Abstract
From biosynthesis to bioremediation, microbes have been engineered to address a variety of biotechnological applications. A promising direction in these endeavors is harnessing the power of designer microbial consortia that consist of multiple populations with well-defined interactions. Consortia can accomplish tasks that are difficult or potentially impossible to achieve using monocultures. Despite their potential, the rules underlying microbial community maintenance and function (i.e. the task the consortium is engineered to carry out) are not well defined, though rapid progress is being made. This limited understanding is in part due to the greater challenges associated with increased complexity when dealing with multi-population interactions. Here, we review key features and design strategies that emerge from the analysis of both natural and engineered microbial communities. These strategies can provide new insights into natural consortia and expand the toolbox available to engineers working to develop novel synthetic consortia.
Introduction
Microbial biotechnology is a growing influence in various global industries because of its capability to produce valuable products like chemicals and pharmaceuticals (Demain 2000). In particular, advances in synthetic biology have demonstrated the potential power of engineered microbes to enable technologies ranging from drug synthesis to addressing the global antibiotic resistance epidemic (Cheng and Lu 2012). Engineered microbial consortia have the potential to advance this further by accomplishing highly complex tasks that are challenging to realize with monocultures, as evidenced by the evolutional development of natural communities. For example, the gut microbiota maintains several roles in metabolism, protection against pathogen invasion, and coordinating immune system response while varying in composition over time and between individuals (Clemente et al. 2012). Similarly, microbial communities have been used for several applications, including synthesizing useful compounds and removing contaminants (Cydzik-Kwiatkowska and Zielinska 2016; Shong et al. 2012).
However, examples of engineered communities often contain only a few members, limiting their utility for applications requiring sophisticated behavior. This limitation is likely because they are significantly more difficult to engineer than clonal populations, with each additional member increasing the size of the interaction matrix geometrically. In this review, we discuss the potential advantages of microbial communities over clonal populations. We then explore design strategies from reviewing both natural and engineered consortia to provide insights towards designing. Finally, we discuss future directions of microbial consortia research. For our discussion, we do not distinguish microbial consortia consisting of members from the same species and those consisting of members from different species, because design strategies to be discussed can be applied to both types.
Division of labor is a key feature of microbial consortia
Division of labor, where different populations execute different tasks, can enables microbial consortia to accomplish tasks at higher efficiency than monocultures. In ecological contexts, division of labor has been defined by both specialization of different cells and cooperation between them to provide an inclusive benefit to the community (West and Cooper 2016). Taking an engineering perspective, we define division of labor as simply the separation of tasks regardless of the ecological and evolutionary consequences. However, when properly constructed, division of labor can facilitate consortia functions in multiple ways.
Improving functionality via specialization
Division of labor can enable rational organization of the pathway by compartmentalizing different steps into different members of the microbial community that offer either a natural or engineered environment benefitting each process (Figure 1A). This separation is necessary when multiple processes cannot coexist in the same cell. For example, feedstocks like lignocellulosic biomass, which contain many simple sugars and other carbon sources, have garnered significant interest for renewable microbial biosynthesis (Isikgor and Becer 2015). However, single populations typically cannot utilize multiple sugars simultaneously due to catabolite repression, instead consuming one sugar at a time in order of preference. In contrast, consortia can more quickly and efficiently utilize such mixtures by metabolizing one carbon source per population. For example, two Escherichia coli strains were engineered such that each population could metabolize only glucose or xylose (Eiteman et al. 2008). The coculture was able to consume a mixture of the sugars more quickly than the single population approach and was robust against varying sugar concentrations (Eiteman et al. 2008). Since then, cocultures where each population specializes in a subset of the entire pathway have been adopted for numerous applications (Table 1). For example, a three-member E. coli consortium, each of which was engineered to utilize one of glucose, xylose, or arabinose, was able to simultaneously metabolize a mixture of all three (Xia et al. 2012). Even individual strains engineered for simultaneous consumption of those same sugars were unable to fully utilize all three sugars or metabolized glucose significantly faster than the others (Bettiga et al. 2009; Hector et al. 2011; Sedlak et al. 2003). Moreover, division of labor in this context allows for more efficient resource utilization, improving overall culture yield (Savage et al. 2007).
Figure 1.
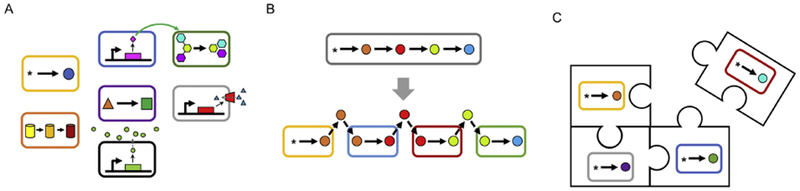
Division of labor, a key feature of microbial consortia, can provide numerous advantages over single populations.
A) Improving functionality via specialization. Division of labor can allow different populations to specialize, permitting sophisticated multifunctionality not possible with monoclonal populations and improving the efficiency of each process
B) Reducing metabolic burden. Division of labor can reduce the metabolic burden experienced by the cells by decreasing the number of enzymes expressed and heterologous parts maintained per population.
C) Modularity. Division of labor can make the system more modular, facilitating easier manipulation and engineering.
Table 1.
Recent achievements in engineering co-cultures for biotechnological applications
| Composition | Final product(s)/process | Application context | Reference |
|---|---|---|---|
| E. coli-E. coli | Ethanol | Biofuels | (L. Wang et al. 2019) |
| Trichoderma reesei-E. coli | Isobutanol | Biofuels | (Minty et al. 2013) |
| E. coli-E. coli | Muconic acid | Biomaterials | (Zhang et al. 2015) |
| E. coli-E. coli | Cadaverine | Biomaterials | (J. Wang et al. 2018b) |
| E. coli-E. coli | Caffeyl alcohol/coniferyl alcohol | Biomaterials/pharmacological activity | (Z. Y. Chen et al. 2017) |
| E. coli-E. coli | Bisdemethoxycurcumin | Pharmacological activity | (Fang et al. 2018) |
| E. coli-E. coli | Rosmarinic acid | Pharmacological activity | (Li et al. 2019) |
| E. coli-S. cerevisiae | Benzylisoquinoline alkaloids | Pharmacological activity | (Minami et al. 2008) |
| Pichia pastoris-P. pastoris | Monacolin J/lovastatin | Anti-hypercholesterolemia | (Liu et al. 2018) |
| E. coli-S. cerevisiae | Oxygenated taxanes | Antitumor | (K. Zhou et al. 2015) |
| E. coli-E. coli | Apigetrin | Antioxidant | (Thuan et al. 2018) |
| Penicillum fuscum-P. camembertiilclavigerum | Berkeleylactone | Macrolide antibiotic | (Stierle et al. 2017) |
| E. coli-E. coli | Naringenin biosensor | Biosensing | (Xiu et al. 2017) |
| Aureobasidium pullans-S. cerevisiae | Fructo-oligosaccharides | Food | (Castro et al. 2019) |
Dividing an entire process, even if the steps are biologically compatible, into a microbial consortium can also improve overall productivity of the system in comparison to a monoculture. A coculture strategy for muconic acid biosynthesis improved specific production of muconic acid from ~100 mg/L/OD in the monoculture to over 800 mg/L/OD (Zhang et al. 2015). In another example, Brenner et. al. engineered a community of two E. coli strains, each containing an orthogonal quorum-sensing module (Brenner et al. 2007). Their consortium coordinated gene expression based on the relative abundances of both populations, generating an output at high density of both populations (Brenner et al. 2007). Moreover, since monocultures of each strain separately displayed minimal gene expression, their system functioned as an AND logic gate, which could be sustained for several days in an engineered biofilm (Brenner et al. 2007). By design, a single cell approach containing both QS modules could not function as an AND gate since cell growth would activate both modules. In other cases, multiple reactions may require the same resource or molecular precursor that is present in low abundance within the host. This constraint can reduce the maximum productivity of the system by reducing access to the limited resource. Moreover, the accumulation of intermediate metabolites (see Reducing metabolic burden) could further decrease pathway productivity via negative feedback (Lindemann et al. 2016). Both limitations can be alleviated by division of labor.
