Abstract
The small ubiquitin-like modifier 2 (SUMO-2) is required for survival when cells are exposed to treatments that induce proteotoxic stress by causing the accumulation of misfolded proteins. Exposure of cells to heat shock or other forms of proteotoxic stress induces the conjugation of SUMO-2 to proteins in the nucleus. Here, we investigated the chromatin landscape of SUMO-2 modifications in response to heat stress. Through chromatin immunoprecipitation assays coupled to high-throughput DNA sequencing and with mRNA sequencing, we showed that in response to heat shock, SUMO-2 accumulated at nucleosome-depleted, active DNA-regulatory elements, which represented binding sites for large protein complexes and were predominantly associated with active genes. However, SUMO did not act as a direct transcriptional repressor or activator of these genes during heat shock. Instead, integration of our results with published proteomics data on heat shock–induced SUMO-2 substrates supports a model in which the conjugation of SUMO-2 to proteins acts as an acute stress response that is required for the stability of protein complexes involved in gene expression and posttranscriptional modification of mRNA. We showed that the conjugation of SUMO-2 to chromatin-associated proteins is an integral component of the proteotoxic stress response, and propose that SUMO-2 fulfills its essential role in cell survival by contributing to the maintenance of protein complex homeostasis.
Introduction
Small ubiquitin-like modifiers (SUMOs) are small proteins that are covalently conjugated to various target proteins and thus influence a broad range of biological functions. Three SUMO paralogs, termed SUMO-1, SUMO-2, and SUMO-3 are present in higher eukaryotes. Based on structural and functional characteristics, SUMO-2 and SUMO-3 are collectively referred to as SUMO-2/3 to distinguish them from SUMO-1. SUMO conjugation involves an E1 activating enzyme SUMO-activating enzyme subunit 1/2 (SAE1/SAE2) and the SUMO E2 conjugating enzyme (UBC9), which catalyzes the formation of an isopeptide bond between the C-terminus of SUMO and the ε-amino group of the target lysine, which is often located within a SUMOylation consensus motif ΨKxE (Ψ, a hydrophobic amino acid; K, lysine; x, any amino acid residue; E, glutamate). A number of E3 ligases, including members of the protein inhibitor of activated STAT protein (PIAS) family, chromobox protein homolog 4 (CBX4), and Ran-binding protein 2 (RanBP2), facilitate the UBC9-dependent conjugation of SUMO to target proteins (1). SUMO conjugation is a highly dynamic process and can be reversed through the action of SUMO proteases (SENPs). By catalyzing the cleavage of SUMO molecules at a specific C-terminal sequence, SENPs are responsible for maturation of SUMO precursor molecules, deconjugation of SUMO from substrates, and depolymerization of SUMO chains (2).
SUMO paralogs display a certain degree of functional redundancy and substantial overlaps in substrate specificities (3). Yet, paralog-specific characteristics have been ascribed to SUMO-1 and SUMO-2/3. SUMO-2 and SUMO3 contain a SUMO consensus modification motif that enables self-modification and the formation of SUMO chains (4). Incorporation of SUMO-1 into these polymers in vivo appears to cap SUMO-2/3 chains (5). Noncovalent binding of SUMO to proteins containing SUMO interaction motifs (SIMs) enables SUMO chains to act as platforms that mediate protein-protein interactions and downstream signaling events (6, 7). SUMO is essential for normal cell function in most eukaryotes (8–12). Although abrogation of UBC9-dependent SUMOylation leads to early embryonic lethality in mice, knockout studies suggest that SUMO-2/3 can functionally compensate for the loss of SUMO-1 (12–14). Modification by SUMO-2/3 and SUMO chain formation can be rapidly induced by proteotoxic stress, including heat and hyperosmotic and oxidative stress, resulting from the accumulation of unfolded or damaged proteins. Heat shock (HS)-induced SUMO conjugation is well-conserved across species and has cytoprotective functions (15–22).
Proteotoxic stress is a threat to cellular homeostasis and is implicated in the development of many age-associated diseases associated with neurodegeneration. The HS response (HSR) enables cells to adapt to and survive proteotoxic insults by coordinating the sensing of protein damage with the actions of cytoprotective response pathways and chaperone networks (23). Its unique hallmark is the marked induction of genes encoding heat shock proteins (HSPs) and chaperones, which preserve cellular protein homeostasis by counteracting protein misfolding, unfolding, and aggregation. The expression of these genes is driven by heat shock transcription factor 1 (HSF1), commonly coined the master regulator of the HSR.
Despite the HS-induced modification of chromatin-associated proteins by SUMO-2/3, the global chromatin-binding profile of SUMO-2 during HS has not been addressed. Here, by combining data from chromatin immunoprecipitation assays coupled to high-throughput DNA sequencing (ChIP-seq) and RNA sequencing (RNA-seq) with previously published proteomics data, we show that in response to HS, SUMO-2 is rapidly conjugated to protein complexes associated with the DNA-regulatory elements of active genes that encode regulators of gene expression and posttranscriptional modification of RNA. Rather than acting as a direct transcriptional repressor or activator during HS, HS-induced SUMO conjugation appears to be an integral component of the proteotoxic stress response. Our data implicate SUMO conjugation as an immediate early mechanism required for the maintenance of protein complex homeostasis in response to protein damage. We suggest that, by complementing the action of HSPs, proteotoxic stress-induced SUMO conjugation is required to tolerate the transient accumulation of damaged and misfolded proteins.
Results
ChIP-seq reveals substantial increases in the binding of SUMO-2 to active DNA-regulatory elements in response to HS
To characterize changes in the association of SUMO-2 with chromatin in response to HS in an unbiased manner, we used chromatin immunoprecipitation (ChIP) coupled to high-throughput sequencing (ChIP-seq) to determine the genome-wide profiles of SUMO-2 in U2OS (human osteosarcoma) cells that were either unstressed or were subjected to HS by shifting the incubation temperature of the cells to 43°C for 30 min (HS cells), when global conjugation of proteins by SUMO-2/3 is maximal (17). SUMO-2–bound chromatin was enriched from crosslinked cell extracts with an antibody with previously confirmed specificity for SUMO-2 (24). Total genomic DNA was sequenced to obtain reference input profiles (fig. S1A). We applied the model-based analysis of ChIP-seq (MACS) peak-calling algorithm (25, 26) to identify sites of statistically significantly increased SUMO-2–binding in HS cells compared to that in untreated cells and input control. The observations described here are based on SUMO-2–binding sites common to two independent ChIP-seq experiments showing a very similar pattern of binding of SUMO-2 to chromatin before and in response to HS (fig. S1B).
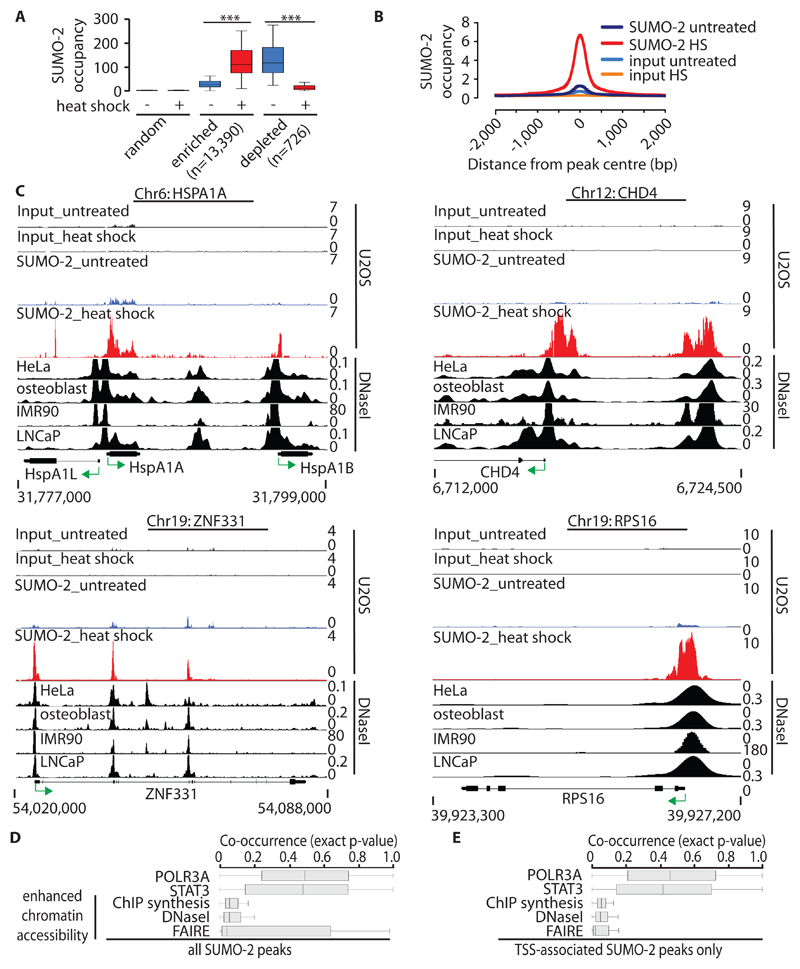
This analysis revealed the HS-induced recruitment of SUMO-2 to chromatin, and indicated that the binding of SUMO-2 to chromatin markedly changed in response to HS at 13390 sites shared between both replicate datasets (Fig. 1A and fig. S1C). These sites represent discrete loci that centered on an average core region of 500 to 1000 bp and were already marked by variable, but low, amounts of chromatin-bound SUMO-2 before HS was induced (Fig. 1, A and B, and fig. S1, C and D). In contrast, we detected only 726 sites shared between both replicate datasets that were depleted of SUMO-2 in response to HS, indicating that, overall, this stress caused a substantial increase in global SUMO-2 binding to chromatin (Fig. 1A, and fig. S1C). Unlike genomic regions bound by SUMO-2 in response to HS, sites depleted of SUMO-2 upon HS did not correlate with the number of genes and regulatory features per chromosome (fig. S2, A to D). Thus, our study focused on genomic loci that showed increased HS-induced binding of SUMO-2. This SUMO-2 ChIP-seq analysis confirmed the previously reported HS-induced recruitment of SUMO-2 to the HSPA1A (HSP70) promoter (27), but also revealed substantial binding of SUMO-2 to the HSPA1A gene body and to the HSPA1B and HSPA1L genes. Statistically significant enrichment of SUMO-2 was also detected at numerous regions for which no HS-induced SUMO-2 binding has been reported to date, including the CHD4, RPS16, and ZNF331 gene loci (Fig. 1C).
