Abstract
A comprehensive laboratory evaluation of the Tetracore RedLine Alert test, a lateral flow immunoassay (LFA) for the rapid presumptive identification of Bacillus anthracis, was conducted at 2 different test sites. The study evaluated the sensitivity of this assay using 16 diverse strains of B. anthracis grown on sheep blood agar (SBA) plates. In addition, 83 clinically relevant microorganisms were tested to assess the specificity of the RedLine Alert test. The results indicated that the RedLine Alert test for the presumptive identification of B. anthracis is highly robust, specific, and sensitive. RedLine Alert is a rapid test that has applicability for use in a clinical setting for ruling-in or ruling-out nonhemolytic colonies of Bacillus spp. grown on SBA medium as presumptive isolates of B. anthracis.
Keywords: Anthrax, Lateral flow assay, Rapid diagnostics, Bacillus anthracis
A comprehensive laboratory evaluation of the Tetracore RedLine Alert test, a lateral flow immunoassay (LFA) for the rapid presumptive identification of Bacillus anthracis, was conducted at 2 different test sites. The study evaluated the sensitivity of this assay using 16 diverse strains of B. anthracis grown on sheep blood agar plates. In addition, 83 clinically relevant microorganisms were tested to assess the specificity of the RedLine Alert test.
Anthrax is an acute infection caused by the gram-positive, rod-shaped, spore-forming facultative anaerobic bacterium Bacillus anthracis.1-5 Cells of B. anthracis are nonmotile, nonhemolytic, encapsulated, and arranged in chains. The vegetative form of this bacterium is not easily transmitted to humans, but the spores, which are the infectious form, are resistant to harsh environmental conditions, including ultraviolet light, ionizing radiation, heat, and various chemicals.4,6-8 Cutaneous anthrax infections make up the vast majority of reported human cases worldwide, accounting for more than 95% of human anthrax cases; if left untreated, up to 20% of cases can be fatal.3,9,10 Oropharyngeal or gastrointestinal anthrax is the second most common form of the disease, characterized by lesions in the oral cavity or stomach/intestinal tract due to consumption of contaminated meat or by swallowing aerosolized spores.11 Lesions cause massive swelling and blockage of the airway, stomach or intestinal perforation, or hemorrhage, and, if left untreated, 25% to 60% of cases are fatal.10,11 Inhalation anthrax, which is the most dangerous form of B. anthracis infection, with untreated fatality rates close to 100%, can be acquired through occupational exposure (eg, mill workers) or from an intentional release of spores, as happened in the anthrax attacks in 2001.3,9,12-16 In recent years, a fourth form, injection/septicemic anthrax, has emerged among intravenous drug users in western Europe.17 The most recent outbreak of injectional anthrax was believed to be the result of contaminated heroin that originated in Pakistan or Iran.18
B. anthracis ranks high on the list of potential agents of bioterrorism, as mortality rates can be significant when illness results from the inhalation of aerosolized spores.19-21 Experts consistently rank anthrax spores as a potential bioweapon when taking into consideration characteristics such as stability and ease of dissemination.20,21 Bioterrorism events and planning scenarios have emphasized the need for rapid and accurate detection and diagnostics to protect public health. Various tests have been developed to detect and/or identify B. anthracis in clinical specimens. Many of these tests have a high sensitivity and/or high specificity, such as real-time PCR, antigen detection tests, gamma phage susceptibility, and direct fluorescent antibody tests.22,23 However, these tests have limitations or high complexity testing.24,25 Because of these limitations, the time to obtain results may be delayed, which can hinder timely and critical treatment decisions.
