Abstract
Background:
Radiotherapy continues to be delivered uniformly without consideration of individual tumor characteristics. To advance toward more precise treatments in radiotherapy, we queried the lung computed tomography (CT)-derived feature space to identify radiation sensitivity parameters that can predict treatment failure and hence guide the individualization of radiotherapy dose.
Methods:
We used a cohort-based registry of 849 patients with cancer in the lung treated with high dose radiotherapy using stereotactic body radiotherapy. We input pre-therapy lung CT images into a multi-task deep neural network, Deep Profiler, to generate an image fingerprint that primarily predicts time to event treatment outcomes and secondarily approximates classical radiomic features. We validated our findings in an independent study population (n = 95). Deep Profiler was combined with clinical variables to derive iGray, an individualized dose that estimates treatment failure probability to be <5%.
Findings:
Radiation treatments in patients with high Deep Profiler scores fail at a significantly higher rate than in those with low scores. The 3-year cumulative incidences of local failure were 20.3% (95% CI: 16.0–24.9) and 5.7% (95% CI: 3.5–8.8), respectively. Deep Profiler independently predicted local failure (hazard ratio 1.65, 95% 1.02–2.66, p = 0.04). Models that included Deep Profiler and clinical variables predicted treatment failures with a concordance index of 0.72 (95% CI: 0.67–0.77), a significant improvement compared to classical radiomics or clinical variables alone (p = <0.001 and <0.001, respectively). Deep Profiler performed well in an external study population (n = 95), accurately predicting treatment failures across diverse clinical settings and CT scanner types (concordance index = 0.77 [95% CI: 0.69–0.92]). iGray had a wide dose range (21.1–277 Gy, BED), suggested dose reductions in 23.3% of patients and can be safely delivered in the majority of cases.
Interpretation:
Our results indicate that there are image-distinct subpopulations that have differential sensitivity to radiotherapy. The image-based deep learning framework proposed herein is the first opportunity to use medical images to individualize radiotherapy dose.
Keywords: artificial intelligence, personalized medicine, precision oncology, tumor heterogeneity
INTRODUCTION
Medical imaging is integral to the management of patients with cancer, with significant roles extending from diagnosis to treatment response monitoring.1 Its ubiquity in clinical practices notwithstanding, its current use remains largely subjective, exemplified by annotations of dimensions delimited by a range of human exactness. Computed tomography (CT) is the most frequently used modality across all cancers and comprises information beyond tumor geometry.2 Information acquired by the scanner is conveyed by a matrix of voxels across thin sections of the body composed of X-ray attenuation values proportional to the density of the incident matter. These values can have a total range of >4,096 intensities. The human eye, on the other hand, resolves a minor proportion of these intensities.3,4 Such limited discriminatory capacity clamors for ‘machine-like’ methods of information extraction and knowledge optimization.
Recent advances in image analysis have allowed for precisely this task. Radiomics permits the extraction of quantitative imaging descriptors or features that could characterize more objective tumor characteristics beyond human detection. This approach converts image data into a high dimensional feature space using a large number of data characterization algorithms.5,6 Some of these features have been shown to capture distinct tumor characteristics and exhibit prognostic power, indicating some value to this approach.7 Limitations to the utility of handcrafted image features, however, are their manual labeling and their inability to conform to a specific task. Manual labeling confines the feature space to elements that humans can learn and lack of deformability is a characteristic of the a priori design of the features, which cannot be modified based on the classification task at hand.
The process of feeding a machine raw data, like CT pixels, and allowing it to discover vectors for classification through the use of multiple layers of features is known as deep learning.8 Compared to natural images, medical images have regulated quality that can reduce noise and therefore make them more useful for deep learning-based approaches.9 However, while medical images can be an ideal source for deep learning, it remains difficult to secure a large quantity of clinically annotated datasets.10 Since classification accuracy is dependent on the size of the initial training datasets, computational methods that seek to optimize model performance are critical.
Cancers are characterized by substantial diversity and the optimal therapeutic approach has been shown to vary on the basis of the genetic features of individual cancers.11,12 Similarly, image-based profiling of tumors may reveal subpopulations that are more or less likely to be sensitive to a particular therapy and therefore guide its delivery. Classifications made by deep learning algorithms have begun to stratify patients on the basis of the type of cancer and genetic alterations.13,14 However, very little progress has been made in the use of deep learning to predict tumor responses to individual anti-cancer therapies.
High dose radiation delivery to the lung via stereotactic body radiotherapy (SBRT) was developed with the intent to effect local tumor control while potentially obviating perioperative or long-term surgical morbidity in patients with early-stage lung cancer or oligometastatic disease to the lung. Despite several prospective clinical trials demonstrating excellent local tumor control rates in medically inoperable patients with lung cancer,15–17 recent studies describe unacceptably high local failure rates in some patient subgroups.18–20 Ongoing and future studies of lung SBRT are likely to be significantly informed by a more accurate and quantitative determination of treatment failure risk and the mitigation of that failure by adjusting radiotherapy dose.
Herein, we incorporated domain-specific information, namely radiomics, in the training signal of a deep neural network and then combined this data with clinical variables to predict the likelihood of treatment success after lung SBRT, a mainstay of treatment for patients with early-stage lung cancer and oligometastatic disease to the lung. Our results signify a new roadmap for deep learning-guided predictions and treatment guidance in the image-replete and highly standardized discipline of radiation oncology.
METHODS
Clinical Characteristics
An institutional review board-approved study (IRB 14–562) was used to identify 1275 patients treated with lung stereotactic body radiotherapy (SBRT). Patients with primary (stage IA-IV) or recurrent lung cancer as well as patients with other cancer types with solitary or oligometastases to the lung were included. Patients without digitally accessible CT image or radiotherapy structure data were excluded from the study. A total of 944 patients met our eligibility criteria. 849 that were treated at the main campus in downtown Cleveland represented the internal study cohort. 95 patients that were treated at eight affiliate regional or national sites (Fairview, Ohio; Hillcrest, OH; Independence, OH; Mansfield, OH; Sandusky, OH; Wooster, OH; Weston, Florida) represented the independent validation cohort.
Patients were treated based on either a pathological or radiographic diagnosis. All primary lung cancer patients were staged using CT of the chest. PET and imaging of the brain (magnetic resonance imaging [MRI] or CT) was employed when clinically indicated. In cases where imaging revealed mediastinal or hilar lymph nodes enlarged by accepted radiographic criteria or where the standardized uptake value (SUV) exceeded a value of 3.0 on PET, pathological mediastinal evaluation with endobronchial ultrasonography-guided sampling (EBUS) was requested.
Radiotherapy was conducted first by patient immobilization with abdominal compression to restrict breathing motion. In cases where motion could not be adequately restricted to less than 1 cm (11.3% of patients), Active Breathing Coordinator (ABC) (Elekta, Stockholm, Sweden) was used. Tumors within a 2-cm expansion of the tracheobronchial tree were categorized as central. A risk-adapted approach for radiation dose delivery was used. Most patients received 50 Gy in five fractions. When the RTOG 0236 trial commenced, eligible patients with peripheral tumors to 60 Gy in three fractions we treated as per protocol while patients with central tumors continued to receive 50 Gy in five fractions. Alternative fractionations were employed for patients enrolled in a clinical trial, if constraints for our standard fractionation schedules could not be met or at the discretion of the treating radiation oncologist. Local failure was defined as radiographic progression within 1 cm of the planning target volume (PTV) to maintain a consistent definition of local/marginal failure in clinical trials of SBRT. Failures within the same lobe of the lung but greater than 1 cm from the PTV of the initial treatment site were defined as lobar failure and were not considered in this analysis. 8.5% of patients received adjuvant chemotherapy. The main indication for adjuvant chemotherapy was a perceived high risk of treatment failure. The recommendation to deliver adjuvant treatments was also influenced by considerations of patient tolerance for additional therapy.
