Abstract
H3K9me3 (methylation of lysine 9 of histone H3) is an epigenetic modification that acts as a repressor mark. Several diseases, including cancers and neurological disorders, have been associated with aberrant changes in H3K9me3 levels. Different tools have been developed to enable detection and quantification of H3K9me3 levels in cells. Most techniques, however, lack live cell compatibility. To address this concern, we have engineered recombinant protein sensors for probing H3K9me3 in situ. A heterodimeric sensor containing a chromodomain and chromo shadow domain from HP1a was found to be optimal in recognizing H3K9me3 and exhibited similar spatial resolution to commercial antibodies. Our sensor offers similar quantitative accuracy in characterizing changes in H3K9me3 compared to antibodies but claims single cell resolution. The sensor was applied to evaluate changes in H3K9me3 responding to environmental chemical atrazine (ATZ). ATZ was found to result in significant reductions in H3K9me3 levels after 24 h of exposure. Its impact on the distribution of H3K9me3 among cell populations was also assessed and found to be distinctive. We foresee the application of our sensors in multiple toxicity and drug-screening applications.
Introduction
DNA in high eukaryotes is packaged into a highly organized chromatin structure. The conventional wisdom suggesting that the underlying genomic sequence encodes the cell fate is fundamentally challenged with the recent revelation of stem cell lineage data.1 This data suggest that distinctive cell types, for example, epithelial and mesenchymal cells, share almost identical genomic information and gain their respective cell identity by differential usage of genomic information, for example, different expression patterns of transcriptional factors and chromatin remodeling regulators.2 For example, during an epithelial mesenchymal transition,3 cell reprogramming occurs first via changes in epigenetic marks, that is, H3K9me3 (tri-methylation of lysine 9 of histone H3).4 Transcription factors, for example, OCT4 and NANOG, essential for maintaining pluripotency, are then epigenetically repressed.5−7 These changes can result in large-scale re-arrangement of chromatin8,9 that subsequently lead to an altered phenotype.
Different chromatin regions, that is, active (transcriptionally “on”) and silenced (transcriptionally “off”) chromatin, are identified via DNA staining (e.g., DAPI, C-, G-, Q-, and N-banding10) in cytohistology. Silenced chromatin, also known as heterochromatin, has a higher DNA density and consists primarily of permanently silenced genes. Methylation of histones, that is, H3K9me3, is a hallmark of heterochromatin.11 Characterizing and understanding H3K9me3 distributions can thus provide crucial insights about heterochromatin organization in cells. Although spatial distribution of H3K9me3 is expected to at least partially correlate with the DNA density, that is, identified via DAPI staining, differences exist. For example, a recent study has shown that an H3K9me3-enriched region of chromosome X can only be weakly stained via DAPI.12 Other than marking of heterochromatin regions, spatial distribution of H3K9me3 also carries important gene regulation information. For example, in the primary hematopoietic stem and progenitor cells, H3K9me3 is preferentially localized to the nuclear periphery with a small number of foci in the center of a nucleus.13 Mature cells lacking differentiation capability, however, exhibit a more pericentric distribution of H3K9me3.13 Aberrant changes in the global level14,15 and spatial distribution15 of H3K9me3 are commonly observed in many diseases, for example, cancer and neurodegenerative diseases. A more direct approach for characterizing H3K9me3 is thus needed for tracking transcriptionally silenced and/or repressed regions, as well as monitoring chromatin re-arrangement and epigenetic re-programming during cell differentiation and disease development.
Limited approaches exist for monitoring H3K9me3 distribution and changes. Immuno-histochemistry, that is, immuno-fluorescence imaging, is commonly used to reveal the distribution of H3K9me3. This approach, however, can only be applied to fixed cells and is further constrained by the availability and quality of commercial antibodies, which is becoming an increasing concern in recent literature studies.16 Several novel probes have been developed in recent years that are live-cell-compatible. Briefly, both Fabs- and Mintbody-based approaches utilize fragments derived from the antibody.17,18 A lengthy development process which involves the production of specific IgG and scFv libraries is typically required.19,20 Although these approaches have been successfully applied to detect several epigenetic changes (e.g., H4K20me and H3K9ac),17,21,22 they have not been used to detect H3K9me3. A FRET-based sensor utilizing a H3K9me3 reader domain [i.e., heterochromatin protein 1a (HP1a)]23 was developed to monitor intracellular H3K9me3 levels. This sensor relies on the use of a reporter histone H3 conjugated to the recognition motif and thus does not provide any information about the spatial distribution of endogenous H3K9me3. Recently, a H3K9me3 sensor based on human HP1β was reported.24 In comparison to other HP1 homologs, HP1β has a relatively high affinity for H3K9me3 (KD = 3 ± 1 μM), while for HP1α and HP1γ, this is of 13 ± 1 and 7 ± 2 μM, respectively.25 Despite its high affinity, image analysis of cells transfected with HP1β-GFP and costained with the H3K9me3 antibody, showed that they do not colocalize with all H3K9me3 foci within the cell.26,27 Recently, it was reported that HP1β is capable of binding to H4K20me3 and thus provides an explanation for its lacking of specificity.28
We thus engineered a H3K9me3 sensor using functional domains derived from HP1a fused with a fluorescent protein. A heterodimer sensor linking chromodomain (CD) (ΔCD) and chromo shadow domain (ΔCSD) was found to be the most effective in colocalizing with H3K9me3. It is recognized that the most optimum sensor design resembles native protein binding motifs but lacks a hinge domain. Comprehensive validation was carried out both in situ and in vitro to confirm their suitability in epigenetic studies which has not been done previously in the literature. The sensor can be used to monitor changes in H3K9me3 in single cells with a comparable spatial resolution to commercial antibodies. We have demonstrated the application of our sensor in monitoring cellular responses to an epigenetic drug (BIX-01294) and an environmental chemical (atrazine, an herbicide).
Results and Discussion
Engineering H3K9me3 Sensors
In this work, we engineered various H3K9me3 sensors derived from CD (ΔCD) and chromo-shadow domain (ΔCSD) of HP1a. All protein sensors were subcloned into a mammalian expression vector and fused to an enhanced green fluorescent protein, allowing us to monitor changes in H3K9me3 in live cells. It is important to note that although we conserved the primary sequence of ΔCD and ΔCSD in our design, the hinge domain was intentionally left out for enhanced selectivity and the probes as constructed vary from the native HP1a sequence. All sensors were successfully transfected via lipofectamine, expressed and enriched in the cell nucleus. NLS–mEGFP lacking the “reader” motif was used as a negative control. Typical images of HEK293T cells transfected with various sensor constructs and the negative control are shown in Figure 1. Additional images of sensor-transfected cells can be found in Figure S1 (Supporting Information). Cells transfected with ΔCSD and ΔCSD−ΔCSD sensors exhibit a diffusive pattern inside the nucleus with no “speckle” or “island” features as we would expect for H3K9me3-rich regions. The same pattern was also observed in cells transfected with the negative control. Sensors containing ΔCD (i.e., ΔCD, ΔCD−ΔCD, and ΔCD−ΔCSD), on the other hand, show distinctive patterns that potentially resemble the distribution of H3K9me3 in HEK293T cells.
Figure 1.
