Abstract
The Plasmodium vivax Duffy binding protein (DBP) is a prime target of the protective immune response and a promising vaccine candidate for P. vivax malaria. Naturally acquired immunity (NAI) protects against malaria in adults residing in infection-endemic regions, and the passive transfer of malarial immunity confers protection. A vaccine that replicates NAI will effectively prevent disease. Here, we report the structures of DBP region II in complex with human-derived, neutralizing monoclonal antibodies obtained from an individual in a malaria-endemic area with naturally acquired immunity. We identified protective epitopes by X-ray crystallography, hydrogen-deuterium exchange mass spectrometry, mutational mapping, and P. vivax invasion studies. These approaches reveal that naturally-acquired human antibodies neutralize P. vivax by targeting the DARC–binding site and dimer interface of P. vivax DBP. Antibody binding is unaffected by polymorphisms in the vicinity of epitopes, suggesting the antibodies have evolved to engage multiple polymorphic variants of DBP. The human antibody epitopes are broadly conserved and are distinct from previously defined epitopes for broadly conserved murine mAbs. A library of globally conserved epitopes of neutralizing human antibodies opens new horizons for rational design of strain-transcending DBP-based vaccines and therapeutics against P. vivax.
Malaria remains a life threatening disease causing high morbidity and mortality1. Despite the long history of human battle against malaria, a viable vaccine is still desperately needed but remains elusive. This is in spite of the recognition that adults in malaria-endemic areas develop relative immunity to malaria infection2–4. Further, passive transfer of gamma globulins isolated from serum of patients exposed to malaria to infected non-immune children less than 5 years of age, demonstrated a significant reduction in parasitemia2,5–7. Defining the structural correlates of naturally acquired immunity is a critical factor for the design of a universal vaccine.
The P. vivax Duffy binding protein (DBP) is the most promising vaccine candidate for P. vivax malaria8–21. During reticulocyte invasion, P. vivax uses a Duffy Binding Like (DBL) domain in DBP also known as region II (DBP-II) to engage the Duffy Antigen Receptor for Chemokines (DARC) on host reticulocytes8–12,14–18. DBP-II binds DARC via receptor-induced ligand dimerization, sandwiching DARC residues 19–30 between two DBP-II molecules17,18. DBP-II is comprised of three subdomains (1 to 3), and subdomain 2 (SD2) is responsible for dimerization and receptor binding that are required to engage DARC17,18. Rabbit and human antibodies that block the DBP:DARC interaction neutralize P. vivax, suggesting a DBP-based vaccine will reduce infection5. However, the successful design of a DBP-II-based vaccine may be limited by strain-specific immune responses due to the polymorphic nature of DBP22,23, and the presence of immunodominant but non-protective epitopes within DBP24,25. Despite the polymorphic nature of DBP, broadly conserved epitopes of three inhibitory murine monoclonal antibodies (mAbs) have been identified in subdomain 3 of DBP-II19. These epitopes are distant from the dimer interface or DARC binding site19. Furthermore, human vaccination with DBP-II elicits antibodies that block the in vitro binding of four alleles of DBP to DARC suggesting broadly-neutralizing epitopes of human antibodies may exist within DBP-II20,21.
The identification of broadly-conserved human neutralizing-antibody epitopes that contribute to naturally acquired immunity is essential for the improved rational design of potent strain-transcending DBP-based vaccines. Here, we present the study of DBP-II with two human neutralizing monoclonal antibodies 053054 and 092096. These human mAbs were produced by sorting individual DBP-II-specific B cells from a Cambodian patient with naturally acquired DBP-II-blocking antibodies and then isolating, sequencing, and cloning the variable regions from human IgG heavy and light chains. Structures of DBP-II antibody complexes were determined by X-ray crystallography, and epitopes further mapped by hydrogen-deuterium exchange mass spectrometry (HDX-MS) and mutational studies. Both antibodies inhibit binding of DBP to red blood cells, and 092096 neutralizes P. vivax in ex vivo experiments. Polysera from patient populations competes with binding of 092096 to DBP. We show that these naturally-acquired human antibodies neutralize P. vivax by targeting the DARC–binding site and dimer interface of P. vivax DBP. This work forms a strong foundation for the rational design of potent strain-transcending DBP-based vaccines against P. vivax.
RESULTS
Isolation of human monoclonal antibodies 053054 and 092096
Igh and Igl PCR products from 98 individual B cells were sequenced from one Cambodian donor, and 16 B cell clonal groups as defined by Ab V heavy chain sequences with the same inferred VH and JH germline sequences, identical CDR3 length, and the same or very similar CDR3 sequences. One or two clones were selected from each group and expressed as full-length IgG1, thereby creating monoclonal antibodies (mAbs). mAbs from eleven clones recognized DBP-II. We selected one mAb from two different clonal groups corresponding to two of the larger clonal groups in terms in the number of DBP-II-specific B cells isolated by single cell sorting. These two mAbs were designated as 092096 and 053054.
Structures of human antibodies 053054 and 092096 in complex with DBP-II
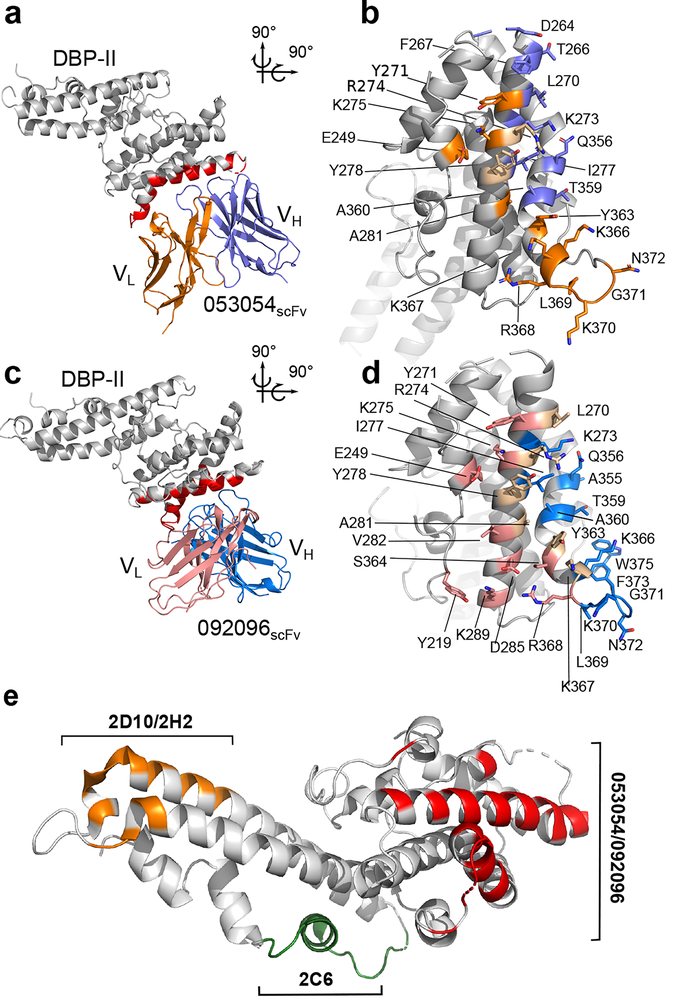
We solved two crystal structures of DBP-II in complex with a single-chain variable fragment (scFv) generated from the human mAbs 053054 and 092096 that were isolated from a Cambodian patient (Fig. 1a,c and Supplementary Table 1). The quality of the electron density maps clearly defined the contact sites of the DBP-II/antibody interface in both structures (Supplementary Fig. 1a,b). Both mAbs bind to the same face of DBP, although the orientation of the heavy and light chains with respect to DBP differs (Supplementary Fig. 2). The interacting residues showed a significant overlap between the two epitopes (Supplementary Table 2).
Figure 1|. Structural definition of human antibody epitopes in DBP.
a, Overall structure and epitope for the 053054 complex. Gray - DBP-II. Dark blue - 053054 heavy chain. Orange - 053054 light chain. Red - epitope. b, orthogonal detailed view of the epitope for 053054 in DBP. Gray - DBP-II. Dark blue – DBP residues contacted by the 053054 heavy chain. Orange – DBP residues contacted by the 053054 light chain. Beige – DBP residues contacted by both heavy and light chains. c, Overall structure and epitope for the 092096 complex. Gray - DBP-II. Light blue - 092096 heavy chain. Pink - 092096 light chain. Red - epitope. d, orthogonal detailed view of the epitope for 092096 in DBP. Gray - DBP-II. Light blue – DBP residues contacted by the 092096 heavy chain. Pink – DBP residues contacted by the 092096 light chain. Beige – DBP residues contacted by both heavy and light chains. e, Comparison of human and murine epitopes19 in DBP-II reveal epitopes are distinct. Orange - epitope of inhibitory murine mAbs 2D10/2H2. Green - epitope of inhibitory murine mAb 2C6. Red- epitopes of neutralizing human mAbs 053054 and 092096.
