In the environment, many microbes form biofilms. However, the interspecies interactions underlying bacterial coexistence within these biofilms remain understudied. Here, we mimic environmentally relevant biofilms by studying a dual-species biofilm formed between Bacillus subtilis and Pantoea agglomerans and subjecting the coculture to chemical and physical stressors that it may experience in the natural world. We determined that both bacteria contribute structural elements to the coculture, which is reflected in its overall viscoelastic behavior. Existence within the coculture can be either beneficial or detrimental depending on the context. Many of the features and determinants of the coculture biofilm appear distinct from those identified in monoculture biofilm studies, highlighting the importance of characterizing multispecies consortia to understand naturally occurring bacterial interactions.
KEYWORDS: Bacillus subtilis, Pantoea agglomerans, antibiotic stress, biofilm, cell-cell interactions, coculture, dual-species, rheology, wrinkles
ABSTRACT
Many microbes coexist within biofilms, or multispecies communities of cells encased in an extracellular matrix. However, little is known about the microbe-microbe interactions relevant for creating these structures. In this study, we explored a striking dual-species biofilm between Bacillus subtilis and Pantoea agglomerans that exhibited characteristics that were not predictable from previous work examining monoculture biofilms. Coculture wrinkle formation required a P. agglomerans exopolysaccharide as well as the B. subtilis amyloid-like protein TasA. Unexpectedly, other B. subtilis matrix components essential for monoculture biofilm formation were not necessary for coculture wrinkling (e.g., the exopolysaccharide EPS, the hydrophobin BslA, and cell chaining). In addition, B. subtilis cell chaining prevented coculture wrinkling, even though chaining was previously associated with more robust monoculture biofilms. We also observed that increasing the relative proportion of P. agglomerans (which forms completely featureless monoculture colonies) increased coculture wrinkling. Using microscopy and rheology, we observed that these two bacteria assemble into an organized layered structure that reflects the physical properties of both monocultures. This partitioning into distinct regions negatively affected the survival of P. agglomerans while also serving as a protective mechanism in the presence of antibiotic stress. Taken together, these data indicate that studying cocultures is a productive avenue to identify novel mechanisms that drive the formation of structured microbial communities.
IMPORTANCE In the environment, many microbes form biofilms. However, the interspecies interactions underlying bacterial coexistence within these biofilms remain understudied. Here, we mimic environmentally relevant biofilms by studying a dual-species biofilm formed between Bacillus subtilis and Pantoea agglomerans and subjecting the coculture to chemical and physical stressors that it may experience in the natural world. We determined that both bacteria contribute structural elements to the coculture, which is reflected in its overall viscoelastic behavior. Existence within the coculture can be either beneficial or detrimental depending on the context. Many of the features and determinants of the coculture biofilm appear distinct from those identified in monoculture biofilm studies, highlighting the importance of characterizing multispecies consortia to understand naturally occurring bacterial interactions.
INTRODUCTION
Microbes often form structured multicellular communities known as biofilms, which are present in varied environments such as on plant roots and medical equipment (1). Biofilms consist of cells encased in an extracellular matrix composed of polysaccharides, proteins, and nucleic acids (2). The extracellular matrix promotes surface adherence, acts as a nutrient reservoir, and protects against physical and chemical stressors such as predation, invasion, fluid flow, osmotic stress, and antibiotics (2–8). The heterogeneity and underlying physical properties of the extracellular matrix and cell-cell interactions result in the formation of organized structured biofilms (9–12).
Most naturally occurring biofilms are composed of multiple bacterial species, and there is evidence that such multispecies biofilms can enhance resistance to antimicrobials (13, 14) and biofilm invasion by planktonic cells (8) as well as lead to synergistic increases in biofilm biomass compared to that of their single-species counterparts (15). Nevertheless, many previous coculture biofilm studies have largely focused on the survival of individual bacterial species within cocultures, rather than on identifying their genetic determinants or physical features. Here, we dissect a coculture biofilm formed between Bacillus subtilis and Pantoea agglomerans, directly exploring the contributions of their extracellular matrix components, the physical properties of the resulting mixed-species biofilm, and how colony structure relates to community function.
B. subtilis is a Gram-positive soil- and plant-associated bacterium that is frequently used as a laboratory model of biofilm formation (16). B. subtilis colonies at the air-agar interface form wrinkled colonies after a few days of growth (17). Under some conditions, the extracellular matrix holds cells together tightly and promotes either vertical buckling (18) or out-of-plane growth (19) that manifests as colony wrinkling. The formation of some B. subtilis biofilm wrinkles has been associated with mechanical stresses (20) resulting from mechanical forces directed at sites of cell death (21) or as a result of self-produced chemical gradients (22). The primary structural components of B. subtilis extracellular matrix are an exopolysaccharide (EPS, encoded by the operon epsA-O), the protein TasA, which assembles into fibers attached to the cell wall and provides structural integrity to the biofilm (23, 24), and the protein BslA, which provides a hydrophobic coat to the surface of the biofilm (25). Although these matrix components are well studied in laboratory biofilms, little is known about whether these or other B. subtilis genes play important roles in biofilm formation within more natural multispecies communities.
P. agglomerans is a soil-dwelling Gram-negative organism in the Enterobacteriaceae family that, like B. subtilis, is present in the rhizosphere and forms biofilms on plants (26). P. agglomerans has been shown to form symplasmata, multicellular aggregates on the surfaces of roots and leaves and within plant tissues (27). The matrix components necessary for biofilm formation in P. agglomerans have not been well characterized. However, the related organism Pantoea stewartii subsp. stewartii produces an EPS, termed stewartan, that is required for biofilm formation and plant host colonization (28).
When B. subtilis and P. agglomerans are grown in coculture on an agar surface, we observed that, in contrast to their monoculture colonies under these conditions, a highly wrinkled biofilm structure forms. Here, we dissect this dual-species interaction by identifying the genetic determinants and functional consequences of this robust coculture colony. Using a combination of genetics, microscopy, and biophysical techniques, we determined that matrix components of both B. subtilis and P. agglomerans (specifically, TasA and exopolysaccharide, respectively) contribute to the coculture biofilm architecture. These data indicate that some B. subtilis components important for monoculture biofilm formation are not necessary for this community behavior. In addition, we determined that contributions from both organisms affect the response of the coculture biofilm to mechanical and chemical perturbations, results that highlight the context-dependent nature of cooperative and competitive coculture interactions and the unique mechanical insights attainable when examining bacterial cocultures.
RESULTS
B. subtilis and P. agglomerans form a striking biofilm in coculture.
Cultures of B. subtilis NCIB3610 and an environmental soil isolate of P. agglomerans were normalized to an optical density at 600 nm (OD600) of 0.5 and spotted individually and in coculture at a 1:1 ratio (by OD) on MSgg, a B. subtilis biofilm-inducing medium. Under these conditions in monoculture, B. subtilis formed a dry, rough flat macrocolony, while P. agglomerans formed a mucoid convex macrocolony (Fig. 1; see also Fig. S1A in the supplemental material). (Note that this study was conducted at 24°C, while most B. subtilis biofilm studies are performed at 30°C, and so we did not observe colony wrinkle formation at 48 h in B. subtilis monoculture.) In coculture, a remarkable structure emerged with pronounced central wrinkles that was distinct from the monoculture colony morphologies (Fig. 1; Fig. S1A).
FIG 1.
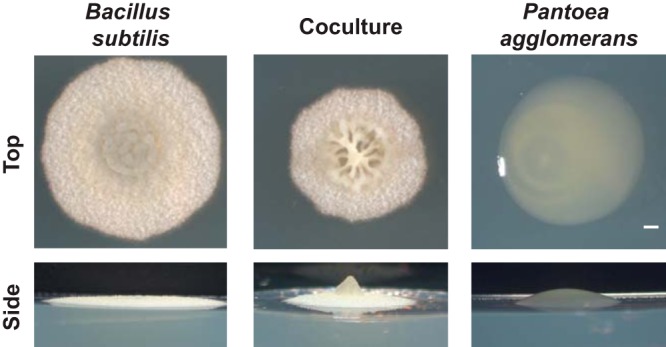
Biofilm architecture of B. subtilis NCIB3610 and environmental soil isolate P. agglomerans in monoculture and coculture grown at 24°C on biofilm-inducing medium MSgg for 48 h. Colony images were taken from the top down to observe wrinkle formation and from the side to visualize the height and 3D structure. Bar, 1 mm.
Related Bacillus species form wrinkled biofilms with P. agglomerans.
We then asked whether other Bacillus species form a similar structure when cocultured with P. agglomerans or whether this pronounced wrinkle formation was unique to these two bacterial strains. We screened a phylogenetically diverse set of Bacillus species in coculture with P. agglomerans and observed wrinkle formation with cocultures involving B. subtilis subsp. spizizenii, Bacillus mojavensis, Bacillus amyloliquefaciens, and Bacillus atrophaeus but not more distantly related Bacillus species (see Fig. S2). Thus, only species within the B. subtilis subclade formed wrinkles when cocultured with P. agglomerans, but not all such representatives did. In addition, there is some correlation between strains that formed a wrinkled coculture biofilm structure and those that possess a gene that codes for a protein homologous to TasA. However, some strains also exhibited a reduced growth rate on MSgg, and so we cannot definitively say whether wrinkle formation and production of a TasA-like protein are related under these culture conditions.
The B. subtilis structural protein TasA is required for the coculture phenotype.
