Abstract
Innate lymphoid cells (ILCs) play important functions in immunity and tissue homeostasis, but their development is poorly understood. Through the use of single-cell approaches, we examined the transcriptional and functional heterogeneity of ILC progenitors and studied the precursor–product relationships that linked the subsets identified. This analysis identified two successive stages of ILC development within TCF-1+ early innate lymphoid progenitors (EILPs), which we named ‘specified EILPs’ and ‘committed EILPs’. Specified EILPs generated dendritic cells, whereas this potential was greatly decreased in committed EILP. TCF-1 was dispensable for the generation of specified EILPs, but required for the generation of committed EILPs. TCF-1 used a pre-existing regulatory landscape established in upstream lymphoid precursors to bind chromatin in EILPs. Our results provide insight into the mechanisms by which TCF-1 promotes developmental progression of ILC precursors, while constraining their dendritic cell lineage potential and enforcing commitment to ILC fate.
Keywords: innate lymphoid cells, ILC, dendritic cells, DC, lineage commitment, TCF-1
Innate lymphoid cells (ILCs) are functionally similar to T cells, but lack somatically recombined antigen receptors. Both early T cell progenitors and ILC progenitors depend on the expression of transcription factors such as TCF-1 (encoded by Tcf7), which might program similar functions in T cells and ILCs during development1–4. Transcription factors specific for early ILCs, such as NFIL3, could imprint the innate features of ILCs. Understanding how these factors act to program the immune effector functions of ILCs requires the identification of the progenitor cells in which they act, and the developmental transitions they control. Early innate lymphoid progenitors (EILPs) are the earliest known specified ILC progenitors1,4. Unlike later ILC progenitors, EILPs are thought to contain uncommitted ILC precursors that can also access the dendritic cell (DC) fate1,4.
Here, we use population level and single-cell approaches to assess the transcriptional and functional heterogeneity within bone marrow (BM) ILC progenitors. We identified two successive stages of development within EILPs. Precursor cells at the earlier stage were specified to the ILC lineage, but expressed transcription factors important for DC development, and accessed the DC fate at steady state in vivo. Precursor cells at the later stage lacked expression of these factors, as well as DC potential, and were committed to the ILC lineage. At this newly characterized developmental bifurcation, we identified key roles for the transcription factor TCF-1 in promoting the developmental progression of ILC precursors and enforcing their commitment to the ILC lineage. Our observations revealed a degree of imprecision during ILC specification prior to commitment, underscoring the complexity of fate transitions in hematopoiesis.
RESULTS
EILPs are transcriptionally heterogeneous
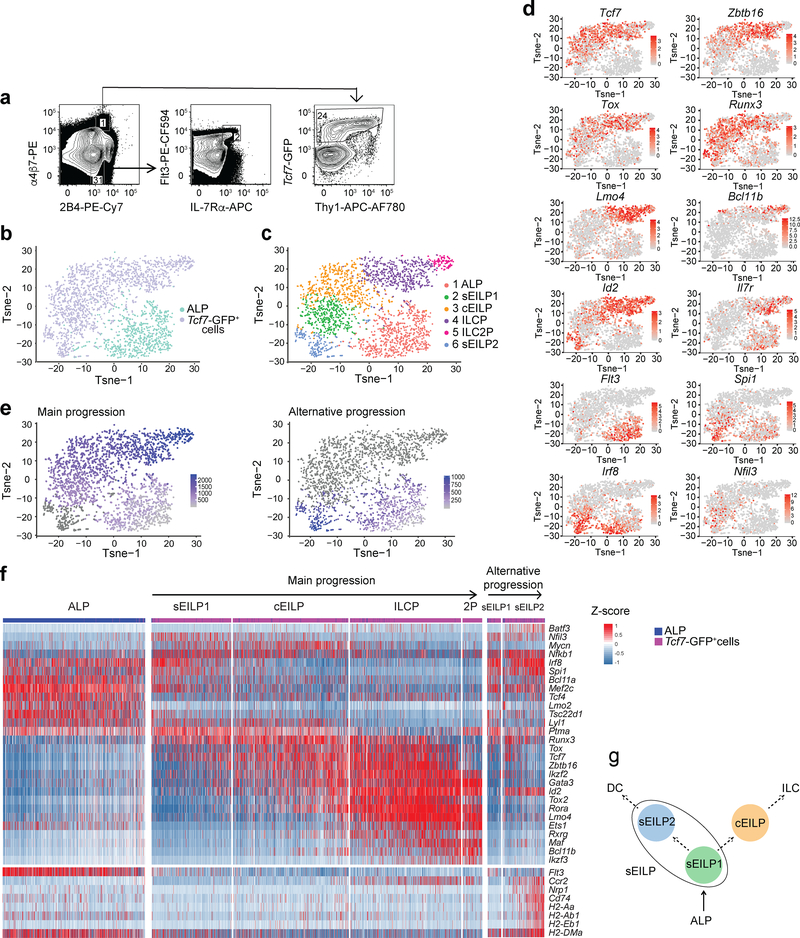
We performed single cell RNA sequencing (scRNA-seq) on Ly-6D−B220−CD19− Mac-1−Gr-1−CD11c−Ter119−NK1.1−CD3ε−CD8α−CD8β−CD4−TCRβ−TCRγδ−(LinILC) Kit+2B4+α4β7+Tcf7-GFP+ cells isolated from the BM of Tcf7EGFP mice, which express EGFP under the control of Tcf7 regulatory elements4. These LinILC–Kit+2B4+α4β7+Tcf7-GFP+ cells included EILPs, ILC precursors (ILCPs) and early ILC2 progenitors (ILC2Ps)1,4–7. We also sequenced LinILC−Kit+2B4+α4β7−Flt3hiIL-7Rα+ all-lymphoid progenitors (ALPs), which are upstream of ILC precursors and are Tcf7-GFP− (Fig. 1a,b)1. After clustering, ALPs formed one cluster (cluster 1), whereas Tcf7-GFP+ cells comprised five clusters (cluster 2–6, Fig. 1c), which we identified based on expression of key transcription factors1,4,8. The Zbtb16hiLmo4hi cluster 4 corresponded to ILCPs; the Zbtb16loBcl11bhi cluster 5 corresponded to ILC2Ps; and clusters 2, 3 and 6 that were Spi1+Irf8+Nfil3+ Zbtb16loId2lo were putative EILP clusters (Fig. 1c,d). Consistently, clusters 2, 3 and 6 had very low expression of the cytokine receptor Il7r compared to all other ILC precursor cells1,4,8.
Figure 1. Single-cell RNA-seq of ILC precursors.
(a) Strategy of isolation of ALP (middle) and Tcf7-GFP+ BM progenitors (right) from Tcf7EGFP/+ mice by flow cytometry. Gated on LinILC− Kit+ cells as shown in Supplementary Fig. 1. Arrows show successive gating and numbers indicate percentages of cells in each gate. (b-e) t-SNE plots showing the ALP (n=786 cells) and Tcf7-GFP+ (n=1799 cells) samples (b), clustering showing known and novel ILC progenitors subsets (c) feature plot showing expression of individual genes differentially expressed by ILC progenitor clusters (d), pseudo-temporal reconstruction of early ILC development showing two plausible progressions (e). The main progression (left) links clusters 1, 2, 3, 4, 5. The alternative progression (right) links clusters 1, 2, 6. The ordering score of individual cells is represented in colors going from light-grey to violet for a given progression. Cells that are not part of the progression are dark grey. (f) Heatmap of expression of transcription factors most differentially expressed between clusters (top), and chosen structural genes (bottom) shown on individual cells ordered along each of the pseudo-time scales from e. The sample (from b), cluster (from c) and developmental progression (from e) in which each cell belongs are indicated above the heatmap. (g) Hypothetical scheme of early ILC development inferred from b-f. Dotted arrows indicate progenitor-successor relationships between EILP subsets and their developmental fate suggested by scRNA-seq analysis. See also Supplementary Table 1.
Next, we examined the developmental relationship between clusters using pseudo-time reconstruction (Fig. 1e)9. A main developmental progression linked ALPs, EILPs, ILCPs and ILC2Ps (clusters 1–5; Fig. 1e). Analysis of differentially expressed transcription factors between clusters showed progressive upregulation or downregulation of factors such as Irf8, Spi1, Tcf7 or Tox along the pseudo-time (Fig. 1f). In an alternative developmental progression, cluster 6 EILPs arose from cluster 2 EILPs (Fig. 1e). Cluster 6 EILPs had high expression of the transcription factors Batf3, Irf8, Spi1, Nfil3 and Id2 (Fig. 1f), which are associated with cDC1 development10 and the DC structural genes Cd74 and MHCII molecules (Fig. 1f and Supplementary Table 1). Cluster 6 EILPs appeared transcriptionally similar to cDC1-committed pre-cDC111,12, although these cDC1 progenitors are CD11c+ and were excluded by the LinILC cocktail. Zbtb46, which is upregulated on committed cDC BM precursors11,12, was not detectably expressed in cluster 6 EILPs (Supplementary Table 1) suggesting these cells represented an early stage of cDC1 priming. ILC-specific transcription factors such as Tox, Tcf7 and Runx3, which are expressed in EILPs and have no described function in DCs and are not expressed in DC progenitors13, were detectable in cluster 6 (Fig. 1f), and their expression was downregulated along the pseudo-temporal progression (Fig. 1f). Based on these results, we designated cluster 2 and cluster 6 as specified EILPs (sEILP), comprising of sEILP1s (cluster 2) and sEILP2s (cluster 6). sEILP1s and sEILP2s highly expressed the cDC genes Flt3, Irf8, Nfil3 and Spi1. Cluster 3 EILPs, which had downregulated the expression of Flt3, Irf8, Nfil3 and Spi1 (Fig. 1f) were designated as committed EILPs (cEILPs). Our analysis placed sEILP1s at a branch point between ILC and cDC1 lineages, indicating that ILC precursors might access cDC1 lineage fate in vivo (Fig. 1g).