The discontinuous conformational epitope for 053054 is composed of amino acids D264-A281 and Q356-N372 belonging to two outer helices of subdomain 2, and E249 of the N-terminal helix in subdomain 1 (Fig. 1a,b and Supplementary Table 2). mAb 092096 binds to the discontinuous conformational epitope comprised of residues L270-K289, A355-W375, E249 and Y219 (Fig. 1c,d and Supplementary Table 2). All complementarity determining regions (CDRs) of both antibodies contact DBP (Supplementary Table 2), and the buried surface area and shape complementarity of the interactions are within standard parameters (Supplementary Fig. 1c,d). Strikingly, these epitopes differ from previously reported epitopes for broadly conserved inhibitory murine mAbs located in SD3 of DBP-II (Fig. 1e). The overlap of human epitopes in DBP coupled with distinct modes of binding indicate multiple avenues for antibody engagement of the surface comprised of residues D264-K289 and Q356-W375 in DBP.
Mechanism of neutralization
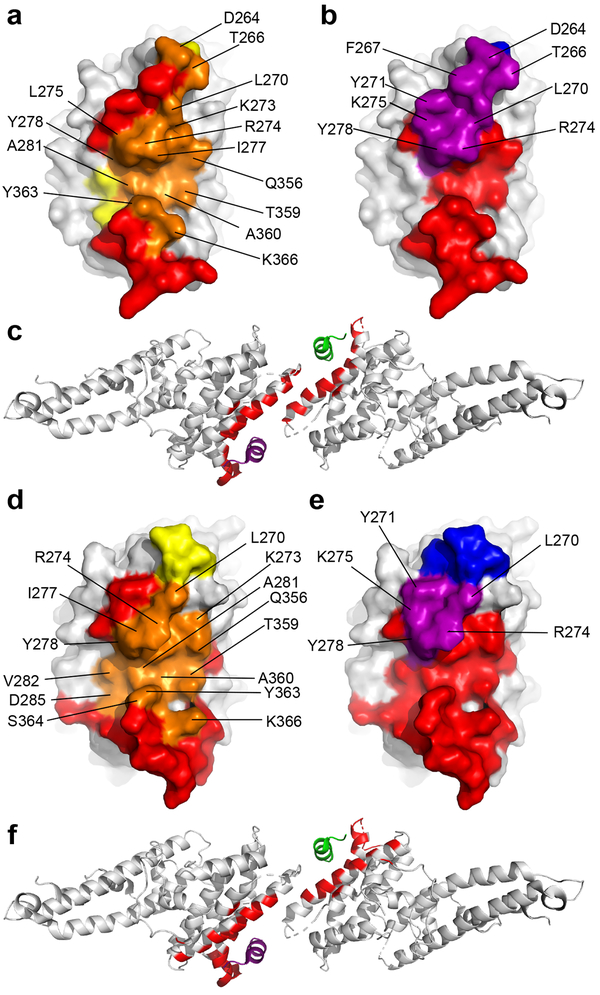
Binding of 053054 or 092096 does not cause conformational changes within DBP-II as there are negligible structure differences between antibody-bound, unbound and DARC-bound DBP (Supplementary Table 3). However, structural comparison of DBP-II/053054 and DBP-II/092096 with DBP-II/DARC complex revealed that mAbs 053054 and 092096 engage residues in the DARC-binding cleft and dimer-interface of DBP-II (Fig. 2). DBP uses residues F261-T266, L270-K289 and Q356-K367 to engage DARC ectodomain and residue range F261-F267, L270-Y278 and E352-Q356 to form a dimer interface18. These segments overlap considerably with the 053054 and 092096 epitopes suggesting that antibody association prevents DBP dimerization and DARC binding (Fig. 2c and 2f).
Figure 2|. Neutralizing mAbs 053054 and 092096 block DARC-binding site and dimer interface.
a, Overlay of the 053054 epitope with the DARC-binding residues in DBP. Orange - overlap between the 053054 epitope and the DARC binding site. Yellow/Orange - DARC binding site. Red/Orange –the 053054 epitope. b, Overlay of 053054 epitope with the dimer interface of DBP. Purple - overlap between the 053054 epitope and the dimer interface. Blue/Purple - dimer interface. Red/Purple –the 053054 epitope. c, Mapping the 053054 epitope on the tetramer structure of DARC-bound DBP reveals the epitope overlaps with the DARC binding residues and dimer interface. Grey – DBP. Green – DARC monomer 1. Purple – DARC monomer 2. Red – epitope of 053054. d, Overlay of the 092096 epitope with the DARC-binding residues in DBP. Orange - overlap between the 092096 epitope and the DARC binding site. Yellow/Orange - DARC binding site. Red/Orange –the 092096 epitope. e, Overlay of 092096 epitope with the dimer interface of DBP. Purple - overlap between the 092096 epitope and the dimer interface. Blue/Purple - dimer interface. Red/Purple –the 092096 epitope. f, Mapping the 092096 epitope on the tetramer structure of DARC-bound DBP reveals the epitope overlaps with the DARC binding residues and dimer interface. Grey – DBP. Green – DARC monomer 1. Purple – DARC monomer 2. Red – epitope of 092096.
Human antibodies 053054 and 092096 block DBP binding to RBCs and neutralize P. vivax invasion ex vivo
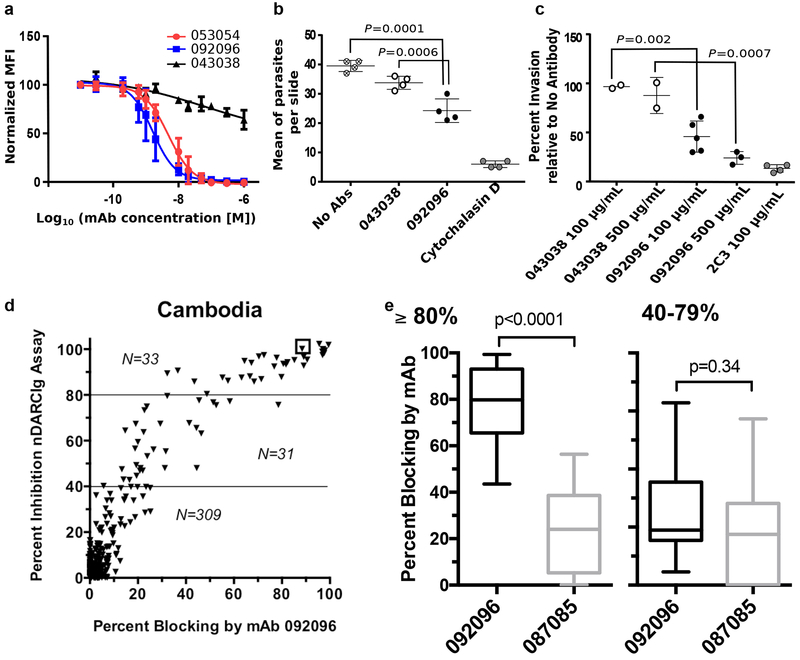
The structural studies suggest that the mAbs 053054 and 092096 directly block DBP binding to DARC on red blood cells and would, therefore, neutralize P. vivax merozoites. To test this hypothesis, we examined inhibition of DBP-II binding to RBCs (Fig. 3a). Both human mAbs completely blocked the binding with IC50 values for 053054 and 092096 as 4.88 ± 1.06 nM and 1.63 ± 1.07 nM, respectively while an isotype control, 043038, could not inhibit binding. 092096 was chosen for its higher potency to test the human mAb’s ability to neutralize P. vivax invasion of human reticulocytes ex vivo. Using P. vivax-infected erythrocytes obtained from Brazilian patients we showed that 092096 neutralizes P. vivax invasion with 43% invasion inhibition at a human mAb concentration of 100 μg/mL compared to no antibody (p = 0.0001) (Fig. 3b). We further performed the P. vivax ex vivo assay with Cambodian clinical isolates to assess the ability of this mAb to neutralize parasites from distinct geographical locations. Compared to no antibody 092096 neutralizes P. vivax invasion with 47.4% neutralization at a human mAb concentration of 100 μg/mL (p =0.002) and 86.6% neutralization at a human mAb concentration of 500 μg/mL (p = 0.0007) (Fig. 3c). Note that the degree of invasion inhibition is fairly consistent for the different parasite isolates. This result indicates a single human mAb that targets the receptor-binding residues and the dimer interface of DBP can neutralize invasion of multiple P. vivax isolates, from distinct geographical locations.
