Abstract
Soybean growers widely use the Resistance to Heterodera glycines 1 (Rhg1) locus to reduce yield losses caused by soybean cyst nematode (SCN). Rhg1 is a tandemly repeated four gene block. Two classes of SCN resistance‐conferring Rhg1 haplotypes are recognized: rhg1‐a (“Peking‐type,” low‐copy number, three or fewer Rhg1 repeats) and rhg1‐b (“PI 88788‐type,” high‐copy number, four or more Rhg1 repeats). The rhg1‐a and rhg1‐b haplotypes encode α‐SNAP (alpha‐Soluble NSF Attachment Protein) variants α‐SNAPRhg1LC and α‐SNAPRhg1HC, respectively, with differing atypical C‐terminal domains, that contribute to SCN resistance. Here we report that rhg1‐a soybean accessions harbor a copia retrotransposon within their Rhg1 Glyma.18G022500 (α‐SNAP‐encoding) gene. We termed this retrotransposon “RAC,” for Rhg1 alpha‐SNAP copia. Soybean carries multiple RAC‐like retrotransposon sequences. The Rhg1 RAC insertion is in the Glyma.18G022500 genes of all true rhg1‐a haplotypes we tested and was not detected in any examined rhg1‐b or Rhg1WT (single‐copy) soybeans. RAC is an intact element residing within intron 1, anti‐sense to the rhg1‐a α‐SNAP open reading frame. RAC has intrinsic promoter activities, but overt impacts of RAC on transgenic α‐SNAPRhg1LC mRNA and protein abundance were not detected. From the native rhg1‐a RAC+ genomic context, elevated α‐SNAPRhg1LC protein abundance was observed in syncytium cells, as was previously observed for α‐SNAPRhg1HC (whose rhg1‐b does not carry RAC). Using a SoySNP50K SNP corresponding with RAC presence, just ~42% of USDA accessions bearing previously identified rhg1‐a SoySNP50K SNP signatures harbor the RAC insertion. Subsequent analysis of several of these putative rhg1‐a accessions lacking RAC revealed that none encoded α‐SNAPRhg1LC, and thus, they are not rhg1‐a. rhg1‐a haplotypes are of rising interest, with Rhg4, for combating SCN populations that exhibit increased virulence against the widely used rhg1‐b resistance. The present study reveals another unexpected structural feature of many Rhg1 loci, and a selectable feature that is predictive of rhg1‐a haplotypes.
Keywords: plant disease resistance, retrotransposon, Rhg1, soybean cyst nematode
1. INTRODUCTION
To thrive in their natural environments, organisms must continually sense and respond to changing conditions, including biotic and abiotic stresses. Transposable elements (TEs) can cause relatively stable variation in numerous plant phenotypes such as flowering time, trichome presence or fruit size (Lisch, 2013). TEs may insertionally disrupt genes, or if TE activity is repressed by epigenetic transcriptional silencing, small interfering RNAs, and chromatin condensation, this can impact the expression of nearby genes (Sigman & Slotkin, 2016). TEs are also increasingly being identified as modulatory factors during periods of host stress (i.e., heat, pathogen) (Ding et al., 2015; Liu, He, Amasino, & Chen, 2004; Xiao, Jiang, Schaffner, Stockinger, & Knaap, 2008). For instance, cis‐regulatory motifs within certain TEs can recruit stress‐responsive host transcriptional factors, thereby influencing nearby host gene expression and potentially conferring a host benefit (Cavrak et al., 2014; Galindo‐Gonzalez, Mhiri, Deyholos, & Grandbastien, 2017; Makarevitch et al., 2015; Matsunaga, Kobayashi, Kato, & Ito, 2012; Matsunaga et al., 2015; McCue & Slotkin, 2012; Negi, Rai, & Suprasanna, 2016; Slotkin & Martienssen, 2007; Woodrow et al., 2010). Some TEs also beneficially modulate the expression of host plant defense or susceptibility genes (Berg et al., 2015; Tsuchiya & Eulgem, 2013). Additionally, numerous studies report TEs lying within or adjacent to putative plant immune genes, however, potential influences on host genes or positive effects are often not apparent (Bhattacharyya, Gonzales, Kraft, & Buzzell, 1997; Henk, Warren, & Innes, 1999; Wawrzynski et al., 2008).
Glycine max (soybean) is an important food and industrial crop (Schmutz et al., 2010). A major pest afflicting global soybean production is the soybean cyst nematode (SCN, Heterodera glycines), which causes yearly U.S. soybean yield losses of over 1 billion USD (Allen et al., 2017; Mitchum, 2016; Niblack, Lambert, & Tylka, 2006). SCN is an obligate parasite that invades host roots and induces individual host cells to form a complex syncytium that serves as the SCN feeding site (Mitchum, 2016; Niblack et al., 2006). SCN feeding depletes available host resources, and a functional syncytium must be maintained for 2–4 weeks for the nematode to complete its lifecycle. Since the unhatched eggs within cysts can remain viable for many years in the field, SCN is difficult to manage and is primarily controlled by growing naturally resistant soybeans (Niblack et al., 2006). Among known soybean loci contributing to SCN resistance, the Rhg1 (Resistance to Heterodera glycines 1) locus found on chromosome 18 provides the strongest protection (Concibido, Diers, & Arelli, 2004). Rhg1 causes the SCN‐induced syncytium to fail a few days after induction, and the soybean PI 88788‐type “rhg1‐b” haplotype is the primary SCN‐resistance locus used in commercially grown soybeans (Concibido et al., 2004; Mitchum, 2016; Niblack et al., 2006).
Soybean Rhg1 is an unusual disease resistance locus that consists of a ~31.2 kb unit that is tandemly repeated as many as 10 times (Cook et al., 2012). Within each 31.2 kb Rhg1 repeat unit are four different Rhg1‐encoded genes: Glyma.18G022400, Glyma.18G022500, Glyma.18G022600, and Glyma.18G022700, none of which have similarity to previously identified resistance genes (Cook et al., 2014, 2012; Lee, Kumar, Diers, & Hudson, 2015). Of the three Rhg1 genes that contribute to SCN resistance, only Glyma.18G022500, an α‐SNAP (alpha‐Soluble NSF Attachment Protein), has amino acid polymorphisms relative to the wild‐type Rhg1 gene alleles present in SCN‐susceptible soybeans (Cook et al., 2014, 2012; Lee et al., 2015). The mRNA transcript abundance of all three resistance‐associated Rhg1 genes is significantly elevated in SCN‐resistant multi‐copy Rhg1 soybeans, relative to SCN‐susceptible single‐copy Rhg1 (WT Rhg1) soybeans (Cook et al., 2014, 2012).
At least two distinct Rhg1 genotype classes exist: “low‐copy Rhg1” (rhg1‐a, sometimes referred to as Rhg1LC, often derived from PI 548402 “Peking”) and “high‐copy Rhg1” (rhg1‐b, sometimes referred to as Rhg1HC, often derived from PI 88788) (Bayless et al., 2018; Brucker, Carlson, Wright, Niblack, & Diers, 2005; Cook et al., 2014, 2012; Niblack et al., 2002). These Rhg1 genotype classes represent two distinct multi‐copy Rhg1 haplotypes that vary most notably by (a) Rhg1 repeat number (a high or low number of Rhg1 repeats) and (b) encoding distinctive resistance‐type α‐SNAP proteins with C‐terminal polymorphisms at a conserved functional site (Bayless et al., 2016; Cook et al., 2014). rhg1‐a resistance is bolstered by an unlinked chromosome 8 locus, Rhg4, whose presence contributes to full‐strength “Peking‐type” SCN resistance (Liu et al., 2012; Meksem et al., 2001). Rhg4 encodes a polymorphic serine hydroxymethyl transferase with altered enzyme kinetics, but the molecular basis of resistance augmentation by Rhg4 is not yet understood (Liu et al., 2012; Mitchum, 2016). Several rhg1‐b and rhg1‐a accessions have been analyzed by whole‐genome sequencing (WGS) studies, and characteristic single nucleotide polymorphism (SNP) signatures predictive of rhg1‐b or rhg1‐a haplotype soybeans have been reported (Cook et al., 2014; Kadam et al., 2016; Lee et al., 2015; Patil et al., 2019; Shi et al., 2015). Additionally, studies by Arelli, Young and others have profiled SCN resistance among thousands of USDA soybean accessions and noted substantial phenotypic variation (e.g., (Anand & Gallo, 1984; Arelli, Sleper, Yue, & Wilcox, 2000; Diers, Skorupska, Rao‐Arelli, & Cianzio, 1997; Hussey, Boerma, Raymer, & Luzzi, 1991; Klepadlo et al., 2018; Vuong et al., 2015; Young, 1995)). However, the influence of all Rhg1 haplotype and/or allelic variation factors on SCN‐resistance expression or plant yield is not yet fully understood.
Several recent studies have deepened our understanding of Rhg1 molecular function and highlight a central role of the SNARE (Soluble NSF Attachment Protein REceptors)‐recycling machinery in SCN resistance (Bayless et al., 2016, 2018; Cook et al., 2014; Lakhssassi et al., 2017; Matsye et al., 2012). α‐SNAP and the ATPase NSF (N‐ethylmaleimide Sensitive Factor) are conserved eukaryotic housekeeping proteins that form the core SNARE‐recycling machinery. They sustain the pool of fusion‐competent SNAREs necessary for new membrane fusion events (Sudhof & Rothman, 2009; Zhao et al., 2015). While most animals encode single NSF and α‐SNAP genes, soybean is a paleopolyploid that encodes two NSF, four or five α‐SNAP and two γ‐SNAP (gamma‐SNAP) genes, respectively. A C‐terminal α‐SNAP domain conserved across all plants and animals recruits NSF to SNARE‐bundles and stimulates the ATPase activity of NSF that powers SNARE‐complex recycling. However, it is this otherwise conserved α‐SNAP C‐terminal region that is atypical among both rhg1‐b‐ and rhg1‐a‐encoded α‐SNAP proteins, and accordingly, both Rhg1 resistance‐type α‐SNAPs are impaired in promoting normal NSF function and instead mediate dosage‐dependent cytotoxicity (Bayless et al., 2016; Cook et al., 2014). The abundance of the atypical rhg1‐b α‐SNAPRhg1HC protein specifically increases in the SCN feeding site and contributes to Rhg1‐mediated collapse of the SCN‐induced syncytium (Bayless et al., 2016).
At least two additional loci associated with SCN resistance are also components of the SNARE‐recycling machinery (Bayless et al., 2018; Lakhssassi et al., 2017). Recently, a specialized allele of NSF, NSFRAN07 (Rhg1‐ associated NSF on chromosome 07), was shown to be necessary for the viability of Rhg1‐containing soybeans (Bayless et al., 2018). Compared with the WT NSFCh07 protein, the NSFRAN07 protein more effectively binds to resistance‐type α‐SNAPs and confers better protection against resistance‐type α‐SNAP‐induced cytotoxicity (Bayless et al., 2018). During the Rhg1‐mediated resistance response, the ratio of Rhg1 resistance‐type to WT α‐SNAPs increases and is apparently an important factor underlying resistance (Bayless et al., 2016, 2018). Two genetic events sharply reduce WT α‐SNAP protein abundance in SCN‐resistant rhg1‐a soybeans (Bayless et al., 2018). First, the wild‐type α‐SNAP‐encoding block at Rhg1 on chromosome 18—a predominant source of total WT α‐SNAP proteins in soybean—is absent from all examined rhg1‐a accessions, thereby diminishing overall WT α‐SNAP protein abundance (Bayless et al., 2018; Cook et al., 2014). Secondly, rhg1‐a lines often carry a null allele of the α‐SNAP encoded on chromosome 11 (Glyma.11G234500)—the other major source of WT α‐SNAP proteins – due to an intronic splice site mutation that causes premature translational termination and loss of protein stability (Bayless et al., 2018; Cook et al., 2014; Lakhssassi et al., 2017; Matsye et al., 2012). Together, the above studies support a paradigm whereby Rhg1 and associated SCN‐resistance loci rewire major components of the soybean SNARE‐recycling machinery. Importantly, soybean accessions that carry rhg1‐a and Rhg4 can resist many of the virulent SCN populations that partially overcome rhg1‐b resistance (Bayless et al., 2016; Brucker et al., 2005; Niblack, Colgrove, Colgrove, & Bond, 2008). Therefore, there is considerable interest in understanding and using rhg1‐a, the subject of the present study, as an alternative to rhg1‐b in commercial soybean cultivars (Brucker et al., 2005; Liu et al., 2012; Yu, Lee, Rosa, Hudson, & Diers, 2016).
Presence/absence variation of TEs at specific loci is common among different soybean accessions, and tens of thousands of non‐reference genome TE insertions occur between cultivated and wild soybean (Tian et al., 2012). Moreover, high TE densities near genomic regions exhibiting structural polymorphisms such as copy number variation are also reported in soybean (McHale et al., 2012). While examining the Rhg1 low‐copy (rhg1‐a) haplotype of soybean accession PI 89772, we uncovered an intact copia retrotransposon within all three copies of the Rhg1‐encoded α‐SNAP genes. We termed this retrotransposon “RAC,” for Rhg1 alpha‐SNAP copia). The RAC element, which is entirely within the first intron of the Glyma.18G022500 (α‐SNAP) gene, appears to be intact and transcribes anti‐sense to the α‐SNAP open reading frame (ORF). BLAST searches revealed similar copia elements across the soybean genome, suggesting why assemblies of Illumina short‐read whole‐genome sequences failed to include this sequence within rhg1‐a assemblies. This α‐SNAP‐RAC insertion was absent from all examined single‐copy Rhg1 (SCN‐susceptible) and high‐copy rhg1‐b (Rhg1HC) accessions. More than half of the USDA accessions with SoySNP50K SNPs preliminarily indicative of a low‐copy rhg1‐a haplotype did not carry RAC, but sub‐sampling among those accessions revealed that they do not encode α‐SNAPRhg1LC and thus are not rhg1‐a. The increasingly important rhg1‐a SCN‐resistant soybean breeding lines do harbor this previously unreported retrotransposon within the α‐SNAPRhg1LC‐encoding gene.
2. MATERIALS AND METHODS
2.1. Transgenic soybean hairy root generation and root culturing
Transgenic soybean roots were produced using Agrobacterium rhizogenes strain “ARqua1” (Quandt, Pühler, & Broer, 1993) and the previously described binary vector pSM101, as in (Cook et al., 2012). Transgenic roots were sub‐cultured in the dark at room temperature on hairy root medium as in (Cook et al., 2012).
2.2. DNA extraction
Soybean genomic DNAs were extracted from expanding trifoliates or root tissues of the respective soybean accessions using standard CTAB methods similar to (Cook et al., 2012).
2.3. Amplification and detection of RAC (Rhg1 alpha‐SNAP copia)
For initial amplification and subcloning of native α‐SNAP‐RAC, approximately 100 ng of CTAB‐extracted gDNA from PI 89772 (rhg1‐a) was PCR‐amplified for 35 cycles using HiFi polymerase (KAPA Biosystems, Wilmington, MA). Primer annealing was at ~70°C for 30 s and extension was at 72°C for 5 min. The resulting α‐SNAP‐RAC amplicon from PI 89772 was separated by agarose gel electrophoresis, gel extracted using a Zymoclean Large Fragment DNA Recovery Kit (Zymo Research) and TA overhang cloned into a pTopo xL vector using the Topo xL PCR Cloning Kit (Life Technologies Corp.), per manufacturer's recommendations. For PCR detection of α‐SNAP‐RAC junctions or WT exon distances, ~25 ng of CTAB‐extracted genomic DNA from each respective accession was amplified using GoTAQ Green (New England Biolabs) for 32 cycles, separated on a 0.8% agarose gel and visualized.
