Most bacteria spend a large part of their life spans attached to surfaces, forming complex multicellular communities called biofilms. Bacteria can colonize virtually any surface, and therefore, they have adapted to bind efficiently in very different environments. In this study, we compare the adhesive holdfasts produced by the freshwater bacterium C. crescentus and a relative, the marine bacterium H. baltica. We show that H. baltica holdfasts have a different morphology and chemical composition and tolerate high ionic strength. Our results show that the H. baltica holdfast is an excellent model to study the effect of ionic strength on adhesion and provides insights into the physicochemical properties required for adhesion in the marine environment.
KEYWORDS: adhesion, bacterial adhesin, Hirschia baltica, holdfast, ionic strength, marine Caulobacterales, polysaccharides
ABSTRACT
Bacterial adhesion is affected by environmental factors, such as ionic strength, pH, temperature, and shear forces. Therefore, marine bacteria must have developed adhesins with different compositions and structures than those of their freshwater counterparts to adapt to their natural environment. The dimorphic alphaproteobacterium Hirschia baltica is a marine budding bacterium in the clade Caulobacterales. H. baltica uses a polar adhesin, the holdfast, located at the cell pole opposite the reproductive stalk, for surface attachment and cell-cell adhesion. The holdfast adhesin has been best characterized in Caulobacter crescentus, a freshwater member of the Caulobacterales, and little is known about holdfast compositions and properties in marine Caulobacterales. Here, we use H. baltica as a model to characterize holdfast properties in marine Caulobacterales. We show that freshwater and marine Caulobacterales use similar genes in holdfast biogenesis and that these genes are highly conserved among the species in the two genera. We determine that H. baltica produces a larger holdfast than C. crescentus and that the holdfasts have different chemical compositions, as they contain N-acetylglucosamine and galactose monosaccharide residues and proteins but lack DNA. Finally, we show that H. baltica holdfasts tolerate higher ionic strength than those of C. crescentus. We conclude that marine Caulobacterales holdfasts have physicochemical properties that maximize binding in high-ionic-strength environments.
IMPORTANCE Most bacteria spend a large part of their life spans attached to surfaces, forming complex multicellular communities called biofilms. Bacteria can colonize virtually any surface, and therefore, they have adapted to bind efficiently in very different environments. In this study, we compare the adhesive holdfasts produced by the freshwater bacterium C. crescentus and a relative, the marine bacterium H. baltica. We show that H. baltica holdfasts have a different morphology and chemical composition and tolerate high ionic strength. Our results show that the H. baltica holdfast is an excellent model to study the effect of ionic strength on adhesion and provides insights into the physicochemical properties required for adhesion in the marine environment.
INTRODUCTION
In their natural environments, bacteria preferentially form surface-associated communities known as biofilms (1). To irreversibly adhere to surfaces and form these complex multicellular communities, bacteria produce strong adhesins, mainly composed of proteins or polysaccharides (2, 3). Bacterial adhesion is affected by different environmental conditions, such as pH, temperature, shear forces, and ionic strength (2, 4–6). In marine environments, bacteria face 500-times-higher ionic strength than in freshwater (7). Therefore, marine bacteria have evolved ways to overcome the effect of ionic strength and bind permanently to surfaces in high-salt environments, such as seas and oceans. Higher ionic strength might affect the structures of adhesins or neutralize their charges, impairing their interaction with surfaces (2).
Caulobacterales are Alphaproteobacteria found in various habitats from oligotrophic aquatic and nutrient-rich soil environments (8, 9). The aquatic Caulobacterales species live in a wide range of environments with different salinity levels, such as pristine fresh river and lake waters, brackish ponds, and marine waters, making them a good model for studying bacterial adhesion in different ionic environments. Caulobacterales species use a polar adhesin structure called a holdfast to adhere permanently to surfaces and form biofilms (8, 10, 11). The holdfast has been primarily studied in Caulobacter crescentus, a freshwater member of the Caulobacterales (2, 3, 12). The C. crescentus holdfast uses both electrostatic and hydrophobic interactions to attach to different surfaces (6). The binding affinity of the C. crescentus holdfast is dramatically impaired in the presence of NaCl (6), yet marine Caulobacterales adhere to surfaces at considerably higher ionic strength, suggesting that their holdfasts have different properties. However, little is known about holdfasts from marine Caulobacterales, and the molecular mechanism used to adhere successfully to surfaces in saline environments is currently unknown.
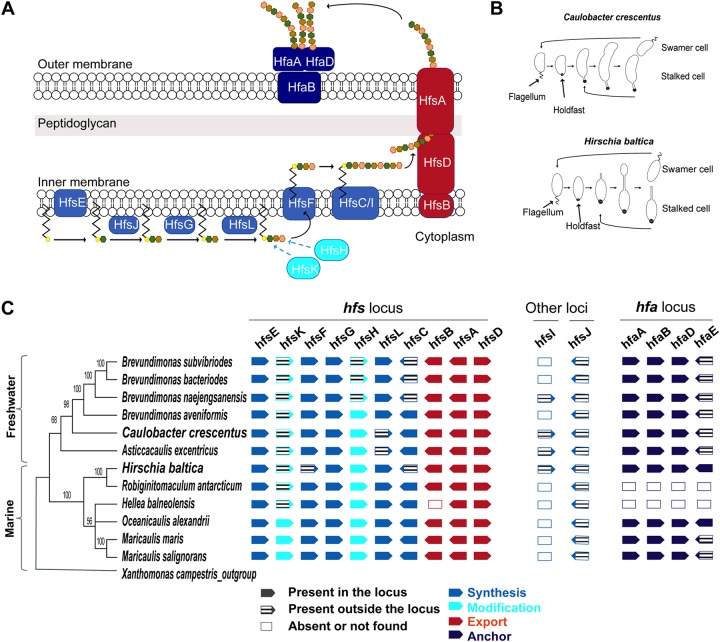
The C. crescentus holdfast is the strongest characterized bioadhesive, with an adhesion force of 70 N/mm2 (13). Despite being identified almost 85 years ago (14), the exact composition and structure of the C. crescentus holdfast remain elusive. Wheat germ agglutinin (WGA) lectin-binding assays show that the holdfast contains N-acetylglucosamine (GlcNAc) residues (10), while other studies suggest that the holdfast is also composed of unidentified peptide and DNA residues (15). The C. crescentus holdfast polysaccharide is produced via a polysaccharide synthesis and export pathway similar to the group I capsular polysaccharide synthesis Wzy/Wzx-dependent pathway in Escherichia coli (16, 17), leading to a model for the holdfast synthesis pathway (Fig. 1A). Holdfast polysaccharide synthesis is hypothesized to be initiated in the cytoplasm by the putative glycosyltransferase HfsE, which is thought to transfer activated sugar phosphate from UDP (UDP-glucose) to an undecaprenyl-phosphate (Und-P) lipid carrier (18). Additional sugar residues, including GlcNAc, are then added to form a repeat unit on the lipid carrier with three glycosyltransferases, HfsG, HfsJ (17), and HfsL (19). The acetyltransferase HfsK (20) and the polysaccharide deacetylase HfsH (30) modify one or more sugar residues. The lipid carrier with the repeat units is transported across the inner membrane into the periplasm by a flippase (HfsF) (17, 22). In the periplasm, the repeat units are polymerized by two polysaccharide polymerases HfsC and HfsI (17). The holdfast polysaccharide chain is then secreted through the export protein complex, composed of HfsA, HfsB, and HfsD (21, 23, 24). Once outside the cell, holdfast polysaccharides are anchored to the cell envelope by the action of holdfast anchor (hfa) proteins: HfaA, HfaB, HfaD, and HfaE (19, 25–27).
FIG 1.
Organization of the holdfast gene cluster in H. baltica. (A) Schematic of holdfast synthesis, modification, secretion, and anchor machineries. Holdfast polysaccharide synthesis is initiated by the glycosyltransferase HfsE, which transfers activated sugar precursors in the cytoplasm to a lipid carrier. Three glycosyltransferases, HfsJ, HfsG, and HfsL, add different sugars to the growing polysaccharide. The acetyltransferase HfsK and the deacetylase HfsH modify one or more sugar residues, and then a flippase, HfsF, transports the lipid carrier into the periplasm. Repeat units are polymerized by polymerases HfsC and HfsI. The polysaccharide is exported outside the cell through the HfsA-HfsB-HfsD complex. The exported polysaccharide is then anchored to the cell body by the secreted proteins HfaA, HfaB, and HfaD. The different colored hexagons represent different sugars. (B) Diagrams of C. crescentus and H. baltica dimorphic cell cycles. A motile swarmer cell differentiates into a stalked cell by shedding its flagellum and synthesizing a holdfast at the same cell pole. C. crescentus stalked cells divide asymmetrically to produce a motile swarmer and a stalked cell (top), and H. baltica reproduces by budding a motile swarmer off the stalk (bottom). (C) Maximum-likelihood phylogeny inferred from 16S rRNA sequences of selected freshwater and marine members of Caulobacterales. The node values represent clade frequencies of 1,000 bootstraps. The genes were identified using reciprocal best-hit analysis on fully sequenced Caulobacterales genomes. Solid gene symbols represent genes within the hfs or hfa loci, while hatched symbols indicate the genes translocated from these loci to a different location in the genome. Empty boxes indicate absent or missing genes.
Hirschia baltica is a marine member of the Caulobacterales isolated from surface water taken from a boat landing in the Kiel Fjord inlet of the Baltic Sea (Germany) (28). H. baltica has a dimorphic life cycle similar to that of C. crescentus (28) but reproduces by budding from the tip of the stalk (Fig. 1B). Newborn swarmer cells are motile by means of a polar flagellum and differentiate into sessile stalked cells after flagellum ejection. The sessile cells produce a holdfast at the same pole as the flagellum and synthesize a stalk at the opposite pole (29). H. baltica cells have been shown to produce holdfasts containing GlcNAc residues, using fluorescent WGA lectin (29, 30). The vast majority of studies on holdfasts have been done using C. crescentus, and therefore, the H. baltica holdfast is poorly understood.