CT Image Dataset
Planning CT images with corresponding physician-designated gross tumor volumes (GTV) were used. Images with contrast were excluded. Four scanners were used in the internal study population, namely three Philips Brilliance CT Big Bore (annotated CT-1, CT-2 and CT-3) and a Philips AcQSim (CT-4). The number of cases scanned on each of the scanners were 499, 244, 40 and 61, respectively; the identity of the scanner could not be definitively determined for 5 cases. The independent validation cohort had CT scanners made by GE, Siemens or Philips and four distinct models were used: GE Medical Systems Discovery ST, Philips Brilliance CT Big Bore, Philips Gemini GXL and a Siemens SOMATOM Definition AS.
Deep Profiler and Multi-task Learning
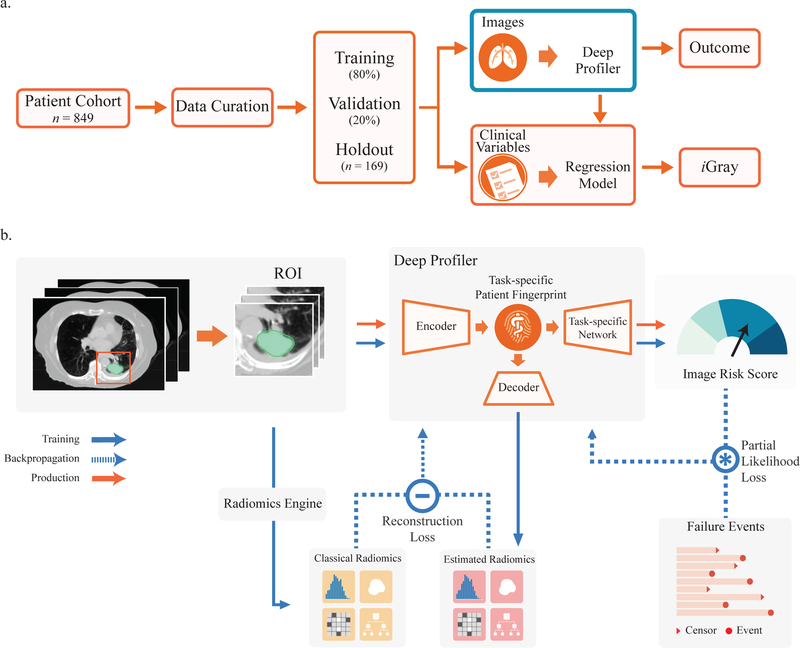
The schema for deriving the Deep Profiler signature is shown in Figure 1a. For a step-by-step protocol for generating Deep Profiler scores and a detailed description of the multi-task learning framework, see Supplementary Methods.
Figure 1.
Study design and neural network architecture. (a) Deep Profiler and iGray are obtained using a 5-fold cross-validation with an 80:20 ratio for training and validation within each fold, respectively. iGray is calculated by combining image and clinical data and using the cumulative incidence function of the regression model. (b) Neural network structure. The neural network consists of three main parts: an encoder for extracting imaging features and building a task-specific fingerprint, a decoder for estimating handcrafted radiomic features and a task-specific network for generating an image signature for therapy outcome prediction. A 3-D convolutional neural network (CNN) was used as an encoder for extracting imaging features. A fully-connected layer was adopted to link the latent fingerprint space with classical radiomics. Another neural network was used to predict the outcome based on latent fingerprint variables. Since therapeutic outcomes contain time-to-event information, we used a proportional hazards model to relate the time that passes before local failure occurs to classify failures.
Classical Radiomics
The 3D handcrafted radiomic features were extracted from GTV encompassing regions of interest (ROI). The handcrafted features can be divided into four groups: (1) intensity, (2) geometry, (3) texture, and (4) wavelet features. The intensity features quantified the first-order statistical distribution of the voxel intensities within the GTV. The geometry features quantified 3D shape characteristics of the tumor. The texture features described spatial distribution of the voxel intensities, thereby quantifying the intratumoral heterogeneity. The intensity and texture features were also computed after applying wavelet transformations to the original image. A total of 365 radiomic features were extracted. A list of all features can be found in Supplementary Table 1. All handcrafted features were extracted using Pyradiomics.21
We examined the performance of handcrafted radiomics to predict local failure. Five-fold cross-validation was also used for this analysis. Given that the number of radiomic features is much larger than the number of failures, either strong feature selection or model regularization was required to prevent overfitting. Feature selection was performed as previously described7. In the training set, we computed the performance of all individual features using C-index, and selected the one best feature from each of the four feature groups. These four features were then combined in a multivariate model for predicting local failure. Parameters estimated from the training were applied to the testing set for performance evaluation. To assess the performance of full handcrafted features, we also designed a multivariate model with Ridge L2 regularization on regression coefficients. Parameters were optimized using the training set and selected based on the performance in the validation set. Similar to the feature selection method, the final performance was evaluated using the testing set.
Clinical Variable Integration and iGray
We also assessed the complementary effect of the image score with other clinical risk factors such as biologically effective dose (BED) and histological subtypes. BED was calculated using an α/β ratio of 10 Gy, modeled as a continuous variable. We assessed the effects of two main histological subtypes, adenocarcinoma and squamous cell carcinoma (SqCC) and modeled them as categorical data. In the presence of the competing risk (death), Fine and Gray regression modeling was used to examine the effect of all factors to the local failure. Univariate analysis was first used to confirm the significance level of each individual factor. All three variables were included in the multivariable model. For directly evaluating the effect of histological subtype between adenocarcinoma and SqCC variable, the model was fitted to a subset of the data (i.e. adenocarcinoma and SqCC patients only).
We used the multivariate regression model with Deep Profiler score and BED to both predict failure and calibrate the radiation dose to modulate the risk of local failure. iGray was defined as the dose that results in a probability of failure of <5% at 24 months and is in units of BED. The calibration was achieved by estimating the cumulative incidence function (CIF) from the regression model. According to the assumptions in Fine and Gray’s model, the predicted CIF can be computed for a subject with covariate vector X as follows
where I0(t) is the estimated baseline CIF, X = (ximg, xBED)T is the covariate vector, and β = (βimg, βBED)T are the regression coefficients for image and BED covariates.
To estimate the feasibility of delivering iGray recommended doses, we permitted prescribed doses up to 180 Gy BED for GTVs that were outside of the central zone per RTOG 023615 and 0618.22 For central tumors, we partitioned the central zone region, which is within a 2 cm radius of large airways or the proximal bronchial tree (PBT), into four equal segments. We assigned a gradient BED schema to tumors from the most proximal to the most distal segments: 108, 132, 149.5 and 168 Gy, respectively. 108 Gy BED (60 Gy in 8 fractions) has been previously shown to be safe for ultra-central tumors.23,24 The use of 132 Gy BED in the next segment is per RTOG 0813, which indicated that the maximal tolerated dose in patients with centrally located tumors is 12 Gy in 5 fractions.25 For tumors from 1–2 cm, minimal to no overlap between the treated volume and the PBT and central organs at risk is expected due to more limited respiratory motion in the central zone and PTV expansions of only ~5 mm. Nevertheless, we used a conservative linear gradient of risk to estimate putative safe doses in these regions. These latter estimates are theoretical as the relationships between dose escalation, a stratified central zone and toxicity have yet to be thoroughly investigated.