2.4. Phylogenetic tree construction
For the RAC‐like nucleotide tree, evolutionary analyses were conducted in MEGA7 (Kumar, Stecher, & Tamura, 2016), and evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura‐Nei model (Tamura, Nei, & Kumar, 2004). The tree with the highest log likelihood (−47751.11) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor‐Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood approach and then selecting the topology with superior log likelihood value. The analysis involved 24 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 4,157 positions in the final dataset.
For the RAC‐polyprotein tree, evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016), and the evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix‐based model (Jones, Taylor, & Thornton, 1992). The tree with the highest log likelihood (−12,400.87) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor‐Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 14 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 644 positions in the final dataset.
2.5. Read‐depth analysis of RAC
Using previously published WGS data (Cook et al., 2014), read depth was computed using the depth program of SAMtools (Li et al., 2009). Depth was averaged in 250 bp intervals on Chromosome 10 from bp 40,650,000–40,690,000 (includes flanking regions of the 99.7% identity RAC‐like element). The copy number of the Chromosome 10 RAC‐like element was then calculated as the ratio of the read coverage per 250 bp from bp 40,672,000–40,675,750, divided by the average read coverage for the flanking regions between bp 40,650,000–40,690,000. Sequencing coverage was visualized using ggplot (Wickham, 2009) within RStudio (RStudio Team, 2015).
2.6. Methylation analysis
McrBC methylation studies were performed similarly to (Cook et al., 2014). Control McrBC reactions contained equivalent amounts of genomic DNA in reaction buffer, but had no added McrBC enzyme. McrBC digestion was performed at 37°C for 90 min, followed by a 20‐min heat inactivation at 65ºC. McrBC‐digested or mock‐treated samples were PCR‐amplified with primers flanking 5′ or 3′ α‐SNAP‐RAC junctions and visualized by agarose gel electrophoresis.
2.7. RNA isolation and cDNA synthesis
Total RNAs were extracted using Trizol (Life Technologies Corp.) or the Direct‐Zol RNA Miniprep Kit (Zymo Research), per manufacturer's instructions. All RNA samples were DNAase treated and quantified using a spectrophotometer. cDNA synthesis was performed using the iScript cDNA Synthesis Kit (Bio‐Rad) according to manufacturer's recommendations using 1.0 µg of purified total RNA.
2.8. qPCR analysis
qPCR was performed with a CFX96 real‐time PCR detection system (Bio‐Rad Laboratories) using SsoFast EvaGreen Supermix (Bio‐Rad Laboratories) as in Cook et al. (2012). Following amplification, a standardized melting curve analysis was performed. Overall cDNA abundances for each sample were normalized using the qPCR signal for reference gene Glyma.18G022300. RAC transcript abundances are presented relative to the mean abundance of RAC transcript for Williams 82 leaf samples.
2.9. RT‐PCR analysis
For RT‐PCR, 31 cycles of amplifications were performed prior to loading PCR product samples for separation and visualization by agarose gel electrophoresis. The number of PCR cycles terminated prior to maximal amplification of product from the most abundant template pool. A primer set complementary to a conserved copia region detected both endogenous RAC‐like transcripts as well as the uniquely tagged RAC transgene. Specific detection of the tagged RAC transgene utilized a primer pair complementary to the engineered region. Transcripts from Glyma.18G022300 or Skp16 served as a control for both cDNA quality and relative transcript abundance.
2.10. Vector construction
Native α‐SNAP‐RAC was PCR‐amplified from pTopo XL subclones using Kapa HiFi polymerase with AvrII and SbfI restriction site overhangs. Following agarose gel purification (cut with XbaI/PstI) (New England Biolabs), gel extractions were performed using the QIAquick Gel Extraction Kit (Qiagen) or the Zymoclean Large Fragment DNA Recovery Kit (Zymo Research). Purified DNA fragments were ligated overnight at 4ºC with T4 DNA ligase (New England Biolabs) per manufacturer's recommendations. To remove RAC from within the native α‐SNAP‐RAC subclone in vector pSM101, the Polymerase Incomplete Primer Extension (PIPE) PCR method was used with Kapa HiFi polymerase (Klock & Lesley, 2009). Similarly, the synonymous tag added within the RAC ORF of native α‐SNAP‐RAC was created using PIPE. Unique nucleotide tag was located ~160 bp downstream of the RAC ATG and maintains an intact ORF. For creating the RAC only vector which assessed inherent RAC transcription, the native RAC ORF with both LTRs (~4.77 kb) was amplified from the initial PI 89772 α‐SNAP‐RAC subclone in pTopoXL using Kapa HiFi. Restriction site overhangs for AvrII and SbfI were incorporated into the primer sequences, and following gel recovery, the PCR amplicon was restriction digested and ligated into a PstI/XbaI cut pSM101 binary vector using T4 DNA ligase (New England Biolabs). For the native α‐SNAP‐RAC with flanking Rhg1 sequence, an 11.1 kb native Rhg1 sequence containing Glyma.18G022400 and Glyma.18G022500 (and ~1 kb downstream of each stop codon), was PCR‐amplified from a previously published fosmid subclone “Fos‐32,” with AvrII and SbfI restriction ends using Kapa HiFi polymerase (Cook et al., 2012). After restriction digestion, this amplified native fragment was ligated into the binary vector pSM101 (digested with PstI and XbaI). This created a native Rhg1 two gene vector of the rhg1‐b type. Then, to make the native rhg1‐a construct, this native rhg1‐b pSM101 vector was used as a scaffold for step‐wise cloning of two different native rhg1‐a fragments amplified from PI 89772 genomic DNA. The first was a 4 kb fragment with an SbfI primer overhang containing Glyma.18G022400 up to an endogenous NruI site at exon 1; the second 11 kb fragment resumed at NruI until ~1.0 kb downstream of the Glyma.18G022500 (α‐SNAPRhg1LC) termination codon and contained an AvrII restriction overhang.
2.11. Immunobloting and antibodies
Affinity‐purified polyclonal rabbit antibodies raised against α‐SNAPRhg1LC and wild‐type α‐SNAP C‐terminus were previously generated and validated (Bayless et al., 2016).
Protein lysates were prepared from ~100 mg of soybean roots that were immediately flash‐frozen in liquid N2. Roots were homogenized in a PowerLyzer 24 (Qiagen) for three cycles of 15 s, with flash‐freezing in‐between each cycle. Protein extraction buffer [50 mM Tris·HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 0.2% Triton X‐100, 10% (vol/vol) glycerol, 1/100 Sigma protease inhibitor cocktail] was then added at a 3:1 volume to mass ratio. Lysates were then centrifuged at 10,000 g for 10 min, and supernatant was added to SDS‐PAGE loading buffer. Immunoblots were performed essentially as in (Bayless et al., 2016; Song, Keppler, Wise, & Bent, 2015; Song, Hyten, et al., 2015). Briefly, immunoblots for α‐SNAPRhg1LC or WT α‐SNAPs were incubated overnight at 4°C in 5% (wt/vol) nonfat dry milk TBS‐T (50 mM Tris, 150 mM NaCl, 0.05% Tween 20) at 1:1,000. NSF immunoblots were performed similarly. Secondary horseradish peroxidase‐conjugated goat anti‐rabbit IgG was added at 1:10,000 and incubated for 1 hr at room temperature on a platform shaker, followed by four washes with TBS‐T. Chemiluminescence signal detection was performed with SuperSignal West Pico or Dura chemiluminescent substrates (Thermo Scientific) and developed using a ChemiDoc MP chemiluminescent imager (Bio‐Rad).
2.12. Immunolabeling and electron microscopy
Immunolabeling was performed as in (Bayless et al., 2016). Transverse sections of ~2 mm long soybean (cv. Forrest) root areas containing syncytia were harvested by hand‐sectioning at 4 dpi. Root sections were fixed in 0.1% glutaraldehyde and 4% (vol/vol) paraformaldehyde in 0.1M sodium phosphate buffer (PB) (pH 7.4) overnight after vacuum infiltration for about 1 hr. After dehydration in ethanol, samples were then embedded in LR White Resin. Ultrathin sections (∼90‐nm) were taken longitudinally along the embedded root pieces using an ultramicrotome (UC‐6; Leica) and mounted on nickel slot grids. For the immunogold labeling procedure, grids were first incubated on drops of 50 mM glycine/PBS for 15 min and then blocked in drops of blocking solutions for goat gold conjugates (Aurion) for 30 min and then equilibrated in 0.1% BSA‐C/PBS (incubation buffer). Grids were then incubated overnight at 4°C with custom α‐SNAPRhg1LC polyclonal antibody (diluted 1:1,000 in incubation buffer), washed five times in incubation buffer, and incubated for 2 hr with goat anti‐rabbit antibody conjugated to 15‐nm gold (Aurion) diluted 1:50 in incubation buffer. After six washes in incubation buffer and two 5‐min washes in PBS, the grids were fixed for 5 min in 2.0% (vol/vol) glutaraldehyde in 0.1 M phosphate buffer, followed by two 5‐min washes in 0.1 M phosphate buffer and five 2‐min washes in water. Images were collected with a MegaView III digital camera on a Philips CM120 transmission electron microscope.
2.13. Oligonucleotide primers
RAC qRT For: GGGTTCGAAATGAATACCTG
RAC qRT Rev: CACGTTCTTCTCATGGATCCTA
RAC Delete PIPE For: CTT CAT CCA CAA TTC TAA TTT ATA TGC TAG
RAC Delete PIPE Rev: GAA TTG TGG ATG AAG TAC GAC AAT CAA C
RAC amplify F: AAAGCCGCCAATTGCTTCAA
RAC amplify R: AGCAATGTGCAGCATCGACA
5′ RAC Junction For: TGGCTCCAAGTATGAAGATGCC
5′ RAC Junction Rev: AACTACAGTGGCTGACCTTCT
3′ RAC Junction For: ACTGTTCATTCAGACCGCGT
3′ RAC Junction Rev: GCAATGTGCAGCATCGACATGGG
WT Junction For: GAGTTTTGAGGTGTCCGATTTCCC
WT Junction Rev: GTGAGCGCAGTCACAAACAAC
5′ Methylation For: TGGCTCCAAGTATGAAGATGCC
5′ Methylation Rev: AACTACAGTGGCTGACCTTCT
3′ Methylation For: ACTGTTCATTCAGACCGCGT
3′ Methylation Rev: GCAATGTGCAGCATCGACATGGG
Skp16 qRT For: GAG CCC AAG ACA TTG CGA GAG
Skp16 qRT Rev: CGG AAG CGG AAG AAC TGA ACC
2570 qRT For: TGA GAT GGG TGG AGC TCA AGA AC
2570 qRT Rev: AGC TTC ATC TGA TTG TGA CAG TGC
For RAC Tag Mut: GCTCTGCTCCTGAGCCCTTGAAAACGGACAGAATGCACGGAG
Rev RAC Tag Mut: GCTCAGGAGCAGAGCCATCTATGAACTCCACTTTATTCTTGGC
RAC Tag Detect For: CAGTCCTAGACTCAACCAATTACC
RAC Tag Detect Rev: CCTTGGCTATACCTGCTCTTTAAATC
For RAC initial TopoXL subclone: GAGATTACATTGGATGATACGGTCGACC
Rev RAC initial TopoXL subclone: AGATAAGATCAGACTCCAGCAACCTC
For RAC Alone subclone AvrII: cctaggGGTGTCCGATTTCCCGATTAATTGAAG
Rev RAC Alone subclone SbfI: cctgcaggCCAACATCAATTTCAAAGTTCGTCACTTTC
LC‐Splice Reverse: AGTAATAACCTCATACTCCTCAAGTT
LC‐Splice Full For: GAGGAGGTTGTTGCTATAACCAATGC
LC‐Splice Isoform For: GAGGAGGAACTGGATCCAACATTTTC
SbfI Native Glyma.18G022400 For: cctgcaggGAGCAGTAGGCTTCTTTGGAACTTG
AvrII Native Glyma.18g022500 Rev:cctaggGTTCCTAAAGTGGCAAACCCTAAGAACAAAG
BglII Native For: AGATCTCCCTGAGAGTATCTTGATTTCAGATCG
BglII Native Rev: AGATCTTTTACGCATATCCGACCTTCAAC
2.14. Accession numbers
The RAC sequence shown in Figure S2 has been deposited as Genbank accession MN340250.
3. RESULTS
3.1. Multiple rhg1‐a haplotypes harbor an intronic copia retrotransposon (RAC) within the Rhg1‐encoded α‐SNAP
The α‐SNAPs encoded by the rhg1‐a and rhg1‐b loci play a key role in SCN resistance (Bayless et al., 2016, 2018; Cook et al., 2014, 2012; Liu et al., 2017). While entire 31.2 kb Rhg1 repeats of rhg1‐b and Rhg1WT haplotypes have been sub‐cloned and characterized (Cook et al., 2012), we sought to study the native genomic rhg1‐a α‐SNAP‐encoding region to investigate potential regulatory differences between rhg1‐a and rhg1‐b. Figure 1a provides a schematic of the 31.2 kb Rhg1 repeat unit with the four Rhg1‐encoded genes, while Figure 1b presents a schematic of the previously published Rhg1 haplotypes (single‐, low‐, and high‐copy) and the respective C‐terminal amino acid polymorphisms of their Rhg1‐encoded α‐SNAP proteins (Cook et al., 2014, 2012). Working from previously generated WGS data, we PCR‐amplified the native genomic rhg1‐a α‐SNAP locus from PI 89772 and, unexpectedly, obtained PCR amplicons ~5 kb larger than WGS‐based estimates (Figure S1a) (Cook et al., 2014; Liu et al., 2017). Overly large amplicons were also obtained using different PI 89772 genomic DNA templates and/or other PCR cycling conditions, and no rhg1‐a α‐SNAP amplicons of expected size were observed (Figure S1a). Sanger DNA sequencing of these unusually sized rhg1‐a α‐SNAP amplicons matched WGS predictions until partway into α‐SNAP intron 1, where a 4.77 kb element was inserted. Immediately following this 4.77 kb insertion, the amplicon sequence again matched WGS predictions (Figure 1c).
Figure 1.