As bacteria have to develop different strategies to adhere to surfaces in a given environment, we hypothesized that H. baltica produces holdfasts with different physicochemical properties because H. baltica’s natural habitat is high-ionic-strength seawater (28), while the freshwater C. crescentus holdfast is highly sensitive to salt (6). Here, we study H. baltica holdfast composition and properties. Using both genetics and bioinformatics analyses, we show that freshwater and marine Caulobacterales use orthologous genes in holdfast biogenesis and that these genes are highly conserved in the two genera. We show that H. baltica produces more holdfast material than C. crescentus and that the holdfasts of the two genera have different chemical compositions and behave differently. In addition to GlcNAc monosaccharides, we show that H. baltica holdfasts contain galactose residues and uncharacterized peptides different than the ones found in C. crescentus holdfasts. Finally, we demonstrate that the H. baltica holdfast tolerates higher ionic strength than that of C. crescentus.
RESULTS
Organization of the holdfast genes in H. baltica.
The genes essential for holdfast synthesis and export in the C. crescentus hfs locus (hfsG, hfsB, hfsA, and hfsD) are conserved in H. baltica (29, 30). To determine if the genomic organization of all the known holdfast-related genes is conserved in both species, we performed reciprocal best-hit analyses using the C. crescentus hfs (holdfast synthesis, modification, and export) and hfa (holdfast anchoring) genes (Fig. 1C). We also extended our analysis to other fully sequenced available Caulobacterales genomes for a more global overview of the organization of these genes in the clade (Fig. 1C). Table 1 gives the locus tag names of all the holdfast-related genes used in this study for C. crescentus CB15 (31), C. crescentus NA1000 (32), and H. baltica IFAM 1418T (29) type strains.
TABLE 1.
Genes involved in holdfast synthesis, modification, and anchoring
| Gene name | Locus tag name |
||
|---|---|---|---|
|
C. crescentus |
H. baltica
IFAM 1418T |
||
| CB15 | NA1000 | ||
| Export apparatus | |||
| hfsA | CC2431 | CCNA_02513 | Hbal_1968 |
| hfsB | CC2430 | CCNA_02512 | Hbal_1967 |
| hfsD | CC2432 | CCNA_02514 | Hbal_1969 |
| Synthesis genes | |||
| hfsC | CC2429 | CCNA_02511 | Hbal_1972 |
| hfsE | CC2425 | CCNA_02507 | Hbal_1963 |
| hfsJ | CC0095 | CCNA_00094 | Hbal_1784 |
| hfsG | CC2427 | CCNA_02509 | Hbal_1964 |
| hfsL | CC2277 | CCNA_02361 | Hbal_1966 |
| hfsI | CC0500 | CCNA_00533 | Hbal_2115 |
| hfsF | CC2426 | CCNA_02508 | Hbal_0100 |
| Modification genes (nonessential) | |||
| hfsH | CC2428 | CCNA_02510 | Hbal_1965 |
| hfsK | CC3689 | CCNA_03803 | Hbal_0069 |
| Anchor genes (nonessential) | |||
| hfaA | CC2628 | CCNA_02711 | Hbal_0652 |
| hfaB | CC2630 | CCNA_02712 | Hbal_0651 |
| hfaD | CC2629 | CCNA_02713 | Hbal_0650 |
| hfaE | CC2639 | CCNA_02722 | Hbal_0649 |
All the genes reported to be involved in holdfast synthesis in C. crescentus are present in the analyzed genomes, with a few rearrangements (Fig. 1C). The general organization of the hfs locus is conserved in all the Caulobacterales genomes analyzed, with the genes encoding proteins essential for holdfast synthesis (the glycosyltransferase gene hfsG and the export genes hfsA, hfsB, and hfsD) and the initiating glycosyltransferase gene hfsE in an organization similar to that in C. crescentus. Some of the genes involved in holdfast synthesis and modification in C. crescentus are not part of the hfs gene cluster (genes encoding the polymerase HfsI [17], the glycotransferases HfsJ [33] and HfsL [19], and N-acetyltransferase HfsK [20]); these genes are also present in H. baltica. Interestingly, in the genomes of the marine Caulobacterales Oceanicaulis alexandrii, Maricaulis maris, and Maricaulis salignorans, all the hfs genes except hfsJ are found in one locus (Fig. 1C). This suggests that the ancestral hfs locus might have contained most of the hfs genes. Most of the genomes analyzed had only one polysaccharide polymerase gene, hfsC, while others had a paralogous polysaccharide polymerase gene, hfsI (Fig. 1C) (17).
Once exported outside the cell by the HfsDAB complex, the holdfast is anchored to the cell envelope by the actions of anchor proteins that have been identified and characterized in C. crescentus HfaA, HfaB, and HfaD (19, 25–27). The organization of the three anchor genes hfaA, hfaB, and hfaD in the hfa locus is conserved in all the analyzed Caulobacterales genomes (Fig. 1C). In C. crescentus and most of the tested Caulobacterales, the recently identified holdfast anchor gene hfaE (19) is not part of the hfaABD operon, while it is present in the hfa locus in both H. baltica and O. alexandrii (Fig. 1C). We could not find orthologs of the hfa genes in the genomes of Robiginitomaculum antarticum and Hellea balneolensis, but this may be due to the incomplete nature of their genome sequences. Alternatively, these species may have a different mechanism to anchor the holdfast to the surface of the cell, as is the case for several other Alphaproteobacteria (34, 35).
Roles of the hfs and hfa genes in H. baltica.
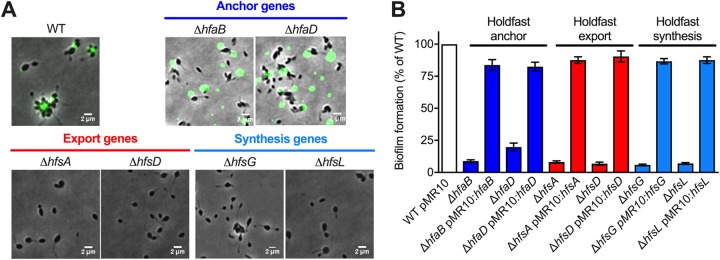
To determine if the genes identified in Fig. 1C are involved in holdfast production and anchoring in H. baltica, we created in-frame deletion mutants of the hfa genes encoding the anchor proteins and the hfs genes shown to be essential for holdfast synthesis in C. crescentus (12). We first monitored the presence of holdfasts in these mutants using fluorescence microscopy with fluorescently labeled WGA lectin (10) (Fig. 2A). We also quantified biofilm formation after 12 h of incubation at room temperature on a plastic surface by normalizing crystal violet staining to the optical density of the cells using 24-well PVC plates (Fig. 2B). All the mutants could be complemented in trans by a replicating plasmid carrying a copy of the deleted gene (Fig. 2B).
FIG 2.
Role of the hfs and hfa genes in H. baltica holdfast production. (A) Representative images showing merged phase and fluorescence channels of different H. baltica WT and mutant strains with holdfasts labeled with WGA-AF488 (green): H. baltica holdfast anchor mutants (ΔhfaB and ΔhfaD), export mutants (ΔhfsA and ΔhfsD), and synthesis mutants (ΔhfsG and ΔhfsL). (B) Quantification of biofilm using crystal violet assay after 12 h for H. baltica hfs and hfa mutants. The data are expressed as averages from 5 independent replicates, and the error bars represent the standard errors.
We first deleted the holdfast anchor genes encoding the HfaB and HfaD proteins. Both H. baltica ΔhfaB and ΔhfaD mutants produced holdfasts, but they failed to anchor them to the cell envelope, resulting in the holdfasts being shed in the medium (Fig. 2A). H. baltica ΔhfaB was not able to permanently attach to surfaces and could not form a biofilm (Fig. 2B). In contrast, H. baltica ΔhfaD mutants were not completely deficient for permanent adhesion, with around 20% biofilm formation compared to the wild type (WT) (Fig. 2B). These results are in agreement with what has been reported for C. crescentus ΔhfaB and ΔhfaD mutants (26), suggesting that the Hfa proteins have similar functions in both organisms.
We then made in-frame deletions of the genes encoding the export proteins HfsA and HfsD. These genes are essential for holdfast production in C. crescentus (23). Deletion of the genes in H. baltica similarly completely abolished holdfast production (Fig. 2A) and surface attachment (Fig. 2B). These results show that deletion of the export genes is sufficient for complete loss of holdfast production and that a holdfast is crucial for surface attachment in H. baltica.
Finally, we made in-frame deletions of the genes encoding the glycosyltransferases HfsG and HfsL, which are essential for holdfast formation in C. crescentus (17, 19). Similarly, H. baltica ΔhfsG and ΔhfsL mutants did not produce holdfasts or form biofilms (Fig. 2A and B).
Effects of modulating hfsL and hfsG expression on H. baltica holdfast properties.
We investigated if varying the expression of the hfsL and hfsG genes could change holdfast synthesis and properties. To achieve this goal, we first engineered a replicating plasmid harboring an inducible promoter suitable for H. baltica. We adapted the system developed for a tightly controlled heavy metal (copper) promoter-inducible system in Hyphomonas neptunium, a marine member of the Caulobacterales closely related to H. baltica (36). Similarly, we used the promoter for the copper-resistant protein operon copAB (Pcu) in H. baltica (copA, hbal_0699, and copB, hbal_0698) (see Fig. S1A, top, in the supplemental material). We first showed that H. baltica can tolerate up to 500 μM CuSO4 without a significant effect on growth (see Fig. S1B and C). We then fused 500 bp upstream of the copAB operon (Pcu) to the lacZ gene and assembled the construct onto the pMR10 replicating plasmid (see Fig. S1A, bottom) to assess Pcu promoter activity, using β-galactosidase as a reporter. We showed that Pcu is a tightly controlled promoter, with a working inducible range of CuSO4 from 10 to 250 μM (see Fig. S1D), concentrations that do not impact H. baltica growth (see Fig. S1B and C).
