Abstract
The ykkC RNA motif was a long-standing orphan riboswitch candidate that has recently been proposed to encompass at least five distinct bacterial riboswitch classes. Most ykkC RNAs belong to the subtype 1 group, which are guanidine-I riboswitches that regulate the expression of guanidine-specific carboxylase and transporter proteins. The remaining ykkC RNAs have been organized into at least four major categories called subtypes 2a through 2d. Subtype 2a RNAs are riboswitches that sense the bacterial alarmone ppGpp, and typically regulate amino acid biosynthesis genes. Subtype 2b riboswitches sense the purine biosynthetic intermediate PRPP, and frequently partner with guanine riboswitches to regulate purine biosynthesis genes. In the current study, we examined ykkC subtype 2c RNAs, which are found upstream of genes encoding hydrolase enzymes that cleave the phosphoanhydride linkages of nucleotide substrates. Subtype 2c representatives mostly recognize adenosine- and cytidine-5ˊ-diphosphate molecules in either their ribose or deoxyribose forms (ADP, dADP, CDP, and dCDP). Other nucleotide-containing compounds, especially nucleoside-5ˊ-triphosphates, are strongly rejected by some members of this putative riboswitch class. High ligand concentrations in vivo are predicted to turn on expression of hydrolase enzymes, which presumably function to balance cellular nucleotide pools. These results further showcase the striking functional diversity derived from the structural scaffold shared among all ykkC motif RNAs, which has been adapted to sense at least five different types of natural ligands. Moreover, riboswitches for nucleoside diphosphates provide additional examples of the numerous partnerships observed between natural RNA aptamers and nucleotide-derived ligands including metabolites, coenzymes and signaling molecules.
Keywords: ADP, aptamer, CDP, HAD hydrolase, NUDIX hydrolase, ykkC RNA motif
Graphical Abstract
INTRODUCTION
A bacterial noncoding RNA (ncRNA) motif called ykkC, originally reported in 2004,1 was proposed to represent the highly-conserved aptamer domain of a riboswitch candidate.1,2 Riboswitches3–6 almost exclusively reside in the 5ˊ-untranslated regions (UTRs) of the bacterial mRNAs whose expression they control. Likewise, ykkC motif RNAs are always found in the 5′ UTRs of mRNAs for certain types of genes, which strongly suggests a regulatory function. Furthermore, the putative aptamer domains represented by the original1 consensus sequence and secondary structure of ykkC motif RNAs are commonly associated with identifiable expression platform structures,7 which interact with specific cellular components to regulate gene expression in a manner dictated by the ligand occupancy state of the aptamer. Thus, ykkC motif RNAs were considered to be excellent ‘orphan’ riboswitch candidates1,2,8 because they were predicted to function as riboswitches, but the natural ligand for any of its numerous representatives that triggered gene regulation remained unknown for over a decade.
Our continued experimental pursuit of ykkC orphan riboswitches was motivated in part by the prospect that the elusive ligand would have an important, but likely unknown or underappreciated, role in bacteria. Representative ykkC motif RNAs are both abundant and widespread in bacteria, suggesting that these riboswitches would sense a ligand that was broadly important to many bacterial species. Another motivating factor was the discovery of two additional RNA motifs, named mini-ykkC9 and ykkC-III,10 that are associated with many of the same types of genes as the original ykkC motif. Like these two additional orphan riboswitch candidates, most of the original ykkC motif RNA representatives are located upstream of genes encoding proteins annotated as urea carboxylases and small multi-drug resistance transporters.1,2,8 This suggested that the ligands for ykkC, mini-ykkC, and ykkC-III would all be solved if the ligand for any individual class could be identified.
Eventually guanidine, a common protein denaturant and chaotropic agent, was found to activate expression of a ykkC riboswitch-lacZ reporter gene fusion construct by testing a library comprised of different growth media conditions.8 Further experiments demonstrated that guanidine, likely in its guanidinium cation form, binds to certain ykkC motif RNAs in vitro. High-resolution x-ray crystal structures11,12 revealed a specific guanidine binding pocket is formed by distal groups of nucleotides within the aptamer domain. Guanidine is toxic to bacteria at high concentrations, and its build-up is sensed by guanidine-responsive RNAs from the original ykkC orphan riboswitch collection, now called guanidine-I riboswitches, to activate the expression of genes whose protein products mitigate toxicity.8 For example, guanidine-I riboswitches most commonly activate the expression of guanidine carboxylases8 (previously annotated as urea carboxylases13) to deplete cellular guanidine levels and diminish its toxic effects. These riboswitches also commonly activate the expression of genes encoding guanidine exporters8,14 (previously annotated as small multi-drug resistance transporters15,16). Likewise, members of both the mini-ykkC and ykkC-III RNA classes were also found to specifically recognize guanidine and thus were renamed guanidine-II17 and guanidine-III18 riboswitches, respectively.
Unlike most other riboswitch classes, validation efforts for the original ykkC motif RNAs were not complete when guanidine was identified as a ligand. Whereas most ykkC motif RNAs specifically recognize guanidine and regulate genes involved in guanidine detoxification, approximately 30% of all RNAs that were originally identified to be part of the ykkC motif collection almost certainly do not bind guanidine.8 This subset of ykkC motif RNAs, termed ykkC subtype 2, carries distinct nucleotide changes in regions of the aptamer that otherwise form the binding pocket for guanidine. Additionally, these variant RNAs associate with genes involved in processes unrelated to guanidine biology. Further analysis of the RNA sequences and consensus models of ykkC subtype 2 RNAs, and the assessment of the diverse types of genes found downstream of these RNAs, revealed that there are probably at least four additional distinct riboswitch classes present within the ykkC subtype 2 collection, termed ykkC subtypes 2a through 2d.19,20
Recently, ykkC subtype 2a RNAs were discovered to recognize guanosine tetraphosphate (ppGpp),19 which is a nearly ubiquitous nucleotide signaling molecule involved in amino acid starvation and other stress responses in bacteria.21–23 Uncharged tRNAs activate synthesis of the ppGpp molecule that then triggers ppGpp riboswitches and other factors in its signaling network in part to activate genes encoding amino acid biosynthesis enzymes.19,24,25 Furthermore, ykkC subtype 2b RNAs were found to sense phosphoribosyl pyrophosphate (PRPP),20 which is essential for the biosynthesis of all purine and pyrimidine nucleotides. PRPP riboswitches sense this ligand to activate the expression of genes encoding enzymes required for the de novo biosynthesis of purines.20 Herein we present evidence that the natural ligands for ykkC subtype 2c RNAs likely include certain nucleoside diphosphate compounds. Thus, only the target ligand for subtype 2d RNAs remains to be identified from among the collection of ligands sensed by members of the original group of ykkC orphan riboswitches.
MATERIALS AND METHODS
Chemicals and Oligonucleotides.
