Abstract
The synthesis of pre-mRNA by RNA polymerase II (Pol II) involves the formation of a transcription initiation complex and a transition to an elongation complex1–4. The large subunit of Pol II contains an intrinsically disordered C-terminal domain (CTD), which is phosphorylated by cyclin-dependent kinases (CDKs) during the initiation-to-elongation transition, thus influencing the CTD’s interaction with different components of the initiation or the RNA splicing apparatus (Fig. 1a)5,6. Recent observations suggest that this model provides only a partial picture of the effects of CTD phosphorylation. Both the transcription initiation machinery and the splicing machinery can form phase-separated condensates containing large numbers of component molecules; hundreds of Pol II and Mediator molecules are concentrated in condensates at super-enhancers7,8 and large numbers of splicing factors are concentrated in nuclear speckles, some of which occur at highly active transcription sites9–12. Here we investigate whether phosphorylation of the CTD regulates its incorporation into phase-separated condensates associated with transcription initiation and splicing. We find that the hypophosphorylated Pol II CTD is incorporated into Mediator condensates and that phosphorylation by regulatory CDKs reduces this incorporation. We also find that the hyperphosphorylated CTD is preferentially incorporated into condensates formed by splicing factors. These results suggest that Pol II CTD phosphorylation drives an exchange from condensates involved in transcription initiation to those involved in RNA processing and implicates phosphorylation as a mechanism to regulate condensate preference.
Previous studies have shown that the hypophosphorylated Pol II CTD can interact with Mediator5,6 and that Pol II and Mediator occur in condensates at super-enhancers7,8 (Fig. 1b, Extended Data Fig. 1a), but have not established whether the CTD contributes to Pol II interactions with Mediator condensates. To investigate whether the Pol II CTD is incorporated into Mediator condensates, we purified the human Mediator complex and recombinant CTD fused to GFP (full length GFP-CTD52 and truncated forms GFP-CTD26 and GFP-CTD10) and measured condensate formation in an in vitro droplet assay. Mediator droplets incorporated and concentrated GFP-CTD52 to a much greater extent than the truncated forms or control GFP (Fig. 1c). We further investigated the interaction of the CTD with Mediator using MED1, the largest subunit of the Mediator complex13. MED1 has proven to be a useful surrogate for the Mediator condensate in previous studies8,14, has an exceptionally large intrinsically disordered region (IDR) that contributes to condensate formation8, and has been shown to associate with Pol II in human cells15. Droplet assays revealed that mCherry-MED1-IDR condensates incorporated and concentrated GFP-CTD52 to a greater extent than its truncated forms or GFP alone (Fig. 1d). The GFP-CTD52/MED1-IDR condensates exhibited liquid-like fusion behavior (Extended Data Fig. 1b) and showed evidence of dynamic internal rearrangement and internal-external exchange of molecules by fluorescence recovery after photobleaching (FRAP; Extended Data Fig. 1c), consistent with liquid-liquid phase-separated condensates16–18. These results are consistent with the idea that the Pol II CTD contributes to Pol II incorporation into Mediator condensates.
Figure 1. The CTD of Pol II is integrated and concentrated in Mediator condensates.
a. A model depicting the transition from transcription initiation to elongation and the role of Pol II CTD phosphorylation in this transition. During initiation, Pol II with a hypophosphorylated CTD interacts with Mediator. CDK7 phosphorylation of the CTD leads to formation of a paused Pol II approximately 50–100 bp downstream of the initiation site, and subsequent CDK9 phosphorylation leads to pause release and elongation1. For simplicity, we show CDK7 and CDK9 phosphorylating the CTD, leading to elongation. During elongation, Pol II with hyperphosphorylated CTD interacts with various RNA splicing factors5,6. The colored compartments surrounding the initiating and elongating polymerases represent initiation and splicing factor condensates, respectively.
b. Representative images exhibiting overlap between immunofluorescence (IF) of MED1 and Pol II with nascent RNA FISH of Nanog and Trim28 in fixed mouse embryonic stem cells (mESCs). The three columns on the right show average RNA FISH signal and average MED1 or Pol II IF signal centered on RNA FISH foci (see methods).
c. Representative images and quantification of partition ratios of droplet experiments measuring full length or truncated CTD incorporation into human Mediator complex droplets. Purified human Mediator complex (~200 nM; see methods) was mixed with 10 μM GFP, GFP-CTD52, or truncated forms of GFP-CTD in droplet formation buffers with 140 mM monovalent salt and 16% Ficoll-400 and visualized on a fluorescence microscope with the indicated filters.
d. Representative images and quantification of partition ratios of droplet experiments measuring full length or truncated CTD incorporation into MED1-IDR droplets. Purified human MED1-IDR fused to mCherry (mCherry-MED1-IDR) at 10 μM was mixed with 3.3 μM GFP, GFP-CTD52, or truncated forms of GFP-CTD in droplet formation buffers with 125 mM NaCl and 16% Ficoll-400.
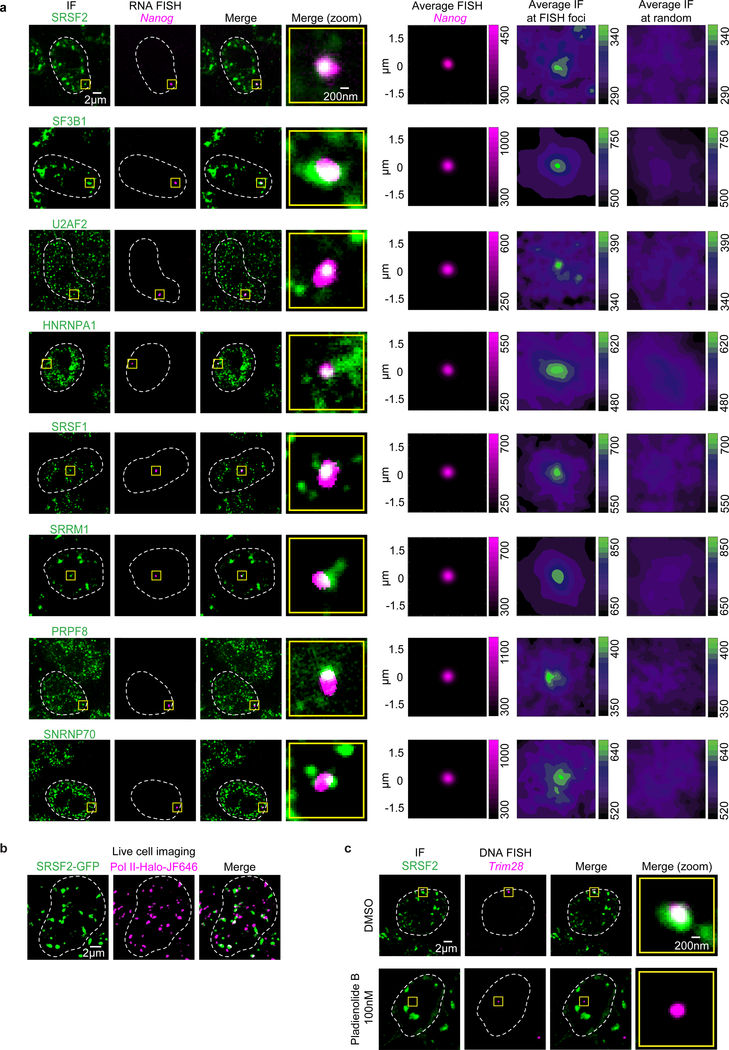
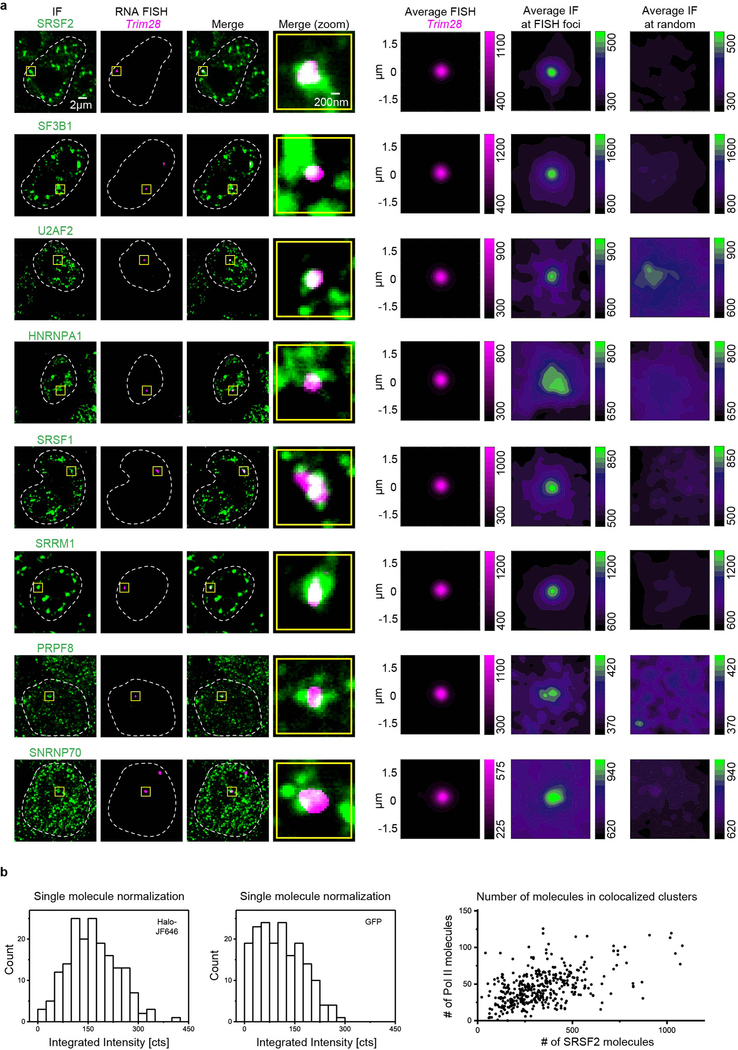
We next sought to determine whether splice factor condensates occur at super-enhancer-associated genes because these genes are transcribed at especially high rates19, RNA splicing can occur co-transcriptionally20–22, and some nuclear speckles have been reported to occur in the vicinity of highly transcribed genes9–12. We selected eight different components of the splicing apparatus and used immunofluorescence (IF) microscopy with concurrent nascent RNA fluorescence in situ hybridization (FISH) for Nanog and Trim28 to determine whether the splicing apparatus occurs in puncta in the vicinity of these super-enhancer-associated genes. The results showed that all eight splicing factors occur in puncta at these two genes (Fig. 2a, Extended Data Fig. 2a). To gain additional insights into splice factor puncta that colocalize with Pol II, mESCs engineered to express endogenously tagged proteins were studied using lattice light sheet imaging in live cells. We previously showed that large numbers of Mediator and Pol II molecules can occur in puncta and that these sometimes colocalize7,8; using a similar approach, we found here that large numbers of SRSF2 molecules occur in puncta and some of these (~15%) overlap with Pol II puncta (Fig. 2b, Extended Data Fig. 2b). Treatment of cells with an inhibitor of splicing, Pladienolide B, which reduced splicing as determined by a splicing reporter (Extended Data Fig. 3a), also reduced the levels of splicing factors, but not RNA polymerase II, in puncta at Trim28 DNA FISH foci (Fig. 2c, Extended Data Fig. 3b, c). This treatment also led to splicing factor incorporation into “mega-speckles” at some distance from the gene (Fig. 2c), a phenomenon observed previously when splicing is inhibited23. These results suggest that functional RNA splicing apparatus is present in condensates at active super-enhancer-associated genes.