sEILPs but not cEILPs have DC lineage potential
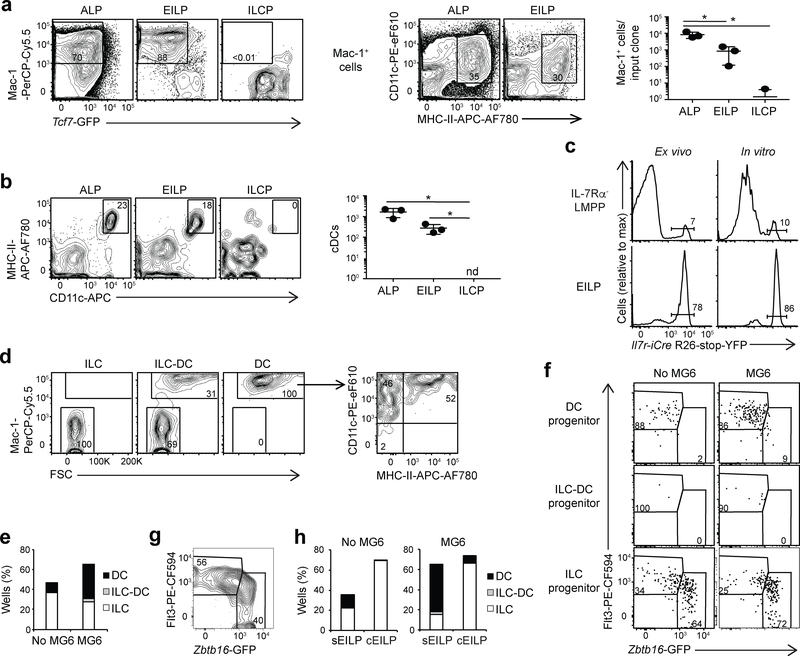
We next characterized the DC potential of EILPs. After 7 days of culture in the presence of OP9 stromal cells and the cytokines SCF, Flt3L and IL-7 (hereafter SF7 conditions), together with GM-CSF and IL-3, which support DC survival and expansion (hereafter SF7-GM3 conditions), LinILC−Kit+2B4+α4β7+Tcf7-GFP+Thy1−EILPs, but not LinILC−Kit+2B4+α4β7+Tcf7-GFP+Thy1+IL-7Rα+ ILCPs, gave rise to Mac-1+CD11c+MHC-II+ cDCs (Fig. 2a). 7 days after transfer into irradiated mice, EILPs, but not ILCPs, differentiated into splenic CD11c+MHC-II+ cDCs (Fig. 2b). To address whether the DC potential of EILPs was due to a small subset of DC precursor cells that co-purified with EILPs, we used Il7r-iCre R26-stop-YFP mice, in which cells that express Il7r permanently express YFP. In these mice, most EILPs1 and ~15% of cDCs14 are Il7r-iCre R26-stop-YFP+. We isolated EILPs and LinILC−KithiSca-1+Flt3hi2B4+α4β7−IL-7Rα− lymphoid-primed multipotential progenitors (LMPPs) from the BM of Il7r-iCre R26-stop-YFP Tcf7EGFP mice and cultured them for 5 days in SF7-GM3 conditions. About 80% of EILPs and EILP-derived Mac-1+ cells were YFP+ (Fig. 2c). In contrast, only 10% of IL-7Rα− LMPPs and IL-7Rα− LMPP-derived Mac-1+ cells were YFP+ (Fig. 2c), suggesting that cDCs develop from EILPs.
Figure 2. Characterization of DC lineage potential of EILPs.
(a) Flow cytometric analysis of cultures from 100 ALPs, EILPs and ILCPs after 7 days in SF7-GM3 condition (left) and quantification of Mac-1+ cell numbers (right). Data are presented as mean ± SD for triplicate wells. (b) Flow cytometric analysis of CD45.2+ splenocytes from wild-type CD45.1+ mice, 7 days after irradiation (800 rads) and injection with 2,000 CD45.2+ ALPs, EILPs, or ILCPs (left) and quantification of CD45.2+ cDC numbers (right). Data are presented as mean ± SEM for n=3 mice per group. (a,b) A two-tailed unpaired Student’s t-test was performed to determine significance. *p<0.05. nd, not detectable. (c) Flow cytometric analysis of IL-7Rα− LMPPs and EILPs from Il7r-Cre R26-stop-YFP Tcf7EGFP/+ mice ex vivo (left), or after 5 days of culture in SF7-GM3 condition (100 cells per well) and gated on Mac-1+ cells (right). (d) Flow cytometric analysis of colonies from single EILPs sorted into 96 well plates and cultured for 10 days in either SF7-GM3 or SF7-GM3-MG6 conditions. Representative profile of individual colonies. Arrows show successive gating. (e) Quantification of wells containing DCs, ILCs, or both lineages as shown in d. (f) Flow cytometric analysis of colonies from single EILPs that were index-sorted from Tcf7YFPZbtb16GFP mice into 96 well plates and cultured for 10 days in either SF7-GM3 or SF7-GM3-MG6 conditions. Profile of EILPs that differentiated into DCs, ILCs, or both, in each condition. (g) Definition of Flt3hiZbtb16-GFPlo sEILPs and Flt3loZbtb16-GFPhi cEILPs deduced from f. Gated on LinILC−Kit+Thy1−2B4+α4β7highTcf7-YFP+ EILP. (h) Quantification of wells containing DCs, ILCs, or both lineages as in e, derived from sEILPs or cEILPs defined in g. (f-h) Data are pooled from 2 experiments. (a-d,f,g) Numbers indicate the percentage of cells in each gate. All data are representative of three independent experiments. See also Supplementary Fig. 2 for the analysis of single EILP differentiation, and Supplementary Fig. 3 for the design of the Tcf7YFP mouse.
To quantify the frequency of DC-competent precursors within EILPs, we sorted single EILPs on OP9 cells in 96-well plates and cultured them for 10 days in SF7-GM3 conditions, or with the additional cytokines M-CSF, G-CSF and IL-6, which promote the differentiation of early T cell precursors into myeloid cells and DCs (hereafter SF7-GM3-MG6 conditions)15. In both conditions, CD45+ EILP-derived colonies contained Mac-1+CD11c+MHC-II+ cDCs (Fig. 2d). Mac-1− cells were ILC lineage cells (Supplementary Fig. 2a–c)1,7. In the SF7-GM3-MG6 cultures, >50% of CD45+ wells contained only Mac-1+ DCs, ~40% contained only Mac-1− ILCs, and <5% contained both ILCs and DCs (Fig. 2d,e). The frequency of Mac-1+ colonies was decreased in SF7-GM3 conditions compared to SF7-GM3-MG6 conditions, but the size of these colonies was comparable in both conditions (Fig. 2e, Supplementary Fig. 2d), suggesting that MG6 cytokines affected the survival of some EILPs. Furthermore, >50% of DC-competent EILPs gave rise to both CD24hi cDC1 and CD172ahi cDC2 in SF7-GM3-MG6 and SF7-GM3 conditions (Supplementary Fig. 2e,f)11,12. Together, these results indicated that a large fraction of EILPs possessed DC potential. However, 40% of EILPs generated exclusively ILCs.
Next, we examined whether transcriptional heterogeneity identified by scRNA-seq in EILPs correlated with heterogeneity identified in differentiation assays. Because the expression of Flt3 and PLZF protein (encoded by Zbtb16) inversely correlates in EILPs1, we tested whether Flt3 and PLZF could be used to distinguish the Flt3hiZbtb16lo sEILPs and Flt3loZbtb16hi cEILPs identified in scRNA-seq. We crossed newly generated Tcf7YFP reporter mice, which express YFP downstream of the Tcf7 gene (Supplementary Fig. 3), with Zbtb16GFP reporter mice7. We used index sorting, which allows a posteriori analyses linking flow cytometric measurements with developmental fate, to sort 1176 single EILPs from Tcf7YFPZbtb16GFP mice into 96-well plates and cultured them in either SF7-GM3 or SF7-GM3-MG6 conditions. Analysis of heterogeneously-expressed factors (Zbtb16-GFP, Tcf7-YFP, Flt3) and cell fate (generation of DCs or ILCs) for single EILPs indicated that almost all DC progenitor cells were Flt3hiZbtb16-GFPlo, whereas ILC progenitors were mostly Flt3loZbtb16-GFPhi (Fig. 2f). Using Flt3 and Zbtb16-GFP, we defined gates that matched the EILP fate (Fig. 2f). These gates identified a Flt3loZbtb16-GFPhi population that gave rise almost exclusively to Mac-1− ILCs in SF7-GM3 and SF7-GM3-MG6 conditions (Fig. 2g,h) and a Flt3hiZbtb16-GFPlo subset that predominantly gave rise to Mac-1+ DCs in SF7-GM3-MG6 conditions, but predominantly gave rise to ILC in SF7-GM3 conditions (Fig. 2g,h).
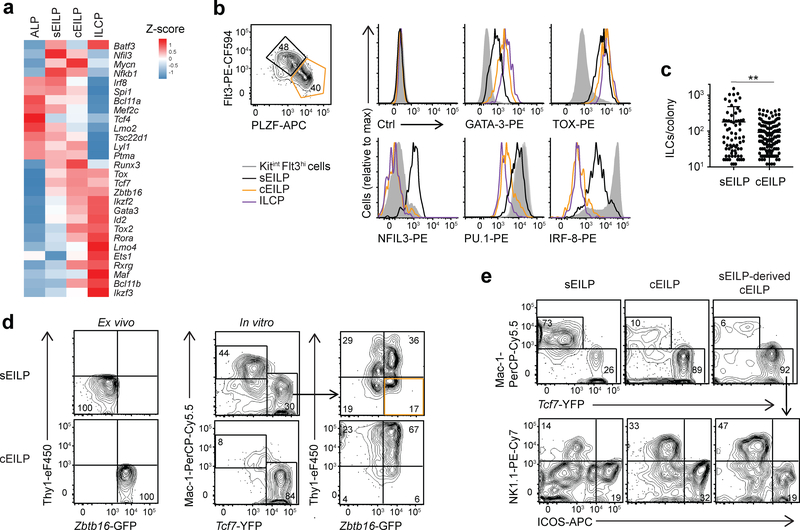
Bulk RNA-sequencing of Flt3hiZbtb16-GFPlo and Flt3loZbtb16-GFPhi EILP subsets (Supplementary Fig. 4a) indicated they were transcriptionally similar to sEILPs and cEILPs respectively (Fig. 3a). We examined expression of the transcription factors GATA-3, TOX, NFIL3, PU.1, IRF-8 in Flt3hiPLZFlo sEILPs and Flt3loPLZFhi cEILPs. Consistent with the bulk RNA-seq and scRNA-seq, GATA-3 expression was upregulated from sEILPs to cEILPs, TOX expression was high in sEILPs and cEILPs, and PU.1 (encoded by Spi1), IRF8 and NFIL3 expression was downregulated from sEILPs to cEILPs (Fig. 3b). Thus, Flt3hiZbtb16-GFPlo and Flt3loZbtb16-GFPhi EILP populations correspond respectively to the sEILP and cEILP populations identified by scRNA-seq.