The genes essential for robust B. subtilis biofilm formation in monoculture are well characterized and include the matrix components EPS (epsA-O), BslA (bslA), and TasA (tasA) and the biofilm signal surfactin (srfA) (29). To determine whether these components also contribute to the coculture biofilm structure, we mixed B. subtilis strains lacking one of these genes with P. agglomerans and grew these cocultures on MSgg to determine whether coculture biofilm architecture was affected. When P. agglomerans was mixed with either the B. subtilis epsA-O, bslA, srfA, or hag (flagellar motility) mutants, the resulting coculture colonies formed wrinkles similar to those formed in wild-type B. subtilis-P. agglomerans cocultures (Fig. 2A; see Fig. S3A). However, the B. subtilis tasA mutant-P. agglomerans coculture no longer formed these distinctive wrinkles (Fig. 2A), indicating that TasA was necessary for the coculture structure.
FIG 2.
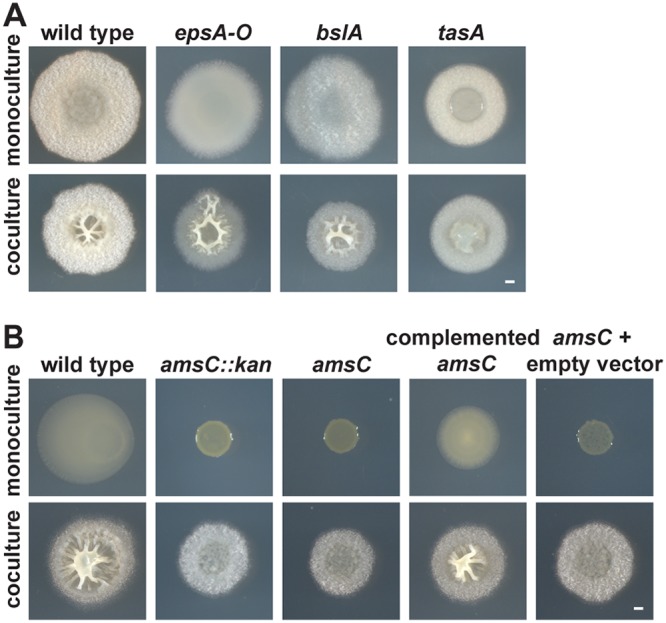
Extracellular matrix components required for coculture biofilm structure. (A) Colony morphology of B. subtilis strains lacking genes encoding essential matrix components (epsA-O, bslA, and tasA) in monoculture (top) and in coculture with P. agglomerans (bottom) grown on MSgg for 48 h. (B) Representative P. agglomerans mutants (transposon insertion amsC::kan, P. agglomerans amsC, P. agglomerans amsC complemented with amsC on a plasmid [pBBR1MCS-amsC], amsC plus pBBR1MCS [empty vector control]) grown in monoculture (top) and coculture with B. subtilis (bottom) in a 1:500 B. subtilis-to-P. agglomerans ratio on MSgg for 48 h. Bars, 1 mm.
P. agglomerans contributes exopolysaccharide to the coculture biofilm.
Under monoculture conditions, B. subtilis typically requires all three biofilm matrix components (epsA-O, bslA, and tasA) to form wrinkles. This raises the question of why neither epsA-O nor bslA appeared necessary for this coculture structure to form. One possibility was that P. agglomerans contributed to this coculture morphology by providing alternative biofilm matrix components. To identify P. agglomerans genes important for the coculture biofilm phenotype, we generated 8,064 transposon mutants in P. agglomerans and screened them to identify those that did not form wrinkles in coculture with B. subtilis. We identified 129 mutants in our primary screen, 40 of which subsequently passed our secondary screen. We have sequenced 12 of these hits, eight of which contained transposons that disrupted genes predicted to be in an EPS biosynthesis cluster, suggesting that the coculture structure requires this P. agglomerans EPS (Fig. 2B; Fig. S3B). The remaining four mutants contained transposon insertions in a diverse set of genes, including a homolog of the transcriptional regulator gene rcsA; crtE, a geranylgeranyl pyrophosphate synthase gene involved in carotenoid synthesis; an unnamed ABC transporter permease gene; and argE, an acetyl ornithine deacetylase gene (Fig. S3B).
Based on the large number of hits observed within the putative EPS biosynthesis gene cluster, we constructed a clean deletion of one of its genes, amsC, which is predicted to encode a glycosyl transferase EpsG family protein. We also constructed an amsC-complemented strain and an empty vector control strain. None of these P. agglomerans variants displayed a growth defect in monoculture in liquid growth (Fig. S3C); nevertheless, we cocultured all of the P. agglomerans variants at a 1:500 (B. subtilis/P. agglomerans) ratio to ensure that the coculture phenotypes were not affected by potential growth defects on solid medium and monitored the CFU of each strain at 48 h (Fig. S3D). When the P. agglomerans amsC mutant was cocultured with B. subtilis, we observed no wrinkles. In contrast, when the amsC complemented strain was cocultured with B. subtilis, it did exhibit coculture wrinkles (Fig. 2B). These data indicate that amsC appears to be both necessary and sufficient for the production of the coculture biofilm structure.
To determine whether the production of this or another putative P. agglomerans component alone was sufficient to induce the formation of a structured coculture biofilm in conjunction with B. subtilis, we examined whether heat-killed P. agglomerans could stimulate coculture colony formation when mixed with B. subtilis. We determined that B. subtilis mixed with heat-killed P. agglomerans resulted in a coculture colony that resembled the B. subtilis monoculture biofilm (Fig. S1B), indicating that active growth or more substantial production of structural components by P. agglomerans was necessary for wrinkle formation in coculture.
B. subtilis-P. agglomerans coculture morphology varies depending upon cell ratios.
Knowing that both B. subtilis and P. agglomerans contributed structural components to the biofilm led us to test the relative contribution of each organism in terms of their viable cell numbers. To explore this question, we altered the initial proportions of the two organisms and monitored the resulting colony morphologies (Fig. 3A; see also Fig. S4A). (Up to this point, a culture of each bacterium had been normalized to an OD600 of 0.5 before being mixed and spotted for coculture growth, which represents a ratio of 1:5 in terms of B. subtilis-P. agglomerans cells.) We quantified the B. subtilis and P. agglomerans CFU present in the original inoculum as well as in the monoculture and coculture colonies over time and saw that in coculture on solid medium, B. subtilis almost always outnumbered P. agglomerans. This was regardless of the initial proportions of cells (Fig. S4B and C) and in spite of the fact that wild-type B. subtilis and P. agglomerans have similar growth rates in liquid culture (Fig. S4D). However, when P. agglomerans cells were initially present at higher cell numbers than B. subtilis, the center of the biofilm became broader and taller with longer wrinkles (Fig. 3A). This supports our interpretation that P. agglomerans contributes to wrinkle formation in coculture.
FIG 3.
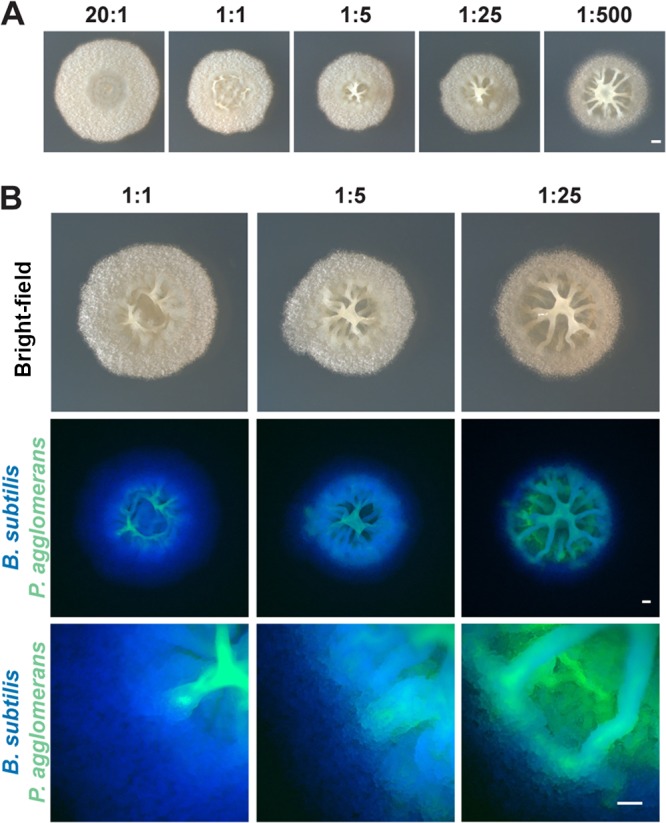
Colony morphology and localization of B. subtilis and P. agglomerans in coculture plated at various initial cell ratios. (A) Colonies of B. subtilis and P. agglomerans in coculture imaged from the top and side at increasing initial ratios of P. agglomerans after growth on MSgg for 48 h. Bar, 1 mm. (B) B. subtilis (false colored blue) and P. agglomerans (false colored green) in coculture at increasing initial ratios of P. agglomerans grown on MSgg after 48 h of growth. Colocalization of two species is indicated in cyan. Coculture images of phase contrast (top) at ×1 magnification, fluorescence (middle) at ×1 magnification, and fluorescence (bottom) at ×8 magnification. Bars, 0.5 mm.
