Abstract
Colletotrichum species are plant pathogens, saprobes, and endophytes on a range of economically important hosts. However, the species occurring on pear remain largely unresolved. To determine the morphology, phylogeny and biology of Colletotrichum species associated with Pyrus plants, a total of 295 samples were collected from cultivated pear species (including P. pyrifolia, P. bretschneideri, and P. communis) from seven major pear-cultivation provinces in China. The pear leaves and fruits affected by anthracnose were sampled and subjected to fungus isolation, resulting in a total of 488 Colletotrichum isolates. Phylogenetic analyses based on six loci (ACT, TUB2, CAL, CHS-1, GAPDH, and ITS) coupled with morphology of 90 representative isolates revealed that they belong to 10 known Colletotrichum species, including C. aenigma, C. citricola, C. conoides, C. fioriniae, C. fructicola, C. gloeosporioides, C. karstii, C. plurivorum, C. siamense, C. wuxiense, and two novel species, described here as C. jinshuiense and C. pyrifoliae. Of these, C. fructicola was the most dominant, occurring on P. pyrifolia and P. bretschneideri in all surveyed provinces except in Shandong, where C. siamense was dominant. In contrast, only C. siamense and C. fioriniae were isolated from P. communis, with the former being dominant. In order to prove Koch’s postulates, pathogenicity tests on pear leaves and fruits revealed a broad diversity in pathogenicity and aggressiveness among the species and isolates, of which C. citricola, C. jinshuiense, C. pyrifoliae, and C. conoides appeared to be organ-specific on either leaves or fruits. This study also represents the first reports of C. citricola, C. conoides, C. karstii, C. plurivorum, C. siamense, and C. wuxiense causing anthracnose on pear.
Keywords: Colletotrichum, multi-gene phylogeny, pathogenicity, Pyrus
INTRODUCTION
Colletotrichum species are important plant pathogens, saprobes, and endophytes, and can infect numerous plant hosts (Cannon et al. 2012, Dean et al. 2012, Diao et al. 2017, Guarnaccia et al. 2017). In recent years, the Colletotrichum species isolated from many host plants, e.g., Camellia sinensis (Theaceae), Capsicum annuum (Solanaceae), Citrus reticulata (Rutaceae), Mangifera indica (Anacardiaceae), and Vitis vinifera (Vitaceae), have been studied at a broad geographical level, which contributed to a better understanding of the genus (Huang et al. 2013, Lima et al. 2013, Vieira et al. 2014, Liu et al. 2015, Yan et al. 2015, Diao et al. 2017, Guarnaccia et al. 2017). Although Pyrus is an important host genus for Colletotrichum spp., the Colletotrichum spp. associated with pear anthracnose remained largely unresolved, with only six individual species identified including C. acutatum, C. aenigma, C. fioriniae, C. fructicola, C. pyricola, and C. salicis (Damm et al. 2012b, Weir et al. 2012). Moreover, previous reports chiefly investigated morphology and ITS sequence data (Wu et al. 2010, Liu et al. 2013b), which is insufficient for distinguishing closely related taxa in several species complexes (Liu et al. 2016a). Additionally, most of the species reported from pear were based on small sample sizes from restricted areas, thus underestimating the species diversity on this host (Damm et al. 2012b, Weir et al. 2012).
In the genus Pyrus, P. bretschneideri, P. communis, P. pyrifolia, P. sinkiangensis, and P. ussuriensis are commercially cultivated (Wu et al. 2013). Of these, P. bretschneideri, P. communis, and P. pyrifolia represent the major cultivated species in China (Zhao et al. 2016). Pear is the third most widespread temperate fruit crop after apple and grape, with the largest production in China (Wu et al. 2013). The pear industry is also one of the most important fruit industries worldwide. Statistical data for 2016 indicated that pear-cultivation area was 1 121 675 ha, yielding 19.5 MT fruit in China, accounting for 70 % of the global pear fruit yield (FAO 2016). Furthermore, Pyrus also originated from the tertiary period (about 65 to 55 M yr ago) in western China, which represents one of the two subcentres for genetic diversity of this genus (Rubtsov 1944, Vavilov 1951, Zeven & Zhukovsky 1975, Wu et al. 2013, Silva et al. 2014).
Characterisation of the Colletotrichum spp. associated with Pyrus plants is expected to provide a better insight into the biology of this important genus. Moreover, pear anthracnose caused by Colletotrichum spp. is an important disease in major pear-cultivation areas of China, occurring in the growth and fruit maturation periods of pear, mainly damaging leaves and fruits. Pear anthracnose has led to substantial economic losses due to excessive fruit rot, or the severe suppression of tree growth. However, a detailed study and knowledge of the Colletotrichum spp. affecting pear production has been lacking in China and is also poorly documented worldwide.
The taxonomy of the genus Colletotrichum has in the past mainly relied on host range and morphological characters (Von Arx 1957, Sutton 1980), which is limited in species resolution (Cai et al. 2009, Hyde et al. 2009, Cannon et al. 2012). Recently, multi-locus phylogenetic analyses together with morphological characteristics have significantly influenced the classification and species concepts in Colletotrichum (Cai et al. 2009, Cannon et al. 2012, Damm et al. 2012a, b, 2013, 2014, 2019, Weir et al. 2012, Liu et al. 2013a, 2014, Vieira et al. 2014, Yan et al. 2015, Guarnaccia et al. 2017). Phylogenetic analyses based on multi-locus DNA sequence data and the application of Genealogical Concordance Phylogenetic Species Recognition (GCPSR) represent an enhanced ability for species resolution (Quaedvlieg et al. 2014, Liu et al. 2016a, Diao et al. 2017), e.g., C. siamense was previously assumed to be a species complex composed of several taxa (Yang et al. 2009, Wikee et al. 2011, Lima et al. 2013, Vieira et al. 2014, Sharma et al. 2015), but was shown to represent a single variable species in the C. gloeosporioides species complex (Weir et al. 2012, Liu et al. 2016a). Based on recent progress, 14 Colletotrichum species complexes and 15 singleton species have been identified (Marin-Felix et al. 2017, Damm et al. 2019).
The aims of the present study were as follows:
identify the prevalence of Colletotrichum spp. associated with Pyrus anthracnose in the major production provinces in China;
validate the taxonomy of the Colletotrichum spp. through morphology, DNA phylogenetic analysis; and
evaluate their pathogenicity by proving Koch’s postulates.
MATERIALS AND METHODS
Sampling and isolation
A survey was conducted in 15 commercial pear orchards and four nurseries (Aug. 2013 to Oct. 2016) in the seven major pear-cultivation provinces (Anhui, Fujian, Hubei, Jiangsu, Jiangxi, Shandong, and Zhejiang) of China. Two kinds of symptoms were observed on fruit, namely 1) bitter rot showing big sunken rot lesions (BrL), 10–35 mm diam, with embedded concentric acervuli, secreting an orange conidial mass under humid conditions (Fig. 1a–c); and 2) tiny black spots (TS) less than 1 mm diam, gradually increasing in number instead of in size during the season (Fig. 1d, e). Three symptom types were observed on leaves, namely 1) big necrotic lesions (BnL); 2) small round spots (SS); and 3) TS. The BnL symptoms were characterised by sunken necrotic lesions 5–10 mm diam, brown in the centre but black along the margin, with black acervuli on the surface, secreting orange conidial tendrils under humid conditions (Fig. 1f). The SS symptoms were characterised by grey-white spots, 3–4 mm diam, circular to subcircular, grey-white in the centre, with a dark-brown margin (Fig. 1g). The TS symptoms were characterised by tiny black spots of less than 1 mm diam, which increased in number instead of in the size, accompanied by chlorosis, yellowing, and ‘green island regions’, resulting in defoliation (Fig. 1h, i).
Fig. 1.

Representative symptoms of pear anthracnose on fruits and leaves in the field. a–c. Symptoms of big sunken rot lesions (BrL; 10–35 mm diam) on fruits of P. pyrifolia (a, b) and P. communis cultivar (cv.) Gyuiot (c); d, e. symptoms of tiny black spots (TS; < 1 mm diam) on young pear fruits of P. pyrifolia cv. Cuiguan and mature pear fruit of P. bretschneideri cv. Huangguan, respectively; f. symptoms of big necrotic lesions (BnL; 5–10 mm diam) on leaves of P. pyrifolia cv. Xiangnan; g. symptoms of small round spots (SS; 3–4 mm diam) on leaves of P. pyrifolia cv. Jinshui No.1; h, i. initial and latter symptoms of TS on P. pyrifolia cv. Cuiguan.
Fruits and leaves showing the symptoms explained above were collected from pear trees of P. pyrifolia cultivars (cvs.) Cuiguan, Guanyangxueli, Hohsui, Huanghua, Huali No.1, Imamuraaki, Jinshui No. 1, Jinshui No. 2, and Xiangnan, P. bretschneideri cvs. Chili, Dangshansuli, Huangguan, Huangxianchangba, and Yali, and P. communis cv. Gyuiot in the surveyed orchards.
Fungi were isolated and linked to symptom types. Diseased tissues (neighbouring the asymptomatic regions) without sporulation were cut into small pieces (4–5 mm2) after surface sterilisation (1 % NaOCl for 45 s, 75 % ethanol for 45 s, washed three times in sterile water and dried on sterilised filter paper; Photita et al. 2005). Excised tissues were placed onto potato dextrose agar (PDA, 20 % diced potato, 2 % glucose, and 1.5 % agar, and distilled water) plates and incubated at 28 °C. For diseased tissues with sporulation, conidia were collected, suspended in sterilised water, diluted to a concentration of 1 × 104 conidia per mL, and spread onto the surface of water agar (WA, 2 % agar, and distilled water) to generate discrete colonies (Choi et al. 1999). Six single colonies of each isolate were picked up with a sterilised needle (insect pin, 0.5 mm diam) and transferred onto PDA plates. Pure cultures were stored in 25 % glycerol at -80 °C until use. Type specimens of new species from this study were deposited in the Mycological Herbarium, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS), and ex-type living cultures were deposited in the China General Microbiological Culture Collection Centre (CGMCC), Beijing, China.
DNA extraction, PCR amplification and sequencing
Mycelial discs were transferred to PDA plates covered with sterile cellophane and incubated at 28 °C in the dark for 5–7 d. Fungal genomic DNA was extracted with cetyltrimethylammonium bromide (CTAB) buffer (2 % w/v CTAB, 1.42 M NaCl, 20 mM EDTA, 100 mM Tris·HCl, pH 8.0, 0.2 % (w/v) β-mercaptoethanol) as previously described (Freeman et al. 1996). Six loci including the 5.8S nuclear ribosomal gene with the two flanking internal transcribed spacers (ITS), a 200-bp intron of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and partial actin (ACT), beta-tubulin (TUB2), chitin synthase (CHS-1), and calmodulin (CAL) genes were amplified using the primer pairs ITS4/ITS5 (White et al. 1990), GDF1/GDR1 (Guerber et al. 2003), ACT-512F/ACT-783R (Carbone & Kohn 1999), T1/Bt2b (Glass & Donaldson 1995, O’Donnell & Cigelnik 1997), CHS-79F/CHS-345R (Carbone & Kohn 1999), and CL1C/CL2C (Weir et al. 2012), respectively.
PCR amplification was conducted as described by Weir et al. (2012) but modified by using an annealing temperature of 56 °C for ITS, 59 °C for ACT and GAPDH, 58 °C for TUB2 and CHS-1, and 57 °C for CAL. PCR amplicons were purified and sequenced at the Sangon Biotech (Shanghai, China) Company, Ltd. Forward and reverse sequences were assembled to obtain a consensus sequence with DNAMAN (v. 9.0; Lynnon Biosoft). Sequences generated in this study were deposited in GenBank (Table 1).
Table 1.
List of 90 representative isolates of 12 Colletotrichum spp. collected from pear in China, with details about host, symptoms, origins, and GenBank accession numbers.
| Species | Isolate No. | Host | Symptoms | Origin | GenBank accession number |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| ITS | GAPDH | CAL | ACT | CHS-1 | TUB2 | |||||
| C. aenigma | PAFQ1 | P. pyrifolia cv. Xiangnan, leaf | BnL | Zhongxiang, Hubei | MG747997 | MG747915 | MG747769 | MG747687 | MG747833 | MG748079 |
| PAFQ5 | P. pyrifolia cv. Huali No.1, leaf | BnL | Zhongxiang, Hubei | MG747998 | MG747916 | MG747770 | MG747688 | MG747834 | MG748080 | |
| PAFQ21 | P. pyrifolia cv. Jinshui No.1, leaf | SS | Wuhan, Hubei | MG747999 | MG747917 | MG747771 | MG747689 | MG747835 | MG748081 | |
| PAFQ23 | P. pyrifolia cv. Jinshui No.1, leaf | SS | Wuhan, Hubei | MG748000 | MG747918 | MG747772 | MG747690 | MG747836 | MG748082 | |
| PAFQ24 | P. pyrifolia cv. Jinshui No.1, leaf | SS | Wuhan, Hubei | MG748001 | MG747919 | MG747773 | MG747691 | MG747837 | MG748083 | |
| PAFQ45 | P. bretschneideri cv. Yali, leaf | BnL | Yancheng, Jiangsu | MG748002 | MG747920 | MG747774 | MG747692 | MG747838 | MG748084 | |
| PAFQ47 | P. bretschneideri cv. Chili, fruit | BrL | Yancheng, Jiangsu | MG748003 | MG747921 | MG747775 | MG747693 | MG747839 | MG748085 | |
| PAFQ64 | P. bretschneideri cv. Huangguan, leaf | BnL | Dangshan, Anhui | MG748004 | MG747922 | MG747776 | MG747694 | MG747840 | MG748086 | |
| PAFQ66 | P. bretschneideri cv. Huangguan, fruit | BrL | Dangshan, Anhui | MG748005 | MG747923 | MG747777 | MG747695 | MG747841 | MG748087 | |
| PAFQ81 | P. pyrifolia cv. Guanyangxueli, leaf | SS | Hangzhou, Zhejiang | MG748006 | MG747924 | MG747778 | MG747696 | MG747842 | MG748088 | |
| PAFQ83 | P. pyrifolia cv. Guanyangxueli, leaf | SS | Hangzhou, Zhejiang | MG748007 | MG747925 | MG747779 | MG747697 | MG747843 | MG748089 | |
| C. citricola | PAFQ13 | P. pyrifolia, leaf | BnL | Wuhan, Hubei | MG748062 | MG747980 | MG747819 | MG747752 | MG747898 | MG748142 |
| C. conoides | PAFQ6 | P. pyrifolia, fruit | BrL | Wuhan, Hubei | MG748008 | MG747926 | MG747780 | MG747698 | MG747844 | MG748090 |
| C. fioriniae | PAFQ8 | P. pyrifolia cv. Jinshui No.1, leaf | SS | Wuhan, Hubei | MG748047 | MG747965 | – | MG747737 | MG747883 | MG748128 |
| PAFQ9 | P. pyrifolia cv. Jinshui No.1, leaf | SS | Wuhan, Hubei | MG748048 | MG747966 | – | MG747738 | MG747884 | – | |
| PAFQ10 | P. pyrifolia cv. Jinshui No.2, leaf | SS | Wuhan, Hubei | MG748049 | MG747967 | – | MG747739 | MG747885 | MG748129 | |
| PAFQ11 | P. pyrifolia cv. Jinshui No.2, leaf | SS | Wuhan, Hubei | MG748050 | MG747968 | – | MG747740 | MG747886 | MG748130 | |
| PAFQ12 | P. pyrifolia cv. Jinshui No.1, leaf | SS | Wuhan, Hubei | MG748051 | MG747969 | – | MG747741 | MG747887 | MG748131 | |
| PAFQ17 | P. pyrifolia, fruit | BrL | Wuhan, Hubei | MG748052 | MG747970 | – | MG747742 | MG747888 | MG748132 | |
| PAFQ18 | P. pyrifolia, fruit | BrL | Wuhan, Hubei | MG748053 | MG747971 | – | MG747743 | MG747889 | MG748133 | |
| PAFQ19 | P. pyrifolia, fruit | BrL | Wuhan, Hubei | MG748054 | MG747972 | – | MG747744 | MG747890 | MG748134 | |
| PAFQ34 | P. pyrifolia cv. Cuiguan, leaf | BnL | Jianning, Fujian | MG748055 | MG747973 | – | MG747745 | MG747891 | MG748135 | |
| PAFQ35 | P. pyrifolia cv. Cuiguan, leaf | BnL | Jianning, Fujian | MG748056 | MG747974 | – | MG747746 | MG747892 | MG748136 | |
| PAFQ36 | P. pyrifolia cv. Cuiguan, leaf | BnL | Jianning, Fujian | MG748057 | MG747975 | – | MG747747 | MG747893 | MG748137 | |
| PAFQ49 | P. pyrifolia, fruit | BrL | Nanjing, Jiangsu | MG748060 | MG747978 | – | MG747750 | MG747896 | MG748140 | |
| PAFQ50 | P. pyrifolia, fruit | BrL | Nanjing, Jiangsu | MG748061 | MG747979 | – | MG747751 | MG747897 | MG748141 | |
| PAFQ55 | P. pyrifolia cv. Cuiguan, leaf | BnL | Jinxi, Jiangxi | MG748058 | MG747976 | – | MG747748 | MG747894 | MG748138 | |
| PAFQ75 | P. communis cv. Gyuiot, fruit | BrL | Yantai, Shandong | MG748059 | MG747977 | – | MG747749 | MG747895 | MG748139 | |
| C. fructicola | PAFQ20 | P. pyrifolia cv. Jinshui No.1, leaf | SS | Wuhan, Hubei | MG748011 | MG747929 | MG747783 | MG747701 | MG747847 | MG748093 |
| PAFQ25 | P. pyrifolia cv. Jinshui No.1, leaf | SS | Wuhan, Hubei | MG748012 | MG747930 | MG747784 | MG747702 | MG747848 | MG748094 | |
| PAFQ31 | P. pyrifolia cv. Cuiguan, leaf | TS | Jianning, Fujian | MG748013 | MG747931 | MG747785 | MG747703 | MG747849 | MG748095 | |
| PAFQ32 | P. pyrifolia cv. Cuiguan, leaf | BnL | Jianning, Fujian | MG748014 | MG747932 | MG747786 | MG747704 | MG747850 | MG748096 | |
| PAFQ33 | P. pyrifolia cv. Cuiguan, leaf | BnL | Jianning, Fujian | MG748015 | MG747933 | MG747787 | MG747705 | MG747851 | MG748097 | |
| PAFQ46 | P. bretschneideri cv. Yali, leaf | BnL | Yancheng, Jiangsu | MG748016 | MG747934 | MG747788 | MG747706 | MG747852 | MG748098 | |
| PAFQ48 | P. bretschneideri cv. Dangshanshuli, fruit | TS | Yancheng, Jiangsu | MG748017 | MG747935 | MG747789 | MG747707 | MG747853 | MG748099 | |
| PAFQ51 | P. pyrifolia cv. Cuiguan, leaf | BnL | Jiangxi | MG748018 | MG747936 | MG747790 | MG747708 | MG747854 | MG748100 | |
| PAFQ57 | P. pyrifolia cv. Cuiguan, leaf | BnL | Jinxi, Jiangxi | MG748019 | MG747937 | MG747791 | MG747709 | MG747855 | MG748101 | |
| PAFQ62 | P. bretschneideri cv. Huangguan, leaf | BnL | Dangshan, Anhui | MG748020 | MG747938 | MG747792 | MG747710 | MG747856 | MG748102 | |
| PAFQ63 | P. bretschneideri cv. Huangguan, leaf | BnL | Dangshan, Anhui | MG748021 | MG747939 | MG747793 | MG747711 | MG747857 | MG748103 | |
| PAFQ77 | P. pyrifolia cv. Guangyangxueli, leaf | BnL | Hangzhou, Zhejiang | MG748023 | MG747941 | MG747795 | MG747713 | MG747859 | MG748105 | |
| PAFQ79 | P. pyrifolia cv. Guanyangxueli, leaf | BnL | Hangzhou, Zhejiang | MG748024 | MG747942 | MG747796 | MG747714 | MG747860 | MG748106 | |
| PAFQ84 | P. pyrifolia cv. Cuiguan, leaf | BnL | Tonglu, Zhejiang | MG748022 | MG747940 | MG747794 | MG747712 | MG747858 | MG748104 | |
| C. gloeosporioides | PAFQ7 | P. bretschneideri cv. Huangxianchangba, leaf | BnL | Wuhan, Hubei | MG748025 | MG747943 | MG747797 | MG747715 | MG747861 | MG748107 |
| PAFQ27 | P. pyrifolia cv. Hohsui, leaf | SS | Wuhan, Hubei | MG748026 | MG747944 | MG747798 | MG747716 | MG747862 | MG748108 | |
| PAFQ29 | P. pyrifolia cv. Hohsui, leaf | SS | Wuhan, Hubei | MG748027 | MG747945 | MG747799 | MG747717 | MG747863 | MG748109 | |
| PAFQ44 | P. bretschneideri cv. Yali, leaf | SS | Yancheng, Jiangsu | MG748028 | MG747946 | MG747800 | MG747718 | MG747864 | MG748110 | |
| C. gloeosporioides (cont.) | PAFQ56 | P. pyrifolia cv. Cuiguan, leaf | BnL | Jinxi, Jiangxi | MG748029 | MG747947 | MG747801 | MG747719 | MG747865 | MG748111 |
| PAFQ58 | P. pyrifolia cv. Cuiguan, leaf | BnL | Jinxi, Jiangxi | MG748030 | MG747948 | MG747802 | MG747720 | MG747866 | MG748112 | |
| PAFQ59 | P. pyrifolia cv. Cuiguan, leaf | BnL | Jinxi, Jiangxi | MG748031 | MG747949 | MG747803 | MG747721 | MG747867 | MG748113 | |
| PAFQ60 | P. pyrifolia cv. Cuiguan, leaf | BnL | Jinxi, Jiangxi | MG748032 | MG747950 | MG747804 | MG747722 | MG747868 | MG748114 | |
| PAFQ61 | P. pyrifolia cv. Huanghua, fruit | BrL | Jinxi, Jiangxi | MG748033 | MG747951 | MG747805 | MG747723 | MG747869 | MG748115 | |
| PAFQ80 | P. pyrifolia cv. Guangyangxueli, leaf | SS | Hangzhou, Zhejiang | MG748035 | MG747953 | MG747807 | MG747725 | MG747871 | MG748117 | |
| PAFQ86 | P. pyrifolia, leaf | BnL | Hangzhou, Zhejiang | MG748034 | MG747952 | MG747806 | MG747724 | MG747870 | MG748116 | |
| C. jinshuiense | PAFQ26, CGMCC 3.18903* | P. pyrifolia cv. Jinshui No.1, leaf | SS | Wuhan, Hubei | MG748077 | MG747995 | – | MG747767 | MG747913 | MG748157 |
| PAFQ26a | P. pyrifolia cv. Jinshui No.1, leaf | SS | Wuhan, Hubei | MG874830 | MG874822 | – | MG874807 | MG874814 | MG874838 | |
| PAFQ26b | P. pyrifolia cv. Jinshui No.1, leaf | SS | Wuhan, Hubei | MG874831 | MG874823 | – | MG874808 | MG874815 | MG874839 | |
| PAFQ26c | P. pyrifolia cv. Jinshui No.1, leaf | SS | Wuhan, Hubei | MG874832 | MG874824 | – | – | MG874816 | MG874840 | |
| PAFQ26d | P. pyrifolia cv. Jinshui No.1, leaf | SS | Wuhan, Hubei | MG874833 | MG874825 | – | MG874809 | MG874817 | MG874841 | |
| C. karstii | PAFQ14 | P. pyrifolia, leaf | BnL | Wuhan, Hubei | MG748063 | MG747981 | MG747820 | MG747753 | MG747899 | MG748143 |
| PAFQ15 | P. pyrifolia, leaf | BnL | Wuhan, Hubei | MG748064 | MG747982 | MG747821 | MG747754 | MG747900 | MG748144 | |
| PAFQ16 | P. pyrifolia, leaf | BnL | Wuhan, Hubei | MG748065 | MG747983 | MG747822 | MG747755 | MG747901 | MG748145 | |
| PAFQ28 | P. pyrifolia cv. Hohsui, leaf | BnL | Wuhan, Hubei | MG748066 | MG747984 | MG747823 | MG747756 | MG747902 | MG748146 | |
| PAFQ37 | P. pyrifolia cv. Cuiguan, leaf | BnL | Jianning, Fujian | MG748067 | MG747985 | MG747824 | MG747757 | MG747903 | MG748147 | |
| PAFQ38 | P. pyrifolia cv. Cuiguan, leaf | BnL | Jianning, Fujian | MG748068 | MG747986 | MG747825 | MG747758 | MG747904 | MG748148 | |
| PAFQ39 | P. pyrifolia cv. Cuiguan, leaf | BnL | Jianning, Fujian | MG748069 | MG747987 | MG747826 | MG747759 | MG747905 | MG748149 | |
| PAFQ40 | P. pyrifolia cv. Huanghua, leaf | BnL | Jianning, Fujian | MG748070 | MG747988 | MG747827 | MG747760 | MG747906 | MG748150 | |
| PAFQ41 | P. pyrifolia cv. Huanghua, leaf | BnL | Jianning, Fujian | MG748071 | MG747989 | MG747828 | MG747761 | MG747907 | MG748151 | |
| PAFQ42 | P. pyrifolia cv. Huanghua, leaf | BnL | Jianning, Fujian | MG748072 | MG747990 | MG747829 | MG747762 | MG747908 | MG748152 | |
| PAFQ43 | P. pyrifolia cv. Huanghua, leaf | BnL | Jianning, Fujian | MG748073 | MG747991 | MG747830 | MG747763 | MG747909 | MG748153 | |
| PAFQ52 | P. pyrifolia cv. Cuiguan, leaf | BnL | Jinxi, Jiangxi | MG748074 | MG747992 | MG747831 | MG747764 | MG747910 | MG748154 | |
| PAFQ82 | P. pyrifolia cv. Guanyangxueli, leaf | BnL | Hangzhou, Zhejiang | MG748075 | MG747993 | MG747832 | MG747765 | MG747911 | MG748155 | |
| C. plurivorum | PAFQ65 | P. bretschneideri cv. Huangguan, leaf | BnL | Dangshan, Anhui | MG748076 | MG747994 | – | MG747766 | MG747912 | MG748156 |
| C. pyrifolia | PAFQ22, CGMCC 3.18902* | P. pyrifolia cv. Jinshui No.1, leaf | SS | Wuhan, Hubei | MG748078 | MG747996 | – | MG747768 | MG747914 | MG748158 |
| PAFQ22a | P. pyrifolia cv. Jinshui No.1, leaf | SS | Wuhan, Hubei | MG874834 | MG874826 | – | MG874810 | MG874818 | MG874842 | |
| PAFQ22b | P. pyrifolia cv. Jinshui No.1, leaf | SS | Wuhan, Hubei | MG874835 | MG874827 | – | MG874811 | MG874819 | MG874843 | |
| PAFQ22c | P. pyrifolia cv. Jinshui No.1, leaf | SS | Wuhan, Hubei | MG874836 | MG874828 | – | MG874812 | MG874820 | MG874844 | |
| PAFQ22d | P. pyrifolia cv. Jinshui No.1, leaf | SS | Wuhan, Hubei | MG874837 | MG874829 | – | MG874813 | MG874821 | MG874845 | |
| C. siamense | PAFQ67 | P. communis cv. Gyuiot, fruit | BrL | Yantai, Shandong | MG748036 | MG747954 | MG747808 | MG747726 | MG747872 | MG748118 |
| PAFQ68 | P. communis cv. Gyuiot, fruit | BrL | Yantai, Shandong | MG748037 | MG747955 | MG747809 | MG747727 | MG747873 | MG748119 | |
| PAFQ69 | P. communis cv. Gyuiot, fruit | BrL | Yantai, Shandong | MG748038 | MG747956 | MG747810 | MG747728 | MG747874 | MG748120 | |
| PAFQ70 | P. communis cv. Gyuiot, fruit | BrL | Yantai, Shandong | MG748039 | MG747957 | MG747811 | MG747729 | MG747875 | MG748121 | |
| PAFQ71 | P. communis cv. Gyuiot, fruit | BrL | Yantai, Shandong | MG748040 | MG747958 | MG747812 | MG747730 | MG747876 | MG748122 | |
| PAFQ72 | P. communis cv. Gyuiot, fruit | BrL | Yantai, Shandong | MG748041 | MG747959 | MG747813 | MG747731 | MG747877 | MG748123 | |
| PAFQ73 | P. communis cv. Gyuiot, fruit | BrL | Yantai, Shandong | MG748042 | MG747960 | MG747814 | MG747732 | MG747878 | MG748124 | |
| PAFQ74 | P. communis cv. Gyuiot, fruit | BrL | Yantai, Shandong | MG748043 | MG747961 | MG747815 | MG747733 | MG747879 | MG748125 | |
| PAFQ76 | P. communis cv. Gyuiot, fruit | BrL | Yantai, Shandong | MG748044 | MG747962 | MG747816 | MG747734 | MG747880 | – | |
| PAFQ78 | P. pyrifolia cv. Guanyangxueli, leaf | BnL | Hangzhou, Zhejiang | MG748046 | MG747964 | MG747818 | MG747736 | MG747882 | MG748127 | |
| PAFQ85 | P. pyrifolia, leaf | BnL | Hangzhou, Zhejiang | MG748045 | MG747963 | MG747817 | MG747735 | MG747881 | MG748126 | |
| C. wuxiense | PAFQ53 | P. pyrifolia cv. Cuiguan, leaf | BnL | Jinxi, Jiangxi | MG748009 | MG747927 | MG747781 | MG747699 | MG747845 | MG748091 |
| PAFQ54 | P. pyrifolia cv. Cuiguan, leaf | BnL | Jinxi, Jiangxi | MG748010 | MG747928 | MG747782 | MG747700 | MG747846 | MG748092 | |
* = Ex-type culture.
