Artificial proteins built from consensus PPR motifs bind the intended RNA in vivo and can be used as RNA affinity tags to purify endogenous RNPs and identify the bound proteins.
Abstract
Pentatricopeptide repeat (PPR) proteins bind RNA via a mechanism that facilitates the customization of sequence specificity. However, natural PPR proteins have irregular features that limit the degree to which their specificity can be predicted and customized. We demonstrate here that artificial PPR proteins built from consensus PPR motifs selectively bind the intended RNA in vivo, and we use this property to develop a new tool for ribonucleoprotein characterization. We show by RNA coimmunoprecipitation sequencing (RIP-seq) that artificial PPR proteins designed to bind the Arabidopsis (Arabidopsis thaliana) chloroplast psbA mRNA bind with high specificity to psbA mRNA in vivo. Analysis of coimmunoprecipitating proteins by mass spectrometry showed the psbA translational activator HCF173 and two RNA binding proteins of unknown function (CP33C and SRRP1) to be highly enriched. RIP-seq revealed that these proteins are bound primarily to psbA RNA in vivo, and precise mapping of the HCF173 and CP33C binding sites placed them in different locations on psbA mRNA. These results demonstrate that artificial PPR proteins can be tailored to bind specific endogenous RNAs in vivo, add to the toolkit for characterizing native ribonucleoproteins, and open the door to other applications that rely on the ability to target a protein to a specified RNA sequence.
INTRODUCTION
The ability to target proteins to specified RNA sequences provides an entrée to diverse approaches for manipulating and analyzing RNA-mediated functions. However, the sequence specificities of most RNA binding proteins are difficult to predict because most RNA binding domains bind short, degenerate sequence motifs and use variable binding modes (reviewed in Helder et al., 2016). In this context, the Pumilio/FBF (PUF) and pentatricopeptide repeat (PPR) protein families have attracted interest due to their unusual mode of RNA recognition (Chen and Varani, 2013; Yagi et al., 2014; Hall, 2016). PUF and PPR proteins have tandem helical repeating units that bind consecutive nucleotides with a specificity that is largely determined by the identities of amino acids at two positions. These amino acid codes have been used to reprogram native proteins to bind new RNA sequences and for the design of artificial proteins with particular sequence specificities (Barkan et al., 2012; Campbell et al., 2014; Coquille et al., 2014; Kindgren et al., 2015; Shen et al., 2015, 2016; Colas des Francs-Small et al., 2018; Miranda et al., 2018; Zhao et al., 2018; Bhat et al., 2019; Yan et al., 2019).
PUF and PPR proteins also differ in important respects. They bind RNA with opposite polarity, and they use distinct amino acid combinations to specify each nucleotide (reviewed in Hall, 2016). PUF proteins comprise a small protein family whose members invariably contain eight repeat motifs (Goldstrohm et al., 2018), whereas the PPR family includes more than 400 members in plants, and the number of PPR motifs per protein ranges from 2 to ∼30 (Lurin et al., 2004). PUF proteins generally localize to the cytoplasm and repress the translation or stability of mRNA ligands (reviewed in Wang et al., 2018), while PPR proteins localize almost exclusively to mitochondria and chloroplasts, where they function in RNA stabilization, translational activation, group II intron splicing, RNA cleavage, and RNA editing (reviewed in Barkan and Small, 2014). The evolutionary malleability of PPR architecture and function suggests that the PPR scaffold may be particularly amenable to tailoring RNA binding affinity, kinetics, and sequence specificity for particular applications.
The PPR code has been used to recode several natural PPR proteins to bind nonnative RNA ligands in vitro (Barkan et al., 2012) and in vivo (Kindgren et al., 2015; Colas des Francs-Small et al., 2018; Rojas et al., 2019). However, the engineering of native PPR proteins is complicated by irregularities in their PPR tracts, which result in variable and unpredictable contributions of their PPR motifs to RNA affinity and specificity (Fujii et al., 2013; Okuda et al., 2014; Miranda et al., 2017; Rojas et al., 2018). By contrast, artificial PPR proteins (aPPRs) built from consensus PPR motifs exhibit predictable sequence specificity in vitro (Coquille et al., 2014; Shen et al., 2015; Miranda et al., 2018; Yan et al., 2019). However, the degree to which such proteins bind selectively to RNAs in vivo has not been reported.
In this work, we advance efforts to engineer PPR proteins by showing that two aPPR proteins bind with high specificity to their intended endogenous RNA target in vivo. At the same time, we demonstrate the utility of aPPRs for a particular application—the purification of specific native ribonucleoprotein particles (RNPs) for identification of the associated proteins. The population of proteins bound to an RNA influences its function and metabolism, but techniques for characterizing RNP-specific proteomes are limited. Thus, our results expand the toolkit for purifying selected RNPs and lay the groundwork for the use of aPPRs in other applications.
RESULTS
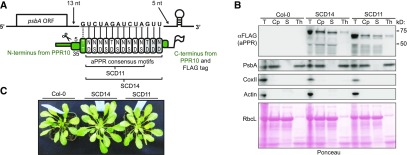
We chose to target aPPRs to the chloroplast psbA mRNA for this proof-of-concept experiment because the psbA mRNA exhibits dynamic changes in translation in response to light, and identification of bound proteins may elucidate the underlying mechanisms (reviewed in Sun and Zerges, 2015; Chotewutmontri and Barkan, 2018). We designed aPPR proteins with either 11 or 14 PPR motifs to bind the psbA 3ʹ untranslated region (UTR) in Arabidopsis (Arabidopis thaliana; Figure 1A; Supplemental Figure 1). We refer to these proteins as SCD11 and SCD14, respectively. Because PPR proteins bind single-stranded RNA, we targeted the proteins to a sequence that is predicted to be unstructured. To avoid disrupting psbA expression, we chose a target sequence that is poorly conserved and that begins sufficiently far from the stop codon that the terminating ribosome and aPPR should not occupy the same nucleotides. We designed the proteins according to the scheme described by Shen et al. (2015), such that a tract of consensus PPR motifs with the appropriate specificity-determining amino acids is embedded within N- and C-terminal segments of the native chloroplast-localized protein PPR10 (Pfalz et al., 2009). We previously reported a comprehensive analysis of the sequence specificity of SCD11 and SCD14 in vitro (Miranda et al., 2018), confirming them to be highly selective for their intended target sequence in vitro while also revealing nuances relevant to prediction of off-target binding. For the in vivo assays described here, the proteins included, in addition, a C-terminal 3xFLAG tag and the N-terminal chloroplast targeting sequence from PPR10, which is cleaved after chloroplast import (Figure 1A; Supplemental Figure 1).