Lateral flow immunochromatographic assays (LFAs) were commercially introduced for pregnancy testing in 1988.26 Simple to use and requiring minimal training,27 these LFAs are ideal for use by first responders and law enforcement officers to test suspicious materials in field settings. LFAs such as the BioThreat Alert® assays have previously been evaluated for the detection of several biothreat agents including orthopoxviruses,28 ricin,29 abrin,30 B. anthracis,31 and Yersinia pestis.32
The purpose of this study was to conduct a comprehensive test and evaluation of the FDA cleared RedLine Alert LFA (Tetracore, Inc., Rockville, MD) (510K Approval No. K030370) for presumptive identification of B. anthracis from nonhemolytic bacillus colonies grown on sheep blood agar (SBA) plates. The RedLine Alert LFA uses a combination of a polyclonal and a monoclonal antibody directed against extractable antigen 1 (EA1). The presence of EA1 in the sample forms a complex with the colloidal gold-labeled monoclonal antibody that migrates along the membrane. An immobilized rabbit capture antibody binds the colloidal gold-labeled antibody-antigen complex to form a colored line in the results window. An internal control line is also present in the results window to ensure that the test has been performed correctly.33,34 The evaluation included the likelihood of false-negative results (assay is negative but the analyte is present at a concentration below the limit of detection, or LOD), false-positive results (assay is positive but the target analyte is not present in the sample), and robustness and reproducibility of an assay that can be incorporated into the Level A protocol for B. anthracis.25 This will enable local hospital and commercial clinical laboratories to obtain a rapid presumptive diagnosis of anthrax infections, including those resulting from an intentional biological attack, so that appropriate notification and clinical intervention can be initiated in a timely manner to save lives.
This study was designed and executed through an interagency collaboration with participation from subject matter experts from the Department of Homeland Security (DHS) Science and Technology Directorate (S&T) Chemical and Biological Defense Division (CBD) and First Responders Group (FRG); Health and Human Services (HHS) Office of the Assistant Secretary for Preparedness and Response/Biomedical Advanced Research and Development Authority (ASPR/BARDA); HHS Centers for Disease Control and Prevention (CDC); HHS Food and Drug Administration (FDA), Office of Laboratory Science and Safety; FDA Center for Food Safety and Applied Nutrition (CSFAN); Department of Justice (DOJ) Federal Bureau of Investigation (FBI); the US Department of Agriculture (USDA); and others.
Materials and Methods
The Level A protocol for the sentinel level clinical laboratory for the presumptive identification of B. anthracis is to inoculate the specimen onto an SBA plate (and chocolate agar, and MacConkey or eosin methylene blue agar, depending on the type of specimen). After incubation at 35°C to 37°C in 5% to 10% CO2 for 18 to 24 hours (growth of B. anthracis may be observed as early as 8 hours), γ-hemolytic (no hemolysis) nonpigmented colonies with a ground glass appearance on the blood agar plate are gram stained. Those consisting of large, gram-positive rods are tested for the presence of catalase and motility. Cultures of γ-hemolytic, nonmotile, catalase positive, and gram-positive rods are considered as presumptive B. anthracis and sent to a CDC Laboratory Response Network (LRN) reference laboratory for confirmatory testing and further characterization.25 B. anthracis can be ruled out if the colonies are α- or β-hemolytic or if the organism is catalase negative and/or motile.25 RedLine Alert test strips (catalog number TC-5123-001) were obtained from Tetracore, Inc., in Rockville, MD. In Phase 1, the sensitivity of the assay was determined using the virulent B. anthracis Ames strain and an inclusivity panel of 15 additional strains. The virulent B. anthracis strains were grown, samples prepared, and 4 replicates tested at the Zoonoses and Select Agent Laboratory, Bacterial Special Pathogens Branch, National Center for Emerging and Zoonotic Infectious Diseases, CDC, Atlanta, GA. In Phase 2, assay specificity was determined using the avirulent Sterne strain of B. anthracis (positive control) and a panel composed of 81 clinically relevant strains. Most of the testing was done at Omni Array Biotechnology, Rockville, MD, by 4 different operators from DHS S&T and FDA CFSAN according to manufacturers' recommendations. Select agent organisms in the clinical panel were grown, samples prepared, and 4 replicates tested at Tetracore, Inc., Rockville, MD.
Phase 1: Inclusivity Panel
All inclusivity strains of B. anthracis (N = 16) representing clades A1.a, A1.b, A3, A3.a, A3.b, A4, B1, B2, and C were typed using Multiple Locus Variable-number Tandem Repeat Analysis (MLVA) and characterized by plasmid profile analysis and 16S typing at CDC (Table 1).
Table 1.