Figure 3|. Human antibodies block DBP binding to RBCs and P. vivax invasion.
a, Inhibition of DBP-II binding to RBCs. The isotype matched mAb 043038 was used as a negative control. Data shown are mean ± SD of three biological replicates. b, Neutralization data for a Brazilian isolate of P.vivax invasion of human reticulocytes by human mAb 092096. Individual data points represent the total number of P. vivax infected cells per 20,000 RBCs and are shown as mean ± 1 SEM, for one P. vivax isolate performed in four biological replicate cultures in one experiment. Statistical differences between 092096 and no invasion inhibitor control (No Abs = medium alone) and non-inhibitory mAb 043038 were analyzed by one-way ANOVA and Dunnett’s test (p =0.0001 and 0.0006, respectively). Cytochalasin D, an actin inhibitor, served as a positive control for the inhibition of invasion. c, Neutralization data for Cambodian isolates of P.vivax invasion of human reticulocytes by human mAb 092096. Each data point represent results from one of five P. vivax isolates and the results presented as mean ± 1 SEM. These data represent results from five separate experiments, each with a different P. vivax isolate. All experiments had no Ab control and 4 out of 5 experiments used mouse anti-DARC monoclonal antibody 2C3 as a positive control and one experiment used heparin as positive control (11% invasion relative to no antibody, not shown in figure). The other controls were performed with only some experiments because of limited number of parasites available for different culture conditions. The isotype matched mAb 043038 serves as an additional negative control. Statistical comparison between non-inhibitory mAb 043038 (n=2) and neutralizling mAb 092096 was performed with one-way ANOVA and Dunnett’s test (p =0.002 for 100 μg/mL mAbs concentration, n=5, p =0.0007 for 500 μg/mL mAbs concentration, n=3). d, Human mAbs compete with epitopes recognized by serum from individuals with high levels of DBPII-specific blocking activity (≥80%) residing in Cambodia. Y-axis indicates the level of blocking activity using a DBPII:DARC binding assay, which measures the overall blocking activity in a serum sample. The x-axis is the percentage of blocking activity in the sample for which a given mAb competed. e, The box and whisker plots (median blocking activity, 25% and 75% interquartile ranges, and 95% confidence intervals) examine the degree to which mAbs 092096 and 087085 compete with serum blocking activity stratified as to whether participants had 80–100% blocking activity (n=33 for both mAbs, p<0.0001) versus lower activity of 40–79% (n=31 for both mAbs, p=0.34). The sample size for the two strata is shown in the scatter plots. Difference in blocking activity was examined using two-tailed Mann-Whitney U test.
Neutralizing epitopes are widely recognized in patient populations
We examined whether the epitope recognized by 092096 is widespread in patient populations exposed to P. vivax (Fig. 3d, e). Functional mAbs found in individuals with high levels (≥80%) of blocking antibody activity to DBPII who resided in P. vivax-endemic regions of Cambodia recognize similar epitopes as 092096 (Fig. 3d). For example, the serum sample at the top right in Fig. 3d (in small box) shows 100% blocking activity of DBP-II binding to DARC N-terminus at a dilution of 1:20, and mAb 092096 (dilution of 1:50) competed for 90% of the blocking activity. The ability of mAbs 092096 and 087086 (a non-blocking mAb to DBPII) to compete with polysera stratified as to whether participants had 80–100% blocking activity versus lower blocking activity of 40–79% (Fig. 3e). mAb 092096 demonstrated clear competition with high-blocking activity polysera. These data demonstrate the epitopes identified here are widely recognized by sera in patient populations.
Mutations and ELISA reveal overlapping binding sites with different binding requirements
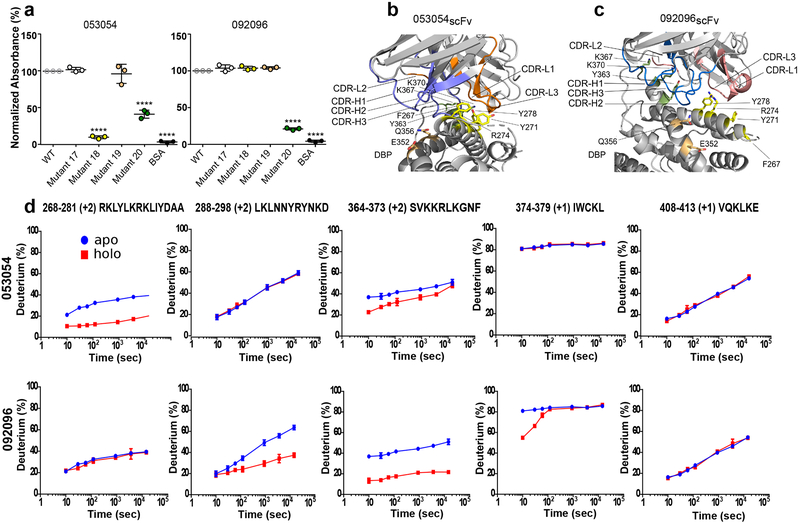
The epitopes were evaluated using surface mutant libraries of DBP-II and ELISA. Both mAbs 053054 and 092096 recognized wild type Sal-1 DBP-II and mutants 17 and 19, with substituted residues outside either epitope, equally well (Fig. 4a, and Supplementary Fig. 3). In contrast, mAb 053054 did not bind to mutants 18 and 20 as these mutants contain mutations in the epitope for 053054. Consistent with overlapping epitopes between the two mAbs, mAb 092096 also lost binding to mutant 20. Mutant 18, however, had no effect on binding to 092096. This divergence in binding to mutant 18 is caused by interaction differences due to the altered orientation of heavy and light chains of 053054 and 092096 with respect to DBP (Fig. 4b,c and Supplementary Fig. 2). Although residues substituted in mutant 18, F267, Y271, K274 and Y278, are bound to the core of 053054 and interact with CDR3 on both the heavy and light chains (Fig. 4b), they only bind to the periphery of 092096 and predominantly interact with its light chain (Fig. 4c). On the contrary, residues Y363, K367 and K370 substituted in mutant 20 interact with the CDR3 on the heavy chain in 092096 and contact the core of 053054 through CDRs 1 and 2 of its light chain. Therefore, this region of DBP is crucial for binding to both mAbs.
Figure 4|. Mutant ELISAs and HDX-MS reveal overlapping but distinct binding modes.
a, Evaluation of 053054 and 092096 binding to DBP Sal1 and four mutants by ELISA. Mutant 17: T257G, D258G, T259S, N260G, F261G, H262S. Mutant 18: F267A, Y271A, R274A, Y278A. Mutant 19: E352A, Q356A. Mutant 20: Y363A, K367A, K370A. Data are shown as mean ± 1 SD, calculated from three independent experiments with three technical replicates each. Statistical differences analyzed by one-way ANOVA and Dunnett’s test (****p < 0.0001). b, c, Position of DBP-II surface mutants tested in (a) with regards to the individual mAbs. The antibodies were structurally aligned in (b) and (c) (see also Supplementary Fig. 2) to show the different locations of the mutations with respect to each complex. DBP-II and antibody colored gray. Mutant 18 – yellow. Mutant 19 – salmon, Mutant 20 - green. d, HDX-MS kinetics for five DBP peptides in the presence (red) or absence (blue) of 053054 or 092096. Peptide sequence and charge state is shown (see also Supplementary Fig. 4 for full peptide coverage of DBP). The HDX data were determined in duplicate, and data are presented as mean +/− SD. For determinations where the deviation is not seen, it is smaller than the size of the data point. The statistical validity of the curves was also assured by making the measurement over seven time points. Given that the purpose of the HDX kinetic plots is to assign binding or no-binding, those assignments were made visually without further statistical analysis.
HDX-MS determination of the epitope for 053054 and 092096
Hydrogen deuterium exchange mass spectrometry (HDX-MS) was used to independently assess the epitopes for mAbs 053054 and 092096 in solution (Fig. 4d). In excellent agreement with the structures, peptide 268–281 is significantly attenuated from exchange upon binding to 053054, whereas no difference is detected for this peptide with 092096. Regions 288–298 and 364–379 show substantial HDX attenuation against 092096 predominantly on peptide 364–373. Peptide 364–373 also responds for 053054 but with less attenuation and converging kinetics indicate weaker binding for 053054. In contrast, peptides 288–298 and 374–379 are silent with 053054 (Fig. 4d). The remaining peptides either show minor differences in deuterium exchange upon antibody binding or no difference including the broadly conserved epitope for murine antibodies 2D10, 2H219 (Fig. 4d, last panel, and Supplementary Fig. 4 and 5).
Polymorphisms within or in the vicinity of the epitopes for 053054 and 092096 do not affect antibody binding to DBP
Having determined the human neutralizing epitopes and the mechanism of antibody neutralization, we examined the DBP variation within epitopes. We first determined antibody affinities for 053054 and 092096 to the reference strain Sal-1 DBP by using bio-layer interferometry (BLI) (Supplementary Table 4 and Supplementary Fig. 6 and 7). The steady state equilibrium dissociation constants were found to be 7.44 ± 0.36 nM for 053054 and 7.63 ± 0.29 nM for 092096, respectively. BLI also provides the association and dissociation rates that can inform the half-life of antibody binding. In both cases, the dissociation rates were remarkably slow, indicating antibody binding leads to a stable, long-lived complex. The slow off rate is consistent with the HDX data that show no convergence of HDX kinetics at long exchange times. These results indicate that the two antibodies have comparable binding affinity.