B. subtilis and P. agglomerans spatially partition within the biofilm.
Next, we wanted to determine where B. subtilis and P. agglomerans cells localized within the colony biofilm. Using constitutively fluorescent B. subtilis and P. agglomerans strains, we saw that B. subtilis cells primarily localized to the outer ring of the colony, while P. agglomerans cells were observed in the center of the colony (Fig. 3B). These results indicate that these two organisms spatially partition themselves into distinct radial regions of the macroscopic biofilm. We were then curious how the coculture structure developed. We monitored the localization of the two species during biofilm development using fluorescence microscopy at the colony level. P. agglomerans appeared to dominate the coculture biofilm at early time points (10 h), with small pockets of B. subtilis present in the center of the structure (see Fig. S5). However, by 18 h, B. subtilis was present in the outer edge of the colony, and it began to envelop the coculture structure by 22 h (Fig. S5).
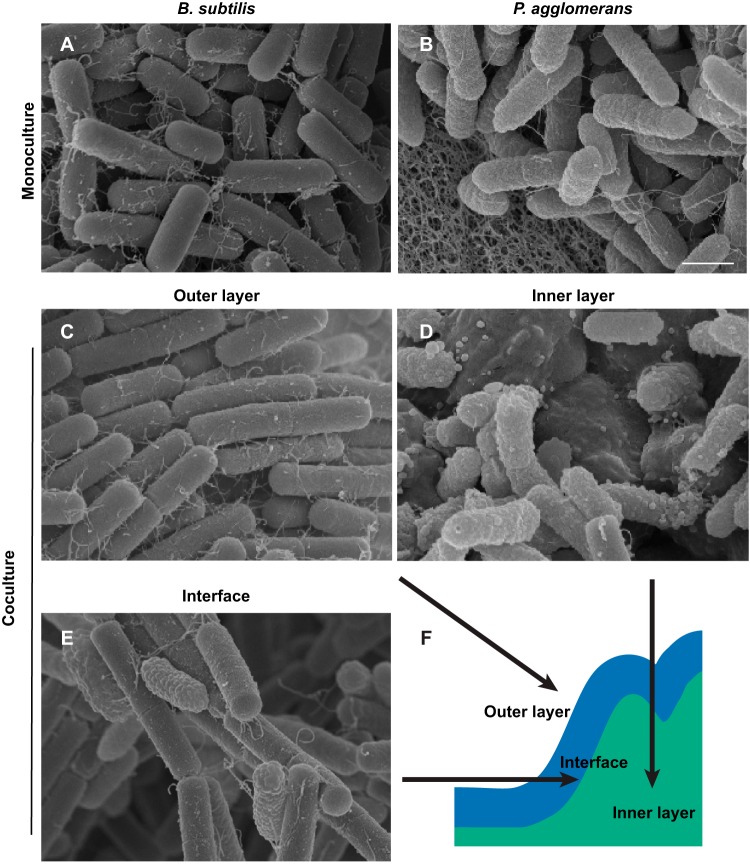
On the basis of this observation, we then wanted to determine how individual cells were physically distributed within the coculture structure. We used scanning electron microscopy (SEM) to visualize the spatial organization of cells in both monoculture and coculture biofilms grown for 24 h. (The large size of the 48-h colonies posed technical challenges for SEM.) There were distinct differences in the cellular morphologies of these two species (apparent in the monoculture micrographs) that allowed us to distinguish them in the coculture biofilms: the cells of B. subtilis appeared smooth with blunt ends (Fig. 4A), while the cells of P. agglomerans appeared wrinkled with rounded ends (Fig. 4B).
FIG 4.
B. subtilis and P. agglomerans cells partition into a layered coculture biofilm. Monoculture and coculture colonies grown on MSgg for 24 h were removed from the agar plate and processed for SEM. (A and B) Cells in B. subtilis monoculture biofilm (A) compared to cells of P. agglomerans in monoculture (B). (C to F) Views of outer (C), inner (D), and interface layers (E) with a schematic of a B. subtilis and P. agglomerans coculture colony (F). Bar, 1 μm.
In the B. subtilis monoculture, cells in the top layer were aligned in long chains that were bundled together tightly (see Fig. S6A and B), as described previously (30). Underneath this top layer, B. subtilis cells were oriented in divergent directions and appeared packed by extracellular material (Fig. S6 and D). In contrast, cells in the top layer of the P. agglomerans monoculture biofilm were disorganized and appeared connected by a fine mesh-like matrix (Fig. S6E and F). Beneath this top layer, the P. agglomerans cells appeared to be bound into multicellular aggregates reminiscent of symplasmata structures (Fig. S6G and H) (27).
In the coculture SEM micrographs, the outer layer of cells resembled B. subtilis monoculture cell chains (Fig. 4C), while the inner layer of the colony resembled the multicellular aggregates of P. agglomerans cells (Fig. 4D). Only at the interface between the outer and inner layers of the coculture colony did we observe physical interactions between B. subtilis and P. agglomerans cells (Fig. 4E).
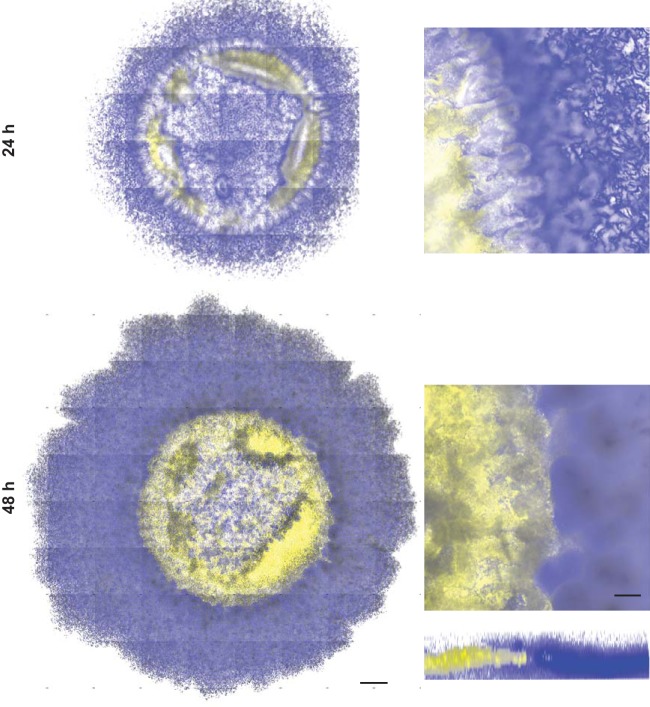
To validate the results observed by SEM, we used confocal laser scanning microscopy (CLSM) to visualize the spatial organization of fluorescently labeled B. subtilis and P. agglomerans in coculture. The fluorescence data indicate that B. subtilis localizes to the outer ring of the coculture biofilm, while P. agglomerans is localized in the center of the structure (Fig. 5; see also Movie S1). Although our CLSM method is unable to image through the entire “height” of the coculture structure, these data, in combination with our SEM images, allow us to conclude that the two organisms vertically segregate within the coculture biofilm structure even though the cultures were initially well mixed.
FIG 5.
B. subtilis and P. agglomerans are spatially organized within the coculture biofilm. Coculture colonies grown on glass coverslips embedded in MSgg agar and imaged at 24 h and 48 h using confocal laser scanning microscopy. B. subtilis PspacC-mTurq (blue) and P. agglomerans PspacC-Ypet (yellow) are shown in coculture colony tile scans (left) and an inset of one tile (right) with a representative three-dimensional (3D) reconstructed biofilm shown at 48 h. Bars, 500 μm and 100 μm for tile scans and individual tiles, respectively.
Rigid B. subtilis cells prevent the formation of the coculture structure.
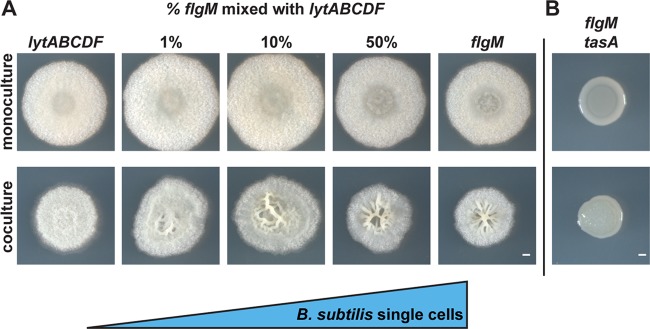
The SEM micrographs indicated that B. subtilis formed a relatively unbroken layer of chained cells on the top of the coculture biofilm. In B. subtilis monoculture, an increase in cell chains has been associated with increased matrix gene expression (31). This raises the question of whether the ability to form cell chains structurally contributed to the wrinkling morphology of the coculture. We tested this using B. subtilis flgM and lytABCDF mutants. FlgM is an antagonist of alternative sigma factor σD, which activates motility; therefore, flgM cells are single motile cells that do not chain (32, 33). Autolysin-related proteins encoded by lytABCDF cleave peptidoglycan connections between mother and daughter cells (34); therefore, lytABCDF mutant cells only exist as nonmotile chains (35). When we cocultured P. agglomerans with B. subtilis lytABCDF, we did not observe wrinkles (Fig. 6A). In contrast, the B. subtilis flgM coculture with P. agglomerans resulted in a more wrinkled structure than with the wild type (Fig. 6A). These data indicate that, counter to expectations, fully chained B. subtilis cells prevent coculture wrinkle formation, while single cells enhance it.
FIG 6.