Saliency Map
To find the voxels of an input volume that contribute the most toward the prediction of treatment failures, we took the derivative of the final partial likelihood loss with respect to the input CT volume and evaluated each volume Xi as . This derivative provides a scalar quantity for each of the voxels in the input volume, indicating the influence of the variation of voxel to the output of the model. The magnitude of these values was projected on the CT image to create a saliency map.
Statistical Analysis
To quantify the predictive performance, the concordance index (C-index) was measured between network output and actual event (local failure) time. The concordance index is a measurement between 0 and 1 that indicates how well the prediction model can order the actual event times - 1 indicates perfect concordance while 0.5 indicates no better concordance than chance. The averaged C-index across all five folds was calculated. The confidence interval was calculated using a bootstrap approach. We calculated cross-validated C-indices based on bootstrap resampling of the testing set and repeated 1000 times. The 2.5th and 97.5th percentile of the bootstrapped C-index distribution was used as an estimation of the 95% confidence interval.
We compared the predictive performance of handcrafted radiomics and our imaging fingerprint using the C-index. The performance of tumor 2D CT size, the maximum 3D diameter and volume were used as comparators. We applied a bootstrap method to compare the significance between different models. For each model, we randomly resampled the testing set and calculated the C-index. This was repeated 100 times for all five folds. Wilcoxon test was used to assess the significance between the C-index distributions of different models.
To further explore the association between the imaging index and failure time, competing risk analysis was performed to estimate the cumulative incidences of local failure. The Kaplan-Meier method is inappropriate for estimating the incidence rate of therapy failure in the presence of death because patient death leads to the censoring of the primary outcome. As mortality is not completely independent from therapy failure, death without evidence of local failure was treated as a competing event. The median score in the training set was computed and then applied as a threshold to stratify patients in the testing set into high and low risk groups. After the cross-validation was complete, each patient was classified into one of the risk groups. Cumulative incidence curves (CICs) were estimated for each group, and Gray’s test was used to determine the significance of difference between two curves.26 Statistical analysis was performed using R 3.2.5.27
Role of the funding source
This work was supported, in part, by Siemens Healthcare. Siemens contributed to data analysis and interpretation and the writing of aspects of the manuscript (see Contributors). The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
RESULTS
Deep Profiler Accurately Predicts Radiation Treatment Failures
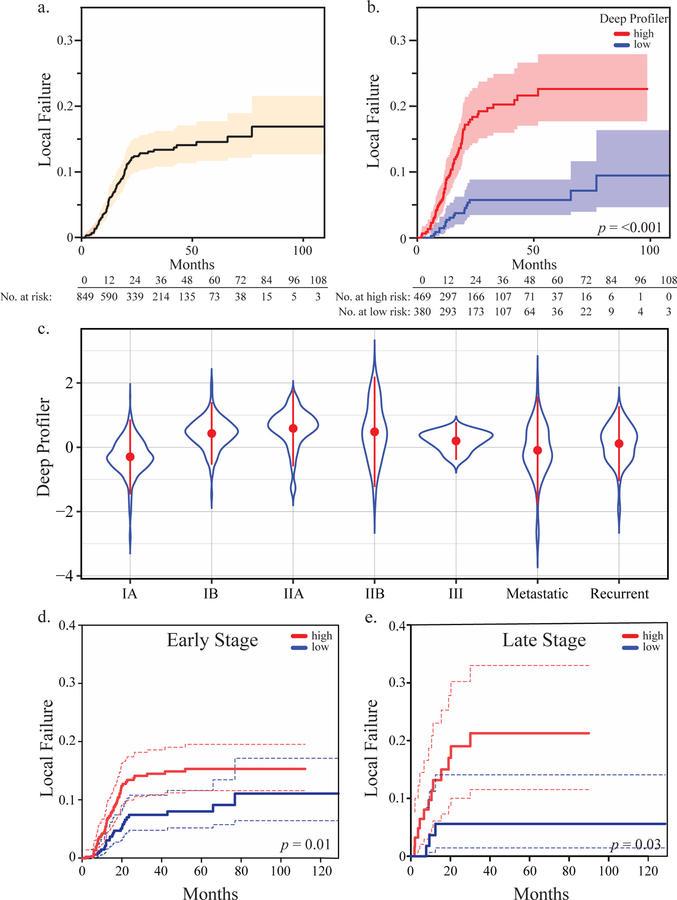
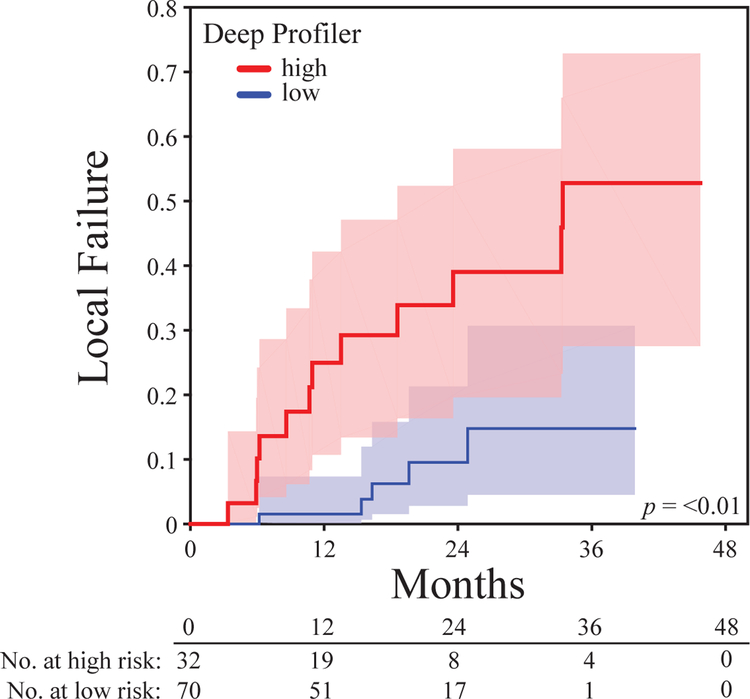
A total of 849 patients met our eligibility criteria. 86.9% of the patients enrolled received definitive treatment for early stage non-small cell lung cancer (Table 1). The cumulative incidence of local failure at 3 years was 13.5% (95% CI: 10.8–16.2). Patients were stratified into high and low risk groups based on a median score cutoff of a neural network derived imaging signature from the training set. This process was repeated for each partition and the results of all five folds were concatenated for statistical analysis. A total of 469 patients were stratified into the high-risk group and 380 patients were in the low risk group. Estimated CICs of local failure for overall population and each risk group are shown in (Figure 2a & b). Gray’s test for equality across Deep Profiler risk groups was significant (p = <0.001). Patients in the low risk group failed radiotherapy at a significantly lower rate than do those in the high-risk group, with 3-year cumulative incidences of local failure of 5.7% (95% CI: 3.5–8.8) and 20.3% (95% CI: 16.0–24.9), respectively.
Table 1.
