Abstract
Background
Postoperative cognitive dysfunction (POCD) affects a large number of post-surgery patients, especially for the elderly. However, the etiology of this neurocognitive disorder is largely unknown. Even if several studies have reported a small number of miRNAs as the essential modulatory factors in POCD, these findings are still rather limited. The aim of current study was to screen the POCD-related miRNAs in the hippocampus tissues and investigate the target genes of differentially expressed miRNAs and their biological functions underlying POCD pathophysiology.
Methods
The miRNA microarray was used to find the abnormal expression of miRNAs in the hippocampus tissues from the POCD model mice to normal mice (Discovery cohort, 3 vs 3). The nominal significant results were validated in an independent sample of hippocampus tissues of 10 mice based on same miRNA microarray (Replication cohort, 5 vs 5). Expression level of the most significantly abnormal miRNA was further validated by real-time quantitative polymerase chain reaction (PCR). To determine the expression pattern among miRNAs and genes and detect the interactions, we conducted a weighted gene co-expression network analysis (WGCNA) in the miRNAs and genes expression data from hippocampus tissue of wild type mice (n = 24). The target genes of miRNAs were predicted using the miRWalk3.0 software. Furthermore, we used the ClueGO software to decipher the pathways network and reveal the biological functions of target genes of miRNAs.
Results
We found that nine miRNAs showed significant associations with POCD in both datasets. Among these miRNAs, mmu-miR-190a-3p was the most significant one. By performing WGCNA analysis, we found 25 co-expression modules, of which mmu-miR-190a-3p was significantly anti-correlated with red module. Moreover, in the red module, 314 genes were significantly enriched in four pathways such as axon guidance and calcium signaling pathway, which are well-documented to be associated with psychiatric disorders and brain development. Also, 169 of the 314 genes were highly correlated with mmu-miR-190a-3p, and four genes (Sphkap, Arhgef25, Tiam1, and Ntrk3) had putative binding sites at 3′-UTR of mmu-miR-190a-3p. Based on protein-protein network analysis, we detected that Tiam1 was a central gene regulated by the mmu-miR-190a-3p.
Conclusions
Taken together, we conclude that mmu-miR-190a-3p is involved in the etiology of POCD and may serve as a novel predictive indicator for POCD.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-6035-0) contains supplementary material, which is available to authorized users.
Keywords: POCD, miR-190a, Neurocognitive disorders, Brain, Animal model
Background
Postoperative cognitive dysfunction (POCD), a commonly-seen postoperative complication especially for elderly patients (age > 65-year-old), is characterized by cognitive impairments in patients underwent major surgery, and associated with high morbidity and mortality [1]. It covers a wide range of cognitive dysfunctions including short or long-term memory loss and attention deficit [2]. POCD was reported to occur in 25.8% of the patients seven days post-surgery, and in 12.7% of the patients 3 months post surgery [3–5]. This indicates that when the early postoperative neurocognitive disorders happen, the human body simultaneously initiates the self-repair mechanism for improving or restoring the cognitive function [6]. However, the mechanisms remain to be fully elucidated. Since POCD often leads to prolonged hospital stays, decreased quality of life, and increased social dependence as well as medical expenses [3], it is important to elucidate its underlying molecular mechanisms for the prevention, diagnosis, and treatment of POCD.
MicroRNAs (miRNAs), which are extensively distributed in eukaryotes, regulate expression of genes by 1) combining with 3-UTR of target gene to inhibit the translation or to degrade target mRNAs at the post-transcription level; and 2) interacting with other target non-coding RNAs, e.g. long non-coding RNA (lncRNA) or circular RNA (circRNA) [7, 8]. A great number of miRNAs enriched in human brain has been shown to express in a developmental stage-specific, tissue-specific, and cell-specific pattern [9] and play crucial roles in development of the neural system and in cognitive process such as learning and memorizing [10, 11]. Recently, multiple lines of evidence have demonstrated that miRNAs are involved in the pathogenesis of several neuropsychiatric diseases [11–15]. For example, based on genome-wide transcription sequencing, Liu et al. demonstrated that miRNAs and lncRNAs were important contributors in the pathogenesis of schizophrenia [14]. Furthermore, multiple aberrantly expressed miRNAs were implicated in the development of Huntington’s disease [13], neural tube defects [16], and Alzheimer’s disease [17]. As for POCD, several miRNAs have been reported to be associated with it [6, 18–20], such as, Yu et al. [6] reported that miR-572 was implicated in the development and restoration of POCD and might serve as a biological marker in early diagnosis of POCD. However, the specific roles and underlying mechanism of miRNAs in POCD are still not yet clear.
Hence, in this study, by focusing on the biology of the dysregulated miRNAs in POCD, we conducted a comprehensive analysis of miRNAs expression profiles in hippocampus tissue of POCD model mice, and made further investigation of the miRNAs’ potential interaction mechanisms with target genes. Our findings not only extend the understanding of miRNAs’ role in the pathogenesis of POCD but also facilitate the identification of the miRNA biomarkers for improving the prognosis of POCD patients.
Methods
Animals
Healthy aged male C57BL/6 mice (n = 22, aged 18 months, weights from 40.2 g to 46.6 g) were purchased from SiBeiFu Experimental Animal Science and Technology Co. Ltd. (Beijing, China. Permit Number: SCXK (Jing) 2016–0002). The mice were individually housed in an air-conditioned room with a temperature of 24 ± 4 °C and 55–65% humidity, under a standard 12 h–12 h light-dark cycle (lights on 6 AM to 6 PM), and had free access to standard food and water. The mice were acclimatized for 1 week before the experiment. The protocol of animal experiment was approved by the Animal Care Committee of the Chinese People’s Liberation Army General Hospital (Beijing, China).
According to the protocol of our research work, all mice were euthanized before we were prepared to obtain hippocampal tissue. When those mice scheduled for obtaining the hippocampal tissue, one by one, they were put into an anesthetic introduction chamber and were anesthetized with isoflurane into a deeper level. The introduction chamber was kept clean to minimize the odor that might distress animals subsequently anesthetized. A rodent anesthesia machine were used (Model: ABM09–002, Reward, Shenzhen) and the anesthetic used was isoflurane. The concentration of the vaporizer was set at 3% and the oxygen flow rate was set at 3 L/min during anesthesia. All mice were deeply anesthetized based on following signs: the slowed rising and falling of chest, no respond to toe pinch, and corneal reflex disappeared. Then, the mice were decapitated by using a guillotine in a uniformly instantaneous manner. The brain was instantly dissected on ice, and the hippocampal tissue was obtained and put into liquid nitrogen. All the mice were decapitated 24 h after operation. The detailed time line of the euthanization of study animals is shown in the Additional file 2: Figure S1.
Tissue sampling
Mice were numbered by weight and randomly divided into two groups: Surgery and Sham, with 11 mice per group. The mice in the Surgery group were exposed to abdominal surgery under local bupivacaine anesthesia (according to the protocol from Xu et al. [21]), whereas those in the Sham group did not suffer from the anesthesia and surgery. At 24 h after the surgery, three mice were randomly selected from each group and sacrificed. The hippocampus tissue was removed immediately and stored in the sterile tube (RNase Free) in liquid nitrogen.
Morris water maze (MWM)
The MWM test, as a hippocampal-dependent test, was applied to evaluate the spatial learning, spatial memory and cognitive flexibility for the mice [22]. The cognitive function of remaining 8 mice in each group were assessed by the MWM experiment. The water maze was a black circular tank (120 cm in diameter and 50 cm in depth) and filled with water of 22 ± 1 °C to a depth of 35 cm. Several visual objects were installed above the pool to help mice identify the direction. The maze was divided into four quadrants, an invisible platform (10 cm in diameter) was placed 1.5 cm blew the water surface in the first quadrant (target quadrant). The whole experiment was performed under a dark and quite environment.
