Abstract
Background
Genome editing tools are important for functional genomics research and biotechnology applications. Recently, the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein-9 (Cas9) system for gene knockout has emerged as the most effective genome-editing tool. It has previously been reported that, in rice plants, knockdown of the Os8N3 gene resulted in enhanced resistance to Xanthomonas oryzae pv. oryzae (Xoo), while displaying abnormal pollen development.
Results
The CRISPR/Cas9 system was employed to knockout rice Os8N3, in order to confer enhanced resistance to Xoo. Analysis of the genotypes and edited Os8N3 in T0, T1, T2, and T3 transgenic rice plants showed that the mutations were transmitted to subsequent generations, and homozygous mutants displayed significantly enhanced resistance to Xoo. Stable transmission of CRISPR/Cas9-mediated Os8N3 gene editing without the transferred DNA (T-DNA) was confirmed by segregation in the T1 generation. With respect to many investigated agronomic traits including pollen development, there was no significant difference between homozygous mutants and non-transgenic control plants under greenhouse growth conditions.
Conclusion
Data from this study indicate that the CRISPR/Cas9-mediated Os8N3 edition can be successfully employed for non-transgenic crop improvements.
Electronic supplementary material
The online version of this article (10.1186/s12284-019-0325-7) contains supplementary material, which is available to authorized users.
Keywords: CRISPR/Cas9, Disease resistance, Os8N3, Rice, xa13, Xanthomonas oryzae pv. oryzae
Background
Rice (Oryza sativa L.) is one of the most important cereal crops in the world, directly feeding more people than any other crop. Bacterial blight, caused by Xanthomonas oryzae pv. oryzae (Xoo), is a prevalent and destructive rice disease that causes serious production loss worldwide (Zhang and Wang 2013). Enhancing rice plants’ resistance to Xoo is known to be an economical and effective approach for managing rice bacterial blight.
Xoo pathogenicity depends on a specific class of virulence factors, called transcription activator-like (TAL) effectors, which mimic plant transcriptional activators (Hutin et al. 2015; Blanvillain-Baufume et al. 2017). The TAL effectors target the host nucleus, where they bind to specific promoter elements of the plant genes and activate their expression, reprogramming the plant transcriptome (Schornack et al. 2013). The genomes of Xanthomonas strains typically contain highly variable numbers of TAL effectors between Asian Xoo (15–26), African Xoo (8–10), and North-American Xoo (0) (Erkes et al. 2017). The rice genes targeted by TAL effectors have been identified as host disease-susceptibility genes, acting as major susceptibility factors during rice and Xoo interactions. In some cases, DNA polymorphisms in the so-called TAL effector binding elements (EBEs), located at the promoter region of the susceptibility gene, lead to no development of the disease (Yang et al. 2006; Hutin et al. 2015). Rice Os8N3 (also known as OsSWEET11), which belongs to the Sugar Will Eventually be Exported Transporters (SWEET) family of sugar transporters, represents one of the susceptibility genes induced by TAL effectors (Yang et al. 2006; Chen 2014). The expression of Os8N3 is induced by strains of Xoo carrying pthXo1, which encodes the TAL effector PthXo1 (Yang et al. 2006; Yuan et al. 2009). PthXo1 from Xoo strain PXO99 directly activates Os8N3 through recognition of TAL EBEs located at the promoter region of Os8N3 (Romer et al. 2010). The recessive resistance gene xa13 occurs as a series of natural alleles of the susceptibility gene Os8N3 (Yang et al. 2006; Yuan et al. 2009). Although it has not been clearly demonstrated, Os8N3 is believed to remove toxic copper from xylem vessels where Xoo multiplies and spreads (Yuan et al. 2010), and make nutrients easily available to Xoo for its growth and virulence to cause disease (Chen et al. 2010; Chen et al. 2012).
Genome editing technologies enable precise modification of DNA sequences in vivo and promise a novel revolution in crop improvement (Sun et al. 2016; Feng et al. 2013). The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein-9 (Cas9) system has revolutionized genome editing and become widely popular because of its specificity, simplicity, and versatility. It allows targeted genome editing in organisms guided by a customizable small noncoding RNA called single guide RNA (sgRNA). Once susceptibility genes targeted by TAL effectors have been identified, the CRISPR/Cas9-mediated genome editing strategy can be employed to create a target mutation in the susceptibility genes. Although it was not edited by the CRISPR/Cas9, Os11N3 (also known as OsSWEET14), the susceptibility gene targeted by AvrXa7 and PthXo3, has been edited by Transcription Activator-Like Effector Nucleases (TALENs) to create bacterial blight-resistant rice through disrupting the EBE site in the promoter region (Li et al. 2012; Blanvillain-Baufume et al. 2017). It can also be applied to negative regulators of disease resistance that have been studied for the last decades (Grand et al. 2012; Wang et al. 2015; Chern et al. 2005). However, to date, only a few examples of improvement of disease resistance using the CRISPR/Cas9 approach have been reported (Wang et al. 2016; Pyott et al. 2016; Peng et al. 2017). For Os8N3, studies on its knockdown rice plants using the gene silencing system and promoter mutations reported that they showed enhanced resistance to Xoo while displaying abnormal pollen development (Yang et al. 2006; Chu et al. 2006). Recently, CRISPR/Cas9-mediated knockout of Os8N3 displayed decreased sucrose concentration in the embryo sacs and defective grain filling, suggesting that Os8N3 plays important role in sucrose transport during early stage of rice grain filling (Ma et al. 2017; Yang et al. 2018).
Here, the CRISPR/Cas9-target mutagenesis of Os8N3 in Kitaake, a Japonica rice cultivar, is reported. The homozygous mutant lines carrying edited Os8N3 displayed significantly enhanced resistance to Xoo with normal pollen development. It was possible to select resistant mutant lines not containing the transferred DNA (T-DNA) by segregation in the T1 generation.
Results
Os8N3 in the rice cultivar Kitaake
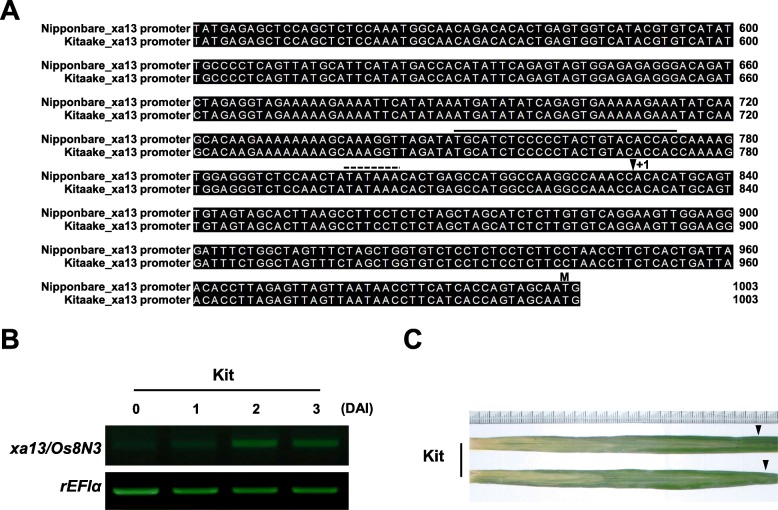
Os8N3 was originally isolated as a susceptibility gene from the rice cultivar Nipponbare (Yang et al. 2006) and later, the EBE in its promoter element bound and activated by TAL effector PthXo1 of PXO99 was determined experimentally (Romer et al. 2010). In this study, rice cultivar Kitaake was investigated to see if it also carries the EBE sequence in the Os8N3 promoter region. Using the Kitaake database (Li et al. 2017), the promoter sequence of the Os8N3 gene, ranging from − 1000 bp to − 1 bp relative to the ATG start codon, was analyzed (Fig. 1a). The putative TATA box (TATAAA) is located at − 32 upstream of the transcription start site (+ 1). The promoter region including PthXo1 EBE (TGCATCTCCCCCTACTGTACACCAC), ranging from − 80 bp to − 56 bp upstream of the transcription start site, displayed 100% identity to Nipponbare (Yang et al. 2006). After inoculation with strain PXO99, Kitaake displayed strong induction of Os8N3 two days after inoculation (DAI) (Fig. 1b) and long water-soaked lesions (approximately 13–14 cm) 12 DAI (Fig. 1c). These results suggest that Kitaake carries a functional susceptible gene Os8N3, whose expression is induced by PXO99 possessing the TAL effector PthXo1.