Adenosine-5′-diphosphate was purchased from Santa Cruz Biotechnology. Diadenosine tetraphosphate (Ap4A), 8-oxo-2′-deoxyadenosine-5′-triphosphate, 2-hydroxy-2′-deoxyadenosine-5′-triphosphate, and 2-hydroxy-2′-deoxyadenosine-5′-triphosphate were purchased from Jena Bioscience. Isoguanosine-5′-diphosphate, 8-oxo-2′-deoxyguanosine-5′-triphosphate, and 8-oxo-adenosine-5′-triphosphate were purchased from TriLink Biotechnologies. All remaining chemicals and synthetic DNA oligonucleotides were purchased from Sigma-Aldrich. [γ−32P] ATP was purchased from PerkinElmer. Enzymes were purchased from New England BioLabs, unless noted otherwise. The complete list of DNA oligonucleotides used in this work is found in Table S1.
Bioinformatic Analyses.
Examples of the ykkC subtype 2c RNA motif (Supplemental File 1) were manually sorted by their downstream gene association as reported earlier19,20 using the previously published alignment of ykkC subtype 2 RNAs.8
RNA Preparation.
Double-stranded DNA (dsDNA) templates for in vitro transcription were produced by extension of overlapping synthetic DNA oligonucleotides (Table S1) using SuperScript™ II Reverse Transcriptase (Thermo Fisher Scientific). Certain oligonucleotides included a 5′ terminal T7 RNA polymerase (T7 RNAP) promoter sequence, enabling in vitro transcription of the resulting dsDNA templates with T7 RNAP. The RNAs were subsequently purified and 5′ 32P-labeled for in-line probing assays as previously described.8,17
In-Line Probing Analysis of RNAs.
In-line probing experiments22,23 were performed as previously described.26,27
RESULTS AND DISCUSSION
Subtype 2c ykkC Motif RNAs are Distinct from Other ykkC Riboswitch Classes.
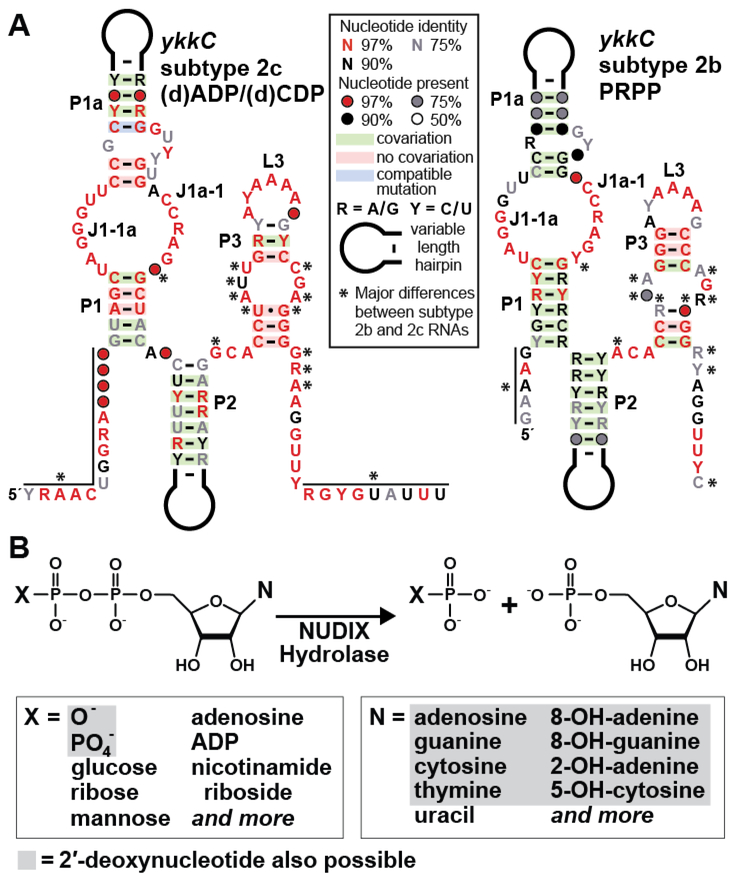
The consensus sequence and secondary structure model for the 46 unique examples of subtype 2c ykkC RNAs (Figure 1A, left) carry sequence and structural features common to all ykkC motif RNAs. Specifically, base-paired stems P1, P1a, P2 and P3 are characteristic of the original ykkC motif RNA class.1,8 Furthermore, numerous nucleotides conserved among all ykkC motif RNAs mostly reside in junctions between stems, such as J1–1a and J1a-1, or in the L3 loop that closes stem P3. These nucleotides are mostly intimately involved in forming the overall structure of each riboswitch aptamer, rather than forming the ligand binding pocket.11,12,28 Thus, we speculate that the general architecture of subtype 2c RNAs will be similar to that seen with other riboswitch aptamers derived from the ykkC motif collection. Another similarity among PRPP, ppGpp (originally ykkC subtype 2a), and ykkC subtype 2c riboswitches is that representatives of these three classes are found exclusively in organisms in the phylum Firmicutes, whereas ykkC subtype 2d RNAs (which sense an undetermined ligand) are found exclusively in organisms in the phyla Proteobacteria and Actinobacteria.15,16
Figure 1.
Distinct features of ykkC subtype 2c riboswitches. (A) Comparison of the consensus sequence and secondary structure models for the 46 unique representatives of ykkC subtype 2c RNAs (left), which sense certain nucleoside diphosphates, and for the 257 unique representatives of the similar ykkC subtype 2b RNAs (right), which sense phosphoribosyl pyrophosphate (PRPP).19 Notable differences (asterisks) include the extended termini of subtype 2c RNAs, and changes to nucleotides known to reside in the binding pockets of other riboswitches uncovered from the original ykkC motif RNA collection.11,12,28 See Supplementary File 1 for sequence alignments of the 46 unique ykkC subtype 2c aptamers. (B) Top: the reaction performed by NUDIX (nucleoside diphosphate linked to x) hydrolase enzymes that are frequently associated with subtype 2c RNAs, but not other ykkC motif RNAs. Bottom: nucleotide and nucleotide-like compounds that are known to act as substrates for certain NUDIX hydrolase enzymes whose expression is predicted to be regulated by ykkC subtype 2c RNAs. Some nucleoside di- and triphosphate substrates can carry a ribose or a deoxyribose moiety (gray shading).
Despite sequence and structural similarities, there are numerous nucleotide positions unique to subtype 2c RNAs that distinguish this subtype from all other ykkC motif RNAs. For example, the region of high sequence conservation for ykkC subtype 2c RNAs (Figure 1A, left) extends well beyond that presented in the original consensus model for ykkC motif RNAs.1,8 The aptamer for the guanidine-I riboswitch class only encompasses the P1 through P3 structural elements,8,11,12 whereas the ppGpp and PRPP riboswitch classes carry additional conserved nucleotide positions upstream (five nucleotides) and downstream (nine nucleotides) to form an additional stem called P0.28 The consensus model for ykkC subtype 2c RNAs extends as many as 14 nucleotides upstream of the P1 stem and as many as 18 nucleotides downstream of the P3 stem.
It is possible that there are base-pairing interactions between these additional conserved nucleotides present in subtype 2c RNAs, but the conservation in this region appears to be distinct from that observed for ppGpp and PRPP riboswitches. Therefore, ykkC subtype 2c RNAs might form a unique substructure by using these 5ˊ and 3ˊ extensions. Another difference is the nucleotide position immediately preceding the P1 stem, which is typically a G residue in ppGpp and PRPP riboswitches,19,20 but is either a U or a G in subtype 2c RNAs. Also, the nucleotide directly preceding the right shoulder of P1 is either a C or a G for subtype 2c RNAs. This is the only subtype for which a G at this position is commonly observed. These and other unique features of subtype 2c RNAs strongly suggest that these RNAs sense and respond to a novel ligand.
