Abstract
Objectives:
Gut bacteria play an essential role during infancy and are strongly influenced by the mode of birth and feeding. A primate model was used to investigate the benefits of exposure to the mother or conversely the negative impact of early nursery rearing on microbial colonization.
Method:
Rectal swabs were obtained from rhesus macaques born vaginally and mother-reared (MR, N=35) or delivered primarily via cesarean-section and human-reared (HR, N=19). Microbiome composition was determined by rRNA gene amplicon sequencing at 2, 4 and 8 weeks of age and KEGG orthologs used to assess influences on functional metabolic pathways in the gut. Growth trajectories and incidence of diarrheic symptoms were evaluated.
Results:
The microbial community structure was different between MR and HR infants with respect to phylogeny and abundance at all 3 ages. When examining dominant phyla, HR infants had a higher Firmicutes-to-Bacteroidetes ratio. At the genus level, breast milk-dependent commensal taxa and adult-typical genera were more abundant in MR infants. This difference resulted in a corresponding shift in the predicted metabolic effects, specifically for microbial genes associated with metabolism and immune function. HR infants had faster growth trajectories (p<0.001), but more diarrheic symptoms by 6 months postnatal (p=0.008).
Conclusions:
MR infants acquired adult-typical microbiota more quickly, and had higher levels of several beneficial commensal taxa. Cesarean-delivered and formula-fed infants had different developmental trajectories of bacterial colonization. Establishment of the gut microbiome was associated with an infant’s growth trajectory, and implicated in the subsequent vulnerability to Campylobacter infections associated with diarrhea in infant monkeys.
Keywords: Breastfeeding, infant nutrition, cesarean section, Bifidobacteria
INTRODUCTION
Birth is an abrupt event transitioning the neonate from relatively sterile uterine conditions into the external world. The full significance of the rapid microbial colonization that begins during delivery and continues postpartum through exposure to microorganisms from the mother and rearing environment is just beginning to be appreciated (1,2). These microbiota have a critical role supporting intestinal homeostasis, stimulating immunity, and influencing host metabolism, and may even contribute to the emergence of different behavioral phenotypes (3–5). Though the foundations of the microbiome can be influenced by prenatal conditions, the community structure of the microbiome is largely impacted by delivery mode and infant diet, especially by decisions about breast- or formula-feeding (6–9). Breast milk contains numerous proteins and prebiotic oligosaccharide substrates, and provides viable bacteria that compete with pathogens for adherence to the intestinal mucus and epithelial surfaces (10,11). It is important to more fully understand the ramifications of delivery mode and parental decisions about infant feeding because, despite recommendations from the World Health Organization, cesarean delivery is common and only 40% of American infants are exclusively breastfed until 6 months of age (12,13).
A nonhuman primate model was employed to investigate the benefits of exposure to the mother and breast milk for microbial colonization. We hypothesized that mother-reared (MR) infants would have a higher abundance of several commensal taxa as compared to infants initially reared in a nursery setting and fed formula (HR) (14,15). Because the microbiome can affect digestive efficiency, host metabolism, and protect against enteric pathogens, we also looked for differences in growth and the later incidence of diarrheic episodes.
METHODS
Subjects.
Fifty-four infant rhesus macaques (Macaca mulatta) were generated from healthy, multiparous mothers at the Harlow Center for Biological Psychology and Wisconsin Primate Research Center. Thirty-five were housed with their mothers and exclusively breast-fed (MR). All were full-term vaginal deliveries. Nineteen were fed formula and reared by humans (HR) for one month in isolate incubators mimicking a Neonatal Intensive Care Unit (NICU) setting, with 13 delivered by cesarean-section (See Text, Supplemental Digital Content 1 for further details of HR husbandry). Infants from both rearing conditions were randomly selected to be representative of the population born between 2016–2018. Mothers and older infants were fed a commercial biscuit diet, supplemented with fruit. Typically, MR infants may first sample biscuits between 2–4 weeks after birth. HR infants were introduced to chow at 2 weeks of age and were progressively weaned from formula by 2 months. All protocols were approved by the Institutional Animal Care and Use Committee and conducted in accordance with federal guidelines.
Rectal swabs were obtained at 2, 4 or 8 weeks of age. Genomic DNA was extracted and quantified using a Qubit 2.0 Fluorometer, and amplicon sequencing performed on Illumina MiSeq (See Text, Supplemental Digital Content 2, for further details of specimen collection, bacterial DNA isolation, and sequence analysis). Infant growth and health were closely monitored. No infants were treated with antibiotics during the sampling period; however, the mothers of 5 MR infants had been administered antibiotics perinatally. Infants were first weighed at a mean 3.65 (± 0.61) days, and then regularly at 2–8 week intervals. Growth was indexed by weight, as well as growth rate between birth and 8 months. Diarrheic symptoms and treatments were recorded. In the majority of cases, to determine if Campylobacter was the cause, plates with selective media were inoculated (Campy CVA Medium and Charcoal Selective Medium), and streaked for isolation. Identification was made using a MALDI-TOF analyzer at the UW Veterinary Clinical Pathology Lab.