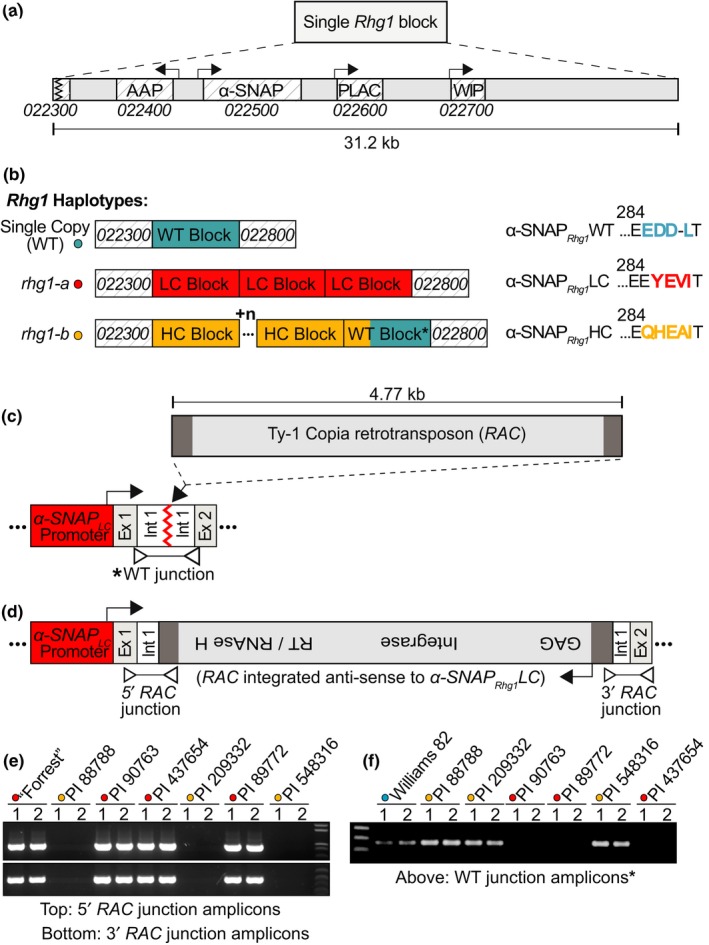
Multiple rhg1‐a haplotypes harbor an intronic copia retrotransposon (RAC) within the Rhg1‐encoded α‐SNAP (Glyma.18g022500). (a) Diagram of a single 31.2 kb Rhg1 block and the four Rhg1‐encoded genes: Glyma.18G022400 (amino acid permease, AAP), Glyma.18G022500 (α‐SNAP), Glyma.18G022600 (PLAC‐domain protein), and Glyma.18G022700 (wound‐inducible protein, WIP). Glyma.18G022300 and Glyma.18G022800 flank Rhg1, but each repeat also includes a truncated 3′ fragment of Glyma.18G022300. (b) Schematic of the three known Rhg1 haplotypes: Rhg1 wild‐type (single‐copy, shown blue), rhg1‐a (low‐copy, shown red), and rhg1‐b (high‐copy, shown orange). Rhg1 α‐SNAP C‐terminal amino acid polymorphisms colored to match Rhg1 block type. (c, d) Model from DNA sequencing of Rhg1 alpha‐SNAP copia (RAC) integration site within the PI 89772 (rhg1‐a) encoded α‐SNAP. The 4.77 kb RAC element (shown gray) is anti‐sense to α‐SNAPRhg1LC and increases predicted overall rhg1-a repeat size to ~36 kb. RAC ORF is intact and encodes a 1438 residue polyprotein. RAC LTRs are shown in dark gray; α‐SNAPRhg1LC promoter are shown in red. “Ex.” and “int.” are α‐SNAPRhg1LC exons and introns, respectively. Connected open triangles indicate PCR products of Figure 1e,f. LTR: long terminal repeat; GAG: group‐specific antigen, RT: reverse transcriptase. (e) Agarose gel showing 5′ and 3′ α‐SNAP‐RAC junction products from the rhg1‐a (low‐copy, red dots) accessions: “Forrest,” PI 90763, PI 437654, PI 89772. No α‐SNAP‐RAC junctions detected from rhg1‐b (high‐copy; orange dots) accessions: PI 88788, PI 209332, or PI 548316. (f) Similar to E and using same template DNA samples, but PCR amplification of a "WT junction" products (Figure 1c) of size similar to the wild‐type α‐SNAP exon 1–2 distance, as in the Williams 82 reference genome
An NCBI nucleotide BLAST of the unknown 4.77 kb region returned hits for conserved features of the Ty‐1 copia retrotransposon superfamily. Notably, the multi‐cistronic ORF of this copia element was fully intact, and both 5′ and 3′ LTRs were present (Long Terminal Repeats; LTRs function as transcriptional promoters and terminators, respectively) (Havecker, Gao, & Voytas, 2004). Subsequently, we named this insert “RAC” for Rhg1 alpha‐SNAP copia. Figure 1d shows the rhg1‐a α‐SNAP‐RAC structure, and Figure S2 provides the complete RAC nucleotide sequence and highlights the α‐SNAPRhg1LC intron 1 sequences directly flanking the RAC integration. The RAC insertion effectively doubles the pre‐spliced α‐SNAPRhg1LC mRNA transcript from 4.70 to 9.47 kb, yet RAC apparently splices out effectively, as all reported cDNAs of mature α‐SNAPRhg1LC transcripts do not contain any RAC sequences (Cook et al., 2014; Lee et al., 2015; Liu et al., 2017). The RAC ORF is uninterrupted and encodes a 1438 residue polyprotein with conserved copia retrotransposon motifs for GAG (group‐specific antigen) protease, integrase, and reverse transcriptase (Havecker et al., 2004; Kanazawa, Liu, Kong, Arase, & Abe, 2009; Peterson‐Burch & Voytas, 2002). These conserved RAC‐polyprotein motifs are highlighted in Figure S3. Intriguingly, RAC integrated just 396 bp downstream of the α‐SNAPRhg1LC start codon, and in an anti‐sense orientation (Figure 1d). The intact LTRs and uninterrupted ORF suggest that RAC integration could have been a relatively recent event and that RAC may remain functional.
PI 89772 (used above) is one of seven rhg1‐a and rhg1‐b soybean accessions used to determine the HG type of SCN populations (Niblack et al., 2002). We subsequently tested for RAC insertions within the Rhg1‐encoded α‐SNAP genes in the other six soybean accessions used in HG type tests. The RAC integration within the PI 89772‐encoded α‐SNAPRhg1LC creates unique 5′ and 3′ sequence junctions within the α‐SNAPRhg1LC intron 1 and substantially increases the distance from α‐SNAP exon 1 to exon 2 from ~400 bp to ~5,000 bp (Figure 1b,c). Therefore, to screen for α‐SNAP‐RAC insertions, we devised PCR assays specific for RAC‐α‐SNAP junctions, or for wild‐type (uninterrupted) “WT junctions” separated by the genomic distances from exon 1 to exon 2 that are annotated in the soybean reference genome (Schmutz et al., 2010), as depicted in Figure 1c,d. Among all rhg1‐a haplotype HG type test accessions (PI 90763, PI 89772, PI 437654 and PI 548402(Peking)‐derived “Forrest”), both 5′ and 3′ α‐SNAP‐RAC junctions were detected (Figure 1d).
3.2. RAC is absent from rhg1‐b and single‐copy Rhg1WT accessions
The α‐SNAP‐RAC junctions were absent from the rhg1‐b accessions tested (PI 88788, PI 548316, PI 209332), which instead gave WT junction PCR products of a size similar to Williams 82 (Figure 1d–f; Figure S1b). Because no SNPs exist at the WT junction primer sites across any of the rhg1‐b repeats (Cook et al., 2014), absence of 5′ RAC junction and 3′ RAC junction PCR products for the rhg1‐b accessions suggests that those accessions do not carry the RAC copia element in the first intron of their α‐SNAP gene. Accession Williams 82 (Wm82, SCN‐susceptible, Rhg1 single‐copy), the source of the soybean reference genome, also gave a product for the WT junction reaction, and no PCR product for a RAC integration within the Rhg1WT (single‐copy) α‐SNAP gene, consistent with the reference genome annotation (Schmutz et al., 2010) (Figure S1b,c). RAC absence from rhg1‐b and WT Rhg1 repeats is also consistent with previous studies that sub‐cloned and Sanger‐sequenced large‐insert genomic fragments spanning entire rhg1‐b and Rhg1WT‐like repeats and noted no unusual insertions (Cook et al., 2012). Although all seven HG type test soybean lines were previously analyzed via WGS, the RAC insertion was evidently omitted from the four rhg1‐a accession assemblies during Illumina short sequence read filtering that excludes repetitive genome elements and hence from subsequent read mapping and assembly to the Williams 82 reference genome, which lacks the RAC insertion (Cook et al., 2014; Lee et al., 2015). The RAC insertion was apparently missed in other studies due to sequencing of post‐splicing Rhg1 α‐SNAP cDNAs (Bayless et al., 2016; Liu et al., 2017).
The WT junction PCR experiment also interrogated if RAC is present within each encoded α‐SNAP gene of all three rhg1‐a repeats. Among all rhg1‐a accessions, no WT exon 1 to exon 2 junctions were detected, while uninterrupted WT junction product distances were present in all rhg1‐b accessions and Wm82 as noted above (Figures 1e,f and S1b,c). Hence RAC is apparently present within the α‐SNAP of each rhg1‐a repeat unit. To independently investigate the same question, previously available Illumina whole‐genome sequence data were queried (Cook et al., 2014). The read depth for RAC was found to be 3–4 fold greater in rhg1‐a accessions relative to the read depth of the flanking DNA regions, or when compared to RAC read depth in rhg1‐b accessions (Figure S4, SI Spreadsheet). Table S1 provides a summary of the Rhg1 haplotype composition, copy number, resistance‐type α‐SNAP allele, and presence of normal versus RAC‐interrupted α‐SNAP among the HG type test accessions and the Wm82 reference genome. Together, these findings indicate that the HG type test rhg1‐a accessions contain the RAC‐α‐SNAP introgression and that their rhg1‐a repeats are ~36.0 kb, as opposed to ~31.2 kb for rhg1‐b repeats and Rhg1WT. Although RAC is integrated in anti‐sense orientation and close to α‐SNAPRhg1LC exon 1, RAC does not eliminate rhg1‐a function because the HG type test accessions are selected owing to their strong SCN resistance, and all have previously been shown to express α‐SNAPRhg1LC mRNA and protein (Bayless et al., 2016, 2018; Cook et al., 2014; Liu et al., 2017). RAC presence within these rhg1‐a lines may even be beneficial.
3.3. RAC is not identical to other copia elements but the RAC‐like copia subfamily is common in soybean
Copia retrotransposons frequently attain high‐copy numbers in plant and animal genomes; therefore, we assessed the abundance of RAC and RAC‐like copia elements in the soybean genome (Du, Grant, et al., 2010; Du, Tian, et al., 2010; Zhao & Ma, 2013). SoyTE, the soybean transposon database, has recorded over 32,000 TEs, including nearly 5,000 intact retrotransposons (Du, Grant, et al., 2010; Du, Tian, et al., 2010). We queried the SoyTE database via Soybase.org using a RAC nucleotide sequence BLASTN search, but no intact or high identity hits were returned. However, a similar BLASTN search against the Wm 82 soybean reference genome at Phytozome.org (Goodstein et al., 2012) returned 146 sequences. These 146 hits spanned all 20 G. max chromosomes and included several intact elements of high nucleotide identity with RAC, as well as numerous short length matches which likely represent fragments of inactive elements (Table S2). We then constructed a nucleotide‐based phylogenetic tree of the soybean RAC family using just one RAC‐family element from each soybean chromosome (Chr) (Figure 2a). This analysis used the element from each chromosome that was most similar to RAC, as well as a previously reported copia retrotransposon (TGMR) residing near the soybean Rps1‐k resistance gene (Bhattacharyya et al., 1997) and the highest RAC‐identity element match from the common bean (Phaseolus vulgaris) genome (Figure 2a). The two RAC‐like elements, from Chr10 and Chr18, had 99.7% and 97.6% nucleotide identity with RAC, respectively (Figure 2a, Table S2). Moreover, the Chr10 element retained an intact ORF and both LTRs. The near‐perfect nucleotide identity with RAC (99.7%; 4464/4477 positions) suggests the Chr10 element as a possible source for the retrotransposition event that created the RAC introgression within α‐SNAPRhg1LC. The above‐noted WGS read‐depth analysis of soybean accessions (Figure S4, SI Spreadsheet) also found that 3 of 10 examined rhg1‐b accessions gave a read depth of zero or close to zero for RAC (with one mismatch allowed), indicating absence of the RAC or a close homolog at the Chr 10 locus in some soybean accessions. Like RAC, the Chr18 element was also integrated anti‐sense within a host gene Glyma.18G268000 (a putative leucine‐rich repeat receptor kinase). We further noted that the Chr20 RAC‐like element (82% identical) was intronically positioned within Glyma.20G250200 (BAR‐domain containing protein) (Table S2). That multiple intact and highly similar RAC‐like elements are in soybean suggests that this retrotransposon family was recently active.
Figure 2.
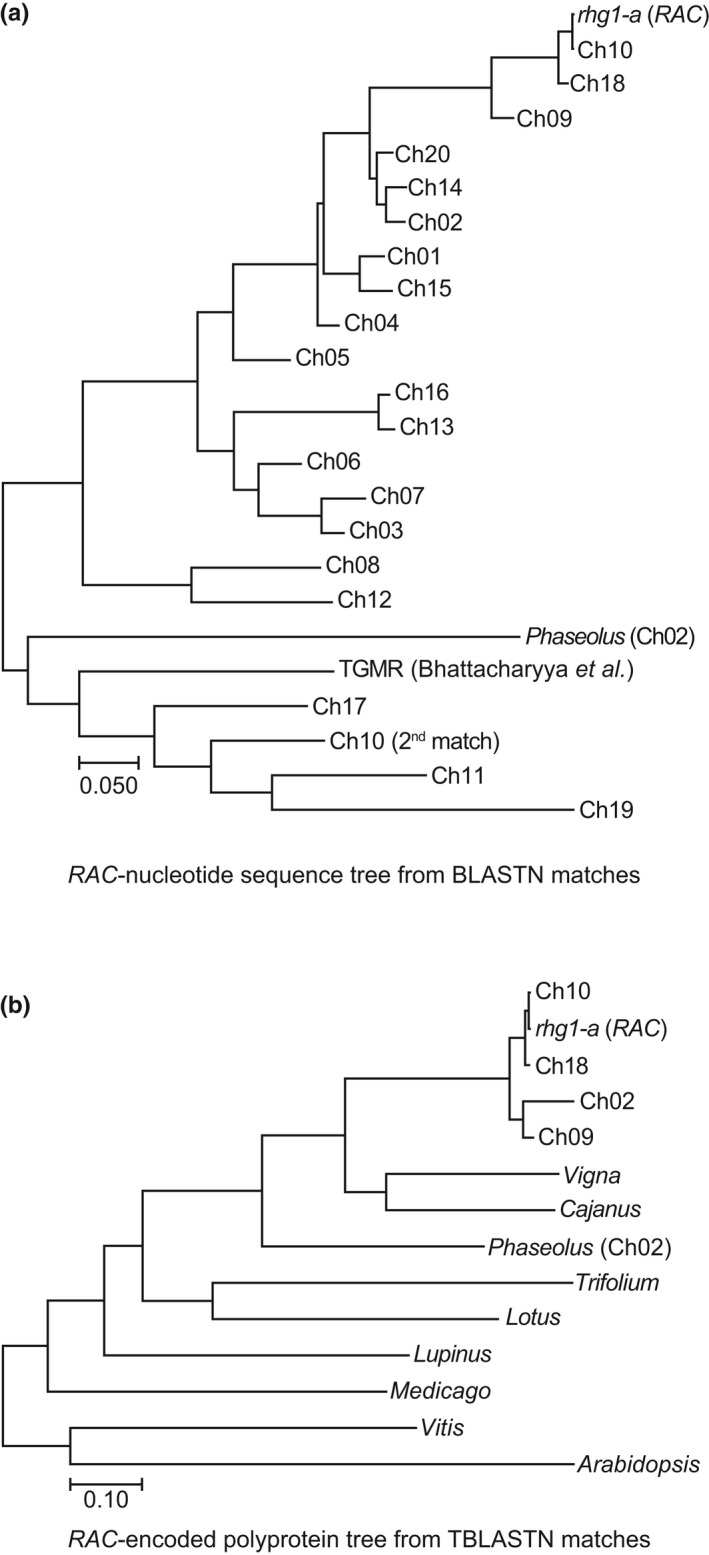
The RAC‐like subfamily of copia retrotransposons is common in soybean and other legumes. (a) Maximum likelihood phylogenetic tree of RAC‐like element nucleotide sequences from soybean. The top hit from each soybean chromosome was included, as was the known soybean retrotransposon “TGMR” and the top RAC‐like match from Phaseolus (common bean). (b) Similar to A; a maximum likelihood tree, but using the RAC‐encoded polyprotein sequences from the four most similar soybean RAC‐like elements, and the most similar element matches from the indicated plant species
Subsequent work examined if RAC‐family elements are present among other plant species. We performed a TBLASTN search of the RAC‐encoded polyprotein at NCBI and obtained numerous hits against multiple species, including Arabidopsis, Cajanus cajans (pigeon pea), Vigna angularis (adzuki bean), P. vulgaris (common bean), Lupinus angustifolius (blue lupin), Medicago truncatula, and clover (Trifolium subterraneum). Similar to Figure 2a, a phylogenetic tree using MEGA (muscle alignment) and the RAC‐family polyprotein sequences of these different plant species was constructed (Kumar et al., 2016) (Figure 2b). Together, these findings demonstrate that the RAC‐like copia members are not only common in G. max, but also in other legumes.