With only 46 representatives, the extent of conservation for some nucleotide positions might not be as meaningful as it is for the other subtypes of which there are many more examples. However, the stems do exhibit ample covariation to support our conclusion that these structures indeed form. Also, most of the highly-conserved nucleotides of subtype 2c RNAs reside in regions known to form both the ligand binding sites and the structural scaffold common to other riboswitches from the ykkC motif collection. This suggests that subtype 2c RNAs employ a similar overall architecture, but form a unique binding pocket for recognition of a different ligand. Based on the presence of a terminator stem downstream of subtype 2c RNAs, which presumably acts as the expression platform for this riboswitch candidate, we infer that the presence of the ligand should turn on expression of the downstream gene, as is also the case for guanidine-I, ppGpp, and PRPP riboswitches.8,19,20 Importantly, among the various ykkC motif RNAs, subtype 2c representatives most closely resemble PRPP (originally ykkC subtype 2b) riboswitches19 (Figure 1A, right). This observation might suggest that the ligand for subtype 2c RNAs might be somewhat similar to PRPP.
Genes Associated with ykkC Subtype 2c RNAs are Nucleotide Hydrolases.
Nearly all ykkC subtype 2c RNAs are located upstream of genes encoding Nucleoside Diphosphate linked to X (NUDIX) hydrolases. NUDIX hydrolases are a broad family of proteins that cleave the phosphoester bond between the α and β phosphate moieties of various nucleotide substrates (Figure 1B).29,30 The founding member of this protein family is MutT, which is an enzyme involved in DNA damage repair whose substrates are primarily 8-oxo-2′-deoxyguanosine di- and triphosphate.30,31 The damaged DNA molecule is cleaved by the NUDIX enzyme to produce the monophosphate version of the nucleotide. Because nucleoside monophosphates cannot be used as a substrate for DNA polymerase, cleavage by the NUDIX hydrolase removes these damaged nucleotides from the cellular nucleotide pool. Similar NUDIX hydrolase family proteins act on the di-and tri-phosphate versions of other types of damaged DNA and RNA nucleosides, including 8-oxoadenosine, 2-hydroxyadenosine, and 5-hydroxycytosine.30,32 NUDIX hydrolase family proteins also act on many other nucleotide-containing compounds, such as the enzyme cofactors NADH and TPP, the signaling molecule Ap4A, and the adenylated sugar ADP-glucose.30,32,33 Some NUDIX hydrolases display broad substrate specificity and act on a range of nucleoside di- and triphosphates. In general, NUDIX hydrolases are considered to be “housekeeping” enzymes that maintain a balance between various nucleotide pools.30,34
In rare instances, ykkC motif RNAs are found upstream of genes encoding Haloacid dehalogenase-like (HAD) hydrolases, which represent another protein superfamily of phosphatases that act on various substrates including nucleotides.35–37 The proteins whose expression is predicted to be regulated by these examples of ykkC RNAs share homology with the Cof-type HAD-IIB hydrolase family, many of which cleave phosphorylated sugar substrates including nucleotides.35,38 The examples of ykkC motif RNAs found upstream of HAD-like hydrolases were assigned to subtype 2c in part because of the similarity in the functions of NUDIX and HAD-like hydrolases. However, the four ykkC motif RNA representatives found upstream of HAD-like hydrolase genes also closely match the consensus sequence for the 42 representatives found upstream of NUDIX hydrolase genes. This observation provides compelling evidence that the orphan riboswitch candidates associated with these two types of nucleotide-processing genes belong to the same subtype 2c candidate riboswitch class.
Representative ykkC Subtype 2c RNAs Bind Nucleoside Diphosphates.
Based on the ligand identities for the ykkC subtype 2a and 2b RNAs, which sense ppGpp and PRPP, respectively,19,20 as well as the known substrates for NUDIX and HAD-like hydrolases,29–38 it seemed very likely that the ligand for subtype 2c would also be a nucleotide-derived molecule. Therefore, we examined various nucleotides and nucleotide derivatives for possible binding by representatives of the ykkC subtype 2c candidate riboswitch class.
We assessed whether any known NUDIX hydrolase substrates might be the ligand for ykkC subtype 2c RNAs by using in-line probing.26,27 This method takes advantage of the instability of RNA internucleotide linkages that cleave via spontaneous phosphoester transfer with rate constants that are dependent on the local structure of the RNA. Specifically, the speed of spontaneous RNA strand scission is greatly decreased for nucleotide positions that are structurally constrained by base-pairing or other interactions, compared to the linkages joining nucleotides that are not actively engaged in forming higher order structures. When the breakdown products are analyzed by denaturing polyacrylamide gel electrophoresis (PAGE), the pattern of fragmentation can be used to examine the structure of the RNA oligonucleotide at single-nucleotide resolution.26 For riboswitch RNAs, some nucleotides in the aptamer domain typically undergo conformational change in response to ligand binding, which yields a corresponding change in the banding pattern that can be quantified to determine ligand binding affinity.
