Abstract
Background
While smoking is known to associate with development of multiple diseases, the underlying mechanisms are still poorly understood. Tobacco smoking can modify the chemical integrity of DNA leading to changes in transcriptional activity, partly through an altered epigenetic state. We aimed to investigate the impact of smoking on lung cells collected from bronchoalveolar lavage (BAL).
Methods
We profiled changes in DNA methylation (5mC) and its oxidised form hydroxymethylation (5hmC) using conventional bisulphite (BS) treatment and oxidative bisulphite treatment with Illumina Infinium MethylationEPIC BeadChip, and examined gene expression by RNA-seq in healthy smokers.
Findings
We identified 1667 total 5mC + 5hmC, 1756 5mC and 67 5hmC differentially methylated positions (DMPs) between smokers and non-smokers (FDR-adjusted P <.05, absolute Δβ >0.15). Both 5mC DMPs and to a lesser extent 5mC + 5hmC were predominantly hypomethylated. In contrast, almost all 5hmC DMPs were hypermethylated, supporting the hypothesis that smoking-associated oxidative stress can lead to DNA demethylation, via the established sequential oxidation of which 5hmC is the first step. While we confirmed differential methylation of previously reported smoking-associated 5mC + 5hmC CpGs using former generations of BeadChips in alveolar macrophages, the large majority of identified DMPs, 5mC + 5hmC (1639/1667), 5mC (1738/1756), and 5hmC (67/67), have not been previously reported. Most of these novel smoking-associating sites are specific to the EPIC BeadChip and, interestingly, many of them are associated to FANTOM5 enhancers. Transcriptional changes affecting 633 transcripts were consistent with DNA methylation profiles and converged to alteration of genes involved in migration, signalling and inflammatory response of immune cells.
Interpretation
Collectively, these findings suggest that tobacco smoke exposure epigenetically modifies BAL cells, possibly involving a continuous active demethylation and subsequent increased activity of inflammatory processes in the lungs.
Fund
The study was supported by the Swedish Research Council, the Swedish Heart-Lung Foundation, the Stockholm County Council (ALF), the King Gustav's and Queen Victoria's Freemasons' Foundation, Knut and Alice Wallenberg Foundation, Neuro Sweden, and the Swedish MS foundation.
Keywords: DNA methylation, DNA hydroxymethylation, Enhancers, EPIC, Epigenetics, Smoking, Oxidative stress, Alveolar macrophages
Abbreviations: BAL, Bronchoalveolar lavage; BS, Bisulphite; DMP, Differentially methylated position; DMR, Differentially methylated region; DVP, Differentially variable position; IPA, Ingenuity Pathway Analysis; oxBS, Oxidative bisulphite; RNA-seq, RNA sequencing; TF, Transcription factor; 5hmC, 5-hydroxymethylcytosine; 5mC, 5-methylcytosine
Research in Context.
Evidence before this study
Tobacco smoking can lead to epigenetic and transcriptional changes in several tissues, and is a well-known risk factor for development of a multiple diseases. There is a growing list of smoking-associated epigenetic biomarkers in blood cells, as well as novel sites in other affected tissues. We searched for articles in PubMed using various combinations of the search terms “DNA methylation”, “DNA hydroxymethylation”, “EPIC”, “850 K”, “Smoking”, and “Alveolar macrophages” (last search April 30th 2019). Two studies on DNA methylation in alveolar macrophages from smokers were identified, one investigating ~25,000 CpG loci (n = 22), and the other ~450,000 (n = 19). These studies have presented evidence of genome-wide changes in total DNA methylation and transcription in alveolar macrophages (lung macrophages), using previously available techniques.
Added value of this study
The present study provides new fundamental information on smoking-associated effects on DNA methylation and transcription in alveolar macrophage-dense BAL cells. Oxidative stress is induced by tobacco smoking, and also links to a DNA demethylation pathway where 5-methylcytosine (5mC), as a first step, is oxidised to 5-hydroxymethylcytosine (5hmC). We investigated smoking-associated DNA methylation and hydroxymethylation changes in bronchoalveolar lavage (BAL) cells, and to the best of our knowledge, this is the first epigenome-wide study identifying differentially hydroxymethylated CpG sites as well as true 5mC sites associated with smoking (n = 35) in any human tissue. It is also the first study on lung cells using the recently released Illumina EPIC BeadChip, covering >850,000 CpG loci. Predominant 5mC hypomethylation in contrast to predominant 5hmC hypermethylation supports the hypothesis of a DNA demethylation process initiated by smoking-induced oxidative stress. Notably, many of the affected loci were located at enhancers. We also investigated transcriptional changes using RNA-seq, and found converging alterations in the transcriptome and methylome, with genes involved in signalling, migration, and inflammatory response of immune cells.
Implications of all the available evidence
The current study provides new insights into understanding the impact of tobacco smoking on human health. We could confirm many differentially methylated sites from previous analyses of alveolar macrophages as well as blood cells. Importantly, we also demonstrate novel findings including differential methylation/hydroxymethylation at specific loci together with transcriptional effects, and find previously unreported pathways. These findings provide information that may be of relevance in inflammatory processes leading up to disease. Further studies are warranted to explore the DNA demethylation pathway in the context of smoking-induced oxidative stress.
Alt-text: Unlabelled Box
1. Introduction
Although tobacco smoking is a well-known toxic agent with serious impact on human health, smoking or exposure to smoke remains common worldwide. Smoke exposure is a risk factor and a common cause of death from cardiovascular disease, chronic obstructive pulmonary disease (COPD) and multiple types of cancer, in particular lung cancer [1]. Tobacco smoking is also a risk factor for autoimmune diseases, most notably rheumatoid arthritis (RA) and multiple sclerosis (MS) for which there is an established gene-environment interaction with specific HLA risk alleles, resulting in a substantially elevated risk for developing diseases [2,3]. Cigarette smoke contains a complex mixture of >6000 chemicals, many of them reaching the lower airways during smoke inhalation [4]. Deep airway alveolar macrophages, which in healthy individuals constitute the majority of bronchoalveolar lavage (BAL) cells, play pivotal roles in clearance of such inhaled particles and debris and initiation of inflammatory response [5]. Overall, both the local pro-inflammatory and oxidative response caused by cigarette smoke inhalation, as well as the systemic impact of smoke exposure can lead to long-term deleterious effects on health [6]. Epigenetic mechanisms have been proposed to mediate some of the impact of cigarette smoke through changes in DNA methylation, which may alter gene expression and contribute to smoking-associated disease [7]. With the development of DNA methylation arrays, the effect of smoking on total DNA methylation has been extensively studied in blood cells from adults, showing marked differences in smokers compared to non-smokers [[7], [8], [9]], which can be even more pronounced in patients, as shown in MS smokers [10]. The effect of tobacco smoking on DNA methylation is also evident in cells from cord blood [11] and blood [12] from new-borns, whose mothers smoked during pregnancy. However, only a few studies have investigated DNA methylation in bronchoalveolar immune cells [13,14], which regulate inflammatory reactions through secretion of modulatory and proinflammatory signal molecules, and are affected directly by tobacco smoke.
DNA methylation plays an important role as transcriptional regulator in many biological contexts, such as cell differentiation, embryogenesis, genomic imprinting, and development of disease [15]. In mammals, DNA methylation is most commonly found as 5-methylcytosine (5mC) in the context of CpG dinucleotides and has been associated with gene regulation, where gene promoter methylation usually exerts a repressive action on transcription [16]. In addition to traditional 5mC, other forms of cytosine DNA modifications have been identified, deriving from a DNA demethylation pathway with sequential oxidation of 5mC by the TET family of enzymes, to 5-hydroxymethylcytosine (5hmC) [17], 5-formylcytosine (5fC), and 5 carboxylcytosine (5caC) [18,19]. Hydroxymethylated cytosine (5hmC), often referred to as the sixth base, is however not only an intermediary step of active demethylation, but also a stable epigenetic mark with unique functional properties such as binding of transcription factors and positive regulation of gene expression [20,21].
Recent studies have demonstrated that the DNA demethylation pathway can be initiated by oxidative stress [22,23]. Briefly, oxidative DNA damage from reactive oxygen species (ROS) leads to formation of 8-oxoguanine (8-oxoG), an oxidised product of guanine, and subsequent oxidation of adjacent 5mC to 5hmC [22]. However, conventional bisulphite (BS) conversion used to detect DNA methylation is not able to distinguish 5mC from 5hmC [24]. Therefore, most of the previously published DNA methylation data are reporting overall signal from a mixture of both modifications. In that context, DNA methylation studies in BAL cells (primarily alveolar macrophages) have identified smoking-associated differentially methylated positions (DMPs) using Infinium HumanMethylation27 BeadChip [13] and Infinium HumanMethylation450 BeadChip [14]. In this study, we hypothesised that oxidative stress caused by smoking [25], might affect not only 5mC but also 5hmC patterns in the lung as part of DNA demethylation. In order to better comprehend the effect of tobacco smoking on pulmonary cells, we examined DNA methylation and hydroxymethylation in healthy smokers and non-smokers using the latest Illumina HumanMethylation EPIC BeadChip, covering over 850 K CpG sites and thus providing a higher coverage compared to previous arrays. Further, we combined DNA methylation analysis with gene expression analysis using RNA-seq in BAL cells from healthy smokers and non-smokers.
2. Material and methods
2.1. Study subjects, bronchoscopy and bronchoalveolar lavage
We obtained 49 BAL samples from healthy volunteers (20 smokers, 29 non-smokers) collected during bronchoscopy as previously described [26]. All subjects gave their written consent, and the study was approved by the Regional Ethical Review Board in Stockholm. None of the subjects had clinically relevant airway infections or allergy symptoms at the time of bronchoscopy, and subjects diagnosed with asthma, COPD, other lung diseases, or other inflammatory conditions were not included in the study. All subjects filled out questionnaires regarding general and pulmonary health, current medications, and previous and current smoking habits, and in addition also underwent dynamic spirometry (Medikro PRO, Aiolos Medical). Individuals with >5 pack years (pack years = (cigarettes smoked per day / 20) x years smoking), or smoking at least 5 cigarettes per day, were defined as smokers. Non-smokers were defined as individuals who has never smoked. All subjects have been included in a previous study [27].
2.2. Differential cell count, DNA and RNA extraction
BAL cells and fluid were separated by centrifugation (400 ×g at 4 °C for 10 min), and the supernatants were immediately stored at -80 °C for later use. Approximately 500.000 BAL cells were collected and resuspended in RPMI 1640 without supplements. Smears for differential cell counts were prepared by cytocentrifugation (Cytospin 2, Shanon Ltd) at 22 g for 3 min and stained with May-Grünwald Giemsa, or toluidine/hematoxylin for mast cell identification. A minimum of 500 cells/sample was counted, to estimate cellular proportions in BAL. Samples with low alveolar macrophage content (< 80%) were not included in the study. Genomic DNA and total RNA were extracted using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen) according to the manufacturer's protocol. The concentration of both DNA and total RNA were quantified using PicoGreen (Invitrogen) and Qubit 3 fluorometer (Invitrogen).
2.3. Total 5mC + 5hmC and 5mC methyl processing
DNA from BAL cells was processed using the TrueMethyl conversion kit (Cambridge Epigenetix) workflow in order to investigate true methylation (5mC) by using oxidative bisulphite treatment (oxBS), and total methylation (5mC + 5hmC) by regular common non-oxidative bisulphite treatment (BS). DNA methylome profiling was carried out using the Infinium HumanMethylationEPIC BeadChip Kit (Illumina), which interrogates over 850,000 CpG sites. Methylation arrays were processed by the National Genomics Infrastructure (NGI), Science for Life Laboratory at Uppsala University. Samples were randomised according to smoking-status, age, gender, and cell-proportion and processed together with technical replicates in one run. DNA methylation β-values from the technical replicates correlated strongly with a Pearson correlation value of 0.992 (P = 2.2 × 10–16). Raw intensity IDAT format files were used for subsequent array analysis.
2.4. Generation of RNA sequencing libraries
RNA integrity number (RIN) measurements were performed on an Agilent Bioanalyzer using the RNA 6000 nano kit (Agilent Technologies). 150 ng of total RNA from each individual was used to generate poly-A selected SMART-seq2 cDNA libraries according to the published protocol [28], with minor adjustments to fit bulk RNA input. Spike-in ERCC (External RNA Controls Consortium) RNA was added at 10.000× dilution and cDNA received 6 rounds of preamplification. Sequence ready libraries was prepared with Illumina Nextera index primers using in-house produced TN5 tagmentation enzyme according to following protocol [29]. All libraries were pool and purified with Ampure XP beads, and sequenced at 125 bp paired-end on an Illumina HiSeq 2500.