Figure 3. Characterization of sEILP and cEILP populations.
(a) RNA-seq analysis of ALPs, sEILPs, cEILPs and ILCPs showing the heat map of expression of transcription factors from Fig. 1f. Data are averaged from two samples for sEILPs and ILCPs or three samples for ALPs and cEILPs as shown in Supplementary Fig. 4a. (b) Flow cytometric analysis of LinILC−Kit+Thy1−2B4+α4β7highTCF-1+ EILPs by intracellular staining. Definition of sEILPs and cEILPs (left), histogram of expression on LinILC−Thy1−2B4+α4β7−KitintFlt3hi cells (shaded grey), sEILPs (black), cEILPs (orange) and ILCPs (purple)(right). (c) Quantification of absolute numbers of ILCs per ILC positive colony from Fig. 2h. Data are presented as mean + SD for n=78 sEILP- and n=143 cEILP-derived colonies. A two-tailed unpaired Student’s t-test with Welsh correction was performed to determine significance. **p<0.01. (d) Flow cytometric analysis of sEILPs and cEILPs isolated from Tcf7YFPZbtb16GFP mice (left) and cultured for two days in SF7 condition (right). (e) Flow cytometric analysis of cultures from in vitro sEILP-derived cEILP population (shown in orange in d) and ex vivo sEILPs and cEILPs after 4 days in SF7 condition. (b,d,e) Arrows show successive gating. Numbers indicate the percentage of cells in each gate. All data are representative of three independent experiments. See also Supplementary Fig. 4a–f for further characterization of sEILP and cEILP populations.
sEILPs differentiate into cEILPs
We next examined the developmental relationship between sEILPs and cEILPs. We examined ILC-positive colonies derived from single sEILP and cEILP cultured for 10 days in SF7-GM3 conditions. Mac-1−ICOS−NK1.1−Tcf7-YFP+ cells with the phenotype of ILC progenitors (Supplementary Fig. 2) were more abundant in sEILP-derived colonies (70%) than cEILP-derived colonies (50%; Supplementary Fig. 4b), suggesting sEILPs were more immature than cEILPs. ILC progeny (Tcf7hiNK1.1+ ILC1s or NK cells, Tcf7loICOS+α4β7lo ILC2s and Tcf7loICOS+α4β7lo ILC3s) were similar between sEILP-derived and cEILP-derived colonies (Supplementary Fig. 4c). However, sEILP-derived ILC colonies were larger than cEILP-derived ILC colonies (Fig. 3c). Furthermore, sEILPs were larger than cEILPs (Supplementary Fig. 4d), and contained a greater fraction of cycling cells (27% of DAPIhi sEILPs versus 5% of cEILPs; Supplementary Fig. 4e), consistent with larger proliferative capacity of more upstream progenitors16.
To test whether sEILPs were upstream of cEILPs, we isolated Flt3hiZbtb16-GFPlo sEILPs and Flt3loZbtb16-GFPhi cEILPs from Tcf7YFPZbtb16GFP mice and cultured them for 2 days in SF7 conditions. Most sEILPs downregulated Tcf7-YFP and upregulated Mac-1 (Fig. 3d), consistent with differentiation into DCs. Most Tcf7-YFP+ sEILP progeny were similar to Zbtb16-GFPhiThy1− cEILPs and Zbtb16-GFPhiThy1+ ILCPs (Fig. 3d). sEILP-derived PLZFloThy1− sEILPs, PLZFhiThy1− cEILPs and PLZFhiThy1+ ILCPs showed transcription factor expression consistent with corresponding ex vivo populations (GATA-3, TOX, NFIL3, PU.1, IRF-8; Supplementary Fig. 4f). Most cEILP-derived cells were Tcf7-YFP+Zbtb16-GFP+Thy1+ ILCPs, and had higher expression of Thy1 than sEILP-derived ILCPs (Fig. 3d). To establish that sEILP-derived cEILPs were similar to ex vivo cEILPs, we isolated Tcf7-YFP+Zbtb16-GFP+Thy1− cEILPs from sEILP SF7 cultures after 2 days, and cultured them for 4 days in SF7 conditions. Only 10% of Mac-1+ DCs were generated in these cultures, similar to ex vivo Flt3loZbtb16hi sEILPs cultured in the same conditions. However, in vitro generated cEILPs and ex vivo cEILPs had similar potential to generate NK1.1+ or ICOS+ ILCs (Fig. 3e). Our results support a linear developmental relationship linking sEILPs, cEILPs and ILCPs.
sEILPs develop into cDC1 precursors in vivo
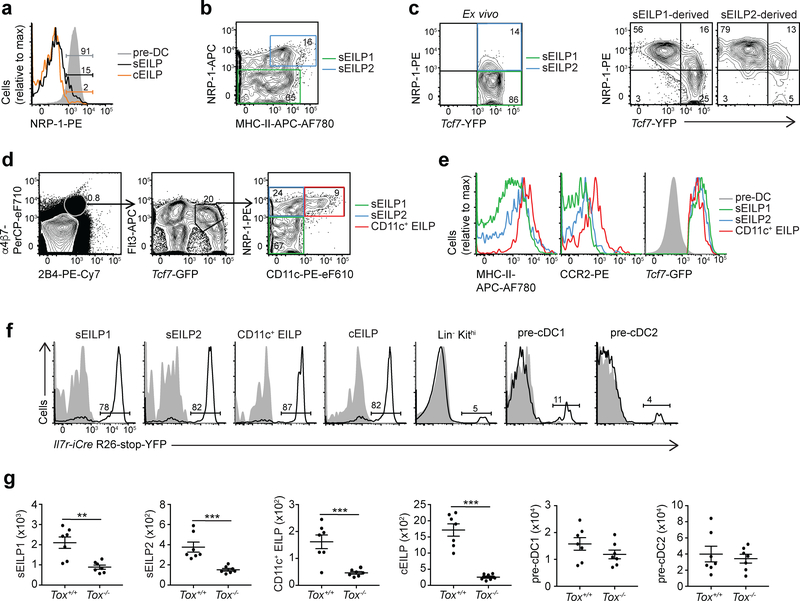
Using scRNA-seq, we identified Nrp1 as encoding a surface marker that could resolve sEILP1s and sEILP2s (Fig. 1f). NRP-1 was homogeneously expressed by LinDC–(B220−CD19−Thy-1−Mac-1−Gr-1−Ter119−NK1.1−CD3ε−CD8α−CD8β−CD4− TCRβ−TCRγδ−)Flt3+CD11c+MHC-IIint/lo pre-DCs (Fig. 4a), as well as a fraction of sEILPs that corresponded to sEILP2s (Fig. 4a). CCR2 and MHC-II11,12, which were upregulated from sEILP1 to sEILP2 at the RNA level (Fig. 1f), were upregulated on NRP-1+ sEILP2s compared to NRP-1− sEILP1s (Fig. 4b). Expression of Tcf7-YFP, PLZF and IRF8 on NRP-1+ sEILP2s was intermediate between NRP-1− sEILP1s and pre-DCs, whereas TOX expression was similar on NRP-1− sEILP1s and NRP-1+ sEILP2s (Supplementary Fig. 4g,h).
Figure 4. Characterization of EILP-derived DC precursor populations.
(a) Flow cytometric analysis of pre-DCs (grey), sEILPs (black) and cEILPs (orange). (b) Flow cytometric analysis of LinILC−Kit+Thy1−2B4+α4β7highTcf7-YFP+ sEILPs defining sEILP1s (green) and sEILP2s (blue). (c) Flow cytometric analysis of sEILP1s and sEILP2s isolated as shown in b (left) and cultured for two days in SF7 condition (right). (d) Flow cytometric analysis of LinDCKit+ BM cells from Tcf7YFP mice showing sEILP1s (green), sEILP2s (blue) and CD11c+ EILPs (red). Arrows show successive gating. (e) Flow cytometric analysis of sEILP1s (green), sEILP2s (blue) and CD11c+ EILPs (red) defined in d, compared to pre-DCs (grey). (f) Flow cytometric analysis of the indicated populations in Il7r-Cre R26-stop-YFP Tcf7EGFP/+ mice (black histogram) compared to Tcf7EGFP/+ control mice (gray histogram). (g) Quantification by flow cytometry of the indicated populations in Tox−/− Tcf7EGFP/+ mice and Tox+/+ Tcf7EGFP/+ littermates. Data are presented as mean ± SEM for n=7 mice per group, pooled from three independent experiments. A two-tailed unpaired Student’s t-test was performed to determine significance. *p<0.05, **p<0.01, ***p<0.005. (a-d,f) Numbers indicate the percentage of cells in each gate. All data are representative of three independent experiments. See Supplementary Fig. 5 for the definition of canonical DC precursors.