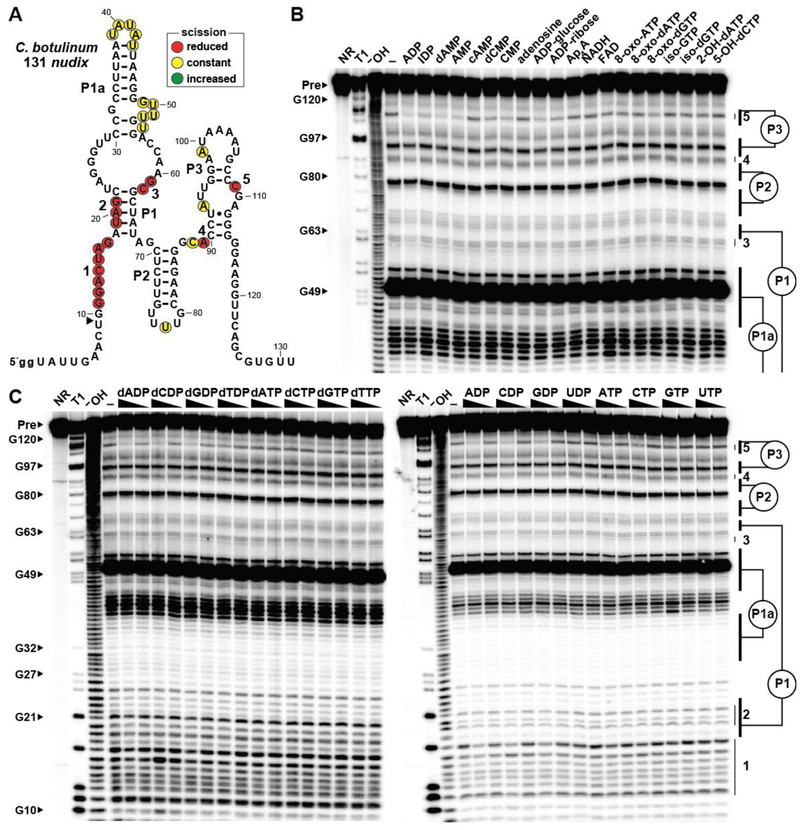
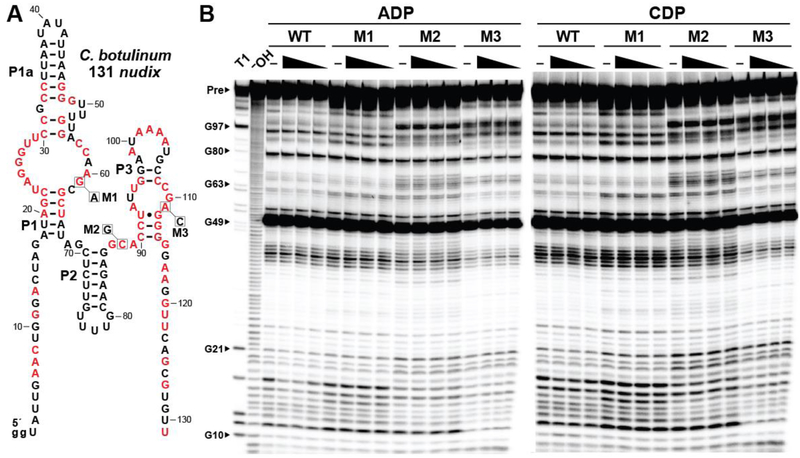
We conducted initial in-line probing assays with 1 mM each of the four common ribonucleoside-5ˊ-diphosphates (NDPs), ribonucleoside-5ˊ-triphosphates (NTPs), their corresponding deoxyribonucleotides (dNDPs and dNTPs), as well as certain ribonucleosides, ribonucleoside monophosphates (NMPs), nucleotide sugars, coenzymes, and oxidatively-modified nucleotides. These analyses were conducted with a representative ykkC subtype 2c RNA comprising 131 nucleotides of the 5′ UTR of a nudix gene in Clostridium botulinum named 131 nudix (Figure 2A). Intriguingly, the 5ˊ 32P-labeled RNA construct exhibits structural modulation in the presence of several nucleotides when tested at 1 mM, including adenosine-5ˊ-diphosphate (ADP) and several related molecules that carry an adenosine moiety (Figure 2B). Similarly, we found that structural modulation is induced by both the ribose and deoxyribose versions of the nucleoside-5ˊ-diphosphates carrying the nucleobases A, C and G (Figure 2C). Inosine-5ˊ-diphosphate is also bound (Figure 2B), suggesting that this particular RNA construct only poorly recognizes the nucleobase.
Figure 2.
Multiple nucleotide compounds are bound by a representative ykkC subtype 2c RNA. (A) Sequence and secondary structure of the 131 nucleotide RNA derived from a nudix gene of C. botulinum. Data collected in B were used to determine regions of reduced or constant scission upon the addition of certain nucleotides to in-line probing assays. The arrowhead demarks the position at the 5′ end of the aptamer where in-line probing data becomes available. Locations of nucleotides beyond position ~70 undergoing structural modulation are estimated, as they cannot be definitively assigned based on the available in-line probing data. Lowercase g notations indicate guanosine nucleotides added to the natural sequence to enable efficient transcription by T7 RNA polymerase. (B) PAGE analysis of the products of in-line probing of 5′ 32P-labeled 131 nudix RNA in the presence of either no ligand (–) or 1 mM of the compound indicated. NR, T1 and –OH represent RNA undergoing no reaction, partial digest with RNase T1 (cleaves after G nucleotides), or partial digest under alkaline conditions (cleaves after all nucleotides), respectively. Bands corresponding to precursor RNA (Pre) and certain G residues are annotated. Annotations 3 through 5, as marked in A, identify prominent regions of the RNA that undergo structural modulation in the presence of certain nucleotide molecules. (C) PAGE analysis of the products of in-line probing of 5′ 32P-labeled 131 nudix RNA in the presence of either no ligand (–), 1 mM, or 100 μM of the nucleoside di- or triphosphate indicated. Annotations are as described for B.
Most riboswitches previously validated are highly selective for their target ligands,6 whereas the binding data described above reveals an unusual lack of selectivity. However, there are several observations that suggest these initial binding results are reflective of the natural function of ykkC subtype 2c riboswitches. For example, the regions of the 131 nudix RNA that undergo structural change in the presence of various (d)NDPs (Figure 2A) are generally consistent with nucleotide positions that form the binding pockets for the guanidine-I,8,11 ppGpp,19,28 and PRPP20,28 riboswitch classes that are also derived from ykkC motif RNAs. This finding is consistent with the hypothesis that the binding pocket for the aptamer present in the 131 nudix construct is undergoing structural reorganization in response to compounds that are at least similar to its natural ligand. Also, compounds that carry certain modifications to the diphosphate are discriminated against by the RNA. Most notably, the addition of a single phosphate moiety causes a substantial reduction in the modulation of band intensities from in-line probing assays (Figure 2C). This suggests that the ligand binding pocket has adapted to reject these natural triphosphate derivatives of NDPs and dNDPs, which can be present at high concentrations in cells. Finally, it seems possible that riboswitches with broadened ligand specificity might be useful for regulating certain NUDIX hydrolase genes because some NUDIX hydrolase enzymes are naturally broad in substrate specificity.29–35
To further evaluate the binding specificity of ykkC subtype 2c RNAs, three additional representative RNA constructs were examined for binding by using the same collection of compounds. The first of these additional representatives is a ykkC subtype 2c RNA construct comprising 129 nucleotides of the 5′ UTR of a nudix mRNA from Salsuginibacillus kocurii (Figure S1A). This 5ˊ 32P-labeled RNA construct, named 129 nudix, exhibits structural modulation only in response to the addition of NDPs and dNDPs carrying either adenine or cytosine as a nucleobase (ADP and CDP) (Figure S1B, C). The 129 nudix RNA strongly discriminates against NTPs, as well as a range of additional nucleotide compounds with more substantial modifications. As explored in greater detail later, it is important to note that some ligands are bound more tightly than others, which ultimately might help determine what compounds naturally serve as the ligands for these riboswitch RNAs.