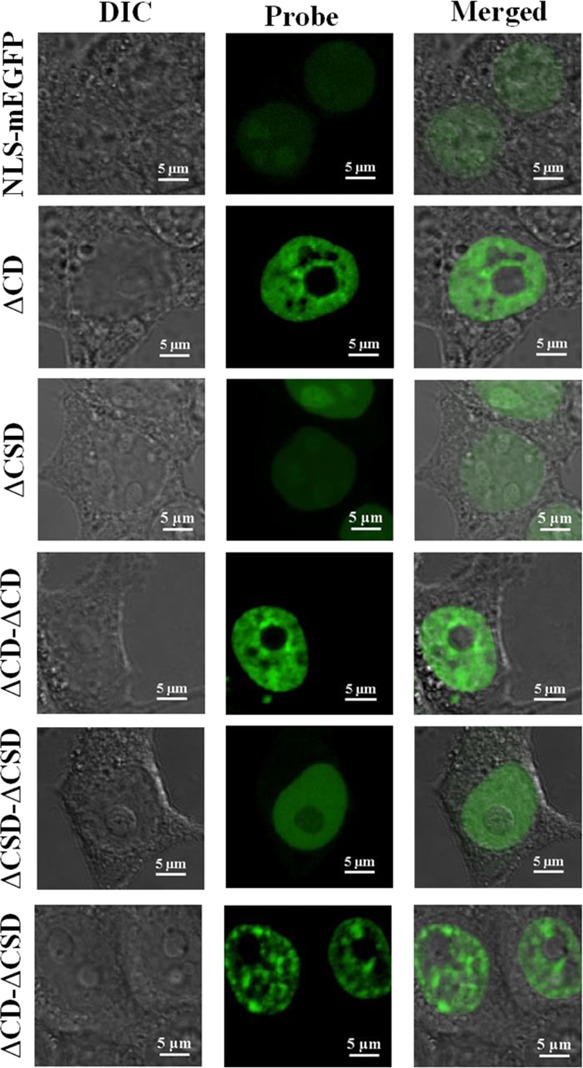
Representative images of HEK293T cells transfected with monomeric and dimeric constructs of H3K9me3 sensors. All cells were imaged 24 h post transfection. All images are 2D confocal slides of cells.
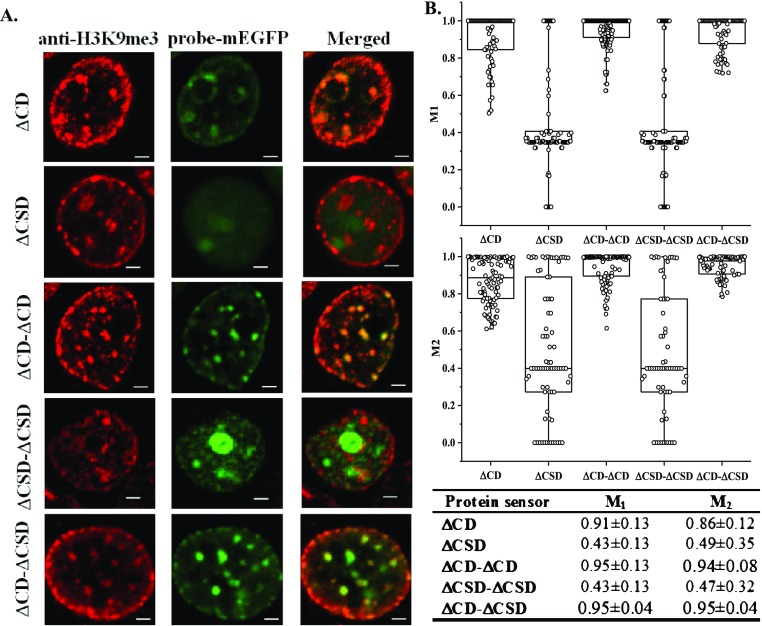
To select among them and identify the sensor that is highly selective in recognizing H3K9me3, we costained transfected cells with an anti-H3K9me3 antibody. Confocal images of costained cells are summarized in Figure 2A [additional images can be found in Figure S2A,B (Supporting Information)]. Mander’s colocalization analysis was performed to assess the degree of colocalization between the antibody and our probes. The results are summarized in Figure 2B. Mander’s coefficients, namely M1 and M2, represent the fraction of the antibody co-localized with our sensors and vice versa, respectively.29 Having Mander’s coefficients close to 1 suggests perfect colocalization. Among all probes, ΔCSD and ΔCSD−ΔCSD do not colocalize with H3K9me3 antibodies. All three probes containing ΔCD exhibit a certain level of colocalization with H3K9me3 antibodies. Among them, ΔCD−ΔCD and ΔCD−ΔCSD sensors have the highest values of M1 and M2. A closer comparison between ΔCD−ΔCD and ΔCD−ΔCSD sensors suggests that ΔCD−ΔCSD exhibits a lower dispersion of M1 and M2 coefficients. We further analyzed the intensity profiles of both probes and compared them to the antibody-stained images as shown in Figure S2C (Supporting Information). Qualitatively, both probes capture a similar trend in intensity changes as the antibody. Between them, ΔCD−ΔCSD exhibits a closer match in the intensity profile. We thus proceeded to evaluate the potential of ΔCD−ΔCSD as a live cell sensor for H3K9me3. Although similar findings have been reported in the biological literature,30−32 our work is the first to apply this engineered design for quantifying the endogenous H3K9me3 modification level.
Figure 2.
(A) HEK293T cells transfected with various protein sensors and co-stained with anti-H3K9me3 antibody (Alexa 564 as secondary). Scale bar = 2 μm. All images are 2D slides of cells. (B) Mander’s correlation analysis was performed between sensor and antibody-stained images. Coefficients of colocalization, namely M1 and M2, were reported in a scattered bar plot and summarized in the table underneath, n = 100.
To verify that the observed fluorescence signal arises from specific binding of our sensors to H3K9me3, we introduced mutations to the CDs of our ΔCD−ΔCSD sensor. Specifically, we mutated the conserved residues at positions 42–43 of our sensor from KW to AA. This mutant has been shown to result in the loss of H3K9me3 binding affinity in the literature.33 The mutant sensor, ΔCD (KW42/43AA)−ΔCSD, can be successfully transfected and translocated into the nucleus but exhibits no H3K9me3-like features as shown in Figure S3 (Supporting Information). This observation confirms that the fluorescent patterns observed in transfected cells, that is, as shown Figures 1 and S1 (Supporting Information), can be attributed to the specific binding of our sensors to H3K9me3 in the cell chromatin.
An in vitro binding assay was performed to characterize the affinity and selectivity of ΔCD−ΔCSD. The affinity measurements were performed using Bio-Layer Interferometry (methods detailed in Supporting Methods (Supporting Information)). Synthetic histone peptides with sequences detailed in Table S1 (Supporting Information) was used as binding substrates. The Kd value of our sensor was found to be ∼0.24 μM and 1.4 mM for H3K9me3 and H3WT peptides, respectively, as shown Figure S4 (Supporting Information). We also tested our sensor using H3K9me2-containing peptides. Although our sensors still exhibit a preference to H3K9me3, it binds to H3K9me2 at a reduced affinity of ∼0.54 μM. Thus, our sensor has a high affinity for H3K9me3 and a high selectivity between H3K9me3 and unmodified H3, although modest binding was also observed for H3K9me2.