Inclusivity Strains of B. anthracis
| S. No. | Strain ID | Clade | Genotype | pX01 | pX02 |
|---|---|---|---|---|---|
| 1 | K8960; A0369; 2011756210 | A1.a | GT7 | Yes | Yes |
| 2 | K1256; A0193; 2000031657 | A1.a | GT10 | Yes | Yes |
| 3 | K9002; A0149; Turkey #32; 2000031650 | A1.b | GT23 | Yes | Yes |
| 4 | K7948; A0264; 2000031659 | A1.b | GT28 | Yes | Yes |
| 5 | K2802; A0248; 2000031652 Ohio ACB | A3 | GT68 | Yes | Yes |
| 6 | K4516; A0376; 2000031654 1015 | A3.a | GT51 | Yes | Yes |
| 7 | AO467; 2002013028 | A3.a | GT91 | Yes | Yes |
| 8 | K7816; Sterne | A3.b | GT59 | Yes | No |
| 9 | K1694; A0462; Ames; 2000031656 | A3.b | GT62 | Yes | Yes |
| 10 | K7222; A0379; 2000031653 SK-102 | A4 | GT69 | Yes | Yes |
| 11 | K4596; A0488; Vollum 1B; 2000031666 | A4 | GT77 | Yes | Yes |
| 12 | K1129; A0337; 2008724774 | A4 | GT74 | Yes | Yes |
| 13 | K8101; A0382; 2008724769 1035 | B1 | GT82 | Yes | Yes |
| 14 | K2762; A0465; 2000031651 RA3 | B2 | GT80 | Yes | Yes |
| 15 | Clade C Wild type; 2002013094 | C | GT133 | Yes | Yes |
| 16 | K5135; A0463; 2000031648 PAK-1 | a | a | Yes | Yes |
Strain K5135 received as GT29, but the allele size for vrrC1 does not match (should be 520 but is an unusual size of 548).
Prior to testing, the inclusivity panel strains were inoculated onto SBA plates and incubated at 37°C under aerobic conditions until visible colonies (2-4 mm in diameter) were observed. A colony was removed using a sterile loop and resuspended in 200 μL of colony isolation buffer (CIB) in a vial. To facilitate transfer, the loop was twirled in the buffer for 5 seconds to dislodge the colony. The tube was vortexed for 5 seconds to resuspend and to evenly distribute the cells in the buffer and then allowed to sit at room temperature for about 2 minutes. Next, the vials were gently vortexed, and 150 μL of the test sample was transferred to the sample well of the RedLine Alert test strip. Four replicates of each strain were tested, and visual readings were noted between 15 and 30 minutes after the addition of the sample.
Phase 2: Clinical Panel
After consultation with subject matter experts, a clinical panel consisting of 81 human pathogens was assembled (Table 2). The panel of bacteria consisted of agents known to cause dermal, pulmonary, gastrointestinal, and septicemic infections.
Table 2.
List of Clinically Relevant Background Organisms (Route of infection – Red color cell)
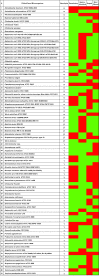 |
This panel was used to evaluate the specificity of the RedLine Alert LFA assay. Strains were grown either on selective medium or SBA by Lawrence Livermore National Laboratory, Livermore, CA, or Tetracore, Inc., Rockville, MD, and shipped to Omni Array, where they were subcultured onto SBA plates and incubated for 16 to 24 hours at 37°C to ensure purity. For long-term storage, individual bacterial seeds were prepared using MICROBANK™ (Pro-Lab, Ontario, Canada), based on manufacturer's guidelines and stored at −80°C. Two days prior to testing, the frozen stocks were retrieved, inoculated onto SBA plates, and incubated overnight at 37°C. The following day, cultures were examined for purity, and a single isolated colony was selected and streaked onto a new SBA plate. On the day of testing, the medium surrounding the colonies was examined. Cultures showing a darkening or discoloration of the medium surrounding the colonies demonstrated alpha-hemolysis. Cultures showing clear halos around and under the colonies exhibited beta-hemolysis. An isolated colony measuring 2 to 4 mm in diameter was removed with a sterile inoculating loop and transferred to a vial containing 200 μL colony isolation buffer. Vials were vortexed for 5 seconds to suspend and evenly distribute the cells in the buffer and allowed to stand at room temperature for at least 2 minutes prior to testing. The vials were gently vortexed, and 150 μL of cell suspension was added to the sample well of each strip. Strips were read visually 15 to 30 minutes following sample addition. Strips on which the control line did not appear were discarded. Each organism was tested by 4 different operators and visual observations recorded.