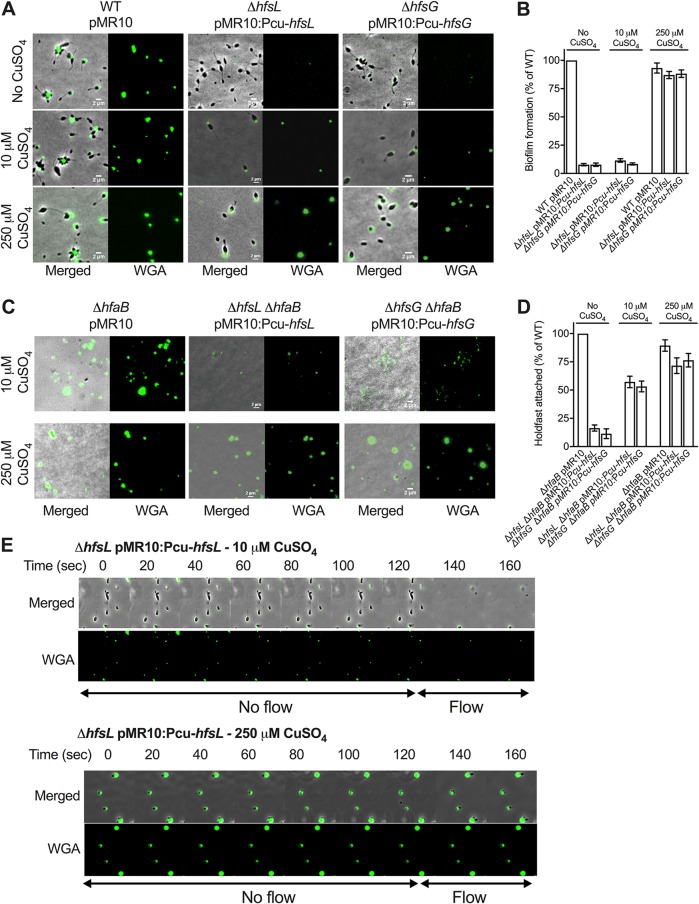
We expressed hfsL or hfsG under the control of the Pcu inducible promoter in H. baltica ΔhfsL and ΔhfsG mutants. In both cases, when gene expression was highly induced (250 μM CuSO4), holdfast size and adhesion were restored to WT levels (Fig. 3A and B). At lower levels of induction (10 μM CuSO4), both complemented strains produced small holdfasts (Fig. 3A) but failed to form biofilms after 12 h (Fig. 3B). To test if these results were due to altered adhesive properties of the smaller holdfasts or if their smaller size did not enable the cells to be retained on the surface, we combined the ΔhfsL and ΔhfsG mutations with an in-frame deletion of the holdfast anchor gene hfaB, resulting in mutants that produced holdfasts shed in the medium upon CuSO4 induction (Fig. 3C). We grew exponential-phase cultures of the double mutants on glass coverslips for 4 h to allow them to attach to the surface. After incubation, the slides were rinsed with distilled H2O (dH2O) to remove all the cells that were unable to anchor their holdfasts to their cell bodies, resulting in coverslips displaying attached holdfasts and no cells (Fig. 3C). At low levels of induction of hfsL or hfsG, shed holdfasts from H. baltica ΔhfaB ΔhfsL and H. baltica ΔhfaB ΔhfsG, though smaller than those from H. baltica ΔhfaB, were still able to efficiently bind to glass slides (Fig. 3C). We determined the numbers of holdfasts attached at different levels of induction of hfsL and hfsG (Fig. 3D). At low induction, the mutants produced 50% of the number of WT holdfasts (Fig. 3D). To visualize how cells with small holdfasts interact with the glass surface, we performed time-lapse microscopy in a microfluidic device, starting with static conditions and adding flow after 2 min to allow the cells to bind to the surface (Fig. 3E). We observed that at low induction of hfsL (10 μM CuSO4), cells efficiently bound to the surface, despite their small holdfasts. However, when the flow was adjusted to 1.4 μl/min in the microfluidic device, generating a drag force of 4 nN, the hfsL mutant cells detached, whereas wild-type cells remained attached. Attached C. crescentus cells have been shown to withstand high drag forces, up to 1 μN (13). These results show that the small holdfasts are not sufficient to withstand high shear forces. At high induction of hfsL (250 μM CuSO4), cells produced bigger holdfasts and were able to bind to the surface and resist the drag force. This result confirms that the smaller holdfasts are still adhesive, but their size is probably not sufficient to allow cells to resist larger drag forces.
FIG 3.
Effect of modulating hfsL and hfsG expression on H. baltica holdfast properties. (A) Representative images showing merged phase and fluorescence channels of H. baltica WT and ΔhfsL and ΔhfsG mutants complemented with copper-inducible promoter constructs and grown in marine broth with 0 μM,10 μM, and 250 μM CuSO4. The holdfasts were labeled with WGA-AF488. (B) Biofilm quantification after 12 h using crystal violet assay of ΔhfsL and ΔhfsG mutants and complementations under copper-inducible promoters in marine broth supplemented with 0 μM, 10 μM, and 250 μM CuSO4. The data are expressed as averages from 6 independent replicates, and the error bars represent the standard errors. (C) Images of WGA-AF488-labeled H. baltica ΔhfaB, H. baltica ΔhfaB ΔhfsL pMR10:PcuhfsL, and H. baltica ΔhfaB ΔhfsG pMR10:PcuhfsG shed holdfasts bound to glass slides. Cells were grown in marine broth with 0 μM, 10 μM, and 250 μM CuSO4 induction for 4 h. (D) Percentages of holdfasts bound to glass slides per field of view at different CuSO4 induction levels shown in panel C. The data are expressed as averages from 5 independent replicates, and the error bars represent the standard errors. (E) Time-lapse montage of H. baltica ΔhfsL pMR10:PcuhfsL induced with 10 μM (top) and 250 μM (bottom) CuSO4 in a microfluidic device, initially with no flow and then with a flow of 1.4 μl/min, generating a drag force of 4 nN, introduced into the microfluidic device. The arrows indicate the times when no flow (first 120 s) and flow (later times) were applied to the device.
H. baltica produces large holdfasts by developmental and surface contact stimulation pathways.
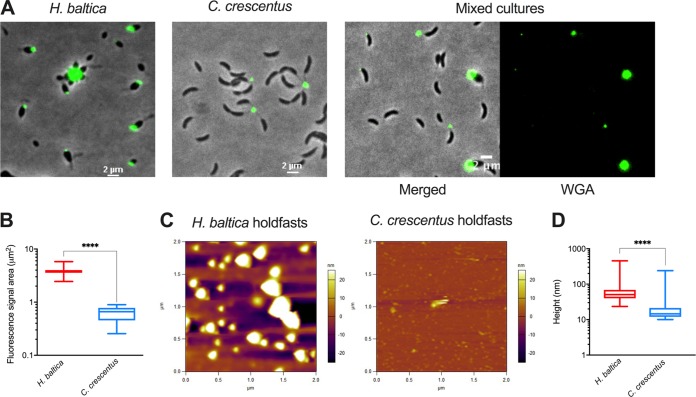
It was previously shown that WGA binds to C. crescentus and H. baltica holdfasts (29, 30). However, side-by-side microscopy imaging using fluorescent WGA suggested that H. baltica holdfasts might be larger than C. crescentus holdfasts (Fig. 4A). To quantify relative holdfast size, we imaged mixed cultures of H. baltica and C. crescentus simultaneously labeled with fluorescent WGA lectin. We measured the area of fluorescent WGA staining on single cells for each strain (Fig. 4A) and determined that, on average, the fluorescence area was 5 times larger for H. baltica holdfasts than for those of C. crescentus (Fig. 4B), whereas the fluorescence intensities were not statistically different. Since WGA binds to GlcNAc residues in the holdfast, either H. baltica holdfasts are larger than those of C. crescentus or H. baltica and C. crescentus holdfasts are similar in size but H. baltica holdfasts contain more GlcNAc residues, yielding an increased fluorescence area from bound WGA. To reliably measure the sizes of holdfasts, we used atomic-force microscopy (AFM) and imaged dry holdfasts deposited on a clean mica surface, free of any stain. The results confirmed that H. baltica produces larger holdfasts than C. crescentus. H. baltica holdfasts had a median height of 68 nm, while C. crescentus produced holdfasts with a median height of 19 nm (Fig. 4C and D), in agreement with previous reports (6, 37).
FIG 4.
H. baltica produces large holdfasts. (A) Images of H. baltica, C. crescentus, and mixed culture with holdfasts labeled with WGA-AF488 (green). (B) Quantification of holdfast size based on WGA-AF488 fluorescence area. The data in the box-and-whisker plots represent 5 independent replicates of 200 holdfasts from each strain. The variance between H. baltica and C. crescentus holdfast fluorescent areas was analyzed using a t test. ****, P < 0.0001. (C) AFM images of dry shed holdfasts from H. baltica ΔhfaB and C. crescentus ΔhfaB deposited on a mica surface. The colors on the scale represent the height of the holdfast relative to the surface. (D) Box-and-whisker plots of holdfast height distribution from AFM images. More than 500 holdfasts were measured in 10 independent images. The variance between H. baltica and C. crescentus holdfast heights was analyzed using a t test. ****, P < 0.0001.
