We read with interest the article by Passamonti et al,1 reporting the performance of two different faecal immunochemical tests (FITs) highlighting the importance of standardisation and validation of screening methodologies. Conventionally, laboratory-based FIT is the preferred approach in testing for occult blood in faeces, which includes colorectal cancer screening programmes.2–4 The potential of preserving stable faecal samples in a widely used FIT buffer for microbiome research would enable prospective microbiome studies in generally healthy subjects undergoing colorectal cancer screening.
For this purpose, we evaluated faecal sample stability in the commonly used OC-Sensor (Eiken Chemical, Tokyo, Japan) under various storage conditions. Faecal samples from five healthy adult individuals were used for the analysis and exposed to 16 different conditions: immediately frozen at −86°C (with FIT (wFIT)/without FIT (woFIT)); immediately frozen at −20°C (wFIT/woFIT); wFIT stored at 4°C for 1, 2, 7, 14 days; stored at 20°C for 1, 2, 7, 14 days; stored at 30°C for 1, 2, 7 days and lastly stored at 4°C for 2 days and at 20°C for additional 2 days. Shotgun metagenomic analysis was performed by Illumina 2500, while taxonomic compositions were determined by 16S rRNA analysis employing Illumina MiSeq and Ion Torrent PGM.
Our first attempt to extract DNA from wFIT samples failed due to low amount of DNA obtained. Thus, lyophilisation for all of the wFIT samples, except for woFIT samples, was applied, increasing the yield of DNA up to 30 times.
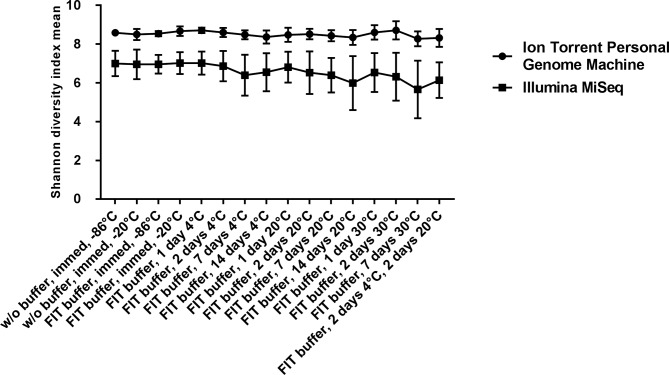
As shown by the figure 1, the Shannon index in both platforms displayed a clear pattern of decreased diversity during prolonged storage. While testing for differences between samples we discovered that woFIT samples, wFIT samples stored at 4°C for 1 and 2 days, wFIT samples stored at 20°C for 1 day from the Illumina MiSeq data significantly differ from wFIT samples stored at 30°C for 7 days (one-way analysis of variance; p<0.05).
Figure 1.
Mean Shannon diversity index values by storage conditions of faecal samples sequenced on Ion Torrent Personal Genome Machine and Illumina MiSeq. Error bars represent SEs. FIT, faecal immunochemical test.
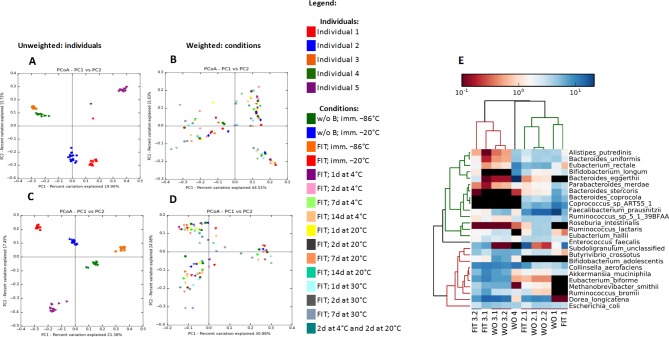
The similarity matrix using unweighted principal coordinate analysis (PCoA, figure 2A,C) demonstrated that samples collected from each individual clustered together and was consistent in data gained from both sequencing platforms, suggesting that the biological effect outweighed the effect of the sample handling. Further, the weighted PCoA (figure 2B,D) displays that native samples tend to cluster closer while samples stored for prolonged periods tend to be located further away from the native ones.
Figure 2.
Beta diversity (panels A–D) analysis of GI tract bacterial communities between the individuals and storage conditions presented in a form of principal coordinate analysis (PCoA) plot of weighted and unweighted UniFrac distances. Panels A and C are coloured according to the individual, panels B and D are coloured according to the conditions that samples were exposed to. Panels A and B represent the UniFrac metrics obtained from sequencing on the Illumina MiSeq platform, whereas panels C and D represent UniFrac metrics obtained from sequencing on the Ion Torrent PGM platform. The results of the metagenomics analysis at the species level are presented within panel E in a form of the heat-map profile for the most abundant entities in the metagenome samples. According to upper dendrogram, there is a consistency within the samples composition as the samples from the same individual are clustering together. Samples WO1, WO2.1, WO2.2, WO3.1, WO3.2, WO4 were without faecal immunochemical test (woFIT) and immediately frozen at −86°C; while samples FIT1, FIT2.1, FIT2.2, FIT3.1, FIT3.2 and FIT4 were with FIT (wFIT) and stored at 4°C for 2 days and then at 20°C for additional 2 days. Samples FIT2.2 and FIT4 failed at the sequencing stage and therefore are not presented within the figure. Both sample groups include technical replicates for individuals 2 and 3 (eg, WO2.1. and WO2.2.; WO3.1 and WO3.2.; FIT2.1 and FIT2.2.; FIT3.1. and FIT3.2.).