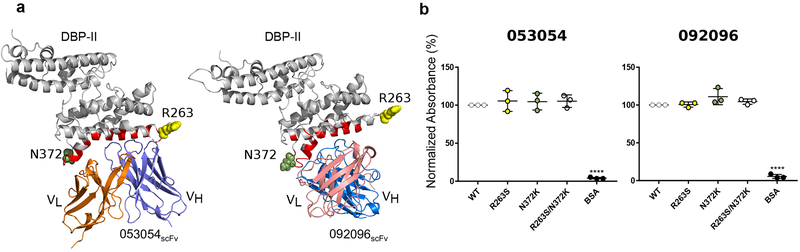
A comparative alignment of 599 DBP sequences from diverse isolates of P. vivax revealed a number of polymorphisms within the epitopes for 053054, 092096, and adjacent residues (Supplementary Table 5). The most common polymorphisms individually and in combination are R263S and N372K. Sal-1, the strain that 053054 and 092096 were isolated and crystalized with, contains R263 and N372 at these positions. R263 is located in a disordered segment at the periphery of the 053054 epitope and therefore is not visible in the crystal structure of the complex (Fig. 5a). R263 is also distant from the epitope of 092096. Nevertheless, to determine the effect of polymorphism within or adjacent to the epitopes on the binding of 053054 and 092096 to DBP, the single mutants and the double mutant were generated and analyzed for the antibody binding by ELISA. All variants retained robust binding to both antibodies (Fig. 5b). We further quantified binding of antibodies to the DBP variants by BLI. Binding affinities showed no difference in equilibrium dissociation constants for the natural DBP variants compared to Sal-I DBP-II (Supplementary Table 4 and Supplementary Fig. 6 and 7). In addition to the most frequent polymorphism, we tested whether the lower frequency polymorphisms L288F, I374M and T359R affected binding by BLI (Supplementary Table 4 and Supplementary Fig. 6 and 7). I374M occurs most frequently in combination with N372K, and T359R occurs most frequently with R263S. Introducing these mutations into DBP also had no effect on antibody affinity or binding parameters. These results demonstrate that the polymorphisms within the epitopes do not affect antibody binding.
Figure 5|. Strain-transcending human mAbs overcome polymorphisms located within the vicinity of their epitopes.
a, Location of polymorphisms R263S and N372K on DBP-II surface shown in DBP-II/053054 and DBP-II/092096 structures. Polymorphic residues 263 and 372 shown in ball representation in yellow and green, respectively. The location of R263 was modeled as this residue is located in a disordered segment that is not visible in either structure. b, ELISA for 053054 and 092096 against Sal-I DBP-II wild type and three DBP-II natural variants, R263S, N372K and double mutant R263S/N372K. Data are shown as mean ± SD, calculated from three independent experiments. Statistical differences between wild type Sal-1 DBP and the mutants were analyzed by one-way ANOVA and Dunnett’s test (****p < 0.0001).
053054 and 092096 heavy and light chain sequences show limited development from the germline genes
The lack of sequence similarity between mAbs 053054 and 092096 (Fig. 6a,b) indicates they do not evolve from a common progenitor. Nevertheless, both mAbs recognize overlapping epitopes in DBP. We analyzed the CDRs of the neutralizing antibodies to DBP to determine how drastic a deviation from germline the paratopes are and how dramatically the antibodies would need to develop. Nucleotide and amino acid sequences were analyzed by NCBI/IgBlast and IMGT/V-Quest to identify the germline V, D and J genes (Fig. 6c–f). The analysis showed that 053054 and 092096 heavy and light chain sequences are very close to the germline genes with a minimal number of junction insertions (Supplementary Table 6). A limited number of somatic hypermutations create direct contacts between the antibodies and DBP: two (T62 and S110) of fourteen contact residues in the 053054 heavy chain; four (E173, H196, T237 and E240) of fifteen contact residues in the 053054 light chain; two (Y108 and F109) of twenty residues in the 092096 heavy chain; and three (R175, P177 and D197) of fourteen residues in the 092096 light chain (Supplementary Table 2). The low number of somatic hypermutations reflects that the mAbs underwent limited affinity maturation to achieve broadly neutralizing activity (Supplementary Table 6).
Figure 6|. Monoclonal antibody maturation.
Sequence identity analysis for 053054 and 092096 mAbs by a, heavy and b, light chain alignment. CDRs 1–3 are marked at the top of the alignment. Similar residues are highlighted gray, identical black. c-f, Amino acid sequence alignment of the variable heavy chain and light chain of the mAbs (c and d) 053054 and (e and f) 092096 against the germline V, D and J gene members with the highest sequence similarity defined by NCBI/IgBlast and IMGT/V-Quest indicated in each alignment. Complementarity determining regions (CDR 1–3) are shown at the top of the alignment. Amino acid mutations are highlighted green. Non-silent somatic hypermutations in CDRs are indicated by black stars. Red triangles mark insertions in the junction region. The secondary structure elements are indicated by blue arrows for β sheets and blue coils for α helices. g, The number of non-silent somatic mutations for CDRs of VH and VL chains of each antibody as well as junction insertions.
DISCUSSION
We found that naturally acquired human antibodies target the DARC-binding site and dimer interface in PvDBP-II. The epitope mapping data are consistent and demonstrate the epitope for mAb 053054 spans residues E249, D264-A281, Q356-N372 of helices belonging to SD2. This epitope overlaps with the epitope for 092096 that comprises residues of Y219, E249, L270-K289 and Q356-W375. These findings result in a clear mechanism for inhibition and neutralization. These human antibodies function by preventing DBP-II binding to the N-terminus of DARC and are neutralizing. Strikingly, we show that a single human mAb 092096 that targets the receptor-binding residues and dimer interface of DBP can neutralize invasion of reticulocytes by multiple P. vivax isolates from distinct geographical locations.
The human mAb epitopes are distinct from broadly neutralizing epitopes of murine mAbs19 and this diversity of epitopes residing in different subdomains of DBP-II is reminiscent of receptor-binding site26 and stem27 epitopes in influenza hemagglutinin. Although 053054 and 092096 mAbs do not share a germ line lineage, they target overlapping epitopes of DBP involved in DARC engagement. Our results suggest the development of neutralizing mAbs can arise from multiple progenitor B cells with straightforward CDR affinity maturation to result in high neutralization and breadth. The epitopes for broadly neutralizing human antibodies presented in this study are, therefore, an excellent platform to develop DBP-II based vaccines.
DBP is highly polymorphic, and this variation may confound vaccine development28,29. Identification of highly conserved epitopes of neutralizing antibodies will have a profound positive effect on the development of a DBP-based vaccine. One caveat of the invasion assays presented here is that the isolates tested were not sequenced to identify the diversity of DBP variants used in the invasion assays. Therefore, we examined the breadth of binding of the antibodies to DBP variants that encompassed polymorphisms within the epitopes. We demonstrate that the most prevalent polymorphisms within or adjacent to the epitopes, as determined by comparative analysis of 599 DBP sequences, were residues N372K, R263S, I374M, L288F, and T359R. As we showed, these polymorphisms individually and in combination do not affect antibody binding to DBP-II. This suggests that human neutralizing antibodies have evolved to compensate for polymorphisms and variation in DBP likely due to repeated exposure to P. vivax, thereby becoming broadly neutralizing antibodies.
A significant challenge for successful vaccination is the need for complex maturation of broadly neutralizing antibodies from their precursors requiring long developmental pathways. For example, broadly neutralizing antibodies that target HIV must undergo dramatic development including multiple rounds of affinity maturation to result in CDRs capable of neutralization breadth and involved extensive lengthening of the CDRs30. This has had a fundamental impact on the immunization scheme necessary to generate suitable broadly neutralizing antibodies for HIV. In contrast, the number of somatic hypermutations seen here reflects that 053054 and 092096 underwent limited affinity maturation for broadly neutralizing activity. This limited development for P. vivax-neutralizing antibodies differs from the extensive development needed for HIV-neutralizing antibodies, and suggests immunization with DBP-based vaccines will likely lead to the ready production of neutralizing mAbs.