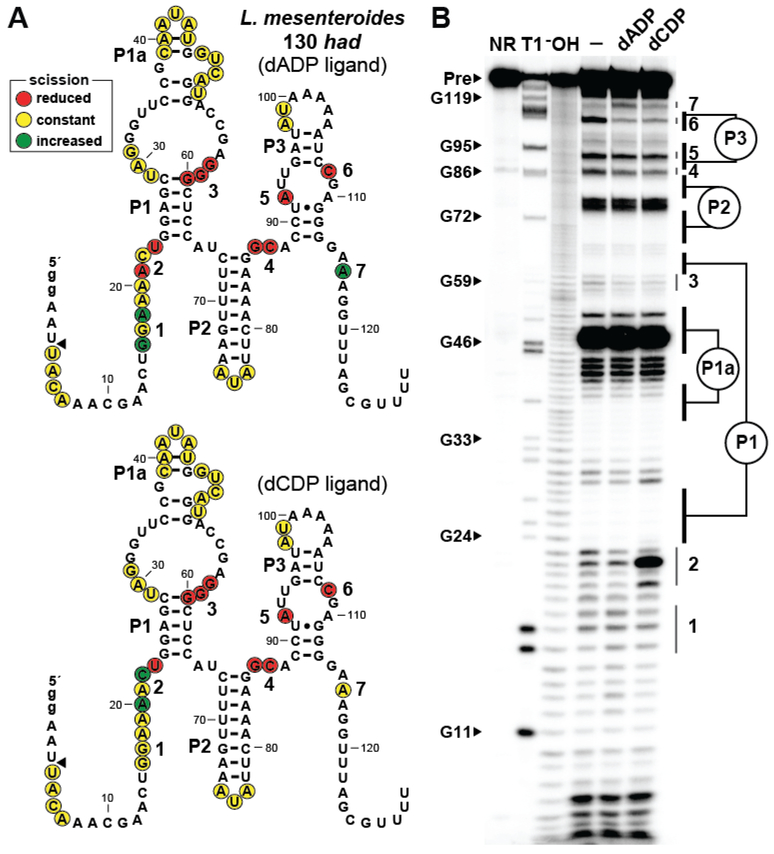
The two other additional constructs examined were a 130 nucleotide RNA derived from the 5′ UTR of a had gene from Leuconostoc mesenteroides called 130 had (Figure 3) and a 126 nucleotide RNA derived from the 5′ UTR of a nudix gene from Bacillus cellulosilyticus called 126 nudix (Figure S2). These two RNA constructs specifically recognize only ADP, CDP, dADP, and dCDP (Figures S3, S4). Importantly, the 130 had RNA, which is predicted to regulate the expression of a HAD-like hydrolase rather than a NUDIX hydrolase, exhibits the same ligand binding preferences as the 129 nudix and the 126 nudix constructs derived from other species as described above. Notably, the 130 had RNA exhibits a different pattern of structural modulation at certain sites during in-line probing analysis depending on whether the aptamer is exposed to dADP (Figure 3A, top) or dCDP (Figure 3A, bottom) (Figure 3B). Similar results are observed for ADP and CDP ligands (Figure S3). Likewise, the 126 nudix RNA construct exhibits distinct structural changes in response to ADP and CDP, particularly in region 2 wherein a C residue experiences a substantial increase in band intensity (Figure S2). These finding suggests that the RNA constructs use binding pockets that adapt somewhat differently to these distinct ligands.
Figure 3.
Adenosine and cytidine nucleotides are differently recognized by ykkC subtype 2c RNAs. (A) Sequence and secondary structure of the 130 nucleotide RNA derived from a had gene of L. mesenteroides. Top: Data collected in B were used to determine regions of reduced, constant, increased scission specific to the addition of dADP (top) or dCDP (bottom). Additional annotations are described in the legend to Figure 2A. (B) PAGE analysis of the products of in-line probing of 5′ 32P-labeled 130 had RNA in the presence of 2 mM dADP or dCDP. Additional annotations are described in the legend to Figure 2B. See Figure S2 for similar results using the 126 nudix construct from B. cellulosilyticus.
Indeed, the unusual differences in banding patterns observed from the in-line probing data for these two RNA constructs might hint at a possible mechanism for nucleobase recognition. As noted above, a C residue at position 22 in 130 had (Figure 3) and position 17 in 126 nudix (Figure S2) appear to become less structured only when the ligands CDP or dCDP are present. In an analogous manner, only the addition of the ligands ADP or dADP causes an increase in the predicted unstructured character of an A residue near position 116 in 130 had and near position 113 in 126 nudix. Perhaps these nucleotide positions that undergo a band intensity increase when ligand is present are being displaced from a structurally important contact to accommodate binding of the ligand’s nucleobase. Future analyses to establish atomic-resolution structural models for these RNAs should resolve the precise mechanisms by which members of the ykkC subtype 2c RNAs bind multiple (d)NDP ligands.
Ligand Binding Affinities for Subtype 2c RNAs.
We next sought to determine the affinity of ykkC subtype 2c RNAs for the various nucleotides that were observed to bind in the initial in-line probing assays described above. Whereas the nucleotide compounds recognized by ykkC subtype 2c RNAs vary slightly depending on the construct tested, all four of the representative RNAs tested display structural modulation in response to (d)ADP and (d)CDP nucleotides. To determine the affinity of ykkC subtype 2c RNAs for adenine- versus cytosine-containing nucleoside diphosphates as well as those carrying ribose versus deoxyribose, each of the four RNA constructs described above were analyzed by in-line probing with ADP, CDP, and dADP over a wide range of ligand concentrations.
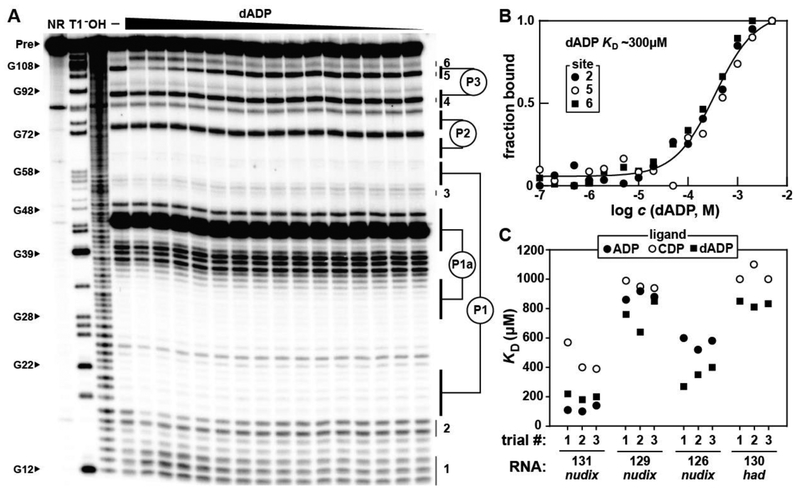
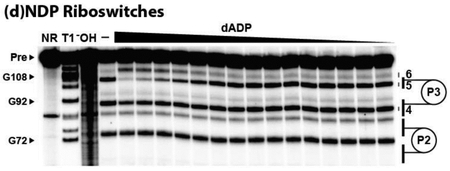
For example, the B. cellulosilyticus 126 nudix RNA (Figure S2) exhibits dose-dependent structural modulation upon the addition of dADP (Figure 4A) in a manner consistent with a one-to-one binding interaction and with a dissociation constant (KD) of ~300 μM (Figure 4B). ADP is bound with an affinity similar to that for dADP, but CDP exhibits a KD that appears to be poorer than 1 mM (Figure 4C, Figure S5). Overall, the four ykkC subtype 2c RNAs examined in this study typically bind dADP best, with KD values ranging between ~200 μM and ~800 μM (Figure 4C, Figures S6-S8). The C. botulinum 131 nudix RNA is the most promiscuous construct (Figure 2), but it also recognizes ADP, CDP, and dADP with the highest affinity of the four RNA constructs that were tested (Figure 4C). It is not obvious to us why 131 nudix accepts a broader range of ligands compared to the other constructs tested. Two notable sequence differences occur at nucleotide positions 16 (G nucleotide) and 62 (C nucleotide) of the 131 nudix, which are U and G nucleotides in the more selective 126 nudix , 129 nudix, and 130 had constructs. These regions undergo structural modulation upon ligand binding, and perhaps are contributing to the unique properties of their ligand-binding pockets.