3.4. RAC is present within only a subclass of the soybean accessions that have the previous SoySNP50K‐predicted rhg1‐a signature
Although we detected RAC in all four rhg1‐a accessions that are used for HG type determination, we sought to determine if the RAC insertion is universal among all rhg1‐a‐containing accessions. Recently, the USDA soybean collection (~20,000 accessions) was genotyped using a 50,000 SNP DNA microarray chip—the SoySNP50K iSelect BeadChip (Song, Keppler, et al., 2015; Song, Hyten, et al., 2015). We searched for and found a SNP on the SoySNP50K chip that detects RAC. Using the SoySNP50K browser at Soybase.org (Soybase.org/snps/), we found a SNP (ss715606985, G to A) that in the Wm82 soybean reference genome was assigned to the Chr10 RAC‐family element (99.7% nucleotide identity with RAC). However, we noted that this ss715606985 SNP is a perfect match to the sequence of RAC within α‐SNAPRhg1LC (Figure S5a). Using this SNP marker for RAC, we then calculated the ss715606985 SNP prevalence among all USDA accessions and found that the SNP is rare—only 390 of 19,645 accessions (~2.0%) were putative RAC+ lines homozygous for the SNP (Figure 3a). The RAC‐SNP (ss715606985) was then directly tested as a marker for the α‐SNAP‐RAC event using the PCR assays described in Figure 1, which test for α‐SNAP‐RAC junctions and normal α‐SNAP exon 1–2 distances. We randomly selected several accessions with SNP signatures of rhg1‐a that were positive or negative for the RAC‐SNP and found that SNP presence correlated perfectly with RAC‐α‐SNAP junction detection, while accessions lacking the RAC‐SNP had normal α‐SNAP exon 1–2 distances indicative of no inserted DNA (Figure 3b).
Figure 3.
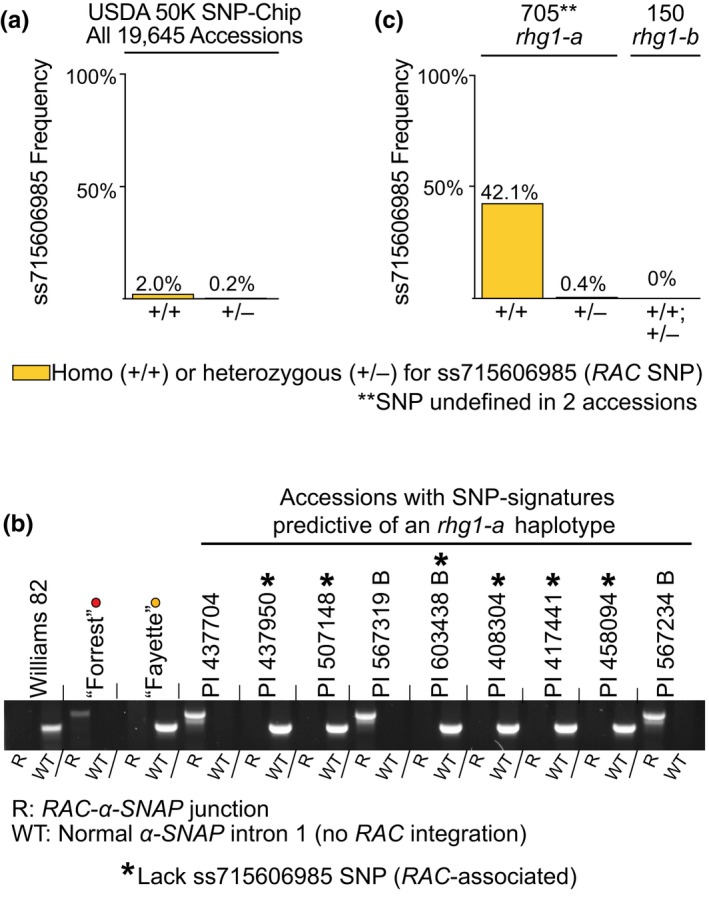
RAC is present within a subclass of rhg1‐a signature soybean accessions. (a) Frequency of RAC‐associated SNP, ss715606985, among 19,645 SoySNP50K‐genotyped USDA soybean accessions. (b) Agarose gel showing PCR detection of α‐SNAP‐RAC junctions or WT α‐SNAP exon 1–2 distances among rhg1‐a signature accessions positive or negative for the RAC‐SNP, ss71560698. Williams 82 (Rhg1WT), “Forrest” (rhg1‐a), and “Fayette” (rhg1‐b) included as controls; Rhg1 haplotypes color coded with dots as in Figure 1. An * denotes an rhg1‐a signature accession lacking the RAC‐SNP. (c) Frequency of RAC‐associated SNP among all USDA Glycine max accessions with consensus SNP signatures of rhg1‐a or rhg1‐b haplotypes
We next examined RAC presence among all G. max USDA accessions with the SoySNP50K SNP signatures of rhg1‐a or rhg1‐b haplotypes, as reported by Lee et al. (Lee et al., 2015). The multi‐SNP SoySNP50K signatures for rhg1‐a and rhg1‐b (Lee et al., 2015) are present in 705 and 150 G. max accessions, respectively, out of 19,645 USDA accessions; these SoySNP50K signatures are provided in Figure S5b. We found that 42% (299 of 705) of accessions with rhg1‐a SNP signatures and 0% (0 of 150) of accessions with rhg1‐b SNP signatures carry the RAC ss715606985 SNP (Figure 3c). That the RAC‐SNP was absent from all rhg1‐b signature accessions is consistent with the PCR screens of Figure 1e,f, which indicated that no rhg1‐b HG type test accession contained RAC‐α‐SNAP junctions. A flowchart is available as Figure S6 that summarizes the above findings and additional work presented below.
Because only 299 of the 390 accessions with the RAC‐SNP had a perfect match rhg1‐a SoySNP50K SNP signature (Figure 3c), we investigated the Rhg1 SNP signature of the remainder. To avoid false positives, the rhg1‐a SNP signature of (Lee et al., 2015) uses 14 SNPs that extend to 11 kb and 54 kb beyond the edges of the ~30 kb Rhg1 repeat. Relaxing the stringency that required 14 perfect matches, we found that 83 of the remaining 91 RAC + accessions carry a perfect match with the four rhg1‐a SNP markers that map within the Rhg1 repeat or within <5 kb of the edge of the Rhg1 repeat. 86 of 91 have only a single reliably called SNP that varies from the rhg1‐a consensus (SI Spreadsheet). Together, the combined findings indicate that the α‐SNAP‐RAC integration is only present within rhg1‐a haplotypes and that RAC retrotransposition may have occurred within a subset of the Rhg1+ population after Rhg1 divergence into the distinctive high‐ and low‐copy haplotype classes.
3.5. The RAC‐SNP allows more accurate prediction of rhg1‐a presence
The above finding that a few hundred of the 705 USDA accessions with the previously identified rhg1‐a SNP signature apparently do not contain RAC was surprising, given that all four of the rhg1‐a HG type test accessions do contain RAC‐α‐SNAP junctions. However, it was possible that these non‐RAC‐containing accessions, despite a consensus SNP signature predicting an rhg1‐a haplotype, might not truly carry an rhg1‐a resistance haplotype. The rhg1‐a and rhg1‐b repeats encode distinct Rhg1 α‐SNAP alleles; thus, we cloned and sequenced the genomic Rhg1 α‐SNAP regions from several non‐RAC rhg1‐a SNP signature accessions and detected coding sequences for either rhg1‐b (α‐SNAPRhg1HC) or Rhg1WT (α‐SNAPRhg1WT) alleles (Figure S5c). None of these accessions encoded α‐SNAPRhg1LC, and thus, they were not rhg1‐a (Figure S5c). These findings indicate that the consensus rhg1‐a SNP signature, while useful, is not a perfect predictor of accessions carrying actual rhg1‐a resistance haplotypes. Rather, combined presence of the ss715606985 SNP for RAC and a near‐consensus rhg1‐a signature is a more accurate predictor of accessions that truly carry rhg1‐a resistance. Additionally, these data suggest that accessions carrying rhg1‐b resistance haplotypes can share the SNP signatures of rhg1‐a accessions. We again refer readers to the flowchart (Figure S6) that summarizes these and other findings.
3.6. RAC presence correlates with a stronger SCN‐resistance profile and the co‐presence of other loci that augment rhg1‐a resistance
rhg1‐a (Rhg1 low‐copy) loci encode unique α‐SNAPRhg1LC alleles, however, robust rhg1‐a resistance requires the co‐presence of Rhg4, and the α‐SNAP Ch11‐IR allele bolsters rhg1‐a resistance further (Bayless et al., 2018; Lakhssassi et al., 2017; Liu et al., 2012; Patil et al., 2019). We sought to compare the SCN‐resistance profiles of the rhg1‐a signature accessions with RAC to those without RAC (which are not true rhg1‐a), to assess how the two groups match what is known about rhg1‐a resistance. Previously, Arelli, Young, and others obtained SCN‐resistance phenotype data, across multiple trials and with various SCN populations, for at least 573 different USDA accessions that are now known to carry the SoySNP50K signatures suggestive of rhg1‐a (Anand, 1984; Arelli et al., 2000; Diers et al., 1997; Hussey et al., 1991; Lee et al., 2015; Young, 1995). The groups all conducted variants of the widely utilized female index assay, allowing comparisons across tests, with the caveat that a few false‐positive and false‐negative results are likely in this dataset for over 500 soybean lines. We used these available SCN‐resistance data from the USDA GRIN database to compare the resistance profiles of rhg1‐a signature accessions which did or did not have the ss715606985 (RAC) SNP signature. Any accession that scored as “R” (resistant) in any single SCN trial was placed into the broad category “R.” Likewise, any accession that scored “MR” (moderately resistant) or “MS” (moderately susceptible) in any trial, with no higher resistance scores in other trials, was placed into those respective categories. Only accessions that scored susceptible (“S”) across all trials were placed into the “S” category. Consistent with previous reports that rhg1‐a accessions possess broad and robust resistance (Concibido et al., 2004; Kadam et al., 2016; Vuong et al., 2015), 91% (51/56) of the accessions in the “R” group were positive for the ss715606985 + RAC‐SNP (Figure 4a). The frequency of RAC presence was substantially lower among the more susceptible phenotypic classes (Figure 4a) while the majority of the ss715606985 ‐ (no RAC) accessions scored either “S” or “MS.” As was noted above, none of the non‐RAC accessions that we examined had rhg1a‐type resistance (Figure 3b,c, Figure S5c). The phenotype scores and relevant SNP markers for all 573 of these SCN‐phenotyped rhg1‐a SNP signature accessions are provided in the SI spreadsheet in the Supplemental Data.
Figure 4.