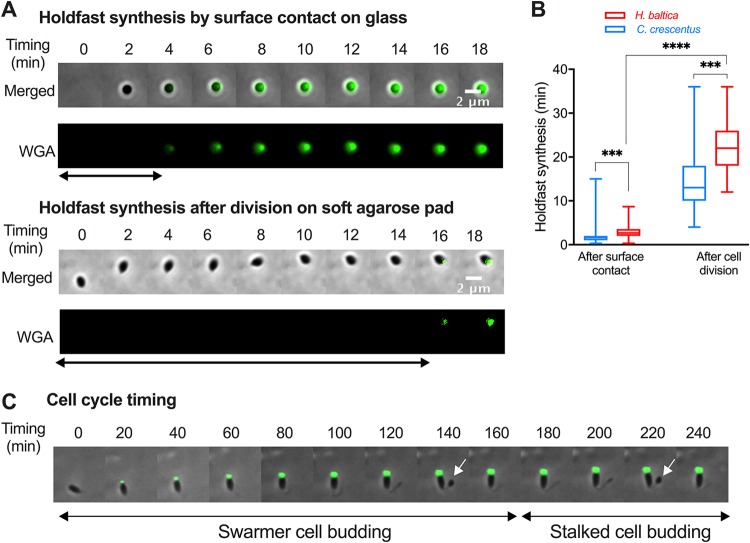
C. crescentus can regulate holdfast synthesis by two distinct pathways, a complex developmental program in a cell cycle-regulated manner or activation upon cell contact with a surface, independent of the cell cycle (38–41). Some Alphaproteobacteria, such as Asticaccaulis biprosthecum (39) and Prosthecomicrobium hirschii (42), are also able to produce holdfasts via developmental and surface-contact-stimulated pathways, while others, like Agrobacterium tumefaciens, produce holdfasts only upon contact with a surface (39, 43). To determine how holdfast production is regulated in H. baltica, we measured the timing of holdfast synthesis in the presence or absence of a hard surface. To test whether H. baltica holdfast production can be stimulated upon contact with a surface, we performed time-lapse microscopy in a microfluidic device where cells were in close proximity to a glass surface, and we tracked single cells as they reached the surface. We observed holdfast production by including fluorescently labeled WGA in the medium, and we recorded the difference between the time when a cell first reached the surface and the time when a holdfast was synthesized (Fig. 5A, top). We observed that H. baltica produces holdfasts within approximately 3 min of surface contact (Fig. 5A and B), showing that surface contact stimulates holdfast synthesis in the species. To assess cell cycle progression and the timing of holdfast synthesis independent of a hard surface, we tracked single cells and monitored cell differentiation and holdfast synthesis by time-lapse microscopy on soft agarose pads containing fluorescent WGA (Fig. 5A, bottom, and Fig. 5B). H. baltica newborn swarmer cells produced holdfasts within 15 to 25 min after budding on an agarose pad (Fig. 5A and B), showing that H. baltica can produce holdfasts through progression of the cell cycle, as part of a developmental pathway. To determine the timing of holdfast production relative to the cell cycle length, we measured the time required for a newborn swarmer cell to complete its first and second budding divisions on agarose pads (Fig. 5C). H. baltica swarmer cells completed their first budding within 160 to 200 min (Fig. 5C). Thus, the holdfast is synthetized within ∼1/10 of the cell cycle, similar to C. crescentus, which synthesizes holdfasts within 8 to 12 min of a 90- to 120-min cell cycle under these conditions (44).
FIG 5.
H. baltica holdfast synthesis is regulated by a developmental pathway and in response to surface contact. (A) Montages of H. baltica holdfast synthesis by a newly budded swarmer cell on a glass surface on a microfluidic device (top) and on soft agarose pads (bottom). The holdfasts were labeled with WGA-AF488 (green). Images shown were acquired every 2 min, and holdfast synthesis timing was processed using MicrobeJ. The arrows indicate the time it took for holdfasts to be detected after surface contact. (B) Box-and-whisker plots representing the quantification of H. baltica holdfast timing via surface contact stimulation and developmental pathways. The data for C. crescentus holdfast synthesis timing were extracted from reference 44. The total number of cells analyzed was 100 for each setup. The variance between H. baltica and C. crescentus holdfast synthesis times was analyzed using a t test. ***, P < 0.001; ****, P < 0.0001. (C) Time-lapse montage of an H. baltica swarmer cell differentiating into a budding stalked cell on an agarose pad containing WGA-AF488 to label the holdfast. Images were collected every 5 min for 3 h. The arrows point to incipient swarmer cells in predivisional cells.
H. baltica holdfasts contain GlcNAc and galactose monosaccharides and proteins.
Holdfasts in diverse Alphaproteobacteria bind to WGA, showing that they contain GlcNAc residues (3). Previous studies using lectin labeling showed that GlcNAc polymers are the main polysaccharides present in C. crescentus holdfasts, while other Caulobacterales strains may have additional monosaccharides in their holdfasts (10). Indeed, WGA lectin (specific to GlcNAc) and Dolichos biflorus agglutinin (specific to N-acetylgalactosamine) both bind Caulobacter henricii holdfasts (10), while Caulobacter subvibrioides holdfasts were shown to interact with D. biflorus agglutinin (specific to N-acetylgalactosamine), concanavalin A (specific to α-mannose), and Ulex europaeus agglutinin (specific to α-fucose) but not WGA (10).
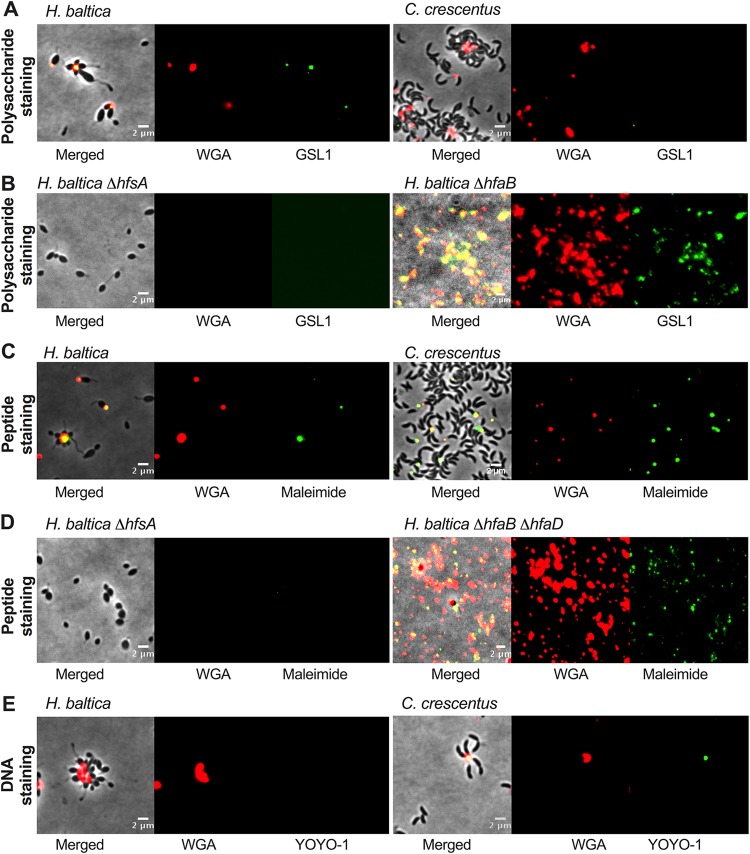
To identify the types of saccharides present in H. baltica holdfasts, we screened a variety of fluorescent lectins to attempt to label H. baltica holdfasts (Table 2; see Table S3 in the supplemental material). Our results indicate that, in addition to binding to WGA, H. baltica holdfasts also bind to Solanum tuberosum potato lectin (STL), Lycopersicon esculentum tomato lectin (LEL), and Datura stramonium lectin 1 (DSL1), all lectins specific to GlcNAc residues (Table 2), confirming that H. baltica holdfasts contain GlcNAc residues. In addition, lectins that specifically recognize α-galactose residues, Griffonia simplicifolia agglutinin 1 (GSL1) and Ricinus communis agglutinin 120 (RCA120) also bind to H. baltica holdfasts (Table 2), while they do not bind to C. crescentus holdfasts (Fig. 6A). Interestingly, soybean agglutinin lectin (45) did not bind to H. baltica holdfasts, showing that these holdfasts contain only galactose and no N-acetylgalactosamine (GalNAc) residues (Table 2). These results show that H. baltica holdfasts have a different sugar composition than Caulobacter holdfasts and contain both GlcNAc and galactose residues. To confirm that the observed galactose-specific binding was holdfast dependent, we labeled H. baltica ΔhfsA and H. baltica ΔhfsG (holdfast-negative strains) and ΔhfaB (a holdfast-shedding strain) mutants with both WGA and GSL1 lectins. None of the lectins labeled the holdfast-deficient ΔhfsA and ΔhfsG mutants, but they labeled shed holdfasts produced by the ΔhfaB mutant (Fig. 6A), confirming that H. baltica holdfasts contain galactose residues.
TABLE 2.
Lectin-binding assay results
| Lectin | Specificity | Presence in holdfasta |
|
|---|---|---|---|
| H. baltica | C. crescentus | ||
| WGA | GlcNAc | + | + |
| L. esculentum tomato lectin | GlcNAc 1-4 | + | +b |
| D. stramonium lectin | GlcNAc 1-4 | +b | − |
| S. tuberosum potato lectin | GlcNAc; prefers trimers and tetramers | + | +b |
| R. communis agglutinin | Galactose | + | − |
| G. simplicifolia lectin 1 | α-GalNAc, α-galactose | + | − |
| Soybean agglutinin | α-GalNAc | − | − |
+, fluorescent signal detected; −, no fluorescent signal detected.
Binding was enhanced on rosettes, with weaker signals on single cells.
FIG 6.
H. baltica holdfasts contain GlcNAc and galactose monosaccharides and proteins. (A to C) Representative images showing merged phase and fluorescence channels on the left and fluorescence channels alone in the middle and on the right. (A) H. baltica and C. crescentus holdfasts were colabeled with WGA-AF594 (red, GlcNAc) and GSL1-AF488 (green, galactose) lectins to stain polysaccharides. (B) H. baltica and C. crescentus holdfasts were colabeled with WGA-AF594 (GlcNAc) lectin and AF488mal to stain peptides. (C) H. baltica and C. crescentus holdfasts were colabeled with WGA-AF594 (GlcNAc) lectin and YOYO-1-AF488 to stain DNA.
C. crescentus holdfasts have been recently shown to contain peptides and DNA residues (15). To test whether H. baltica holdfasts contain proteins, we attempted to label putative cysteines in the holdfasts using a fluorescent maleimide dye (Alexa Fluor 488 maleimide [AF488mal]). As for C. crescentus holdfasts, H. baltica holdfasts could be stained with AF488mal, showing that these holdfasts possess molecules with free, accessible thiols, suggesting the presence of peptides containing cysteines (Fig. 6B). The staining was holdfast specific, as AF488mal did not label the holdfast-deficient ΔhfsA and ΔhfsG mutants (Fig. 6B). It has been shown that in C. crescentus, holdfast labeling by AF488mal was specific to holdfasts attached to cells, as shed holdfasts from a holdfast anchor mutant were not labeled, suggesting that the cysteine-containing HfaD in cell-anchored holdfasts is responsible for the labeling of those holdfasts with AF488mal (15). In H. baltica, both the anchor proteins HfaB and HfaD contain cysteines. In order to test whether AF488mal interacts with HfaB or HfaD, we stained shed holdfasts produced by an H. baltica ΔhfaB ΔhfaD double mutant and could detect staining (Fig. 6B). This is in stark contrast with C. crescentus holdfasts, which react with AF488mal only when attached to WT cells (15, 44). This result shows that the holdfast compositions in the two microorganisms are different.