Cytotoxicity of Protein Sensors
To evaluate the potential cytotoxicity of the selected protein sensor, we assessed the effects of transfected probes on cell growth and viability. We followed transfected cells for a total duration of ∼60 h post-transfection. At least 92% of transfected cells in the control remain viable within our observation window and maintain a normal growth pattern [Figure S5 (Supporting Information)]. The proliferation rate of transfected cells (doubling time of 19.4 ± 0.5 h) was slightly lowered compared to untransfected ones (doubling time of 18.6 ± 0.5 h). The difference, however, was found not statistically significant (p > 0.05). In addition, cells transfected with either ΔCD−ΔCSD or its mutant, ΔCD (KW42/43AA)−ΔCSD, present a similar morphology [Figures 1 and S3 (Supporting Information)], suggesting that the specific binding of our sensor minimally disrupted the cell morphology.
Track H3K9me3 in Live Cells
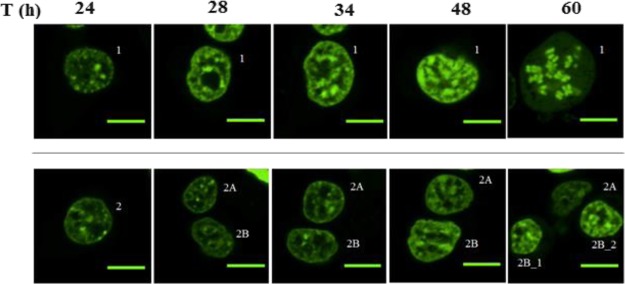
Single-cell images of transfected cells were collected for ∼60 h after transfection. Grid slides were used to ensure that we image the same area at different time points, following a procedure that we detailed in our previous publication34 and briefly illustrated in Figure S6 (Supporting Information). The time course of two individual cells (marked as 1 and 2) is shown in Figure 3. Cell 1 (Figure 3, top row) undergoes division within 48 and 60 h of observation with significant alterations in chromatin organization. Cell 2 (Figure 3, bottom row) undergoes division twice, once between 24 and 28 h and then between 48 and 60 h of observation. Resulting cells are identified as (2A and 2B, and 2B_1 and 2B_2). The observed changes in sensor distributions are consistent with the expected H3K9me3 distribution changes along the cell cycle.35 Thus, daughter cells from the transfected ones were able to continuously express H3K9me3 sensors and highlight features in H3K9me3. The transfected cells were then able to retain the fluorescence signal for >60 h, enabling live-cell tracking.
Figure 3.
Two cells transfected with ΔCD−ΔCSD probes and tracked for 60 h. Cell images were collected at 60× at areas specified in Figure S6 (Supporting Information). Scale bar = 10 μm.
Quantifying Changes in H3K9me3 Using Fluorescence Imaging
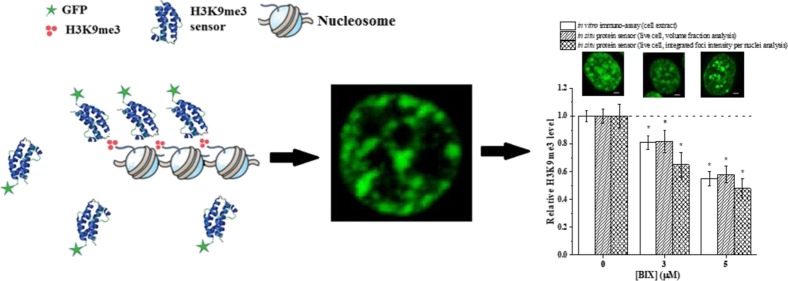
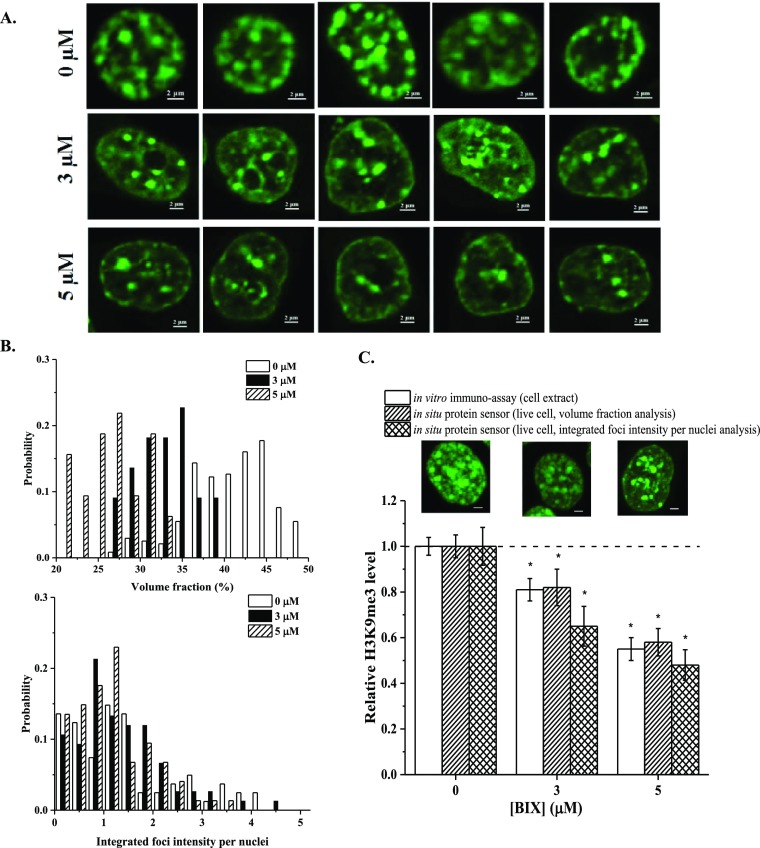
So far, we have demonstrated that we can track H3K9me3 in live cells. We then proceeded to determine the quantitative accuracy of our H3K9me3 sensor. To do that, HEK293T cells were treated with BIX-01294 to reduce H3K9me3 levels. Transfected cells undergoing treatments were imaged and then lysed. Cell extracts were analyzed using an immunoassay, as shown in Figure S7A (Supporting Information) to reveal relative changes in H3K9me3 [Figure S7B (Supporting Information)]. Meanwhile, fluorescent images of cells (Figure 4A) were analyzed for the same purpose. We used two approaches to analyze the collected fluorescent images, namely, a volume fraction (volume %) analysis and a foci intensity analysis. Volume fraction analysis utilizes the 3D stacks of the collected images. This analysis characterizes the relative abundance of H3K9me3 within a nucleus and is thus expected to be a good reflection of cellular H3K9me3 levels. Similar analysis was used previously in the literature.36 A foci intensity analysis uses 2D projection of all stacks as shown in Figure S8 (Supporting Information). We integrated the intensity of all foci within a cell nucleus to obtain the integrated foci intensity per nuclei (IFIN). The IFIN is equivalent to the fluorescence intensity above the diffusive background and is thus expected to correlate with the number of probes binding specifically to its target (i.e., H3K9me3) and reflect the overall H3K9me3 level of individual cells. The probability distribution of volume fraction and IFIN is plotted in Figure 4B. The probability histogram for the volume % shows a normal distribution shape. Conversely, for the integrated foci intensity, the probability histogram is right-skewed. We also observed potential narrowing in distribution and the rise of a subpopulation of cells with much lower H3K9me3 levels that do not exist in untreated cell populations. This type of information is otherwise inaccessible using population-based experiments. The average values of volume % and IFIN were compared to the untreated group to determine relative changes in H3K9me3. The calculated numbers were then compared to the results from our immunoassays as shown in Figure 4C. At 3 μM of BIX, the reduction in H3K9me3 was found to be 19% (immuno-assay), 18% (volume %), and 35% (IFIN). At 5 μM of BIX, the reduction was found to be 45% (immuno-assay), 42% (volume %), and 52% (IFIN), respectively. Despite some observed differences among varying analysis techniques, they are not found to be statistically different (p > 0.05). The observed changes in H3K9me3 are in close accordance with the literature reports.37,38 We thus consider both analysis approaches, namely, a volume fraction (volume %) and an IFIN analyses, being valid choices to quantify changes in H3K9me3 levels using our H3K9me3 live-cell sensors with a single-cell resolution.