Figure 2. Splicing factor condensates occur at active super-enhancer-driven genes.
a. Representative images exhibiting overlap between IF of splicing factors SRSF2, SF3B1, U2AF2, HNRNPA1, SRSF1, SRRM1, PRPF8, or SNRNP70 with nascent RNA FISH of Nanog in fixed mESCs. The rightmost column shows average IF signal for splicing factors centered on randomly selected nuclear positions (see methods).
b. Representative lattice light sheet images of live mESCs engineered to express GFP tagged SRSF2 and Halo-JF646 tagged Pol II. Maximum intensity projection after background subtraction.
c. Representative images exhibiting overlap or absence of overlap between IF of SRSF2 and DNA FISH of Trim28 in mESCs treated with DMSO or splicing inhibitor Pladienolide B for 4 hrs.
Actively transcribed genes may become associated with nuclear speckles or obtain splicing apparatus stored in speckles9–12, which are thought to be phase-separated16. In live cell imaging, we found that the SRSF2 puncta exhibited features of liquid-like condensates: they all showed evidence of dynamic internal rearrangement and internal-external exchange of molecules by FRAP (Extended Data Fig. 4a–c), were sensitive to treatment with 1,6-hexanediol (Extended Data Fig. 4d), and some would occasionally fuse (Extended Data Fig. 4e). These results are consistent with previous reports regarding speckle behavior9 and suggest that the SRSF2-containing puncta that come into contact with active super-enhancer-associated genes are liquid-like condensates. At highly transcribed genes such as those driven by super-enhancers, large numbers of Pol II molecules may be engaged in transcription elongation7,8,17, and these might serve to recruit into condensates some portion of the apparatus otherwise located in speckles.
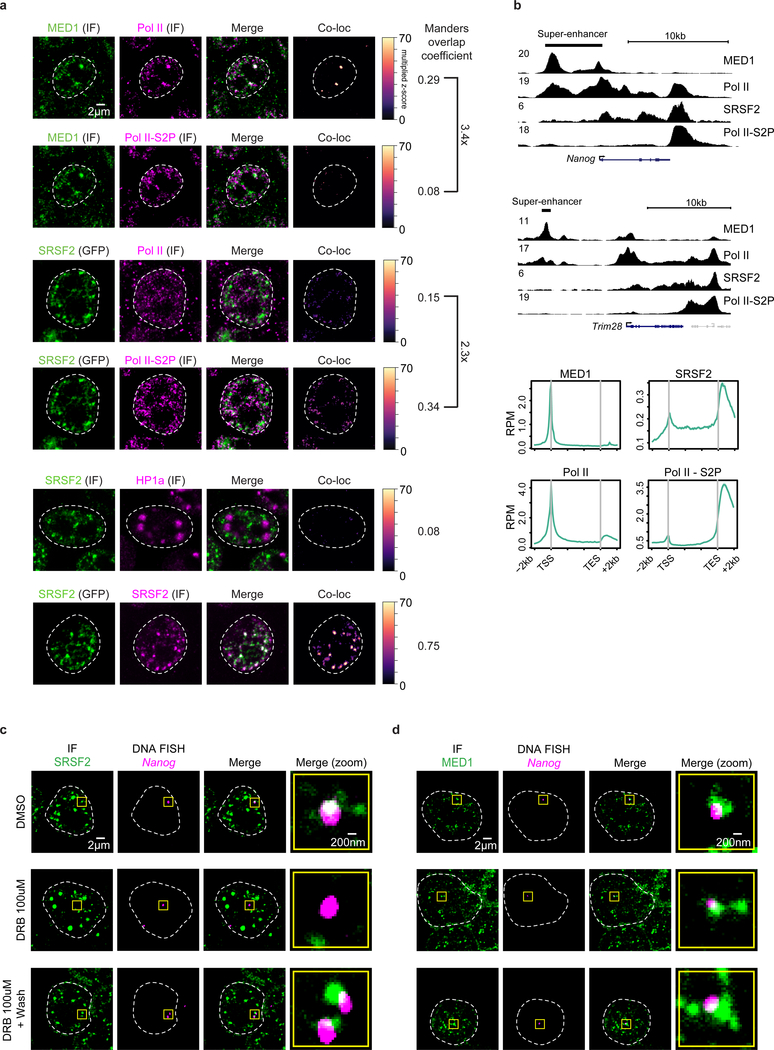
We next investigated whether hypophosphorylated Pol II tends to be associated with MED1 condensates whereas hyperphosphorylated Pol II tends to be associated with SRSF2 condensates. Using immunofluorescence and antibodies against hypophosphorylated or serine 2 phosphorylated CTD, we confirmed this prediction: MED1 puncta more frequently overlapped with signals for the hypophosphorylated CTD whereas SRSF2 puncta more frequently overlapped with signals for the serine 2 phosphorylated CTD (Fig. 3a). A control experiment showed that there was essentially no overlap between SRSF2 puncta and the heterochromatin protein HP1a, and strong overlap of SRSF2 puncta visualized using independent methods (Fig. 3a). An independent experimental approach using ChIP-seq with antibodies against MED1, SRSF2, and the two phosphoforms of Pol II also confirmed that MED1 tends to occupy super-enhancers and promoters together with Pol II containing hypophosphorylated CTD, whereas SRSF2 is observed across the transcription unit and is prominent at the ends of genes together with Pol II containing serine 2 phosphorylated CTD (Fig. 3b).
Figure 3. Pol II partitioning in transcriptional and splicing factor condensates.
a. First four rows: IF imaging using antibodies for the hypophosphorylated and serine 2 phosphorylated (S2P) Pol II CTD, coupled with IF for MED1 or direct visualization of SRSF2 in the GFP-SRSF2 mESCs. Last two rows: IF for SRSF2 coupled to IF for HP1a or direct visualization of SRSF2 in the GFP-SRSF2 mESCs. The “Co-loc” column highlights overlapped pixels for each factor in an example z-slice, and the Manders’ overlap coefficient gives a relative score for the degree of overlap from multiple cells and images (see methods). For each experimental comparison, one coverslip of cells was stained for the indicated factors and 5 independent fields were imaged and analyzed.
b. Top: Representative ChIP-seq tracks of MED1, SRSF2 and the hypophosphorylated or serine 2 phosphorylated forms of Pol II in mESCs. Y-axis in reads per million (RPM). Bottom: Metagene plots of average ChIP-seq RPM for the same factors across gene bodies (see methods). ChIP-seq was performed once for each factor with approximately 100 million cells.
c. Representative images exhibiting overlap or lack of overlap between IF of SRSF2 and DNA FISH of Nanog in mESCs treated with DMSO for 2 hrs, DRB for 2 hrs, or DRB for 2 hrs followed by a 2 hr washout.
d. Representative images exhibiting overlap between IF of MED1 and DNA FISH of Nanog in mESCs treated as in panel c.