2.5. Normalisation of 5mC + 5hmC and 5mC methyl data
Raw IDAT files were imported and processed in R software (version 3.5.2) using minfi (version 1.28.4) [30,31] and ChAMP (version 2.12.4) [32] packages. BS and oxBS samples from the same individual were run on the same array. We compared two different normalisation strategies, stratified quantile normalisation (SQN) and subset-quantile within array normalisation (SWAN) [33] and obtained similar results. Final normalisation was done with SQN, and BS-treated (5mC + 5hmC) and oxBS-treated (5mC) samples were processed separately.
Methylation signals were computed as β-values, ranging from 0 to 1. β-values from BS-treated samples represent the total methylation, including both 5hmC and 5mC. The β-values from oxBS-treated samples represent true methylation, and only include 5mC. Samples with probe coverage <95% were removed, as well as probes with a detection P value >.01 in >5% of samples. Further, sex chromosome, cross-reactive and SNP-related probes were removed [34], resulting in 43 samples with 764,958 total 5mC + 5hmC probes and 36 samples with 736,336 true 5mC probes. Finally, only probes and samples overlapping between the datasets were kept, resulting in 35 samples with both 5mC and 5mC + 5hmC readings of 735,794 probes (Supplementary Fig. 1).
2.6. Genomic annotation of CpG sites
Genomic regions were annotated using HumanMethylationEPIC probe annotations through the ChAMP [32] Bioconductor package. The following categories were used as locations in relation to gene: TSS1500 (200 to 1500 nucleotides, nt, upstream of transcription start site, TSS); TSS200 (up to 200 nt upstream of TSS); 5′ UTR (5′ untranslated region); 1st exon; Body (gene body); ExonBnd (exon boundaries), IGR (intergenic regions), and 3′ UTR (3′ untranslated region). Annotations related to CpG islands (CGIs) were divided into following categories: CGIs, CGI shores (flanking shore regions, <2 kb up- and downstream of CGIs), CGI shelfs (2–4 kb up- and downstream of CGIs), and open sea (non-CGI-related sites). Venn diagrams were created using the R package VennDiagram (version 1.6.20) [35].
2.7. Differential methylation and variability analysis
DMP analysis of 5mC + 5hmC and 5mC were done on M-values after transformation from β-values (M = log2(β/(1-β)) as previously recommended [36]. We used a linear model (limma) with the empirical Bayes approach with non-smokers as the reference group and adjusted for covariates (age and sex). A probe was considered significantly differentially methylated if the methylation difference (β-values) between the smokers and non-smokers were at least 15% with a FDR-adjusted (Benjamini-Hochberg) P value <.05. For hydroxymethyl (5hmC) DMP finding, normalised 5mC (oxBS) β-values were subtracted from normalised 5mC + 5hmC β-values to calculate the hydroxymethyl level (Δβ) at each probe. Similarly to previous analysis we used limma with empirical Bayes approach adjusting for age and sex. Additionally, since oxBS treatment is known to introduce negative β-values when subtracting 5mC from 5mC + 5hmC, we performed a second limma where negative β-values were set to a value close to zero (1 × 10−7). We combined the two methods and only 5hmC DMPs that overlapped between the generated lists were considered significant to limit false positives. The RUVm method [37] was used to generate a comparison dataset where unwanted variation was removed. Instead of using known covariates, which can be limited by inaccurate measurements, this method only assume the presence of hidden covariates. A linear regression model with ebayes was run on smoking status yielding statistically non-significant CpGs (FDR-adjusted P value >.5) that were treated as negative control probes, i.e. not associated with smoking. Next, RUVfit was run using RUV-inverse function, and then adjusted through RUVadj (FDR-adjusted P value <.05).
In addition to the single site DMP analysis, we applied the DMRcate package (version 1.18.0) [38] with default settings to detect differentially methylated regions (DMRs) between smokers and non-smokers. We fitted a linear model of methylation values at each probe as a function of smoking status and adjusting for covariates (age, sex). DMRs were defined as those with Stouffer-transformed limma-derived FDR-adjusted P values <.05. Identified DMRs were also filtered using cut-off values of mean absolute Δβ >15%.
Analysis of differential variability was applied to identify smoking-associated DVPs (differential variability positions). DiffVar first uses an empirical Bayes Levene-type algorithm to calculate absolute deviations from respective group means, then uses moderated t-test to compare the distribution of deviations between the groups, assuming that the variability is driven by a range of outliers and not just a few. Age and sex were included as covariates, and DVPs were defined as sites with and FDR-adjusted P value <.05.
A significant DMP associated with the gene body of ITSN1 (cg11650372) was selected based on significance and Δβ and was validated both with oxidative and non-oxidative BS pyrosequencing. Primers were designed using the PyroMark Assay Design 2.0 software and afterwards optimised for best annealing temperature (Supplementary Table 1). 1 μL of oxidised and non-oxidised converted DNA (10 ng) was used for PCR amplification with the PyroMark PCR kit (Qiagen). The product was used together with streptavidin Sepharose high-performance beads (GE Healthcare), the respective sequencing primer, and PyroMark Gold Q96 reagent kit (Qiagen), for pyrosequencing with the PSQ 96 system (Qiagen). Data were analysed on the PyroMark Q96 software.
2.8. Identification of cell-type specific signal and transcription factor analysis with eFORGE analyses
In order to account for differences in cell-type composition in BAL, and identify if they might influence the observed methylation differences, we performed analyses with eFORGE (experimentally derived Functional element Overlap analysis of ReGions from EWAS) [39]. As input, eFORGE accepts a minimum of 20 and a maximum of 1000 CpG sites, in our case including the top 1000 DMPs for total methyl and 5mC, and 67 CpG sites for 5hmC. We examined enrichments for DNase I hypersensitive sites (DHSs) and histone marks (H3K27me3, H3K36me3, H3K4me3, H3K9me3, and H3K4me1). Default settings were used when running the analysis. An in-house pipeline for eFORGE-TF analysis was used to investigate TF motif associations. TF motifs were linked back to a TF gene list and analysed with PANTHER pathway analysis.
2.9. Differential expression analysis
RNA sequencing reads were quality filtered and trimmed for adapters using TrimGalore (version 0.6.0) at default parameters. Afterwards the filtered reads were processed using the pseudoalignment-based Kallisto algorithm (version 0.45.0) with GENCODE v24 comprehensive transcript set as reference transcriptome. For downstream analysis, only samples with a RIN value above 7 were included, and only genes with >10 normalised read counts were kept. In total 23 samples passed these criteria, 10 smokers and 13 non-smokers, which were used for differential expression analysis using DESeq2 package in R. We adjusted for the covariates sex and age and only considered genes with a BH-adjusted (FDR) P value <.05 as significant, together with an absolute log2 fold change threshold >1.
2.10. Gene ontology analyses
Gene ontology (GO) analysis was performed using Ingenuity Pathway Analysis (IPA) (Qiagen) on the annotated genes from 5mC + 5hmC and 5mC DMP genes (FDR-adjusted P < .05, absolute Δβ >0.15; 928 and 938 genes, respectively), differentially expressed genes (adj. P value <.05, logFC >1, 633 genes) as well as dysregulated genes that harbour 5mC + 5hmC DMPs (70 genes), applying unbiased parameters for all criteria including tissues selection. GO analysis was also performed on annotated genes from 5hmC DMP genes (nominal P < .001, absolute Δβ >0.05; 983 genes). Right-tailed Fisher's exact test was used to calculate P values, with P < .05 considered statistically significant. We confirmed IPA findings using over-representation analysis (www.webgestalt.org) [40] on differentially expressed genes and annotated 5mC + 5hmC DMPs genes. Comparison between findings from gene expression and DNA methylation analyses was performed using REVIGO tool [41] based on multidimensional scaling of overrepresented GO terms with semantic similarities. STRING network was generated using STRING database version 10.5 with a minimum level of confidence >0.4.
2.11. Statistical analysis
Statistical methods used for genome-wide DNA methylation and gene expression analyses are detailed in the previous sections. For feature-specific distribution analysis of β-values, we used Wilcoxon rank sum test with Bonferroni adjusted P values for multiple comparison. For enrichment and depletion analysis of differential methylation, we used Pearson's Chi-squared test on contingency tables of count data, and adjusted for multiple comparison using Bonferroni. Correlation analysis between β-values and normalised gene counts was performed using Pearson method, and the Spearman method for correlation analysis between β-values and macrophage fractions. Power analysis (post-hoc) was performed using the pwrEWAS package [42], a tool designed specifically for power analysis of EWAS studies. Input parameters were as follows; 735.000 CpGs, 1700 target CpGs, minimum Δβ detection limit 0.05, FDR-adjusted P value <.05, and adult PBMC as the closest available tissue.
2.12. Data availability
DNA methylation data from this study is available in Gene Expression Omnibus (GEO) database under accession number GSE133062, and RNA-seq data is available upon request.
3. Results
3.1. Characteristics of study participants
Our study included a total of 49 healthy volunteers who all underwent bronchoscopy with bronchoalveolar lavage (BAL), spirometry, and clinical assessment. Quality checks and filtering described in Supplementary Fig. 1 resulted in 35 DNA methylation samples (14 smokers and 21 non-smokers) and 23 RNA-seq samples (10 smokers and 13 non-smokers) for further analyses. Description of the cohort and cell counts is shown in Table 1. Smokers and non-smokers were similar in age (median 28 vs 24 years, P = ns) and both included a larger proportion of females with non-significant difference between groups (P = ns). Smokers displayed a significant increase in BAL fluid cell concentration (P < .001), as well as an altered cellular composition compared to non-smokers (Table 1). More specifically, alveolar macrophages proportion was higher (P < .001) and lymphocytes proportion lower (P < .001) in smokers compared to non-smokers while other cellular compartments remained unchanged. Altogether, the composition of BAL fluid is consistent with previous analyses of smoking and non-smoking healthy volunteers [43].
Table 1.
Characterisation of healthy individuals included in our study.
| All subjects | Non-smokers | Smokers | |
|---|---|---|---|
| Subject | 35 | 21 | 14 |
| Sex (male/female) | 13/22 | 9/12 | 4/10 |
| Age (years) | 25.0 [22.0–29.0] | 24.0 [22.0–28.0] | 28.0 [24.3–29.8] |
| Cigarettes/day | 12.5 [10.0–19.8] | N/A | 12.5 [10.0–19.8] |
| Pack years | 6.0 [5.0–9.5] | N/A | 6.0 [5.0–9.5] |
| FEV1, % predicted | 103.0 [97.5–110.5] | 103.0 [98.0–107.0] | 105.0 [94.8–110.8] |
| FVC, % predicted | 109.0 [102.0–117.0] | 108.0 [102.0–117.0] | 110.5 [103.0–115.3] |
| FEV/FVC % | 81.0 [78.0–83.5] | 82.0 [79.0–83.0] | 80.0 [76.5–84.5] |
| BALF cell concentration (x106/L) | 119.6 [75.4–254.9] | 76.8 [72.2–105.4] | 300.6 [219.0–357.5]*** |
| BAL recovery (%) | 71.0 [62.5–77.5] | 74.0 [70.0–79.0] | 64.0 [58.0–69.5]** |
| BAL macrophages (%) | 93.4 [88.9–96.1] | 90.8 [85.8–93.4] | 96.5 [93.8–97.4]*** |
| BAL lymphocytes (%) | 4.8 [2.8–9.7] | 7.3 [4.2–12.0] | 2.2 [1.8–3.45]*** |
| BAL neutrophils (%) | 0.8 [0.4–1.3] | 1.0 [0.6–2.0] | 0.7 [0.4–1.15] |
| BAL eosinophils (%) | 0.2 [0–0.4] | 0.2 [0–0.3] | 0.1 [0–0.4] |
| BAL basophils (%) | 0 [0–0] | 0 [0–0] | 0 [0–0] |
| BAL mast cells (%) | 1.0 [0–2.5] | 0 [0–3.0] | 1.0 [0.3–2.0] |
| BAL CD4/CD8 ratio | 1.6 [1.1–2.7] | 1.9 [1.4–3.0] | 1.1 [0.8–1.8] |
Basic characterisation of individuals included in our cohort. Data represent n or median [25th–75th percentile]. Pack years: (cigarettes smoked per day / 20) x years smoking); FEV1: forced expiratory volume in 1 s; FVC: forced vital capacity; BALF: bronchoalveolar lavage fluid; BAL: bronchoalveolar lavage; N/A: not applicable. Statistics calculated using Chi-square test for male/female ratio and Mann-Whitney U test for other comparisons: * P < .05 compared to non-smokers, ** P < .01 compared to non-smokers, *** P < .001 compared to non-smokers.