Fig. 1. Genome-wide analysis of SUMO-2 chromatin occupancy during HS.
(A) SUMO-2 occupancy at enriched, depleted, and control sites in untreated U2OS cells and in U2OS cells subjected to HS (dataset 2). The y-axis represents RPKM (reads per kb per million mapped reads). Boxplots (64) are shown without outliers. ***P ≤ 10-16 by pairwise Wilcoxon signed-rank test. (B) Profile of HS-induced SUMO-2 peaks (dataset 2). Alignment of sequencing reads at 13,390 HS-induced SUMO-2–binding sites. The y-axis represents reads per base per million reads. (C) Alignment of HS-induced SUMO-2–binding sites associated with HSPA1A, CHD4, RPS16, and ZNF331 in U2OS cells with DHSs from untreated HeLa-S3 cells, osteoblasts, IMR90 cells, and LNCaP cells (from ENCODE). Green arrows indicate the direction of transcription. Chr, chromosome. (D) Co-occurrence of HS-induced SUMO-2 peaks (n = 13390) and sites of enhanced chromatin accessibility (from ENCODE) 5 kb either side of the SUMO-2 peaks calculated by IntervalStats. POLR3A and STAT3 (from ENCODE) are shown as negative controls. Enhanced chromatin accessibility datasets are derived from untreated HeLa-S3 cells. FAIRE, formaldehyde-assisted isolation of regulatory elements; DNaseI, DNaseI-seq; ChIP synthesis, compilation of transcription factor–binding sites identified by ChIP-seq experiments for various transcription factors. (E) Co-occurrence of HS-induced TSS-associated SUMO-2 peaks only (n = 7325 peaks; TSS ± 2 kb). Details are as described for (D).
Under HS conditions, SUMO-2 is covalently attached to a large set of nuclear substrates that are enriched for transcription factors and chromatin-binding proteins (17). To relate HS-induced SUMO-2 ChIP-seq peaks to regulatory factor–binding sites in chromatin, the number of observed SUMO-2 peaks per chromosome was compared to the number of expected peaks. This revealed that the number of observed SUMO-2 peaks correlated with genes and regulatory sites in chromatin (fig. S2, A and B). Second, SUMO-2 ChIP-seq signals were compared to sites of transcription factor–binding and enhanced chromatin accessibility from unstressed cells (ENCODE, see Materials and Methods for GEO accession numbers). Regions of enhanced sensitivity to nucleases, such as DNaseI, correspond to nucleosome-depleted regions. These nuclease-hypersensitive sites mark active cis-regulatory elements, including transcription start sites (TSSs), enhancers, insulators, and silencers. The region of maximal nuclease sensitivity directly correlates with maximal regulatory factor occupancy (28). Analysis of this data (29) revealed the co-occurrence of SUMO-2–enriched peaks at regions of enhanced chromatin accessibility as defined by DNaseI-hypersensitivity, formaldehyde-assisted isolation of DNA regulatory elements (FAIRE), and transcription factor–binding site data (ChIP synthesis) (Fig. 1, D and E). This co-location of SUMO-2 peaks and DNaseI–hypersensitive sites (DHSs) was observed for individual sites associated with the HSPA1A, CHD4, ZNF331, and RPS16 loci (Fig. 1C). Together, these observations imply that SUMO-2 covalently modifies substrate proteins that occupy active DNA-regulatory elements during HS.
SUMO-2 localizes to DNA-regulatory elements predominantly associated with genes and an active chromatin environment
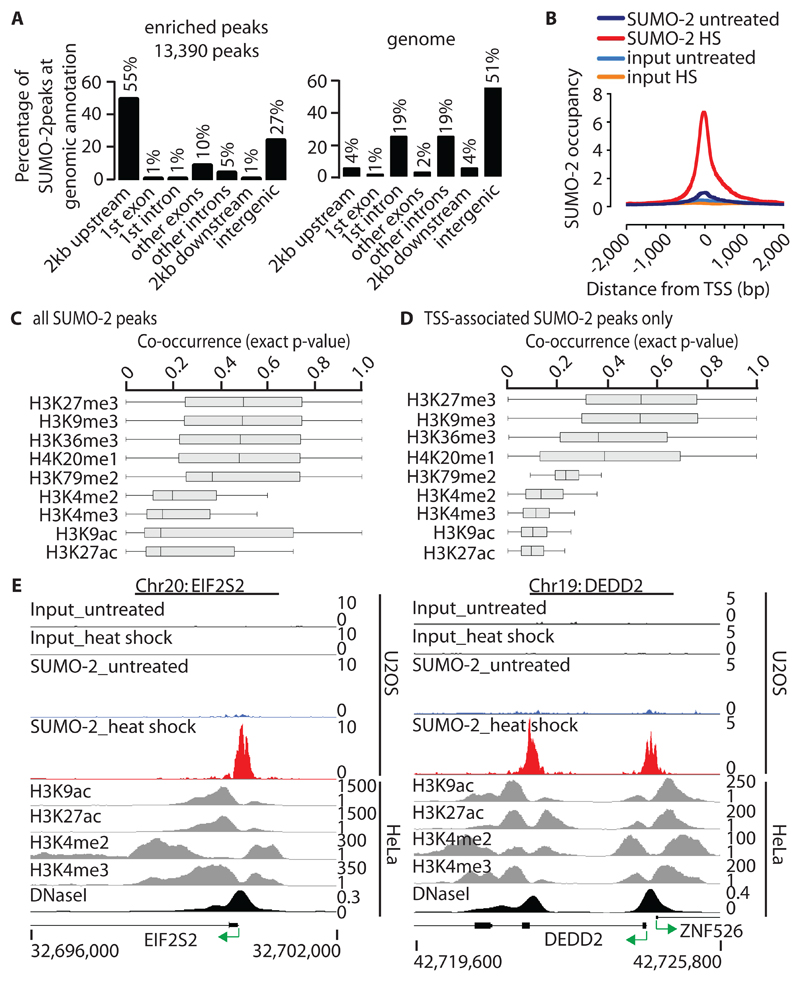
More than two-thirds (73%) of all SUMO-2–enriched sites are located within protein-coding genes or their proximal regions spanning 2 kb upstream or downstream (9721 peaks, 8292 genes) (Fig. 2A). Whereas most (55%) SUMO-2 peaks map to the promoter-proximal region, only 1% localize to the 2-kb region downstream of genes. Of the remaining SUMO-2 peaks, 17% are located within gene bodies, and less than a third (27%) are found in the intergenic region (> 2 kb from any protein-coding gene), possibly representing distal gene-regulatory elements (Fig. 2A and fig. S3, A to G) (30, 31).
Fig. 2. Genomic distribution of HS-induced SUMO-2 peaks and their relationship to active histone marks.
(A) Left: Location of HS-induced SUMO-2–binding sites relative to the annotated protein-coding genome, including 2 kb upstream from the TSS, 2 kb downstream from the end of the gene, and intergenic sites (> 2 kb from any gene). Right: The distribution of annotations across the human genome is shown for comparison. (B) Alignment of HS-induced SUMO-2 peaks to the TSSs of protein-coding genes (dataset 2). A total of 7325 HS-induced SUMO-2 peaks map to a region spanning 2 kb either side of TSSs (n = 6290 peaks). The y-axis represents reads per base per million reads. (C) Co-occurrence of HS-induced SUMO-2 peaks (n = 13390) and histone modification marks within a region spanning 5 kb either side of a SUMO-2 peak as calculated by IntervalStats. Histone modification data sets are derived from untreated HeLa-S3 cells. (D) Co-occurrence of HS-induced TSS-associated SUMO-2 peaks only (n = 7325 peaks; TSS ± 2 kb). Details are as described for (C). (E) Alignment of HS-induced SUMO-2 peaks with active histone modification marks at sites associated with EIF2S2 (left) and DEDD2 (right). See (C) for histone datasets.
In agreement with the observed HS-induced recruitment of SUMO-2 to the promoter-proximal region of genes, 7325 HS-induced SUMO-2 peaks map to gene start sites (± 2 kb of the gene start position; 6290 genes) (Fig. 2B and fig. S4) (30, 31). Concordantly, comparison of SUMO-2 peaks with ENCODE transcription factor–binding datasets showed a strong co-occurrence of SUMO-2 and several transcription- and chromatin-binding factors (fig. S5, A and B). For example, SUMO-2 peaks corresponded to binding sites for general transcriptional regulators and chromatin-remodelers, including GTF2F1, TBP, and CHD2, whereas binding sites for the RNA polymerase III subunit POLR3A or the inflammatory response specific transcription factor Signal transducer and activator of transcription 3 (STAT3) did not co-locate with SUMO-2 peaks. Comparing the co-location of transcription factor–binding sites with either all genome-wide SUMO-2 peaks or SUMO-2 peaks found at the TSSs of protein-coding genes revealed that co-location was more pronounced at TSS-associated SUMO-2 peaks (fig. S5, A and B). Comparison of the genome-wide distribution of SUMO-2–binding sites with ChIP-seq profiles for several histone modifications (ENCODE) revealed that, during HS, SUMO-2 targeted a genomic environment that was already enriched for active histone marks, including acetylated Lys27 (K27) of histone H3 (H3K27ac), H3K9ac, trimethylated K4 of histone H3 (H3K4me3), and dimethylated K4 of histone H3 (H3K4me2), before HS was induced (Fig. 2, C to E). Repressive histone modifications, including H3K27me3 and H3K9me3, were excluded from SUMO-2–occupied regions (Fig. 2, C and D). Thus, SUMO-2 was enriched at DHSs that correspond to regions that were depleted of all histone marks and were flanked on either side by regions enriched in active histone marks. This is exemplified by the genomic landscape at the EIF2S2, DEDD2 and ZNF526 genes (Fig. 2E). These data suggest that HS-induced modification by SUMO-2 targets chromatin-regulatory factors embedded in or recruited to a chromatin environment that is marked by active histone modifications before HS occurs.