Figure 1.
Overview of Artificial PPR Proteins Designed to Bind the psbA 3ʹ UTR.
(A) Protein design. SCD11 and SCD14 were designed to bind the indicated 14- or 11 (underlined)-nucleotide (nt) sequence in the 3ʹ UTR of the psbA mRNA in Arabidopsis. The targeted sequence begins 13 nucleotides downstream of the stop codon and ends five (SCD14) or eight (SCD11) nucleotides upstream of the 3ʹ-terminal stem-loop in the psbA mRNA. SCD14 and SCD11 contain 13 and 10 consensus PPR motifs, respectively, flanked by sequences from PPR10 (green). The motifs that are found in SCD14, but not in SCD11, are marked in gray. The specificity-determining amino acids (Barkan et al., 2012) (positions 5 and 35 according to the nomenclature in Yin et al., 2013) are indicated, and each repeat is aligned with its nucleotide ligand. The PPR10-derived sequence at the N terminus includes a chloroplast targeting sequence and PPR10’s first PPR motif, which has a noncanonical specificity code (dotted line). The targeting sequence is cleaved after import into the chloroplast (scissors). Both proteins contain a C-terminal 3xFLAG tag. ORF, open reading frame.
(B) Immunoblots demonstrating chloroplast-localization of SCD11 and SCD14. Chloroplasts (Cp) were isolated from total leaf (T) of wild-type (Col-0) and transgenic Arabidopsis plants and fractionated to generate thylakoid membrane (Th) and soluble (S) fractions. Aliquots representing an equivalent amount of starting material were probed to detect markers for cytosol (Actin), mitochondria (Cox II), and thylakoid membranes (PsbA). The aPPR proteins were detected with anti-FLAG antibody. The Ponceau S–stained filter is shown below to demonstrate the partitioning of the chloroplast stromal protein RbcL (the large subunit of Rubisco).
(C) Visible phenotype of transgenic Arabidopsis plants expressing SCD14 and SCD11. Col-0 is the wild-type progenitor of the transgenic lines.
Immunoblot analysis of leaf and chloroplast fractions from transgenic Arabidopsis plants expressing SCD11 and SCD14 confirmed that they localize to chloroplasts (Figure 1B). Both proteins were found predominantly in the soluble fraction, as expected given that they lack transmembrane segments or thylakoid targeting signals. Laddering beneath the band corresponding to each full-length protein suggests that these artificial proteins are prone to proteolysis in vivo. The transgenic plants were phenotypically normal (Figure 1C) and had normal levels of PsbA protein (Figure 1B), indicating that the aPPRs did not disrupt psbA expression or have off-target effects that compromised plant growth.
SCD11 and SCD14 Bind with High Specificity to psbA RNA in Vivo
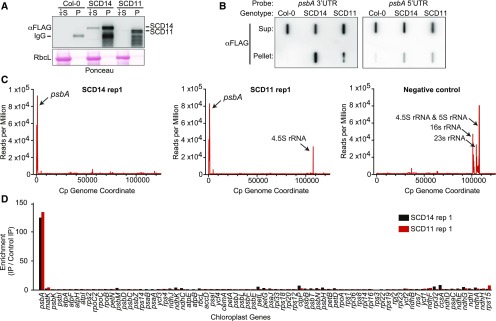
To identify RNAs that are bound to SCD11 and SCD14 in vivo, we isolated chloroplasts from the transgenic plants, used anti-FLAG antibody to immunoprecipitate each protein from stromal extract (Figure 2A), and purified RNA from the coimmunoprecipitates. Slot-blot hybridizations showed that psbA RNA was highly enriched in immunoprecipitates from the transgenic lines in comparison to the wild-type (Columbia [Col]-0) progenitor (Figure 2B). Furthermore, RNA from the 3ʹ UTR was more highly enriched than that from the 5ʹ UTR (Figure 2B), consistent with the binding of the aPPRs to the 3ʹ UTR, as intended.
Figure 2.
RIP-Seq Analysis of RNAs That Coimmunoprecipitate with SCD14 and SCD11.
(A) Immunoprecipitation of SCD14 and SCD11. Stromal extracts from transgenic Arabidopsis expressing the indicated protein or from the Col-0 progenitor were used for immunoprecipitation with anti-FLAG antibody. The pellet (P) and supernatant (S) fractions were analyzed by immunoblot analysis using anti-FLAG antibody. An excerpt of the Ponceau S–stained filter is shown to illustrate the abundance of the large subunit of Rubisco (RbcL), which serves as a loading control. An equal proportion of each pellet fraction was analyzed; one-fourth that proportion of each supernatant was analyzed to avoid overloading the lane.
(B) Slot-blot hybridization analysis of RNAs that coimmunoprecipitate with SCD14 and SCD11. RNA was extracted from the immunoprecipitations analyzed in (A) and applied to nylon membrane via a slot-blot manifold. The same proportion of all samples was analyzed to illustrate the partitioning of psbA RNA between the pellet and supernatant (Sup) fractions. Replicate blots were hybridized with oligonucleotide probes specific for the psbA 5ʹ UTR or 3ʹ UTR.
(C) Plastome-wide view of RNAs that coimmunoprecipitate with SCD11 and SCD14. Results are plotted as the sequence coverage in consecutive 10-nucleotide windows along the chloroplast genome (accession NC_000932.1) per million reads mapped to the chloroplast genome. The negative control used an antibody that does not detect proteins in Arabidopsis together with extract from the SCD14 line. Data for replicate experiments are shown in Supplemental Figure 2A. Read counts for all RIP-seq experiments are provided in Supplemental Data Set 1.
(D) Enrichment of RNA from each protein-coding chloroplast gene in SCD11 and SCD14 coimmunoprecipitates. The ratio of normalized reads/gene in the experimental versus control immunoprecipitations (IP) is shown for replicate 1. Data for replicate experiments are shown in Supplemental Figure 2A.