If the formation or maintenance of splicing factor condensates is dependent on CTD phosphorylation, we would expect that inhibition of CTD phosphorylation in cells would prevent the formation of splicing factor condensates at super-enhancer-driven genes. Indeed, inhibition of CTD phosphorylation by the CDK inhibitor D-ribofuranosylbenzimidazole (DRB) caused a marked reduction in the occupancy of multiple splicing factor condensate components (SRSF2, SRSF1, SF3B1, U2AF2, PRPF8) at Nanog or Trim28 DNA FISH foci, and a washout of the drug led to a partial reestablishment of most of these splicing factors within 2 hours (Fig. 3c, Extended Data Fig. 5a, d). In contrast, DRB treatment had minor effects on Mediator and Pol II condensates (Fig. 3d, Extended Data Fig. 5b–d). These DRB treatment and washout results suggest that CTD phosphorylation is necessary for formation of splicing factor condensates at these genes in vivo, although it is possible that altered phosphorylation of other CDK substrates may contribute to these observations.
The transition of Pol II from initiation to elongation is accompanied by phosphorylation of the CTD heptapeptide repeat by CDK7 and CDK924,25. Phosphorylation of the CTD has been shown to affect its interaction with hydrogels formed by the low-complexity domains of FET (FUS/EWS/TAF15) proteins26, suggesting that phosphorylation may affect the condensate interacting properties of the CTD. We investigated whether phosphorylation of the CTD by CDK7 or CDK9 would affect its incorporation into Mediator condensates. We found that CTD phosphorylation by either CDK7 or CDK9 (Extended Data Fig. 6a, b) caused a reduction in CTD incorporation into Mediator droplets (Fig. 4a). Similarly, CTD phosphorylation caused a reduction in CTD incorporation into MED1-IDR droplets (Extended Data Fig. 6c, d). These results are consistent with the model that Pol II CTD phosphorylation causes eviction from a Mediator condensate.
Figure 4. CTD phosphorylation promotes a condensate preference switch between Mediator and splicing factor condensates.
a. Representative images and quantification of partition ratios of droplet experiments measuring CTD incorporation into Mediator droplets. Purified human Mediator complex (~200 nM; see methods) was mixed with 10 μM GFP, GFP-CTD52 or GFP-CTD52 phosphorylated with CDK7 or CDK9 in droplet formation buffers with 140 mM monovalent salt and 16% Ficoll-400.
b. Representative images and quantification of partition ratios of droplet experiments measuring CTD incorporation into SRSF2 droplets. Purified human SRSF2 fused to mCherry (mCherry-SRSF2) at 2.5 μM was mixed with 3.3 μM GFP, GFP-CTD52 or GFP-CTD52 phosphorylated with CDK7 or CDK9 in droplet formation buffers with 120 mM NaCl and 10% PEG-8000.
c. Representative images and quantification of partition ratios of droplet experiments measuring CTD incorporation into SRSF1 droplets. Same conditions as in panel b.
The observation that CTD phosphorylation is necessary for splicing factor condensate formation at highly transcribed genes (Fig. 3c) suggests that CTD phosphorylation might enhance its partitioning into condensates formed by splicing components. To investigate this idea, we first selected four human splicing factors (SRSF2, SRSF1, U2AF2 and hnRNPA1) as surrogates for the more complex splicing factor condensates and explored their condensate forming properties. Each of the four purified human proteins, fused to mCherry, formed phase-separated droplets (Extended Data Fig. 7a, b). SRSF2 is among the multiple proteins involved in pre-mRNA splicing that contain serine:arginine (SR) dipeptide repeats and has an especially large SR-rich domain27, so we used SRSF2 as a core component to study whether it could concentrate the other three factors into heterotypic droplets. Indeed, all of these factors could form binary heterotypic droplets with SRSF2 (Extended Data Fig. 7c). We then asked whether CTD phosphorylation influences its incorporation into splicing factor condensates in vitro using recombinant SRSF1 and SRSF2. The results showed that unphosphorylated CTD was not efficiently incorporated into SRSF1 or SRSF2 droplets, whereas CDK7 or CDK9-phosphorylated CTD was incorporated and concentrated in both SRSF1 and SRSF2 droplets (Fig. 4b, c, Extended Data Fig. 8a–c). The ability of SRSF2 to incorporate phosphorylated CTD was dependent on CTD length (Extended Data Fig. 8d), as expected for a high valency condensate interaction16–18 and consistent with models where CTD truncation leads to splicing defects28. We conclude that phosphorylation of the Pol II CTD leads to a switch in its preference for interactions between Mediator and SR-protein-containing condensates.
Our results indicate that Pol II CTD phosphorylation alters its condensate partitioning behavior and may thus drive an exchange of Pol II from condensates involved in transcription initiation to those involved in RNA splicing at super-enhancer-associated genes. This model is consistent with evidence from previous studies that large clusters of Pol II can fuse with Mediator condensates in cells7, that phosphorylation dissolves CTD-mediated Pol II clusters29, that CDK9/Cyclin T can interact with the CTD through a phase separation mechanism30, that Pol II is no longer associated with Mediator during transcription elongation13, and that nuclear speckles containing splicing factors can be observed at loci with high transcriptional activity9–12. Previous studies have shown that the CTD can interact with components of the transcription initiation apparatus and RNA processing machinery in a phosphoform-specific manner5,6, but did not explore the possibility that these components occur in condensates or that phosphorylation of the Pol II CTD alters its partitioning behavior between these condensates. Our results reveal that Mediator and splicing factor condensates occur at the same super-enhancer driven genes and suggest that the transition of Pol II from interactions with components involved in initiation to those involved in splicing can be mediated in part through a CTD phosphorylation regulated change in condensate partitioning. These results also suggest that phosphorylation may be among the mechanisms that regulate condensate partitioning of proteins in processes where protein function involves eviction from one condensate and migration to another.
Methods
Cell culture
V6.5 murine embryonic stem cells (mESCs) were a gift from the Jaenisch lab. Cells were grown on 0.2% gelatinized (Sigma, G1890) tissue culture plates in 2i media, DMEM-F12 (Life Technologies, 11320082), 0.5X B27 supplement (Life Technologies, 17504044), 0.5X N2 supplement (Life Technologies, 17502048), an extra 0.5 mM L-glutamine (Gibco, 25030–081), 0.1 mM beta-mercaptoethanol (Sigma, M7522), 1% Penicillin Streptomycin (Life Technologies, 15140163), 1X nonessential amino acids (Gibco, 11140–050), 1000 U/ml LIF (Chemico, ESG1107), 1 μM PD0325901 (Stemgent, 04–0006-10), 3 μM CHIR99021 (Stemgent, 04–0004-10). Cells were grown at 37°C with 5% CO2 in a humidified incubator. For confocal imaging, cells were grown on glass coverslips (Carolina Biological Supply, 633029), coated with 5 μg/mL of poly-L-ornithine (Sigma Aldrich, P4957) for at least 30 min at 37°C and with 5 μg/ml of Laminin (Corning, 354232) for 2–16 hrs at 37 °C. For passaging, cells were washed in PBS (Life Technologies, AM9625), 1000 U/mL LIF. TrypLE Express Enzyme (Life Technologies, 12604021) was used to detach cells from plates. TrypLE was quenched with FBS/LIF-media (DMEM K/O (Gibco, 10829–018), 1X nonessential amino acids, 1% Penicillin Streptomycin, 2 mM L-Glutamine, 0.1 mM beta-mercaptoethanol and 15% Fetal Bovine Serum, FBS, (Sigma Aldrich, F4135).
Western blot
Purified phosphorylated CTD was mixed in 1X XT buffer (Bio-Rad) and run on 3–8% Criterion™ XT Tris-acetate Precast Gels (Bio-Rad) at 100 V until the dye front reached the end of the gel. Protein was then wet transferred to a 0.45 μm PVDF membrane (Millipore, IPVH00010) in ice-cold transfer buffer (25 mM Tris, 192 mM glycine, 10% methanol) at 250 mA for 2 hours at 4°C. After transfer, the membrane was blocked with 5% non-fat milk in TBS for 1 hour at room temperature, with shaking. The membrane was then incubated with a 1:5,000 dilution of anti-GFP (Abcam #ab290) antibodies in 5% non-fat milk in TBST overnight at 4°C, with shaking. The membrane was washed three times with TBST for 10 min at room temperature with shaking. The membrane was incubated with 1:10,000 secondary antibodies (GE health) for 1 hr at RT and washed three times in TBST for 5 mins. Membranes were developed with Femto ECL substrate (Thermo Scientific, 34095) and imaged using a CCD camera.