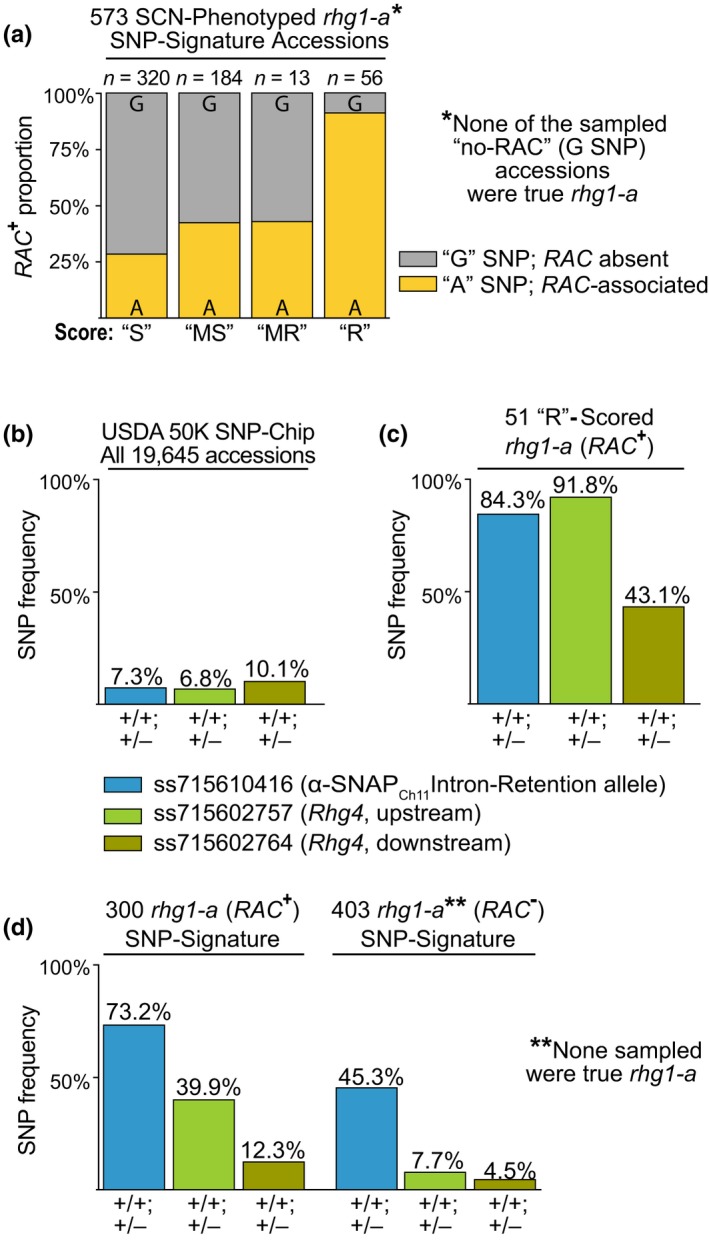
RAC presence correlates with a stronger SCN‐resistance profile and the co‐presence of other loci that augment rhg1‐a resistance. (a) Proportion of RAC + (ss715606985 A SNP) versus RAC ‐ (G SNP) accessions among 573 SCN‐phenotyped soybeans with consensus SoySNP50K SNP signatures predictive of rhg1‐a. *Note that none of the sampled RAC ‐ (G SNP) accessions had rhg1‐a (none encoded α‐SNAPRhg1LC). “S”: susceptible in all trials, “MS”: moderately susceptible in at least one trial, “MR”: moderately resistant in at least one trial, “R”: resistant in at least one trial. Fisher's Exact Test pairwise comparisons: “R‐MR” (p = 2.6E‐4), “R‐MS” (p = 2.3E‐11), “R‐S” (p = 2.2E‐16), “MR‐MS” (p = 1.0), “MR‐S” (p = .25), “MS‐S” (p = 2.4E‐3). (b) Frequency of SNPs associated with Rhg4 (ss715602757, ss715602764) or the Chromosome 11‐encoded α‐SNAP intron‐retention (α‐SNAPCh11IR) allele, ss71559743 among 19,645 USDA accessions. (c) Frequency of the Rhg4 and α‐SNAPCh11IR associated SNPs among the 51 “Resistant” scored RAC + rhg1‐a signature accessions. (d) Frequency of the Rhg4 and α‐SNAPCh11‐IR associated SNPs among all RAC + (300) or RAC ‐ (403) USDA G. max accessions with consensus SNP signatures predictive of rhg1‐a (705 total; two accessions undefined for ss715606985 SNP)
In the above analysis (Figure 4a), some of the RAC+ (ss715606985+) accessions, which are rhg1‐a, had scored as “S” or “MS.” This seemed likely to be because they lack a resistance‐conferring allele at Rhg4 and/or the resistance‐enhancing allele of the Chr 11‐encoded α‐SNAP (α‐SNAPCh11‐IR) (Bayless et al., 2018; Lakhssassi et al., 2017; Liu et al., 2012). Accordingly, we investigated if the SCN‐resistance phenotype scores also correlated with co‐presence of those loci. None of the SoySNP50K markers resides within the Rhg4 gene but we noted that the two SoySNP50K SNPs that most closely flank the Rhg4 locus are rare among USDA accessions (Figure 4b; ss715602757, ss715602764), and one or both of these SNPs are present in the Rhg4‐containing HG type test lines. We also used a SNP, ss715610416, previously associated with the Chr11 α‐SNAP intron‐retention allele (α‐SNAPCh11‐IR) (Bayless et al., 2018). Among the 51 RAC + accessions with an SCN‐=resistance score of “R,” we found that SNPs associated with both α‐SNAPCh11‐IR and Rhg4 were enriched ~10‐fold, as compared to the entire USDA collection (Figure 4b,c). Additionally, among the 705 USDA accessions with SoySNP50K signatures predictive of rhg1‐a, we found that the Rhg4 and α‐SNAPCh11‐IR SNPs were enriched among the RAC + accessions relative to the RAC – accessions (Figure 4d). Thus, the heightened SCN resistance of the RAC‐positive (ss715606985+) rhg1‐a signature accessions is consistent with previous reports, and as expected, the resistance is associated with the co‐presence of additional loci like Rhg4 and α‐SNAPCh11‐IR (Bayless et al., 2018; Kadam et al., 2016; Lakhssassi et al., 2017; Patil et al., 2019; Vuong et al., 2015).
3.7. The rhg1‐a RAC element has intrinsic transcriptional activity
While the RAC‐SNP apparently identifies true rhg1‐a resistance sources, possible impacts of the RAC element itself on α‐SNAPRhg1LC expression remained to be explored. Typically, eukaryotic cells silence TEs using small RNA‐directed DNA methylation pathways, and this can also silence adjacent genes (Kim & Zilberman, 2014; McCue, Nuthikattu, Reeder, & Slotkin, 2012). Since the α‐SNAPRhg1LC mRNA transcript and protein are readily detected in RAC‐containing soybeans, RAC does not, at least constitutively, eliminate α‐SNAPRhg1LC expression (Bayless et al., 2016, 2018; Cook et al., 2014; Liu et al., 2017). Nonetheless, we examined DNA methylation at the rhg1‐a α‐SNAP‐RAC junction, as well as transcriptional activity of the rhg1‐a RAC element. The restriction enzyme McrBC cleaves only methylated DNA, so potentially methylated DNA regions may be assessed via McrBC digestion and subsequent attempted PCR across areas of interest. After genomic DNAs from “Forrest” and “Peking” (PI 548402) were treated with McrBC, both the 5′ and 3′ borders of the α‐SNAP‐RAC did not PCR amplify relative to the mock‐treated controls, indicating the presence of methylated cytosines at the α‐SNAP‐RAC junctions (Figure 5a).
Figure 5.
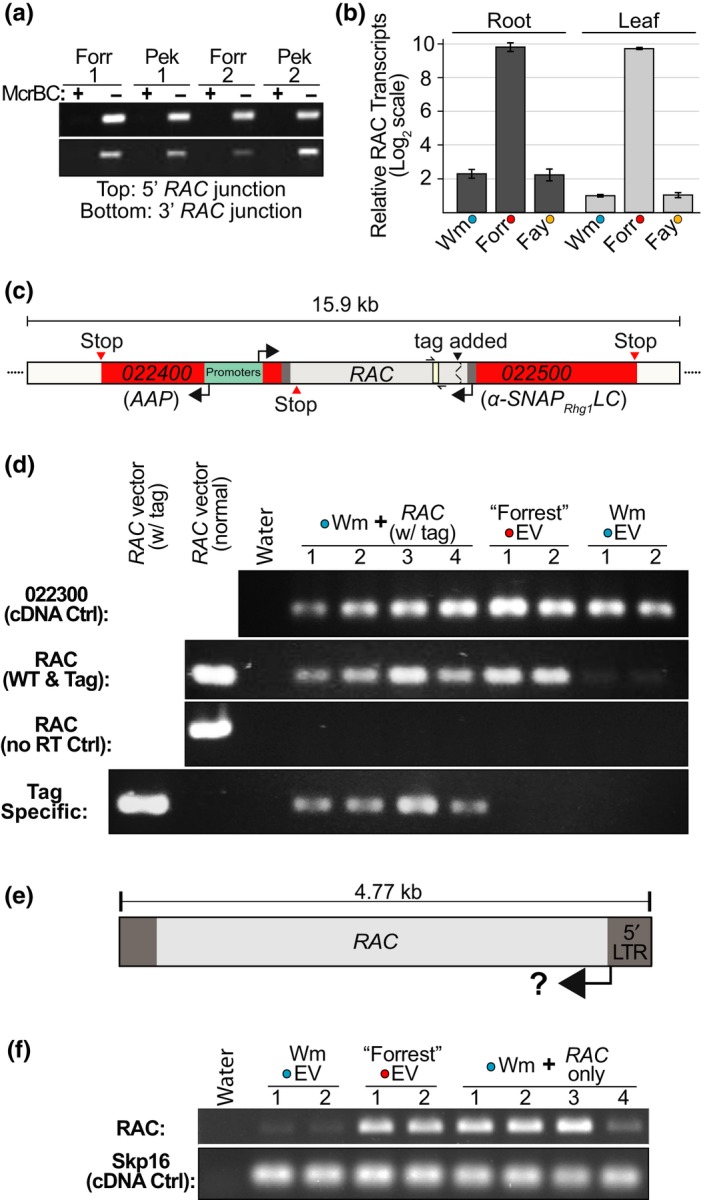
The rhg1‐a RAC element is methylated but has intrinsic transcriptional activity. (a) Agarose gel showing PCR amplicons for α‐SNAP‐RAC regions from McrBC‐treated (+) or mock‐treated (−) genomic DNAs from “Forrest” (Forr) or “Peking” (Pek, PI 548402) roots. (b) qPCR analysis of mRNA transcript abundance for RAC and similar RAC‐like elements, in leaf or root tissues of Williams 82 (Wm; Rhg1WT), “Forrest” (Forr; rhg1‐a) or “Fayette” (Fay; rhg1‐b). Colored dots indicate Rhg1 haplotype as in Figure 1. Normalized RAC transcript abundances are presented relative to the mean abundance of RAC transcript for Williams 82 leaf samples. Y‐axis uses log2 scale. (c) Schematic showing unique nucleotide tag addition to an otherwise native α‐SNAP‐RAC cassette. This construct contains native flanking Rhg1 sequence including Glyma.18G022400 (transcribes from the bidirectional α‐SNAP promoter) and 1.8 kb upstream, as well as 4.7 kb of downstream RAC flanking sequence (~1.0 kb after the α‐SNAPRhg1LC termination codon). The RAC region detected and amplified via qPCR or RT‐PCR is colored ivory and flanked by half‐arrows. (d) Agarose gel of RT‐PCR cDNAs of “Forrest” or Wm 82 transgenic roots transformed with an empty vector (EV) or the native tagged α‐SNAP‐RAC construct. Tag primers amplify only the modified α‐SNAP‐RAC while the normal RAC primer set amplifies both endogenous RAC‐like transcripts as well as the tagged α‐SNAP‐RAC transgene. Glyma.18G022300 mRNA transcript used as a cDNA quality and loading control; no RT (reverse transcriptase) ctrl verifies absence of amplifiable genomic DNA. (e) Schematic showing the sub‐cloned 4.77 kb RAC expression cassette tested in F. (f) Like D, but “Forrest” or Wm 82 roots transformed with empty vector or the 4.77 kb RAC element (all flanking Rhg1 sequence context removed)
Because RAC has both LTRs and an intact ORF, we tested for transcription of RAC in the rhg1‐a soybean genotype “Forrest” as compared to “Fayette” (rhg1‐b) and Wm82 (Rhg1WT). Fayette and Wm82 do not carry the Rhg1 α‐SNAP‐RAC but do carry other RAC‐like copia elements that match the qPCR primers used. qPCR analysis of cDNAs from root or leaf tissues indicated that mRNA transcripts from RAC or RAC‐like sequences were ~200‐fold higher in “Forrest” than in Wm82 or “Fayette” (rhg1‐b) (Figure 5b). This suggested but did not firmly demonstrate that the Rhg1‐embedded RAC is the primary source of the detected transcript, because RAC has high nucleotide identity with other RAC‐like elements (Figure 2a) whose activity may also vary between accessions.
We conducted additional tests for transcription of α‐SNAP‐RAC by transforming Wm82 roots with a ~15 kb cloned segment of native α‐SNAP‐RAC genomic DNA (including the upstream Glyma.18G024400 Rhg1 gene, which shares the same bidirectional promoter; depicted in Figure 5c). Importantly, we engineered this otherwise native α‐SNAP‐RAC cassette with a unique nucleotide tag to distinguish between transgene‐derived transcripts versus transcripts from other RAC‐family elements in the genome (Figure 5c). Low abundance of RAC transcripts in Wm82 roots relative to “Forrest” roots had been documented (Figure 5b), so RT‐PCR of Wm82 readily visualized RAC‐specific transcript expression from the transgenically introduced construct. In control roots, sharp contrasts in RAC expression were again observed between “Forrest” roots and Wm82 roots transformed with empty vector (Figure 5d). But Wm82 roots transformed with the uniquely tagged α‐SNAP‐RAC transgene had substantially elevated RAC transcripts compared with isogenic Wm82 controls, as indicated by a primer pair that amplifies all RAC sequences (native or tagged), and by a primer pair that amplifies only the uniquely tagged α‐SNAP‐RAC transcript (Figure 5d). Controls using template samples prepared without reverse transcriptase verified successful DNAase treatment of cDNA preparations (Figure 5d). We further tested the activity of the RAC promoter itself by constructing a native 4.77 kb RAC element cassette divorced from the flanking Rhg1 DNA (Figure 5e), which we then transformed into Wm82. Similar to Figure 5d, the native 4.77 kb RAC transgene substantially increased RAC transcript abundance in Wm82 roots, relative to empty vector controls (Figure 5f). Together, these findings demonstrate that presence of the rhg1‐a locus RAC can substantially elevate RAC mRNA transcripts and that RAC itself possesses intrinsic promoter activity. The findings suggest that the high RAC transcript abundance observed in “Forrest” (rhg1‐a), but not “Fayette” (rhg1‐b) or Wm 82 (single‐copy Rhg1), is likely to be derived from the rhg1‐a locus RAC insertion. These findings also support the possibility that RAC may retain the potential to promote transposition.
3.8. α‐SNAPRhg1LC protein is expressed despite RAC presence
TEs can influence the expression of host genes. Because RAC is present in rhg1‐a accessions previously chosen for use in agricultural breeding due to their strong SCN resistance, RAC presence may benefit rhg1‐a‐containing soybeans. In light of RAC’s anti‐sense orientation and close proximity to the Rhg1 α‐SNAPRhg1LC promoter, we sought to examine if RAC influences α‐SNAPRhg1LC protein expression. We were not able to compare expression of α‐SNAPRhg1LC between native rhg1‐a loci that do or do not contain RAC, because no rhg1‐a accessions without RAC have been identified and deleting RAC from all Rhg1 repeats of a rhg1‐a accession would not be trivial. Therefore, we left intact or removed the 4.77 kb RAC insertion from the native α‐SNAP‐RAC construct used for Figure 5 (Figure 6a) and then examined α‐SNAPRhg1LC protein abundance in transgenic Wm82 roots carrying the respective constructs. Immunoblotting was conducted using previously described α‐SNAPRhg1LC‐specific and WT α‐SNAP‐specific antibodies (Bayless et al., 2016). The results of a representative experiment are shown in Figure 6b. Across multiple experiments containing independently transformed roots, the constitutive expression of α‐SNAPRhg1LC protein was highly variable, regardless of RAC presence/absence. However, with respect to constitutive expression of α‐SNAPRhg1LC protein we observed no requirement for RAC nor any obvious detrimental impact of RAC (Figure 6b).
Figure 6.