To gain a comprehensive view of the RNAs bound to each protein, we sequenced the coimmunoprecipitating RNA (RIP-seq) in two replicate experiments. Comparison of these RNAs to those from immunoprecipitations with an antibody that does not recognize proteins in Arabidopsis showed that the psbA RNA was strongly enriched in the SCD11 and SCD14 immunoprecipitates (Figure 2C; Supplemental Figure 2A; Supplemental Data Set 1). The enrichment of RNAs mapping to each chloroplast protein-coding gene was highly reproducible between the replicate experiments (Figure 2D; Supplemental Figure 2A); RNA from the psbA gene was enriched more than 100-fold, whereas RNA from most genes showed no enrichment. This establishes that aPPR proteins can bind specifically to an intended RNA target in vivo. The 4.5S rRNA was also enriched in the SCD11 immunoprecipitates. However, this may be artifactual because it was not enriched in SCD14 immunoprecipitates, and 4.5S rRNA is an unlikely ligand for an aPPR protein due to the fact that it is highly structured and largely embedded in the ribosome.
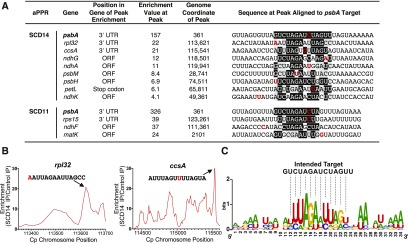
Reproducible low-level enrichment of RNA from several genes other than psbA suggested a small degree of off-target binding, particularly by SCD14 (Figure 2D; Supplemental Figure 2A). All genes whose RNAs showed greater than threefold enrichment in either the SCD14 or SCD11 coimmunoprecipitates are listed in Figure 3A. Most of these contain local peaks of enrichment harboring sequences resembling the intended binding site in the psbA 3ʹ UTR (Figure 3B; Supplemental Figure 3). A sequence logo representing the frequency of nucleotides at each position in SCD14’s off-target sequence set closely resembles SCD14’s intended binding site (Figure 3C).
Figure 3.
Analysis of Off-Target Binding by SCD14 and SCD11.
(A) Genes whose transcripts were enriched more than threefold in both replicate SCD14 and SCD11 coimmunoprecipitates. The position of peak enrichment within each gene (see [B] and Supplemental Figure 3) and the magnitude of enrichment at that peak (average of replicates) are indicated, together with sequence motifs within the peak that resemble the intended binding site for SCD14 and SCD11. Matches to the intended binding site in the psbA 3ʹ UTR are shaded in black. Transition mismatches are shaded in gray. The nucleotide at the peak of enrichment (see profiles in Figure 3B and Supplemental Figure 3) is marked in red. ORF, open reading frame.
(B) Local sequence enrichment profile for the off-target sites in rpl32 and ccsA. The regions shown span the open reading frame and UTRs of the indicated gene. The displayed sequences overlap the point of maximum enrichment (nucleotide at peak marked in red) and resemble the intended binding site of SCD14 (see [A]). Analogous plots for other genes listed in (A) are shown in Supplemental Figure 3. IP, immunoprecipitate.
(C) Sequence logo derived from the off-target sites of SCD14. Sequences listed in (A) were weighted according to the degree to which RNA from the corresponding region was enriched in the SCD14 immunoprecipitation and analyzed with WebLogo (Crooks et al., 2004).
Identification of Known and Novel psbA RNA Binding Proteins in SCD14 and SCD11 Coimmunoprecipitates
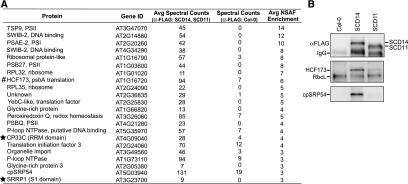
Results above demonstrated that psbA RNA is, by far, the most highly enriched mRNA in SCD14 and SCD11 coimmunoprecipitates. To identify native proteins that are bound to psbA mRNA in vivo, we analyzed the SCD11 and SCD14 coimmunoprecipitates by mass spectrometry. Approximately 400 different proteins were identified in at least one of the immunoprecipitates (Supplemental Data Set 2). The enrichment of each protein was calculated with respect to its representation in an anti-FLAG immunoprecipitate from the nontransgenic host line (Col-0). Approximately 50 proteins were enriched at least twofold in either the SCD11 or SCD14 immunoprecipitation (Supplemental Data Set 2). Proteins whose enrichment averaged at least threefold in the two experiments are listed in Figure 4A. This protein set included several proteins that are known to associate with psbA mRNA: HCF173, which activates psbA translation (Schult et al., 2007); cpSRP54, which binds cotranslationally to PsbA (Nilsson and van Wijk, 2002); and various ribosomal proteins and general translation factors. Immunoblot analysis of anti-FLAG coimmunoprecipitates confirmed that HCF173 and cpSRP54 coimmunoprecipitate with the aPPR proteins from extracts of the transgenic plants (Figure 4B). The differing efficiency with which these proteins were coprecipitated from the two lines may be due to the higher abundance of SCD14 in the stromal extracts (Figure 1B), or to differing degrees of RNA degradation in the two preparations, as RNA cleavage will separate the bound aPPRs from proteins bound elsewhere on the RNA.
Figure 4.
Highly-Enriched Proteins in SCD11 and SCD14 Coimmunoprecipitates.
(A) Proteins whose average (Avg) enrichment from lines expressing SCD11 or SCD14 in comparison to the Col-0 progenitor was three or greater. Stars mark two RNA binding proteins of unknown function. HCF173 (hashtag) is known to associate with psbA mRNA and to activate its translation (Schult et al., 2007; Williams-Carrier et al., 2019). The full data set is provided in Supplemental Data Set 2, which includes a more complete explanation of the data analysis.
(B) Immunoblot validation of two proteins identified by MS analysis. Chloroplast stroma from plants expressing SCD11 or SCD14, or from the Col-0 progenitor was used for immunoprecipitation with anti-FLAG antibody. Replicate immunoblots were probed with anti-FLAG antibody to detect SCD11 and SCD14 (top), HCF173 (middle), or cpSRP54 (bottom). The HCF173 blot was initially probed to detect RbcL (the large subunit of Rubisco), an abundant protein that typically contaminates immunoprecipitates; this serves as an internal standard.