Figure 4.
Nucleoside diphosphate binding by various ykkC subtype 2c RNAs. (A) PAGE analysis of the products of in-line probing using the 5′ 32P-labeled 126 nudix RNA derived from B. cellulosilyticus (Figure S2) in the presence of various dADP concentrations. In-line probing experiments contained either no ligand (–) or dADP ranging from 5 mM to 100 nM decreasing in ~1/3 log units. Annotations are described in the legend to Figure 2B. (B) Plot of the fraction of RNA bound to dADP, as determined by the normalized fraction of RNA scission at regions 2, 5, and 6 as a function of the logarithm of dADP concentration (c). The dissociation constant (KD) was estimated by a sigmoidal fit of the average of the data points from the indicated regions of RNA structural modulation. Data shown are representative of at least three separate in-line probing experiments. (C) Plot of KD values for the 131 nudix from C. botulinum, 129 nudix from S. kocurii, 126 nudix from B. cellulosilyticus, and 130 had from L. mesenteroides ykkC subtype 2c RNAs with ADP, CDP, and dADP. Each RNA construct was tested with each ligand in three separate trials and all values are reported. Representative PAGE images for each RNA construct with each ligand can be found in panel A for 126 nudix with dADP, and Figures S3, S5-S7 for all remaining data points. In-line probing data for the 126 nudix RNA with CDP (Figure S5A) and for the 130 had RNA with ADP (Figure S8A) indicate that the KD values for these RNA-ligand interactions are poorer than 1 mM, but likely better than 10 mM.
Regardless, it seems reasonable to speculate that the natural ligand for most members of the ykkC subtype 2c riboswitch class is dADP, although we cannot yet exclude the possibility that the riboswitch aptamers are intentionally promiscuous to match the substrate specificities of the enzymes whose expression they control. It also seems possible that some of the riboswitches in the subtype 2c collection have evolved to favor somewhat different nucleotide derivatives, again to correspond to the natural enzymatic specificities of their associated enzymes.
Conserved Nucleotides Characteristic of the ykkC Motif Consensus are Required for ADP and CDP Binding by Subtype 2c RNAs.
To ensure that the modulation observed is caused by a specific interaction between the aptamer and ligand molecule, we examined a series of three mutant 131 nudix RNA constructs (M1-M3), wherein each carries a single nucleotide change to an otherwise strictly conserved position (Figure 5A). These mutant RNAs are all unable to recognize ADP or CDP, even at a concentration of 1 mM (Figure 5B), whereas the wild-type (WT) construct performs as previously observed (Figure 2B).
Figure 5.
Mutation of conserved nucleotides disrupts ligand binding. (A) Sequence and secondary structure of the 131 nudix RNA from C. botulinum. Red letters denote nucleotides where this RNA representative matches the highly conserved (>97%) positions of the ykkC subtype 2C consensus sequence. Positions that were altered to produce each mutant RNA are identified with boxes, annotated with the nucleotide identity of each mutation, and labeled M1 through M3. (B) PAGE analysis of in-line probing assays with 5ˊ 32P-labeled WT and mutated versions of the 131 nudix RNA in the absence (–) or presence of 1, 0.5, or 0.1 mM ADP or CDP. Additional annotations are as described in the legend to Figure 2B.
Given the general similarity between the various riboswitch aptamers derived from the original ykkC motif collection,1,2,8,19,20 we assessed whether subtype 2c RNAs are unique in their ability to recognize nucleoside diphosphates. A representative ppGpp aptamer (Figure S9A), which was previously determined to bind ppGpp with a KD of ~10 nM,19 does not exhibit any structural modulation in the presence of 1 mM ADP (Figure S9B). Moreover, a ykkC subtype 2c construct, 126 nudix, does not recognize the ligands for any of the other riboswitch classes encompassed by the original ykkC motif collection (Figure S9C). These findings strongly support our conclusion that subtype 2c RNAs function as a distinct class of riboswitches that bind to certain NDP and dNDP ligands, of which dADP is particularly favored.
Speculation on the Natural Ligand for ykkC Subtype 2c Riboswitch Class.
Given that ykkC subtype 2c aptamers recognize multiple nucleotide ligands in our binding assays, the true natural ligand is not yet readily apparent. Indeed, subtype 2c RNAs might have diverse natural ligands, given the well-established substrate heterogeneity of NUDIX hydrolases. The current binding data generated with four RNA constructs from different species suggests the ligand generally conforms to Xpp(d)M, where “X” is a chemical moiety attached to the 5′ β-phosphate of a nucleotide molecule, “pp” are the α and β 5′ phosphates, “(d)” indicates that the sugar of the nucleotide can be either ribose or deoxyribose, and “M” indicates the presence of either an adenine or cytosine nucleobase. The “X” of the ligand could just simply be hydrogen (or a negative charge), making the ligand a canonical nucleoside diphosphate. X appears unlikely to be phosphate, glucose, or ribose because NTPs, ADP-glucose, and ADP-ribose are not recognized at all when tested at 1 mM by three of the four representative subtype 2c RNAs examined in this study (Figures S1, S3, S4). However, we cannot rule out the possibility that another substituent at position X is present in the preferred natural ligand for this riboswitch class.
The nucleobases most consistently recognized by ykkC subtype 2c riboswitches are adenine and cytosine. However, some NUDIX hydrolases act on nucleotide substrates with oxidatively-damaged nucleobases, such as 2-hydroxyadenine, 8-oxoadenine, and 5-hydroxycytosine.29–31 Although we did not observe any RNA structural modulation with these nucleotide derivatives, they were only commercially available in the 5′ triphosphate form. Members of this riboswitch class strongly discriminate against nucleotides with a γ 5′ phosphate, and thus we cannot completely exclude the possibility that one of these damaged nucleobase molecules might be preferentially recognized by subtype 2c RNAs in its nucleoside diphosphate form.
Nevertheless, we do not favor the hypothesis that an oxidatively-damaged nucleotide is the natural ligand for this riboswitch class, based on the observation that (d)NTPs are strongly rejected as ligands. If the function of ykkC subtype 2c RNAs is to act as receptors for damaged nucleotides that subsequently activate expression of phosphatase enzymes that degrade damaged nucleotides, then it does not seem likely that the riboswitch would exclude these modified compounds in their triphosphate form. Some other riboswitch classes, such as those for ppGpp (which also binds pppGpp)19 and ZTP (which also binds ZMP),39 strongly bind their target ligands in multiple phosphorylation states. Given this precedence, we would expect the ykkC subtype 2c riboswitch class to recognize damaged (d)NDPs and (d)NTPs with similar affinity if these were the true natural ligand for this riboswitch class.
The existing data for representatives such as 130 had and 126 nudix fit best with the hypothesis that ykkC subtype 2c RNAs represent a riboswitch class that naturally responds to both (d)ADP and (d)CDP. To achieve this broad specificity, a portion of the binding pocket might recognize the sugar and diphosphate moieties of the ligand, while a separate portion of the aptamer recognizes each nucleobase. The sugar-diphosphate portion of the binding pocket could be provided by the aptamer nucleotides corresponding to the binding site for this same chemical moiety that is present in the ligands for ppGpp19,28 (ykkC subtype 2a) and PRPP20,28 (ykkC subtype 2b) aptamers. The nucleobase-sensing portion of the aptamer could be located in the additional nucleotides that flank the P1-P3 stems, based on the modulation observed by in-line probing (Figures 2–4). These regions might form a distinct substructure into which the nucleobase portion of the ligand (either adenine for ADP or cytosine for CDP) can insert and replace the structural role of a nucleotide residue of the same identity within the aptamer.