SUMO-2 targets active genes associated with gene expression and the posttranscriptional modification of RNA
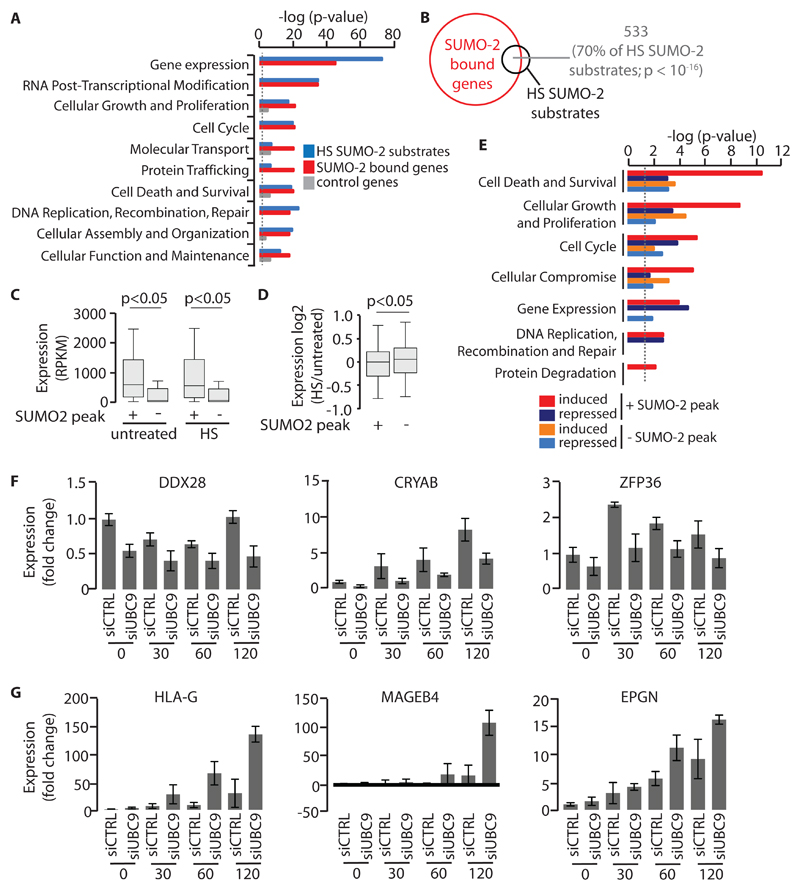
To understand the biological functions of proteins encoded by SUMO-2 target genes during HS, we subjected our list of 8292 SUMO-2–bound protein-coding genes (gene ± 2 kb) to functional analysis. This demonstrated that HS-induced modification by SUMO-2 targeted an extensive set of actively transcribed genes. Most statistically significantly enriched were those genes that encode factors involved in gene expression and posttranscriptional modification of RNA (Fig. 3A). Previous proteomics studies revealed that SUMO-2 substrates were particularly enriched for factors associated with biological processes almost identical to those associated with the SUMO-2 target genes during HS (16–18). To assess this apparent similarity more directly, we compared our HS TAP-SUMO-2 (SUMO-2 fused to a tandem affinity protein tag) substrate dataset, which lists cellular targets of HS–induced SUMOylation identified by a quantitative proteomics approach (17), with our ChIP-seq dataset of genes that were bound by SUMO-2 in response to HS. This comparative analysis showed that the two most statistically significantly enriched biological processes shared by both datasets are “gene expression” and “RNA posttranscriptional modification” (Fig. 3A). Indeed, of the individual HS-induced SUMO-2 substrates identified by proteomics, 70% of the genes encoding these proteins were marked by SUMO-2 modification after HS, suggesting that SUMO-2 conjugation modulated gene expression and RNA-modification pathways at multiple levels during HS (Fig. 3B).
Fig. 3. Relationship between HS-induced SUMO-2 occupancy and gene expression.
(A) Functional analysis of HS TAP-SUMO-2 substrates (n = 755 HS TAP-SUMO-2 substrates) (17) and genes associated with HS SUMO-2 peaks (n = 8036 genes associated with HS SUMO-2 peaks). A random gene set (n = 2500 genes) not associated with HS-induced SUMO-2 peaks served as a control. The dotted line indicates P = 0.05. (B) Venn diagram showing overlap between HS TAP-SUMO-2 substrates (n = 755 HS TAP-SUMO-2 substrates) and SUMO-2–bound genes (n = 8036). (C) The relative abundances of mRNAs of genes that were either bound or not bound by SUMO-2 in untreated U2OS cells or in U2OS cells subjected to HS for 4 hours were measured by RNA-seq. Boxplots and P values were derived as described in Fig. 1A. Data are from three biological replicates. (D) Differential expression analysis of SUMO-2 target genes and non-SUMO-2 target genes in response to HS. Boxplots and P values were derived as described in Fig. 1A. Data are from three biological replicates. (E) Molecular and cellular function analysis of SUMO-2 target genes and non-SUMO-2 target genes upon HS. Dotted line denotes P = 0.05. (F) Effect of the siRNA-mediated depletion of Ubc9 on SUMO-2 target gene expression after HS. Cells were transfected with control siRNA or Ubc9-specific siRNA. Ninety-six hours later, cells were left untreated or were subjected to HS for the indicated times before newly synthesized RNAs were labeled with 4-thiouridine and the indicated RNAs were quantified. (G) Effect of the siRNA-mediated depletion of Ubc9 on non-SUMO-2 target gene expression after HS. Cells were transfected with control siRNA or Ubc9-specific siRNA. Ninety-six hours later, cells were left untreated or were subjected to HS for the indicated times before newly synthesized RNAs were labeled with 4-thiouridine and the indicated RNAs were quantified. Data in (F) and (G) are means ± SEM of three independent biological replicates.
SUMO-2 is required to maintain expression of its target genes
To assess the effect on gene expression of the binding of SUMO-2 to active DNA-regulatory elements, we used RNA-seq to measure mRNA abundance in cells before HS was induced or 4 hours afterwards. This analysis showed that genes associated with HS-induced SUMO-2 peaks were generally more highly expressed than were genes that did not recruit SUMO-2 (hereafter referred to as non-SUMO-2 target genes) (Fig. 3C). To address whether the recruitment of SUMO-2 to active promoters was involved in their transcriptional repression or activation during HS, we performed differential expression analysis between SUMO-2 target genes and non-SUMO-2 target genes (Fig. 3D). This analysis revealed that, similar to non-SUMO-2 target genes, SUMO-2 target genes could be categorized into groups of genes that were induced or repressed, or whose mRNA abundance was not altered during HS. To characterize the regulation and function of SUMO-2–bound genes, genes with a False Discovery Rate (FDR) ≤ 0.01 were grouped according to their differential expression and SUMO-2 binding status in response to HS. The top 25% of genes in each category, as judged by fold-change in gene expression after HS, were then subjected to functional analysis (Fig. 3E). This revealed that HS-repressed SUMO-2 target genes were linked with biological processes, including “gene expression” and “cell cycle,” whereas HS-induced SUMO-2 target genes were particularly associated with terms such as “cell death and survival” and “cellular growth and proliferation.” Accordingly, SUMO-2 target genes include many that encode stress-inducible regulators of cell death and survival, as well as chaperones, and they share HSF1 as a common upstream regulator (Fig. 3F and fig. S6, A to D and fig. S7A). These data suggest that HS-induced modification by SUMO-2 preferentially targets either genes that are highly active in unstressed cells or those whose expression is induced upon HS. These findings also imply that the recruitment of SUMO-2 to active DNA-regulatory elements does not determine whether an associated gene is activated or repressed during HS.
To test the role of SUMO conjugation on gene expression during HS, the expression of UBC9, which encodes the SUMO-conjugating enzyme UBC9, was inhibited by short inhibitory RNA (siRNA), and newly synthesized mRNA was measured in unstressed cells and in cells subjected to HS for 30 min or 1 or 2 hours. Based on our ChIP-seq and RNA-seq data, we compared SUMO-2–bound genes with non-SUMO-2–bound genes. Quantification of 4-thiouridine–labelled newly synthesized RNA showed that UBC9 was required for the active expression of a number of SUMO-2–bound genes in unstressed cells (Fig. 3F and fig. S7A). Abrogation of HS-induced SUMO-2 conjugation led to a reduction in the expression of many SUMO-2 target genes, including DDX28 and the HS-inducible CRYAB and HspA1A genes (Fig. 3F and fig. S7A). In contrast, expression of most of the tested HS-induced non-SUMO-2 target genes, including HLA-G, MAGEB4, and EPGN, was generally not impaired, but rather increased in the absence of SUMO conjugation (Fig. 3G and fig. S7B).
Because HSF1 can be SUMOylated, it was important to exclude the possibility that impaired HSF1 activation accounted for the reduced expression of SUMO-2 target genes in UBC9-depleted cells. However, neither the HSF1 protein amount nor its HS-induced activation, as measured by Western blotting analysis of its phosphorylation on Ser326 (32) and the appearance of a high molecular mass species of HSF1, were substantially affected by depletion of UBC9 (fig. S7C). Furthermore, impaired SUMOylation of HSF1 would be expected to result in increased transcriptional activity of HSF1, rather than having an inhibitory effect (33). These results suggest that SUMO conjugation is required to maintain the maximal expression of SUMO-2 target genes during HS by regulating the maintenance of gene-associated transcription-regulatory protein complexes.