During the experiment, mice were released into the water facing the wall of the tank from one of the four quadrants. The mice were trained to find the hidden platform and climb onto it within 60 s. The animals were allowed to stay on the platform for at least 10 s after each trial. If the mice were unable to find the platform in 60 s, it was then placed on the platform for 10 s. After that, the mice were put back to the cage and the second mouse was tested on trial 1. This rotation was repeated until all animals completed trial 1. Subsequently, the process was repeated for subsequent trials until 4 trials completed per day for 5 consecutive days. At 6th day, the platform was removed, and the mice were sent to evaluate their reference memory by being released from the third quadrant. The swimming speed, platform-site crossing numbers, dwelling time in the target quadrant and the escape latency were recorded.
RNA extraction
Total miRNAs were extracted from the hippocampal tissue (n = 3 from each group) by the miRcutemiRNA kit (TIANGEN, DP501). The nanodrop was utilized to determine the miRNA concentration of each sample according to the optical absorption at 260 nm and the gel electrophoresis was used to detect the miRNA integrity.
Microarray and statistical analysis
MiRNA microarray (Affymetrix miRNA 4.0) was conducted by PREMEDICAL Co. Ltd. (Beijing, China). The microarray was utilized to find the aberrant expression of miRNAs from the POCD model mice to normal mice. Fluorescent signals were transformed from picture signal to digital data based on the degree of fluorescent for each probe, and then the data were saved as. DAT files by AGCC software (Affymetrix Genechip Command Console Software). The differential expression analyses were performed by using Transcriptome Analysis Console (v 4.0) and using an FDR correction for multiple testing. Considering that none of the identified miRNAs reached the threshold of adjusted significance, we chose the threshold of a nominal P value of < 0.05 for further replication.
The miRNA expression profile data of replication cohort were downloaded from the Gene Expression Omnibus (GEO) database (Accession number: GSE95070; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE95070), which were deposited by Wei et al. [19] from the Chaoyang Hospital. With regard to this dataset, hippocampus tissues of 10 mice (5 per group) were dissected, and the different miRNA expression levels between two groups were detected by Affymetrix miRNA 4.0 as well. Similarly, the differential expression analysis was performed by using the Transcriptome Analysis Console (v 4.0) with same parameter.
Quantitative real-time PCR
Expression level of the most significantly aberrant miRNA, mmu-miR-190a-3p, was validated by using real-time PCR assay. Reverse transcription reaction was performed with M-MLV Reverse Transcriptase kit (Takara Code: D2639A) based on the manufacturers’ protocol. Real-time PCR was performed with SYBR Premix Ex Taq kit (Takara Code: DRR041A). The miRNA expression level was evaluated relative to the expression of U6 of the 2 -ΔΔCt. The primers for miRNA mmu-miR-190a-3p are listed in Additional file 1: Table S1.
Statistical analysis
Data were analyzed using GraphPad PRISM (version 6; GraphPad Prism Software, Inc. San Diego, CA, USA). Measurements of dwelling time, number of grossing, escape latency, and speed in MWM test among preoperative and postoperative mice were analyzed by using Student’s t-test. qRT-PCR data were also analyzed with Student’s t-test. A P value of < 0.05 was considered statistically significant.
WGCNA analysis
The genes and miRNAs expression data of GSE73507 were acquired from GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73507). The GSE73507 dataset was designed to gain insight into the relationship between the CAG repeat length and the Huntington disease [23]. By excluding the expression data from other brain regions and mutant mice, we only downloaded the mRNAs and miRNAs expression data from hippocampus tissue of wild type mice for our analysis (n = 24). Because of recent progress of alignment and mapping approaches, which are capable of detecting the transcriptome profiles in a more accurate and effective way compared with former tools [24]. The RNA and miRNA sequencing data were aligned and mapped to the GRCm38 version of mice genome using Hisat2 (v 2.1.0) and StringTie (v 1.3.4) [25], and the miRNA expression data by miRDeep2 (v 2.0.0.7) [26]. A total of 16,425 mRNAs and 1057 miRNAs were correctly mapped onto the mouse genome. Threshold for filtering out genes expressed at low levels was set to greater than 1 of the average fpkm. After the filtering process, 13,241 mRNAs and 546 miRNAs were included for WGCNA analysis.
The R package of WGCNA was used to construct the network modules of highly correlated transcripts subsets [27]. This approach aims to find the gene pairs with similar expression patterns and highly topological overlap, and it represents a valuable tool for identifying promising target genes and understanding the pathology of complex disorders [28]. In order to provide a comprehensive expression pattern among the mRNAs and miRNAs and detect the interaction of the transcripts, we performed our co-expression analysis by combined the mRNA and miRNA dataset together. First, we constructed a weighted network according to the gene pair correlations among all the mRNAs and miRNAs; second, by using the default parameters to assess the network interconnection, 25 specific modules were hierarchically clustered. These module sizes were from 50 to 17,500 genes. In the network we only showed a connection of the corresponding topological overlap is above a threshold of 0.05 with mmu-miR-190a-3p in the red module (n = 169). The visual gene-gene network plot was displayed by using the Cytoscape version 3.5.1 (https://www.cytoscape.org/) [29].
Concordance between the highly correlated mRNAs with mmu-miR-190a-3p and those not involved in the module genes were assayed by performing density plots. We compared the distribution of Pearson correlation coefficients of the 169 potential interaction targets of mmu-miR-190a-3p to a control distribution of non-predicted targets which consisted of all other mRNAs that we mapped. The mRNAs which were possibly modulated by mmu-miR-190a-3p displayed more significant negative correlation compared with the control.
Pathway analysis
ClueGO (v. 2.3.4), a plug-in software of Cytoscape, was used to decipher the pathways network and determine their biological functions for the candidate genes [30]. The potential biological functions of each gene set were annotated using the pathway profiles of Kyoto Encyclopedia of Genes and Genomes (KEGG) [31].
Target prediction
We conducted further analysis to screen the most potential regulated genes by mmu-miR-190a-3p. We first employed the miRWalk3.0 (http://mirwalk.umm.uni-heidelberg.de/) to predict the target genes regulated by mmu-miR-190a-3p [32]. By using a stringent standard to obtain reliable targets (Additional file 1: Table S2), we set the parameters for target prediction in the miRwalk3.0 as following: 1) Binding over than 0.9; 2) Energy less than − 16; and 3) Accessibility less than 0.05. Second, using these predicted targets for mmu-miR-190a-3p to overlap with the WGCNA results of those highly correlated genes. Finally, we conducted a protein-protein interaction network analysis for those overlapped genes according to the STRING v 10.5 under default parameters (https://string-db.org/cgi/input.pl). By combining the gene-gene interaction result, prediction targets and the protein-protein interaction analysis, we attempted to find the high degree receivable genes regulated by mmu-miR-190a-3p.
Results
Morris water maze test
Our MWM test showed that the dwelling time (p = 0.002) and the number of crossings (p = 0.049) of mice in Surgery group were significantly decreased than those in Sham group (Table 1). The swimming speed did not differ between the two groups (p = 0.660), however, the escape latency increased significantly in the Surgery group (p < 0.001). These results demonstrated that the lower ability to find the platform was not a result from the motor ability, but derived from the spatial learning memory impairment and the cognitive impairment. This indicates that the spatial learning ability of the mice in the Surgery group was impaired notably and a POCD mouse model has been developed.
Table 1.