BrL: big sunken rot lesions; BnL: big necrotic lesions; SS: small round spots; TS: tiny black spots.
Phylogenetic analyses
Multiple sequences of concatenated ACT, TUB2, CAL, CHS-1, GAPDH and ITS sequences were aligned using MAFFT v. 7 (Katoh & Standley 2013) with default settings, and if necessary, manually adjusted in MEGA v. 7.0.1 (Kumar et al. 2016). Bayesian inference (BI) was used to construct phylogenies using MrBayes v. 3.1.2 (Ronquist & Huelsenbeck 2003). MrModeltest v. 2.3 (Nylander 2004) was used to carry out statistical selection of best-fit models of nucleotide substitution using the corrected Akaike information criterion (AIC) (Table 2). Two analyses of four Markov Chain Monte Carlo (MCMC) chains were conducted from random trees with 1 × 107 generations for the C. gloeosporioides species complex, 3 × 106 for the C. dematium species complex and the related reference species involved in the same phylogenetic tree, and 2 × 106 generations for C. acutatum and C. boninense species complexes. The analyses were sampled every 1 000 generations, which were stopped once the average standard deviation of split frequencies was below 0.01. Convergence of all parameters was checked using the internal diagnostics of the standard deviation of split frequencies and performance scale reduction factors (PSRF), and then externally with Tracer v. 1.6 (Rambaut et al. 2013). The first 25 % of trees were discarded as the burn-in phase of each analysis and posterior probabilities determined from the remaining trees. Additionally, maximum parsimony analyses (MP) were performed on the multi-locus alignment using PAUP (Phylogenetic Analysis Using Parsimony) v. 4.0b10 (Swofford 2002). Phylogenetic trees were generated using the heuristic search option with Tree Bisection Reconnection (TBR) branch swapping and 1 000 random sequence additions. Maxtrees were set up to 5 000, branches of zero length collapsed, and all multiple parsimonious trees were saved. Clade stability was assessed using a bootstrap analysis with 1 000 replicates. Afterwards, tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI) were calculated. Furthermore, maximum likelihood (ML) analyses were implemented on the multi-locus alignments using the RaxmlGUI v. 1.3.1 (Silvestro & Michalak 2012). Clade stability was assessed using bootstrap analyses with 1 000 replicates. A general time reversible model (GTR) was applied with an invgamma-distributed rate variation. Phylogenetic trees were visualised in FigTree v. 1.4.2 (Rambaut 2014). The alignments and phylogenetic trees were deposited in TreeBASE (study 22264).
Table 2.
Nucleotide substitution models used in the phylogenetic analyses.
| Gene | Gloeosporioides clade | Acutatum clade | Boninense clade | Dematium clade and other taxa |
|---|---|---|---|---|
| ITS | GTR+I+G | GTR+I | SYM+I+G | GTR+I+G |
| ACT | GTR+G | HKY+G | HKY+G | HKY+I+G |
| GAPDH | HKY+G | GTR+G | HKY+I | HKY+I+G |
| TUB2 | SYM+G | GTR+G | HKY+I | HKY+I+G |
| CHS-1 | K80+I | SYM+G | GTR+I | GTR+I+G |
| CAL | GTR+I+G | HKY+I |
For the phylogenetically close but not clearly delimited species, sequences were analysed using the GCPSR model by performing a pairwise homoplasy index (PHI) test as described by Quaedvlieg et al. (2014). The PHI test was performed in SplitsTree 4 (Huson 1998, Huson & Kloepper 2005, Huson & Bryant 2006) to determine the recombination level within phylogenetically closely related species using a six-locus concatenated dataset (ACT, TUB2, CAL, CHS-1, GAPDH, and ITS). If the resulting pairwise homoplasy index was below a 0.05 threshold (Ôw < 0.05), it was indicative of significant recombination in the dataset. The relationship between closely related species was visualised by constructing a splits graph.
Morphological analysis
Morphological and cultural features were characterised according to Yan et al. (2015). Briefly, mycelial discs (5 mm diam) were taken from the growing edge of 5-d-old cultures in triplicate, transferred on PDA, oatmeal agar (OA; Crous et al. 2009) and synthetic nutrient-poor agar medium (SNA; Nirenberg 1976), and incubated in the dark at 28 °C. Colony diameters were measured daily for 5 d to calculate their mycelial growth rates (mm/d). The shape, colour and density of colonies were recorded after 6 d. Moreover, the shape, colour and size of sporocarps, conidia, conidiophores, asci and ascospores were observed using light microscopy (Nikon Eclipse 90i or Olympus BX63, Japan), and 50 conidia or ascospores were measured to determine their sizes unless no or less spores were produced. Conidial appressoria were induced by dropping a conidial suspension (106 conidia/mL; 50 μL) on a concavity slide, placed inside plates containing moistened filter papers with distilled sterile water, and then incubated at 25 °C in the dark. After incubating for 24 to 48 h, the sizes of 30 conidial appressoria formed at the ends of germ tubes were measured (Yang et al. 2009).
Prevalence
To determine the prevalence of Colletotrichum species in sampled provinces, the Pyrus spp. and pear organ (leaf or fruit) involved were established. The Isolation Rate (RI) was calculated for each species with the formula, RI % = (NS / NI) × 100, where NS was the number of isolates from the same species, and NI was the total number of isolates from each sample-collected province, Pyrus sp. or pear organ (Vieira et al. 2014, Wang et al. 2016). The overall RI was calculated using the NI value equal to the total number of isolates obtained from pear plants.
Pathogenicity tests
Representative Colletotrichum isolates were selected for pathogenicity tests with a spore suspension on detached leaves (approx. 4-wk-old) of P. pyriforia cv. Cuiguan in eight replicates as previously described (Cai et al. 2009). Briefly, tender healthy-looking leaves were collected, washed three times with sterile water, and air-dried on sterilised filter paper. The leaves are inoculated using the wound/drop and non-wound/drop inoculation methods (Lin et al. 2002, Kanchana-udomkan et al. 2004, Than et al. 2008). For the wound/drop method, an aliquot of 6 μL of spore suspension (1.0 × 106 conidia or ascospores per mL) was dropped on the left side of a leaf after wounding once by pin-pricking with a sterilised needle (insect pin, 0.5 mm diam), and sterile water on the right side of the same leaf in parallel as control. For non-wound/drop method, the spore suspension was dropped on the left side of a leaf without being unwounded, and sterile water on the right side of the same leaf in parallel as control. The infection rates were calculated using the formula (infection rate = the number of infected leaves or fruits/the number of inoculated leaves or fruits) at 14 d post inoculation (dpi) (Huang et al. 2013).
Additionally, pathogenicity was also determined on detached mature pear fruits of P. bretschneideri cv. Huangguan in triplicate as previously described (Cai et al. 2009). Briefly, healthy fruits were surface-sterilised with 1 % sodium hypochlorite for 5 min, washed three times with sterile water, and air-dried. Wound/drop and non-wound/drop inoculation methods were also used (Lin et al. 2002, Kanchana-udomkan et al. 2004, Than et al. 2008). For the wound/drop method, an aliquot of 6 μL of spore suspension (1 × 106 conidia or ascospores per mL) was dropped on the fruits after wounding three times by pin-pricking with a sterilised needle (5 mm deep). For the non-wound/drop method, the same spore suspension was also directly dropped on the surface of unwounded pear fruits. Sterile water was dropped on the fruit in parallel as control. Symptom development under wounded conditions was evaluated by determining the mean lesion lengths at 10 dpi. Symptom development on fruits was studied by determining the infection rates at 30 dpi using the aforementioned formula.
After inoculation, the detached leaves and fruits were put on plastic trays, covered with plastic wrap to maintain a 99 % relative humidity, and incubated at 25 °C with a 12/12 h light/dark photoperiod. Pathogens were re-isolated from the resulting lesions and identified as described above. The pathogenicity tests were repeated once.
RESULTS
Colletotrichum isolates associated with pear anthracnose
A total of 295 pear samples (249 leaves and 46 fruits) affected by pear anthracnose, including BrL and TS on fruits, and BnL, SS, and TS on leaves were collected for fungal isolation, resulting in a total of 488 Colletotrichum isolates identified based on morphology and ITS sequence data. A total of 90 representative isolates were chosen for further analyses based on their morphology (colony shape, colour, and conidial morphology), ITS sequence data, symptom type, origin, and host cultivar involved (Table 1).
Multi-locus phylogenetic analyses
The 90 representative isolates (Table 1) together with 181 reference isolates from previously described species (Table 3) were subjected to multi-locus phylogenetic analyses with concatenated ACT, TUB2, CAL, CHS-1, GAPDH, and ITS sequences for those belonging to the C. gloeosporioides and C. boninense species complexes, or with concatenated ACT, TUB2, CHS-1, GAPDH, and ITS sequences for other species of which no CAL sequences are available. The results showed that isolates clustered together with 12 species in five Colletotrichum species complexes, including gloeosporioides (50 isolates), acutatum (15), boninense (14), dematium (5), and orchidearum (1), and one singleton species (5) (Fig. 2–5).
Table 3.
List of isolates of the Colletotrichum species used in this study, with details about host/substrate, country, and GenBank accession numbers.
| Species | Culturex | Host/Substrate | Country | GenBank accession number |
|||||
|---|---|---|---|---|---|---|---|---|---|
| ITS | GAPDH | CAL | ACT | CHS-1 | TUB2 | ||||
| C. abscissum | COAD 1877* | Citrus sinensis cv. Pera | Brazil | KP843126 | KP843129 | – | KP843141 | KP843132 | KP843135 |
| C. acerbum | CBS 128530* | Malus domestica | New Zealand | JQ948459 | JQ948790 | – | JQ949780 | JQ949120 | JQ950110 |
| C. acutatum | CBS 112996* | Carica papaya | Australia | JQ005776 | JQ948677 | – | JQ005839 | JQ005797 | JQ005860 |
| C. aenigma | ICMP 18608* | Persea americana | Israel | JX010244 | JX010044 | JX009683 | JX009443 | JX009774 | JX010389 |
| ICMP 18686 | Pyrus pyrifolia | Japan | JX010243 | JX009913 | JX009684 | JX009519 | JX009789 | JX010390 | |
| C. aeschynomenes | ICMP 17673* | Aeschynomene virginica | USA | JX010176 | JX009930 | JX009721 | JX009483 | JX009799 | JX010392 |
| C. agaves | CBS 118190 | Agave striate | Mexico | DQ286221 | – | – | – | – | – |
| C. alatae | CBS 304.67* | Dioscorea alata | India | JX010190 | JX009990 | JX009738 | JX009471 | JX009837 | JX010383 |
| C. alienum | ICMP 12071* | Malus domestica | New Zealand | JX010251 | JX010028 | JX009654 | JX009572 | JX009882 | JX010411 |
| C. annellatum | CBS 129826* | Hevea brasiliensis, leaf | Colombia | JQ005222 | JQ005309 | JQ005743 | JQ005570 | JQ005396 | JQ005656 |
| C. anthrisci | CBS 125334* | Anthriscus sylvestris,dead stem | Netherlands | GU227845 | GU228237 | – | GU227943 | GU228335 | GU228139 |
| CBS 125335 | Anthriscus sylvestris,dead stem | Netherlands | GU227846 | GU228238 | – | GU227944 | GU228336 | GU228140 | |
| C. aotearoa | ICMP 18537* | Coprosma sp. | New Zealand | JX010205 | JX010005 | JX009611 | JX009564 | JX009853 | JX010420 |
| C. asianum | ICMP 18580* | Coffea arabica | Thailand | FJ972612 | JX010053 | FJ917506 | JX009584 | JX009867 | JX010406 |
| C. australe | CBS 116478* | Trachycarpus fortunei | South Africa | JQ948455 | JQ948786 | – | JQ949776 | JQ949116 | JQ950106 |
| C. beeveri | CBS 128527* | Brachyglottis repanda | New Zealand | JQ005171 | JQ005258 | JQ005692 | JQ005519 | JQ005345 | JQ005605 |
| C. boninense | CBS 123755* | Crinum asiaticum var. sinicum | Japan | JQ005153 | JQ005240 | JQ005674 | JQ005501 | JQ005327 | JQ005588 |
| CBS 128506 | Solanum lycopersicum, fruit rot | New Zealand | JQ005157 | JQ005244 | JQ005678 | JQ005505 | JQ005331 | JQ005591 | |
| C. brasiliense | CBS 128501* | Passiflora edulis, fruit anthracnose | Brazil | JQ005235 | JQ005322 | JQ005756 | JQ005583 | JQ005409 | JQ005669 |
| C. brassicicola | CBS 101059* | Brassica oleracea, leaf spot | New Zealand | JQ005172 | JQ005259 | JQ005693 | JQ005520 | JQ005346 | JQ005606 |
| C. brevisporum | BCC 38876* | Neoregalia sp. | Thailand | JN050238 | JN050238 | – | JN050216 | KF687760 | JN050244 |
| C. brisbanense | CBS 292.67* | Capsicum annuum | Australia | JQ948291 | JQ948621 | – | JQ949612 | JQ948952 | JQ949942 |
| C. cairnsense | BRIP 63642* | Capsicum annuum | Australia | KU923672 | KU923704 | – | KU923716 | KU923710 | KU923688 |
| C. camelliae-japonicae | CGMCC 3.18118* | Camellia japonica | Japan | KX853165 | KX893584 | – | KX893576 | – | KX893580 |
| CGMCC 3.18117 | Camellia japonica | Japan | KX853164 | KX893583 | – | KX893575 | – | KX893579 | |
| C. carthami | SAPA100011* | Carthamus tinctorium | Japan | AB696998 | – | – | – | – | AB696992 |
| C. cattleyicola | CBS 170.49* | Cattleya sp. | Belgium | MG600758 | MG600819 | – | MG600963 | MG600866 | MG601025 |
| C. chlorophyti | IMI 103806* | Chlorophytum sp. | India | GU227894 | GU228286 | – | GU227992 | GU228384 | GU228188 |
| C. chrysanthemi | IMI 364540 | Chrysanthemum coronarium, leaf spot | China | JQ948273 | JQ948603 | – | JQ949594 | JQ948934 | JQ949924 |
| C. circinans | CBS 221.81* | Allium cepa | Serbia | GU227855 | GU228247 | – | GU227953 | GU228345 | GU228149 |
| C. citricola | CBS 134228* | Citrus unshiu | China | KC293576 | KC293736 | KC293696 | KC293616 | KC293696 | KC293656 |
| CBS 134229 | Citrus unshiu | China | KC293577 | KC293737 | KC293697 | KC293617 | KC293793 | KC293657 | |
| CBS 134230 | Citrus unshiu | China | KC293578 | KC293738 | KC293698 | KC293618 | KC293794 | KC293658 | |
| C. clidemiae | ICMP 18658* | Clidemia hirta | USA, Hawaii | JX010265 | JX009989 | JX009645 | JX009537 | JX009877 | JX010438 |
| C. cliviicola | CBS 125375* | Clivia miniata | China | JX519223 | JX546611 | – | JX519240 | JX519232 | JX519249 |
| CSSS1 | Clivia miniata | China | GU109479 | GU085867 | – | GU085861 | GU085865 | GU085869 | |
| CSSS2 | Clivia miniata | China | GU109480 | GU085868 | – | GU085862 | GU085866 | GU085870 | |
| C. colombiense | CBS 129818* | Passiflora edulis, leaf | Colombia | JQ005174 | JQ005261 | JQ005695 | JQ005522 | JQ005348 | JQ005608 |
| C. conoides | CGMCC 3.17615* | Capsicum annuum | China | KP890168 | KP890162 | KP890150 | KP890144 | KP890156 | KP890174 |
| CAUG33 | Capsicum annuum | China | KP890169 | KP890163 | KP890151 | KP890145 | KP890157 | KP890175 | |
| CAUG34 | Capsicum annuum | China | KP890170 | KP890164 | KP890152 | KP890146 | KP890158 | KP890176 | |
| C. constrictum | CBS 128504* | Citrus limon, fruit rot | New Zealand | JQ005238 | JQ005325 | JQ005759 | JQ005586 | JQ005412 | JQ005672 |
| C. cordylinicola | ICMP 18579* | Cordyline fruticosa | Thailand | JX010226 | JX009975 | HM470238 | HM470235 | JX009864 | JX010440 |
| C. cosmi | CBS 853.73* | Cosmos sp., seed | Netherlands | JQ948274 | JQ948604 | – | JQ949595 | JQ948935 | JQ949925 |
| C. costaricense | CBS 330.75* | Coffea arabica, cv. Typica, berry | Costa Rica | JQ948180 | JQ948510 | – | JQ949501 | JQ948841 | JQ949831 |
| C. curcumae | IMI 288937* | Curcuma longa | India | GU227893 | GU228285 | – | GU227991 | GU228383 | GU228187 |
| C. cuscutae | IMI 304802* | Cuscuta sp. | Dominica | JQ948195 | JQ948525 | – | JQ949516 | JQ948856 | JQ949846 |
| C. cymbidiicola | IMI 347923* | Cymbidium sp., leaf lesion | Australia | JQ005166 | JQ005253 | JQ005687 | JQ005514 | JQ005340 | JQ005600 |
| C. dacrycarpi | CBS 130241* | Dacrycarpus dacrydioides, leaf endophyte | New Zealand | JQ005236 | JQ005323 | JQ005757 | JQ005584 | JQ005410 | JQ005670 |
| C. dematium | CBS 125.25* | Eryngium campestre,dead leaf | France | GU227819 | GU228211 | – | GU227917 | GU228309 | GU228113 |
| CBS 123728 | Genista tinctoria, leaf spot | Czech Republic | GU227822 | GU228214 | – | GU227920 | GU228312 | GU228116 | |
| C. dracaenophilum | CBS 118199* | Dracaena sp. | China | JX519222 | JX546707 | – | JX519238 | JX519230 | JX519247 |
| C. euphorbiae | CBS 134725* | Euphorbia sp. | South Africa | KF777146 | KF777131 | – | KF777125 | KF777128 | KF777247 |
| C. fioriniae | CBS 125396 | Malus domestica, fruit lesion | USA | JQ948299 | JQ948629 | – | JQ949620 | JQ948960 | JQ949950 |
| IMI 324996 | Malus pumila | USA | JQ948301 | JQ948631 | – | JQ949622 | JQ948962 | JQ949952 | |
| CBS 126526 | Primula sp., leaf spots | Netherlands | JQ948323 | JQ948653 | – | JQ949644 | JQ948984 | JQ949974 | |