The findings from this study of DBP will inform neutralization of and vaccine development for an important family of parasite invasion ligands. DBP is a member of the Erythrocyte Binding like (EBL) family of Plasmodium red cell invasion proteins that includes EBA-17531–37, EBA-14038–44, EBA-18143,45–47, and EBL-148. EBL family members have either a single or double tandemly arranged Duffy Binding Like (DBL) domains that Plasmodium parasites use for receptor-binding and attachment to the host erythrocytes. This study has established that a single monoclonal antibody that targets the receptor binding residues of DBP can neutralize P. vivax parasites, and underscores the importance of targeting the functional receptor-binding residues of other EBL ligands. In addition, DBP and EBA-175 undergo receptor-induced dimerization required for invasion17,18,34,35. mAbs 053054 and 092096 also block the dimer interface of DBP suggesting that antibody disruption of oligomeric interfaces in antigens could be a general method for neutralization. Defining the full complement of mechanisms for neutralizing DBL domains using antibodies will aid in the direct design of highly efficient immunogens against malaria. Although additional receptor-ligand interactions have been identified with proposed roles in P. vivax malaria49,50, a P. vivax vaccine will likely include DBP given the central role of the DBP:DARC interaction in infection.
In conclusion, we studied two human neutralizing monoclonal antibodies 053054 and 092096 that target P. vivax DBP isolated from an individual with naturally acquired immunity to P. vivax and with high levels of blocking antibodies to DBP. These two monoclonal antibodies arose from different clonal groups, bind overlapping epitopes in DBP, and compete with mAbs found extensively in multiple patients with blocking antibody activity to DBP. We characterized their interaction with DBP, confirmed their high neutralizing potential, structurally defined their epitopes and proposed a mechanism for neutralization. This work extends our knowledge of the function of parasitic ligands and methods to leverage the immune response for neutralization. These naturally acquired human antibodies appear to have a short pathway for development with broadly neutralizing capacity. Monoclonal Abs to DBP-II following immunization of mice recognized a distinct set of epitopes that do not target the receptor binding domain. Therefore, current vaccination regiments may not favor targeting the receptor-binding domain and dimer interface of DBP to generate antibodies capable of blocking parasite invasion. Furthermore, while naturally acquired immunity can result in the desired neutralizing antibodies described here, naturally acquired immunity requires repeated exposure to the parasite and develops over the lifetime of a patient. The structure definition of conformational epitopes provides a solid foundation for focusing the immune response to the neutralizing human antibody epitopes identified through immunogen design and structural vaccinology. This work enables the design of immunogens that primarily elicit antibodies to the neutralizing epitopes identified here and to avoid or reduce targeting immunodominant but non-protective epitopes within DBP. Improved immunogens will aid in the design of successful and efficient DBP-based vaccines for malaria.
ONLINE METHODS
Cell staining and sorting of antigen-specific memory B cells
Blood samples were obtained from Cambodians, Brazilians and Papua New Guineans residing Pv endemic areas2,51–53. Samples were screened for blocking Abs to PvDBPII (see below). A subset of Cambodian and Brazilian adults with high levels of blocking Abs to PvDBPII (>80% binding inhibitory activity at a titer 1:10 or higher) donated up to 200 ml of peripheral venous blood. Peripheral blood mononuclear cells (PBMC) were from prepared from the venous blood and cryopreserved. Institutional review boards from the United States National Institutes of Health (NIAID protocol #08-N094, Clinicaltrials.gov NCT00663546), Cambodian Ministry of Health, University Hospital of the University of Sao Paulo (1025/10), National Human Research Ethics Committee of the Ministry of Health of Brazil (551/2010), Medical Research Advisory Counsel of Papua New Guinea (PNGIMR No 1409, PNG MRAC No. 1400, and UH IRB No. 04–14-19) and University Hospitals of Cleveland Medical Center approved the protocols. Written informed consent was obtained from all study participants or their parents/guardians.
Single cells were identified and sorted according to previously described techniques54 from cryopreserved PBMC without activation. B cells from cryopreserved PBMC were enriched using immunomagnetic positive selection with anti-CD19 magnetic MACS beads (MiltenyiBiotec). Cells were washed at least twice with 5 ml FACS buffer with 3 mM EDTA and adjusted to a cell density of 1–2×106 cells/ml. Cells were stained with mouse anti-human CD20 (PE-Cy5.5, Invitrogen) and anti-human IgG Abs (PE-Cy7 clone G18–145; Becton, Dickinson and Co., BD) along with DBPII- or TTCF-prepared tetramers using streptavidin coupled with allophycocyanin (BD) and SYTOX Green Dead Cell Stain (Invitrogen) to gate out dead cells. Stained CD19+ cells were sorted on a BD FACSAria II based on size and complexity and individual DBPII- or TTCF-specific CD20+, IgG+ memory B cells were single cell sorted directly into 4μl mRNA extraction buffer on a cooled 96-well metal block. After cells were collected, plates were frozen immediately on dry ice and stored at −80°C until further processing.
cDNA synthesis
The 96-well plates with single cells were thawed on ice; a cold volume of 7 μl containing 300ng random hexamers (Qiagen Operon), 12U Rnasin (Promega) and 0.9% NP-40 (Thermo Scientific Pierce) was added to each well. After thorough pipetting and rinsing, wells were capped, centrifuged at 4°C, heated to 68°C in a thermal cycler for 5 min and placed on ice for at least 1 min. Reverse transcription was performed with the addition of 7 μl containing 3.6 μl 5X reverse transcriptase buffer, 10 U RNAsin (Promega), 62 U Superscript III RT (Invitrogen), 0.62 μl dNTPs 25 mM each (Omega Bio-Tek) and 1.25 μl 0.1 M DTT (Sigma). All wells were capped, the plate placed in a cold rack and vortexed for 10 sec before centrifugation at 300xg. Thermal cycler conditions for reverse transcription were as follows: 42°C 5 min, 25°C 10 min, 50°C 60 min, 94°C 5 min and 4°C hold. When completed, 10 μl of nuclease-free PCR water was added to each well.
Ig gene amplification
Immediately following cDNA synthesis, IgG genes (Igg) were amplified in a total of 20 μl per well for the first round of nested PCR for IgG heavy chain (Iggh), IgG kappa (Iggκ) and IgG lambda (Iggλ), utilizing primers (Supplemental Table 1) as previously described55. In brief, a master mix was prepared consisting of 15.58 μl water, 2 μl 10X HotStar PCR buffer (Qiagen), 0.065 μl 5’ primer mix, 0.065 μl 3’ primer, 0.2 μl dNTP solution and 0.09 μl HotStarTaq per well, to which 2 μl cDNA from individual sorted B cells were added and Igg amplified under the following conditions: thermal cycle PCR at 94°C for 15 min; 50 cycles at 94°C for 30 sec, then 58°C (Iggh and Iggκ) or 60°C (Iggλ) for 30 sec, then 72°C for 55 sec; then one cycle at 72°C for 10 min. Second round of nested PCR for Iggh, Iggκ and Iggλ utilized 2 μl of first-round PCR product with second-round primers55 and the same master mix protocol, with the following conditions: thermal cycle PCR at 94°C for 15 min; 50 cycles at 94°C for 30 sec, then 58°C (Iggh and Iggκ) or 60°C (Iggλ) for 30 sec, then 72°C for 45 sec; then one cycle at 72°C for 10 min. The PCR product generated was purified and sequenced, with V(D)J genes determined using IMGT/V-Quest56.
Specific V(D)J region amplification and cloning
Primers specific with restriction enzyme sites for V and J regions were used to amplify the first-round PCR product to generate a fragment for cloning based on previously described primers55. PCR product was purified, restriction enzyme digested, cloned into Iggh, Iggκ or Iggλ expression vectors and chemically transformed into 5 μl aliquots of TOP10 E. coli cells (Thermo Fisher Scientific). Successful transformants were screened by PCR amplification utilizing a vector-specific primer paired with an insert-specific primer, sequenced and compared to the second-round PCR product sequence.
Definition of clonal groups
Clonal groups were based on heavy chain nucleotide sequences. Any PCR product with >0.8% nucleotide sequences with a Phred score <20 was excluded. From PCR-amplified sequences, we determined heavy chain alleles using IMGT/V-QUEST (http://www.imgt.org, international ImMunoGeneTic information system). A clonal group was defined by Ab V heavy chain sequences with the same inferred VH and JH germline sequences, identical CDR3 length, and the same or very similar CDR3 sequences (CDR, complementarity-determining region), i.e. >72% similarity of each CDR3 amino acid sequence. Clonal grouping was determined using Sequence Manipulation Suite: Ident and Sim57 using Ab-specific clusters as previously defined58.
