Significance
The avocado is a nutritious, economically important fruit species that occupies an unresolved position near the earliest evolutionary branchings of flowering plants. Our nuclear genome sequences of Mexican and Hass variety avocados inform ancient evolutionary relationships and genome doublings and the admixed nature of Hass and provide a look at how pathogen interactions have shaped the avocado’s more recent genomic evolutionary history.
Keywords: avocado genome, angiosperm phylogeny, genome duplications, Phytophthora, genome evolution
Abstract
The avocado, Persea americana, is a fruit crop of immense importance to Mexican agriculture with an increasing demand worldwide. Avocado lies in the anciently diverged magnoliid clade of angiosperms, which has a controversial phylogenetic position relative to eudicots and monocots. We sequenced the nuclear genomes of the Mexican avocado race, P. americana var. drymifolia, and the most commercially popular hybrid cultivar, Hass, and anchored the latter to chromosomes using a genetic map. Resequencing of Guatemalan and West Indian varieties revealed that ∼39% of the Hass genome represents Guatemalan source regions introgressed into a Mexican race background. Some introgressed blocks are extremely large, consistent with the recent origin of the cultivar. The avocado lineage experienced 2 lineage-specific polyploidy events during its evolutionary history. Although gene-tree/species-tree phylogenomic results are inconclusive, syntenic ortholog distances to other species place avocado as sister to the enormous monocot and eudicot lineages combined. Duplicate genes descending from polyploidy augmented the transcription factor diversity of avocado, while tandem duplicates enhanced the secondary metabolism of the species. Phenylpropanoid biosynthesis, known to be elicited by Colletotrichum (anthracnose) pathogen infection in avocado, is one enriched function among tandems. Furthermore, transcriptome data show that tandem duplicates are significantly up- and down-regulated in response to anthracnose infection, whereas polyploid duplicates are not, supporting the general view that collections of tandem duplicates contribute evolutionarily recent “tuning knobs” in the genome adaptive landscapes of given species.
The avocado, Persea americana, is a commercially important tree fruit species in the Lauraceae family, otherwise known for the spices cinnamon, bay leaves, and sassafras (gumbo filé) (1). Lauraceae is contained within the early diverging magnoliid lineage of angiosperms, which at about 11,000 total species is minuscule in comparison to the dominant eudicot and monocot flowering plant lineages, comprising about 285,000 species combined (2). Avocados are a vital crop for Mexico, from which almost 50% of all avocado exports originate, valued at about $2.5 billion US dollars.* Although the avocado has an ancient cultivation history in Mexico and Central to South America (5), its extreme worldwide popularity as an oily, nutty-flavored fruit with highly beneficial nutritional properties dates mainly from the early 20th century (6). Cultivated avocados occur in 3 landraces with possibly independent cultivation origins that reflect their current distribution: the Mexican, Guatemalan, and West Indian varieties (6). The principal industrial avocado cultivar is known as Hass, after the grower who first patented it in 1935. Hass represents a hybrid between the Guatemalan and Mexican races, but its precise breeding history is unknown (6, 7). Here, we generate and analyze the complete genome sequences of a Hass individual and a representative of the highland Mexican landrace, Persea americana var. drymifolia. We also study genome resequencing data for other Mexican individuals, as well as Guatemalan and West Indian accessions. We use these data to study the admixed origin of the Hass cultivar and demonstrate its racewise parentage more precisely. We evaluate the phylogenetic origin of avocado among angiosperms and provide information on avocado’s unique polyploid ancestry. The adaptive landscape of the avocado genome in terms of its duplicate gene functional diversity was also explored. We further evaluate gene expression patterns during the defense response of Hass avocado to anthracnose disease and how this is partitioned by gene duplication mechanisms.
Results and Discussion
Plant Material, Genome Assembly, and Annotation.
Due to growing market demand, 90% of cultivated avocado corresponds to the cultivar Hass, which in Mexico is commonly grafted on Mexican race (P. americana var. drymifolia) rootstock (6). This practice makes it possible to maintain high productivity as the indigenous race is well-adapted to Mexican highland soils. The Hass cultivar and Mexican race were chosen to generate reference genomes (SI Appendix, section 1). Additionally, to explore the genetic diversity available in avocado, we resequenced representative individuals from the 3 avocado botanical varieties (vars. drymifolia, guatemalensis, and americana), including the disease-resistant rootstock Velvick (8), an additional Hass specimen and the early flowering/fruiting Hass somatic mutant Carmen Hass cultivar [otherwise known as Mendez No. 1 (6, 9)], as well as wild avocados of the West Indian variety (P. americana var. costaricensis). Persea shiedeana (the edible coyo), a species relatively closely related to P. americana (10), was also included (SI Appendix, Tables S1 and S2).
De novo and evidence-directed annotation revealed a similar number of protein coding genes in each genome: 22,441 from the Mexican race and 24,616 from Hass (Table 1, SI Appendix, section 2, and Datasets S2 and S3). We next used the Benchmarking Universal Single-Copy Orthologs (BUSCO) software to estimate the presence of 1,440 conserved embryophyte single-copy genes (11) in the annotations, leading to estimated completeness percentages of 85% and 86.3% for Hass and Mexican avocado, respectively (Table 1). The Mexican race was sequenced using the short-read, high-coverage Illumina sequencing platform, while the Hass genome was sequenced using the long-read Pacific Biosciences sequencing technology. Given the similar BUSCO scores, we used the larger Hass genome assembly for downstream single-nucleotide polymorphism (SNP) calling, as PacBio technology lowers the probability of contig misassembly and permits incorporation of substantially more repetitive DNA sequence and genes lying within it into the assembled genome, which might have otherwise been missed.
Table 1.
General statistics of the avocado assemblies and their annotations
| P. americana assembly | ||
| Metrics | Var. drymifolia | Hass cultivar |
| No. of contigs | 99,957 | 8,135 |
| Total length of contigs, bp | 668,137,248 | 912,697,600 |
| No. of scaffolds | 42,722 | — |
| Total length of scaffolds, bp | 823,419,498 | — |
| Longest contig/scaffold, bp | 254,240/4,610,966 | 2,811,280/– |
| Mean contig/scaffold length, bp | 6,684/19,274 | 112,194/– |
| N50 contig/scaffold length, bp | 11,724/323,854 | 296,371/– |
| Assembly in scaffolded contigs, % | 87.6 | 0 |
| Assembly in unscaffolded contigs, % | 12.4 | 100 |
| Protein coding genes (% BUSCO completeness) | 22,441 (86.3%) | 24,616 (85%) |
We also anchored the Hass genome to an avocado genetic map (12). Two large mapping populations of 1,339 trees were genotyped with 5,050 SNP markers from transcribed genes, and the resulting map was used to order the Hass scaffolds into 12 linkage groups, matching the avocado haploid chromosome number (see, e.g., chromosome 4, Fig. 2A). The total length of the anchored genome accounts for 46.2% of the Hass assembly and represents 915 scaffolds, 361 of which could be oriented (SI Appendix, section 1.5).