3.2. Genomic profiling of total 5mC + 5hmC and 5mC reveals feature-specific presence of hydroxymethyl in BAL cells
We determined genome-wide total 5mC + 5hmC and 5mC CpG levels in BAL cells from smokers and non-smokers using the Illumina HumanMethylationEPIC BeadChip. The array covers over 850 K CpG sites with improved coverage of regulatory elements compared to its 27 K and 450 K predecessors, including 58% of Functional Annotation of the Mammalian Genome 5 (FANTOM5) enhancers. Total methylation (5mC + 5hmC) was quantified using BS-treated (bisulphite-treated) DNA samples and represents a combination of 5mC + 5hmC signals. In order to decipher the contribution of 5mC from 5hmC signals we performed oxidative BS (oxBS) treatment in parallel to BS treatment prior to array hybridisation. After QC and filtering steps (Supplementary Fig. 1), we retained 35 subjects for further analysis, with overlapping oxBS and BS data and 735,794 probes in common.
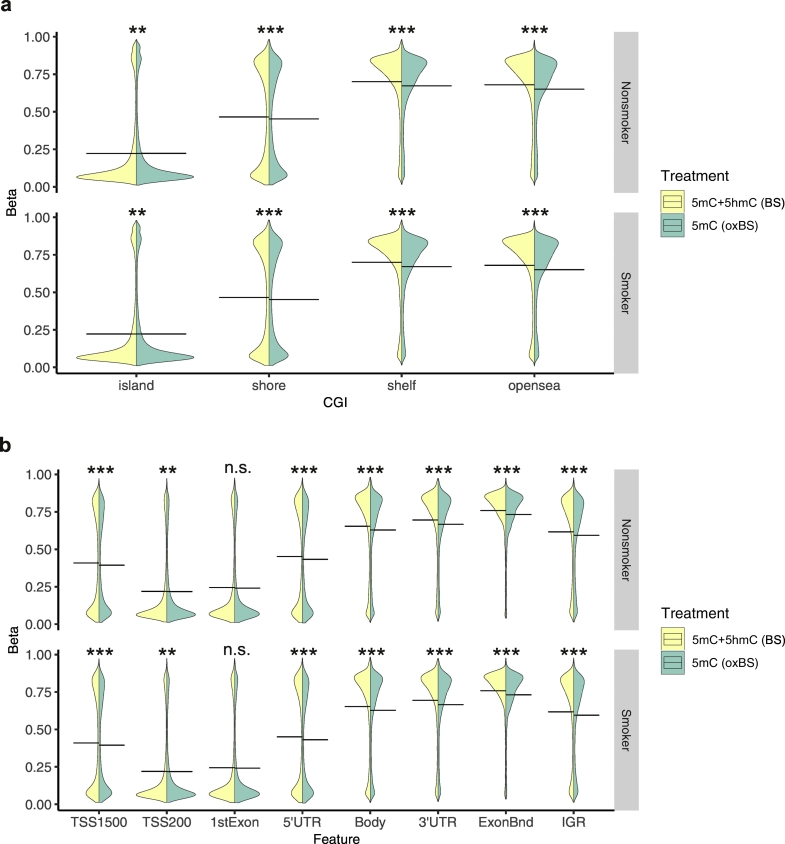
As an exploratory first step, we examined distributions of all 735,794 probes focusing on the genomic features related to CpG island (CGI) (Fig. 1a), and gene location (Fig. 1b). BAL cells from both smokers and non-smokers had a significantly higher 5mC + 5hmC than 5mC for probes located in open sea, shelfs and shore regions (Wilcoxon rank sum test, P < .001). The probe distribution was also significantly different between smokers and non-smokers in open sea (P < .001) and shelf regions (5mC + 5hmC P < .01, 5mC P < .05; Supplementary Fig. 2a). Since the subtraction of true 5mC values from 5mC + 5hmC values results in 5hmC values, our data suggest a genome-wide presence of 5hmC in BAL cells across CGI-related features, most likely outside CGI as the typically low CGI methylation levels is less permissive to demethylation. Similar to reports describing other peripheral tissues [44], we observed relatively low overall levels of 5hmC in BAL cells.
Fig. 1.
Feature-specific differences in β-value distribution between total 5mC + 5hmC and true 5mC. Violin plots representing the distribution of β-values from 735,794 probes after SQN normalisation, plotted as densities in relation to CpG islands (a) and genomic features (b). Yellow plots represent standard bisulphite-treated samples with a combination of both 5mC and 5hmC (total BS methyl). Green plots represent oxidative bisulphite-treated samples with 5mC (true 5mC, oxBS). Plots representing smoker densities are shown in upper rows (a-b) and non-smoker plots in lower rows (a-b). Differences in distributions between 5mC and 5mC + 5hmC was tested using Wilcoxon signed-rank test was considered significant with a P value <.05. * P < .05; ** P < .01; *** P < .001; n.s = not significant. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Exploration of DNA methylation signals throughout the genome revealed significant differences in distribution between 5mC + 5hmC and 5mC signals (Fig. 1b). No significant difference was observed between 5mC + 5hmC and 5mC signal in 1st exon, displaying low range of methylation distribution in both smokers and non-smokers. Overall, distribution profiles at promoter regions (defined here as TSS200 + TSS1500) indicated low methylation. Smokers and non-smokers displayed relatively similar 5mC + 5hmC and 5mC signal distribution across all genomic regions, but with significant differences in gene bodies and IGR (P < .001; Supplementary Fig. 2b). Thus, global differences between total 5mC + 5hmC and true 5mC (oxBS) methylation profiles indicate presence of 5hmC in BAL cells.
3.3. Site and feature-specific changes in total 5mC + 5hmC, 5mC, and 5hmC in BAL cells from smokers
To determine genome-wide differences in total 5mC + 5hmC and true DNA methylation (5mC) between smokers and non-smokers in BAL cells, we applied a linear regression model with age and sex included as covariates and with a Benjamini-Hochberg (FDR) adjusted level of significance of P < .05. We set a stringent delta beta (Δβ) cut-off of 15% for differential methylation, which was substantially higher than the difference in content of alveolar macrophages in BAL of smokers (median = 96.5% compared to median = 90.8% for non-smokers), to exclude potential DMPs that would be caused by a difference in cell proportions alone. Analysis of 5mC + 5hmC and 5mC data from BAL cells revealed 1667 total 5mC + 5hmC DMPs (Table 2; Supplementary Table 2) and 1713 5mC DMPs (Table 2; Supplementary Table 3) associated with smoking (FDR-adjusted P <0.05, absolute Δβ >0.15). 5hmC was quantified by subtracting 5mC β-values from 5mC + 5hmC β-values (735,794) in 35 overlapping BS and oxBS samples from smokers and non-smokers. As expected, 5hmC methylation values ranged at lower level and a minor fraction of probes displayed slightly negative values. We therefore set an absolute Δβ threshold of >0.05 when calling 5hmC and found 67 significant 5hmC DMPs (Table 2; Supplementary Table 4) after correcting for multiple testing (FDR-adjusted P < .05). The most significant 5mC + 5hmC, 5mC, and 5hmC DMPs are shown in Supplementary Fig. 3.
Table 2.
Summary of the top 10 significant smoking-associated DMPs of total 5mC + 5hmC (BS), true 5mC (oxBS), and 5hmC.
| Probe | adj.P.Val | Chr | Position | Gene | Delta Beta |
Island Relation |
Feature | Enh | 450 k chip |
Blood | AMs 450 k |
DMP overlap |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5mC + 5hmC DMPs | ||||||||||||
| cg04857037 | 3.77E-12 | 1 | 26,136,971 | SEPN1 | −0.24 | OpenSea | Body | 5mC | ||||
| cg04308301 | 4.68E-12 | 8 | 131,454,686 | ASAP1 | −0.21 | N_Shore | 5’UTR | * | 5mC | |||
| cg26721868 | 8.01E-12 | 19 | 4,567,641 | −0.38 | S_Shore | IGR | * | 5mC | ||||
| cg02233197 | 9.41E-11 | 15 | 51,390,542 | TNFAIP8L3 | −0.30 | S_Shelf | Body | * | * | 5mC | ||
| cg04135110 | 1.72E-10 | 5 | 346,695 | AHRR | 0.19 | S_Shelf | Body | * | * | 5mC | ||
| cg01596674 | 2.01E-10 | 9 | 130,342,290 | FAM129B | −0.22 | OpenSea | TSS1500 | * | 5mC | |||
| cg12617080 | 2.95E-10 | 1 | 156,509,844 | IQGAP3 | −0.25 | OpenSea | Body | * | 5mC | |||
| cg01668352 | 3.27E-10 | 12 | 64,482,597 | SRGAP1 | −0.30 | OpenSea | Body | * | * | * | 5mC | |
| cg21513724 | 3.27E-10 | 10 | 105,409,153 | SH3PXD2A | −0.23 | OpenSea | Body | * | 5mC | |||
| cg14223856 | 5.35E-10 | 9 | 139,508,740 | −0.32 | OpenSea | IGR | * | * | 5mC | |||
| 5mC DMPs | ||||||||||||
| cg02233197 | 2.39E-09 | 15 | 51,390,542 | TNFAIP8L3 | −0.29 | S_Shelf | Body | * | * | BS | ||
| cg26721868 | 2.39E-09 | 19 | 4,567,641 | −0.26 | S_Shore | IGR | * | BS | ||||
| cg14223856 | 5.30E-09 | 9 | 139,508,740 | −0.40 | OpenSea | IGR | * | * | BS | |||
| cg10655682 | 3.49E-08 | 19 | 4,567,177 | −0.34 | S_Shore | IGR | * | BS | ||||
| cg11180972 | 3.49E-08 | 12 | 105,066,468 | CHST11 | −0.29 | OpenSea | Body | * | BS | |||
| cg25711726 | 3.49E-08 | 11 | 124,949,155 | SLC37A2 | −0.34 | OpenSea | ExonBnd | BS | ||||
| cg07457727 | 3.49E-08 | 8 | 131,451,983 | −0.30 | N_Shelf | IGR | * | * | BS | |||
| cg09552070 | 3.49E-08 | 17 | 3,704,607 | ITGAE | −0.22 | OpenSea | TSS200 | BS | ||||
| cg04135110 | 4.88E-08 | 5 | 346,695 | AHRR | 0.20 | S_Shelf | Body | * | * | BS | ||
| cg10360854 | 5.81E-08 | 11 | 124,949,180 | SLC37A2 | −0.28 | OpenSea | Body | * | * | BS | ||
| 5hmC DMPs | ||||||||||||
| cg00456797 | 1.36E-03 | 2 | 207,233,872 | 0.16 | OpenSea | IGR | ||||||
| cg20991802 | 8.13E-03 | 22 | 17,724,817 | 0.12 | OpenSea | IGR | ||||||
| cg13638884 | 8.13E-03 | 10 | 125,223,731 | 0.12 | OpenSea | IGR | BS | |||||
| cg11385411 | 1.23E-02 | 1 | 184,133,807 | 0.11 | OpenSea | IGR | * | |||||
| cg13631605 | 1.23E-02 | 6 | 160,405,572 | IGF2R | 0.12 | OpenSea | Body | |||||
| cg09550697 | 1.23E-02 | 8 | 145,012,068 | PLEC1 | 0.15 | S_Shelf | Body | * | * | 5mC | ||
| cg13388131 | 1.23E-02 | 11 | 62,211,493 | AHNAK | 0.12 | N_Shore | Body | |||||
| cg19800026 | 1.23E-02 | 5 | 14,492,945 | TRIO | 0.18 | OpenSea | Body | * | * | BS | ||
| cg01025883 | 1.23E-02 | 16 | 23,867,088 | PRKCB | 0.12 | OpenSea | Body | * | ||||
| cg27058773 | 1.58E-02 | 17 | 66,309,601 | ARSG | 0.17 | OpenSea | Body | * | ||||
Probe: Illumina probe ID; adj.P.Val: Benjamini-Hochberg corrected P value (FDR); Chr: Chromosome; Gene: UCSC gene name; Delta Beta: difference in mean β-values between smokers and non-smokers; Island Relation: Relation to CpG Island; Feature: Gene feature; Enh: Identified enhancer in FANTOM5 consortium; 450 k chip: Present on Infinium 450 k BeadChip; Blood: Significant in blood cell DNA methylation smoke signature as previously reported [7]; AMs 450 k; Significant in alveolar macrophages in smokers as previously reported [14]; * indicates presence in database, on chip, or in related studies; DMP overlap: Overlapping with BS (total 5mC + 5hmC), 5mC or 5hmC DMPs.