Sequence and Statistical Analysis.
Significance was tested by implementations in Quantitative Insights into Microbial Ecology (QIIME) (16), PAST (PAleontological STatistics) (17), and R Statistics, including ANOVA, Analysis of Similarity (ANOSIM), nonparametric t-tests (Kruskal-Wallis), and adjustments for False Discovery Rates (FDR). Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) (18) was utilized to correct for variable 16S copy number. Taxonomic analyses were restricted to more abundant taxa present at a minimum 1% composition in total observations across samples and are presented at phylum and genus levels. To examine diversity indices, Faith’s Phylogenetic Diversity (PD), Chao1 Index, and Observed Species were generated to examine richness (alpha diversity), and weighted UniFrac dissimilarity matrices examined for differences in microbial community structure (beta diversity) while accounting for both abundance and phylogenetic relatedness. Repeated measures ANOVA examined developmental changes in microbial composition, followed by pairwise comparisons adjusted for multiplicity.
Results are presented in two ways: first comparing the MR and HR infants at the 3 age points. Serial changes in community structure for the MR infants are then described (See Text, Supplemental Digital Content 3).
Using GreenGenes 13_5 (19) assigned OTUs, PICRUSt was employed to predict microbial metagenomes from the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthologs in order to evaluate whether shifts in community structure influenced the functional potential of bacterial communities. Finally, linear discriminant analysis (LDA) effect size algorithm from LEfSe (20) identified microbial taxa that distinguished the two rearing conditions.
RESULTS
Analysis 1: Community structure.
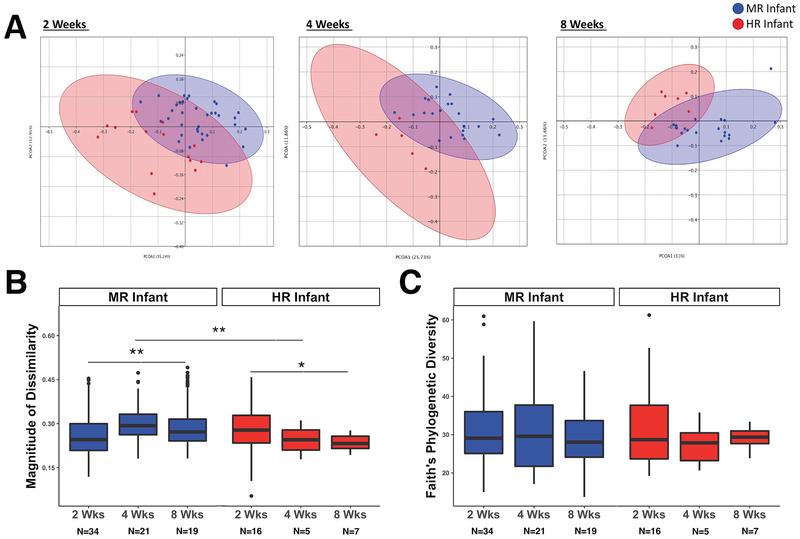
102 rectal swabs were collected from 35 MR infants and 19 HR infants at 2, 4 and 8 weeks of age (See Table, Supplemental Digital Content 4, for more details). The microbial diversity and composition for the 5 MR infants born to mothers administered perinatal antibiotics did not differ significantly from MR infants without antibiotic exposure. Rearing condition had a significant effect on the gut microbial community structure. Principle Coordinates Analysis for phylogenetic and abundance data revealed a distinctive clustering based on rearing evident at each age point (Figure 1A), and permutational multivariate analysis of variance (PERMANOVA) indicated divergent microbiota at 2, 4 and 8 weeks (p=0.001, p=0.001, p=0.002, respectively). Unpaired t-test comparisons of the mean dissimilarity distances for the microbial profiles indicated that sample-to-sample phylogenetic distances were larger in HR infants by 2 weeks of age, (t=2.99; p<0.001), but the dissimilarity in community structure then shifted and was greater in MR infants at 4 and 8 weeks (t=3.29; p<0.01, t=3.60; p<0.005; Figure 1B). Overall, divergence of the microbial community structure increased in MR infants across the first 2 months of life, while decreasing among HR infants (p<0.001 and p=0.011, respectively). Averaged OTU counts for HR infants were higher than for MR infants (444.62 versus 404.89). However, there was not a large difference in the within-group pairwise phylogenetic distances (PD index; Figure 1C), sample diversity (Chao1), or total species richness (Observed OTUs) at any age point age (See Table, Supplemental Digital Content 5, for alpha diversity metric results).
Figure 1.
Microbiome Diversity Indices across the first two months of life. A) Analyses of beta diversity based on weighted UniFrac distances between samples. Principal coordinates analysis with ellipses representing 95% confidence intervals shows clear separation of microbial profiles between MR and HR infants at 2, 4, and 8 weeks. B) Inter-individual distances illustrate the magnitude of dissimilarity in the profiles, which was significantly larger in HR infants at 2 weeks of age, but was lower than the microbial diversity across all MR infants at 4 and 8 weeks of age. C) Further categorization based on the phylogenetic analogue of taxon richness revealed no effect of rearing condition at any age point. Values are: box, median; whiskers, 25 and 75% quartiles; lines, 1.5 times the interquartile range. Outliers are illustrated by circles.