MAb expression and purification
Two plasmids that included coding sequences for full-length IgG1 heavy and light chains were transfected into HEK293-H cells using polyethyleneimine (PEI). Five hundred micrograms PEI were incubated for 25 min at room temperature with 250μg of each plasmid and then added to the HEK293-H at a density of 1×106 cells/ml in total volume of 500ml54,55. Transfected cells were adapted for growth in Freestyle 293 serum-free expression medium (Gibco, Thermo Fisher Scientific) under suspension conditions of 37°C and 8.5% CO2. Cells were centrifuged 96 h post transfection and culture medium was harvested, filtered through a 0.22 μm filter and supernatants concentrated 20 times using a 50 kDa cut-off Vivaflow 50 System (Vivasciences). One volume of IgG binding buffer (Thermo Scientific Pierce) was added and IgG was purified on a Protein A HP HiTrap column (GE Healthcare) eluted with IgG elution buffer (Thermo Scientific Pierce) and neutralized with 1M Tris pH 9.0. Proteins were concentrated and buffer exchanged with PBS using Amicon Ultra4 10kDa. Protein concentrations were determined on Nanodrop (Thermo Fisher Scientific) and sample purity was analyzed by SDS-PAGE.
Protein expression and purification
Sal-1 DBP-II WT and DBP-II mutants were prepared as described17–19. scFvs were produced by expression and refolding from E. coli. The light chain variable region was linked to the heavy chain variable region by using a (GGGGS)3 linker, cloned into the pET28(a+) expression vector using restriction sites NcoI/XhoI and expressed in E. coli. Inclusion bodies were solubilized in 6 M guanidinium hydrochloride overnight at 4 °C and refolded with 400 mM L-Arginine, 2 mM reduced glutathione and 0.2 mM oxidized glutathione in 50 mM Tris, pH 8.0, 10 mM EDTA and 0.1 mM PMSF at 4 °C for 24 h. Refolded scFv were concentrated in Amicon®Stirred Cells using Biomax® 10 kDa Ultracentrifugation Discs and purified by size exclusion chromatography on GF200 (GE Healthcare) into 10 mM HEPES pH 7.4, 100 mM NaCl.
Protein crystallization and data collection
Purified DBP-II and 053054-scFv or 092096-scFv were mixed in a 1.2:1 (antigen:antibody) molar ratio and incubated at room temperature for 30 minutes. Complexes was purified by size exclusion chromatography (GF200 - GE Healthcare) in 10 mM HEPES pH 7.4, 100 mM NaCl. Crystals were grown by hanging-drop vapor diffusion by mixing 1 μL of complex at 7 mg/mL for DBP-II/053054-scFv or 10 mg/mL for DBP-II/092096-scFv with 2 μL of reservoir (0.2M ammonium sulfate, 0.1 M sodium citrate pH 5.6, 25% w/v PEG 4,000 for DBP-II/053054-scFv or 12% w/v PEG 6,000, 0.1 M MES, pH 6.0 for DBP-II/092096-scFv). Crystals were flash frozen with 30% ethylene glycol as cryoprotectant in liquid nitrogen. Data for DBP-II/053054-scFv were collected at the beamline 4.2.2 of the Advanced Light Source. Data for DBP-II/092096-scFv were collected at the beamline 23-ID-D (GM/CA) of Advanced Photon Source. Both data sets were processed with XDS59.
Structure solution and analysis
The DBP-II/053054-scFv and DBP-II/092096-scFv structures were solved by molecular replacement in PHASER60 by using PDB 4NUV and scFv domains modeled by the PIGS server61 as search models. Refinement was performed using PHENIX62 and COOT63. Summary of diffraction data and refinement statistics are given in Supplementary Table 1. The quality of the models was assessed using the MolProbity server64. All structures have good Ramachandran statistics with no outliers. 94.89 % and 5.11% of residues for DBP-II/053054-scFv or 93.80 % and 6.20% of residues for DBP-II/092096-scFv in favored and allowed regions of Ramachandran plot, respectively. Interaction interfaces were determined using PDBePISA65. Software used in the project was installed and configured by SBGrid66.
Hydrogen-Deuterium Exchange Mass Spectrometry
Before HDX footprinting experiments, 10 μM PvDBP-II was incubated with 12 μM of mAb in PBS for 30 min at 25 °C. The apo-state sample was 10 μM PvDBP in PBS. Continuous HDX on apo- and holo-state samples were performed after diluting into D2O buffer at the ratio of 1:5 and incubate for 10, 30, 60, 120, 900, 3,600, and 14,400 s, as described previously19 followed by on-line pepsin digestion and reversed-phase HPLC separation. Peptides were analyzed by a Thermo LTQ-FT mass spectrometer (Thermo Scientific). Deuterium uptakes of various time points were analyzed using HDX Workbench (Scripps, FL).
ELISA
The ELISAs were performed as previously described36. Briefly, BSA, Sal1 DBP-II, and DBP-II mutants were coated on plates overnight at 4 °C, washed with PBS/Tween-20, and blocked with 2% BSA in PBS/Tween-20 for 1 h at room temperature. The plates were washed with PBS/Tween-20 and then incubated with each mAb (053054, 092096) in a concentration 250ng/ml for 1 h at room temperature. The plates were washed with PBS/Tween-20 and incubated with an anti-human antibody conjugated to HRP for 30 min at room temperature. After a final wash step with PBS/Tween-20 ELISA, substrate TMB was added to the plates. The reaction was quenched by addition of 0.2 M sulfuric acid, and the absorbance at 450 nm was measured using a POLARstar Omega (BMG Labtech) plate reader.
Biotinylation of BirA-Tagged Sal-1 DBP-II
BirA-tagged Sal-1 DBP-II was buffer exchanged into Phosphate buffered saline. Then 50 μL of BiomixA (Avidity), 50 μL of BiomixB (Avidity), and 10 μL of 5 mM D-biotin (Avidity) were added to the protein along with BirA ligase, followed by overnight incubation at 4 °C. The biotinylation was confirmed by Western Blot using Streptavidin HRP-conjugate (Thermo Scientific). Before use, the reaction mix was buffer-exchanged into PBS.
Biolayer Interferometry Binding Assays
The binding affinity of purified Sal1 DBP-II protein with human mAbs was monitored by BLI conducting on an Octet-Red96 device (Pall ForteBio) by using Streptavidin (SA) biosensors (ForteBio). The biotinylated DBP-II was loaded onto biosensors until saturation, typically 1 μM for 15 min, in 10 mM HEPES (pH 7.4), 150 mM NaCl, 3 mM EDTA, and 0.005% P20 surfactant with 3% BSA. The mAb scFvs (analyte) were applied at two-fold serial dilution concentrations (100 nM to 1.675 nM). Association and dissociation were measured at 25 °C for all mAbs. The real-time data were analyzed using Biaevaluation 4.1 (GE Healthcare). Steady-state equilibrium concentration curves were fitted using a 1:1 binding model.
Fluorescence Activated Cell Sorting Assay
Biotinylated purified DBP-II protein in concentration of 10 nM was incubated with ten different concentrations of 053054 or 092096 (from 0.03 nM to 1 μM interval of concentrations) for 1 h at RT. Red blood cells were added and incubated for an additional 1 h at RT. To detect bound DBP Alexa Fluor 488 Streptavidin conjugate (Fisher) was added to the mix and incubated at RT for 1 h followed by washing twice with PBS. FITC labeling was measured by flow cytometry. An IC50 value was calculated in GraphPad Prism from three independent biological replicates. IC50 curve for the isotype control mAbs 043038 was plotted as a negative control.
P. vivax invasion assay
P. vivax ex vivo assays were independently performed with clinical isolates from Brazil and Cambodia. Cryopreserved P. vivax infected erythrocytes (iRBC) obtained from Brazilian donors with acute vivax malaria were thawed, enriched using Percoll density-gradients, immediately re-suspended in 100 μl IMDM plus 10% human AB serum with GlutaMax (1:100) to a hematocrit of 6% and cultured with a gas mix of 5% O2, 5% CO2, 90% N2 at 37 °C until established. The cultures were performed in quadruplicate from a single isolate of P. vivax and incubated with 092096 or isotype control comprising anti-TT monoclonal antibody 043038 at a concentration of 100 μg ml−1 and examined for parasite viability and maturation three times: after 20 h of initial incubation (initial growth from ring stages into early trophozoites, since trophozoites and schizont stages of the parasites do not survive cryopreservation), after additional incubation for 38 h, at which time most of the parasite have matured to schizont stages prior to merozoite release, parasite cultures were supplemented with fresh blood cells enriched for reticulocytes to ~2.5–3.0% at a ratio of 1:1 and additional incubation for 24 h to allow for new erythrocyte invasion. For examination, two smear slides were prepared for each culture well and stained with Giemsa. Rings and early trophozoites per 20,000 RBC were counted blindly three times. Cytochalasin D (5 μg/mL), a cell-permeable fungal toxin that inhibits merozoite invasion of erythrocytes, was used as a positive invasion inhibitor control.