As expected, DNA methylation QQ plots (Supplementary Fig. 4) show inflated observed –log10 P values, both after adjusting for covariates (sex+age) and when removing unwanted variables using RUVm [37]. This is not uncommon in EWAS studies [45,46] where alterations in the phenotype also may reflect other underlying factors, unlike genome-wide association studies where a genetic variant is closely linked to a phenotype. In order to investigate statistical power, we performed a post-hoc analysis using a power analysis tool designed specifically for EWAS studies (pwrEWAS). With a sample size of 35 and an effect size (absolute Δβ) of 0.15, the estimated statistical power of the current study is >0.8 (Supplementary Fig. 5), which is a common cut-off for well-powered studies.
Regarding cell composition across individuals, we observed a high proportion of macrophages across our samples (median > 90%). However, BAL from smokers is known to present an increased macrophage fraction and decreased lymphocyte fraction [43], which was also the case in our cohort. We hypotesise that correction for cell type composition might mask some of the impact of smoke exposure since the smoking effect will also be adjusted due to multicollinearity. To test this, we compared our findings with 5mC + 5hmC, 5mC, 5hmC DMP analyses using cell-type as an additional covariate in the linear model while applying the same criteria for significance (FDR-adjusted P < .05) and effect size (5mC + 5hmC and 5mC: absolute Δβ >0.15; 5hmC: absolute Δβ >0.05). Spearman's correlation coefficients (Rho) indicated that there was no overall strong correlation between cell type and DMP β-values (Supplementary Fig. 6, Rho and P values has been added to Supplementary Table 2–4). Additionally, the FDR-adjusted P values from the cell type-adjusted and unadjusted DMP lists strongly correlated (Supplementary Fig. 7). Given the high proportion of macrophages across our samples, it is likely that our DNA methylation changes are specific to this particular cell type. To address this, we performed eFORGE analyses [39] (based on DNase I hypersensitive sites or DHSs) applied to the top DMPs of total 5mC + 5hmC (1000 probes), 5mC (1000 probes), and 5hmC (67 probes). Since alveolar macrophages are not part of the Epigenome Roadmap [47], we based the overlaps on peripheral blood cell types including monocytes. As expected, eFORGE results (Supplementary Fig. 8) showed that monocytes (primary monocytes in peripheral blood) were by far the most enriched cell type among our DMPs (5mC + 5hmC q-value = 1.77 × 10−152; 5mC q-value = 1.04 × 10−153; 5hmC q-value = 1.77 × 10−13), demonstrating co-localisation of monocyte DHS sites and smoking-associated DMPs. In contrast, we did not see any significant enrichment of T cells (primary T cells from peripheral blood) among our DMPs (5mC + 5hmC q-value = ns; 5mC q-value = ns; 5hmC q-value = ns), further validating that our findings were indeed macrophage-specific.
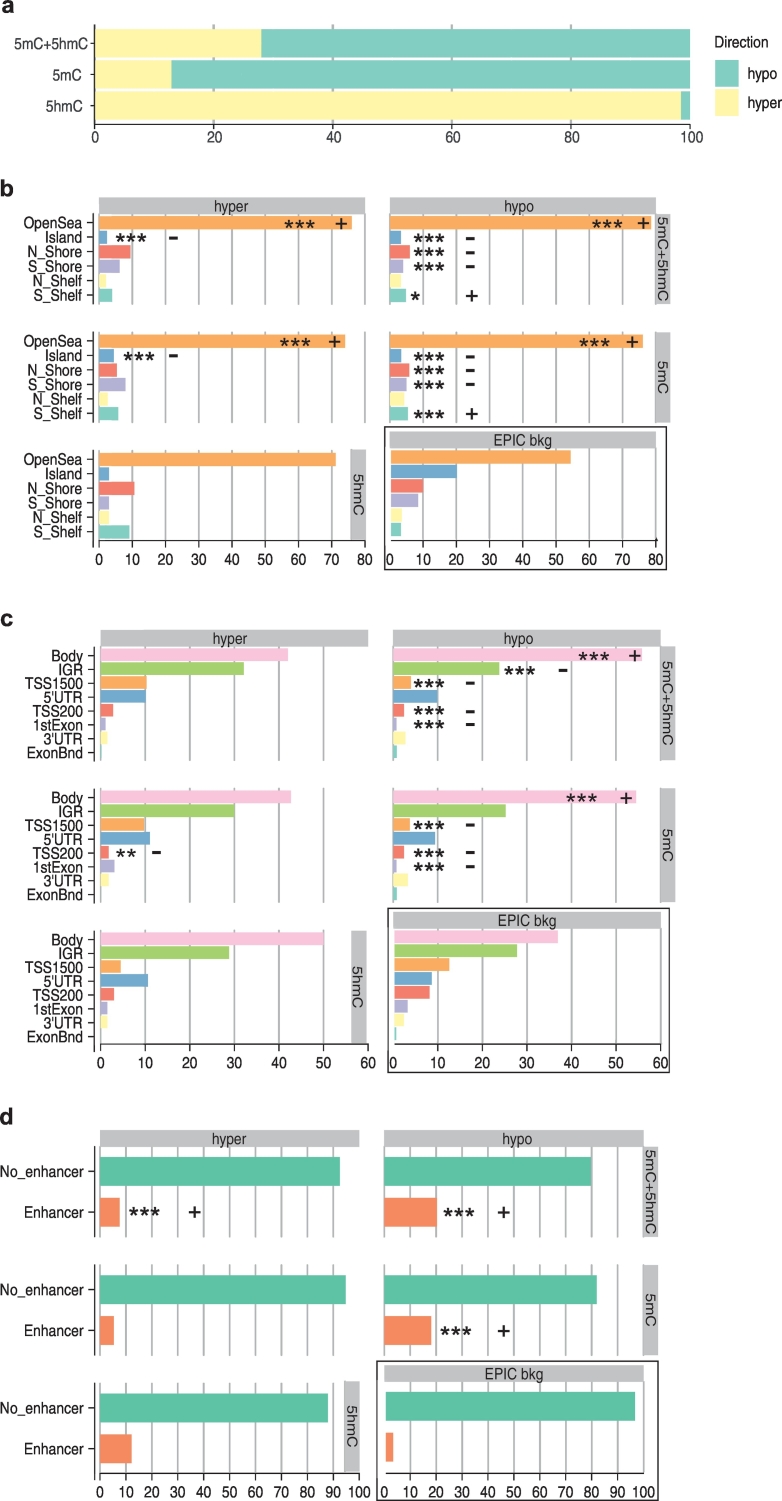
The majority of the significant DMPs were hypomethylated in smokers compared to non-smokers (Fig. 2a), but to a larger extent in 5mC (87%) than in 5mC + 5hmC (72%), suggesting that a fraction of the 5mC + 5hmC DMPs display 5hmC modification. This is confirmed by the predominant hypermethylation in 5hmC DMPs (Fig. 2a). Next, we stratified the significant DMPs into CGI (Fig. 2b) and gene-related features (Fig. 2c), and performed enrichment analysis relative to the EPIC background (735,794) probe distribution. In summary, analysis of significant DMPs revealed that both 5mC + 5hmC and 5mC hypo- and hypermethylated changes, caused by smoking, predominantly occur in open seas and CGI (P < .001). Hypermethylated 5hmC sites (66 DMPs) were also found more often in open sea and under-represented at CGI, without however, reaching significance level. Hypomethylated 5mC + 5hmC and 5mC DMPs were enriched in gene bodies (Fig. 2c), and depleted in TSS1500, TSS200, and 1st Exon (P < .001). Hypermethylated 5hmC sites were not significantly changed, but followed the same directionality as hypomethylated 5mC, with enrichment at gene bodies and depletion at TSS1500 and TSS200. Interestingly, we found a striking enrichment of both among 5mC + 5hmC and 5mC DMPs in FANTOM5 enhancers (Fig. 2d). This is the case especially for hypomethylated DMPs, with enrichment from 3.3% of total EPIC probes (background) to 20.3% of significant 5mC + 5hmC DMPs (244 DMPs, P < .001) and 18.1% of 5mC DMPs (276 DMPs, P < .001).
Fig. 2.
Smoking-associated DMPs are predominantly hypomethylated in 5mC and total 5mC + 5hmC with enrichment in gene bodies, non-CGI context and enhancer sites. Horizontal bar plots (a-d) illustrating relative frequencies of DMPs associated with smoking. (a) Relative frequencies of hypermethylated and hypomethylated 5mC + 5hmC, 5mC, and 5hmC DMPs. Percentage of hypermethylated and hypomethylated 5mC + 5hmC, 5mC and 5hmC across (b) CGI-related features (CpG islands, shores, shelves, open sea), (c) gene features (TSS1500, TSS200, 1stExon, 5’UTR, Body, 3’UTR, ExonBnd, IGR), and (d) enhancers. The distribution of all EPIC array probes included in our analysis are shown as EPIC bkg (background) for comparison (b-d). Enrichment/depletion analysis was performed using Chi-square test on frequencies, adjusting P values for multiple testing (Bonferroni). * P < .05 compared to non-smokers, ** P < .01 compared to non-smokers, *** P < .001 compared to non-smokers.
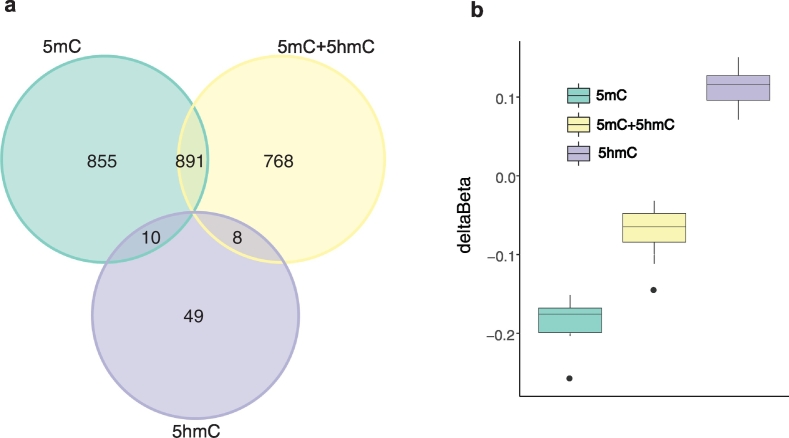
Interestingly, only 53% of the true 5mC DMPs (891) overlapped with 5mC + 5hmC DMPs (Fig. 3a), suggesting that relevant changes might be undetected by conventional BS methylation analysis. The 10 overlapping 5mC and 5hmC DMPs (Fig. 3a-b) showed hypomethylated 5mC and hypermethylated 5hmC, further supporting involvement of the DNA demethylation pathway.
Fig. 3.
Hypomethylation and hyperhydroxymethylation in smokers. Overlapping total 5mC + 5hmC (yellow), 5mC (green) and 5hmC (purple) with adjusted P value <.05 (absolute Δβ threshold: 5mC + 5hmC and 5mC >0.15; 5hmC >0.05). (a) Venn diagram illustrating number of DMPs and overlaps between 5mC + 5hmC, 5mC, and 5hmC. (b) Boxplot showing 5mC + 5hmC, 5mC and 5hmC Δβ for the 10 overlapping DMPs between 5mC and 5hmC. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Predominant hypomethylation could be further confirmed at region level. In order to do so, we investigated clusters of neighbouring probes for potential differential methylation between smokers and non-smokers, using again a linear model adjusting for age and sex. We focused on differentially methylated regions (DMRs) with clusters of at least two consecutive differentially methylated CpGs and identified 60 5mC + 5hmC and 83 5mC DMRs associated with smoking status at genome-wide level (FDR-adjusted P < .05) (Supplementary Table 5 and Supplementary Table 6). In accordance with DMPs analysis, most DMRs were found hypomethylated (90% and 92% of 5mC + 5hmC and 5mC DMRs, respectively).
For technical validation of the EPIC methylation array, we selected a DMP mapping to ITSN1 (cg11650372), displaying strong hypermethylation in smokers compared to non-smokers. BS and oxBS pyrosequencing confirmed differential methylation in accordance with the EPIC data (BS methyl and 5mC; P value <.0001; Supplementary Fig. 9).
3.4. Known and novel smoking-associated DMPs
In order to compare our findings with previously reported DMPs generated using BS-treatment alone, we focused on the 1667 DMPs from total 5mC + 5hmC signals. Strikingly, out of the identified total 1667 DMPs, a substantial fraction (63.8%, 1063 DMPs) were covered only by the EPIC chip (hence absent from previous platforms) (Supplementary Table 2). Among the remaining 604 overlapping probes, 18% (110/604) are known smoking-associated sites from genome-wide studies in blood [7]. In alveolar macrophages, 60% (18/30) of previously reported DMPs [14] were also found significant in our cohort. Seven of our DMPs overlapped with smoking-associated DMPs in both blood cells and alveolar macrophages and map to the SRGAP1 (cg01668352), AHRR (cg25648203), SUSD4 (cg25466245), DAB2 (cg17576603), FGR (cg11254522), CD80 (cg13458803) and RYBP (cg09006487) genes, respectively.