B. subtilis cells in chains prevent formation of the wrinkled coculture phenotype while single cells enhance it. (A) Colonies of B. subtilis lytABCDF (chained cells) cultured with increasing amounts of B. subtilis flgM (single cells) and spotted in monoculture (top) and in coculture with P. agglomerans (bottom) on MSgg and imaged after 48 h of growth. (B) B. subtilis flgM tasA double mutant grown in monoculture (top) and in coculture with P. agglomerans (bottom) on MSgg for 48 h. Bars, 1 mm.
To determine what proportion of single cells are necessary for wrinkle formation, we mixed various ratios of B. subtilis flgM and lytABCDF cells before combining them with P. agglomerans for coculture growth. As the proportion of B. subtilis flgM cells in the coculture colony was increased, we observed larger magnitude wrinkle formation; only a small percentage of single cells appear necessary for wrinkles to form (Fig. 6A). Even though the colony morphology differs when P. agglomerans is cocultured with B. subtilis flgM and lytABCDF mutants, we still observed spatial partitioning (see Fig. S7A). In addition, B. subtilis flgM and lytABCDF mutants and P. agglomerans were present at similar numbers to the wild-type coculture (Fig. S7B). These data indicate that the coculture wrinkles require the presence of single B. subtilis cells, while an excess of B. subtilis chains may result in an overly rigid cover that suppresses wrinkle formation.
We then predicted that, in the absence of cell chains, TasA might be providing the necessary structure to B. subtilis cells to generate a wrinkled colony. To test this, we generated a flgM tasA double mutant and cocultured it with P. agglomerans. Even though the flgM mutant exhibited prominent wrinkle formation, we did not observe the characteristic wrinkled coculture biofilm structure when TasA was eliminated in the double mutant (Fig. 6B), suggesting that TasA was an essential biofilm component for holding the unchained flgM cells together and allowing the coculture wrinkles to form.
Mechanical properties of the coculture biofilm reflect those of the monocultures.
Having identified the genetic determinants of this coculture morphology, we next used bulk rheology (36) to measure the mechanical properties of both the monoculture and coculture biofilms. This technique allows us to quantify the response of a material to known applied forces and thereby measure the shear storage modulus (G′, associated with elasticity) and shear loss modulus (G″, associated with viscosity) of a viscoelastic material. Because biofilms are viscoelastic, they not only store elastic energy but also flow irreversibly rather than returning to their initial state (37–39). Due to the differences in matrix compositions described above, we hypothesized that the mechanical properties of the monocultures and coculture would differ.
Monoculture and coculture biofilms were grown to a 50-mm diameter for 48 h on polycarbonate filters and then transferred to the bottom plate of the rheometer, along with the polycarbonate (the filters allowed us to keep the colony structures intact during rheometer loading). Our data indicated that monoculture B. subtilis has a storage modulus larger than its loss modulus, reflecting its more elastic (solid-like) nature (Fig. 7A). At low (biologically relevant) frequencies, the P. agglomerans biofilm was more viscous than elastic, which we would expect from its ability to “flow” at the macrocolony level (Fig. 7A). In coculture, the elastic behavior dominated over the viscous behavior (Fig. 7A). The data we obtained are consistent with a Reuss model (see Materials and Methods), which predicts a storage modulus G′co of 356 ± 188 Pa (compared to a measured value of 160 ± 100 Pa) and a loss modulus G″co of 576 ± 283 Pa (compared to a measured value of 121 ± 70 Pa). For this model, we assume both B. subtilis and P. agglomerans are approximately incompressible and therefore have a Poisson ratio of 1/2 (40). One possible reason for the underprediction by the model is that there could be slip present between the various components of the biofilm.
FIG 7.
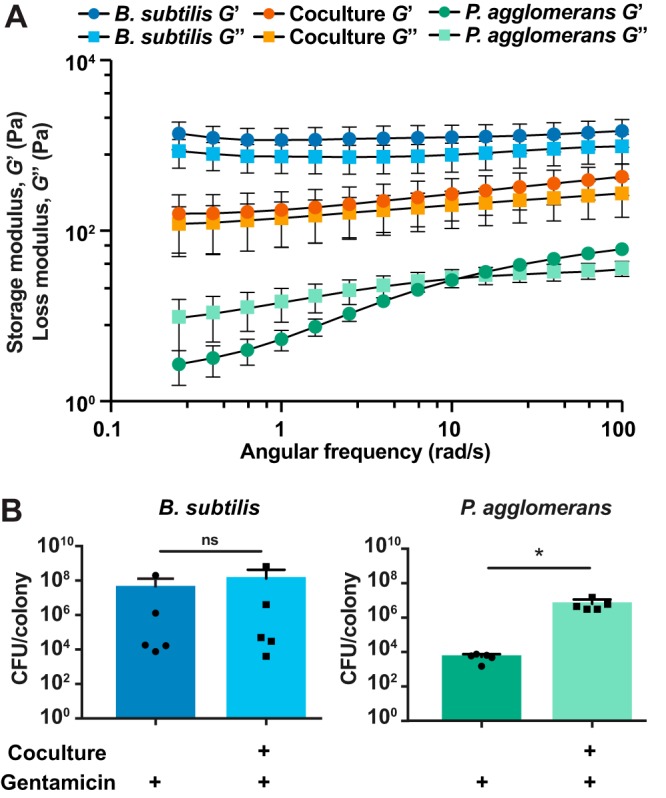
Physical properties of each organism impact the coculture structure and function. (A) Storage modulus (G′, elastic component) and loss modulus (G″, viscous component) of B. subtilis and P. agglomerans biofilms in monoculture and coculture measured over a range of frequencies after 48 h of growth on MSgg (n = 5). Error bars represent standard errors of means. (B) CFU per colony counts of B. subtilis and P. agglomerans in monoculture and coculture biofilms treated with gentamicin for 24 h after 24 h of prior growth. Error bars indicate standard deviations. *, P < 0.05. Statistical significance was determined by a two-tailed t test.
Coculture confers antibiotic resistance to cells within the coculture.
We next asked whether the coculture conferred any functional benefits to the individual bacteria. To test this, we applied antibiotic stress (in the form of gentamicin) to both monoculture and coculture colonies after 24 h of growth. Biofilms were harvested at 48 h and CFU per colony were quantified (Fig. 7B). When gentamicin was spotted onto the top of the colony, B. subtilis cell numbers were significantly reduced when treated in monoculture or coculture compared to that in untreated colonies (Fig. 7B; see also Fig. S8). In contrast, growth within the coculture protected P. agglomerans from antibiotic killing: P. agglomerans was killed when exposed to gentamicin in monoculture (Fig. 7B), but significantly more cells survived when treated in the coculture (Fig. 7B). These data indicated that the coculture provides protection to some constituent cells when facing gentamicin stress.
DISCUSSION
Although most natural biofilms are composed of multispecies communities, much of what we have learned about biofilm formation is from single-species studies. Here, we add to a growing body of literature exploring the genetic determinants and physical properties of bacterial cocultures by examining a dual-species biofilm formed between B. subtilis and P. agglomerans. We determined that these two bacteria produce a wrinkled biofilm in coculture and that this structure requires three components: (i) unchained single B. subtilis cells, (ii) the B. subtilis matrix protein TasA, and (iii) a P. agglomerans exopolysaccharide. Our rheology measurements indicate that the mechanical properties of the monoculture biofilms differ by several orders of magnitude, while the coculture moduli are indicative of mechanical contributions from both B. subtilis and P. agglomerans on the basis of a simple composite theory model. Rheology studies have been performed on monoculture B. subtilis colonies (41). However, as far as we are aware, this is the first examination of the mechanical properties of a coculture colony biofilm, since previous rheology studies on mixed-species biofilms have focused on submerged flow cell systems (40). We also showed that, depending on the measures evaluated, this dual-species interaction can be both detrimental (fewer P. agglomerans cells overall survive in coculture) and beneficial (the coculture structure protected P. agglomerans from environmental assaults such as antibiotics).
Both B. subtilis and P. agglomerans contribute matrix components to the coculture biofilm structure. TasA, the only characterized B. subtilis extracellular matrix component essential for this coculture phenotype, provides structural integrity to B. subtilis biofilms by anchoring cells to one another (42), and tasA colonies that are lacking these connections have a reduced ability to expand (30). The disruption of these cell-cell interactions in tasA strains may decrease the storage modulus of a B. subtilis biofilm, which may shorten the wrinkle wavelength and amplitude (43). In contrast to the requirement for TasA, we showed that this coculture structure forms even with a B. subtilis eps mutant. The necessity of a P. agglomerans exopolysaccharide component suggests the intriguing idea that the P. agglomerans EPS may be functionally complementing the EPS that B. subtilis typically produces. Although previous studies have detailed enhanced metabolic cooperation (44), antibiotic resistance (14), and spatial organization (45) in mixed-species communities, to our knowledge, the ability to share an exopolysaccharide in an interspecies interaction has not been shown. Going forward, it would thus be particularly interesting to determine the spatial organization of specific biofilm matrix components within this mixed-species structure using established methods (46) to obtain insights into how B. subtilis and P. agglomerans each contribute to the dual-species structure.