For the assay with Cambodian isolates, cryopreserved iRBCs obtained from Cambodian patients with acute P.vivax malaria were thawed and cultured in IMDM medium (Gibco) supplemented with 0.5% Albumax II (Gibco), 2.5% heat-inactivated human serum, 25 mM HEPES (Gibco), 20 μg/ml gentamicin (Sigma) and 0.2 mM hypoxanthine (C-C Pro) for ~24 or ~48h until a majority of schizont stage parasites were noted. The schizont-infected erythrocytes were enriched using Percoll-KCl as described67, then mixed at a ratio of 1 erythrocyte to 1 with reticulocytes enriched from cord blood and labeled with CellTrace Far Red dye. The cultures were incubated for ~8h in a final volume of 100 μl in 96 well plates or 20 μl in 384 well plates, in presence of the human monoclonal antibodies 092096 or anti-TT MoAb 043038 while a positive invasion inhibitor control used the mouse anti-DARC monoclonal antibody 2C368 at 100 μg/ml. Cells were stained with DNA stain Hoechst 33342 post invasion and examined by flow cytometry. Reticulocytes which were Hoechst 33342 and Far Red positive were scored as new invasion events. Invasion of reticulocytes ranged from 0.19% to 6.52% (media alone) for the six experiments, each of which corresponded to one of six P. vivax isolates.
DBP-II patient population ELISAs and binding inhibition of DBPII to DARC fusion protein
To assess Ab blocking activity, plasma from Pv-exposed individuals was incubated with DBP-II at specified concentrations. We then measured levels of binding inhibition of DBP-II to a fusion protein containing amino acids 1–60 from the DARC N-terminal region fused to the Fc region of human IgG (nDARCIg)5,69. Pooled plasma samples with high blocking activity from Papua New Guinea or Brazil served as positive controls. Diluent alone or pooled samples from North Americans not exposed to malaria were used for negative controls. Percent inhibition was calculated by (1-(optical density [OD] of test sample/OD of negative control) x100).
Competition experiments
For assays measuring competition between mAbs and naturally-acquired Abs, plates were coated with mAbs at 0.5 μg/ml per well and remained overnight at 4 °C, then washed with PBST and blocked with 3% BSA in PBST for 2 h at room temperature. Serum from naturally immune subjects, at a dilution of 1/50, was pre-incubated with biotinylated DBP-II Sal I (20 ng/ml) for 20 min at room temperature. The serum mixture was added to plates and incubated 1 h at room temperature. Detection of DBP-II was achieved using High Sensitivity Streptavidin-HRP (Thermo Fisher Scientific) as described for competition assays between individual mAbs.
DBP epitope conservation analysis
A total of 599 sequences representing global variation in DBP-II70 was obtained from GenBank (accession codes XM_001608337.1, DQ156519, AF289480–AF289483, AF289635–AF289653, AF291096, AY970837–AY970925, AF469515–AF469602, U50575–U50590, DQ156513, DQ156515, DQ156522–DQ156523, AF215737–AF215738, AF220657, AF220659–AF220667, FJ491142–FJ491241, EF219451, EF368159–EF368180, EF379127–EF379132, EF379134, GU143914–GU144013, DQ156520, EU812839–EU812960, and EU860428–EU860438), aligned using ClustalW71, and inspected in JalView72. All 599 sequences contained complete coverage for the 053054 and 092096 epitopes.
Antibody evolution analysis
To compare heavy and light chains of 053054 and 092096 with germlines we used IMGT/V-Quest, an integrated software program for immunoglobulin and T cell receptor V–J and V–D–J rearrangement analysis73.
Statistical analyses
Analysis of ELISA data was performed using GraphPad Prism version 6.03 (GraphPad Software).
DATA AND SOFTWARE AVAILABILITY
Atomic coordinates and structural factors have been deposited in the Protein Data Bank with accession codes 6OAN and 6OAO (https://www.rcsb.org/structure/6OAN and https://www.rcsb.org/structure/6OAO).
Supplementary Material
ACKNOWLEDGMENTS
Funding: This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, the Extramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health (R56 AI080792 to NHT; contract HHSN272201400018C to NHT, JHA, CLK and MLG; R01 AI064478 to JHA, NHT and CLK; and P41 GM103422 to MLG), the Veterans Affairs Research Service (BX001350 to CLK) and the Burroughs Wellcome Fund (to NHT). Fundação de Amparo à Pesquisa do Estado de São Paulo, Brazil (FAPESP, 2009/52729–9 to MUF). VCN is supported by a scholarship from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) of Brazil, which also provides a senior researcher scholarship to MUF. We thank Y. Colin for the gift of the murine 2C3 anti-DARC. We thank J. Nix and ALS Beamline 4.2.2 supported by contract DE-AC02–05CH11231, and the Facility of the Rheumatic Diseases Core Center under award number P30AR048335. We also thank GM/CA-CAT beamlines 23-ID-D at the Advanced Photon Source, Argonne National Laboratory. Our special thanks to Dr. Christopher Nelson for assistance in the analysis of BLI data. We also thank Dr. Audrey Odom-John for the use of a plate reader, and J. Patrick Gorres for his assistance in preparing this manuscript for publication.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
SUPPLEMENTARY INFORMATION
Supplementary Information is available online.
REFERENCES
- 1.Guerra CA et al. The international limits and population at risk of Plasmodium vivax transmission in 2009. PLoS Negl Trop Dis 4, e774 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King CL et al. Naturally acquired Duffy-binding protein-specific binding inhibitory antibodies confer protection from blood-stage Plasmodium vivax infection. Proc Natl Acad Sci U S A 105, 8363–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chootong P et al. Mapping epitopes of the Plasmodium vivax Duffy binding protein with naturally acquired inhibitory antibodies. Infect Immun 78, 1089–95 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole-Tobian JL et al. Age-acquired immunity to a Plasmodium vivax invasion ligand, the duffy binding protein. J Infect Dis 186, 531–9 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Grimberg BT et al. Plasmodium vivax invasion of human erythrocytes inhibited by antibodies directed against the Duffy binding protein. PLoS Med 4, e337 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michon P, Fraser T & Adams JH Naturally acquired and vaccine-elicited antibodies block erythrocyte cytoadherence of the Plasmodium vivax Duffy binding protein. Infect Immun 68, 3164–71 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGregor IA The passive transfer of human malarial immunity. Am J Trop Med Hyg 13, SUPPL 237–9 (1964). [DOI] [PubMed] [Google Scholar]
- 8.Miller LH, Mason SJ, Dvorak JA, McGinniss MH & Rothman IK Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science 189, 561–3 (1975). [DOI] [PubMed] [Google Scholar]
- 9.Miller LH, Mason SJ, Clyde DF & McGinniss MH The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N. Engl. J. Med. 295, 302–4 (1976). [DOI] [PubMed] [Google Scholar]
- 10.Wertheimer SP & Barnwell JW Plasmodium vivax interaction with the human Duffy blood group glycoprotein: identification of a parasite receptor-like protein. Exp Parasitol 69, 340–50 (1989). [DOI] [PubMed] [Google Scholar]
- 11.Adams JH et al. The Duffy receptor family of Plasmodium knowlesi is located within the micronemes of invasive malaria merozoites. Cell 63, 141–53 (1990). [DOI] [PubMed] [Google Scholar]
- 12.Adams JH et al. A family of erythrocyte binding proteins of malaria parasites. Proc Natl Acad Sci U S A 89, 7085–9 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chitnis CE & Miller LH Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J Exp Med 180, 497–506 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chitnis CE, Chaudhuri A, Horuk R, Pogo AO & Miller LH The domain on the Duffy blood group antigen for binding Plasmodium vivax and P. knowlesi malarial parasites to erythrocytes. J Exp Med 184, 1531–6 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ranjan A & Chitnis CE Mapping regions containing binding residues within functional domains of Plasmodium vivax and Plasmodium knowlesi erythrocyte-binding proteins. Proc Natl Acad Sci U S A 96, 14067–72 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.VanBuskirk KM, Sevova E & Adams JH Conserved residues in the Plasmodium vivax Duffy-binding protein ligand domain are critical for erythrocyte receptor recognition. Proc Natl Acad Sci U S A 101, 15754–9 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batchelor JD, Zahm JA & Tolia NH Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nature Structural & Molecular Biology 18, 908–U67 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batchelor JD et al. Red Blood Cell Invasion by Plasmodium vivax: Structural Basis for DBP Engagement of DARC. Plos Pathogens 10(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen E et al. Broadly neutralizing epitopes in the Plasmodium vivax vaccine candidate Duffy Binding Protein. Proc Natl Acad Sci U S A 113, 6277–82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Payne RO et al. Human vaccination against Plasmodium vivax Duffy-binding protein induces strain-transcending antibodies. JCI Insight 2(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh K et al. Malaria vaccine candidate based on Duffy-binding protein elicits strain transcending functional antibodies in a Phase I trial. NPJ Vaccines 3, 48 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cole-Tobian JL et al. Strain-specific duffy binding protein antibodies correlate with protection against infection with homologous compared to heterologous plasmodium vivax strains in Papua New Guinean children. Infect Immun 77, 4009–17 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuboi T et al. Natural variation within the principal adhesion domain of the Plasmodium vivax duffy binding protein. Infect Immun 62, 5581–6 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ntumngia FB & Adams JH Design and immunogenicity of a novel synthetic antigen based on the ligand domain of the Plasmodium vivax duffy binding protein. Clin Vaccine Immunol 19, 30–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen E, Salinas ND, Ntumngia FB, Adams JH & Tolia NH Structural analysis of the synthetic Duffy Binding Protein (DBP) antigen DEKnull relevant for Plasmodium vivax malaria vaccine design. PLoS Negl Trop Dis 9, e0003644 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt AG et al. Viral receptor-binding site antibodies with diverse germline origins. Cell 161, 1026–1034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ekiert DC et al. Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246–51 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ceravolo IP et al. Naturally acquired inhibitory antibodies to Plasmodium vivax Duffy binding protein are short-lived and allele-specific following a single malaria infection. Clin Exp Immunol 156, 502–10 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.VanBuskirk KM et al. Antigenic drift in the ligand domain of Plasmodium vivax duffy binding protein confers resistance to inhibitory antibodies. J Infect Dis 190, 1556–62 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Verkoczy L, Kelsoe G, Moody MA & Haynes BF Role of immune mechanisms in induction of HIV-1 broadly neutralizing antibodies. Curr Opin Immunol 23, 383–90 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orlandi PA, Sim BK, Chulay JD & Haynes JD Characterization of the 175-kilodalton erythrocyte binding antigen of Plasmodium falciparum. Mol Biochem Parasitol 40, 285–94 (1990). [DOI] [PubMed] [Google Scholar]
- 32.Klotz FW et al. Binding of Plasmodium falciparum 175-kilodalton erythrocyte binding antigen and invasion of murine erythrocytes requires N-acetylneuraminic acid but not its O-acetylated form. Mol Biochem Parasitol 51, 49–54 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sim BK, Chitnis CE, Wasniowska K, Hadley TJ & Miller LH Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science 264, 1941–4 (1994). [DOI] [PubMed] [Google Scholar]
- 34.Tolia N, Enemark E, Sim B & Joshua-Tor L Structural basis for the EBA-175 erythrocyte invasion pathway of the malaria parasite Plasmodium falciparum (vol 122, pg 183, 2005). Cell 122, 183 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Chen E, Paing MM, Salinas N, Sim BKL & Tolia NH Structural and Functional Basis for Inhibition of Erythrocyte Invasion by Antibodies that Target Plasmodium falciparum EBA-175. Plos Pathogens 9(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salinas ND, Paing MM & Tolia NH Critical Glycosylated Residues in Exon Three of Erythrocyte Glycophorin A Engage Plasmodium falciparum EBA-175 and Define Receptor Specificity. Mbio 5(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salinas ND & Tolia NH A quantitative assay for binding and inhibition of Plasmodium falciparum Erythrocyte Binding Antigen 175 reveals high affinity binding depends on both DBL domains. Protein Expression and Purification 95, 188–194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayer DC, Kaneko O, Hudson-Taylor DE, Reid ME & Miller LH Characterization of a Plasmodium falciparum erythrocyte-binding protein paralogous to EBA-175. Proc Natl Acad Sci U S A 98, 5222–7 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narum DL, Fuhrmann SR, Luu T & Sim BK A novel Plasmodium falciparum erythrocyte binding protein-2 (EBP2/BAEBL) involved in erythrocyte receptor binding. Mol Biochem Parasitol 119, 159–68 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Lobo CA, Rodriguez M, Reid M & Lustigman S Glycophorin C is the receptor for the Plasmodium falciparum erythrocyte binding ligand PfEBP-2 (baebl). Blood 101, 4628–31 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Lin DH, Malpede BM, Batchelor JD & Tolia NH Crystal and solution structures of Plasmodium falciparum erythrocyte-binding antigen 140 reveal determinants of receptor specificity during erythrocyte invasion. J Biol Chem 287, 36830–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malpede BM, Lin DH & Tolia NH Molecular basis for sialic acid-dependent receptor recognition by the Plasmodium falciparum invasion protein erythrocyte-binding antigen-140/BAEBL. J Biol Chem 288, 12406–15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopaticki S et al. Reticulocyte and erythrocyte binding-like proteins function cooperatively in invasion of human erythrocytes by malaria parasites. Infect Immun 79, 1107–17 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Persson KE et al. Erythrocyte-binding antigens of Plasmodium falciparum are targets of human inhibitory antibodies and function to evade naturally acquired immunity. J Immunol 191, 785–94 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilberger TW et al. A novel erythrocyte binding antigen-175 paralogue from Plasmodium falciparum defines a new trypsin-resistant receptor on human erythrocytes. J Biol Chem 278, 14480–6 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Mayer DC et al. Polymorphism in the Plasmodium falciparum erythrocyte-binding ligand JESEBL/EBA-181 alters its receptor specificity. Proc Natl Acad Sci U S A 101, 2518–23 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maier AG, Baum J, Smith B, Conway DJ & Cowman AF Polymorphisms in erythrocyte binding antigens 140 and 181 affect function and binding but not receptor specificity in Plasmodium falciparum. Infect Immun 77, 1689–99 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mayer DC et al. Glycophorin B is the erythrocyte receptor of Plasmodium falciparum erythrocyte-binding ligand, EBL-1. Proc Natl Acad Sci U S A 106, 5348–52 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ntumngia FB et al. A Novel Erythrocyte Binding Protein of Plasmodium vivax Suggests an Alternate Invasion Pathway into Duffy-Positive Reticulocytes. MBio 7(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gruszczyk J et al. Transferrin receptor 1 is a reticulocyte-specific receptor for. Science 359, 48–55 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orjuela-Sanchez P et al. Higher microsatellite diversity in Plasmodium vivax than in sympatric Plasmodium falciparum populations in Pursat, Western Cambodia. Exp Parasitol 134, 318–26 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hostetler JB et al. Independent Origin and Global Distribution of Distinct Plasmodium vivax Duffy Binding Protein Gene Duplications. PLoS Negl Trop Dis 10, e0005091 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicolete VC, Frischmann S, Barbosa S, King CL & Ferreira MU Naturally Acquired Binding-Inhibitory Antibodies to Plasmodium vivax Duffy Binding Protein and Clinical Immunity to Malaria in Rural Amazonians. J Infect Dis 214, 1539–1546 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wardemann H & Kofer J Expression cloning of human B cell immunoglobulins. Methods Mol Biol 971, 93–111 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Tiller T et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods 329, 112–24 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giudicelli V, Brochet X & Lefranc MP IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb Protoc 2011, 695–715 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Stothard P The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28, 1102, 1104 (2000). [DOI] [PubMed] [Google Scholar]
- 58.Mirsky A, Kazandjian L & Anisimova M Antibody-specific model of amino acid substitution for immunological inferences from alignments of antibody sequences. Mol Biol Evol 32, 806–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kabsch W Xds. Acta Crystallogr D Biol Crystallogr 66, 125–32 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCoy AJ et al. Phaser crystallographic software. J Appl Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marcatili P, Rosi A & Tramontano A PIGS: automatic prediction of antibody structures. Bioinformatics 24, 1953–4 (2008). [DOI] [PubMed] [Google Scholar]
- 62.Adams PD et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58, 1948–54 (2002). [DOI] [PubMed] [Google Scholar]
- 63.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–32 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Davis IW et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35, W375–83 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krissinel E & Henrick K Inference of macromolecular assemblies from crystalline state. J Mol Biol 372, 774–97 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Morin A et al. Collaboration gets the most out of software. Elife 2, e01456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rangel GW et al. Enhanced Ex Vivo Plasmodium vivax Intraerythrocytic Enrichment and Maturation for Rapid and Sensitive Parasite Growth Assays. Antimicrob Agents Chemother 62(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Russell B et al. A reliable ex vivo invasion assay of human reticulocytes by Plasmodium vivax. Blood 118, e74–81 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choe H et al. Sulphated tyrosines mediate association of chemokines and Plasmodium vivax Duffy binding protein with the Duffy antigen/receptor for chemokines (DARC). Mol Microbiol 55, 1413–22 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Nóbrega de Sousa T, Carvalho LH & Alves de Brito CF Worldwide genetic variability of the Duffy binding protein: insights into Plasmodium vivax vaccine development. PLoS One 6, e22944 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Larkin MA et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–8 (2007). [DOI] [PubMed] [Google Scholar]
- 72.Waterhouse AM, Procter JB, Martin DM, Clamp M & Barton GJ Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–91 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brochet X, Lefranc MP & Giudicelli V IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res 36, W503–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structural factors have been deposited in the Protein Data Bank with accession codes 6OAN and 6OAO (https://www.rcsb.org/structure/6OAN and https://www.rcsb.org/structure/6OAO).