Phase 3: Informational Panel
Bacillus cereus biovar anthracis strains CA and CI were obtained from the Center for Biological Threats and Special Pathogens, Robert Koch Institute, Berlin Germany.35 Strains were grown and suspensions prepared for testing as described above for the inclusivity panel (Table 3).
Table 3.
Informational Panel Organisms (Route of infection – Red color cell)
Biosafety Considerations
The virulent B. anthracis and B. cereus biovar anthracis strains used in this study were handled with appropriate biosafety conditions at the CDC according to institutional biosafety guidelines. Virulent B. anthracis strains were handled in a BSL-3 laboratory. All other organisms, including low-risk bacterial strains, were handled, processed, and tested under safety protocols in accordance with Biosafety in Microbiological and Biomedical Laboratories (BMBL).36 To minimize risk of aerosols, cultures were handled using BSL-2 practices that also required personal protective equipment and procedures such as gowning, use of gloves, protective eyewear, and working in a certified Class II biosafety cabinet (BSC). All work areas before and after the testing were cleansed with 10% bleach, while disposal of stock cultures or biomedical waste was done in accordance with institutional guidelines.
Statistical Analysis
The performance of the lateral flow assay was assessed by calculating its sensitivity, specificity, and accuracy based on the results from all the testing done in this study. Sensitivity is defined as the proportion of true positives that are correctly identified by the test and is calculated as:
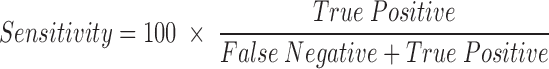 |
Specificity is defined as the proportion of true negatives that are correctly identified by the test and is calculated as:
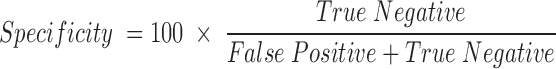 |
Accuracy is defined as the proportion of true positives and true negatives correctly identified by the test, the overall probability that the test correctly classifies the presence of the analyte (Bacillus anthracis in this instance), and is calculated as:
 |
Sensitivity, specificity, and accuracy from the visual results of the lateral flow assay were calculated using MedCalc Statistical Software version 18.11.3 (MedCalc Software bvba, Ostend, Belgium; http://www.medcalc.org; 2019).
Results
RedLine Alert test results of all 3 phases of testing—Phase 1, inclusivity panel; Phase 2, clinical panel; and Phase 3, informational panel—microorganisms are shown in Table 4. Number of samples tested as positive controls and negative controls in each phase are also given in the table.
Table 4.
RedLine Alert test results of 3 phase testing comprising B. anthracis Phase 1 inclusivity panel, Phase 2 clinical panel, and Phase 3 informational panel microorganisms
| Description | No. of Samples Tested | No. of Positive Controls Tested | No. of Negative Controls Tested | Total |
|---|---|---|---|---|
| Phase 1: Inclusivity panel testing | 64 | 16 | 48 | 128 |
| Phase 2: Clinical panel testing | 324 | 20 | 44 | 388 |
| Phase 3: Informational panel testing | 8 | 4 | 4 | 16 |
| Total | 396 | 40 | 96 | 532 |
Phase 1: Inclusivity Panel Testing
Four replicates of 16 geographically diverse B. anthracis strains that comprised the inclusivity panel were tested. A combined total of 64 tests were performed in this phase. B. anthracis Sterne strain was used as a positive control, and the manufacturer-supplied CIB was used as a negative control. Forty-eight negative controls and 16 positive controls were also tested in Phase 1 in addition to the inclusivity panel organism testing. All samples containing B. anthracis produced positive visual results; no reactivity was observed for any of the negative controls.
Phase 2: Clinical Panel Testing
The clinical panel used in the current study consisted of 40 (49.4%) γ-hemolytic organisms, 8 (9.9%) β-hemolytic microorganisms, and 33 (40.7%) α-hemolytic organisms (Table 2). As in Phase 1 testing, a combined total of 324 tests were performed in Phase 2 along with 44 negative controls and 20 positive controls.
Testing of the clinical panel by 4 operators yielded negative results for 80 of 81 (98.76%) organisms shown in Table 2 by visual inspection. The only positive result occurred with a β-hemolytic Bacillus cereus strain. When evaluating the assay using data from all organisms regardless of hemolytic activity, the sensitivity of the RedLine Alert assay was 100% and its specificity was 98.76% (Table 5). The sensitivity and specificity of the assay using results from only the nonhemolytic organisms in the clinical panel (40 out of 81) were 100% and 100%, respectively (Table 6). The data show that the assay is robust and does not cross-react with other clinically relevant organisms tested based on the testing algorithm. Nevertheless, the manufacturer's product disclosure notes that the RedLine Alert test may cross-react with B. cereus and B. thuringiensis; however, these organisms are β-hemolytic, so the predefined algorithm would exclude them for testing.