The pattern of HS-induced SUMO-2 conjugation on chromatin suggests that SUMO is an integral component of the proteotoxic stress response
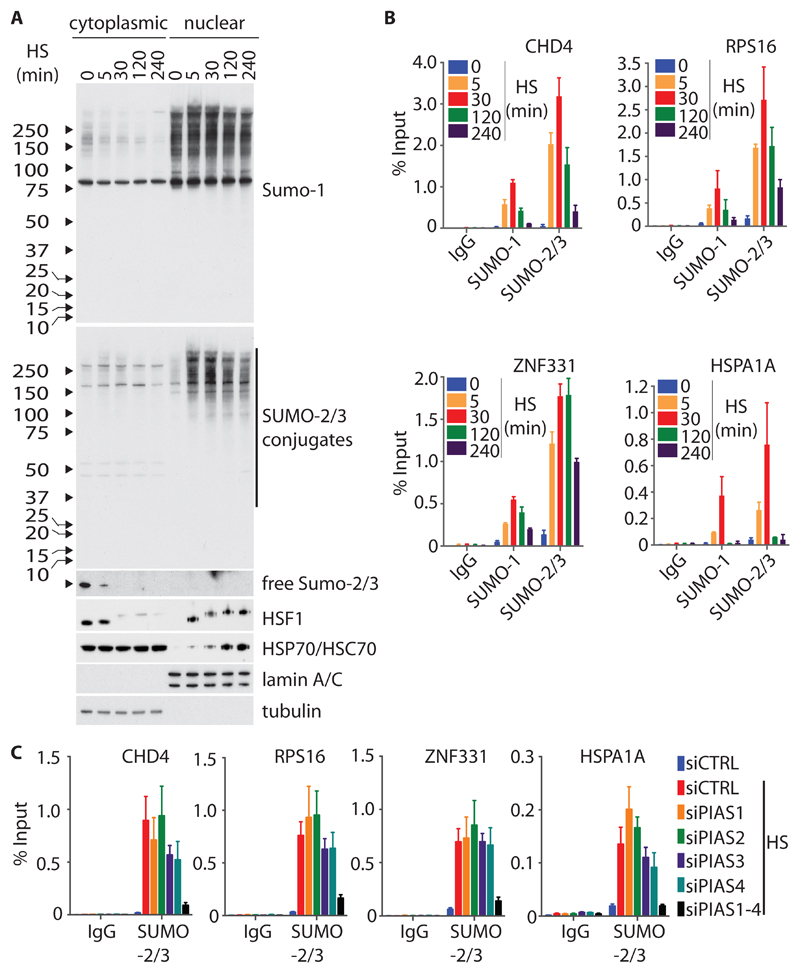
Because the HS-induced conjugation of SUMO-2 to proteins did not appear to act as a direct transcriptional repressor or activator of target genes, we investigated the possibility that SUMO-2 conjugation is an integral component of the HSR that regulates protein homeostasis of chromatin-associated regulatory complexes. A direct link between SUMO-2 conjugation and the HSR is supported by the observation that SUMOylation represents an immediate event after an increase in temperature (17, 20). Accordingly, when we characterized the dynamics of the recruitment of SUMO-2 to chromatin, we noted that SUMO-2 occupancy at DHSs in response to HS mirrored the temporal pattern of global SUMO-2 conjugation in cells (Fig. 4, A and B). Within 5 min of HS induction, SUMO-2 and SUMO-1 were enriched at all of the DHSs that were examined (Fig. 4B). After reaching a maximal occupancy after approximately 30 of HS, the amounts of chromatin-bound SUMO-2 and SUMO-1 began to decrease (Fig. 4B). Despite a uniform pattern of SUMO recruitment across DHSs, the clearance of SUMO from the HSPA1A promoter appeared to follow faster kinetics than were observed for other loci (Fig. 4B). We suggest that this difference is based on faster recycling rates of transcription regulatory protein complexes linked to high transcriptional activity at the HSPA1A locus. Overall, these observations support the idea that the SUMO modification status during HS is controlled by a central mechanism that acts on a system-wide scale and is interlinked with the HSR. ChIP experiments with an antibody raised against PIAS1 (fig. S8, A and B) revealed that the kinetics of the HS-induced recruitment to chromatin of the SUMO E3 ligase PIAS1 was similar to that of SUMO (fig. S8C), suggesting that at least a fraction of SUMO modification occurs directly on chromatin. Whereas single depletion of individual PIAS proteins (PIAS1 to PIAS4) did not result in a major reduction in the amount of SUMO-2 at DHSs, simultaneous ablation of all four PIAS isoforms abolished the recruitment of SUMO-2 to DHSs (Fig. 4C).
Fig. 4. Kinetics of the HS-induced recruitment of SUMO-2 to chromatin.
(A) Time-course of the HS-induced conjugation of SUMO-2 to proteins. U2OS cells were left untreated or were subjected to HS for the indicated times, fractionated into cytoplasmic and nuclear extracts, and then analyzed by Western blotting with antibodies against the indicated targets. Western blots are representative of two independent experiments. (B) Time-course of the HS-induced recruitment of SUMO-2 to chromatin. U2OS cells were left untreated or were subjected to HS for the indicated times. SUMO-1 and SUMO-2 chromatin occupancy was analyzed by ChIP-RT-qPCR with primers specific for the regulatory regions of the indicated genes. Data are means ± SD of at least two independent experiments and were analyzed by GraphPad Prism software. (C) Effect of the depletion of PIAS proteins on the HS-induced recruitment of SUMO-2 to chromatin. U2OS cells were transfected with non-targeting siRNA (siCTRL), siRNAs specific for individual PIAS proteins, or a pool thereof. Transfections were repeated 48 hours after the initial transfection. Ninety-six hours after the initial transfection, the cells were left untreated or were subjected to HS for 30 min. The extent of recruitment of SUMO-2 to chromatin was analyzed as described for (B). Data are means ± SEM of six biological replicates derived from three independent experiments and were analyzed by GraphPad Prism software.
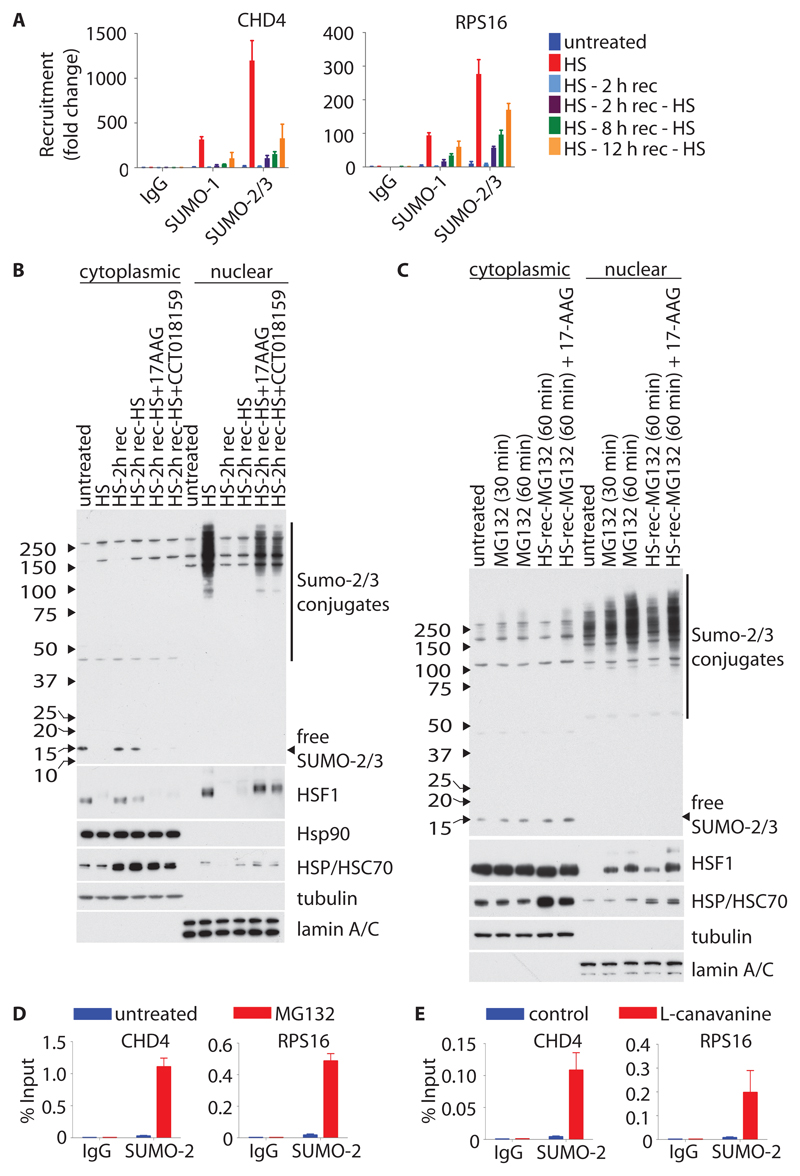
To investigate the temporal dynamics of HS-induced SUMOylation, we compared the recruitment of SUMO to DHSs both in response to HS and after a 2-hour recovery period at 37°C. After recovery from HS, the amounts of chromatin-associated SUMO-2 and SUMO-1 were reduced to those before treatment; however, exposure of cells to a second round of HS after a recovery period of 2, 8, or 12 hours led to impaired recruitment of SUMO-2 and SUMO-1 (Fig. 5A and fig. S9A). To establish the cellular compartments that provided the pool of free SUMO-2 that was used for HS-induced conjugation, cells were subjected to HS and recovery as described earlier, but before analysis, the cells were fractionated into cytoplasmic and nuclear extracts (Fig. 5B). An inverse relationship was observed between the pattern of binding of SUMO-2 to chromatin (Fig. 5A and fig. S9A) and the cytoplasmic pool of monomeric SUMO-2, which was rapidly depleted because SUMO-2 was conjugated to nuclear substrates during HS (Fig. 5B). After a 2-hour recovery period, when the amounts of nuclear proteins conjugated with SUMO-2 declined, the cytoplasmic pool of monomeric SUMO-2 was restored (Fig. 5B).
Fig. 5. The HS-induced recruitment of SUMO to chromatin is part of the proteotoxic stress response.
(A) U2OS cells were left untreated or were subjected to HS for 30 min and allowed to recover (rec) at 37°C for the indicated times before being exposed to a second round of HS for 30 min (HS-rec-HS). The recruitment of SUMO-1 and SUMO-2 to chromatin was measured by ChIP-RT-qPCR analysis with primers specific for the regulatory regions of the indicated genes. Data are means ± SD of two biological replicates from two independent experiments. (B) SUMO conjugation during HS and recovery. U2OS cells were treated as described in (A), and where indicated, the HSP90 inhibitors 17-AAG or CCT018159 were added at the start of the recovery period. Cells were then fractionated into cytoplasmic and nuclear extracts before being analyzed by Western blotting with antibodies against the indicated targets. Western blots are representative of two independent experiments. (C) U2OS cells were left untreated or were treated with proteasome inhibitor MG132 for indicated times. Where indicated, cells were subjected to HS (30 min) and a two-hour recovery period before the MG132 was added (HS-rec-MG132). 17-AAG was added at the start of the recovery period. Cells were then fractionated into cytoplasmic and nuclear extracts before being analyzed by Western blotting with antibodies against the indicated targets. Western blots are representative of two independent experiments. (D) The recruitment of SUMO-2 to chromatin in response to MG132-induced proteotoxic stress was determined as described in (A). (E) The recruitment of SUMO-2 to chromatin in response to L-canavanine-induced proteotoxic stress was determined as described in (A). Data in (D) and (E) are means ± SD of two biological replicates from two independent experiments.