Fig. 1.
Os8N3 is a susceptibility gene for Xoo strain PXO99 in rice cultivar Kitaake. a Promoters containing a PthXo1 EBE (upper line) from Nipponbare and Kitaake displayed 100% identity to each other. The putative TATA box is shown by a dashed line. The transcription start site is represented by a vertical arrowhead noted as + 1. The translational initiating ATG codon is shown as ‘M’. b Expression of Os8N3 is elevated after inoculation with Xoo strain PXO99 in Kitaake. Rice elongation factor 1α (rEF1α) was used as an internal control. c Kitaake exhibited a susceptible phenotype with long water-soaked lesions after inoculation with PXO99. The lesions were photographed 12 days after inoculation (DAI) and arrowheads indicated the end of the lesion
CRISPR/Cas9 design for xa13/Os8N3 editing
In monocot plants, the rice U3 small nuclear RNA promoter (OsU3) is generally used to express sgRNA (Belhaj et al. 2013). Recently, the efficiency of mutations targeted by sgRNAs driven by different small nuclear RNA promoters including OsU3, OsU6a, OsU6b, and OsU6c, were compared in an Indica cultivar 93–11 (Ma et al. 2015b). OsU6a was slightly more efficient in driving genome editing than the other promoters. It has also been reported that U6 promoters derived from the target plants function better than heterologous U6 promoters (Sun et al. 2015). Therefore, it was decided to use the OsU6a promoter isolated from the Japonica cultivar Kitaake. The OsU6a promoter amplified from Kitaake contains five single-nucleotide substitutions and one 5-bp deletion compared with one from Indica cultivar 93–11 (Additional file 1: Figure S1). The Arabidopsis U6 promoter in the CRISPR/Cas9 vector, pHAtC (Kim et al. 2016), was replaced with the Kitaake OsU6a promoter, and the resulting OsU6a::pHAtC was used for rice CRISPR/Cas9-mediated target mutagenesis.
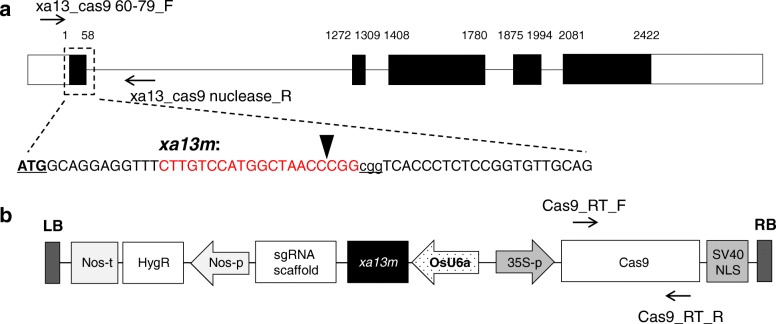
To design a CRISPR/Cas9 that targets the Os8N3 gene, a 20-bp nucleotide sequence (xa13m) in the first exon of Os8N3 was chosen as the target site (Fig. 2a). The xa13m targeting sequence and protospacer adjacent motif (PAM) sequence are represented in red and in underlined lower-case letters, respectively. The predicted Cas9 cleavage site (vertical arrowhead) in the coding region of the gene was 31 bp downstream from the ATG initiation codon. The recombinant binary plasmid, OsU6a::xa13m-sgRNA/pHAtC, carrying xa13m-sgRNA targeting the Os8N3 gene under the control of the OsU6a promoter, was then constructed based on the OsU6a::pHAtC (Fig. 2b).
Fig. 2.
Schematic representation of CRISPR/Cas9-mediated targeted mutagenesis in the rice Os8N3 gene. a Schematic diagram of Os8N3 gene and xa13m targeting sequence. Rice Os8N3 contains five exons, represented by black rectangles, and the untranslated region portion, represented by white rectangles. The enlarged area indicated by the black broken line shows the coding sequence and position of the first exon of Os8N3. The 20-bp sgRNA targeting sequence (xa13m) and protospacer adjacent motif (PAM) sequence are shown in red and in underlined lower-case letters, respectively. The vertical arrowhead indicates an expected cleavage site. The underlined bold ATG indicates a translation initiation codon. b T-DNA region of the recombinant OsU6a::xa13m-sgRNA/pHAtC vector carrying xa13m-sgRNA under the control of the OsU6a promoter. Expression of Cas9 is driven by the Cauliflower mosaic virus 35S (CaMV35S) promoter; expression of the xa13m-sgRNA is driven by the OsU6a promoter; expression of hygromycin (HPT) is driven by the nopalin synthase (NOS) promoter; NLS: nuclear localization signal of Simian virus 40 (SV40) large T antigen; nos-t: gene terminator; LB and RB: left and right border, respectively. Primers used in the PCR are indicated by black arrows
CRISPR/Cas9-mediated targeted mutagenesis of xa13/Os8N3
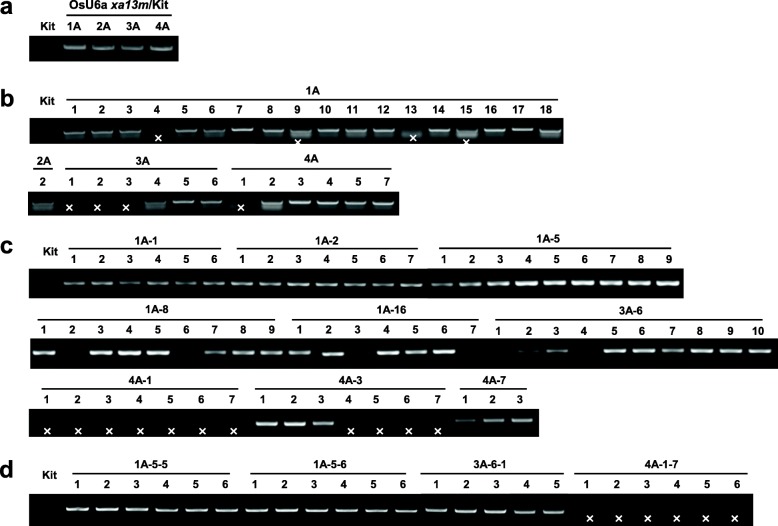
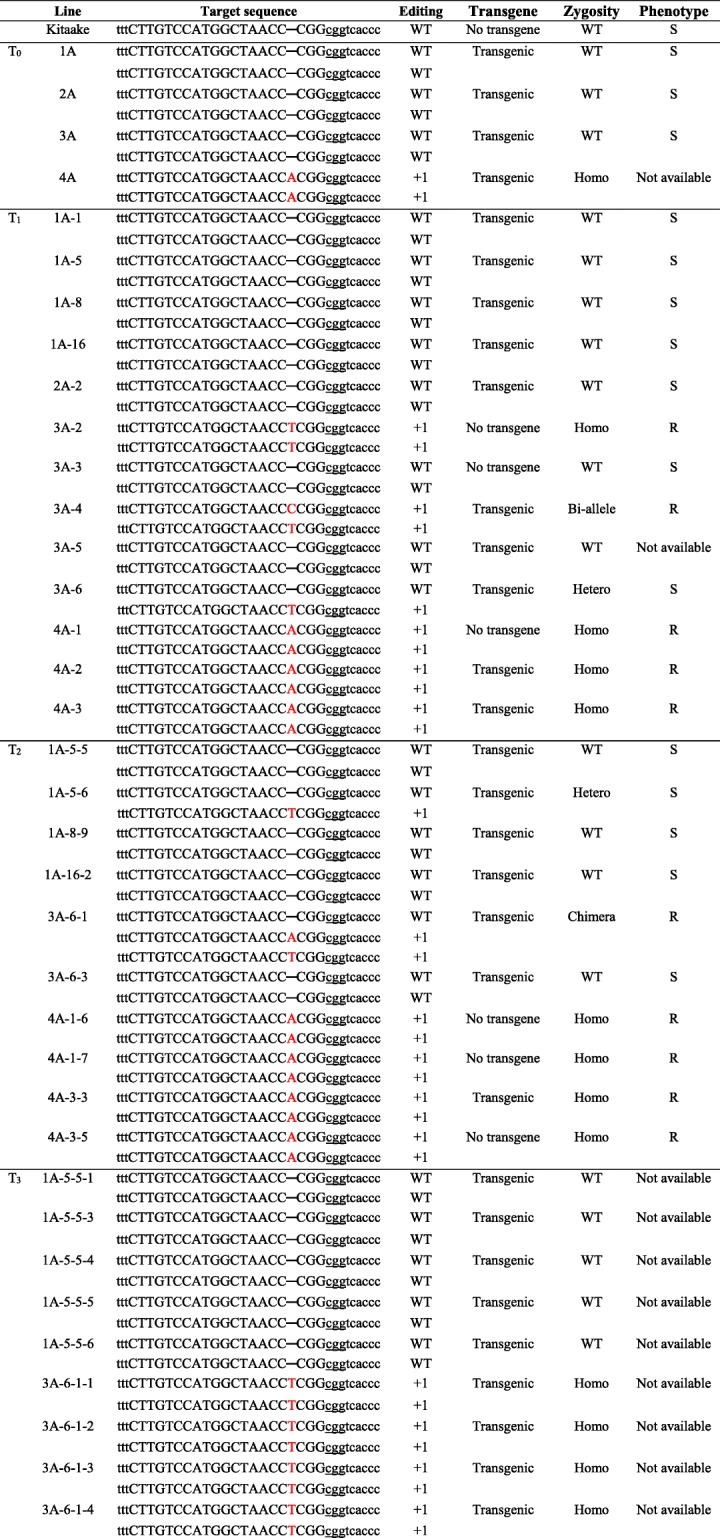
