LETTER
The plasmid-mediated quinolone resistance (PMQR) gene qnrE1 has been limited to South America to date; in 2007, it was identified for the first time in Klebsiella pneumoniae from a human infection in Argentina (1). A study in Brazil reported the identification of qnrE1 in K. pneumoniae isolated from an infected parrot in 2016 (2). The presence of this gene was also confirmed in Salmonella enterica serovar Enteritidis, Newport, and Infantis strains isolated in 2011, 2013, and 2014, respectively, from humans in Brazil (3).
During a retrospective genomic surveillance study in our laboratory to examine the presence of the qnrE1 gene, 43 S. enterica serovar Typhimurium strains obtained in 2000 to 2015 from the Brazilian agricultural food sector, including strains isolated from poultry, swine, and turkey, were directly sequenced using short-read whole-genome sequencing. Eight (18.6%) S. Typhimurium isolates from broiler chicken (n = 6) and swine (n = 2) samples, collected in São Paulo and Parana states, respectively, were positive for the qnrE1 gene. These isolates were found to be genetically unrelated by pulsed-field gel electrophoresis (PFGE) using XbaI restriction (4), since isolates were clustered in two pulsotypes, designated A (A1, A2, and A3) and B (B1, B2, and B3) (see Fig. S1 in the supplemental material). Subcluster A1 included S. Typhimurium strains (STy2 and STy13) obtained from swine samples, whereas subclusters A2 (STy08) and A3 (STy011) were composed of S. Typhimurium strains obtained from broiler chicken samples. Additionally, isolates from cluster A were recovered from different geographical locations, in different periods (Fig. S1). Subclusters B1 (STy06 and STy07), B2 (STy013), and B3 (STy05) included Salmonella strains recovered from broiler chicken samples collected in São Paulo state in 2015 (Fig. S1).
All qnrE1-positive S. Typhimurium strains analyzed in this study shared a genetic environment with high levels of identity (>99%), composed of inverted repeat left (IRL)-ISEcp1-inverted repeat right (IRR)-qnrE1-araJ-ahp (alkyl hydroperoxidase-encoding gene) (4,659 bp in size) (Fig. 1). Among these isolates, we identified the presence of the qnrE1 gene in a S. Typhimurium strain isolated from a pig liver sample in 2000, 7 and 11 years earlier than previously reported in Argentina (1) and Brazil (3), respectively.
