362. doi: 10.1016/j.tips.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Duncan MC, Linington RG, Auerbuch V. 2012. Chemical inhibitors of the type three secretion system: disarming bacterial pathogens. Antimicrob Agents Chemother 56:5433–5441. doi: 10.1128/AAC.00975-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Charro N, Mota LJ. 2015. Approaches targeting the type III secretion system to treat or prevent bacterial infections. Expert Opin Drug Discov 10:373–387. doi: 10.1517/17460441.2015.1019860. [DOI] [PubMed] [Google Scholar]
- 75.Fuqua C, Winans SC, Greenberg EP. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol 50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 76.Hughes DT, Sperandio V. 2008. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol 6:111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim J, Kim J, Kim Y, Oh S, Song M, Choe JH, Whang KY, Kim KH, Oh S. 2018. Influences of quorum-quenching probiotic bacteria on the gut microbial community and immune function in weaning pigs. Anim Sci J 89:412–422. doi: 10.1111/asj.12954. [DOI] [PubMed] [Google Scholar]
- 78.Thompson JA, Oliveira RA, Djukovic A, Ubeda C, Xavier KB. 2015. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep 10:1861–1871. doi: 10.1016/j.celrep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 79.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 80.Winter SE, Lopez CA, Baumler AJ. 2013. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep 14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hughes ER, Winter MG, Duerkop BA, Spiga L, Furtado de Carvalho T, Zhu W, Gillis CC, Buttner L, Smoot MP, Behrendt CL, Cherry S, Santos RL, Hooper LV, Winter SE. 2017. Microbial respiration and formate oxidation as metabolic signatures of inflammation-associated dysbiosis. Cell Host Microbe 21:208–219. doi: 10.1016/j.chom.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu W, Winter MG, Byndloss MX, Spiga L, Duerkop BA, Hughes ER, Büttner L, de Lima Romão E, Behrendt CL, Lopez CA, Sifuentes-Dominguez L, Huff-Hardy K, Wilson RP, Gillis CC, Tükel Ç, Koh AY, Burstein E, Hooper LV, Bäumler AJ, Winter SE. 2018. Precision editing of the gut microbiota ameliorates colitis. Nature 553:208–211. doi: 10.1038/nature25172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cummings JH. 1983. Fermentation in the human large intestine: evidence and implications for health. Lancet 321:1206. doi: 10.1016/S0140-6736(83)92478-9. [DOI] [PubMed] [Google Scholar]
- 84.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. 2013. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kamada N, Chen GY, Inohara N, Nunez G. 2013. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lawley TD, Walker AW. 2013. Intestinal colonization resistance. Immunology 138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jacobson A, Lam L, Rajendram M, Tamburini F, Honeycutt J, Pham T, Van Treuren W, Pruss K, Stabler SR, Lugo K, Bouley DM, Vilches-Moure JG, Smith M, Sonnenburg JL, Bhatt AS, Huang KC, Monack D. 2018. A gut commensal-produced metabolite mediates colonization resistance to salmonella infection. Cell Host Microbe 24:296–307.e7. doi: 10.1016/j.chom.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.De la Cruz MA, Morgan JK, Ares MA, Yáñez-Santos JA, Riordan JT, Girón JA. 2016. The two-component system CpxRA negatively regulates the locus of enterocyte effacement of enterohemorrhagic Escherichia coli involving sigma(32) and Lon protease. Front Cell Infect Microbiol 6:11. doi: 10.3389/fcimb.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kumar A, Sperandio V. 2019. Indole signaling at the host-microbiota-pathogen interface. mBio 10:e01031-19. doi: 10.1128/mBio.01031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rivera-Chavez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, Xu G, Velazquez EM, Lebrilla CB, Winter SE, Baumler AJ. 2016. Depletion of butyrate-producing clostridia from the gut microbiota drives an aerobic luminal expansion of salmonella. Cell Host Microbe 19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lopez CA, Miller BM, Rivera-Chavez F, Velazquez EM, Byndloss MX, Chavez-Arroyo A, Lokken KL, Tsolis RM, Winter SE, Baumler AJ. 2016. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science 353:1249. doi: 10.1126/science.aag3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smanski MJ, Zhou H, Claesen J, Shen B, Fischbach MA, Voigt CA. 2016. Synthetic biology to access and expand nature's chemical diversity. Nat Rev Microbiol 14:135–149. doi: 10.1038/nrmicro.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ericsson AC, Franklin CL. 2015. Manipulating the gut microbiota: methods and challenges. ILAR J 56:205–217. doi: 10.1093/ilar/ilv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marino S, Baxter NT, Huffnagle GB, Petrosino JF, Schloss PD. 2014. Mathematical modeling of primary succession of murine intestinal microbiota. Proc Natl Acad Sci U S A 111:439–444. doi: 10.1073/pnas.1311322111. [DOI] [PMC free article] [PubMed] [Google Scholar]