Figure 4.
(A) Typical images of HEK293T cells transfected with ΔCD−ΔCSD sensors and treated with different concentrations of BIX-01294. (B) Probability distribution of volume % and IFIN under varying BIX-01294 concentrations. (C) Relative changes in mean volume % and IFIN of transfected cells under varying [BIX-01294] and a comparison with findings from immuno-assays. n ≥ 3 form immuno-assays. n ≥ 90 for images-based assays. *: p < 0.05 denoting statistical difference among compared groups. All images are 2D projections of Z-stacks. Scale bar = 2 μm.
Atrazine Effect on H3K9me3
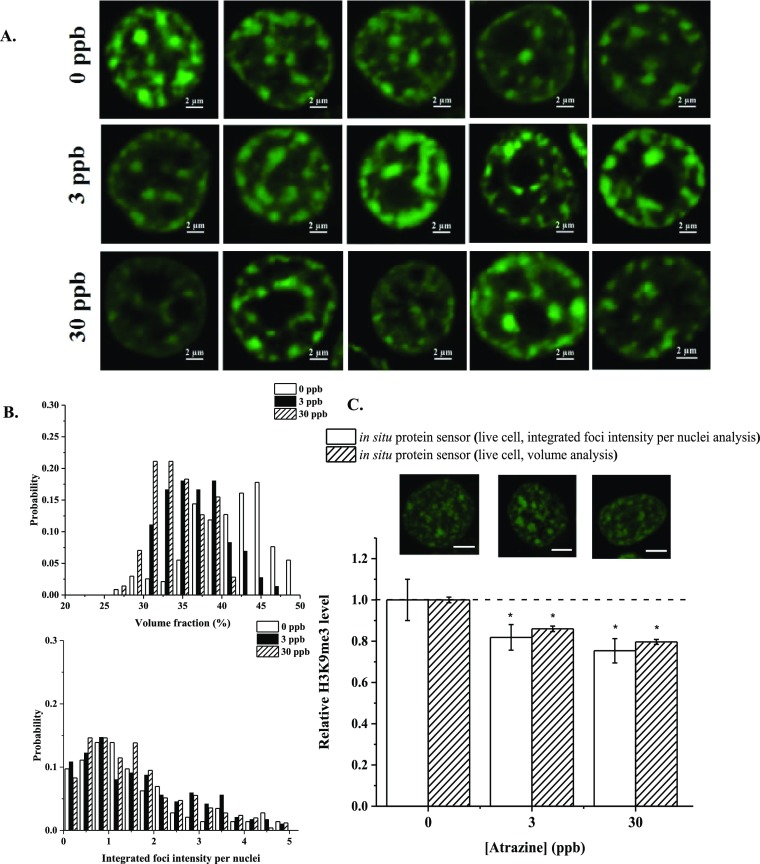
With a viable single-cell tool in hand, we proceeded to demonstrate the application of our probes in monitoring cellular response to an environmental chemical. For our applications, we demonstrated how cells respond to an organic compound (atrazine, ATZ) via changes in H3K9me3. HEK293T cells were treated with 3 and 30 ppb of ATZ for 24 h. Typical images of cells treated with ATZ can be found in Figure 5A. Histograms of volume % and IFIN under different treatment conditions are shown in Figure 5B. Our image-based analysis suggests that H3K9me3 levels were reduced by ∼14–18 and 20–25% with 3 and 30 ppb of ATZ treatments, respectively (Figure 5C). The probability histogram for the volume % shows a normal distribution shape, while for the integrated foci intensity, the probability histogram is right-skewed. A leftward shift was observed under high ATZ concentrations. These results reveal that ATZ exposure can significantly lower H3K9me3 in HEK293T cells. To the best of our knowledge, no previous report has evaluated the effect of ATZ on H3K9me3 levels using human cells.
Figure 5.
(A) Typical images of HEK293T cells transfected with ΔCD−ΔCSD sensors and treated with different concentrations of Atrazine (ATZ). (B) Probability distribution of volume % and IFIN under varying ATZ concentrations. (C) Relative changes in mean volume % and IFIN of transfected cells under varying [ATZ] and a comparison with findings from immuno-assays. n ≥ 3 form immuno-assays. n ≥ 90 for images-based assays. *: p < 0.05 denoting statistical difference among compared groups. All images are 2D projections of Z-stacks. Scale bar = 2 μm.
Demonstration of in Vitro Applicability of ΔCD−ΔCSD–mEGFP
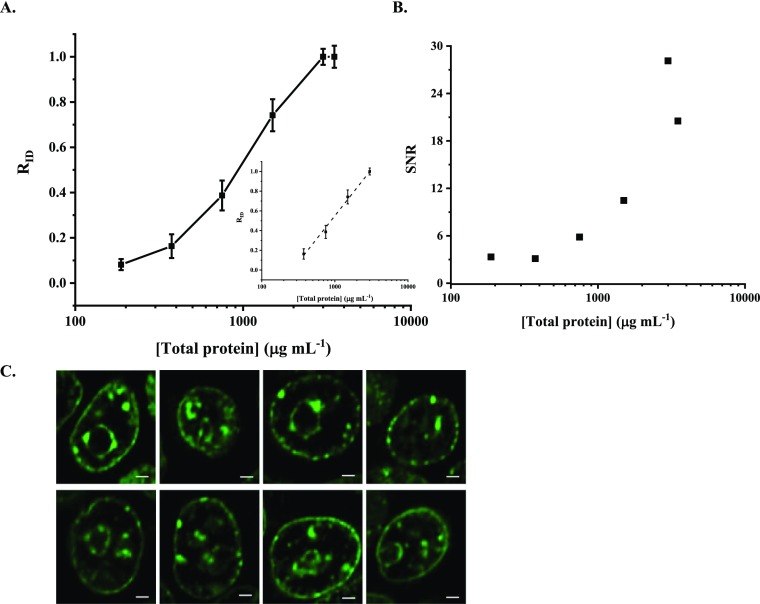
Compared to commercial antibodies, recombinant protein sensors are advantageous because of their lower cost and high reproducibility. We used recombinant ΔCD−ΔCSD–mEGFP as a replacement for commercial antibodies and demonstrated its application in dot blotting and fixed cell staining. To start, we prepared nuclear cell extracts (NCEs) from HEK293T. Dilution series of NCE were prepared, dotted on a membrane, bound to the protein sensor, and imaged to quantify changes in H3K9me3 concentrations. Representative images of dot blots are shown in Figure S9A (Supporting Information). Images were then analyzed to reveal the linear range of the probe as shown in Figure S9B (Supporting Information) and Figure 6. Figures 6A and S9B describe the correlation between the intensity and protein concentrations and RID with protein concentrations, respectively. A close-to-linear relation was observed (see Figure 6A inset) in the protein concentrations ranging from 375 to 3000 μg mL–1 (R2 = 0.995 and a slop of 0.94 ± 0.04). The signal to noise ratio (SNR) within this probing range is consistently higher than 3 (Figure 6B), suggesting good signal quality based on the current standard.39,40
Figure 6.