After Kitaake was transformed with OsU6a::xa13m-sgRNA/pHAtC using Agrobacterium-mediated transformation, four independent transgenic Kitaake plants (OsU6a xa13m/Kit T0, 1A, 2A, 3A, and 4A) were generated. The putative transgenic plants were subjected to polymerase chain reaction (PCR)-based selection using the Cas9-specific primers, Cas9_RT_F and Cas9_RT-R (Fig. 2b), and all of them generated a Cas9-specific 400-bp amplicon (Fig. 3a). To further investigate CRISPR/Cas9-targeted mutagenesis of Os8N3, the target-containing amplicons obtained from all PCR-positive transgenic plants were directly sequenced and analyzed by decoding via the Degenerate Sequence Decoding method (Liu et al. 2015; Ma et al. 2015a). Rice plants are diploid with two copies of each gene, one copy on each chromosome of a chromosome pair. Therefore, when CRISPR/Cas9 is inserted into the genome and begins to function, one or both copies of the target gene Os8N3 can be cleaved and mutated, generating five possible genotypes in the transgenic plants: homozygote, biallele, heterozygote, chimera, and wild type (WT). In four T0 transgenic plants, there was only one homozygous mutation, 1-bp insertion (+A), in 4A, whereas no target sequence changes could be detected in the other plants (T0 in Table 1 and Additional file 2: Figure S2).
Fig. 3.
Generation of transgenic rice plants carrying the Cas9 transgene with a sgRNA targeting the Os8N3 gene. Genotyping was performed using the specific primers for Cas9, Cas9_RT_F and Cas9_RT_R (see Fig. 2b), from four independently transformed plants and their progenies (OsU6a xa13m/Kit T0, T1, T2, and T3 generations). Genomic DNAs were extracted from Kit (Kitaake) and OsU6a xa13m/Kit T0 (a), T1 (b), T2 (c), and T3 (d). ‘ × ’ indicates PCR negative
Table 1.
Transmission and segregation of CRISPR/Cas9-mediated target mutagenesis from T0, T1, T2, and T3 of the OsU6a xa13m/Kit transgenic plant. The recovered mutated alleles of the xa13/Os8N3 gene in the OsU6a xa13m/Kit transgenic plant are shown below the Kitaake sequence. Nucleotide sequences at the target sites are shown in black capital letters and black dashes. PAM motifs are underlined. Red capital letters indicate the inserted nucleotide. The genotype of the mutation is indicated at the right of each sequence. WT indicates the nucleotide sequences identical to the Os8N3 gene in Kitaake plants. “+” indicates the insertion of the indicated number of nucleotides. No transgene: PCR negative for Cas9 gene; Transgenic: PCR positive for Cas9 gene; S: susceptible to PXO99; R: resistant to PXO99; Not available: inoculation data are not available
Inheritance of Os8N3 mutations and enhanced resistance to Xoo
To determine if and how the CRISPR/Cas9-targeted mutagenesis of Os8N3 by OsU6a::xa13m-sgRNA/pHAtC was transmitted to the next generation, all OsU6a xa13m/Kit T0 transgenic plants were self-pollinated and the targeted Os8N3 of some T1 transgenic plants was directly sequenced and analyzed (Fig. 3b, Table 1, and Additional file 3: Figure S3). The homozygous mutated T0 line (4A) produced homozygous mutated T1 progeny (4A-1, 4A-2, and 4A-3) and did not display additional different mutations. There was no mutation observed in the sequenced T1 progenies of the WT 1A, and 2A lines. However, new targeted sequence changes were detected in the T1 progeny of the WT 3A line. Previously, sequencing results indicated a putative WT genotype of the targeted Os8N3 in the T0 3A line, whereas three (3A-2, 3A-4, and 3A-6) out of the five sequenced T1 progenies of the WT 3A line displayed a 1-bp insertion (Table 1): 3A-2 was homozygous; 3A-4 was bi-allelic; and 3A-6 was heterozygous.
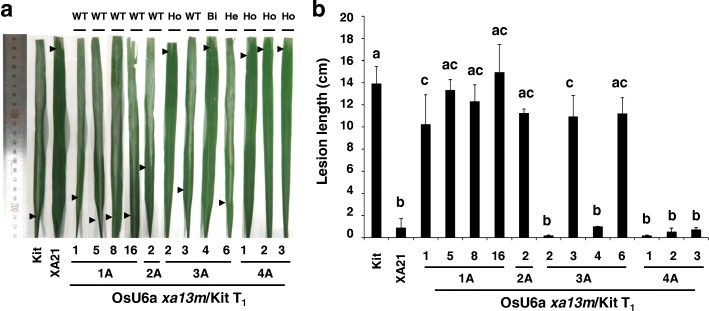
To characterize the bacterial blight resistance phenotype of the mutant lines, T1 lines (progeny of OsU6a xa13m/Kit 1A, 2A, 3A, and 4A) with different types of allelic mutations were inoculated with PXO99 at the eight-week stage (Fig. 4a). Kitaake and transgenic Kitaake carrying Xa21 (XA21), driven by the ubiquitin promoter, were used as the susceptible and resistant control for PXO99, respectively (Park et al. 2010). As expected, while the XA21 plant was highly resistant, displaying short lesions, the inoculated leaves of the Kitaake plants developed long water-soaked lesions typical of bacterial blight disease. Homozygous (OsU6a xa13m/Kit 3A-2, 4A-1, 4A-2, and 4A-3) and bi-allelic (3A-4) xa13 mutant plants displayed a robust resistance phenotype compared with heterozygous (3A-6) mutant and Kitaake control plants (T1 in Table 1 and Fig. 4a). The differences were further evaluated by quantification of the lesion lengths and significance analysis using Tukey’s HSD test (Fig. 4b). Homozygous and bi-allelic mutant plants displaying a resistance phenotype showed no significant differences in lesion lengths compared with the XA21 plants. These results indicated that the homozygous and bi-allelic mutant lines were significantly different from Kitaake and heterozygous mutant plants, and that CRISPR/Cas9-mediated mutagenesis in both Os8N3 alleles conferred robust resistance to PXO99.
Fig. 4.