Notably absent from the set of proteins detected in the SCD11 and SCD14 immunoprecipitates (Supplemental Data Set 2) is the pentatricopeptide repeat protein LPE1, which was reported to bind the psbA 5ʹ UTR and recruit HCF173 for psbA translation (Jin et al., 2018). However, this role for LPE1 was recently called into question (Williams-Carrier et al., 2019). The failure to detect LPE1 in the aPPR coimmunoprecipitates despite the strong enrichment of HCF173 supports the revised view that LPE1 neither binds psbA RNA nor activates its translation.
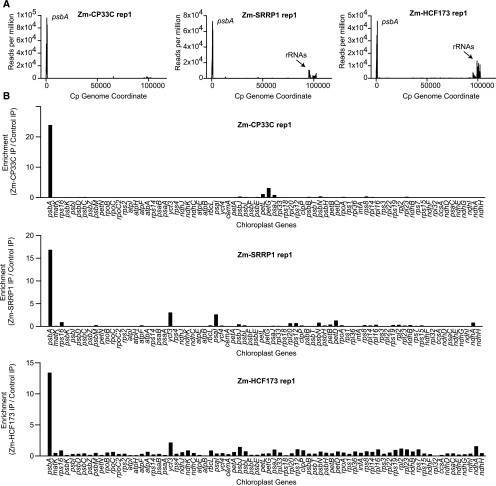
Two predicted RNA binding proteins of unknown function were strongly enriched in the aPPR coimmunoprecipitates (Figure 4A): SRRP1 (AT3G23700) and CP33C (AT4G09040). SRRP1 has two S1 RNA binding domains and was proposed to harbor RNA chaperone activity based on in vitro and Escherichia coli complementation data (Gu et al., 2015). CP33C has two RRM RNA binding domains (Ruwe et al., 2011), but functional studies have not been reported. To determine whether these proteins associate with psbA RNA in vivo, we performed RIP-seq using antibodies generated against the Zea mays (maize) orthologs (sequences shown in Supplemental Figure 4A). At the same time, we performed RIP-seq with antibody to the maize ortholog of HCF173 (Zm-HCF173); HCF173 was previously shown to coimmunoprecipitate with psbA RNA, but limited information was provided about interactions with other RNAs (Schult et al., 2007). Because the Zm-CP33C antibodies did not detect the Arabidopsis ortholog, we used maize stroma for this set of RIP-seq assays. The psbA RNA was highly enriched in each coimmunoprecipitate and was the only RNA to be strongly enriched in each case (Figure 5; replicates in Supplemental Figure 2B). Chloroplast rRNAs were reproducibly enriched in the Zm-HCF173 and Zm-SRRP1 immunoprecipitates, but not in the Zm-CP33C immunoprecipitates, suggesting that HCF173 and SRRP1, but not CP33C, associate with ribosomes and/or with psbA RNA that is undergoing translation. Several other mRNAs were reproducibly enriched in each immunoprecipitate, but to a much smaller degree (Figure 5B; Supplemental Figure 2B). These results validate the utility of the aPPR-affinity tag approach to identify proteins that associate with a specific RNA-of-interest in vivo.
Figure 5.
RIP-Seq Analysis of Zm-HCF173, Zm-CP33C, and Zm-SRRP1.
Maize chloroplast stroma was used for immunoprecipitations with antibodies to the maize orthologs of HCF173, CP33C, and SRRP1, and the coimmunoprecipitated RNA was analyzed by deep sequencing.
(A) Average sequence coverage in consecutive 10-nucleotide windows along the chloroplast genome, per million reads mapped to the chloroplast genome (NCBI NC_001666).
(B) Ratio of normalized reads/gene in the experimental immunoprecipitations (IP) versus a control using antibody to AtpB (subunit of the chloroplast ATP synthase). Analogous plots for replicate experiments are shown in Supplemental Figure 2B. Read counts are provided in Supplemental Data Set 1, which also includes data for tRNAs. Immunoblots demonstrating immunoprecipitation of each protein are shown in Supplemental Figure 5.
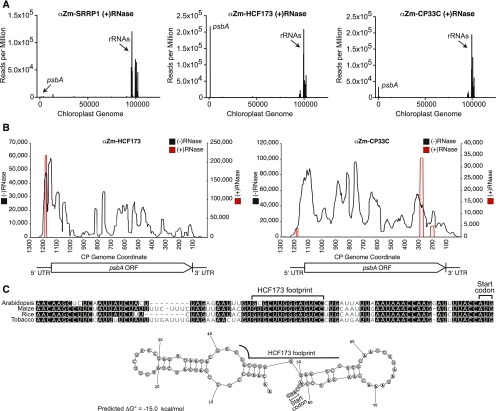
High-Resolution RIP-Seq Pinpoints Binding Sites for HCF173 and CP33C on psbA mRNA
The precise location of an RNA binding protein on RNA can be informative with regard to its functions and mechanisms. With that in mind, we modified the RIP-seq protocol by adding a ribonuclease I digestion step prior to antibody addition, aiming to limit the span of RNA that coimmunoprecipitates due to proximity to the binding site. The RNase treatment all but eliminated psbA RNA from the SRRP1 coimmunoprecipitation (Figure 6A), suggesting that the Zm-SRRP1 binding site is too short or of too low an affinity to be recovered with this method. However, psbA sequences remained highly enriched in the Zm-HCF173 and Zm-CP33C coimmunoprecipitates (Figure 6A). The apparent increase in rRNA abundance in the +RNase immunoprecipitates likely reflects a decrease in the balance of true RNA ligand (psbA RNA) to contaminants (rRNA), as we aimed to obtain a similar number sequence reads in all experiments. A higher resolution view of these data showed that both antibodies immunoprecipitated well-defined, specific fragments of psbA RNA (Figure 6B). The sequences that coimmunoprecipitated with CP33C mapped toward the end of the open reading frame (Figure 6B; Supplemental Figure 4B). The sequence that coimmunoprecipitated with HCF173 mapped in the 5ʹ UTR (Figure 6B, red) and coincided with a conserved sequence patch (Figure 6C). We refer to this as the HCF173 footprint, but we recognize that HCF173’s interaction with this sequence could be mediated by another protein. Interestingly, this sequence spans a junction between two predicted conserved RNA hairpins (Figure 6C). The downstream hairpin intrudes on the footprint of the initiating ribosome (Chotewutmontri and Barkan, 2016) and is therefore predicted to inhibit translation initiation (Scharff et al., 2011). Association of HCF173 with the site defined here would likely prevent formation of this hairpin, providing a plausible mechanism for HCF173’s translation activation function.