When viable B. subtilis and P. agglomerans cells were quantified from the coculture, our data indicated that growth in coculture was detrimental to P. agglomerans survival. Upon treatment of the coculture with gentamicin, however, P. agglomerans survived better than in monoculture, indicating that under these conditions, P. agglomerans benefited from growth in coculture. The notion that competition and cooperation are context dependent is supported by other diverse studies, such as those examining bacterial interactions mediated by kin discrimination (47), cellular positioning (48), and antibiotic production (49). Functionally, our results suggest that the “outer layer” of B. subtilis cells that forms over the coculture colony may be acting as a protective shield over the internal P. agglomerans cells. Our SEM micrographs suggest that the B. subtilis layer is at least 60 to 80 μm thick, and measurements of oxygen levels in the colonies of other bacterial species indicate that anoxic zones are present at a 60-μm depth. While the position of B. subtilis in the outer layer of the biofilm may provide B. subtilis cells with access to oxygen (50), allowing them to dominate the coculture in terms of cell numbers, we speculate that the thick B. subtilis layer may cut off P. agglomerans’ access to oxygen. This may indirectly protect P. agglomerans from antibiotics by forcing them into an anaerobic state, where growth is reduced (51); if the antibiotic requires cellular activity, the reduction of P. agglomerans growth would then render the antibiotic ineffective (52). Other groups have similarly observed increased resistance to antibiotics in multispecies biofilms (13), although no structural mechanism for this resistance was implied in those studies.
The mechanisms governing the spatial segregation of B. subtilis and P. agglomerans within the coculture colony are not yet clear. There is evidence in the literature that pattern formation emerges when microbes compete for resources in dense communities (53). The partitioning we observe could also simply be due to cellular morphology. For instance, previous studies suggest that cells in some biofilms may partition based on nutrient availability (54) or based on cell shape, which resulted in shorter cells lying on top of longer cells (55). However, we observed the opposite segregation patterns (i.e., the longer chained B. subtilis cells gravitated toward the outside structure). This difference could simply be due to the consequences of coculturing two physiologically distinct organisms (rather than shape-altered variants of the same bacterial species that were previously examined) (55). It would therefore be informative to utilize newly developed microscopy techniques that enable single-cell imaging of bacterial colony formation (56) to visualize how B. subtilis and P. agglomerans cells orient and assemble themselves during coculture biofilm formation.
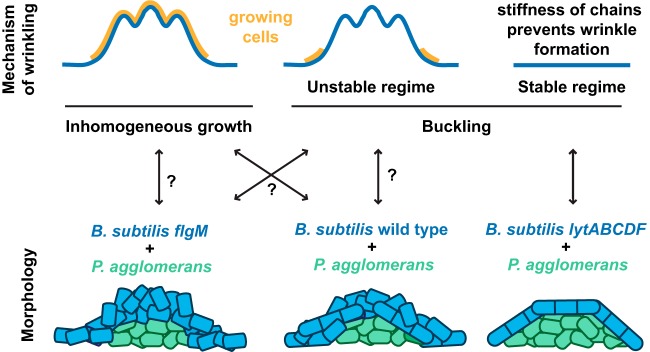
The observed spatial segregation of the B. subtilis and P. agglomerans coculture informed our interpretation of how this colony responded to mechanical stress. In this coculture, we observed an upper shell composed of an elastic behavior-dominated (G′ > G″) B. subtilis layer above a central viscous behavior-dominated (G″ > G′) “droplet” of P. agglomerans (Fig. 4, 5, and 7). This elastic shell of B. subtilis extends outwards, past the central droplet of P. agglomerans and directly onto the agar substrate. This B. subtilis shell appears to prevent the viscous behavior-dominated P. agglomerans from pushing upward through the shell or growing outward onto the agar. Because wrinkle formation only occurs for some bacteria, we can use these observations to test theories about the mechanisms underlying wrinkle formation. Wrinkle formation in B. subtilis biofilms has been proposed to occur through one of the two main mechanisms (18, 19, 21, 57, 58) shown schematically in Fig. 8. The first mechanism, inhomogeneous growth, applies in any circumstance where the outer material is growing faster than the inner material and leads to wrinkles within the growing regions. Under the second mechanism, buckling, there must be compressive forces that cause the material to buckle after it has already grown; buckling arises when cells encounter spatial constraints (19) or adhere to the agar (18).
FIG 8.
Schematic images illustrating possible mechanisms of wrinkling and the spatial organization of B. subtilis and P. agglomerans within the biofilm. (Top) Inhomogeneous growth occurs when the upper layer of the material grows faster than the interior, leading to wrinkling during growth. Buckling occurs in response to compressive stresses on materials that have already formed. Wrinkles occur only within unstable buckling regimes, when the compressive stresses are above a modulus-dependent threshold; this threshold, along with its wavelengths and amplitude, depends on both the geometry and properties of the thin sheet and its substrate (71, 72). In the stable buckling regime, the material is too stiff for the applied stresses and no buckling occurs. (Bottom) The colony morphology of each coculture is shown with the B. subtilis strain in blue (forming an elastic shell) and the P. agglomerans strain in green (forming a central “droplet” of more viscous material).
In the case of the B. subtilis wild-type or flgM cocultures, either the inhomogeneous growth or buckling mechanism could be invoked to explain the observed wrinkles (Fig. 8). Inhomogeneous growth could arise from, for example, B. subtilis cells in the outer shell having greater access to oxygen and nutrients due to their proximity to both the air and agar (11). In this situation, as B. subtilis reproduces, it would be easy for the B. subtilis cells to grow upward into the free space above the colony (rather than downward into the area occupied by P. agglomerans), leading to wrinkles throughout the central growing region (Fig. 8). We could also interpret the wild-type and flgM coculture morphologies in light of the buckling mechanism (Fig. 8). For instance, if only the B. subtilis cells with direct access to the agar are able to reproduce because of their access to nutrients, then their growth could generate a compressive (inward) stress, causing cells in the center of the colony to buckle into wrinkles (Fig. 8). Distinguishing between these wrinkling mechanisms is challenging based on morphology alone and, ideally, would be determined using controlled experiments where the wavelengths and amplitudes of wrinkled samples were correlated with their dimensions and moduli.
In contrast, when the B. subtilis lytABCDF mutant is cocultured with P. agglomerans, we observe no wrinkles. This could be because B. subtilis is nonmotile. These colonies contained packed, constitutively chained B. subtilis cells (Fig. 6A). We propose that this dense structure provides additional rigidity (59) that places this coculture within the stable buckling regime, where the B. subtilis shell is too stiff to wrinkle under the applied stresses (Fig. 8). Thus, by examining different B. subtilis strains in coculture with P. agglomerans, we speculate that we have been able to observe the shift either between two buckling regimes (unstable and stable) or between the two proposed wrinkling mechanisms (inhomogeneous growth and buckling) (Fig. 8).
Exploring the features of natural biofilms is critical to obtaining a mechanistic understanding of multispecies communities. Here, we studied isolates of B. subtilis and P. agglomerans that inhabit the same habitat and could potentially interact in the environment and subjected their biofilms to chemical and physical stressors they may experience in the natural world. Our work indicates that this interspecies interaction results in a biofilm structure with distinct extracellular matrix requirements, physical properties, and responses to antibiotic treatment from those observed for either B. subtilis or P. agglomerans when grown in isolation. Critically, these coculture biofilm characteristics are not predictable from studying monocultures alone. Thus, investigating coculture biofilms may provide us with insights into the underlying cell-cell interactions that drive the formation of structured communities in the environment as well as provide opportunities to develop models that are applicable to more complex microbial communities.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used in this study are listed in Table S1 in the supplemental material. We routinely cultured B. subtilis and P. agglomerans strains on lysogeny broth (LB)-Lennox medium (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl, 1.5% agar) at 30°C for 16 to 18 h with antibiotics as necessary. TY broth consisted of LB supplemented with 10 mM MgSO4 and 100 μM MnSO4 after autoclaving. Coculture assays were performed on MSgg medium (5 mM potassium phosphate [pH 7], 100 mM morpholinepropanesulfonic acid [MOPS; pH 7], 2 mM MgCl2, 700 μM CaCl2, 50 μM MnCl2, 50 μM FeCl3, 1 μM ZnCl2, 2 μM thiamine, 0.5% glycerol, 0.5% glutamate) with 1.5% agar. Antibiotics (final concentrations) were used as follows unless noted otherwise: MLS (1 μg/ml erythromycin, 25 μg/ml lincomycin), spectinomycin (100 μg/ml), kanamycin (15 μg/ml), tetracycline (10 μg/ml), and chloramphenicol (5 μg/ml). Escherichia coli strains were cultured in LB-Miller medium (10 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter NaCl, 1.5% agar). Antibiotics (final concentrations) were used as follows unless noted otherwise: kanamycin (50 μg/ml), chloramphenicol (50 μg/ml), and carbenicillin (50 μg/ml).
Coculture and secondary screens.
Strains were grown overnight and suspended to an OD600 of 0.5 in LB-Lennox broth. For monoculture colonies, 1 μl of each culture was spotted on MSgg agar. For coculture colonies, the two cultures were mixed at a 1:1 ratio by OD600 measurement, and 1 μl of the mixed culture was spotted on MSgg agar. Plates were incubated at 24°C, and colonies were imaged or harvested at times noted in the text and figures.
Colony morphology phase contrast and fluorescence imaging.
Macrocolony biofilm images were taken using a Zeiss SteREO Discovery.V8 dissecting stereomicroscope (Zeiss, Oberkochen, Germany) with a 1 × 0.63 lens objective. To obtain a colony side-profile view, a razor blade was used to excise an agar block surrounding the colony. Agar blocks including the colonies were removed from the plate and mounted at 90° on a microscopy slide for imaging.
Scanning electron microscopy.