Baseline Characteristics of the Study Population
| Characteristics | |
|---|---|
| No. patients | 849 |
| Follow-up, months | 20.93 (11.03–37.97) |
| Age | 74.1 (67.6–80.7) |
| Sex | |
| Female | 440 (51.8%) |
| Male | 409 (48.2%) |
| Treated tumor size, cm | 2.3 (1.6–3.4) |
| Overall stage | |
| I | 645 (76%) |
| II | 81 (9.5%) |
| III | 8 (0.9%) |
| IV | 74 (8.7%) |
| Recurrent | 41 (4.8%) |
| Histology | |
| Adeno | 255 (30.0%) |
| SqCC | 248 (29.2%) |
| NSCLC (NOS) | 47 (5.5%) |
| Neuroendocrine | 14 (1.7%) |
| Other | 22 (2.6%) |
| Non-diagnostic biopsy | 74 (8.7%) |
| No biopsy | 189 (22.3%) |
| Indications for treatment | |
| Definitive | 738 (86.9%) |
| Salvage | 52 (6.0%) |
| Oligometastatic | 50 (5.9%) |
| Other | 9 (1.1%) |
| Total dose, Gy | 50 (30–60) |
| No. fractions | 5 (1–10) |
| BED, Gy | 100 (39–180) |
Continuous variables are represented as medians with inter-quartile ranges, with the exception of total dose, no. of fraction and BED, Gy, which are represented by median and range.
Abbreviations: Adeno, adenocarcinoma; BED, biologically effective dose; CT, computed tomography; IQR, inter-quartile range; NOS, not otherwise specified; SqCC, squamous cell carcinoma; SUV, standardized uptake values.
Figure 2.
Deep Profiler predicts local failure. Estimated cumulative incidence curves for local failure in the overall population (a) or apportioned by the median Deep Profiler score based on each cross-validation step followed by concatenation (b). (c) Violin plot of Deep Profiler scores by tumor overall stage. The mean (point) and standard deviations (point range) are shown. (d) Estimated cumulative incidence curves for local failure apportioned by the median Deep Profiler score for early stage (IA-IIB) (e) or late stage tumors (III, IV and recurrent) (f). Gray’s test was used to test for equality across Deep Profiler risk groups.
To determine the clinical setting(s) in which Deep Profiler can be most predictive of local failure, we examined the impact of tumor stage on Deep Profiler and its prediction accuracy. Scores varied based on tumor stage, with IA tumors having the lowest mean score (Figure 2c). Despite differences in the mean scores of Deep Profiler across some stages of disease, there was significant variation within and across individual stages. These results suggested that information beyond tumor stage was learned by Deep Profiler. Consistent with this observation, Deep Profiler predicted local failure in patients with early- or late-stage cancers (Figure 2d & e).
To assess the influence of possible variation in the types of treatments delivered or CT image acquisition, we assessed the impact of motion management, the use of adjuvant chemotherapy and CT scanner type on Deep Profiler. Scores were not significantly different based on the type of motion management used for treatment (ABC versus abdominal compression, p = 0.353) (Supplementary Figure 1). Scores were, however, significantly higher in patients that received adjuvant chemotherapy (p = <0.001), although there was significant overlap across the two groups (Supplementary Figure 2). This data is consistent with the physician-directed recommendation of adjuvant chemotherapy on the basis of variables that are perceived to lead to a higher risk of treatment failure28 and suggests that Deep Profiler could potentially inform these recommendations. Lastly, scores obtained from the two most frequently used CT scanners (both of which are Philips Big Bore had similar accuracy in predicting local failures (Supplementary Figure 3).
Deep Learning Outperforms Classical Radiomics
We compared the performance of our neural network-derived imaging index to two-dimensional (2D) CT size, maximum 3D CT size, 3D tumor volume and classical radiomic features. Our learning-based framework is superior to classical radiomics features, which were in turn superior to tumor volume followed by 2D size values (Table 2). The superiority of Deep Profiler indicated that features beyond tumor size, which has been previously shown to be associated with local failure after high-dose radiotherapy to the lung,29 can be identified using our deep learning algorithm.
Table 2.
Prognostic performance of different models
| Model | Concordance Index | Confidence Interval | v. Deep Profiler p-value |
|---|---|---|---|
| 2D CT Size | 0.610 | [0.545, 0.672] | 2.05×10−32 |
| Max 3D diameter | 0.655 | [0.600, 0.712] | 1.06×10−20 |
| 3D Volume | 0.669 | [0.611, 0.726] | 9.78×10−14 |
| Classical Radiomics (feature selection) | 0.651 | [0.600, 0.710] | 4.23×10−25 |
| Classical Radiomics (regularization) | 0.680 | [0.625, 0.739] | 1.18×10−10 |
| Deep Profiler | 0.711 | [0.660, 0.767] | - |
On univariate analysis, a higher image-based risk score (Deep Profiler), lower radiation dose and histological subtype were associated with an increased risk of local failure (Supplementary Table 2). On multivariate analyses, all three factors remained significantly associated with local failure (Table 3). The multivariable models that included Deep Profiler and clinical variables predicted treatment failures with a C-index of 0.72 (95% CI: 0.67–0.77), which was a significant improvement when compared to classical radiomics (p = <0.001) or 3D volume (p = <0.001). These results indicated that an image-based score can provide complementary information to the clinical established variables of histological subtype and radiation dose18.
Table 3.
Predictors of Local Failure in Multivariable Analysis
| HR | 95% L | 90% U | p-value | |
|---|---|---|---|---|
| Deep Profiler signature | 1.645 | 1.016 | 2.663 | 0.042 |
| BED (continuous) | 0.978 | 0.969 | 0.987 | 0.026 |
| Adeno vs. SqCC | 0.494 | 0.281 | 0.868 | 0.029 |
| Adeno vs. Others | 0.515 | 0.286 | 0.927 | 0.027 |
iGray and Personalized Radiation Dose Delivery
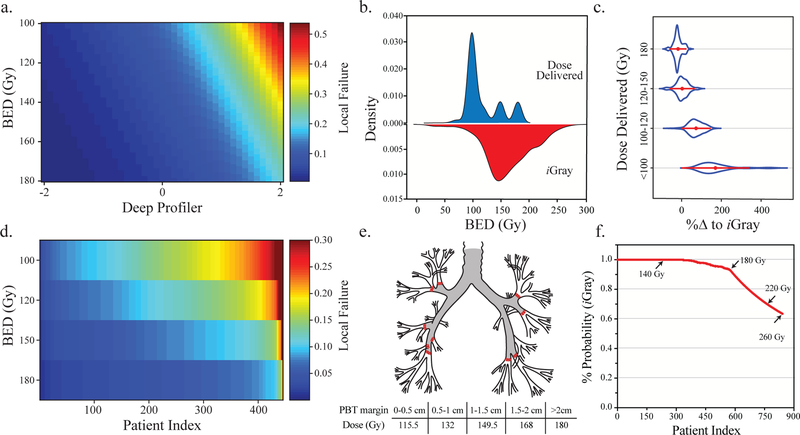
We posited that treatment failures can be mitigated by higher radiation doses and that we can model this relationship for the purpose of guiding dose individualization. First, we built a Fine and Gray’s regression model using the imaging signature and dose of radiation. This enabled us to model the risk of local recurrence by tuning the dose of radiation accordingly. Importantly, the type of treatment delivered is the only mutable variable identified in the model; tumor size, CT image features and histology are fixed. Using this model, we calculated the probability of local failure at 24-months after treatment as a function of radiation dose. Our results indicated that local failure can be significantly reduced as a function of radiation dose (Figure 3a).