To probe for the presence of DNA in H. baltica holdfasts, we labeled holdfasts with the fluorescent DNA dye YOYO-1, which binds to double-stranded DNA molecules. As previously reported, C. crescentus holdfasts were labeled with YOYO-1 (15). However, YOYO-1 failed to label H. baltica holdfasts (Fig. 6C), suggesting that H. baltica holdfasts do not contain DNA. It has been previously shown that, in C. crescentus, extracellular DNA (eDNA) released during C. crescentus cell lysis binds specifically to C. crescentus holdfasts, preventing adhesion to surfaces and biofilm formation (46), and it has been hypothesized that this could be due to a specific interaction between the DNA present in the holdfast and eDNA (15). We showed above that H. baltica holdfasts were devoid of DNA, so we tested whether eDNA could inhibit H. baltica binding. We performed short-term adhesion assays in the presence of H. baltica and C. crescentus eDNAs (see Fig. S2A in the supplemental material). When C. crescentus eDNA was present, the number of C. crescentus cells attached to the glass slide after 60 min was dramatically decreased compared to when H. baltica eDNA was added and to the control (no DNA addition) (see Fig. S2A), confirming previous studies that showed that, in C. crescentus, eDNA inhibition was specific for C. crescentus eDNA (46). However, H. baltica adhesion was not impaired by the presence of eDNA, from itself or from C. crescentus (see Fig. S2A). We also performed long-term biofilm assays in the presence of eDNA and showed that H. baltica biofilm formation is not impaired by the presence of eDNA in the medium after 24 h of incubation (see Fig. S2B).
Taking these results together, we showed that the H. baltica holdfast is different than that of C. crescentus: it is larger and contains GlcNAc, galactose, and peptide residues but is devoid of DNA.
H. baltica holdfasts tolerate high ionic strength.
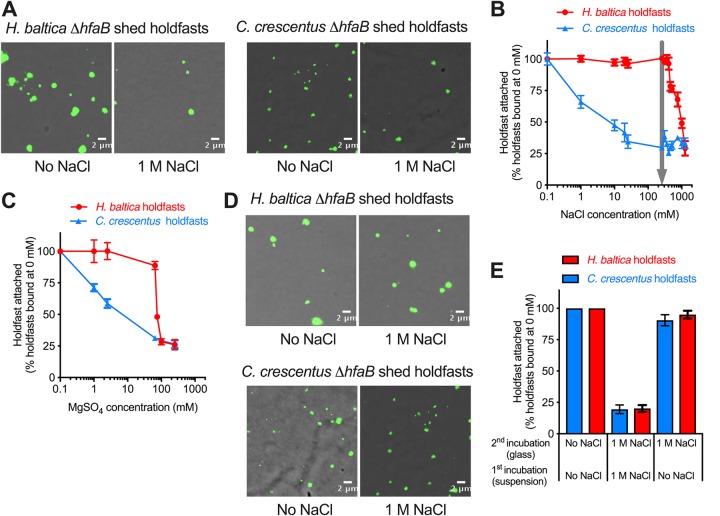
It has been shown that C. crescentus holdfasts are very sensitive to ionic strength, as the efficiency of purified-holdfast binding to glass decreased by 50% with the addition of 10 mM NaCl (6). C. crescentus is a freshwater bacterium and has probably evolved without selective pressure to bind under high ionic strength. This compelled us to investigate how purified holdfasts from H. baltica are affected by ionic strength. We first used NaCl to study the effects of ionic strength on holdfast binding, since it is the most abundant ionic element in marine water and it has been used in many studies to assess the effect of ionic strength on bacterial adhesins (6, 47–49). Holdfasts from both species were purified from a holdfast-shedding mutant during 2 h of growth in peptone yeast extract (PYE) following growth in optimal medium. We quantified purified-holdfast binding to glass at different NaCl concentrations, using fluorescent WGA, and plotted the relative numbers of holdfasts per field of view bound to glass at different concentrations of NaCl (Fig. 7A and B). Our results confirmed that the C. crescentus holdfast is very sensitive to NaCl, as only 50% of holdfasts could bind to glass when 10 mM NaCl was added (Fig. 7B). However, H. baltica holdfasts tolerated up to 500 mM NaCl without any effect on surface binding (Fig. 7B). There was a 50% decrease in H. baltica holdfast binding at 600 mM (Fig. 7B), showing that H. baltica holdfasts are more than 50 times more resistant to NaCl than those of C. crescentus. H. baltica was originally isolated from the Baltic Sea, which has 250 mM NaCl (Fig. 7B, arrow) (28), and at that NaCl concentration, the binding efficiency of H. baltica holdfasts was still at its maximum. Interestingly, H. baltica holdfasts still bound efficiently at low ionic strength. We observed similar results using different concentrations of MgSO4 (Fig. 7C): H. baltica holdfasts were 50 times more resistant to MgSO4 than those of C. crescentus, showing that the binding inhibition is not specific to NaCl but is rather dependent on ionic strength. The salt dose-response curves are noticeably different for the two types of holdfasts: while C. crescentus holdfast binding slowly decreases as the salt concentration increases, the binding efficiency of H. baltica holdfasts remains steady up to 500 mM NaCl or 250 mM MgSO4 and sharply decreases at high salt concentrations (Fig. 7B and C). This shows that the two holdfasts behave differently under increased ionic strength, suggesting different properties.
FIG 7.
H. baltica holdfasts tolerate higher ionic strength than C. crescentus holdfasts. (A) Images of WGA-AF488-labeled H. baltica ΔhfaB and C. crescentus ΔhfaB shed holdfasts bound to glass slides and incubated in different concentration of NaCl for 4 h. (B) Percentages of holdfasts bound per field of view at different concentrations of NaCl. The number of holdfasts bound per field of view at 0 M NaCl was standardized to 100%. The arrow indicates the ionic strength of marine broth and the Baltic Sea (250 mM), from which H. baltica was isolated. The data are expressed as averages from 6 independent replicates, and the error bars represent the standard errors. (C) Percentages of holdfasts bound per field of view at different concentrations of MgSO4. The number of holdfasts bound per field of view at 0 M MgSO4 was standardized to 100%. The data are expressed as averages from 4 independent replicates, and the error bars represent the standard errors. (D) Images of WGA-AF488-labeled holdfasts already bound to a glass surface and incubated in 0 mM NaCl and 1 M NaCl for 12 h. (E) Percentages of holdfasts bound per field of view at 0 M or 1 M NaCl. The first incubation was done by adding 0 M or 1 M NaCl to a holdfast suspension spotted on a glass slide. After a 12 h of incubation, the second incubation was done, after washing off unbound holdfasts, by adding 0 M or 1 M NaCl directly to the holdfasts attached to the glass slide and incubating them for another 12 h. The number of holdfasts bound per field of view at 0 M NaCl was standardized to 100%. The data are expressed as averages from 5 independent replicates, and the error bars represent the standard errors.
Our results show that, in H. baltica, initial holdfast binding to glass did not change for NaCl concentrations up to 500 mM and then drastically decreased to reach around 25% of holdfasts attached at 1 M NaCl (Fig. 7B). To test whether high ionic strength could remove holdfasts previously attached to the glass surface, we first incubated purified holdfasts for 4 h without any salt added and then added 1 M NaCl for 12 h to the bound holdfasts (Fig. 7D). Bound holdfasts from H. baltica and C. crescentus were not dislodged from the glass surface (Fig. 7D and E), indicating that while high ionic strength inhibits holdfasts from binding to a surface, it cannot dislodge bound holdfasts from a glass surface (Fig. 7E).
DISCUSSION
Different bacterial species harbor an adhesive holdfast and use it to attach to surfaces (2, 3, 9, 50, 51). They represent an extremely diverse group in terms of their physiologies and the natural environments they inhabit (soil, freshwater, and marine environments). They have evolved the ability to adhere to surfaces with vastly different compositions under varying environmental conditions (salinity, pH, temperature, etc.). Holdfast chemical properties have been mainly studied in the model organism C. crescentus CB15, a freshwater member of the Caulobacterales (6, 10, 13, 15, 19, 20, 30, 37, 52). Little is known about holdfast properties and composition in Caulobacterales isolated from habitats other than oligotrophic freshwater environments. In this study, we used H. baltica as a model species living in a marine environment and found that it has a holdfast tailored for adhesion under high-salinity conditions. We show that holdfasts in H. baltica are different than those of C. crescentus: they are larger, have a different chemical composition, and have a high tolerance for ionic strength.
The bioinformatics analysis of holdfast genes indicated that the hfs and hfa loci are highly conserved among Caulobacterales, with some reshuffling of the genes (Fig. 1C). The arrangement of the holdfast genes in the hfs and hfa loci appears to be ancestral, while the relocation of some of the genes is a recent event that could affect their levels of expression (53). Through deletion and complementation of important hfs and hfa genes, we confirmed that holdfast biogenesis and anchoring to the cell body in H. baltica use genes similar to those identified in C. crescentus (2, 19) (Fig. 2).
We showed that the two glycosyltransferase genes hfsL and hfsG are essential for holdfast production and that their expression level modulates the amount of sugar monosaccharides added to holdfast polysaccharides. Small holdfasts with fewer polysaccharides bind to glass, but not strongly enough to support cell adhesion to glass in flow (Fig. 3). This phenomenon could be due to the smaller surface contact area of the small holdfasts being insufficient to resist drag and shear forces during the washing steps of our assays or to a change in holdfast structure or composition due to the lower expression of the glycosyltransferases HfsL and HfsG. More studies on the roles of HfsL and HfsG will help us to determine if these enzymes play important roles in specific physicochemical properties of H. baltica holdfasts.
In C. crescentus, the growing holdfast polysaccharide repeat units are thought to be modified by the acetyltransferase HfsK (20) and the polysaccharide deacetylase HfsH (21) (Fig. 1A). These two enzymes are not essential for holdfast production in C. crescentus, but they modify the adhesiveness and cohesiveness of the holdfasts. C. crescentus ΔhfsH and ΔhfsK mutants produced thread-like holdfasts with weaker adhesion (20, 30). In addition, fully acetylated purified holdfasts from the C. crescentus ΔhfsH mutant were not affected by ionic strength (6), suggesting that holdfast modification can modulate salt tolerance. Our future work will determine how holdfast modification impacts H. baltica holdfast tolerance for high ionic strength and the possible roles of HfsH and HfsK.
