Significance
Accurate description of protein structure and function is a fundamental step toward understanding biological life and highly relevant in the development of therapeutics. Although greatly improved, experimental protein structure determination is still low-throughput and costly, especially for membrane proteins. As such, computational structure prediction is often resorted. Predicting the structure of a protein without similar structures in the Protein Data Bank is very challenging and usually needs a large amount of computing power. This paper shows that by using a powerful deep learning technique, even with only a personal computer we can predict new folds much more accurately than ever before. This method also works well on membrane protein folding.
Keywords: protein folding, deep learning, protein contact prediction, protein distance prediction, direct coupling analysis
Abstract
Direct coupling analysis (DCA) for protein folding has made very good progress, but it is not effective for proteins that lack many sequence homologs, even coupled with time-consuming conformation sampling with fragments. We show that we can accurately predict interresidue distance distribution of a protein by deep learning, even for proteins with ∼60 sequence homologs. Using only the geometric constraints given by the resulting distance matrix we may construct 3D models without involving extensive conformation sampling. Our method successfully folded 21 of the 37 CASP12 hard targets with a median family size of 58 effective sequence homologs within 4 h on a Linux computer of 20 central processing units. In contrast, DCA-predicted contacts cannot be used to fold any of these hard targets in the absence of extensive conformation sampling, and the best CASP12 group folded only 11 of them by integrating DCA-predicted contacts into fragment-based conformation sampling. Rigorous experimental validation in CASP13 shows that our distance-based folding server successfully folded 17 of 32 hard targets (with a median family size of 36 sequence homologs) and obtained 70% precision on the top L/5 long-range predicted contacts. The latest experimental validation in CAMEO shows that our server predicted correct folds for 2 membrane proteins while all of the other servers failed. These results demonstrate that it is now feasible to predict correct fold for many more proteins lack of similar structures in the Protein Data Bank even on a personal computer.
Computational structure prediction of proteins without detectable homology to experimentally solved structures is a very challenging problem. Even after decades of research, progress on this problem has been slow, and many methods require considerable computational resources, even for relatively small proteins. Nevertheless, in recent years good progress has been achieved thanks to accurate contact prediction enabled by direct coupling analysis (DCA) (1–9) and deep convolutional neural networks (DCNN) (10–16). As such, contact-assisted protein folding has gained a lot of attention and contact prediction has garnered considerable research effort.
We have developed the CASP12- and CASP13-winning method RaptorX-Contact (10) that uses deep and fully convolutional residual neural network (ResNet) to predict contacts. ResNet is one type of DCNN (17) but is much more powerful than traditional DCNN. RaptorX-Contact has good accuracy even for some proteins with only dozens of sequence homologs. The precision of RaptorX-Contact decreases more slowly than DCA when more predicted contacts are evaluated, especially when the protein under study has few sequence homologs (10). As reported in refs. 10 and 12, without extensive fragment-based conformation sampling, the 3D models constructed from contacts predicted by RaptorX-Contact have much better quality than those built from contacts predicted by DCA methods such as CCMpred (6) and the CASP11 winner MetaPSICOV (18). RaptorX-Contact also works well for membrane proteins (MPs) even trained by soluble proteins (12) and for complex contact prediction even trained by single-chain proteins (19).
Both our ResNet and DCA are global prediction methods because they predict the contact/distance score or probability of one residue pair by considering its correlation with other residue pairs at distant sequence positions, which is the key to the significant improvement in contact prediction. In principle, when many convolutional layers are used, it is possible to capture correlation between any two residue pairs across the whole contact/distance matrix. However, ResNet differs from DCA in that 1) ResNet can capture higher-order residue correlation (e.g., structure motifs) while DCA mainly focuses on pairwise relationships, 2) ResNet tries to learn the global context of a contact matrix, and 3) existing DCA methods are roughly linear models with tens of millions of parameters estimated from a single protein family, while ResNet is a nonlinear model with parameters estimated from thousands of protein families. Deep learning (DL) models such as CMAPpro (20) and Deep Belief Networks (DBN) (21) were used for contact prediction before, but ResNet is a DL method that greatly outperforms shallow methods such as MetaPSICOV (18). Different from ResNet and DCA, DBN and MetaPSICOV are local prediction methods, as they predict the label (i.e., contact or distance) of 1 residue pair without considering the labels of others. This is one of the major reasons why DBN and MetaPSICOV underperformed RaptorX-Contact. Inspired by the success of RaptorX-Contact, many CASP13 predictors have employed fully ResNet or DCNN (13, 15, 22), as shown in the CASP13 abstract book. Notably, the Cheng group, who developed DBN, has switched to DCNN for contact prediction (13). The Peng group, who employed traditional DCNN in CASP12 (16), has switched to ResNet in CASP13.
Although contact prediction is drawing considerable attention, here we study distance prediction and treat contact prediction as a by-product. The rationale for this decision is 2-fold. The distance matrix contains finer-grained information than the contact matrix and provides more physical constraints of a protein structure (e.g., distance is metric but contact is not). Because of this, a distance matrix can determine a protein structure (except mirror image) much more accurately than a contact matrix. Trained by distance instead of contact matrices, ResNet may learn more about the intrinsic properties of a protein structure and thus greatly reduce the conformation space and improve folding accuracy. Further, different from DCA that aims to predict only a small number of contacts to assist time-consuming conformation sampling, we would like to predict the whole distance matrix and then directly construct protein 3D models without extensive conformation sampling. By doing so, we significantly reduce running time needed for protein folding, especially for a large protein. As we use many more distance restraints to construct 3D models, the impact of individual distance prediction error may be reduced (by the law of large numbers). In contrast, contact-assisted conformation sampling may be misguided by several wrongly predicted contacts and needs a long time to generate a good conformation for a large protein.
Distance prediction is not totally new (23–26). We have employed a probabilistic neural network to predict interresidue distance distribution, converted it to protein-specific distance-based statistical potential (27), and studied its folding simulation (28). Recently, we showed that protein-specific distance potential derived from deep ResNet may improve by a large margin protein threading with remote templates (29). In addition to distance prediction, we predicted secondary structure and backbone torsion angles by deep ResNet. By feeding these predicted restraints to CNS (30), we are able to quickly construct accurate 3D models, as evidenced by self-benchmark on 37 CASP12 hard targets (14) and 41 Continuous Automated Model Evaluation (CAMEO) hard targets (10) as well as rigorous experimental validation in CASP13 and CAMEO.