Figure 3.
Ameliorating treatment failure by adjusting the radiation dose. (a) Heatmap demonstrating the relationship between Deep Profiler score (x-axis), iGray in biologically effective dose (BED) units (y-axis) and the probability of local failure (right legend). As iGray dose increases, the risk of failure decreases and vice versa. (b) The kernel density estimation of the actual dose of radiation delivered and iGray. (c) Violin plots of the distribution for the percent change in dose delivered required to achieve iGray stratified by the four treatment categories (y-axis). (d) The probability of local failure was calculated for patients receiving the most common treatment regimen of 100 Gy BED after incremental increases in radiation dose. The overall rate of local failure is significantly diminished at the highest doses. (e) A depiction of the proximal bronchial tree (PBT). A proposed theoretical dose scheme to avoid proximal airway toxicity is shown in the bottom (see Methods). (f) The cumulative relative frequency of feasibly delivering iGray per the scheme in (e). Some iGray doses are shown (arrows) to highlight the range in which a substantial decrement in feasibility occurs.
We then calculated the patient-specific dose that reduces the probability of treatment failure to <5%, iGray, for each patient using a permuted holdout set design. The kernel densities of dose delivered compared to iGray showed significant overlap (Figure 3b). The range of iGray was wider (21.15–277.1 Gy) with greater standard deviation (40.6 v. 30 Gy). The percent dose difference required to achieve iGray for each patient was calculated and its distributions were plotted for a function of each dose delivery interval (Figure 3c). These results indicated that iGray is likely to be feasible in a majority of patients receiving high-dose radiotherapy to the lung.
To assess the feasibility of delivering iGray dose recommendations, we first estimated the impact of incremental dose increases on the probability of local failure in patients who received a BED of 100 Gy, the most frequent treatment dose in the cohort (n = 445). We used our model to estimate local failure probabilities at 24-months (Figure 3d) and showed that even incremental increases in the dose delivered to these patients can significantly reduce treatment failure probability. To generate an estimate of the extent of feasibility in all patients in our cohort, we used a gradient dose scheme that is extrapolated from previous dose escalation studies to avoid airway toxicity in based on the proximity of the tumor to the proximal bronchial tree (Figure 3e and Methods). Using this scheme, the cumulative relative frequency of safely achieving iGray is 63.5% (Figure 3f).
Model Accuracy and Scalability
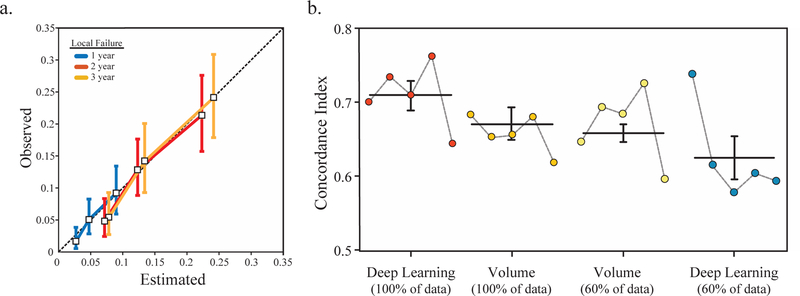
To examine the agreement between the observed outcomes and the multivariate model with the iGray and BED, we calculated calibration curves. A calibration curve was obtained by plotting the average predicted probability at 1, 2, or 3 years after radiation treatment against CIC estimates of the actual outcome (Figure 4a). Our results indicated that our model accurately predicts treatment outcomes.
Figure 4.
Model performance and the scalability of the deep neural network. (a) The calibration curves for the multivariable model that includes iGray and BED demonstrated strong agreement between the estimated and observed 1-, 2-, and 3-year outcomes. Vertical bars represent 95% confidence intervals for observed local failure probabilities. (b) Comparison of model performance using the full data set and 60% of the dataset demonstrates the superiority and the potential scalability of the deep learning algorithm. Horizontal lines and error bars represent the mean C-indices and standard errors across five folds.
In addition, we sought to determine the impact of dataset size on prediction accuracy. To achieve this, we randomly selected 60% of the patients in the dataset and calculated the concordance indices using our deep learning platform and a classical clinical risk factor (i.e. volume) and compared it to our analyses using 100% of the patients. Our results indicated that whereas volume appears to reach a plateau in accuracy, our framework’s performance is significantly higher with increases in sample size (Figure 4b). These results establish the scalability of deep learning in our dataset and suggest that improvements in accuracy are more likely using deep learning-based approaches compared to tumor volume measurements with dataset growth.
Voxel Saliency and Tumor Volume
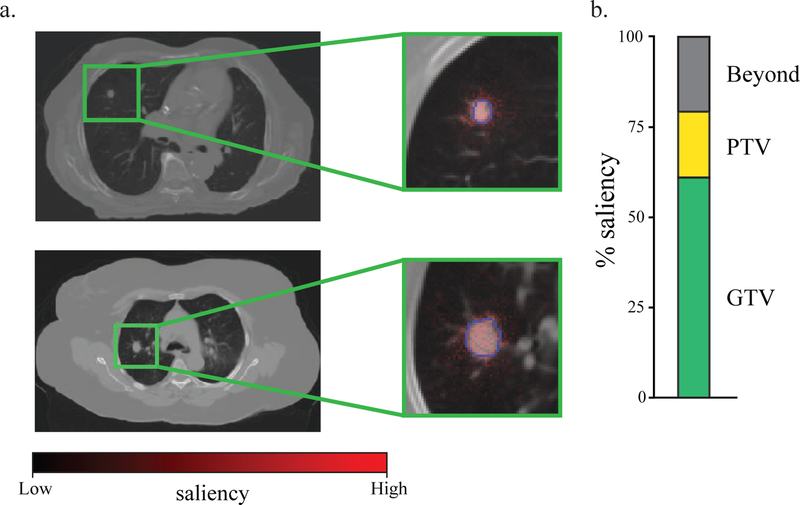
To determine the significance of each voxel on treatment failure, we calculated a saliency map for each tumor. Saliency projects a weight in heatmap form to each voxel in the image and this weight reflects the importance of that voxel on the image risk score (Figure 5a). Critically, the majority of the most salient voxels were within the GTVs and PTVs across all tumors, indicating that the gross tumor and the peri-tumoral region are the most relevant voxels to the model (Figure 5b). This saliency detection method is also a critical examination of the spatial sampling methodology of cropping to a 64 × 64 × 32 sub-volume encompassing the tumor, indicating that the volume is sufficiently large to encompass the most salient voxels but not too broad resulting in a classifier that fails to understand despite having high accuracy (e.g. spurious voxel associations).30 Lastly, there were a number of salient voxels outside of the GTV (37.8%) and PTV (20.5%) across the dataset. The role of these outlying salient voxels in marginal treatment failures remains unclear.
Figure 5.
Tumor saliency extends beyond the physician delimited tumor volume. (a) A saliency map represents an overlay of heatmap color (red=high saliency; black=low saliency) on voxels in the CT image region of interest. Blue contour lines represent the delineation of tumor volume. Two patient examples are shown. (b) The proportion of voxels with saliency >0.20 within the gross tumor volume (GTV) (green), the planning target volume (PTV) or beyond both volumes (gray) are shown.