After 24 h of growth on MSgg, plates were vapor fixed by inverting plates over 10 ml of 8% paraformaldehyde and 12.5% glutaraldehyde for 24 h (60). Agar blocks containing whole and longitudinally sectioned colonies were harvested; for longitudinal colony sections, a scalpel was used to slice the colony in half. The blocks were immersed in 1% osmium tetroxide in 0.05 M sodium cacodylate buffer (pH 7.4) solution for 1 h, followed by 3 washes in sodium cacodylate buffer (pH 7.4). The samples were placed in baskets submerged in 30% ethanol and further dehydrated by transferring through increasing concentrations of ethanol (50%, 75%, 90%, and 100% twice) for 20 min each step. The colonies were transferred in 100% ethanol to a critical-point dryer and dried using liquid carbon dioxide as the transitional solvent. Agar blocks were mounted on aluminum stubs, and for longitudinal sections, both halves were mounted on their sides to view the cut edges. The samples were sputter coated with gold-palladium alloy (60 Au/40 Pd) to a thickness of 10 nm. Images were taken using a Zeiss Supra 25 field emission SEM with an in-lens secondary electron detector at an accelerating voltage of 5 kV in high-vacuum mode.
Confocal laser scanning microscopy.
Autoclaved glass 22-mm by 22-mm coverslips were placed on an MSgg agar plate and then coated with an additional 1 ml of molten MSgg agar medium. After solidification, coculture colonies for CLSM were grown as described earlier. At 24 h or 48 h, the glass coverslip was then gently removed from the agar with forceps to keep the colony intact, and the bottom coverslip surface was cleaned and then placed upright on the ZEISS 780 microscope. Individual images were taken at ×100 magnification with tile scans and Z-stacks as composites.
Transposon mutagenesis library construction, screen, and identification.
P. agglomerans was mutagenized by mating with E. coli containing the pKMW3 vector (61) on medium supplemented with diaminopimelic acid (DAP; 300 μM) at 30°C overnight. P. agglomerans mutants were harvested, suspended in LB-Lennox broth with 20% glycerol, and stored in a 96-well plate format at −80°C. From frozen stocks, small volumes of P. agglomerans mutants were inoculated in LB-Lennox broth using a 96-prong replica plater (model number 140500; Boekel Scientific, Feasterville, PA) and grown overnight at 30°C. After overnight growth, ∼1 μl P. agglomerans mutant cultures was spotted on MSgg in a Nunc OmniTray using the 96-prong replica plater. Before the P. agglomerans spots dried, B. subtilis cells were spotted on top of the P. agglomerans mutant cells using the 96-prong replica plater. Plates were incubated at 24°C for 48 h and hits were identified visually. For validation, hits were tested in coculture with B. subtilis as described above. We identified genes containing the transposon insertion by a rescue cloning method. Briefly, genomic DNA of transposon mutants was purified using the Qiagen DNA Easy blood and tissue kit (Hilden, Germany) and digested with AfeI, AgeI, or BamHI restriction enzymes from New England BioLabs (Ipswich, MA). The digested DNA was concentrated and ligated with T4 ligase buffer (New England BioLabs). The resulting plasmid was transformed into E. coli [F− Δlac169 rpoS(Am) robA1 creC510 hsdR514 endA recA1 uidA(ΔMluI)::pir-116] cells, and transformants were selected on LB medium supplemented with kanamycin. The plasmid was purified from transformants and sent for sequencing using primer ES579 (62) and primer ES630 (this study) (see Table S1), which annealed to the transposon in order to sequence the gene containing the insertion bidirectionally (61).
Construction of Pantoea agglomerans mutants.
A two-step allelic-exchange method was employed to generate clean gene deletion mutants in P. agglomerans. To make the amsC gene deletion, a 500-bp fragment upstream of the amsC gene was amplified from P. agglomerans with primers ES880 and ES881 (see Table S1) and a 500-bp fragment downstream of the amsC gene was amplified with primers ES882 and ES883. Both fragments were cloned into pSR47S (63) predigested with SalI and BamHI using an E5510 Gibson assembly cloning kit (NEB), resulting in pSR47S-amsC. The resulting plasmid was transformed into E. coli PIR1, and transformants were selected for kanamycin resistance. Kanamycin-resistant transformants were checked by colony PCR for the inserts, and the plasmids from the positive hits were sequenced using primers up- and downstream of the pSR47S multiple cloning site primer ES896 and primer ES897 (Table S1). P. agglomerans amsC transconjugants were generated by electroporating pSR47S-amsC into P. agglomerans electrocompetent cells, and mutants were selected for kanamycin resistance. To select for the second crossover event, the resulting P. agglomerans transconjugant colonies were struck out on 5% and 10% sucrose (vol/vol) 1× LB agar. The resulting colonies were then screened by colony PCR for deletion of amsC using primer ES871 and primer ES872, up- and downstream of the amsC gene. P. agglomerans-positive hits were then confirmed to be kanamycin sensitive and PCR checked using primer ES915 and primer ES916 (Table S1). Complementation of the mutant was achieved by amplifying the amsC gene and the 300-bp region upstream of the gene using primer ES898 and primer ES900 from P. agglomerans. The gene was then cloned into pBBR1MCS (64) predigested with SacI and HindIII using an E5510 Gibson assembly master mix (NEB). The resulting plasmid, pBBR1MCS-amsC, was transformed into E. coli TOP10 electrocompetent cells and transformants were selected for chloramphenicol resistance. Chloramphenicol-resistant transformants were checked by colony PCR for the insert, and the plasmids from the positive hits were sequenced using the primers to amplify the insert, primer ES898 and primer ES900 (Table S1). The complementing plasmid was introduced into P. agglomerans amsC mutant by electroporation, and strains harboring the plasmid were selected for chloramphenicol resistance. Growth of newly constructed strains was monitored in shaking liquid culture using a Synergy H1 plate reader (BioTek, Winooski, VT).
Pantoea agglomerans genomic sequencing.
Genomic DNA was extracted from P. agglomerans using a standard phenol-chloroform extraction protocol. Library preparation and genome sequencing were performed by the University of North Carolina School of Medicine High-Throughput Sequencing Facility (HTSF) using the Illumina MiSeq platform. The quality control of the sequence data was determined using FastQC (65). To assemble the genome, adaptor sequence removal, trimming, error correction, and de novo assembly were performed using the A5-miseq-pipeline (66), and the assembly was analyzed with QUAST (67).
Phylogenetic tree construction.
16S rRNA sequences from NCBI for the Bacillus strains (see Table S1) were trimmed and assembled using DNASTAR Lasergene 14. We constructed a phylogenetic tree in ClustalX using Paenibacillus polymyxa as the outgroup.
TasA homology.
The amino acid identity between TasA in Bacillus subtilis subsp. subtilis strain NCIB 168 and the other bacilli shown in Fig. S2 (see Table S1 for strain details) was determined by inputting the 168 TasA amino acid sequence (CDS NP_390342.1) into NCBI Protein BLAST and searching the genome sequences available for those strains or their species.
Rheology.
To grow cultures suitable for use in a rheometer, a Whatman nucleopore polycarbonate filter (5 cm, 0.3 μm pore; Maidstone, United Kingdom) was placed on top of an MSgg plate using sterile forceps. One hundred microliters of each monoculture and coculture at an OD600 of 0.5 was spread on top of the membrane. After incubation for 48 h, the polycarbonate membrane and biofilm were transferred to the bottom plate of the Anton Paar MCR 302 rheometer (Graz, Austria) using sterile forceps. The measurement was performed in a standard cone-plate geometry; the top plate is a cone of diameter 5 cm and the cone angle was 1°. We measured the complex shear modulus (G′, G″) using a frequency sweep from 0.25 rad/s to 100 rad/s at constant 5% strain and a gap size of 0.102 mm.
To understand how the coculture moduli are influenced by the components of B. subtilis and P. agglomerans, we applied the rule of bonded mixtures for calculating the moduli of composite materials. The bonded mixture model can be understood by considering the composite material to be composed of two incompressible homogeneous materials; two relevant models are the Voigt model and the Reuss model. The Voigt model arises from considering a sample under uniaxial strain: this is analogous to connecting two springs of differing stiffness in parallel. As such, the two values of shear modulus add in proportion to the fraction f that is present: Gco = fGB + (1 – f)Gp. Here, G is the storage or loss shear modulus, with the subscripts denoting coculture (co), B. subtilis (B), and P. agglomerans (P). The Reuss model, in contrast, arises from considering a sample under uniaxial stress: this is analogous to connecting two springs of differing stiffness in series. As such, the two inverse values of shear modulus add in proportion to the fraction f that is present: Gco = (f/GB + [1 – f]/Gp)−1. Incompressible homogeneous materials comprising a more complicated configuration, with a mix of material elements connected in series and in parallel, would result in shear moduli lying between the Voigt upper bound and the Reuss lower bound.
Construction of B. subtilis flgM::tet.
The flgM::tet insertion-deletion allele was generated by long-flanking-homology PCR using primer pairs ES981/ES982 and ES983/ES984 (Table S1), and DNA containing a tetracycline drug resistance gene (pDG1515) was used as a template for marker replacement (68, 69).
SPP1 phage transduction.