In response to HS, HSF1 is released from HSP70- and HSP90-mediated inhibition in the cytoplasm and translocates to the nucleus to drive the expression of HSP-encoding genes, including HSPA1A (34, 35). When the extent of SUMO-2 conjugation was maximal after 30 min of HS, the amounts of HSP70 protein remained similar to those in unstimulated cells (Fig. 5B). However, after a 2- or 8-hour recovery period from HS, when SUMO-2–conjugated proteins had declined to background amounts, an increase in the amount of HSP70 protein was apparent (Fig. 5B and fig. S9B). Consistently, SUMO-2 conjugation was fully restored only after a recovery period of 24 hours, which resulted in a concomitant decrease in the cellular amounts of HSP70 (fig. S9B).
An alternative approach to investigating the link between the HSR and SUMO conjugation was provided by pharmacological inhibition of the chaperone function of HSP90. Cells were treated with two structurally distinct HSP90 inhibitors, 17-AAG or CCT018159 (36, 37). Both inhibitors relieved the block on SUMO-2 conjugation caused by the initial round of HS and substantially restored the amounts of nuclear SUMO-conjugates that were attained during the second round of HS (Fig. 5B). To reveal whether this effect was HS-specific or represented a general regulatory mechanism of proteotoxic stress–induced SUMO conjugation, proteotoxic stress was induced by treating the cells with MG132, which disrupts cellular protein homeostasis by blocking the proteasomal degradation of incorrectly folded proteins. Similar to the effects of HS, MG132 caused an increase in the conjugation of SUMO-2 to nuclear substrates (Fig. 5C), and, with the exception of the HSPA1A promoter, led to substantial recruitment of SUMO-2 to the same genomic loci that were targeted by SUMO-2 during HS (Fig. 5D and fig. S9C). The lack of robust SUMO-2 occupancy at the HSPA1A promoter correlated with weak activation of HSF1 in response to MG132 as judged by the translocation of HSF1 to the nucleus and the impaired mobility of the protein in an SDS-PAGE gel (Fig. 5, B and C). Additionally, MG132-induced SUMO-2 conjugation was inhibited by a preceding round of HS. As was observed for HS, this block in MG132-induced SUMO-2 conjugation was relieved by pharmacological inhibition of HSP90 (Fig. 5C). Furthermore, L-canavanine, a non-proteinogenic L-arginine analog that disrupts protein structure upon its incorporation, also led to the recruitment of SUMO-2 to chromatin (Fig. 5E and fig. S9D). When comparing the dynamics of HSF1 nucleo-cytoplasmic translocation, and thus HSF1 activation, with those of SUMO-2 conjugation in response to HS and MG132, we noted that the temporal and subcellular activation pattern of HSF1 correlated with that of SUMO-2 conjugation (Fig. 5, B and C), suggesting that SUMO-2 conjugation also forms an integral component of the HSR and is regulated as such.
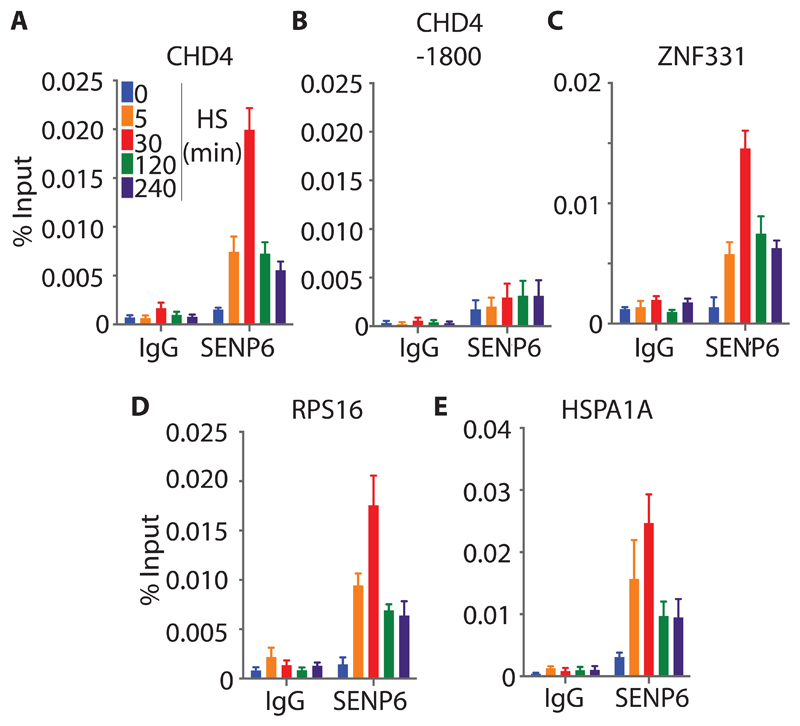
Consistent with the observation that the recruitment of SUMO-2 to chromatin was impaired in response to a second round of HS (Fig. 5A and fig. S9A), SUMO-2 remained almost entirely in its cytoplasmic, monomeric form, suggesting that SUMO was rapidly deconjugated from its substrates upon removal of the stress stimulus, and that further conjugation was inhibited (Fig. 5B). Concordantly, the SUMO protease SENP6 was also recruited to active DNA-regulatory elements upon HS (Fig. 6, A to E).
Fig. 6. The HS-induced recruitment of SENP6 to chromatin.
(A to E) Time-course analysis of the HS-induced recruitment of SENP6 to chromatin. U2OS cells were left untreated or were subjected to HS for the indicated times. The recruitment of SENP6 to chromatin was analyzed by ChIP and RT-qPCR with primers specific for the regulatory regions of the indicated genes. Primers specific for a region 1800 bp upstream of the CHD4 regulatory region (CHD4-1800) were used as control. Data in (A) to (E) are means ± SEM of four biological replicates from two independent experiments and were analyzed by GraphPad Prism.
SUMO-2 targets large protein complexes during HS
Because SUMO-2 conjugation appeared to form a central part of the proteotoxic stress response, it seemed reasonable to suggest that SUMO itself might maintain the integrity of the regulatory complexes that occupied active DNA-regulatory elements. To test this hypothesis, we used ChIP analysis to assess the HS-induced recruitment of SUMO-2 to DNA-regulatory elements in cells individually depleted of SUMO-2 substrates likely to localize to TSSs (17), such as HSF1 and the E3 ubiquitin-protein ligase ubiquitin-like PHD and RING finger domain–containing protein 1 (UHRF1). To study the recruitment to chromatin of UHRF1, we raised an antibody against this protein and validated its specificity (fig. S10, A and B). Both HSF1 and UHRF1 are known SUMO-2 substrates and are specifically localized to active DNA regulatory elements of the CHD4 promoter, but not to a control site 1800 bp upstream, upon exposure to HS (fig. S10, C to F) (17). Whereas depletion of HSF1 led to the reduced binding of HSF1 to the promotors of CHD4, ZNF331, and RPS16 (fig. S10, C, G, and H), the recruitment of SUMO-2 was not affected during HS (fig. S10, I to K). Similarly, the amounts of SUMO-2 at the CHD4 promoter were unaltered in cells depleted of UHRF1, although the amounts of UHRF1 were reduced (fig. S10, B and L). These data suggest that the pool of SUMO-2 recruited to DHSs upon HS was not the result of one single factor being SUMOylated at these sites or of one single SUMO-2–modified protein being recruited. Instead, the data suggest that SUMO-2 recruitment was a combination of the HS-induced modification by SUMO-2 of multiple substrates, for example components of multiprotein complexes. During HS, most cellular substrates of SUMO associated with the insoluble, nuclear fraction (fig. S11A). SUMOlyated proteins were released from this fraction by increasing the salt concentration, suggesting that SUMO was associated with large protein complexes upon HS (38). Salt-induced release of SUMO conjugates from this insoluble material required pretreatment with benzonase or DNaseI, suggesting that the SUMOylated protein complexes were tightly associated with DNA and chromatin (fig. S11, A and B).
Discussion
SUMO conjugation is implicated as an important stress response mechanism (15–17, 19, 20). Here, we showed that SUMO conjugation acts as a dynamic and integral component of the HSR. Upon HS, both SUMO-2 and SUMO-1 were conjugated to components of large nuclear protein complexes that are involved in the regulation of active gene expression and posttranscriptional modification of RNA in either proliferating cells or during HS. Integration of our ChIP-seq and RNA-seq datasets revealed that a substantial proportion of these SUMO-modified complexes was associated with transcription factor–binding sites located close to the TSS of many highly expressed genes. These DNA regulatory elements were nucleosome-depleted and are flanked on either side by stretches of active histone marks, implying that chromatin-binding factors, rather than histones, were the targets for SUMO modification. Other forms of protein-damaging stimuli induced a similar recruitment of SUMO to chromatin, implying that SUMO conjugation is not HS-specific, but is a general response to proteotoxic stress.
In concurrence with a study describing the dynamic enrichment of SUMO at predominantly active genes in WI38 human fibroblasts grown under normal conditions, our data reveal that HS-induced SUMO-2 target genes were also enriched for fundamental cellular processes such as gene expression and cell cycle (39). However, whereas the SUMO pathway has a predominantly repressive function on the transcription of many genes that encode factors that regulate growth and proliferation in fibroblasts, our data suggest that SUMO is required for maximal expression of many of its target genes during HS. In particular, the association of SUMO-2 with HS-induced genes encoding survival factors offers a mechanism by which SUMO contributes to increased survival rates of cells after proteotoxic stress.