Reducing metabolic burden
Maintaining heterologous components such as enzymes and plasmids can impose a significant metabolic burden on the host. Specifically, gene expression can funnel away host resources, including nucleotides, amino acids, and ATP, that are subsequently not available for cell growth or maintenance (G. Wu et al. 2016). This burden reduces the fitness of the culture, which can result in low productivity of the culture or promote loss-of-function mutations (G. Wu et al. 2016). For particularly complex systems, a single host containing all the components may not grow at all due to the severe burden.
Splitting the responsibility of carrying out the engineered function among different populations via division of labor can reduce the burden experienced by each (Figure 1B). This strategy can improve system performance by improving the fitness of individual populations, thus increasing product yield over a single population (Tsoi et al. 2018). This increase occurs because the reduction in metabolic burden outweighs the lower productivity due to metabolite transport (Tsoi et al. 2018). As a result, division of labor can enable more complex pathways to be engineered in consortia that may not be optimal or functional in single populations (Jones et al. 2017). Indeed, many examples of engineered consortia feature longer pathways, fewer enzymes per population, and/or improved performance compared to their monoculture counterparts (Atsumi et al. 2008; Minty et al. 2013; Zhang et al. 2015). For example, a consortium of E. coli-Saccharomyces cerevisiae was engineered to produce scoulerine from dopamine in seven steps (Minami et al. 2008). A S. cerevisiae monoculture was engineered to produce the same product but utilized a four-step pathway starting from the intermediate norlaudanosoline (Hawkins and Smolke 2008).
Conceptually, the microbial consortia approach provides two potential advantages, though direct evaluation has yet to be carried out. First, dopamine is both cheaper and easier to acquire, making it the more ideal substrate for scalable, industrial applications. Thus, the consortium approach may be preferable since dopamine was not a suitable substrate in the single-cell system due to its low conversion efficiency to S-reticuline (Hawkins and Smolke 2008). Second, the Saccharomyces cerevisiae strain in the single-cell approach was tasked with both intermediate (S-reticuline) and benzylisoquinoline alkaloid (BIA) conversion (Hawkins and Smolke 2008). In contrast, in the consortium approach S. cererisiaewas only tasked with BIA conversion (Minami et al. 2008). As a result, the consortium approach decreased the number of heterologous enzymes expressed, which would reduce the metabolic burden experienced by the yeast strain.
Reducing engineering complexity
Compartmentalizing cellular processes into different populations can increase the modularity of the system. Specifically, individual modules can be tuned, modified, or replaced in a plug-and-play manner independently without affecting other functions (Figure 1C). As a result, depending on its complexity, a consortium utilizing division of labor may be simpler to manipulate than a single cell, since each individual population would contain only a subset of overall complexity. This level of controllability is a fundamental principle and feature of computation, as circuits and machines are simply specific assemblies of discrete parts that can be changed without reconstructing the entire system. For example, a consortium-based approach to sensing organophosphorus pesticides (OPs) used a combination of two E. coli strains (Khatun et al. 2018). The first strain converted the pesticide to p-nitrophenol, which induced expression of β-galactosidase in the second strain after complexing with a transcriptional regulator (Khatun et al. 2018). The authors noted that the division of labor approach allowed for easy modulation of its two components by varying the relative abundance of each population (Khatun et al. 2018). As a result, the consortium was able to detect ethyl-paraoxon at up to 200-fold higher sensitivity than a single-cell based whole-cell approach and up to 400-fold higher sensitivity for other pesticides (Khatun et al. 2018). Other examples of biosensors have also utilized a co-culture strategy to improve modularity and ease of manipulation (Xiu et al. 2017).
Strategies for engineering robust microbial consortia
Given the potential benefits of communities over clonal populations, how may we successfully engineer them to take advantage of their properties? To do so, we must understand the ways in which multiple populations can coexist as it is the most important requirement for stable consortia. Thus, we highlight numerous examples of natural and engineered microbial consortia from the literature and categorize their interactions into different mechanisms for coexistence. We propose that a thorough understanding of these mechanisms can then serve as design strategies for engineering novel microbial communities (Table 1). We note these strategies are not mutually exclusive—indeed, many examples have utilized multiple strategies together to ensure stable coexistence. Additionally, other antagonistic interactions may still result in competitive exclusion between populations in spite of using one or multiple design strategies proposed below. If so, a constraint on these design strategies is the need to ensure sufficient cooperation between members to achieve coexistence. Depending on the context, individual strategies may be more effective at achieving coexistence compared to others, or multiple strategies in conjunction may be necessary.
Resource partitioning
To avoid competitive exclusion, different species typically occupy different ecological niches in nature, a process termed niche differentiation. A common mechanism of niche differentiation is resource partitioning, wherein each community member depends on a different nutrient or set of nutrients such that no two populations directly compete with one another for the same metabolites (Figure 2). These separate pools of nutrients can be inherently present in the environment, provided by different members of the community, or segregated via physical barriers (see Symbiosis and Spatial organization for discussion on the latter two, respectively). In well-mixed systems, implementing resource partitioning in microbial consortia is typically achieved by engineering the metabolic profiles of the members such each member is orthogonal for at least one primary metabolite, typically the preferred carbon source (see Improving functionality via specialization). By ensuring that each member has exclusive access to a key nutrient, resource partitioning reduces competition for resources between populations, which would otherwise result in a monoculture due to interspecific competition (Ley et al. 2006). Indeed, stochastic niche theory also predicts that the probability of a successful consortia assembly with new species depends on its metabolic requirements (Tilman 2004). Specifically, each newly established member reduced the available resources in the environment, decreasing probability of a subsequent “invader” being able to invade the community (Tilman 2004). Thus, successful communities with several coexisting members were composed of metabolically distinct members that occupied different niches (Tilman 2004).
Figure 2.
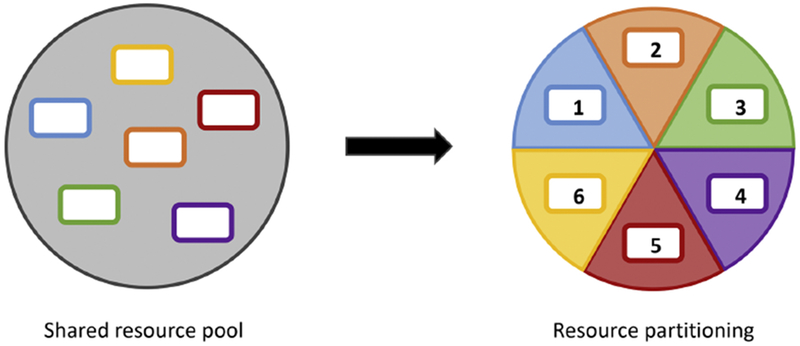
Resource partitioning can maintain coexistence by reducing competition between different species for resources.