CRISPR/Cas9-mediated mutagenesis in both Os8N3 alleles conferred enhanced resistance to Xoo. a Bacterial blight resistance phenotypes of the xa13 mutant rice lines (T1). Rice plants 12 DAI with Xoo. From left to right: Kitaake (Kit), transgenic line (XA21, 7A-8) carrying Xa21 driven by the ubiquitin promoter, and transgenic lines (OsU6a xa13m/Kit, T1) carrying the OsU6a::xa13m-sgRNA/pHAtC construct. Arrowheads indicated the end of the lesion. WT; wild type: Ho; homozygous: Bi; bi-allelic: He; Heterozygous. b Lesion lengths measured 12 DAI in Kitaake, XA21, and OsU6a xa13m/Kit T1. Error bars in the graph represent standard error of at least three leaves from each plant. Letters indicate a significant difference at P < 0.050 by Tukey’s HSD test
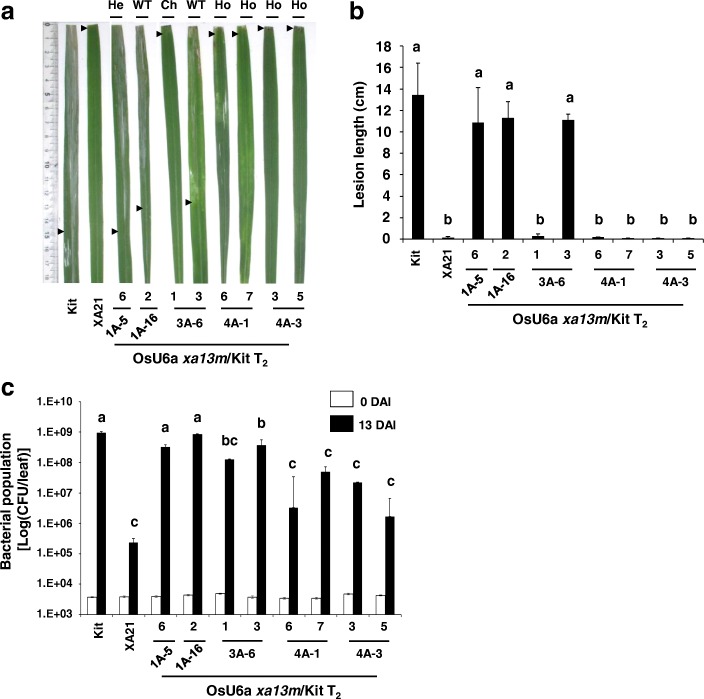
To further investigate the inheritance of targeted mutations in later generations, the genotypes of several OsU6a xa13m/Kit T2 plants were analyzed and inoculated with PXO99. New allelic mutation was detected in the T2 progeny of WT 1A-5. Although all sequenced T0 and T1 generations of the 1A line carry WT Os8N3, T2 progeny (1A-5-6) of the 1A line displayed a heterozygous 1-bp insertion (+T) mutation (Table 1 and Additional file 4: Figure S4). Heterozygous mutated 3A-6 (+T) produced chimera 3A-6-1 with three distinct alleles detected at the target site, displaying additional different mutations (+A). All T1 plants derived from the homozygous T0 mutant plant (4A) and T2 plants derived from homozygous T1 mutant plants (4A-1 and 4A-3) were homozygous for the same mutations (Table 1). All homozygous mutant lines (4A-1-6, 4A-1-7, 4A-3-3, and 4A-3-5) and chimera (3A-6-1) displayed significantly short lesion lengths (Fig. 5a and b) and low bacterial populations compared with the heterozygous mutant (1A-5–6) and Kitaake plants (Fig. 5c). These results indicate that the mutations in these homozygous mutant lines and enhanced resistance to PXO99 were stably transmitted to the next generation.
Fig. 5.
Homozygous mutants in both Os8N3 alleles displayed enhanced resistance to Xoo. Transgenic Kitaake plants targeting xa13 (OsU6a xa13m/Kit T2) display enhanced resistance to Xoo. a Inoculation results for mutant rice lines 12 DAI with Xoo. From left to right: Kitaake (Kit), transgenic line (XA21, 7A-8) carrying Xa21 driven by the ubiquitin promoter, and transgenic lines (OsU6a xa13m/Kit, T2) carrying the OsU6a::xa13m-sgRNA/pHAtc construct. Arrowheads indicated the end of the lesion. He; Heterozygous; WT; wild type: Ch; chimeric: Ho; homozygous. b Lesion lengths measured 12 DAI in Kitaake, XA21, and OsU6a xa13m/Kit T2. Error bars in the graph represent standard error of at least three leaves from each plant. Letters indicate a significant difference at P < 0.050 by Tukey’s HSD test. c Bacterial population in Kitaake, XA21, and OsU6a xa13m/Kit T2 plants 0 and 12 DAI, determined by the number of CFU per inoculated leaf. Error bars represent standard deviation from at least three technical replicates. Letters indicate a significant difference at P < 0.050 by Tukey’s HSD test
Main agronomic traits in xa13 mutants
To determine whether mutations in the Os8N3 gene affect agronomic traits, two independent homozygous mutant lines (T3) were analyzed by measuring their plant height, flag leaf length/width, the number of productive panicles, and panicle length (Table 2, Additional file 5: Figure S5 and Additional file 6: Figure S6). Tukey’s HSD test indicated that the mutant lines displayed no significant difference to Kitaake, in terms of the investigated agronomic traits, under our greenhouse conditions.
Table 2.
Analysis of the agronomic traits of T3 mutant lines
| Plant height (cm) | Flag leaf length (cm) | Flag leaf width (mm) | No. of productive panicles | Panicle length (cm) | |
|---|---|---|---|---|---|
| Kitaake | 69.8 ± 4.1a | 27.8 ± 4.9a | 11.7 ± 0.9a | 3.0 ± 0.0a | 11.1 ± 2.1a |
| Progeny of 3A-6-1 | 65.6 ± 6.4a | 26.9 ± 4.0a | 12.7 ± 0.5a | 3.0 ± 0.0a | 12.1 ± 1.4a |
| Progeny of 4A-1-7 | 65.7 ± 7.9a | 29.1 ± 4.8a | 12.0 ± 0.4a | 2.5 ± 1.1a | 12.0 ± 2.2a |
The results shown are from more than three homozygous mutants of each mutant line, and are represented as the mean ± SE. The values marked with the same letter (a) are non-significantly different (P < 0.050, Tukey’s HSD test)
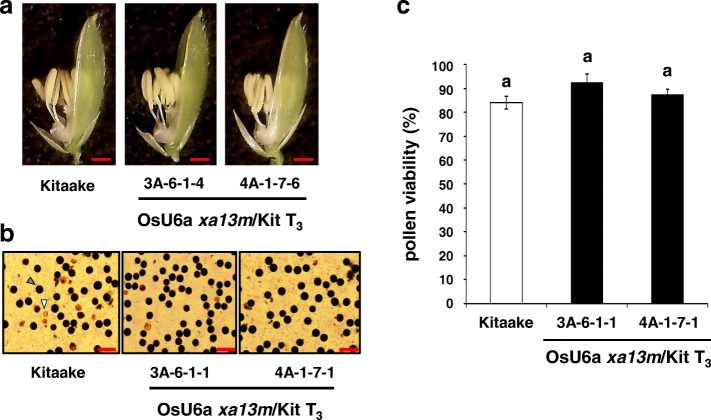
Previously, Os8N3 knockdown transgenic plants displayed abnormal pollen development (Yang et al. 2006; Chu et al. 2006). To investigate whether Os8N3 knockout mutations affect pollen development, their pollen developments were assessed (Fig. 6). The phenotypical analysis showed that two independent homozygous T3 mutant lines (3A-6-1-4 and 4A-1-7-6) exhibited normal golden yellow anthers (Fig. 6a). In addition, pollen grains from Kitaake and two independent homozygous T3 mutant lines (3A-6-1-1 and 4A-1-7-1) were stained with iodine potassium iodide (I2-KI) (Fig. 6b). Dark-stained pollen grains (black in color) were considered viable and those that were lightly stained (yellow in color) were considered sterile. Homozygous mutants (3A-6-1-1 and 4A-1-7-1) displayed similar pollen viabilities to Kitaake, under our greenhouse conditions (Fig. 6c). The seed-setting rates and grain fillings were further analyzed in the Os8N3 knockout mutant lines (Additional file 7: Figure S7). Although, under greenhouse conditions, the caryopses from two independent homozygous mutants (3A-6-1-1 and 4A-1-7-1) were slightly wrinkled as they matured (Additional file 7: Figure S7c), no significant alteration in the seed-setting rate was observed between progeny of two homozygous mutants (3A-6-1 and 4A-1-7) and Kitaake plants (Additional file 7: Figure S7a and S7b).
Fig. 6.