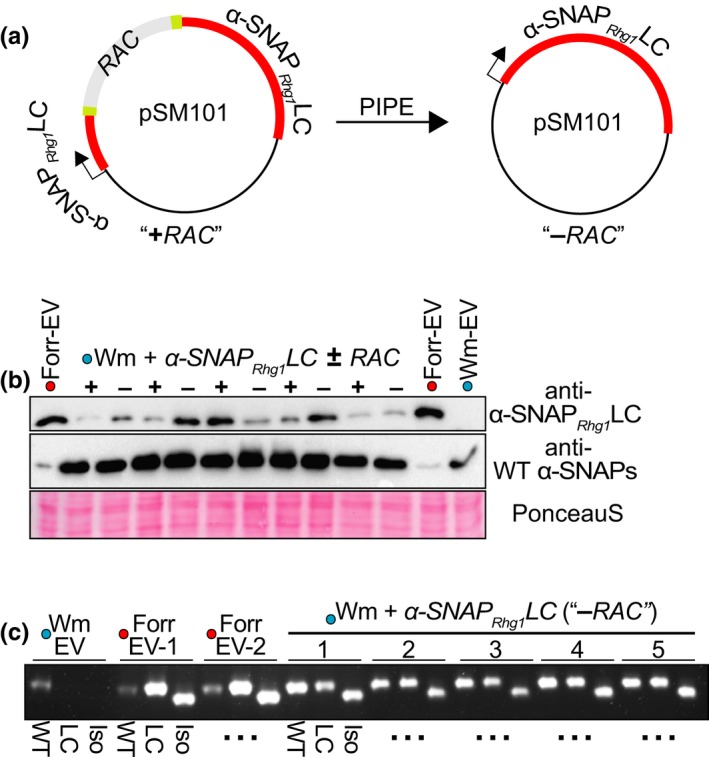
α‐SNAPRhg1LC protein is expressed despite RAC presence. (a) Schematic showing PIPE‐mediated removal of RAC from the native α‐SNAP‐RAC construct, pSM101. (b) Immunoblots of independent “Forrest” or Wm82 transgenic root lysates using previously described antibodies for α‐SNAPRhg1LC or WT α‐SNAP proteins. “+” denotes α‐SNAP‐RAC transformation, “−” indicates transformation with α‐SNAPRhg1LC (RAC removed), and EV is transformed with empty vector. Ponceau S staining serves as a loading control. (c) Agarose gel showing RT‐PCR amplification of mature α‐SNAPRhg1LC transcript isoforms from roots of Wm 82 or “Forrest” transformed with α‐SNAP‐RAC (+), or a native α‐SNAPRhg1LC cassette with RAC removed (−), or an empty vector control. WT refers to primers specific for WT α‐SNAP transcripts, LC detects full‐length α‐SNAPRhg1LC transcripts, while “Iso” amplifies a previously described α‐SNAPRhg1LC alternative transcript isoform that splices out 36 bp (Cook et al., 2014). "...": same WT‐LC‐Iso pattern
Previously, we reported that native α‐SNAPRhg1LC mRNA transcripts include an alternative splice product (Bayless et al., 2016; Cook et al., 2014). Because TEs can influence host mRNA splicing (Krom, Recla, & Ramakrishna, 2008), we also examined how RAC influenced splicing of the known α‐SNAPRhg1LC alternative transcript. As above, we generated transgenic roots of Wm82 containing either a RAC + α‐SNAPRhg1LC native genomic segment or a version with RAC precisely deleted (Figure 6a). We then generated cDNAs and performed RT‐PCR with primer sets specific for either the full‐length or shorter splice isoform. As shown by agarose gel electrophoresis, RAC presence was not required for alternate splicing of this α‐SNAPRhg1LC isoform (Figure 6c).
3.9. α‐SNAPRhg1LC hyperaccumulates at SCN infection sites
We also examined infection‐associated α‐SNAPRhg1LC protein expression in non‐transgenic soybean roots that carry the native rhg1‐a locus. We previously reported that during rhg1‐b‐mediated SCN resistance, α‐SNAPRhg1HC abundance is elevated ~12‐fold within syncytial cells (SCN feeding sites) relative to adjacent non‐syncytial cells (Bayless et al., 2016). To test whether the RAC‐containing rhg1‐a follows a similar expression pattern during the resistance response, the present study examined α‐SNAPRhg1LC abundance at SCN infection sites in soybean variety “Forrest” using SDS‐PAGE and immunoblots. Using the aforementioned α‐SNAPRhg1LC‐specific antibody, we detected increased α‐SNAPRhg1LC accumulation within tissues enriched for SCN feeding sites, while expression was barely detectable in mock‐treated roots (Figure 7a). As previously reported, NSF proteins were also increased in SCN‐infested roots, albeit less prominently (Figure 7a) (Bayless et al., 2016). Thus, even in RAC presence, rhg1‐a haplotypes drive an expression pattern of α‐SNAPRhg1LC similar to that observed for rhg1‐b and α‐SNAPRhg1HC.
Figure 7.
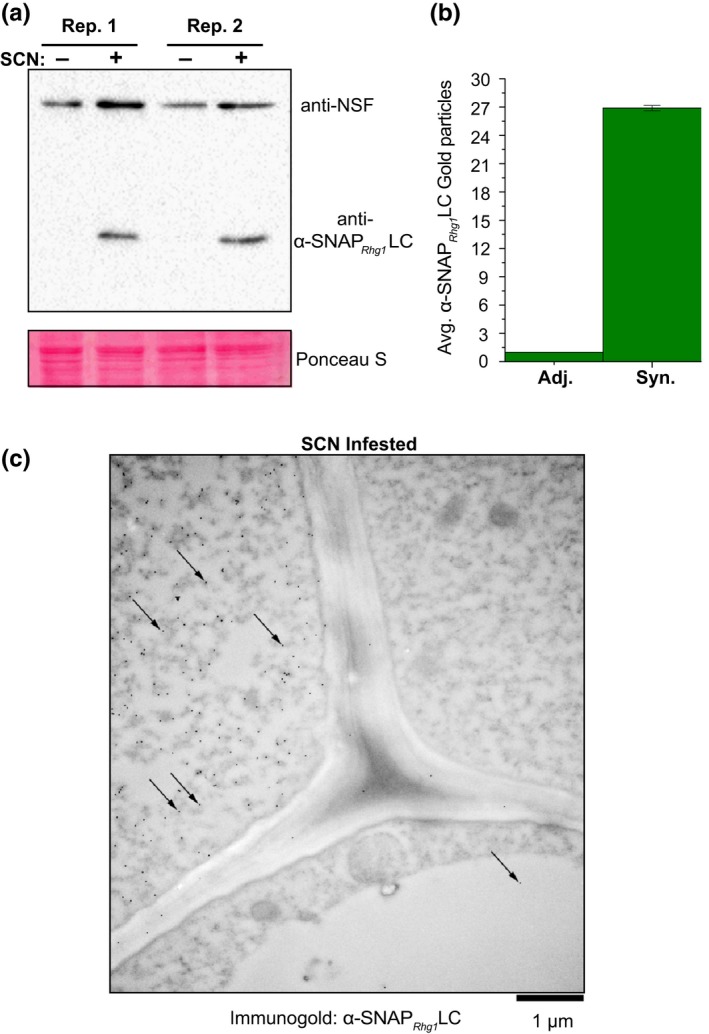
α‐SNAPRhg1LC hyperaccumulates at SCN infection sites in low‐copy rhg1‐a soybean accession “Forrest.” (a) Immunoblot of non‐transgenic “Forrest” root samples from SCN‐infested root regions (SCN +) harvested 4 days after SCN infection, or similar regions from mock‐inoculated controls (SCN −). Blot was probed simultaneously with anti‐α‐SNAPRhg1LC and anti‐NSF polyclonal antibodies. Ponceau S staining before blotting served as a loading control. (b) Representative electron microscope image (7 dpi) showing anti‐α‐SNAPRhg1LC immunogold signal in SCN‐associated syncytium cells from “Forrest” roots. Arrows highlight only some of the 15 nm immunogold particle dots. Frequent α‐SNAPRhg1LC signal was observed in syncytium cells (upper left, “Syn”) but rare in adjacent cells (upper right and bottom, “Adj.”). CW, cell wall; M, mitochondrion; Vac, vacuole. Bar = 1 µm. (c) Mean and SEM of α‐SNAPRhg1LC gold particle abundance in syncytia, normalized to the count from adjacent cells in the same image. Anti‐α‐SNAPRhg1LC immunogold particles were counted for one 9 µm2 area within cells having syncytium morphology and in a region with the highest observable signal in directly adjacent cells with normal root cell morphology (large central vacuole). Data are for 23 images (11 and 12 root sections, respectively, from two experiments), for root sections 7 days after inoculation
To more precisely locate the α‐SNAPRhg1LC increases, SCN‐infested root sections were imaged using transmission electron microscopy and immunogold labeling of bound α‐SNAPRhg1LC‐specific antibody. Syncytium‐specific accumulation of α‐SNAPRhg1LC protein was observed and quantified in root sections taken 7 days after SCN inoculation (Figure 7b,c). The average increase of immunogold particles per equal area of adjacent non‐syncytial root cells (cells still carrying a large central vacuole) was ~25‐fold (Figure 7c). In control experiments, EM sections from mock‐inoculated roots (no SCN) exhibited no immunogold signal above background (Figure S7a). Similarly, no immunogold signal above background was observed when secondary antibody and all other reagents were used but the primary antibody was omitted (Figure S7b). The specificity of the antibody for α‐SNAPRhg1LC protein was previously demonstrated (signal for recombinant α‐SNAPRhg1LC protein or total protein from roots with rhg1‐a, no signal for α‐SNAPRhg1HC protein or total protein from roots with rhg1‐b or Rhg1WT) (Bayless et al., 2016). The above results, similar to the previously observed ~12‐fold increase reported for the α‐SNAPRhg1HC in syncytia from rhg1‐b roots, indicate that α‐SNAPRhg1LC protein abundance is also elevated within syncytia upon SCN infection (Bayless et al., 2016). Collectively, these findings demonstrate that while a potentially active retrotransposon (RAC) has integrated within the important rhg1‐a α‐SNAPRhg1LC resistance gene, and its presence correlates with rhg1‐a haplotypes preferred for SCN‐resistance breeding, no negative impacts of RAC on α‐SNAPRhg1LC mRNA or protein expression were detected.
4. DISCUSSION
Rhg1 is the principal SCN‐resistance locus in commercially grown soybeans. The increasing occurrence of SCN populations that at least partially overcome the overwhelmingly utilized “PI 88788‐type” rhg1‐b resistance source is an important concern for soybean breeders and growers (McCarville, Marett, Mullaney, Gebhart, & Tylka, 2017), (www.thescncoalition.com). Alternating use of different Rhg1 haplotypes should help bolster and preserve resistance against these virulent SCN populations (Brucker et al., 2005; Niblack et al., 2008)(www.thescncoalition.com). In this study we report that the other Rhg1 haplotype available for SCN control, rhg1‐a (also known as “Peking‐type” Rhg1), carries a distinct genetic structure. rhg1‐a unexpectedly contains an intact and transcriptionally active retrotransposon within an intron of the key Rhg1 α‐SNAP resistance gene in each repeat. The “Hartwig‐type” SCN resistance from PI 437654 also carries the rhg1‐a haplotype and also carries the RAC retrotransposon within the Rhg1 α‐SNAP genes.
Transposons have been coopted for the service of defense responses in both plants and animals (Huang et al., 2016; Tsuchiya & Eulgem, 2013). V(D)J recombination, which underlies the remarkable diversity of vertebrate adaptive immunity, apparently derives from a domesticated RAG‐family transposase (Huang et al., 2016). RAC has inherent transcriptional activity and is positioned anti‐sense within the first intron of the rhg1‐a α‐SNAP gene. It is unclear if RAC impacts rhg1‐a function (discussed below). However, this study revealed the utility of RAC and the RAC‐associated ss715606985 (G to A) SNP in more accurately identifying SCN resistance‐conferring rhg1‐a germplasm. Among the 19,645 USDA soybean accessions genotyped using the SoySNP50K iSelect BeadChip (Song, Keppler, et al., 2015; Song, Hyten, et al., 2015), a few hundred accessions with a rhg1a‐type SNP signature apparently do not actually carry a rhg1‐a locus. All of the rhg1‐a signature soybeans we examined that do not carry RAC encoded rhg1‐b or Rhg1WT α‐SNAP alleles. Conversely, all examined rhg1‐a signature soybeans with RAC carried the rhg1‐a α‐SNAP allele.
Active retrotransposon families are abundant in soybean (Wawrzynski et al., 2008). RAC has similarities to a copia element near a Phytophthora sojae resistance locus identified by Bhattacharyya et al., however, SoyTE database searches returned no highly similar RAC‐family TEs (Bhattacharyya et al., 1997; Du, Grant, et al., 2010; Du, Tian, et al., 2010). Although WGS studies previously examined the HG type test soybean accessions, the RAC insertion within α‐SNAPRhg1LC was apparently omitted during the filtering steps of DNA sequence read mapping and assembly (Cook et al., 2014; Liu et al., 2017). Our findings revealed multiple RAC‐family elements in soybean, and this abundance of RAC‐family elements likely led to RAC omission from previous rhg1‐a sequence assemblies. It is intriguing that RAC, at least from PI 89772, has inherent transcriptional activity and an intact ORF encoding conserved functional motifs—features of an autonomous element. Additionally, RAC’s near‐perfect identity with the Chr10 element supports that RAC‐family retrotransposons were recently active in soybean.
Host silencing of TEs can establish cis‐regulatory networks where the expression of nearby host genes may also be impacted (Lisch & Bennetzen, 2011; McCue et al., 2012; McCue & Slotkin, 2012). In some cases, biotic stresses can influence transposon methylation, and subsequently, alter the expression of host genes near transposons (Dowen et al., 2012). We noted DNA methylation at the α‐SNAP‐RAC junctions. However, we also found evidence that RAC is transcriptionally active. In addition, the Rhg1 α‐SNAP gene that contains RAC successfully increases expression of the α‐SNAPRhg1LC protein during SCN resistance, similarly to that observed for α‐SNAPRhg1HC. Future analyses of small RNAs may provide evidence of differential silencing of RAC or α‐SNAPRhg1LC. Cyst nematode infection of Arabidopsis has been reported to trigger the hypomethyation and activation of certain TEs, and moreover, many of these TEs reside near host genes whose expression is altered during syncytium establishment (Hewezi et al., 2017; Piya, Bennett, Rambani, & Hewezi, 2017). Thus, it remains an intriguing hypothesis that RAC may influence the epigenetic landscape of α‐SNAPRhg1LC or the overall ~36 kb rhg1‐a repeat during infection, particular stresses, developmental stages or in specific tissues. Moreover, small RNAs deriving from the other RAC‐like elements in the soybean genome could modulate α‐SNAPRhg1LC expression in trans (McCue et al., 2012; McCue, Nuthikattu, & Slotkin, 2013; Slotkin & Martienssen, 2007). Our BLAST searches revealed at least two other RAC‐like elements positioned intronically or adjacent to putative host defense and/or developmental genes. Future studies may interrogate if RAC, and/or other endogenous retrotransposons, impacts the regulation of host defense gene networks in soybean.
