Abstract
The aim of this study was to provide a comprehensive analysis of the plant-specific B3 domain-containing transcription factors (TFs) in chickpea. Scanning of the chickpea genome resulted in the identification of 51 B3 domain-containing TFs that were located on seven out of eight chickpea chromosomes. Based on the presence of additional domains other than the B3 domain, the candidates were classified into four subfamilies, i.e., ARF (24), REM (19), LAV (6) and RAV (2). Phylogenetic analysis classified them into four groups in which members of the same group had similar intron–exon organization and motif composition. Genome duplication analysis of the candidate B3 genes revealed an event of segmental duplication that was instrumental in the expansion of the B3 gene family. Ka/Ks analysis showed that the B3 gene family was under purifying selection. Further, chickpea B3 genes showed maximum orthology with Medicago followed by soybean and Arabidopsis. Promoter analyses of the B3 genes led to the identification of several tissue-specific and stress-responsive cis-regulatory elements. Expression profiling of the candidate B3 genes using publicly available RNA-seq data of several chickpea tissues indicated their putative role in plant development and abiotic stress response. These findings were further validated by real-time expression analysis. Overall, this study provides a comprehensive analysis of the B3 domain-containing proteins in chickpea that would aid in devising strategies for crop manipulation in chickpea.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1875-5) contains supplementary material, which is available to authorized users.
Keywords: Chickpea, B3 domain-containing protein, Phylogeny, Expression analysis
Introduction
Transcription factors (TFs) are critical regulatory elements that bind to the specific DNA sequences present over the promoter regions of their target genes (Franco-Zorrilla et al. 2014). With the availability of whole genome sequence, the numbers of candidate transcription factors with implicated regulatory roles from different species are continuously increasing in numerous transcription factor databases. At present, a total of 320,370 TFs from 165 species are submitted in the plant transcription factor database (Jin et al. 2017). Amongst them, the B3 domain-containing transcription factors are plant-specific TFs that play key roles in various facets of plant development. In addition, these TFs are instrumental for plant adaptation and survival during various abiotic and biotic stresses (Sasnauskas et al. 2018; Swaminathan et al. 2008). The members of this family have also been found to be involved in various hormone-related signal transduction pathways, including abscisic acid (ABA), ethylene, cytokinin and jasmonates (JAs) (Romanel et al. 2009; Swaminathan et al. 2008). All proteins, encoded by B3 genes contain a highly conserved region of ~ 110 aa that encodes a B3 domain. A typical B3 domain usually contains seven β strands arranged like an open barrel and two alpha helices (Romanel et al. 2009). The B3 domain was initially recognized in the VIVIPAROUS (VP1) gene of Zea mays (Suzuki et al. 1997). Members of this superfamily can be further classified into four subfamilies based on the occurrence of domains other than B3 domain such as LAV [Leafy Cotyledon2 (LEC2) Abscisic Acid Insensitive3 (ABI3)-VAL], RAV (related to ABI3 and VP1), auxin response factor (ARF) and reproductive meristem (REM) (Swaminathan et al. 2008). B3 family members from different crop species have been identified utilizing whole genome sequences that include 92, 77, 55 and 88 B3 proteins from Arabidopsis, rice, maize and poplar, respectively (Peng and Weselake 2013; Romanel et al. 2009). Different binding sites of the B3 domain have been reported in various studies while analyzing B3 members of distinct families. The B3 domain of LAV family members binds to RY elements (CATGCA) in the promoter regions of seed-specific genes (Reidt et al. 2000; Tsukagoshi et al. 2005). The members of the ARF family possess N-terminal B3 domains that recognize TGTCTC motifs (Yamasaki et al. 2013). Similarly, the N-terminal region of RAV family proteins harbors an AP2 domain that can bind to the CAACA sequence, and C-terminal region containing B3 domain that recognizes the CACCTG motif (Magnani et al. 2004).
Three well-known B3 family members that belong to AFL network (ABI3, FUS3 and LEC2) under LAV subfamily have been characterized in Arabidopsis (Devic and Roscoe 2016: Kim et al. 2013). Their roles in seed development, particularly during storage and maturation stage, have been established using the loss-of-function mutants of the corresponding genes. Similarly, some members of RAV subfamily such as Tempranillo1 (TEM1), (TEM2) and NAGATHA genes have been shown to play prominent roles during flower and leaf development (Castillejo and Pelaz 2008; Gu et al. 2017). A large number of studies identifying the ARF family members on a genome-wide scale in various plants have been done. In such studies, 23, 24, 31 and 51 ARF TFs have been reported in Arabidopsis, Medicago, Brassica and soybean, respectively (Li et al. 2016; Mun et al. 2012; Shen et al. 2015; Van Ha et al. 2013). Role of several ARF genes such as MONOPTEROUS (ARF5), ETTIN (ARF3), ARF6, ARF7 and ARF8 have been established in embryo patterning, carpel development, floral organ maturation, lateral root formation and development of parthenocarpic fruit, respectively (Li et al. 2016; Vidaurre et al. 2007; Zhang et al. 2018).
Chickpea is one of the most important legume crops in the world, especially for human consumption as its seeds are good sources of protein, carbohydrate and minerals (Pradhan et al. 2014). As chickpea root nodules contribute to nitrogen fixation it offers benefits to cereal-based cropping systems through crop rotation (Kant et al. 2016). The available whole genome sequences of chickpea have facilitated identification of different members of various gene families (Jain et al. 2013; Varshney et al. 2013). In chickpea, genome-wide analyses of various transcription factor families such as NAC (Ha et al. 2014), AP2 (Agarwal et al. 2016), Aux/IAA (Singh and Jain 2015), and C3H (Pradhan et al. 2017) has been done soon after the availability of its whole genome sequence. However, the B3 superfamily that contains several members known to regulate various developmental processes remains largely unexplored in chickpea.
To this effect, in this study, identification of the members of B3 superfamily was undertaken at a genome-wide scale. The phylogenetic relationships of the identified members with homologs from Medicago and Arabidopsis were studied. In addition, these genes were subjected to gene structure analysis, promoter sequence analysis, and evolutionary analysis. Apart from this, expression profiling of the B3 members using the available RNA-seq data of various chickpea tissues and under stress conditions has also been carried out. Furthermore, quantitative real-time PCR (qRT-PCR) of selected B3 genes was done to validate the in silico expression patterns of the B3 genes. Additionally, expression profiling of selected B3 members was examined in response to treatment with plant hormones.
Materials and methods
Plant materials, tissue collection and treatment
Cicer arietinum cv. ICCV2 was grown at National Institute of Plant Genome Research, India, and used for harvesting of developing seeds. Field-grown plants were observed for flowering. The individual flowers were tagged at the first day of anthesis and subsequently used for collecting seeds at different time points, i.e., 10, 20, 30 and 40 days after anthesis (DAA) in three biological replicates. For vegetative tissues collection, the plants were maintained in a growth chamber at a plant growth facility at NIPGR under optimal conditions with 16/8 h of light/dark at 22 °C and 64% relative humidity. Leaf and root tissue were collected from 10-day-old seedlings, flash-frozen in liquid N2 and used for RNA isolation.
To understand the response of chickpea B3 genes under various stress treatments, 10-day-old seedlings were subjected to four different stresses: dehydration, desiccation, cold and salinity. Initially, chickpea seeds were germinated and grown in sterilized soilrite soaked with half-strength MS medium for 10 days in controlled growth conditions. For dehydration stress and salt stress, 10-day-old seedlings were transferred to the solutions of 20% PEG4000 and 150 mM NaCl in half-strength MS medium. For desiccation stress, seedlings were kept between the blotting sheets. The seedlings were kept at 4 °C to evoke cold stress. Shoots were harvested from stressed samples after 0 (control), 3, 6, 12 and 24 h. For hormonal treatment, 10-day-old seedlings were transferred to ½ MS solution containing ABA (100 µM) and IBA (10 µM) hormones for 0 (control), 3, 6, 12, 24 h. Seedlings that were transferred to 1/2 MS solution without any additives were used as 0 h control for normalization. Shoots of six seedlings were bulked and used for RNA isolation.
Identification of B3 domain-containing proteins from the chickpea genome
To identify the B3 domain-containing proteins from the chickpea whole genome (Varshney et al. 2013), HMM profile of B3 domain (PF02362) was downloaded from the Pfam database (http://www.pfam.xfam.org/) and used to carry out HMM search (HMMER v3.1) against all predicted proteins of kabuli chickpea (Varshney et al. 2013). The identified proteins were retrieved and scanned using InterProScan v5.0 for the presence of specific B3 domain. Further, the sequences were checked for redundancy and subjected to Blast (https://www.blast2go.com/) analysis in order to assign annotations. Sequences of the B3 domain-containing proteins of other plant species, i.e., Arabidopsis, Medicago and G. max were downloaded from Phytozome v10.