Indeed, this strategy is often featured in natural consortia because a single source of a nutrient typically cannot sustain the diversity of species in a community. For instance, carbon heterogeneity in soil was correlated with noncompetitive diversity since populations could avoid competition by specialization (J. Zhou et al. 2002). In another example, a microbial consortium within Olavius alganmsis was shown to metabolize waste products from their host, some of which were shared amongst multiple clusters and some of which were uniquely consumed by particular members (Woyke et al. 2006). Not only does the resource partitioning benefit the different species, especially during periods of nutrient variation, but it also benefits the host, which does not have a digestive nor an excretory system (Woyke et al. 2006). Finally, Wilson et. al. evaluated the coexistence between Pseudomonas syringae and several other strains (Wilson and Lindow 1994). They found that coexistence between species pairs was inversely proportional to the similarity of their respective carbon utilization profiles (Wilson and Lindow 1994). As a result, several species exhibited greater coexistence with P. syringae than a genetically similar P. syringae variant (Wilson and Lindow 1994). As a result, resource partitioning between ecologically different strains reduced the degree of competition between them for resources, enabling more robust coexistence.
Resource partitioning is also commonly used to design synthetic consortia since it not only maintains coexistence between populations also often improves overall output. For example, Eiteman et al. utilized a co-culture of E. coli strains, each of which could only consume one of either glucose or xylose, to produce lactate through simultaneous metabolism of a mixture of both sugars (Eiteman et al. 2009). Similarly, multiple co-culture systems in metabolic engineering also utilized multiple feedstocks, such as glucose and glycerol, to produce compounds of interest (Ahmadi et al. 2016; T. Chen et al. 2019; J. Wang et al. 2018a). Many of the previous studies discussed in Improving functionality via specialization also feature different members of their respective consortia specializing in different carbon sources (Eiteman et al. 2008; Xia et al. 2012; Zhang et al. 2015).
Chemical symbiotic relationships
Chemical interactions between populations can incentivize and stabilize coexistence by tying together the survival of different species. The chemicals exchanged are typically metabolites, which can directly influence growth via cellular metabolism, or small molecules, which can induce gene expression to promote survival via cell-to-cell signaling (Figure 3A). For example, one of the most common symbiotic interactions in natural microbial communities is commensalism in which one population benefits another population but receives no harm or benefit in return. Under the right conditions, commensalism can promote coexistence since a “sturdy” population can support the growth of a lower fitness one that would otherwise be unable to survive (Gudelj et al. 2016). Indeed, many predicted two-species commensal relationships were able to sustain growth of both populations, including both same-species and different-species pairs (Klitgord and Segre 2010). Interestingly, the authors found that E. coli could form commensal relationships with many different species in various environments, specifically serving as the provider, possibly due to its ability to survive on various carbon sources and capability to export numerous byproducts (Klitgord and Segre 2010). Given that E. coli is the standard for microbial engineering, this supports the viability of commensalism as a design strategy for engineering consortia.
Figure 3.
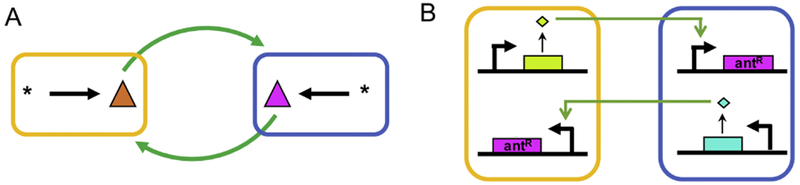
Chemical interactions can stabilize coexistence between different populations.
A) Metabolic symbiosis such as cross-feeding in which different populations produce and share nutrients with one another can outweigh competition and ensure survival of all populations.
B) Cell-cell communication via small molecules can also ensure coexistence. For example, cells could mediate survival of other populations via gene circuits.
Indeed, several engineered consortia have been constructed that utilize commensalism to ensure coexistence between different populations (Kong et al. 2018). For example, Weber et. al. engineered an E. coli-mammalian cell commensal, mutualistic, and predator-prey relationships using cell to cell communication via acetaldehyde (Weber et al. 2007). In their commensal system, E. coli produced acetaldehyde, which induced transcription of neomycin phosphotransferase in mammalian cells when cocultured (Weber et al. 2007). As a result, E. coli enabled survival of the mammalian cells in the presence of neomycin while its own growth was unaffected by the mammalian cells (Weber et al. 2007). Similarly, Brenner et. al. engineered a commensal E. coli-E. coli consortium that cooperate via quorum sensing in a biofilm (Brenner and Arnold 2011). Specifically, one population is unable to synthesize key metabolites for survival unless the second population is present at sufficiently high density (Brenner and Arnold 2011). While this population relies on the first to form the biofilm, this relationship is not mutualistic because the second population can survive independently.
Another symbiotic relationship that can maintain coexistence is mutualism, in which both populations depend on each other for survival. One of the most common methods whereby mutualism is realized is cross-feeding, in which each population relies on metabolites produced by the other population for survival (Bernstein et al. 2012; Harcombe et al. 2014; Shou et al. 2007). An E. coli-S. cereiisiae consortium engineered to produce paclitaxel precursors cooperatively utilized different resources (K. Zhou et al. 2015). Specifically, E. coli consumed xylose and generated the byproduct acetate, which both inhibited the growth of E. coli and served as the sole carbon source of S. cerevisiae. Compared to a previous design in which both populations consumed fed glucose, this approach stabilized the co-culture and doubled the final yield (K. Zhou et al. 2015). Additionally, Mee et. al. engineered E. coli mutants and constructed increasingly complex syntrophic communities to analyze synergistic interactions in microbial communities (Mee et al. 2014). They found that not only did cocultures outgrow the individual strains separately, but also three-member cultures performed even better (Mee et al. 2014). In another example, Pande et. al. engineered several different E. coli variants that were auxotrophic for one amino acid but overproduced a different amino acid (Pande et al. 2014). Coculturing the complementary pairs of strains resulted in increased amino acid production and subsequently higher growth rates over wild-type E. coli via a division of metabolic labor between the individual populations (Pande et al. 2014).
Non-metabolite chemicals can also be used to achieve mutualistic systems (Figure 3B). For example, Hu et al. created an engineered mutualistic system whose population dynamics were modulated via quorum sensing gene circuits (Hu et al. 2010). Specifically, expression of antibiotic resistance genes was regulated by cell density such that modulating antibiotic concentration and initial cell densities could produce various symbiotic interactions, including both mutualism and the previously mentioned commensalism (Hu et al. 2010). From the previous commensalism example, Weber et al. also engineered a mutualistic E. coli-mammalian cell relationship utilizing the same acetylaldehyde signaling design (Weber et al. 2007). In this case, E. coli was sensitive to ampicillin, and acetyladehyde induced both neomycin resistance and production and secretion of beta lactamase in the mammalian population (Weber et al. 2007). As a result, the populations rescue one another from antibiotic-induced death, and neither population was able to grow in the absence of the other (Weber et al. 2007). Similarly, Kong et. al. constructed several two-population symbiotic consortia, including predator-prey, commensal, and mutualistic systems (Kong et al. 2018). In the mutualism case, one population produced a nisin precursor while the second population converted that precursor to active nisin (Kong et al. 2018). Nisin then induced tetracycline resistance in both populations, rescuing them in tetracycline-supplemented media (Kong et al. 2018). Each population was unable to grow by itself but both grew in coculture (Kong et al. 2018). Finally, Wu et. al. engineered two mutualistic E. coli populations which rescued each other from CcdB (toxin)-induced cell death (F. Wu et al. 2019). Specifically, each partner produced a unique quorum sensing molecule, which at sufficiently high density induced expression of the antitoxin CcdA in the opposing population (F. Wu et al. 2019). As a result, either both populations coexisted or the whole system collapsed (F. Wu et al. 2019).