Pollen viability of the homozygous xa13 mutants. a Anthers in mature spikelets of Kitaake, homozygous mutant (T3, 3A-6-1-4), and homozygous mutant (T3, 4A-1-7-6). Scale bars, 1 mm. b Representative images of pollen viability tests from Kitaake and homozygous mutants (T3, 3A-6-1-1 and 4A-1-7-1). Viable pollen grains are stained dark (gray arrow) and sterile pollen grains are stained light yellow (white arrow). Scale bars, 100 μm. c Statistical analysis of pollen viability of Kitaake, homozygous mutants (T3, 3A-6-1-1 and 4A-1-7-1) lines. Pollen viability percentage was calculated relative to the total pollen counted in three microscopic images
Selection of transgene-free mutant rice lines
To select rice plants harboring the mutation in Os8N3 but without the T-DNA of the OsU6a::xa13m-sgRNA/pHAtC construct, PCR and phenotypic analysis for the OsU6a xa13m/Kit T0, T1, and T2 plants was performed. Thirty-one segregating T1 plants were analyzed and six of them (19.35%) did not generate a Cas9-specific amplicon from the T-DNA (Fig. 3b). Similarly, PCR analysis also failed to detect the T-DNA in 11 out of the 65 segregating T2 plants (16.92%) derived from nine T1 plants (1A-1, 1A-2, 1A-5, 1A-8, 1A-16, 3A-6, 4A-1, 4A-3, and 4A-7) (Fig. 3c). Notably, the 4A-1 plant was a Cas9-free homozygous mutant harboring the desired xa13/Os8N3 modifications (Fig. 3b and Fig. 4, and Additional file 3: Figure S3 and Additional file 4: Figure S4). None of the seven T2 plants derived from the T1 mutant plant 4A-1 generated the Cas9-specific amplicon (Fig. 3c). Two (4A-1-6 and 4A-1-7) out of the seven carried a 1-bp insertion (+A) and displayed significantly enhanced resistance to PXO99 (Fig. 5), which has also been observed in their parent (4A-1) (Fig. 4). The T3 plant (4A-1-7-1) not generating the Cas9-specific amplicon carried the same Os8N3 modification observed in the T2 mutant plant 4A-1-7 (Fig. 3d and Additional file 4: Figure S4 and Additional file 5: Figure S5). These results indicate that T-DNA-free mutant plants carrying the desired gene modifications can be acquired through genetic segregation in T1, T2, and T3 generations.
Discussion
The CRISPR/Cas9 system has been widely used to provide new avenues in crop improvements in rice, tomato, wheat, and maize (Xu et al. 2015; Feng et al. 2013; Wang et al. 2016; Ito et al. 2015; Wang et al. 2014; Zhou et al. 2014). In this study, OsU6a::pHAtC, which replaced the Arabidopsis U6 promoter in the pHAtC vector (Kim et al. 2016) with the OsU6a promoter of Kitaake, was constructed for rice CRISPR/Cas9-mediated target mutagenesis. Using the OsU6a::pHAtC, targeted mutagenesis in the recessive resistance gene, Os8N3, was generated.
One xa13 mutant line 4A (T0) from four independent transgenic OsU6a xa13m/Kit plants carrying OsU6a::xa13m-sgRNA/pHAtC was obtained. However, new targeted sequence changes were continuously detected in the transgenic OsU6a xa13m/Kit plants in subsequent generations. For example, two additional independent mutant lines (progenies of 3A and 1A-5) were identified in the T1 and T2 generations, respectively. Except for line 2A, which was lost in T1, all available lines in T2 were successfully mutated at the target sequence. Because the CRISPR/Cas9 system has been shown to be active in heterozygous and chimeric plants (Xu et al. 2015; Zhou et al. 2014), it is possible for the WT allele to be continuously modified in subsequent generations. Therefore, non-mutated transgenic plants, in which the OsU6a::xa13m-sgRNA/pHAtC construct remained active, continually cleaved the target site for generations, resulting in new mutations. Multiple mutations were also detected at the target site in the T2 mutant plant 3A-6-1. Because 3A-6 was heterozygous, the presence of a chimeric mutation may result from delayed cleavage in the primary embryogenic cell of 3A-6-1. This chimeric mutation by the CRISPR/Cas9 system is likely a common phenomenon and has been reported in many plant species including rice (Xu et al. 2015; Feng et al. 2013; Wang et al. 2016), Arabidopsis (Feng et al. 2014), and tomato (Ito et al. 2015).
Regarding all examined agronomic traits, there was no significant difference between T3 homozygous mutants and Kitaake plants under greenhouse growth conditions. The homozygous mutant plants had a similar height, flag leaf length and width, number of productive panicles, panicle length, and pollen viability to Kitaake plants. It has been previously reported that Os8N3 is expressed at a high level in panicles and anthers during pollen development (Chu et al. 2006; Yang et al. 2006). Consistent with these observations, although detailed molecular mechanisms have not been elucidated, Os8N3-silenced rice plants displayed reduced fertility, and most pollen grains were defective (Chu et al. 2006; Yang et al. 2006). Therefore, Os8N3, conferring disease resistance by expressional loss-of-function in rice, has been considered an essential constituent for pollen development. However, in this study, homozygous mutants in both Os8N3 alleles were generated, and the mutations were stably transmitted to later generations, T3. The homozygous T3 mutant plants had normal pollen development, and most pollen grains were well preserved, in comparison with ones from Kitaake plants.
Thus far, it has been believed that Os8N3 plays roles in both copper and sugar transport, indicating its complex function in copper/sugar metabolism and signaling (Chen et al. 2010; Chen 2014; Yuan et al. 2010). However, no one dissected the molecular connection between Xoo resistance by copper/sugar metabolism and pollen development. Among the different in vivo functions of xa13/Os8N3, knockout mutation, in particular, displayed enhanced resistance against Xoo without affecting pollen development. It is not yet understood why OsU6a xa13m/Kit mutant lines did not display the sterile phenotype previously observed in Os8N3-knockdown rice plants (Chu et al. 2006; Yang et al. 2006). Because frameshift mutations of Os8N3 in OsU6a xa13m/Kit lines are located at the very beginning of the Os8N3 polypeptide, it is very unlikely that the mutated polypeptide is functional. Lack of a functional Os8N3 protein in the mutant lines was also supported by a robust resistant phenotype of the homozygous mutant lines, but not heterozygous or Kitaake plants. Therefore, it is possible that there is a novel gene genetically compensating essential pollen development directly or indirectly in homozygous OsU6a xa13m/Kit mutant lines. Genetic compensation was recently proposed to explain increasing numbers of studies revealing phenotypic differences between knockouts and knockdowns in plants (Gao et al. 2015; Braun et al. 2008; Chen et al. 2014) and animals (Young et al. 2009; De Souza et al. 2006; Daude et al. 2012; McJunkin et al. 2011; Law and Sargent 2014; Evers et al. 2016; Karakas et al. 2007; Morgens et al. 2016; Kok et al. 2015; Rossi et al. 2015). For example, similar to Os8N3, there have been studies on Arabidopsis auxin-binding protein 1 (ABP1) that revealed phenotypic differences between knockouts and knockdowns (Gao et al. 2015; Braun et al. 2008; Chen et al. 2014). Inducible abp1 knockdown lines showed defects in shoot and root growth, cell remodeling, or clathrin-mediated endocytosis of PIN auxin efflux carriers (Braun et al. 2008; Paque et al. 2014; Robert et al. 2010). However, abp1 knockout mutants generated by CRISPR/Cas9 are indistinguishable from wild type plants at every developmental stage analyzed (Gao et al. 2015). Although one possible explanation for the difference is off-target effects of ABP1 antisense RNA, it is not yet understood how independent abp1 knockdown lines, which generate fundamentally different approaches for functional down-regulation of the ABP1 gene, display indistinguishable morphological defect phenotypes (Michalko et al. 2016). Recently, genetic compensation was studied in depth on zebrafish (Rossi et al. 2015). While knockdown of zebrafish EGF-like domain 7 (egfl7), an endothelial extracellular matrix gene, leads to severe vascular defects, most egfl7 mutants display no obvious defects (Rossi et al. 2015). Elastin microfibril interfacer (Emilin) genes were proposed as compensating genes in the edgl7 knockout mutants (Rossi et al. 2015). Supporting this hypothesis, Os8N3 mutants showed increased expressions of several SWEET genes such as OsSWEET3a, OsSWEET6b, OsSWEET13, and OsSWEET15 (Ma et al. 2017; Yang et al. 2018) and double mutants of Os8N3 and OsSWEET15 displayed much more wrinkled grain morphology, compared with single Os8N3 mutant (Yang et al. 2018). These reports suggest that some of SWEET genes are able to at least partially compensate for the lack of Os8N3. Currently, we are trying to identify candidate genes that compensate for xa13/Os8N3 in the pollen development pathway without affecting Xoo resistance in homozygous mutant lines.