Results
Approach Summary.
We use a DL network very similar to that described in ref. 10 to predict the Euclidean distance distribution of 2 atoms (of different residues) in a protein to be folded (Fig. 1). Our DL network consists of one 1D deep ResNet, one 2D deep dilated ResNet, and one Softmax layer. The 1D ResNet captures sequential context of 1 residue (or sequence motifs) and the 2D ResNet captures pairwise context of a residue pair (or structure motifs). A dilated 2D convolutional operation (31) is used to capture a broader pairwise context with fewer parameters. The 1D ResNet consists of 6 to 7 convolutional layers and the same number of instance normalization layers and ReLU activation layers. The kernel size of the 1D convolutional operation is 15. The 2D dilated ResNet is much more important, consisting of 30 to 40 residual blocks, each having 2 convolutional layers, 2 instance normalization layers, and 2 ReLU activation layers. The contact and distance prediction accuracy may further improve slightly when 50 to 60 residual blocks are used for the 2D ResNet. We use 5 × 5 as the kernel size of the 2D convolutional operation. Note that the convolutional operation is applied to the whole protein sequence and matrix. In the case that the matrix is too big to fit into the limited graphics processing unit (GPU) memory, a submatrix of 300 × 300 or 400 × 400 is randomly sampled. As mentioned by us before (11), we have tested a few other slightly different network architectures such as dense deep network, wide ResNet, and adding LSTM onto the 2D ResNet but have not observed any significant performance gain.
Fig. 1.
The overall deep network architecture for protein distance prediction.
We discretize interatom distance into 25 bins: <4.5 Å, 4.5 to 5 Å, 5 to 5.5 Å, …, 15 to 15.5 Å, 15.5 to 16 Å, and >16 Å and treat each bin as a classification label. Our DL model for distance prediction is trained using a procedure similar to that described in ref. 10. By summing up the predicted probability values of the first 8 distance labels (corresponding to distance ≤8 Å), our distance-based DL model can be used for contact prediction and has 3 to 4% better long-range prediction accuracy than the DL model directly trained from contact matrices.
In addition to Cβ–Cβ, we also trained separate DL models to predict distance distribution for 4 other atom pairs: Cα–Cα, Cα–Cg, Cg–Cg, and N–O, where Cg is the first CG atom in an amino acid. When CG does not exist, OG or SG is used. The predicted distance of these 5 atom pairs is used together to fold a protein, which on average yields slightly better models than using Cβ–Cβ distance alone. We also trained a 1D ResNet to predict backbone torsion angles from sequence profile, which is used together with predicted distance to build 3D models.
In CASP13 we registered RaptorX-Contact for contact prediction and distance-based ab initio folding and RaptorX-DeepModeller for distance-based folding of a target based upon its alignment to weakly similar templates. In addition to features used by RaptorX-Contact, RaptorX-DeepModeller employed a few alignment-based input features including amino acid and profile similarity, secondary structure similarity, and an initial distance matrix extracted from template. In this paper we mainly focus on RaptorX-Contact.
Distance-Based Folding Outperforms Contact-Based Folding.
On the 37 CASP12 FM (free-modeling) targets, our distance-based ab initio folding method works much better than 3 contact-based folding methods (i.e., our own contact-based method and CCMpred- and MetaPSICOV-based) and 4 top CASP12 groups, Baker-server (32), Baker-human (33), Zhang-server (34), and Zhang-human (35) (Table 1, Fig. 2A, and Dataset S1). The top CASP12 groups folded some hard targets using contacts predicted by DCA and/or shallow machine learning methods. Zhang-human also extracted information from CASP12 server predictions. The 3D models predicted by our distance-based folding method for CASP12 FM targets have average TMscore of 0.466 and 0.476, respectively, when the top 1 and the best of the top 5 models are evaluated. When all 5 models are considered, our distance-based folding method can predict correct folds (TMscore >0.5) for 21 CASP12 FM targets, much better than our contact-based method and the best CASP12 human groups. With only predicted secondary structure and contacts predicted by MetaPSICOV and CCMpred, we may generate correct folds for 2 and 0 targets, respectively. That is, contacts predicted by CCMpred and MetaPSICOV alone are insufficient for 3D modeling.
Table 1.
Modeling accuracy of selected methods on CASP12 hard targets
| Method | Top 1 | Top 5 | #OK |
| This work | 0.466 | 0.476 | 21 |
| Our contact | 0.354 | 0.397 | 10 |
| CCMpred | 0.216 | 0.235 | 0 |
| MetaPSICOV | 0.262 | 0.289 | 2 |
| Baker-server | 0.326 | 0.370 | 9 |
| Zhang-server | 0.347 | 0.404 | 10 |
| Baker-human | 0.392 | 0.422 | 11 |
| Zhang-human | 0.375 | 0.420 | 11 |
Columns “Top 1” and “Top 5” list the average TMscore of the top 1 and the best of top 5 models, respectively. Column “#OK” lists the number of models with TMscore > 0.5.
Fig. 2.
Distance prediction and folding results on the 37 CASP12 FM and 41 CAMEO hard targets. (A) Quality of distance- vs. contact-based 3D models predicted by our method. (B) Distance-based 3D model quality vs. logarithm of Meff. (C) Cβ–Cβ distance prediction error vs. logarithm of Meff. (D) Distance-based 3D model quality vs. Cβ–Cβ distance prediction error. Here model quality or quality of a model denotes the quality of a predicted 3D model measured by TMscore.
The folding results on the CAMEO data are consistent with that on the CASP12 data (Table 2, Fig. 2A, and Dataset S1). When the first and the best of the top 5 models are evaluated, our distance-based 3D models have average TMscores of 0.551 and 0.577, respectively, about 10% better than our contact-based models, which have average TMscores of 0.504 and 0.524, respectively, and much better than the models built from contacts generated by CCMpred and MetaPSICOV. Our distance- and contact-based folding methods predicted correct folds for 30 and 23 of 41 CAMEO targets, respectively.
Table 2.
Modeling accuracy of selected methods on CAMEO hard targets
| Method | Top 1 | Top 5 | #OK |
| This work | 0.551 | 0.577 | 30 |
| Our contact | 0.507 | 0.525 | 23 |
| CCMpred | 0.292 | 0.316 | 4 |
| MetaPSICOV | 0.365 | 0.392 | 8 |
See Table 1 for explanation.