To determine whether sEILP1s and sEILP2s represented two successive stages of development towards DCs, we isolated Flt3hiZbtb16-GFPloNRP-1− sEILP1s and Flt3hiZbtb16-GFPloNRP-1+MHC-II+ sEILP2s from Tcf7YFPZbtb16GFP mice and examined their fate after 2 days of culture in SF7 conditions. 16% of sEILP1-derived cells were phenotypically similar to Tcf7-YFP+NRP-1+ sEILP2s (Fig. 4c). The remaining cells were Tcf7-YFP−NRP-1+ (Fig. 4c) and had upregulated the expression of the DC markers Mac-1, MHCII and CD11c (Supplementary Fig. 4i), consistent with DC differentiation. Conversely, 82% of sEILP2 progeny were NRP-1+ and were mostly DCs (Fig. 4c, Supplementary Fig. 4i). More Tcf7− sEILP2 progeny expressed Mac-1, MHC-II and CD11c compared to Tcf7− sEILP1 progeny, and at higher levels (Fig. 4c and Supplementary Fig. 4i). Both sEILP1s and sEILP2s gave rise to Mac-1+ DCs after culture for 10 days in SF7-GM3 and SF7-GM3-MG6 conditions; however, sEILP1-derived DC colonies were larger than sEILP2-derived DC colonies (Supplementary Fig. 4j,k), consistent with larger proliferative capacity of more upstream progenitors16. These results indicate that sEILP1s transitioned through the sEILP2 stage, prior to DC differentiation.
To determine whether sEILPs differentiated into DCs in vivo at steady state, we analyzed LinDC-Kit+2B4+α4β7+Tcf7-GFPloFlt3hi sEILPs for CD11c expression (Fig. 4d). This analysis identified a population of CD11c+ cells that was phenotypically similar to sEILPs (CD11c+ EILP; Fig. 4d). Expression of NRP-1 (Fig. 4d), MHC-II and CCR2 proteins (Fig. 4e) was low on sEILP1s, intermediate on sEILP2s, and high on CD11c+ EILPs. Tcf7-GFP was lower on CD11c+ EILPs than on sEILP1s and sEILP2s, but clearly positive compared to pre-DCs (Fig. 4e). Further characterization of CD11c+ EILPs using surface markers described on pre-DC populations (CD24, CCR2, Ly-6D, Ly-6C, Siglec-H)11,12 showed that CD11c+ EILPs were phenotypically similar to pre-cDC1s (Supplementary Fig. 5c).
To test whether CD11c+ EILPs derived from EILPs, we analyzed these cells in Il7r-iCre R26-stop-YFP mice crossed with the Tcf7EGFP mice. More than 80% of CD11c+ EILPs were Il7r-iCre R26-stop-YFP+, similarly to sEILPs (Fig. 4f)1. In contrast, only ~10% of canonical DC precursors pre-cDC1 and pre-cDC2, and even fewer Lin−Kithi upstream hematopoietic progenitor cells were Il7r-iCre R26-stop-YFP+ (Fig. 4f). In Tox−/− mice, CD11c+ EILP and sEILP absolute numbers were 3–4-fold reduced compared to Tox+/+ littermates (Fig. 4g)1, whereas canonical DC precursors such as pre-cDC1s and pre-cDC2s were present in normal numbers (Fig. 4g). These results indicate that CD11c+ EILPs derive from EILPs.
Although EILPs are 10-fold less abundant than pre-cDC1 and pre-cDC2 (Fig. 4g), we examined whether ILC precursors contribute to DC generation in vivo. In Tcf7EGFPIl7r-iCre R26-stop-YFP mice, Tcf7-GFP+Il7r-iCre R26-stop-YFP+ cells represented <5% of pre-cDC1 in vivo, and were almost undetectable within other DC precursor populations (Supplementary Fig. 5e). These cells likely include CD11c+ EILPs, and other putative EILP-derived DC lineage cells. Furthermore, Tox−/− LinILC−KithiSca-1+ hematopoietic progenitor cells, which are deficient in EILP generation1, generated CD8α+Mac-1lo cDC1s normally in long-term competitive BM chimeras (Supplementary Fig. 5f), suggesting EILPs were unlikely to contribute significantly to DC development in vivo. Our results establish that EILPs generate cDC1 precursors in vivo at steady state, and support a linear developmental relationship linking sEILP1s, sEILP2s and the pre-cDC1 subset CD11c+ EILPs.
TCF-1-deficient sEILPs do not generate cEILPs
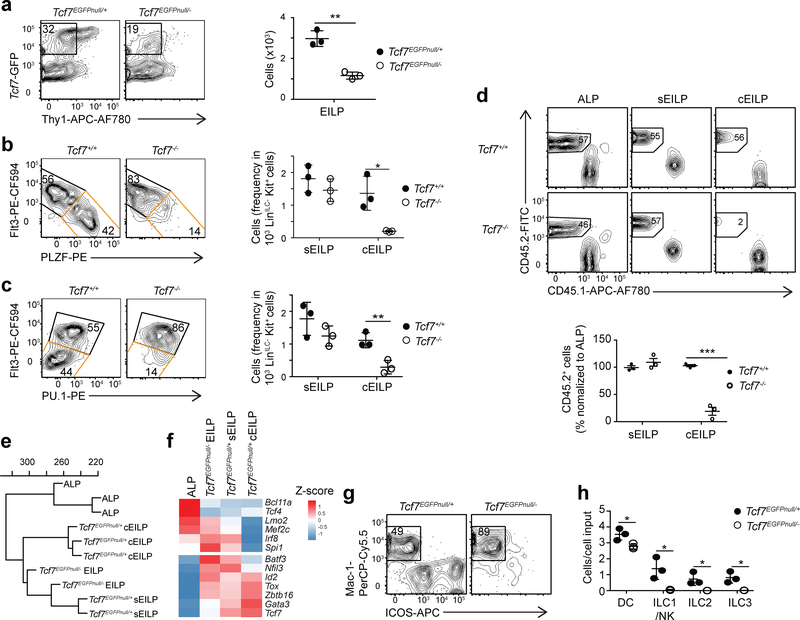
Next, we investigated if TCF-1 regulated the transition from sEILP1s to cEILPs. Deletion of exon 2 of Tcf7 and the surrounding floxed region in Tcf7EGFP mice generated mice with a Tcf7EGFPnull allele (Supplementary Fig. 6a), which were crossed to Tcf7−/− mice to obtain Tcf7EGFPnull/− mice. Tcf7EGFPnull/− mice reported Tcf7 expression like Tcf7EGFP mice, but lacked TCF-1 protein (Supplementary Fig. 6a–d). LinILC−Tcf7-GFP+ cells were detectable in Tcf7EGFPnull/− BM (Supplementary Fig. 6e)2 and were α4β7+2B4+ (Supplementary Fig. 6f). Numbers of EILPs in Tcf7EGFPnull/− mice were reduced 2-fold compared to wild-type mice (Fig. 5a)2 and Tcf7EGFPnull/− EILPs were Flt3hi (Supplementary Fig. 6f), suggesting that ILC development was arrested at the sEILP stage. In the absence of TCF-1 protein, we used TOX expression to quantify the number of EILPs in Tcf7−/− mice (Supplementary Fig. 6g, h), and used LinILC-depleted BM cells, which allowed reliable detection of TOX by intracellular staining. Absolute numbers of LinILC−Kit+ were similar in Tcf7−/− and Tcf7+/+ BM (Supplementary Fig. 6h), thus the frequency of LinILC−Kit+2B4+α4β7+Thy1−TOX+ EILPs could be quantified in LinILC−Kit+ cells. Within TOX+ EILP, expression of Flt3 in combination with either PLZF (Fig. 5b) or PU.1 (Fig. 5c) was used to separate Flt3hiPU.1hiPLZFlo sEILP from Flt3loPU.1loPLZFhi cEILP. Both strategies showed that the numbers of sEILP were not significantly affected, whereas the numbers of cEILP were greatly reduced in Tcf7−/− mice compared to Tcf7+/+ littermates (Fig. 5b,c). This defect was similar when ILC precursors developed in the presence of competitor wild-type cells in lethally irradiated CD45.1+ mice reconstituted with a mix of CD45.2+ LinILC− BM cells from Tcf7−/− or wild-type littermate controls, and CD45.1+ LinILC− BM cells (Fig. 5d), confirming TCF-1 was required cell-autonomously for the generation of cEILPs.
Figure 5. Developmental arrest at sEILP stage in the absence of TCF-1.
(a) Flow cytometric analysis of LinILC−Kit+2B4+α4β7+ BM cells from Tcf7EGFPnull/+ and Tcf7EGFPnull/− littermate mice (left) and quantification of EILP numbers (right). (b,c) Flow cytometric analysis of TOX+ EILPs defined in Supplementary Fig. 6g, in Tcf7−/− and Tcf7+/+ littermate mice. sEILP and cEILP gates are shown in black and orange respectively (left). Quantification of frequencies of sEILPs and cEILPs within LinILC−Kit+ BM cells (right). (a-c) Data are presented as mean ± SEM for n=3 mice per group. (d) Flow cytometric analysis of BM cells from CD45.1+ mice that were lethally irradiated (850 rads), injected with Tcf7−/− or wild-type littermate CD45.2+LinILC− BM cells mixed with CD45.1+LinILC− BM cells, and reconstituted for 10–15 weeks. Data are presented as mean ± SEM for n=3 mice per group. (e) RNA-seq analysis of the indicated samples. Hierarchical clustering using complete linkage calculated from Euclidian distances. (f) Heatmap of expression of selected transcription factors from RNA-seq analysis. Data are averaged from two or three samples shown in e. (g) Flow cytometric analysis of cultures from EILPs isolated from Tcf7EGFPnull/− or Tcf7EGFPnull/+ littermate mice after 4 days in SF7 condition (100 cells per well). Profile of the derived population. (h) Quantification of DC and ILC numbers as in g. Data are presented as mean ± SD for n=3 triplicate wells. (a-d,g) Numbers indicate the percentage of cells in each gate. (a-d,g-h) Data are representative of three independent experiments. (a-d,h) A two-tailed unpaired Student’s t-test was performed to determine significance. *p<0.05, **p<0.01. See also Supplementary Fig. 6 for description of the Tcf7EGFPnull mouse strain and Supplementary Table 2 for additional RNA-seq data.
Tcf7EGFPnull/− EILPs were transcriptionally similar to wild-type sEILPs (Fig. 5e), consistent with developmental arrest of Tcf7EGFPnull/− EILPs at the sEILP stage. Genes dynamically regulated between ALPs and sEILPs, such as Nfil3, Id2 and Tox, were similarly regulated between ALP and Tcf7EGFPnull/− EILPs (Fig. 5f, Supplementary Fig. 6i and Supplementary Table 2), indicating early ILC lineage specification was largely unaffected by loss of TCF-1. Consistently, Tcf7EGFPnull/− EILPs cultured for 4 days in SF7 conditions were still able to generate DCs (Fig. 5g,h). These observations indicated that TCF-1 was dispensable for initial ILC specification, but was required for progression toward the ILC lineage.