Deep Profiler Generalizes to Independent Populations
We sought to measure the accuracy of Deep Profiler using a different but plausibly related independent population of patients who received SBRT to the lung. A total of 95 patients with 102 tumors (metachronous and/or synchronous treatments were included) from eight affiliate treatment centers met our eligibility criteria. Differences in baseline patient characteristic compared to our internal study population included a shorter median time to follow-up (p < 0.01), smaller tumors (p < 0.001) and lower radiation doses (p < 0.01) (Table 4). These results indicated sufficient differences between the internal and independent validation cohorts to allow for an assessment of both the reproducibility and transportability of the model.31
Table 4.
Baseline Characteristics of the External Validation Population
| Characteristics | P value | |
|---|---|---|
| No. patients / tumors | 95 / 102 | |
| Follow-up, months | 16.4 (11.4–24.6) | <0.01 |
| Treated tumor size, cm | 1.8 (1.3–2.7) | <0.001 |
| Overall stage | 0.29 | |
| I | 78 (76.5%) | |
| II | 10 (9.8%) | |
| III | 0 (0 %) | |
| IV | 5 (4.9%) | |
| Recurrent | 9 (8.8%) | |
| Histology | 0.21 | |
| Adeno | 43 (42.1%) | |
| SqCC | 24 (23.5%) | |
| NSCLC (NOS) | 8 (7.8%) | |
| Neuroendocrine | 1 (0.9%) | |
| Other | 1 (0.9%) | |
| Non-diagnostic biopsy | 6 (5.9%) | |
| No biopsy | 19 (18.6%) | |
| Indications for treatment | 0.52 | |
| Definitive | 88 (86.2%) | |
| Salvage | 9 (8.8%) | |
| Oligometastatic | 5 (4.9%) | |
| Other | 0 (0%) | |
| Total dose, Gy | 50 (34–60) | 0.02 |
| No. fractions | 5 (1–5) | <0.01 |
| BED, Gy | 100 (72–180) | <0.01 |
| CT Simulator Type | NA | |
| GE Medical Systems Discovery ST | 14 (13.7%) | |
| Philips Brilliance CT Big Bore | 22 (21.6%) | |
| Philips Gemini GXL | 17 (16.7%) | |
| Siemens SOMATOM Definition AS | 49 (48.0%) |
Continuous variables are represented as medians with inter-quartile ranges, with the exception of total dose, no. of fraction and BED, Gy, which are represented by median and range.
Abbreviations: Adeno, adenocarcinoma; BED, biologically effective dose; CT, computed tomography; IQR, inter-quartile range; NOS, not otherwise specified; SqCC, squamous cell carcinoma; SUV, standardized uptake values.
p values were calculated using Pearson’s Chi-squared test for categorical variables and by the non-parametric test of medians for continuous variables. *p value of < 0.05 was considered statistically significant.
The cumulative incidence of local failure in this population at 2 years was 19.7% (95% CI: 10.9–30.4). Patients were stratified into high and low risk groups based on a median Deep Profiler score from a training dataset that included all 849 patients in our internal study population. Estimated CICs of local failure for overall population and each risk group are shown in (Figure 6). Gray’s test for equality across Deep Profiler risk groups was significant (p = 0.002). Patients in the low risk group failed radiotherapy at a significantly lower rate than do those in the high-risk group, with 2-year cumulative incidences of local failure of 9.5% (95% CI: 2.8–21.3) and 39% (95% CI: 19.6–58.1), respectively. Deep Profiler predicted treatment failure with a C-index of 0.77 (95% CI: 0.66–0.92), which was calculated based on bootstrap resampling of the external dataset and repeated 1000x. These results indicate that Deep Profiler can predict treatment failures accurately across diverse clinical settings and distinct CT simulator scanners (Table 4).
Figure 6.
Deep Profiler accurately predicts local failure in an external study population. Estimated cumulative incidence curves for local failure apportioned by the median Deep Profiler score derived from the internal study population.
DISCUSSION
Accurate estimates of the probability of treatment success for individual patients can significantly improve clinical outcomes. In this study, we show that the clinical responses of cancer to radiotherapy vary in a manner not fully explained by clinical and histopathological variables alone and that CT image-based features contribute to this variance. The most important message in our study is that predictive features can be learned from CT images and contribute to the individualization of radiation dose.
Quantitative image analyses to date have not been used to personalize cancer treatment delivery.32–34 To address these limitations, we trained a machine to learn the multi-dimensional feature space in a large cohort of patients with cancer in the lung that was treated with a wide dose range of radiation. In unconstrained machine learning algorithms of imaging data, the machine predominantly seeks relationships between multi-dimensional inputs and outcomes data. However, most clinically available datasets have generally been smaller and more limited than the datasets other disciplines may use to tune their predictive algorithms, and the quality and completeness of existing outcomes data could be key barrier to these approaches. To address these limitations, we used a multi-task approach that takes advantage of established image-based radiomic features to partially delimit and inform the neural network. We demonstrated that this approach, Deep Profiler, is superior to deep learning or classical radiomics alone. We also demonstrated that Deep Profiler can accurately predict treatment failures in varied clinical settings.
To limit spurious voxel associations with our predicted outcome, we incorporated prior knowledge by using the physician delimited tumor volume as part of the input into our network. Although manual annotation could bias the feature extractions, we showed that the voxels that are most deterministic for treatment failure localize within the physician contoured volumes (GTV or PTV). Conversely, some salient voxels localized to the peri-tumoral regions or tumor margin. Since classical radiomics approaches disregard image information outside of the GTV, the identification of these salient voxels is an additional advantage of this approach. The potential association of these voxels with marginal recurrences remains to be explored. To the extent that marginal salient voxels are predictive of local failures, automatic contouring of tumor saliency maps could represent a leap toward more accurate tumor volume delineation and informed inhomogeneous dose delivery.35,36
Our results have several additional clinical implications. First, there are image-distinct subpopulations that demonstrate differential sensitivity to radiotherapy. We showed that Deep Profiler has a wide range of values and is significantly associated with treatment failures across varied clinical settings, including plausibly related populations. These populations, in part, included distinct stages of disease, CT simulation scanners, motion management techniques, linear accelerators, radiation oncologists and therapists, geographies (local, regional and national) and longitudinal periods. Second, we provide an integrated method that uses image and, importantly, established clinical variables to individualize radiation dose. Moreover, iGray uses the clinically validated linear quadratic model,37 is empirically derived in that no assumptions are made regarding individual tumor radiosensitivity (α and β in the tumor toxicity isoeffect remain constant) and its output is directly clinically actionable by recommending a dose that can be achieved using several treatment schedules. Third, our prediction accuracy is evolvable. A critical feature of neural network-based prediction is the potential for substantial improvements in accuracy with scale. As our dataset increases in sample size and/or is augmented by integration into large data sharing collaborations, the network is expected to substantially improve in prediction accuracy. This is in contrast with other computational methods like classical radiomics, whose accuracy appears to plateau in the early phases of dataset growth (see Figure 4). Another important element of the evolvability of our model is the eventual stratification of the dataset into more homogeneous populations based on variables such as cancer subtype, clinical stage, use of systemic adjuvant therapy, et cetera. This is particularly compelling considering the demonstrated preliminary efficacy of SBRT in patients with oligometastatic disease from distinct cancer types.38,39 The use of SBRT in varied clinical settings will result in larger and more diverse datasets that are more amenable to data partitions, and therefore improved model accuracy.