Dependency on Multiple Sequence Alignment and Direct Coupling Score.
Our distance-based model quality is correlated (coefficient ∼0.6) with logarithm of Meff (i.e., the multiple sequence alignment [MSA] depth) (Fig. 2B). When Meff > 55 or ln(Meff) > 4, there is a good chance that our predicted 3D models have a correct fold. Our distance-based folding method can fold 8 out of 21 CASP12 FM targets with Meff ≤ 100: T0862-D1, T0863-D1, T0869-D1, T0870-D1, T0894-D1, T0898-D1, T0904-D1, and T0915-D1. Meanwhile, 5 of them have 3D models with TMscores >0.6. In contrast, Zhang-Server, Zhang-Human, Baker-Server, and Baker-Human predicted models with TMscores >0.6 for only 2, 1, 0, and 0 targets with Meff ≤100, respectively (Dataset S1).
To evaluate the importance of direct coupling score produced by CCMpred, we trained our ResNet without using it but keeping all other features including raw and average product correction (APC)-corrected mutual information (MI). On the CASP12 FM targets, this ResNet has top L, L/2, L/5, and L/10 long-range contact precision 36.3%, 48.4%, 61.9% and 66.7%, respectively, where L is the sequence length. The average TMscores of the top 1 and best of the top 5 models built from distance predicted by this ResNet are 0.400 and 0.411, respectively. By contrast, when this direct coupling score is used, the top L, L/2, L/5, and L/10 long-range contact precision is 43.1%, 56.9%, 66.8%, and 73.7%, respectively. That is, without this score, the long-range contact precision and 3D model quality drop by ∼7% and 0.066, respectively. The performance does not decrease too much since many targets have a small Meff and thus this direct coupling score is not much better than MI.
Distance Prediction Error and Implications on 3D Modeling.
We only consider the pairs of atoms with sequence separation of at least 12 residues and predicted distance ≤15 Å. We calculated the average quality of predicted distance on each CASP12 or CAMEO target (Dataset S1) and the average quality of each dataset (SI Appendix, Table S1). Distance prediction error is correlated (coefficient ∼−0.53) with logarithm of Meff (Fig. 2C). When Meff > 55 or ln(Meff) > 4, Cβ–Cβ distance prediction error is likely to be less than 4 Å. Three-dimensional modeling quality is strongly correlated (coefficient ∼−0.80) with distance prediction error (Fig. 2D), which implies that as long as distance prediction is accurate CNS is able to build good 3D models. When distance error is 8 Å, the resultant 3D models have very bad quality.
Rigorous Blind Test in CASP13.
RaptorX-Contact was officially ranked first among 46 human and server contact predictors, in terms of a combination of many metrics. In terms of F1 of top L/5 long-range predicted contacts, the top 5 groups are RaptorX-Contact (0.233), TripletRes (0.213), ResTriplet (0.208), RRMD (0.192), and TripletRes_AT (0.191). DeepMind did not submit contact prediction, according to its presentation at the seventh Critical Assessment of Prediction of Interactions meeting; AlphaFold’s F1 values on the top L/5, L/2, and L predicted long-range contacts are 0.227, 0.369, and 0.419, respectively, slightly better than ours (0.233, 0.362, and 0.411). On 12 FM/TBM targets, AlphaFold’s F1 values are 31.4, 48.7, and 55.1, better than ours (28.7, 43.2, and 51.7). This could be due to the difference of training data. AlphaFold used a larger training set (Cath S35 as of 16 March 2018) and a much deeper ResNet. Cath S35 contains proteins with <35% sequence identity and thus has a better coverage for FM/TBM targets than PDB25 used by us. See SI Appendix for a more detailed study of the impact of training sets on contact prediction (SI Appendix, Table S4). To the best of our knowledge, all these top-performing groups used deep ResNet. As a control, MetaPSICOV ran by the CASP13 organizers has top L/5 long-range precision = 25.16% and F1 = 0.078, respectively, and a DCA method GaussDCA has precision = 21.757% and F1 = 0.067, respectively.
Table 3 summarizes the average quality of predicted distance on the FM targets (Dataset S2). For most targets, the Cβ–Cβ distance prediction error is less than 4 Å (Fig. 3A) and its correlation with ln(Meff) is not very strong (coefficient −0.45). RaptorX-Contact predicted distance well for quite a few targets such as T0969-D1 and T0957s2-D1 (SI Appendix, Fig. S1) but did badly on T0953s1 and T0989-D1, both of which have ln(Meff) around 4. T0969-D1 has MSA depth >1,000, but T0957s2-D1 has only a shallow MSA. T0953s1 has 72 residues, but only 34 long-range residue pairs with native distance <15 Å, which is much smaller than typical. While estimating distance bounds from predicted distribution, we assumed each target had about 7L long-range Cβ–Cβ pairs with distance <15 Å, which resulted in a big prediction error on T0953s1 (SI Appendix, Fig. S1). T0989 is a 2-domain target. Its first domain has much better coevolution signal than the second one. We did not split T0989 into 2 domains in CASP13, which resulted in many more Cβ–Cβ pairs in D1 being assumed to have distance <15 Å and thus led to a big prediction error (SI Appendix, Fig. S1). When T0989-D1 is predicted independently, its Cβ–Cβ distance error is only 4.89 Å.
Table 3.
Quality of predicted distance on the 32 CASP13 FM targets
| Atom | AbsE, Å | RelE | Prec | Recall | F1 |
| CbCb | 3.76 | 0.259 | 0.678 | 0.540 | 0.588 |
| CgCg | 4.02 | 0.278 | 0.656 | 0.532 | 0.573 |
| CaCg | 3.84 | 0.262 | 0.671 | 0.512 | 0.567 |
| CaCa | 3.84 | 0.253 | 0.666 | 0.532 | 0.577 |
| NO | 3.75 | 0.253 | 0.674 | 0.505 | 0.566 |
Columns “AbsE” and “RelE” are the absolute and relative error. “Prec” represents precision.