Immunofluorescence with RNA FISH
Coverslips were coated at 37°C with 5 μg/mL poly-L-ornithine (Sigma-Aldrich, P4957) for 30 minutes and 5 μg/mL of Laminin (Corning, 354232) for 2 hrs. Cells were plated on the pre-coated cover slips and grown for 24 hours followed by fixation using 4% paraformaldehyde, PFA, (VWR, BT140770) in PBS for 10 minutes. After washing cells three times in PBS, the coverslips were put into a humidifying chamber or stored at 4°C in PBS. Permeabilization of cells was performed using 0.5% triton X100 (Sigma Aldrich, X100) in PBS for 10 minutes followed by three PBS washes. Cells were blocked with 4% IgG-free Bovine Serum Albumin, BSA, (VWR, 102643–516) for 30 minutes. Cells were then incubated with the indicated primary antibody at a concentration of 1:500 in PBS for 4–16 hrs. Antibodies used for IF in this study include SRSF2 (Abcam ab11826), MED1 (Abcam ab64965), Pol II-CTD (Abcam ab817), Pol II-CTD-S2 (Millipore 04–1571), SF3B1 (Sigma HPA050275), U2AF2 (Abcam ab37530), HNRNPA1 (Abcam ab5832), SRSF1 (Santa Cruz 33652), SRRM1 (Abcam ab221061), PRPF8 (Santa Cruz 55533), SNRNP70 (Sigma HPA043516), and HP1a (Abcam ab203432). SRSF2, MED1, Pol II-CTD, U2AF2, HNRNPA1, SRSF1, and SRRM1 antibodies were validated in house by siRNA knockdown. Pol II-CTD-S2 antibody was validated in house by treatment of cells with DRB. SF3B1 and SNRNP70 antibodies were validated by The Cell Atlas and meet the “enhanced” validation criteria. HP1a antibody was knockout validated by Abcam. Cells were washed with PBS three times followed by incubation with secondary antibody at a concentration of 1:500 in PBS for 1 hour. After washing twice with PBS, cells were fixed using 4% paraformaldehyde, PFA, (VWR, BT140770) in PBS for 10 minutes. After two washes of PBS, Wash buffer A (20% Stellaris RNA FISH Wash Buffer A (Biosearch Technologies, Inc., SMF-WA1–60), 10% Deionized Formamide (EMD Millipore, S4117)) in RNase-free water (Life Technologies, AM9932) was added to cells and incubated for 5 minutes. 12.5 μM RNA probe in Hybridization buffer (90% Stellaris RNA FISH Hybridization Buffer (Biosearch Technologies, SMF HB1–10) and 10% Deionized Formamide) was added to cells and incubated overnight at 37°C. After washing with Wash buffer A for 30 minutes at 37°C, the nuclei were stained in 20 μm/mL Hoechst 33258 (Life Technologies, H3569) for 5 minutes, followed by a 5 minute wash in Wash buffer B (Biosearch Technologies, SMFWB1–20). Cells were washed once in water followed by mounting the coverslip onto glass slides with Vectashield (VWR, 101098–042) and finally sealing the cover slip with nail polish (Electron Microscopy Science Nm, 72180). Images were acquired on the RPI Spinning Disk confocal microscope with 100x objective using MetaMorph acquisition software and a Hammamatsu ORCA-ER CCD camera (W.M. Keck Microscopy Facility, MIT). Images were post-processed using Fiji Is Just ImageJ (FIJI). RNA FISH probes were custom designed and generated by Agilent to target Nanog and Trim28 intronic regions to visualize nascent RNA.
Cell line generation
V6.5 murine embryonic stem cells were a gift from the Jaenisch lab, and have been verified using short tandom repeat (STR) analysis and tested negative for mycoplasma contamination. Other cell lines in this manuscript were generated from these V6.5 cells. CRISPR/Cas9 was used to generate endogenously-mEGFP-tagged SRSF2 mESC, and endogenously-Halo-tagged RPB1/mEGPF-tagged SRSF2 mESC. Oligos coding for guide RNAs targeting the N-terminus were cloned into a px330 vector expressing Cas9 and mCherry (gift from R. Jaenisch). The sequence that was targeted for SRSF2 was 5’-CGTAGCTCATGGCTGCGAAG-3’. The sequence that was targeted for RPB1 was 5’-TGCCTCGCCATGCACGGGGG-3’. Repair templates were cloned into a pUC19 vector (NEB) containing mEGFP, a GS linker, and 800 bp homology arms flanking the insert. 500K mouse ESCs were transfected with 1.25 μg px330 vector and 1.25 μg repair templates using Lipofectamine-3000 (Thermofisher). Cells were sorted 2 days after transfection for mCherry and 1 week after first sort for mEGFP. 50K cell were serially diluted in a 6 well plate and colonies were picked 4 days after seeding into a 96 well plate. 2–4 days after colony picking, cells were passaged into 3 plates. 1 plate was used for genotyping and the other 2 were frozen down at −80°C in 10% DMSO, 10% FBS and 80% 1x DMEM. The primer pairs that were used for genotyping were the following:
mEGFP-SRSF2
FW - 5’ TTTGGCGGGCTTTCTAACTGC 3’
RV - 5’ CGGTAGGTCAGGTTGTCCAC 3’
Halo-RPB1
FW - 5’ GAGCCCTAGCGTCAACAACT 3’
RV - 5’ CCTCTGGTATCAGCTCCCCT 3’
A clone with heterozygous mEGFP-SRSF2 was subsequently passaged for all assays. A clone of homozygous Halo-RPB1 and heterozygous mEGFP-SRSF2 were used for live cell lattice light sheet imaging.
Live cell imaging of GFP-SRSF2 cell line
Cells were grown on glass dishes (Mattek Corporation P35G-1.5–20-C) coated with 5 μg/mL of poly-L-ornithine (Sigma-Aldrich, P4957) for 30 min at 37°C and with 5 μg/mL of Laminin (Corning, 354232) for 2–16 hrs at 37°C. Before imaging cells, culture media was replaced with phenol red-free 2i media and imaged using the Andor Revolution Spinning Disk Confocal. Raw Andor images were processed using FIJI/ImageJ.
Fluorescence recovery after photobleaching in live cells
FRAP was performed on an Andor Revolution Spinning Disk Confocal with 488 nm laser. Bleaching was performed using 100% laser power with 30 microseconds dwell time for 5 cycles and images were collected every 500 milliseconds. Fluorescence intensity at the bleached spot, a control unbleached spot, and background was measured using the FIJI plugin FRAP Profiler. Background intensity was subtracted and values are reported relative to the unbleached spot to control for photobleaching during image acquisition.
Immunofluorescence with DNA FISH
Immunofluorescence was performed as previously described in the “Immunofluorescence with RNA FISH” section. After incubating the cells with the secondary antibodies, cells were washed three times in PBS for 5min at RT, fixed with 4% PFA in PBS for 10min and washed three times in PBS. Cells were incubated in 70% ethanol, 85% ethanol and then 100% ethanol for 1 minute at RT. Probe hybridization mixture was made by mixing 7 mL of FISH Hybridization Buffer (Agilent G9400A), 1 mL of FISH probe (see below for region) and 2 mL of water. 5 mL of mixture was added on a slide and the coverslip was placed on top (cell-side toward the hybridization mixture). Coverslips were sealed using rubber cement. Once rubber cement solidified, genomic DNA and probes were denatured at 78°C for 5 minutes and slides were incubated at 37°C in the dark overnight. The coverslip was removed from the slide and incubated in pre-warmed Wash buffer 1 (Agilent G9401A) at 73°C for 2 minutes and in Wash Buffer 2 (Agilent, G9402A) for 1 minute at room temperature. Slides were air dried and nuclei were stained with Hoechst in PBS for 5 minutes at RT. Coverslips were washed three times in PBS, mounted on slides using Vectashield and sealed with nail polish. Images were acquired on an RPI Spinning Disk confocal microscope with a 100x objective using MetaMorph acquisition software and a Hammamatsu ORCA-ER CCD camera (W.M. Keck Microscopy Facility, MIT). Images were post-processed using FIJI. DNA FISH probes were custom designed and generated by Agilent to target Nanog and Trim28 super-enhancers.
Nanog
Design Input Region – mm9
chr6 122605249 – 122705248
Design Region – mm9
chr6: 122605985–122705394
Trim28
Design Input Region – mm9
Chr7:13551990–13651989
Design Region – mm9
chr7:13552132–13651971
Drug treatments
V6.5 mESCs were grown in 24 well plates on coated glass coverslips as described previously. Cells were treated with drugs or vehicle (Pladienolide B at 100nM, DRB at 100uM, or DMSO at 0.1%), followed by fixation with 4% PFA in PBS. For DRB washout experiments, cells were treated with DRB for 2 hours, gently washed twice with fresh 2i media, and left to recover in 2i media containing 0.1% DMSO for 2 hrs before fixation.
Co-immunofluorescence co-localization analysis
For analysis of co-localization data (Figure 3a), custom Python scripts were written to process and analyze 3D image data gathered in IF and DAPI channels. Nuclei were detected by Otsu thresholding, and a mask of nuclei were applied to the IF channels. Manual minimal thresholds were called for the IF channels. Manders coefficients were then calculated for masked IF channels A and B in 3D with the following formulas: M1 = IA[IB > 0] / ∑IA, M2 = IB[IA > 0] / ∑IB (see Bolte and Cordelieres. 2006)31. Manders coefficients were calculated for at least 3 images per sample and then averaged. To generate the heatmaps of co-localization in Figure 3a, a representative z-slice was selected for each dataset. Each channel was standardized by subtracting the mean and dividing by the standard deviation of the slice to generate a per-pixel z-score. The z-scores between channels were then multiplied and a heatmap was generated from multiplied z-scores using the Python package matplotlib with the magma heatmap. All heatmaps are displayed on the same scale (vmax = 70). This method highlights areas where both channels have overlapping pixels.