Results of Morris Water Maze test among preoperative and postoperative mice
| Sample ID | Dwelling Time | Number of Crossing | Escape Latency | Speed |
|---|---|---|---|---|
| CTRL1 | 33.1 | 1.0 | 36.3 | 0.2 |
| CTRL2 | 16.9 | 2.0 | 36.0 | 0.2 |
| CTRL3 | 26.3 | 1.0 | 60.0 | 0.2 |
| CTRL4 | 23.5 | 2.0 | 60.0 | 0.2 |
| CTRL5 | 36.3 | 3.0 | 47.9 | 0.2 |
| CTRL6 | 16.2 | 3.0 | 31.9 | 2.1 |
| CTRL7 | 25.9 | 2.0 | 46.9 | 0.2 |
| CTRL8 | 26.3 | 2.0 | 43.1 | 0.2 |
| CASE1 | 26.0 | 2.0 | 60.0 | 0.3 |
| CASE2 | 15.4 | 2.0 | 55.0 | 0.2 |
| CASE3 | 13.7 | 1.0 | 60.0 | 0.2 |
| CASE4 | 4.8 | 0.0 | 60.0 | 1.3 |
| CASE5 | 30.6 | 2.0 | 60.0 | 0.2 |
| CASE6 | 12.3 | 1.0 | 60.0 | 0.2 |
| CASE7 | 17.1 | 1.0 | 56.3 | 0.2 |
| CASE8 | 18.0 | 1.0 | 53.6 | 0.2 |
| p Value | 0.044 | 0.0596 | 0.005 | 0.754 |
Note: CTRL represents preoperative mice, CASE represents postoperative mice
Differential expression analysis
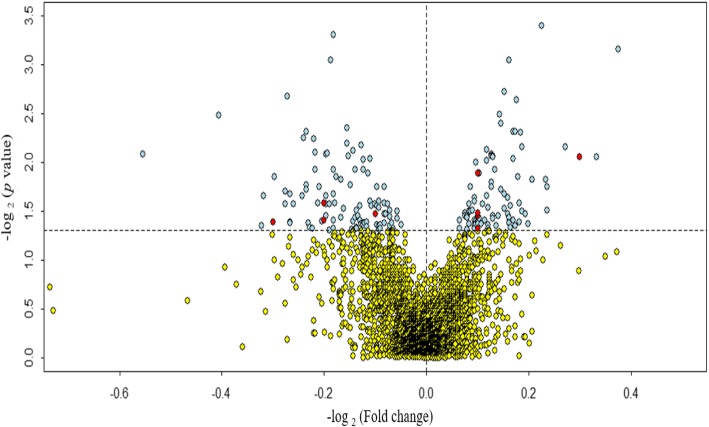
In order to assess the effects of aberrantly expressed miRNAs in hippocampus tissue of POCD mice brain, we performed differential miRNAs expression analysis of six samples. A total of 3162 mice miRNAs have been mapped. Compared with the mice in the Sham group, the microarray analysis results showed that 83 miRNAs were up-regulation and 103 miRNAs were down-regulation (p < 0.05) (Fig. 1).
Fig. 1.
Volcano plot of miRNA expression in discovery cohort. Plotted along the x-axis is the mean of log2 fold-change, along the y-axis the negative logarithm of the log2 p values. Light blue demonstrates the miRNAs with significant p value (p < 0.05), and red denotes these significant miRNAs validated by the replication cohort. The horizontal line represents the threshold of significant p value
Further, we performed a replication analysis with the same procedure by using a published miRNA microarray dataset (i.e., GSE95070 dataset). In this miRNAs expression profile, we detected 85 significantly up-regulation miRNAs and 103 significantly down-regulation miRNAs (p < 0.05) (Additional file 2: Figure S2). By comparing the analysis results from these two datasets, we found that five up-regulation miRNAs and four down-regulation miRNAs, which were significantly changed in both datasets with the same direction (Table 2).
Table 2.
The replicated differential expressed miRNAs in hippocampus tissues
| miRNA | Chr | Start | End | Discovery Cohort | Replication Cohort | ||
|---|---|---|---|---|---|---|---|
| Log2 (FC) | p Value | Log2 (FC) | p Value | ||||
| mmu-miR-190a-3p | chr9 | 67,236,664 | 67,236,685 | 2.4 | 7.0E-04 | 1.9 | 2.4E-03 |
| mmu-mir-7648 | chr15 | 90,224,360 | 90,224,412 | 1.3 | 1.3E-02 | 1.1 | 2.3E-02 |
| mmu-mir-1907 | chr15 | 50,889,025 | 50,889,114 | 1.2 | 3.3E-02 | 1.4 | 2.1E-02 |
| mmu-miR-184-5p | chr9 | 89,802,302 | 89,802,323 | 1.2 | 3.5E-02 | 1.2 | 3.9E-02 |
| mmu-miR-6999-5p | chr2 | 91,944,908 | 91,944,930 | 1.2 | 4.8E-02 | 1.3 | 3.6E-02 |
| mmu-miR-496a-3p | chr12 | 109,739,165 | 109,739,186 | −1.5 | 2.6E-02 | −1.1 | 3.3E-02 |
| mmu-miR-592-3p | chr6 | 27,936,672 | 27,936,693 | −1.9 | 4.1E-02 | −2.3 | 6.6E-05 |
| mmu-miR-6389 | chr7 | 57,581,070 | 57,581,090 | −1.5 | 3.9E-02 | −1.2 | 5.0E-02 |
| mmu-miR-6939-5p | chr12 | 112,659,327 | 112,659,347 | −1.2 | 3.4E-02 | −1.4 | 4.0E-02 |
Notes: 1) Chr: Chromosome; 2) FC: Fold Change
Among of these replicated targets, mmu-miR-190a-3p was the most significantly one in the Surgery group (i.e., POCD model mice) compared to the mice of the Sham group. For mmu-miR-190a-3p, it was significantly increased in the mice with POCD (Discovery sample: log2(FC) = 2.4, p = 7.0 × 10− 4; Replication sample: log2(FC) = 1.9, p = 2.4 × 10− 3). Real-time quantitative PCR was performed to technically validate the expression pattern of this miRNA in our discovery sample. Consistently, the result showed that the mmu-miR-190a-3p was significantly increased in the hippocampus tissue of mice with POCD (log2(FC) = 1.8, p = 0.041; see Additional file 2: Figure S3).
Co-expression network analysis
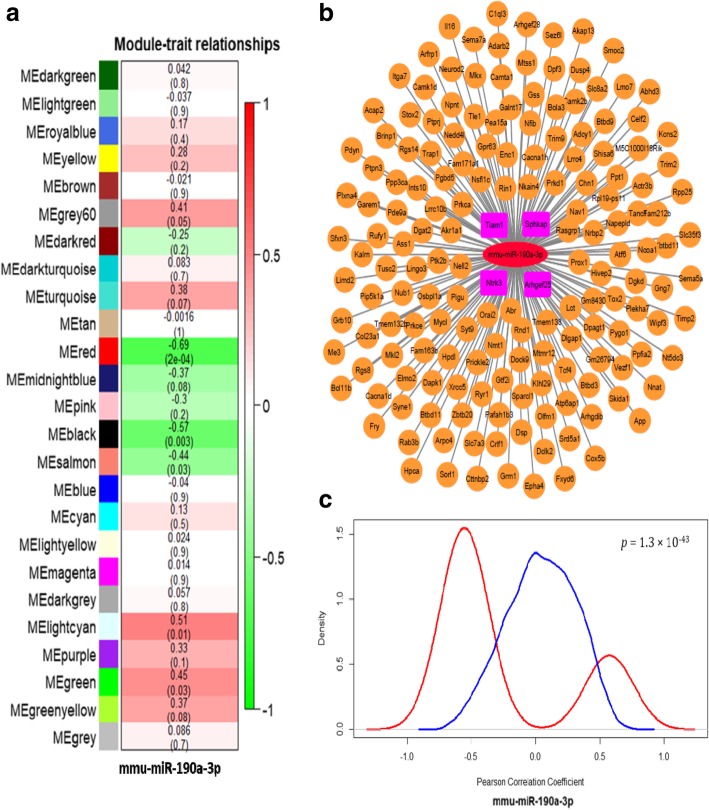
To determine the relevant POCD pathways modulated by target miRNAs, we investigated the orchestrating of the transcriptome in the hippocampus of mice brain by performing weighted gene co-expression network analysis to the 24 specimens as described in Materials and Methods. We detected 25 modules with highly significant co-expression patterns (Additional file 2: Figure S4). Because the mmu-miR-190a-3p was significantly highly expressed in the POCD model mice brain and one of the most significant miRNA in two samples (Table 2), we paid special attention on this miRNA in this communication. In addition, as showed in the Fig. 2a, mmu-miR-190a-3p was significantly inverse-correlated with the red module.