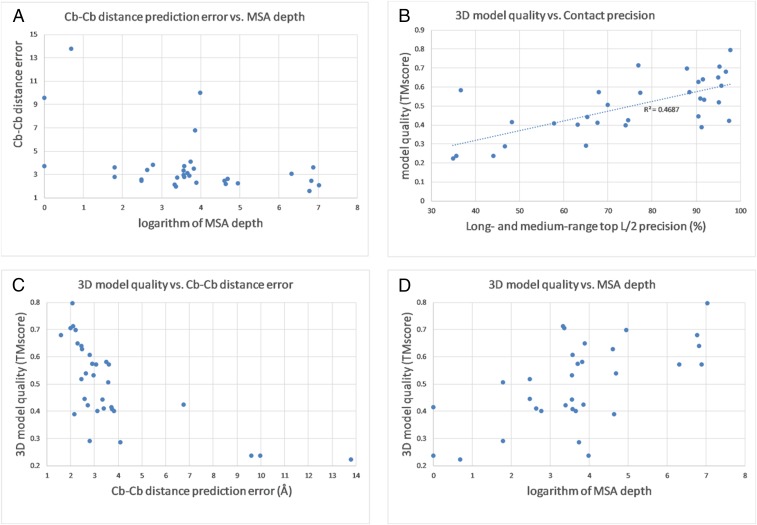
Fig. 3.
Distance prediction and folding results of RaptorX-Contact on the 32 CASP13 FM targets. (A) Cβ–Cβ distance prediction error vs. logarithm of Meff. (B) Three-dimensional model quality vs. contact precision. (C) Three-dimensional model quality vs. Cβ–Cβ distance prediction error. (D) Three-dimensional model quality vs. logarithm of Meff.
Table 4 shows the modeling accuracy of top 2 human and 6 server groups for the CASP13 FM targets. Our 2 distance-based folding servers are only slightly worse than Zhang’s 2 servers, which used ResNet-predicted contacts to guide folding simulation. If we merge our 2 servers into a single group, the best models have an average TMscore = 0.5264. Similarly, if Zhang-Server and QUARK are merged, the best models produced by the Zhang group have an average TMscore = 0.5348. Robetta underperformed the top 4 servers by a large margin, possibly because it did not use DL to predict contacts or distance. Although AlphaFold and RaptorX-Contact have similar contact prediction performance, AlphaFold did much better in 3D modeling. AlphaFold predicted much better 3D models for quite a few targets (e.g., T0968s2-D1, T0980s1-D1, T0986s2-D1, T0990-D1, T1015s1-D1, T1017s2-D1, and T1021s3-D2), although the Zhang group or RaptorX also predicted correct folds for them. On several FM targets (T0950-D1, T0960-D2, and T0963-D2) the Zhang group or RaptorX predicted better 3D models than AlphaFold, however.
Table 4.
Folding results of selected methods on CASP13 FM targets
| Top 1 model | Best of top 5 model | |||||||
| Methods | rmsd, Å | TM | GDT | #OK | rmsd, Å | TM | GDT | #OK |
| AlphaFold | 9.05 | 0.583 | 0.516 | 20 | 7.85 | 0.625 | 0.561 | 23 |
| Zhang-human | 8.93 | 0.521 | 0.462 | 18 | 7.87 | 0.558 | 0.487 | 20 |
| Zhang-server | 9.92 | 0.487 | 0.422 | 16 | 8.97 | 0.524 | 0.453 | 20 |
| Zhang-QUARK | 9.10 | 0.490 | 0.426 | 16 | 8.85 | 0.514 | 0.442 | 19 |
| RX-DeepModeller | 10.64 | 0.471 | 0.406 | 16 | 9.79 | 0.501 | 0.431 | 17 |
| RX-Contact | 10.92 | 0.474 | 0.409 | 15 | 10.09 | 0.498 | 0.427 | 17 |
| Robetta Server | 13.64 | 0.390 | 0.339 | 7 | 13.02 | 0.430 | 0.372 | 10 |
| RX-TBM | 12.49 | 0.402 | 0.345 | 7 | 11.87 | 0.420 | 0.358 | 9 |
The scores are calculated by us and may be slightly different from the official. AlphaFold and Zhang-human are human groups and the others are server groups. Zhang-human did not submit models for T0950-D1, so its evaluation is based upon 31 targets.
The quality (TMscore) of RaptorX-Contact 3D models is correlated (coefficient ∼0.68) with top L/2 long- and medium-range contact precision (Fig. 3B) and Cβ–Cβ distance prediction error (Fig. 3C). When Meff > 55 or ln(Meff) > 4, RaptorX-Contact is likely to predict a correct fold (TMscore > 0.5). When ln(Meff) is between 3 and 4, RaptorX-Contact may predict correct folds for half of the targets. Here we use the HHblits MSA depth reported by CASP13 as Meff. RaptorX-Contact predicted very good 3D models for T0969-D1 (354 residues), T0953s2-D2 (127 residues), and T0957s2-D1 (155 residues). T0969-D1 has >1,000 sequence homologs, but the other two target domains have only ∼30 sequence homologs. The 3D model for T0969-D1 has TMscore = 0.8 and rmsd = 5.1 Å and the models for T0953s2-D2 and T0957s-D1 have TMscore > 0.7 and rmsd = 3 to 4 Å (SI Appendix, Fig. S2).
DL Can Predict New Folds and Its Dependency on Target-Training Similarity.
Some CASP13 FM targets may have weakly similar experimental structures in the Protein Data Bank (PDB), although they are hard to detect by sequence (profile) information. Because of this, there are 2 possible reasons why DL performed better than previous methods. One is that DL is just a better fold recognition method and the other is that DL indeed can predict 3D models better than what can be copied from individual training proteins. To address this, we analyze the FM targets and their models from a few different perspectives (Dataset S3).
First, we use our structure alignment tool DeepAlign (36) to calculate target-training structure similarity, that is, the structure similarity in terms of TMscore between target experimental structures and their most similar training protein structures. Some targets indeed have similar structures in our training set (Fig. 4A). However, the quality (in terms of TMscore) of RaptorX-Contact first models is weakly and negatively correlated (correlation coefficient = −0.199 and trendline R2 = 0.0396) with target-training structure similarity. RaptorX-Contact first models for 11 targets have quality (i.e., TMscore) higher than their target-training structure similarity (SI Appendix, Table S2). Meanwhile, 6 of them have MSA depth <200 and 8 of them have no similar folds in the training set at all (i.e., TMscore <0.5). That is, for these targets RaptorX-Contact can predict 3D models much better than what can be copied from their most similar training protein structures. Second, 26 of the 32 RaptorX-Contact first models have TMscore <0.51 with all training protein structures (Fig. 4B), which means they are not very similar to any training proteins, that is, not simply copied from individual training proteins. Eleven of these 26 models have quality much better than 0.51 (Dataset S3). We have also employed another tool, TMalign, to calculate the structure similarity of two structures/models, which does not change the conclusion drawn here. See SI Appendix for a more detailed description.