The exact composition and structure of the holdfast in the model organism C. crescentus are still unknown. Lectin-binding assays and lysozyme treatment support GlcNAc as one of the important components in holdfasts (10, 37). Treating C. crescentus holdfasts with proteinase K and DNase I affects their structure and force of adhesion, suggesting that they contain peptide and DNA residues (15). In this work, we identified different components present in H. baltica holdfasts: these holdfasts contain galactose monosaccharides in addition to GlcNAc (Fig. 6A). In the different hfs mutants generated in this study, galactose monosaccharides were not detected on the cell pole (Fig. 6A), suggesting that GlcNAc and galactose are parts of the same polysaccharide or secreted by the same proteins. Shed holdfasts from H. baltica ΔhfaB contain both GlcNAc and galactose (Fig. 6A), implying that they are both anchored to the cell envelope with the same anchor proteins. H. baltica holdfasts are void of DNA, a stark contrast to those of C. crescentus (Fig. 6C). In addition, H. baltica holdfasts could be successfully stained with a fluorescent maleimide dye, which suggests the presence of a protein or peptide with a cysteine residue (54). The maleimide dye stains only cells with a holdfast and interacts with holdfasts without the presence of cells, indicating that the reactive molecules are an intrinsic part of H. baltica holdfasts (Fig. 6B), another notable difference from C. crescentus holdfasts, where maleimide dye interacts only with holdfasts attached to cells (15). In aggregate, our results suggest that the two holdfasts from H. baltica and C. crescentus have different compositions.
Bacterial adhesins have been shown to use electrostatic and hydrophobic interactions to attach to surfaces (6). Electrostatic interactions are impaired in high-ionic-strength environments, like seawater, with 600 mM NaCl (7). The C. crescentus holdfast uses both ionic and hydrophobic interactions, and its binding is impaired in the presence of NaCl in the medium (6). We have shown that H. baltica holdfasts tolerate high ionic strength compared to C. crescentus (Fig. 7A to C). Marine Caulobacterales face a higher-ionic-strength environment than the freshwater bacteria; therefore, it is vital that marine Caulobacterales produce holdfasts that are more tolerant of ionic strength and strongly adhere in saline environments. Holdfasts do not efficiently bind at 1 M NaCl, but holdfasts already attached to a surface cannot be removed by adding 1 M NaCl (Fig. 7D), suggesting that the binding inhibition at 1 M NaCl takes place during the initial stage of surface interaction. These results imply that holdfasts interact with surfaces initially by using electrostatic interactions before a permanent molecular bond is formed (6, 55). The differences in ionic tolerance between freshwater and marine Caulobacterales indicate that there are significant differences in physicochemical properties between the two types of holdfasts. Holdfast structure and binding properties could depend on the types and the amounts of sugars polymerized in the holdfast polysaccharides that are specialized to interact with different surfaces (56).
In conclusion, we have shown that H. baltica produces holdfasts with different binding and physicochemical properties than C. crescentus holdfasts. This suggests that there are additional holdfast-related genes or regulators that have not been identified. A careful genetic screen of H. baltica will provide more insights into holdfast production and the underlying mechanisms yielding enhanced adhesion at high ionic strength.
MATERIALS AND METHODS
Identification of orthologous holdfast genes and phylogenetic analysis.
C. crescentus holdfast genes were used to find bidirectional best hits (BBH) on Caulobacterales genomes. The putative genes were selected for an E value of >10−4 and a sequence identity of >30%. The phylogenic tree was built using 16S rRNA sequences of the selected Caulobacterales. Sequences were aligned using MUSCLE software (47). The aligned sequences were used to construct a maximum-likelihood phylogeny using MEGA6 software (57). The LG+G+I models and analysis of 1,000 bootstraps were used to generate the node values for each clade.
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table S1 in the supplemental material. H. baltica strains were grown in marine medium (Difco marine broth/agar; reference 2216), except when studying the effect of ionic strength on holdfast binding, when they were grown in PYE medium (8) supplemented with 0 or 1.5% NaCl or MgSO4. C. crescentus was grown in PYE medium. Both H. baltica and C. crescentus strains were grown at 30°C. When appropriate, kanamycin (Kan) was added at 5 μg/ml in liquid and 20 μg/ml on agarose plates. H. baltica strains with copper-inducible promoters were grown in marine broth supplemented with 0 to 250 μM CuSO4. E. coli strains were grown in Luria-Bertani medium (31) at 37°C with no antibiotics or with 30 μg/ml Kan in liquid or 25 μg/ml on agarose plates when needed.
Strain construction.
All the plasmids and primers used in this study are listed in Tables S1 and S2 in the supplemental material, respectively. In-frame deletion mutants were obtained by double homologous recombination as previously described (58), using suicide plasmids transformed into the H. baltica host strains by mating or electroporation (59). Briefly, genomic DNA was used as the template to PCR amplify 500-bp fragments from upstream and downstream regions of the gene to be deleted. The pNPTS139 plasmid was cut using EcoRV-HF endonuclease from New England Biolabs (NEB). The primers used to amplify 500 bp upstream and downstream of the gene were designed to have 25 bp overlapping for isothermal assembly (60) using the New England Biolabs NEBuilder tools for Gibson assembly into plasmid pNPTS139. Then, pNPTS139-based constructs were transformed into an α-select E. coli strain and introduced into the host H. baltica by mating or electroporation (59). The two-step selection for homologous recombination was carried out using sucrose resistance and kanamycin sensitivity (61).
For gene complementation, the pMR10 plasmid was cut with EcoRV-HF, and 500 bp of the promoter and the gene was ligated into plasmid pMR10 using NEBuilder tools. The pMR10-based constructs were transformed into an α-select E. coli strain and introduced into the H. baltica host by mating or electroporation, followed by Kan selection. The plasmid constructs and mutants were confirmed by sequencing.
Holdfast labeling using fluorescently labeled lectins.
Alexa Fluor (AF)-conjugated lectins (Vector Laboratories) (Table 2; see Table S3) were added to 100 μl of exponential-phase culture to a final concentration of 0.5 μg/ml and incubated at room temperature for 5 min; 3 μl of the labeled culture was spotted on a glass cover slide and covered with a 1.5% (wt/vol) SeaKem LE agarose (Lonza) pad in water and visualized by epifluorescence microscopy. Holdfasts were imaged by epifluorescence microscopy using an inverted Nikon Ti-E microscope with a Plan Apo 60× objective, a green fluorescent protein (GFP)/DsRed filter cube, an Andor iXon3 DU885 EM charge-coupled device (CCD) camera, and Nikon NIS Elements imaging software with a 200-ms exposure time. Images were processed in ImageJ (45).
Short-term and biofilm binding assays.
Short-term and biofilm binding assays were performed as previously described (30) with the following modifications. For short-term binding, exponential cultures (optical density at 600 nm [OD600] = 0.6 to 0.8) were diluted to an OD600 of 0.4 in fresh marine broth, added to 24-well plates (1 ml per well), and incubated with shaking (100 rpm) at room temperature for 4 h. For biofilm assays, overnight cultures were diluted to an OD600 of 0.10, added to 24-well plates (1 ml per well), and incubated at room temperature for 12 h with shaking (100 rpm). In both setups, OD600 values were measured before the wells were rinsed with dH2O to remove nonattached bacteria, stained using 0.1% crystal violet (CV), and rinsed again with dH2O to remove excess CV. The CV was dissolved in 10% (vol/vol) acetic acid and quantified by measuring the absorbance at 600 nm (A600). The biofilm formation was normalized to the A600/OD600 and expressed as a percentage of the WT value.
hfsL and hfsG expression using copper-inducible promoters.
Strains bearing copper-inducible plasmids were inoculated from freshly grown colonies into 5 ml marine broth containing 5 μg/ml Kan and incubated with shaking (200 rpm) at 30°C overnight. The overnight cultures were diluted in the same culture medium to an OD600 of 0.10 and incubated until an OD600 of 0.4 was reached. When needed, copper sulfate dissolved in marine broth was added to a final concentration of 0 to 250 μM. The induced cultures and controls were added to a 24-well plate (1 ml per well) and incubated with shaking (100 rpm) at room temperature for 4 to 8 h. Then, OD600 values were measured before the wells were rinsed with dH2O to remove nonattached bacteria, stained using 0.1% CV, and rinsed again with dH2O to remove excess CV. The CV was dissolved in 10% (vol/vol) acetic acid and quantified by measuring the A600. The biofilm formation was normalized to the A600/OD600 and expressed as a percentage of the WT value.
Visualization of holdfasts attached to a glass surface.
Visualization of holdfast binding to glass surfaces was performed as described previously (30) with the following modifications. H. baltica and C. crescentus strains grown to exponential phase (OD600 = 0.2 to 0.6) were incubated on washed glass coverslips at room temperature in a saturated humidity chamber for 4 to 8 h. After incubation, the slides were rinsed with dH2O to remove unbound cells, and the holdfasts were labeled using 50 μl of fluorescent Alexa Fluor 488 (AF488)- or AF594-conjugated lectins (Molecular Probes or Vector Laboratories) (Table 2) at a final concentration of 0.5 μg/ml. Then, the slides were rinsed with dH2O and topped with a glass coverslip. The holdfasts were imaged by epifluorescence microscopy using an inverted Nikon Ti-E microscope with a Plan Apo 60× objective, a GFP/DsRed filter cube, an Andor iXon3 DU885 EM CCD camera, and Nikon NIS Elements imaging software with a 200-ms exposure time. Images were processed in ImageJ (45).
Atomic-force microscopy.
AFM imaging was performed using the tapping mode on a Cypher AFM (Asylum Research) at 20°C, as described previously (6, 22), with the following modifications. H. baltica ΔhfaB and C. crescentus ΔhfaB grown to exponential phase were diluted and spotted on freshly cleaved mica. Samples were grown overnight at room temperature in a humid chamber. The samples were then rinsed with sterile dH2O to remove unbound cells and debris and air dried. AFM topographic images of dried holdfasts attached to the mica surface were obtained using a silicon Olympus AC160TS cantilever (resonance frequency = 300 kHz; spring constant = 26 N/m). Forty images of 4 independent replicates were obtained. Holdfast height was determined using the built-in image analysis function of the Igor Pro/Asylum Research AFM software.