Fig. 2.
The mmu-miR-190a-3p involved module in the hippocampal tissue. (a) Clustering dendrograms of genes according to the topological overlap to distinguish the subgroup and used same color to assign highly correlated modules. As the result, 25 co-expression modules were constructed and painted with different colors. Pearson correlation analysis was used to determine whether co-expression modules are correlated with mmu-miR-190a-3p. Red color indicates the module was positively correlated with mmu-miR-190a-3p, and green demonstrates negative correlation. (b) Overview of candidate genes modified by the mmu-miR-190a-3p. Nodes are defined as the target genes modulated by the mmu-miR-190a-3p which was representing by red ellipse. The 4 pink rectangular represented the prediction results from miRwalk3.0 databases. (c) Distribution of Pearson correlation coefficients of predicted mmu-miR-190a-30-mRNA pairs. We determined the correlation between the expression of that miRNA and the expression of a predicted mRNA targets in all samples by calculating a Pearson correlation coefficient. This process was repeated for all targets and the red line indicated the distribution of these coefficients as a density plot. The blue line as the control curve represented the distribution of the coefficient of others unrelated mRNA with mmu-miR-190a-3p. A lift shift of the red line comparing with the red line demonstrated that the potential targets for mmu-miR-190a-3p are preferentially more negative correlated with targets gene than random
A total of 314 genes and 5 miRNAs were grouped in the red module (Additional file 1: Table S3). Further, by conducting the pathway analysis of the genes in red module, we found four significant enrichment pathways (Fig. 3). The most significant pathway was axon guidance (p = 1.9 × 10− 10), and the second to fourth pathways were calcium signaling pathway (p = 7.7 × 10− 08), Fc gamma R-mediated phagocytosis (p = 1.2 × 10− 2), and Fat digestion and absorption (p = 5.0 × 10− 02), respectively. Among these four significant pathways, the axon guidance, calcium signaling pathway and Fc gamma R-mediated phagocytosis have been widely identified to be related with the psychiatric disorders and brain development [33–38].
Fig. 3.
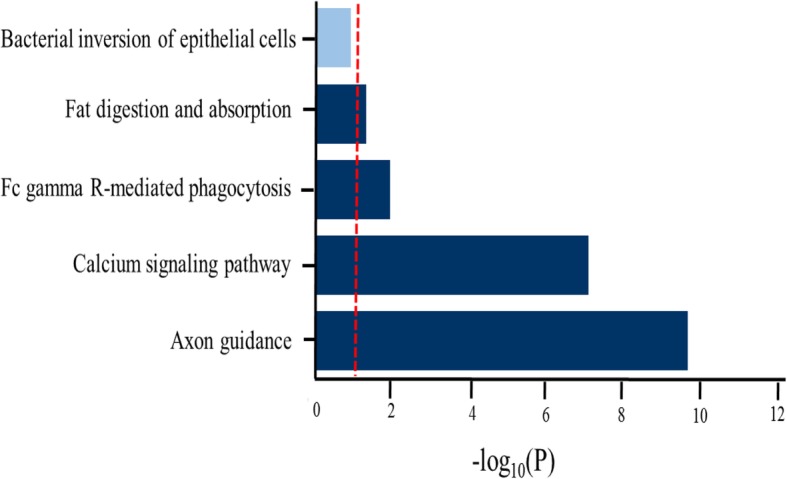
KEGG pathway enrichment analysis for genes in red module. The red vertical line indicates the threshold of the significant pathway enrichment p value after FDR correction. Dark blue color of horizontal bars represents the significantly enriched pathway, and the light blue color represents the non-significantly enriched pathway
In addition, we refined 169 genes from red module with a highly connected with mmu-miR-190a-3p (Fig. 2b). To determine whether these highly correlated genes have critical roles in psychiatric disorders, we performed similar pathway-based enrichment analysis of these 169 genes and found several significant enriched pathways (Additional file 2: Figure S5), which was in line with the results based on all genes in red module. These findings indicated that the mmu-miR-190a-3p acted as a key regulator to red module genes, which might be involved in the pathology of psychiatry diseases. Since miRNAs negatively altered the expression of its target genes [39], we expect a left shift in the predicted target genes distribution toward the negative correlations compared with the background. To explore this possibility, we compared the Pearson correlation coefficients for mmu-miR-190a-3p with its predicted targets. Indeed, we found a significantly left shift of these target genes compared with randomly selected non-predicted targets (p = 1.3 × 10− 43; Fig. 2c).
As a result, we found that 4 of 169 correlated genes (Sphkap, Arhgef25, Tiam1, and Ntrk3) involved putative binding sites at 3′-UTR and CDS regions of mmu-miR-190a-3p (Additional file 1: Table S2). Further, we performed correlation analysis with mmu-miR-190a-3p and 4 targets genes. As shown in Fig. 4, the mmu-miR-190a-3p was significantly negative correlated with the expression levels of Sphkap (R2 = 0.35, p = 1.3 × 10− 3), Tiam1(R2 = 0.19, p = 2.03 × 10− 2) and Ntrk3 (R2 = 0.43, p = 2.9 × 10− 4), respectively. As for Arhgef25, we observed a marginally significant correlation with the mmu-miR-190a-3p (R2 = 0.12, p = 5.2 × 10− 2). Importantly, as showed in Fig. 5, the evidence from protein-protein interaction analysis displayed that Tiam1 as a hub gene was possibly dysregulated by mmu-miR-190a-3p.
Fig. 4.