Horizontal gene transfer
Fiorizontal gene transfer (HGT), the non-genealogical transfer of genetic material between microbial species, has been implicated as a primary driving force of microbial evolution (Boto 2010; Gogarten et al. 2002). These mobile genes typically encode functions such as new metabolic capabilities, antibiotic resistance, and virulence that are advantageous for the host under certain conditions (Dutta and Pan 2002). Shuffling these genes can promote diversification and specialization in a microbial community by forming various phenotypes that can coexist in different ecological niches (Polz et al. 2013). For example, HGT in Vibrionaceae populations drove specialization of different populations into three different categories—pioneers, harvesters, and scavengers—that metabolized different alginate polysaccharides (Hehemann et al. 2016). There these populations coexisted symbiotically, as the pioneers degraded insoluble alginate polymer into smaller oligomers that could be consumed by harvesters, which were less efficient at degrading the polymer, and scavengers, which could not use the polymer at all (Hehemann et al. 2016). This level of diversification is possible because HGT can drive acquisition and loss of physiologically novel traits that may not be possible alone with mutations, which occur less often and are not guaranteed to have the desired phenotypic consequence (Dutta and Pan 2002).
Regarding consortia in particular, HGT is a valuable trait for maintaining robust coexistence between populations. For example, HGT can promote coexistence in communities by maintaining cooperative traits (Figure 4). Consider the synthesis of a public good, which is beneficial to a community but also metabolically costly, making the community vulnerable to destabilization by cheater populations that receive the public good without producing any themselves (Nogueira et al. 2009). HGT can prevent loss of a cooperative behavior by co-transferring cellular addiction systems with the public good, converting cheaters into cooperative producers (Nogueira et al. 2009). Additionally, HGT can improve both the stability and robustness of coexistence against environmental perturbation. In this case, HGT expands the range of parameters and initial conditions that lead to coexistence between a cooperator and a cheater (Fan et al. 2018). Similarly, another study found uptake of DNA from the environment was positively correlated with coexistence between cells containing an extrachromosomal plasmid and those not containing it (Mao and Lu 2016). Conversely, coexistence in microbial consortia can be disrupted by suppressing HGT (Lopatkin et al. 2017). For example, conjugation has been shown to maintain plasmids-carrying cells in a population even if the plasmid itself does not provide a selective advantage (Lopatkin et al. 2017). However, plasmid conjugation can be suppressed to eliminate the plasmid-carrying population from the community through competitive exclusion, leaving a monoculture of empty cells (Lopatkin et al. 2017).
Figure 4.
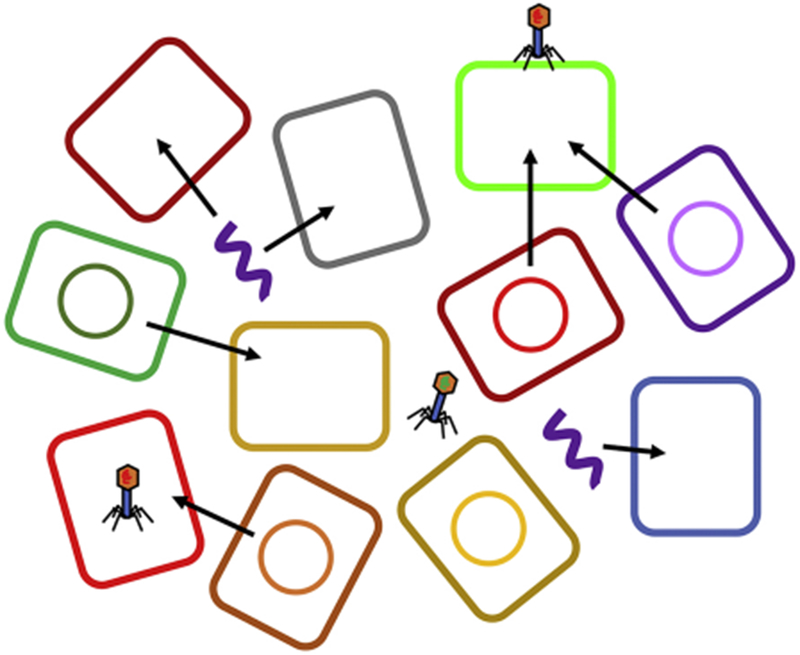
Horizontal gene transfer (HGT) can maintain diversity and modulate coexistence between different populations. For example, HGT can maintain genetic diversity in a population, promoting diversification and specialization of different species into different niches and thus coexistence.
Higher-order interactions
For certain communities, knowing the pairwise interactions between members can enable reliable predictions of overall behavior. However, for other communities, it might be critical to consider higher-order interactions (HOIs) to ensure stable coexistence and function. HOIs describe the non-additive interactions between populations when one population modulates the interaction between two other populations (Figure 5) (Mayfield and Stouffer 2017). Depending on the number of species and interactions between them, HOI networks can be highly sophisticated. As a result, the role of HOIs in consortia and the extent to which they modulate coexistence is likely to be system-specific (Layeghifard et al. 2017). Some studies have found that HOIs play a lesser role in determining community dynamics compared with mono-species growth parameters and pairwise interactions (Venturelli et al. 2018). Instead, pairwise negative feedback loops created by mixtures of positive and negative interactions could enable robust coexistence alone by normalizing relative fitness levels between populations (Venturelli et al. 2018). Similarly, pairwise interactions alone could predict metabolic dynamics between a four-species community of environmental isolates (Guo and Boedicker 2016). In contrast, other studies have found that HOIs do impact community dynamics. For example, Mee et al. engineered two- and three-strain cocultures in which each population was auxotrophic for a different amino acid (Mee et al. 2014). Certain three-member combinations exhibited positive synergy such that the triplet consortia grew significantly more than expected based on pairwise cultures (Mee et al. 2014).
Figure 5.
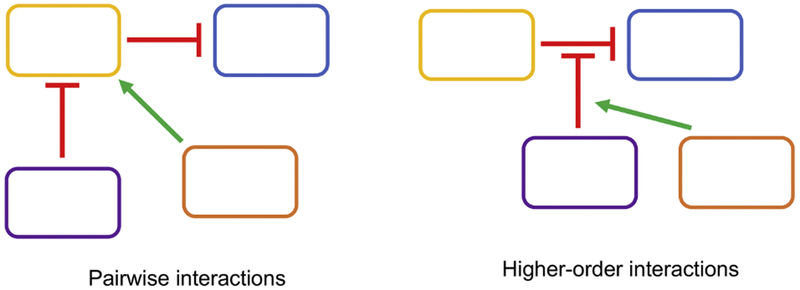
Pairwise interactions only describe the direct interactions between populations (left), while Higher-order interactions describe when an interaction between two populations is modulated by one or more other populations (right). High-order interactions can ensure coexistence in complex microbial communities. In this case, two additional species could decrease the negative interaction between two antagonistic populations but otherwise have no direct competition with them, promoting coexistence between all four populations.