*, p<.01; **, p<.005.
Taxonomic Abundance.
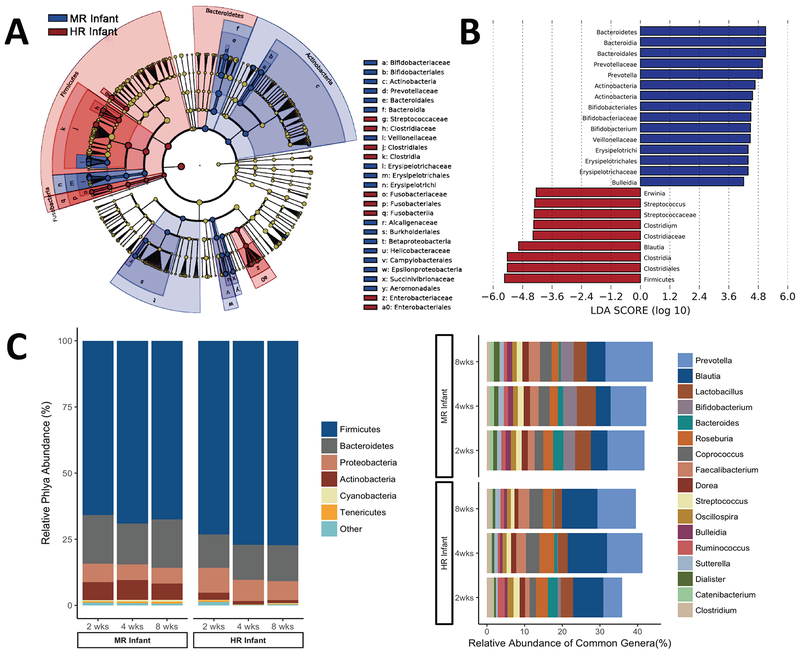
Differences were evident at all taxonomic levels, but most prominent at the phylum and genus levels (Figure 2A,B). Similar to humans, the most abundant microbes were members of the gram-positive Firmicutes and gram-negative Bacteroidetes phyla. However, the Firmicutes-to-Bacteroidetes ratio was impacted by rearing, with Firmicutes being relatively more abundant in HR infants at 2, 4 and 8 weeks of age (p<0.001, p=0.05, p<0.001, respectively; Figure 2C). Within Bacteroidetes, Prevotella was consistently the most abundant genus in MR infants, comprising an average 9.9% compared to only 5% in HR infants at 2 weeks of age (p=0.03). The highest Prevotella levels were seen in MR infants at 8 weeks (13%), an abundance similar to adult monkeys.
Figure 2.
Rearing environment predicted differences in taxonomic composition of gut microbiota. A) Taxonomic cladogram plotted from LEfSe analysis of 16S sequences from all time points. (Blue) MR infant enriched taxa; (Red) HR infant enriched taxa. Brightness of each dot is proportional to its effect size. B) Histogram of Linear Discriminant Analysis scores computed for features that distinguish between MR and HR infants which estimates the effect size of each differentially abundant feature (all LDA scores >4.0). C) Stacked bar graphs show abundance of phyla and genera based on pooled data from each rearing condition. Phyla comprising less than 1% abundance were grouped into the ‘other’ category. At the Phylum level, MR infants consistently had a greater abundance of Firmicutes, as well as increased colonization by commensal genera stimulated by breast milk constituents. At the level of genera, only taxa comprising >1% total abundance is included.
Conversely, the genus Blautia (within Firmicutes) was more enriched in HR infants at 4 and 8 weeks of age (10.5% vs. 3.9%, p=0.07; 9.4% vs. 4.9%, p=0.02, respectively). HR infants also had a lower relative abundance of the phyla Actinobacteria (Figure 2B), reflecting a significantly lower abundance of the genus Bifidobacterium at 2, 4, and 8 weeks of age (p<0.001, p=0.007, p=0.002, respectively). Moreover, Bifidobacteria diminished over time in HR infants; constituting only .2% and .15% at 4 and 8 weeks as compared to 1% at 2 weeks (p=0.018). Lactobacilli were also lower in HR than MR infants, but this difference was only significant at 8 weeks (p=0.017) and was small (2% vs. 3.5% abundance). At 2 weeks, the proportion of Clostridium was twice as high in HR compared to MR infants (1.7% vs. .8%), but the abundance decreased with age (p=0.056). The carriage rate, or percentage of infants in which the typically pathogenic Staphylococcus was detected was also higher among HR as compared to MR infants at 2 weeks (75% and 47%, χ2=3.48, p=0.063), but differences in prevalence diminished by 4 weeks of age (p=0.34).
Predicted shifts in functional pathways.