In vitro application of the recombinant protein sensor ΔCD−ΔCSD–mEGFP. Relative ID (RID) (A) and signal to noise ratio (SNR) (B) are plotted against protein concentrations in HEK293T NCEs. (A) Inset shows the linear detection range. (C) Images of fixed HEK293T cells stained with recombinant ΔCD−ΔCSD–mEGFP.
Staining fixed HEK293T cells with the recombinant protein sensor ΔCD−ΔCSD–mEGFP was carried out using a similar protocol to conventional immunostaining. Figure 6C shows representative images of fixed cells stained with the protein sensor. Like antibody-stained images (see also Figures 2A and S2), characteristic features such as isolated islands, distinctive foci, nuclear periphery, and nucleolus rings are clearly visible in the stained cells. These observations agree with what has been reported in the literature, where heterochromatin, in which H3K9me3 has been recognized as an epigenetic hallmark,11 is mainly positioned at the nuclear periphery, interacting with the nuclear lamina and presenting around nucleoli.41,42 The isolated islands and foci rich in H3K9me3 have been associated with the heterochromatin responsible for cell senescence43,44 and pericentromeric regions.45,46
To this end, we have developed a sensor to probe H3K9me3 levels in situ and in vitro. The engineered sensors provide spatial resolution and quantitative accuracy comparable to commercial antibodies but offer the flexibility of live-cell tracking and lower costs. The probes were applied to understand how epigenetic drugs and environmental chemicals can affect H3K9me3 changes at a single cell level. The probes can be easily extended to study other epigenetic drugs and chemicals in situ.
Materials and Methods
Development of H3K9me3 Sensors
Lysine methylation in histones is typically recognized by epigenetic reader domains such as chromo-, chromobarrel- or tudor domains.47 Among them, the CD of HP1 has high affinities for H3K9me3 (HP1α (human): KD = 13 ± 3 μM; and HP1a (Drosophila): KD = 4 ± 1 μM) and high selectivity [KD > 500 μM for unmodified H3 for HP1α (human) and 1 mM for HP1a (Drosophila)].25,48,49 A survey among different HP1 orthologs suggests that the CD found in Drosophila melanogaster (HP1a) has the highest reported affinity and selectivity for H3K9me3.48 We thus chose to work with CDs from HP1a. HP1a is a multidomain protein containing a CD and a CSD connected by a flexible hinge.50 Although CD is considered to be primarily responsible for recognizing H3K9me3,25,51 CSD has been reported to enhance the target recognition by stabilizing the neighboring interactions.30−32 The hinge domain, however, has been shown to interact with ligands like RNA and DNA and non-histone proteins like deacetylases,32 thus potentially conferring off-target effects. Based on this, we designed protein sensors using the truncated CD (ΔCD, containing residues 24–82 from HP1a UniProtKB-P05205) and/or CSD (ΔCSD, containing residues 147–205 from HP1a UniProtKB-P05205) lacking the original hinge connector as shown in Figure S10. The flexible hinge was not included in our design to minimize off-target interactions. Multimeric domain designs were used to enhance the performance of our sensor, as we have demonstrated previously.34 The detailed primary structure of designed sensors is included in Table S2 (Supporting Information). Glycine linkers are used for connecting two functional domains.
Gene fragments encoding the monomeric sensor, that is, ΔCD and ΔCSD, were custom-synthesized (GenScript). Dimeric probes were cloned using compatible restriction enzyme pairs (i.e., EcoRI, MfeI, and SalI), as we have described in our previous work.34 The probes were subcloned into pET-21b (+) and pRK5 vectors for expressing in bacteria and human cells, respectively. The encoding fragment was verified using gel electrophoresis [Figure S11 (Supporting Information)] and DNA sequencing. Recombinant protein sensors were expressed using Escherichia coli host strains [detailed in Supporting Methods (Supporting Information)].
Mammalian Cell Culture
Human embryonic kidney 293T (HEK293T) cells were cultured as described previously.34 Briefly, cells were cultured in a Dulbecco modified Eagle medium supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) of a penicillin–streptomycin solution (Gibco, CA, US) and incubated at 37 °C with 5% CO2. Cells (3 × 104 cells per well) were seeded onto μslide 8-well and grid-500 coverslips (Ibidi, WI, US) for live-cell imaging and single cell tracking, respectively.
BIX-01294 (Sigma, MO, US) is a potent histone methyltransferase (HMTase) inhibitor (IC50 = 1.9 μM for G9a and IC50 = 38 μM for GLP)52,53 for reducing cellular H3K9me3 levels.54 Cells were treated with BIX-01294 at a concentration of 0, 3, or 5 μM for 24 h to obtain cell population with different H3K9me3 levels. The exposure dose and duration were selected because they were found effective in altering histone methylation levels while cell viability remains minimally perturbed.55 The relative changes in H3K9me3 were also individually characterized using NCEs via immunoassays [approach detailed in Supporting Methods (Supporting Information)].
Fluorescent Microscopy
Cells were imaged using a Nikon Eclipse Ti-2 inverted microscope using 63×/1.49 NA oil objective. Images of cells were collected using Nikon EZ-C1 software. To validate our engineered protein sensor, sensor-transfected cells were fixed, followed by immunostaining with the H3K9me3 antibody as we have described previously.34 The anti-H3K9me3 antibody (ab8898, Abcam, CA, US) was used as a primary and an Alexa 568 coupled to the goat anti rabbit antibody was used as a secondary (ab175471, Abcam, CA, US). All collected images were analyzed using ImageJ (NIH) and/or CellProfiler (BROAD Institute).56
Selection of the Environmental Chemical Treatment
Atrazine (ATZ, 2-chloro-4-ethylamino-6-isopropylamino-1,3,5-triazine) is an herbicide widely used in the United States.57 ATZ can affect endogenous hormone signaling pathways and is thus a potential endocrine disrupting chemical.58−60 ATZ exposure has been affiliated with DNA hypomethylation in common carp (Cyprinus carpio L.).61,62 Down regulation of H3K4me3 have also been observed in a mice study.63 Little is known about how ATZ may affect H3K9me3. We treated HEK293T cells with 3 and 30 ppb of ATZ for 24 h. The treatment dose was selected because exposure to these doses has shown to alter the transcriptome of genes associated with the reproductive system function and development in zebrafish embryos, while minimal mortality and morphological changes were observed.64 ATZ of up to 5.936 ppb in community water systems has been reported in a recent study conducted at Indiana, USA.65 To investigate the ATZ effect on H3K9me3, cells were treated with ATZ and imaged similar to what we have described for BIX-01294 in the previous section.
In Vitro Characterization of the Selected Protein Sensor
To characterize the sensing properties of selected protein sensors in vitro, they were expressed as recombinant proteins in an E. coli system and purified individually as described in the Supporting Information. Briefly, the protein sensors, ΔCD−ΔCSD and ΔCD−ΔCSD–mEGFP, were subcloned into a pET-21b(+) vector with a His-tag on its C-terminus. The quality of the purified protein was verified using SDS-PAGE and Western blot (Figure S12, Supporting Information). ΔCD−ΔCSD was used to characterize the binding affinity and selectivity of the probe via Bio-Layer Interferometry. ΔCD−ΔCSD–mEGFP was used to probe changes in H3K9me3 levels of cell extracts using dot blots. The blotting membranes were imaged and analyzed to reveal their integrated density (ID) similar as described in the literature.66 We defined the signal as the relative ID (RID) following eq 1 and the SNR following (eq 2)
| 1 |
| 2 |
where IDS, IDB, and IDSS are the integrated intensity of a sample spot, the background, and the saturation level, respectively. X̅S and SS stand for the mean and standard deviation of RID, respectively. Based on the RID definition, it accounts for the measured fluorescence intensity emitted by the protein sensor corrected for run-to-run variations. RID and SNR will thus be used to monitor the accuracy of our dot blot assays. To further validate the feasibility of our selected protein sensors, fixed cells were stained with purified ΔCD−ΔCSD–mEGFP [see Supporting Methods (Supporting Information)], qualitatively analyzed, and compared to conventional immunostaining.