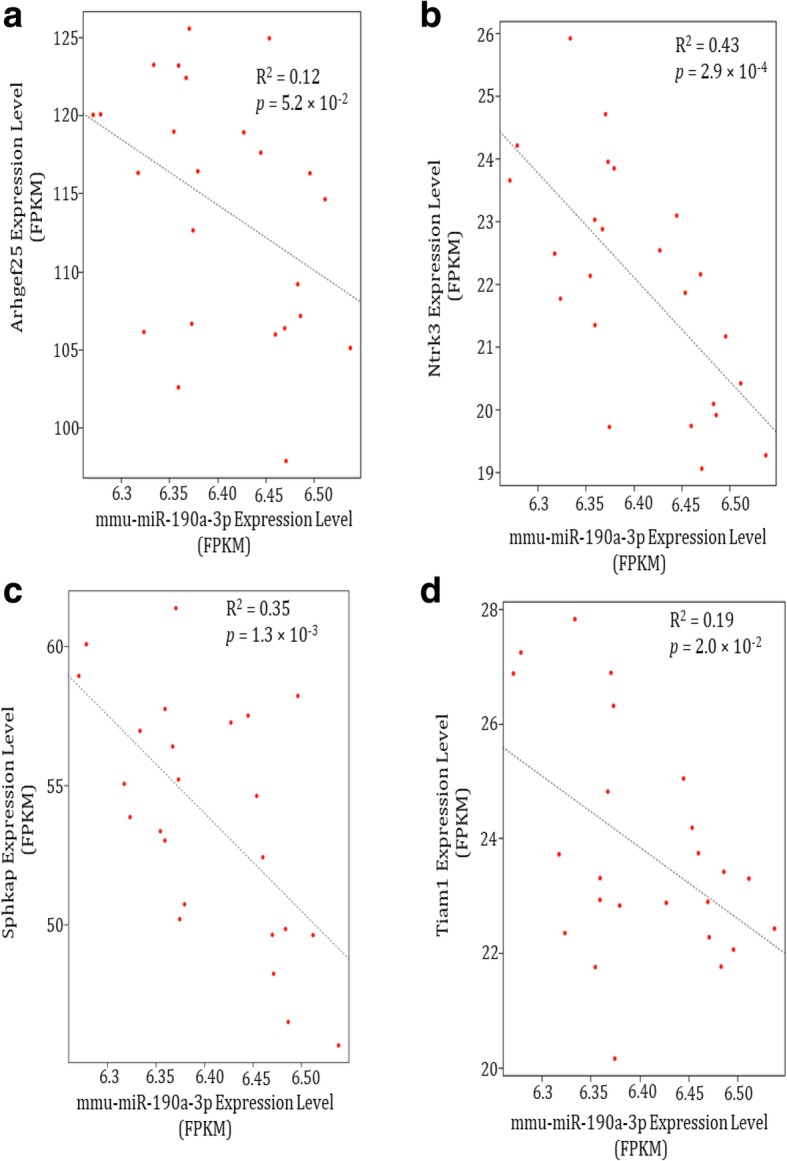
Prediction binding targets show anti-correlation with mmu-miR-190a-3p. The target mRNAs expression level (FPKM) of: a) Arhgef25; b) Ntrk3; c) Sphkap and d) Tiam1 are inversely correlated with the expression level of mmu-miR-190a-3p
Fig. 5.
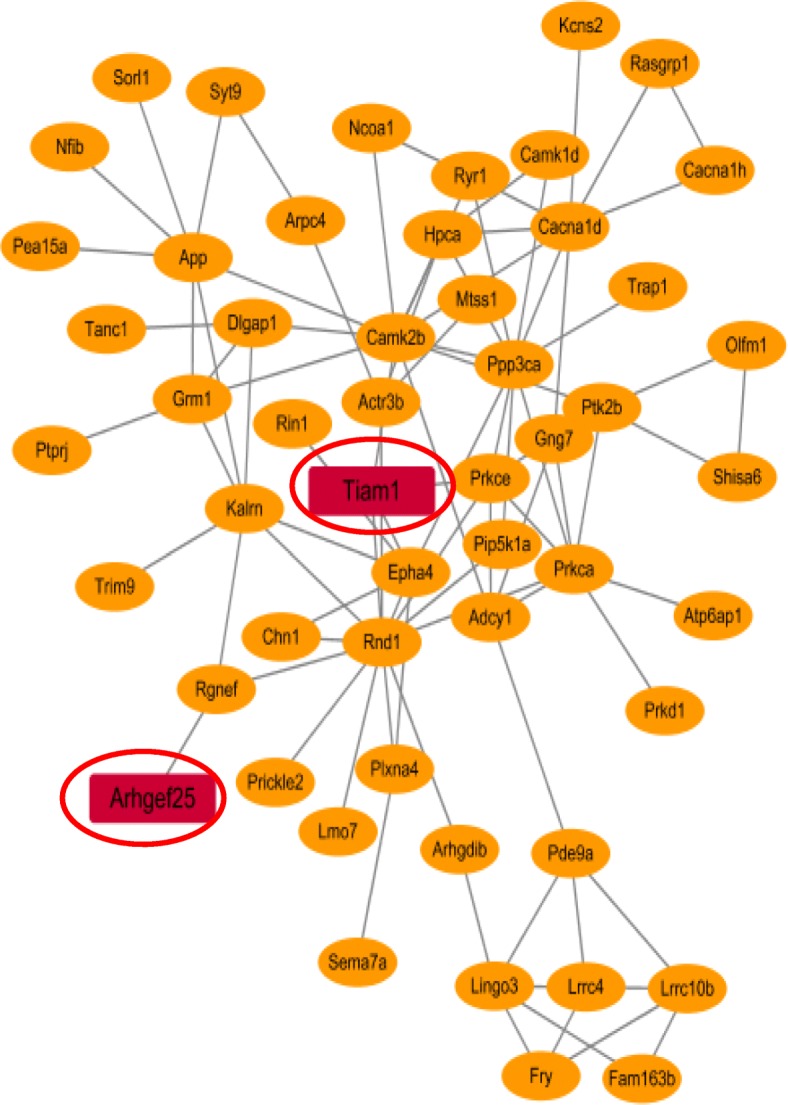
Protein-protein interaction network for mmu-miR-190a-3p potential regulated genes in mice hippocampus tissue. The genes overlapped with prediction targets by mmu-miR-190a-3p were highlighted as red rectangular
Discussion
In this study, we examined dysregulated miRNAs in the hippocampus of POCD mice and further analyzed the potential mechanisms of the target miRNAs in this neurocognitive disorder. Firstly, based on two independent miRNA microarray datasets, we identified 9 significant dysregulated miRNAs in the hippocampus tissues of POCD mice. Among of these identified miRNAs, mmu-miR-190a-3p was the most significantly differential expressed one in both datasets, and this had been validated by using the qRT-PCR. Secondly, we used a sequence-based dataset of both mRNA and miRNA data to perform co-expression analysis, which revealed 25 co-expression modules. Interestingly, the co-expression red module including mmu-miR-190a-3p was of significant enrichment in the neuron-related pathways and immunology pathways, which have been well-documented to play crucial roles in the psychiatry diseases [33–38]. Finally, by using the database of miRwalk3.0 to conduct miRNA targets prediction analysis, we found Tiam1, a Rac-specific guanine nucleotide exchange factor (GEF) required high level expression in development of spine and synapse, might be repressed by mmu-miR-190a-3p in the POCD patients.
Previous studies have reported that several miRNAs were significantly associated with POCD [6, 18–20], which provide supportive evidence of the important role of miRNAs in POCD. Consistently, some of those reported miRNAs were replicated in our current study. For example, Chen and colleagues [20] reported that mmu-miR-146a showed increased expression in the BV-2 microglial cells stimulated with LPS and hippocampal tissues of mice with POCD. For the mmu-miR-146a, we observed that it was marginally significantly associated with POCD (Log2 (FC) = 1.1; P = 0.054). In addition, Shan et al. illuminated that the expression of mmu-miR-30a was significantly decreased in the mPFC and hippocampus of aged rats compared with control and was involved in the expression regulation of several innate immune system related genes [40]. Such a finding was validated by our study, where we found that the mmu-miR-30a decreased the expression level in the POCD model mice compared with control (Log2 (FC) = − 1.2; P = 0.046).
Furthermore, we also performed an integrative analysis to refine a collection of putative mRNA-miRNA interaction pairs for mmu-miR-190a-3p. By applying to a powerful approach, which was combining expression data of miRNAs and mRNAs while performing the co-expression analysis based on Pearson correlation, we provided a new clue to detect the biological network relationship between miRNA and mRNA [41–44]. Because the primary function of miRNAs was to inhibit expression of target genes [39], it was reasonable to consider that there were more significantly negative correlation coefficient between mmu-miR-190a-3p and its target genes than the background genes. Consistently, we found that the target genes of mmu-miR-190a-3p were significantly shifted to negative part whereas the background genes were always presented a normal distribution. According to the prediction results from miRwalk3.0, we filtered out the genes with no binding sites to mmu-miR-190a-3p, which lead to the identification of likely four target genes for mmu-miR-190a-3p. Also, based on the protein-protein interaction analysis, two of four putative genes, Tiam1 and Arhgef25, were involved in the interaction network. Interestingly, Tiam1, as a hub gene located on the central of network map, might be modified by abnormal expression of mmu-miR-190a-3p and played important roles in the pathogenesis of POCD.