| CBS 124958 | Pyrus sp., fruit rot | USA | JQ948306 | JQ948636 | – | JQ949627 | JQ948967 | JQ949957 | |
| IMI 504882 | Fragaria × ananassa | New Zealand | KT153562 | KT153552 | – | KT153542 | KT153547 | KT153567 | |
| CBS 129938 | Malus domestica | USA | JQ948296 | JQ948626 | – | JQ949617 | JQ948957 | JQ949947 | |
| CBS 119292 | Vaccinium sp., fruit | New Zealand | JQ948313 | JQ948643 | – | JQ949634 | JQ948974 | JQ949964 | |
| CBS 129930 | Malus domestica | New Zealand | JQ948304 | JQ948634 | – | JQ949625 | JQ948965 | JQ949955 | |
| ATCC 28992 | Malus domestica | USA | JQ948297 | JQ948627 | – | JQ949618 | JQ948958 | JQ949948 | |
| C. fructi | CBS 346.37* | Malus sylvestris, fruit | USA | GU227844 | GU228236 | – | GU227942 | GU228334 | GU228138 |
| C. fructicola | ICMP 18581* | Coffea arabica | Thailand | JX010165 | JX010033 | FJ917508 | FJ907426 | JX009866 | JX010405 |
| ICMP 18613 | Limonium sinuatum | Israel | JX010167 | JX009998 | JX009675 | JX009491 | JX009772 | JX010388 | |
| ICMP 18645 | Theobroma cacao | Panama | JX010172 | JX009992 | JX009666 | JX009543 | JX009873 | JX010408 | |
| ICMP 18727 | Fragaria × ananassa | USA | JX010179 | JX010035 | JX009682 | JX009565 | JX009812 | JX010394 | |
| ICMP 18120 | Dioscorea alata | Nigeria | JX010182 | JX010041 | JX009670 | JX009436 | JX009844 | JX010401 | |
| C. fructicola (syn. C. ignotum) | ICMP 18646* | Tetragastris panamensis | Panama | JX010173 | JX010032 | JX009674 | JX009581 | JX009874 | JX010409 |
| C. fructicola (syn. Glomerella cingulata var. minor) | ICMP 17921* | Ficus edulis | Germany | JX010181 | JX009923 | JX009671 | JX009495 | JX009839 | JX010400 |
| C. fructivorum | CBS 133125* | Vaccinium macrocarpon | USA | JX145145 | – | – | – | – | JX145196 |
| CBS 133135 | Rhexia virginica | USA | JX145133 | – | – | – | – | JX145184 | |
| C. gloeosporioides | IMI 356878* | Citrus sinensis | Italy | JX010152 | JX010056 | JX009731 | JX009531 | JX009818 | JX010445 |
| ICMP 12939 | Citrus sp. | New Zealand | JX010149 | JX009931 | JX009728 | JX009462 | JX009747 | – | |
| ICMP 18695 | Citrus sp. | USA | JX010153 | JX009979 | JX009735 | JX009494 | JX009779 | – | |
| ICMP 18694 | Mangifera indica | South Africa | JX010155 | JX009980 | JX009729 | JX009481 | JX009796 | – | |
| C. gloeosporioides (syn. Gloeosporium pedemontanum) | ICMP 19121* | Citrus limon | Italy | JX010148 | JX010054 | JX009745 | JX009558 | JX009903 | – |
| C. godetiae | CBS 133.44* | Clarkia hybrida | Denmark | JQ948402 | JQ948733 | – | JQ949723 | JQ949063 | JQ950053 |
| C. hebeiense | JZB330024 | Vitis vinifera cv. Cabernet Sauvignon | China | KF156873 | KF377505 | – | KF377542 | – | – |
| CGMCC 3.17464* | Vitis vinifera cv. Cabernet Sauvignon | China | KF156863 | KF377495 | – | KF377532 | KF289008 | KF288975 | |
| C. hemerocallidis | CDLG5* | Hemerocallis fulva var. kwanso | China | JQ400005 | JQ400012 | – | JQ399991 | JQ399998 | JQ400019 |
| C. hippeastri | CBS 125376* | Hippeastrum vittatum, leaf | China | JQ005231 | JQ005318 | JQ005752 | JQ005579 | JQ005405 | JQ005665 |
| C. horii | ICMP 10492* | Diospyros kaki | Japan | GQ329690 | GQ329681 | JX009604 | JX009438 | JX009752 | JX010450 |
| C. insertae | MFLU 15-1895* | Parthenocissus inserta | Russia | KX618686 | KX618684 | – | KX618682 | KX618683 | KX618685 |
| C. jasminigenum | MFLUCC 10-0273 | Jasminum sambac | Vietnam | HM131513 | HM131499 | – | HM131508 | – | HM153770 |
| C. jiangxiense | CGMCC 3.17362 | Camellia sinensis, endophyte | China | KJ955198 | KJ954899 | KJ954749 | KJ954469 | – | KJ955345 |
| CGMCC 3.17363* | Camellia sinensis, pathogen | China | KJ955201 | KJ954902 | KJ954752 | KJ954471 | – | KJ955348 | |
| C. johnstonii | CBS 128532* | Solanum lycopersicum, fruit rot | New Zealand | JQ948444 | JQ948775 | – | JQ949765 | JQ949105 | JQ950095 |
| C. kahawae subsp. ciggaro | ICMP 18539* | Olea europaea | Australia | JX010230 | JX009966 | JX009635 | JX009523 | JX009800 | JX010434 |
| ICMP 18534 | Kunzea ericoides | New Zealand | JX010227 | JX009904 | JX009634 | JX009473 | JX009765 | JX010427 | |
| ICMP 12952 | Persea americana | New Zealand | JX010214 | JX009971 | JX009648 | JX009431 | JX009757 | JX010426 | |
| C. kahawae subsp. kahawae | IMI 319418* | Coffea arabica | Kenya | JX010231 | JX010012 | JX009642 | JX009452 | JX009813 | JX010444 |
| C. kahawae subsp. kahawae (cont.) | ICMP 17905 | Coffea arabica | Cameroon | JX010232 | JX010046 | JX009644 | JX009561 | JX009816 | JX010431 |
| ICMP 17915 | Coffea arabica | Angola | JX010234 | JX010040 | JX009638 | JX009474 | JX009829 | JX010435 | |
| C. karstii | CBS 113087 | Malus sp. | USA | JQ005181 | JQ005268 | JQ005702 | JQ005529 | JQ005355 | JQ005615 |
| CBS 128524 | Citrullus lanatus, rotten fruit | New Zealand | JQ005195 | JQ005282 | JQ005716 | JQ005543 | JQ005369 | JQ005629 | |
| CBS 128551 | Citrus sp. | New Zealand | JQ005208 | JQ005295 | JQ005729 | JQ005556 | JQ005382 | JQ005642 | |
| CBS 129832 | Musa sp. | Mexico | JQ005177 | JQ005264 | JQ005698 | JQ005525 | JQ005351 | JQ005611 | |
| CBS 129824 | Musa AAA, fruit | Colombia | JQ005215 | JQ005302 | JQ005736 | JQ005563 | JQ005389 | JQ005649 | |
| CBS 128552 | Synsepalum dulcificum, leaves | Taiwan | JQ005188 | JQ005275 | JQ005709 | JQ005536 | JQ005362 | JQ005622 | |
| C. kinghornii | CBS 198.35* | Phormium sp. | UK | JQ948454 | JQ948785 | – | JQ949775 | JQ949115 | JQ950105 |
| C. laticiphilum | CBS 112989* | Hevea brasiliensis | India | JQ948289 | JQ948619 | – | JQ949610 | JQ948950 | JQ949940 |
| C. ledebouriae | CBS 141284* | Ledebouria floridunda | South Africa | KX228254 | – | – | KX228357 | – | – |
| C. liaoningense | CGMCC 3.17616* | Capsicum sp. | China | KP890104 | KP890135 | – | KP890097 | KP890127 | KP890111 |
| C. lindemuthianum | CBS 144.31* | Phaseolus vulgaris | Germany | JQ005779 | JX546712 | – | JQ005842 | JQ005800 | JQ005863 |
| C. lineola | CBS 125337* | Apiaceae, dead stem | Czech Republic | GU227829 | GU228221 | – | GU227927 | GU228319 | GU228123 |
| CBS 124.25 | Trillium sp., leaf spot | Czech Republic | GU227836 | GU228228 | – | GU227934 | GU228326 | GU228130 | |
| C. lupini | CBS 109225* | Lupinus albus | Ulkraine | JQ948155 | JQ948485 | – | JQ949476 | JQ948816 | JQ949806 |
| C. magnum | CBS 519.97* | Citrullus lanatus | USA | MG600769 | MG600829 | – | MG600973 | MG600875 | MG601036 |
| C. menispermi | MFLU 14-0625* | Menispermum dauricum | Russia | KU242357 | KU242356 | – | KU242353 | KU242355 | KU242354 |
| C. musae | CBS 116870* | Musa sp. | USA | JX010146 | JX010050 | JX009742 | JX009433 | JX009896 | HQ596280 |
| C. musicola | CBS 132885* | Musa sp. | Mexico | MG600736 | MG600798 | – | MG600942 | MG600853 | MG601003 |
| C. neosansevieriae | CBS 139918* | Sansevieria trifasciata | South Africa | KR476747 | KR476791 | – | KR476790 | – | KR476797 |
| C. novae-zelandiae | CBS 128505* | Capsicum annuum, fruit rot | New Zealand | JQ005228 | JQ005315 | JQ005749 | JQ005576 | JQ005402 | JQ005662 |
| C. nupharicola | CBS 470.96* | Nuphar lutea subsp. Polysepala | USA | JX010187 | JX009972 | JX009663 | JX009437 | JX009835 | JX010398 |
| C. nymphaeae | CBS 515.78* | Nymphaea alba | Netherlands | JQ948197 | JQ948527 | – | JQ949518 | JQ948858 | JQ949848 |
| C. oncidii | CBS 129828* | Oncidium sp., leaf | Germany | JQ005169 | JQ005256 | JQ005690 | JQ005517 | JQ005343 | JQ005603 |
| C. orbiculare | CBS 514.97 | Cucumis sativus | Japan | JQ005778 | KF178491 | – | JQ005841 | JQ005799 | JQ005862 |
| C. orchidearum | CBS 135131* | Dendrobium nobile | Netherlands | MG600738 | MG600800 | – | MG600944 | MG600855 | MG601005 |
| C. orchidophilum | CBS 632.80* | Dendrobium sp. | USA | JQ948151 | JQ948481 | – | JQ949472 | JQ948812 | JQ949802 |
| C. paranaense | CBS 134729* | Malus domestica | Brazil, Parana | KC204992 | KC205026 | – | KC205077 | KC205043 | KC205060 |
| C. parsonsiae | CBS 128525 | Parsonsia capsularis, leaf endophyte | New Zealand | JQ005233 | JQ005320 | JQ005754 | JQ005581 | JQ005407 | JQ005667 |
| C. paxtonii | IMI 165753* | Musa sp. | Saint Lucia | JQ948285 | JQ948615 | – | JQ949606 | JQ948946 | JQ949936 |
| C. petchii | CBS 378.94* | Dracaena marginata, spotted leaves | Italy | JQ005223 | JQ005310 | JQ005744 | JQ005571 | JQ005397 | JQ005657 |
| C. phormii | CBS 118194* | Phormium sp. | Germany | JQ948446 | JQ948777 | – | JQ949767 | JQ949107 | JQ950097 |
| C. phyllanthi | CBS 175.67* | Phyllanthus acidus, anthracnose | India | JQ005221 | JQ005308 | JQ005742 | JQ005569 | JQ005395 | JQ005655 |
| C. piperis | IMI 71397* | Piper nigrum | Malaysia | MG600760 | MG600820 | – | MG600964 | MG600867 | MG601027 |
| C. plurivorum | CBS 125474* | Coffea sp. | Vietnam | MG600718 | MG600781 | – | MG600925 | MG600841 | MG600985 |
| CBS 125473 | Coffea sp. | Vietnam | MG600717 | MG600780 | – | MG600924 | MG600840 | MG600984 | |
| CGMCC 3.17358 | Camellia sinensis, endophyte | China | KJ955215 | KJ954916 | – | KJ954483 | – | KJ955361 | |
| CMM 3742 | Mangifera indica | Brazil | KC702980 | KC702941 | – | KC702908 | KC598100 | KC992327 | |
| LJTJ30 | Capsicum annuum | China | KP748221 | KP823800 | – | KP823741 | – | KP823853 | |
| MAFF 243073 | Amorphophallus rivieri | Japan | MG600730 | MG600793 | – | MG600936 | MG600847 | MG600997 | |
| MAFF 305790 | Musa sp. | Japan | MG600726 | MG600789 | – | MG600932 | MG600845 | MG600993 | |
| C. psidii | CBS 145.29* | Psidium sp. | Italy | JX010219 | JX009967 | JX009743 | JX009515 | JX009901 | JX010443 |
| C. pyricola | CBS 128531* | Pyrus communis, fruit rot | New Zealand | JQ948445 | JQ948776 | – | JQ949766 | JQ949106 | JQ950096 |
| C. queenslandicum | ICMP 1778* | Carica papaya | Australia | JX010276 | JX009934 | JX009691 | JX009447 | JX009899 | JX010414 |
| C. quinquefoliae | MFLU 14-0626* | Parthenocissus quinquefolia | Russia | KU236391 | KU236390 | – | KU236389 | – | KU236392 |
| C. rhexiae | CBS 133134* | Rhexia virginica | USA | JX145128 | – | – | – | – | JX145179 |
| C. rhexiae (cont.) | CBS 133132 | Vaccinium macrocarpon | USA | JX145157 | – | – | – | – | JX145209 |
| C. rhombiforme | CBS 129953* | Olea europaea | Portugal | JQ948457 | JQ948788 | – | JQ949778 | JQ949118 | JQ950108 |
| C. salicis | CBS 607.94* | Salix sp., leaf, spot | Netherlands | JQ948460 | JQ948791 | JQ949781 | JQ949121 | JQ950111 | – |
| C. salsolae | ICMP 19051* | Salsola tragus | Hungary | JX010242 | JX009916 | JX009696 | JX009562 | JX009863 | JX010403 |
| C. sansevieriae | MAFF 239721* | Sansevieria trifasciata | Japan | AB212991 | – | – | – | – | – |
| C. sedi | MFLUCC 14-1002* | Sedum sp. | Russia | KM974758 | KM974755 | – | KM974756 | KM974754 | KM974757 |
| C. siamense | ICMP 18578* | Coffea arabica | Thailand | JX010171 | JX009924 | FJ917505 | FJ907423 | JX009865 | JX010404 |
| ICMP 12567 | Persea americana | Australia | JX010250 | JX009940 | JX009697 | JX009541 | JX009761 | JX010387 | |
| ICMP 18574 | Pistacia vera | Australia | JX010270 | JX010002 | JX009707 | JX009535 | JX009798 | JX010391 | |
| ICMP 18121 | Dioscorea rotundata | Nigeria | JX010245 | JX009942 | JX009715 | JX009460 | JX009845 | JX010402 | |
| ICMP 17795 | Malus domestica | USA | JX010162 | JX010051 | JX009703 | JX009506 | JX009805 | JX010393 | |
| C. siamense (syn. C. hymenocallidis) | ICMP 18642* | Hymenocallis americana | China | JX010278 | JX010019 | JX009709 | GQ856775 | GQ856730 | JX010410 |
| C. siamense (syn. C. jasmini-sambac) | ICMP 19118* | Jasminum sambac | Vietnam | HM131511 | HM131497 | JX009713 | HM131507 | JX009895 | JX010415 |
| C. simmondsii | CBS 122122* | Carica papaya | Australia | JQ948276 | JQ948606 | – | JQ949597 | JQ948937 | JQ949927 |
| C. sloanei | IMI 364297* | Theobroma cacao, leaf | Malaysia | JQ948287 | JQ948617 | – | JQ949608 | JQ948948 | JQ949938 |
| C. sojae | ATCC 62257* | Glycine max | USA | MG600749 | MG600810 | – | MG600954 | MG600860 | MG601016 |
| CGMCC 3.15171 | Bletilla ochracea | China | HM751813 | KC843501 | – | KC843550 | – | KC244161 | |
| C. sonchicola | JZB330117 | Sonchus sp. | Italy | KY962756 | KY962753 | – | KY962747 | KY962750 | – |
| MFLUCC 17-1300 | Sonchus sp. | Italy | KY962758 | KY962755 | – | KY962749 | KY962752 | – | |
| C. spinaciae | CBS 128.57 | Spinacia oleracea | Netherlands | GU227847 | GU228239 | – | GU227945 | GU228337 | GU228141 |
| C. sydowii | CBS 135819 | Sambucus sp. | China, Taiwan | KY263783 | KY263785 | – | KY263791 | KY263787 | KY263793 |
| C. tamarilloi | CBS 129814* | Solanum betaceum, fruit, anthracnose | Colombia | JQ948184 | JQ948514 | – | JQ949505 | JQ948845 | JQ949835 |
| C. temperatum | CBS 133122* | Vaccinium macrocarpon | USA | JX145159 | – | – | – | – | JX145211 |
| CBS 133120 | Vaccinium macrocarpon | USA | JX145135 | – | – | – | – | JX145186 | |
| C. theobromicola | CBS 124945* | Theobroma cacao | Panama | JX010294 | JX010006 | JX009591 | JX009444 | JX009869 | JX010447 |
| C. ti | ICMP 4832* | Cordyline sp. | New Zealand | JX010269 | JX009952 | JX009649 | JX009520 | JX009898 | JX010442 |
| C. torulosum | CBS 128544* | Solanum melongena | New Zealand | JQ005164 | JQ005251 | JQ005685 | JQ005512 | JQ005338 | JQ005598 |
| C. tropicale | CBS 124949* | Theobroma cacao | Panama | JX010264 | JX010007 | JX009719 | JX009489 | JX009870 | JX010407 |
| C. tropicicola | BCC 38877* | Citrus maxima | Thailand | JN050240 | JN050229 | – | JN050218 | – | JN050246 |
| MFLUCC100167 | Paphiopedilum bellatolum | Thailand | JN050241 | JN050230 | – | JN050219 | – | JN050247 | |
| C. truncatum | CBS 151.35* | Phaseolus lunatus | USA | GU227862 | GU228254 | – | GU227960 | GU228352 | GU228156 |
| C. viniferum | GZAAS 5.08601* | Vitis vinifera cv. Shuijing | China | JN412804 | JN412798 | JQ309639 | JN412795 | – | JN412813 |
| C. vittalense | CBS 181.82* | Theobroma cacao | India | MG600734 | MG600796 | – | MG600940 | MG600851 | MG601001 |
| C. walleri | CBS 125472* | Coffea sp., leaf tissue | Vietnam | JQ948275 | JQ948605 | – | JQ949596 | JQ948936 | JQ949926 |
| C. wuxiense | CGMCC 3.17894* | Camellia sinensis | China | KU251591 | KU252045 | KU251833 | KU251672 | KU251939 | KU252200 |
| JS1A44 | Camellia sinensis | China | KU251592 | KU252046 | KU251834 | KU251673 | KU251940 | KU252201 | |
| C. xanthorrhoeae | ICMP 17903* | Xanthorrhoea preissii | Australia | JX010261 | JX009927 | JX009653 | JX009478 | JX009823 | JX010448 |
| C. yunnanense | CBS 132135* | Buxus sp. | China | JX546804 | JX546706 | – | JX519239 | JX519231 | JX519248 |
| Colletotrichum sp. | CGMCC 3.15172 | Bletilla ochracea | China | HM751816 | KC843522 | – | KC843547 | – | KC244162 |
| Q026 | Rubus glaucus | Colombia | JN715839 | KC860013 | – | KC859970 | KC859995 | KC860039 | |
| Glomerella cingulata ‘f. sp. camelliae’ | ICMP 10643 | Camellia × williamsii | UK | JX010224 | JX009908 | JX009630 | JX009540 | JX009891 | JX010436 |
| Monilochaetes infuscans | CBS 869.96* | Ipomoea batatas | South Africa | JQ005780 | JX546612 | – | JQ005843 | JQ005801 | JQ005864 |
x ATCC: American Type Culture Collection; BCC: BIOTEC Culture Collection, National Center for Genetic Engineering and Biotechnology (BIOTEC), Khlong Luang, Pathumthani, Thailand; BRIP: Plant Pathology Herbarium, Department of Employment, Economic, Development and Innovation, Queensland, Australia; CBS: Culture collection of the Centraalbureau voor Schimmelcultures, Fungal Biodiversity Centre, Utrecht, The Netherlands; CGMCC: China General Microbiological Culture Collection; CMM: Culture Collection of Phytopathogenic Fungi Prof. Maria Menezes, Federal Rural University of Pernambuco, Brazil; COAD: Coleção Octávio Almeida Drummond, Viçosa, Brazil; GZAAS: Guizhou Academy of Agricultural Sciences Herbarium, China; ICMP: International Collection of Microorganisms from Plants, Auckland, New Zealand; IMI: Culture collection of CABI Europe UK Centre, Egham, UK; MAFF: MAFF Genebank Project, Ministry of Agriculture, Forestry and Fisheries, Tsukuba, Japan; MFLU: Herbarium of Mae Fah Luang University, Chiang Rai, Thailand; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand.
* = ex-type culture.
Fig. 2.
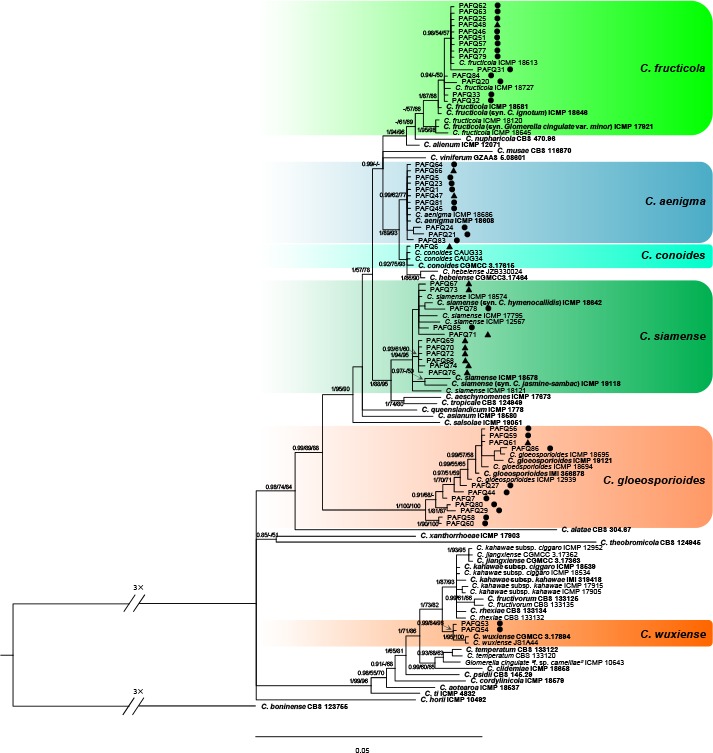
A Bayesian inference phylogenetic tree of 111 isolates in the C. gloeosporioides species complex. The species C. boninense (CBS 123755) was selected as an outgroup. The tree was built using concatenated sequences of the ACT, TUB2, CAL, CHS-1, GAPDH, and ITS genes. Bayesian posterior probability (PP ≥ 0.90), MP bootstrap support values (ML ≥ 50 %), and RAxML bootstrap support values (ML ≥ 50 %) were shown at the nodes (PP/MP/ML). Ex-type isolates are in bold. Coloured blocks indicate clades containing isolates from Pyrus spp. in this study; circles indicate isolates isolated from leaves, triangles indicate isolates isolated from fruits. The scale bar indicates 0.05 expected changes per site.