An image-based framework for the personalization of radiotherapy dose can substantially alter the clinical radiotherapy paradigm. The radiation oncologist is advantaged because the dose of radiation delivered can be calibrated on the basis the risk of treatment failure, which itself is a continuum. This largely mitigates binary decisions of “to treat or not to treat” and instead permits the adjustment of radiation treatments to prevent treatment failures. iGray can assist in the design of image-stratified, radiotherapy-based trials. In this role, it can guide the evolution of radiotherapy toward dose delivery strategies that are calibrated on the basis of individual predictions of tumor control probability.
There are several characteristics of Deep Profiler and iGray that suggest a lower implementation barrier. Due to the strict requirement for the acquisition of radiation planning CT images for radiotherapy, each radiation treatment center is likely to have an extensive CT dataset that could be utilized for model development and implementation. Combined with the automated feature algorithms of scalable deep learning-based prediction platforms, this represents an accessible opportunity to directly improve medical-decision support across broad cancer patient populations receiving radiotherapy.
The strengths of our study include the large number of patients evaluated, the completeness of the dataset, the use of a carefully annotated radiotherapy specific outcome (local failure) rather than surrogate of treatment failure (e.g. progression-free survival, cancer-specific mortality or overall survival) and the use of readily implementable and highly tractable image-based score as a backbone for our analyses. The limitations of our study include the following: we cannot fully account for all potential causes of bias, there is explicit population heterogeneity in our datasets (e.g. clinical stage, radiation dose, CT scanners, motion management, et cetera), the independent validation cohort is limited in size and we do not account for normal tissue toxicity. These limitations can, in part, be addressed with the incorporation of new datasets and emerging tools that predict lung toxicity,40 respectively.
In summary, we combined clinical variables with deformable radiomic features through the deep learning of CT imaging-based features to individualize radiation dose delivery using a clinically meaningful unit, the Gray (iGray). This framework could be readily implemented for pretreatment risk stratification and risk-adapted dose optimization in clinical trials and, ultimately, in everyday clinical practices that use radiotherapy.
Supplementary Material
Supplementary Figure 1. Deep Profiler scores were independent of motion management technique. Violin plot with the mean (point) and standard deviation (point range) are shown. A difference between the two means was measured by a Welch two sample t-test and was not significant (p = 0.353).
Supplementary Figure 2. Deep Profiler scores were higher in patients that received adjuvant chemotherapy. Violin plot with the mean (point) and standard deviation (point range) are shown. A difference between the two means was measured by a Welch two sample t-test and was significant (p = <0.001).
Supplementary Figure 3. Deep Profiler predicted local failure using images from at least two distinct CT scanners. Estimated cumulative incidence curves for local failure using scanner CT-1 (a) or CT-2 (b). CT-1 and CT-2 were used non-contemporaneously. CT-1 was used from 2009–2017 and CT-2 was used from 2017–2019. Both are Philips Big Bore scanners. Sample sizes were 499 and 244, respectively. The sample size for the other two scanners used in the study, CT-3 and CT-4, were 40 and 61, respectively. The image sets from these scanners were not sufficiently powered to assess predictive accuracy (1-β = 0.24 and 0.35 for an effect size | r | = 0.2 and α = 0.05, respectively).
RESEARCH IN CONTEXT.
Evidence before this study
CT images comprise voxel intensities that, to the extent that they are discernable by the treating physician, can guide the manual delineation of tumor volumes. They do not, however, currently contribute to the individualization of radiation dose prescriptions. Extraction and analysis of rigidly defined radiomic features has been used to transform medical imaging data into quantifiable variables used to predict survival, other failure modes and response to therapeutic agents.
Added value of this study
To our knowledge, this study is the first to implement a deep neural network using deformable multi-tasking with the ability to create new radiomic features to predict the risk of failure for patients treated with radiotherapy. This study also represents an innovation in personalized medicine by the projection of an optimized radiation dose, iGray. The cohort of patients evaluated represents one of the largest datasets of chest CT images heretofore used in outcome prediction analysis.
Implications of all the available evidence
Accurate estimates of the likelihood of response to treatments coupled with optimized dose delivery can significantly improve clinical outcomes and limit toxicity for patients treated with radiotherapy. Our framework could provide readily implementable treatment strategy guidance for under-resourced medical facilities and populations. The ability of the neural network, Deep Profiler, to generate new predictive features represents a major advance in radiomics and artificial intelligence (AI). Augmenting this impact, Deep Profiler’s prediction accuracy is scalable in that it will improve as our dataset increases in sample number via natural growth, federated datasets and data partitions into more homogenous populations.
Acknowledgments
DECLARATION OF INTERESTS
B.L., A.K., N.M., L.L. and M.E.A. are named inventors in a patent pending for the use of Deep Profiler and iGray to personalize radiotherapy dose. M.E.A. receives grant support, travel support and honoraria from Bayer AG. M.E.A. receives grant support from Siemens Healthcare. M.E.A. is also supported by NIH KL2 TR0002547, NIH R37 CA222294, the American Lung Association and VeloSano.
Footnotes
Publisher's Disclaimer: DISCLAIMER
Publisher's Disclaimer: Deep Profiler and iGray are based on research and are not currently commercially available. Due to regulatory reasons, their future availability cannot be guaranteed.
DATA AVAILABILITY
The datasets analyzed during the current study will be available from the corresponding author (abazeem@ccf.org) at the time of publication. Per institutional policy, the datasets are designated limited access. Upon receiving access, the investigator may only use them for the purposes outlined in the request to the data provider and redistribution of the data is prohibited.