TCF-1-deficient EILPs are diverted toward the DC lineage
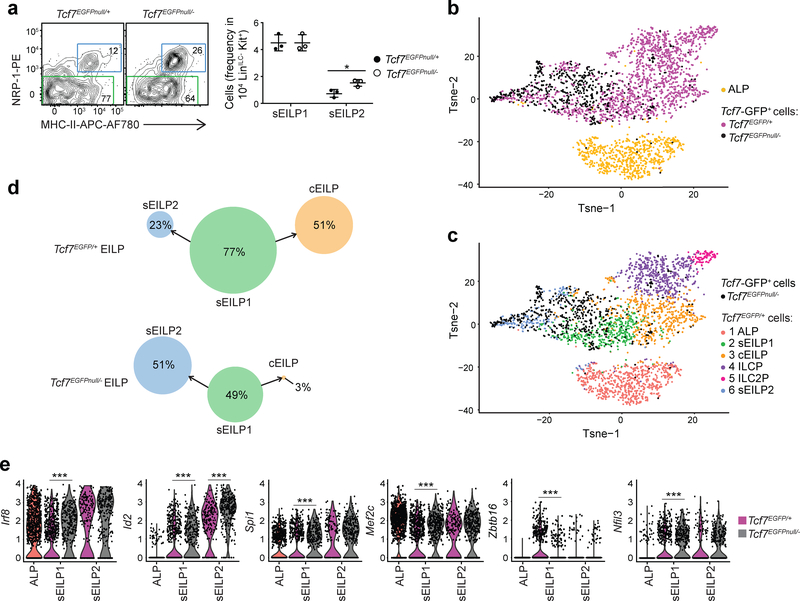
Although developmental arrest at the sEILP stage in TCF-1-deficient mice predicted a 2-fold higher frequency of DC-competent sEILPs in Tcf7EGFPnull/− EILPs, Tcf7EGFPnull/− EILPs cultured for 4 days in SF7 conditions gave rise to fewer DCs than wild-type EILPs (Fig. 5g,h). Expression of Batf3 and Id2, which was higher on sEILP2s compared to sEILP1s (Fig. 1f), was 2-fold higher on Tcf7EGFPnull/− EILPs compared to wild-type sEILP (Fig. 5f, Supplementary Table 2), suggesting that a larger fraction of sEILPs might be primed toward the DC lineage in the absence of TCF-1. The number of NRP-1− sEILP1s was similar, whereas the number of NRP-1+MHC-II+ sEILP2s was increased 2-fold in Tcf7EGFPnull/− mice compared to wild-type mice (Fig. 6a). In single cell assays, Tcf7EGFPnull/− sEILPs generated fewer Mac-1+ DCs, than wild-type sEILPs (Supplementary Fig. 7a,b), indicating that Tcf7EGFPnull/− sEILPs had functional properties similar to sEILP2.
Figure 6. Commitment failure and lineage diversion in the absence of TCF-1.
(a) Flow cytometric analysis of LinILC−Kit+2B4+α4β7+Tcf7-GFP+Flt3+ sEILPs from Tcf7EGFPnull/+ and Tcf7EGFPnull/− littermate mice, showing sEILP1s (green) and sEILP2s (blue)(left). Quantification of percentages of sEILP1s and sEILP2s within LinILC−Kit+ BM cells (right). Data are representative of three independent experiments and presented as mean ± SEM for n=3 mice per group. A two-tailed unpaired Student’s t-test was performed to determine significance. *p<0.05. (b,c) scRNA-seq analysis of Tcf7-GFP+ BM progenitors isolated from Tcf7EGFPnull/− mice and compared to wild-type ALPs and Tcf7-GFP+ BM progenitors from Fig. 1. t-SNE plots showing: ALP (n=786 cells), Tcf7EGFP/+ (n=1799 cells) and Tcf7EGFPnull/− (n=594 cells) samples (b), similarity of Tcf7EGFPnull/− Tcf7-GFP+ BM progenitors (black) with the wild-type ALP and Tcf7-GFP+ BM progenitor subsets colored by clusters as defined in Fig. 1c (c). (d) Quantification of Tcf7EGFPnull/− and Tcf7EGFP/+ cell number in each cluster from c, calculated as percentage of sEILP. The arrows show the developmental relationships linking clusters. The size of the circle that symbolizes each cluster is relative to cell number. (e) Expression of the indicated genes by n=752 ALPs (red), n= 270 Tcf7EGFPnull/− sEILP1s, and n=276 Tcf7EGFPnull/− sEILP2s (black), and n=615 Tcf7EGFP/+ sEILP1s and n=187 Tcf7EGFP/+ sEILP2s (pink) from scRNA-seq analysis. A two-sided Wilcoxon rank-sum test was used to determine the significance of gene expression differences between Tcf7EGFPnull/– and Tcf7EGFP/+ cells for a given subset. ***p<0.005. See also Supplementary Fig. 7, and Supplementary Table 3 and 4.
We performed scRNA-seq on LinILC−Kit+2B4+α4β7+Tcf7-GFP+ BM progenitors from Tcf7EGFPnull/− mice, and merged the data with Tcf7EGFP/+ LinILC−Kit+2B4+α4β7+Tcf7-GFP+ BM progenitors and ALP scRNA-seq data (Fig. 6b). In t-SNE analysis, only ~5% Tcf7EGFPnull/− cells overlapped with wild-type cEILP, ILCPs or ILC2Ps (Fig. 6c). Relative quantification of Tcf7EGFPnull/− and Tcf7EGFP/+ cells in each cluster showed that 51% of Tcf7EGFPnull/− sEILPs corresponded to sEILP2, whereas only 23% of wild-type sEILPs were sEILP2s (Fig. 6d). Transcription factors Irf8, Id2, Nfil3, Spi1 and Mef2c were upregulated, and Zbtb16 was downregulated in Tcf7EGFPnull/− sEILP1s compared to wild-type sEILP1s (Fig. 6e and Supplementary Table 3). These altered expression patterns were most obvious for sEILP1s on the main progression (Supplementary Fig. 7c,d), suggesting they preceded diversion toward the DC lineage (Supplementary Fig. 7e and Supplementary Table 4). These observations indicated that, in the absence of TCF-1, sEILP1s adopted a cDC1 progenitor-like transcriptional profile and were diverted towards the DC fate.
TCF-1 enhances expression of ILC genes and represses DC genes
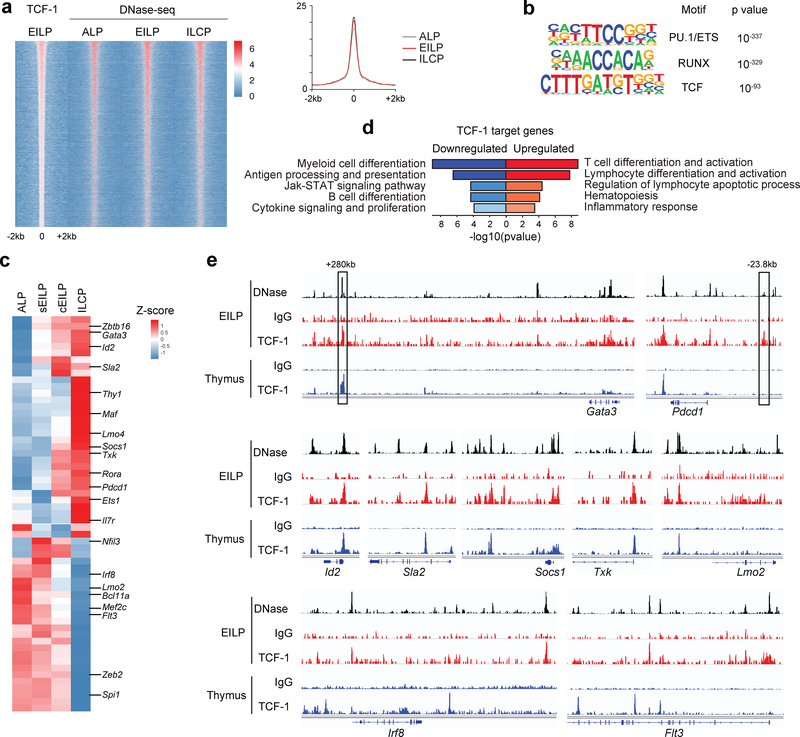
To identify TCF-1 gene targets during ILC development, we first inferred regulatory interactions between genes using LEAP (lag-based expression association for pseudotime series) algorithm17. We used the main progression to identify genes whose expression was correlated during ILC development, allowing a gap in correlation to take into account delays between the expression of a controller and its targets (Supplementary Fig. 7f, Supplementary Table 5). To determine which of the putative TCF-1 target genes identified were directly regulated by TCF-1, we characterized TCF-1 binding in EILPs using ChIC-seq18, and identified open chromatin regions in ALPs, EILPs and ILCPs using DNase-seq. We identified 9649 TCF-1 peaks genome-wide in EILPs (Supplementary Table 6). 99% of these peaks were located in regions of open chromatin in ALPs (Fig. 7a, Supplementary Fig. 7g). Regions bound by TCF-1 showed significant enrichment for TCF motifs, as well as RUNX and PU.1 motifs (Fig. 7b).
Figure 7. Identification of TCF-1 gene targets during early ILC development.
(a) TCF-1 ChIC-seq analysis in EILPs and DNase-seq analysis in ALPs, EILPs, ILCPs. Heat map centered on TCF-1 binding sites in EILPs (± 2kb) (n=9549 peaks) (left), and quantification of DNase I hypersensitivity enrichment (right). (b) Known transcription factor motif analysis (HOMER) of TCF-1 binding sites from a (n=9549 peaks). Chosen enriched motifs and associated p values were calculated based on the cumulative binomial distribution. (c) RNA-seq analysis averaged from 3 ALP samples, 2 sEILP samples, 3 cEILP samples and 2 ILCP samples, showing the expression of TCF-1 gene targets. (d) Biological process enrichment analysis on genes upregulated (n=31 genes, red), or downregulated by TCF-1 (n=28 genes, blue). Most significant biological processes and associated p values calculated based on the accumulative hypergeometric distribution. (e) Traces of DNase-seq in EILPs (black), IgG or TCF-1 ChIC-seq in EILPs (blue), and IgG or TCF-1 ChIP-seq in thymocytes (blue). Previously described enhancers are indicated by black boxes and their distance relative to the TSS is indicated20,21. See also Supplementary Fig. 7, and Supplementary Tables 5, 6, and 7.