A riboswitch class specific for both (d)ADP and (d)CDP that promotes the expression of nucleotide hydrolase enzymes could be useful for cells to monitor the phosphorylation status of nucleotide pools in the cell. It appears that subtype 2c RNAs do not consistently discriminate between rNDPs and dNDPs, and perhaps this ligand ambiguity is exploited to sense nucleoside diphosphates in both their 2ˊ-ribose and 2ˊ-deoxyribose forms. Intriguingly, some members of this riboswitch class sense (d)NDP molecules that carry at least one purine and at least one pyrimidine, which are generated through separate biosynthesis pathways. This dual recognition capability could allow bacteria to simultaneously monitor the phosphorylation state of their purine and pyrimidine nucleotide pools to maintain the balance between the different nucleotide forms in the cell.
The in vitro binding affinity of ykkC subtype 2c RNAs appears to be relatively poor compared to most riboswitch classes. However, there are several pertinent factors to consider when evaluating these affinities, including the typical in vivo concentration of the natural ligand, the concentration of this ligand at which it becomes necessary to trigger its corresponding riboswitch class, and whether the riboswitch aptamer reaches thermodynamic equilibrium with its ligand or if the riboswitch is kinetically driven.40,41 For example, the glutamine riboswitch class42,43 displays a poor in vitro affinity of ~1 mM for its cognate ligand glutamine.42 However, the concentration of this amino acid has been measured at nearly 4 mM in Escherichia coli cells.44 The affinities measured for ykkC subtype 2c RNAs for their (d)NDP ligands are generally in the range of 0.1 to 1 mM. ADP has been found to exist in cells at a similar range of concentrations.44–46 Perhaps, under circumstances in which the concentration of ADP substantially exceeds its typical concentration, it would be beneficial for the cell to express a nucleotide hydrolase enzyme that helps maintain the (d)NDP concentrations within this lower range.
Concluding Remarks.
The functions of ykkC motif RNAs and the various riboswitch classes they represent have revealed several interesting insights into the properties of RNA molecules and the biological processes they control. Previously, we have reported that subtype 1 ykkC motif RNAs selectively respond to guanidine, which revealed numerous genes involved in overcoming the toxic effects of this nitrogen-rich compound.8 Most recently, we reported that subtype 2a and 2b ykkC motif RNAs function as riboswitches for ppGpp19 and PRPP,20 respectively. In the present study, we provide evidence that subtype 2c ykkC motif RNAs bind most tightly to (d)ADP and (d)CDP nucleotides. At this time we cannot be certain that the natural ligand for ykkC subtype 2c riboswitches is (d)ADP or (d)CDP. The true ligand for this riboswitch class could be a nucleotide molecule with additional modifications, which could even be a nucleotide compound currently unknown to biology. It seems reasonable to conclude that the ligand molecule is likely to conform to the Xpp(d)M consensus structure as discussed above. Alternatively, the ligand ambiguity we observe in vitro might parallel the substrate recognition patterns typical of some NUDIX hydrolase enzymes, which also exhibit broader specificities for certain nucleoside diphosphate substrates.
Another group of natural RNAs that display a broad specificity for multiple putative ligands that are chemically similar is the azaaromatic riboswitch class.47 Representatives of this riboswitch class, whose members were first collectively called the yjdF RNA motif,10 exhibit unusually broad ligand binding specificity for various azaaromatic compounds.47 It has yet to be definitively determined whether there is a single compound that is the natural cognate ligand for this riboswitch class or if yjdF RNAs act as a riboswitch for a variety of planar, azaaromatic molecules. The existence of another riboswitch class comprised of the ykkC subtype 2c RNAs that might respond to more than one ligand supports the hypothesis that binding sites made of RNA have the structural ability to carry out more challenging molecular recognition tasks. Regardless, this nucleoside diphosphate riboswitch class once again exemplifies the amazing diversity of the ykkC motif scaffold, and adds to the collection of natural RNA aptamers that recognize nucleotide-derived ligand molecules.6
Finally, we note that there is at least one more collection of ykkC motif RNA variants that constitute an unsolved orphan riboswitch class. Subtype 2d RNAs, which most closely resemble the consensus model for guanidine-I riboswitches (ykkC subtype 1 RNAs), have yet to be experimentally linked to a ligand. We speculate that the ligand is unlikely to be another nucleotide or nucleotide-like compound, because these RNAs lack some of the aptamer nucleotides that form the sugar-phosphate binding pocket of subtypes 2a,24 2b,24 and probably 2c. Also, the nature of the transporter proteins whose expression is regulated by this candidate riboswitch class indicate that subtype 2d RNAs might sense a smaller molecule or ion that is not a nucleotide derivative. Currently subtype 2d RNAs represent the longest known orphan riboswitch candidate, and thus present an intriguing scientific puzzle to solve.
Supplementary Material
Supplemental File 1: Alignment of ykkC subtype 2c RNAs (.sto)
ACKNOWLEDGEMENTS
We thank Shira Stav, Adam Roth, Narasimhan Sudarsan, and other members of the Breaker laboratory for helpful discussions.
Funding
M.E.S. and H.S. were supported by an NIH Cellular and Molecular Biology Training Grant (T32GM007223). This work was also supported by NIH grants to R.R.B. (GM022778 and AI136794). R.R.B. is also supported by the Howard Hughes Medical Institute.