Alternatively, the extent to which pairwise interactions or HOIs modulate coexistence may depend on the size of the community. Bairey et al. found that pairwise interactions were destabilized as the species number increased, whereas four-dimensional HOIs were destabilized as the species number decreased (three-dimensional HOI stability did not change with community size) (Bairey et al. 2016). As a result, communities comprising solely pairwise interactions experienced member extinction beyond a certain threshold, setting an upper bound on member diversity (Bairey et al. 2016). In contrast, communities comprising solely four-dimensional HOIs experienced member extinction below a different threshold, setting a lower bound on member diversity (Bairey et al. 2016). Thus, while we would expect HOIs to play a large role in natural consortia with high species diversity, HOIs may play a lesser role in the stability of engineered consortia comprising a small number of populations. Indeed, other studies have reached similar conclusions regarding HOI importance and community size. For example, Friedman et al. evaluated the predictive power of interactions between subsets of individual members in assembling complex consortia (Friedman et al. 2017). Similar to the study from Bairey et al., they found that the predictive power of their approach depended on the size of the community being designed (Friedman et al. 2017). Specifically, survival of three-species consortia could be predicted from pairwise interactions alone, but seven- and eight-species consortia required information from both pairwise and triple-population interactions to predict accurately (Friedman et al. 2017).
Despite their ambiguous role in predicting community behavior, HOIs have contributed to engineering stable microbial communities in certain cases by facilitating coexistence between members. For example, in a three-member consortium a third population could attenuate the negative interaction between two antagonistic populations (Kelsic et al. 2015). The consortium can also be stable even if the third species is antagonistic to the first two species so long as each population modulates the inhibitory interactions between the remaining two members (Kelsic et al. 2015). These results can extend up to five-population network topologies where all populations compete with one another for resources (Grilli et al. 2017). Alternatively, the presence of an additional population could synergize with an existing community, resulting in a more productive syntrophic consortium. Embree et al. found that, within their syntrophic three-, four-, and five-member consortia, amino acid auxotrophies created complex interdependencies between members that promoted stability and robustness by allowing for metabolic redundancy among community members (Embree et al. 2015). Ultimately, the extent to which a HOI will maintain coexistence in a community will likely depend on the network structure and the level of resource partitioning between members. To this end, mathematical modeling can help predict the degree to which higher order interactions influence the consortium in question, informing engineers on how to address them.
Spatial organization
In a well-mixed condition without forces to promote coexistence, competitive exclusion prevents populations from forming stable microbial consortia (Velazquez et al. 2014; Venters et al. 2017). In nature, microbial communities are typically distributed heterogeneously into different niches with spatially defined structures (Brenner et al. 2007; Lindemann et al. 2016). For example, spatial localization has an important role in the biofilm formation and is tightly related to the functionality of the gut microbiota (Venters et al. 2017; Venturelli et al. 2018). Spatial organization can enable stable coexistence within a microbial consortia by reducing competition for resources via niche differentiation, ensuring survival of the members with lower fitness, and/or improving overall resilience to environmental stresses (Johns et al. 2016). To this end, researchers have used various physical segregation techniques, including microfluidic devices, bioprinting, molding/encapsulation, and other fabrication techniques, to study how the spatial organization affects the interactions among multi-species and the functionality of the whole communities (Figure 6).
Figure 6.
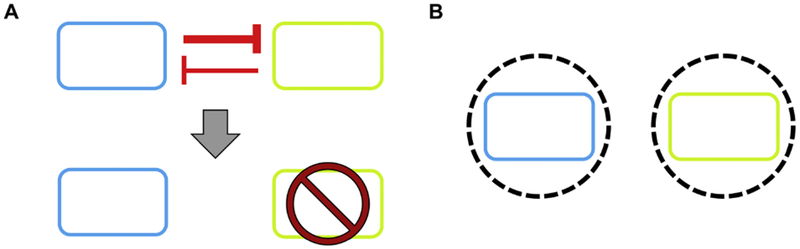
Spatial organization can separate different populations into distinct niches to maintain coexistence.
A) In a well-mixed condition without any stabilizing forces, competition for resources will result in competitive exclusion of the least fit population.
B) By segregating antagonistic populations into their own micro-environments, competition is reduced to maintain stable coexistence of the two populations.
By controlling the “distance” between two different populations, spatial organization can modulate the interaction strength between them. For example, Kim et al. fabricated a microfluidic device to study spatial-dependent interactions within a three-member syntrophic soil consortium that could not coexist in a well-mixed culture due to resource competition (Kim et al. 2008). The authors found that stable coexistence between the three populations occurred at intermediate separation distances of 0.6–1.2 mm where, according to their modeling, the combined consumption rate of the shared nutrients would be comparable to their production rates (Kim et al. 2008). Without spatial segregation, the consortium recapitulated the well-mixed condition, whereas, at a separation distance of 1.8 mm, the syntrophic exchange of nutrients was too low to sustain coexistence (Kim et al. 2008). Similarly, Hynes et al. used bioprinting of microbes to evaluate the distance-dependency of metabolite sharing in an engineered syntrophic consortium of E. coli and S. cerevisiae (Hynes et al. 2018). Growth of both species decreased with increasing separation distance, supporting the notion that spatial organization modulated the strength of cross-feeding between the two populations (Hynes et al. 2018). Moreover, the placement of a third species affected the growth of the consortium depending on the location of the new population relative to the two original members (Hynes et al. 2018). Song et al. also investigated the impact of spatial organization, in the form of habitat partitioning and cellular motility, on biodiversity in a synthetic predator-prey consortium (Song et al. 2009). They found that not only did the spatial segregation between the two populations modulate their coexistence, but also increasing cellular motility (i.e. reducing effective partitioning between populations) decreased biodiversity at intermediate segregation distances (Song et al. 2009). Finally, Borer et al. observed spatial segregation and coexistence of an intermixed community of Pseudomonas putida and Pseudomonas veronii when placed on glass-etched micrometric pore networks that mimicked resource gradients in soil (Borer et al. 2018). In contrast, a well-mixed culture of the two populations resulted in a monoculture of one of the species depending on the availability of oxygen (Borer et al. 2018).
Spatial organization can also promote stable coexistence of microbial consortia by providing beneficial traits. For example, Connell et al. developed a microscopic three-dimensional (3D) printing strategy which can arrange multiple bacterial species in a spatially segregated manner (Connell et al. 2013). Specifically, their technique enables nested arrangements in which one species forms a “shell” around another “core” population (Connell et al. 2013). This configuration allowed Pseudomonas aeruginosa to confer antibiotic resistance to Staphylococcus aureus by surrounding and shielding S. aureus from β-lactam antibiotics (Connell et al. 2013). In another study, Kim et al. evaluated the impact of spatial organization on the metabolic capabilities of a consortium of Sphingobium chlorcphenolicum and Ralstonia metallidurans (Kim et al. 2011). This community was also organized into a core-shell structure with S. chlorophenolicum as the core and R. metallidurans as the shell (Kim et al. 2011). In a well-mixed condition, the consortium was unable to degrade a mixture of pentachlorophenol (PCP) and mercuric ion (Hg(II)) because S. chlorophenolicum was sensitive to Hg(II) (Kim et al. 2011). When organized into the core-shell structure, however, the community was able to remove both chemical pollutants because R. metallidurans protected S. chlorophenolicum from toxic Hg(II) (Kim et al. 2011). In both cases, the spatial organization of two populations could be considered a form of cooperative behavior similar to the chemical symbiotic relationships previously mentioned. It is unclear if the “core” populations provided any fitness benefit to the “shell” populations in return or whether a mutual benefit is necessary to maintain this structure in vivo.