Chromosomal positions, duplication events and prediction of Ka/Ks values
The GFF file of kabuli chickpea genome was used to identify the start positions of the B3 genes and assign chromosomal locations on the eight chickpea chromosomes. Their locations were visualized using the MapChart software. Duplication events that occurred in the B3 genes were predicted using MCScanX (Wang et al. 2012). Ka/Ks values were calculated among orthologous and paralogous genes using a PAL2NAL web server (Suyama et al. 2006).
Analysis of phylogeny, gene structure, conserved domains and motifs
Full-length amino acid sequences were used for phylogenetic tree construction. Sequences were aligned using Clustal W. The phylogenetic trees were constructed using the neighbor-joining method on MEGA6 (Tamura et al. 2013) with a number of bootstrap replications as 1000. Domain analysis was carried out using the InterProScan (http://www.ebi.ac.uk/interpro/) web tool and CD search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The Pfam database was used to examine the schemata of various domains present over the amino acid sequences (Finn et al. 2016). MEME v4.12.0 (Bailey et al. 2009) was used to search for conserved motifs in the candidate protein sequences. The maximum number of motifs was set as 10. Genomic organization of chickpea B3 genes was visualized using Gene Structure Display Server (GSDS) v2.0 (Guo et al. 2007). The intron–exon information of B3 genes in GFF3 file format was used as input.
Synteny analysis
To perform synteny analysis, the chickpea B3 gene sequences were aligned to the genomes of Medicago, soybean and Arabidopsis (Phytozome v10) using BLASTn with an E value cut-off of 1E−05. Mapped locations of B3 genes were utilized for comparison. Circos software (v0.61) (Krzywinski et al. 2009) was used for diagrammatical visualization of synteny.
Promoter analysis
For in silico promoter analysis, the 2 kb sequences upstream of the ATG start site of B3 genes were retrieved from chickpea genome (Varshney et al. 2013) and analyzed using PLACE database (Higo et al. 1998).
Expression profiling using RNA-seq data
The expression pattern of the B3 genes was examined using the available RNA-seq data of different chickpea tissues: leaf, root, flower-bud, and pod (SRX048833, SRX048832, SRX048834, and SRX208035) and seed tissues (SRX125162). Further, the expression pattern of B3 genes was also analyzed under three abiotic stresses by using the available RNA-seq data of chickpea seedlings under desiccation, salinity and cold stresses (SRP034839). Filtered reads from all tissues were mapped onto the identified B3 genes using Burrows–Wheeler Aligner with default parameters (Li and Durbin 2009). Mapped reads were used for measuring RPKM values. Heat maps were generated using log2 normalized values in the MeV software (Howe et al. 2011).
RNA extraction and qRT-PCR analysis
Total RNA from different chickpea tissues were extracted using lithium chloride (Choudhary et al. 2009). RNA yield and purity were assessed by measuring 260/280 and 260/230 ratio using NanoDropTM 8000 (Thermo Fisher Scientific). Only the RNA samples with a 260/280 ratio of ~ 2.0 and 260/230 ratio from 2.0 to 2.3 were used for the analysis. The integrity of RNA samples was also assessed by agarose gel electrophoresis. Primer pairs were designed employing the Primer Express software (Applied Biosystems, USA). All primer sequences are listed in Online Resource 1. cDNA synthesis and qRT-PCR was carried out according to the protocol as described in Verma and Bhatia (2019). Statistical significance was determined using the Student’s two-tailed t test. Genes that had ≥ 2-fold expression change (as compared to control) with P ≤ 0.05 were considered as differentially expressed/up-regulated or having significantly higher expression.
Results
Identification of B3 domain-containing proteins
To identify B3 domain-containing proteins, HMM search was carried out using the HMM profile of the B3 domain against chickpea protein dataset (Varshney et al. 2013). This resulted in the identification of 51 full-length B3 domain-containing proteins that were further confirmed by the presence of at least one B3 domain in their amino acid sequences. Domains present in chickpea B3 proteins and their positions are listed in Online Resource 2. The corresponding genes of identified proteins were designated as CarB3_1–CarB3_51 according to the position on the chickpea chromosomes 1–8. Further, these genes were used to carry out Blast search against the NCBI database using the Blast2GO tool to identify their annotation description which is listed in Table 1. These genes were classified into four subfamilies based on the presence of additional domains other than the B3 domain. Based on the presence of a conserved region (Auxin_resp), 24 proteins were classified as ARF proteins. In addition to the B3 domain and Auxin_resp, some of them also contained Aux/IAA domain (PF02309). Those containing one or more than one B3 domain were classified as REM (19 nos.), and proteins containing an additional AP2 domain were classified as RAV proteins (2 nos.). In Arabidopsis, the LAV subfamily consists of two subgroups: the LEC2-ABI3 subgroup and the VAL subgroup. Based on the presence of an additional zf-CW domain, chickpea B3 proteins were classified into VAL subgroup. The three well-known members namely ABI3, FUS3 and LEC2 were classified into LEC2-ABI3 subgroup. A schematic representation of the domain composition of different subfamilies is depicted in Online Resource 3. The 51 identified CarB3 proteins from chickpea ranged from 145 (CarB3_43) to 1090 (CarB3_13) amino acids in length, with a relative molecular mass of 16.19 kDa (CarB3_43) to 123.34 kDa (CarB3_13) and protein pIs from 5.03 (CarB3_37) to 9.91 (CarB3_36) (Table 1).
Table 1.
Characteristics of chickpea B3 proteins
| Gene name | Chickpea_id | Chromosome/scaffold | Predicted protein | Family | NCBI annotation | ||
|---|---|---|---|---|---|---|---|
| Length (aa) | Mw (Da) | PI | |||||
| CarB3_1 | Ca_25633 | scaffold151 | 322 | 37,072.85 | 7.78 | REM | b3 domain-containing protein at5g06250-like |
| CarB3_2 | Ca_24123 | scaffold470 | 382 | 41,763.25 | 8.99 | RAV | ap2 erf and b3 domain-containing transcription repressor tem1-like |
| CarB3_3 | Ca_23185 | scaffold682 | 907 | 100,349.05 | 6.33 | VAL | b3 domain-containing transcription repressor val2-like isoform x1 |
| CarB3_4 | Ca_21518 | scaffold1281 | 794 | 88,689.71 | 5.71 | LAV/ABI3 | b3 domain-containing transcription factor abi3-like isoform x1 |
| CarB3_5 | Ca_11175 | scaffold1301_1 | 449 | 49,576.73 | 6.18 | ARF | Auxin response factor 17-like |
| CarB3_6 | Ca_00244 | Ca1 | 854 | 94,533.74 | 6.72 | VAL | b3 domain-containing protein os07g0563300-like |
| CarB3_7 | Ca_00347 | Ca1 | 699 | 76,248.34 | 6.13 | ARF | Auxin response factor 3 |
| CarB3_8 | Ca_00467 | Ca1 | 915 | 101,575.86 | 6.22 | ARF | Auxin response factor 6 |
| CarB3_9 | Ca_02541 | Ca1 | 719 | 79,444.74 | 8.56 | ARF | Auxin response factor 18-like |