Footnotes
ASSOCIATED CONTENT
Supporting Information
REFERENCES
- (1).Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, Wickiser JK, and Breaker RR (2004) New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. U.S.A 101, 6421–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Meyer MM, Hammond MC, Salinas Y, Roth A, Sudarsan N, and Breaker RR (2011) Challenges of ligand identification for riboswitch candidates. RNA Biol. 8, 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Roth A, and Breaker RR (2009) The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem 78, 305–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Serganov A, and Nudler E (2013) A decade of riboswitches. Cell 152, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Sherwood AV, and Henkin TM (2016) Riboswitch-mediated gene regulation: Novel RNA architectures dictate gene expression responses. Annu. Rev. Microbiol 70, 361–374. [DOI] [PubMed] [Google Scholar]
- (6).McCown PJ, Corbino KA, Stav S, Sherlock ME, and Breaker RR (2017) Riboswitch diversity and distribution. RNA 23, 995–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Breaker RR (2012) Riboswitches and the RNA World. Cold Spring Harb. Perspect. Biol 4, pii: a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Nelson JW, Atilho RM, Sherlock ME, Stockbridge RB, and Breaker RR (2017) Metabolism of free guanidine in bacteria is regulated by a widespread riboswitch class. Mol Cell 65, 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, Neph S, Tompa M, Ruzzo WL, and Breaker RR (2007) Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res 35, 4809–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, and Breaker RR (2010) Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol 11, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Reiss CW, Xiong Y, and Strobel SA (2017) Structural basis for ligand binding to the guanidine-I riboswitch. Structure 25, 195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Battaglia RA, Price IR, and Ke A (2017) Structural basis for guanidine sensing by the ykkC family of riboswitches. RNA 23, 578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Kanamori T, Kanou N, Atomi H, and Imanaka T (2004) Enzymatic characterization of a prokaryotic urea carboxylase. J Bacteriol 186, 2532–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Kermani AA, Macdonald CB, Gundepudi R, and Stockbridge RB (2018) Guanidinium export is the primal function of SMR family transporters. Proc Natl Acad Sci USA 115, 3060–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Jack DL, Storms ML, Tchieu JH, Paulsen IT, and Saier MH Jr. (2000) A broad-specificity multidrug efflux pump requiring a pair of homologous SMR-type proteins. J Bacteriol 182, 2311–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Schuldiner S (2009) EmrE, a model for studying evolution and mechanism of ion-coupled transporters. Biochim Biophys Acta 1794, 748–762. [DOI] [PubMed] [Google Scholar]
- (17).Sherlock ME, Malkowski SN, and Breaker RR (2017) Biochemical validation of a second guanidine riboswitch class. Biochemistry 56, 352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Sherlock ME, and Breaker RR (2017) Biochemical validation of a third guanidine riboswitch class in bacteria. Biochemistry 56, 359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Sherlock ME, Sudarsan N, and Breaker RR (2018) Riboswitches for the alarmone ppGpp expand the collection of RNA-based signaling systems. Proc. Natl. Acad. Sci. U.S.A 115, 6052–6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Sherlock ME, Sudarsan N, Stav S, and Breaker RR (2018) Tandem riboswitches form a natural Boolean logic gate to control purine metabolism in bacteria. eLife 7, e33908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Cashel M (1969) The control of ribonucleic acid synthesis in Escherichia coli. J Biol Chem 244, 3133–3141. [PubMed] [Google Scholar]
- (22).Dalebroux ZD, and Swanson MS (2012) ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol 10, 203–212. [DOI] [PubMed] [Google Scholar]
- (23).Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, and Gerdes K (2015) Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13, 298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, and Conway T (2008) The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 68, 1128–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Shivers RP, and Sonenshein AL (2004) Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol Micriobiol 53, 599–611. [DOI] [PubMed] [Google Scholar]
- (26).Soukup GA, and Breaker RR (1999) Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5, 1308–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Regulski EE, and Breaker RR (2008) In-line probing analysis of riboswitches. Methods Mol Biol 419, 53–67. [DOI] [PubMed] [Google Scholar]
- (28).Knappenberger AJ, Reiss CW, and Strobel SA (2018) Structures of two aptamers with differing ligand specificity reveal ruggedness in the functional landscape of RNA. eLife 7, pii: e36381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).McLennan AG (2006) The Nudix hydrolase superfamily. Cell Mol Life Sci 63, 123–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Bessman MJ, Frick DN, and O’Handley SF (1996) The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem 271, 25059–25062. [DOI] [PubMed] [Google Scholar]
- (31).Takagi Y, Setoyama D, Ito R, Kamiya H, Yamagata Y, and Sekiguchi M (2012) Human MTH3 (NUDT18) protein hydrolyzes oxidized forms of guanosine and deoxyguanosine diphosphates: comparison with MTH1 and MTH2. J Biol Chem 287, 21541–21549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Nguyen VN, Park A, Xu A, Srouji JR, Brenner SE, and Kirsch JF (2016) Substrate specificity characterization for eight putative nudix hydrolases. Evaluation of criteria for substrate identification within the Nudix family. Proteins 84, 1810–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Muñoz FJ, Baroja-Fernández E, Morán-Zorzano MT, Alonso-Casajús N, and Pozueta-Romero J (2006) Cloning, expression and characterization of a Nudix hydrolase that catalyzes the hydrolytic breakdown of ADP-glucose linked to starch biosynthesis in Arabidopsis thaliana. Plant Cell Physiol 47, 926–934. [DOI] [PubMed] [Google Scholar]
- (34).Fisher DI, Cartwright JL, Harashima H, Kamiya H, and McLennan AG (2004) Characterization of a Nudix hydrolase from Deinococcus radiodurans with a marked specificity for (deoxy)ribonucleoside 5’-diphosphates. BMC Biochem 5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Kuznetsova E, Proudfoot M, Gonzalez CF, Brown G, Omelchenko MV, Borozan I, Carmel L, Wolf YI, Mori H, Savchenko AV, Arrowsmith CH, Koonin EV, Edwards AM, and Yakunin AF (2006) Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J Biol Chem 281, 36149–36161. [DOI] [PubMed] [Google Scholar]
- (36).Lu Z, Dunaway-Mariano D, and Allen KN (2005) HAD superfamily phosphotransferase substrate diversification: structure and function analysis of HAD subclass IIB sugar phosphatase BT4131. Biochemistry 44, 8684–8696. [DOI] [PubMed] [Google Scholar]
- (37).Caparrós-Martín JA, McCarthy-Suárez I, and Culiáñez-Macià FA (2013) HAD hydrolase function unveiled by substrate screening: enzymatic characterization of Arabidopsis thaliana subclass I phosphosugar phosphatase AtSgpp. Planta 237, 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Kuznetsova E, Nocek B, Brown G, Makarova KS, Flick R, Wolf YI, Khusnutdinova A, Evdokimova E, Jin K, Tan K, Hanson AD, Hasnain G, Zallot R, de Crécy-Lagard V, Babu M, Savchenko A, Joachimiak A, Edwards AM, Koonin EV, and Yakunin AF (2015) Functional diversity of haloacid dehalogenase superfamily phosphatases from Saccharomyces cerevisiae: biochemical, structural, and evolutionary insights. J Biol Chem 290, 18678–18698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Kim PB, Nelson JW, and Breaker RR (2015) An ancient riboswitch class in bacteria regulates purine biosynthesis and one-carbon metabolism. Mol Cell 57, 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Wickiser JK, Winkler WC, Breaker RR, and Crothers DM (2005) The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell 18, 49–60. [DOI] [PubMed] [Google Scholar]
- (41).Wickiser JK, Cheah MT, Breaker RR, and Crothers DM (2005) The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry 44, 13404–13414. [DOI] [PubMed] [Google Scholar]
- (42).Ames TD, and Breaker RR (2011) Bacterial aptamers that selectively bind glutamine. RNA Biol 8, 82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Ren A, Xue Y, Peselis A, Serganov A, Al-Hashimi HM, and Patel DJ (2015) Structural and dynamic basis for low affinity-high selectivity binding of L-glutamine by the glutamine riboswitch. Cell Rep 13, 1800–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, and Rabinowitz JD (2009) Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol 5, 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Bochner BR, and Ames BN (1982) Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem 257, 9759–9769. [PubMed] [Google Scholar]
- (46).Buckstein MH, He J, and Rubin H (2008) Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J Bacteriol 190, 718–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Li S, Hwang XY, Stav S, and Breaker RR (2016) The yjdF riboswitch candidate regulates gene expression by binding diverse azaaromatic compounds. RNA 22, 530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental File 1: Alignment of ykkC subtype 2c RNAs (.sto)