PICRUSt predictions indicated the different community structures would be accompanied by significant shifts in several metabolic pathways, including environmental and genetic information processing, cellular processes, and metabolism (Table 1). At Level 2 of the KEGG pathway analysis, functional predictions for HR infants indicated pathway increases related to energy, carbohydrate and lipid metabolism and xenobiotic biodegradation and metabolism. Conversely, MR infants evinced increases in genes mapping to glycan biosynthesis and pathways associated with metabolism of vitamins, terpenoids and polyketides, and amino acids. KEGG predictions also projected that genes related to environmental adaption and immunity would be downregulated in HR infants, while biochemical pathways implicated in human infectious disease would be enhanced.
TABLE 1.
Significant differences predicted for metabolic pathways associated with the microbiota of Mother and Human-Reared infants*
| KEGG Prediction | Test Statistic | p | FDR-P | MR Mean | HR Mean | Difference |
|---|---|---|---|---|---|---|
| Environmental Information Processing | ||||||
| Signal transduction | 20.56 | 0.000 | 0.000 | 213,446.61 | 278,115.08 | 64,668.47 |
| Membrane transport | 11.39 | 0.001 | 0.003 | 1,858,851.12 | 2,091,898.12 | 233,047.00 |
| Cellular Processes | ||||||
| Cell motility | 17.61 | 0.000 | 0.001 | 264,134.76 | 357,417.04 | 93,282.28 |
| Cellular processes AND signaling | 16.81 | 0.000 | 0.001 | 631,089.28 | 699,712.36 | 68,623.08 |
| Cell growth and death | 5.99 | 0.014 | 0.019 | 86,701.01 | 78,069.84 | 8,631.17 |
| Organismal Systems | ||||||
| Excretory system | 16.41 | 0.000 | 0.001 | 1,625.78 | 2,517.24 | 891.46 |
| Environmental adaptation | 12.44 | 0.000 | 0.002 | 23,713.73 | 26,061.76 | 2,348.03 |
| Immune system | 11.01 | 0.001 | 0.003 | 14,732.84 | 13,770.64 | 962.20 |
| Nervous system | 7.32 | 0.007 | 0.011 | 16,079.82 | 15,362.52 | 717.30 |
| Circulatory system | 6.50 | 0.011 | 0.015 | 1,110.34 | 1,263.84 | 153.50 |
| Human Diseases | ||||||
| Infectious diseases | 14.82 | 0.000 | 0.001 | 62,751.11 | 65,904.16 | 3,153.05 |
| Neurodegenerative diseases | 5.16 | 0.023 | 0.029 | 19,478.16 | 19,588.24 | 110.08 |
| Metabolic diseases | 5.10 | 0.024 | 0.029 | 18,132.22 | 15,709.72 | 2,422.50 |
| Metabolism | ||||||
| Lipid metabolism | 13.43 | 0.000 | 0.002 | 425,769.61 | 456,215.64 | 30,446.03 |
| Energy metabolism | 11.72 | 0.001 | 0.003 | 948,442.86 | 958,682.16 | 10,239.30 |
| Metabolism of cofactors and vitamins | 10.38 | 0.001 | 0.003 | 710,506.43 | 689,044.68 | 21,461.75 |
| Carbohydrate metabolism | 9.92 | 0.002 | 0.004 | 1,673,406.01 | 1,740,094.60 | 66,688.59 |
| Metabolism of terpenoids and polyketides | 9.61 | 0.002 | 0.005 | 273,118.54 | 264,845.20 | 8,273.34 |
| Metabolism of other amino acids | 9.37 | 0.002 | 0.005 | 251,763.42 | 242,611.20 | 9,152.22 |
| Glycan biosynthesis and metabolism | 9.22 | 0.002 | 0.005 | 426,098.57 | 366,852.00 | 59,246.57 |
| Amino acid metabolism | 8.78 | 0.003 | 0.006 | 1,542,558.74 | 1,521,755.72 | 20,803.02 |
| Xenobiotics biodegradation and metabolism | 8.64 | 0.003 | 0.006 | 248,805.08 | 256,821.84 | 8,016.76 |
| Nucleotide metabolism | 6.56 | 0.010 | 0.015 | 698,936.45 | 630,544.24 | 68,392.21 |
| Biosynthesis of other secondary metabolites | 6.03 | 0.014 | 0.019 | 153,533.15 | 141,316.84 | 12,216.31 |
| Genetic Information Processing | 9.56 | 0.002 | 0.005 | 433,820.50 | 423,507.68 | 10,312.82 |
| Transcription | 11.33 | 0.001 | 0.003 | 424,546.05 | 467,090.48 | 42,544.43 |
| Folding, sorting and degradation | 10.53 | 0.001 | 0.003 | 405,204.04 | 389,156.56 | 16,047.48 |
| Replication and repair | 6.94 | 0.008 | 0.013 | 1,514,143.88 | 1,380,323.64 | 133,820.24 |
| Translation | 5.84 | 0.016 | 0.020 | 980,135.35 | 884,161.76 | 95,973.59 |
KEGG predictions are representative of all time points.
Infant Growth and Health Trajectories.