FIG 1.
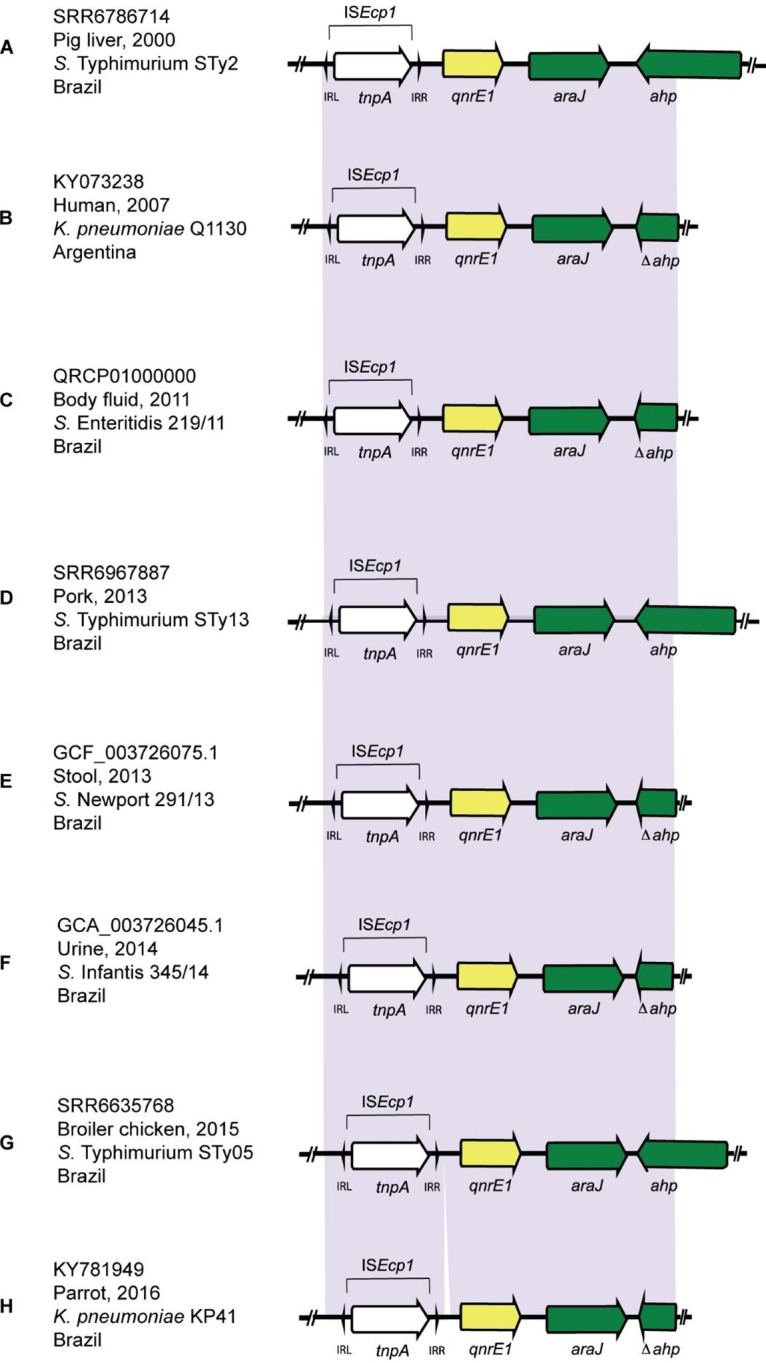
Chronological comparison between genetic contexts of qnrE1 genes carried by S. enterica serovars (A, C, D, E, F, and G) and K. pneumoniae (B and H) strains isolated in South America. Genes and shotgun sequences were extracted from the GenBank database. Arrows indicate the positions and directions of the genes; Δ indicates truncated genes. Regions with >99% identity are indicated with purple shading. (A, D, and G) Strains contain ahp genes of 1,035 bp. (B) Strain contains an ahp gene of 438 bp. (C, E, F, and H) Strains contain ahp genes of 327 bp.
Furthermore, the downstream region of the qnrE1 genetic platform, as well as the upstream region, was found to be slightly different from that in previous surveys (1–3). First, as reported previously, pKp1130 (GenBank accession no. KY073238) possesses a 5′-truncated ahp gene that is 438 bp in size, while plasmid pKp41M (GenBank accession no. KY781949) and the 219/11 (GenBank accession no. QRCP01000000), 345/14 (GenBank accession no. GCA_003726045.1), and 291/13 (GenBank accession no. RDPW00000000.1) isolates have a different 5′-truncated ahp that is 327 bp in size. Therefore, while our isolates present a complete 1,035-bp ahp gene, compared to the reference sequence of the ahp gene from S. Typhimurium strain CFSAN033917 (GenBank accession no. KYE07389.1), deletions of 597 bp and 708 bp in the ahp gene could be observed, as reported previously for K. pneumoniae and Salmonella isolates, respectively (1, 3). Another difference within this genetic platform was that, although an IRR sequence was identified downstream of ISEcp1 in our isolates, an alternative IRR could not be found at the right edge, in contrast to previous observations for K. pneumoniae (1). Moreover, pKp41M (GenBank accession no. KY781949) contained an additional 152 bp in the upstream region of the qnrE1 gene (Fig. 1H). These findings provide new insights into this genetic environment and suggest that the emergence of qnrE1 was much earlier than previously thought.