Fig. 5.
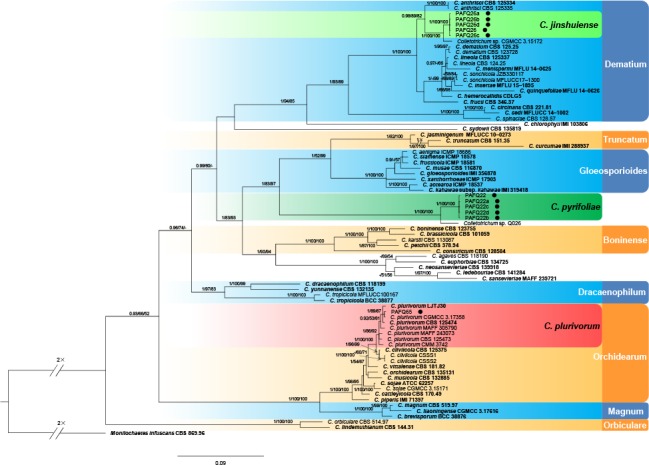
Phylogenetic tree generated by Bayesian inference based on concatenated sequences of the ACT, CHS-1, GAPDH, ITS, and TUB genes. Monilochaetes infuscans (CBS 869.96) was selected as an outgroup. Bayesian posterior probability (PP ≥ 0.90), MP bootstrap support values (ML ≥ 50 %), and RAxML bootstrap support values (ML ≥ 50 %) were shown at the nodes (PP/MP/ML). Ex-type isolates are in bold. Coloured blocks are used to indicate clades containing isolates from Pyrus spp. in this study; circles indicate isolates isolated from leaves. The scale bar indicates 0.09 expected changes per site.
In the phylogenetic tree constructed for the isolates in the C. gloeosporioides species complex, 50 isolates clustered in six clades corresponding to C. fructicola (14 isolates), C. aenigma (11), C. siamense (11), C. gloeosporioides (11), C. wuxiense (2), and C. conoides (1) (Fig. 2). For the isolates in the C. acutatum species complex, 13 isolates grouped in subclade II of C. fioriniae (Bayesian posterior probabilities value 1/PAUP bootstrap support value 97/RAxML bootstrap support value 100) as defined in a previous study (Damm et al. 2012b), while two isolates (PAFQ49 and PAFQ50) formed a further subclade, which is designated as subclade III (Fig. 3). For isolates in the C. boninense species complex, 13 isolates clustered with C. karstii, and one with C. citricola (Fig. 4). For the remaining 11 isolates, PAFQ65 clustered with C. plurivorum (1/86/92), while five isolates formed a distinct clade (1/100/100) as sister to Colletotrichum sp. isolate CGMCC 3.15172 in the C. dematium species complex. In addition, the remaining five isolates, which formed a distinct clade (1/100/100), clustered distantly from any known Colletotrichum species complex (Fig. 5).
Fig. 3.
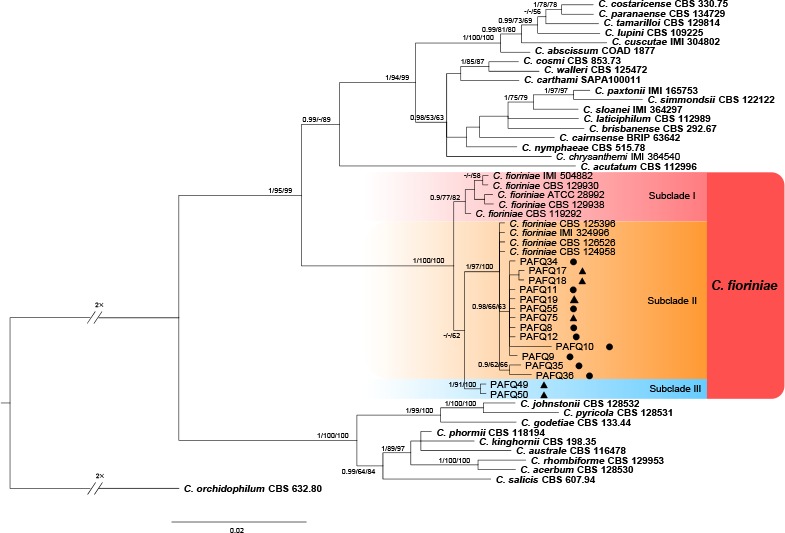
A Bayesian inference phylogenetic tree of 51 isolates in the C. acutatum species complex. The species C. orchidophilum (CBS 632.80) was selected as an outgroup. The tree was built using concatenated sequences of the ACT, TUB2, CHS-1, GAPDH, and ITS genes. Bayesian posterior probability (PP ≥ 0.90), MP bootstrap support values (ML ≥ 50 %), and RAxML bootstrap support values (ML ≥ 50 %) were shown at the nodes (PP/MP/ML). Ex-type isolates are in bold. Coloured blocks indicate clades containing isolates from Pyrus spp. in this study; circles indicate isolates isolated from leaves, triangles indicate isolates isolated from fruits. The scale bar indicates 0.02 expected changes per site.
Fig. 4.
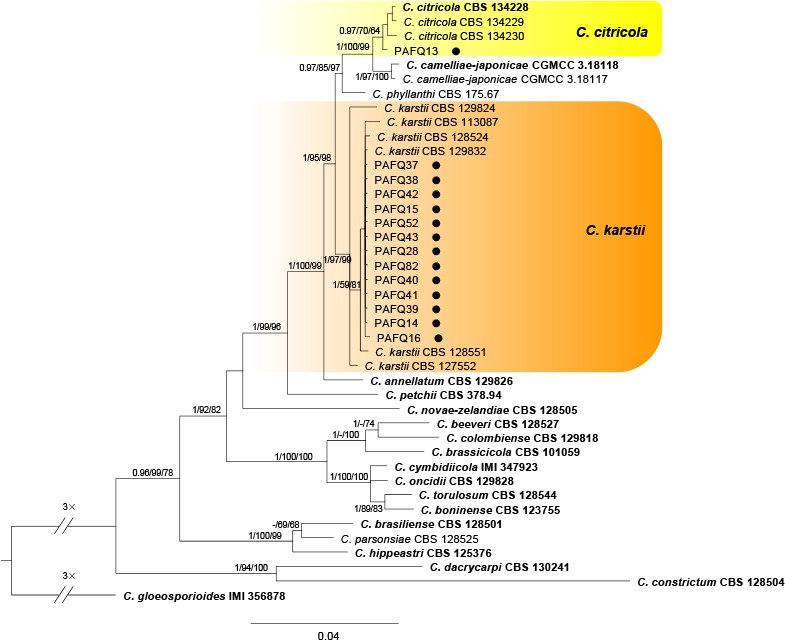
A Bayesian inference phylogenetic tree of 41 isolates in the C. boninense species complex. The species C. gloeosporioides (IMI 356878) was selected as an outgroup. The tree was built using concatenated sequences of the ACT, TUB2, CAL, CHS-1, GAPDH, and ITS genes. Bayesian posterior probability (PP ≥ 0.90), MP bootstrap support values (ML ≥ 50 %), and RAxML bootstrap support values (ML ≥ 50 %) were shown at the nodes (PP/MP/ML). Ex-type isolates are in bold. Coloured blocks indicate clades containing isolates from Pyrus spp. in this study; circles indicate isolates isolated from leaves. The scale bar indicates 0.04 expected changes per site.
To exclude the possibility that species delimitation might be interfered by recombination among the genes used for phylogenetic analyses, the multi-locus (ACT, TUB2, CHS-1, GAPDH, and ITS) concatenated datasets were subjected to two PHI tests (Fig. 6) to determine the recombination level within phylogenetically closely related species. The results showed that no significant recombination events were observed between C. jinshuiense and phylogenetically related isolates or species (Colletotrichum sp. isolate CGMCC 3.15172, C. anthrisci and C. fructi) (Fig. 6a), and between C. pyrifoliae and phylogenetically related isolates or species (Colletotrichum sp. isolate Q026, C. boninense and C. kahawae) (Fig. 6b).
Fig. 6.
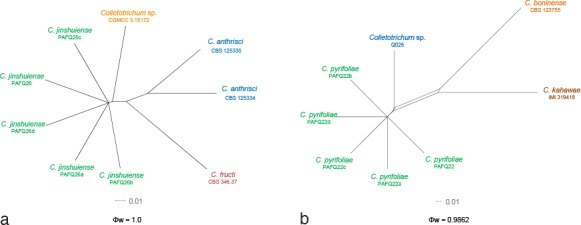
The result of the pairwise homoplasy index (PHI) tests of closely related species using both LogDet transformation and splits decomposition. a, b. The PHI of C. jinshuiense (a) or C. pyrifoliae (b) and their phylogenetically related isolates or species, respectively. PHI test value (Φw) < 0.05 indicate significant recombination within the dataset.
Taxonomy
Based on morphology and multi-locus sequence data, the 90 isolates were assigned to 12 Colletotrichum spp. Of these, two species proved to represent new taxa that are described below. Six species are reported from pear for the first time. Eight species formed sexual morphs in vitro.
Colletotrichum aenigma B.S. Weir & P.R. Johnst., Stud. Mycol. 73: 135. 2012. — Fig. 7
Fig. 7.
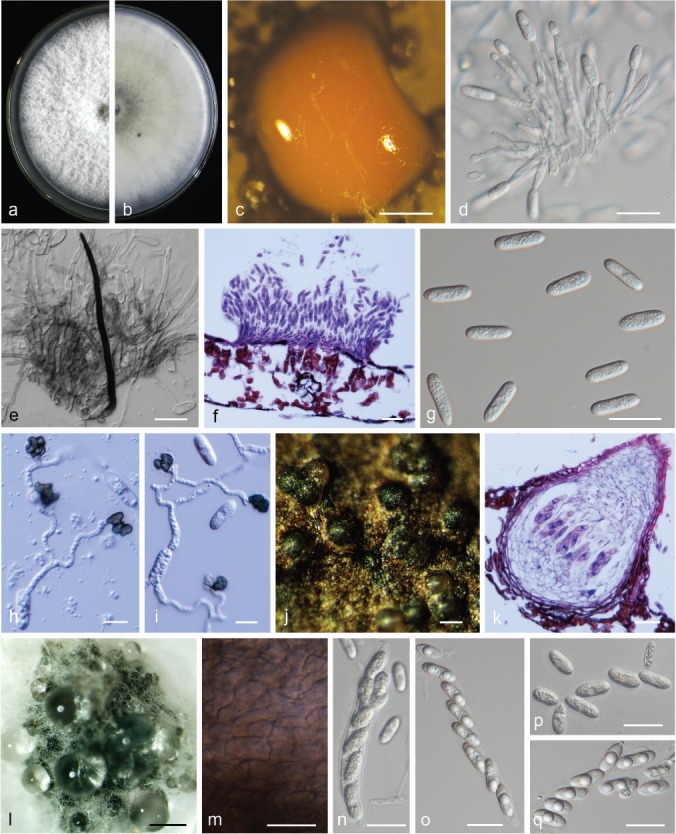
Colletotrichum aenigma. a, b. Front and back view, respectively, of 6-d-old PDA culture; c. conidiomata; d. conidiophores; e. seta; f. section view of acervulus produced on pear leaf (P. pyrifolia cv. Cuiguan); g. conidia; h, i. appressoria; j. ascomata produced on pear leaf (P. bretschneideri cv. Dangshansuli); k. section view of ascoma produced on pear leaf (P. pyrifolia cv. Cuiguan); l. ascomata; m. outer surface of peridium; n, o. asci; p, q. ascospores (a–c, i–m. isolate PAFQ1; d–h. isolate PAFQ47; n, p. isolate PAFQ3; o, q. isolate PAFQ2; a–e, g, l–q produced on PDA agar medium). — Scale bars: c, l = 500 μm; d–g, k, m–q = 20 μm; h, i =10 μm; j = 100 μm.
Description & Illustration — Weir et al. (2012), Wang et al. (2016).
Materials examined. CHINA, Hubei Province, Zhongxiang City, on leaves of P. pyrifolia cv. Xiangnan, 1 Sept. 2015, M. Fu (culture PAFQ1); ibid., on leaves of P. pyrifolia cv. Huanghua, 1 Sept. 2015, M. Fu (PAFQ3); ibid., on leaves of P. pyrifolia cv. Huali No.1, 1 Sept. 2015, M. Fu (PAFQ5); Jiangsu Province, Yancheng City, on fruits of P. bretschneideri cv. Renli, 1 Sept. 2015, M. Fu (PAFQ47); ibid., on leaves of P. bretschneideri cv. Yali, 1 Sept. 2015, M. Fu (PAFQ45); Zhejiang Province, Hangzhou City, on leaves of P. pyrifolia cv. Guanyangxueli, 18 Aug. 2016, M. Fu (PAFQ81); Anhui Province, Dangshan County, on fruits of P. bretschneideri cv. Huangguan, 4 Aug. 2016, M. Fu (PAFQ66).
Notes — A total of 40 isolates were collected. Colletotrichum aenigma has been reported to cause anthracnose diseases of P. pyrifolia from Japan (Weir et al. 2012), and P. communis from Italy (Schena et al. 2014). This is the first report of C. aenigma causing anthracnose on P. bretschneideri and on Pyrus in China.
Colletotrichum citricola F. Huang et al., Fung. Diversity 61: 67. 2013. — Fig. 8
Fig. 8.
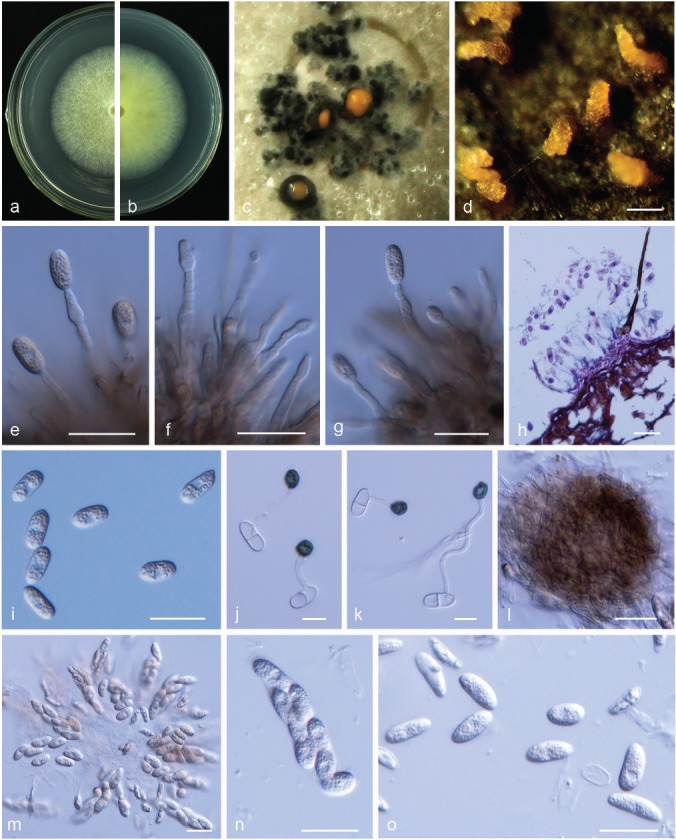
Colletotrichum citricola. a, b. Front and back view, respectively of 6-d-old PDA culture; c, d. conidiomata; e–g. conidiophores; h. section view of acervulus produced on pear leaf (P. pyrifolia cv. Cuiguan); i. conidia; j, k. appressoria; l. ascoma; m, n. asci; o. ascospores (a–o. isolate PAFQ13; a–c, e–g, i, l–o. produced on PDA agar medium, d. produced on pear leaf (P. bretschneideri cv. Dangshansuli)). — Scale bars: d = 100 μm; e–i, l–o = 20 μm; j, k = 10 μm.
Description & Illustration — Huang et al. (2013).
Materials examined. CHINA, Hubei Province, Wuhan City, on leaves of P. pyrifolia, 1 Sept. 2015, P.F. Zhang (culture PAFQ13).
Notes — Colletotrichum citricola was first reported as a saprobe from Citrus unshiu in China (Huang et al. 2013). Isolate PAFQ13 was isolated from pear leaves, and clustered together with the ex-type culture of C. citricola (CBS 134228) in the multi-locus phylogenetic tree (Fig. 4). This is the first report of C. citricola causing anthracnose on P. pyrifolia.
Ascospores of the isolate PAFQ13 (13.5–20 × 5–8 μm, mean ± SD = 17.4 ± 1.4 × 7.1 ± 0.7 μm) are slightly larger than those of the ex-type isolate CBS 134228 (12.8–18.4 × 5.3–6.7 μm, mean = 15.8 × 6.1 μm) of C. citricola. Setae were observed in the acervuli formed on pear leaves, being brown, smooth-walled, 2-septate, 41–84 μm long, base rounded, 6 μm diam, tip more or less acute.
Colletotrichum conoides Y.Z. Diao et al., Persoonia 38: 27. 2017. — Fig. 9
Fig. 9.
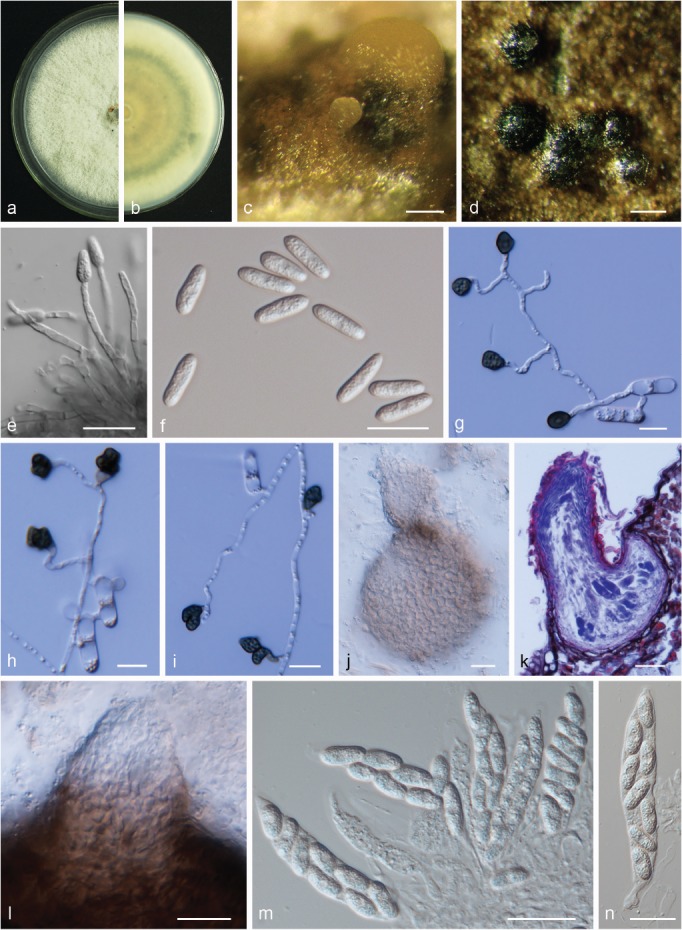
Colletotrichum conoides. a, b. Front and back view, respectively, of 6-d-old PDA culture; c. conidiomata; d. ascomata produced on pear leaf (P. bretschneideri cv. Dangshansuli); e. conidiophores; f. conidia; g–i. appressoria; j. ascoma; k. section view of ascoma produced on pear leaf (P. pyrifolia cv. Cuiguan); l. neck of ascoma; m, n. asci (a–n. isolate PAFQ6; a–c, e, f, j, l–n. produced on PDA agar medium). — Scale bars: c, d = 100 μm; e, f, j–n = 20 μm; g–i =10 μm.
Sexual morph developed on PDA. Ascomata ovoid to obpyriform, light to dark brown, 77–180 × 69–159 μm, ostiolate. Asci cylindrical to clavate, 59.5–99 × 13.5–18.5 μm, 8-spored. Ascospores hyaline, smooth-walled, aseptate, cylindrical, sometimes slightly curved, both sides rounded, contents granular, 12.5–21 × 5.5–7.5 μm, mean ± SD = 15.9 ± 1.3 × 6.8 ± 0.5 μm, L/W ratio = 2.3.
Asexual morph developed on PDA. Conidiophores hyaline, smooth-walled, septate, branched. Conidiogenous cells hyaline, cylindrical to clavate, 18–34.5 × 2–3 μm. Conidia hyaline, aseptate, smooth-walled, cylindrical, both ends round or one end slightly acute, usually broader towards one side, contents granular, 16–20 × 4.5–6 μm, mean ± SD = 18.4 ± 0.8 × 5.6 ± 0.3 μm, L/W ratio = 3.3. Appressoria dark brown, irregular, but often square to ellipsoid in outline, the margin lobate, 7–12.5 × 5–8.5 μm, mean ± SD = 9.7 ± 1.3 × 6.9 ± 1.1 μm, L/W ratio = 1.4.
Culture characteristics — Colonies on PDA flat with entire margin, aerial mycelium white, cottony, dense; reverse light grey in the centre and pale white margin, olivaceous coloured pigments formed in the shape of a concentric ring pattern; colony diam 77–78 mm in 5 d. Conidia in mass orange.
Materials examined. CHINA, Hubei Province, Wuhan City, on fruits of P. pyrifolia, 1 Sept. 2015, M. Fu (culture PAFQ6).
Notes — Colletotrichum conoides was first reported on Capsicum annuum (chili) from China (Diao et al. 2017). In the present study, one isolate (PAFQ6) from pear fruit clustered together with the ex-type culture of C. conoides (CGMCC 3.17615) in the multi-locus phylogenetic tree (Fig. 2). This is the first report of C. conoides to cause anthracnose on P. pyrifolia and the first description of its sexual morph.
Conidia of the isolate PAFQ6 (16–20 × 4.5–6 μm, mean ± SD = 18.4 ± 0.8 × 5.6 ± 0.3 μm) are longer than those of the ex-type isolate CGMCC 3.17615 (13–17.5 × 5–6.5 μm, mean = 15.9 × 5.9 μm) of C. conoides.
Colletotrichum fioriniae (Marcelino & Gouli) Pennycook,
Mycotaxon 132: 150. 2017. — Fig. 10
Fig. 10.
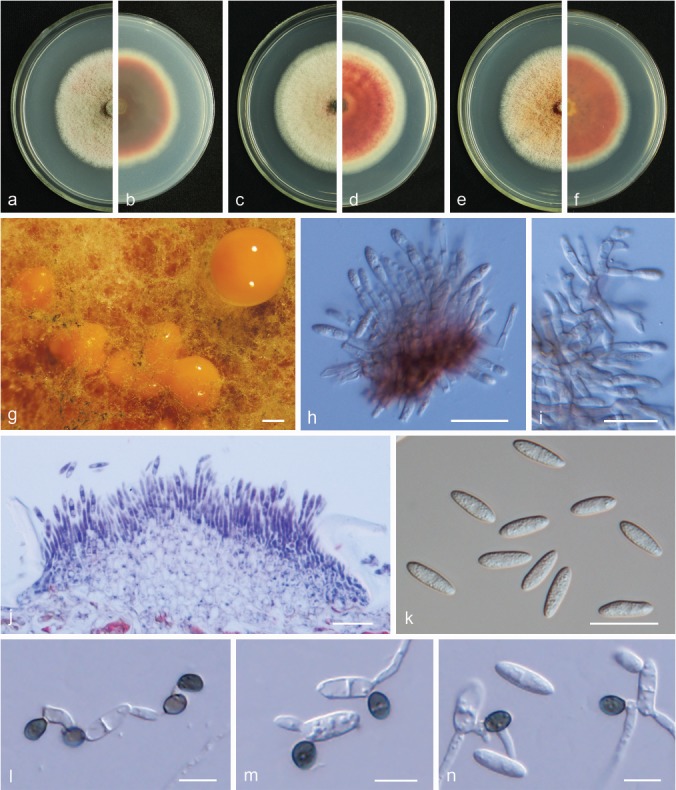
Colletotrichum fioriniae. a, c, e. Front views of 6-d-old PDA culture; b, d, f. back views of 6-d-old PDA culture; g. conidiomata; h, i. conidiophores; j. section view of acervulus produced on pear fruit (P. bretschneideri cv. Huangguan); k. conidia; l–n. appressoria (a, b, g–l. isolate PAFQ8, c, d, m. isolate PAFQ36, e, f, n. isolate PAFQ49; a–i, k produced on PDA agar medium). — Scale bars: g = 400 μm; h–k = 20 μm; l–n = 10 μm.