| CarB3_10 | Ca_02516 | Ca1 | 742 | 83,157.89 | 8.01 | ARF | Auxin response factor-like protein |
| CarB3_11 | Ca_19289 | Ca1 | 830 | 92,157.49 | 5.7 | ARF | Auxin response factor 19-like |
| CarB3_12 | Ca_15659 | Ca2 | 773 | 85,244.41 | 6.25 | REM | b3 domain-containing transcription repressor val1 |
| CarB3_13 | Ca_15677 | Ca2 | 1090 | 123,348.24 | 9.45 | REM | Metal tolerance protein 4-like |
| CarB3_14 | Ca_15694 | Ca2 | 833 | 92,562.88 | 5.89 | ARF | Auxin response factor 8-like |
| CarB3_15 | Ca_14329 | Ca2 | 504 | 55,425.74 | 6.63 | ARF | Auxin response factor |
| CarB3_16 | Ca_23296 | Ca3 | 460 | 51,878.1 | 5.9 | ARF | Auxin response factor 9-like |
| CarB3_17 | Ca_09486 | Ca3 | 390 | 44,059.38 | 5.75 | REM | b3 domain-containing transcription repressor val2-like isoform x2 |
| CarB3_18 | Ca_05976 | Ca3 | 327 | 36,606.97 | 5.81 | LAV/FUS3 | b3 domain-containing transcription factor fus3-like |
| CarB3_19 | Ca_00826 | Ca3 | 277 | 31,998.24 | 6.46 | REM | b3 domain-containing protein at2g36080-like |
| CarB3_20 | Ca_01368 | Ca3 | 413 | 47,875.65 | 5.84 | REM | b3 domain-containing transcription factor nga1-like |
| CarB3_21 | Ca_08436 | Ca4 | 384 | 41,588.05 | 8.95 | RAV | ap2 erf and b3 domain-containing transcription repressor tem1-like |
| CarB3_22 | Ca_08488 | Ca4 | 611 | 67,927.19 | 7.58 | ARF | Auxin response factor 18-like |
| CarB3_23 | Ca_17136 | Ca4 | 692 | 77,443.39 | 7.05 | ARF | Auxin response factor 18-like isoform x2 |
| CarB3_24 | Ca_14825 | Ca4 | 434 | 49,413.7 | 8.84 | REM | b3 domain-containing transcription factor vrn1 |
| CarB3_25 | Ca_09188 | Ca4 | 212 | 24,737.4 | 9.46 | REM | e1 protein |
| CarB3_26 | Ca_10857 | Ca4 | 557 | 62,256.9 | 8.68 | REM | b3 domain-containing protein os01g0723500-like |
| CarB3_27 | Ca_10856 | Ca4 | 232 | 26,737.25 | 8.14 | REM | Plant-specific b3-dna-binding domain protein |
| CarB3_28 | Ca_10855 | Ca4 | 436 | 48,925.86 | 6.4 | REM | b3 domain-containing protein rem16-like |
| CarB3_29 | Ca_10794 | Ca4 | 279 | 31,246.94 | 8.24 | ARF | Auxin response factor 5-like |
| CarB3_30 | Ca_10789 | Ca4 | 291 | 31,955.22 | 6.86 | REM | Auxin response factor 5-like |
| CarB3_31 | Ca_10748 | Ca4 | 917 | 102,506.7 | 5.55 | ARF | Auxin response factor 5 |
| CarB3_32 | Ca_18739 | Ca5 | 239 | 27,640.46 | 9.42 | REM | b3 dna-binding domain protein |
| CarB3_33 | Ca_18738 | Ca5 | 251 | 29,086.52 | 9.6 | REM | b3 dna-binding domain protein |
| CarB3_34 | Ca_08924 | Ca5 | 827 | 91,419.93 | 8.33 | VAL | High-level expression of sugar-inducible gene isoform 1 |
| CarB3_35 | Ca_08872 | Ca5 | 897 | 99,653.76 | 5.67 | ARF | Auxin response factor 8-like |
| CarB3_36 | Ca_01536 | Ca5 | 227 | 25,934.93 | 9.91 | REM | b3 domain-containing protein at5g42700-like |
| CarB3_37 | Ca_07410 | Ca5 | 476 | 54,204.83 | 5.03 | REM | b3 domain-containing protein os02g0598200-like isoform x1 |
| CarB3_38 | Ca_05876 | Ca6 | 1089 | 120,334.39 | 6.18 | ARF | Auxin response factor 19-like isoform x1 |
| CarB3_39 | Ca_05681 | Ca6 | 817 | 91,497.01 | 6.24 | ARF | Auxin response factor 2 |
| CarB3_40 | Ca_05025 | Ca6 | 917 | 102,746.01 | 6.17 | ARF | Auxin response factor 6 |
| CarB3_41 | Ca_06323 | Ca6 | 293 | 34,209.52 | 5.84 | LAV/LEC2 | b3 domain-containing transcription factor lec2 |
| CarB3_42 | Ca_06352 | Ca6 | 723 | 80,098.44 | 6.32 | ARF | Auxin response factor 3 |
| CarB3_43 | Ca_06552 | Ca6 | 145 | 16,195.76 | 6.82 | REM | b3 domain-containing protein os04g0386900-like |
| CarB3_44 | Ca_14590 | Ca6 | 807 | 89,813.72 | 6.53 | ARF | Auxin response factor 4 |
| CarB3_45 | Ca_26121 | Ca6 | 706 | 78,577.19 | 7.03 | ARF | Auxin response factor 18 |
| CarB3_46 | Ca_21948 | Ca6 | 687 | 76,186.72 | 6.39 | ARF | Auxin response factor 9-like |
| CarB3_47 | Ca_13671 | Ca6 | 431 | 48,587.08 | 9.03 | REM | b3 domain-containing transcription factor vrn1-like isoform x1 |
| CarB3_48 | Ca_03128 | Ca7 | 990 | 110,777.92 | 6.33 | ARF | Auxin response factor 19-like |
| CarB3_49 | Ca_09273 | Ca7 | 435 | 49,459.13 | 8.37 | REM | b3 domain-containing protein os01g0234100-like isoform x1 |
| CarB3_50 | Ca_17624 | Ca7 | 571 | 64,731.54 | 7.28 | ARF | Auxin response factor 18-like isoform x2 |
| CarB3_51 | Ca_17636 | Ca7 | 679 | 76,102.9 | 6.38 | ARF | Auxin response factor 18-like isoform x1 |
Chromosomal locations and duplication analysis
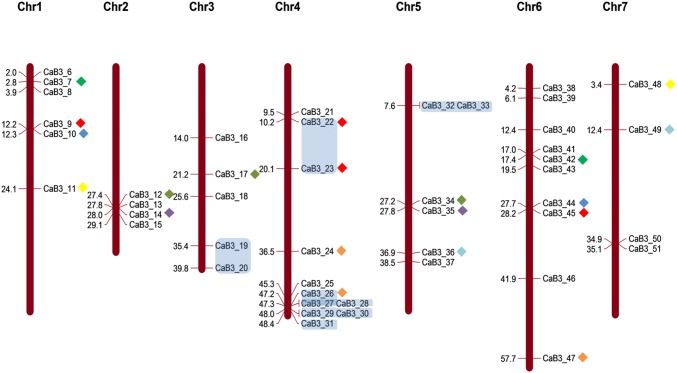
Out of 51 CarB3 genes, 46 could be mapped on all chickpea chromosomes except chromosome 8 whereas five genes, i.e., CarB3_1, 2, 3, 4, and 5 were located on scaffolds 151, 470, 682, 1281 and 1301, respectively. Chromosomal locations of the chickpea B3 genes revealed that chromosome 4 and 6 had the highest number of B3 genes (11 and 10, respectively) followed by chromosome 2 (4) and chromosome 7 (4) (Fig. 1). Investigation of duplication events using MCScanX led to the discovery of 21 pairs of paralogous B3 genes distributed on different chromosomes. Seven pairs of B3 genes were identified as tandem gene duplications and 14 gene pairs were identified as segmental duplications (Fig. 1; Online Resource 4).
Fig. 1.
Chromosomal distribution of CarB3 members on eight chickpea chromosomes. Genes under colored blocks are tandemly duplicated genes and genes with symbols are segmentally duplicated genes (Online Resource 4)
Analysis of phylogeny, gene structure and motifs
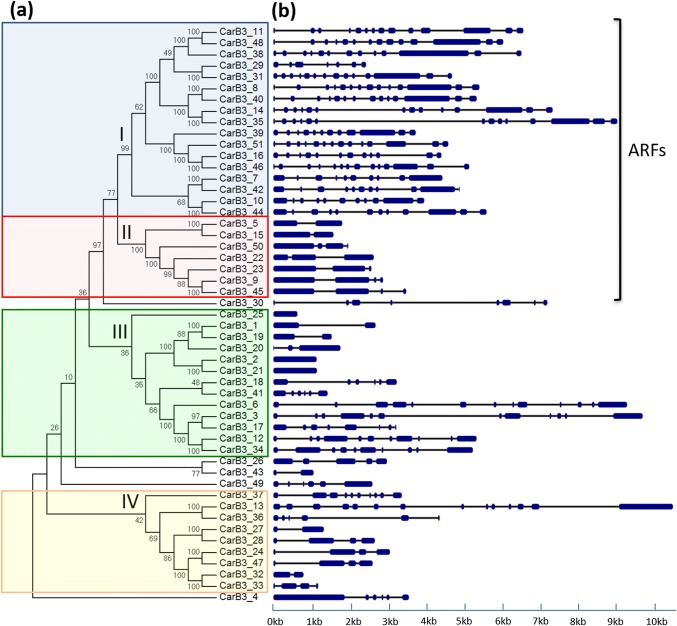
To gain insights into the diversity of chickpea B3 proteins, a phylogenetic tree was created using the NJ method. Multiple sequence alignment of full-length amino acid sequences of all 51 B3 proteins was utilized for phylogenetic tree construction. According to the phylogenetic analysis, chickpea B3 proteins were classified into four independent groups designated I–IV (Fig. 2a). A total of 19 sister pairs were found among the 51 B3 proteins, and 16 of them exhibited high bootstrap value (≥ 99%). Five proteins, i.e., CarB3_30, CarB3_26, CarB3_43, CarB3_49 and CarB3_4 were quite diverse and could not be grouped into any of the major groups.
Fig. 2.
a Phylogenetic classification of CarB3 members. The CarB3 members were divided into four groups based on their clustering pattern. b Exon–intron organization of the CarB3 genes. Exons and introns are represented by blue boxes and grey lines, respectively
The phylogenetic relationships were further strengthened by the prediction of conserved domains present in the 51 CarB3 proteins (Online Research 2). Interestingly, members of the same group contained similar types of domains. For example, all 24 members encoding ARF-type proteins were clustered in group I and group II as they possessed an Aux_resp domain in addition to the B3 domain in their protein sequences. Remaining B3 proteins were clustered into group III and IV with some being exceptions (CarB3_30, 26, 43 and 39). Group III contained 13 B3 proteins including 6 proteins of REM family, 2 proteins of RAV family, 3 proteins of VAL family and, LEC2 and FUS3. Notably, all members of group IV except for CarB3_13, 36, and 27 contained more than one B3 domains in their protein sequences.