Fig. 4.
Novelty of RaptorX-Contact 3D models. (A) RaptorX-Contact first model quality vs. target-training structure similarity. (B) Structure similarity between RaptorX-Contact models and training proteins. (C) First model quality of RaptorX-Contact vs. CNFpred. (D) First model quality of RaptorX-Contact vs. RaptorX-TBM.
We further compare RaptorX-Contact with 2 in-house threading methods, CNFpred (36, 37) and RaptorX-TBM. CNFpred integrates sequence profile, secondary structure, and solvent accessibility via a machine learning approach to build sequence–template alignments, which are then fed into MODELER (38) to build 3D models. CNFpred is more sensitive than HHpred (39), although both mainly rely on sequence profile similarity. RaptorX-TBM used our new threading program DeepThreader (29) to build sequence–template alignments and then Rosetta-CM (40) to build 3D models from alignment. DeepThreader greatly outperforms previous threading methods by integrating CNFpred with distance-based statistical potential converted from distance distribution predicted by RaptorX-Contact. Although mainly a template-based method, RaptorX-TBM performed well on the CASP13 FM targets, only second to Zhang’s 2 servers and our 2 servers that applied contact or distance-based folding (Table 4).
CNFpred and DeepThreader used PDB90 as the template database while RaptorX-Contact was trained by PDB25. Although PDB90 has a larger coverage, in terms of TMscore of the first 3D models, RaptorX-Contact exceeded CNFpred by 0.236 (Fig. 4C) and RaptorX-TBM by 0.072 (Fig. 4D). RaptorX-Contact and CNFpred models have weakly correlated quality (correlation coefficient = 0.431 and trendline R2 = 0.1868) since they use the same set of sequence profile, predicted secondary structure, and solvent accessibility. The correlation coefficient and R2 may further drop to 0.351 and 0.1234, respectively, if we exclude two targets, T0957s2-D1 and T0975-D1, for which CNFpred generates 3D models with TMscore >0.4 (Dataset S3). That is, RaptorX-Contact model quality is only weakly correlated with target-PDB sequence profile similarity. RaptorX-Contact and RaptorX-TBM model quality has higher correlation coefficient (0.665) and trendline R2 (0.4419) because they used the same set of predicted distance information and sequence profile. In summary, RaptorX-Contact can do much better than a very good fold recognition method and its modeling accuracy is only weakly correlated with sequence profile similarity.
Distance Violation for Model Quality Assessment.
In the absence of experimental structures, although a sophisticated method could be developed to assess the quality of a 3D model, we find out that the quality of a model is well correlated with distance violation (produced by CNS) of this model with respect to the set of distance restraints predicted by RaptorX-Contact (Fig. 5 and Dataset S4). When distance violation is less than 4 Å, there is a good chance that the resultant 3D model has a TMscore >0.5. On the CASP12 FM targets, the correlation coefficient between model quality (TMscore) and distance violation is −0.795. On the CASP13 FM targets, the correlation coefficient is slightly worse (−0.707), possibly because for CASP13 distance violation is calculated on domains predicted by us while model quality is calculated on official domains. In contrast, for CASP12 targets the official domain definitions are used for both prediction and evaluation. Note that the quality of our 3D models has a stronger correlation with distance violation than with logarithm of MSA depth, which is ∼0.6 on both CASP12 and CASP13 FM targets (Datasets S1 and S2).
Fig. 5.
Relationship between RaptorX-Contact model quality and distance violation.
DL Can Fold MPs.
Since mid-September 2018 we have tested RaptorX-Contact (CAMEO ID: server 60) in CAMEO (https://www.cameo3d.org/), an online blind benchmark currently testing ∼40 servers including Robetta, RaptorX, Swiss-Model, and HHpred. There are no hard MP targets in CASP13. We show that RaptorX-Contact predicted correct folds for two hard MP targets (CAMEO-3D IDs: 2018-11-03_00000053_1 and 2018-11-17_00000062_1). The first MP target (PDB ID code 6bhpC) has 200 residues and Meff = 229. RaptorX-Contact predicted a 3D model with TMscore = 0.68 and rmsd = 5.65 Å (SI Appendix, Fig. S2), while the other servers failed to predict a correct fold (SI Appendix, Fig. S3). The second MP target (PDB ID code 6a2jA) has 309 residues and a few thousand sequence homologs. RaptorX-Contact predicted a 3D model with TMscore = 0.73 and rmsd = 5.51 Å (SI Appendix, Fig. S2), while the other servers predicted 3D models with TMscore ≤0.4 (SI Appendix, Fig. S4). The most similar training proteins have TMscore ∼0.45 with the experimental structure of 6a2jA. That is, 6a2jA does not have a similar fold in our training set. For 6bhpC, 7 training proteins, 4qn1A, 5gheA, 4p79A, 4k1pA, 5fnnA, 2nrjA, and 4phoA, have TMscore >0.6 (0.655, 0.633, 0.627, 0.627, 0.626, 0.623, and 0.604) with its experimental structure. Its RaptorX-Contact best model has TMscore 0.489, 0.534, 0.533, 0.541, 0.537, 0.531, and 0.506 with these proteins, respectively. That is, the RaptorX-Contact model for 6bhpC is not simply copied from one of these training proteins.
Information Content of Contacts Predicted by DL.
One may argue that DL has better precision than DCA simply because DL fills in redundant contacts in a secondary structure pair while DCA does not. This is not true for some targets. For 4 CASP13 FM targets, T0975-D1 (MSA depth >500), T0950-D1, T0957s2-D1, and T0980s1-D1, even when top n predicted medium- and long-range contacts are considered (where n is the number of native contacts) CCMpred failed to predict any native contacts in the circled regions but RaptorX-Contact predicted many (Fig. 6). By the way, the most similar training proteins for T0975-D1 and T0980s1-D2 have TMscore 0.448 and 0.476, respectively, with their experimental structures. Even for targets with a deep MSA, CCMpred generates too many false positives spreading over the whole contact matrix (SI Appendix, Fig. S5).