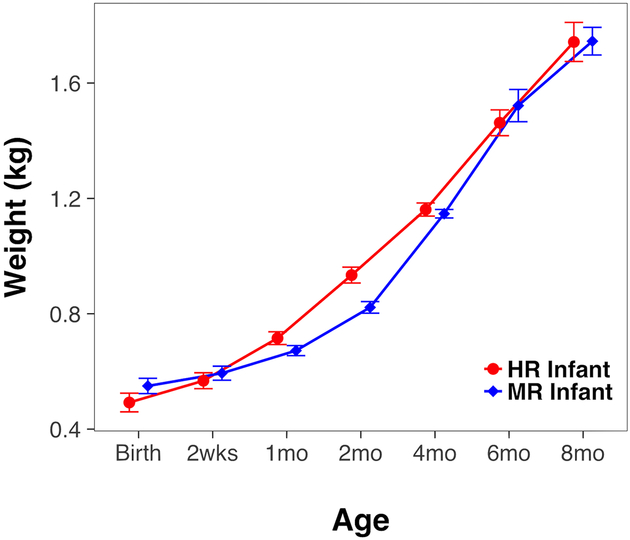
Growth curve analysis was used to analyze weight gain through 8 months of age. The overall growth trajectory of HR infants was faster [F(1,78.77)=14.52, p<0.001; Figure 3]. Despite having similar birth weights (p=0.22), HR infants weighed an average 188 g more at 2 months [F(1, 51)=38.76, p<0.001] and 88 g more at 4 months [F(1, 51)=7.60, p<0.01]. Weight differences diminished by 6 months after infants had transitioned to solid food. The phylogenetic richness of the microbial community structure (PD index) at 8 weeks tended to be positively associated with infant weight at the time of sampling (r=.350, p=0.08), and was significantly correlated with subsequent weights at 4, 6, and 8 months of age (r=.433, p=0.027; r=.408, p=0.043; and r=.418, p=0.034, respectively). More microbial richness at 8 weeks was also associated with a faster growth trajectory (r=.404, p=0.041). When comparing gut health between rearing conditions, more incubator-reared and formula-fed infants were treated for both acute and chronic diarrhea by 6 months of age (7 of 19 HR vs. 3 of 35 MR, χ2=6.53, p=0.011; See Figure, Supplemental Digital Content 6). Differences in vulnerability to enteric pathogens, identified as C. jejuni in 70% of cultured stool, continued to persist after infants were weaned to solids (6 of 19 HR vs. 2 of 35 MR, χ2=4.69, p=0.03).
Figure 3.
Infant growth differed by rearing condition. In keeping with formula-feeding and a unique gut microbial structure, HR infants grew significantly faster until 6 months of age, despite similar birth weights in both rearing conditions. All infants were consuming solid food by 4 months of age when growth rates converged.
DISCUSSION
Our findings concur with prior research demonstrating that bacterial colonization is strongly affected by early rearing conditions. Vaginally-born infants were colonized by microbial populations more closely related to the maternal reproductive tract and gut, and dominated by Prevotella and Lactobacillus (8,21). This natural colonization was perturbed if delivered by cesarean-section and instead the infants show a delayed colonization by Bacteroides and Bifidobacterium and may have been enriched in skin microbiota, including Staphylococcus, thus resembling the gut microbial composition of human infants exposed to a NICU (22–24).
Breastfeeding is a second major factor influencing the establishment of the microbiome during the neonatal period. It favored the growth of commensal Bifidobacteria and Lactobacillus, whereas the formula-fed infant monkeys had an overrepresentation of Clostridium (25–27). However, while Bifidobacteria species can comprise nearly 81% of the gut microbiota in vaginally-born and breastfed human infants (7), levels were considerably lower in the monkey (28). This species difference may be due to the higher concentrations of Bifidobacterium-promoting oligosaccharides present in human milk. The ratio of oligosaccharides to lactose is 1:1.26 in humans, whereas it is only 1:6 in the rhesus monkey (29,30). We expected a higher abundance of Lactobacilli in MR infants due to vertical transmission and stimulation from prebiotic constituents in breast milk. However, other studies have found that this effect is not always clearly evident (8,31). In addition, the absence of a larger difference when compared to Lactobacilli abundance in the HR infant may reflect the recent addition of prebiotic glycans to the particular formula they consumed. Fructooligosaccharides, including those present in Similac Sensitive® formula, can significantly increase both Lactobacillus and Bifidobacterium. However, Lactobacilli growth is less dependent on oligosaccharides (32) and a dose-dependent stimulating effect on growth has been seen only for Bifidobacteria (33). The effect of early feeding regimens on Lactobacilli may also be delayed. A prior study on nursery-reared rhesus infants fed formula with prebiotics indicated they ultimately had substantially lower levels of Lactobacillus, but not until after they were weaned onto solid foods (26).