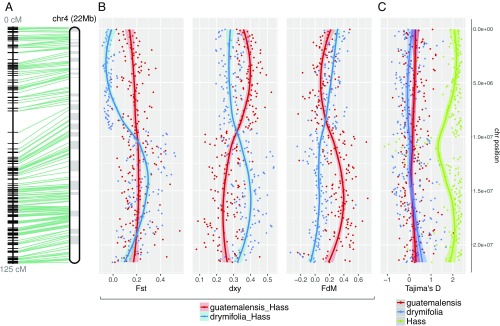
Fig. 2.
SNP diversity analysis reveals the hybrid genomic background of Hass avocado. (A) Twenty-two megabases (Mb) of anchored DNA on chromosome 4 exemplify the hybrid nature of Hass, in which genomic introgression from the Guatemalan avocado race (var. guatemalensis) occurred into a Mexican (var. drymifolia) genetic background. (B) While in the top chromosome arm the blue trend line shows a low differentiation index (FST) between Hass and the Mexican subpopulation as well as a high introgression signal from var. drymifolia into Hass, these signals should not be misinterpreted as introgression events, since the absolute genetic divergence between both sets of accessions does not vary along the chromosome. The lower arm of the chromosome, however, has inverted trends, where our estimators describe an elevated introgression signal from var. guatemalensis into Hass, as confirmed by the decay in (red trend line), and higher FST between Hass and Mexican accessions. (C) No evidence for selective sweeps or domestication signatures were identified; Mexican and Guatemalan subpopulations displayed neutral D values while Hass maintained extreme D values at the theoretical upper limit of the estimator (∼2). Such positive values reflect a “bottlenecked” origin with clonal expansion after the very recent foundation of the cultivar only a few decades ago. Each dot in the plots corresponds to statistics for SNP data in nonoverlapping 100-kb windows (confidence interval of 0.90 for graphical smoothed conditional means). Apparent centromeric regions are located at around 10 Mb, where FST, and intersect and Tajima’s D for Hass decreases.
SNPs, Population Structure, and the Parentage of Hass.
To study avocado from a population genomic perspective, we resequenced accessions of different races and cultivars and mapped the reads against the Hass reference genome assembly (SI Appendix, Table S2). The estimated depth of coverage ranged from 3.3 to 39×, with breadth of coverage between 70 and 92% (SI Appendix, section 3.2). Given the uneven sequencing coverage, we used ANGSD to call SNPs across the entire (unanchored) genome assembly, followed by a stringent pruning based on per-site depth, minor allele frequencies, and linkage disequilibrium, that resulted in 179,029 high-quality SNP variants. Phylogenetic, principal component, and identity-by-state (IBS) analyses derived from this dataset (Fig. 1A and SI Appendix, section 4) cluster the samples belonging to the Hass cultivar and Guatemalan variety into 2 groups as expected according to their genetic background. Principal component analysis of genome-wide SNPs showed relative uniformity in Costa Rican/West Indian/Guatemalan group but strong heterogeneity within the Mexican subpopulation, wherein the unusual accession Tiny Charly is a divergent sample (SI Appendix, Fig. S15). SNPhylo (13) results reflected the poor fit of the SNP data to a bifurcating tree by embedding the hybrid Hass within an otherwise Mexican clade, 1 known parent of this hybrid cultivar (SI Appendix, Fig. S13). Furthermore, in that lineage’s sister group, Guatemalan accessions were derived within an otherwise Costa Rican/West Indian lineage, suggesting an admixed origin involving Guatemalan and other sources. Phylogenetic patterns generated from chromosome-wise SNP subsets (based only on contigs anchored to chromosomes) recapitulated these relationships for 7 of avocado’s 12 chromosomes, whereas 1 chromosome supported Hass to be sister to the Costa Rican/West Indian lineage, perhaps reflective of chromosomal differences in the admixture proportions of this hybrid cultivar (SI Appendix, Fig. S14 and Dataset S6). Furthermore, IBS clustering placed Hass intermediate between the Guatemalan and Mexican subpopulations, agreeing with the hybrid nature of this variety (SI Appendix, Fig. S16).
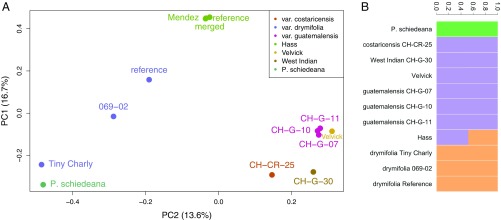
Fig. 1.
Population genomic structure of avocado. (A) Principal component analysis (PCA) of genome-wide SNPs reveals population groupings among races and varieties. The Guatemalan and West Indian/Costa Rican accessions are closely related, while the Mexican (P. americana var. drymifolia) specimens are more diverse, with the unusual individual Tiny Charly drawn toward the outgroup species P. schiedeana by PC2. Hass and its sport Mendez are tightly clustered and intermediate between Mexican and Guatemalan and West Indian/Costa Rican on PC2. (B) NGSAdmix analysis reveals similar population structure at K = 3. The P. schiedeana outgroup is distinct, and the Hass reference genome is revealed to be admixed between Guatemalan–West Indian and Mexican source populations, the Mexican source clearly contributing greater than 50%.
To account for further evidence of admixture in the Hass reference genome, we used NGSAdmix (14) modeling different possible numbers of source populations (K = 1 to 6) (SI Appendix, section 5 and Fig. S17). The Akaike information criterion (AIC) indicated K = 1 as the preferred model, reflecting poor population structuring within avocado as a whole. However, since we know Hass is admixed a priori, we chose the smallest (most parsimonious) K for which Hass admixture appears (K = 3). This criterion predicts the following 3 populations: 1) P. schiedeana, 2) the West Indian plus the Guatemalan varieties, and the 3) Mexican accessions (Fig. 1B). Combining the IBS and NGSAdmix observations, we specifically calculated the contribution of Guatemalan and Mexican backgrounds into the Hass subpopulation. EIGMIX (15) revealed that the greatest admixture proportion, 61%, stemmed from the Mexican race (SI Appendix, Fig. S18 and Table S8).