REFERENCES
- 1.Morin O, Vallieres M, Jochems A, et al. A Deep Look Into the Future of Quantitative Imaging in Oncology: A Statement of Working Principles and Proposal for Change. Int J Radiat Oncol 2018; 102(4): 1074–82. [DOI] [PubMed] [Google Scholar]
- 2.Mathematics and Physics of Emerging Biomedical Imaging. Washington (DC); 1996. [PubMed] [Google Scholar]
- 3.Barten PGJ. Physical Model for the Contrast Sensitivity of the Human Eye. P Soc Photo-Opt Ins 1992; 1666: 57–72. [Google Scholar]
- 4.Barten PGJ. Contrast sensitivity of the human eye and its effects on image quality [doctoral]. Bellingham, WA: Technische Universiteit Eindhoven; 1999. [Google Scholar]
- 5.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nature Reviews Clinical Oncology 2017; 14(12): 749–62. [DOI] [PubMed] [Google Scholar]
- 6.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: Extracting more information from medical images using advanced feature analysis. European Journal of Cancer 2012; 48(4): 441–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aerts HJWL, Velazquez ER, Leijenaar RTH, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach (vol 5, pg 4006, 2014). Nature Communications 2014; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521(7553): 436–44. [DOI] [PubMed] [Google Scholar]
- 9.Samei E, Rowberg A, Avraham E, Cornelius C. Toward clinically relevant standardization of image quality. J Digit Imaging 2004; 17(4): 271–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Budin-Ljosne I, Burton P, Isaeva J, et al. DataSHIELD: An Ethically Robust Solution to Multiple-Site Individual-Level Data Analysis. Public Health Genom 2015; 18(2): 87–96. [DOI] [PubMed] [Google Scholar]
- 11.Weinstein JN, Collisson EA, Mills GB, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013; 45(10): 1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yard BD, Adams DJ, Chie EK, et al. A genetic basis for the variation in the vulnerability of cancer to DNA damage. Nat Commun 2016; 7: 11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nature medicine 2018; 24(10): 1559–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Causey JL, Zhang JY, Ma SQ, et al. Highly accurate model for prediction of lung nodule malignancy with CT scans. Scientific reports 2018; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Timmerman R, Paulus R, Galvin J, et al. Stereotactic body radiation therapy for inoperable early stage lung cancer. Jama 2010; 303(11): 1070–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Timmerman RD, Hu C, Michalski J, et al. Long-term Results of RTOG 0236: A Phase II Trial of Stereotactic Body Radiation Therapy (SBRT) in the Treatment of Patients with Medically Inoperable Stage I Non-Small Cell Lung Cancer In: Int J Radiat Oncol Biol Phys; 2014. p. S30. [Google Scholar]
- 17.Videtic GM, Hu C, Singh AK, et al. A Randomized Phase 2 Study Comparing 2 Stereotactic Body Radiation Therapy Schedules for Medically Inoperable Patients With Stage I Peripheral Non-Small Cell Lung Cancer: NRG Oncology RTOG 0915 (NCCTG N0927). Int J Radiat Oncol Biol Phys 2015; 93(4): 757–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woody NM, Stephans KL, Andrews M, et al. A Histologic Basis for the Efficacy of SBRT to the lung. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 2017; 12(3): 510–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horner-Rieber J, Bernhardt D, Dern J, et al. Histology of non-small cell lung cancer predicts the response to stereotactic body radiotherapy. Radiother Oncol 2017; 125(2): 317–24. [DOI] [PubMed] [Google Scholar]
- 20.Baine MJ, Verma V, Schonewolf CA, Lin C, Simone CB. Histology significantly affects recurrence and survival following SBRT for early stage non-small cell lung cancer. Lung cancer 2018; 118: 20–6. [DOI] [PubMed] [Google Scholar]
- 21.van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Research 2017; 77(21): E104–E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Timmerman RD, Paulus R, Pass HI, et al. Stereotactic Body Radiation Therapy for Operable Early-Stage Lung Cancer: Findings From the NRG Oncology RTOG 0618 Trial. JAMA Oncol 2018; 4(9): 1263–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murrell DH, Laba JM, Erickson A, Millman B, Palma DA, Louie AV. Stereotactic ablative radiotherapy for ultra-central lung tumors: prioritize target coverage or organs at risk? Radiat Oncol 2018; 13(1): 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura T, Nagata Y, Harada H, et al. Phase I study of stereotactic body radiation therapy for centrally located stage IA non-small cell lung cancer (JROSG10–1). International journal of clinical oncology 2017; 22(5): 849–56. [DOI] [PubMed] [Google Scholar]
- 25.Bezjak A, Paulus R, Gaspar LE, et al. Safety and Efficacy of a Five-Fraction Stereotactic Body Radiotherapy Schedule for Centrally Located Non-Small-Cell Lung Cancer: NRG Oncology/RTOG 0813 Trial. J Clin Oncol 2019: Jco1800622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scrucca L, Santucci A, Aversa F. Regression modeling of competing risk using R: an in depth guide for clinicians. Bone marrow transplantation 2010; 45(9): 1388–95. [DOI] [PubMed] [Google Scholar]
- 27.Team RDC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2018. [Google Scholar]
- 28.Ernani V, Appiah AK, Marr A, et al. Adjuvant Systemic Therapy in Patients With Early-Stage NSCLC Treated With Stereotactic Body Radiation Therapy. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 2019; 14(3): 475–81. [DOI] [PubMed] [Google Scholar]
- 29.Allibhai Z, Taremi M, Bezjak A, et al. The Impact of Tumor Size on Outcomes After Stereotactic Body Radiation Therapy for Medically Inoperable Early-Stage Non-Small Cell Lung Cancer. Int J Radiat Oncol 2013; 87(5): 1064–70. [DOI] [PubMed] [Google Scholar]
- 30.Ribeiro MT, Singh S, Guestrin C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. San Francisco, California, USA: ACM; 2016. p. 1135–44. [Google Scholar]
- 31.Debray TP, Vergouwe Y, Koffijberg H, Nieboer D, Steyerberg EW, Moons KG. A new framework to enhance the interpretation of external validation studies of clinical prediction models. J Clin Epidemiol 2015; 68(3): 279–89. [DOI] [PubMed] [Google Scholar]
- 32.Huynh E, Coroller TP, Narayan V, et al. CT-based radiomic analysis of stereotactic body radiation therapy patients with lung cancer. Radiother Oncol 2016; 120(2): 258–66. [DOI] [PubMed] [Google Scholar]
- 33.Coroller TP, Grossmann P, Hou Y, et al. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol 2015; 114(3): 345–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parmar C, Leijenaar RT, Grossmann P, et al. Radiomic feature clusters and prognostic signatures specific for Lung and Head & Neck cancer. Scientific reports 2015; 5: 11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hosny A, Parmar C, Coroller TP, et al. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLoS Med 2018; 15(11): e1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang ZL, Wang XG, Wang JS, Liu WY, Wang JD. Weakly-Supervised Semantic Segmentation Network with Deep Seeded Region Growing. Proc Cvpr Ieee 2018: 7014–23. [Google Scholar]
- 37.Brenner DJ. The linear-quadratic model is an appropriate methodology for determining isoeffective doses at large doses per fraction. Semin Radiat Oncol 2008; 18(4): 234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palma DA, Olson R, Harrow S, et al. Stereotactic ablative radiotherapy versus standard of care palliative treatment in patients with oligometastatic cancers (SABR-COMET): a randomised, phase 2, open-label trial. Lancet 2019. [DOI] [PubMed] [Google Scholar]
- 39.Gomez DR, Blumenschein GR Jr., Lee JJ, et al. Local consolidative therapy versus maintenance therapy or observation for patients with oligometastatic non-small-cell lung cancer without progression after first-line systemic therapy: a multicentre, randomised, controlled, phase 2 study. Lancet Oncol 2016; 17(12): 1672–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cunliffe A, Armato SG, Castillo R, Pham N, Guerrero T, Al-Hallaq HA. Lung Texture in Serial Thoracic Computed Tomography Scans: Correlation of Radiomics-based Features With Radiation Therapy Dose and Radiation Pneumonitis Development. Int J Radiat Oncol 2015; 91(5): 1048–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Deep Profiler scores were independent of motion management technique. Violin plot with the mean (point) and standard deviation (point range) are shown. A difference between the two means was measured by a Welch two sample t-test and was not significant (p = 0.353).
Supplementary Figure 2. Deep Profiler scores were higher in patients that received adjuvant chemotherapy. Violin plot with the mean (point) and standard deviation (point range) are shown. A difference between the two means was measured by a Welch two sample t-test and was significant (p = <0.001).
Supplementary Figure 3. Deep Profiler predicted local failure using images from at least two distinct CT scanners. Estimated cumulative incidence curves for local failure using scanner CT-1 (a) or CT-2 (b). CT-1 and CT-2 were used non-contemporaneously. CT-1 was used from 2009–2017 and CT-2 was used from 2017–2019. Both are Philips Big Bore scanners. Sample sizes were 499 and 244, respectively. The sample size for the other two scanners used in the study, CT-3 and CT-4, were 40 and 61, respectively. The image sets from these scanners were not sufficiently powered to assess predictive accuracy (1-β = 0.24 and 0.35 for an effect size | r | = 0.2 and α = 0.05, respectively).