Despite significant differences in overall composition and individual taxa, we did not see large differences in phylogenetic richness between the two rearing conditions. However, phylogenetic diversity was predictive of weight gain in both conditions as the infants began to consume solid foods. Other studies have found diminished diversity in HR infant monkeys at weaning and during the transition to solids (26), so it is possible that a diminished community richness only becomes apparent later. Our HR infants did, however, evince a higher ratio of Firmicutes-to-Bacteroidetes, due in part to an abundance of Blautia, a profile associated with more efficient absorption of calories and metabolic disorders in humans (14,15,34). This difference resulted in a corresponding shift in predicted metabolic pathways based on functions attributed to these microbes, including increased genes associated with carbohydrate and lipid metabolism. KEGG predictions were substantiated by larger and faster weight gains in HR infants. A differential effect of early rearing on growth rate has been reported previously for nursery-reared monkeys as well as in formula-fed human infants (28,35). Though this difference was diminished after weaning, rapid weight gain during infancy has been correlated with later risk for adult obesity, dyslipidemia, and insulin resistance in humans (36).
In addition to influencing host metabolism, gut bacteria can sustain infant health by providing protection against pathogens (3,4). When clinical records were reviewed at both 6 and 12 months, the HR monkeys exhibited more diarrheic symptoms with verified C. jejuni infections during and subsequent to the transition to solid foods, confirming the benefits of breastfeeding for providing sustained protection against enteric pathogens (37). C. jejuni is associated with gut dysbiosis and the abundance of Campylobacter is predictive of the overall microbial composition, even in asymptomatic infant and juvenile monkeys (38–40). Further demonstrating the importance of maternal influences, a prior study documented that a high-fat maternal diet prior to birth had a protracted effect on the abundance of Campylobacter present without symptoms in infant monkeys (9). Previous studies have also documented differences in the immune responses of MR and HR monkeys (28,41), which are likely associated with the protective qualities of lactic acid bacteria and Bifidobacterium, commensal strains that were more abundant in MR infants and known to enhance intestinal epithelial barrier function (42,43). Oligosaccharides in breast milk can selectively stimulate the propagation of these microbes, as seen in the predicted upregulation of genes involved in the metabolism of host glycans in the MR infants (32,44). Through these mechanisms, breast milk can be protective against diarrheal disease. Specifically, it has been shown that probiotics can discourage the growth and intestinal adhesion of C. jejuni, Clostridium, and other pathogenic organisms (45). Moreover, Prevotella, the predominant genera in MR monkeys, may also have a protective role because it contributes to the production of fermentation enzymes responsible for short-chain fatty acids (SCFA) (46). These enzymes and SCFA are critically important for the regulation of immune responses. Collectively, these findings reaffirm the view that there is a critical early window for initiating the trajectory to gut health.
While our findings concur with many prior studies, limitations should be acknowledged. The HR condition differed in more than one way from the MR condition, because it involved both formula-feeding and early rearing in an incubator. However, both factors can co-occur in human infants. Infants born premature or delivered through cesarean-section are more likely to be admitted to the NICU and their mothers are less likely to breastfeed, or to delay breastfeeding initiation (47,48). Many of the HR infants were being reared in this manner because the biological mother had a cesarean delivery. There were repeated attempts to reunite the HR infant with its mother during the first week of life, a practice that could be considered to mimic the bacterial exposure that might occur during the ‘skin-to-skin contact’ encouraged for preterm NICU infants (49). Future research will have to more selectively vary the dietary and social variables that differed between the two rearing conditions. Ours was designed to discern more maximal microbial differences that might occur in the absence of the mother and breast milk. For this type of investigation, a monkey is preferable to rodent models because breast milk composition and gut maturity are different in species with altricial neonates (29,30). Though ours is one of the larger studies to examine factors influencing the infant microbiome in monkeys, the sample size did limit statistical power. It precluded stratified analyses to identify interactions between delivery mode and feeding method. We were also underpowered to conduct a serial analysis of the changing microbiome in individual HR infants, but did have a sufficient number of MR infants for repeated measures analysis (see Text, Supplemental Digital Content 3). Finally, the gut microbiome is not the only outcome known to differ between MR and HR monkeys; the mother also stimulates neural and behavioral development (50).
In summary, breastfed infant rhesus exposed continuously to maternal sources of bacteria more quickly acquired microbiota typical of adults and had higher levels of several beneficial symbionts. The findings concur with the view that there is a biological expectancy that a mother will be present to provide a sustained microbial inoculation. Many clinical, personal and economic factors contribute to obstetrical decisions about delivery mode and parental decisions about feeding regimens after birth, but the influence on the infant’s gut microbiome should be taken into consideration. While the improved composition of formula, including prebiotic factors, now allow it to more closely resemble mother’s milk, we still need to advance our understanding of its prebiotic functions. The initial community structure of the infant’s microbiome can have long-term metabolic and physiologic effects influencing the developmental trajectory to adult health in animals and humans.
Supplementary Material
Supplemental Digital Content 1. Text detailing husbandry of human-reared infant. pdf
Supplemental Digital Content 2. Text detailing specimen collection, isolation of bacterial DNA, and sequence analysis. pdf
Supplemental Digital Content 3. Text describing serial changes in the community structure for the MR infants. pdf
Supplemental Digital Content 4. Table with descriptive details of infant recruitment. pdf
Supplemental Digital Content 5. Table with results of alpha diversity indices. pdf
Figure, Supplemental Digital Content 6 Prevalence of diarrheic symptoms. HR infants were more likely to be treated for both acute and chronic diarrhea by 6 months of age and the increased susceptibility to enteric pathogens continued during the 6-month period following weaning. Clinical stool cultures were run on 73% of infants exhibiting symptoms; Campylobacter jejuni was most common pathogen identified.