To identify TCF-1 target genes, we identified genes predicted to be regulated by TCF-1 (Supplementary Table 5) and showed TCF-1 binding in open chromatin regions in their vicinity (Supplementary Table 6). We excluded genes that were properly regulated in the absence of TCF-1 in scRNA-seq or bulk RNA-seq (Supplementary Tables 2 and 3). This analysis identified 59 genes that received direct regulatory inputs from TCF-1 in EILPs (Fig. 7c, Supplementary Table 7). Downregulated genes were enriched for genes involved in myeloid cell differentiation (Fig. 7d), some of which (Flt3, Nfil3, Mef2c, Spi1 and Irf8) were upregulated in Tcf7EGFPnull/− sEILP1 cells in the main trajectory compared to wild-type sEILP1s (Supplementary Fig. 7h, Supplementary Table 4), and were downregulated from sEILP1s to cEILPs in wild-type ILC precursors (Fig. 1f). On the other hand, genes upregulated by TCF-1 were enriched for genes involved in lymphocyte differentiation and activation (Fig. 7d) such as Id2 or Gata3, which play important functions in EILPs1,4 (Fig. 7c), indicating that TCF-1 promoted ILC differentiation. Genes important at later stages of development and ILC maturation, such as Il7r, Ets1 or Pdcd1 (encoding PD1) were also predicted TCF-1 targets (Fig. 7d). A few genes predicted to be positively regulated by TCF-1 (Zbtb16, Ccl5 and Rora) were upregulated between ALPs and sEILP1s (Supplementary Table 1) and had lower expression in Tcf7EGFPnull/− sEILP1s compared to wild-type sEILP1s (Supplementary Fig. 7e,h).
Comparison of TCF-1 binding in EILPs and T lineage cells19 revealed that several TCF-1 binding regions were shared, including the downstream Gata3 enhancer important for early T cell development (Tce1)20, and the Pdcd1 enhancer active in exhausted T cells21 (Fig. 7e). TCF-1 contributes to the regulation of these enhancers in T cells20,21, indicating that TCF-1 can regulate similar loci in T cells and ILCs. Additional genes expressed in both T cells and ILCs, such as Id2, Socs1, Sla2 and Txk had similar patterns of TCF-1 binding in both lineages, whereas other genes such as Irf8, Lmo2 and Flt3 showed distinct patterns (Fig. 7e). Our analysis indicated that TCF-1 promoted ILC development by positively regulating ILC-specific genes; and enforced ILC commitment through the repression of genes shared by sEILP1s and DCs (Supplementary Fig. 7i).
DISCUSSION
In this study, we used population level and minimally biased single-cell approaches to identify transcriptional and functional heterogeneity of ILC progenitor cells, and understand developmental relationships between them. Our work confirmed a linear developmental path between ALPs, EILPs, ILCPs and ILC2Ps1,8. We identified two successive stages of ILC development within EILPs which we called sEILP1 and cEILP, and delineated a commitment checkpoint during early ILC development. We also identified a developmental bifurcation toward the DC fate at the sEILP1 stage at steady state in vivo. We further defined a role for the transcription factor TCF-1 in controlling these developmental transitions. We find that TCF-1 was dispensable for initial ILC specification and generation of sEILP1s, but required for the development of cEILPs. Mechanistically, TCF-1 enforced commitment to the ILC lineage by providing positive regulatory inputs to key ILC genes and by repressing expression of genes key for DC development that were expressed at early steps of ILC specification and were downregulated during ILC commitment.
Many genes bound and upregulated by TCF-1 during early ILC development also have important functions in T cells, which raises the possibility that TCF-1 regulate these genes in both lineages. Consistently, key enhancers described to receive regulatory inputs from TCF-1 in T cells, including the distal Gata3 and Pdcd1 enhancer, were bound by TCF-1 in ILC precursors. We found that chromatin bound by TCF-1 in EILPs was almost entirely accessible in upstream ALPs. This finding indicates that TCF-1 binding is largely dictated by the regulatory landscape established prior to T cell and ILC specification, in precursors of these lineages. TCF-1 binding sites showed significant enrichment for binding motifs for RUNX and PU.1 transcription factors. Hence, RUNX1 and PU.1 may mediate chromatin opening to allow TCF-1 binding22,23, or these factors may serve to recruit TCF-1 to its binding sites during T cell and ILC specification24. Our findings raise the possibility that TCF-1 regulates a shared set of core genes in T cell and ILC, which coordinates the acquisition of functional similarities between the two lineages1.
Although the observation that ILC progenitors had the ability to develop into DCs was similar to DC lineage potentials reported for other uncommitted lymphoid progenitors, such as ETP25 and ALPs26, our study provided evidence that some lymphoid precursors actually fulfilled this potential in vivo at steady state, and that DC development occurred from non-canonical DC precursors. Because sEILPs and other lymphoid precursors do not appear to contribute significantly to DC generation, nor generate distinct lineages of DCs27,28, such a pathway may not be important in physiological conditions. We speculate that the residual DC development described here is the result of the imprecision of molecular mechanisms that underly early lymphoid specification, and a consequence of the transcriptional similarities between early innate lymphoid progenitors and DC progenitors10. More generally, because of the complexity of specification mechanisms, other developmental processes may be similarly prone to imprecisions29,30.
In summary, our work revealed that EILPs are heterogeneous, and that innate cell lineage specification prior to commitment occurred with a degree of imprecision and co-opted transcriptional programs permissive for DC development. Our work revealed that a residual DC development accompanied early ILC specification in vivo, and identified mechanisms by which TCF-1 restrained this alternative potential and mediated commitment to the ILC lineage.
METHODS
Mice
B6-Ly5.2 (CD45.1) and CMV-Cre mice were from the Jackson Laboratory. Tcf7EGFP4, Zbtb16GFP7, Tcf7−/−31, Tox−/−32, Il7r-iCre R26-stop-YFP14 mice have previously been described. Tcf7YFP mice were generated as followed: the targeting vector was generated by BAC recombineering: First a 10538 nt long mouse genomic Tcf7 fragment starting with atgtatatatgtgccccttcct…. and ending with ….cagggttacaaagctgggt was inserted into the BAC retrieval vector PL253 (NCI at Frederick) by BAC recombineering. Second, a synthetic DNA construct was designed: It started with acacaagattcctcttg…. of Tcf7 intron 9 followed by the beginning of exon 10, which was fused at the C terminus of its …..FLPMTVL amino acid sequence to the P2A ribosomal skipping peptide. This was followed by the YFP sequence and the rest of Tcf7 exon10 starting at taggctgtccccggtccccagc….. and the synthetic construct ended with the Tcf7 sequence ….agaaactctttttgcgcc. After synthesis (Blueheron, Inc.) this 2251 nt long construct was inserted into the EcoRI and KpnI sites of PL452 (NCI at Frederick). In addition, a ~400 nt Tcf7 genomic arm located upstream of the ….acacaagattcctcttg sequence of intron 9 was cloned into the multiple cloning site of PL452 adjacent to its loxP site resulting in a ‘mini targeting vector’ containing the P2A-YFP construct and both arms of homology. This ‘mini vector’ was recombined into the above pL253 Tcf7 genomic vector by a second round of BAC recombineering resulting in the final targeting vector. This final plasmid contains in this order an upstream arm of 4259 nt, a 1888 nt long floxed pgk neo cassette, the 218 nt long 3’ terminal end of Tcf7 intron 9, exon 10 with the P2A ribosomal skipping peptide inserted after the ..FLPMTVL sequence (total of 310 nt), the YFP sequence (720 nt), the additional downstream part of exon10 (1159 nt) and additional 4039 nt intronic sequence (total downstream arm is 5198 nt). C57Bl6N ES cells (strain EAP6, developed by Tobias Raabe, Jean Richa, Klaus Kaestner and Eric Pierce as members of the Penn Knockout Mouse Project) were electroporated with this vector, selected by G418 (geneticin) and 196 resulting ES clones were screened by Southern blotting, resulting in several positive clones. After ascertaining correct karyotype by chromosome counting, independent positive ES cell clones were injected into C57BL/6 blastocysts at the Penn Transgenic and Chimeric Mouse Facility, yielding 100% C57BL/6 background male chimeras, which were bred to B6 females. Tcf7YFP mice are described in Supplementary Fig. 3. Tcf7EGFPnull mice were generated by breeding the Tcf7EGFP mice4 with CMV-Cre mice and are described in Supplementary Fig. 6. Mice used were 6–10 weeks old and of either sex. Animal procedures were approved by relevant NIH Animal Care and Use Committees.