Conclusion
Much of the research towards engineering stable microbial consortia has focused on addressing coexistence between different species or strains. To this end, numerous strategies have been employed to promote cooperation or reduce the competition for resources between populations. However, another key aspect of consortia stability, especially for industrial applications, is the compositional and functional robustness of the communities against perturbations over time. This includes both their ability to maintain themselves in the presence of a perturbation, termed resistance, as well as their ability to return to their original condition after being perturbed, termed resilience (Shade et al. 2012). While such robustness is often cited as a core advantage of microbial consortia, different communities display diverse levels of robustness against various perturbations such as changes in nutrient availability, changes in environmental conditions, and invasion by exploitative “cheater” populations (Lindemann et al. 2016; Mello et al. 2016; Stump et al. 2018; Valentin-Vargas et al. 2014).
For instance, consortia may be more robust to perturbations than monocultures due to their species diversity and cooperation between members within the community. Specifically, environmental perturbations may favor proliferation of specific populations, which can in turn sustain coexistence of the entire community. As a result, temporary propagation of these “hardier” species can buffer the entire community from collapsing and can even maintain the community’s functional stability by taking over tasks from the lower fitness ones. Indeed, this notion is supported by multiple anecdotal examples. In the gut microbiota, while perturbations due to changes in diet can change community composition, the micro flora remains functionally stable (Lozupone et al. 2012). Engineered consortia, even those composed of simpler interaction networks, can also display robustness against changes in the environment. A consortium of cross-feeding S. cerevisiae strains was able to coexist for a wide range of parameters and survived through several population bottlenecks (Shou et al. 2007).
Microbial communities may also be more robust against invasion by non-cooperating populations such as cheaters. Cheaters can arise in a population from loss-of-function mutations or by external invasion and steal resources from functioning cells without paying the same fitness cost. While monocultures can be exploited by cheaters due to their higher relative fitness, consortia can stably maintain coexistence and prevent proliferation of these external populations (Stump et al. 2018). For example, the gut microbiota is responsible for colonization resistance against opportunistic pathogens, which can exploit the gut for nutrients. Moreover, successful invasion by pathogens is more likely when the gut microbiota is destabilized such as by antibiotic treatment. Similarly, a model consortium of three species cooperating via antibiotic degradation was resistant to invasion by cheating populations unless they had a significant relative growth advantage (Kelsic et al. 2015). Microbial communities can resist cheater invasion via various mechanisms, including occupying all available niches, punishing cheaters that arise, selecting for the group via quorum sensing, and spatial organization (Stump et al. 2018; Tilman 2004).
However, the robustness of microbial communities for biotechnological applications remains a fundamental, unresolved question. Engineered consortia in particular can display significantly different behavior depending on their starting conditions, underlining the need for methods to better control and accurately measure them. For example, while many natural mutualistic systems have shown susceptibility to cheater invasion, cheaters may not be able to completely take over and sometimes can actually benefit the system (Ferriere et al. 2002; Genini et al. 2010). Additionally, the increased complexity of a microbial community may destabilize the community, reducing its intrinsic stability and its ability to withstand perturbations. Instead of promoting cooperation, additional populations may instead result in more antagonistic pairwise or HOIs among the consortium, opening up niches for foreign invaders to eventually exploit the unstable coexistence. We speculate that whether a community is more or less robust depends on the structure of the community as well as the environmental conditions surrounding it. To this end, future work is needed to probe and quantify the factors influencing consortia stability in diverse natural or engineered consortia. Doing so may produce key design principles that can help guide engineering design of novel communities for industrial and/or medical applications.
Table 2.
Design strategies to maintain coexistence in engineered microbial consortia
| Design strategy | Coexistence mechanisms | Examples |
|---|---|---|
| Resource partitioning | Reducing competition for resources | E. coli-E. coli coculture producing lactate from both glucose and xylose (Eiteman et al. 2009) |
| Chemical symbiosis | Engineering commensalism via non-metabolite molecules | Acetaldehyde-mediated coexistence of E. coli- mammalian cell consortium (Weber et al. 2007) |
| Engineering metabolic syntrophy | Pairing auxotrophic E. coli strains that cross-fed amino acids (Pande et al. 2014) | |
| Engineering mutualism via non-metabolite molecules | Cooperative, nisin-mediated survival of a Lactococcus lactis-L. lactis consortium (Kong et al. 2018) | |
| Horizontal gene transfer | Promoting specialization | HGT-induced specialization of Vibrionaceae populations into pioneers, harvesters, and scavengers (Hehemann etal. 2016) |
| Higher-order interactions | Modulating interaction strength between members | Complex interdependences within a methanogenic community created by amino acid auxotrophies (Embree et al. 2015) |
| Spatial organization | Modulating interaction strength between members | Spatial organization modulates the strength of cross-feeding between two members of a syntrophic consortium (Kim et al. 2008; Song et al. 2009) |
| Conferring beneficial/cooperative traits | P. aeruginosa surrounding and shielding S. aureus from β-lactam antibiotics in a core-shell structure (Connell et al. 2013) | |
Acknowledgements
We thank F. Wu for her insightful comments and suggestions. Research in the You lab related to the review has been partially supported by the National Science Foundation, the National Institutes of Health, the Office of Naval Research, the Army Research Office, and a David and Lucile Packard Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmadi MK, et al. (2016), ‘E. coli metabolic engineering for gram scale production of a plant-based anti-inflammatory agent’, Metab Eng, 38, 382–88. [DOI] [PubMed] [Google Scholar]
- Atsumi S, Hanai T, and Liao JC (2008), ‘Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels’, Nature, 451 (7174), 86–9. [DOI] [PubMed] [Google Scholar]
- Bairey E, Kelsic ED, and Kishony R (2016), ‘High-order species interactions shape ecosystem diversity’, Nat Commun, 7, 12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein HC, Paulson SD, and Carlson RP (2012), ‘Synthetic Escherichia coli consortia engineered for syntrophy demonstrate enhanced biomass productivity’, J Biotechnol, 157 (1), 159–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettiga M, et al. (2009), ‘Arabinose and xylose fermentation by recombinant Saccharomyces cerevisiae expressing a fungal pentose utilization pathway’, Microb Cell Fact, 8, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer B, Tecon R, and Or D (2018), ‘Spatial organization of bacterial populations in response to oxygen and carbon counter-gradients in pore networks’, Nat Commun, 9 (1), 769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boto L (2010), ‘Horizontal gene transfer in evolution: facts and challenges’, Proc Biol Sci, 277 (1683), 819–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner K and Arnold FH (2011), ‘Self-organization, layered structure, and aggregation enhance persistence of a synthetic biofilm consortium’, PLoS One, 6 (2), e16791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner K, et al. (2007), ‘Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium’, Proc Natl Acad Sci U S A, 104 (44), 17300–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro CC, et al. (2019), ‘Microbial co-culturing strategies for fructo-oligosaccharide production’, N Biotechnol, 51, 1–7. [DOI] [PubMed] [Google Scholar]