Figure 6.
High-Resolution RIP-Seq Analysis of HCF173 and CP33C Pinpointing Interaction Sites in the psbA mRNA.
Experiments were performed as in Figure 5 except that the stromal extract was briefly treated with RNase I prior to antibody addition.
(A) Average sequence coverage in consecutive 10-nucleotide windows along the chloroplast genome, per million reads mapped to the chloroplast genome (NCBI NC_001666).
(B) Sequence coverage along the psbA mRNA, with [red, (+) RNase] or without [black, (−) RNase] a pretreatment of stroma with RNase I. The number of reads per million mapped to the chloroplast genome (y axes) is shown according to position along the chloroplast genome (x axes). The sequences of the RNase-resistant peaks are shown in (C) (HCF173) and Supplemental Figure 4B (CP33C).
(C) Site of HCF173 interaction in the psbA 5ʹ UTR. The RNase-resistant sequence that coimmunoprecipitates with HCF173 (HCF173 footprint) is marked on a multiple sequence alignment of the psbA 5ʹ UTR from Arabidopsis, maize, tobacco (Nicotiana tabacum), and rice (Oryza sativa). A conserved secondary structure was predicted by Dynalign (Fu et al., 2014) using the maize and Arabidopsis sequences as input; the prediction for maize is shown below.
DISCUSSION
PPR proteins have attracted interest as potential scaffolds for the development of designer RNA binding proteins (Filipovska and Rackham, 2011; Yagi et al., 2014; Hall, 2016). However, the feasibility of designing PPR proteins to bind specifically to a wide variety of RNA sequences, and of predicting the landscape of RNA occupancy in vivo, remains to be established. Results presented here advance these efforts by demonstrating that artificial proteins built from consensus PPR motifs bind with high specificity to the intended RNA target in vivo, that the affinity of these interactions is sufficient for RNA-tagging applications, and that the landscape of low-level off-target binding correlates well with data from in vitro experiments.
As is true of all RNA binding proteins, the sequence specificity of PPR proteins is not absolute: the population of bound RNAs will inevitably be influenced by protein concentration, salt conditions, RNA structure, and competition from other RNAs. As expected, we observed a small degree of off-target binding by our artificial proteins. Encouragingly, however, features of these off-target interactions (Figure 3) closely resemble those from our prior in vitro bind-n-seq analyses of the same proteins (Miranda et al., 2018). For example, SCD14 was more permissive to mismatches in the binding site than was SCD11, nucleotide selectivity dropped off toward the C terminus, pyrimidine binding motifs discriminated poorly between U and C, and purine binding motifs (especially those aligning with AGA at positions 5 through 7 of the binding site) were the most highly selective. Additionally, sequences flanking the off-target sites are AU-rich (see logo in Figure 3C), implying that off-target binding in vivo is concentrated in regions of low RNA structure. This is consistent with our finding that binding of a model PPR protein in vitro is inhibited even by very weak RNA structures (McDermott et al., 2018). The fact that in vitro bind-n-seq data were highly predictive of the in vivo interaction landscapes of our artificial PPR proteins indicates that bind-n-seq analysis is a fruitful precursor to in vivo applications.
In addition, we show that the capability of aPPRs to bind predictably to RNA in vivo can be used to characterize proteomes associated with specific RNAs. The panoply of proteins that bind an RNA determine many aspects of its function and metabolism. Although excellent approaches are available for identifying RNAs bound to a protein of interest, the identification of proteins bound to particular RNAs remains problematic. Several methods for the RNA-centric purification of RNPs have been reported previously. Some of these rely on the insertion of an RNA affinity tag into the target RNA (e.g., Butter et al., 2009; Panchapakesan et al., 2017; Ramanathan et al., 2018). However, insertions can alter RNA functionalities, modification of endogenous genes is technically challenging in some experimental systems (such as organelles), and expression of ectopic modified genes can disrupt the balance of trans-factors to their cis-targets. These limitations are addressed by assays that purify untagged RNPs by coupling in vivo crosslinking with postlysis antisense oligonucleotide purification (e.g., Chu et al., 2015; McHugh et al., 2015; Rogell et al., 2017; Spiniello et al., 2018). However, UV crosslinking is inefficient and is practical only with cultured cells or lysates. Formaldehyde crosslinking provides an alternative, but it is prone to capturing both transient and stable interactions. Recently, a type VI–related CRISPR-Cas system was engineered to bind and modify the splicing of endogenous RNAs (Konermann et al., 2018). Whether this system can achieve the degree of RNA occupancy needed for use in affinity purification approaches remains to be determined.
Designer PPR affinity tags add to this toolkit by binding unmodified RNPs within intact tissues and allowing their purification under non denaturing conditions. Given the successes in using designer PUF proteins to modify the expression of specific mRNAs (Wang et al., 2009; Campbell et al., 2014) and to track untagged RNAs in vivo (Yoshimura and Ozawa, 2016), they may also be useful as affinity tags for RNP purification. However, the greater flexibility in repeat tract length with the PPR scaffold (Hall, 2016) may facilitate customization of RNA binding affinity, kinetics, and specificity for this application.
Our approach uncovered two RNA binding proteins of unknown function that associate primarily with psbA RNA in vivo. It seems likely that these proteins influence psbA expression, and elucidation of their functions will be an interesting area for future investigation. However, there are several important caveats relevant to the general applicability of our approach. First, the psbA mRNA is highly abundant, and analysis of proteomes associated with less abundant mRNAs is likely to be more challenging. Second, PPR proteins in nature function almost exclusively inside mitochondria and chloroplasts, and it is as yet unclear how they will perform in the nuclear-cytosolic compartment. Thus, an important next step is to test this approach on a cytosolic RNA target. Additionally, our finding that SCD14 and SCD11 are prone to proteolysis in vivo warrants exploration of alternative consensus designs that are more robust to the in vivo protease milieu.