Although based on ∼46% anchoring of scaffolds to chromosomes, we investigated chromosomewise signatures of admixture in the Hass genome (SI Appendix, section 5.2). We calculated the , , and estimators of introgression and divergence (SI Appendix, Fig. S19 and Dataset S7) according to Martin et al. (16) in nonoverlapping 100-kb windows, controlling the directionality of gene flow from the Guatemalan race into Hass versus the Mexican race into Hass, setting P. schiedeana as the outgroup and leaving Tiny Charly out of the Mexican subpopulation to avoid the bias this divergent accession could introduce into calculations (Fig. 2B). Genomic regions that behave as outliers can be distinguished as introgressed from ancestral variation if the absolute genetic distance is also reduced between donor (P3) and receptor (P2). In the presence of gene flow, genomic windows coalesce more recently than the lineage split, so the magnitude of reduction in P2 − P3is greater than in the case where recombination and hybridization are absent. We evaluated several cutoffs (Q50, 75, and 90; SI Appendix, Fig. S20) and observed a remarkable reduction of genetic divergence in the scenario where gene flow occurs from the Guatemalan race into Hass. Considering those blocks with > 0.174 (Q50), < 0.113 (mean divergence between subpopulations) and < 0.114 (Q50), we were able to define 840 high-confidence regions of Guatemalan origin across the 12 chromosomes (Fig. 2, SI Appendix, section 5.2 and Figs. S21 and S23–S34). Chromosome 4 illustrates these analyses well, demonstrating that a huge Guatemalan block, which could encompass an entire chromosome arm, is present in the Hass genome (Fig. 2A). The length of this Guatemalan-derived block, uninterrupted by recombination, reflects the extremely recent hybrid origin of the cultivar.
We also calculated the level of nucleotide diversity [π (17, 18)] in each population (Mexican, Guatemalan, and Hass), the FST index (19) to determine regions of high differentiation between varieties, and Tajima’s D (20) in order to evaluate any deviations from neutral evolution (SI Appendix, section 5.3 and Dataset S7). We observed that Hass has the lowest nucleotide diversity (π = 0.06) and very high Tajima’s D in all chromosomes (genomic average of 1.5), as expected for individuals derived from a recent founder event and clonally propagated; these values contrast with the low, positive Tajima’s D values in the Mexican and Guatemalan populations (genomic averages of 0.19 and 0.11, respectively; SI Appendix, Fig. S22). In the case of chromosome 4, the FST index between the Mexican race and Hass corroborates our previous conclusions on admixture, showing that approximately half of the chromosome corresponds to the Mexican background whereas the other half has its origin in the Guatemalan population (Fig. 2B).
Whole-Genome Duplication History.
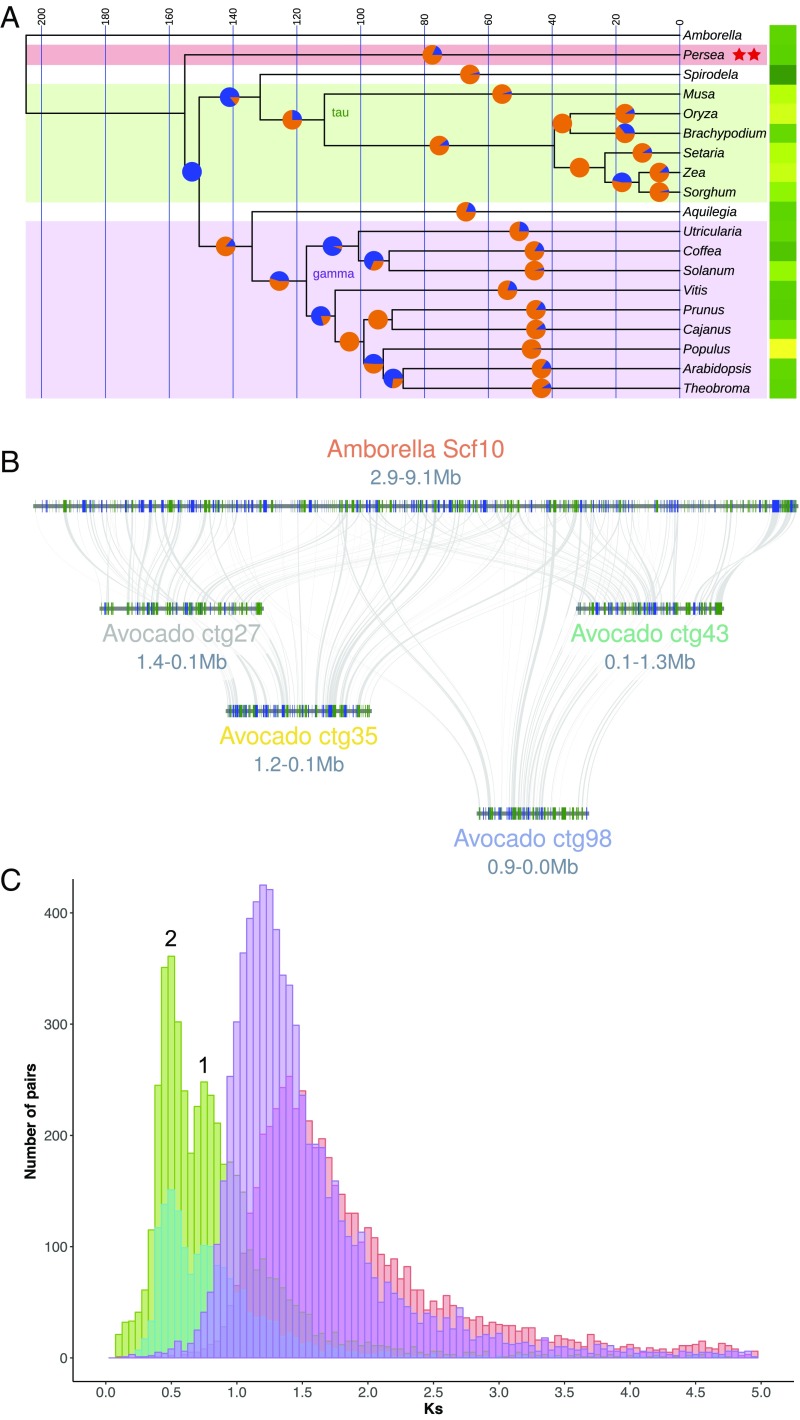
Next, we investigated P. americana genome structural history and relatedness of the species to other major groups of angiosperms (SI Appendix, section 6). We used the CoGe SynMap tool (21) to examine avocado self:self and avocado:Amborella synteny. Amborella, the single living representative of the sister lineage to all other angiosperms, is known to show a 1:3 syntenic block relationship compared with Vitis (grapevine) and its ancient hexaploid structure shared with all core eudicots (22–25). As such, the Amborella genome displays no additional whole-genome duplications (WGDs) since the angiosperm last common ancestor. Consequently, paralogous syntenic blocks discovered within self:self and avocado:Amborella plots could reflect WGDs unique to the avocado lineage that occurred since last common ancestry with Amborella. Using these approaches, we discovered that the avocado genomes show evidence for 2 ancient polyploidy events (Fig. 3 A–C). We investigated the relative timing of these events with respect to the gamma hexaploidy (23, 24) and species splits using Ks density plots of orthologous and paralogous gene pairs between avocado, Amborella, and Vitis (SI Appendix, section 6.2). Avocado polyploidy events are lineage-specific as both postdate the divergence of the avocado lineage from common ancestry with either Amborella or Vitis (Fig. 3C and SI Appendix, Fig. S36). Considerable fractionation (alternative deletion of duplicated genes between polyploid subgenomes) since these 2 polyploidy events is observable in blockwise relationships of about 4:1 (Fig. 3B and SI Appendix, Fig. S35). The blockwise relationship of 4:1 for avocado:Amborella suggests, though does not prove, that the 2 events were WGDs and not triplications. Further quantitative analysis using well-conserved orthologous syntenic “superblocks” (26) between avocado and 15 other angiosperms strongly supported the conclusion that the most recent polyploidy event in avocado was a WGD and not a triplication, as in the ancient gamma hexaploidy event (SI Appendix, section 6.3).