The majority of excitatory synapses existed in micron-sized dendritic protrusions of the mammalian brain, which can change along with the variation of synaptic activity in terms of the alternation in number and shape [45]. Tiam1 is widely expressed in the developing central nervous system [46]. Tiam1 is one of the important Rac-specific GEFs which coordinate activation of the GTPase Rac1 with other GEFs in many partitioning-defective (PAR) processes within spines [47]. The PAR complex is an essential determinant of cellular polarity [48, 49]. This complex included Par3, Par6, and atypical protein kinase C, which modulates polarized processes, like axon specification, neuronal migration, and development of spine and synapse [50–52]. Par3 combines with Tiam1 together to exert vital effects on cellular polarity by spatially restricting Rac1’s activation and localization in cyto-skeletal remodeling [47]. Due the loss of Tiam1’s function in synapses, the Rac1’s activation and spinogenesis cannot be formed in neurons from glutamate/NMDA receptor signaling and ephrinB/EphB receptor signaling [53, 54]. If the Par3 were knock-down and Tiam1 were delocalized in the neurons, it would result in Rac1’s dysregulation and synaptic digenesis [52]. In addition, Tiam1 activates and recruits to the membranes after exposure to amyloid beta peptide in hippocampal neurons, and this might exert influence to the pathology of Alzheimer disease [55, 56]. A study reported that Tiam1 knockdown declined hippocampal neuronal vulnerability to oxygen/glucose depravation [57]. Conclusively, these results demonstrated that the Tiam1 is an important gene for the hippocampal neurons’ development, and for the etiology of several psychiatry diseases. We showed here that increasing expression levels of mmu-miR-190a-3p might inhibit the activation of the Tiam1/Rac1 pathway in the hippocampus after surgery.
There were a few of limitations in this study. First, due to the limited number of sample size and low expression level of miRNA in brain, some weak signals of miRNAs were difficulty to be observed. To obtain more reliable expression signals, we used both the public data from GEO database and RT-PCR to validate the significantly differential expressed miRNAs. Second, although we employed various robust bioinformatics methods to find the binding targets of mmu-miR-190a-3p, such as the expression of Trim1gene which may be restrained by mmu-miR-190a-3p in the hippocampus of POCD, we did not confirm these gene-miRNA interactions experimentally, which should be the research focus of future study.
Conclusions
To sum up, we provided robust evidence supporting that mmu-miR-190a-3p has a strong positive correlation with the incidence of POCD. The pathway analysis displayed that these mmu-miR-190a-3p targeted genes were enriched in the psychiatry-related pathways and immune system-related pathways. Among of these target genes, Tiam1 was inhibited by mmu-miR-190a-3p in the hippocampus tissue of POCD model mice. These results demonstrated that mmu-miR190a-3p probably serve as an important regulator to inhibit target genes expression in the hippocampus tissue of POCD patients. More molecular-based studies are warranted to explore the underlying biological mechanisms of target genes by mmu-miR-190a-3p in the pathogenesis of POCD.
Additional files
Table S1. The primer sequence of the validated miRNA. i.e., the primer sequence of mmu-miR-190a-3p used for real-time PCR assay. Table S2. Predictionbinding sites for mmu-miR-190a-3p. The data were predicted by using miRwalk 3.0 for obtaining reliable targets for mmu-miR-190a-3p. Table S3. Genes and miRNAs involved in red module. This red module were generated by using our co-expression weighted network analysis by combined the 314 mRNA and 5 miRNAs data. (DOCX 49 kb)
Figure S1. The detailed time line of the euthanization of study animals. According to the protocol, all mice were euthanized before obtaining hippocampal tissue. All mice were deeply anesthetized based on several signs (see Methods part). Then the mice were decapitated 24 h after operation, brains were instantly dissected on ice, and the hippocampal tissues were obtained and stored in liquid nitrogen. Figure S2. Volcano plot of miRNA expression in replication cohort. This dataset (GSE95070) was downloaded from the GEO database. Figure S3. The results of qRT-PCR for technical replication. Expression level of the most significantly mmu-miR-190a-3p was validated by using real-time PCR assay. Reverse transcription reaction was performed with M-MLV Reverse Transcriptase kit (Takara Code: D2639A) based on the manufacturers’ protocol. Figure S4. WGCNA module-based analysis for genes and miRNAs expression data. The genes and miRNAs expression data of GSE73507 were acquired from GEO database. After the filtering process, 13,241 mRNAs and 546 miRNAs from hippocampus tissue of wild type mice (n = 24) were included for WGCNA analysis. Figure S5. KEGG pathway analyses for mmu-miR-190a-3p highly related genes in red module. We refined 169 genes from red module with a highly connected with mmu-miR-190a-3p (Fig. 2b) and used ClueGO (v. 2.3.4) to decipher the pathways and determine their biological functions. (PPTX 118 kb)
Acknowledgements
The authors would like to extend their sincere thanks to Professor Wang Yong’an from the Institute of Pharmacology and Toxicology, Academy of Military Medical Sciences, for the support and help in MWM equipment provision and tissue storage.
Abbreviations
- AGCC
Affymetrix Genechip Command Console
- circRNA
circular RNA
- GEO
Gene expression omnibus
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- lncRNA
long non-coding RNA
- miRNA
microRNA
- MWM
Morris water maze
- PCR
Polymerase chain reaction
- POCD
Postoperative cognitive dysfunction
- WGCNA
Weighted gene co-expression network analysis
Authors’ contributions
QL, JL, YY, KN managed the literature searches and data analysis. AH, YZ, YG, HL performed laboratory experiments. QL and AH wrote the first draft of the manuscript. YM, and JC conceived the study and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This project was supported by the National Natural Science Foundation of China (NO. 81541114, NO. 81771129), the Hainan Province Natural Science Foundation of China (NO. 817353), and the China Postdoctoral Science Foundation (2018 M630667). The funders had no role in the designing and conducting of this study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets generated and/or analysed during the current investigation are available from the corresponding author until it becomes available from a public repository.
Ethics approval and consent to participant
The protocol of animal experiment was approved by the Animal Care Committee of the Chinese People’s Liberation Army General Hospital (Beijing, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qiang Liu and Aisheng Hou contributed equally to this work.
Contributor Information
Qiang Liu, Email: sxndlq@sina.com.
Aisheng Hou, Email: houaisheng301@163.com.
Yongyi Zhang, Email: freshstart301@163.com.
Ying Guo, Email: 301guoying301@sina.com.
Jingjing Li, Email: jingjinglea@163.com.
Yinghao Yao, Email: bioyao15@163.com.
Kaimeng Niu, Email: niukaimeng47@163.com.
Hao Li, Email: lihao301@126.com.
Yunlong Ma, Email: glb-biotech@zju.edu.cn.
Jiangbei Cao, Email: cjb2000@sina.com.