The most significant 5mC + 5hmC DMP map to the antioxidant-producing SEPN1 gene (cg04857037, adjusted P value = 3.77 × 10−12), and is specific to the EPIC bead array (Table 2; Supplementary Table 2). The most significant 5mC DMP (Table 2; Supplementary Table 3) was annotated to TNFAIP8L3 (cg02233197, adjusted P value = 2.39Ex10−9), and has been previously reported in alveolar macrophages [14], while the most significant 5hmC DMP (cg00456797, adjusted P value = 1.85 × 10−3) is intergenic and specific to the EPIC bead array (Table 2; Supplementary Table 4). Overall, 30% of the 5mC + 5hmC, 50% of the 5mC and 70% of 5hmC top 10 DMPs were specific to the EPIC chip.
3.5. Differential variability of total 5mC + 5hmC and 5mC associated to smoking
Next, we tested whether DNA methylation variability was significantly associated with cigarette smoking. Group-wise analyses of differential variability on M-values revealed 19 significant 5mC + 5hmC differentially variable positions (DVPs) (Supplementary Table 7) and 3 significant 5mC DVPs (Supplementary Table 8) with an FDR-adjusted P < .05. The commonly known smoking-associated AHRR CpG site cg05575921 was among the top 5mC + 5hmC DVPs, together with CpG site in the bidirectional promoter of oncogene BRCA1 and the NBR2 gene (cg20760063). Out of the 3 significant 5mC DVPs, 2 were annotated to the 3’UTR region of NFYA (cg06671660, cg09580153).
3.6. Changes in methylation correlate with transcriptional differences
To determine whether the total DNA methylation profiles were associated with differentially expressed genes, RNA was simultaneously extracted from BAL cells from the same individuals and subjected to RNA sequencing (RNA-seq). After filtering reads passing quality threshold (RIN >7, total read count >10), a total of 633 genes were differentially expressed between smokers and non-smokers with an adjusted P value <.05 and absolute log2-fold change >1 (Supplementary Table 9).
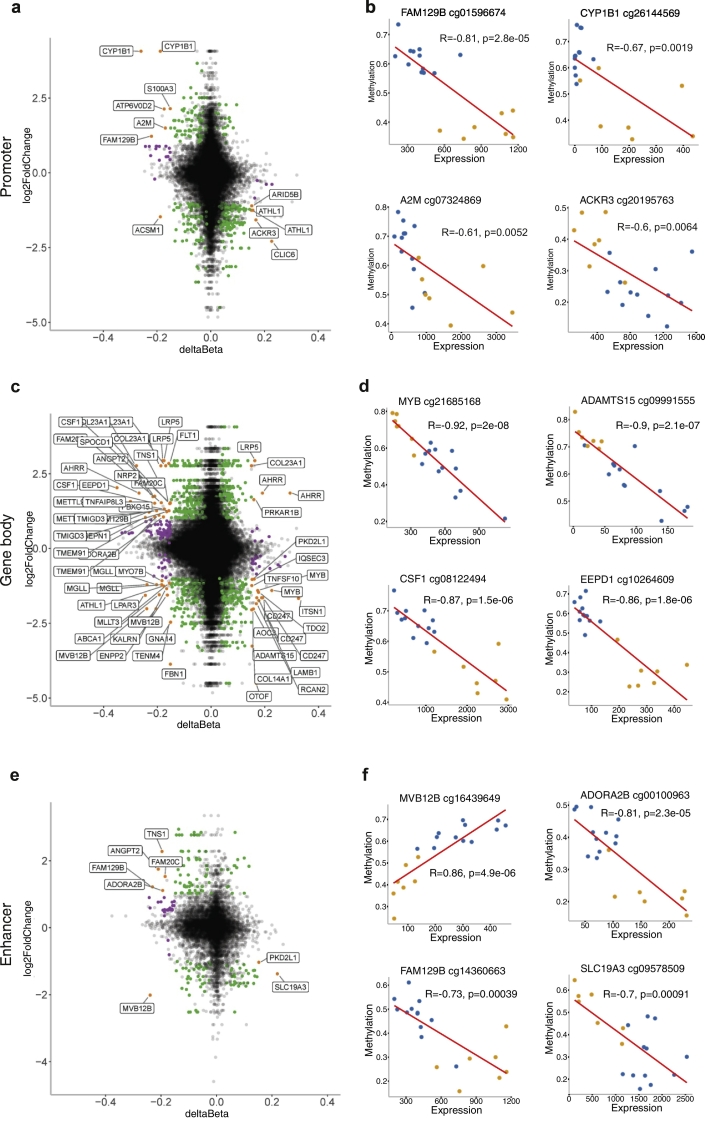
Since DNA methylation in promoters and gene bodies often correlate with gene regulation, and given the considerable evidence of interactions between enhancers and promoters with subsequent transcriptional regulation [48], we sought to investigate associations between methylation and expression specifically in these regions. We thus focused on all probes annotated to promoter regions (defined here as TSS200 or TSS1500), gene bodies, and enhancer sites. Comparison of 5mC + 5hmC and gene expression changes in smokers vs non-smokers revealed 60 differentially expressed genes, with 90 DMPs annotated to them. A large fraction, 75.5% (68/90), of these 5mC + 5hmC β-values correlated with gene expression (Pearson correlation, R > 0.5) (Supplementary Table 10). More specifically, in promoters, a total of 10 differentially expressed genes also showed 5mC + 5hmC changes (12 DMPs) (Fig. 4a). 66.7% (8/12 DMPs) of these genes displayed a negative correlation (R > 0.5) between DNA promoter methylation and gene expression, as is often the case with promoter methylation. Smoking associated with hypomethylation and upregulation of 5 genes, including FAM129B, CYP1B1, and A2M (Fig. 4b), along with hypermethylation and downregulation of 4 genes, such as ACKR3 and ATHL1. An additional 45 genes showed significant differences in both gene body total DNA methylation (63 5mC + 5hmC DMPs) outside of promoter region, and gene expression (Fig. 4c-d), including MYB, EEPD1, FLT1, and the antioxidant-producing SEPN1. Interestingly, gene bodies include both hypo- and hypermethylation as well as over- and under-expression, supporting a mechanistic difference between promoters and gene bodies. Enhancer-associated DMPs negatively correlated with gene expression and were mostly located at gene bodies (87.5%, 7/8) including the MVB12B, ADORA2B, and FAM129B genes (Fig. 4e-f). Overall, 5mC methylation and gene expression associations largely overlapped with 5mC + 5hmC, but included additional genes such as IL4I1, NFIA (Supplementary Fig. 10a-c), and selectin L-producing SELL. A few 5hmC DMPs were also differentially expressed, including XYLT1, SH3RG1, and CDA (Supplementary Fig. 10d-e).
Fig. 4.
Correlation of promoter, gene body, and enhancer methylation with gene expression. Plots showing genes with differences in both DNA methylation and expression in promoters (a-b), gene body (c-d), and enhancers (e-f) of smokers-associated DMPs and genes. Scatterplots of mean gene expression values (log2 fold change) and total methyl (5mC + 5hmC) mean Δβ in promoter (a), gene body (c), and enhancer (e). Orange dots represent genes with significant smoking-associated changes in both total 5mC + 5hmC (>0.15 absolute Δβ) and gene expression (log2 fold change >1), green dots represent significant expression but not methylation, and purple dots represent changes in 5mC + 5hmC but not gene expression (b,d,f). Correlation plots of selected genes showing both methylation (5mC + 5hmC, β-values) and gene expression (normalised gene count), with Pearson correlation coefficient and P value (b,d,f). Smokers (n = 7) are represented by yellow dots, and (n = 12) non-smokers by blue dots. Promoters are represented by TSS200 and TSS1500 CpG sites. Enhancer sites are present at various genomic features and can overlap with both promoter and gene body sites. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.7. DNA methylation and expression changes associate with genes involved in immune cells migration and activation
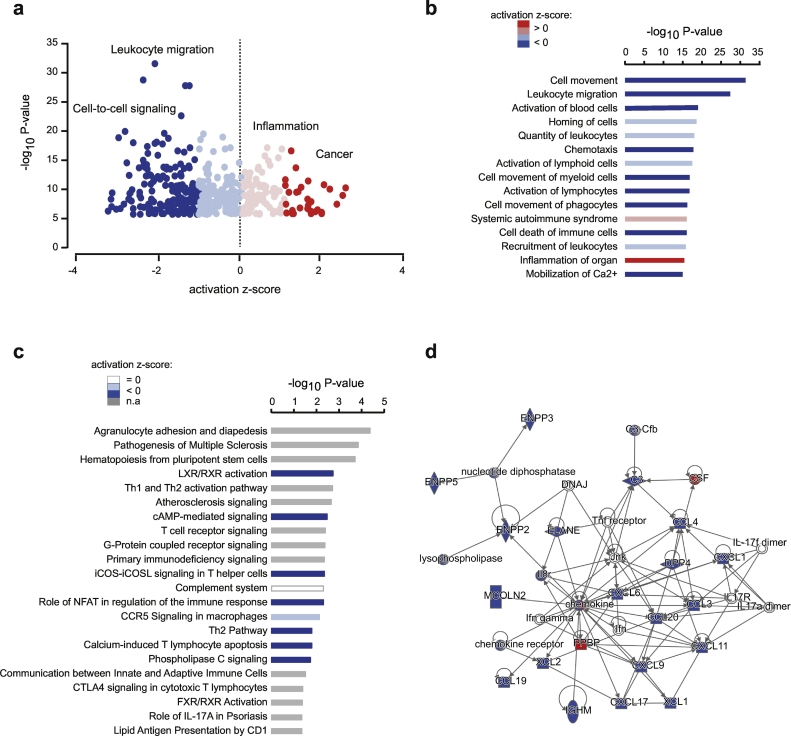
To gain insight into biological functions associated with transcriptional changes in BAL cells from smokers, we performed Ingenuity Pathway analysis (IPA) on the 633 differentially expressed genes (adj. P value <.05, log2-fold change >1). Enriched biological functions associated with transcriptional changes showed inhibition of processes such as cellular movement and cell-to-cell signalling while inflammatory response and cancer-related processes were predicted to be activated (Fig. 5a). This is exemplified by the most significant biological functions implicating decreased activity of immune cell chemotaxis, movement and activation (Fig. 5b). The most enriched canonical pathways (Fig. 5c) showed alteration of LXR/RXR activation pathway involved in lipid metabolism and inflammation (Fig. 5d), adhesion and diapedesis of mononuclear leukocytes pathway, a key event in the process of inflammation, and complement system pathway bridging the innate and acquired immune systems, including cell killing, clearance of immune complexes and apoptotic cells and activation of inflammation.
Fig. 5.
Functional annotations of differentially expressed genes. All (a) and top (b) biological processes and diseases associated with differentially expressed genes obtained with Ingenuity Pathway Analysis (IPA). Top canonical pathways (c) associated with differentially expressed genes obtained with IPA. (d) Schematic representation of the top gene interaction network obtained with IPA, with downregulated and upregulated genes depicted in blue and red, respectively. (a-c) Significance is represented as –log10P value and colours indicate predicted activation z-score, with decreased, no effect, and increased activation in blue, white and red colours respectively. n.a. prediction not available. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
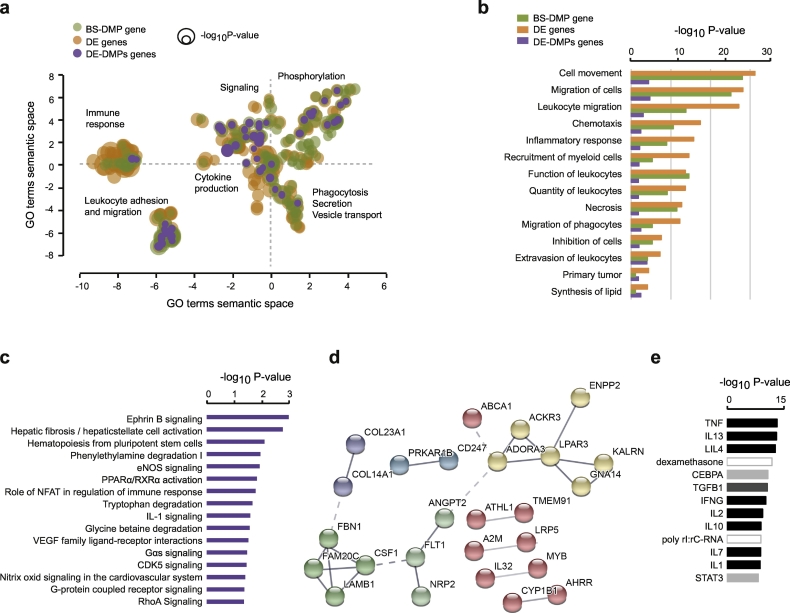
Interestingly, biological processes (GO terms) associated with 5mC + 5hmC DMP-annotated genes strongly overlap with the differentially expressed genes, and differentially expressed genes harbouring DMPs also clusters with them (Fig. 6a). Top biological functions for both differentially methylated and expressed genes converge to immune-related processes such as leukocyte recruitment, migration and adhesion (Fig. 6a-b). Of note, functions and pathways of genes annotated by novel EPIC smoking-associated DMPs converge to similar GO terms as the ones from genes annotated with known (450 K) smoking-associated DMPs (Supplementary Fig. 11). IPA analysis of dysregulated genes affected by 5mC + 5hmC DMPs shows enrichment of canonical pathways related to haematopoiesis, angiogenesis, immune/oxidative pathways and metabolism (Fig. 6c), cores genes segregating into functional groups of interconnected genes (Fig. 6d). Among them, AHRR and CYP1B1 are known genes involved in AHR detoxification pathway. Likewise, genes showing DNA methylation and transcriptional changes have common upstream regulators, most of which are cytokines typically found in an inflammatory milieu, such as the pro-inflammatory TNF, IFN-γ and IL-1 and the anti-inflammatory IL-4 and IL-10 cytokines. Thus, smoking-associated changes in DNA methylation and gene expression alter genes important for immune functions of BAL cells.