Fig. 3.
Phylogenomic and whole-genome duplication history of avocado. (A) An ultrametric time tree based on universally present single-copy protein sequences depicts 1 of 3 common resolutions of Persea (Magnoliidae) relationships to other flowering plants. This topology, showing avocado sister to monocots plus eudicots, mirrors phylogenetic relationships derived from syntenic distances. Here, the split time between the last common ancestor of avocado and the monocot/eudicot crown group is less than 4 million y. Pie charts at 50% positions on branches show proportions of gene gains (orange) versus losses (blue) as determined by BadiRate’s birth–death–innovation model. Yellow–green (greater–lesser) heat map to the right of the tree depicts relative numbers of genes in the modern genomes. Syntenic analysis revealed 2 independent WGD events (red stars) during avocado’s evolutionary history. (B) Hass avocado (bottom 4 genomic blocks) shows 4:1 intercalated syntenic relationships with Amborella (upper block). (C) Syntenic homologs in avocado show a bimodal Ks distribution suggestive of 2 polyploidy events (numbered 1 and 2; cyan: Hass:Hass paralogs; green: Hass:drymifolia homologs) following the split between magnoliids and Amborella (red syntenic homologs). These events postdate the species split between Vitis and avocado (purple syntenic homologs) and so are independent of the gamma triplication that underlies Vitis.
Phylogenomic Analysis of Avocado’s Placement among Angiosperms.
To further corroborate the placement of these 2 polyploidy events as specific to the avocado lineage, we generated phylogenetic trees of representatives of the major angiosperm lineages using 2 data forms: coding sequence alignments and modal distances within large collections of syntenic orthologs between species pairs (SI Appendix, section 7).
Single-copy gene families [presumed unambiguous orthologs, those that returned to single copy following duplicate deletions after the various polyploidy events in flowering plant history (27)] were retrieved from orthogroup classification of 19 angiosperm proteomes, including those of avocado, Amborella, and representatives of monocots and eudicots (SI Appendix, section 3.1 and Dataset S4). Phylogenetic trees based on 176 stringently filtered single-copy gene alignments (SI Appendix, section 7.1 and Datasets S8 and S9) gave different results for amino acid versus inferred codon data. Based on protein sequences, avocado was resolved as sister to monocots plus eudicots (i.e., branching before their divergence from each other; cf. refs. 28 and 29), whereas from coding sequences avocado was placed as sister to monocots only (cf. ref. 30) (SI Appendix, Figs. S40 and S41, respectively). In a different analysis we included Gnetum (a gymnosperm) and Selaginella (a nonseed plant) in orthogroup classification to generate a rooted species tree from all gene trees (4,694) that contained one or more (i.e., paralogous) gene copies from all species (SI Appendix, section 7.2). Here, avocado was resolved as sister to eudicots only (SI Appendix, Fig. S42), a result similarly found in transcriptome-based analyses of large numbers of species (26, 31). In an altogether different approach (32, 33), we generated a neighbor-joining tree based on modal dissimilarity scores from thousands of syntenically validated ortholog pairs generated by the SynMap function on the CoGe platform (21) (SI Appendix, section 7.3). Here, avocado was again placed as sister to monocots plus eudicots, as in Fig. 3A (SI Appendix, Fig. S44).
Apparently, the early branching orders of the angiosperms are extremely difficult to determine using protein coding sequences. This problem is due in part to sequence parallelism/reversal over deep time, limitations in taxon sampling (including unknown extinctions), biases in sequence-based ortholog versus paralog determination, but clearly also to the relatively coincident branching times of the species involved (see figure 6 of ref. 34 and also below). Rapid species divergences can lead to real gene-tree/species-tree discordances through enhanced occurrence of incomplete lineage sorting (ILS), wherein polymorphic allele states in ancestral populations do not have enough time to fix according to the species tree (35–37).