References
- 1.Shoair OA, Grasso Ii MP, Lahaye LA, Daniel R, Biddle CJ, Slattum PW. Incidence and risk factors for postoperative cognitive dysfunction in older adults undergoing major noncardiac surgery: a prospective study. J Anaesthesiol Clin Pharmacol. 2015;31(1):30–36. doi: 10.4103/0970-9185.150530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patron E, Messerotti Benvenuti S, Zanatta P, Polesel E, Palomba D. Preexisting depressive symptoms are associated with long-term cognitive decline in patients after cardiac surgery. Gen Hosp Psychiatry. 2013;35(5):472–479. doi: 10.1016/j.genhosppsych.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Steinmetz J, Christensen KB, Lund T, Lohse N, Rasmussen LS, Group I Long-term consequences of postoperative cognitive dysfunction. Anesthesiology. 2009;110(3):548–555. doi: 10.1097/ALN.0b013e318195b569. [DOI] [PubMed] [Google Scholar]
- 4.Engelhard K, Werner C. Postoperative cognitive dysfunction in geriatric patients. Anasthesiologie, Intensivmedizin, Notfallmedizin, Schmerztherapie : AINS. 2008;43(9):606–614. doi: 10.1055/s-0028-1090023. [DOI] [PubMed] [Google Scholar]
- 5.Biedler A, Juckenhofel S, Larsen R, Radtke F, Stotz A, Warmann J, Braune E, Dyttkowitz A, Henning F, Strickmann B, et al. Postoperative cognition disorders in elderly patients. The results of the “international study of postoperative cognitive dysfunction” ISPOCD 1. Anaesthesist. 1999;48(12):884–895. doi: 10.1007/s001010050802. [DOI] [PubMed] [Google Scholar]
- 6.Yu X, Liu S, Li J, Fan X, Chen Y, Bi X, Liu S, Deng X. MicroRNA-572 improves early post-operative cognitive dysfunction by down-regulating neural cell adhesion molecule 1. PLoS One. 2015;10(2):e0118511. doi: 10.1371/journal.pone.0118511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murchison EP, Hannon GJ. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr Opin Cell Biol. 2004;16(3):223–229. doi: 10.1016/j.ceb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 8.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao YS, Pak TR. microRNAs and the adolescent brain: filling the knowledge gap. Neurosci Biobehav Rev. 2016;70:313–322. doi: 10.1016/j.neubiorev.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F, Wei G, Bai Y, Li Y, Huang F, Lin J, Hou Q, Deng R, Zhou JH, Zhang SX, et al. MicroRNA-574 is involved in cognitive impairment in 5-month-old APP/PS1 mice through regulation of neuritin. Brain Res. 1627;2015:177–188. doi: 10.1016/j.brainres.2015.09.022. [DOI] [PubMed] [Google Scholar]
- 11.Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466(7310):1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kocerha J, Faghihi MA, Lopez-Toledano MA, Huang J, Ramsey AJ, Caron MG, Sales N, Willoughby D, Elmen J, Hansen HF, et al. MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc Natl Acad Sci U S A. 2009;106(9):3507–3512. doi: 10.1073/pnas.0805854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoss AG, Kartha VK, Dong X, Latourelle JC, Dumitriu A, Hadzi TC, Macdonald ME, Gusella JF, Akbarian S, Chen JF, et al. MicroRNAs located in the Hox gene clusters are implicated in huntington's disease pathogenesis. PLoS Genet. 2014;10(2):e1004188. doi: 10.1371/journal.pgen.1004188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Chang X, Hahn CG, Gur RE, Sleiman PAM, Hakonarson H. Non-coding RNA dysregulation in the amygdala region of schizophrenia patients contributes to the pathogenesis of the disease. Transl Psychiatry. 2018;8(1):44. doi: 10.1038/s41398-017-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gokbuget D, Pereira JA, Bachofner S, Marchais A, Ciaudo C, Stoffel M, Schulte JH, Suter U. The Lin28/let-7 axis is critical for myelination in the peripheral nervous system. Nat Commun. 2015;6:8584. doi: 10.1038/ncomms9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu H, Li H, Zhang L, Luan H, Huang T, Wang L, Fan Y, Zhang Y, Liu X, Wang W, et al. Diagnostic role of microRNA expression profile in the serum of pregnant women with fetuses with neural tube defects. J Neurochem. 2012;122(3):641–649. doi: 10.1111/j.1471-4159.2012.07812.x. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Wang ZH, Wu YY, Tang H, Tan L, Wang X, Gao XY, Xiong YS, Liu D, Wang JZ, et al. Melatonin attenuates scopolamine-induced memory/synaptic disorder by rescuing EPACs/miR-124/Egr1 pathway. Mol Neurobiol. 2013;47(1):373–381. doi: 10.1007/s12035-012-8355-9. [DOI] [PubMed] [Google Scholar]
- 18.Wu C, Wang R, Li X, Chen J. Preoperative serum MicroRNA-155 expression independently predicts postoperative cognitive dysfunction after laparoscopic surgery for Colon Cancer. Med Sci Monit. 2016;22:4503–4508. doi: 10.12659/MSM.898397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei C, Luo T, Zou S, Zhou X, Shen W, Ji X, Li Q, Wu A. Differentially expressed lncRNAs and miRNAs with associated ceRNA networks in aged mice with postoperative cognitive dysfunction. Oncotarget. 2017;8(34):55901–55914. doi: 10.18632/oncotarget.18362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L, Dong R, Lu Y, Zhou Y, Li K, Zhang Z, Peng M. MicroRNA-146a protects against cognitive decline induced by surgical trauma by suppressing hippocampal neuroinflammation in mice. Brain Behav Immun. 2019;78:188–201. doi: 10.1016/j.bbi.2019.01.020. [DOI] [PubMed] [Google Scholar]
- 21.Xu Z, Dong Y, Wang H, Culley DJ, Marcantonio ER, Crosby G, Tanzi RE, Zhang Y, Xie Z. Age-dependent postoperative cognitive impairment and Alzheimer-related neuropathology in mice. Sci Rep. 2014;4:3766. doi: 10.1038/srep03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris RG, Garrud P, Rawlins JN, O'Keefe J. Place navigation impaired in rats with hippocampal lesions. Nature. 1982;297(5868):681–683. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- 23.Langfelder P, Cantle JP, Chatzopoulou D, Wang N, Gao F, Al-Ramahi I, Lu XH, Ramos EM, El-Zein K, Zhao Y, et al. Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice. Nat Neurosci. 2016;19(4):623–633. doi: 10.1038/nn.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahraeian SME, Mohiyuddin M, Sebra R, Tilgner H, Afshar PT, Au KF, Bani Asadi N, Gerstein MB, Wong WH, Snyder MP, et al. Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat Commun. 2017;8(1):59. doi: 10.1038/s41467-017-00050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedlander MR, Mackowiak SD, Li N, Chen W. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40(1):37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11(11):1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nature methods. 2015;12(8):697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 33.Kim JM, Park SK, Yang JJ, Shin ES, Lee JY, Yun JY, Kim JS, Park SS, Jeon BS. SNPs in axon guidance pathway genes and susceptibility for Parkinson's disease in the Korean population. J Hum Genet. 2011;56(2):125–129. doi: 10.1038/jhg.2010.130. [DOI] [PubMed] [Google Scholar]