Description & Illustration — Damm et al. (2012b).
Materials examined. CHINA, Hubei Province, Wuhan City, on leaves of P. pyrifolia cv. Jinshui No. 1, 1 Sept. 2015, M. Fu (cultures PAFQ8 and PAFQ9); ibid., on fruits of P. pyrifolia, 1 Aug. 2016, M. Fu (PAFQ17); Fujian Province, Jianning County, on leaves of P. pyrifolia cv. Cuiguan, 1 Apr. 2016, M. Fu (PAFQ35, PAFQ36); Jiangxi Province, Jinxi County, on leaves of P. pyrifolia cv. Cuiguan, 23 July 2016, M. Fu (PAFQ55); Shandong Province, Yantai City, on fruits of P. communis cv. Gyuiot, 27 Aug. 2016, M. Fu (PAFQ75); Jiangsu Province, Nanjing City, on leaves of P. pyrifolia, 20 Aug. 2016, M. Fu (PAFQ49).
Notes — Colletotrichum fioriniae was first reported on Persea americana and Acacia acuminata from Australia (Shivas & Tan 2009) and also caused fruit rot on Pyrus sp. in the USA (Damm et al. 2012b). In the study of Damm et al. (2012b), isolates clustered in two subclades, here designated as I and II. In the current study, an additional subclade (III) was detected (Fig. 3), which differs from subclade I in 2–3 bp in ACT, 1 bp in CHS, 1 bp in GAPDH, and 1 bp in TUB2, and subclade II in 3 bp in CHS, 4 bp in GAPDH, and 2 bp in TUB2.
Colletotrichum fructicola Prihast. et al., Fung. Diversity 39: 96. 2009. — Fig. 11
Fig. 11.
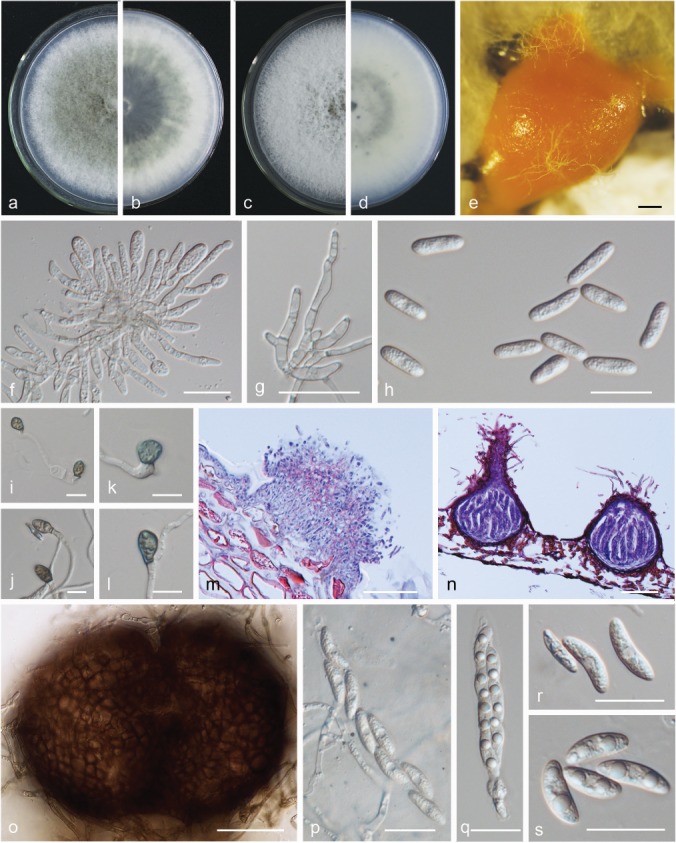
Colletotrichum fructicola. a, c. Front views of 6-d-old PDA culture; b, d. back views of 6-d-old PDA culture; e. conidiomata; f, g. conidiophores; h. conidia; i–l. appressoria; m. section view of acervulus produced on pear fruit (P. bretschneideri cv. Huangguan); n. section view of ascomata produced on pear leaf (P. pyrifolia cv. Cuiguan); o. ascomata; p, q. asci; r, s. ascospores (a, b, h–l, o, q, r. isolate PAFQ31, c–e, m, n. isolate PAFQ32, p, s. isolate PAFQ48, f, g. isolate PAFQ30; a–h, o–s produced on PDA agar medium). — Scale bars: e = 500 μm; f–h, p–s = 20 μm; i–l = 10 μm; m–o = 50 μm.
Description & Illustration — Prihastuti et al. (2009).
Materials examined. CHINA, Fujian Province, Jianning County, on leaves of P. pyrifolia cv. Cuiguan, Apr. 2014, P.F. Zhang (cultures PAFQ30 and PAFQ31); ibid., 1 Sept. 2015, M. Fu (PAFQ32, PAFQ33); Jiangxi Province, Jinxi County, on leaves of P. pyrifolia cv. Cuiguan, 23 July 2016, M. Fu (PAFQ88); Hubei Province, Wuhan City, on leaves of P. pyrifolia cv. Jingshui, 1 Aug. 2016, M. Fu (PAFQ20, PAFQ25); Zhejiang Province, Hangzhou City, on leaves of P. pyrifolia cv. Guanyangxueli, 18 Aug. 2016, M. Fu (PAFQ79); ibid., Tonglu County, on leaves of P. pyrifolia cv. Cuiguan, 18 Aug. 2016, M. Fu (PAFQ84); Jiangsu Province, Yancheng City, on fruits of P. bretschneideri cv. Dangshanshuli, 1 Sept. 2015, M. Fu (PAFQ48); ibid., on leaves of P. bretschneideri cv. Yali, 1 Sept. 2015, M. Fu (PAFQ46); Anhui Province, Dangshan County, on leaves of P. bretschneideri cv. Huangguan, 4 Aug. 2016, M. Fu (PAFQ62); ibid., on fruits of P. bretschneideri cv. Huangguan, 4 Aug. 2016, M. Fu (PAFQ90).
Notes — Colletotrichum fructicola was first reported on Coffea arabica in Thailand (Prihastuti et al. 2009), and subsequently reported on Pyrus pyrifolia in Japan (Weir et al. 2012), Citrus reticulata in China (Huang et al. 2013), Pyrus bretschneideri in China (Li et al. 2013), and other plants (e.g., Lima et al. 2013, Liu et al. 2015, Diao et al. 2017). The species was identified as responsible for pear anthracnose, causing TS symptoms on P. pyrifolia leaves (Zhang et al. 2015) and P. bretschneideri fruits in China (Jiang et al. 2014).
Colletotrichum gloeosporioides (Penz.) Penz. & Sacc., Atti Reale Ist. Veneto Sci. Lett. Arti., ser. 6, 2: 670. 1884. — Fig. 12
Fig. 12.
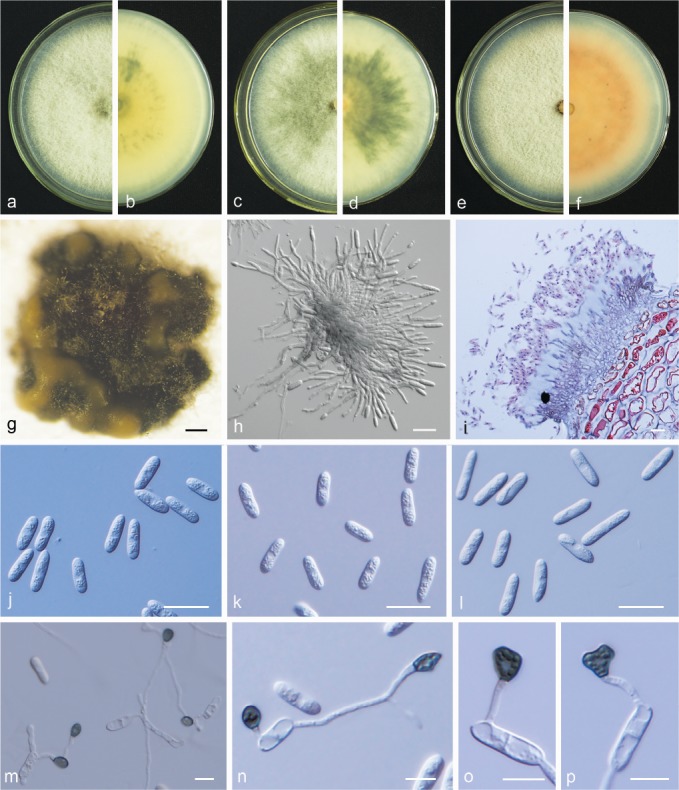
Colletotrichum gloeosporioides. a, c, e. Front views of 6-d-old PDA culture; b, d, f. back views of 6-d-old PDA culture; g. conidiomata; h. conidiophores; i. section view of acervulus produced on pear fruit (P. bretschneideri cv. Huangguan); j–l. conidia; m–p. appressoria (a, b, j, m. isolate PAFQ80, c, d, k, n. isolate PAFQ7, e–i, l, o, p. isolate PAFQ56; a–h, j–l produced on PDA agar medium). — Scale bars: g = 200 μm; h–l = 20 μm; m–p = 10 μm.
Description & Illustration — Cannon et al. (2008), Liu et al. (2015).
Materials examined. CHINA, Jiangxi Province, Jinxi County, on leaves of P. pyrifolia cv. Cuiguan, 23 July 2016, M. Fu (culture PAFQ56); ibid., on fruits of P. pyrifolia cv. Huanghua, 23 July 2016, M. Fu (PAFQ61); Hubei Province, Wuhan City, on leaves of P. pyrifolia cv. Hohsui, 1 Aug. 2016, M. Fu (PAFQ27); ibid., on leaves of P. bretschneideri cv. Huangxianchangba, 1 Sept. 2016, M. Fu (PAFQ7); Jiangsu Province, Yancheng City, on leaves of P. bretschneideri cv. Yali, 1 Sept. 2015, M. Fu (PAFQ44); Zhejiang Province, Hangzhou City, on leaves of P. pyrifolia cv. Guanyangxueli, 18 Aug. 2016, M. Fu (PAFQ80); ibid., on leaves of Pyrus sp., 18 Sept. 2016, M. Fu (PAFQ86).
Notes — Although C. gloeosporioides has been identified as responsible for pear anthracnose in China, these identifications were chiefly based on morphology and/or ITS sequence data (Wu et al. 2010, Liu et al. 2013b). In this study, 20 isolates of C. gloeosporioides isolated from fruits and leaves of pear were identified as C. gloeosporioides based on multi-loci phylogenetic analyses and confirmed as responsible for pear anthracnose following Koch’s postulates.
Colletotrichum jinshuiense M. Fu & G.P. Wang, sp. nov. — MycoBank MB824216; Fig. 13
Fig. 13.

Colletotrichum jinshuiense. a, b. Front and back view, respectively, of 6-d-old PDA culture; c. acervuli produced on pear leaf (P. bretschneideri cv. Dangshansuli); d. acervuli produced on pear fruit; e, f. section view of acervulus produced on pear leaf and fruit, respectively; g, h. conidiophores; i. setae; j, k. conidia; l, m. appressoria (a–m. isolate PAFQ26; a, b. produced on PDA agar medium; c, e, j, l. from pear leaf (P. pyrifoliae cv. Cuiguan), d, f–i, k–m. from pear fruit (P. bretschneideri cv. Huangguan)). — Scale bars: c = 200 μm; d = 100 μm; e–k = 20 μm; l, m = 10 μm.
Etymology. Referring to the host variety (P. pyrofolia cv. Jinshui) from which the fungus was isolated.
Sexual morph not observed. Asexual morph on pear leaves and fruit. Conidiomata acervular, conidiophores and setae formed from a brown stroma. Setae dark brown to black, opaque, tip acute, base cylindrical, 1–4-septate, 59–363 (on leaf surface) and 70–272 μm long (on fruit surface). Conidiophores pale brown to hyaline, simple to 2-septate, unbranched. Conidiogenous cells (on fruit surface) hyaline, smooth-walled, cylindrical, 12.5–27 × 3.5–4.5 μm, opening 1–2 μm. Conidia, hyaline, smooth-walled, aseptate, curved, base subtruncate, apex acute, contents with 1–2 guttules, on leaf surface: 25–29.5 × 3.5–4.5 μm, mean ± SD = 27.1 ± 1.7 × 4.0 ± 0.3 μm, L/W ratio = 6.8; on fruit surface: 21–30.5 × 3–4.5 μm, mean ± SD = 24.4 ± 2.1 × 4.0 ± 0.3 μm, L/W ratio = 6.2. Appressoria pale brown, smooth-walled, ellipsoidal to clavate, 8–17 × 5–7.5 μm, mean ± SD = 10.7 ± 1.7 × 6.0 ± 0.5 μm, L/W ratio = 1.8.
Culture characteristics — Colonies on PDA flat with entire margin, aerial mycelium sparse, cottony, surface pale grey-black with white margin; reverse black to dark grey-green in centre with white margin. Colony diam 56–57 mm in 5 d. Conidia in mass not observed on PDA or SNA.
Materials examined. CHINA, Hubei Province, Wuhan City, on leaves of P. pyrifolia cv. Jinshui, 1 Aug. 2016, M. Fu (holotype HMAS 247824, culture ex-type CGMCC 3.18903 = PAFQ26); ibid., culture PAFQ26a, PAFQ26b, PAFQ26c, and PAFQ26d.
Notes — Isolates of C. jinshuiense are phylogenetically closely related to Colletotrichum sp. isolate CGMCC 3.15172 (Fig. 5), which was reported as an endophytic Colletotrichum species from Bletilla ochracea (Orchidaceae) in China (Tao et al. 2013), whereas they are different in GAPDH (94.98 %), and TUB2 (98.12 %). Furthermore, the PHI test (Φw = 1) did not detect recombination between these isolates and Colletotrichum sp. isolate CGMCC 3.15172 (Fig. 6a). In this study, C. jinshuiense clustered in the C. dematium species complex, which is often associated with herbaceous plants (Damm et al. 2009). The asexual and sexual morphs of C. jinshuiense were not observed on PDA or SNA, while they easily developed on pear fruit and leaves, indicating that pear tissue plays an important part in the epidemiology and life cycle of C. jinshuiense.
Colletotrichum karstii Yan L. Yang et al., Cryptog. Mycol. 32: 241. 2011. — Fig. 14
Fig. 14.
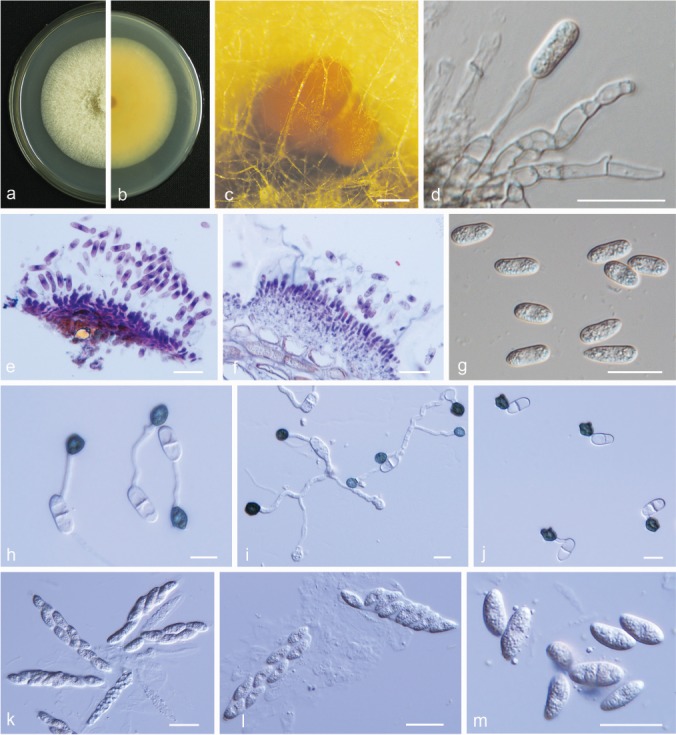
Colletotrichum karstii. a, b. Front and back view, respectively, of 6-d-old PDA culture; c. conidiomata; d. conidiophores; e, f. section view of acervulus produced on pear leaf (P. pyrifolia cv. Cuiguan) and fruit (P. bretschneideri cv. Huangguan), respectively; g. conidia; h–j. appressoria; k, l. asci; m. ascospores (a–h. isolate PAFQ14, i, k–m. isolate PAFQ40, j isolate PAFQ52; a–d, g, k–m produced on PDA agar medium). — Scale bars: c = 200 μm; d–g, k–m = 20 μm; h–j = 10 μm.
Description & Illustration — Yang et al. (2011).
Materials examined. CHINA, Hubei Province, Wuhan City, on leaves of P. pyrifolia, 1 Sept. 2015, P.F. Zhang (culture PAFQ14); ibid., on leaves of P. pyrifolia cv. Hohsui, 1 Aug. 2016, M. Fu (PAFQ28); Fujian Province, Jianning County, on leaves of P. pyrifolia cv. Cuiguan, 20 Oct. 2016, M. Fu (PAFQ40); Zhejiang Province, Hangzhou City, on leaves of P. pyrifolia cv. Guanyangxueli, 18 Aug. 2016, M. Fu (PAFQ82); Jiangxi Province, Jinxi County, on leaves of P. pyrifolia cv. Cuiguan, 23 July 2016, M. Fu (PAFQ52).
Notes — Colletotrichum karstii was first reported on Vanda sp. in China (Yang et al. 2011) and is diverse in its geographical distribution and host range (Damm et al. 2012a). In this study, 19 isolates of Colletotrichum were identified as belonging to this species, and this is the first report of C. karstii causing anthracnose of P. pyrifolia.
Conidia of the ex-type (GZAAS 090006, 12–19.5 × (5–)6–7.5 μm, mean ± SD = 15.4 ±1.3 × 6.5 ± 0.5 μm) of C. karstii are slightly smaller than that of isolate PAFQ82 (12.5–21 × 5–8 μm, mean ± SD = 16.8 ± 1.6 × 7.2 ± 0.6 μm), but larger than that of isolate PAFQ40 (12.5–16 × 5.5–7.5 μm, mean ± SD = 13.6 ± 0.8 × 6.5 ± 0.4 μm) and isolate PAFQ52 (11.5–16 × 5.5–7.5 μm, mean ± SD = 13.9 ± 1.0 × 6.8 ± 0.3 μm).
Colletotrichum plurivorum Damm et al., Stud. Mycol. 92: 31. 2019. — Fig. 15
Fig. 15.
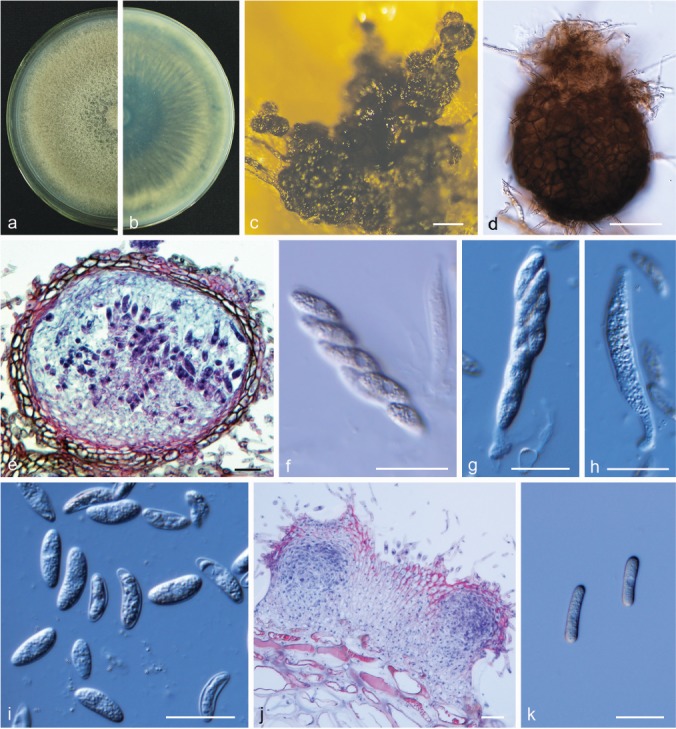
Colletotrichum plurivorum. a, b. Front and back view, respectively, of 6-d-old PDA culture; c, d. ascomata; e. section of ascoma; f, g. asci; h. immature ascus; i. ascospores; j. section view of acervulus produced on pear fruit (P. bretschneideri cv. Huangguan); k. conidia (a–k. isolate PAFQ65; a–i. produced on PDA agar medium, j, k. from pear fruits). — Scale bars: c = 200 μm; d = 50 μm; e–k = 20 μm.
Description & Illustration — Damm et al. (2019).
Materials examined. CHINA, Anhui Province, Dangshan County, on leaves of P. bretschneideri cv. Huangguan, 4 Aug. 2016, M. Fu (culture PAFQ65).
Notes — Colletotrichum plurivorum was first reported as C. sichuanensis from fruits of Capsicum annuum in China (Liu et al. 2016b), further regarded as a synonym of C. cliviicola (as C. cliviae) (Douanla-Meli et al. 2018), but later distinguished from the latter by Damm et al. (2019). In this study, isolate PAFQ65 was isolated from pear leaves and clustered together with the ex-type culture of C. plurivorum (CBS 125474) in the multi-locus phylogenetic tree. This is the first report of C. plurivorum associated with anthracnose in P. bretschneideri. Notably, isolate PAFQ65 rapidly developed the sexual morph on PDA, but the asexual morph was not observed on PDA.
Colletotrichum pyrifoliae M. Fu & G.P. Wang, sp. nov. — MycoBank MB824217; Fig. 16
Fig. 16.
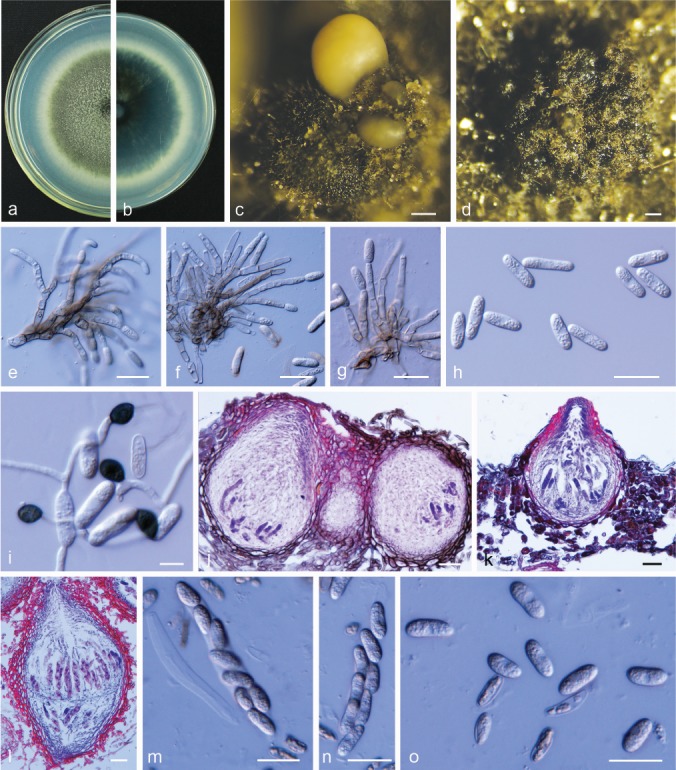
Colletotrichum pyrifoliae. a, b. Front and back view, respectively, of 6-d-old PDA culture; c. conidiomata; d. ascomata; e–g. conidiophores; h. conidia; i. appressoria; j, k. section view of ascomata produced on pear fruit (P. bretschneideri cv. Huangguan) and leaf (P. pyrifolia cv. Cuiguan), respectively; l. section view of ascoma; m, n. asci; o. ascospores (a–o. isolate PAFQ22; a–e, h, l–o. produced on PDA, f. produced on OA, g. produced on SNA). — Scale bars: c, d = 200 μm; e–h, j–o = 20 μm; i = 10 μm.