Another interesting observation is that, for both human and parrot infected hosts reported in Brazil, the occurrence of qnrE1 has been associated with carriage of blaCTX-M-8 and blaOXA-9 β-lactamase genes, conferring a broad spectrum and a narrow spectrum, respectively, of activity against cephalosporins (2, 3, 5). This finding may suggest an evolutionary mechanism of adaptation, most likely driven by selective pressure through the use of cephalosporins in infected hosts such as parrots (2) and humans (3), particularly in Brazil, since all S. Typhimurium strains analyzed in this study were isolated from healthy animals and did not carry these genes.
A limitation of this study was that plasmids harboring the qnrE1 gene were not reconstructed, because of the limitations of short-read sequencing (6). However, the findings demonstrated in this study substantially aid in the ongoing development of this new genetic platform; it has not escaped our attention that further studies are necessary to fully comprehend the epidemiology of S. Typhimurium strains carrying the qnrE1 gene and their potential to be spread throughout the food chain.
In summary, we present evidence that qnrE1 genes have been circulating in South America since at least 2000, which is much earlier than reported previously. Therefore, since qnrE1-carrying Enterobacteriaceae strains are established in Latin America (1–3), we demonstrate the need for continuous surveillance programs to track this and other antimicrobial resistance genes important in therapeutic efficacy. Further, continuous surveillance can aid in the prevention of wide dissemination of this gene following the development of mitigation strategies. It is likely that, unless continuous surveillance is established, we will continue to underestimate the prevalence of these and other antimicrobial resistance genes at the environment-animal-human interface.
Data availability.
Sequence data were deposited as part of the GenomeTrakr Project. The genome sequences of the S. Typhimurium strains have been deposited in DDBJ/ENA/GenBank with BioSample accession numbers SAMN08606827, SAMN08874445, SAMN08386803, SAMN07279711, SAMN08606818, SAMN08874450, SAMN08386758, and SAMN07283712.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by research grants from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (Food Research Center grant 2013/07914-8 and grant 2016/03044-7 to D.F.M.). The project was partially developed during the time of D.F.M. as a visiting scholar at North Carolina State University, under a fellowship grant from the FAPESP (grant 2017/15967-5). N.L. is a research grant fellow of CNPq.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00571-19.
REFERENCES
- 1.Albornoz E, Tijet N, De Belder D, Gomez S, Martino F, Corso A, Melano RG, Petroni A. 2017. qnrE1, a member of a new family of plasmid-located quinolone resistance genes, originated from the chromosome of Enterobacter species. Antimicrob Agents Chemother 61:e02555-16. doi: 10.1128/10.1128/AAC.02555-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cunha MPV, Davies YM, Cerdeira L, Dropa M, Lincopan N, Knöbl T. 2017. Complete DNA sequence of an IncM1 plasmid bearing the novel qnrE1 plasmid-mediated quinolone resistance variant and blaCTX-M-8 from Klebsiella pneumoniae sequence type 147. Antimicrob Agents Chemother 61:e00592-17. doi: 10.1128/AAC.00592-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soares FB, Camargo CH, Cunha MPV, de Almeida EA, Bertani AMJ, Carvalho E, de Paiva JB, Fernandes SA, Tiba-Casas MR. 2019. Co-occurrence of qnrE1 and blaCTX-M-8 in IncM1 transferable plasmids contributing to MDR in different Salmonella serotypes. J Antimicrob Chemother 74:1155–1156. doi: 10.1093/jac/dky516. [DOI] [PubMed] [Google Scholar]
- 4.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 5.Antunes NT, Fisher JF. 2014. Acquired class D β-lactamases. Antibiotics (Basel) 3:398–434. doi: 10.3390/antibiotics3030398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arredondo-Alonso S, Willems RJ, van Schaik W, Schürch AC. 2017. On the (im)possibility of reconstructing plasmids from whole-genome short-read sequencing data. Microb Genom 3:e000128. doi: 10.1099/mgen.0.000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data were deposited as part of the GenomeTrakr Project. The genome sequences of the S. Typhimurium strains have been deposited in DDBJ/ENA/GenBank with BioSample accession numbers SAMN08606827, SAMN08874445, SAMN08386803, SAMN07279711, SAMN08606818, SAMN08874450, SAMN08386758, and SAMN07283712.