Statistical Analysis
Experimental assays were conducted using biological triplicates. Ensemble data are presented as average and standard deviation. Mander’s colocalization coefficients (M1 and M2) were used to assess similarity in binding patterns. Coste’s thresholding was used for background correction. Pairwise statistical difference was analyzed using an ANOVA, followed by the Duncan’s multiple-range test. Differences between groups were considered significant when p < 0.05, on OriginPro (OriginLab Corp, Northampton, MA). Comparisons between distributions where the distribution was non-normal were made using the Wilcoxon Rank Sum test via the Wilcoxon test function as implemented in R (Version 3.4.0).
Acknowledgments
O.S. and A.M. would like to thank Dr. James A. Schaber from the Bindley Bioscience Imaging Facility for helping with cell imaging. This work was supported by US Army Medical Research [W81XWH-14-1-0012], National Science Foundation [CBET-1512285 & CBET-1705560], and Indiana Clinical and Translational Science Institute. Administrative Department of Science, Technology and Innovation from Colombia and Fulbright Colombia [529 to O.F.S]. The authors gratefully acknowledge the support from the Purdue University Center for Cancer Research, NIH grant P30 CA023168.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.9b01413.
Detail experimental procedures for in vitro experiments; representative images of HEK293T cells transfected with A. monomeric and B. dimericchromodomain protein sensors; representative images of transfected HEK293T cells with A. monomeric and B dimeric sensors and costained with the anti-H3K9me3 antibody and an Alexa 564 secondary antibody; representative images of HEK293T cells transfected with the mutated protein sensor ΔCD(KW42/43AA)-ΔCSD; affinity measurements via biolayer interferometry (BLI). A.Comparison of binding curves of ΔCD-ΔCSDwith wild-type H3 and H3K9me3 protein; growth curve (○, untransfected and□, transfected) and viability (■: transfected relative to untransfected) of cells; selection of a defined area using cells transfected with the ΔCD-ΔCSD protein sensor over successive time points; effect of BIX-01294 on global H3K9me3 of HEK293T cells after 48 h of exposure; 2D images of a selected HEK293T cell transfected with ΔCD-ΔCSD in a z-stack; in vitro quantification of H3K9me3 via quantitative dot blot using the protein sensor ΔCD-ΔCSD-mEGFP; sequences of synthetic H3 peptides; and detailed amino acid sequences of various sensor designs (PDF)
Author Contributions
O.F.S and C.Y. designed the experiments. O.F.S. and A.M performed the experiments. A.M. performed statistical analysis. O.F.S., A.M., A.M., J.L., and C.Y aided in interpreting the results. J.L. provided the material source. All the authors provided critical feedback and helped shape the research and manuscript. All the authors approved the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Hemberger M.; Dean W.; Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nat. Rev. Mol. Cell Biol. 2009, 10, 526–537. 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- Mani S. A.; Guo W.; Liao M.-J.; Eaton E. N.; Ayyanan A.; Zhou A. Y.; Brooks M.; Reinhard F.; Zhang C. C.; Shipitsin M.; Campbell L. L.; Polyak K.; Brisken C.; Yang J.; Weinberg R. A. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S.; Xu J.; Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald O. G.; Wu H.; Timp W.; Doi A.; Feinberg A. P. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat. Struct. Mol. Biol. 2011, 18, 867–874. 10.1038/nsmb.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.-Y.; Pu M.-T.; Hirasawa R.; Li B.-Z.; Huang Y.-N.; Zeng R.; Jing N.-H.; Chen T.; Li E.; Sasaki H.; Xu G.-L. Synergistic Function of DNA Methyltransferases Dnmt3a and Dnmt3b in the Methylation of Oct4 and Nanog. Mol. Cell. Biol. 2007, 27, 8748–8759. 10.1128/mcb.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.; Vodyanik M. A.; Smuga-Otto K.; Antosiewicz-Bourget J.; Frane J. L.; Tian S.; Nie J.; Jonsdottir G. A.; Ruotti V.; Stewart R.; Slukvin I. I.; Thomson J. A. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science 2007, 318, 1917–1920. 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Spivakov M.; Fisher A. G. Epigenetic signatures of stem-cell identity. Nat. Rev. Genet. 2007, 8, 263–271. 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- Lanctôt C.; Cheutin T.; Cremer M.; Cavalli G.; Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 2007, 8, 104–115. 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- Cremer M.; Küpper K.; Wagler B.; Wizelman L.; Hase J. v.; Weiland Y.; Kreja L.; Diebold J.; Speicher M. R.; Cremer T. Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei. J. Cell Biol. 2003, 162, 809–820. 10.1083/jcb.200304096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. S.; Punina E. O.; Rodionov A. V. Chromosome Cpd (pi/dapi)-and Cma/dapi-banding Patterns in Allium cepa L. Russ. J. Genet. 2002, 38, 392–398. 10.1023/a:1015250219322. [DOI] [PubMed] [Google Scholar]
- Saksouk N.; Simboeck E.; Déjardin J. Constitutive heterochromatin formation and transcription in mammals. Epigenet. Chromatin 2015, 8, 1–17. 10.1186/1756-8935-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessler J. B.; Andersen E. C.; Villeneuve A. M. Differential Localization and Independent Acquisition of the H3K9me2 and H3K9me3 Chromatin Modifications in the Caenorhabditis elegans Adult Germ Line. PLoS Genet. 2010, 6, e1000830 10.1371/journal.pgen.1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarte F.; Sousae R.; Cinquin B.; Martin E. W.; Krietsch J.; Sanchez G.; Inman M.; Tsang H.; Warr M.; Passegué E.; Larabell C. A.; Forsberg E. C. Progressive Chromatin Condensation and H3K9 Methylation Regulate the Differentiation of Embryonic and Hematopoietic Stem Cells. Stem Cell Rep. 2015, 5, 728–740. 10.1016/j.stemcr.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slee R. B.; Steiner C. M.; Herbert B.-S.; Vance G. H.; Hickey R. J.; Schwarz T.; Christan S.; Radovich M.; Schneider B. P.; Schindelhauer D.; Grimes B. R. Cancer-associated alteration of pericentromeric heterochromatin may contribute to chromosome instability. Oncogene 2012, 31, 3244–3253. 10.1038/onc.2011.502. [DOI] [PubMed] [Google Scholar]