Fig. 6.
Contacts predicted by CCMpred (upper right triangle) and RaptorX-Contact (lower left triangle) on T0950-D1, T0957s2-D1, T0975-D1, and T0980s1-D1. Native, correctly predicted, and incorrectly predicted contacts are displayed in gray, red, and green, respectively. Top n medium- and long-range predicted contacts are displayed where n is the number of native contacts.
To evaluate information content of predicted contacts, we employ 2 strategies (Dataset S5). First, we employ entropy score to estimate the utility of predicted contacts for 3D modeling. A set of widely dispersed contacts may be more useful for protein modeling than the same number of contacts confined in a small contact submatrix. Entropy score was introduced in CASP12 to assess contact prediction (14). It is calculated on only correctly predicted contacts and ignores the impact of wrongly predicted contacts. As reported by CASP13, when top 10, L/5, and L/2 long-range contacts are considered, RaptorX-Contact has entropy score 0.311, 0.643, and 1.255, respectively, much larger than GaussDCA (41) (the only DCA method blindly tested in CASP13), which has entropy score 0.151, 0.332, and 0.553, respectively. On most targets RaptorX-Contact has better entropy score than GaussDCA (SI Appendix, Fig. S6). Among the 5 CASP13 FM targets (T0969-D1, T0975-D1, T1000-D2, T1021S3-D1, and T1022s1-D1) with MSA depth >500, RaptorX-Contact has entropy score 0.198, 0.763, and 1.687, respectively, while GaussDCA has entropy score 0.251, 0.80, and 1.365, respectively, when top 10, L/5, and L/2 long-range contacts are evaluated. That is, when only a small number of predicted contacts are considered, GaussDCA is slightly better for these targets, but when more are considered, RaptorX-Contact is better. By the way, the entropy score of RaptorX-Contact is negatively and weakly correlated with target-training structure similarity (Dataset S5). This further confirms that RaptorX-Contact predicts contacts not by copying from structurally the most similar training proteins.
In addition to entropy score, we calculate the ratio of native contacts covered by the correct ones among top n predicted contacts where n is the number of native medium- and long-range contacts (Dataset S5). We say 1 native contact (of residues i and j) is covered by a correctly predicted contact (of residues k and l) if their city-block distance (i.e., |i − k| + |j − l|) is no more than a given cutoff c. When c = 0, the coverage is just the recall. If RaptorX-Contact simply predicts redundant contacts, it shall have similar coverage as DCA when c is big. Besides GaussDCA, we also evaluate CCMpred’s contact prediction generated by us during CASP13. At the same c, the advantage of RaptorX-Contact over CCMpred and GaussDCA is very large, although it is decreasing along with increasing c (SI Appendix, Table S3). At c = 6, RaptorX-Contact, CCMpred, and GaussDCA have coverage ratio 0.726, 0.534, and 0.307, respectively. Of the 5 targets with MSA depth >500, RaptorX-Contact has a much larger coverage on T0969-D1 and T0975-D1 than CCMpred and GaussDCA. To exceed RaptorX-Contact’s coverage at c = 0, c = 4 is needed for CCMpred and c > 6 for GaussDCA. To reach RaptorX-Contact’s coverage at c = 1, c = 6 is needed for CCMpred. RaptorX-Contact coverage ratio is not correlated with target-training structure similarity at all (Dataset S5), which again confirms that RaptorX-Contact predicts contacts not by copying from the most similar training protein structures.
Running Time.
Our algorithm runs very fast, consisting of 3 main steps: 1) generating multiple sequence alignments and input features, 2) predicting angles and distance, and 3) folding by CNS. On average it takes minutes for the first step, seconds for the second step, and 10 min to a few hours for the third step on 20 central processing units (CPUs). By running the third step in parallel, it took in total ∼13 h to fold the 41 CAMEO targets and ∼4 h to fold the 37 CASP12 targets.
Discussion
We have shown that we can predict an interatom distance distribution well by deep ResNet and that even without extensive fragment-based conformation sampling predicted distance can be used to fold many more proteins than ever before. Our method works for both soluble and membrane proteins and the modeling accuracy is not correlated with target-training structure similarity and only weakly correlated with sequence profile similarity. For some FM targets our method indeed can predict 3D models with quality score much higher than target-training structure similarity. Our method also runs very fast, taking from 10 min to a few hours to generate 200 decoys on 20 CPUs. That is, it is now feasible to fold a protein on a personal computer equipped with a GPU card.
We do not evaluate the accuracy of secondary structure and torsion angle prediction because 1) although in CASP13 we employed deep ResNet to predict them, their accuracy is similar to what we have reported before (42, 43) and 2) secondary structure and torsion angles are much less important than distance for protein folding. Without predicted torsion angles, the 3D model quality decreases by ∼0.008 in terms of average TMscore. Nevertheless, predicted torsion angles may help reduce mirror images.
We only reported the folding results when all of the 5 types of atom pairs are used. In fact, using only Cβ–Cβ, our method can generate slightly worse 3D models than using all 5 types of atom pairs because their distance is highly correlated. Among the 5 types of atom pairs, Cβ–Cβ is the most informative. Nevertheless, using all 5 types of atom pairs can help reduce noise and may improve side-chain packing.
We have also experimented with discretizing the distance into 12 bins (i.e., bin width = 1 Å) and 52 bins (i.e., bin width = 0.25 Å). Using 25 bins and 52 bins has similar accuracy, better than using 12 bins. Instead of using a discrete representation of distance, we may predict a real-valued distance matrix by assuming that distance has a log-normal distribution (44) and revising our method to predict its mean and variance. CNS can easily take the predicted mean and variance as distance restraints to build 3D models. In the future, we will study whether 3D modeling accuracy can be further improved by using the whole real-valued distance matrix, especially for the determination of domain orientation of a multidomain protein.
In CASP13 AlphaFold also employed deep ResNet to predict interresidue distance distribution (which is similar to our work) and then converted this distribution into distance-based statistical potential for energy minimization (see refs. 27 and 29) for how to implement this). In CASP13 we employed such a distance potential for protein threading (i.e., RaptorX-TBM) but did not get a chance to integrate it into RaptorX-Contact. AlphaFold used both fragment-based sampling and fragment-free gradient descent to minimize distance-based potential and then employed Rosetta to refine models and pack side-chain atoms. By contrast, RaptorX-Contact did not have these steps, which may be the reason why RaptorX-Contact generated worse 3D models than AlphaFold. Nevertheless, AlphaFold’s performance further confirms that deep ResNet, which was first developed by us for contact/distance prediction, allows us to fold proteins without time-consuming conformation sampling. Recently an end-to-end DL method was proposed to directly predict a protein structure from sequence profile (45). The idea is unique and attractive, but rigorous test results are needed to show its effectiveness.