METHODS
Development of Transgenic Lines
Genes for SCD14 and SCD11 were codon optimized for Arabidopsis (Arabidopsis thaliana) and assembled by PCR from several overlapping synthetic DNA fragments (IDT). The nucleotide and protein sequences are provided in Supplemental Figure 1. They were designed with the PPR nucleotide specificity codes described previously (Barkan et al., 2012; Miranda et al., 2018) and are summarized in Figure 1A. The PPR-encoding genes were inserted into a modified form of pCambia1300 harboring the Superpromoter (Lee et al., 2007) to drive transgene expression (inserted at the XbaI site of pCambia1300) and encoding a 3xFLAG tag at the C terminus of the inserted open reading frame (a gift from Jie Shen and Zhizhong Gong, China Agricultural University). The plasmids were used to transform Arabidopsis (ecotype Col-0) using the floral dip method (Zhang et al., 2006). Lines were screened by immunoblotting for aPPR expression, and those with the highest expression were used for further experiments. An additional transgenic line was developed using the MCD14 protein design we reported previously (Miranda et al., 2018); however, MCD14 transgenic lines failed to express the protein.
Plant Growth
Arabidopsis seeds were sterilized by incubation for 10 min in a solution containing 1% (v/v) bleach and 0.1% (w/v) SDS, followed by a 70% (v/v) ethanol wash. The seeds were then washed three times with sterile water. Seeds were plated and grown in tissue culture dishes containing Murashige and Skoog agar medium: 4.33 g/L Murashige and Skoog basal salt medium (Sigma-Aldrich), 2% Suc, and 0.3% Phytagel, pH 5.7. Transgenic plants were selected by the addition of 50 µg/mL hygromycin to the growth medium. Plants used for chloroplast isolation and immunoprecipitation assays were grown in a growth chamber in diurnal cycles (under 10 h of light at 120 µmol photons m−2 s−1, 14 h of dark, 22°C) for 14 d. Arabidopsis was grown using cool-white, high-output fluorescent lamps (F48T12/CW/HO, Sylvania).
Maize (Zea mays) was grown and used for the preparation of stromal extracts using methods described previously (Schmitz-Linneweber et al., 2005). Maize insertion alleles for Zm-cp33C (GRMZM2G023591) and Zm-srrp1 (GRMZM2G016084) were obtained from the UniformMu collection: the GRMZM2G023591 mutant corresponds to line (mu1032521, UFMu-02565) and the GRMZM2G016084 mutant corresponds to line (mu1076060, UFMu-09028). Positions of the insertions are diagrammed in Supplemental Figure 5.
Chloroplast Isolation and Fractionation
Maize chloroplast stroma for use in RIP-seq assays was prepared as described previously (Ostheimer et al., 2003). Arabidopsis chloroplast stroma was prepared from chloroplasts isolated from the aerial portion of 2-week-old seedlings (40 g of tissue) as described previously (Kunst, 1998), with the following modifications: seedlings were not placed in ice water before homogenization, sorbitol concentration in the homogenization buffer was reduced to 0.33 M, and plants were homogenized in a blender using three 5-s bursts. Purified chloroplasts were resuspended and lysed in hypotonic lysis buffer (30 mM HEPES-KOH, pH 8.0, 10 mM MgOAc⋅4H2O, 60 mM KOAc, 2 mM DTT, 2 µg/mL aprotinin, 2 µg/mL leupeptin, 1 µg/mL pepstatin A, and 0.8 mM phenylmethylsulfonyl fluoride), using a minimal buffer volume. Lysed chloroplasts were centrifuged for 40 min at 18,000g and 4°C in a tabletop microcentrifuge to pellet membranes and particulates. The supernatant was removed, and the pellet was resuspended in hypotonic lysis buffer and centrifuged again under the same conditions. Supernatants were combined, aliquoted, and frozen at −80°C. The thylakoid membranes (pellet fraction) were aliquoted and frozen at −80°C.
Antibodies, SDS-PAGE, and Immunoblot Analysis
SDS-PAGE and immunoblot analyses were performed as described previously (Barkan, 1998). A mouse monoclonal anti-FLAG M2 antibody was purchased from Sigma-Aldrich (F1804-1 MG, lot SLBW3851). The SRP54 antibody was a generous gift of Masato Nakai (Osaka University). Cytochrome oxidase subunit II and actin antisera were purchased from Agrisera (AS04 053A and AS13 2640, respectively). Polyclonal antibodies were raised in rabbits to recombinant fragments of the maize orthologs of HCF173, AT4G09040/CP33C, and AT3G23700/SRRP1; these correspond to maize genes GRMZM2G397247, GRMZM2G023591, and GRMZM2G016084, respectively (seehttp://cas-pogs.uoregon.edu for evidence of orthology). The amino acids used for the HCF173, S1, and RRM protein antigens and evidence for the specificity of the resulting antisera are shown in Supplemental Figure 4; Supplemental Figure 5. Maize CRP1 antibody (Fisk et al., 1999) was used as the negative control for immunoprecipitations from Arabidopsis extract because it does not interact with proteins in Arabidopsis. Antibodies used for immunoprecipitations were affinity purified against their antigen prior to use.
Coimmunoprecipitation Experiments
Immunoprecipitation for analysis of proteins by mass spectrometry was performed as described previously (Watkins et al., 2007), with minor modifications. In brief, experiments used Arabidopsis stromal extract, anti-FLAG antibodies were crosslinked to magnetic Protein A/G beads (Pierce), the beads were prewashed in communoprecipitation buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% (v/v) Nonidet P-40, and 5 µg/mL aprotinin), and the antibody-crosslinked beads were titrated to determine the amount required to deplete the aPPR from the stromal extract. Stromal extract (400 μL at 6 mg protein/mL) was supplemented with RNAsin (Promega) to a concentration of 1 unit/μL and precleared by centrifugation for 10 min at 18,000g at 4°C. The supernatant was removed to a new tube, antibody-bound beads were added, and the mixture was incubated at 4°C for 1 h while rotating. Beads were captured with a magnet (Invitrogen), and the supernatant was removed. The beads (pellets) were washed three times with communoprecipitation buffer and then twice with 50 mM ammonium bicarbonate, pH 7.5. Proteins were digested on the beads with trypsin (Promega mass spectrometry grade at 25 ng/μL in 50 mM ammonium bicarbonate, pH 7.5) overnight at 25°C while shaking. Beads were captured, and the supernatant was transferred to a new tube. This step was repeated five times to ensure the removal of all beads. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed by the University of California–Davis Proteomics Core Facility, where the data were analyzed using Scaffold2 (Proteome Software). Protein enrichment was calculated by dividing the average normalized spectral abundance factor (NSAF) values (Zhang et al., 2010) from the two aPPR lines by NSAF values from the control immunoprecipitation using extract of Col-0 plants. To avoid division by zero, a correction term of 0.001 was added to each NSAF value in the control; therefore, the actual enrichment of proteins that were not detected in the control is under-estimated. The data are shown in Supplemental Data Set 2.