- Mansuroglu Z.; Benhelli-Mokrani H.; Marcato V.; Sultan A.; Violet M.; Chauderlier A.; Delattre L.; Loyens A.; Talahari S.; Bégard S. Loss of Tau protein affects the structure, transcription and repair of neuronal pericentromeric heterochromatin. Sci. Rep. 2016, 6, 33047. 10.1038/srep33047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhofer T. A.; Minoda A.; Klugman S.; Lee K.; Kolasinska-Zwierz P.; Alekseyenko A. A.; Cheung M.-S.; Day D. S.; Gadel S.; Gorchakov A. A.; Gu T.; Kharchenko P. V.; Kuan S.; Latorre I.; Linder-Basso D.; Luu Y.; Ngo Q.; Perry M.; Rechtsteiner A.; Riddle N. C.; Schwartz Y. B.; Shanower G. A.; Vielle A.; Ahringer J.; Elgin S. C. R.; Kuroda M. I.; Pirrotta V.; Ren B.; Strome S.; Park P. J.; Karpen G. H.; Hawkins R. D.; Lieb J. D. An assessment of histone-modification antibody quality. Nat. Struct. Mol. Biol. 2011, 18, 91–93. 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Takanaka Y.; Yamagata K.; Wakayama T.; Stasevich T. J.; Kainuma T.; Tsurimoto T.; Tachibana M.; Shinkai Y.; Kurumizaka H.; Nozaki N.; Kimura H. Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling. Nucleic Acids Res. 2011, 39, 6475. 10.1093/nar/gkr343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y.; Mukai M.; Ueda J.; Muraki M.; Stasevich T. J.; Horikoshi N.; Kujirai T.; Kita H.; Kimura T.; Hira S. Genetically encoded system to track histone modification in vivo. Sci. Rep. 2013, 3, 2436. 10.1038/srep02436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (US) Committee on Methods of Producing Monoclonal Antibodies . Summary of Advantages and Disadvantages of In Vitro and In Vivo Methods; National Academic Press: Washington D.C., 1999; pp 22–24. [Google Scholar]
- Kimura H.; Sato Y.; Zhang J.; Schultz C.. Histone Modification Sensors in Living Cells. In Optical Probes in Biology; CRC Press: Florida, 2015; pp 317−334. [Google Scholar]
- Sato Y.; Kujirai T.; Arai R.; Asakawa H.; Ohtsuki C.; Horikoshi N.; Yamagata K.; Ueda J.; Nagase T.; Haraguchi T.; Hiraoka Y.; Kimura A.; Kurumizaka H.; Kimura H. A Genetically Encoded Probe for Live-Cell Imaging of H4K20 Monomethylation. J. Mol. Biol. 2016, 428, 3885–3902. 10.1016/j.jmb.2016.08.010. [DOI] [PubMed] [Google Scholar]
- Suzuki M.; Takagi C.; Miura S.; Sakane Y.; Suzuki M.; Sakuma T.; Sakamoto N.; Endo T.; Kamei Y.; Sato Y. In vivo tracking of histone H3 lysine 9 acetylation in Xenopus laevis during tail regeneration. Genes Cells 2016, 21, 358. 10.1111/gtc.12349. [DOI] [PubMed] [Google Scholar]
- Lin C.-W.; Jao C. Y.; Ting A. Y. Genetically Encoded Fluorescent Reporters of Histone Methylation in Living Cells. J. Am. Chem. Soc. 2004, 126, 5982–5983. 10.1021/ja038854h. [DOI] [PubMed] [Google Scholar]
- Lungu C.; Pinter S.; Broche J.; Rathert P.; Jeltsch A. Modular fluorescence complementation sensors for live cell detection of epigenetic signals at endogenous genomic sites. Nat. Commun. 2017, 8, 649. 10.1038/s41467-017-00457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W.; Tseng B. S.; Dormann H. L.; Ueberheide B. M.; Garcia B. A.; Shabanowitz J.; Hunt D. F.; Funabiki H.; Allis C. D. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 2005, 438, 1116–1122. 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- Dialynas G. K.; Makatsori D.; Kourmouli N.; Theodoropoulos P. A.; McLean K.; Terjung S.; Singh P. B.; Georgatos S. D. Methylation-independent Binding to Histone H3 and Cell Cycle-dependent Incorporation of HP1β into Heterochromatin. J. Biol. Chem. 2006, 281, 14350–14360. 10.1074/jbc.m600558200. [DOI] [PubMed] [Google Scholar]
- Dialynas G. K.; Terjung S.; Brown J. P.; Aucott R. L.; Baron-Luhr B.; Singh P. B.; Georgatos S. D. Plasticity of HP1 proteins in mammalian cells. J. Cell Sci. 2007, 120, 3415–3424. 10.1242/jcs.012914. [DOI] [PubMed] [Google Scholar]
- Bosch-Presegué L.; Raurell-Vila H.; Thackray J. K.; González J.; Casal C.; Kane-Goldsmith N.; Vizoso M.; Brown J. P.; Gómez A.; Ausió J.; Zimmermann T.; Esteller M.; Schotta G.; Singh P. B.; Serrano L.; Vaquero A. Mammalian HP1 Isoforms Have Specific Roles in Heterochromatin Structure and Organization. Cell Rep. 2017, 21, 2048–2057. 10.1016/j.celrep.2017.10.092. [DOI] [PubMed] [Google Scholar]
- Dunn K. W.; Kamocka M. M.; McDonald J. H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 2011, 300, C723–C742. 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richart A. N.; Brunner C. I. W.; Stott K.; Murzina N. V.; Thomas J. O. Characterization of Chromoshadow Domain-mediated Binding of Heterochromatin Protein 1α (HP1α) to Histone H3. J. Biol. Chem. 2012, 287, 18730–18737. 10.1074/jbc.m111.337204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowieson N. P.; Partridge J. F.; Allshire R. C.; McLaughlin P. J. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr. Biol. 2000, 10, 517–525. 10.1016/s0960-9822(00)00467-x. [DOI] [PubMed] [Google Scholar]
- Smothers J. F.; Henikoff S. The Hinge and Chromo Shadow Domain Impart Distinct Targeting of HP1-Like Proteins. Mol. Cell. Biol. 2001, 21, 2555–2569. 10.1128/mcb.21.7.2555-2569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A. J.; Zegerman P.; Partridge J. F.; Miska E. A.; Thomas J. O.; Allshire R. C.; Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 2001, 410, 120. 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Sanchez O. F.; Mendonca A.; Carneiro A. D.; Yuan C. Engineering Recombinant Protein Sensors for Quantifying Histone Acetylation. ACS Sens. 2017, 2, 426. 10.1021/acssensors.7b00026. [DOI] [PubMed] [Google Scholar]
- Park J.-A.; Kim A.-J.; Kang Y.; Jung Y.-J.; Kim H. K.; Kim K.-C. Deacetylation and Methylation at Histone H3 Lysine 9 (H3K9) Coordinate Chromosome Condensation during Cell Cycle Progression. Mol. Cells 2011, 31, 343–349. 10.1007/s10059-011-0044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I.-H.; Nagel J.; Otten S.; Knerr B.; Eils R.; Rohr K.; Dietzel S. Quantitative comparison of DNA detection by GFP-lac repressor tagging, fluorescence in situ hybridization and immunostaining. BMC Biotechnol. 2007, 7, 92. 10.1186/1472-6750-7-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punga T.; Bühler M. Long intronic GAA repeats causing Friedreich ataxia impede transcription elongation. EMBO Mol. Med. 2010, 2, 120–129. 10.1002/emmm.201000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navakauskienė R.; Mori M.; Christodoulou M.; Zentelytė A.; Botta B.; Dalla Via L.; Ricci F.; Damia G.; Passarella D.; Zilio C. Histone demethylating agents as potential S-adenosyl-L-methionine-competitors. MedChemComm 2016, 7, 1245–1255. 10.1039/c6md00170j. [DOI] [Google Scholar]
- Lu X.; Dong X.; Zhang K.; Han X.; Fang X.; Zhang Y. A gold nanorods-based fluorescent biosensor for the detection of hepatitis B virus DNA based on fluorescence resonance energy transfer. Analyst 2013, 138, 642–650. 10.1039/c2an36099c. [DOI] [PubMed] [Google Scholar]