- Chen T, et al. (2019), ‘Advances in heterologous biosynthesis of plant and fungal natural products by modular co-culture engineering’, Biotechnol Lett, 41 (1), 27–34. [DOI] [PubMed] [Google Scholar]
- Chen ZY, et al. (2017), ‘Metabolic engineering of Escherichia coli for microbial synthesis of monolignols’, Metabolic Engineering, 39, 102–09. [DOI] [PubMed] [Google Scholar]
- Cheng Allen A. and Lu Timothy K. (2012), ‘Synthetic Biology: An Emerging Engineering Discipline’, Annual Review of Biomedical Engineering, 14 (1), 155–78. [DOI] [PubMed] [Google Scholar]
- Clemente JC, et al. (2012), ‘The impact of the gut microbiota on human health: an integrative view’, Cell, 148 (6), 1258–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell JL, et al. (2013), ‘3D printing of microscopic bacterial communities’, Proc Natl Acad Sci U S A, 110 (46), 18380–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cydzik-Kwiatkowska A and Zielinska M (2016), ‘Bacterial communities in full-scale wastewater treatment systems’, World J Microbiol Biotechnol, 32 (4), 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demain AL (2000), ‘Microbial biotechnology’, Trends in Biotechnology, 18 (1), 26–31. [DOI] [PubMed] [Google Scholar]
- Dutta C and Pan A (2002), ‘Horizontal gene transfer and bacterial diversity’, J Biosci, 27 (1 Suppl 1), 27–33. [DOI] [PubMed] [Google Scholar]
- Eiteman MA, Lee SA, and Altman E (2008), ‘A co-fermentation strategy to consume sugar mixtures effectively’, J Biol Eng, 2, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiteman MA, et al. (2009), ‘A substrate-selective co-fermentation strategy with Escherichia coli produces lactate by simultaneously consuming xylose and glucose’, Biotechnol Bioeng, 102 (3), 822–7. [DOI] [PubMed] [Google Scholar]
- Embree M, et al. (2015), ‘Networks of energetic and metabolic interactions define dynamics in microbial communities’, Proc Natl Acad Sci U S A, 112 (50), 15450–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, et al. (2018), ‘Horizontal gene transfer can help maintain the equilibrium of microbial communities’, J Theor Biol, 454, 53–59. [DOI] [PubMed] [Google Scholar]
- Fang Z, et al. (2018), ‘Engineering Escherichia coli Co-Cultures for Production of Curcuminoids From Glucose’, Biotechnology Journal, 13 (5). [DOI] [PubMed] [Google Scholar]
- Ferriere R, et al. (2002), ‘Cheating and the evolutionary stability of mutualisms’, Proc Biol Sci, 269 (1493), 773–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Higgins LM, and Gore J (2017), ‘Community structure follows simple assembly rules in microbial microcosms’, Nat Ecol Evol, 1 (5), 109. [DOI] [PubMed] [Google Scholar]
- Genini J, et al. (2010), ‘Cheaters in mutualism networks’, Biol Lett, 6 (4), 494–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten JP, Doolittle WF, and Lawrence JG (2002), ‘Prokaryotic evolution in light of gene transfer’, Mol Biol Evol, 19 (12), 2226–38. [DOI] [PubMed] [Google Scholar]
- Grilli J, et al. (2017), ‘Higher-order interactions stabilize dynamics in competitive network models’, Nature, 548 (7666), 210–13. [DOI] [PubMed] [Google Scholar]
- Gudelj I, et al. (2016), ‘Stability of Cross-Feeding Polymorphisms in Microbial Communities’, PLoS Comput Biol, 12 (12), e1005269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X and Boedicker JQ (2016), ‘The Contribution of High-Order Metabolic Interactions to the Global Activity of a Four-Species Microbial Community’, PLoS Comput Biol, 12 (9), e1005079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcombe WR, et al. (2014), ‘Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics’, Cell Rep, 7 (4), 1104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins KM and Smolke CD (2008), ‘Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae’, Nat Chem Biol, 4 (9), 564–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector RE, et al. (2011), ‘Engineering industrial Saccharomyces cerevisiae strains for xylose fermentation and comparison for switchgrass conversion’, J Ind Microbiol Biotechnol, 38 (9), 1193–202. [DOI] [PubMed] [Google Scholar]
- Hehemann JH, et al. (2016), ‘Adaptive radiation by waves of gene transfer leads to fine-scale resource partitioning in marine microbes’, Nat Commun, 7, 12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, et al. (2010), ‘An environment-sensitive synthetic microbial ecosystem’, PLoS One, 5 (5), e10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes William F, et al. (2018), ‘Bioprinting microbial communities to examine interspecies interactions in time and space’, Biomedical Physics & Engineering Express, 4 (5). [Google Scholar]
- Isikgor Furkan H. and Becer C. Remzi (2015), ‘Lignocellulosic biomass: a sustainable platform for the production of bio-based chemicals and polymers’, Polymer Chemistry, 6 (25), 4497–559. [Google Scholar]
- Johns NI, et al. (2016), ‘Principles for designing synthetic microbial communities’, Current Opinion in Microbiology, 31, 146–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JA, et al. (2017), ‘Complete Biosynthesis of Anthocyanins Using E. coli Polycultures’, MBio, 8 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsic ED, et al. (2015), ‘Counteraction of antibiotic production and degradation stabilizes microbial communities’, Nature, 521 (7553), 516–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatun MA, et al. (2018), ‘Bacterial Consortium-Based Sensing System for Detecting Organophosphorus Pesticides’, Analytical Chemistry, 90 (17), 10577–84. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Du WB, and Ismagilov RF (2011), ‘Complex function by design using spatially pre-structured synthetic microbial communities: degradation of pentachlorophenol in the presence of Hg(II)’, Integrative Biology, 3 (2), 126–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, et al. (2008), ‘Defined spatial structure stabilizes a synthetic multispecies bacterial community’, Proc Natl Acad Sci U S A, 105 (47), 18188–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klitgord N and Segre D (2010), ‘Environments that induce synthetic microbial ecosystems’, PLoS Comput Biol, 6 (11), e1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, et al. (2018), ‘Designing microbial consortia with defined social interactions’, Nat Chem Biol, 14 (8), 821–29. [DOI] [PubMed] [Google Scholar]
- Layeghifard M, Hwang DM, and Guttman DS (2017), ‘Disentangling Interactions in the Microbiome: A Network Perspective’, Trends Microbiol, 25 (3), 217–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, and Gordon JI (2006), ‘Ecological and evolutionary forces shaping microbial diversity in the human intestine’, Cell, 124 (4), 837–48. [DOI] [PubMed] [Google Scholar]
- Li Z, Wang X, and Zhang H (2019), ‘Balancing the non-linear rosmarinic acid biosynthetic pathway by modular co-culture engineering’, Metab Eng. [DOI] [PubMed] [Google Scholar]
- Lindemann SR, et al. (2016), ‘Engineering microbial consortia for controllable outputs’, ISME J, 10 (9), 2077–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YQ, et al. (2018), ‘Engineered monoculture and co-culture of methylotrophic yeast for de novo production of monacolin J and lovastatin from methanol’, Metabolic Engineering, 45, 189–99. [DOI] [PubMed] [Google Scholar]