Fig. 6.
Functional annotations of differentially methylated and expressed genes. (a) Multidimensional scaling of GO Biological processes terms associated with differentially methylated (BS-DMP: total 5mC + 5hmC) (green), differentially expressed (DE) (orange) genes according to semantic similarities. Findings from annotated DE genes that harbour BS-DMPs are depicted in purple. GO terms were obtained using over-representation analysis and visualization was generated by REVIGO. (b) Top common biological processes and diseases associated with differentially methylated (BS-DMP) (green) and DE (orange) genes obtained with Ingenuity Pathway Analysis (IPA), with IPA findings from genes displaying both BS-DMP (FDR < 0.05, Δβ > 0.15) and transcriptional change in purple. (c) Top canonical pathways associated to both DE- and DMP-genes. (d) Representation of the genes network obtained with IPA on both DE- and DMP-genes and visualised using STRING. Grey line gradient and thickness indicates the strength of data support (darker thick grey representing stronger confidence) and colours represent different cluster (Markov Clustering set at 6). (e) Top upstream regulators for both differentially methylated and expressed genes, with colours depicting different classes of regulators. Significance is represented as –log10P value. DE, differentially expressed, BS-DMPs, bisulphite-generated differentially methylated positions (5mC + 5hmC). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Overall, we found a major overlap of 5mC + 5hmC and 5mC (Supplementary Table 11), suggesting that 5mC accounts for a substantial fraction of the biological terms for pathway analysis. Regarding 5hmC as a separate modification, there was not enough significant DMPs at FDR-adjusted P value <.05 to perform pathway analysis. As an exploratory step, we performed analysis on DMPs at an unadjusted P value <.001 (1659 DMPs, Supplementary Table 11). Even though these findings should be interpreted with caution, they reveal that the large majority (80%) of canonical pathways associated to candidate 5hmC DMPs overlap with 5mC DMPs.
3.8. Transcription factor analysis
The monocyte-specific signature of the DMPs (Supplementary Fig. 8), indicates that these probes point to regulatory elements present in cells from the monocyte/macrophage line rather than T cells or B cells. DHSs can include several classes of regulatory elements, including promoters, enhancers, and CTCF-binding sites. In order to refine our previous associations, we performed further analyses with chromatin states (defined by the Epigenomics Roadmap Project [47]) on the same 5mC + 5hmC and 5mC DMPs and found monocyte/macrophage enhancers as the top category, suggesting that the DHS enrichment is driven mainly by overlap with monocyte/macrophage enhancers. To gain more insight into the molecular processes involved in the observed smoking response signature, we performed eFORGE-TF (transcription factor) analysis and identified several significant TF motif associations with both the 5mC + 5hmC and 5mC DMPs. Similar TF motifs were found for 5mC + 5hmC (Supplementary Fig. 12; Supplementary Table 12) and 5mC DMPs (Supplementary Fig. 13; Supplementary Table 13). Using PANTHER pathway analysis, genes associated with these TF motifs revealed the platelet derived growth factor (PDGF) signalling pathway as the top enriched pathway (Fold enrichment: 14.55, P value: 2.74 × 10−2). Thus, eFORGE-TF and PANTHER analyses of differentially methylated changes in BAL macrophages link smoking with the PDGF signalling pathway.
4. Discussion
We have investigated DNA modifications and gene expression in alveolar macrophage-enriched bronchoalveolar lavage cells from healthy smokers and non-smokers. This is, to our knowledge, the first report of smoking-associated genome-wide methylation data in pulmonary cells that includes true 5mC and 5hmC as separate modifications, especially in combination with the EPIC BeadChip. We identified a large number of smoking-associated DMPs (5mC + 5hmC, 1667; 5mC, 1713; 5hmC, 67), many of them located in enhancers. Our findings suggest that smoking-associated differences may include DNA demethylation of 5mC with a 5hmC intermediate, an interpretation based on the detected opposing hypomethylated 5mC and hypermethylated 5hmC data. Importantly, we could correlate our DNA methylation data with gene expression, and associate tobacco smoking with genes involved in cancer, immune cell migration, and activation pathways. The observed smoking-associated effect on DNA methylation and gene expression in BAL cells is likely to have implications on disease risk of several pathologies such as cancer, COPD, and autoimmune diseases.
In this study, we do not only show that tobacco smoking induce alterations in 5mC + 5hmC and gene expression in BAL cells from healthy individuals, but also report significant changes in 5hmC. It has been well-established that oxidative stress (ROS) underlying smoke-related lung inflammation, results in DNA damage and cytotoxic events [49]. For example, the oxidative damage biomarker 8-oxoG, is significantly elevated in lung tissue of smokers and also correlates with number of cigarettes smoked per day [25]. The DNA demethylation system catalysed by the TET enzymes also appears to be regulated by oxidative state, where oxidative stress may lead to demethylation initiated by TET enzymes [22]. This may induce formation of 5hmC but could importantly also lead to active continuous oxidation to formyl and carboxyl, which would actually lower the amount of 5hmC. Since 8-oxoG is essential for initiation of oxidative stress-induced DNA demethylation [22] and is abundant in smokers [25,49], we would expect to see many hypomethylated 5mC DMPs in smokers compared to nonsmokers. Interestingly, nearly all of the 67 identified 5hmC DMPs were hypermethylated while most of the 5mC + 5hmC and 5mC top DMPs, DVPs and DMRs were hypomethylated. This association supports the hypothesis that smoking can promote demethylation by oxidation of 5mC into 5hmC, a modification that can also exert effect as a stable mark. Future studies are warranted to decipher the mechanisms underlying the methylation profiles observed in BAL cells from smokers in general, and the contribution of oxidative stress-related 8-oxoG in such processes.
DNA demethylation of 5mC into 5hmC may also lead to functional effects, since these modifications have distinct properties such as different affinity to transcription factors [23]. In contrast to 5mC, which is able to bind transcriptional repressors, 5hmC can inhibit this binding and thereby counteract the repressive effect of 5mC [21]. While 5mC is often associated with gene repression, 5hmC facilitates transcription by contributing to an open chromatin state [21]. This has been widely studied in adult neurons and during embryogenesis where there is a relatively high abundance of 5hmC. For example, in mouse embryonic stem cells, the majority of 5hmC are located in gene bodies, and associated with moderate CpG density [50]. In our study, 5hmC is mainly located in highly methylated regions such as 3’UTR, 5’UTR, gene bodies, exon boundaries, and intergenic regions, and this is also where most smoking-associated 5hmC DMPs are observed. It should also be noted that while the hydroxymethylation results of the current study are highly interesting and novel, they also need to be replicated in future studies.
CpG sites associated with the aryl hydrocarbon receptor repressor (AHRR) gene often appear as top significantly differentially methylated in studies on the effect of smoking [7,10,13]. This is the case in our cohort as well, where three 5mC + 5hmC and six 5mC DMPs were annotated to AHRR, along with one 5mC + 5hmC DVP. The aryl hydrocarbon pathway regulates cytochrome P450 family members, metabolic enzymes involved in the elimination of xenobiotics that may have entered the body through i.e. the lungs. One group of xenobiotics includes the polycyclic aromatic hydrocarbons (PAH), which are increased in the lung tissue of tobacco smokers [51]. Many of these PAHs are carcinogenic and are, after metabolic activation, able to bind to DNA bases and form DNA adducts that in turn can lead to mutations in oncogenes and tumour suppressor genes [51]. The AhR-dependent cytochrome P450 family member 1B1 (CYP1B1), another differentially methylated gene in our study, is highly induced by PAHs, and responsible for metabolising many xenobiotics, including metabolic activation of PAH [52]. In addition, CYP1B1 also metabolizes many physiological compounds, including steroid hormones such as testosterone and oestrogen. Noteworthy, CYP1B1 is involved in both hydroxylation and demethylation of oestrogen products, and the rapid oxidation of 4-hydroxylated oestrogens to quinones [53,54]. This redox recycling leads to ROS production and oxidative DNA damage, which may lead to DNA adduct formation and carcinogenesis [54,55].
In our study, we found several hypomethylated promoter DMPs in genes from smokers that associate with increased gene expression, one of them being CYP1B1. Previous studies of smokers have reported upregulated CYP1B1 expression in the oral mucosa [56] and bronchial airway epithelium [57]. Due to the direct contact between alveolar macrophages and lung tissue, the hypomethylated promoter of CYP1B1 in combination with the increased expression, may have a significant role in the activation of procarcinogens and biotransformation to carcinogens. Functional phagocytic capacity is highly relevant for alveolar macrophages and this was impaired in a study on CYP1B1-deficient mice, suggesting a role for CYP1B1 in clearance of debris [58], and an attempt to increase this capacity as a response to smoking. Genome-wide epigenetic studies have emphasised that the location of DNA methylation in relation to the gene, influences the direction of gene expression. For example, DNA methylation occurring in promoter regions of genes, is well known to repress gene expression [59]. In gene bodies on the other hand, DNA methylation does not inhibit transcription and might even be positively correlated with expression [15]. Interestingly, it has been shown that AhR can bind to the promoter region (TSS1500) of CYP1B1 [60], the same region we found hypomethylated both at DMP and DMR level in our study.
Enhancer–promoter interaction through spatial chromatin organisation is a fundamental part of transcriptional regulation [48]. Enhancers are located at various distances from promoters and contain binding sites for sequence-specific transcription factors, and compared to promoters, enhancer usage varies vastly across cell types [61]. Tissue-resident macrophages, such as alveolar macrophages, exhibit distinct enhancer landscapes that are influenced by the tissue microenvironment in which they reside [61]. In the present study, we observed a large enrichment of hypomethylated DMPs annotated to enhancers, suggesting that these sites are highly affected by smoking exposure. Investigation of transcription factor motifs enriched for our DMPs identified the PDGF signalling pathway, a mediator of vascular remodelling that may be implicated in cigarette smoke-induced pulmonary artery hypertension [62].
Pathway analysis of transcriptional changes in our study revealed biological functions highly relevant to well-known smoking-related disease processes. We identified inhibition of biological processes such as cellular movement, migration and adhesion and cell-to-cell signalling, while inflammatory response and cancer-related processes showed increased activity. Interestingly, the most enriched canonical pathway is related to LXR/RXR activation with involvement in lipid metabolism and inflammation. In support of this, our highly significant DMP and gene expression top hit EEPD1 has recently been identified as a LXR target gene, proposed to promote cellular cholesterol efflux in macrophages, by controlling ABCA1 activity and protein levels [63]. Additionally EEPD1 has recently been characterised as a protein being recruited to stalled replication forks during replication stress, where it promotes restart of the replication fork [64]. Since smoking is known to damage DNA, and thereby create barriers for DNA replication, the upregulated EEPD1 expression seen in BAL cells could therefore be a response to the replication fork stress.
By combining methylome and transcriptome data from BAL cells, our study provides new insights into the biological impact of smoking locally in the lungs and possibly systemically in peripheral immune functions. Our findings confirm previously reported DNA methylation and gene expression results from alveolar macrophages, but also reveal new smoking-associated signatures. These novel targets especially map to regions that have not been covered with the previous methodologies, thus increasing and refining our knowledge of the molecular mechanisms underlying the effect of smoking.