Protein purification
Human cDNA was cloned into a modified version of a T7 pET expression vector. The base vector was engineered to include a 5’ 6xHIS followed by either mEGFP or mCherry and a 14 amino acid linker sequence “GAPGSAGSAAGGSG.” NEBuilder® HiFi DNA Assembly Master Mix (NEB E2621S) was used to insert these sequences (generated by PCR) in-frame with the linker amino acids. For MED1-IDR, the inserted sequence encodes residues 948 to 1574 of the full length MED1 protein. Vector expressing mEGFP alone contains the linker sequence followed by a STOP codon. All expression constructs were sequenced to ensure sequence identity.
For protein expression, plasmids were transformed into LOBSTR cells (gift of Chessman Lab). A fresh bacterial colony was inoculated into LB media containing kanamycin and chloramphenicol and grown overnight at 37°C. Cells containing CTD constructs were diluted 1:30 in 500ml room temperature LB with freshly added kanamycin and chloramphenicol and grown 1.5 hours at 16°C. IPTG was added to 1 mM and growth continued for 20 hours. Cells were collected and stored frozen at −80°C. Cells containing all other constructs were treated in a similar manner except they were grown for 5 hours at 37°C after IPTG induction.
For wildtype (GFP-CTD52, full-length CTD with 52 heptapeptide repeats) and mutant (GFP-CTD26, C-terminal 26 repeats; GFP-CTD10, C-terminal 10 repeats) CTD and hnRNPA1, pellets of 500 ml of cells were resuspended in 15 ml of Buffer A (50 mM Tris pH7.5, 500 mM NaCl) with cOmplete protease inhibitors, (Roche,11873580001) and sonicated (ten cycles of 15 seconds on, 60 seconds off). The lysates were cleared by centrifugation at 12,000 xg for 30 minutes and added to 1 ml of Ni-NTA agarose (Invitrogen, R901–15) that had been pre-equilibrated with 10 volumes of the same buffer. and rotated at 4°C for 1.5 hours. The slurry was centrifuged at 3,000 rpm for 10 minutes in a Thermo Legend XTR swinging bucket rotor. The resin pellets were washed 2 X with 5 ml of Buffer A followed by centrifugation as above. Protein was eluted 3 X with 2 ml of buffer A plus 250 mM imidazole. For each cycle the elution buffer was added and rotated at least 10 minutes at 4C and centrifuged as above. Elutes were analyzed on a 12% acrylamide gel stained with Coomassie. Fractions containing protein of the expected size were pooled, diluted 1:1 with the 250 mM imidazole buffer and dialyzed against two changes of buffer containing 50 mM Tris 7.5, 12 5mM NaCl, 10% glycerol and 1 mM DTT at 4°C. Protein concentration was measured by Thermo BCA Protein Assay Kit – Reducing Agent Compatible.
For SRSF2, SRSF1 and U2AF2, pellets of 500ml of cells were resuspended in 15 ml of denaturing buffer (50 mM Tris 7.5, 300 mM NaCl, 10 mM imidazole, 8 M Urea) with cOmplete protease inhibitors, sonicated and the cleared as above. The lysates were added to 1 ml of Ni-NTA agarose that had been pre-equilibrated with 10 volumes of the same denaturing buffer. Tubes containing this agarose lysate slurry were rotated for 1.5 hours at room temperature. The slurry was centrifuged and washed 2 X as above. Protein was eluted with 3 X 2ml of denaturing buffer containing 250 mM imidazole. Fractions containing protein of the expected size were diluted 1:1 and dialyzed vs 50 mM Tris pH 7.5, 500 mM NaCl, 1 mM DTT with 4 M Urea, followed by the same buffer containing 2M Urea and lastly 2 changes of buffer with 10% Glycerol and no Urea. Any precipitate after dialysis was removed by centrifugation at 3,000 rpm for 10 minutes and concentration determined assayed by BCA.
Purification of Mediator
The Mediator samples were purified as previously described32 with modifications. Prior to affinity purification, the P0.5M/QFT fraction was concentrated, to 12 mg/mL, by ammonium sulfate precipitation (35%). The pellet was resuspended in pH 7.9 buffer containing 20 mM KCl, 20 mM HEPES, 0.1 mM EDTA, 2 mM MgCl2, 20% glycerol and then dialyzed against pH 7.9 buffer containing 0.15 M KCl, 20 mM HEPES, 0.1 mM EDTA, 20% glycerol and 0.02% NP-40 prior to the affinity purification step. Affinity purification was carried out as described32, eluted material was loaded onto a 2.2 mL centrifuge tube containing 2 mL 0.15M KCl HEMG (20 mM HEPES, 0.1 mM EDTA, 2 mM MgCl2, 10% glycerol) and centrifuged at 50K RPM for 4h at 4ºC. This served to remove excess free GST-VP16 and to concentrate the Mediator in the final fraction. Prior to droplet assays, purified Mediator was further concentrated using Microcon-30kDa Centrifugal Filter Unit with Ultracel-30 membrane (Millipore MRCF0R030) to reach ~650 nM of Mediator complex. Concentrated Mediator was added to the droplet assay to a final concentration of ~200 nM with 10 μM indicated GFP-tagged protein. Droplet reactions contained 16% Ficoll-400 and 140 mM salt.
Chromatin immunoprecipitation sequencing (ChIP-seq)
mES were grown to 80% confluence in 2i media. About 1% formaldehyde in PBS was used for crosslinking of cells for 15 minutes, followed by quenching with Glycine at a final concentration of 125mM on ice. Cells were washed with cold PBS and harvested by scraping cells in cold PBS. Collected cells were pelleted at 1000 g for 3 minutes at 4°C, flash frozen in liquid nitrogen and stored at −80°C. All buffers contained freshly prepared cOmplete protease inhibitors (Roche, 11873580001). For ChIPs using phospho-specific antibodies, all buffers contained freshly prepared PhosSTOP phosphatase inhibitor cocktail (Roche, 4906837001). Frozen crosslinked cells were thawed on ice and then resuspended in LB1 (50 mM Hepes-KOH, pH7.9, 140 mM NaCl, 1 mM EDTA 0.5 mL 0.5 M, 10% glycerol, 0.5% NP-40, 1% Triton X-100, 1x protease inhibitor) and incubated for 20 minutes rotating at 4°C. Cells were pelleted for 5 minutes at 1350 g, resuspended in LB2 (10 mM Tris pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1x protease inhibitor) and incubated for 5 minutes rotating at 4°C. Pellets were resuspended in LB3 (10 mM Tris pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% sodium-deoxycholate, 0.5% sodium lauroyl sarcosinate, 1% Triton X-100, 1x protease inhibitor) at a concentration of 30–50 million cells/ml. Cells were sonicated using Covaris S220 for 12 minutes (Duty cycle: 5%, intensity: 4, cycles per burst: 200). Sonicated material was clarified by spinning at 20000 xg for 30 minutes at 4°C. The supernatant is the soluble chromatin used for the ChIP. Dynabeads pre-blocked with 0.5% BSA were incubated with indicated antibodies for 2 hours. Chromatin was added to antibody-bead complex and incubated rotating overnight at 4°C. Beads were washed three times each with Wash buffer 1 (50 mM Hepes pH7.5, 500 mM NaCl, 1 mM EDTA, 1 mM EGTA,1% Triton, 0.1% NaDoc, 0.1% SDS) and Wash Buffer 2 (20 mM Tris pH 8, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% NaDoc) at 4°C, followed by washing one time with TE at room temperature. Chromatin was eluted by adding Elution buffer (50 mM, Tris pH 8.0, 10 mM EDTA, 1% sodium dodecyl sulfate) to the beads and incubated shaking at 60°C for 30 minutes. Reversal of crosslinking was performed overnight at 58°C. RNaseA was added and incubated for 1 hour at 50°C for RNA removal. Proteinase K was added and incubated for 1 hour at 60°C for protein removal. DNA was purified using Qiagen PCR purification kit, as per manufacturer’s instructions, and eluted in 50 μL 10 mM Tris-HCl, pH 8.5, which was used for quantitation and ChIP library preparation. ChIP Libraries were prepared with the Swift Biosciences Accel-NGS® 2S Plus DNA Library Kit according to kit instructions with an additional size selection step on the PippinHT system from Sage Science. Following library prep, ChIP libraries were run on a 2% gel on the PippinHT with a size collection window of 200–600 bases. Final libraries were quantified by qPCR with the KAPA Library Quantification kit from Roche and sequenced in single-read mode for 40 bases on an Illumina HiSeq 2500.
ChIP-Seq data were aligned to the mm9 version of the mouse reference genome using bowtie with parameters –k 1 –m 1 –best and –l set to read length. Wiggle files for display of read coverage in bins were created using MACS with parameters –w –S – space=50 –nomodel –shiftsize=200, and read counts per bin were normalized to the millions of mapped reads used to make the wiggle file. Reads-per-million-normalized wiggle files were displayed in the UCSC genome browser. Metagene plots were made using ngs.plot33 (v2.61) using default parameters. Top 20% of expressed genes were calculated from a published RNA-seq dataset (GSE112807)8.
SRSF2 and Ser2-P Pol II ChIP-seqs were generated in this study using antibodies against SRSF2 (Abcam ab11826) and Pol II Ser2 phospho CTD (Millipore 04–1571), whereas MED1 and total Pol II ChIP-seqs were previously published (GSE112808)8.