Table 5.
2 x 2 Contingency table and statistical analysis assessing the accuracy of RedLine Alert Assay based on the data from Phase 1 testing and all clinical panel microorganisms in Phase 2
| Result | Anthrax Positive | Anthrax Negative | Total |
|---|---|---|---|
| Red Line Alert Test positive | 64 | 0 | 64 |
| Red Line Alert Test negative | 4 | 320 | 324 |
| Total | 68 | 320 | 388 |
| Parameter | Percentage | Confidence Interval | |
|---|---|---|---|
| Sensitivity | 100.00% | 94.40% to 100.00% | |
| Specificity | 98.77% | 96.87% to 99.66% | |
| Accuracy | 98.97% | 97.38% to 99.72% | |
| Area under the curve | 0.99 | (0.99 to 1.00) | |
| Negative predictive value | 100.00% | — | |
Table 6.
2 x 2 Contingency table and statistical analysis assessing the accuracy of RedLine Alert assay based on the data from Phase 1 testing and Phase 2 testing of only nonhemolytic strains
| Result | Anthrax Positive | Anthrax Negative | Total |
|---|---|---|---|
| Red Line Alert Test positive | 64 | 0 | 64 |
| Red Line Alert Test negative | 0 | 160 | 160 |
| Total | 64 | 160 | 224 |
| Parameter | Percentage | Confidence Interval | |
|---|---|---|---|
| Sensitivity | 100.00% | 94.40% to 100.000% | |
| Specificity | 100.00% | 97.66% to 100.000% | |
| Accuracy | 100.00% | 98.34% to 100.00% | |
| Area under the curve | 1.00 | (0.98 to 1.00) | |
| Negative predictive value | 100.00% | — | |
Phase 3: Informational Panel Testing
B. cereus biovar anthracis is an emerging pathogen that has caused fatal anthrax-like infections in nonhuman primates such as chimpanzees and gorillas in West Africa.35 Human infections with this organism have not been reported but cannot be ruled out due to the paucity of clinical laboratories in areas where this organism has been found. These organisms possess some phenotypic properties of B. anthracis (γ-hemolytic, colony morphology), which may make them difficult to identify in a clinical laboratory. However, B. cereus biovar anthracis is motile, which would rule out B. anthracis prior to testing with the RedLine Alert assay. In a study limited by the availability of strains, B. cereus biovar anthracis strains CA and CI were tested 4 times each; 4 replicates of negative controls and 4 replicates of positive controls were also tested during the informational panel testing. Strain CA gave negative results, but strain CI gave positive results in the RedLine Alert test. Results of the informational panel testing were not included in the sensitivity, specificity, and accuracy calculations shown in Table 5. While these results are interesting, additional strains need to be tested before we can state with certainty that this organism can occasionally produce false-positive test results if the manufacturer's algorithm is not followed.
Analytical Sensitivity, Specificity, and Accuracy
Calculation of the analytical sensitivity, specificity, and accuracy of the RedLine Alert test is shown in Table 5 using the results from testing organisms in the inclusivity panel and the clinical panel; Table 6 uses the results from testing of clinically relevant nonhemolytic organisms (40) from the clinical panel and the inclusivity panel. Results of positive controls and negative controls tested in each phase were not included in the statistical analysis and calculations.
Of the 388 RedLine Alert tests performed in this study, 64 tests were positive (ie, “B. anthracis present in sample”), while 320 were negative (ie, “B. anthracis absent from sample”), and 1 negative sample “Bacillus cereus” yielded a false-positive result which equates to 4 tests. Based on these data, the assay sensitivity was calculated to be 100% and specificity at 98.76%, with an area under the curve value of 0.99. Since the test is FDA cleared for use in a clinical setting for testing unidentified γ-hemolytic bacterial colonies for presence of B. anthracis, analysis using data from only the γ-hemolytic organisms produced a sensitivity and specificity of 100% and 100%, respectively, with an area under the curve value of 1.