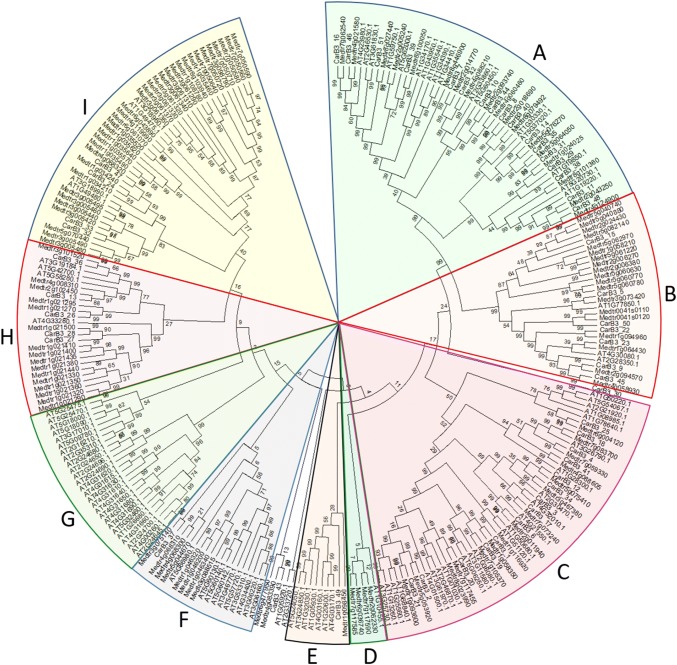
Further, the phylogenetic relationships between CarB3 proteins and the members of the B3 domain-containing proteins of Medicago and Arabidopsis were analyzed. For this, B3 protein sequences from Arabidopsis and Medicago were downloaded from phytozome v10. These sequences were scanned for redundancy and the presence of the B3 domain. After redundancy removal and domain confirmation, a total of 98 and 116 proteins were identified as B3 domain-containing proteins from Arabidopsis and Medicago, respectively. A phylogenetic tree was constructed utilizing the full-length B3 protein sequences of chickpea, Arabidopsis and Medicago. The phylogenetic tree comprised 265 B3 proteins from Arabidopsis (98), Medicago (116) and chickpea (51). As shown in Fig. 3, the phylogenetic tree could be divided into 9 well-supported groups designated as A to I. Almost all groups contained at least one CarB3 protein except for the group G, where only B3 proteins of Arabidopsis were clustered. It was observed that all members of chickpea B3 proteins had very close phylogenetic relationship with their homologs from Medicago except for CarB3_3 which showed a close relationship with the gene of Arabidopsis (AT4G32010.1).
Fig. 3.
A combined NJ-based phylogenetic tree based on B3 sequences of chickpea, Medicago and Arabidopsis
Exon–intron structural diversity is one of the possible contributors to the evolution of multigene families. To better understand the structural diversity and classification of chickpea B3 genes, their exon/intron organization was analyzed. It was observed that 48 CarB3 genes possessed 1–13 intron(s) in their genomic sequences, whereas only 3 CarB3 genes were found lacking introns (Fig. 2b). Remarkably, members of the same group exhibited nearly a similar exon/intron organization. For example, members of the group I contained 11–13 introns, whereas members of group II had 1–3 introns only. Notably, members of group III contained the 3 intron-less genes. It was observed that most of the sister gene pairs in the same group showed a conserved intron/exon structure. However, differences in the structural organization and the number of exons and introns were also observed in some of the sister gene pairs. For example, CarB3_29 contained 6 introns in its genomic sequence, whereas its close homolog, CarB3_31 contained 13 introns. Likewise, CarB3_13 was found to have 13 introns, whereas its closest homolog, CarB3_36 contained 5 introns.
Conserved motifs and their distribution in the 51 B3 proteins were identified using MEME tool. A total of 10 conserved motifs were identified in the B3 proteins (Online Resource 5). The identified motifs varied in length from 22 to 50 amino acids (Online Resource 6). Motif I typically appeared in all proteins except CarB3_27 and 33. This motif was observed as highly conserved amongst all motifs and was present in the region encoding B3 domain. Composition and number of motifs further validated the phylogenetic analysis, as members belonging to the same group displayed similar motif organization. Members of group I and II shared maximum identical motifs at the N-terminal region of proteins, whereas two motifs, i.e., motifs 8 and 9 were conserved in the C-terminal region. In group IV, only motifs 1 and 7 appeared in most members.
Synteny analysis
In order to recognize the orthologous relationship and to determine the conserved order of B3 genes on chromosomes between chickpea and other sequenced genomes including Medicago, soybean and Arabidopsis, a comparative analysis was carried out. Sequences of B3 genes were aligned to genomes of Medicago, soybean and Arabidopsis. The comparative analysis revealed that 43 B3 genes of chickpea found homologs in the M. truncatula genome, 38 members found homologs in soybean genome and 11 B3 genes found homologs in Arabidopsis genome (Online Resource 7). Ka/Ks ratio was calculated to determine the nature of selection (neutral, diversifying or purifying) of the B3 gene family. Ka/Ks ratio was calculated for paralogous and orthologous genes. The results indicated that the B3 genes were under purifying selection as all gene pairs had the Ka/Ks ratio < 1 (Online Resource 8).
Promoter analysis
A promoter sequence largely determines the spatiotemporal expression pattern of genes. In addition, the cis-regulatory elements present over the promoter sequence often provide binding sites for transcription factors and other accessory proteins which in turn can activate or repress the expression of gene. In order to identify the important cis-regulatory elements present over the promoters of B3 genes, 2 kb upstream sequences from ATG start site of each B3 gene were analyzed using PLACE database. In silico analysis of promoter regions of all B3 genes revealed the presence of several different cis-elements in the upstream region of the genes (Online Resource 9). cis-elements such as AUXREPSIAA4, AUXRETGA2GMGH3, BBOXSITE1STPAT, CARGATCONSENSUS, CONSERVED11NTZMATP1 and LREBOXIPCCHS1 were unique to CarB3_26, CarB3_21, CarB3_38, CarB3_15, CarB3_8 and CarB3_10, respectively. It was observed that one or more motifs related to seed-specific expression were also present in promoter regions of B3 genes. These motifs included AACA2SSEEDPROTBANAPA, GCN4OSGLUB1, NAPINMOTIFBN, SEF1MOTIF, SEF3MOTIFGM and RYREPEATBNNAPA motifs which are characteristically found in promoters of genes encoding seed storage proteins. Apart from this, cis-elements related to dehydration-response (CBFHV, ACGTATERD1, DRECRTCOREAT, MYB1AT, and MYB2CONSENSUSAT) and flower development (CARGATCONSENSUS, WUSATAG) were also present in many B3 gene promoters.
In silico expression analysis and validation through qRT-PCR
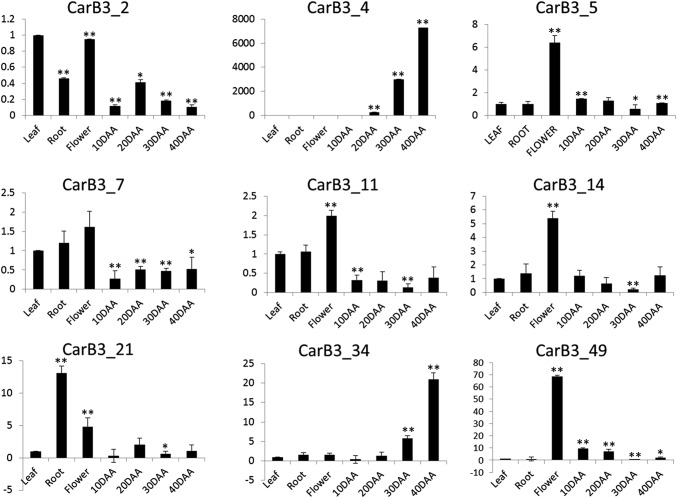
All 51 Car_B3 genes were examined for their in silico expression pattern using RNA-seq data from different chickpea tissues: leaf, root, flower-bud, pod and 4 stages of seed development (Fig. 4). The analysis showed that 12 genes exhibited preferential expression in at least one of the seed tissues (Fig. 4). For instance, four B3 genes such as CarB3_19, 9, 37 and 12 were expressed specifically in 10 DAA seeds, and three genes such as CarB3_18, 42, and 22 were expressed predominantly in 20 DAA seeds. In addition, CarB3_47 had an almost equal expression in 10, 20 and 30 DAA seeds. One B3 gene, CarB3_31 was observed having higher expression in early stages of seed development (10 and 20 DAA) whereas two B3 genes, CarB3_34 and CarB3_4 were found to be expressed higher in late stages (30 and 40 DAA). Four genes (CarB3_17, 27, 15, and 50) were found to have no expression in any of the seed tissues. However, some B3 genes such as CarB3_14, 8, 11, 46 and 16 exhibited lower expression in seed tissue when compared to other tissues. It was observed that CarB3_7, 8, 40, 35 had high expression in flower bud. Several B3 genes (60.08%) were found to have no expression in leaf tissue (Fig. 4). Only one B3 gene, i.e., CarB3_2 exhibited higher expression in leaf tissue. Two B3 genes, i.e., CarB3_21 and 24 were seen to display extensively high expression in root tissue (Fig. 4). Some of the members were selected for validation through qRT-PCR. Significant correlation between in silico data and real-time PCR data was observed that validated the in silico prediction of the CarB3 gene expression in different tissues (Fig. 5).