- Lopatkin AJ, et al. (2017), ‘Persistence and reversal of plasmid-mediated antibiotic resistance’, Nat Commun, 8 (1), 1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, et al. (2012), ‘Diversity, stability and resilience of the human gut microbiota’, Nature, 489 (7415), 220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J and Lu T (2016), ‘Population-Dynamic Modeling of Bacterial Horizontal Gene Transfer by Natural Transformation’, Biophys J, 110 (1), 258–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield MM and Stouffer DB (2017), ‘Higher-order interactions capture unexplained complexity in diverse communities’, Nat Ecol Evol, 1 (3), 62. [DOI] [PubMed] [Google Scholar]
- Mee MT, et al. (2014), ‘Syntrophic exchange in synthetic microbial communities’, Proc Natl Acad Sci U S A, 111 (20), E2149–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello BL, et al. (2016), ‘Nutrient availability shapes the microbial community structure in sugarcane bagasse compost-derived consortia’, Sci Rep, 6, 38781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami H, et al. (2008), ‘Microbial production of plant benzylisoquinoline alkaloids’, Proc Natl Acad Sci U S A, 105 (21), 7393–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minty JJ, et al. (2013), ‘Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass’, Proc Natl Acad Sci U S A, 110 (36), 14592–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira T, et al. (2009), ‘Horizontal gene transfer of the secretome drives the evolution of bacterial cooperation and virulence’, Curr Biol, 19 (20), 1683–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande S, et al. (2014), ‘Fitness and stability of obligate cross-feeding interactions that emerge upon gene loss in bacteria’, ISME J, 8 (5), 953–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polz MF, Alm EJ, and Hanage WP (2013), ‘Horizontal gene transfer and the evolution of bacterial and archaeal population structure’, Trends Genet, 29 (3), 170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage VM, Webb CT, and Norberg J (2007), ‘A general multi-trait-based framework for studying the effects of biodiversity on ecosystem functioning’, J Theor Biol, 247 (2), 213–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlak M, Edenberg HJ, and Ho NWY (2003), ‘DNA microarray analysis of the expression of the genes encoding the major enzymes in ethanol production during glucose and xylose co-fermentation by metabolically engineered Saccharomyces yeast’, Enzyme and Microbial Technology, 33 (1), 19–28. [Google Scholar]
- Shade A, et al. (2012), ‘Fundamentals of microbial community resistance and resilience’, Front Microbiol, 3, 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shong J, Jimenez Diaz MR, and Collins CH (2012), ‘Towards synthetic microbial consortia for bioprocessing’, Curr Opin Biotechnol, 23 (5), 798–802. [DOI] [PubMed] [Google Scholar]
- Shou W, Ram S, and Vilar JM (2007), ‘Synthetic cooperation in engineered yeast populations’, Proc Natl Acad Sci U S A, 104 (6), 1877–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, et al. (2009), ‘Spatiotemporal modulation of biodiversity in a synthetic chemical-mediated ecosystem’, Nat Chem Biol, 5 (12), 929–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierle AA, et al. (2017), ‘The Berkeleylactones, Antibiotic Macrolides from Fungal Coculture’, J Nat Prod, 80 (4), 1150–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump SM, Johnson EC, and Klausmeier CA (2018), ‘Local interactions and self-organized spatial patterns stabilize microbial cross-feeding against cheaters’, J R Soc Interface, 15 (140). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuan NH, et al. (2018), ‘Engineering co-culture system for production of apigetrin in Escherichia coli’, Journal of Industrial Microbiology & Biotechnology, 45 (3), 175–85. [DOI] [PubMed] [Google Scholar]
- Tilman D (2004), ‘Niche tradeoffs, neutrality, and community structure: a stochastic theory of resource competition, invasion, and community assembly’, Proc Natl Acad Sci U S A, 101 (30), 10854–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi R, et al. (2018), ‘Metabolic division of labor in microbial systems’, Proc Natl Acad Sci U S A, 115 (10), 2526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Vargas A, et al. (2014), ‘Environmental factors influencing the structural dynamics of soil microbial communities during assisted phytostabilization of acid-generating mine tailings: a mesocosm experiment’, Sci Total Environ, 500–501, 314–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez J, Garrahan JP, and Eichhorn MP (2014), ‘Spatial Complementarity and the Coexistence of Species’, PLoS One, 9 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters M, et al. (2017), ‘Effects of Spatial Localization on Microbial Consortia Growth’, Plos One, 12 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturelli OS, et al. (2018), ‘Deciphering microbial interactions in synthetic human gut microbiome communities’, Mol Syst Biol, 14 (6), e8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. (2018a), ‘A Novel Process for Cadaverine Bio-Production Using a Consortium of Two Engineered Escherichia coli’, Front Microbiol, 9, 1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. (2018b), ‘A Novel Process for Cadaverine Bio-Production Using a Consortium of Two Engineered Escherichia coli’, Frontiers in Microbiology, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, et al. (2019), ‘Simultaneous fermentation of biomass-derived sugars to ethanol by a co culture of an engineered Escherichia coli and Saccharomyces cerevisiae’, Bioresource Technology, 273, 269–76. [DOI] [PubMed] [Google Scholar]
- Weber W, Daoud-El Baba M, and Fussenegger M (2007), ‘Synthetic ecosystems based on airborne inter- and intrakingdom communication’, Proc Natl Acad Sci U S A, 104 (25), 10435–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SA and Cooper GA (2016), ‘Division of labour in microorganisms: an evolutionary perspective’, Nat Rev Microbiol, 14 (11), 716–23. [DOI] [PubMed] [Google Scholar]
- Wilson M and Lindow SE (1994), ‘Coexistence among Epiphytic Bacterial Populations Mediated through Nutritional Resource Partitioning’, Appl Environ Microbiol, 60 (12), 4468–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyke T, et al. (2006), ‘Symbiosis insights through metagenomic analysis of a microbial consortium’, Nature, 443 (7114), 950–5. [DOI] [PubMed] [Google Scholar]
- Wu Feilun, et al. (2019), ‘A unifying framework for interpreting and predicting mutualistic systems’, Nature Communications, 10 (1), 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, et al. (2016), ‘Metabolic Burden: Cornerstones in Synthetic Biology and Metabolic Engineering Applications’, Trends Biotechnol, 34 (8), 652–64. [DOI] [PubMed] [Google Scholar]
- Xia T, Eiteman MA, and Altman E (2012), ‘Simultaneous utilization of glucose, xylose and arabinose in the presence of acetate by a consortium of Escherichia coli strains’, Microb Cell Fact, 11, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiu Y, et al. (2017), ‘Naringenin-responsive riboswitch-based fluorescent biosensor module for Escherichia coli co-cultures’, Biotechnol Bioeng, 114 (10), 2235–44. [DOI] [PubMed] [Google Scholar]
- Zhang H, et al. (2015), ‘Engineering Escherichia coli coculture systems for the production of biochemical products’, Proc Natl Acad Sci U S A, 112 (27), 8266–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, et al. (2002), ‘Spatial and resource factors influencing high microbial diversity in soil’, Appl Environ Microbiol, 68 (1), 326–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, et al. (2015), ‘Distributing a metabolic pathway among a microbial consortium enhances production of natural products’, Nat Biotechnol, 33 (4), 377–83. [DOI] [PMC free article] [PubMed] [Google Scholar]