In an experimental approach to the problem, we further investigated the possible role of ILS using gene family turnover analysis as incorporated in BadiRate (38) (SI Appendix, section 7.4). Trees with the 3 alternative placements of avocado were converted into time-calibrated ultrametric trees, and the likelihoods of duplicate gains versus losses were evaluated under 4 different branch models (SI Appendix, section 7.4). The AIC clearly favored free-rates (FR) models, supporting heterogeneous rates of multigene family evolution across lineages (Dataset S10). Interestingly, such uneven rates of gene turnover cannot be entirely explained by lineage-specific WGD/whole-genome triplication (WGT) events, given that FR models fit multigene family data better than WGD/WGT models alone. Additionally, allowing turnover rates to vary in each short branch (<10 My) also improved likelihood and AIC values, although the fit was still worse than under the FR model (Dataset S10). That FR models fit gene count data significantly better could be explained by their flexibility to accommodate variation that is not explicitly accounted for by current turnover models, such as gene copy variation within species. Intraspecific variation, segregating in an ancestral population, can be inherited differently by 2 splitting lineages, which will thus start diverging with a significant fraction of differentiation. This predicts that divergence will be inflated for short branches, and that this bias will become negligible as divergence times increase, because its relative contribution to the total divergence tends to be comparatively small over time (39, 40). We observe a correlation between turnover rates and branch lengths at the multigene family level (SI Appendix, Fig. S46), suggesting pervasive copy number variation (CNV) in the ancestral species, possibly exacerbated by WGD and subsequent fractionation processes. Short phylogenetic branches, representing rapid speciation events, increase the incidence of ILS in phylogeny reconstruction since extinctions of alternative duplicate copies within ancestral populations (e.g., unfixed CNVs) further break up branches that are nearly time-coincident already (41). According to BadiRate estimates, the temporal impact of ILS on turnover rates extended well beyond 10 Mya, a time frame exceeding the branch length of the lineage that existed immediately prior to avocado divergence from other species, which varied in age from only 7.4 to as little as 3.8 My (Fig. 3A and SI Appendix, Table S11 and Figs. S46 and S47). This implies that the 3 different placements of avocado among angiosperms may be impossible to discriminate among for purely biological reasons (cf. ref. 42). Yet, 1 of the 3 different tree topologies was preferred based on AIC contrasts under the FR model: the topology wherein magnoliids are sister to monocots plus eudicots (Fig. 3A and SI Appendix, Fig. S47).
Functional Enrichments in Duplicate Gene Space.
Duplicate gene collections within plant genomes mainly derive from 2 processes, local and ongoing tandem duplication events, many of which may be recent, and global and often ancient polyploidy events wherein entire gene complements are duplicated (43). Subfunctionalization and/or neofunctionalization of duplicate gene copies (44) results in retained descendants of duplication events that have differentially escaped the otherwise usual fate of duplicates—pseudogenization—through functional divergence. Tandem duplication is problematic for genes that are part of dosage-sensitive transcriptional regulatory networks, or for genes that code for parts of multiprotein complexes (45). Such functions are more likely to remain among the surviving duplicate complements stemming from precisely dosage-balancing polyploidy events (46). On the other hand, dosage-responsive functions such as secondary metabolism (including biochemical pathway addition) are among those most likely to survive as sub- or neofunctionalized tandem duplicates (45). These patterns have been repeatedly observed among plant genomes, wherein secondary metabolic function is most prevalent among tandems, and transcriptional function is enriched among polyploid duplicates (e.g., refs. 46 and 47). The avocado genome provides no exception to this rule; we identified precisely these overrepresentation patterns among Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) categories for these different classes of gene duplicates, separated using the CoGe platform (SI Appendix, section 8). Among 2,433 total polyploid duplicates, “regulation of transcription, DNA-templated” was significantly overrepresented by 352 genes (Dataset S12). Enriched functions among tandem duplicates was highly illustrative of the secondary metabolic landscape particular to avocado (Dataset S12). We show that “phenylpropanoid biosynthesis” and closely related KEGG pathways (Dataset S13) are significantly enriched among tandem duplicates (P = 2.08e-08; Fisher’s exact test, Bonferroni-corrected). This functional enrichment in a long-lived tree may have evolved in response to pathogen infection, including Colletotrichum (anthracnose) and Phytophthora cinnamomi (avocado root rot), both of which are reported to activate the phenylpropanoid biosynthetic pathways in avocado (48–50). Several GO functional enrichments among avocado tandems (for example, “1,3-beta-d-glucan synthase activity” and “regulation of cell shape”; P = 1.64e-05 and 0.00258, respectively; Fisher’s exact test, Bonferroni-corrected) relate to callose synthase activity (51, 52), a recently discovered avocado defense mechanism against P. cinnamomi (53). Other significantly enriched GOs include “phenylpropanoid metabolic process,” “lignin biosynthetic process,” and “UDP-glycosyltransferase activity” (P = 0.00142, 7.36e-07 and 5.16e-07, respectively; Fisher’s exact test, Bonferroni-corrected), categories directly or closely related to phenylpropanoid biosynthesis (54, 55). The lignin functional enrichment, for example, includes diverse tandemly duplicated genes involved in many pathway-interrelated processes, including homologs of both biosynthetic and regulatory genes encoding HYDROXYCINNAMOYL-COA SHIKIMATE/QUINATE HYDROXYCINNAMOYL TRANSFERASE (HCT), CINNAMYL ALCOHOL DEHYDROGENASE 5 (CAD5), LACCASE 17 (LAC17), CAFFEATE O-METHYLTRANSFERASE 1 (COMT1), PEROXIDASE 52 (PRX52), NAC DOMAIN CONTAINING PROTEIN 12 (NAC012), and NAC SECONDARY WALL THICKENING PROMOTING FACTOR 1 (NST1). As could be expected from the above, the GOs “defense response” and “defense response to fungus” are significantly enriched among tandem duplicates (P = 0.000165 and 0.0167, respectively; Fisher’s exact test, Bonferroni-corrected), as has been discovered for other plant genomes, and involving many different gene families and responses. Tandem O-methyltransferases homologous to COMT1 may also contribute to synthesis of the phenylpropanoid derivative and insecticide estragole (56), which is largely responsible for the anise-like leaf scent and fruit taste of many avocado cultivars, particularly of the Mexican race (57). Another relevant enriched GO category among tandems is “ethylene-activated signaling pathway” (P = 0.000463; Fisher’s exact test, Bonferroni-corrected), which annotates many different transcription factor duplicates. Ethylene signaling factors such as ERF1 (represented by 2 homologs) are heavily involved in pathogen-induced responses, including to infection by Colletotrichum and other necrotrophic fungi (58–61). Also identified are 3 homologs of EIN3, a transcription factor that initiates downstream ethylene responses, including fruit ripening (62). Avocado fruit matures on the tree in a process that involves ethylene synthesis and signaling, while it does not ripen until harvested—a desirable trait that allows growers to delay harvesting for several months (63).
Given the ancient derivation of avocado’s retained polyploid duplicates, most tandem duplicates in the genome are expected to be of more recent origin, having been generated by ongoing gene birth–death–innovation processes that operate in all eukaryotic genomes. As such, sub- or neofunctionalized tandem duplicates that survive the usual fate of duplicated genes—pseudogenization—should be enriched in functions that fine-tune a given species’ recent selective environment. In the case of avocado, response to fungal pathogens is precisely reflected in its tandemly duplicated gene complement.
Differential Expression of Tandem versus Polyploid Duplicates.