RNA FISH average image analysis
For analysis of RNA FISH with immunofluorescence, custom Python scripts were written to process and analyze 3D image data gathered in FISH and IF channels. Nuclear stains were blurred with a Gaussian filter (sigma = 2.0), maximally projected in the z plane, and clustered into 2 clusters (nuclei and background) by K-means. FISH foci were either manually called with ImageJ or automatically called using the scipy ndimage package. For automatic detection, an intensity threshold (mean + 3*standard deviation) was applied to the FISH channel. The ndimage find_objects function was then used to call contiguous FISH foci in 3D. These FISH foci were then filtered by various criteria, including size (minimum 100 voxels), circularity of a max z-projection (; 0.7), and being present in a nucleus (determined by nuclear mask described above). For manual calling, FISH foci were identified in maximum z-projections of the FISH channel, and the x and y coordinates were used as reference points to guide the automatic detection described above. The FISH foci were then centered in a 3D-box (length size l = 3.0 μm). The IF signal centered at FISH foci for each FISH and IF pair are then combined and an average intensity projection is calculated, providing averaged data for IF signal intensity within a l x l square centered at FISH foci. As a control, this same process was carried out for IF signal centered at an equal number of randomly selected nuclear positions. These average intensity projections were then used to generate 2D contour maps of the signal intensity. Contour plots are generated using the matplotlib python package. For the contour plots, the intensity-color ranges presented were customized across a linear range of colors (n! = 15). For the FISH channel, black to magenta was used. For the IF channel, we used chroma.js (an online color generator) to generate colors across 15 bins, with the key transition colors chosen as black, blue-violet, medium-blue, lime. This was done to ensure that the reader’s eye could more readily detect the contrast in signal. The generated colormap was employed to 15 evenly spaced intensity bins for all IF plots. The averaged IF centered at FISH or at randomly selected nuclear locations are plotted using the same color scale, set to include the minimum and maximum signal from each plot.
Lattice light sheet microscopy
For lattice light sheet microscopy cells were plated on coated coverslip 24 hrs before imaging. Before imaging cells were incubated for 20 min with 250 nM Halo-JF646 ligand34 and washed in growth medium for 20 min. Dual-color stacks were acquired with 100 ms exposure time and 340 nm effective z-spacing. Light sheet data was processed (deskewed, deconvolved, chromatic aberration correction) using LLSpy35 and analyzed using custom MATLAB scripts. Foci were localized in 3D following a two-step procedure. First, background was subtracted by subtracting a median filtered image from each slice in a z-stack, and intensity peaks were detected using the MTT algorithm36. Foci were then identified as peaks that were found in at least 4 subsequent z-slices within a 100nm radius in x-y. A 3D Gaussian peak function was fitted to the intensity distribution to obtain 3D center coordinates for foci of SRSF2 and Pol II. Detection of foci was performed on deconvolved, background-subtracted data, whereas subsequent quantitative analysis of foci brightness was performed on deskewed, background-subtracted data. To estimate the number of fluorescently tagged proteins in foci cells were fixed in 4% PFA for 10 mins, washed 3x in PBS, and imaged on the lattice light sheet microscope until bleached almost entirely. A single plane was then imaged continuously for 1000 frames to detect single emitter signals. The apparent brightness of Halo-JF646 (n = 204) or GFP (n = 236) single emitters was determined by calculating the integrated intensity above background37 (Extended Data Fig. 2b). We note that signals are close to the noise floor of the camera, and the integrated intensity measure can sometimes (~10% of the cases) yield negative values when nearby emitters lead to overestimation of the local background intensity. For the subsequent analysis we excluded emitters with negative integrated intensity measures. We used the mean estimates obtained for the remaining single emitters to normalize values obtained by the same metric for foci of SRSF2-GFP, and Halo-JF646-Pol II in the z slice closest to the 3D center position, taking into account higher laser power densities that were used to detect single emitters. To assess co-localization we paired Pol II-Halo foci with their nearest neighbor in 3D space. We note that axial resolution of the imaging method is considerably lower than xy-resolution. We therefore call only those foci co-localized with center coordinates separated by less than the optical resolution of 300 nm laterally (xy), and 900nm axially (z).
Splicing reporter assay
The splicing reporter assay was performed as described in Younis et al. (2010)38. Briefly, mESCs were transfected with a plasmid encoding luciferase with an intervening intron (Addgene 62858) or a plasmid encoding luciferase with no intervening intron (Addgene 62857). Cells were then treated with DMSO or 100 nM Pladienolide B for 4 hrs, at which point they were lysed and assayed for luciferase activity. Relative splicing levels in each condition were determined by normalizing the luciferase activity detected in cells transfected with the intron containing plasmid to the luciferase activity detected in cells transfected with the intronless plasmid.
FISH-IF overlap analysis
DNA FISH spots were identified as described in “RNA FISH average image analysis”. Images of the spots with the corresponding IF channel for all conditions were randomized and blindly scored for FISH-IF overlap (at least 25% of the FISH spot overlapping with an IF puncta) or no overlap. Overlap scores for each condition were then tallied and compared. For presentation, the FISH foci overlap with IF was indexed with the DMSO condition set to 1.
In vitro droplet assay
Recombinant GFP or mCherry fusion proteins were concentrated and desalted to an appropriate protein concentration and 125–500 mM NaCl using Amicon Ultra centrifugal filters (30K MWCO, Millipore). Recombinant proteins were added to solutions at varying concentrations with 120–125 mM final salt and 16% Ficoll-400 or 10% PEG-8000 as crowding agent in droplet formation buffer (50 mM Tris-HCl pH 7.5, 10% glycerol, 1 mM DTT) as described in figure legends. The protein solution was incubated for 1 hour and loaded onto a homemade chamber comprising a glass slide with a coverslip attached by two parallel strips of double-sided tape. Slides were then imaged with the Andor confocal microscope with a 150x objective. Unless indicated, images presented are of droplets settled on the glass coverslip. For FRAP of in vitro droplets, 2 pulses of laser (20% power) at a 20 us dwell time were applied to the droplet, and recovery was imaged on the Andor microscope every 1s for the indicated time periods. For CDK7 or CDK9 mediated CTD phosphorylation, commercially available active CDK7/MAT1/CCNH (CAK complex; Millipore 14–476) or CDK9/Cyclin T1 (Millipore 14–685) was used to phosphorylate GFP-CTD52 in kinase reaction buffer (20 mM MOPs-NaOH pH 7.0, 1 mM EDTA, 0.001% NP-40, 2.5 % glycerol, 0.05% beta-mercaptoethanol, 10 mM MgAc, 10 μM ATP) at room temperature for 3 hours. The CTD to enzyme ratio is ~1 μM CTD to ~5 ng/ul CDK7 or CDK9.
In vitro droplet quantification
To analyze in vitro droplet experiments, custom Python scripts using the scikit-image package were written to identify droplets and characterize their size, shape, and intensity. Droplets were segmented from average images of captured channels on various criteria: (1) an intensity threshold three standard deviations above the mean of the image, (2) size thresholds (20 pixel minimum droplet size), (3) and a minimum circularity () of 0.8 (1 being a perfect circle). After segmentation, mean intensity for each droplet was calculated while excluding pixels near the phase interface and background-corrected by subtracting intensity of dark images of droplet formation buffer only (see Banani et al Cell 2016)39. Hundreds of droplets identified in typically 10 independent fields of view were quantified. The mean intensity within the droplets (C-in) and in the bulk (C-out) were calculated for each channel. The partition ratio was computed as (C-in)/(C-out). The box plots show the distributions of all droplets. Each dot represents an individual droplet. The measured datasets for partition ratio versus the protein concentration in Extended Data Figure 7b were fitted by the logistic equation (see Wang et al Cell 2018)40:
Where f is the partition ratio and x is the corresponding protein concentration.
Statistics and reproducibility
For all immunofluorescence and fluorescence in situ hybridization experiments, one coverslip of cells was stained for the indicated factors and at least eight independent imaging fields were acquired, which typically contained 50–200 FISH foci. The exact number of FISH foci analyzed, and the fraction of those foci that overlap with IF puncta and relevant comparative statistics for experiments where these comparisons were made are as follows:
Fig. 1b/Extended Data Fig. 1a: 86 Nanog foci, 131 Trim28 foci.
Fig. 2a: Nanog FISH foci counts for each IF experiment- SRSF2: 97, SF3B1: 122, U2AF2: 74, HNRNPA1: 88, SRSF1: 109, SRRM1: 137, PRPF8: 103, SNRNP70: 119.
Fig. 2c/Extended Data Fig. 3b, c: The numbers of overlapped foci for each factor in the DMSO and Pladienolide B condition, and the p-values associated with each DMSO vs. Pladienolide B comparison are as follows: (SRSF2, 31/61, 19/125, <0.0001; SF3B1, 29/61, 30/126, 0.0014; Pol II, 16/71, 15/65, >0.9999).