Conclusions
In summary, the CRISPR/Cas9 system was highly efficient in generating Os8N3 gene editing in rice. Mutant lines harboring the desired modification in Os8N3 but without the T-DNA of the OsU6a::xa13m-sgRNA/pHAtC were obtained. T-DNA-free homozygous mutant lines displayed significantly enhanced resistance to Xoo and normal pollen development. This study provides a successful example of improving bacterial blast resistance using CRISPR/Cas9 technology.
Materials and methods
Plant and pathogen materials
Rice cultivar Kitaake (Oryza sativa L. ssp. Japonica) was generously provided by Prof. Pamela Ronald (University of California Davis, USA). Rice plants in this study were maintained in the greenhouse facility at Sejong University in Korea. Xoo strain PXO99 was used in this study. PXO99 was cultured in peptone sucrose agar media (PSA: peptone 10.0 g/L, sucrose 1.0 g/L, L-glutamic acid 1.0 g/L, and agar 16.0 g/L) containing 15.0 mg/L cephalexin at 28 °C for two days (Bai et al. 2000).
Vector construction
The Gateway™ destination vector, pHAtC binary vector (Kim et al. 2016), was used to construct OsU6a::pHAtC carrying the OsU6a promoter to express sgRNA. A 472-bp DNA fragment containing the OsU6a promoter (Ma et al. 2015c) was amplified from the genomic DNA of Kitaake using primers, EcoRI_OsU6a_F (5′-GGAATTCTTTTTTCCTGTAGTTTTCCCAC-3′) and XhoI_OsU6a_R (5′-GCTCGAGACACCTGCCTCCAATCCGGCAGCCAAGCCAGCACCC-3′). The PCR product was cloned into the pGEM®-T Easy Vector according to the manufacturer’s instructions (Promega, USA), and the insert was confirmed by Sanger sequencing. The OsU6a promoter was cut out from the pGEM®-T Easy Vector using EcoRI + XhoI and cloned into the pHAtC, generating an OsU6a::pHAtC vector.
Cloning of sgRNA expression vector
The OsU6a::xa13m-sgRNA/pHAtC vector expressing sgRNA for xa13/Os8N3 (xa13m-sgRNA) was constructed according to the method previously described (Kim et al. 2016). Briefly, the target sequence (xa13m) for Os8N3 editing of Kitaake was designed by the CRISPR-RGEN Tools website (http://rgenome.ibs.re.kr) (Park et al. 2015). The sgRNA templates (xa13m) for Os8N3 were annealed using two primers, 5′-GATTGCTTGTCCATGGCTAACCCGG-3′ and 5′- AAACCCGGGTTAGCCATGGACAAGC-3′, and cloned into AarI-digested OsU6a::pHAtC. Construction of the sgRNA expression vector, OsU6a::xa13m-sgRNA/pHAtC, and its flanking sequences were confirmed by Sanger sequencing.
Rice transformations
Rice transformations were carried out as previously described (Chern et al. 2005). Agrobacterium tumefaciens strain LBA4404 was used to infect callus tissue induced from Kitaake seeds. Transformants carrying OsU6a::xa13m-sgRNA/pHAtC constructs were selected using hygromycin. Transgenic Kitaake plants overexpressing xa13m-sgRNA (OsU6a xa13m/Kit) were confirmed by PCR using Cas9-specific primers, Cas9_RT_F (5′-CGAGCTGACCAAGGTGAAGT-3′) and Cas9_RT_R (5′-CGTTGATAAGCTTGCGGCTC-3′).
Expression
For reverse transcription polymerase chain reaction (RT-PCR) analysis of Cas9 and sgxa13 transgenes, total RNA was extracted from fully expanded leaves of OsU6a xa13m/Kit plants using TRIzol reagent (Invitrogen, USA). First-strand cDNA was synthesized using quantified RNA (5 μg of total RNA). Expression of Cas9 was confirmed by RT-PCR using Cas9_RT_F and Cas9_RT_R. Meanwhile, the rEFla cDNA fragment was amplified as a control using specific primers, rEF1a1048F (5′-ACTGCCACACCTCCCACATTG-3′) and rEF1a1552R (5′-CAACAGTCGAAGGGCAATAATAAGTC-3′).
Identification of mutant transgenic plants
Rice genomic DNA was extracted from Kitaake leaves and transgenic OsU6a xa13m/Kit plants. All transgenic hygromycin-resistant T0 plants were analyzed by PCR using the Cas9-specific primers, Cas9_RT_F and Cas9_RT_R. Subsequently, the DNA fragment across the xa13 target site was amplified from the genomic DNAs of all PCR-positive plants using xa13-specific primers, xa13_cas9 60-79_F (5′-TCTGGCTAGTTTCTAGCTGG-3′) and xa13_cas9 nuclease_R (5′-TGCATGAGCTGAAGCTAGGG-3′). The PCR amplicons were then directly sequenced using primer xa13_cas9 60-79_F. The sequencing chromatograms with superimposed peaks of bi-allelic and heterozygous mutations were decoded using the Degenerate Sequence Decoding method (http://skl.scau.edu.cn/dsdecode/) (Liu et al. 2015; Ma et al. 2015a).
Xoo inoculation and determination of bacterial populations
For Xoo inoculation, Kitaake, XA21, and transgenic OsU6a xa13m/Kit plants were grown in a greenhouse normally until they reached the eight-week stage, unless otherwise stated. PXO99 was used to inoculate rice plants using the scissors dip method (Song et al. 1995; Chern et al. 2005). For lesion length measurements, at least three inoculated leaves were measured to calculate the average and standard deviation 12 days after inoculation (DAI). Representative leaves were photographed 12 DAI. For Xoo colony counts from inoculated leaves 0 and 12 DAI, 20 cm of leaf tissue from the top, including lesions and tissue showing no lesions, was ground up and resuspended in 10 ml water to harvest bacteria. The extract was diluted accordingly and plated out on PSA plates containing 15.0 mg/L cephalexin. Plates were incubated at 28 °C for two days, and then colony forming units (CFU) were counted. Statistical analysis was performed using Tukey’s HSD test.
Pollen viability tests
Pollen viability was evaluated as previously described (Chhun et al. 2007). Before flowering, six anthers from Kitaake and transgenic OsU6a xa13m/Kit plants were removed and crushed into a fine powder. Pollen grains were stained with 10 μl I2-KI solution (1% I2, 3% KI) and 1 μl of stained pollen grains was harvested to observe fertile and infertile pollen under a light microscope. Dark-stained pollen grains were considered viable and the percentage of pollen viability was calculated relative to the total pollen counted in five microscopic images. Seed viability represents the percentage of spikelets that set seed per total number. Statistical analysis was performed using Tukey’s HSD test.
Additional files
Figure S1. Sequence comparison of OsU6a promoters from Japonica cultivar Kitaake and Indica cultivar 93–11. (PDF 72 kb)
Figure S2. Sequencing chromatogram at the target site of Os8N3 in the CRISPR/Cas9-induced plants (OsU6a xa13m/Kit T0). The vertical arrowhead indicates an expected cleavage site. (PDF 105 kb)
Figure S3. Sequencing chromatogram at the target site of Os8N3 in the CRISPR/Cas9-induced plants (OsU6a xa13m/Kit T1). The vertical arrowhead indicates an expected cleavage site. (PDF 232 kb)
Figure S4. Sequencing chromatogram at the target site of Os8N3 in the CRISPR/Cas9-induced plants (OsU6a xa13m/Kit T2). The vertical arrowhead indicates an expected cleavage site. (PDF 188 kb)
Figure S5. Sequencing chromatogram at the target site of Os8N3 in the CRISPR/Cas9-induced plants (OsU6a xa13m/Kit T3). The vertical arrowhead indicates an expected cleavage site. (PDF 196 kb)
Figure S6. Gross morphology of Kitaake and two homozygous Os8N3 mutant lines, T3 progeny of 3A-6-1 and 4A-1-7. (PDF 72 kb)
Figure S7. Seed-setting rates of homozygous xa13 mutants. a Representative panicles from Kitaake, homozygous mutant (T3, 3A-6-1-2), and homozygous mutant (T3, 4A-1-7-4). b Seed-setting rates of Kitaake, homozygous mutant (progeny of 3A-6-1), and homozygous mutant (progeny of 4A-1-7). c Mature caryopses of Kitaake, homozygous mutant (T3, 3A-6-1-1), and homozygous mutant (T3, 4A-1-7-1). Scale bars, 2.5 mm. (PDF 78 kb)
Acknowledgments
We thank Hyesu Jo for technical assistance in generating transgenic plants.