Etymology. Referring to the host species and host organ from which the fungus was isolated.
Sexual morph developed on PDA. Ascomata formed on PDA after 20–22 d, semi-immersed in the agar medium, pyriform to subglobose, dark brown, 78–212 × 75–160 μm, ostiolate. Asci fasciculate, clavate, 66–92 × 11–20 μm, 8-spored. Ascospores hyaline, smooth-walled, aseptate, cylindrical with rounded ends, straight, rarely slightly curved, contents granular, 11.5–20.5 × 4.5–7 μm, mean ± SD = 16.8 ± 1.6 × 6.4 ± 0.5 μm, L/W ratio = 2.6.
Asexual morph developed on PDA. Vegetative hyphae 2–6.5 μm diam, hyaline, smooth-walled, septate, branched. Setae not observed. Conidiophores hyaline to pale brown, smooth-walled, septate and branched. Conidiogenous cells hyaline to pale brown, cylindrical to clavate, 15–32 × 3–5 μm, opening 1.5–2.5 μm. Conidia hyaline, smooth-walled, aseptate, cylindrical, both ends rounded, contents granular, 14–23 × 5.5–7 μm, mean ± SD = 18.1 ± 1.8 × 6.4 ± 0.4 μm, L/W ratio = 2.9. Appressoria dark-brown, elliptical, 7–12 × 6–8 μm, mean ± SD = 8.8 ± 1.0 × 6.9 ± 0.5 μm, L/W ratio = 1.3.
Asexual morph developed on OA. Setae not observed. Conidiophores hyaline to pale brown, smooth-walled, septate and branched. Conidiogenous cells hyaline to pale brown, cylindrical to clavate, 8–23 × 4–5 μm. Conidia hyaline, smooth-walled, aseptate, cylindrical, both ends rounded, contents granular, 15.5–21.5 × 5–6.5 μm, mean ± SD = 17.8 ± 1.3 × 5.7 ± 0.4 μm, L/W ratio = 3.1.
Asexual morph developed on SNA. Setae not observed. Conidiophores hyaline to pale brown, smooth-walled, septate and branched. Conidiogenous cells hyaline to pale brown, cylindrical to clavate, 12–24.5 × 4–6 μm. Conidia hyaline, smooth-walled, aseptate, cylindrical, both ends rounded, contents granular, 16–22 × 5–6.5 μm, mean ± SD = 18.5 ± 1.3 × 5.6 ± 0.3 μm, L/W ratio = 3.3.
Culture characteristics — Colonies on PDA flat with entire margin, aerial mycelium sparse, cottony in the centre, surface grey-green with white margin; reverse dark grey-green with white margin; colony diam 48–50 mm in 5 d. Conidia in mass pale yellow.
Materials examined. CHINA, Hubei Province, Wuhan City, on leaves of P. pyrifolia cv. Jinshui, 1 Aug. 2016, M. Fu (holotype HMAS 247825, culture ex-type CGMCC 3.18902 = PAFQ22); ibid., PAFQ22a, PAFQ22b, PAFQ22c, and PAFQ22d.
Notes — Colletotrichum pyrifoliae is phylogenetically closely related to Colletotrichum sp. isolate Q026 (Fig. 5), which was reported to be associated with anthracnose of Rubus glaucus in Colombia (Afanador-Kafuri et al. 2014). However, C. pyrifoliae differs from the latter in ACT (with 95.62 % sequence identity), CHS-1 (96.47 %), GAPDH (93.01 %), ITS (99.25 %), and TUB2 (96.41 %) sequences. Moreover, isolates of C. pyrifoliae have larger conidia (PAFQ22, 14–23 × 5.5–7 μm, mean ± SD = 18.1 ± 1.8 × 6.4 ± 0.4 μm) than those of Colletotrichum sp. isolate Q026 (mean = 10.4 × 2.9 μm). The PHI test (Φw = 0.9862) detected no significant recombination between the isolates and Colletotrichum sp. isolate Q026 (Fig. 6b). Colletotrichum pyrifoliae is a singleton species, which grouped neither with the C. gloeosporioides nor the C. boninense species complexes (Fig. 5).
Colletotrichum siamense Prihast. et al., Fung. Diversity 39: 98. 2009. — Fig. 17
Fig. 17.
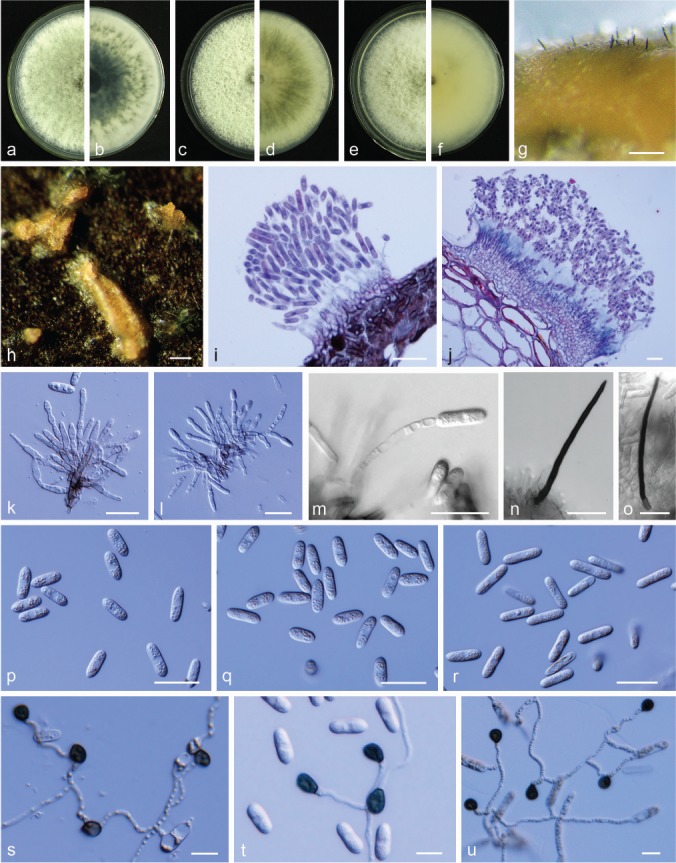
Colletotrichum siamense. a, c, e. Front views of 6-d-old PDA culture; b, d, f. back views of 6-d-old PDA culture; g, h. conidiomata; i, j. section view of acervulus produced on pear leaf (P. pyrifolia cv. Cuiguan) and fruit (P. bretschneideri cv. Huangguan), respectively; k–m. conidiophores; n, o. setae; p–r. conidia; s–u. appressoria (a, b, k, p, s. from PAFQ67, c, d, g, h, j, l, n, q, t. from PAFQ74, e, f, i, m, o, r, u. from PAFQ78; a–g, k–r. produced on PDA, h. produced on pear leaf (P. bretschneideri cv. Dangshansuli)). — Scale bars: g, h = 100 μm; i–r = 20 μm; s–u = 10 μm.
Description & Illustration — Prihastuti et al. (2009).
Materials examined. CHINA, Shandong Province, Yantai City, on fruits of P. communis cv. Gyuiot, 27 Aug. 2016, M. Fu (cultures PAFQ67, PAFQ68, PAFQ71, PAFQ73, PAFQ74); Zhejiang Province, Hangzhou City, on leaves of P. pyrifolia cv. Guanyangxueli, 18 Aug. 2016, M. Fu (PAFQ78); ibid., on leaves of P. pyrifolia cv. Cuiguan, 18 Aug. 2016, M. Fu (PAFQ85).
Notes — Colletotrichum siamense was first reported on Coffea arabica in Thailand (Prihastuti et al. 2009) and subsequently reported on a wide range of hosts (e.g., Yang et al. 2009, Wikee et al. 2011, Weir et al. 2012, Wang et al. 2016, Liu et al. 2016b). Notably, this is the first report and characterisation of C. siamense causing anthracnose on P. pyrifolia and P. communis.
The isolates of C. siamense were divided into three groups (I–III) in this study according to morphology. Group I colonies (13 isolates, representative isolate PAFQ67) flat, grey-green with white margin; reverse dark green to black in the centre and pale white margin, sporadic pigment at the margin. Group II colonies (25 isolates, representative isolate PAFQ74) flat, surface white; reverse pale yellow in the centre and pale white margin, sometimes grey radial pigment produced. Group III colonies (1 isolate, representative isolate PAFQ78) convex, surface pale white in the centre and white margin; reverse pale yellow in the centre and pale white margin, sometimes grey pigment produced. Moreover, these isolates have similar appressorial sizes but different conidium sizes among the three colony types. Of these, conidium sizes of the type III isolates (PAFQ78, 15–21 μm, mean lengths ± SD = 17.4 ± 1.1 μm) were longer than those of type I (12–19 μm, mean lengths from 15.5 ± 1.0 to 16.0 ± 1.2 μm) and II (12–17.5 μm, mean lengths from 14.7 ± 1.0 to 15.1 ± 0.9 μm) isolates (Table 4 and Fig. 17p–r). Setae were observed in isolates PAFQ78 and PAFQ74 on PDA, and setae were dark brown to black, opaque, tip acute, base cylindrical, 3-septate, 67–95 μm long.
Table 4.
The sizes of conidia, appresoria and ascospores of the representative isolates of Colletotrichum spp. obtained in this study.
| Conidia |
Appresoria |
Ascospores) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Species and strain number | Length (μm)x | Width (μm)y | Means ± SD of conidia sizez | Length (μm)x | Width (μm)y | Means ± SD of appresoria sizez | Length (μm)x | Width (μm)y | Means ± SD of ascospores sizez | Growth rate (mm/d) |
| C. aenigma | ||||||||||
| PAFQ1 | 15.5–20 | 5–6.5 | 17.2 ± 1.0 × 5.6 ± 0.3 | 7.5–15.5 | 6–11 | 10.5 ± 1.8 × 8.0 ± 1.2 | 13.5–22 | 6–8 | 18.0 ± 1.7 × 6.9 ± 0.5 | 8.2 |
| PAFQ3 | 14.5–20 | 5.5–7.5 | 17.1 ± 1.1 × 6.6 ± 0.4 | / | / | / | 14.5–20.5 | 5–8 | 17.5 ± 1.6 × 6.5 ± 0.6 | 3.7 |
| PAFQ5 | 16–21.5 | 5.5–7.5 | 18.5 ± 1.1 × 6.7 ± 0.5 | 7.5–11 | 5–9.5 | 9.2 ± 1.1 × 7.1 ± 1.1 | 14.5–19 | 4–8 | 16.7 ± 1.1 × 6.1 ± 0.8 | 6.9 |
| PAFQ47 | 15–19 | 5.5–7 | 16.9 ± 0.9 × 6.3 ± 0.3 | 8–11.5 | 5.5–9 | 9.4 ± 1.0 × 7.3 ± 0.9 | 12.5–19.5 | 5–8 | 15.7 ± 1.6 × 6.6 ± 0.8 | 7.9 |
| PAFQ66 | 14.5–18 | 5.5–6.5 | 16.0 ± 0.7 × 5.8 ± 0.3 | 6–11.5 | 6–11.5 | 9.0 ± 1.3 × 7.6 ± 1.1 | 15–20 | 5.5–8.5 | 17.1 ± 1.1 × 6.5 ± 0.6 | 7.5 |
| PAFQ81 | 15–19 | 5–6 | 17.1 ± 0.9 × 5.8 ± 0.3 | 5.5–11 | 5.5–8 | 8.8 ± 1.2 × 6.7 ± 0.8 | 14.5–21 | 5.5–8 | 18.0 ± 1.6 × 6.7 ± 0.5 | 7.5 |
| C. citricola | ||||||||||
| PAFQ13 | 12.5–17 | 6–8 | 14.4 ± 1.0 × 7.1 ± 0.4 | 7–9.5 | 5.5–7.5 | 8.2 ± 0.6 × 6.7 ± 0.5 | 13.5–20 | 5–8 | 17.4 ± 1.4 × 7.1 ± 0.7 | 4.4 |
| C. conoides | ||||||||||
| PAFQ6 | 16–20 | 4.5–6 | 18.4 ± 0.8 × 5.6 ± 0.3 | 7–12.5 | 5–8.5 | 9.7 ± 1.3 × 6.9 ± 1.1 | 12.5–21 | 5.5–7.5 | 15.9 ± 1.3 × 6.8 ± 0.5 | 7.8 |
| C. fioriniae | ||||||||||
| PAFQ8 | 13.5–16 | 4.5–5.5 | 15.8 ± 1.0 × 5.6 ± 0.3 | 5.5–9 | 3.5–6 | 7.1 ± 0.9 × 4.9 ± 0.5 | / | / | / | 3.5 |
| PAFQ17 | 13–15 | 4–5 | 15.2 ± 1.2 × 5.1 ± 0.5 | 5.5–8.5 | 3.5–6 | 7.1 ± 0.6 × 5.2 ± 0.5 | / | / | / | 4.3 |
| PAFQ36 | 11.5–14 | 4.5–5 | 14.2 ± 1.2 × 5.3 ± 0.4 | 5.5–8.5 | 4.5–6 | 7.2 ± 0.7 × 5.3 ± 0.4 | / | / | / | 4.7 |
| PAFQ49 | 13–16 | 4.5–5.5 | 16.1 ± 1.3 × 5.7 ± 0.4 | 6.5–10 | 4.5–6.5 | 7.7 ± 0.7 × 5.4 ± 0.5 | / | / | / | 4.6 |
| PAFQ55 | 12.5–16.5 | 4–5 | 16.3 ± 1.4 × 5.0 ± 0.4 | 6–9 | 4.5–6.5 | 7.3 ± 0.7 × 5.3 ± 0.5 | / | / | / | 4.6 |
| PAFQ75 | 13–15.5 | 4.5–5.5 | 15.4 ± 1.3 × 5.4 ± 0.3 | 6.5–10.5 | 4–7 | 7.8 ± 1.0 × 5.2 ± 0.6 | / | / | / | 4.4 |
| C. fructicola | ||||||||||
| PAFQ30 | 14.5–19 | 5–7.5 | 17.1 ± 1.1 × 6.4 ± 0.6 | 6.5–13 | 5–8.5 | 8.5 ± 1.7 × 6.7 ± 0.9 | 15.5–24 | 4–6 | 18.8 ± 1.9 × 5.4 ± 0.5 | 7 |
| PAFQ31 | 14.5–20 | 5–7.5 | 17.1 ± 1.5 × 6.1 ± 0.6 | 8–12.5 | 6–9.5 | 9.9 ± 1.2 × 7.2 ± 0.9 | 14–22 | 3.5–6 | 17.1 ± 1.9 × 4.6 ± 0.6 | 7.6 |
| PAFQ32 | 13–17.5 | 5.5–7 | 15.1 ± 1.0 × 6.5 ± 0.4 | 8–14.5 | 6–9.5 | 10.9 ± 1.5 × 7.5 ± 0.9 | 12.5–22.5 | 4–6 | 17.1 ± 1.9 × 4.9 ± 0.5 | 7.3 |
| PAFQ48 | 13.5–16.5 | 4–6 | 15.0 ± 0.7 × 5.1 ± 0.4 | 7–10 | 5.5–8 | 8.2 ± 0.8 × 6.7 ± 0.7 | 14.5–25.5 | 4.5–7 | 18.3 ± 1.9 × 5.4 ± 0.5 | 7.8 |
| PAFQ77 | 13.5–19.5 | 4–6 | 16.2 ± 1.5 × 5.3 ± 0.4 | 6.5–13 | 5–7 | 9.5 ± 1.5 × 6.0 ± 0.5 | 12.5–18.5 | 3.5–6 | 15.5 ± 1.5 × 4.9 ± 0.7 | 6.6 |
| PAFQ84 | 14–19 | 4.5–6 | 16.1 ± 1.1 × 5.4 ± 0.4 | 6.5–14 | 5–7 | 7.8 ± 1.4 × 6.0 ± 0.5 | / | / | / | 7.9 |
| C. gloeosporioides | ||||||||||
| PAFQ7 | 16–22.5 | 5–7.5 | 18.0 ± 1.4 × 6.1 ± 0.6 | 7–10.5 | 5–7 | 8.4 ± 0.8 × 6.1 ± 0.5 | / | / | / | 7.9 |
| PAFQ44 | 11.5–21 | 4–6 | 16.6 ± 1.7 × 5.5 ± 0.4 | 7.5–12.5 | 5.5–8.5 | 9.0 ± 1.2 × 7.0 ± 0.7 | / | / | / | 8.3 |
| PAFQ56 | 16–32 | 4.5–6.5 | 21.5 ± 4.1 × 5.5 ± 0.4 | 6–10.5 | 5–9 | 8.3 ± 1.0 × 6.6 ± 0.8 | / | / | / | 7 |
| PAFQ61 | 15.5–22.5 | 5–6.5 | 17.7 ± 1.6 × 5.6 ± 0.3 | 6.5–10 | 4.5–7.5 | 8.2 ± 0.8 × 6.3 ± 0.7 | / | / | / | 7.4 |
| PAFQ80 | 15–21 | 5–6.5 | 16.9 ± 1.1 × 5.9 ± 0.3 | 6.5–11 | 5–6.5 | 7.8 ± 0.9 × 5.9 ± 0.4 | / | / | / | 7.4 |
| PAFQ86 | 14–18 | 5–6.5 | 16.1 ± 0.9 × 5.8 ± 0.3 | 7–11.5 | 5–7.5 | 9.0 ± 1.3 × 6.4 ± 0.6 | / | / | / | 7.1 |
| C. jinshuiense | ||||||||||
| PAFQ26 | 21–30.5 α | 3–4.5 α | 24.4 ± 2.1 × 4.0 ± 0.3 α | 8–17 | 5–7.5 | 10.7 ± 1.7 × 6.0 ± 0.5 | / | / | / | 5.6 |
| C. karstii | ||||||||||
| PAFQ14 | 12.5–18 | 5.5–8 | 15.8 ± 1.0 × 7.2 ± 0.5 | 6.5–10 | 5.5–7.5 | 8.3 ± 0.8 × 6.4 ± 0.5 | / | / | / | 4.3 |
| PAFQ28 | 12.5–18.5 | 6–8 | 15.5 ± 1.4 × 6.8 ± 0.5 | 6.5–10 | 5–8.5 | 8.4 ± 0.7 × 6.9 ± 0.7 | / | / | / | 5.2 |
| PAFQ40 | 12.5–16 | 5.5–7.5 | 13.6 ± 0.8 × 6.5 ± 0.4 | 6.5–9.5 | 6–8.5 | 8.0 ± 0.7 × 7.3 ± 0.6 | 14–19 | 5–8 | 16.4 ± 1.1 × 6.8 ± 0.7 | 5.3 |
| PAFQ52 | 11.5–16 | 5.5–7.5 | 13.9 ± 1.0 × 6.8 ± 0.3 | 7–10.5 | 5–8 | 8.8 ± 0.7 × 6.8 ± 0.8 | / | / | / | 5.3 |
| PAFQ82 | 12.5–21 | 5–8 | 16.8 ± 1.6 × 7.2 ± 0.6 | 8–14 | 5–9.5 | 10.5 ± 1.4 × 7.2 ± 1.0 | / | / | / | 4.4 |
| C. plurivorum | ||||||||||
| PAFQ65 | 14–24 α | 4.5–7 α | 18.1 ± 2.1 × 5.6 ± 0.7 α | / | / | / | 15–20.5 | 4.5–6 | 18.2 ± 1.6 × 5.4 ± 0.4 | 7.2 |
| C. pyrifoliae | ||||||||||
| PAFQ22 | 14–23 | 5.5–7 | 18.1 ± 1.8 × 6.4 ± 0.4 | 7–12 | 6–8 | 8.8 ± 1.0 × 6.9 ± 0.5 | 11.5–20.5 | 4.5–7 | 16.8 ± 1.6 × 6.4 ± 0.5 | 4.9 |
| C. siamense | ||||||||||
| PAFQ67 | 12–18 | 5–6.5 | 15.5 ± 1.0 × 5.6 ± 0.3 | 6–10.5 | 4.5–8.5 | 8.1 ± 1.3 × 6.2 ± 0.7 | / | / | / | 7.9 |
| PAFQ68 | 12.5–17.5 | 5.5–7 | 14.7 ± 1.0 × 5.8 ± 0.4 | 5.5–10.5 | 5.5–7.5 | 8.0 ± 1.1 × 6.3 ± 0.6 | / | / | / | 8.2 |
| PAFQ71 | 13–19 | 4.5–6.5 | 15.8 ± 1.1 × 5.3 ± 0.4 | 5.5–9.5 | 5–6.5 | 7.7 ± 1.0 × 5.8 ± 0.4 | / | / | / | 7.7 |
| PAFQ73 | 13.5–19 | 4–6 | 16.0 ± 1.2 × 5.6 ± 0.4 | 6.5–8.5 | 4.5–6.5 | 7.4 ± 1.0 × 5.7 ± 0.4 | / | / | / | / |
| PAFQ74 | 13–17.5 | 4.5–6.5 | 15.1 ± 0.9 × 5.7 ± 0.3 | 6–9 | 4.5–6.5 | 7.8 ± 0.6 × 5.7 ± 0.5 | / | / | / | 7.8 |
| PAFQ78 | 15–21 | 4–6 | 17.4 ± 1.1 × 5.4 ± 0.5 | 6.5–12 | 5.5–9 | 9.0 ± 1.2 × 6.7 ± 0.8 | / | / | / | 7.6 |
| PAFQ85 | 14–20 | 4.5–5.9 | 15.9 ± 1.1 × 5.4 ± 0.3 | 5.5–10 | 4.5–6.5 | 7.8 ± 1.0 × 5.8 ± 0.5 | / | / | / | 8.3 |
| PAFQ91 | 12–17.5 | 5–7 | 15.0 ± 1.1 × 5.9 ± 0.4 | 6.5–10 | 4–7 | 7.8 ± 1.2 × 5.9 ± 0.5 | / | / | / | / |
| C. wuxiense | ||||||||||
| PAFQ53 | 11.5–17 | 4.5–6.5 | 14.9 ± 1.3 × 5.3 ± 0.3 | 6.5–12 | 5.5–11 | 9.4 ± 1.1 × 7.1 ± 1.4 | 14–20 β | 4–6.5 β | 17.2 ± 1.3 × 5.0 ± 0.5 β | 7.1 |
| PAFQ54 | 13–18 | 4.5–6 | 15.0 ± 1.3 × 5.1 ± 0.4 | / | / | / | 13–21 β | 4.5–6 β | 17.7 ± 1.5 × 5.2 ± 0.4 β | 7 |
x Numbers indicate minimum and maximum sizes for length of 50 conidia, ascospores and 30 appresoria recorded from the representative strains of Colletotrichum spp. obtained in this study. Significance at P = 0.05 level.
y Numbers indicate minimum and maximum sizes for width of 50 conidia, ascospores and 30 appresoria recorded from the representative strains of Colletotrichum spp. obtained in this study. Significance at P = 0.05 level.
z Numbers indicate mean conidia, appresoria, ascospores sizes of each representative strain calculated by the statistical analysis. Data were analyzed with SPSS Statistics 21.0 (WinWrap® Basic; http://www.winwrap.com) by one-way ANOVA, and means were compared using Duncan’s test at a significance level of P = 0.05. SD: standard deviation.
/ Appresoria, ascospores or data of growth rate were absent.
α Conidia induced on fruit.
β Ascospores induced on SNA medium.
Colletotrichum wuxiense Y.C. Wang et al., Sci. Rep. 6: 8. 2016. — Fig. 18
Fig. 18.