Fig. 3c, d/Extended Data Fig. 5a–d: The numbers of overlapped foci for each factor in the DMSO, DRB, and washout condition, and the p-values associated with the DMSO vs. DRB and DRB vs. washout comparison are as follows: (SRSF2 Nanog, 40/91, 11/146, 19/78, <0.0001, 0.0008; Pol II Nanog, 33/114, 23/92, 36/160, 0.6368, 0.6467; MED1 Nanog, 28/89, 32/133, 27/84, 0.2804, 0.2122; SRSF2 Trim28, 26/71, 5/111, 12/92, <0.0001, 0.0403; SRSF1 Trim28, 19/36, 12/55, 22/73, 0.0347, 0.3189; SF3B1 Trim28, 48/91, 14/63, 27/99, 0.0002, 0.5788; U2AF2 Trim28, 21/42, 17/50, 27/78, 0.1406, >0.9999; PRPF8 Trim28, 15/80, 6/69, 17/102, 0.0996, 0.1721; MED1 Trim28, 19/73, 30/96, 21/91, 0.4971, 0.251; Pol II Trim28, 25/102, 22/93, 21/78, >0.9999, 0.7238).
Extended Data Fig. 2a: Trim28 FISH foci counts for each IF experiment - SRSF2: 115, SF3B1: 151, U2AF2: 104, HNRNPA1: 90, SRSF1: 145, SRRM1: 127, PRPF8: 175, SNRNP70: 157.
For lattice light sheet imaging, the number of images acquired and data points plotted are as follows:
Fig. 2b: full image dataset comprises 102 cells from 10 independent fields of view.
Extended Data Fig. 2b: 288 colocalized condensates were plotted in the scatter plot.
For all in vitro droplet experiments, one slide of droplet mix was imaged and at least seven independent fields of view were acquired, which typically contained ~100–1000 droplets. In all cases where the droplet data is quantified and displayed as a box plot, the box in the boxplot extends from the 25th to 75th percentiles, the line in the middle of the box is plotted at the median, the whiskers represent the range within 1.5x interquartile, and each dot represents an individual droplet. The exact number of fields and droplets analyzed are as follows:
Fig. 1c (fields/droplets): GFP, 10/540; GFP-CTD52, 7/842; GFP-CTD26, 10/879; GFP-CTD10 10/1293.
Fig. 1d (fields/droplets): GFP 11/159, GFP-CTD52 10/175, GFP-CTD26 11/207, GFP-CTD10 10/206.
Fig. 4a (fields/droplets): GFP 10/114, CTD 7/420, P-CTD(CDK7) 8/342, P-CTD(CDK9) 8/464.
Fig. 4b (fields/droplets): GFP 10/385, CTD 10/328, P-CTD(CDK7) 10/294, P-CTD(CDK9) 12/361.
Fig. 4c (fields/droplets): GFP 10/400, CTD 10/369, P-CTD(CDK7) 10/314, P-CTD(CDK9) 15/513.
Extended Data Fig. 1b: One fusion event was captured from one field.
Extended Data Fig. 6c, d (fields/droplets): 16% Ficoll, GFP 21/231, CTD 21/289, P-CTD(CDK7) 10/134, P-CTD(CDK9) 11/144; 10% PEG, GFP 24/147, CTD 21/227, P-CTD(CDK7) 10/106, P-CTD(CDK9) 10/83.
Extended Data Fig. 7a, b: The number of fields acquired are as follows, SRSF2: 5μM 10, 2.5μM 10, 1.25μM 10, 0.625μM 10, 0.313μM 10, 0.156μM 11, 0.078μM 12, 0.039μM 12, 0.0195μM 10, 0.0098μM 10, 0.0049μM 10, 0.0024μM 10, 0.0012μM 10, 0.0006μM 10; SRSF1: 5μM 10, 2.5μM 10, 1.25μM 10, 0.625μM 10, 0.313μM 10, 0.156μM 10, 0.078μM 10, 0.039μM 10, 0.0195μM 10, 0.0098μM 10, 0.0049μM 10, 0.0024μM 11, 0.0012μM 11, 0.0006μM 10; U2AF2: 10 for each sample; hnRNPA1: 10 for each sample; mCherry: 10 for each sample.
Extended Data Fig. 7c: 10 independent fields of view were acquired for each sample.
Extended Data Fig. 8a, b: same as Fig. 4b, c.
Extended Data Fig. 8c (fields/droplets): GFP 10/365, CTD 10/321, P-CTD(CDK7) 10/325, P-CTD(CDK9) 10/313.
Extended Data Fig. 8d: (fields/droplets): GFP 10/423, CTD 11/437, P-CTD(CDK7) 10/412, P-CTD(CDK9) 10/381.
Code availability
All custom code used in this study is available upon request.
Data availability
Datasets generated in this study have been deposited in the Gene Expression Omnibus under accession number GSE120656.
Extended Data
Extended Data Figure 1. CTD partitioning in Mediator condensates.
a. Average IF signal for MED1 and Pol II centered on randomly selected nuclear positions.
b. Images of a fusion event between two full-length CTD/MED1-IDR droplets. GFP-CTD52 at 10 μM was mixed with 10 μM mCherry-MED1-IDR in droplet formation buffer with 125 mM NaCl and 16% Ficoll-400.
c. Top: representative images of FRAP of heterotypic droplets of mCherry-MED1-IDR and GFP-CTD52. Droplet formation conditions are the same as in b. Bottom: Quantification of the fraction of fluorescence recovery following photobleaching of mCherry-MED1-IDR and GFP-CTD52. Data represent mean +/− SD (n = 3).
Extended Data Figure 2. Splicing factors at Trim28, SRSF2/Pol II molecule quantification.
a. Representative images exhibiting overlap between IF of splicing factors SRSF2, SF3B1, U2AF2, HNRNPA1, SRSF1, SRRM1, PRPF8, or SNRNP70 with nascent RNA FISH of Trim28 in fixed mESCs.
b. Left: Histogram of the integrated intensity of single Halo-JF646 (n=178) and single GFP emitters (n=177). Mean values of 164.8 +/− 5.6 cts (mean +/− s.e.m.) and 108.6 +/− 5.1 (mean +/− s.e.m.) were used to normalize the integrated intensity of Pol II-Halo-JF646 and SRSF2-GFP, respectively. Right: Scatter plot depicting estimated numbers of Pol II and SRSF2 molecules in colocalizing Pol II and SRSF2 puncta (see methods).
Extended Data Figure 3. Splicing inhibition and splicing factor condensates.
a. Left: diagram depicting the splicing reporter used to measure splicing inhibition following treatment with Pladienolide B. Right: relative levels of splicing in cells treated with DMSO vs. cells treated with Pladienolide B for 4 hrs. The mean of 3 biological replicates (each replicate shown as dot) with standard deviations is plotted. See methods for details.
b. Representative images exhibiting overlap or absence of overlap between IF of SF3B1 or Pol II and DNA FISH of Trim28 in mESCs treated with either DMSO or splicing inhibitor Pladienolide B for 4 hours.
c. Fraction of overlap of FISH foci with IF puncta in cells treated with Pladienolide B relative to cells treated with DMSO. A star above the drug treated bar indicates a p-value of <0.05 from a two-tailed Chi-squared test comparing the number of overlapping and non-overlapping FISH spots in the DMSO vs. drug condition. See methods for details.
Extended Data Figure 4. Liquid-like properties of SRSF2 condensates.
a. Live cell imaging of GFP-SRSF2 mESCs. Ten independent fields from one plate of cells were imaged.
b. Representative images of fluorescence recovery after photobleaching (FRAP) experiments performed on the GFP-SRSF2 mESC cell line.
c. Quantification of experiment depicted in b. Data represent means +/− SD (n = 9).
d. Representative images of live cells before and after treatment with 10% 1,6-hexanediol for ~17 minutes. Five independent fields from one plate of cells were imaged before and after treatment.
e. Examples of fusion events occurring between SRSF2 puncta in the GFP-SRSF2 cell line. Two fields from two independent plates of cells were imaged over a two hour time course, and each showed at least one fusion event.
Extended Data Figure 5. DRB treatment effects on splicing factor and transcriptional condensates.
a. Representative images exhibiting overlap or lack of overlap between IF of SRSF2 and DNA FISH of Trim28 in mESCs treated with DMSO for 2 hrs, DRB for 2 hrs, or DRB for 2 hrs followed by a 2 hr DRB washout.
b. Representative images exhibiting overlap between IF of MED1 and DNA FISH of Trim28 in mESCs treated with DMSO for 2 hrs, DRB for 2 hrs, or DRB for 2 hrs followed by a 2 hr DRB washout.
c. Representative images exhibiting overlap between IF of Pol II and DNA FISH of Nanog or Trim28 in mESCs treated with DMSO for 2 hrs, DRB for 2 hrs, or DRB for 2 hrs followed by a 2 hr DRB washout.
d. Fraction of overlap of FISH foci with IF puncta in cells treated with DRB or DRB followed by washout relative to cells treated with DMSO. A star above the DRB treated bar indicates a p-value of <0.05 from a two-tailed Chi-squared test comparing the number of overlapping and non-overlapping FISH spots in the DMSO vs. DRB condition. A star over the washout bar indicates a p-value of <0.05 from a two-tailed Chi-squared test comparing the DRB vs. washout condition. Splicing factors tested showed significantly decreased or trended towards decreased overlap with FISH spots in the DRB condition compared to DMSO, while MED1 and Pol II exhibited limited changes. See methods for details.