Following our prediction that many tandem duplicates fixed in the avocado genome may have evolved under relatively recent pathogen pressure, we examined differential expression of Hass genes after treatment with the anthracnose causal agent (64) (SI Appendix, section 9). Hass transcriptome reads for untreated control versus pathogen-treated were mapped to Hass gene models using Kallisto (65), normalized to transcript-per-million values and thresholded by identifying genes with treatment/control log2 fold change outside of the [2, −2] interval. Tandems were significantly enriched among up-regulated (P = 3.536e-09; Fisher’s exact test) and down-regulated genes (P = 7.274e-07), whereas polyploid duplicates did not show enrichment (SI Appendix, section 9). We interpret these results to indicate that tandem duplicates are the most dynamic component of the avocado duplicate gene space under pathogen treatment.
We also examined functional enrichments for up- versus down-regulated tandem duplicates (Dataset S16). The only significantly enriched category was xyloglucan:xyloglucosyl transferase activity (P = 0.038984; Fisher’s exact test, Bonferroni-corrected). Among genes with this annotation were 4 homologs of XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 22 [XTH22; also known as TOUCH 4 (TCH4) (66)]. XTH22 and similar genes encoding cell-wall–modifying proteins have been shown to up- or down-regulate after Citrus Huanglongbing infection (67), whitefly infestation (65), and herbivore (68) or mechanical stimulation (69), the latter provoking a Botrytis-protective response. In a different pathogen response, up-regulation of XTH22 occurs in concert with pectin digestion in Pseudomonas-sensitive Arabidopsis lines that overexpress IDA-LIKE 6 [IDL6 (70)].
Conclusions
Our genomes of Mexican and Hass avocados provide the requisite resources for genome-wide association studies to identify important traits among natural avocado genetic diversity present in Mesoamerica, to develop genome-assisted breeding and genetic modification efforts crucial for the improvement of this long-life-cycle crop, to fight threatening avocado diseases, and to optimize growth and desirable phenotypic traits. We anchored almost half of the sequenced Hass genome to a genetic map, providing linkage information for genetic variation on 12 chromosomes. We resequenced 10 genomes representing small populations of Guatemalan, West Indian, Mexican, and Hass-related cultivars—and the genome of the closely related species P. schiedeana—in order to call SNPs and study genetic diversity among these chromosomes. Analyses of admixture and introgression clearly highlighted the hybrid origin of Hass avocado, pointed to its Mexican and Guatemalan progenitor races, and showed Hass to contain Guatemalan introgression in approximately one-third of its genome. Introgressed blocks of chromosome arm size matched expectation based on Hass’s recent (20th century) origin. We uncovered 2 ancient polyploidy events that occurred in the lineage leading to avocado and conclude that these were independent from genome duplications or triplications known to have occurred in other angiosperm clades. We contributed to solving the problem of magnoliid phylogenetic relationships to other major angiosperm clades by showing that thousands of syntenic orthologs among 14 species support an arrangement wherein the magnoliid clade branched off before the split between monocots and eudicots. However, this resolution is tentative, with coding sequence phylogenomics inconclusive and gene family birth/death analysis suggesting appreciable duplicate gene turnover—and therefore enhanced possibility for ILS—during what appears to have been a nearly coincident radiation of the major angiosperm clades. We also studied the adaptive landscape of the avocado genome through functional enrichment analyses of its mechanistically distinct duplicate gene collections, that is, tandem versus polyploid duplicates. Tandem duplicates were enriched with many potentially important metabolic responses that may include relatively recent adaptation against fungal pathogens. In contrast, ancient polyploid duplicates, which originated in 2 distinct waves, were enriched with transcriptional regulatory functions reflective of core physiological and developmental processes. We discovered that tandem duplicates were more dynamically transcribed following anthracnose infection, and that some of the up-regulated genes could be related to defense responses. In sum, our work paves the way for genomics-assisted avocado improvement (1).
Data Availability.
Bioproject: PRJNA508502. Biosamples: SAMN10523735, SAMN10523720, SAMN10523736, SAMN10523738, SAMN10523739, SAMN10523746, SAMN10523747, SAMN10523748; SAMN10523749, SAMN10523750, SAMN10523752, SAMN10523753, and SAMN10523756. SRA submission: SUB4878870. Whole Genome Shotgun projects have been deposited at DDBJ/ENA/GenBank under the accession nos. SDXN00000000 and SDSS00000000. The versions described in this paper are versions SDXN01000000 and SDSS01000000 (P. americana var. drymifolia and P. americana cultivar Hass, respectively). The genome assemblies and annotations are available at https://genomevolution.org/CoGe/SearchResults.pl?s=29305&p=genome and https://genomevolution.org/coge/SearchResults.pl?s=29302&p=genome (P. americana var. drymifolia and P. americana cultivar Hass, respectively).
Materials and Methods
P. americana var. drymifolia was obtained from the germplasm bank of the Instituto Nacional de Investigaciones Forestales y Agropecuarias in Uruapan and the Hass and Carmen Hass cultivars were collected from a commercial orchard in Tingambato, both in Michoacán, Mexico. The remaining resequenced accessions were obtained from the Fundación Salvador Sánchez Colín germplasm bank located at La Cruz Experimental Center at Coatepec Harinas in the state of Mexico. The Velvick rootstock was provided by the University of Queensland, Australia. DNA was extracted from young leaves of single individuals for all cultivars sequenced. For the reference genome of the Hass cultivar, high-quality megabase-sized DNA was submitted to the National Center for Genome Resources for PacBio single-molecule real-time sequencing. A P. americana var. drymifolia reference individual was sequenced using different fragment-size libraries (∼0.5, 1, 3, 5, or 8 kb). For detailed information about assembly, annotation, and other bioinformatic analysis see SI Appendix.
Supplementary Material
Acknowledgments
This project was funded in large part by Grant 00126261from the Secretaria de Agrícultura, Ganadería, Recursos Pesqueros y Alimentos/Consejo Nacional de Ciencia y Tecnología sectorial program to L.H.-E.; Grant 05-2018 from the Governor University Research Initiative program from the state of Texas; Howard Hughes Medical Institute Grant 55005946 to L.H.-E.; Grants 0922742 and 1442190 to V.A.A., N.M., and A.H. from the National Science Foundation; Horticulture Innovation Australia Ltd; and the Australian Bureau of Agricultural and Resource Economics and Sciences. We thank the Fundación Salvador Sánchez Colín–Centro de Investigaciones Científicas y Tecnológicas de Aguacate en el Estado de México, S.C. for providing avocado specimens. We also thank Araceli Fernandez and Emanuel Villafan, administrators of the high-performance computing systems at Laboratorio Nacional de Genomica para la Biodiversidad and Instituto Nacional de Ecología, respectively.
Footnotes
The authors declare no conflict of interest.