Fig. 4.
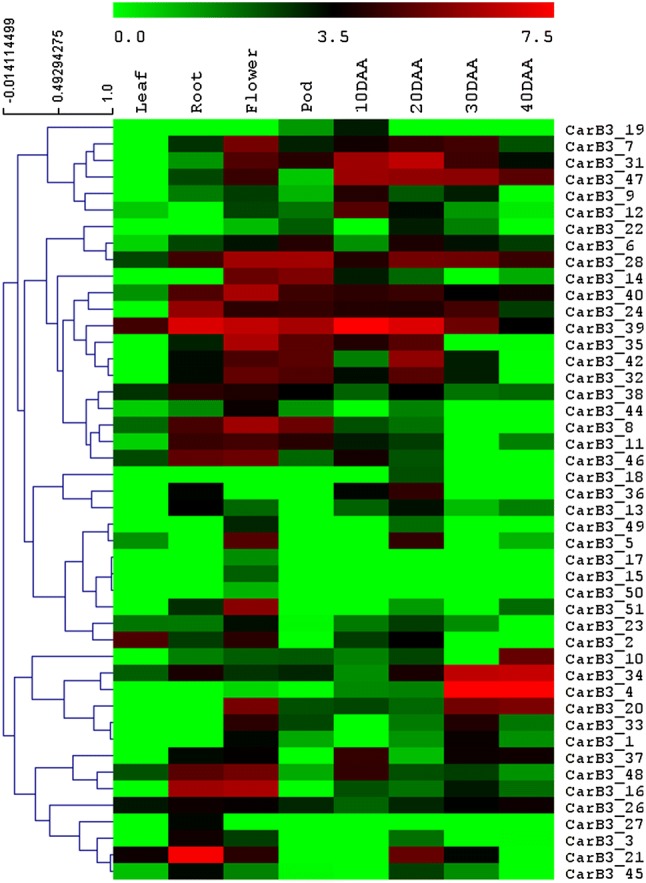
Hierarchical clustering of CarB3 genes based on digital expression analysis in different chickpea tissues. Scale bar represents the log2 normalized RPKM values. Color gradient denotes level of expression; red being high and green being low
Fig. 5.
Expression analysis of nine selected CarB3 genes in different chickpea tissues through qRT-PCR. Values on Y-axis denote relative expression values. DAA, days after anthesis. Error bars indicate standard deviation (± SD) of three biological replicates each calculated from three technical replicates. Asterisks indicate statistically significant difference between control (leaf) and other tissues (t test, *P < 0.05, **P < 0.01)
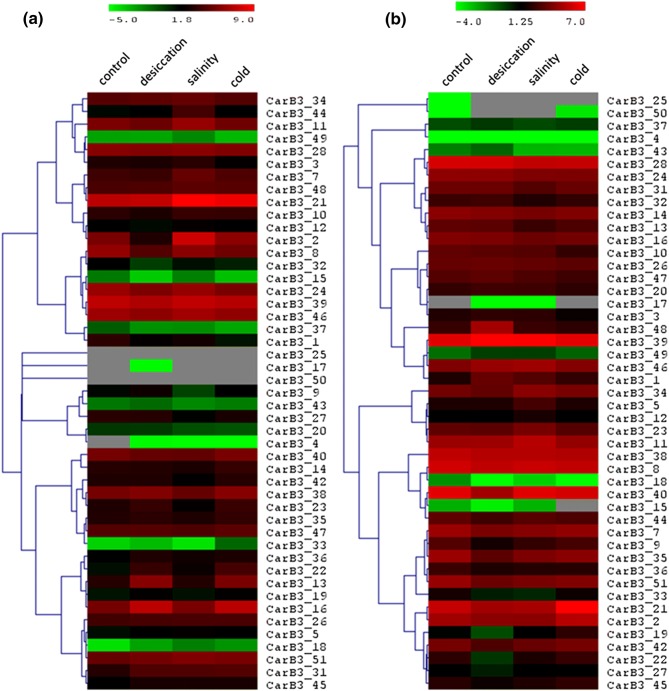
Further, the in silico expression patterns of B3 genes were analyzed under three abiotic stresses: desiccation, salinity and cold by utilizing the RNA-seq reads (SRP034839) of chickpea stressed tissues. Majority of the genes exhibited differential expression in stressed root tissues when compared to control tissue (Fig. 6a). Two genes such as CarB3_31 and CarB3_4 were found to be up-regulated in all three stress conditions. In addition, three B3 genes such as CarB3_7, 44 and 2 were expressed more in salinity stress. None of these B3 genes were detected having expression specifically in cold-stressed root tissue. Two B3 genes such as CarB3_13 and 16 were up-regulated in both desiccation and cold stresses in root tissue. Most of the B3 genes (CarB3_6, 8, 20, 36, 38, 26, 24, 14, 16, 45) were observed having less than twofold difference (almost similar expression) in expression in stressed shoot tissue when compared to the control shoot tissue (Fig. 6b). Two B3 genes such as CarB3_48 and 1 were found to be up-regulated in desiccated shoot tissue. Two B3 genes, CarB3_2 and CarB3_21, were expressed more in cold stress. Two B3 genes, i.e., CarB3_39 and 17 were found having similar expression in both desiccation and salinity stress (Fig. 6b).
Fig. 6.
Heat map showing the digital expression of CarB3 genes in different stress conditions: desiccation, salinity, cold. a Control and stressed root tissues. b Control and stressed shoot tissues. Clustering method: hierarchical clustering. Color gradient denotes level of expression; red being high and green being low. Grey color depicts no expression
qRT-PCR analysis of selected members under different stresses and hormonal treatments
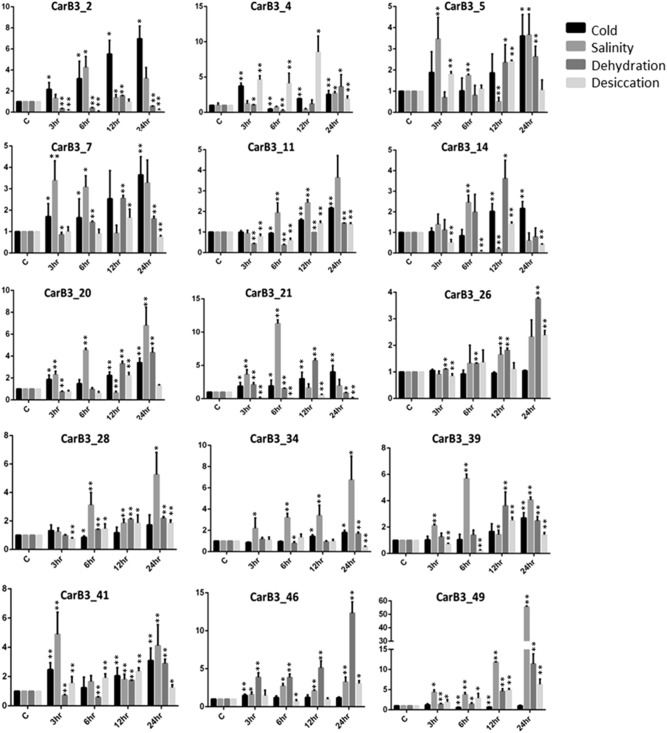
Further, we selected some B3 members on the basis of their in silico expression profiles in different tissues and analyzed their expression through real-time PCR under four stress conditions (salinity, cold, dehydration and desiccation) at different time points. Three B3 genes (CarB3_2, 5 and 7) were found to be up-regulated in cold stress. They showed a steady increase in expression at 12 and 24 h after treatment. However, genes such as CarB3_49, 26, 46 were found having expression similar to the control tissue in cold stress (Fig. 7). Seven B3 genes (CarB3_49, 39, 11, 34, 41, 21 and 28) were up-regulated significantly in response to salinity stress. Amongst them, CarB3_49, 11, 28 and 34 were up-regulated at 24 h after treatment, whereas CarB3_39, 21 were up-regulated at 6 h after treatment. Another gene, CarB3_41 was up-regulated at 3 h and 24 h after salinity treatment. Gene, CarB3_7 showed up-regulation in response to cold and salinity stress but did not show any activity in desiccation stress. CarB3_4 and CarB3_46 were up-regulated more in desiccation stress and dehydration stress, respectively (Fig. 7).