The following are the supplementary data related to this article.
Description of inclusion criteria and filtering steps of study subjects. Schematic representing criteria for inclusion of samples and probes in 5mC + 5hmC, 5mC, and 5hmC analysis, as well as RNA-Seq.
Feature-specific differences in β-value distribution between smokers and non-smokers. Violin plots representing the distribution of β-values from 735,794 probes after SQN normalisation, plotted as densities in relation to CpG islands (a) and genomic features (b). Green plots represent smokers and orange plots represent non-smokers. Plots representing total methyl (5mC + 5hmC) densities are shown in upper rows (a-b) and 5mC plots in lower rows (a-b). Differences in distributions between smokers and non-smokers was tested using Wilcoxon signed-rank test was considered significant with a P value <.05. * P < .05; ** P < .01; *** P < .001; n.s. = not significant.
Significant smoking-associated changes at CpG level in total 5mC + 5hmC, 5mC and 5hmC. (a-i) β-values of of 14 smokers (orange) and 21 non-smokers (green). Each plot represents a single CpG site with gene annotation (if the site is annotated to a gene). Top significant hypomethylated DMPs in total 5mC + 5hmC (a-c) and 5mC (d-f) in smokers. Top significant hypermethylated 5hmC DMPs (g-i) in smokers.
Quantile-quantile (QQ) plots of DNA methylation changes between smokers and non-smokers. The QQ plots shows observed versus expected −log10 P values for association at all investigated loci in 5mC + 5hmC (a-b), 5mC (c-d) and 5hmC (e-f). The left column (a, c, e) shows the QQ plots from linear model testing with ebayes using covariates age + sex, and the right column (b, d, f) shows the QQ plots after removal of unwanted variables using RUVm.
Power curve as a function of sample size by effect size (absolute Δβ). The plot shows the average power (y-axis) by total sample size (x-axis), ranging from 20 to 50 patients with increments of 10, and by target Δβ ranging from 0.05 to 0.3. The following input parameters were used: 735.000 CpGs, 1700 target CpGs, minimum deltabeta detection limit 0.05, FDR-adjusted P value <.05, and adult PBMC as the closest available tissue.
Histograms of Spearman correlation rank Rho-values when correlating β-values and macrophage fractions. Plotted frequencies (y-axis) of Rho-values (x-axis) when DMP β-values were tested for correlation with the macrophage fraction of BAL fluid (a-f). Total 5mC + 5hmC DMPs Rho-values in non-smokers and smokers (a). True 5mC DMPs Rho-values in non-smokers and smokers (b). 5hmC DMPs Rho-values in non-smokers and smokers (c).
Correlation of adjusted P values of DMPs before and after cell-type correction. Plots of “BH”-adjusted P values from DMPs before and after cell type correction. Adjusted P values from cell type-corrected DMPs on the y-axis and non-cell type-corrected DMP P values on the x-axis in total BS (a) and true 5mC (b).
Annotation of DMPs with respect to DHSs. The enrichment –log10 binomial P-values for DHSs in blood cell types was obtained with eFORGE, using default settings. Dots are colored according to their FDR-adjusted q-value, and significant enrichments are labeled in pink (q < 0.05) and red (q < 0.01), while blue represent no significance (q > 0.05). PB: Peripheral blood; CB: Cord blood.
Oxidative BS and BS pyrosequencing validation of ITSN1.****: P < .0001.
Association of 5mC and 5hmC promoter, gene body, and enhancer methylation with gene expression. Scatterplots of mean gene expression values (log2 fold change) and 5mC mean Δβ in promoter (a), gene body (b), and enhancer (c); mean gene expression and 5hmC mean Δβ in promoter (d), gene body (e), and enhancer (f). Orange dots represent genes with significant smoking-associated changes in both total 5mC + 5hmC (>0.15 absolute Δβ) and gene expression (log2 fold change >1), green dots represent significant expression but not methylation, and purple dots represent changes in 5mC + 5hmC but not gene expression. Promoters are represented by TSS200 and TSS1500 CpG sites. Enhancer sites are present at various genomic features and can overlap with both promoter and gene body sites.
Functional annotations of 450 K and novel EPIC smoking-associated DMPs. (a) Smoking-associated 5mC + 5hmC DMPs were separated into 450 K and novel EPIC annotated genes. (b) Multidimensional scaling of GO Biological processes terms from smoking-associated 450 K (red) and novel EPIC (blue) 5mC + 5hmC DMP annotated genes. GO terms were obtained using over-representation analysis and visualization was generated by REVIGO. (c) Top canonical pathways from IPA associated with all smoking associated 5mC + 5hmC DMPs with red and blue colours representing the contribution of 450 K and novel EPIC annotated genes. Significance is represented as –log10P-value.
Transcription factor association of 5mC + 5hmC DMPs by eFORGE-TF. The enrichment –log10 binomial P-values for transcription factors are shown on the y-axis, and transcription factors on the x-axis. Dots are colored according to their FDR-adjusted q-value, and significant enrichments are labeled in pink (q < 0.05) and red (q < 0.01), while blue dots represent no significance (q > 0.05).
Transcription factor association 5mC DMPs by eFORGE-TF. The enrichment –log10 binomial P-values for transcription factors are shown on the y-axis, and transcription factors on the x-axis. Dots are colored according to their FDR-adjusted q-value, and significant enrichments are labeled in pink (q < 0.05) and red (q < 0.01), while blue dots represent no significance (q > 0.05).
Supplementary material 1
Supplementary material 2
Acknowledgments
Acknowledgments
We thank all the volunteers contributing to the study. We would also like to thank Heléne Blomqvist, Margitha Dahl, and Gunnel de Forest for recruitment and management of volunteers, and Benita Engvall for sample handling. We also thank the Uppsala Multidisciplinary Centre for Advanced Computational Science (UPPMAX; Uppsala University) for computational resources.
Funding sources
This study was supported by the Swedish Research Council (MH-2012-13; 2016–01209), the Swedish Heart-Lung Foundation (20120448; 20140666; 20150559; 20160300), the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institutet, The King Gustav's and Queen Victoria's Freemasons' Foundation, Knut and Alice Wallenberg Foundation (grant number KAW 201.0148), Neuro Sweden, and the Swedish MS foundation. The funders had no role in the design of the study, data collection, data analysis, interpretation, or the writing of this report. The corresponding authors had access to all the data in the study and had final responsibility for the decision to submit for publication.
Declaration of interests
The authors declare that they have no competing interests.
Author contributions
M.V.R., M.H-J., J.Ö. and T.J.E. conceptualised and designed the study. M.V.R. conducted DNA methylation experiments and M.H-J extracted DNA/RNA and performed RNA-seq experiments. M.V.R. performed the main data and statistical analysis, and M.N. and L.K contributed with input and interpretation. M.H-J and Y.L. performed RNA-seq analysis and L.K. conducted IPA analysis. L.K.S. and L.S. performed pyrosequencing analysis. C.B. performed transcription factor analysis. J.W., J.G., B.B., designed and/or contributed to the collection of patient samples. S.K. performed BAL and characterised study subjects. M.J., M.A., J.Ö., and T.J.E. supervised the work in each contributing research group. M.V.R. and T.J.E. initiated the study and wrote the manuscript with contribution from L.K., M.N, M.H-J., and M.J. All authors read and approved the final manuscript.
Contributor Information
Mikael V. Ringh, Email: mikael.ringh@ki.se.
Tomas J. Ekström, Email: tomas.ekstrom@ki.se.
References
- 1.Ezzati M., Lopez A.D. Estimates of global mortality attributable to smoking in 2000. Lancet. 2003;362(9387):847–852. doi: 10.1016/S0140-6736(03)14338-3. [DOI] [PubMed] [Google Scholar]
- 2.Kallberg H., Ding B., Padyukov L., Bengtsson C., Ronnelid J., Klareskog L. Smoking is a major preventable risk factor for rheumatoid arthritis: estimations of risks after various exposures to cigarette smoke. Ann. Rheum. Dis. 2011;70(3):508–511. doi: 10.1136/ard.2009.120899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hedstrom A.K., Sundqvist E., Baarnhielm M., Nordin N., Hillert J., Kockum I. Smoking and two human leukocyte antigen genes interact to increase the risk for multiple sclerosis. Brain. 2011;134:653–664. doi: 10.1093/brain/awq371. Pt 3. [DOI] [PubMed] [Google Scholar]
- 4.Rodgman A., Perfetti T.A. 2nd Edition ed. CRC Press; Boca Raton: 2013. The chemical components of tobacco and tobacco smoke. [Google Scholar]
- 5.Hussell T., Bell T.J. Alveolar macrophages: plasticity in a tissue-specific context. Nat. Rev. Immunol. 2014;14(2):81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 6.Lee J., Taneja V., Vassallo R. Cigarette smoking and inflammation: cellular and molecular mechanisms. J. Dent. Res. 2012;91(2):142–149. doi: 10.1177/0022034511421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joehanes R., Just A.C., Marioni R.E., Pilling L.C., Reynolds L.M., Mandaviya P.R. Epigenetic signatures of cigarette smoking. Circ. Cardiovasc. Genet. 2016;9(5):436–447. doi: 10.1161/CIRCGENETICS.116.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breitling L.P., Yang R., Korn B., Burwinkel B., Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am. J. Hum. Genet. 2011;88(4):450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambatipudi S., Cuenin C., Hernandez-Vargas H., Ghantous A., Le Calvez-Kelm F., Kaaks R. Tobacco smoking-associated genome-wide DNA methylation changes in the EPIC study. Epigenomics. 2016;8(5):599–618. doi: 10.2217/epi-2016-0001. [DOI] [PubMed] [Google Scholar]
- 10.Marabita F., Almgren M., Sjoholm L.K., Kular L., Liu Y., James T. Smoking induces DNA methylation changes in multiple sclerosis patients with exposure-response relationship. Sci. Rep. 2017;7(1) doi: 10.1038/s41598-017-14788-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joubert B.R., Haberg S.E., Nilsen R.M., Wang X., Vollset S.E., Murphy S.K. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ. Health Perspect. 2012;120(10):1425–1431. doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joubert B.R., Felix J.F., Yousefi P., Bakulski K.M., Just A.C., Breton C. DNA methylation in Newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am. J. Hum. Genet. 2016;98(4):680–696. doi: 10.1016/j.ajhg.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Philibert R.A., Sears R.A., Powers L.S., Nash E., Bair T., Gerke A.K. Coordinated DNA methylation and gene expression changes in smoker alveolar macrophages: specific effects on VEGF receptor 1 expression. J. Leukoc. Biol. 2012;92(3):621–631. doi: 10.1189/jlb.1211632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monick M.M., Beach S.R., Plume J., Sears R., Gerrard M., Brody G.H. Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2012;159B(2):141–151. doi: 10.1002/ajmg.b.32021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 16.Li E., Zhang Y. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 2014;6(5) doi: 10.1101/cshperspect.a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin S.G., Wu X., Li A.X., Pfeifer G.P. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011;39(12):5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendonca A., Chang E.H., Liu W., Yuan C. Hydroxymethylation of DNA influences nucleosomal conformation and stability in vitro. Biochim. Biophys. Acta. 2014;1839(11):1323–1329. doi: 10.1016/j.bbagrm.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 22.Zhou X., Zhuang Z., Wang W., He L., Wu H., Cao Y. OGG1 is essential in oxidative stress induced DNA demethylation. Cell. Signal. 2016;28(9):1163–1171. doi: 10.1016/j.cellsig.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 23.Menezo Y.J., Silvestris E., Dale B., Elder K. Oxidative stress and alterations in DNA methylation: two sides of the same coin in reproduction. Reprod. BioMed. Online. 2016;33(6):668–683. doi: 10.1016/j.rbmo.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Nestor C., Ruzov A., Meehan R., Dunican D. Enzymatic approaches and bisulfite sequencing cannot distinguish between 5-methylcytosine and 5-hydroxymethylcytosine in DNA. Biotechniques. 2010;48(4):317–319. doi: 10.2144/000113403. [DOI] [PubMed] [Google Scholar]
- 25.Asami S., Manabe H., Miyake J., Tsurudome Y., Hirano T., Yamaguchi R. Cigarette smoking induces an increase in oxidative DNA damage, 8-hydroxydeoxyguanosine, in a central site of the human lung. Carcinogenesis. 1997;18(9):1763–1766. doi: 10.1093/carcin/18.9.1763. [DOI] [PubMed] [Google Scholar]