The currently available picture of Rhg1 haplotype evolution is incomplete and has been dominated by study of lines that are the product of ongoing selection for the most effective SCN resistance (e.g., modern 10‐copy rhg1‐b and 3‐copy rhg1‐a haplotypes). The finding of RAC in all copies of the rhg1‐a repeat, and in all confirmed rhg1‐a haplotypes that were tested to date, suggests but does not confirm that RAC plays an adaptive role in those haplotypes. The sequence of the Rhg1 repeat junction is identical between rhg1‐b and rhg1‐a haplotypes, as are many SNPs not present in the Williams 82 soybean reference genome, providing evidence of the shared evolutionary origin of rhg1‐b and rhg1‐a (Cook et al., 2012). The finding to date of RAC only in rhg1‐a haplotypes suggests that this retroelement probably inserted in Rhg1 after the divergence of rhg1‐b and rhg1‐a. However, it is also possible that the RAC retroelement was ancestrally present but then purged from the progenitors of current rhg1‐b accessions. A correlation has been demonstrated between Rhg1 copy number and SCN resistance, and rhg1‐a in the absence of Rhg4 confers only partial SCN resistance (Cook et al., 2014; Kandoth et al., 2017; Lee, Diers, & Hudson, 2016; Liu et al., 2012; Yu et al., 2016). Yet there are no known instances of rhg1‐a accessions with an Rhg1 copy number above three. The RAC may allow increased and/or more tightly regulated expression of rhg1‐a. Alternatively, it is possible that absence of RAC is advantageous in allowing increased copy number of Rhg1, but that too is only a hypothesis, raised by the present work and in need of future testing. Additional questions about Rhg1 locus evolution remain that have functional implications for the efficacy of SCN resistance. For example, we know of no rhg1‐a haplotypes that carry an α‐SNAPRhg1WT‐encoding Rhg1 repeat, which all examined rhg1‐b haplotypes do contain. Might RAC acquisition have influenced this absence of the WT Rhg1 repeat? Do any α‐SNAPRhg1LC‐expressing accessions exist that do not carry the RAC integration? The USDA soybean collection contains numerous accessions that are positive for NSFRAN07 but which carry Rhg1 copy numbers below 3 or 10, or have slight deviations from consensus rhg1‐a or rhg1‐b SNP signatures. Intensive study of these accessions may shed further light on Rhg1 haplotype evolution and, moreover, may facilitate the discovery of new and agriculturally useful Rhg1 alleles.
The finding that popular rhg1‐a breeding sources contain an intact retrotransposon within α‐SNAPRhg1LC was surprising, given that these accessions have previously been sequenced multiple times by different groups (Cook et al., 2014; Liu et al., 2017; Patil et al., 2019). The RAC‐SNP is rare among all USDA accessions, and approximately 700 of 19,645 USDA accessions carry a SoySNP50K signature predictive of an rhg1‐a haplotype. However, ~400 of these putative rhg1‐a accessions do not carry the RAC‐SNP and all of the non‐RAC putative rhg1‐a accessions that we sampled did not encode α‐SNAPRhg1LC, indicating that they are not true rhg1‐a. Accordingly, most of these non‐RAC accessions scored phenotypically as SCN‐susceptible. Taken together with our PCR assays showing perfect correlation of the RAC‐SNP with RAC presence, our findings indicate that the RAC‐SNP successfully identifies true rhg1‐a loci (i.e., those that encode the α‐SNAPRhg1LC protein) which, in combination with Rhg4 and other loci, confers strong SCN resistance.
Correlation of SNP data with previously published SCN‐resistance phenotype data recorded in the GRIN database indicated that the vast majority of “R” scoring rhg1‐a signature accessions were RAC +. Many of the 705 accessions postulated (using the earlier SoySNP50K SNP signature) to be lines that carry rhg1‐a turned out to carry rhg1‐b, which would explain their resistance to SCN. The large majority of the subset that are not positive for RAC were scored as SCN‐susceptible or moderately susceptible. Some RAC + lines also were scored as SCN‐susceptible or moderately susceptible, but most of these are apparently due to the absence of a resistance‐associated Rhg4 and/or the Chr 11 α‐SNAP intron‐retention allele (α‐SNAPCh11‐IR), consistent with the established contributions of those loci to SCN resistance (Kandoth et al., 2017; Meksem et al., 2001; Yu et al., 2016). Among the “R” scoring RAC+ accessions, SNPs genetically linked to Rhg4 and the Chr 11 α‐SNAP intron‐retention allele were substantially elevated. The five lines from the GRIN database that had the SoySNP50K rhg1‐a SNP signature and scored as “R” (SCN‐resistant) despite lacking RAC merit future investigation. They may be a combination of plants mis‐scored as “R,” plants that unexpectedly carry rhg1‐b or rhg1‐b‐derived variants despite their SNP signature, plants with novel rhg1 alleles, and/or plants that carry combinations of other SCN‐resistance QTLs including possible novel QTLs.
Potential modulation of α‐SNAPRhg1LC expression by RAC, either during the SCN‐resistance response or in certain developmental or stress situations, could benefit rhg1‐a‐containing soybeans that have significantly depleted WT α‐SNAP proteins. α‐SNAPs, together with NSF, carry out essential eukaryotic housekeeping functions by maintaining SNARE proteins for vesicle trafficking. Notably, among true rhg1‐a accessions, the abundance of wild‐type (WT) α‐SNAP proteins is sharply diminished, as compared to rhg1‐b‐ or SCN‐susceptible soybeans (Bayless et al., 2018). Moreover, α‐SNAPRhg1LC protein was shown to be cytotoxic in Nicotiana benthamiana while WT α‐SNAP co‐expression alleviated this toxicity (Bayless et al., 2016). The more recent discovery of the NSFRAN07 allele as a requisite for the viability of Rhg1 soybeans further underscores the necessity of the SNARE‐recycling machinery for overall plant health (Bayless et al., 2018). The intronic copia element within the Arabidopsis RPP7 (Recognition of Peronospora Parasitica 7) gene serves as an example of a retroelement with immunomodulatory function, having been shown to modulate RPP7 transcript splicing and expression (Tsuchiya & Eulgem, 2013). However, we did not detect any influence of RAC on constitutive α‐SNAPRhg1LC protein expression from transgenes delivered to roots. We also did not detect a functional influence of RAC in its native rhg1‐a haplotype context, insofar as that α‐SNAPRhg1LC protein abundance is successfully elevated in syncytia similarly to the reported syncytium elevation of the α‐SNAPRhg1HC protein of rhg1‐b haplotypes, which do not carry RAC (Bayless et al., 2016). Hence although we did not find any true rhg1‐a soybeans without RAC integrations, it remains possible that RAC integration was a neutral event that confers no host advantage or disadvantage.
SCN causes the most yield loss of any disease for U.S. soybean farmers, and rhg1‐a offers a potential solution to SCN populations that overcome commonly used rhg1‐b resistance sources (Allen, 2017; Brucker et al., 2005; Niblack et al., 2008). Findings continue to emerge that further characterize different sources of SCN resistance, including exciting new findings regarding copy number variation at Rhg4 (Patil et al., 2019). An attractive overall hypothesis for future study of RAC is that, in the presence of SCN or other stresses, RAC provides an additional regulatory layer to optimize the SCN‐resistance response mediated by rhg1‐a and Rhg4, and/or promotes plant health in the absence of SCN. By revealing the existence of RAC within the important rhg1‐a haplotype, the present study provides a marker for finding such soybeans and expands our knowledge regarding the genetic structure and divergence of the agriculturally valuable Rhg1 source of SCN resistance.
CONFLICT OF INTEREST
The authors declare no conflict of interest associated with the work described in this manuscript.
AUTHOR CONTRIBUTIONS
A.M.B., R.W.Z., S.H., and A.F.B designed the research; A.M.B., R.W.Z., S.H., D.J.G., and K.K.A. performed the research; all authors analyzed the data and contributed to writing the paper that was drafted primarily by A.M.B.
Supporting information
ACKNOWLEDGMENTS
This work was supported by USDA‐NIFA‐AFRI award 2014‐67013‐21775 and United Soybean Board award 1920‐172‐0122‐B to A.F.B. The work was also supported by the National Science Foundation Graduate Research Fellowship under Grant No. (DGE‐1256259) to A.M.B. We thank anonymous reviewers for their comments on a previous version of this manuscript (submitted for review December 2018).
Bayless AM, Zapotocny RW, Han S, Grunwald DJ, Amundson KK, Bent AF. The rhg1‐a (Rhg1 low‐copy) nematode resistance source harbors a copia‐family retrotransposon within the Rhg1‐encoded α‐SNAP gene. Plant Direct. 2019;3:1–19. 10.1002/pld3.164
Adam M. Bayless and Ryan W. Zapotocny contributed similarly.
This manuscript was previously deposited as a preprint at https://www.biorxiv.org/content/10.1101/653568v1
REFERENCES
- Allen, T. W. , Bradley, C. A. , Sisson, A. J. , Byamukama, E. , Chilvers, M. I. , Coker, C. M. , … Wrather, J. A. (2017). Soybean yield loss estimates due to diseases in the United States and Ontario, Canada, from 2010 to 2014. Plant Health Progress, 18(1), 19–27. 10.1094/PHP-RS-16-0066 [DOI] [Google Scholar]
- Anand, S. C. , & Gallo, K. M. (1984) Identification of additional soybean germ plasm with resistance to race 3 of the soybean cyst nematode. Plant Disease, 68(7), 593– 10.1094/PD-69-593 [DOI] [Google Scholar]
- Arelli, P. R. , Sleper, D. A. , Yue, P. , & Wilcox, J. A. (2000). Soybean reaction to races 1 and 2 of Heterodera glycines. Contribution of the Missouri Agric. Exp. Stn. Journal Series No. 12,751. Crop Science, 40, 824–826. 10.2135/cropsci2000.403824x [DOI] [Google Scholar]
- Bayless, A. M. , Smith, J. M. , Song, J. , McMinn, P. H. , Teillet, A. , August, B. K. , & Bent, A. F. (2016). Disease resistance through impairment of alpha‐SNAP‐NSF interaction and vesicular trafficking by soybean Rhg1 . Proceedings of the National Academy of Sciences of the United States of America, 113, E7375–E7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless, A. M. , Zapotocny, R. W. , Grunwald, D. J. , Amundson, K. K. , Diers, B. W. , & Bent, A. F. (2018). An atypical N‐ethylmaleimide sensitive factor enables the viability of nematode‐resistant Rhg1 soybeans. Proceedings of the National Academy of Sciences of the United States of America, 115, E4512–E4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg, J. A. , Appiano, M. , Santillan Martinez, M. , Hermans, F. W. , Vriezen, W. H. , Visser, R. G. , … Schouten, H. J. (2015). A transposable element insertion in the susceptibility gene CsaMLO8 results in hypocotyl resistance to powdery mildew in cucumber. BMC Plant Biology, 15, 243 10.1186/s12870-015-0635-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya, M. K. , Gonzales, R. A. , Kraft, M. , & Buzzell, R. I. (1997). A copia‐like retrotransposon Tgmr closely linked to the Rps1‐k allele that confers race‐specific resistance of soybean to Phytophthora sojae . Plant Molecular Biology, 34, 255–264. [DOI] [PubMed] [Google Scholar]
- Brucker, E. , Carlson, S. , Wright, E. , Niblack, T. , & Diers, B. (2005). Rhg1 alleles from soybean PI 437654 and PI 88788 respond differentially to isolates of Heterodera glycines in the greenhouse. TAG. Theoretical and Applied Genetics, 111, 44–49. [DOI] [PubMed] [Google Scholar]
- Cavrak, V. V. , Lettner, N. , Jamge, S. , Kosarewicz, A. , Bayer, L. M. , & Mittelsten Scheid, O. (2014). How a retrotransposon exploits the plant's heat stress response for its activation. PLoS Genetics, 10, e1004115 10.1371/journal.pgen.1004115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concibido, V. C. , Diers, B. W. , & Arelli, P. R. (2004). A decade of QTL mapping for cyst nematode resistance in soybean. Crop Science, 44, 1121–1131. 10.2135/cropsci2004.1121 [DOI] [Google Scholar]
- Cook, D. E. , Bayless, A. M. , Wang, K. , Guo, X. , Song, Q. , Jiang, J. , & Bent, A. F. (2014). Distinct copy number, coding sequence, and locus methylation patterns underlie Rhg1‐mediated soybean resistance tosoybean cyst nematode. Plant Physiology, 165, 630–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, D. E. , Lee, T. G. , Guo, X. , Melito, S. , Wang, K. , Bayless, A. M. , … Bent, A. F. (2012). Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science, 338, 1206–1209. 10.1126/science.1228746 [DOI] [PubMed] [Google Scholar]
- Diers, B. W. , Skorupska, H. T. , Rao‐Arelli, A. P. , & Cianzio, S. R. (1997). Genetic relationships among soybean plant introductions with resistance to soybean cyst nematodes. Crop Science, 37, 1966–1972. 10.2135/cropsci1997.0011183X003700060048x [DOI] [Google Scholar]
- Ding, M. , Ye, W. , Lin, L. , He, S. , Du, X. , Chen, A. , … Rong, J. (2015). The hairless stem phenotype of cotton (Gossypium barbadense) is linked to a copia‐like retrotransposon insertion in a homeodomain‐leucine zipper gene (HD1). Genetics, 201, 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen, R. H. , Pelizzola, M. , Schmitz, R. J. , Lister, R. , Dowen, J. M. , Nery, J. R. , … Ecker, J. R. (2012). Widespread dynamic DNA methylation in response to biotic stress. Proceedings of the National Academy of Sciences of the United States of America, 109, E2183–2191. 10.1073/pnas.1209329109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, J. , Grant, D. , Tian, Z. , Nelson, R. T. , Zhu, L. , Shoemaker, R. C. , & Ma, J. (2010). SoyTEdb: A comprehensive database of transposable elements in the soybean genome. BMC Genomics, 11, 113 10.1186/1471-2164-11-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, J. , Tian, Z. , Hans, C. S. , Laten, H. M. , Cannon, S. B. , Jackson, S. A. , … Ma, J. (2010). Evolutionary conservation, diversity and specificity of LTR‐retrotransposons in flowering plants: Insights from genome‐wide analysis and multi‐specific comparison. The Plant Journal, 63, 584–598. 10.1111/j.1365-313X.2010.04263.x [DOI] [PubMed] [Google Scholar]
- Galindo‐Gonzalez, L. , Mhiri, C. , Deyholos, M. K. , & Grandbastien, M. A. (2017). LTR‐retrotransposons in plants: Engines of evolution. Gene, 626, 14–25. 10.1016/j.gene.2017.04.051 [DOI] [PubMed] [Google Scholar]