Immunoprecipitations for RIP-seq analysis were performed similarly, except that antibodies were not crosslinked to the beads, the Arabidopsis experiments used 200 μL of extract, and the maize experiments used 70 μL of stromal extract at ∼10 mg protein/mL and did not include RNAsin. Antibody to maize CRP1 was used as a negative control for the aPPR RIP-seq assays; this antibody does not recognize proteins in Arabidopsis chloroplasts. The aPPR RIP-seq experiments were performed two times, using the same stromal extracts. Antibody to AtpB (subunit of the chloroplast ATP synthase) was used as the negative control for the RIP-seq assays using maize stroma. The HCF173, CP33C, and SRRP1 RIP-seq experiments were each performed twice, using antibodies from different rabbits. For high-resolution RIP-seq, stromal extract was pretreated with 1 U/μL RNase I (Ambion) at 25°C for 10 min. The sample was placed on ice and the remainder of the procedure was as described for standard RIP-seq.
Analysis of Coimmunoprecipitated RNA by RIP-Seq and Slot-Blot Hybridization
An equal volume of TESS buffer (10 mM Tris, pH 7.5, 1 mM EDTA, 150 mM NaCl, and 0.2% (w/v) SDS) supplemented with Proteinase K (0.2 μg/µL) was added to the supernatant and pellet fractions and incubated for 30 min at 37°C. RNA was then purified by phenol-chloroform extraction and ethanol precipitation, resuspended in water, and quantified by Qubit. The RNA was used directly for slot-blot hybridizations as described previously by Schmitz-Linneweber et al. (2005), or phosphorylated (T4 polynucleotide kinase; New England Biolabs) and processed for sequencing using the NEXTflex Small RNA-Seq Kit 3 (catalog no. NOVA-5132-06; BIOO Scientific). For Arabidopsis, 50 ng of pellet RNA was used as the input for sequencing libraries. The maize experiments used 20 ng of pellet RNA for library preparation and included an RNA fragmentation step: RNA was fragmented by incubation at 95°C for 4 min in 40 mM Tris-acetate, pH 8, 100 mM KOAc, and 30 mM Mg(OAc)2. The reaction was stopped by the addition of EDTA to a final concentration of 50 mM, the RNA was ethanol precipitated in the presence of 1.5 µg of GlycoBlue (Thermo Fisher Scientific), and phosphorylated (T4 polynucleotide kinase; New England Biolabs) prior to library preparation. RNase I-RIP-seq experiments did not include an RNA fragmentation step. Libraries were gel purified to enrich for inserts between 15 and 100 nucleotides. Libraries were sequenced at the University of Oregon Genomics and Cell Characterization Core Facility, with read lengths of 75 or 100 nucleotides. Sequencing data were processed as described in Chotewutmontri and Barkan (2018) except that all read lengths were included and reads were aligned only to the chloroplast genome. Read counts for RIP-seq experiments are summarized in Supplemental Data Set 1. Enrichment values were calculated as the normalized abundance of reads mapping to each gene (including UTRs) in an experimental sample relative to the control.
Accession Numbers
The gene identification numbers for genes mentioned in this study are as follows: HCF173, AT1G16720; Zm-HCF173, Zm00001d014716_T002 (B73 v4) or GRMZM2G397247_T03 (B73 v3); SRRP1, AT3G23700; Zm-SRRP1, GRMZM2G016084_T02 (B73 v3) or Zm00001d034828 (B73 v4); CP33C, AT4G09040; Zm-CP33C, GRMZM2G023591_T01 (B73 v4) or Zm00001d031258_T005 (Bs3 v4).
Supplemental Data
Supplemental Figure 1. Sequences of SCD14 and SCD11
Supplemental Figure 2. Replicate RIP-seq data for SCD14, SCD11, HCF173, SRRP1, and CP33C
Supplemental Figure 3. High-resolution views of RNA enrichment along genes listed in Figure 3A.
Supplemental Figure 4. Maize CP33C and SRRP1 antigens and CP33C RNA footprint.
Supplemental Figure 5. Additional information to support HCF173, CP33C, and SRRP1 RIP-seq.
Supplemental Data Set 1. Read counts and RPKM values for RIP-seq experiments.
Supplemental Data Set 2. Proteins found in SCD11 and SCD14 coimmunoprecipitates as detected by LC-MS/MS.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Jie Shen (Chinese Academy of Sciences) and Zhizhong Gong (China Agricultural University) for advice and for their gift of the pCAMBIA1300 vector modified to encode a FLAG tag. We also thank Masato Nakai (Osaka University) for the gift of cpSRP54 antibody; Carolyn Brewster, Margarita Rojas, and Susan Belcher (University of Oregon) for technical assistance; the UniformMu project (University of Florida) for maize insertion lines; and the University of California–Davis Proteomics Core for LC-MS/MS proteomic analyses. This work was supported by National Science Foundation (grant MCB-1616016 to A.B.) and National Institutes of Health Training Grant (T32-GM007759 to J.J.M.).