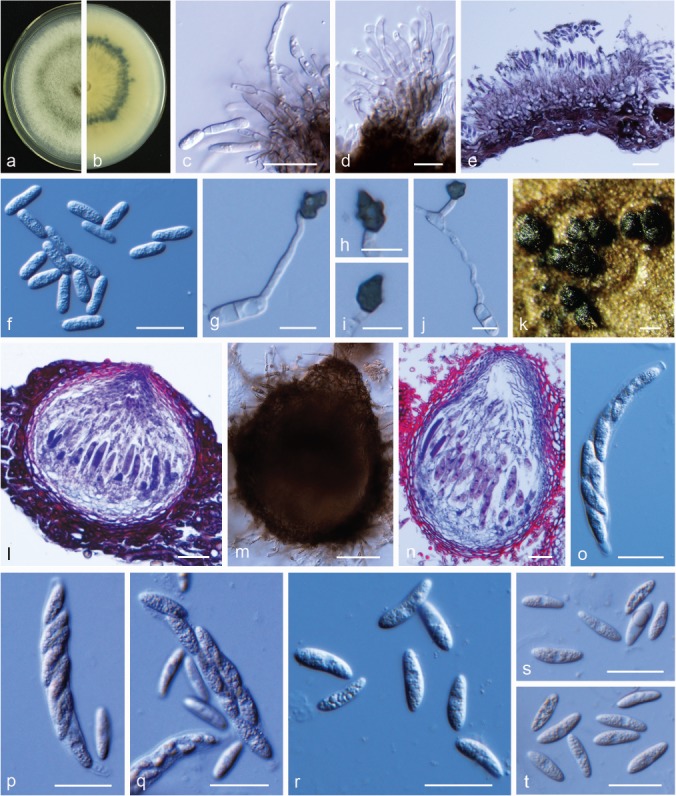
Colletotrichum wuxiense. a, b. Front and back view, respectively, of 6-d-old PDA culture; c, d. conidiophores; e. section view of acervulus produced on pear leaf; f. conidia; g–j. appressoria; k. ascomata; l. section view of ascoma produced on pear fruit; m. ascoma produced on PDA; n. section view of ascoma; o–q. asci; r–t. ascospores (a–l, n, o, q–s. isolate PAFQ53, m, p, t. isolate PAFQ54; a–f, m–t. produced on PDA agar medium, m, n, p, q, s, t. produced on SNA agar medium). — Scale bars: c–f, l, n–t = 20 μm; g–j = 10 μm; k = 100 μm; m = 50 μm.
Sexual morph on SNA. Ascomata developed on SNA after 18–22 d, immersed or semi-immersed in the agar medium, subglobose to pyriform, dark brown, 88–249 × 88–224 μm, ostiolate. Asci clavate, 43–91 × 9–13 μm, 8-spored. Ascospores hyaline, smooth-walled, aseptate, fusiform, slightly curved, rarely straight, rounded ends, contents granular, sometimes with 1–3 guttules, 14–20 × 4–6.5 μm, mean ± SD = 17.2 ± 1.3 × 5.0 ± 0.5 μm, L/W ratio = 3.4.
Sexual morph developed on PDA. Ascomata pyriform to subglobose, dark brown, 74–139 × 64–127 μm, ostiolate. Asci clavate, 57–96 × 12–16 μm, 8-spored. Ascospores hyaline, smooth-walled, aseptate, fusoid, slightly curved, straight with round ends, contents granular, 15.5–22 × 5–6.5 μm, mean ± SD = 18.37 ± 1.39 × 5.80 ± 0.44 μm, L/W ratio = 3.2.
Asexual morph developed on PDA. Vegetative hyphae 1.5–4.5 μm diam, hyaline, smooth-walled, septate, branched. Setae not observed. Conidiophores hyaline to pale brown, smooth-walled, septate and branched. Conidiogenous cells hyaline to pale brown, cylindrical, 8.5–28 × 2.5–4 μm. Conidia hyaline, smooth-walled, aseptate, cylindrical, both ends rounded or one end slightly acute, contents granular or guttulate, 11.5–17 × 4.5–6.5 μm, mean ± SD = 14.9 ± 1.3 × 5.3 ± 0.3 μm, L/W ratio = 2.8. Appressoria dark-brown, irregular in shape or bullet-shaped with an acute tip, lobed, 6.5–12 × 5.5–11 μm, mean ± SD = 9.4 ± 1.1 × 7.1 ± 1.4 μm, L/W ratio = 1.3.
Culture characteristics — Colonies on PDA convex with entire margin, aerial mycelium dense, surface greenish in the centre, with white margin; reverse pale yellow with white margin, and a dark green concentric ring in the middle of the colony. Colony diam 70–71 mm in 5 d. Conidia in mass orange.
Materials examined. CHINA, Jiangxi Province, Jinxi County, on leaves of P. pyrifolia cv. Cuiguan, 23 July 2016, M. Fu (cultures PAFQ53 and PAFQ54).
Notes — According to the results obtained in the multi-locus phylogenetic analyses (Fig. 2), two isolates (PAFQ53, PAFQ54) from pear leaves clustered together with the ex-type culture of C. wuxiense (CGMCC 3.17894), which was initially reported on Camellia sinensis in China (Wang et al. 2016). Notably, the conidium sizes of C. wuxiense isolates in this study (PAFQ53: 11.5–17 × 4.5–6.5 μm, mean ± SD = 14.9 ± 1.3 × 5.3 ± 0.3 μm; PAFQ54: 13–18 × 4.5–6 μm, mean ± SD = 15.0 ± 1.3 × 5.1 ± 0.4 μm) were smaller than those of the ex-type culture of C. wuxiense (CGMCC 3.17894: 16.5–23 × 4.5–6.5 μm, mean ± SE = 19.0 ± 1.4 × 5.6 ± 0.5 μm). This is the first report of C. wuxiense to cause anthracnose on P. pyrifolia and the first description of its sexual morph.
Prevalence of Colletotrichum species
Analyses of the prevalence of 12 Colletotrichum species revealed that C. fructicola isolates (298 isolates, 61.1 % of the total isolates) were predominantly isolated from six provinces (Anhui, Fujian, Hubei, Jiangsu, Jiangxi, and Zhejiang), followed by C. fioriniae (52 isolates, 10.7 %, isolated from Anhui, Fujian, Hubei, Jiangsu, Jiangxi, and Shandong), C. siamense (43 isolates, 8.8 %, isolated from Shandong and Zhejiang), C. aenigma (40 isolates, 8.2 %, isolated from Anhui, Hubei, Jiangsu, and Zhejiang), C. gloeosporioides (20 isolates, 4.1 %, isolated from Hubei, Jiangsu, Jiangxi, and Zhejiang), and C. karstii (19 isolates, 3.9 %, isolated from Fujian, Hubei, Jiangxi, and Zhejiang) (Fig. 19a, b). The remaining six species account for 3.2 % of the isolates (Fig. 19a, b). These results revealed that C. fructicola is the most dominant species on pear in China; C. aenigma, C. fioriniae, C. gloeosporioides, C. karstii, and C. siamense were less dominant and C. citricola, C. conoides, C. jinshuiense, C. plurivorum, C. pyrifoliae, and C. wuxiense the least dominant species. Moreover, C. fructicola isolates causing black spot symptoms were mainly detected in the Yangtze valley regions in the Fujian, Hubei, Jiangsu, Jiangxi, and Zhejiang provinces.
Fig. 19.
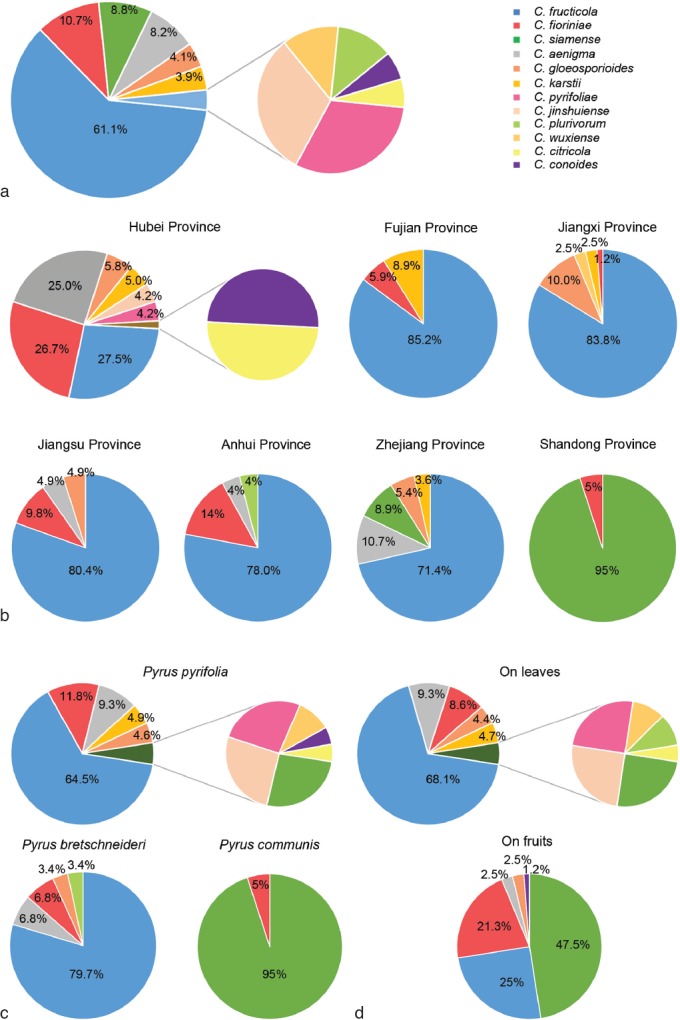
The prevalence of Colletotrichum species isolated from pear. a. Overall isolation rate (%) of Colletotrichum species; b–d. isolation rate (%) of Colletotrichum species from each sampled province (b), Pyrus spp. (c), and pear organs (d), respectively.
Analyses of the isolation rate of these Colletotrichum species in each of the sampled provinces revealed that C. fructicola was dominantly isolated in Fujian, Jiangxi, Jiangsu, Anhui, and Zhejiang provinces, accounting for 85.2 %, 83.8 %, 80.4 %, 78 %, and 71.4 % of the obtained isolates, respectively. Isolates of each other species accounted for less than 15 % (Fig. 19b). However, in the Shandong province, C. siamense isolates were dominantly isolated, accounting for 95 % of the total isolates from this province; in the Hubei province, C. fructicola, C. fioriniae, and C. aenigma isolates were commonly isolated, accounting for 27.5 %, 26.7 %, and 25.0 %, respectively, of the total isolates from this province (Fig. 19b).
Analyses of the isolation rate of these Colletotrichum species from each of the sampled pear species revealed that C. fructicola isolates were dominant on P. pyrifolia and P. bretschneideri, accounting for 64.5 % and 79.7 % of the total isolates, respectively, followed by C. fioriniae (11.8 %), C. aenigma (9.3 %), C. karstii (4.9 %), and C. gloeosporioides (4.6 %) from P. pyrifolia, and C. fioriniae (6.8 %), C. aenigma (6.8 %), C. plurivorum (3.4 %), and C. gloeosporioides (3.4 %) from P. bretschneideri. The remaining species (C. citricola, C. conoides, C. jinshuiense, C. pyrifoliae, C. siamense, and C. wuxiense) were isolated in a low incidence of less than 5.0 % from P. pyrifolia. Only C. siamense and C. fioriniae were isolated from P. communis, with the former accounting for an incidence of 95 % and the latter for 5 % (Fig. 19c). Analyses of the incidence of these Colletotrichum species from the leaves and fruits revealed that C. aenigma, C. fructicola, C. gloeosporioides, C. fioriniae, and C. siamense were isolated from both leaves and fruits, while C. citricola, C. jinshuiense, C. karstii, C. plurivorum, and C. pyrifoliae were isolated only from leaves, and C. conoides only from fruits (Fig. 19d).
Pathogenicity
Thirteen representative Colletotrichum isolates (one from each species except two from C. fructicola related to two different symptom types) were selected to prove Koch’s postulates with a spore suspension on detached leaves of P. pyriforia cv. Cuiguan. Under unwounded conditions, only C. fructicola (isolate PAFQ31) and C. siamense (isolate PAFQ78) were pathogenic to leaves by inducing lesions on the leaf tissues (Fig. 20). Of these, isolate PAFQ31 caused TS symptoms at 8 dpi (Fig. 20b2) and isolate PAFQ78 caused extended BnL symptoms at 14 dpi (Fig. 20b5). Under wounded conditions inoculated at 14 dpi, all the species were pathogenic to leaves, but with obviously varied infection rates depending on the species/isolates (Table 5), with the least 2/16 infection rates for C. plurivorum (isolate PAFQ65) to 16/16 for C. fructicola (isolate PAFQ31). In the case of successful infection, all species started to induce small dark-brown to black necrotic lesions at 6 dpi but 10 dpi for C. citricola (isolate PAFQ13). The small lesions quickly expanded into large dark-brown to black lesions, with the lesion lengths varying among the species (Fig. 20c1–c13) and formed concentric rings of acervuli on the leaf tissues and exuded an orange conidia mass (6–10 dpi) at 25 °C under 99 % relative humidity. It is worth to mention that C. fructicola isolate PAFQ31 isolated from a leaf showing TS symptoms in the field induced similar symptoms around the BnL on inoculated leaves (Fig. 20c2), while another C. fructicola isolate PAFQ32 from a leaf showing BnL symptoms induced big black lesions only (Fig. 20c3). Moreover, C. conoides isolate PAFQ6, which was only isolated from pear fruits, also caused BnL symptoms on pear leaves (Fig. 20c7). No lesions were induced in the control fruits inoculated with sterile water.
Fig. 20.
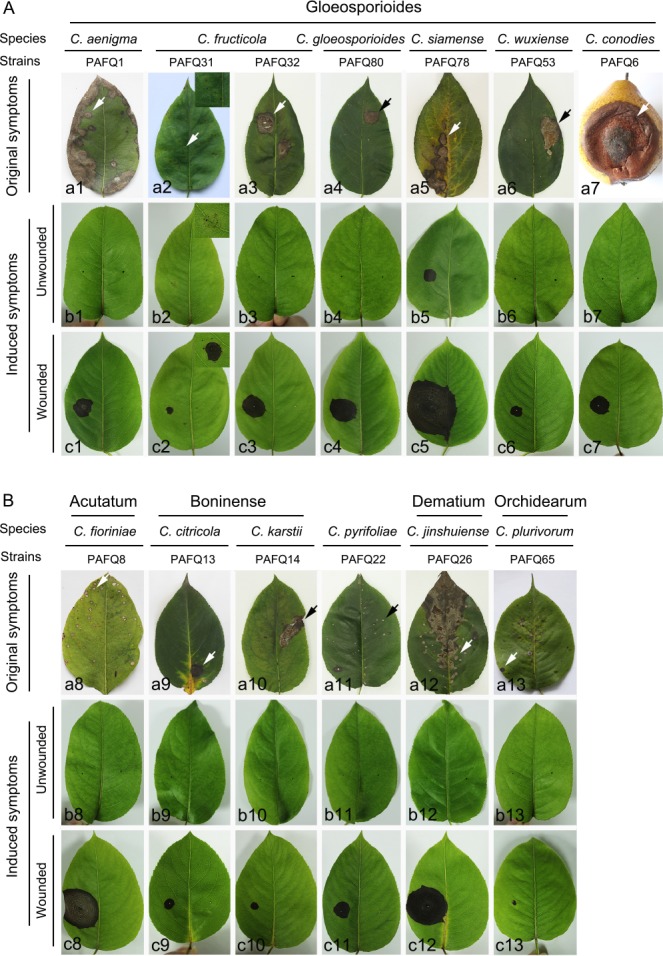
Representative symptoms of pear leaves (P. pyrifolia cv. Cuiguan) induced by inoculation of spore suspensions of 12 Colletotrichum spp. under unwounded and wounded conditions. The symptoms caused by these species were photographed at 14 dpi (except for b2, c2, c3 at 8 dpi). A, B. The symptoms induced by the isolates/species belonging to the C. gloeosporioides complex (A) and other complexes or singleton species (B), respectively. The inoculation was conducted by dropping 1 × 106 spores (conidia or ascospores) per mL on detached about four-weeks-old leaves of P. pyrifolia cv. Cuiguan in eight replicates after wounded by pin-pricking each leaf for one time with a sterilized needle (wounded) or kept unwounded (unwounded). Under unwounded conditions, inoculated positions are indicated with blue spots.
Table 5.
Infection rates of Colletotrichum spp. inoculated on leaves of P. pyrifolia cv. Cuiguan.
| Species | Strain | Origin | Infection rate |
|---|---|---|---|
| C. aenigma | PAFQ1 | Leaf | 14/16 |
| C. citricola | PAFQ13 | Leaf | 7/16 |
| C. conoides | PAFQ6 | Fruit | 6/16 |
| C. fioriniae | PAFQ8 | Leaf | 15/16 |
| C. fructicola | PAFQ31 | Leaf | 16/16 |
| PAFQ32 | Leaf | 10/16 | |
| C. gloeosporioides | PAFQ80 | Leaf | 9/16 |
| C. jinshuiense | PAFQ26 | Leaf | 9/16 |
| C. karstii | PAFQ14 | Leaf | 7/16 |
| C. plurivorum | PAFQ65 | Leaf | 2/16 |
| C. pyrifoliae | PAFQ22 | Leaf | 10/16 |
| C. siamense | PAFQ78 | Leaf | 12/16 |
| C. wuxiense | PAFQ53 | Leaf | 7/16 |
| control | H2O | 0 |
Pathogenicity was also accessed on detached pear fruits of P. bretschneideri cv. Huangguan. Under unwounded conditions, all the isolates isolated from the fruits were pathogenic to the fruits at 30 dpi, with infection rates ranging from 2/6 for C. fioriniae (PAFQ19) to 5/6 for C. gloeosporioides (PAFQ61) (Table 6). These isolates started to induce small brown or dark brown lesions at different time points post inoculation, i.e., at 28–30 dpi for C. aenigma, C. conoides, and C. fioriniae, 18–22 dpi for C. gloeosporioides, and 6–8 dpi for C. siamense. The small lesions expanded to large brown or dark brown lesions over time and formed concentric rings of acervuli at 4–6 dpi, which exuded an orange conidium mass (Fig. 21b1, b4–b6, b8). For the isolates isolated from pear leaves, only C. fructicola isolates (PAFQ31 and PAFQ32) were pathogenic to the inoculated fruits, with infection rates of 6/6 for isolate PAFQ31 and 5/6 for isolate PAFQ32 (Table 6). It is worth to note that C. fructicola isolates PAFQ31 and PAFQ32 induced black spots (Fig. 21b2) and fruit rot symptoms (Fig. 21b3) at 30 dpi, respectively, similar to those in sizes on the leaves observed in the field. The remaining six species isolated from pear leaves induced no visual fruit symptoms (Fig. 21b7, b9–b13). Under wounded conditions, all species were pathogenic to pear fruits at 10 dpi, but with obviously varying aggressiveness among species (Fig. 21c1–c13 and Fig. 22). Of these, the isolates of the C. gloeosporioides species complex induced significantly longer lesions (40–62.5 mm) than those induced by C. fioriniae (20–22 mm), C. citricola (3 mm), C. karstii (31–32 mm), C. pyrifoliae (20.5 mm), and C. jinshuiense (24.5 mm) (Fig. 22). No lesions were induced in the control fruits inoculated with sterile water.
Table 6.
Infection rates of Colletotrichum spp. inoculated on the fruits of P. bretschneideri cv. Huangguan.
| Species | Strain | Origin | Infection rate |
|---|---|---|---|
| C. aenigma | PAFQ66 | Fruit | 4/6 |
| C. citricola | PAFQ13 | Leaf | 0/6 |
| C. conoides | PAFQ6 | Fruit | 3/6 |
| C. fioriniae | PAFQ19 | Fruit | 2/6 |
| C. fructicola | PAFQ31 | Leaf | 6/6 |
| PAFQ32 | Leaf | 5/6 | |
| C. gloeosporioides | PAFQ61 | Fruit | 5/6 |
| C. jinshuiense | PAFQ26 | Leaf | 0/6 |
| C. karstii | PAFQ14 | Leaf | 0/6 |
| C. plurivorum | PAFQ65 | Leaf | 0/6 |
| C. pyrifoliae | PAFQ22 | Leaf | 0/6 |
| C. siamense | PAFQ74 | Fruit | 4/6 |
| C. wuxiense | PAFQ53 | Leaf | 0/6 |
| control | H2O | 0 |
Fig. 21.
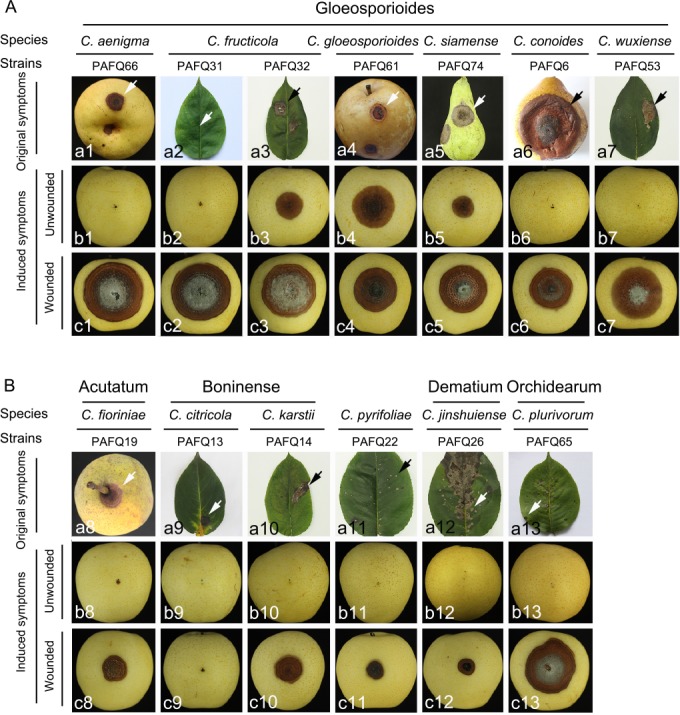
Representative symptoms of pear fruits (P. bretschneideri cv. Huangguan) induced by inoculation with spore suspensions of 12 Colletotrichum spp. under unwounded and wounded conditions. The symptoms under unwounded conditions were photographed at 30 dpi, whereas these under the wounded at 10 dpi. A, B. The symptoms induced by the isolates/species belonging to the C. gloeosporioides complex (A) and other complexes or singleton species (B), respectively. The inoculation was conducted by dropping 1 × 106 spores (conidia or ascospores) per mL on detached fruits in triplicate after wounded by pin-pricking each position for three times with a sterilized needle (wounded) or kept unwounded (unwounded). Under unwounded conditions, inoculated positions are indicated with blue spots.
Fig. 22.
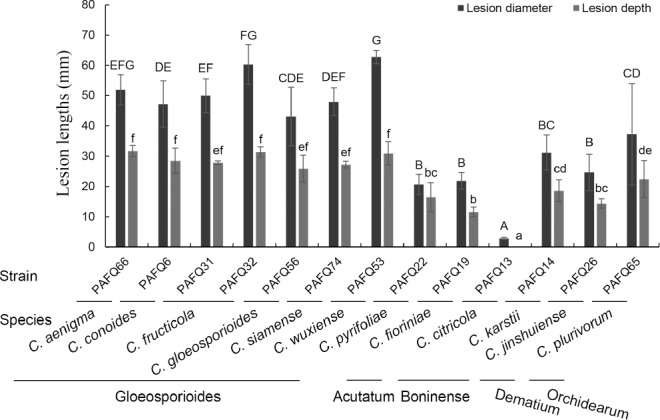
Lesion lengths and depths on wounded pear fruits (P. bretschneideri cv. Huangguan) at 10 dpi induced by conidial suspensions of 13 representative isolates of 12 Colletotrichum spp. The involved isolates and their belonging are indicated at the bottom of the bars. Data were analysed with SPSS Statistics 21.0 (WinWrap Basic; http://www.winwrap.com) by one-way analysis of variance, and means were compared using Duncan’s test at a significance level of P = 0.05. Letters over the error bars indicate the significant difference at the P = 0.05 level.
From the diseased leaf and fruit tissues, fungi were further isolated from the lesions neighbouring the asymptomatic regions. These results showed that the obtained colonies matched the original ones used for inoculation regarding their morphology and ITS sequence data.