- 34.Lesnick TG, Papapetropoulos S, Mash DC, Ffrench-Mullen J, Shehadeh L, de Andrade M, Henley JR, Rocca WA, Ahlskog JE, Maraganore DM. A genomic pathway approach to a complex disease: axon guidance and Parkinson disease. PLoS Genet. 2007;3(6):e98. doi: 10.1371/journal.pgen.0030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rapti G, Li C, Shan A, Lu Y, Shaham S. Glia initiate brain assembly through noncanonical Chimaerin-Furin axon guidance in C. elegans. Nat Neurosci. 2017;20(10):1350–1360. doi: 10.1038/nn.4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quan Y, Moller T, Weinstein JR. Regulation of Fcgamma receptors and immunoglobulin G-mediated phagocytosis in mouse microglia. Neurosci Lett. 2009;464(1):29–33. doi: 10.1016/j.neulet.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puram SV, Riccio A, Koirala S, Ikeuchi Y, Kim AH, Corfas G, Bonni A. A TRPC5-regulated calcium signaling pathway controls dendrite patterning in the mammalian brain. Genes Dev. 2011;25(24):2659–2673. doi: 10.1101/gad.174060.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haustein MD, Kracun S, Lu XH, Shih T, Jackson-Weaver O, Tong X, Xu J, Yang XW, O'Dell TJ, Marvin JS, et al. Conditions and constraints for astrocyte calcium signaling in the hippocampal mossy fiber pathway. Neuron. 2014;82(2):413–429. doi: 10.1016/j.neuron.2014.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 40.Shan L, Ma D, Zhang C, Xiong W, Zhang Y. miRNAs may regulate GABAergic transmission associated genes in aged rats with anesthetics-induced recognition and working memory dysfunction. Brain Res. 1670;2017:191–200. doi: 10.1016/j.brainres.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 41.Saddic LA, Chang TW, Sigurdsson MI, Heydarpour M, Raby BA, Shernan SK, Aranki SF, Body SC, Muehlschlegel JD. Integrated microRNA and mRNA responses to acute human left ventricular ischemia. Physiol Genomics. 2015;47(10):455–462. doi: 10.1152/physiolgenomics.00049.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muniategui A, Pey J, Planes FJ, Rubio A. Joint analysis of miRNA and mRNA expression data. Brief Bioinform. 2013;14(3):263–278. doi: 10.1093/bib/bbs028. [DOI] [PubMed] [Google Scholar]
- 43.Ruike Y, Ichimura A, Tsuchiya S, Shimizu K, Kunimoto R, Okuno Y, Tsujimoto G. Global correlation analysis for micro-RNA and mRNA expression profiles in human cell lines. J Hum Genet. 2008;53(6):515–523. doi: 10.1007/s10038-008-0279-x. [DOI] [PubMed] [Google Scholar]
- 44.Zhu M, Yi M, Kim CH, Deng C, Li Y, Medina D, Stephens RM, Green JE. Integrated miRNA and mRNA expression profiling of mouse mammary tumor models identifies miRNA signatures associated with mammary tumor lineage. Genome Biol. 2011;12(8):R77. doi: 10.1186/gb-2011-12-8-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller MB, Yan Y, Eipper BA, Mains RE. Neuronal rho GEFs in synaptic physiology and behavior. Neuroscientist. 2013;19(3):255–273. doi: 10.1177/1073858413475486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ehler E, van Leeuwen F, Collard JG, Salinas PC. Expression of Tiam-1 in the developing brain suggests a role for the Tiam-1-Rac signaling pathway in cell migration and neurite outgrowth. Mol Cell Neurosci. 1997;9(1):1–12. doi: 10.1006/mcne.1997.0602. [DOI] [PubMed] [Google Scholar]
- 47.Mertens AE, Pegtel DM, Collard JG. Tiam1 takes PARt in cell polarity. Trends Cell Biol. 2006;16(6):308–316. doi: 10.1016/j.tcb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 48.Ohno S. Intercellular junctions and cellular polarity: the PAR-aPKC complex, a conserved core cassette playing fundamental roles in cell polarity. Curr Opin Cell Biol. 2001;13(5):641–648. doi: 10.1016/s0955-0674(00)00264-7. [DOI] [PubMed] [Google Scholar]
- 49.Welchman DP, Mathies LD, Ahringer J. Similar requirements for CDC-42 and the PAR-3/PAR-6/PKC-3 complex in diverse cell types. Dev Biol. 2007;305(1):347–357. doi: 10.1016/j.ydbio.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi SH, Jan LY, Jan YN. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 2003;112(1):63–75. doi: 10.1016/s0092-8674(02)01249-7. [DOI] [PubMed] [Google Scholar]
- 51.Spindler SR, Hartenstein V. Bazooka mediates secondary axon morphology in Drosophila brain lineages. Neural Dev. 2011;6:16. doi: 10.1186/1749-8104-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang H, Macara IG. The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nat Cell Biol. 2006;8(3):227–237. doi: 10.1038/ncb1368. [DOI] [PubMed] [Google Scholar]
- 53.Tolias KF, Bikoff JB, Burette A, Paradis S, Harrar D, Tavazoie S, Weinberg RJ, Greenberg ME. The Rac1-GEF Tiam1 couples the NMDA receptor to the activity-dependent development of dendritic arbors and spines. Neuron. 2005;45(4):525–538. doi: 10.1016/j.neuron.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 54.Tolias KF, Bikoff JB, Kane CG, Tolias CS, Hu L, Greenberg ME. The Rac1 guanine nucleotide exchange factor Tiam1 mediates EphB receptor-dependent dendritic spine development. Proc Natl Acad Sci U S A. 2007;104(17):7265–7270. doi: 10.1073/pnas.0702044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma QL, Yang F, Calon F, Ubeda OJ, Hansen JE, Weisbart RH, Beech W, Frautschy SA, Cole GM. p21-activated kinase-aberrant activation and translocation in Alzheimer disease pathogenesis. J Biol Chem. 2008;283(20):14132–14143. doi: 10.1074/jbc.M708034200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mendoza-Naranjo A, Gonzalez-Billault C, Maccioni RB. Abeta1-42 stimulates actin polymerization in hippocampal neurons through Rac1 and Cdc42 rho GTPases. J Cell Sci. 2007;120(Pt 2):279–288. doi: 10.1242/jcs.03323. [DOI] [PubMed] [Google Scholar]
- 57.Blanco-Suarez E, Fiuza M, Liu X, Chakkarapani E, Hanley JG. Differential Tiam1/Rac1 activation in hippocampal and cortical neurons mediates differential spine shrinkage in response to oxygen/glucose deprivation. J Cereb Blood Flow Metab. 2014;34(12):1898–1906. doi: 10.1038/jcbfm.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The primer sequence of the validated miRNA. i.e., the primer sequence of mmu-miR-190a-3p used for real-time PCR assay. Table S2. Predictionbinding sites for mmu-miR-190a-3p. The data were predicted by using miRwalk 3.0 for obtaining reliable targets for mmu-miR-190a-3p. Table S3. Genes and miRNAs involved in red module. This red module were generated by using our co-expression weighted network analysis by combined the 314 mRNA and 5 miRNAs data. (DOCX 49 kb)
Figure S1. The detailed time line of the euthanization of study animals. According to the protocol, all mice were euthanized before obtaining hippocampal tissue. All mice were deeply anesthetized based on several signs (see Methods part). Then the mice were decapitated 24 h after operation, brains were instantly dissected on ice, and the hippocampal tissues were obtained and stored in liquid nitrogen. Figure S2. Volcano plot of miRNA expression in replication cohort. This dataset (GSE95070) was downloaded from the GEO database. Figure S3. The results of qRT-PCR for technical replication. Expression level of the most significantly mmu-miR-190a-3p was validated by using real-time PCR assay. Reverse transcription reaction was performed with M-MLV Reverse Transcriptase kit (Takara Code: D2639A) based on the manufacturers’ protocol. Figure S4. WGCNA module-based analysis for genes and miRNAs expression data. The genes and miRNAs expression data of GSE73507 were acquired from GEO database. After the filtering process, 13,241 mRNAs and 546 miRNAs from hippocampus tissue of wild type mice (n = 24) were included for WGCNA analysis. Figure S5. KEGG pathway analyses for mmu-miR-190a-3p highly related genes in red module. We refined 169 genes from red module with a highly connected with mmu-miR-190a-3p (Fig. 2b) and used ClueGO (v. 2.3.4) to decipher the pathways and determine their biological functions. (PPTX 118 kb)
Data Availability Statement
The datasets generated and/or analysed during the current investigation are available from the corresponding author until it becomes available from a public repository.