Holdfast synthesis timing by time-lapse microscopy on agarose pads.
H. baltica holdfast synthesis timing was observed in live cells on agarose pads by time-lapse microscopy, as described previously (40), with some modifications. A 1-μl aliquot of exponential-phase cells (OD600 = 0.4 to 0.8) was placed on top of a pad containing 0.8% agarose in marine broth with 0.5 μg/ml WGA-AF488. A coverslip was placed on top of the agarose pad and sealed with VALAP (petrolatum, lanolin, and paraffin wax). Time-lapse microscopy images were taken every 2 min for 4 h using an inverted Nikon Ti-E microscope and a Plan Apo 60× objective, a GFP/DsRed filter cube, and an Andor iXon3 DU885 EM CCD camera. Time-lapse movies were visualized in ImageJ (45) to manually assess the timing of a swarmer cell producing a holdfast (lectin detection) after budding. The time difference between holdfast synthesis and budding was determined using MicrobeJ (62).
Holdfast synthesis timing by time-lapse microscopy in microfluidic devices.
The holdfast synthesis timing experiment was performed as previously described (41) with the following modifications. Cell cultures were grown to mid-exponential phase (OD600 = 0.4 to 0.6), and 200 μl of culture was diluted in 800 μl fresh marine broth in the presence of 0.5 μg/ml WGA-AF488 for holdfast labeling. One milliliter of the cell culture was flushed into a microfluidic device containing a 10-μm-high linear chamber fabricated in polydimethylsiloxane (PDMS) as described previously (40). After injection of the cells into the microfluidic chamber, the flow rate was adjusted so that attachment could be observed under static conditions or at a low flow rate of 1.4 μl/min. The drag force generated by the flow in the microfluidic device was calculated as previously described by Persat and colleagues (63).
Time-lapse microscopy was performed using an inverted Nikon Ti-E microscope and a Plan Apo 60× objective, a GFP/DsRed filter cube, an Andor iXon3 DU885 EM CCD camera, and Nikon NIS Elements imaging software. Time-lapse videos were collected for strains over a period of 3 h at 20-s intervals. Cell attachment was detected at the glass-liquid interface within the microfluidic chamber using phase-contrast microscopy, while holdfast synthesis was detected using fluorescence microscopy. Cells that hit the surface and attached permanently via their holdfasts during this 3-h period were analyzed for the timing of holdfast synthesis. The time difference between holdfast synthesis and cell surface contact was determined using MicrobeJ (62) and define as holdfast delay. Cells that were present on the surface at the start of the time-lapse experiment were not analyzed.
Holdfast labeling using fluorescently labeled maleimide and YOYO-1.
Alexa Fluor (AF488mal)-conjugated maleimide C5 (ThermoFisher Scientific) was added to 100 μl of exponential-phase culture to a final concentration of 0.5 μg/ml and incubated at room temperature for 5 min. Similarly, YOYO-1 (a fluorescent DNA stain; Molecular Probes) was added to 100 μl of exponential-phase culture to a final concentration of 0.5 μg/ml and incubated at room temperature for 5 min; 3 μl of the labeled culture was spotted on a glass cover slide, covered with a 1.5% (wt/vol) agarose pad in water, and visualized by epifluorescence microscopy. Holdfasts were imaged by epifluorescence microscopy using an inverted Nikon Ti-E microscope with a Plan Apo 60× objective, a GFP/DsRed filter cube, an Andor iXon3 DU885 EM CCD camera, and Nikon NIS Elements imaging software with a 200-ms exposure time. Images were processed in ImageJ (45).
Effect of ionic strength on holdfast binding.
Purified holdfasts attached to a surface at different ionic strengths were visualized as described previously (6) with a few modifications. Briefly, H. baltica ΔhfaB and C. crescentus ΔhfaB cells were grown to late exponential phase (OD600 = 0.6 to 0.8) in PYE plus 1.5% NaCl and plain PYE, respectively. The cells were pelleted by centrifugation for 30 min at 4,000 × g, resuspended in PYE, and incubated for 2 h at 30°C to produce shed holdfasts. Then, the cells were again pelleted by centrifugation, and 100 μl of supernatant containing free holdfasts shed by the cells was mixed with PYE-NaCl to make a final concentration of 0 to 1,000 mM NaCl. Fifty microliters of the mixture was incubated on washed glass coverslips at room temperature in a saturated humidity chamber for 4 to 12 h. After incubation, the slides were rinsed with dH2O to remove unbound material, and labeled holdfasts were visualized with Alexa Fluor lectins (Vector Laboratories). The holdfasts were imaged by epifluorescence microscopy using an inverted Nikon Ti-E microscope with a Plan Apo 60× objective, a GFP/DsRed filter cube, an Andor iXon3 DU885 EM CCD camera, and Nikon NIS Elements imaging software with a 200-ms exposure time. Images were processed in ImageJ (45). The number of holdfasts bound per field of view was determined using MicrobeJ (62).
Supplementary Material
ACKNOWLEDGMENTS
We thank Bogdan Dragnea, Department of Chemistry, Indiana University, for use of his AFM and facilities for analysis of the shed holdfasts. We thank the members of the Brun laboratory for comments on the manuscript.
This work was supported by National Institutes of Health grants R01GM102841 and R35GM122556 to Y.V.B. and a fellowship from the Department of Biology, Indiana University, to N.K.C. Y.V.B holds a Canada 150 Research Chair in Bacterial Cell Biology.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00061-19.
REFERENCES
- 1.Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. 1987. Bacterial biofilms in nature and disease. Annu Rev Microbiol 41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 2.Berne C, Ellison CK, Ducret A, Brun YV. 2018. Bacterial adhesion at the single-cell level. Nat Rev Microbiol 16:616–627. doi: 10.1038/s41579-018-0057-5. [DOI] [PubMed] [Google Scholar]
- 3.Berne C, Ducret A, Hardy GG, Brun YV. 2015. Adhesins involved in attachment to abiotic surfaces by Gram-negative bacteria. Microbiol Spectr 3:MB-0018-2015. doi: 10.1128/microbiolspec.MB-0018-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donlan RM. 2002. Biofilms: microbial life on surfaces. Emerg Infect Dis 8:881. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abu‐Lail N, Camesano T. 2003. Polysaccharide properties probed with atomic force microscopy. J Microsc 212:217–238. doi: 10.1111/j.1365-2818.2003.01261.x. [DOI] [PubMed] [Google Scholar]
- 6.Berne C, Ma X, Licata NA, Neves BR, Setayeshgar S, Brun YV, Dragnea B. 2013. Physiochemical properties of Caulobacter crescentus holdfast: a localized bacterial adhesive. J Phys Chem B 117:10492–10503. doi: 10.1021/jp405802e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garrels R, Thompson M. 1962. A chemical model for sea water at 25 degrees C and one atmosphere total pressure. Am J Sci 260:57–66. doi: 10.2475/ajs.260.1.57. [DOI] [Google Scholar]
- 8.Poindexter JS. 1964. Biological properties and classification of the Caulobacter group. Bacteriol Rev 28:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilhelm R. 2018. Following the terrestrial tracks of Caulobacter—redefining the ecology of a reputed aquatic oligotroph. ISME J 12:3025–3037. doi: 10.1038/s41396-018-0257-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merker RI, Smit J. 1988. Characterization of the adhesive holdfast of marine and freshwater caulobacters. Appl Environ Microbiol 54:2078–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ong CJ, Wong M, Smit J. 1990. Attachment of the adhesive holdfast organelle to the cellular stalk of Caulobacter crescentus. J Bacteriol 172:1448–1456. doi: 10.1128/jb.172.3.1448-1456.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown PJ, Hardy GG, Trimble MJ, Brun YV. 2008. Complex regulatory pathways coordinate cell-cycle progression and development in Caulobacter crescentus. Adv Microb Physiol 54:1–101. doi: 10.1016/S0065-2911(08)00001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsang PH, Li G, Brun YV, Freund LB, Tang JX. 2006. Adhesion of single bacterial cells in the micronewton range. Proc Natl Acad Sci U S A 103:5764–5768. doi: 10.1073/pnas.0601705103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henrici AT, Johnson DE. 1935. Studies of freshwater bacteria: II. Stalked bacteria, a new order of schizomycetes 1. J Bacteriol 30:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernando-Pérez M, Setayeshgar S, Hou Y, Temam R, Brun YV, Dragnea B, Berne C. 2018. Layered structure and complex mechanochemistry underlie strength and versatility in a bacterial adhesive. mBio 9:e02359-17. doi: 10.1128/mBio.02359-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuthbertson L, Mainprize IL, Naismith JH, Whitfield C. 2009. Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in gram-negative bacteria. Microbiol Mol Biol Rev 73:155–177. doi: 10.1128/MMBR.00024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toh E, Kurtz HD, Brun YV. 2008. Characterization of the Caulobacter crescentus holdfast polysaccharide biosynthesis pathway reveals significant redundancy in the initiating glycosyltransferase and polymerase steps. J Bacteriol 190:7219–7231. doi: 10.1128/JB.01003-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel KB, Toh E, Fernandez XB, Hanuszkiewicz A, Hardy GG, Brun YV, Bernards MA, Valvano MA. 2012. Functional characterization of UDP-glucose:undecaprenyl-phosphate glucose-1-phosphate transferases of Escherichia coli and Caulobacter crescentus. J Bacteriol 194:2646–2657. doi: 10.1128/JB.06052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hershey DM, Fiebig A, Crosson S. 2019. A genome-wide analysis of adhesion in Caulobacter crescentus identifies new regulatory and biosynthetic components for holdfast assembly. mBio 10:e02273-18. doi: 10.1128/mBio.02273-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sprecher KS, Hug I, Nesper J, Potthoff E, Mahi M-A, Sangermani M, Kaever V, Schwede T, Vorholt J, Jenal U. 2017. Cohesive properties of the Caulobacter crescentus holdfast adhesin are regulated by a novel c-di-GMP effector protein. mBio 8:e00294-17. doi: 10.1128/mBio.00294-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Javens J, Wan Z, Hardy GG, Brun YV. 2013. Bypassing the need for subcellular localization of a polysaccharide export-anchor complex by overexpressing its protein subunits. Mol Microbiol 89:350–371. doi: 10.1111/mmi.12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardy GG, Toh E, Berne C, Brun YV. 2018. Mutations in sugar-nucleotide synthesis genes restore holdfast polysaccharide anchoring to Caulobacter crescentus holdfast anchor mutants. J Bacteriol 200:e00597-17. doi: 10.1128/JB.00597-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith CS, Hinz A, Bodenmiller D, Larson DE, Brun YV. 2003. Identification of genes required for synthesis of the adhesive holdfast in Caulobacter crescentus. J Bacteriol 185:1432–1442. doi: 10.1128/JB.185.4.1432-1442.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurtz HD Jr, Smith J. 1994. The Caulobacter crescentus holdfast: identification of holdfast attachment complex genes. FEMS Microbiol Lett 116:175–182. doi: 10.1111/j.1574-6968.1994.tb06697.x. [DOI] [PubMed] [Google Scholar]