Extended Data Figure 6. CTD in vitro phosphorylation and partitioning in MED1-IDR droplets.
a. Western blot showing phosphorylation of CTD by CDK7. Equal amounts of unphosphorylated and phosphorylated GFP-CTD52 (see methods) were run on a 3–8% SDS PAGE gel and analyzed by western blot using anti-GFP antibodies. Similar results were obtained in two biological replicates. For gel source data, see Supplementary Figure 1.
b. Western blot showing phosphorylation of CTD by CDK9. Similar results were obtained in two biological replicates. For gel source data, see Supplementary Figure 1.
c. Representative images of droplet experiments measuring CTD incorporation into MED1-IDR droplets. Purified human MED1-IDR fused to mCherry (mCherry-MED1-IDR) at 10 μM was mixed with 3.3 μM GFP, GFP-CTD52, GFP-CTD52 phosphorylated with CDK7 or CDK9 in droplet formation buffers with 125 mM NaCl and 16% Ficoll-400 or 10% PEG-8000.
d. Partition ratios of GFP and GFP-CTD in MED1-IDR droplets, and partition ratios of mCherry-MED1-IDR from experiments depicted in c.
Extended Data Figure 7. Splicing factors form droplets in vitro.
a. Representative images of droplet formation by mCherry-SRSF2, SRSF1, U2AF2 and hnRNPA1 with increasing protein concentrations. All assays were performed in the presence of 120 mM NaCl and 10% PEG-8000.
b. Quantification of the partition ratios from the experiments depicted in a.
c. Representative images of heterotypic droplets formed between SRSF2 and other splicing factors, including SRSF1, U2AF2 and hnRNPA1. Purified human SRSF2 fused to GFP (GFP-SRSF2) at 2.5 μM was mixed with 2.5 μM purified mCherry-fused to human SRSF2, SRSF1, U2AF2 or hnRNPA1 in droplet formation buffers with 120mM NaCl and 10% PEG-8000.
Extended Data Figure 8. CTD partitioning in SR-protein droplets.
a. Quantification of the partition ratios of SRSF2 from experiments depicted in Fig. 4b.
b. Quantification of the partition ratios of SRSF1 from experiments depicted in Fig. 4c.
c. Representative images of droplet experiments measuring CTD incorporation into SRSF2 droplets. Same reagents and conditions were used as in Fig. 4b except that 16% Ficoll-400 was used as a crowding agent.
d. Representative images and quantification of partition ratios of droplet experiments measuring phosphorylated full length or truncated CTD incorporation into SRSF2 droplets. Droplet formation conditions are the same as described in Fig. 4b.
Supplementary Material
Acknowledgements
We thank Isaac A. Klein, Jurian Schuijers, Charles H. Li, Eliot L. Coffey and other members in the Young lab for helpful discussions, Wendy Salmon of the W.M Keck Microscopy Facility, Tom Volkert, Jennifer Love and Sumeet Gupta of the Whitehead Genomics Core facility for technical assistance, Michael Stubna for help with droplet and biochemical assays, and Steven L. McKnight, Ilmin Kwon, and Masato Kato for CTD constructs. The work was supported by NIH grant GM123511 (R.A.Y.), NSF grant PHY1743900 (R.A.Y. and P.A.S.), NIH GM117370 and GM110064 (D.J.T.), NIH grant R01-GM034277 (P.A.S.), Cancer Research Institute Irvington Fellowship (Y.E.G.), Damon Runyon Cancer Research Foundation Fellowship (2309-17) (B.R.S.), Hope Funds for Cancer Research fellowship (B.J.A.), Swedish Research Council Postdoctoral Fellowship (VR 2017-00372) (A.B.), DFG Postdoctoral Fellowship (SP 1680/1-1) (J.H.S.), the German Research Foundation DFG DE 3069/1-1 (T-M.D.), NIH T32 GM008759 (C.B.F.).
Footnotes
Supplementary information
Uncropped gel images can be found in Supplementary Figure 1.
References
- 1.Adelman K & Lis JT Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 13, 720–731, doi: 10.1038/nrg3293 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harlen KM & Churchman LS The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat Rev Mol Cell Biol 18, 263–273, doi: 10.1038/nrm.2017.10 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Levine M, Cattoglio C & Tjian R Looping back to leap forward: transcription enters a new era. Cell 157, 13–25, doi: 10.1016/j.cell.2014.02.009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sainsbury S, Bernecky C & Cramer P Structural basis of transcription initiation by RNA polymerase II. Nat Rev Mol Cell Biol 16, 129–143, doi: 10.1038/nrm3952 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Eick D & Geyer M The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev 113, 8456–8490, doi: 10.1021/cr400071f (2013). [DOI] [PubMed] [Google Scholar]
- 6.Ebmeier CC et al. Human TFIIH Kinase CDK7 Regulates Transcription-Associated Chromatin Modifications. Cell Rep 20, 1173–1186, doi: 10.1016/j.celrep.2017.07.021 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho WK et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415, doi: 10.1126/science.aar4199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabari BR et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, doi: 10.1126/science.aar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spector DL & Lamond AI Nuclear speckles. Cold Spring Harb Perspect Biol 3, doi: 10.1101/cshperspect.a000646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y et al. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J Cell Biol, doi: 10.1083/jcb.201807108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quinodoz SA et al. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell 174, 744–757 e724, doi: 10.1016/j.cell.2018.05.024 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall LL, Smith KP, Byron M & Lawrence JB Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol 288, 664–675, doi: 10.1002/ar.a.20336 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen BL & Taatjes DJ The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol 16, 155–166, doi: 10.1038/nrm3951 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boija A et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 175, 1842–1855 e1816, doi: 10.1016/j.cell.2018.10.042 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X et al. MED1/TRAP220 exists predominantly in a TRAP/ Mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription. Mol Cell 19, 89–100, doi: 10.1016/j.molcel.2005.05.015 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Banani SF, Lee HO, Hyman AA & Rosen MK Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18, 285–298, doi: 10.1038/nrm.2017.7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hnisz D, Shrinivas K, Young RA, Chakraborty AK & Sharp PA A Phase Separation Model for Transcriptional Control. Cell 169, 13–23, doi: 10.1016/j.cell.2017.02.007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin Y & Brangwynne CP Liquid phase condensation in cell physiology and disease. Science 357, doi: 10.1126/science.aaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Hnisz D et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947, doi: 10.1016/j.cell.2013.09.053 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braunschweig U, Gueroussov S, Plocik AM, Graveley BR & Blencowe BJ Dynamic integration of splicing within gene regulatory pathways. Cell 152, 1252–1269, doi: 10.1016/j.cell.2013.02.034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herzel L, Ottoz DSM, Alpert T & Neugebauer KM Splicing and transcription touch base: co-transcriptional spliceosome assembly and function. Nat Rev Mol Cell Biol 18, 637–650, doi: 10.1038/nrm.2017.63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsin JP & Manley JL The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev 26, 2119–2137, doi: 10.1101/gad.200303.112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotake Y et al. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol 3, 570–575, doi: 10.1038/nchembio.2007.16 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Akhtar MS et al. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell 34, 387–393, doi: 10.1016/j.molcel.2009.04.016 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Czudnochowski N, Bosken CA & Geyer M Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat Commun 3, 842, doi: 10.1038/ncomms1846 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Kwon I et al. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060, doi: 10.1016/j.cell.2013.10.033 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long JC & Caceres JF The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417, 15–27, doi: 10.1042/BJ20081501 (2009). [DOI] [PubMed] [Google Scholar]
- 28.McCracken S et al. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385, 357–361, doi: 10.1038/385357a0 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Boehning M et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat Struct Mol Biol, doi: 10.1038/s41594-018-0112-y (2018). [DOI] [PubMed] [Google Scholar]
- 30.Lu H et al. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323, doi: 10.1038/s41586-018-0174-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods references
- 31.Bolte S & Cordelieres FP A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224, 213–232, doi: 10.1111/j.1365-2818.2006.01706.x (2006). [DOI] [PubMed] [Google Scholar]
- 32.Meyer KD et al. Cooperative activity of cdk8 and GCN5L within Mediator directs tandem phosphoacetylation of histone H3. EMBO J 27, 1447–1457, doi: 10.1038/emboj.2008.78 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen L, Shao N, Liu X & Nestler E ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15, 284, doi: 10.1186/1471-2164-15-284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grimm JB et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Methods 12, 244–250, 243 p following 250, doi: 10.1038/nmeth.3256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.tlambert03/LLSpy: v0.3.8. (Zenodo, 2018). [Google Scholar]
- 36.Serge A, Bertaux N, Rigneault H & Marguet D Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat Methods 5, 687–694, doi: 10.1038/nmeth.1233 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Verdaasdonk JS, Lawrimore J & Bloom K Determining absolute protein numbers by quantitative fluorescence microscopy. Methods Cell Biol 123, 347–365, doi: 10.1016/B978-0-12-420138-5.00019-7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Younis I et al. Rapid-response splicing reporter screens identify differential regulators of constitutive and alternative splicing. Mol Cell Biol 30, 1718–1728, doi: 10.1128/MCB.01301-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banani SF et al. Compositional Control of Phase-Separated Cellular Bodies. Cell 166, 651–663, doi: 10.1016/j.cell.2016.06.010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 174, 688–699 e616, doi: 10.1016/j.cell.2018.06.006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets generated in this study have been deposited in the Gene Expression Omnibus under accession number GSE120656.