Antibodies and flow cytometry
BM cell suspensions were incubated with a mix of purified rat, mouse and hamster IgG before addition of specific antibodies. Antibodies specific for Ly-6D (49H4), B220 (RA3–6B3), CD19 (1D3), Mac-1 (M1/70), Gr-1 (8C5), CD11c (N418), Ter119 (TER119), NK1.1 (PK136), CD3ε (2C11), CD8α (53–6.72), CD8β (H35–17.2), CD4 (GK1.5), TCRβ (H57), TCRγδ (GL-3), MHC-II (M5/114.15.2), CD205 (205yekta), Kit (2B8), Sca-1 (D7), Thy-1.2 (53–2.1), α4β7 (DATK32), IL-7Rα (A7R34), ICOS (C398.4A), 2B4 (eBio244F4), CD24 (M1/69), CD172a (P84), Ly-6C (HK1.4), Siglec-H (eBio440c), CD25 (PC61.5), CD45.1 (A20), CD45.2 (104), TOX (TXRX10), PLZF (Mags.21F7), GATA-3 (TWAJ), NFIL3 (S2M-E19), IRF-8 (V3GYWCH) were from eBioscience, anti-NRP-1 (3E12), CD122 (TM-β1), and PU.1 (7C2C34) were from Biolegend, anti-Flt3 (A2F10) was from BD, anti-CCR2 (475301) was from R&D, and anti-TCF-1 (C63D9 and C46C7) were from Cell Signaling. The ILC lineage ‘cocktail’ (LinILC) is a mix of the following antibodies: anti-Ly-6D, B220, CD19, Mac-1, Gr-1, CD11c, Ter119, NK1.1, CD3ε, CD8α, CD8β, CD4, TCRβ and TCRγδ. The DC lineage ‘cocktail’ (LinDC) is a mix of the following antibodies: anti-B220, CD19, Thy-1, Mac-1, Gr-1, Ter119, NK1.1, CD3ε, CD8α, CD8β, CD4, TCRβ and TCRγδ. TOX, PLZF, GATA-3, NFIL3, PU.1, IRF-8 and TCF-1 expression were detected by intracellular staining using eBioscience’s transcription factor staining buffer set according to the manufacturer’s instructions. The antibody C63D9 was used for TCF-1 detection unless specified otherwise. Live/dead discrimination was performed by staining with DAPI or LIVE/DEAD Fixable Blue (Invitrogen). Samples were acquired using an LSRFortessa flow cytometer (BD) and analyzed using FlowJo software (Tree Star). All analyses are presented on singlet live cells as showed in Supplementary Fig. 1. GFP/YFP separation was achieved using the filters 509/21, 505LP and 530/30, 525LP. BM progenitors were sorted using an Aria flow cytometer (BD).
BM progenitor definition, isolation and culture
Unless specified otherwise, BM progenitors were defined and isolated by flow cytometric sort as following: ALPs (LinILC−Kit+2B4+α4β7−Flt3hiIL-7Rα+, see Supplementary Fig. 1 and Fig. 1a). EILPs (LinILC−Kit+2B4+α4β7+Tcf7+Thy1−) and ILCPs (LinILC−Kit+2B4+α4β7+Tcf7+Thy1+IL-7Rα+) were isolated from Tcf7EGFP or Tcf7YFP mice. IL-7Rα− LMPPs (LinILC−KithiSca-1+Flt3hi2B4+α4β7−IL-7Rα−). EILPs were further separated in Tcf7YFPZbtb16GFP mice as Flt3hiZbtb16-GFPlo sEILPs and Flt3loZbtb16-GFPhi cEILPs as shown in Fig. 2g, or by intracellular staining as Flt3hiPLZFlo sEILPs and Flt3loPLZFhi cEILPs as shown in Fig. 3b. Alternatively, sEILPs could be defined without Zbtb16 expression, as Flt3hiTcf7lo as show in Fig. 4d. sEILP1s and sEILP2s were separated using NRP-1 and MHC-II expression on sEILPs as shown in Fig. 4b. Canonical precursors were divided into subsets using previously described markers33,34. Pre-DC are defined as LinDC-Flt3+CD11c+MHC-IIint/lo, and pre-cDC are Siglec-H− pre-DC (Supplementary Fig. 5a). Pre-cDC can be further separated into Kit+Ly-6C− pre-cDC1 and Kit−Ly-6C+ pre-cDC2 (Supplementary Fig. 5b). BM progenitors were cultured on irradiated OP9 stromal layers in α-MEM media supplemented with 20% FBS, glutamine, penicillin and streptomycin. All cultures were supplemented with SCF, Flt3-L and IL-7 at 30ng/mL, called SF7 condition. GM-CSF and IL-3 (10ng/mL) were added in SF7-GM3 cytokine condition, and M-CSF, G-CSF, and IL-6 (10ng/mL) were additionally used in SF7-GM3-MG6 cytokine condition. All cytokines were purchased from PeproTech. CD45.2+ cells were considered for analysis of hematopoietic progeny.
Single-cell RNA-seq and analysis
B cells isolated from spleen were used to spike in the scRNA-seq samples. scRNA-seq libraries were prepared using the Chromium Single Cell 3’ kits, according to the manufacturer’s instructions (v2 chemistry, 10x Genomics). The obtained libraries were sequenced with a NextSeq (v2 chemistry, Illumina). Primary analysis was performed with the Cellranger v2.0.1 software using the default parameters. Median number of UMI counts ranged between 2,894 – 5,309 per cell. Cells with low UMI counts are determined by the 10x Genomics Cell Ranger Algorithm (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/algorithms/overview). After estimating the maximum total UMI count (or m) as the 99th percentile of the top 3000 barcodes, those whose UMI count exceeds m/10 are called cells. The data were normalized, scaled, and cell cycle scores and mitochondrial percentages were regressed using the Seurat package35. The Seurat’s PCAFast function was run and the first 13 principal components were used in subsequent analysis. Cells were clustered using Seurat’s FindClusters function. To visualize the data, t-distributed stochastic neighbor embedding (t-SNE) plots were generated using the Seurat’s RunTSNE function. B cells spiked in all samples formed their own cluster and overlapped on the t-SNE plot, thus confirming the validity of the normalization across samples. Small contaminant populations were identified based on signature gene expression of mature populations from BM13. Such cells formed their own clusters, and appeared distant from the main progenitor populations on t-SNE. B cells and contaminant cells that represented 5% of the data were electronically removed prior to subsequent analysis. Seurat’s FindClusters function was used to cluster the remaining data using default perplexity. A range of resolutions were tested, and a resolution of 0.65 was selected to capture known ILC progenitor groups and new subgroups without adding subdivisions within relatively homogenous ALPs. Differentially expressed genes between clusters were identified using Seurat’s FindMarkers function. Only genes showing expression in at least 10% of cells in one of the groups compared were considered. A two-sided Wilcoxon rank-sum test was used to determine significance. Genes significantly different between clusters are shown in Supplementary Table 1. Pseudo-time reconstruction analysis was performed on the first 5 principal components using the TSCAN package9. Violin plots shown in Figures were made using Seurat’s log transformed normalized data. Correlation network analysis along the scRNA-seq pseudo-time was performed using the LEAP package applied to the most variable genes expressed by at least 10% of the cells (1094 genes)17. A lag of 1 third of the pseudo-time was allowed between correlated genes. Correlations >= 0.2 were considered for further analysis (FDR < 5X10−5) and are shown in Supplementary Table 5. Network visualization was done using Cytoscape (Supplementary Fig. 7a)36. Additional analysis and visualization was done using R37.
Bulk RNA-seq and analysis
RNA was extracted using RNeasy plus micro kit (Qiagen) according to the manufacturer’s instructions. Quality control was performed by bioanalyzer (Agilent), and RNA samples with a RIN > 9 were subsequently used. mRNA sequencing libraries were prepared using the SMARTer Ultra Low Input RNA Kit v3 (Clontech) and Nextera XT DNA library preparation kit (Illumina). Paired-end sequence reads of 126bp were generated by a HiSeq2500 sequencer (Illumina). The raw RNA-Seq FASTQ reads were aligned to mouse genome (mm10) using STAR (STAR_2.5.2b) on 2-pass mode with mouse gencode (release M12) gtf38. Genes were subsequently counted using Rsubread39 and further analyzed for gene expression changes and statistics using limma-voom40 with quantile normalization and batch correction using ComBat41. The gene and sample-specific normalization factors were then used to correct counts and to generate bigwig files. Visualization was done using R37.
DNase-seq, ChIC-seq and analysis
DNase-seq: DNase-seq assays were performed as described42. Briefly, 300 cells from each cell type were collected by FACS sorting. 0.3 unit of DNase I (Roche, 04716728001) was added to each cell type and incubated at 37 °C for 5 minutes. Reactions were stopped by adding 80 μl of stop buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl, 0.15% SDS, 10 mM EDTA) containing 1 μl of 20 mg/ml proteinase K. Samples were incubated at 55 °C overnight, and DNA was purified by phenol–chloroform extraction followed by precipitation with ethanol in the presence of 20 μg glycogen. DNA was further processed for library production.
Small cell number ChIC-seq (Small cell number chromatin immunocleavage sequencing): ChIC-seq was performed as described18. Cells were fixed by adding 1:15 volume of 16%w/v formaldehyde solution (Thermo Fisher Scientific) and incubating at RT for 10 minutes. Reaction was terminated by adding 1:10 volume of 1.25 M glycine and incubating on ice for 5 minutes. The fixed cells were washed by ice cold PBS twice, and collected by centrifugation. 1ml RIPA buffer (10 mM Tris-Cl, 1 mM EDTA, 0.1% sodium deoxycholate, 0.2% sodium dodecyl sulfate, 1% Triton X-100) was added to each sorted 500 ALP, EILP, and ILCP cells, and incubated at room temperature for 30 minutes. Cells were rinsed with 500 μl binding buffer (10 mM Tris-Cl, 1 mM EDTA, 150 mM sodium chloride, 0.1% Triton X-100) twice and re-suspended in 50 μl binding buffer. To prepare anti-TCF-1 bound PA-MNase (Ab+PA-MNase), anti-TCF-1 (C46C7, Cell Signaling) and the PA-MNase at a molecular ratio of 1:2 were pre-incubated at 4 °C for 30 minutes in 50 μl binding buffer. TCF1+PA-MNase were add to 50 μl binding buffer re-suspended cells and incubated one hour at 4 °C with rotation. Cells were washed using 200 μl wash buffer (10 mM Tris-Cl, 1 mM EDTA, 150 mM sodium chloride, 0.1%w/v SDS, 0.1%w/v sodium deoxycholate and 1%v/v Triton X-100) three times and pelleted by centrifugation at 600g for 2 minutes. Next, cells were rinsed using 200 μl rinsing buffer (10 mM Tris-Cl and 10 mM sodium chloride, 0.1%v/v Triton X-100). The MNase digestion was initiated by re-suspending the rinsed cells in 40 μl reaction solution buffer (10 mM Tris-Cl, 10 mM sodium chloride, 0.1%v/v Triton X-100, 2 mM CaCl2) and incubating at 37 °C for 3 minutes. The reaction was stopped by adding 80 μl stop buffer (20 mM Tris-Cl (pH 8.0), 10 mMethylenedioxy-bis-(ethylenenitrilo)-tetraacetic acid (EGTA), 20 mM sodium chloride, 0.2%w/v SDS) and 1 μl proteinase K (SigmaAldrich), then incubating at 65 °C for overnight. DNAs were purified by using phenol-chloroform extraction and ethanol precipitation. The purified DNA was further processed for library production.