- 25.Cole JL, Hardy GG, Bodenmiller D, Toh E, Hinz A, Brun YV. 2003. The HfaB and HfaD adhesion proteins of Caulobacter crescentus are localized in the stalk. Mol Microbiol 49:1671–1683. doi: 10.1046/j.1365-2958.2003.03664.x. [DOI] [PubMed] [Google Scholar]
- 26.Hardy GG, Allen RC, Toh E, Long M, Brown PJ, Cole‐Tobian JL, Brun YV. 2010. A localized multimeric anchor attaches the Caulobacter holdfast to the cell pole. Mol Microbiol 76:409–427. doi: 10.1111/j.1365-2958.2010.07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurtz H, Smith J. 1992. Analysis of a Caulobacter crescentus gene cluster involved in attachment of the holdfast to the cell. J Bacteriol 174:687–694. doi: 10.1128/jb.174.3.687-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlesner H, Bartels C, Sittig M, Dorsch M, Stackebrandt E. 1990. Taxonomic and phylogenetic studies on a new taxon of budding, hyphal proteobacteria, Hirschia baltica gen. nov., sp. nov. Int J Syst Evol Microbiol 40:443–451. doi: 10.1099/00207713-40-4-443. [DOI] [PubMed] [Google Scholar]
- 29.Chertkov O, Brown PJB, Kysela DT, de Pedro MA, Lucas S, Copeland A, Lapidus A, Del Rio TG, Tice H, Bruce D, Goodwin L, Pitluck S, Detter JC, Han C, Larimer F, Chang Y-J, Jeffries CD, Land M, Hauser L, Kyrpides NC, Ivanova N, Ovchinnikova G, Tindall BJ, Göker M, Klenk H-P, Brun YV. 2011. Complete genome sequence of Hirschia baltica type strain (IFAM 1418 T). Stand Genomic Sci 5:287. doi: 10.4056/sigs.2205004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan Z, Brown PJ, Elliott EN, Brun YV. 2013. The adhesive and cohesive properties of a bacterial polysaccharide adhesin are modulated by a deacetylase. Mol Microbiol 88:486–500. doi: 10.1111/mmi.12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nierman WC, Feldblyum TV, Laub MT, Paulsen IT, Nelson KE, Eisen J, Heidelberg JF, Alley MRK, Ohta N, Maddock JR, Potocka I, Nelson WC, Newton A, Stephens C, Phadke ND, Ely B, DeBoy RT, Dodson RJ, Durkin AS, Gwinn ML, Haft DH, Kolonay JF, Smit J, Craven MB, Khouri H, Shetty J, Berry K, Utterback T, Tran K, Wolf A, Vamathevan J, Ermolaeva M, White O, Salzberg SL, Venter JC, Shapiro L, Fraser CM. 2001. Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci U S A 98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marks ME, Castro-Rojas CM, Teiling C, Du L, Kapatral V, Walunas TL, Crosson S. 2010. The genetic basis of laboratory adaptation in Caulobacter crescentus. J Bacteriol 192:3678–3688. doi: 10.1128/JB.00255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fiebig A, Herrou J, Fumeaux C, Radhakrishnan SK, Viollier PH, Crosson S. 2014. A cell cycle and nutritional checkpoint controlling bacterial surface adhesion. PLoS Genet 10:e1004101. doi: 10.1371/journal.pgen.1004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fritts RK, LaSarre B, Stoner AM, Posto AL, McKinlay JB. 2017. A Rhizobiales-specific unipolar polysaccharide adhesin contributes to Rhodopseudomonas palustris biofilm formation across diverse photoheterotrophic conditions. Appl Environ Microbiol 83:e03035-16. doi: 10.1128/AEM.03035-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson MA, Onyeziri MC, Fuqua C. 2018. Function and regulation of Agrobacterium tumefaciens cell surface structures that promote attachment. Curr Top Microbiol Immunol 418:143–184. doi: 10.1007/82_2018_96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung A, Eisheuer S, Cserti E, Leicht O, Strobel W, Möll A, Schlimpert S, Kühn J, Thanbichler M. 2015. Molecular toolbox for genetic manipulation of the stalked budding bacterium Hyphomonas neptunium. Appl Environ Microbiol 81:736–744. doi: 10.1128/AEM.03104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li G, Smith CS, Brun YV, Tang JX. 2005. The elastic properties of the Caulobacter crescentus adhesive holdfast are dependent on oligomers of N-acetylglucosamine. J Bacteriol 187:257–265. doi: 10.1128/JB.187.1.257-265.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levi A, Jenal U. 2006. Holdfast formation in motile swarmer cells optimizes surface attachment during Caulobacter crescentus development. J Bacteriol 188:5315–5318. doi: 10.1128/JB.01725-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li G, Brown PJ, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. 2012. Surface contact stimulates the just‐in‐time deployment of bacterial adhesins. Mol Microbiol 83:41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffman MD, Zucker LI, Brown PJ, Kysela DT, Brun YV, Jacobson SC. 2015. Timescales and frequencies of reversible and irreversible adhesion events of single bacterial cells. Anal Chem 87:12032–12039. doi: 10.1021/acs.analchem.5b02087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ellison CK, Kan J, Dillard RS, Kysela DT, Ducret A, Berne C, Hampton CM, Ke Z, Wright ER, Biais N, Dalia AB, Brun YV. 2017. Obstruction of pilus retraction stimulates bacterial surface sensing. Science 358:535–538. doi: 10.1126/science.aan5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams M, Hoffman MD, Daniel JJ, Madren SM, Dhroso A, Korkin D, Givan SA, Jacobson SC, Brown PJ. 2016. Short-stalked Prosthecomicrobium hirschii cells have a Caulobacter-like cell cycle. J Bacteriol 198:1149–1159. doi: 10.1128/JB.00896-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomlinson AD, Fuqua C. 2009. Mechanisms and regulation of polar surface attachment in Agrobacterium tumefaciens. Curr Opin Microbiol 12:708–714. doi: 10.1016/j.mib.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berne C, Ellison CK, Agarwal R, Severin GB, Fiebig A, Morton RI III, Waters CM, Brun YV. 2018. Feedback regulation of Caulobacter crescentus holdfast synthesis by flagellum assembly via the holdfast inhibitor HfiA. Mol Microbiol 110:219–238. doi: 10.1111/mmi.14099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berne C, Kysela DT, Brun YV. 2010. A bacterial extracellular DNA inhibits settling of motile progeny cells within a biofilm. Mol Microbiol 77:815–829. doi: 10.1111/j.1365-2958.2010.07267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otto K, Elwing H, Hermansson M. 1999. Effect of ionic strength on initial interactions of Escherichia coli with surfaces, studied on-line by a novel quartz crystal microbalance technique. J Bacteriol 181:5210–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zita A, Hermansson M. 1994. Effects of ionic strength on bacterial adhesion and stability of flocs in a wastewater activated sludge system. Appl Environ Microbiol 60:3041–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dang H, Lovell CR. 2016. Microbial surface colonization and biofilm development in marine environments. Microbiol Mol Biol Rev 80:91–138. doi: 10.1128/MMBR.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohari B, Thompson MA, Trinidad JC, Setayeshgar S, Fuqua C. 2018. Multiple flagellin proteins have distinct and synergistic roles in Agrobacterium tumefaciens motility. J Bacteriol 200:e00327-18. doi: 10.1128/JB.00327-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li G, Brun YV, Tang JX. 2013. Holdfast spreading and thickening during Caulobacter crescentus attachment to surfaces. BMC Microbiol 13:139. doi: 10.1186/1471-2180-13-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arber W. 2000. Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol Rev 24:1–7. doi: 10.1111/j.1574-6976.2000.tb00529.x. [DOI] [PubMed] [Google Scholar]
- 54.Kim Y, Ho SO, Gassman NR, Korlann Y, Landorf EV, Collart FR, Weiss S. 2008. Efficient site-specific labeling of proteins via cysteines. Bioconjug Chem 19:786–791. doi: 10.1021/bc7002499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karatan E, Watnick P. 2009. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev 73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhosle N, Suci P, Baty A, Weiner R, Geesey G. 1998. Influence of divalent cations and pH on adsorption of a bacterial polysaccharide adhesin. J Colloid Interface Sci 205:89–96. doi: 10.1006/jcis.1998.5597. [DOI] [PubMed] [Google Scholar]
- 57.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ried JL, Collmer A. 1987. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 57:239–246. doi: 10.1016/0378-1119(87)90127-2. [DOI] [PubMed] [Google Scholar]
- 59.Ely B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol 204:372–384. doi: 10.1016/0076-6879(91)04019-K. [DOI] [PubMed] [Google Scholar]
- 60.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 61.Link AJ, Phillips D, Church GM. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol 179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ducret A, Quardokus EM, Brun YV. 2016. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol 1:16077. doi: 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Persat A, Nadell CD, Kim MK, Ingremeau F, Siryaporn A, Drescher K, Wingreen NS, Bassler BL, Gitai Z, Stone HA. 2015. The mechanical world of bacteria. Cell 161:988–997. doi: 10.1016/j.cell.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.