Fig. 7.
Expression analysis of 15 selected CarB3 genes in response to different stress conditions: cold, salinity, dehydration, desiccation at different time points through qRT-PCR. Values on Y-axis denote relative expression values. Error bars indicate standard deviation (± SD) of three biological replicates each calculated from three technical replicates. Asterisks indicate statically significant difference from the control (t test, *P < 0.05, **P < 0.01)
To analyze the response of CarB3 genes under ABA and auxin treatment, qRT-PCR was carried out for selected B3 members of chickpea. ABA treatment caused a marked change in the expression of all selected B3 genes (Online Resource 10). Maximum genes (CarB3_4, 49, 2, 39, 14, 21, 7 and 20) were found to be expressed significantly higher at 12 h after ABA treatment. Three B3 genes (CarB3_5, 41 and 46) maintained the up-regulation at 12 and 24 h after treatment. In response to auxin treatment, many genes were up-regulated at two or more than two time points. For example, CarB3_5 was up-regulated at 3, 6 and 24 h after treatment and CarB3_39 was up-regulated at 3, 12 and 24 h after treatment (Online Resource 11). CarB3_28 showed up-regulation at 12 h and 24 h after treatment. One B3 gene, CarB3_49 was highly expressed (> 2-fold) at 24 h after treatment. Two genes (CarB3_21 and 46) had more than twofold expression at 3 h after treatment when compared to tissue of 0 h treatment. No significant changes were observed for three genes in the expression in response to auxin treatment (Online Resource 11).
Discussion
The entire process of plant development is well orchestrated by several transcription factors (TFs) acting in a combinatorial manner. Transcription factor families have been extensively identified (Davuluri et al. 2003; Jin et al. 2017), amongst them the plant-specific B3 TF family has special significance owing to its role in regulation of a variety of pathways involved in plant development and defence responses (Romanel et al. 2009; Swaminathan et al. 2008). In addition, B3 TFs play crucial roles in several hormone-related signal transduction pathways (Swaminathan et al. 2008). In the present study, the available whole genome sequence of chickpea (Varshney et al. 2013), facilitated identification of the complete set of 51 B3 domain-containing proteins from the 28,269 annotated proteins of chickpea. However, in the plant transcription factor database, 48 B3 transcription factors were reported from Cicer arietinum genome, which is comparable with our identification. The complete set of 51 proteins were analyzed at the molecular and structural level that revealed diversity in their lengths, domain patterns and isoelectric points suggesting diverse roles for transcripts in regulating a wide range of biological processes in different phases of chickpea plant development. This is the first report in chickpea where the B3 proteins were identified and characterized at the molecular level.
The identified 51 B3 domain-containing proteins were classified into four subfamilies, i.e., ARF, LAV, RAV and REM based on the classification by Swaminathan et al. 2008. However, a few reports classified them into five distinct subfamilies, where they sub-categorized LAV as ABI3/VP1 and HIS (Romanel et al. 2009). In the present study, we classified some of the CarB3 proteins lacking additional B3 domain into REM family, similar to the previous reports (Swaminathan et al. 2008, Romanel et al. 2009). In the CarB3 superfamily, ARF was the largest subfamily in chickpea similar to the case of soybean, where 54 ARFs were reported to form the largest subfamily (Peng and Weselake 2013; Zhang et al. 2011). In contrast, ABI3-VP1 shared the largest proportion of B3 proteins in Arabidopsis, B. rapa and cocoa (Peng and Weselake 2013). In chickpea, LEC2-ABI3/VP1 subgroup includes three well-known members namely ABI3, FUS3 and LEC2. Their role in different stages of seed development particularly in maturation stage has been established by analyzing the loss-of-function mutants of corresponding genes in Arabidopsis (Kim et al. 2013; Luerßen et al. 1998; Nambara et al. 1995).
Phylogenetic analysis of different gene families from various crop species has illustrated that the members of the same clade represent similar motif structure, domain organization and intron–exon structure thereby suggesting their conserved function and origin from a common ancestor (Bhattacharjee et al. 2015; Li et al. 2017; Singh et al. 2014; Tang et al. 2016). In the present study, 51 chickpea B3 proteins were clustered into four clades each of which was well supported by the similar gene organization and presence of conserved motifs (Fig. 2, Online Resource 5). Analysis of the intron–exon structure of the B3 genes revealed significant variations in the number and length of introns, suggesting that these genes might have experienced intron loss or intron gain during the course of evolution. Three intron-less B3 genes were observed in chickpea (Fig. 2). These genes might have lost introns to evolve efficient and speedy processes of transcription regulation (Deutsch and Long 1999). Protein sequence motif is a short stretch of amino acids which is considered as a structural unit required for proper folding of the proteins (Bailey et al. 2015). Within the chickpea B3 proteins, significantly high level of conservation was observed amongst identified motifs (Online Resource 5). Interestingly, members of the same clade in the phylogenetic tree were found to contain similar motifs and also exhibited similar motif organization which further validated the phylogenetic analysis. Conservation of motifs among the members of the same clade has been observed in various gene families in chickpea (Pradhan et al. 2014; Singh and Jain 2015; Singh et al. 2014). Furthermore, such kind of identity in motif patterns reflects the structural and functional redundancy in the members of the same group. Domain analysis is an imperative aspect of structural genomics that provides information about the evolution, function and structure of a protein. Each B3 protein contained at least one B3 domain which was highly conserved among all the B3 proteins. Notably, members of the same group in the phylogenetic tree appeared to contain similar numbers and types of domains in addition to the B3 domain, suggesting structural conservation amongst the members of the same group. A combined phylogenetic tree comprising B3 proteins of Arabidopsis, Medicago and chickpea was also constructed (Fig. 3). The tree topology indicated that the majority of orthologous genes could be grouped into the same clade. Notably, all chickpea ARF and REM genes clustered together with their orthologous genes in similar groups indicating their functional and structural conservation. For instance, all chickpea ARFs were clustered together with their orthologs of Arabidopsis and Medicago in group A and B. Likewise, orthologous genes of ABI3, FUS3 and LEC2 were clustered in group C which further supported their functional conservation in the regulation of seed development (Nambara et al. 1995, 2000; To et al. 2006).
Gene duplication events (tandem and segmental) are the major contributors to the expansion of a gene family (Cannon et al. 2004). With regard to the duplication of chickpea B3 genes, it was clear that these genes primarily evolved through segmental duplication since their level was greater than the tandem duplications (Fig. 1). Even at the whole genome level, it had been demonstrated that overall chickpea gene families expanded by undergoing segmental duplications (Jain et al. 2013). Notably, duplicated genes were found to be located as sister pairs in the phylogenetic tree, suggesting their evolutionary closeness. Furthermore, high sequence similarity between the duplicated genes indicated their redundant function in the common biological processes. However, due to long-term evolution, functional diversification may exist in the duplicated genes.
Synteny analysis provides valuable information about the gene order, evolutionary history and conservation among multiple genomes (Salse et al. 2002). A comparative analysis between chickpea and other genomes, i.e., Medicago, soybean and Arabidopsis was carried out by utilizing the information of physically mapped B3 genes (Online Resource 7). Chickpea and Medicago were observed to be sharing maximum similarity and orthologous conservation as compared to Arabidopsis and soybean genome. The close proximity of chickpea to Medicago has repeatedly been demonstrated in various studies carried out for different gene families in chickpea (Gupta et al. 2015; Pradhan et al. 2017). And, it is also supported by the taxonomy of legumes where both chickpea and Medicago occur in the Galegoid clade (Jain et al. 2013).
Selection is the key to the creation of new variations in a gene pool. During evolution, selection occurs by selecting advantageous mutations and removing deleterious variations. In genetics, the ratio of non-synonymous (Ka) and synonymous (Ks) substitutions is used to determine the nature of selection (neutral, diversifying or purifying) acting on a set of homologous protein-coding genes. A Ka/Ks ratio of 1 designates neutral selection, Ka/Ks > 1 indicates positive (diversifying) selection and Ka/Ks < 1 implies that amino acids evolve primarily through negative (purifying) selection (Zhou et al. 2010). In the present study, the Ka/Ks ratio was used to determine the rate of evolution in homologous protein-coding B3 genes. It was observed that the B3 genes are under purifying selection as all gene pairs had the Ka/Ks ratio < 1 (Online Resource 8). Purifying selection maintains long-term stability by eliminating deleterious mutations (Sironi et al. 2015). Therefore, it may be said that the importance of B3 gene family in chickpea plant development has necessitated the maintenance and conservation of its members.