Data deposition: Bioproject: PRJNA508502. Biosamples: SAMN10523735, SAMN10523720, SAMN10523736, SAMN10523738, SAMN10523739, SAMN10523746, SAMN10523747, SAMN10523748, SAMN10523749, SAMN10523750, SAMN10523752, SAMN10523753, and SAMN10523756. SRA submission: SUB4878870. Whole Genome Shotgun projects have been deposited at DDBJ/ENA/GenBank (accession nos. SDXN00000000 and SDSS00000000). The versions described in this paper are version SDXN01000000 and SDSS01000000 (P. americana var. drymifolia and P. americana cultivar Hass, respectively). The genome assemblies and annotations are available at https://genomevolution.org/CoGe/SearchResults.pl?s=29305&p=genome and https://genomevolution.org/coge/SearchResults.pl?s=29302&p=genome (P. americana var. drymifolia and P. americana cultivar Hass, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1822129116/-/DCSupplemental.
References
- 1.Chanderbali A. S., et al. , Persea americana (avocado): Bringing ancient flowers to fruit in the genomics era. BioEssays 30, 386–396 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Christenhusz M. J., Byng J. W., The number of known plants species in the world and its annual increase. Phytotaxa 261, 201–217 (2016). [Google Scholar]
- 3.Statista Research Department, Avocado industry–Statistics & facts. https://www.statista.com/topics/3108/avocadoindustry/. Accessed 29 July 2019.
- 4.Fresh Plaza, Mexico: Avocado exports generate 2.5 billion dollars. https://www.freshplaza.com/article/178230/Mexico-Avocado-exportsgenerate-2.5-billion-dollars/. Accessed 29 July 2019.
- 5.Galindo-Tovar M. E., Arzate-Fernández A. M., Ogata-Aguilar N., Landero-Torres I., The avocado (Persea americana, Lauraceae) crop in Mesoamerica: 10,000 years of history. Harv. Pap. Bot. 12, 325–334 (2007). [Google Scholar]
- 6.Schaffer B. A., Wolstenholme B. N., Whiley A. W., The Avocado: Botany, Production and Uses (CABI, 2013). [Google Scholar]
- 7.Chen H., Morrell P. L., Ashworth V. E., de la Cruz M., Clegg M. T., Tracing the geographic origins of major avocado cultivars. J. Hered. 100, 56–65 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Willingham S., et al. , Rootstock influences postharvest anthracnose development in‘Hass’ avocado. Aust. J. Agric. Res. 52, 1017–1022 (2001). [Google Scholar]
- 9.Illsley-Granich C., Brokaw R., Ochoa-Ascencio S., “Hass Carmen, a precocious flowering avocado tree” in Proceedings VII World Avocado Congress (2011), pp. 5–9.
- 10.Furnier G., Cummings M., Clegg M., Evolution of the avocados as revealed by DNA restriction fragment variation. J. Hered. 81, 183–188 (1990). [Google Scholar]
- 11.Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Kuhn D., et al. , Application of genomic tools to avocado (Persea americana) breeding: SNP discovery for genotyping and germplasm characterization. Sci. Hortic. (Amsterdam) 246, 1–11 (2019). [Google Scholar]
- 13.Lee T. H., Guo H., Wang X., Kim C., Paterson A. H., SNPhylo: A pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics 15, 162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skotte L., Korneliussen T. S., Albrechtsen A., Estimating individual admixture proportions from next generation sequencing data. Genetics 195, 693–702 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng X., Weir B. S., Eigenanalysis of SNP data with an identity by descent interpretation. Theor. Popul. Biol. 107, 65–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin S. H., Davey J. W., Jiggins C. D., Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol. Biol. Evol. 32, 244–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nei M., Li W.-H., Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U.S.A. 76, 5269–5273 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watterson G. A., On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7, 256–276 (1975). [DOI] [PubMed] [Google Scholar]
- 19.Holsinger K. E., Weir B. S., Genetics in geographically structured populations: Defining, estimating and interpreting F(ST). Nat. Rev. Genet. 10, 639–650 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tajima F., Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyons E., Pedersen B., Kane J., Freeling M., The value of nonmodel genomes and an example using SynMap within CoGe to dissect the hexaploidy that predates the rosids. Trop. Plant Biol. 1, 181–190 (2008). [Google Scholar]
- 22.Albert V. A., et al. ; Amborella Genome Project , The Amborella genome and the evolution of flowering plants. Science 342, 1241089 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Tang H., et al. , Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 18, 1944–1954 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiao Y., et al. , A genome triplication associated with early diversification of the core eudicots. Genome Biol. 13, R3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaillon O., et al. , The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Zheng C., Santos Muñoz D., Albert V. A., Sankoff D., Syntenic block overlap multiplicities with a panel of reference genomes provide a signature of ancient polyploidization events. BMC Genomics 16 (suppl. 10), S8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng C., Chen E., Albert V. A., Lyons E., Sankoff D., Ancient eudicot hexaploidy meets ancestral eurosid gene order. BMC Genomics 14 (suppl. 7), S3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore M. J., Bell C. D., Soltis P. S., Soltis D. E., Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc. Natl. Acad. Sci. U.S.A. 104, 19363–19368 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soltis D. E., et al. , Angiosperm phylogeny: 17 genes, 640 taxa. Am. J. Bot. 98, 704–730 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Soltis D. E., et al. , Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences. Bot. J. Linn. Soc. 133, 381–461 (2000). [Google Scholar]
- 31.Wickett N. J., et al. , Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. U.S.A. 111, E4859–E4868 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sankoff D., Zheng C., Lyons E., Tang H., “The trees in the peaks” in International Conference on Algorithms for Computational Biology, Holmes I., Martin-Vide C., Vega-Rodríguez M. A., Eds. (Springer, 2016), pp. 3–14. [Google Scholar]
- 33.Sankoff D., et al. , Models for similarity distributions of syntenic homologs and applications to phylogenomics. IEEE/ACM Trans. Comput. Biol. Bioinformatics 10.1109/TCBB.2018.2849377 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Zeng L., et al. , Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat. Commun. 5, 4956 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hobolth A., Christensen O. F., Mailund T., Schierup M. H., Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden Markov model. PLoS Genet. 3, e7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pollard D. A., Iyer V. N., Moses A. M., Eisen M. B., Widespread discordance of gene trees with species tree in Drosophila: Evidence for incomplete lineage sorting. PLoS Genet. 2, e173 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitfield J. B., Lockhart P. J., Deciphering ancient rapid radiations. Trends Ecol. Evol. 22, 258–265 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Librado P., Vieira F. G., Rozas J., BadiRate: Estimating family turnover rates by likelihood-based methods. Bioinformatics 28, 279–281 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Charlesworth D., Don’t Forget the Ancestral Polymorphisms (Nature Publishing Group, 2010). [DOI] [PubMed] [Google Scholar]