- 26.Olsen H.H., Grunewald J., Tornling G., Skold C.M., Eklund A. Bronchoalveolar lavage results are independent of season, age, gender and collection site. PLoS One. 2012;7(8) doi: 10.1371/journal.pone.0043644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ockinger J., Hagemann-Jensen M., Kullberg S., Engvall B., Eklund A., Grunewald J. T-cell activation and HLA-regulated response to smoking in the deep airways of patients with multiple sclerosis. Clin. Immunol. 2016;169:114–120. doi: 10.1016/j.clim.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Picelli S., Faridani O.R., Bjorklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9(1):171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 29.Picelli S., Bjorklund A.K., Reinius B., Sagasser S., Winberg G., Sandberg R. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 2014;24(12):2033–2040. doi: 10.1101/gr.177881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fortin J.P., Triche T.J., Jr., Hansen K.D. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33(4):558–560. doi: 10.1093/bioinformatics/btw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian Y., Morris T.J., Webster A.P., Yang Z., Beck S., Feber A. ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics. 2017;33(24):3982–3984. doi: 10.1093/bioinformatics/btx513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maksimovic J., Gordon L., Oshlack A. SWAN: subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13(6):R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pidsley R., Zotenko E., Peters T.J., Lawrence M.G., Risbridger G.P., Molloy P. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17(1):208. doi: 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H., Boutros P.C. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011;12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du P., Zhang X., Huang C.C., Jafari N., Kibbe W.A., Hou L. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maksimovic J., Gagnon-Bartsch J.A., Speed T.P., Oshlack A. Removing unwanted variation in a differential methylation analysis of Illumina HumanMethylation450 array data. Nucleic Acids Res. 2015;43(16):e106. doi: 10.1093/nar/gkv526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peters T.J., Buckley M.J., Statham A.L., Pidsley R., Samaras K., R V.L. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin. 2015;8:6. doi: 10.1186/1756-8935-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Breeze C.E., Paul D.S., van Dongen J., Butcher L.M., Ambrose J.C., Barrett J.E. eFORGE: a tool for identifying cell type-specific signal in Epigenomic data. Cell Rep. 2016;17(8):2137–2150. doi: 10.1016/j.celrep.2016.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang B., Kirov S., Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33(Web Server):W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Supek F., Bosnjak M., Skunca N., Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6(7) doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graw S., Henn R., Thompson J.A., Koestler D.C. pwrEWAS: a user-friendly tool for comprehensive power estimation for epigenome wide association studies (EWAS) BMC Bioinformatics. 2019;20(1):218. doi: 10.1186/s12859-019-2804-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karimi R., Tornling G., Grunewald J., Eklund A., Skold C.M. Cell recovery in bronchoalveolar lavage fluid in smokers is dependent on cumulative smoking history. PLoS One. 2012;7(3) doi: 10.1371/journal.pone.0034232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Globisch D., Munzel M., Muller M., Michalakis S., Wagner M., Koch S. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5(12) doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rask-Andersen M., Martinsson D., Ahsan M., Enroth S., Ek W.E., Gyllensten U. Epigenome-wide association study reveals differential DNA methylation in individuals with a history of myocardial infarction. Hum. Mol. Genet. 2016;25(21):4739–4748. doi: 10.1093/hmg/ddw302. [DOI] [PubMed] [Google Scholar]
- 46.Liang L., Willis-Owen S.A.G., Laprise C., Wong K.C.C., Davies G.A., Hudson T.J. An epigenome-wide association study of total serum immunoglobulin E concentration. Nature. 2015;520(7549):670–674. doi: 10.1038/nature14125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roadmap Epigenomics C., Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mora A., Sandve G.K., Gabrielsen O.S., Eskeland R. In the loop: promoter-enhancer interactions and bioinformatics. Brief. Bioinform. 2016;17(6):980–995. doi: 10.1093/bib/bbv097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schieber M., Chandel N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014;24(10):R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu H., D'Alessio A.C., Ito S., Wang Z., Cui K., Zhao K. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25(7):679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldman R., Enewold L., Pellizzari E., Beach J.B., Bowman E.D., Krishnan S.S. Smoking increases carcinogenic polycyclic aromatic hydrocarbons in human lung tissue. Cancer Res. 2001;61(17):6367–6371. [PubMed] [Google Scholar]
- 52.Shimada T., Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004;95(1):1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han X., Liehr J.G. Microsome-mediated 8-hydroxylation of guanine bases of DNA by steroid estrogens: correlation of DNA damage by free radicals with metabolic activation to quinones. Carcinogenesis. 1995;16(10):2571–2574. doi: 10.1093/carcin/16.10.2571. [DOI] [PubMed] [Google Scholar]
- 54.Parl F.F., Egan K.M., Li C., Crooke P.S. Estrogen exposure, metabolism, and enzyme variants in a model for breast cancer risk prediction. Cancer Informat. 2009;7:109–121. doi: 10.4137/cin.s2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.D'Uva G., Baci D., Albini A., Noonan D.M. Cancer chemoprevention revisited: cytochrome P450 family 1B1 as a target in the tumor and the microenvironment. Cancer Treat. Rev. 2018;63:1–18. doi: 10.1016/j.ctrv.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 56.Gumus Z.H., Du B., Kacker A., Boyle J.O., Bocker J.M., Mukherjee P. Effects of tobacco smoke on gene expression and cellular pathways in a cellular model of oral leukoplakia. Cancer Prev. Res. (Phila.) 2008;1(2):100–111. doi: 10.1158/1940-6207.CAPR-08-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steiling K., Lenburg M.E., Spira A. Airway gene expression in chronic obstructive pulmonary disease. Proc. Am. Thorac. Soc. 2009;6(8):697–700. doi: 10.1513/pats.200907-076DP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ward J.M., Nikolov N.P., Tschetter J.R., Kopp J.B., Gonzalez F.J., Kimura S. Progressive glomerulonephritis and histiocytic sarcoma associated with macrophage functional defects in CYP1B1-deficient mice. Toxicol. Pathol. 2004;32(6):710–718. doi: 10.1080/01926230490885706. [DOI] [PubMed] [Google Scholar]
- 59.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 60.Yang X., Solomon S., Fraser L.R., Trombino A.F., Liu D., Sonenshein G.E. Constitutive regulation of CYP1B1 by the aryl hydrocarbon receptor (AhR) in pre-malignant and malignant mammary tissue. J. Cell. Biochem. 2008;104(2):402–417. doi: 10.1002/jcb.21630. [DOI] [PubMed] [Google Scholar]
- 61.Lavin Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159(6):1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xing A.P., Hu X.Y., Shi Y.W., Du Y.C. Implication of PDGF signaling in cigarette smoke-induced pulmonary arterial hypertension in rat. Inhal. Toxicol. 2012;24(8):468–475. doi: 10.3109/08958378.2012.688885. [DOI] [PubMed] [Google Scholar]
- 63.Nelson J.K., Koenis D.S., Scheij S., Cook E.C., Moeton M., Santos A. EEPD1 is a novel LXR target gene in macrophages which regulates ABCA1 abundance and cholesterol efflux. Arterioscler. Thromb. Vasc. Biol. 2017;37(3):423–432. doi: 10.1161/ATVBAHA.116.308434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu Y., Lee S.H., Williamson E.A., Reinert B.L., Cho J.H., Xia F. EEPD1 rescues stressed replication forks and maintains genome stability by promoting end resection and homologous recombination repair. PLoS Genet. 2015;11(12) doi: 10.1371/journal.pgen.1005675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of inclusion criteria and filtering steps of study subjects. Schematic representing criteria for inclusion of samples and probes in 5mC + 5hmC, 5mC, and 5hmC analysis, as well as RNA-Seq.
Feature-specific differences in β-value distribution between smokers and non-smokers. Violin plots representing the distribution of β-values from 735,794 probes after SQN normalisation, plotted as densities in relation to CpG islands (a) and genomic features (b). Green plots represent smokers and orange plots represent non-smokers. Plots representing total methyl (5mC + 5hmC) densities are shown in upper rows (a-b) and 5mC plots in lower rows (a-b). Differences in distributions between smokers and non-smokers was tested using Wilcoxon signed-rank test was considered significant with a P value <.05. * P < .05; ** P < .01; *** P < .001; n.s. = not significant.
Significant smoking-associated changes at CpG level in total 5mC + 5hmC, 5mC and 5hmC. (a-i) β-values of of 14 smokers (orange) and 21 non-smokers (green). Each plot represents a single CpG site with gene annotation (if the site is annotated to a gene). Top significant hypomethylated DMPs in total 5mC + 5hmC (a-c) and 5mC (d-f) in smokers. Top significant hypermethylated 5hmC DMPs (g-i) in smokers.
Quantile-quantile (QQ) plots of DNA methylation changes between smokers and non-smokers. The QQ plots shows observed versus expected −log10 P values for association at all investigated loci in 5mC + 5hmC (a-b), 5mC (c-d) and 5hmC (e-f). The left column (a, c, e) shows the QQ plots from linear model testing with ebayes using covariates age + sex, and the right column (b, d, f) shows the QQ plots after removal of unwanted variables using RUVm.
Power curve as a function of sample size by effect size (absolute Δβ). The plot shows the average power (y-axis) by total sample size (x-axis), ranging from 20 to 50 patients with increments of 10, and by target Δβ ranging from 0.05 to 0.3. The following input parameters were used: 735.000 CpGs, 1700 target CpGs, minimum deltabeta detection limit 0.05, FDR-adjusted P value <.05, and adult PBMC as the closest available tissue.
Histograms of Spearman correlation rank Rho-values when correlating β-values and macrophage fractions. Plotted frequencies (y-axis) of Rho-values (x-axis) when DMP β-values were tested for correlation with the macrophage fraction of BAL fluid (a-f). Total 5mC + 5hmC DMPs Rho-values in non-smokers and smokers (a). True 5mC DMPs Rho-values in non-smokers and smokers (b). 5hmC DMPs Rho-values in non-smokers and smokers (c).
Correlation of adjusted P values of DMPs before and after cell-type correction. Plots of “BH”-adjusted P values from DMPs before and after cell type correction. Adjusted P values from cell type-corrected DMPs on the y-axis and non-cell type-corrected DMP P values on the x-axis in total BS (a) and true 5mC (b).
Annotation of DMPs with respect to DHSs. The enrichment –log10 binomial P-values for DHSs in blood cell types was obtained with eFORGE, using default settings. Dots are colored according to their FDR-adjusted q-value, and significant enrichments are labeled in pink (q < 0.05) and red (q < 0.01), while blue represent no significance (q > 0.05). PB: Peripheral blood; CB: Cord blood.
Oxidative BS and BS pyrosequencing validation of ITSN1.****: P < .0001.
Association of 5mC and 5hmC promoter, gene body, and enhancer methylation with gene expression. Scatterplots of mean gene expression values (log2 fold change) and 5mC mean Δβ in promoter (a), gene body (b), and enhancer (c); mean gene expression and 5hmC mean Δβ in promoter (d), gene body (e), and enhancer (f). Orange dots represent genes with significant smoking-associated changes in both total 5mC + 5hmC (>0.15 absolute Δβ) and gene expression (log2 fold change >1), green dots represent significant expression but not methylation, and purple dots represent changes in 5mC + 5hmC but not gene expression. Promoters are represented by TSS200 and TSS1500 CpG sites. Enhancer sites are present at various genomic features and can overlap with both promoter and gene body sites.
Functional annotations of 450 K and novel EPIC smoking-associated DMPs. (a) Smoking-associated 5mC + 5hmC DMPs were separated into 450 K and novel EPIC annotated genes. (b) Multidimensional scaling of GO Biological processes terms from smoking-associated 450 K (red) and novel EPIC (blue) 5mC + 5hmC DMP annotated genes. GO terms were obtained using over-representation analysis and visualization was generated by REVIGO. (c) Top canonical pathways from IPA associated with all smoking associated 5mC + 5hmC DMPs with red and blue colours representing the contribution of 450 K and novel EPIC annotated genes. Significance is represented as –log10P-value.
Transcription factor association of 5mC + 5hmC DMPs by eFORGE-TF. The enrichment –log10 binomial P-values for transcription factors are shown on the y-axis, and transcription factors on the x-axis. Dots are colored according to their FDR-adjusted q-value, and significant enrichments are labeled in pink (q < 0.05) and red (q < 0.01), while blue dots represent no significance (q > 0.05).
Transcription factor association 5mC DMPs by eFORGE-TF. The enrichment –log10 binomial P-values for transcription factors are shown on the y-axis, and transcription factors on the x-axis. Dots are colored according to their FDR-adjusted q-value, and significant enrichments are labeled in pink (q < 0.05) and red (q < 0.01), while blue dots represent no significance (q > 0.05).
Supplementary material 1
Supplementary material 2
Data Availability Statement
DNA methylation data from this study is available in Gene Expression Omnibus (GEO) database under accession number GSE133062, and RNA-seq data is available upon request.