A promoter sequence harbors specific DNA elements that provide binding sites to proteins, especially transcription factors (Bulyk 2004) and serve as the key factors for controlling gene transcription and regulated expression. Although B3 proteins themselves are the transcription factors, they may be under the control of other transcription factors. To gain insights into their regulatory functions, the promoter sequences of the chickpea B3 genes were analyzed. This highlighted the presence of several cis-regulatory elements, of which some were common among the B3 genes, whereas some promoters contained unique motifs (Online Resource 9). Several tissue-specific cis-regulatory elements were found in the promoter regions of many B3 genes which strongly indicated their involvement in various developmental pathways of chickpea development including leaf, flower, nodule, and seed tissues. For example, genes that had “WUSATAg” in their promoter sequences were found to be expressed highly in flower tissue. Similarly, CarB3_34 was observed to have strong seed-specific expression and its promoter was seen to contain several seed-specific cis-regulatory elements. Further, several cis-regulatory elements responsible for stress-responsive expression such as ABRE, DRE and/or LTRE were also observed in many of the B3 genes, suggesting their stress-responsive regulation. In addition, regulatory elements for plant hormone responses were also detected in the promoters of B3 genes in chickpea thereby endorsing the fact that CarB3 genes may play critical roles in hormonal crosstalk as well.
Expression levels of genes provide useful information about their function during tissue-specific events and under different stress conditions. Therefore, members of CarB3 family were investigated for their in silico expression pattern in various chickpea tissues that was further validated through real-time PCR analysis (Figs. 4, 5). Differential expression (> 2-fold) for most of the CarB3 genes was observed amongst different chickpea tissues indicating the potential role of B3 genes in various developmental pathways. In the present study, CarB3 members encoding ABI3, FUS3, VAL1 and NGA1 were observed having seed-specific expression which is consistent with the study on B3 genes in Arabidopsis (Peng and Weselake 2013). Moreover, in the transcriptome analysis carried out for chickpea seed tissues, the B3 members were found to be over-represented which strongly indicates the involvement of B3 TFs in the process of chickpea seed development (Pradhan et al. 2014). Similarly, VRN1 encoding CarB3 gene was seen to have significantly higher expression in flower tissue. The role of VRN1 in flowering time control and vernalization has been demonstrated in Arabidopsis (Levy et al. 2002). Furthermore, some ARFs were found to be expressed significantly in root tissue suggesting their active involvement in root development related pathways. For instance, CarB3 that encodes ARF19 had the maximum expression in chickpea root tissue and its role in lateral root formation has been established in Arabidopsis (Okushima et al. 2007).
Abiotic stresses adversely affect plant growth and development (Quan et al. 2008). Understanding the complex molecular mechanisms underlying stress conditions is a major challenge. In the present study, digital expression analysis of chickpea B3 genes was carried out using publicly available RNA-seq data of stressed chickpea tissues which was further validated in four stress conditions at different time periods using real-time PCR analysis (Fig. 7). Many CarB3 genes displayed significant differential expression in one or more stressed tissues at different time points suggesting that they might play crucial roles during stress (Fig. 6). For example, CarB3_4, encoding, ABI3, was found to be up-regulated in desiccation stress in both digital and real-time expression analysis. ABI3 has been reported to be an important regulator in the acquisition of desiccation tolerance in Arabidopsis (Ooms et al. 1993). Similarly, the expression of CarB3_49 was found to be up-regulated in salinity stress, both in real-time as well as in digital expression data of salinity stressed shoot tissue (Figs. 6, 7). In short, several CarB3 genes were found to have differential expression during various stresses which further necessitates a better investigation of the molecular and genetic regulation of chickpea B3 genes in abiotic stress responses.
Hormones such as ABA and auxin are known to regulate abiotic stress-related pathways as well as various aspects of plant growth and development. Genes involved in auxin and ABA-mediated signaling pathways have been identified and analyzed in different plant species (Gao et al. 2016; Gu et al. 2012; Kazan 2013; Li et al. 2018; Park et al. 2011; Rabbani et al. 2003; Tiwari et al. 2003). To investigate the involvement of CarB3 genes in hormonal signaling, expression of selected B3 members in response to ABA and IBA treatment was analyzed. Most of the analyzed genes were found to respond differentially at different time points to ABA, suggesting ABA specific regulation of these genes under stress conditions (Online Resource 10). Similarly, many analyzed CarB3 genes responded to exogenous auxin treatment indicating that they may play diverse roles in many developmental processes under hormonal regulation (Online Resource 11).
This study identifies genes encoding B3 domain-containing proteins in chickpea and provides an in-depth analysis of their phylogenetic relationships, promoter sequences, and expression patterns in various chickpea tissues and under different abiotic stress conditions. The findings would serve as an excellent foundation for deepening the understanding of B3 proteins in chickpea under different developmental and physiological processes, including environmental stress responses. This will further help in channelizing directional efforts towards their functional characterization with the overall aim of improving crop chickpea.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was funded by the Department of Biotechnology, Ministry of Science and Technology, Government of India, under the Challenge Programme on Chickpea Functional Genomics (Grant number: BT/AGR/CG-Phase II/01/2014). SV acknowledges the award of a research fellowship from the Department of Biotechnology, Govt. of India. The authors are thankful to DBT-eLibrary Consortium (DeLCON) for providing access to e-resources.
Author contributions
SV and SB were involved in the designing and execution of the work. SV majorly conducted all the experiments, lab work, analyzed data, and prepared the manuscript draft. SB corrected the manuscript and gave the final approval for the version to be published.
Compliance with ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
References
- Agarwal G, et al. Genome-wide dissection of AP2/ERF and HSP90 gene families in five legumes and expression profiles in chickpea and pigeonpea. Plant Biotechnol J. 2016;14:1563–1577. doi: 10.1111/pbi.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Johnson J, Grant CE, Noble WS. The MEME suite. Nucleic Acids Res. 2015;43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee A, Ghangal R, Garg R, Jain M. Genome-wide analysis of homeobox gene family in legumes: identification, gene duplication and expression profiling. PLoS One. 2015 doi: 10.1371/journal.pone.0119198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulyk ML. Computational prediction of transcription-factor binding site locations. Genome Biol. 2004;5:201. doi: 10.1186/gb-2003-5-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C, Pelaz S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol. 2008;18:1338–1343. doi: 10.1016/j.cub.2008.07.075. [DOI] [PubMed] [Google Scholar]
- Choudhary S, Sethy NK, Shokeen B, Bhatia S. Development of chickpea EST–SSR markers and analysis of allelic variation across related species. Theor Appl Genet. 2009;118:591–608. doi: 10.1007/s00122-008-0923-z. [DOI] [PubMed] [Google Scholar]
- Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, Grotewold E. AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinform. 2003;4:25. doi: 10.1186/1471-2105-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch M, Long M. Intron-exon structures of eukaryotic model organisms. Nucleic Acids Res. 1999;27:3219–3228. doi: 10.1093/nar/27.15.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devic M, Roscoe T. Seed maturation: simplification of control networks in plants. Plant Sci. 2016;252:335–346. doi: 10.1016/j.plantsci.2016.08.012. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci USA. 2014;111:2367–2372. doi: 10.1073/pnas.1316278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, et al. Genome-wide survey of Aux/IAA gene family members in potato (Solanum tuberosum): identification, expression analysis, and evaluation of their roles in tuber development. Biochem Biophys Res Commun. 2016;471(2):320–327. doi: 10.1016/j.bbrc.2016.02.013. [DOI] [PubMed] [Google Scholar]
- Gu H, et al. Identification and characterization of the LEA family gene CarLEA4 from chickpea (Cicer arietinum L.) Mol Biol Rep. 2012;39(4):3565–3572. doi: 10.1007/s11033-011-1130-6. [DOI] [PubMed] [Google Scholar]
- Gu C, et al. Multiple regulatory roles of AP2/ERF transcription factor in angiosperm. Bot Stud. 2017;58(1):6. doi: 10.1186/s40529-016-0159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display server. Yi chuan Hereditas. 2007;29:1023–1026. doi: 10.1360/yc-007-1023. [DOI] [PubMed] [Google Scholar]
- Gupta S, Garg V, Kant C, Bhatia S. Genome-wide survey and expression analysis of F-box genes in chickpea. BMC Genom. 2015 doi: 10.1186/s12864-015-1293-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CV, et al. Genome-wide identification and expression analysis of the CaNAC family members in chickpea during development, dehydration and ABA treatments. PLoS One. 2014;9:e114107. doi: 10.1371/journal.pone.0114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Higo H. PLACE: a database of plant cis-acting regulatory DNA elements. Nucleic Acids Res. 1998;26:358–359. doi: 10.1093/nar/26.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe EA, Sinha R, Schlauch D, Quackenbush J. RNA-seq analysis in MeV. Bioinformatics (Oxford, England) 2011;27:3209–3210. doi: 10.1093/bioinformatics/btr490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, et al. A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.) Plant J. 2013;74:715–729. doi: 10.1111/tpj.12173. [DOI] [PubMed] [Google Scholar]