Phage transduction was carried out as previously described (70). Briefly, B. subtilis donor strain was grown at 37°C in TY broth until the culture reached an OD600 of 1.0. Cells were infected with SPP1 phage stock and plated on 0.5% TY soft top agar, overlaid on TY 1.5% agar plates, and incubated at 37°C for 8 to 16 h. B. subtilis donor phage plaques were collected and pelleted using a clinical centrifuge. Three hundred microliters of supernatant was used to infect B. subtilis recipient cells, and then the cell lysate was plated on LB-Lennox with 10 mM citrate and antibiotic to which the donor strain was resistant. Plates were incubated at 37°C for 12 to 24 h. Three colonies were picked from each phage transduction and struck on LB-Lennox plates with antibiotic. After growth, strains were restruck two more times on LB-Lennox plates with antibiotic. Cells were spotted on MSgg and incubated at 30°C to ensure growth, which indicates the cells have a 3610 rather than a 168 background (which is an amino acid auxotroph). Specifically, to construct ES148 (hag::tet PtapA-yfp), a previously constructed 168 mutant containing hag::tet (HV1145, kindly provided by Hera Vlamakis) was transduced into B. subtilis 3610 containing PtapA-yfp. B. subtilis hag::tet PtapA-yfp colonies were selected on Lennox plates containing tetracycline and citrate. To construct B. subtilis tasA::kan PtapA-yfp, B. subtilis tasA::kan eps::tet phage was transduced into a PtapA-yfp-containing B. subtilis recipient strain. B. subtilis tasA::kan PtapA-yfp colonies were selected on Lennox supplemented with kanamycin and citrate. To construct B. subtilis flgM tasA, we infected B. subtilis flgM::tet with B. subtilis tasA::kan eps::tet phage and plated cells on Lennox with kanamycin and citrate.
Construction of a constitutively fluorescent B. subtilis strain.
A plasmid containing mTurquoise2 (mTurq) was generated using primer ES395 and primer ES315 (see Table S1) to amplify mTurq from GL-FP-31. The fragment was inserted in an already constructed plasmid pEA003 [amyE::PspacC-cfp(camR)] and cut with HindIII and BamHI. Upon construction, the linearized plasmid was transformed into B. subtilis 168 cells grown to stationary phase. Cells were plated on Lennox-chloramphenicol to select for transformants. B. subtilis PspacC-mTurq phage was used to infect the B. subtilis wild type and the flgM and lytABCDF mutants. Cells were plated on Lennox plates with chloramphenicol and citrate to select for B. subtilis cells that contained PspacC-mTurq.
Construction of a constitutively fluorescent P. agglomerans strain.
A plasmid containing Ypet driven by a constitutive promoter PspacC was constructed using ES429 and ES430 (Table S1) primers to amplify Ypet. The product was inserted into pES036 contained in strain ES739 and digested with BamHI and HindIII. The resulting plasmid was transformed into P. agglomerans competent cells using an Eppendorf Electroporator 2510 (Eppendorf, Hamburg, Germany) at 1,800 V. Transformed cells were recovered by the addition of SOC (2% tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, and 20 mM glucose) and incubated with rolling at 37°C for 1 h. Recovered cells were plated on LB-Miller agar plates with carbenicillin.
Antibiotic treatment of biofilms.
Monoculture and coculture cultures were spotted on MSgg as previously described. After 24 h of incubation at 24°C, the colonies were spotted with 10 μl of 500 μg/ml gentamicin and returned to incubation at 24°C. After 48 h of total growth, colonies were harvested to quantify CFU per colony.
Data availability.
This whole-genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number SIJO00000000 (data may be downloaded at https://www.ncbi.nlm.nih.gov/Traces/wgs/SIJO01?display=contigs). The version described in this paper is version SIJO01000000.
Supplementary Material
ACKNOWLEDGMENTS
We thank Daniel Kearns (Indiana University) for providing B. subtilis flgM and lytABCDF strains, Ethan Garner (Harvard University) for the codon-optimized mTurq sequence (GL-FP-31) used in our B. subtilis strain construction, Susanna Harris for assistance with imaging biofilms by confocal laser scanning microscopy, Howard Hughes Medical Institute for funding for the Harvard University undergraduate project lab that led to the isolation of P. agglomerans, Kriti Sharma and Jamie Winshell for constructing B. subtilis PspacC-mTurq and PspacC-Ypet, Lews Caro for assistance in screening the P. agglomerans transposon mutant library, Vicky Madden, Kristen White, and the Microscopy Services Laboratory in the Department of Pathology and Laboratory Medicine at UNC at Chapel Hill for assistance with the colony sample preparation for SEM and use of a Zeiss Supra 25 FESEM, and the UNC Biology Department Microscopy Core for use of a Zeiss-780 confocal laser scanning microscope.
This work was supported by research funds provided by the National Institutes of Health (GM112981 to E.A.S.) and the National Science Foundation (INSPIRE IOS-1343020 to E.A.S. and DMR-1608097 to K.E.D.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00670-18.
REFERENCES
- 1.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 2.Flemming H-C, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 3.Mah TF, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 4.Hobley L, Harkins C, MacPhee CE, Stanley-Wall NR. 2015. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol Rev 39:649–669. doi: 10.1093/femsre/fuv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matz C, Kjelleberg S. 2005. Off the hook–how bacteria survive protozoan grazing. Trends Microbiol 13:302–307. doi: 10.1016/j.tim.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Stewart PS. 2012. Mini-review: convection around biofilms. Biofouling 28:187–198. doi: 10.1080/08927014.2012.662641. [DOI] [PubMed] [Google Scholar]
- 7.Cooley BJ, Thatcher TW, Hashmi SM, L'Her G, Le HH, Hurwitz DA, Provenzano D, Touhami A, Gordon VD. 2013. The extracellular polysaccharide Pel makes the attachment of P. aeruginosa to surfaces symmetric and short-ranged. Soft Matter 9:3871–3876. doi: 10.1039/c3sm27638d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nadell CD, Drescher K, Wingreen NS, Bassler BL. 2015. Extracellular matrix structure governs invasion resistance in bacterial biofilms. ISME J 9:1700–1709. doi: 10.1038/ismej.2014.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Persat A, Nadell CD, Kim MK, Ingremeau F, Siryaporn A, Drescher K, Wingreen NS, Bassler BL, Gitai Z, Stone HA. 2015. The mechanical world of bacteria. Cell 161:988–997. doi: 10.1016/j.cell.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nadell CD, Drescher K, Foster KR. 2016. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol 14:589–600. doi: 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- 11.Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat Rev Microbiol 6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 12.Volfson D, Cookson S, Hasty J, Tsimring LS. 2008. Biomechanical ordering of dense cell populations. Proc Natl Acad Sci U S A 105:15346–15351. doi: 10.1073/pnas.0706805105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burmølle M, Webb JS, Rao D, Hansen LH, Sørensen SJ, Kjelleberg S. 2006. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl Environ Microbiol 72:3916–3923. doi: 10.1128/AEM.03022-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee KWK, Periasamy S, Mukherjee M, Xie C, Kjelleberg S, Rice SA. 2014. Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J 8:894–907. doi: 10.1038/ismej.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren D, Madsen JS, Sørensen SJ, Burmølle M. 2015. High prevalence of biofilm synergy among bacterial soil isolates in cocultures indicates bacterial interspecific cooperation. ISME J 9:81–89. doi: 10.1038/ismej.2014.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A 98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ben Amar M, Wu M. 2014. Patterns in biofilms: from contour undulations to fold focussing. Europhys Lett 108:38003. doi: 10.1209/0295-5075/108/38003. [DOI] [Google Scholar]
- 19.Trejo M, Douarche C, Bailleux V, Poulard C, Mariot S, Regeard C, Raspaud E. 2013. Elasticity and wrinkled morphology of Bacillus subtilis pellicles. Proc Natl Acad Sci U S A 110:2011–2016. doi: 10.1073/pnas.1217178110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh P, Ben-Jacob E, Levine H. 2013. Modeling cell-death patterning during biofilm formation. Phys Biol 10:066006. doi: 10.1088/1478-3975/10/6/066006. [DOI] [PubMed] [Google Scholar]
- 21.Asally M, Kittisopikul M, Rue P, Du Y, Hu Z, Cagatay T, Robinson AB, Lu H, Garcia-Ojalvo J, Suel GM. 2012. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proc Natl Acad Sci U S A 109:18891–18896. doi: 10.1073/pnas.1212429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trinschek S, John K, Lecuyer S, Thiele U. 2017. Continuous versus arrested spreading of biofilms at solid-gas interfaces: the role of surface forces. Phys Rev Lett 119:078003. doi: 10.1103/PhysRevLett.119.078003. [DOI] [PubMed] [Google Scholar]
- 23.Branda SS, Chu F, Kearns DB, Losick R, Kolter R. 2006. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol 59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- 24.Romero D, Aguilar C, Losick R, Kolter R. 2010. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A 107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi K, Iwano M. 2012. BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol Microbiol 85:51–66. doi: 10.1111/j.1365-2958.2012.08094.x. [DOI] [PubMed] [Google Scholar]
- 26.Walterson AM, Stavrinides J. 2015. Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol Rev 39:968–984. doi: 10.1093/femsre/fuv027. [DOI] [PubMed] [Google Scholar]