Abbreviations
- CRISPR/Cas9
Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein-9 nuclease
- EBE
Effector binding element
- TAL effector
Transcription activator-like effector
- Xoo PR6
Xanthomonas oryzae pv. oryzae Philippine Race 6
Authors’ contributions
YAK, HM and CJP conceived and designed the experiments. YAK and HM performed the experiments and analyzed the data. YAK, HM, and CJP wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Basic Research Program of National Research Foundation of Korea (NRF-2017R1D1A1B03032215), funded by the Ministry of Science and ICT.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
The manuscript has been approved by all authors.
Competing interests
The authors declare that they have no competing interests.
Footnotes
The original version of this article was revised as all PCR bands in Fig. 3a did not appear in the published version.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Young-Ah Kim and Hyeran Moon contributed equally to this work.
Change history
9/13/2019
It was highlighted that in the original article (Kim, 2019).
Contributor Information
Young-Ah Kim, Email: eco10730@naver.com.
Hyeran Moon, Email: kasiya1221@gmail.com.
Chang-Jin Park, Phone: +82 2-3408-4378, Email: cjpark@sejong.ac.kr.
References
- Bai J, Choi SH, Ponciano G, Leung H, Leach JE. Xanthomonas oryzae pv. Oryzae avirulence genes contribute differently and specifically to pathogen aggressiveness. Mol Plant-Microbe Interact. 2000;13(12):1322–1329. doi: 10.1094/MPMI.2000.13.12.1322. [DOI] [PubMed] [Google Scholar]
- Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V. Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods. 2013;9(1):39. doi: 10.1186/1746-4811-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanvillain-Baufume S, Reschke M, Sole M, Auguy F, Doucoure H, Szurek B, Meynard D, Portefaix M, Cunnac S, Guiderdoni E, Boch J, Koebnik R. Targeted promoter editing for rice resistance to Xanthomonas oryzae pv. oryzae reveals differential activities for SWEET14-inducing TAL effectors. Plant Biotechnol J. 2017;15(3):306–317. doi: 10.1111/pbi.12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun N, Wyrzykowska J, Muller P, David K, Couch D, Perrot-Rechenmann C, Fleming AJ. Conditional repression of AUXIN BINDING PROTEIN1 reveals that it coordinates cell division and cell expansion during postembryonic shoot development in Arabidopsis and tobacco. Plant Cell. 2008;20(10):2746–2762. doi: 10.1105/tpc.108.059048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ. SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 2014;201(4):1150–1155. doi: 10.1111/nph.12445. [DOI] [PubMed] [Google Scholar]
- Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, Chermak D, Antony G, White FF, Somerville SC, Mudgett MB, Frommer WB. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468(7323):527–532. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR, Frommer WB. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science. 2012;335(6065):207–211. doi: 10.1126/science.1213351. [DOI] [PubMed] [Google Scholar]
- Chen X, Grandont L, Li H, Hauschild R, Paque S, Abuzeineh A, Rakusova H, Benkova E, Perrot-Rechenmann C, Friml J. Inhibition of cell expansion by rapid ABP1-mediated auxin effect on microtubules. Nature. 2014;516(7529):90–93. doi: 10.1038/nature13889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern M, Canlas PE, Fitzgerald HA, Ronald PC. Rice NRR, a negative regulator of disease resistance, interacts with Arabidopsis NPR1 and rice NH1. Plant J. 2005;43(5):623–635. doi: 10.1111/j.1365-313X.2005.02485.x. [DOI] [PubMed] [Google Scholar]
- Chhun T, Aya K, Asano K, Yamamoto E, Morinaka Y, Watanabe M, Kitano H, Ashikari M, Matsuoka M, Ueguchi-Tanaka M. Gibberellin regulates pollen viability and pollen tube growth in rice. Plant Cell. 2007;19(12):3876–3888. doi: 10.1105/tpc.107.054759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Z, Yuan M, Yao J, Ge X, Yuan B, Xu C, Li X, Fu B, Li Z, Bennetzen JL, Zhang Q, Wang S. Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 2006;20(10):1250–1255. doi: 10.1101/gad.1416306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daude N, Wohlgemuth S, Brown R, Pitstick R, Gapeshina H, Yang J, Carlson GA, Westaway D. Knockout of the prion protein (PrP)-like Sprn gene does not produce embryonic lethality in combination with PrP(C)-deficiency. Proc Natl Acad Sci U S A. 2012;109(23):9035–9040. doi: 10.1073/pnas.1202130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza AT, Dai X, Spencer AG, Reppen T, Menzie A, Roesch PL, He Y, Caguyong MJ, Bloomer S, Herweijer H, Wolff JA, Hagstrom JE, Lewis DL, Linsley PS, Ulrich RG. Transcriptional and phenotypic comparisons of Ppara knockout and siRNA knockdown mice. Nucleic Acids Res. 2006;34(16):4486–4494. doi: 10.1093/nar/gkl609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkes A, Reschke M, Boch J, Grau J. Evolution of transcription activator-like effectors in Xanthomonas oryzae. Genome Biol Evol. 2017;9(6):1599–1615. doi: 10.1093/gbe/evx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers B, Jastrzebski K, Heijmans JP, Grernrum W, Beijersbergen RL, Bernards R. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat Biotechnol. 2016;34(6):631–633. doi: 10.1038/nbt.3536. [DOI] [PubMed] [Google Scholar]
- Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, Wang Z, Zhang Z, Zheng R, Yang L, Zeng L, Liu X, Zhu JK. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci U S A. 2014;111(12):4632–4637. doi: 10.1073/pnas.1400822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, Cao F, Zhu S, Zhang F, Mao Y, Zhu JK. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23(10):1229–1232. doi: 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci U S A. 2015;112(7):2275–2280. doi: 10.1073/pnas.1500365112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grand X, Espinoza R, Michel C, Cros S, Chalvon V, Jacobs J, Morel JB. Identification of positive and negative regulators of disease resistance to rice blast fungus using constitutive gene expression patterns. Plant Biotechnol J. 2012;10(7):840–850. doi: 10.1111/j.1467-7652.2012.00703.x. [DOI] [PubMed] [Google Scholar]
- Hutin M, Perez-Quintero AL, Lopez C, Szurek B. MorTAL Kombat: the story of defense against TAL effectors through loss-of-susceptibility. Front Plant Sci. 2015;6:535. doi: 10.3389/fpls.2015.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Nishizawa-Yokoi A, Endo M, Mikami M, Toki S. CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem Biophys Res Commun. 2015;467(1):76–82. doi: 10.1016/j.bbrc.2015.09.117. [DOI] [PubMed] [Google Scholar]
- Karakas B, Weeraratna AT, Abukhdeir AM, Konishi H, Gustin JP, Vitolo MI, Bachman KE, Park BH. P21 gene knock down does not identify genetic effectors seen with gene knock out. Cancer Biol Ther. 2007;6(7):1025–1030. doi: 10.4161/cbt.6.7.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim ST, Ryu J, Choi MK, Kweon J, Kang BC, Ahn HM, Bae S, Kim J, Kim JS, Kim SG. A simple, flexible and high-throughput cloning system for plant genome editing via CRISPR-Cas system. J Integr Plant Biol. 2016;58(8):705–712. doi: 10.1111/jipb.12474. [DOI] [PubMed] [Google Scholar]
- Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, Kirchmaier BC, Peterson-Maduro J, Kourkoulis G, Male I, DeSantis DF, Sheppard-Tindell S, Ebarasi L, Betsholtz C, Schulte-Merker S, Wolfe SA, Lawson ND. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell. 2015;32(1):97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law SH, Sargent TD. The serine-threonine protein kinase PAK4 is dispensable in zebrafish: identification of a morpholino-generated pseudophenotype. PLoS One. 2014;9(6):e100268. doi: 10.1371/journal.pone.0100268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Jain R, Chern M, Pham NT, Martin JA, Wei T, Schackwitz WS, Lipzen AM, Duong PQ, Jones KC, Jiang L, Ruan D, Bauer D, Peng Y, Barry KW, Schmutz J, Ronald PC. The sequences of 1504 mutants in the model Rice variety Kitaake facilitate rapid functional genomic studies. Plant Cell. 2017;29(6):1218–1231. doi: 10.1105/tpc.17.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Liu B, Spalding MH, Weeks DP, Yang B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol. 2012;30(5):390–392. doi: 10.1038/nbt.2199. [DOI] [PubMed] [Google Scholar]