- Goodstein, D. M. , Shu, S. , Howson, R. , Neupane, R. , Hayes, R. D. , Fazo, J. , … Rokhsar, D. S. (2012). Phytozome: A comparative platform for green plant genomics. Nucleic Acids Research, 40, D1178–1186. 10.1093/nar/gkr944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker, E. R. , Gao, X. , & Voytas, D. F. (2004). The diversity of LTR retrotransposons. Genome Biology, 5, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henk, A. D. , Warren, R. F. , & Innes, R. W. (1999). A new Ac‐like transposon of Arabidopsis is associated with a deletion of the RPS5 disease resistance gene. Genetics, 151, 1581–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi, T. , Lane, T. , Piya, S. , Rambani, A. , Rice, J. H. , & Staton, M. (2017). Cyst nematode parasitism induces dynamic changes in the root epigenome. Plant Physiology, 174, 405–420. 10.1104/pp.16.01948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S. , Tao, X. , Yuan, S. , Zhang, Y. , Li, P. , Beilinson, H. A. , … Xu, A. (2016). Discovery of an active RAG transposon illuminates the origins of V(D)J recombination. Cell, 166, 102–114. 10.1016/j.cell.2016.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey, R. S. , Boerma, H. R. , Raymer, P. L. , & Luzzi, B. M. (1991). Resistance in soybean cultivars from maturity groups V‐VIII to soybean cyst and root‐knot nematodes. Journal of Nematology, 23, 576–583. [PMC free article] [PubMed] [Google Scholar]
- Jones, D. T. , Taylor, W. R. , & Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Computer Applications in the Biosciences, 8, 275–282. 10.1093/bioinformatics/8.3.275 [DOI] [PubMed] [Google Scholar]
- Kadam, S. , Vuong, T. D. , Qiu, D. , Meinhardt, C. G. , Song, L. , Deshmukh, R. , … Nguyen, H. T. (2016). Genomic‐assisted phylogenetic analysis and marker development for next generation soybean cyst nematode resistance breeding. Plant Science, 242, 342–350. 10.1016/j.plantsci.2015.08.015 [DOI] [PubMed] [Google Scholar]
- Kanazawa, A. , Liu, B. , Kong, F. , Arase, S. , & Abe, J. (2009). Adaptive evolution involving gene duplication and insertion of a novel Ty1/copia‐like retrotransposon in soybean. Journal of Molecular Evolution, 69, 164–175. 10.1007/s00239-009-9262-1 [DOI] [PubMed] [Google Scholar]
- Kandoth, P. K. , Liu, S. , Prenger, E. , Ludwig, A. , Lakhssassi, N. , Heinz, R. , … Mitchum, M. G. (2017). Systematic mutagenesis of serine hydroxymethyltransferase reveals an essential role in nematode resistance. Plant Physiology, 175, 1370–1380. 10.1104/pp.17.00553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M. Y. , & Zilberman, D. (2014). DNA methylation as a system of plant genomic immunity. Trends in Plant Science, 19, 320–326. 10.1016/j.tplants.2014.01.014 [DOI] [PubMed] [Google Scholar]
- Klepadlo, M. , Meinhardt, C. G. , Vuong, T. D. , Patil, G. , Bachleda, N. , Ye, H. , … Nguyen, H. T. (2018). Evaluation of soybean germplasm for resistance to multiple nematode species: Heterodera glycines, Meloidogyne incognita, and Rotylenchulus reniformis . Crop Science, 58, 2511–2522. [Google Scholar]
- Klock, H. E. , & Lesley, S. A. (2009). The polymerase incomplete primer extension (PIPE) method applied to high‐throughput cloning and site‐directed mutagenesis. Methods in Molecular Biology, 498, 91–103. [DOI] [PubMed] [Google Scholar]
- Krom, N. , Recla, J. , & Ramakrishna, W. (2008). Analysis of genes associated with retrotransposons in the rice genome. Genetica, 134, 297–310. 10.1007/s10709-007-9237-3 [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhssassi, N. , Liu, S. , Bekal, S. , Zhou, Z. , Colantonio, V. , Lambert, K. , … Meksem, K. (2017). Characterization of the soluble NSF attachment protein gene family identifies two members involved in additive resistance to a plant pathogen. Scientific Reports, 7, 45226 10.1038/srep45226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T. G. , Diers, B. W. , & Hudson, M. E. (2016). An efficient method for measuring copy number variation applied to improvement of nematode resistance in soybean. The Plant Journal, 88, 143–153. 10.1111/tpj.13240 [DOI] [PubMed] [Google Scholar]
- Lee, T. G. , Kumar, I. , Diers, B. W. , & Hudson, M. E. (2015). Evolution and selection of Rhg1, a copy‐number variant nematode‐resistance locus. Molecular Ecology, 24, 1774–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. , Fennell, T. , Ruan, J. , Homer, N. , … 1000 Genome Project Data Processing Subgroup (2009). The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch, D. (2013). How important are transposons for plant evolution? Nature Reviews Genetics, 14, 49–61. 10.1038/nrg3374 [DOI] [PubMed] [Google Scholar]
- Lisch, D. , & Bennetzen, J. L. (2011). Transposable element origins of epigenetic gene regulation. Current Opinion in Plant Biology, 14, 156–161. 10.1016/j.pbi.2011.01.003 [DOI] [PubMed] [Google Scholar]
- Liu, J. , He, Y. , Amasino, R. , & Chen, X. (2004). siRNAs targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis. Genes & Development, 18, 2873–2878. 10.1101/gad.1217304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Kandoth, P. K. , Lakhssassi, N. , Kang, J. , Colantonio, V. , Heinz, R. , … Meksem, K. (2017). The soybean GmSNAP18 gene underlies two types of resistance to soybean cyst nematode. Nature Communications, 8, 14822 10.1038/ncomms14822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Kandoth, P. K. , Warren, S. D. , Yeckel, G. , Heinz, R. , Alden, J. , … Meksem, K. (2012). A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature, 492, 256–260. 10.1038/nature11651 [DOI] [PubMed] [Google Scholar]
- Makarevitch, I. , Waters, A. J. , West, P. T. , Stitzer, M. , Hirsch, C. N. , Ross‐Ibarra, J. , & Springer, N. M. (2015). Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genetics, 11, e1004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga, W. , Kobayashi, A. , Kato, A. , & Ito, H. (2012). The effects of heat induction and the siRNA biogenesis pathway on the transgenerational transposition of ONSEN, a copia‐like retrotransposon in Arabidopsis thaliana . Plant and Cell Physiology, 53, 824–833. 10.1093/pcp/pcr179 [DOI] [PubMed] [Google Scholar]
- Matsunaga, W. , Ohama, N. , Tanabe, N. , Masuta, Y. , Masuda, S. , Mitani, N. , … Ito, H. (2015). A small RNA mediated regulation of a stress‐activated retrotransposon and the tissue specific transposition during the reproductive period in Arabidopsis. Frontiers in Plant Science, 6, 48 10.3389/fpls.2015.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsye, P. D. , Lawrence, G. W. , Youssef, R. M. , Kim, K. H. , Lawrence, K. S. , Matthews, B. F. , & Klink, V. P. (2012). The expression of a naturally occurring, truncated allele of an alpha‐SNAP gene suppresses plant parasitic nematode infection. Plant Molecular Biology, 80, 131–155. [DOI] [PubMed] [Google Scholar]
- McCarville, M. T. , Marett, C. C. , Mullaney, M. P. , Gebhart, G. D. , & Tylka, G. L. (2017). Increase in soybean cyst nematode virulence and reproduction on resistant soybean varieties in Iowa from 2001 to 2015 and the effects on soybean yields. Plant Health Progress, 18, 146–155. 10.1094/PHP-RS-16-0062 [DOI] [Google Scholar]
- McCue, A. D. , Nuthikattu, S. , Reeder, S. H. , & Slotkin, R. K. (2012). Gene expression and stress response mediated by the epigenetic regulation of a transposable element small RNA. PLoS Genetics, 8, e1002474 10.1371/journal.pgen.1002474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCue, A. D. , Nuthikattu, S. , & Slotkin, R. K. (2013). Genome‐wide identification of genes regulated in trans by transposable element small interfering RNAs. RNA Biology, 10, 1379–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCue, A. D. , & Slotkin, R. K. (2012). Transposable element small RNAs as regulators of gene expression. Trends in Genetics, 28, 616–623. 10.1016/j.tig.2012.09.001 [DOI] [PubMed] [Google Scholar]
- McHale, L. K. , Haun, W. J. , Xu, W. W. , Bhaskar, P. B. , Anderson, J. E. , Hyten, D. L. , … Stupar, R. M. (2012). Structural variants in the soybean genome localize to clusters of biotic stress‐response genes. Plant Physiology, 159, 1295–1308. 10.1104/pp.112.194605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meksem, K. , Pantazopoulos, P. , Njiti, V. , Hyten, L. , Arelli, P. , & Lightfoot, D. (2001). ’Forrest’resistance to the soybean cyst nematode is bigenic: Saturation mapping of the Rhg1and Rhg4 loci. Theoretical and Applied Genetics, 103, 710–717. 10.1007/s001220100597 [DOI] [Google Scholar]
- Mitchum, M. G. (2016). Soybean resistance to the soybean cyst nematode heterodera glycines: An update. Phytopathology, 106, 1444–1450. [DOI] [PubMed] [Google Scholar]
- Negi, P. , Rai, A. N. , & Suprasanna, P. (2016). Moving through the stressed genome: Emerging regulatory roles for transposons in plant stress response. Frontiers in Plant Science, 7, 1448 10.3389/fpls.2016.01448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niblack, T. L. , Arelli, P. R. , Noel, G. R. , Opperman, C. H. , Orf, J. H. , Schmitt, D. P. , … Tylka, G. L. (2002). A revised classification scheme for genetically diverse populations of Heterodera glycines . Journal of Nematology, 34, 279–288. [PMC free article] [PubMed] [Google Scholar]
- Niblack, T. , Colgrove, A. , Colgrove, K. , & Bond, J. (2008). Shift in virulence of soybean cyst nematode is associated with use of resistance from PI 88788. Plant Health Progress, 9(1), 29– 10.1094/PHP-2008-0118-01-RS [DOI] [Google Scholar]
- Niblack, T. L. , Lambert, K. N. , & Tylka, G. L. (2006). A model plant pathogen from the kingdom Animalia: Heterodera glycines, the soybean cyst nematode. Annual Review of Phytopathology, 44, 283–303. [DOI] [PubMed] [Google Scholar]
- Patil, G. B. , Lakhssassi, N. , Wan, J. , Song, L. , Zhou, Z. , Klepadlo, M. , … Nguyen, H. T. (2019). Whole genome re‐sequencing reveals the impact of the interaction of copy number variants of the rhg‐1 and Rhg4 genes on broad‐based resistance to soybean cyst nematode. Plant Biotechnology Journal, 17, 1595‐1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson‐Burch, B. D. , & Voytas, D. F. (2002). Genes of the pseudoviridae (Ty1/copia retrotransposons). Molecular Biology and Evolution, 19, 1832–1845. 10.1093/oxfordjournals.molbev.a004008 [DOI] [PubMed] [Google Scholar]
- Piya, S. , Bennett, M. , Rambani, A. , & Hewezi, T. (2017). Transcriptional activity of transposable elements may contribute to gene expression changes in the syncytium formed by cyst nematode in arabidopsis roots. Plant Signaling & Behavior, 12, e1362521 10.1080/15592324.2017.1362521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt, H. J. , Pühler, A. , & Broer, I. (1993). Transgenic root nodules of Vicia hirsuta: A fast and efficient system for the study of gene expression in indeterminate‐type nodules. MPMI‐Molecular Plant Microbe Interactions, 6, 699–706. [Google Scholar]
- RStudio Team (2015). RStudio: Integrated Development for R. Boston, MA: RStudio, Inc. Retrieved from http://www.rstudio.com/ [Google Scholar]
- Schmutz, J. , Cannon, S. B. , Schlueter, J. , Ma, J. , Mitros, T. , Nelson, W. , … Jackson, S. A. (2010). Genome sequence of the palaeopolyploid soybean. Nature, 463, 178–183. 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- Shi, Z. , Liu, S. , Noe, J. , Arelli, P. , Meksem, K. , & Li, Z. (2015). SNP identification and marker assay development for high‐throughput selection of soybean cyst nematode resistance. BMC Genomics, 16, 314 10.1186/s12864-015-1531-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigman, M. J. , & Slotkin, R. K. (2016). The first rule of plant transposable element silencing: Location, location, location. The Plant Cell, 28, 304–313. 10.1105/tpc.15.00869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin, R. K. , & Martienssen, R. (2007). Transposable elements and the epigenetic regulation of the genome. Nature Reviews Genetics, 8, 272–285. 10.1038/nrg2072 [DOI] [PubMed] [Google Scholar]
- Song, J. , Keppler, B. D. , Wise, R. R. , & Bent, A. F. (2015). PARP2 Is the predominant poly(ADP‐ribose) polymerase in Arabidopsis DNA damage and immune responses. PLoS Genetics, 11, e1005200 10.1371/journal.pgen.1005200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Q. , Hyten, D. L. , Jia, G. , Quigley, C. V. , Fickus, E. W. , Nelson, R. L. , & Cregan, P. B. (2015) Fingerprinting soybean germplasm and its utility in genomic research. G3 (Bethesda), 5, 1999–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof, T. C. , & Rothman, J. E. (2009). Membrane fusion: Grappling with SNARE and SM proteins. Science, 323, 474–477. 10.1126/science.1161748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Nei, M. , & Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor‐joining method. Proceedings of the National Academy of Sciences, 101, 11030–11035. 10.1073/pnas.0404206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Z. , Zhao, M. , She, M. , Du, J. , Cannon, S. B. , Liu, X. , … Ma, J. (2012). Genome‐wide characterization of nonreference transposons reveals evolutionary propensities of transposons in soybean. The Plant Cell, 24, 4422–4436. 10.1105/tpc.112.103630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, T. , & Eulgem, T. (2013). An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proceedings of the National Academy of Sciences, 110, E3535–3543. 10.1073/pnas.1312545110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong, T. D. , Sonah, H. , Meinhardt, C. G. , Deshmukh, R. , Kadam, S. , Nelson, R. L. , … Nguyen, H. T. (2015). Genetic architecture of cyst nematode resistance revealed by genome‐wide association study in soybean. BMC Genomics, 16, 593 10.1186/s12864-015-1811-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzynski, A. , Ashfield, T. , Chen, N. W. , Mammadov, J. , Nguyen, A. , Podicheti, R. , … Innes, R. W. (2008). Replication of nonautonomous retroelements in soybean appears to be both recent and common. Plant Physiology, 148, 1760–1771. 10.1104/pp.108.127910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. (2009). Ggplot2: Elegant graphics for data analysis. New York, NY: Springer. [Google Scholar]
- Woodrow, P. , Pontecorvo, G. , Fantaccione, S. , Fuggi, A. , Kafantaris, I. , Parisi, D. , & Carillo, P. (2010). Polymorphism of a new Ty1‐copia retrotransposon in durum wheat under salt and light stresses. Theoretical and Applied Genetics, 121, 311–322. 10.1007/s00122-010-1311-z [DOI] [PubMed] [Google Scholar]
- Xiao, H. , Jiang, N. , Schaffner, E. , Stockinger, E. J. , & van der Knaap, E. (2008). A retrotransposon‐mediated gene duplication underlies morphological variation of tomato fruit. Science, 319, 1527–1530. 10.1126/science.1153040 [DOI] [PubMed] [Google Scholar]
- Young, L. D. (1995). Soybean germplasm resistant to Races 3, 5, or 14 of soybean cyst nematode. Crop Science, 35, 895–896. 10.2135/cropsci1995.0011183X003500030044x [DOI] [Google Scholar]
- Yu, N. , Lee, T. G. , Rosa, D. P. , Hudson, M. , & Diers, B. W. (2016). Impact of Rhg1 copy number, type, and interaction with Rhg4 on resistance to Heterodera glycines in soybean. Theoretical and Applied Genetics, 129, 2403–2412. 10.1007/s00122-016-2779-y [DOI] [PubMed] [Google Scholar]
- Zhao, M. , & Ma, J. (2013). Co‐evolution of plant LTR‐retrotransposons and their host genomes. Protein Cell, 4, 493–501. 10.1007/s13238-013-3037-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M. , Wu, S. , Zhou, Q. , Vivona, S. , Cipriano, D. J. , Cheng, Y. , & Brunger, A. T. (2015). Mechanistic insights into the recycling machine of the SNARE complex. Nature, 518, 61–67. 10.1038/nature14148 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