Library preparation: Libraries were prepared according to Illumina’s instructions. Briefly, DNA was end-repaired using a combination of T4 DNA polymerase, E. coli DNA Pol I large fragment and T4 polynucleotide kinase. The blunt, phosphorylated ends were treated with Klenow fragment (3’ to 5’ exo minus) and dATP to yield a protruding 3- ‘A’ base for ligation of Illumina’s adapters which have a single ‘T’ base overhang at the 3’ end. The ligation reaction was performed by adding 2 μM Illumina’s adapters to each sample and incubated at RT one hour. After adapter ligation DNA was PCR amplified with Illumina primers for 16 cycles and library fragments from 180bp to 300bp were isolated from an agarose gel. The purified DNA was captured on an Illumina flow cell for cluster generation. Libraries were sequenced on Hi-seq3000 following the manufacturer’s protocols.
Analysis: Sequencing reads were aligned against mm9 reference genome using Bowtie 243 with default parameters. Reads from technical replicates and biological replicates were combined for peak calling. Duplicated reads were removed from further analysis. TCF1 peaks were called using MACS244 with default parameters. Motif discovery (Fig. 7b), peak file annotation (Supplementary Table 6) and additional analysis was done with HOMER (version 4.10.1)22. Biological process enrichment and visualization was done with Metascape (Fig. 7d)45. Additional visualization was done using R37.
Statistics
Statistical analysis was performed on groups with limited variance using Excel or Prism. Differences between groups of mice or wells were determined by a two-tailed unpaired Student’s t-test. A Welsh correction was applied for groups of unequal SD. p < 0.05 was considered significant. Sample sizes were empirically determined, no samples or animals were excluded from the analysis, no randomization or blinding was used.
Data availability statement
The accession number for the raw data of the RNA-seq is GSE113767. The accession number for the raw data of the DNase-seq and ChIC-seq is GSE128483. All other relevant data are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We thank H.-R. Rodewald for sharing Il7r-iCre mice and J. Richa from the Transgenic and Chimeric Mouse Facility, University of Pennsylvania, for injection of Tcf7YFP ES cells into mouse blastocysts. We thank T. Ciucci, R. Bosselut, J. Chen, the CCR Sequencing facility, the CCR flow cytometry core facility, and the DNA sequencing facility of the University of Pennsylvania for technical support. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Footnotes
REFERENCES
- 1.Harly C, Cam M, Kaye J, Bhandoola A. Development and differentiation of early innate lymphoid progenitors. The Journal of experimental medicine 2018, 215(1): 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeevan-Raj B, Gehrig J, Charmoy M, Chennupati V, Grandclément C, Angelino P, et al. The Transcription Factor Tcf1 Contributes to Normal NK Cell Development and Function by Limiting the Expression of Granzymes. Cell Report 2017, 3(20): 613–626. [DOI] [PubMed] [Google Scholar]
- 3.Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature 2011, 476(7358): 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Q, Li F, Harly C, Xing S, Ye L, Xia X, et al. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nature immunology 2015, 16(10): 1044–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity 2012, 37(4): 634–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klose CS, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, et al. Differentiation of Type 1 ILCs from a Common Progenitor to All Helper-like Innate Lymphoid Cell Lineages. Cell 2014, 157(2): 340–356. [DOI] [PubMed] [Google Scholar]
- 7.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature 2014, 508(7496): 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu Y, Tsang JC, Wang C, Clare S, Wang J, Chen X, et al. Single-cell RNA-seq identifies a PD-1hi ILC progenitor and defines its development pathway. Nature 2016, 539(7627): 102–106. [DOI] [PubMed] [Google Scholar]
- 9.Ji Z, Ji H. TSCAN: Pseudo-time reconstruction and evaluation in single-cell RNA-seq analysis. Nucleic Acids Res 2016, 44(13): e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy TL, Grajales-Reyes GE, Wu X, Tussiwand R, Briseno CG, Iwata A, et al. Transcriptional Control of Dendritic Cell Development. Annu Rev Immunol 2016, 34: 93–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grajales-Reyes GE, Iwata A, Albring J, Wu X, Tussiwand R, Kc W, et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8alpha(+) conventional DC clonogenic progenitor. Nature immunology 2015, 16(7): 708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlitzer A, Sivakamasundari V, Chen J, Sumatoh HR, Schreuder J, Lum J, et al. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nature immunology 2015, 16(7): 718–728. [DOI] [PubMed] [Google Scholar]
- 13.Heng TS, Painter MW, Immunological Genome Project C. The Immunological Genome Project: networks of gene expression in immune cells. Nature immunology 2008, 9(10): 1091–1094. [DOI] [PubMed] [Google Scholar]
- 14.Schlenner SM, Madan V, Busch K, Tietz A, Laufle C, Costa C, et al. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity 2010, 32(3): 426–436. [DOI] [PubMed] [Google Scholar]
- 15.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature 2008, 452(7188): 764–767. [DOI] [PubMed] [Google Scholar]
- 16.Morrison SJ, Wandycz AM, Hemmati HD, Wright DE, Weissman IL. Identification of a lineage of multipotent hematopoietic progenitors. Development 1997, 124(10): 1929–1939. [DOI] [PubMed] [Google Scholar]
- 17.Specht AT, Li J. LEAP: constructing gene co-expression networks for single-cell RNA-sequencing data using pseudotime ordering. Bioinformatics 2017, 33(5): 764–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ku WL, Nakamura K, Gao W, Cui K, Hu G, Tang Q, et al. Single-cell chromatin immunocleavage sequencing (scChIC-seq) to profile histone modification. Nature methods 2019, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Zhang JA, Dose M, Kueh HY, Mosadeghi R, Gounari F, et al. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood 2013, 122(6): 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohmura S, Mizuno S, Oishi H, Ku CJ, Hermann M, Hosoya T, et al. Lineage-affiliated transcription factors bind the Gata3 Tce1 enhancer to mediate lineage-specific programs. J Clin Invest 2016, 126(3): 865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sen DR, Kaminski J, Barnitz RA, Kurachi M, Gerdemann U, Yates KB, et al. The epigenetic landscape of T cell exhaustion. Science 2016, 354(6316): 1165–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell 2010, 38(4): 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang D, Diao H, Getzler AJ, Rogal W, Frederick MA, Milner J, et al. The Transcription Factor Runx3 Establishes Chromatin Accessibility of cis-Regulatory Landscapes that Drive Memory Cytotoxic T Lymphocyte Formation. Immunity 2018, 48(4): 659–674 e656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinke FC, Yu S, Zhou X, He B, Yang W, Zhou B, et al. TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4(+) T cell fate and interact with Runx3 to silence Cd4 in CD8(+) T cells. Nature immunology 2014, 15(7): 646–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feyerabend TB, Terszowski G, Tietz A, Blum C, Luche H, Gossler A, et al. Deletion of Notch1 converts pro-T cells to dendritic cells and promotes thymic B cells by cell-extrinsic and cell-intrinsic mechanisms. Immunity 2009, 30(1): 67–79. [DOI] [PubMed] [Google Scholar]
- 26.Wu L, D’Amico A, Hochrein H, O’Keeffe M, Shortman K, Lucas K. Development of thymic and splenic dendritic cell populations from different hemopoietic precursors. Blood 2001, 98(12): 3376–3382. [DOI] [PubMed] [Google Scholar]
- 27.Shigematsu H, Reizis B, Iwasaki H, Mizuno S, Hu D, Traver D, et al. Plasmacytoid dendritic cells activate lymphoid-specific genetic programs irrespective of their cellular origin. Immunity 2004, 21(1): 43–53. [DOI] [PubMed] [Google Scholar]
- 28.Ishikawa F, Niiro H, Iino T, Yoshida S, Saito N, Onohara S, et al. The developmental program of human dendritic cells is operated independently of conventional myeloid and lymphoid pathways. Blood 2007, 110(10): 3591–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothenberg EV, Kueh HY, Yui MA, Zhang JA. Hematopoiesis and T-cell specification as a model developmental system. Immunol Rev 2016, 271(1): 72–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farrell JA, Wang Y, Riesenfeld SJ, Shekhar K, Regev A, Schier AF. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 2018, 360(6392). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, et al. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature 1995, 374(6517): 70–74. [DOI] [PubMed] [Google Scholar]
- 32.Aliahmad P, Kaye J. Development of all CD4 T lineages requires nuclear factor TOX. The Journal of experimental medicine 2008, 205(1): 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naik SH, Sathe P, Park HY, Metcalf D, Proietto AI, Dakic A, et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nature immunology 2007, 8(11): 1217–1226. [DOI] [PubMed] [Google Scholar]
- 34.Onai N, Obata-Onai A, Schmid MA, Ohteki T, Jarrossay D, Manz MG. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nature immunology 2007, 8(11): 1207–1216. [DOI] [PubMed] [Google Scholar]
- 35.Satija R, Butler A, Hoffman P. Seurat: Tools for Single Cell Genomics. R package version 2.3.0. https://CRAN.R-project.org/package=Seurat. 2018.
- 36.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research 2003, 13(11): 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Team RC. R: A Language and environment for Statistical Computing (R Foundation for Statistical Computing; ). 2014. [Google Scholar]
- 38.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29(1): 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 2013, 41(10): e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015, 43(7): e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leek JT, Johnson WE, Parker HS, Fertig EJ, Jaffe AE, Storey JD, et al. sva: Surrogate Variable Analysis. R package version 3.26.0. 2017.
- 42.Cooper J, Ding Y, Song J, Zhao K. Genome-wide mapping of DNase I hypersensitive sites in rare cell populations using single-cell DNase sequencing. Nat Protoc 2017, 12(11): 2342–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods 2012, 9(4): 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome biology 2008, 9(9): R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, et al. Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell host & microbe 2015, 18(6): 723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the raw data of the RNA-seq is GSE113767. The accession number for the raw data of the DNase-seq and ChIC-seq is GSE128483. All other relevant data are available from the corresponding author upon request.