Supplemental Digital Content 6. Figure that illustrates the prevalence of diarrheic symptoms during the first 6 months of age and for 6 months following weaning. tiff
What Is Known:
Bacterial colonization plays a major role in the development of postnatal gut function, with effects on gut health and host metabolism.
Breastfeeding and vaginal delivery are among the most influential factors affecting the establishment of the gut microbiome in young infants.
What Is New:
The microbiome of human-reared infant monkeys is characterized by a higher ratio of Firmicutes-to-Bacteroidetes, a profile associated with metabolic disorders in humans.
The stimulatory effect of breast milk on commensal propagation was more evident for Bifidobacteria than Lactobacilli.
The gut microbiome during early infancy is predictive of future risk for Campylobacter jejuni infections in rhesus monkeys.
Acknowledgments
Acknowledgements are due to the staff of the WNPRC who provided specimens from the HR infants (P51OD011106) and to Alexandra Proctor for help with DNA extractions and advice on the 16S sequence analysis. All bacterial DNA was extracted and processed by the laboratory of GP and ML at ISU.
Source of Funding:
This research was supported by a grant from the National Institute of Mental Health investigating the significance of the gut microbiome for gut health and the gut-brain axis during infancy (MH104198).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
REFERENCES
- 1.Lozupone CA, Stombaugh JI, Gordon JI, et al. Diversity, stability and resilience of the human gut microbiota. Nature 2012;489:220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faith JJJ, Guruge JLJ, Charbonneau M, et al. The long-term stability of the human gut microbiota. Science 2013;341:1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gomez de Agüero M, Ganal-Vonarburg SC, Fuhrer T, et al. The maternal microbiota drives early postnatal innate immune development. Science 2016;351:1296–1302. [DOI] [PubMed] [Google Scholar]
- 4.Cussotto S, Sandhu KV, Dinan TG, et al. The neuroendocrinology of the microbiota-gut-brain axis: A behavioural perspective. Front Neuroendocrinol 2018. [DOI] [PubMed] [Google Scholar]
- 5.Bäckhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci 2004;101:15718–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Penders J, Thijs C, Vink C, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 2006;118:511–21. [DOI] [PubMed] [Google Scholar]
- 7.Martin R, Makino H, Yavuz AC, et al. Early-Life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One 2016;11:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madan JC, Hoen AG, Lundigren SN, et al. Effects of Cesarean delivery and formula supplementation on the intestinal microbiome of six-week old infants. JAMA Pediatr 2016;170:212–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, et al. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat Commun 2014;5:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sela DA, Mills DA. The marriage of nutrigenomics with the microbiome: the case of infant-associated bifidobacteria and milk. Am J Clin Nutr 2014;99:697S–703S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernández L, Langa S, Martín V, et al. The human milk microbiota: Origin and potential roles in health and disease. Pharmacol Res 2013;69:1–10. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization. WHO Statement on Caesarean Section Rates. Geneva: World Health Organization; 2015. (WHO/RHR/15.02). [Google Scholar]
- 13.Cai X, Wardlaw T, Brown DW. Global trends in exclusive breastfeeding. Int Breastfeed J 2012;7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koliada A, Syzenko G, Moseiko V, et al. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol 2017;17:4–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammer Ø, Harper D, Ryan PD. PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electron 2001;4:1–9. [Google Scholar]
- 18.Langille MGI, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013;31:814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006;72:5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci 2010;107:11971–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med 2016;February:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakobsson HE, Abrahamsson TR, Jenmalm MC, et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by Caesarean section. Gut 2014;63:559–66. [DOI] [PubMed] [Google Scholar]
- 24.Bokulich NA, Chung J, Battaglia T, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med 2016;8:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azad MB, Konya T, Maughan H, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ 2013;185:385–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ardeshir A, Narayan NR, Méndez-lagares G, et al. Breast-fed and bottle-fed infant rhesus macaques develop distinct gut microbiotas and immune systems. Sci Transl Med 2014;6:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bezirtzoglou E, Tsiotsias A, Welling GW. Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe 2011;17:478–82. [DOI] [PubMed] [Google Scholar]
- 28.O’Sullivan A, He X, McNiven EMS, et al. Early diet impacts infant rhesus gut microbiome, immunity, and metabolism. J Proteome Res 2013;12:2833–45. [DOI] [PubMed] [Google Scholar]