Our work reveals SUMO conjugation as an integral component of the HSR and proteotoxic stress response, and suggests a model in which HS-induced SUMO conjugation targets large, chromatin-associated protein complexes to maintain their homeostasis during proteotoxic stress. Evidence in support of this model is based on three observations. First, our data suggest a direct link between SUMO conjugation and the HSR. The almost identical set of activating stimuli (15, 16, 20, 40, 41) and the fact that the dynamics of proteotoxic stress–induced SUMO conjugation resemble those of HSF1 activation imply that both pathways shared a common upstream regulatory mechanism. Similar to HSF1 activation, proteotoxic stress-induced SUMO conjugation and recruitment of SUMO to chromatin were impaired in cells preconditioned by a priming HS, suggesting that this initial HS induced a common inhibitor of both pathways. In addition to our finding that the priming HS event led to increased amounts of HSP70 protein, which negatively correlated with SUMO conjugation, overexpression of HSP70 in plants results in a block in HS-induced SUMOylation (20). Further reinforced by the observation that pharmacological inhibition of a different chaperone, HSP90, restored proteotoxic stress–induced SUMO conjugation in cells that had been preconditioned by a priming HS, we propose a model in which proteotoxic stress–induced SUMOylation is dependent on HSP70 and HSP90. The pool of monomeric SUMO-2 is likely to be located in the cytoplasm in unstressed cells because cellular fractionation experiments showed a predominantly cytoplasmic localization of unconjugated SUMO-2. This observation is unlikely to be the consequence of monomeric SUMO-2 leaking out of the nucleus during the fractionation procedure, because this would result in the free SUMO being evenly distributed between nuclear and cytoplasmic compartments. In analogy to the “chaperone titration model” that was suggested for HSF1 regulation (42), we thus suggest that SUMO-2 is also negatively controlled by a repressive, cytoplasmic heteroprotein complex consisting of the HSP70 and HSP90 chaperones. Proteotoxic stress–induced accumulation of misfolded proteins stimulates the release of SUMO-2 and HSF1 from this inhibitory complex while these chaperones are redirected towards sites of protein damage. However, excessive amounts of HSP present in response to an initial HS event would lead to a block in SUMO conjugation and HSF1 activation upon a second proteotoxic stress stimulus. Therefore, conjugation of SUMO-2 to nuclear substrates appears to require two separate events: release from HSP70- and HSP90-dependent cytoplasmic retention and the accumulation of unfolded proteins in the nucleus as an additional trigger.
Second, numerous observations have implicated SUMO in the maintenance of protein complex stability under conditions of compromised protein homeostasis. Several studies focusing on the identification of SUMO substrates in yeast and human cells demonstrated that the SUMO pathway preferentially targets components of macromolecular protein complexes for modification (16, 17, 43–46). This function of SUMO is required in various biological pathways, including DNA damage repair (45), ribosome assembly (47, 48), and the establishment of protein complexes required for dosage compensation, the epigenetic process responsible for the normalization of gene expression that results from unequal copy numbers of sex chromosomes, in C. elegans (49). Proteotoxic stress-induced SUMO substrates are enriched for a diverse set of transcription factors and chromatin remodelers (16–18, 46). These factors are typical components of the large protein complexes commonly associated with active DNA-regulatory elements that are also sites of HS-induced SUMO recruitment. Stabilization of pre-existing, chromatin-bound regulatory complexes, at TSSs for example, during proteotoxic stress is crucial, because their deposition and replacement is often confined to a narrow window of the cell cycle, such as ongoing DNA replication during S-phase (50, 51). Thus, we propose that the SUMO-mediated maintenance of protein complex homeostasis is essential for maintaining the regulatory status quo of gene promoters and other SUMO-associated DNA-regulatory elements. This role of SUMO in protein complex stability is exemplified by the requirement for SUMO modification in Promyelocytic leukemia protein (PML) nuclear body formation (52). We propose that SUMO modulates protein complex homeostasis by exhibiting chaperone-like functions. There are several mechanisms that SUMO might use to achieve these functions. Comparable to the stabilizing effect of HS-induced, PARP-dependent ADP-ribose polymers on the HSP70 locus (53), SUMO could exert a stabilizing effect on protein assemblies and act as a scaffold or glue by potentiating physical interactions between protein complex components (45). For example, one SUMO-modified protein can interact with another non-SUMOylated protein through just SUMO-SIM interactions. According to this model, the scaffolding effect of SUMOylation would not be restricted to covalently modified SUMO substrates or proteins already in direct contact with each other, but would also support indirect interactions between distant complex components, an effect that could even be potentiated by simultaneous modification of some proteins at multiple sites (46). Consequently, only a small number of proteins within a complex need to be SUMOylated under these circumstances. Additionally, the SUMO scaffold can be rapidly disassembled through the action of SUMO-specific proteases, for example, to enable protein complexes to resume their normal functions in cells recovering from a protein-damaging insult. Similar to the function suggested for polyphosphate chains (54), HS-induced poly-SUMO chain formation (17, 21) could also be required for the maintenance of protein solubility, thereby antagonizing the irreversible aggregation of complex components and promoting the maintenance of a refolding-competent state. This proposal is supported by a study that described the N-terminal tail of SUMO as an entropic bristle whose disordered structure exerts a solubilizing effect on associated or modified proteins by increasing their soluble surface areas and limiting contacts between aggregation-prone protein folds (55). Therefore, as an acute and reversible protein modification, SUMO conjugation would be a powerful mechanism to prevent protein aggregation and would enable adaptation to stress without the requirement for de novo RNA or protein synthesis. Finally, further evidence supporting a direct involvement of SUMO in the proteotoxic stress response is contributed by studies in plants and cultured human cells, which demonstrated a requirement for SUMO for cell survival in response to hyperthermic stress (17, 19).
Materials and Methods
Cell culture and treatments
U2OS cells were cultured according to standard procedures. To induce HS, cells were incubated at 43°C for the times indicated in the legends. To induce proteotoxic stress through the incorporation of L-canavanine, cells were cultured for 12 hours in Dulbecco’s Modified Eagle Medium (DMEM) in which L-arginine was replaced with L-canavanine. To inhibit proteasomes, cells were incubated in the presence of 50 μM MG132 for the times indicated in the legends. To inhibit HSP90, cells were incubated with 1 μM 17-AAG [17-(Allylamino)-17-demethoxygeldanamycin) or 10 μM CCT018159 as indicated. DMSO was used as vehicle control.
Cell lysis and fractionation
To generate whole-cell lysates, cells were lysed directly in 2 x Laemmli sample buffer. To fractionate cells into cytoplasmic and nuclear lysates, 0.5 to 1 x 107 U2OS cells were washed in phosphate-buffered saline (PBS) and scraped into ice-cold PBS containing 100 mM iodoacetamide. Cell pellets were resuspended in approximately 300 µl of ice-cold buffer A [10 mM Hepes (pH 7.8), 0.08% NP-40, 10 mM KCl, 1.5 mM MgCl2, protease inhibitors, 100 mM iodoacetamide]. All buffers were supplemented with RNasin (20 U/ml, Promega) when nuclei were prepared for treatment with DNaseI. After incubation on ice for 15 min, cells were gently syringed 20 times through a 19G needle, and nuclei were separated from the cytoplasmic fraction by centrifugation at 500 g. Nuclei were washed twice in buffer A without iodoacetamide before lysis in SDS lysis buffer. Equal amounts of lysates were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting. To separate nuclear contents into soluble, chromatin-associated, and insoluble, “matrix-associated” fractions, nuclei were treated with benzonase (125 to 250 U/ml for 1 to 3 x 106 nuclei, Fermentas) or DNaseI (500 U/ml for 1 to 3 x 106 nuclei, Roche) for 1 hour at 4°C or room temperature, respectively. For salt-extraction of nuclear material, benzonase- or DNaseI-treated nuclei were incubated for 3 min with 0.3 or 0.5 M NaCl, respectively. Soluble and insoluble fractions were separated by centrifugation at 17,000g for 5 min at 4°C.
siRNA
siRNA was purchased from Dharmacon. Cells were transfected with siRNAs according to established protocols for Lipofectamine RNAiMAX (Invitrogen).
Metabolic labelling, purification, and quantitation of newly synthesized RNA
Metabolic labelling, biotinylation, and purification of newly synthesized RNA were performed as described previously (56) with the following modifications. U2OS cells were transfected with non-targeting siRNA or UBC9-specific siRNA and incubated for 96 hours. For metabolic labelling of RNA for a one-hour, single time point (fig. S7, A and B), U2OS cells were left untreated or were subjected to HS at 43°C for 5 min. 4-thiouridine (500 μM, 4SU, Sigma) was added directly to the tissue culture medium and the cells were incubated at 37°C or 43°C for a further 55 min. Total cellular RNA was isolated with the RNeasy Mini kit (Qiagen). Biotinylation of 4SU-labeled RNA was performed by incubating 80 to 100 μg of total RNA at a final concentration of 100 ng/μl in a buffer containing EZ-Link HPDP-Biotin [N-[6-(Biotinamido)hexyl]-3'-(2'-pyridyldithio)propionamide, 0.2 mg/ml, Thermo], 10 mM Tris (pH 7.4), and 1 mM EDTA for 90 min at room temperature. Unbound HPDP-Biotin was removed by two rounds of extraction with chloroform:isoamylalcohol (24:1, Sigma) in Phase-lock-gel (heavy) tubes (Eppendorf). RNA was precipitated by the addition of a 1/10 volume of 5 M NaCl and an equal volume of isopropanol, followed by centrifugation at 20,000g for 20 min. After two washes with 75% ethanol, the RNA pellet was resuspended in RNase-free water, denatured by incubation at 65°C for 10 min, and cooled on ice. Biotinylated RNA was captured with Dynabeads MyOne Streptavidin C1 (Invitrogen) that were prepared according to the manufacturer’s instructions. For RNA capture, RNA was incubated with an equivalent of 100 µl bead solution in a buffer containing 5 mM Tris (pH 7.4), 0.5 mM EDTA, and 1 M NaCl for 15 min by rotation at room temperature. Bead-RNA complexes were washed three times with 1 ml of wash buffer [10 mM Tris (pH 7.4), 10 mM EDTA, 1 M NaCl, 0.1% Tween20] at 65°C, followed by three washes with wash buffer at room temperature. Newly synthesized RNA was eluted twice by incubation with 100 μl of 100 mM DTT for 5 min, and recovered with the RNeasy MinElute Spin kit (Qiagen). The concentration of purified RNA was measured with a Nanodrop spectrophotometer. Equal amounts of labelled RNA were reverse-transcribed into complementary DNA (cDNA) with qScript cDNA mastermix (Quanta). The abundances of the cDNAs of interest were measured by RT-qPCR analysis with gene-specific primer sets and taking into consideration the dilution factor derived from the adjustment of the RNA concentration for reverse transcription. For time-course experiments (Fig. 3, F and G), cells were exposed to HS for 5, 30, or 90 min before being incubated in the presence of 4-thiouridine for an additional 30 min at 43°C to obtain HS time points of 30, 60, and 120 min, respectively.