Discussion
B. anthracis is high on the list of potential bioterrorism agents because of its ease of dissemination, stability and hardiness of the spore in the environment, high morbidity and mortality rate, and past development as a biowarfare agent. One key to saving lives is rapid diagnosis and clinical intervention with appropriate antibiotics. The current Level A protocol used in clinical laboratories in the United States involves the identification of gram-positive, rod-shaped bacteria, which grow as γ-hemolytic (nonhemolytic) colonies on SBA plates, produce catalase, and are nonmotile.25 However, there are some drawbacks to relying solely on these methods. Laboratory personnel require additional training and certification to accurately identify these uncommon agents, using the appropriate biosafety practices, and to package and transfer the agents to a nearby LRN laboratory for secondary analysis and confirmation.24 The ability to rapidly identify B. anthracis in a clinical laboratory will greatly benefit the workflow and support timely therapeutic decisions.
The results of this extensive evaluation led us to conclude that the FDA-cleared RedLine Alert is a highly robust, sensitive, and specific test for the presumptive identification of B. anthracis from γ-hemolytic (nonhemolytic) colonies on SBA. It is important to emphasize that a motility test should be performed to differentiate B. anthracis (nonmotile) from B. cereus biovar anthracis (motile), as some strains of the latter organism may give a positive result with the RedLine Alert test.
The diagnostic methods used for the presumptive identification of B. anthracis by Level A LRN laboratories include culture on 5% SBA plates to determine colony morphology and gram stain characteristics, hemolysis (B. anthracis is γ-hemolytic), motility (B. anthracis is nonmotile), and microscopic observation for spores.37 The RedLine Alert test can reduce the time required for a presumptive identification of B. anthracis in the absence of a confirmed outbreak or biological attack. During an outbreak or biological attack, the assay can be used to confirm the presence of B. anthracis in order that appropriate antibiotic treatment can be started as quickly as possible.
This test may also support timely diagnosis in resource-limited endemic regions where there is a high prevalence of anthrax. For use in a low-prevalence region such as the United States, this test can serve as an important diagnostic tool in the event of an outbreak or biological attack. The 2001 anthrax incident in the United States prompted an extensive review of how clinical and public health laboratories should respond to bioterrorism. The LRN, which is capable of detecting, confirming, and reporting potential bioterrorism agents, was established prior to 2001.24 The LRN consists of 3 tiers of laboratories: (1) sentinel laboratories (Level A), which use Level A protocols developed by the American Society for Microbiology (ASM)—these laboratories generally function as a first line of defense for detecting possible outbreaks or infections caused by biothreat agents and alerting state and federal agencies;25 (2) reference laboratories, which are generally public health laboratories that perform confirmatory tests to produce high-confidence test results for threat analysis and intervention by public health and public safety authorities; and (3) national laboratories (eg, CDC) for definitive characterization.24 Figure 1 indicates that up to 31 hours may elapse between the time a symptomatic patient arrives at a clinic, an initial diagnosis is made, and presumptive and confirmatory testing are completed. Therefore, it is vital that a simple, rapid method be incorporated to rule in the presence of B. anthracis in a clinical sample as quickly as possible so that proper treatment can be started. Equally important, the 100% Negative Predictive Value (NPV, Table 6) of the RedLine Alert assay suggests that it can be used to rapidly rule out γ-hemolytic colonies of gram-positive bacilli as potential B. anthracis.
Figure 1.
Concept of operation for use of RedLine Alert™ test
The RedLine Alert kit is a very simple, easy-to-use, objective test for the presumptive identification of B. anthracis. This test can reduce the diagnostic time by several hours. Additionally, the RedLine Alert lateral flow assay has been approved by the FDA since 2003 for identifying B. anthracis in specimens from symptomatic patients cultured on SBA. However, it is critical that non–select agent registered laboratories (such as most hospital and commercial labs) promptly notify the CDC Select Agent Program and forward all samples and specimens to the closest LRN laboratory for further characterization to avoid violating the select agent regulation prohibiting possession of a select agent by an unregistered laboratory. Once testing has been completed and cultures/specimens forwarded to an LRN laboratory, remaining samples associated with the patient should be destroyed by the unregistered laboratory using appropriate methods within the time period stated in the select agent regulations.