- Jin J, Tian F, Yang DC, Meng YQ, Kong L, Luo J, Gao G. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017;45:D1040–d1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant C, Pradhan S, Bhatia S. Dissecting the root nodule transcriptome of chickpea (Cicer arietinum L.) PLoS One. 2016;11:e0157908. doi: 10.1371/journal.pone.0157908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K. Auxin and the integration of environmental signals into plant root development. Ann Bot. 2013;112:1655–1665. doi: 10.1093/aob/mct229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HU, Jung S-J, Lee K-R, Kim EH, Lee S-M, Roh KH, Kim J-B. Ectopic overexpression of castor bean LEAFY COTYLEDON2 (LEC2) in Arabidopsis triggers the expression of genes that encode regulators of seed maturation and oil body proteins in vegetative tissues. FEBS Open Bio. 2013;4:25–32. doi: 10.1016/j.fob.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: anesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy YY, Mesnage S, Mylne JS, Gendall AR, Dean C. Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science. 2002;297:243–246. doi: 10.1126/science.1072147. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics (Oxford, England) 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SB, Xie ZZ, Hu CG, Zhang JZ. A review of auxin response factors (ARFs) in plants. Front Plant Sci. 2016 doi: 10.3389/fpls.2016.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, et al. Genome-wide analysis of WRKY gene family in the sesame genome and identification of the WRKY genes involved in responses to abiotic stresses. BMC Plant Biol. 2017;17:152. doi: 10.1186/s12870-017-1099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wu M, Liu H, Gao Y, Xiang Y. Systematic identification and expression pattern analysis of Aux/IAA and ARF gene families in moso bamboo (Phyllostachys edulis) Plant Physiol Biochem. 2018;130:431–444. doi: 10.1016/j.plaphy.2018.07.033. [DOI] [PubMed] [Google Scholar]
- Luerßen H, Kirik V, Herrmann P, Miséra S. FUSCA3 encodes a protein with a conserved VP1/ABI3-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J. 1998;15:755–764. doi: 10.1046/j.1365-313X.1998.00259.x. [DOI] [PubMed] [Google Scholar]
- Magnani E, Sjolander K, Hake S. From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. Plant Cell. 2004;16:2265–2277. doi: 10.1105/tpc.104.023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun JH, Yu HJ, Shin JY, Hwang HJ, Chung H. Auxin response factor gene family in Brassica rapa: genomic organization, divergence, expression, and evolution. Mol Genet Genom. 2012;287(10):765–784. doi: 10.1007/s00438-012-0718-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, McCourt P, Naito S. A regulatory role for the ABI3 gene in the establishment of embryo maturation in Arabidopsis thaliana. Development. 1995;121:629–636. [Google Scholar]
- Nambara E, Hayama R, Tsuchiya Y, Nishimura M, Kawaide H, Kamiya Y, Naito S. The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Dev Biol. 2000;220:412–423. doi: 10.1006/dbio.2000.9632. [DOI] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms JJ, Leon-Kloosterziel KM, Bartels D, Koornneef M, Karssen CM. Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana (a comparative study using abscisic acid-insensitive abi3 mutants) Plant Physiol. 1993;102:1185–1191. doi: 10.1104/pp.102.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MY, Kang JY, Kim SY. Overexpression of AtMYB52 confers ABA hypersensitivity and drought tolerance. Mol Cells. 2011;31(5):447–454. doi: 10.1007/s10059-011-0300-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng FY, Weselake RJ. Genome-wide identification and analysis of the B3 superfamily of transcription factors in Brassicaceae and major crop plants. Theor Appl Genet. 2013;126:1305–1319. doi: 10.1007/s00122-013-2054-4. [DOI] [PubMed] [Google Scholar]
- Pradhan S, Bandhiwal N, Shah N, Kant C, Gaur R, Bhatia S. Global transcriptome analysis of developing chickpea (Cicer arietinum L.) seeds. Front Plant Sci. 2014 doi: 10.3389/fpls.2014.00698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan S, Kant C, Verma S, Bhatia S. Genome-wide analysis of the CCCH zinc finger family identifies tissue specific and stress responsive candidates in chickpea (Cicer arietinum L.) PLoS One. 2017;12:e0180469. doi: 10.1371/journal.pone.0180469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan LJ, Zhang B, Shi WW, Li HY. Hydrogen peroxide in plants: a versatile molecule of the reactive oxygen species network. J Integr Plant Biol. 2008;50:2–18. doi: 10.1111/j.1744-7909.2007.00599.x. [DOI] [PubMed] [Google Scholar]
- Rabbani MA, et al. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stress and abscisic acid application using cDNA microarray and RNA gel-blot analysis. Plant Physiol. 2003;133(4):1755–1767. doi: 10.1104/pp.103.025742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidt W, et al. Gene regulation during late embryogenesis: the RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J. 2000;21:401–408. doi: 10.1046/j.1365-313x.2000.00686.x. [DOI] [PubMed] [Google Scholar]
- Romanel EAC, Schrago CG, Couñago RM, Russo CAM, Alves-Ferreira M. Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS One. 2009;4:e5791. doi: 10.1371/journal.pone.0005791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salse J, Piégu B, Cooke R, Delseny M. Synteny between Arabidopsis thaliana and rice at the genome level: a tool to identify conservation in the ongoing rice genome sequencing project. Nucleic Acids Res. 2002;30:2316–2328. doi: 10.1093/nar/30.11.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasnauskas G, Manakova E, Lapėnas K, Kauneckaitė K, Siksnys V. DNA recognition by Arabidopsis transcription factors ABI 3 and NGA 1. FEBS J. 2018;285(21):4041–4059. doi: 10.1111/febs.14649. [DOI] [PubMed] [Google Scholar]
- Shen C, et al. Genome-wide identification and expression analysis of auxin response factor gene family in Medicago truncatula. Front Plant Sci. 2015;6:73. doi: 10.3389/fpls.2015.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh VK, Jain M. Genome-wide survey and comprehensive expression profiling of Aux/IAA gene family in chickpea and soybean. Front Plant Sci. 2015 doi: 10.3389/fpls.2015.00918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh VK, Jain M, Garg R. Genome-wide analysis and expression profiling suggest diverse roles of GH3 genes during development and abiotic stress responses in legumes. Front Plant Sci. 2014;5:789. doi: 10.3389/fpls.2014.00789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi M, Cagliani R, Forni D, Clerici M. Evolutionary insights into host–pathogen interactions from mammalian sequence data. Nat Rev Genet. 2015;16:224–236. doi: 10.1038/nrg3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, McCarty DR. The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell. 1997;9:799–807. doi: 10.1105/tpc.9.5.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan K, Peterson K, Jack T. The plant B3 superfamily. Trends Plant Sci. 2008;13:647–655. doi: 10.1016/j.tplants.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang R, Zhu W, Song X, Lin X, Cai J, Wang M, Yang Q. Genome-wide identification and function analyses of heat shock transcription factors in potato. Front Plant Sci. 2016;7:490. doi: 10.3389/fpls.2016.00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T. The role of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15(2):533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F. A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell. 2006;18:1642–1651. doi: 10.1105/tpc.105.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H, Saijo T, Shibata D, Morikami A, Nakamura K. Analysis of a sugar response mutant of Arabidopsis identified a novel B3 domain protein that functions as an active transcriptional repressor. Plant Physiol. 2005;138:675–685. doi: 10.1104/pp.104.057752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ha C, et al. The auxin response factor transcription factor family in soybean: genome-wide identification and expression analysis during development and water stress. DNA Res. 2013;20(5):511–524. doi: 10.1093/dnares/dst207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney RK, et al. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat Biotechnol. 2013;31:240–246. doi: 10.1038/nbt.2491. [DOI] [PubMed] [Google Scholar]
- Verma S, Bhatia S. Analysis of genes encoding seed storage proteins (SSPs) in chickpea (Cicer arietinum L.) reveals co-expressing transcription factors and a seed-specific promoter. Funct Integr Genom. 2019;19(3):373–390. doi: 10.1007/s10142-018-0650-8. [DOI] [PubMed] [Google Scholar]
- Vidaurre DP, Ploense S, Krogan NT, Berleth T. AMP1 and MP antagonistically regulate embryo and meristem development in Arabidopsis. Development. 2007;134:2561–2567. doi: 10.1242/dev.006759. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Seki M, Shinozaki K, Yokoyama S. DNA-binding domains of plant-specific transcription factors: structure, function, and evolution. Trends Plant Sci. 2013;18(5):267–276. doi: 10.1016/j.tplants.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Zhang H, Jin J, Tang L, Zhao Y, Gu X, Gao G, Luo J. PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 2011;39:D1114–D1117. doi: 10.1093/nar/gkq1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, et al. AUXIN RESPONSE FACTOR3 regulates floral meristem determinacy by repressing cytokinin biosynthesis and signaling. Plant Cell. 2018;30(2):324–346. doi: 10.1105/tpc.17.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Gu W, Wilke CO. Detecting positive and purifying selection at synonymous sites in yeast and worm. Mol Biol Evol. 2010;27:1912–1922. doi: 10.1093/molbev/msq077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.