Antibodies
The antibodies used in this study are listed in table S1. Antigen affinity-purified sheep anti-SUMO-1, sheep anti-SUMO-2 and sheep anti-SENP6 antibodies have been described previously (6, 57). Antibodies against mouse PIAS1 and human UHRF1 were raised in sheep against bacterially produced recombinant proteins. Antibodies were antigen affinity purified and used in ChIP experiments described in figures S8C and S10, B, E and F, respectively. To validate the antibodies, PIAS1 and UHRF1 expression was ablated by a pool of siRNA (Dharmacon) and extracts from these cells were compared to control cells by Western blotting (fig. S8A and fig. S10A) and ChIP analysis (fig. S8B and fig. S10B).
ChIP
ChIP assays were performed according to the Upstate protocol (17-295) with the following modifications. Approximately 1 to 2 × 106 U2OS cells were used per reaction, crosslinked by adding formaldehyde directly to the cell culture medium to a final concentration of 1%, and incubated at room temperature for 10 min. Formaldehyde was quenched by the addition of 0.125 M glycine for 5 min. DNA was sheared to fragments of approximately 200 to 500 bp by sonication for 15 cycles (7.5 min total sonication time) at the high setting at 4°C with a Bioruptor (Diagenode). Cleared lysates were diluted 10-fold in dilution buffer [1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris (pH 8.1)]. Immunoprecipitation was performed by overnight incubation with 1 μg of specific antibody or control immunoglobulin G (IgG), followed by incubation with protein G Dynabeads (Life Technologies) for 1 hour. Eluates were incubated with RNase A (0.1 mg/ml, Fermentas) for 30 min at 37°C before removal of crosslinks by overnight incubation at 65°C in the presence of 0.2 M sodium chloride, followed by digestion with proteinase K (0.25 mg/ml, Roche) for 1 hour at 55°C. DNA was purified with a polymerase chain reaction (PCR) purification kit (Qiagen). Enrichment of chromatin-binding factors was assessed by real-time, quantitative PCR (RTqPCR on an ABI7500 real-time PCR machine with the specific primers listed in table S2.
ChIP-seq
Two independent SUMO-2 enrichment ChIP-seq data sets were generated. The first set using protein G sepharose 4B (Sigma; dataset 1) included untreated and HS samples for which SUMO-2 ChIP and input (whole-cell lysates) samples were run. The second set using protein G Dynabeads (Life Technologies; dataset 2) consisted of untreated and HS SUMO-2 ChIP samples only. ChIP assays were performed as described above with the following adjustments. Approximately 3.5 to 4.5 × 107 U2OS cells were used per treatment. Crosslinked cell pellets were flash-frozen in liquid nitrogen and stored at -80°C. For the experiment with protein G sepharose 4B beads, frozen cell pellets were lysed in 2.1 ml of lysis buffer. DNA was sheared by sonication of 300-μl volumes for 14 cycles (7.5 min total sonication time) at the high setting at 4°C with a Bioruptor (Diagenode). To capture protein-DNA complexes, a fraction corresponding to 90% of the cleared and diluted lysates was incubated overnight with 21 µg of anti–SUMO-2 antibody (Life Technologies, 51-9100) and subsequently with bovine serum albumin (BSA)-blocked protein G sepharose 4B beads (Sigma) for 1 hour with a final bead bed volume of 150 µl. Bead-bound, protein-DNA complexes were washed twice in 14 ml of each wash buffer and 14 ml of TE buffer and eluted into 2.5 ml of elution buffer. An equal volume of TE buffer was added to the eluates before incubation with RNase A (0.2 mg/ml, Fermentas) at 37°C for 2 hours. Samples were incubated overnight at 65°C in the presence of 0.2 M sodium chloride, followed by digestion with proteinase K (0.25 mg/ml, Roche) for 1 hour at 55°C. DNA was purified with a PCR purification kit (Qiagen). To derive genomic DNA to be used as an input reference for SUMO-2 ChIP-seq, a fraction corresponding to the sonicated, diluted, and cleared whole-cell extracts that had been prepared for SUMO-2 ChIP was removed. These input samples were treated to remove RNA and reverse crosslinking, and DNA was extracted as described earlier. When protein G Dynabeads were used, frozen cell pellets were lysed in 1.8 ml of lysis buffer. After sonication for 15 cycles (7.5 min total sonication time), lysates were incubated with 13 µg of anti–SUMO-2 antibody and subsequently with protein G Dynabeads (Life Technologies) for 1 hour with an equivalent of 300 µl of bead solution. Bead-bound protein-DNA complexes were washed in 13 ml of each wash buffer and 13 ml of TE buffer and eluted into 2.3 ml of elution buffer. Salmon sperm DNA was not used at any stage throughout the entire protocol.
ChIP-seq library construction and next generation sequencing
Library construction and sequencing were performed at Genotypic (Bangalore, India). Libraries were constructed with ~10 ng of purified DNA according to a modification of the Illumina ChIP-Seq library protocol. Briefly, DNA was subjected to end-repair and adaptor ligation (Illumina ChIP Seq Library preparation kit). Adapter-ligated fragments were enriched by PCR amplification, and libraries were size-selected by 2% low-melting agarose gel electrophoresis with subsequent gel purification with the MinElute gel extraction kit (Qiagen). To validate the quantity and quality of the libraries, aliquots were analyzed on a Nanodrop spectrophotometer and High Sensitivity Bioanalyzer ChIP (Agilent), respectively. Libraries were sequenced on an Illumina Genome Analyser IIx with either 54-bp (for experiments with protein G sepharose 4B) or 36-bp single-end (for experiments with protein G Dynabeads) sequencing.
Sequence alignment and peak assignment
Sequence alignment was performed with the Subread aligner included in the R-bioconductor (58) package Rsubread (59) against the GRCh37 human reference obtained from Ensemble. Regions of SUMO-2 enrichment and depletion were identified with MACS2 software (25, 26) with an FDR cut-off of 0.01. To combine the datasets and derive a set of high-confidence enriched and depleted peaks, the following procedure was followed. SUMO-2 ChIP samples where run against the corresponding input sample to detect SUMO-2–enriched regions, regions assigned as enriched for SUMO-2 in the HS samples, but not in the untreated samples, were termed “SUMO-2–enriched peaks,” and regions assigned as enriched in the untreated, but not in the HS samples, were termed “SUMO-2–depleted peaks.” Regions assigned as “enriched peaks” in both independent datasets were termed “high-confidence enriched peaks” and similarly, regions assigned as “depleted peaks” in both datasets were termed “high-confidence depleted peaks. These high-confidence regions were further validated with MACS2 to call enriched and depleted regions between the HS SUMO ChIP and the untreated SUMO ChIP datasets with its model-free setting. These two sets of high-confidence peaks were used for subsequent analysis.
Genomic features
Datasets for the ChIP-seq of histone modifications, DNaseI-seq, and FAIRE-seq were retrieved from ENCODE (https://genome.ucsc.edu/ENCODE/) and are associated with the following GEO accession numbers: H3K4me2 (Hela-S3, GSM733734), H3K4me3 (Hela-S3, GSM733682), H3K9ac (Hela-S3, GSM733756), H3K27ac (HeLa-S3, GSM733684), H3K27me3 (Hela-S3, GSM733696), H3K79me2 (Hela-S3, GSM733669), H3K36me3 (Hela-S3, GSM733711), H4K20me1 (Hela-S3, GSM733689), H3K9me3 (U2OS, GSM788078), DNaseI HeLa-S3 (GSM816643), DNaseI IMR90 (GSM468792), DNaseI Osteoblast (GSM816654), DNaseI LNCaP (GSM816637), FAIRE-seq (Hela-S3, GSM864348), and ChIP synthesis (Hela-S3, GSM1002653). Data tracks were loaded and displayed in the Integrated Genome Browser (http://bioviz.org/igb/). Co-occurance of SUMO-2 peaks with gene transcript annotations and annotated genomic regulatory features was performed with the R-bioconductor package ChIPpeakAnno (60) based on the coordinates of Ensembl hg19 transcripts and regulatory features. We assigned peaks to annotations with the following hierarchy: 2 kb upstream of the TSS > first exon > other exon > first intron > other intron > 2 kb downstream of the transcript end > intergenic (> 2 kb from any transcript). To analyze the statistical significance of the co-occurance of SUMO-2 peaks and histone marks, open chromatin, or transcription factor–binding sites, we compared our SUMO-2 ChIPseq data to respective datasets obtained from ENCODE (61) using IntervalStats (29). IntervalStats generates an exact P value that represents the probability that the query feature (the SUMO-2 peak) would be as closely or more closely associated with the reference feature (for example, a DHS) within the specified region of interest (for example, a 4-kb window surrounding a TSS) if its location was selected by random chance. This is calculated by dividing the number of places as close or closer to the reference feature by the number of all possible locations. In contrast to a classical P value, it is an exact P value, because all possible locations are calculated rather than applying a subsampling approach of an assumed statistical distribution. An exact P value is generated for each query feature against all target features in each domain, and a distribution of exact P values is generated describing the co-occurance of the feature rather than a single P value.
RNA-seq
U2OS cells were left untreated or subjected to heat shock at 43°C for 4 hours. Extraction of total RNA from three replicate samples was performed with the RNeasy mini kit (Qiagen) according to the manufacturer’s instructions. Library preparation and transcriptome sequencing were performed at the Beijing Genomics Institute (BGI). Briefly, to generate a sequencing library suitable for transcriptome analysis, mRNA was purified, fragmented, and reverse-transcribed into cDNA according to the Illumina protocol. The resulting cDNAs were then ligated to adapters, PCR-amplified, size-selected (200-bp), and validated with a 2100 Bioanalyser (Agilent) and an ABI StepOnePlus Real-Time PCR System. Libraries were subjected to 91-bp, paired-end sequencing on a HiSeq2000 instrument (Illumina). Sequence alignment was performed using tophat2 (62) against the GRCh37 human reference from Ensembl. Reads were assigned to transcripts with featureCounts from the Rsubread software package (59), and differential expression analysis was performed with the R-bioconductor package DESeq (63).