- 27.Tecon R, Leveau JHJ. 2016. Symplasmata are a clonal, conditional, and reversible type of bacterial multicellularity. Sci Rep 6:31914. doi: 10.1038/srep31914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dolph PJ, Majerczak DR, Coplin DL. 1988. Characterization of a gene cluster for exopolysaccharide biosynthesis and virulence in Erwinia stewartii. J Bacteriol 170:865–871. doi: 10.1128/jb.170.2.865-871.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.López D, Fischbach M, Chu F, Losick R, Kolter R. 2009. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci U S A 106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Gestel J, Vlamakis H, Kolter R. 2015. From cell differentiation to cell collectives: Bacillus subtilis uses division of labor to migrate. PLoS Biol 13:e1002141. doi: 10.1371/journal.pbio.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chai Y, Kolter R, Losick R. 2010. Reversal of an epigenetic switch governing cell chaining in Bacillus subtilis by protein instability. Mol Microbiol 78:218–229. doi: 10.1111/j.1365-2958.2010.07335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caramori T, Barill D, Nessi C, Sacchi L, Galizzi A. 1996. Role of FlgM in σD-dependent gene expression in Bacillus subtilis. J Bacteriol 178:3113–3118. doi: 10.1128/jb.178.11.3113-3118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Courtney CR, Cozy LM, Kearns DB. 2012. Molecular characterization of the flagellar hook in Bacillus subtilis. J Bacteriol 194:4619–4629. doi: 10.1128/JB.00444-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith TJ, Blackman SA, Foster SJ. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249–262. doi: 10.1099/00221287-146-2-249. [DOI] [PubMed] [Google Scholar]
- 35.Cozy LM, Kearns DB. 2010. Gene position in a long operon governs motility development in Bacillus subtilis. Mol Microbiol 76:273–285. doi: 10.1111/j.1365-2958.2010.07112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon VD, Davis-Fields M, Kovach K, Rodesney CA. 2017. Biofilms and mechanics: a review of experimental techniques and findings. J Phys D Appl Phys 50:223002. doi: 10.1088/1361-6463/aa6b83. [DOI] [Google Scholar]
- 37.Peterson BW, He Y, Ren Y, Zerdoum A, Libera MR, Sharma PK, van Winkelhoff A-J, Neut D, Stoodley P, van Der Mei HC, Busscher HJ. 2015. Viscoelasticity of biofilms and their recalcitrance to mechanical and chemical challenges. FEMS Microbiol Rev 39:234–245. doi: 10.1093/femsre/fuu008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korstgens V, Flemming H-C, Wingender J, Borchard W. 2001. Uniaxial compression measurement device for investigation of the mechanical stability of biofilms. J Microbiol Methods 46:9–17. doi: 10.1016/S0167-7012(01)00248-2. [DOI] [PubMed] [Google Scholar]
- 39.Shaw T, Winston M, Rupp CJ, Klapper I, Stoodley P. 2004. Commonality of elastic relaxation times in biofilms. Phys Rev Lett 93:098102. doi: 10.1103/PhysRevLett.93.098102. [DOI] [PubMed] [Google Scholar]
- 40.Towler BW, Rupp CJ, Cunningham AB, Stoodley P. 2003. Viscoelastic properties of a mixed culture biofilm from rheometer creep analysis. Biofouling 19:279–285. doi: 10.1080/0892701031000152470. [DOI] [PubMed] [Google Scholar]
- 41.Kesel S, Grumbein S, Gümperlein I, Tallawi M, Marel A-K, Lieleg O, Opitz M. 2016. Direct comparison of physical properties of Bacillus subtilis NCIB 3610 and B-1. Appl Environ Microbiol 82:2424–2432. doi: 10.1128/AEM.03957-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romero D, Vlamakis H, Losick R, Kolter R. 2011. An accessory protein required for anchoring and assembly of amyloid fibers in B. subtilis biofilms. Mol Microbiol 80:1155–1168. doi: 10.1111/j.1365-2958.2011.07653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerda E, Mahadevan L. 2003. Geometry and physics of wrinkling. Phys Rev Lett 90:074302. doi: 10.1103/PhysRevLett.90.074302. [DOI] [PubMed] [Google Scholar]
- 44.Breugelmans P, Barken KB, Tolker-Nielsen T, Hofkens J, Dejonghe W, Springael D. 2008. Architecture and spatial organization in a triple-species bacterial biofilm synergistically degrading the phenylurea herbicide linuron. FEMS Microbiol Ecol 64:271–282. doi: 10.1111/j.1574-6941.2008.00470.x. [DOI] [PubMed] [Google Scholar]
- 45.Xiao J, Klein MI, Falsetta ML, Lu B, Delahunty CM, Yates JR III, Heydorn A, Koo H. 2012. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog 8:e1002623. doi: 10.1371/journal.ppat.1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berk V, Fong JCN, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, Yildiz FH, Chu S. 2012. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science 337:236–239. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lyons NA, Kolter R. 2017. Bacillus subtilis protects public goods by extending kin discrimination to closely related species. mBio 8:e00723-17. doi: 10.1128/mBio.00723-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frost I, Smith WPJ, Mitri S, Millan AS, Davit Y, Osborne JM, Pitt-Francis JM, MacLean CR, Foster KR. 2018. Cooperation, competition and antibiotic resistance in bacterial colonies. ISME J 12:1582–1593. doi: 10.1038/s41396-018-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cordero OX, Wildschutte H, Kirkup B, Proehl S, Ngo L, Hussain F, Le Roux F, Mincer T, Polz MF. 2012. Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science 337:1228–1231. doi: 10.1126/science.1219385. [DOI] [PubMed] [Google Scholar]
- 50.Kim W, Racimo F, Schluter J, Levy SB, Foster KR. 2014. Importance of positioning for microbial evolution. Proc Natl Acad Sci U S A 111:E1639–E1647. doi: 10.1073/pnas.1323632111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dietrich LEP, Okegbe C, Price-Whelan A, Sakhtah H, Hunter RC, Newman DK. 2013. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J Bacteriol 195:1371–1380. doi: 10.1128/JB.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hancock REW. 1981. Aminoglycoside uptake and mode of action–with special reference to streptomycin and gentamicin. I. Antagonists and mutants. J Antimicrob Chemother 8:249–276. doi: 10.1093/jac/8.4.249. [DOI] [PubMed] [Google Scholar]
- 53.Mitri S, Clarke E, Foster KR. 2016. Resource limitation drives spatial organization in microbial groups. ISME J 10:1471–1482. doi: 10.1038/ismej.2015.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Serra DO, Richter AM, Klauck G, Mika F, Hengge R. 2013. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. mBio 4:e00103-13. doi: 10.1128/mBio.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith WPJ, Davit Y, Osborne JM, Kim W, Foster KR, Pitt-Francis JM. 2017. Cell morphology drives spatial patterning in microbial communities. Proc Natl Acad Sci U S A 114:E280–E286. doi: 10.1073/pnas.1613007114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan J, Sharo AG, Stone HA, Wingreen NS, Bassler BL. 2016. Vibrio cholerae biofilm growth program and architecture revealed by single-cell live imaging. Proc Natl Acad Sci U S A 113:E5337–E5343. doi: 10.1073/pnas.1611494113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilking JN, Angelini TE, Seminara A, Brenner MP, Weitz DA. 2011. Biofilms as complex fluids. MRS Bull 36:385–391. doi: 10.1557/mrs.2011.71. [DOI] [Google Scholar]
- 58.Wilking JN, Zaburdaev V, De Volder M, Losick R, Brenner MP, Weitz DA. 2013. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proc Natl Acad Sci U S A 110:848–852. doi: 10.1073/pnas.1216376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu AJ, Nagel SR. 2010. The jamming transition and the marginally jammed solid. Annu Rev Condens Matter Phys 1:347–369. doi: 10.1146/annurev-conmatphys-070909-104045. [DOI] [Google Scholar]
- 60.Jones B, Young R, Mahenthiralingam E, Stickler DJ. 2004. Ultrastructure of Proteus mirabilis swarmer cell rafts and role of swarming in catheter-associated urinary tract infection. Infect Immun 72:3941–3950. doi: 10.1128/IAI.72.7.3941-3950.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wetmore KM, Price MN, Waters RJ, Lamson JS, He J, Hoover CA, Blow MJ, Bristow J, Butland G, Arkin AP, Deutschbauer A. 2015. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. mBio 6:e00306-15. doi: 10.1128/mBio.00306-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bouhenni R, Gehrke A, Saffarini D. 2005. Identification of genes involved in cytochrome c biogenesis in Shewanella oneidensis, using a modified mariner transposon. Appl Environ Microbiol 71:4935–4937. doi: 10.1128/AEM.71.8.4935-4937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Merriam JJ, Mathur R, Maxfield-Boumil R, Isberg RR. 1997. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect Immun 65:2497–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kovach ME, Phillips RW, Elzer PH, Roop RM, Peterson KM. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800–802. [PubMed] [Google Scholar]
- 65.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 66.Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 67.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guérout-Fleury A-M, Shazand K, Frandsen N, Stragier P. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 1:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 69.Wach A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259–265. doi:. [DOI] [PubMed] [Google Scholar]
- 70.Yasbin RE, Young FE. 1974. Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol 14:1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kundukad B, Seviour T, Liang Y, Rice SA, Kjelleberg S, Doyle PS. 2016. Mechanical properties of the superficial biofilm layer determine the architecture of biofilms. Soft Matter 12:5718–5726. doi: 10.1039/c6sm00687f. [DOI] [PubMed] [Google Scholar]
- 72.Dervaux J, Ben Amar M. 2008. Morphogenesis of growing soft tissues. Phys Rev Lett 101:068101. doi: 10.1103/PhysRevLett.101.068101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This whole-genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number SIJO00000000 (data may be downloaded at https://www.ncbi.nlm.nih.gov/Traces/wgs/SIJO01?display=contigs). The version described in this paper is version SIJO01000000.