DISCUSSION
In this study we employed morphological and multi-locus phylogenetic analyses to identify the species associated with pear anthracnose, and pathogenicity tests to confirm Koch’s postulates. We revealed 12 species belonging to five Colletotrichum species complexes, including gloeosporioides (C. aenigma, C. conoides, C. fructicola, C. gloeosporioides, C. siamense, and C. wuxiense), acutatum (C. fioriniae), boninense (C. citricola and C. karstii), dematium (C. jinshuiense), orchidearum (C. plurivorum), and one singleton species (C. pyrifoliae). Of these, C. conoides, C. siamense, C. wuxiense, C. citricola, C. karstii, and C. plurivorum were confirmed to be responsible for pear anthracnose for the first time. More importantly, this study differentiated two new species responsible for pear anthracnose, namely C. jinshuiense and C. pyrifoliae.
Corresponding to the taxonomic classification determined by multi-locus phylogenetic analyses, most Colletotrichum species also exhibited characteristic morphological characters, including their colony colours, the density of aerial mycelium, and shapes and sizes of conidia, ascospores, appressoria and setae (Fig. 7–18). Most of these features have been used to delimit species in previous studies (Damm et al. 2012a, b, 2014, Liu et al. 2013a, 2015, Hou et al. 2016, Guarnaccia et al. 2017). It is worth to note that the Colletotrichum species associated with pear anthracnose secreted pigments that differed in colour among species and isolates. Moreover, these species also differed in their ability to form a sexual morph in culture. For example, C. gloeosporioides, C. siamense, C. fioriniae, and C. jinshuiense produced no ascospores under the culture conditions employed. Additionally, C. citricola and C. jinshuiense produced setae on the host tissues, but C. aenigma and C. siamense did so on PDA. Importantly, the macro- and micro-morphologies of the Colletotrichum species isolated from pear showed differences compared with those from other plants. For example, most of the C. gloeosporioides isolates (e.g., PAFQ56, PAFQ61, and PAFQ7; 15.5–32 μm) from pear had longer conidia than those from tea (11–15.5 μm) (Liu et al. 2015) and citrus (11.3–14.7 μm) (Huang et al. 2013); and most of C. fructicola isolates (PAFQ30, PAFQ31, and PAFQ84; 14.0–20 × 4.5–7.5 μm) from pear had larger conidia than those from coffee (9.7–14 × 3–4.3 μm) (Prihastuti et al. 2009).
The prevalence of a Colletotrichum species associated with pear anthracnose is closely related to the sampling area, Pyrus sp. and plant organ. For example, C. fructicola is the most prevalent species in most pear-growing regions in China studied, and most frequently isolated from P. pyrifolia and P. bretschneideri in all the sampled areas except for the Shandong province, where C. siamense was most frequently isolated and prevalent on P. communis. Geographical preference was also found for C. aenigma and C. fioriniae, which were mainly isolated in the Hubei province. However, C. jinshuiense, C. pyrifoliae, C. wuxiense, C. plurivorum, C. conoides, and C. citricola showed low prevalence and restricted distribution. Moreover, a high species diversity was observed in the Hubei province as compared to the Fujian and Shandong provinces. It is worth to note that C. acutatum, C. pyricola, and C. salicis were not detected in this study although they were linked to pear anthracnose in New Zealand (Damm et al. 2012b).
In previous reports the pathogenicity of most of the identified Colletotrichum species associated with pear anthracnose, including C. aenigma, C. fructicola, C. acutatum, C. fioriniae, C. pyricola, and C. salicis (Damm et al. 2012b, Weir et al. 2012, Jiang et al. 2014, Schena et al. 2014, Zhang et al. 2015), remained unresolved. Here, pathogenicity tests were conducted in order to confirm Koch’s postulates for all the isolated species to clarify their pathogenicity. From these data it was revealed that the Colletotrichum species/isolates showed broad diversities in their pathogenicity and aggressiveness. Notably, C. fructicola caused TS symptoms on leaves and fruits under unwounded conditions, while it caused rot symptoms on fruits or necrosis lesions on leaves under wounded conditions; the BnL symptoms on leaves could also be induced by C. fructicola isolates, if these isolates were isolated from leaves showing BnL symptoms, indicating C. fructicola to have two pathogenic types. Other species including C. aenigma, C. citricola, C. wuxiense, C. gloeosporioides, C. karstii, and C. siamense are also related to the leaf BnL symptoms; C. fioriniae, C. fructicola, C. aenigma, C. gloeosporioides, C. pyrifoliae, and C. jinshuiense are related to leaf SS symptoms; and C. aenigma, C. fioriniae, C. gloeosporioides, C. siamense, and C. conoides are related to fruit BrL symptoms. Notably, many isolates caused obvious lesions on fruits (or leaves) under wounded conditions but not under unwounded conditions. This phenomenon is related to the quiescent infection of these species, which is an important feature of Colletotrichum spp. and always occurs at the immature fruit stage, progressively developing to rot as the fruits ripen (Peres et al. 2005, Alkan et al. 2015, De Silva et al. 2017). Previous results indicated that wounding can break the quiescent infection and enhance the infectivity of C. fructicola, leading to more rapid rot of young and mature fruits (Jiang et al. 2014). It is worth to note that although the 12 species obtained in this study can infect pear fruits under wounded conditions, those isolated from pear leaves (C. citricola, C. jinshuiense, C. karstii, C. plurivorum, C. pyrifoliae, and C. wuxiense) showed no pathogenicity to pear fruits (P. bretschneideri cv. Huangguan) under unwounded conditions up to 30 dpi. These results revealed a clear organ specificity for the pathogenicity of some Colletotrichum isolates. Some studies also provide clues that some isolates of Glomerella cingulata, C. gloeosporioides and C. acutatum, are host organ specific; they mainly infected the leaves instead of causing bitter rot on apple and pear fruit (Yano et al. 2004, González et al. 2006, Tashiro et al. 2012). Additionally, most of the isolates belonging to the C. gloeosporioides species complex showed higher aggressiveness than those of C. fioriniae, C. citricola, and C. pyrifoliae (Fig. 22).
Previous studies revealed that C. fructicola caused anthracnose on many plants, e.g., Citrus reticulata (Huang et al. 2013), Capsicum sp. (Diao et al. 2017), Camellia sinensis (Liu et al. 2015), Mangifera indica (Lima et al. 2013), and Malus sp. (Munir et al. 2016), resulting in lesions rather than TS symptoms. Therefore, it is interesting that C. fructicola causes TS symptoms on pear. Colletotrichum aenigma was reported on P. pyrifolia in Japan (Weir et al. 2012) and P. communis in Italy (Schena et al. 2014) without mention about the infected organs and induced symptoms. This is the first report of C. aenigma to induce pear anthracnose of P. bretschneideri (on fruits and leaves) and P. pyrifoliae (on leaves) in China (Fig. 19c, d), with a dominant incidence on the latter. Colletotrichum fioriniae was reported causing leaf spots on Cinnamomum subavenium and Juglans regia in China (Sun et al. 2012, Zhu et al. 2015), Salvia leucantha in Italy (Garibaldi et al. 2016), and bitter rot on Pyrus sp. in the USA and Croatia (Damm et al. 2012b, Ivic et al. 2013) and P. communis in France (Da Lio et al. 2017). This is the first report of C. fioriniae in China, which caused pear bitter rot and was associated with pear leaf spot on P. pyrifolia, P. bretschneideri, and P. communis. Colletotrichum citricola was first reported on Citrus unchiu in China, where it was a saprobe on leaves (Huang et al. 2013), but this is the first report of C. citricola on P. pyrifolia, where it was found to cause anthracnose on pear leaves.
This study provides the first systematic investigation, morphological, molecular and biological characterisation of Colletotrichum spp. associated with Pyrus plants, and represents the first reports of C. citricola, C. conoides, C. karstii, C. plurivorum, C. siamense, and C. wuxiense, together with the novel species, causing anthracnose on pear. This study also reveals taxonomic, morphological and biological diversity of Colletotrichum spp. associated with different Pyrus spp. in China in respect to tissue type, geographical location and climate, contributing useful information to help understand the ecology of the Colletotrichum spp. involved in pear anthracnose.
Acknowledgments
This work was financially supported by the earmarked fund for Pear Modern Agro-Industry Technology Research System (CARS-28-15) of the Chinese Ministry of Agriculture and the Fundamental Research Funds for the Central Universities (no. 2662016PY107). The authors would like to thank Dr Lei Cai for critical comments on the manuscript, Dr Fang Liu for technical assistance in the pairwise homoplasy index tests, Dr Xiushi Song for technical assistance in microscopy, and Dr Fangluan Gao for help with the phylogenetic analyses.
REFERENCES
- Afanador-Kafuri L, González A, Gañán L, et al. 2014. Characterization of the Colletotrichum species causing anthracnose in Andean blackberry in Colombia. Plant Disease 98: 1503–1513. [DOI] [PubMed] [Google Scholar]
- Alkan N, Friedlander G, Ment D, et al. 2015. Simultaneous transcriptome analysis of Colletotrichum gloeosporioides and tomato fruit pathosystem reveals novel fungal pathogenicity and fruit defence strategies. New Phytologist 205: 801–815. [DOI] [PubMed] [Google Scholar]
- Cai L, Hyde KD, Taylor PWJ, et al. 2009. A polyphasic approach for studying Colletotrichum. Fungal Diversity 39: 183–204. [Google Scholar]
- Cannon PF, Buddie AG, Bridge PD. 2008. The typification of Colletotrichum gloeosporioides. Mycotaxon 104: 189–204. [Google Scholar]
- Cannon PF, Damm U, Johnston PR, et al. 2012. Colletotrichum – current status and future directions. Studies in Mycology 73: 181–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- Choi YW, Hyde KD, Ho WH. 1999. Single spore isolation of fungi. Fungal Diversity 3: 29–38. [Google Scholar]
- Crous PW, Verkleij GJM, Groenewald JZ, et al. (eds). 2009. Fungal Biodiversity. CBS Laboratory Manual Series 1. Centraalbureau voor Schimmelcultures, Utrecht, Netherlands. [Google Scholar]
- Da Lio D, Baroncelli R, Weill A, et al. 2017. First report of pear bitter rot caused by Colletotrichum fioriniae in France. Plant Disease 101: 1319. [Google Scholar]
- Damm U, Cannon PF, Liu F, et al. 2013. The Colletotrichum orbiculare species complex: important pathogens of field crops and weeds. Fungal Diversity 61: 29–59. [Google Scholar]
- Damm U, Cannon PF, Woudenberg JHC, et al. 2012a. The Colletotrichum boninense species complex. Studies in Mycology 73: 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Cannon PF, Woudenberg JHC, et al. 2012b. The Colletotrichum acutatum species complex. Studies in Mycology 73: 37–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, O’Connell RJ, Groenewald JZ, et al. 2014. The Colletotrichum destructivum species complex – hemibiotrophic pathogens of forage and field crops. Studies in Mycology 79: 49–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Sato T, Alizadeh A, et al. 2019. The Colletotrichum dracaenophilum, C. magnum and C. orchidearum species complexes. Studies in Mycology 92: 1–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Woudenberg JHC, Cannon PF, et al. 2009. Colletotrichum species with curved conidia from herbaceous hosts. Fungal Diversity 39: 45–87. [Google Scholar]
- De Silva DD, Crous PW, Ades PK, et al. 2017. Life styles of Colletotrichum species and implications for plant biosecurity. Fungal Biology Reviews 31: 155–168. [Google Scholar]
- Dean R, Van Kan JAL, Pretorius ZA, et al. 2012. The top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology 13: 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao YZ, Zhang C, Liu F, et al. 2017. Colletotrichum species causing anthracnose disease of chili in China. Persoonia 38: 20–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douanla-Meli C, Unger JG, Langer E. 2018. Multi-approach analysis of the diversity in Colletotrichum cliviae sensu lato. Antonie van Leeuwenhoek 111: 423–435. [DOI] [PubMed] [Google Scholar]
- FAO – Food and Agricultural Organization of the United Nations, China. 2016. Pear fruits fresh and processed: annual statistics. http://www.fao.org/faostat/en/#data/QC. [Google Scholar]
- Freeman S, Katan T, Shabi E. 1996. Characterization of Colletotrichum gloeosporioides isolates from avocado and almond fruits with molecular and pathogenicity tests. Applied and Environmental Microbiology 62: 1014–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garibaldi A, Gilardi G, Franco-Ortega SF, et al. 2016. First report of leaf spot caused by Colletotrichum fioriniae on Mexican bush sage (Salvia leucantha) in Italy. Plant Disease 100: 654. [Google Scholar]
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González E, Sutton TB, Correll JC. 2006. Clarification of the etiology of Glomerella leaf spot and bitter rot of apple caused by Colletotrichum spp. based on morphology and genetic, molecular, and pathogenicity tests. Phytopathology 96: 982–992. [DOI] [PubMed] [Google Scholar]
- Guarnaccia V, Groenewald JZ, Polizzi G, et al. 2017. High species diversity in Colletotrichum associated with citrus diseases in Europe. Persoonia 39: 32–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerber JC, Liu B, Correll JC, et al. 2003. Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns, mtDNA and intron RFLPs, and mating compatibility. Mycologia 95: 872–895. [PubMed] [Google Scholar]
- Hou LW, Liu F, Duan WJ, et al. 2016. Colletotrichum aracearum and C. camelliae-japonicae, two holomorphic new species from China and Japan. Mycosphere 7: 1111–1123. [Google Scholar]
- Huang F, Chen GQ, Hou X, et al. 2013. Colletotrichum species associated with cultivated citrus in China. Fungal Diversity 61: 61–74. [Google Scholar]
- Huson DH. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14: 68–73. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 23: 254–267. [DOI] [PubMed] [Google Scholar]
- Huson DH, Kloepper TH. 2005. Computing recombination networks from binary sequences. Bioinformatics 21: 159–165. [DOI] [PubMed] [Google Scholar]
- Hyde KD, Cai L, McKenzie EHC, et al. 2009. Colletotrichum: a catalogue of confusion. Fungal Diversity 39: 1–17. [Google Scholar]
- Ivic D, Voncina D, Sever Z, et al. 2013. Identification of Colletotrichum species causing bitter rot of apple and pear in Croatia. Journal of Phytopathology 161: 284–286. [Google Scholar]
- Jiang JJ, Zhai HY, Li HN, et al. 2014. Identification and characterization of Colletotrichum fructicola causing black spots on young fruits related to bitter rot of pear (Pyrus bretschneideri Rehd.) in China. Crop Protection 58: 41–48. [Google Scholar]
- Kanchana-udomkan C, Taylor PWJ, Mongkolporn O. 2004. Development of a bioassay to study anthracnose infection of Capsicum chinense Jacq. fruit caused by Colletotrichum capsici. Thai Journal of Agricultural Science 37: 293–297. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HN, Jiang JJ, Hong N, et al. 2013. First report of Colletotrichum fructicola causing bitter rot of pear (Pyrus bretschneideri) in China. Plant Disease 97: 1000. [DOI] [PubMed] [Google Scholar]
- Lima NB, De A. Batista MV, De Morais MA, Jr, et al. 2013. Five Colletotrichum species are responsible for mango anthracnose in northeastern Brazil. Fungal Diversity 61: 75–88. [Google Scholar]
- Lin Q, Kanchana-udomkan C, Jaunet T, et al. 2002. Genetic analysis of resistance to pepper anthracnose caused by Colletotrichum capsici. Thai Journal of Agricultural Science 35: 259–264. [Google Scholar]
- Liu F, Cai L, Crous PW, et al. 2014. The Colletotrichum gigasporum species complex. Persoonia 33: 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Damm U, Cai L, et al. 2013a. Species of the Colletotrichum gloeosporioides complex associated with anthracnose diseases of Proteaceae. Fungal Diversity 61: 89–105. [Google Scholar]
- Liu F, Tang G, Zheng X, et al. 2016b. Molecular and phenotypic characterization of Colletotrichum species associated with anthracnose disease in peppers from Sichuan Province, China. Scientific Reports 6: 32761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wang M, Damm U, et al. 2016a. Species boundaries in plant pathogenic fungi: a Colletotrichum case study. BMC Evolutionary Biology 16: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Weir BS, Damm U, et al. 2015. Unravelling Colletotrichum species associated with Camellia: employing ApMat and GS loci to resolve species in the C. gloeosporioides complex. Persoonia 35: 63–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LZ, Chen ZY, Qian GL, et al. 2013b. Isolation, identification and biological characteristics of Colletotrichum gloeosporioides in pear. Jiangsu Journal of Agricultural Sciences 29: 60–64. [Google Scholar]
- Marin-Felix Y, Groenewald JZ, Cai L, et al. 2017. Genera of phytopathogenic fungi: GOPHY 1. Studies in Mycology 86: 99–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir M, Amsden B, Dixon E, et al. 2016. Characterization of Colletotrichum species causing bitter rot of apple in Kentucky orchards. Plant Disease 100: 2194–2203. [DOI] [PubMed] [Google Scholar]
- Nirenberg HI. 1976. Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem 169: 1–117. [Google Scholar]
- Nylander JAA. 2004. MrModelTest v. 2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- O’Donnell K, Cigelnik E. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. [DOI] [PubMed] [Google Scholar]
- Peres NA, Timmer LW, Adaskaveg JE, et al. 2005. Life styles of Colletotrichum acutatum. Plant Disease 89: 784–796. [DOI] [PubMed] [Google Scholar]
- Photita W, Taylor PWJ, Ford R, et al. 2005. Morphological and molecular characterization of Colletotrichum species from herbaceous plants in Thailand. Fungal Diversity 18: 117–133. [Google Scholar]
- Prihastuti H, Cai L, Chen H, et al. 2009. Characterization of Colletotrichum species associated with coffee berries in northern Thailand. Fungal Diversity 39: 89–109. [Google Scholar]
- Quaedvlieg W, Binder M, Groenewald JZ, et al. 2014. Introducing the consolidated species concept to resolve species in the Teratosphaeriaceae. Persoonia 33: 1–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. 2014. FigTree v. 1.4.2. Institute of Evolutionary Biology, University of Edinburgh; http://tree.bio.ed.ac.uk/software/figtree/. [Google Scholar]
- Rambaut A, Suchard M, Drummond AJ. 2013. Tracer v 1.6. Institute of Evolutionary Biology, University of Edinburgh; http://tree.bio.ed.ac.uk/software/tracer/. [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rubtsov GA. 1944. Geographical distribution of the genus Pyrus and trends and factors in its evolution. American Naturalist 78: 358–366. [Google Scholar]
- Schena L, Mosca S, Cacciola SO, et al. 2014. Species of the Colletotrichum gloeosporioides and C. boninense complexes associated with olive anthracnose. Plant Pathology 63: 437–446. [Google Scholar]
- Sharma G, Pinnaka AK, Shenoy BD. 2015. Resolving the Colletotrichum siamense species complex using ApMat marker. Fungal Diversity 71: 247–264. [Google Scholar]
- Shivas RG, Tan YP. 2009. A taxonomic re-assessment of Colletotrichum acutatum, introducing C. fioriniae comb. et stat. nov. and C. simmondsii sp. nov. Fungal Diversity 39: 111–122. [Google Scholar]
- Silva GJ, Souza TM, Barbieri RL, et al. 2014. Origin, domestication, and dispersing of pear (Pyrus spp.). Advances in Agriculture 2014: 1–8. [Google Scholar]
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution 12: 335–337. [Google Scholar]
- Sun W, Su YY, Cai L, et al. 2012. First report of leaf disease on Cinnamomum subavenium caused by Colletotrichum fioriniae in China. Plant Disease 96: 143. [DOI] [PubMed] [Google Scholar]
- Sutton BC. 1980. The coelomycetes. Fungi imperfecti with pycnidia, acervuli and stromata. Commonwealth Mycological Institute, Kew, Surrey, England. [Google Scholar]
- Swofford D. 2002. PAUP 4.0 b10: Phylogenetic analysis using parsimony (*and other methods). Computer programme. Sinauer Associates, Sunderland, MA, USA. [Google Scholar]
- Tao G, Liu ZY, Liu F, et al. 2013. Endophytic Colletotrichum species from Bletilla ochracea (Orchidaceae), with descriptions of seven new species. Fungal Diversity 61: 139–164. [Google Scholar]
- Tashiro N, Manabe K, Ide Y. 2012. Emergence and frequency of highly benzimidazole-resistant Colletotrichum gloeosporioides, pathogen of Japanese pear anthracnose, after discontinued use of benzimidazole. Journal of General Plant Pathology 78: 221–226. [Google Scholar]
- Than PP, Jeewon R, Hyde KD, et al. 2008. Characterization and pathogenicity of Colletotrichum species associated with anthracnose on chilli (Capsicum spp.) in Thailand. Plant Pathology 57: 562–572. [Google Scholar]
- Vavilov NI. 1951. The origin, variation, immunity and breeding of cultivated plants. Soil Science 72: 482. [Google Scholar]
- Vieira WAS, Michereff SJ, De Morais MA, et al. 2014. Endophytic species of Colletotrichum associated with mango in northeastern Brazil. Fungal Diversity 67: 181–202. [Google Scholar]
- Von Arx JA. 1957. Die Arten der Gattung Colletotrichum Cda. Phytopathologische Zeitschrift 29: 413–468. [Google Scholar]
- Wang YC, Hao XY, Wang L, et al. 2016. Diverse Colletotrichum species cause anthracnose of tea plants (Camellia sinensis (L.) O. Kuntze) in China. Scientific Reports 6: 35287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Johnston PR, Damm U. 2012. The Colletotrichum gloeosporioides species complex. Studies in Mycology 73: 115–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al. (eds), PCR protocols: a guide to methods and applications: 315–322. Academic Press, San Diego, California. [Google Scholar]
- Wikee S, Cai L, Pairin N, et al. 2011. Colletotrichum species from Jasmine (Jasminum sambac). Fungal Diversity 46: 171–182. [Google Scholar]
- Wu J, Wang ZW, Shi ZB, et al. 2013. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Research 23: 396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LQ, Zhu LW, Heng W, et al. 2010. Identification of Dangshan pear anthracnose pathogen and screening fungicides against it. Scientia Agricultura Sinica 43: 3750–3758. [Google Scholar]
- Yan JY, Jayawardena MMRS, Goonasekara ID, et al. 2015. Diverse species of Colletotrichum associated with grapevine anthracnose in China. Fungal Diversity 71: 233–246. [Google Scholar]
- Yang YL, Cai L, Yu ZN, et al. 2011. Colletotrichum species on Orchidaceae in southwest China. Cryptogamie Mycologie 32: 229–253. [Google Scholar]
- Yang YL, Liu ZY, Cai L, et al. 2009. Colletotrichum anthracnose of Amaryllidaceae. Fungal Diversity 39: 123–146. [Google Scholar]
- Yano K, Ishll H, Fukaya M, et al. 2004. Anthracnose on Japanese pear caused by intermediately benzimidazole-resistant strains of Colletotrichum gloeosporioides (Glomerella cingulata). Japanese Journal of Phytopathology 70: 314–319. [Google Scholar]
- Zeven AC, Zhukovsky PM. 1975. Dictionary of cultivated plants and their centers of diversity: 62–63. Center for Agricultural Publishing and Documentation, Wageningen, The Netherlands. [Google Scholar]
- Zhang PF, Zhai LF, Zhang XK, et al. 2015. Characterization of Colletotrichum fructicola, a new causal agent of leaf black spot disease of sandy pear (Pyrus pyrifolia). European Journal of Plant Pathology 143: 651–662. [Google Scholar]
- Zhao DY, Xu K, Yuan JC, et al. 2016. Analysis on the current situation of production and sales of world pear’s main country of origin and its development. China Fruits 2: 94–100. [Google Scholar]
- Zhu YZ, Liao WJ, Zou DX, et al. 2015. First report of leaf spot disease on walnut caused by Colletotrichum fioriniae in China. Plant Disease 99: 289. [DOI] [PubMed] [Google Scholar]