Materials and Methods
Feature Generation.
To ensure a fair comparison with the results in refs. 10 and 11 and the CASP12 groups, we used the same MSAs and protein features as described in refs. 10 and 11 for the CASP12 and CAMEO targets and the same MSAs and protein features as described in ref. 10 for the training proteins. That is, for each test protein we generated four MSAs by running HHblits with 3 iterations and E-value set to 0.001 and 1, respectively, to search the UniProt20 library released in November 2015 and February 2016, respectively. Since the sequence databases were created before CASP12 started in May 2016, the comparison with the CASP12 groups is fair. For CASP13, we generated MSAs (and other sequence features) using the UniClust30 library (46) created in October 2017 and the UniRef sequence database (47) created early in 2018. From each individual MSA, we derived both sequential and pairwise features. Sequential features include sequence profile and secondary structure as well as solvent accessibility predicted by RaptorX-Property (42). Pairwise features include raw and APC-corrected MI, pairwise contact potential, and interresidue coupling score generated by CCMpred (6). In summary, 1 test protein has 4 sets of input features and accordingly 4 predicted distance matrices, which are then averaged to obtain the final prediction. In CASP13, we did not make use of metagenomic sequence databases, which have been reported to be useful for some proteins (48).
Predict Secondary Structure and Torsion Angles.
We employed a 1D deep ResNet of 19 convolutional layers to predict 3-state secondary structure and backbone torsion angles and for each residue (Fig. 1). Two types of input features are used: position-specific scoring matrix generated by HHblits (49) and primary sequence represented as a 20 × L binary matrix where L is sequence length. For secondary structure, logistic regression is used in the last layer to predict the probability of 3 secondary structure types. For torsion angles we do not use logistic regression, but directly predict the below distribution:
| [1] |
In Eq. 1, are the mean, are the variance, and is the correlation. That is, our deep ResNet outputs the mean and variance of the torsion angles at each residue. We use maximum likelihood to train the network for secondary structure and angle prediction, that is, maximizing the probability (defined by logistic regression or Eq. 1) of the observed properties of our training proteins. Note that the predicted mean and variance for angles is residue-specific. Our method for angle prediction is different from many existing ones, which usually discretize angles and formulate it as a classification problem. The same set of training proteins (created in 2016) for distance prediction is used to train our DL models for secondary structure and angle prediction.
Folding by Predicted Distance, Secondary Structure, and Torsion Angles.
Given a protein to be folded, we first predict its interatom distance matrix, secondary structure, and backbone torsion angles then convert them into CNS restraints for 3D model building (30). CNS is a software program for experimental protein structure determination. Given a matrix corresponding to the distance probability distribution for each atom type, we pick 7L (L is sequence length) of the residue pairs with the highest predicted likelihood (probability) having distance < 15 Å and assume their probability of having distance >16 Å is 0. From the predicted distance probability distribution, we may estimate the mean distance and SD (denoted as m and s, respectively) of one atom pair, and then use m − s and m + s as its distance lower and upper bounds. We used the same method as CONFOLD (50) to derive hydrogen-bond restraints from predicted alpha helices. CONFOLD derived backbone torsion angles from predicted secondary structure, but we use the mean degree and variance predicted by our 1D deep ResNet as torsion angle restraints.
For each protein, we run CNS to generate 200 possible 3D decoys and then choose 5 with the least violation of distance restraints as the final models. CNS uses distance geometry to build 3D models from distance restraints and thus can generate a 3D model within seconds. CNS first builds an initial 3D model from predicted distance by heuristic embedding, which may not have physically plausible bond length and angles. Then CNS runs simulated annealing to refine the bond length and angles. For a pair of atoms, the distance violation is calculated as the absolute difference between their Euclidean distance in the 3D model and the corresponding value in the predicted distance restraint. The distance violation of a predicted 3D model is the mean violation of all of the atom pairs included in the distance restraint set.
Data Availability.
See Datasets S6 and S7 for the training proteins used in CASP13 and the 41 CAMEO hard targets, respectively. The CASP12 and CASP13 targets are available at http://predictioncenter.org/. Our stand-alone code is available at https://github.com/j3xugit/RaptorX-Contact. Our web server is available at http://raptorx.uchicago.edu/AbInitioFolding/.
Supplementary Material
Acknowledgments
This work is supported by NIH grant R01GM089753 and NSF grant DBI-1564955. I thank Dr. Sheng Wang for feature generation and Mr. Matthew Mcpartlon for proofreading.
Footnotes
The author declares no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Stand-alone code related to this paper is available at https://github.com/j3xugit/RaptorX-Contact. Our web server is available at http://raptorx.uchicago.edu/AbInitioFolding/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821309116/-/DCSupplemental.