Comparing observed operational taxonomic units (OTUs) between samples that were stored within various storage conditions, we were unable to identify any specific OTU cluster across all taxonomical levels that would significantly differ (Kruskal-Wallis test) between any storage condition. Nevertheless, we found that the relative abundances of Gram-negative bacteria tended to decrease while the abundances for most of the Gram-positive bacteria tended to increase over time. However, it is possible that even within native samples certain bacterial OTUs could fluctuate regardless of the sequencing platform employed.
Shotgun sequencing was performed on 12 samples (figure 2E) from four individuals: six woFIT samples (WO1–4) that were immediately stored at −86°C and six wFIT samples from three individuals (FIT1–4), stored for 2 days at 4°C and for 2 days at 20°C to imitate the typical sample treatment procedure when the material is being transported to a central laboratory by standard mail delivery. In addition to access the reproducibility of acquired data, technical replicates in the form of DNA that was extracted from independently collected samples from individuals 2 and 3 were included within this analysis. However, two wFIT samples (eg, FIT2.2 and FIT4) failed during the sequencing stage and therefore were excluded from further analysis. A similarity matrix was built with relative abundances to calculate the PCoA. Figure 2E illustrates the similarity between species for two types of sampling showing that results are not biased to the sampling procedures. Results indicate that sample similarities tended towards samples from the same subject rather than the storage method.
In this study, we sequenced microbiome samples employing two different sequencing platforms—Ion Torrent PGM that allows to analyse one variable region (V3) and to verify acquired results and also Illumina MiSeq platform that enables the analysis of two variable regions (V3–V4) at a time. Analysing absolute OTU frequencies within the samples, we did not find a significant difference between immediately frozen samples, which coincide with previous studies.5–8 Although previous study9 has found lower FIT stability measures, we observed some limitations as they extracted DNA from a small proportion of sample, while in our study design lyophilisation procedure was included allowing to extract DNA from whole volume. However, we did not include technical replicates for 16S rRNA analysis. Another critical aspect in FIT-oriented studies is the variety of available FIT test tubes in the market. Each FIT buffer from various manufacturers might contain slightly different ingredients and concentrations,10 which seems to be a trade secret and might alter the microbial composition. In our study, employed OC-Sensor is a reliable and convenient sampling device that can be used in large-scale microbiome studies on a population level even outside colorectal cancer screening programmes.
Footnotes
ML and LE contributed equally.
DG and SS contributed equally.
Contributors: Study concept and design was created by ML and LE. Data collection was performed by DG, HW, IR-S, RH, IK, IE, GS. Bioinformatics analysis performed by DG, SS, IS, RP. DG and SS wrote the manuscript. Funding was obtained by LE, DF, JK and ML. All authors approved the final version for publication.
Funding: The work was supported by the ERDF project ‘Long-term effects of H. pylori eradication on gastrointestinal tract microbiome and development of screening system for detection of extended-spectrum beta-lactamase coding genes within faeces samples’ (project no: 1.1.1.1/16/A/272) and in part by the Swedish Research Council (project no: K2013-56X-10848) and the Söderbergs Foundation.
Competing interests: None declared.
Patient consent: Obtained.
Ethics approval: Biomedical Ethics Committee of the Riga East University Hospital Support Foundation, approval no: 13-A/13.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Passamonti B, Malaspina M, Fraser CG, et al. A comparative effectiveness trial of two faecal immunochemical tests for haemoglobin (FIT). Assessment of test performance and adherence in a single round of a population-based screening programme for colorectal cancer. Gut 2018;67:485–96. 10.1136/gutjnl-2016-312716 [DOI] [PubMed] [Google Scholar]
- 2. Directorate-General for Health and Consumers (European Commission) EAfHaC, World Health Organization. European guidelines for quality assurance in colorectal cancer srceening and diagnosis. Luxemborug, 2010. [Google Scholar]
- 3. Young GP, Symonds EL, Allison JE, et al. Advances in fecal occult blood tests: the FIT revolution. Dig Dis Sci 2015;60:609–22. 10.1007/s10620-014-3445-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Robertson DJ, Lee JK, Boland CR, et al. Recommendations on fecal immunochemical testing to screen for colorectal neoplasia: a consensus statement by the us multi-society task force on colorectal cancer. Am J Gastroenterol 2017;112:37–53. 10.1038/ajg.2016.492 [DOI] [PubMed] [Google Scholar]
- 5. Hale VL, Tan CL, Knight R, et al. Effect of preservation method on spider monkey (Ateles geoffroyi) fecal microbiota over 8 weeks. J Microbiol Methods 2015;113:16–26. 10.1016/j.mimet.2015.03.021 [DOI] [PubMed] [Google Scholar]
- 6. Carroll IM, Ringel-Kulka T, Siddle JP, et al. Characterization of the fecal microbiota using high-throughput sequencing reveals a stable microbial community during storage. PLoS One 2012;7:e46953 10.1371/journal.pone.0046953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vlčková K, Mrázek J, Kopečný J, et al. Evaluation of different storage methods to characterize the fecal bacterial communities of captive western lowland gorillas (Gorilla gorilla gorilla). J Microbiol Methods 2012;91:45–51. 10.1016/j.mimet.2012.07.015 [DOI] [PubMed] [Google Scholar]
- 8. Cardona S, Eck A, Cassellas M, et al. Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol 2012;12:158 10.1186/1471-2180-12-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vogtmann E, Chen J, Kibriya MG, et al. Comparison of fecal collection methods for microbiota studies in Bangladesh. Appl Environ Microbiol 2017;83:e00361–17. 10.1128/AEM.00361-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grow MA, Shah VD. Fecal sample immunoassay method testing for hemoglobin. European Patent Office., 1993. [Google Scholar]