- Wang W.; Chen C.; Qian M.; Zhao X. S. Aptamer biosensor for protein detection using gold nanoparticles. Anal. Biochem. 2008, 373, 213–219. 10.1016/j.ab.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Kalverda B.; Röling M. D.; Fornerod M. Chromatin organization in relation to the nuclear periphery. FEBS Lett. 2008, 582, 2017–2022. 10.1016/j.febslet.2008.04.015. [DOI] [PubMed] [Google Scholar]
- Padeken J.; Heun P. Nucleolus and nuclear periphery: Velcro for heterochromatin. Curr. Opin. Cell Biol. 2014, 28, 54–60. 10.1016/j.ceb.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Zhang R.; Chen W.; Adams P. D. Molecular dissection of formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 2007, 27, 2343–2358. 10.1128/mcb.02019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black J. C.; Whetstine J. R. Chromatin landscape: Methylation beyond transcription. Epigenetics 2011, 6, 9–15. 10.4161/epi.6.1.13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksouk N.; Simboeck E.; Déjardin J. Constitutive heterochromatin formation and transcription in mammals. Epigenet. Chromatin 2015, 8, 3. 10.1186/1756-8935-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déjardin J. Switching between Epigenetic States at Pericentromeric Heterochromatin. Trends Genet. 2015, 31, 661–672. 10.1016/j.tig.2015.09.003. [DOI] [PubMed] [Google Scholar]
- Greer E. L.; Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes R. M.; Wiggins K. R.; Khorasanizadeh S.; Waters M. L. Recognition of trimethyllysine by a chromodomain is not driven by the hydrophobic effect. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 11184–11188. 10.1073/pnas.0610850104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W.; Wang Y.; Jacobs S. A.; Kim Y.; Allis C. D.; Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003, 17, 1870–1881. 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomberk G.; Wallrath L.; Urrutia R. The Heterochromatin Protein 1 family. Genome Biol. 2006, 7, 228. 10.1186/gb-2006-7-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P. R.; Nietlispach D.; Mott H. R.; Callaghan J.; Bannister A.; Kouzarides T.; Murzin A. G.; Murzina N. V.; Laue E. D. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 2002, 416, 103–107. 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- Chang Y.; Zhang X.; Horton J. R.; Upadhyay A. K.; Spannhoff A.; Liu J.; Snyder J. P.; Bedford M. T.; Cheng X. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nat. Struct. Mol. Biol. 2009, 16, 312–317. 10.1038/nsmb.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek S.; O’Sullivan R. J.; August E. M.; Hickey E. R.; Zhang Q.; Teodoro M. L.; Rea S.; Mechtler K.; Kowalski J. A.; Homon C. A.; Kelly T. A.; Jenuwein T. Reversal of H3K9me2 by a Small-Molecule Inhibitor for the G9a Histone Methyltransferase. Mol. Cell 2007, 25, 473–481. 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Coward W. R.; Feghali-Bostwick C. A.; Jenkins G.; Knox A. J.; Pang L. A central role for G9a and EZH2 in the epigenetic silencing of cyclooxygenase-2 in idiopathic pulmonary fibrosis. FASEB J. 2014, 28, 3183–3196. 10.1096/fj.13-241760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backliwal G.; Hildinger M.; Kuettel I.; Delegrange F.; Hacker D. L.; Wurm F. M. Valproic acid: a viable alternative to sodium butyrate for enhancing protein expression in mammalian cell cultures. Biotechnol. Bioeng. 2008, 101, 182–189. 10.1002/bit.21882. [DOI] [PubMed] [Google Scholar]
- Jones T. R.; Kang I.; Wheeler D. B.; Lindquist R. A.; Papallo A.; Sabatini D. M.; Golland P.; Carpenter A. E. CellProfiler Analyst: data exploration and analysis software for complex image-based screens. BMC Bioinf. 2008, 9, 482. 10.1186/1471-2105-9-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USEPA Atrazine Chemical Summary. https://archive.epa.gov/region5/teach/web/pdf/atrazine_summary.pdf (accessed Feb 11, 2017).
- Bisson M.; Hontela A. Cytotoxic and endocrine-disrupting potential of atrazine, diazinon, endosulfan, and mancozeb in adrenocortical steroidogenic cells of rainbow trout exposed in vitro. Toxicol. Appl. Pharmacol. 2002, 180, 110–117. 10.1006/taap.2002.9377. [DOI] [PubMed] [Google Scholar]
- Giusi G.; Facciolo R. M.; Canonaco M.; Alleva E.; Belloni V.; Dessi’-Fulgheri F.; Santucci D. The endocrine disruptor atrazine accounts for a dimorphic somatostatinergic neuronal expression pattern in mice. Toxicol. Sci. 2006, 89, 257–264. 10.1093/toxsci/kfj012. [DOI] [PubMed] [Google Scholar]
- Jin Y.; Wang L.; Chen G.; Lin X.; Miao W.; Fu Z. Exposure of mice to atrazine and its metabolite diaminochlorotriazine elicits oxidative stress and endocrine disruption. Environ. Toxicol. Pharmacol. 2014, 37, 782–790. 10.1016/j.etap.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Wang C.; Zhang Z.; Yao H.; Zhao F.; Wang L.; Wang X.; Xing H.; Xu S. Effects of atrazine and chlorpyrifos on DNA methylation in the liver, kidney and gill of the common carp (Cyprinus carpio L.). Ecotoxicol. Environ. Saf. 2014, 108, 142–151. 10.1016/j.ecoenv.2014.06.011. [DOI] [PubMed] [Google Scholar]
- Xing H.; Wang C.; Wu H.; Chen D.; Li S.; Xu S. Effects of atrazine and chlorpyrifos on DNA methylation in the brain and gonad of the common carp. Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol. 2015, 168, 11–19. 10.1016/j.cbpc.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Hao C.; Gely-Pernot A.; Kervarrec C.; Boudjema M.; Becker E.; Khil P.; Tevosian S.; Jégou B.; Smagulova F. Exposure to the widely used herbicide atrazine results in deregulation of global tissue-specific RNA transcription in the third generation and is associated with a global decrease of histone trimethylation in mice. Nucleic Acids Res. 2016, 44, 9784. 10.1093/nar/gkw840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G. J.; Sepúlveda M. S.; Peterson S. M.; Lewis S. S.; Freeman J. L. Transcriptome Alterations Following Developmental Atrazine Exposure in Zebrafish Are Associated with Disruption of Neuroendocrine and Reproductive System Function, Cell Cycle, and Carcinogenesis. Toxicol. Sci. 2013, 132, 458–466. 10.1093/toxsci/kft017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Acuña H.; Frankenberger J.; Hahn L.; Carbajo C. Drinking-water herbicide exposure in Indiana and prevalence of small-for-gestational-age and preterm delivery. Environ. Health Perspect. 2009, 117, 1619. 10.1289/ehp.0900784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen L.; Chingwaru W.; Sgorbati B.; Biavati B.; Cencic A. Gut health promoting activity of new putative probiotic/protective Lactobacillus spp. strains: A functional study in the small intestinal cell model. Int. J. Food Microbiol. 2009, 135, 288–294. 10.1016/j.ijfoodmicro.2009.08.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.