- Liu W, Xie X, Ma X, Li J, Chen J, Liu YG. DSDecode: a web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol Plant. 2015;8(9):1431–1433. doi: 10.1016/j.molp.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Ma L, Zhang D, Miao Q, Yang J, Xuan Y, Hu Y. Essential role of sugar transporter OsSWEET11 during the early stage of Rice grain filling. Plant Cell Physiol. 2017;58(5):863–873. doi: 10.1093/pcp/pcx040. [DOI] [PubMed] [Google Scholar]
- Ma X, Chen L, Zhu Q, Chen Y, Liu YG. Rapid decoding of sequence-specific nuclease-induced heterozygous and Biallelic mutations by direct sequencing of PCR products. Mol Plant. 2015;8(8):1285–1287. doi: 10.1016/j.molp.2015.02.012. [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu YG. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;8(8):1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- McJunkin K, Mazurek A, Premsrirut PK, Zuber J, Dow LE, Simon J, Stillman B, Lowe SW. Reversible suppression of an essential gene in adult mice using transgenic RNA interference. Proc Natl Acad Sci U S A. 2011;108(17):7113–7118. doi: 10.1073/pnas.1104097108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalko J, Glanc M, Perrot-Rechenmann C, Friml J. Strong morphological defects in conditional Arabidopsis abp1 knock-down mutants generated in absence of functional ABP1 protein. F1000Res. 2016;5:86. doi: 10.12688/f1000research.7654.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgens DW, Deans RM, Li A, Bassik MC. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol. 2016;34(6):634–636. doi: 10.1038/nbt.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paque S, Mouille G, Grandont L, Alabadi D, Gaertner C, Goyallon A, Muller P, Primard-Brisset C, Sormani R, Blazquez MA, Perrot-Rechenmann C. AUXIN BINDING PROTEIN1 links cell wall remodeling, auxin signaling, and cell expansion in arabidopsis. Plant Cell. 2014;26(1):280–295. doi: 10.1105/tpc.113.120048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CJ, Lee SW, Chern M, Sharma R, Canlas PE, Song MY, Jeon JS, Ronald PC. Ectopic expression of rice Xa21 overcomes developmentally controlled resistance to Xanthomonas oryzae pv. oryzae. Plant Sci. 2010;179(5):466–471. doi: 10.1016/j.plantsci.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Bae S, Kim JS. Cas-designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics (Oxford, England) 2015;31(24):4014–4016. doi: 10.1093/bioinformatics/btv537. [DOI] [PubMed] [Google Scholar]
- Peng A, Chen S, Lei T, Xu L, He Y, Wu L, Yao L, Zou X. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol J. 2017;15(12):1509–1519. doi: 10.1111/pbi.12733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyott DE, Sheehan E, Molnar A. Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol Plant Pathol. 2016;17(8):1276–1288. doi: 10.1111/mpp.12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S, Kleine-Vehn J, Barbez E, Sauer M, Paciorek T, Baster P, Vanneste S, Zhang J, Simon S, Covanova M, Hayashi K, Dhonukshe P, Yang Z, Bednarek SY, Jones AM, Luschnig C, Aniento F, Zazimalova E, Friml J. ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell. 2010;143(1):111–121. doi: 10.1016/j.cell.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer P, Recht S, Strauss T, Elsaesser J, Schornack S, Boch J, Wang S, Lahaye T. Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. New Phytol. 2010;187(4):1048–1057. doi: 10.1111/j.1469-8137.2010.03217.x. [DOI] [PubMed] [Google Scholar]
- Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, Stainier DY. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524(7564):230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- Schornack S, Moscou MJ, Ward ER, Horvath DM. Engineering plant disease resistance based on TAL effectors. Annu Rev Phytopathol. 2013;51:383–406. doi: 10.1146/annurev-phyto-082712-102255. [DOI] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald P. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270(5243):1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Sun X, Hu Z, Chen R, Jiang Q, Song G, Zhang H, Xi Y. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci Rep. 2015;5:10342. doi: 10.1038/srep10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Li J, Xia L. Precise genome modification via sequence-specific nucleases-mediated gene targeting for crop improvement. Front Plant Sci. 2016;7:1928. doi: 10.3389/fpls.2016.01928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Lin R, Feng J, Chen W, Qiu D, Xu S. TaNAC1 acts as a negative regulator of stripe rust resistance in wheat, enhances susceptibility to Pseudomonas syringae, and promotes lateral root development in transgenic Arabidopsis thaliana. Front Plant Sci. 2015;6:108. doi: 10.3389/fpls.2015.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wang C, Liu P, Lei C, Hao W, Gao Y, Liu YG, Zhao K. Enhanced Rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One. 2016;11(4):e0154027. doi: 10.1371/journal.pone.0154027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32(9):947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- Xu RF, Li H, Qin RY, Li J, Qiu CH, Yang YC, Ma H, Li L, Wei PC, Yang JB. Generation of inheritable and "transgene clean" targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci Rep. 2015;5:11491. doi: 10.1038/srep11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Sugio A, White FF. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc Natl Acad Sci U S A. 2006;103(27):10503–10508. doi: 10.1073/pnas.0604088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Luo D, Yang B, Frommer WB, Eom JS. SWEET11 and 15 as key players in seed filling in rice. New Phytol. 2018;218(2):604–615. doi: 10.1111/nph.15004. [DOI] [PubMed] [Google Scholar]
- Young R, Passet B, Vilotte M, Cribiu EP, Beringue V, Le Provost F, Laude H, Vilotte JL. The prion or the related Shadoo protein is required for early mouse embryogenesis. FEBS Lett. 2009;583(19):3296–3300. doi: 10.1016/j.febslet.2009.09.027. [DOI] [PubMed] [Google Scholar]
- Yuan M, Chu Z, Li X, Xu C, Wang S. Pathogen-induced expressional loss of function is the key factor in race-specific bacterial resistance conferred by a recessive R gene xa13 in rice. Plant Cell Physiol. 2009;50(5):947–955. doi: 10.1093/pcp/pcp046. [DOI] [PubMed] [Google Scholar]
- Yuan M, Chu Z, Li X, Xu C, Wang S. The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. Plant Cell. 2010;22(9):3164–3176. doi: 10.1105/tpc.110.078022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang S. Rice versus Xanthomonas oryzae pv. Oryzae: a unique pathosystem. Curr Opin Plant Biol. 2013;16(2):188–195. doi: 10.1016/j.pbi.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Zhou H, Liu B, Weeks DP, Spalding MH, Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014;42(17):10903–10914. doi: 10.1093/nar/gku806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sequence comparison of OsU6a promoters from Japonica cultivar Kitaake and Indica cultivar 93–11. (PDF 72 kb)
Figure S2. Sequencing chromatogram at the target site of Os8N3 in the CRISPR/Cas9-induced plants (OsU6a xa13m/Kit T0). The vertical arrowhead indicates an expected cleavage site. (PDF 105 kb)
Figure S3. Sequencing chromatogram at the target site of Os8N3 in the CRISPR/Cas9-induced plants (OsU6a xa13m/Kit T1). The vertical arrowhead indicates an expected cleavage site. (PDF 232 kb)
Figure S4. Sequencing chromatogram at the target site of Os8N3 in the CRISPR/Cas9-induced plants (OsU6a xa13m/Kit T2). The vertical arrowhead indicates an expected cleavage site. (PDF 188 kb)
Figure S5. Sequencing chromatogram at the target site of Os8N3 in the CRISPR/Cas9-induced plants (OsU6a xa13m/Kit T3). The vertical arrowhead indicates an expected cleavage site. (PDF 196 kb)
Figure S6. Gross morphology of Kitaake and two homozygous Os8N3 mutant lines, T3 progeny of 3A-6-1 and 4A-1-7. (PDF 72 kb)
Figure S7. Seed-setting rates of homozygous xa13 mutants. a Representative panicles from Kitaake, homozygous mutant (T3, 3A-6-1-2), and homozygous mutant (T3, 4A-1-7-4). b Seed-setting rates of Kitaake, homozygous mutant (progeny of 3A-6-1), and homozygous mutant (progeny of 4A-1-7). c Mature caryopses of Kitaake, homozygous mutant (T3, 3A-6-1-1), and homozygous mutant (T3, 4A-1-7-1). Scale bars, 2.5 mm. (PDF 78 kb)
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.