- 40.Peterson G. I., Masel J., Quantitative prediction of molecular clock and ka/ks at short timescales. Mol. Biol. Evol. 26, 2595–2603 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rasmussen M. D., Kellis M., Unified modeling of gene duplication, loss, and coalescence using a locus tree. Genome Res. 22, 755–765 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suh A., Smeds L., Ellegren H., The dynamics of incomplete lineage sorting across the ancient adaptive radiation of neoavian birds. PLoS Biol. 13, e1002224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lynch M., The Origins of Genome Architecture (Sinauer Associates, Sunderland, MA, 2007), vol. 98. [Google Scholar]
- 44.Force A., et al. , Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freeling M., Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 60, 433–453 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Cheng F., et al. , Gene retention, fractionation and subgenome differences in polyploid plants. Nat. Plants 4, 258–268 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Salojärvi J., et al. , Genome sequencing and population genomic analyses provide insights into the adaptive landscape of silver birch. Nat. Genet. 49, 904–912 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Beno-Moualem D., Prusky D., Early events during quiescent infection development by Colletotrichum gloeosporioides in unripe avocado fruits. Phytopathology 90, 553–559 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Acosta-Muñiz C. H., et al. , Identification of avocado (Persea americana) root proteins induced by infection with the oomycete Phytophthora cinnamomi using a proteomic approach. Physiol. Plant. 144, 59–72 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Engelbrecht J., Van den Berg N., Expression of defence-related genes against Phytophthora cinnamomi in five avocado rootstocks. S. Afr. J. Sci. 109, 1–8 (2013). [Google Scholar]
- 51.Luna E., et al. , Callose deposition: A multifaceted plant defense response. Mol. Plant Microbe Interact. 24, 183–193 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Eshraghi L., et al. , Phosphite primed defence responses and enhanced expression of defence genes in Arabidopsis thaliana infected with Phytophthora cinnamomi. Plant Pathol. 60, 1086–1095 (2011). [Google Scholar]
- 53.Berg N. v. d., Christie J. B., Aveling T. A. S., Engelbrecht J., Callose and β-1,3-glucanase inhibit Phytophthora cinnamomi in a resistant avocado rootstock. Plant Pathol. 67, 1150–1160 (2018). [Google Scholar]
- 54.Le Roy J., Huss B., Creach A., Hawkins S., Neutelings G., Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front. Plant Sci. 7, 735 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonawitz N. D., Chapple C., The genetics of lignin biosynthesis: Connecting genotype to phenotype. Annu. Rev. Genet. 44, 337–363 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Chang C. L., Cho I. K., Li Q. X., Insecticidal activity of basil oil, trans-anethole, estragole, and linalool to adult fruit flies of Ceratitis capitata, Bactrocera dorsalis, and Bactrocera cucurbitae. J. Econ. Entomol. 102, 203–209 (2009). [DOI] [PubMed] [Google Scholar]
- 57.King J. R., Knight R. J., Occurrence and assay of estragole in the leaves of various avocado cultivars. J. Agric. Food Chem. 35, 842–844 (1987). [Google Scholar]
- 58.Zhu X., et al. , The wheat ethylene response factor transcription factor pathogen-induced ERF1 mediates host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant Physiol. 164, 1499–1514 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xing L., et al. , Overexpression of ERF1-V from Haynaldia villosa can enhance the resistance of wheat to powdery mildew and increase the tolerance to salt and drought stresses. Front. Plant Sci. 8, 1948 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Diniz I., et al. , A first insight into the involvement of phytohormones pathways in coffee resistance and susceptibility to Colletotrichum kahawae. PLoS One 12, e0178159 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Broekaert W. F., Delauré S. L., De Bolle M. F., Cammue B. P., The role of ethylene in host-pathogen interactions. Annu. Rev. Phytopathol. 44, 393–416 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Lü P., et al. , Genome encode analyses reveal the basis of convergent evolution of fleshy fruit ripening. Nat. Plants 4, 784–791 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Yahia E. M., Woolf A. B., “Avocado (Persea americana Mill.)” in Postharvest Biology and Technology of Tropical and Subtropical Fruits: Açai to Citrus, Yahia E., Ed. (Woodhead Publishing, 2011), pp. 125–186e. [Google Scholar]
- 64.Xoca-Orozco L.-Á., et al. , Transcriptomic analysis of avocado hass (Persea americana Mill) in the interaction system fruit-chitosan-Colletotrichum. Front. Plant Sci. 8, 956 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bray N. L., Pimentel H., Melsted P., Pachter L., Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Xu W., et al. , Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 7, 1555–1567 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu Y., et al. , Comparative transcriptome analysis unveils the tolerance mechanisms of Citrus hystrix in response to ‘Candidatus Liberibacter asiaticus’ infection. PLoS One 12, e0189229 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davila Olivas N. H., et al. , Effect of prior drought and pathogen stress on Arabidopsis transcriptome changes to caterpillar herbivory. New Phytol. 210, 1344–1356 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Tomas-Grau R. H., et al. , Soft mechanical stimulation induces a defense response against Botrytis cinerea in strawberry. Plant Cell Rep. 37, 239–250 (2018). [DOI] [PubMed] [Google Scholar]
- 70.Wang X., et al. , IDL6-HAE/HSL2 impacts pectin degradation and resistance to Pseudomonas syringae pv tomato DC3000 in Arabidopsis leaves. Plant J. 89, 250–263 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bioproject: PRJNA508502. Biosamples: SAMN10523735, SAMN10523720, SAMN10523736, SAMN10523738, SAMN10523739, SAMN10523746, SAMN10523747, SAMN10523748; SAMN10523749, SAMN10523750, SAMN10523752, SAMN10523753, and SAMN10523756. SRA submission: SUB4878870. Whole Genome Shotgun projects have been deposited at DDBJ/ENA/GenBank under the accession nos. SDXN00000000 and SDSS00000000. The versions described in this paper are versions SDXN01000000 and SDSS01000000 (P. americana var. drymifolia and P. americana cultivar Hass, respectively). The genome assemblies and annotations are available at https://genomevolution.org/CoGe/SearchResults.pl?s=29305&p=genome and https://genomevolution.org/coge/SearchResults.pl?s=29302&p=genome (P. americana var. drymifolia and P. americana cultivar Hass, respectively).