Acknowledgments
This work was funded by US Department of Homeland Security Science and Technology Directorate Contract #HSHQDC-12-C-00071. The article reflects the views of the authors and should not be construed to represent the views or policies of DHS or HHS.
References
- 1. Bouzianas DG. Medical countermeasures to protect humans from anthrax bioterrorism. Trends Microbiol 2009;17(11):522-528 [DOI] [PubMed] [Google Scholar]
- 2. Brossier F, Mock M. Toxins of Bacillus anthracis. Toxicon 2001;39(11):1747-1755 [DOI] [PubMed] [Google Scholar]
- 3. Jamie WE. Anthrax: diagnosis, treatment, prevention. Prim Care Update Ob Gyns 2002;9:117-121 [Google Scholar]
- 4. Koehler TM. Bacillus anthracis physiology and genetics. Mol Aspects Med 2009;30(6):386-396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Helgason E, Okstad OA, Caugant DA, et al. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl Environ Microbiol 2000;66(6):2627-2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pilo P, Frey J. Bacillus anthracis: molecular taxonomy, population genetics, phylogeny and patho-evolution. Infect Genet Evol 2011;11(6):1218-1224 [DOI] [PubMed] [Google Scholar]
- 7. Dragon DC, Rennie RP. The ecology of anthrax spores: tough but not invincible. Can Vet J 1995;36(5):295-301 [PMC free article] [PubMed] [Google Scholar]
- 8. Fraise AP, Maillard JY, Sattar S, eds. Russell, Hugo and Ayliffe's Principles and Practice of Disinfection, Preservation and Sterilization. 5th ed. Wiley-Blackwell; 2012 [Google Scholar]
- 9. Balali-Mood M, Moshiri M, Etemad L. Medical aspects of bio-terrorism. Toxicon 2013;69:131-142 [DOI] [PubMed] [Google Scholar]
- 10. Cavallo JD, Ramisse F, Girardet M, Vaissaire J, Mock M, Hernandez E. Antibiotic susceptibilities of 96 isolates of Bacillus anthracis isolated in France between 1994 and 2000. Antimicrob Agents Chemother 2002;46(7):2307-2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cote CK, Welkos SL, Bozue J. Key aspects of the molecular and cellular basis of inhalational anthrax. Microbes Infect 2011;13(14-15):1146-1155 [DOI] [PubMed] [Google Scholar]
- 12. Beatty ME, Ashford DA, Griffin PM, Tauxe RV, Sobel J. Gastrointestinal anthrax: review of the literature. Arch Intern Med 2003;163(20):2527-2531 [DOI] [PubMed] [Google Scholar]
- 13. Centers for Disease Control and Prevention (CDC). Update: investigation of bioterrorism-related anthrax, 2001. MMWR Morb Mortal Wkly Rep 2001;50(45):1008-1010 [PubMed] [Google Scholar]
- 14. Friedlander AM, Welkos SL, Pitt ML, et al. Postexposure prophylaxis against experimental inhalation anthrax. J Infect Dis 1993;167(5):1239-1243 [DOI] [PubMed] [Google Scholar]
- 15. Hanna PC, Ireland JA. Understanding Bacillus anthracis pathogenesis. Trends Microbiol 1999;7(5):180-182 [DOI] [PubMed] [Google Scholar]
- 16. Holty JE, Bravata DM, Liu H, Olshen RA, McDonald KM, Owens DK. Systematic review: a century of inhalational anthrax cases from 1900 to 2005. Ann Intern Med 2006;144(4):270-280 [DOI] [PubMed] [Google Scholar]
- 17. Hanczaruk M, Reischl U, Holzmann T, et al. Injectional anthrax in heroin users, Europe, 2000-2012. Emerg Infect Dis 2014;20(2):322-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Price EP, Seymour ML, Sarovich DS, et al. Molecular epidemiologic investigation of an anthrax outbreak among heroin users, Europe. Emerg Infect Dis 2012;18(8):1307-1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rao SS, Mohan KV, Atreya CD. Detection technologies for Bacillus anthracis: prospects and challenges. J Microbiol Methods 2010;82(1):1-10 [DOI] [PubMed] [Google Scholar]
- 20. Inglesby TV, O'Toole T, Henderson DA, et al. Anthrax as a biological weapon, 2002: updated recommendations for management. JAMA 2002;287(17):2236-2252 [DOI] [PubMed] [Google Scholar]
- 21. Morse SA, Kellogg RB, Perry S, et al. Detecting biothreat agents: the Laboratory Response Network. ASM News 2003;69(9):433-437 [Google Scholar]