References
- 1.Marks D. S., et al. , Protein 3D structure computed from evolutionary sequence variation. PLoS One 6, e28766 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marks D. S., Hopf T. A., Sander C., Protein structure prediction from sequence variation. Nat. Biotechnol. 30, 1072–1080 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morcos F., et al. , Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl. Acad. Sci. U.S.A. 108, E1293–E1301 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Juan D., Pazos F., Valencia A., Emerging methods in protein co-evolution. Nat. Rev. Genet. 14, 249–261 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Jones D. T., Buchan D. W., Cozzetto D., Pontil M., PSICOV: Precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics 28, 184–190 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Seemayer S., Gruber M., Söding J., CCMpred–Fast and precise prediction of protein residue-residue contacts from correlated mutations. Bioinformatics 30, 3128–3130 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamisetty H., Ovchinnikov S., Baker D., Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proc. Natl. Acad. Sci. U.S.A. 110, 15674–15679 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma J., Wang S., Wang Z., Xu J., Protein contact prediction by integrating joint evolutionary coupling analysis and supervised learning. Bioinformatics 31, 3506–3513 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weigt M., White R. A., Szurmant H., Hoch J. A., Hwa T., Identification of direct residue contacts in protein-protein interaction by message passing. Proc. Natl. Acad. Sci. U.S.A. 106, 67–72 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang S., Sun S., Li Z., Zhang R., Xu J., Accurate de novo prediction of protein contact map by ultra-deep learning model. PLoS Comput. Biol. 13, e1005324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S., Sun S., Xu J., Analysis of deep learning methods for blind protein contact prediction in CASP12. Proteins 86 (suppl. 1), 67–77 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S., Li Z., Yu Y., Xu J., Folding membrane proteins by deep transfer learning. Cell Syst. 5, 202–211.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adhikari B., Hou J., Cheng J., DNCON2: Improved protein contact prediction using two-level deep convolutional neural networks. Bioinformatics 34, 1466–1472 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaarschmidt J., Monastyrskyy B., Kryshtafovych A., Bonvin A. M. J. J., Assessment of contact predictions in CASP12: Co-evolution and deep learning coming of age. Proteins 86 (suppl. 1), 51–66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanson J., Paliwal K., Litfin T., Yang Y., Zhou Y., Accurate prediction of protein contact maps by coupling residual two-dimensional bidirectional long short-term memory with convolutional neural networks. Bioinformatics 34, 4039–4045 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Liu Y., Palmedo P., Ye Q., Berger B., Peng J., Enhancing evolutionary couplings with deep convolutional neural networks. Cell Syst. 6, 65–74.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He K., Zhang X., Ren S., Sun J., Deep residual learning for image recognition. arXiv:1512.03385 (10 December 2015).
- 18.Jones D. T., Singh T., Kosciolek T., Tetchner S., MetaPSICOV: Combining coevolution methods for accurate prediction of contacts and long range hydrogen bonding in proteins. Bioinformatics 31, 999–1006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng H., et al. , ComplexContact: A web server for inter-protein contact prediction using deep learning. Nucleic Acids Res. 46, W432–W437 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Lena P., Nagata K., Baldi P., Deep architectures for protein contact map prediction. Bioinformatics 28, 2449–2457 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eickholt J., Cheng J., Predicting protein residue-residue contacts using deep networks and boosting. Bioinformatics 28, 3066–3072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones D. T., Kandathil S. M., High precision in protein contact prediction using fully convolutional neural networks and minimal sequence features. Bioinformatics 34, 3308–3315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aszódi A., Gradwell M. J., Taylor W. R., Global fold determination from a small number of distance restraints. J. Mol. Biol. 251, 308–326 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Kloczkowski A., et al. , Distance matrix-based approach to protein structure prediction. J. Struct. Funct. Genomics 10, 67–81 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pietal M. J., Bujnicki J. M., Kozlowski L. P., GDFuzz3D: A method for protein 3D structure reconstruction from contact maps, based on a non-euclidean distance function. Bioinformatics 31, 3499–3505 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Kukic P., et al. , Toward an accurate prediction of inter-residue distances in proteins using 2D recursive neural networks. BMC Bioinformatics 15, 6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao F., Xu J., A position-specific distance-dependent statistical potential for protein structure and functional study. Structure 20, 1118–1126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z., “Knowledge-based machine learning methods for macromolecular 3D structure prediction,” PhD thesis, Toyota Technological Institute at Chicago, Chicago (2016).
- 29.Zhu J. W., Wang S., Bu D. B., Xu J. B., Protein threading using residue co-variation and deep learning. Bioinformatics 34, 263–273 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brunger A. T., Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2, 2728–2733 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Yu F., Koltun V., Multi-scale context aggregation by dilated convolutions. arXiv:1511.07122v3 [cs.CV] (30 April 2016).
- 32.Kim D. E., Chivian D., Baker D., Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 32, W526–W531 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ovchinnikov S., Park H., Kim D. E., DiMaio F., Baker D., Protein structure prediction using Rosetta in CASP12. Proteins 86 (suppl. 1), 113–121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy A., Kucukural A., Zhang Y., I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C., Mortuza S. M., He B., Wang Y., Zhang Y., Template-based and free modeling of I-TASSER and QUARK pipelines using predicted contact maps in CASP12. Proteins 86 (suppl. 1), 136–151 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S., Ma J., Peng J., Xu J., Protein structure alignment beyond spatial proximity. Sci. Rep. 3, 1448 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma J., Peng J., Wang S., Xu J., A conditional neural fields model for protein threading. Bioinformatics 28, i59–i66 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eswar N., et al. , Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 31, 3375–3380 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Söding J., Biegert A., Lupas A. N., The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song Y., et al. , High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baldassi C., et al. , Fast and accurate multivariate Gaussian modeling of protein families: Predicting residue contacts and protein-interaction partners. PLoS One 9, e92721 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S., Peng J., Ma J. Z., Xu J. B., Protein secondary structure prediction using deep convolutional neural fields. Sci. Rep. 6, 18962 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao Y. J., Wang S., Deng M. H., Xu J. B., RaptorX-angle: Real-value prediction of protein backbone dihedral angles through a hybrid method of clustering and deep learning. BMC Bioinformatics 19 (suppl. 4), 100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rieping W., Habeck M., Nilges M., Modeling errors in NOE data with a log-normal distribution improves the quality of NMR structures. J. Am. Chem. Soc. 127, 16026–16027 (2005). [DOI] [PubMed] [Google Scholar]
- 45.AlQuraishi M., End-to-end differentiable learning of protein structure. Cell Systems 8, 292–301.es (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mirdita M., et al. , Uniclust databases of clustered and deeply annotated protein sequences and alignments. Nucleic Acids Res. 45, D170–D176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzek B. E., Wang Y., Huang H., McGarvey P. B., Wu C. H.; UniProt Consortium , UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31, 926–932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ovchinnikov S., et al. , Protein structure determination using metagenome sequence data. Science 355, 294–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Remmert M., Biegert A., Hauser A., Söding J., HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 9, 173–175 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Adhikari B., Bhattacharya D., Cao R., Cheng J., CONFOLD: Residue-residue contact-guided ab initio protein folding. Proteins 83, 1436–1449 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
See Datasets S6 and S7 for the training proteins used in CASP13 and the 41 CAMEO hard targets, respectively. The CASP12 and CASP13 targets are available at http://predictioncenter.org/. Our stand-alone code is available at https://github.com/j3xugit/RaptorX-Contact. Our web server is available at http://raptorx.uchicago.edu/AbInitioFolding/.