- 29.Beck KL, Weber D, Phinney BS, et al. Comparative proteomics of human and macaque milk reveals species-specific nutrition during postnatal development. J Proteome Res 2015;14:2143–57. [DOI] [PubMed] [Google Scholar]
- 30.O’Sullivan A, He X, McNiven EMSS, et al. Metabolomic phenotyping validates the infant rhesus monkey as a model of human infant metabolism. J Pediatr Gastroenterol Nutr 2013;56:355–363. [DOI] [PubMed] [Google Scholar]
- 31.Fallani M, Young D, Scott J, et al. Intestinal microbiota of 6-week-old infants across Europe: Geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr 2010;51:77–84. [DOI] [PubMed] [Google Scholar]
- 32.Macfarlane GT, Steed H, Macfarlane S. Bacterial metabolism and health-related effects of galacto-oligosaccharides and other prebiotics. J Appl Microbiol 2008;104:305–44. [DOI] [PubMed] [Google Scholar]
- 33.Moro G, Minoli I, Mosca M, et al. Dosage-related bifidogenic effects of galacto- and fructooligosaccharides in formula-fed term infants. J Pediatr Gastroenterol Nutr 2002;34:291–5. [DOI] [PubMed] [Google Scholar]
- 34.Kasai C, Sugimoto K, Moritani I, et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol 2015;15:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomez-Gallego C, Garcia-Mantrana I, Salminen S, et al. The human milk microbiome and factors influencing its composition and activity. Semin Fetal Neonatal Med 2016;21:400–5. [DOI] [PubMed] [Google Scholar]
- 36.Ekelund U, Ong KK, Linné Y, et al. Association of weight gain in infancy and early childhood with metabolic risk in young adults. J Clin Endocrinol Metab 2007;92:98–103. [DOI] [PubMed] [Google Scholar]
- 37.Morrow A, Ruiz-Palacios G, Altaye M, et al. Human milk oligosaccharides are associated with protection against diarrhea in breast-fed infants. J Pediatr 2004;145:297–303. [DOI] [PubMed] [Google Scholar]
- 38.Kampmann C, Dicksved J, Engstrand L, Rautelin H. Composition of human faecal microbiota in resistance to Campylobacter infection. Clin Microbiol Infect 2016;22:61.e1–8. [DOI] [PubMed] [Google Scholar]
- 39.Connerton IF, Cummings NJ, O’Kane PM, Lafontaine GM, Fish NM, Ghaffar N, et al. The effect of the timing of exposure to Campylobacter jejuni on the gut microbiome and inflammatory responses of broiler chickens. Microbiome 2018;6:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey MT, Coe CL. Maternal separation disrupts the integrity of the intestinal microflora in infant rhesus monkeys. Dev Psychobiol 1999;35:146–55. [PubMed] [Google Scholar]
- 41.Lubach GR, Coe CL, Ershler WB. Effects of early rearing environment on immune-responses of infant rhesus-monkeys. Brain Behav Immun 1995;9:31–46. [DOI] [PubMed] [Google Scholar]
- 42.Gueimonde M, Laitinen K, Salminen S, et al. Breast milk: a source of bifidobacteria for infant gut development and maturation? Neonatology 2007;92:64-. [DOI] [PubMed] [Google Scholar]
- 43.Ohland CL, Macnaughton WK. Probiotic bacteria and intestinal epithelial barrier function. Am J Gastrointest Liver Physiol 2010;298:G807–19. [DOI] [PubMed] [Google Scholar]
- 44.Marcobal A, Barboza M, Froehlich J, et al. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem 2011;76:1358–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ekmekciu I, Fiebiger U, Stingl K, et al. Amelioration of intestinal and systemic sequelae of murine Campylobacter jejuni infection by probiotic VSL#3 treatment. Gut Pathog 2017;9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci 2010;107:14691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kamath B, Todd JK, Glazner JE, et al. Neonatal outcomes after elective cesarean delivery. Obstet Gynecol 2009;113:1231–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hobbs AJ, Mannion CA, McDonald SW, et al. The impact of caesarean section on breastfeeding initiation, duration and difficulties in the first four months postpartum. BMC Pregnancy Childbirth 2016;16:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feldman R, Rosenthal Z, Eidelman AI. Maternal-preterm skin-to-skin contact enhances child physiologic organization and cognitive control across the first 10 years of life. Biol Psychiatry 2014;75:56–64. [DOI] [PubMed] [Google Scholar]
- 50.Rommeck I, Capitanio JP, Strand SC, et al. Early social experience affects behavioral and physiological responsiveness to stressful conditions in infant rhesus macaques (Macaca mulatta). Am J Primatol 2011;73:692–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Digital Content 1. Text detailing husbandry of human-reared infant. pdf
Supplemental Digital Content 2. Text detailing specimen collection, isolation of bacterial DNA, and sequence analysis. pdf
Supplemental Digital Content 3. Text describing serial changes in the community structure for the MR infants. pdf
Supplemental Digital Content 4. Table with descriptive details of infant recruitment. pdf
Supplemental Digital Content 5. Table with results of alpha diversity indices. pdf
Figure, Supplemental Digital Content 6 Prevalence of diarrheic symptoms. HR infants were more likely to be treated for both acute and chronic diarrhea by 6 months of age and the increased susceptibility to enteric pathogens continued during the 6-month period following weaning. Clinical stool cultures were run on 73% of infants exhibiting symptoms; Campylobacter jejuni was most common pathogen identified.
Supplemental Digital Content 6. Figure that illustrates the prevalence of diarrheic symptoms during the first 6 months of age and for 6 months following weaning. tiff