Gene ontology analysis
Comparative analysis and analysis of the biological functions of genes and proteins was performed with IPA Ingenuity software.
Supplementary Material
Acknowledgments
We thank M. Tatham for helpful discussions, as well as E. Jaffray and J. Bett for generation and purification of the antibodies specific for PIAS1 and UHRF1.
Funding: This work was supported by the MRC and The Wellcome Trust. G.J.B. acknowledges support from Wellcome Trust Strategic Grants (100476/Z/12/Z and 097945/Z/11/Z). R.T.H. holds a Wellcome Trust Senior Investigator Award (098391/Z/12/Z).
Footnotes
Author contributions: A.S. designed and conducted the experiments and data analysis, processed data, and interpreted results; P.S. conducted bioinformatics and statistical analysis and processed the data; G.J.B. advised about bioinformatics analysis; R.T.H. conceived the project and interpreted the results; and A.S. and R.T.H. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: ChIP-seq and RNA-seq data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession numbers E-MTAB-2368 and E-MTAB-2349, respectively.
References
- 1.Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J. 2010;428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hay RT. SUMO-specific proteases: a twist in the tail. Trends Cell Biol. 2007;17:370–376. doi: 10.1016/j.tcb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Barysch SV, Dittner C, Flotho A, Becker J, Melchior F. Identification and analysis of endogenous SUMO1 and SUMO2/3 targets in mammalian cells and tissues using monoclonal antibodies. Nat Protoc. 2014;9:896–909. doi: 10.1038/nprot.2014.053. [DOI] [PubMed] [Google Scholar]
- 4.Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 5.Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, Hay RT, Lamond A, Mann M, Vertegaal AC. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol Cell Proteomics. 2008;7:132–144. doi: 10.1074/mcp.M700173-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 7.Kerscher O. SUMO junction-what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–555. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. Embo J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka K, Nishide J, Okazaki K, Kato H, Niwa O, Nakagawa T, Matsuda H, Kawamukai M, Murakami Y. Characterization of a fission yeast SUMO-1 homologue, pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol Cell Biol. 1999;19:8660–8672. doi: 10.1128/mcb.19.12.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saracco SA, Miller MJ, Kurepa J, Vierstra RD. Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 2007;145:119–134. doi: 10.1104/pp.107.102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9:769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Evdokimov E, Sharma P, Lockett SJ, Lualdi M, Kuehn MR. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J Cell Sci. 2008;121:4106–4113. doi: 10.1242/jcs.038570. [DOI] [PubMed] [Google Scholar]
- 14.Zhang FP, Mikkonen L, Toppari J, Palvimo JJ, Thesleff I, Janne OA. Sumo-1 function is dispensable in normal mouse development. Mol Cell Biol. 2008;28:5381–5390. doi: 10.1128/MCB.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 16.Tatham MH, Matic I, Mann M, Hay RT. Comparative proteomic analysis identifies a role for SUMO in protein quality control. Sci Signal. 2011;4:rs4. doi: 10.1126/scisignal.2001484. [DOI] [PubMed] [Google Scholar]
- 17.Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 18.Miller MJ, Scalf M, Rytz TC, Hubler SL, Smith LM, Vierstra RD. Quantitative proteomics reveals factors regulating RNA biology as dynamic targets of stress-induced SUMOylation in Arabidopsis. Mol Cell Proteomics. 2013;12:449–463. doi: 10.1074/mcp.M112.025056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoo CY, Miura K, Jin JB, Lee J, Park HC, Salt DE, Yun DJ, Bressan RA, Hasegawa PM. SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol. 2006;142:1548–1558. doi: 10.1104/pp.106.088831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung DY, Vierstra RD. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem. 2003;278:6862–6872. doi: 10.1074/jbc.M209694200. [DOI] [PubMed] [Google Scholar]
- 21.Bruderer R, Tatham MH, Plechanovova A, Matic I, Garg AK, Hay RT. Purification and identification of endogenous polySUMO conjugates. EMBO Rep. 2011;12:142–148. doi: 10.1038/embor.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewicki MC, Srikumar T, Johnson E, Raught B. The S. cerevisiae SUMO stress response is a conjugation-deconjugation cycle that targets the transcription machinery. Journal of proteomics. 2014 doi: 10.1016/j.jprot.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 23.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin Y, Seifert A, Chua JS, Maure JF, Golebiowski F, Hay RT. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 2012;26:1196–1208. doi: 10.1101/gad.189274.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin N, Schwamborn K, Schreiber V, Werner A, Guillier C, Zhang XD, Bischof O, Seeler JS, Dejean A. PARP-1 transcriptional activity is regulated by sumoylation upon heat shock. Embo J. 2009;28:3534–3548. doi: 10.1038/emboj.2009.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chikina MD, Troyanskaya OG. An effective statistical evaluation of ChIPseq dataset similarity. Bioinformatics. 2012;28:607–613. doi: 10.1093/bioinformatics/bts009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crawford GE, Holt IE, Whittle J, Webb BD, Tai D, Davis S, Margulies EH, Chen Y, Bernat JA, Ginsburg D, Zhou D, et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome research. 2006;16:123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Graf S, Huss M, Keefe D, Liu Z, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou SD, Prince T, Gong J, Calderwood SK. mTOR is essential for the proteotoxic stress response, HSF1 activation and heat shock protein synthesis. PLoS One. 2012;7:e39679. doi: 10.1371/journal.pone.0039679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci U S A. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 35.Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 36.Cheung KM, Matthews TP, James K, Rowlands MG, Boxall KJ, Sharp SY, Maloney A, Roe SM, Prodromou C, Pearl LH, Aherne GW, et al. The identification, synthesis, protein crystal structure and in vitro biochemical evaluation of a new 3,4-diarylpyrazole class of Hsp90 inhibitors. Bioorg Med Chem Lett. 2005;15:3338–3343. doi: 10.1016/j.bmcl.2005.05.046. [DOI] [PubMed] [Google Scholar]
- 37.Schulte TW, Neckers LM. The benzoquinone ansamycin 17-allylamino-17-demethoxygeldanamycin binds to HSP90 and shares important biologic activities with geldanamycin. Cancer Chemother Pharmacol. 1998;42:273–279. doi: 10.1007/s002800050817. [DOI] [PubMed] [Google Scholar]
- 38.Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 2009;19:460–469. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neyret-Kahn H, Benhamed M, Ye T, Le Gras S, Cossec JC, Lapaquette P, Bischof O, Ouspenskaia M, Dasso M, Seeler J, Davidson I, et al. Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res. 2013;23:1563–1579. doi: 10.1101/gr.154872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11:545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pirkkala L, Alastalo TP, Zuo X, Benjamin IJ, Sistonen L. Disruption of heat shock factor 1 reveals an essential role in the ubiquitin proteolytic pathway. Mol Cell Biol. 2000;20:2670–2675. doi: 10.1128/mcb.20.8.2670-2675.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voellmy R, Boellmann F. Chaperone regulation of the heat shock protein response. Adv Exp Med Biol. 2007;594:89–99. doi: 10.1007/978-0-387-39975-1_9. [DOI] [PubMed] [Google Scholar]
- 43.Panse VG, Hardeland U, Werner T, Kuster B, Hurt E. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem. 2004;279:41346–41351. doi: 10.1074/jbc.M407950200. [DOI] [PubMed] [Google Scholar]
- 44.Wohlschlegel JA, Johnson ES, Reed SI, Yates JR., 3rd Global analysis of protein sumoylation in Saccharomyces cerevisiae. J Biol Chem. 2004;279:45662–45668. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- 45.Psakhye I, Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 2012;151:807–820. doi: 10.1016/j.cell.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 46.Tammsalu T, Matic I, Jaffray EG, Ibrahim AF, Tatham MH, Hay RT. Proteome-wide identification of SUMO2 modification sites. Sci Signal. 2014;7:rs2. doi: 10.1126/scisignal.2005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panse VG, Kressler D, Pauli A, Petfalski E, Gnadig M, Tollervey D, Hurt E. Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic. 2006;7:1311–1321. doi: 10.1111/j.1600-0854.2006.00471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finkbeiner E, Haindl M, Muller S. The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. Embo J. 2011;30:1067–1078. doi: 10.1038/emboj.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pferdehirt RR, Meyer BJ. SUMOylation is essential for sex-specific assembly and function of the Caenorhabditis elegans dosage compensation complex on X chromosomes. Proc Natl Acad Sci U S A. 2013;110:E3810–E3819. doi: 10.1073/pnas.1315793110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan J, Enge M, Whitington T, Dave K, Liu J, Sur I, Schmierer B, Jolma A, Kivioja T, Taipale M, Taipale J. Transcription Factor Binding in Human Cells Occurs in Dense Clusters Formed around Cohesin Anchor Sites. Cell. 2013;154:801–813. doi: 10.1016/j.cell.2013.07.034. [DOI] [PubMed] [Google Scholar]
- 51.Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol. 2013;20:14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zobeck KL, Buckley MS, Zipfel WR, Lis JT. Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells. Mol Cell. 2010;40:965–975. doi: 10.1016/j.molcel.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gray MJ, Wholey WY, Wagner NO, Cremers CM, Mueller-Schickert A, Hock NT, Krieger AG, Smith EM, Bender RA, Bardwell JC, Jakob U. Polyphosphate Is a Primordial Chaperone. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grana-Montes R, Marinelli P, Reverter D, Ventura S. N-Terminal Protein Tails Act as Aggregation Protective Entropic Bristles: The SUMO Case. Biomacromolecules. 2014 doi: 10.1021/bm401776z. [DOI] [PubMed] [Google Scholar]
- 56.Dolken L, Ruzsics Z, Radle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, Koszinowski UH. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA. 2008;14:1959–1972. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hattersley N, Shen L, Jaffray EG, Hay RT. The SUMO protease SENP6 is a direct regulator of PML nuclear bodies. Molecular Biology of the Cell. 2011;22:78–90. doi: 10.1091/mbc.E10-06-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic acids research. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu LJ, Gazin C, Lawson ND, Pages H, Lin SM, Lapointe DS, Green MR. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, Lee BT, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic acids research. 2013;41:D56–63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McGill R, Tukey JW, Larsen WA. Variations of box plots. The American Statistician. 1978;32:12–16. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.