- 22. Tatti KM, Greer P, White E, et al. Morphologic, immunologic, and molecular methods to detect Bacillus anthracis in formalin-fixed tissues. Appl Immunohistochem Mol Morphol 2006;14(2):234-243 [DOI] [PubMed] [Google Scholar]
- 23. Hoffmaster AR, Fitzgerald CC, Ribot E, Mayer LW, Popovic T. Molecular subtyping of Bacillus anthracis and the 2001 bioterrorism-associated anthrax outbreak, United States. Emerg Infect Dis 2002;8(10):1111-1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wagar E. Bioterrorism and the role of the clinical microbiology laboratory. Clin Microbiol Rev 2016;29(1):175-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Buchan BW, Mahlen SD, Relich RF. Sentinel Level Clinical Laboratory Guidelines for Suspected Agents of Bioterrorism and Emerging Infectious Diseases. Washington, DC: American Society for Microbiology; 2018. https://www.asm.org/ASM/media/Policy-and-Advocacy/Biosafety_Sentinel_Guideline_October_2018_FINAL.pdf Accessed June12, 2019 [Google Scholar]
- 26. Gubala V, Harris LF, Ricco AJ, Tan MX, Williams DE. Point of care diagnostics: status and future. Anal Chem 2012;84(2):487-515 [DOI] [PubMed] [Google Scholar]
- 27. Andreotti PE, Ludwig GV, Peruski AH, Tuite JJ, Morse SS, Peruski LF., Jr Immunoassay of infectious agents. Biotechniques 2003;35(4):850-859 [DOI] [PubMed] [Google Scholar]
- 28. Townsend MB, MacNeil A, Reynolds MG, et al. Evaluation of the Tetracore Orthopox BioThreat® antigen detection assay using laboratory grown orthopoxviruses and rash illness clinical specimens. J Virol Methods 2013;187(1):37-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hodge DR, Prentice KW, Ramage JG, et al. Comprehensive laboratory evaluation of a highly specific lateral flow assay for the presumptive identification of ricin in suspicious white powders and environmental samples. Biosecur Bioterror 2013;11(4):237-250 [DOI] [PubMed] [Google Scholar]
- 30. Ramage JG, Prentice KW, Morse SA, et al. Comprehensive laboratory evaluation of a specific lateral flow assay for the presumptive identification of abrin in suspicious white powders and environmental samples. Biosecur Bioterror 2014;12(1):49-62 [DOI] [PubMed] [Google Scholar]
- 31. Ramage JG, Prentice KW, DePalma L, et al. Comprehensive laboratory evaluation of a highly specific lateral flow assay for the presumptive identification of Bacillus anthracis spores in suspicious white powders and environmental samples. Health Secur 2016;14(5):351-365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tomaso H, Thullier P, Seibold E, et al. Comparison of hand-held test kits, immunofluorescence microscopy, enzyme-linked immunosorbent assay, and flow cytometric analysis for rapid presumptive identification of Yersinia pestis. J Clin Microbiol 2007;45(10):3404-3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mangold BL, Aldrich JL. Spore Specific Antibodies. Tetracore, United States patent 7,772,374 B2. 2010
- 34. Cohen MN, Robison RA, Venkateswaran N, et al. Evaluation of a lateral flow assay kit for the detection of Bacillus anthracis and its application in laboratory diagnosis of anthrax. Bacillus ACT Annual meeting; 2015; New Delhi, India [Google Scholar]
- 35. Klee SR, Ozel M, Appel B, et al. Characterization of Bacillus anthracis-like bacteria isolated from wild great apes from Cote d'Ivoire and Cameroon. J Bacteriol 2006;188(15):5333-5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chosewood LC, Wilson DE, eds. Biosafety in Microbiological and Biomedical Laboratories. 5th ed. Washington, DC: US Department of Health and Human Services; 2009. https://www.cdc.gov/labs/pdf/CDC-BiosafetyMicrobiologicalBiomedicalLaboratories-2009-P.PDF Accessed June12, 2019 [Google Scholar]
- 37. Centers for Disease Control and Prevention. Anthrax. Reviewed January 31, 2017. http://www.cdc.gov/anthrax Accessed June12, 2019