AUTHOR CONTRIBUTIONS
A.B. and J.J.M. conceived the project and designed the experiments. J.J.M., R.W.-C., and K.P.W. performed the experiments. All authors analyzed the data. A.B. and J.J.M. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Barkan A. (1998). Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 297: 38–57. [Google Scholar]
- Barkan A., Small I. (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65: 415–442. [DOI] [PubMed] [Google Scholar]
- Barkan A., Rojas M., Fujii S., Yap A., Chong Y.S., Bond C.S., Small I. (2012). A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8: e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat V.D., McCann K.L., Wang Y., Fonseca D.R., Shukla T., Alexander J.C., Qiu C., Wickens M., Lo T.W., Tanaka Hall T.M., Campbell Z.T. (2019). Engineering a conserved RNA regulatory protein repurposes its biological function in vivo. eLife 8: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butter F., Scheibe M., Mörl M., Mann M. (2009). Unbiased RNA-protein interaction screen by quantitative proteomics. Proc. Natl. Acad. Sci. USA 106: 10626–10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell Z.T., Valley C.T., Wickens M. (2014). A protein-RNA specificity code enables targeted activation of an endogenous human transcript. Nat. Struct. Mol. Biol. 21: 732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Varani G. (2013). Engineering RNA-binding proteins for biology. FEBS J. 280: 3734–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P., Barkan A. (2016). Dynamics of chloroplast translation during chloroplast differentiation in maize. PLoS Genet. 12: e1006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P., Barkan A. (2018). Multilevel effects of light on ribosome dynamics in chloroplasts program genome-wide and psbA-specific changes in translation. PLoS Genet. 14: e1007555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C., Zhang Q.C., da Rocha S.T., Flynn R.A., Bharadwaj M., Calabrese J.M., Magnuson T., Heard E., Chang H.Y. (2015). Systematic discovery of Xist RNA binding proteins. Cell 161: 404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas des Francs-Small C., Vincis Pereira Sanglard L., Small I. (2018). Targeted cleavage of nad6 mRNA induced by a modified pentatricopeptide repeat protein in plant mitochondria. Commun. Biol 1: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquille S., Filipovska A., Chia T., Rajappa L., Lingford J.P., Razif M.F., Thore S., Rackham O. (2014). An artificial PPR scaffold for programmable RNA recognition. Nat. Commun. 5: 5729. [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. (2004). WebLogo: A sequence logo generator. Genome Res. 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovska A., Rackham O. (2011). Designer RNA-binding proteins: New tools for manipulating the transcriptome. RNA Biol. 8: 978–983. [DOI] [PubMed] [Google Scholar]
- Fisk D.G., Walker M.B., Barkan A. (1999). Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 18: 2621–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Sharma G., Mathews D.H. (2014). Dynalign II: Common secondary structure prediction for RNA homologs with domain insertions. Nucleic Acids Res. 42: 13939–13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S., Sato N., Shikanai T. (2013). Mutagenesis of individual pentatricopeptide repeat motifs affects RNA binding activity and reveals functional partitioning of Arabidopsis PROTON gradient regulation3. Plant Cell 25: 3079–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm A.C., Hall T.M.T., McKenney K.M. (2018). Post-transcriptional regulatory functions of mammalian Pumilio proteins. Trends Genet. 34: 972–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L., Jung H.J., Kim B.M., Xu T., Lee K., Kim Y.O., Kang H. (2015). A chloroplast-localized S1 domain-containing protein SRRP1 plays a role in Arabidopsis seedling growth in the presence of ABA. J. Plant Physiol. 189: 34–41. [DOI] [PubMed] [Google Scholar]
- Hall T.M. (2016). De-coding and re-coding RNA recognition by PUF and PPR repeat proteins. Curr. Opin. Struct. Biol. 36: 116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helder S., Blythe A.J., Bond C.S., Mackay J.P. (2016). Determinants of affinity and specificity in RNA-binding proteins. Curr. Opin. Struct. Biol. 38: 83–91. [DOI] [PubMed] [Google Scholar]
- Jin H., Fu M., Duan Z., Duan S., Li M., Dong X., Liu B., Feng D., Wang J., Peng L., Wang H.B. (2018). LOW PHOTOSYNTHETIC EFFICIENCY 1 is required for light-regulated photosystem II biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 115: E6075–E6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P., Yap A., Bond C.S., Small I. (2015). Predictable alteration of sequence recognition by RNA editing factors from Arabidopsis. Plant Cell 27: 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S., Lotfy P., Brideau N.J., Oki J., Shokhirev M.N., Hsu P.D. (2018). Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173: 665–676.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L. (1998). Preparation of physiologically active chloroplasts from Arabidopsis. Methods Mol. Biol. 82: 43–48. [DOI] [PubMed] [Google Scholar]
- Lee L.Y., Kononov M.E., Bassuner B., Frame B.R., Wang K., Gelvin S.B. (2007). Novel plant transformation vectors containing the superpromoter. Plant Physiol. 145: 1294–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott J.J., Civic B., Barkan A. (2018). Effects of RNA structure and salt concentration on the affinity and kinetics of interactions between pentatricopeptide repeat proteins and their RNA ligands. PLoS One 13: e0209713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh C.A., et al. (2015). The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521: 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda R.G., Rojas M., Montgomery M.P., Gribbin K.P., Barkan A. (2017). RNA-binding specificity landscape of the pentatricopeptide repeat protein PPR10. RNA 23: 586–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda R.G., McDermott J.J., Barkan A. (2018). RNA-binding specificity landscapes of designer pentatricopeptide repeat proteins elucidate principles of PPR-RNA interactions. Nucleic Acids Res. 46: 2613–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R., van Wijk K.J. (2002). Transient interaction of cpSRP54 with elongating nascent chains of the chloroplast-encoded D1 protein; ‘cpSRP54 caught in the act’. FEBS Lett. 524: 127–133. [DOI] [PubMed] [Google Scholar]
- Okuda K., Shoki H., Arai M., Shikanai T., Small I., Nakamura T. (2014). Quantitative analysis of motifs contributing to the interaction between PLS-subfamily members and their target RNA sequences in plastid RNA editing. Plant J. 80: 870–882. [DOI] [PubMed] [Google Scholar]
- Ostheimer G.J., Williams-Carrier R., Belcher S., Osborne E., Gierke J., Barkan A. (2003). Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J. 22: 3919–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchapakesan S.S.S., Ferguson M.L., Hayden E.J., Chen X., Hoskins A.A., Unrau P.J. (2017). Ribonucleoprotein purification and characterization using RNA Mango. RNA 23: 1592–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J., Bayraktar O.A., Prikryl J., Barkan A. (2009). Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 28: 2042–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan M., et al. (2018). RNA-protein interaction detection in living cells. Nat. Methods 15: 207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogell B., Fischer B., Rettel M., Krijgsveld J., Castello A., Hentze M.W. (2017). Specific RNP capture with antisense LNA/DNA mixmers. RNA 23: 1290–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas M., Ruwe H., Miranda R.G., Zoschke R., Hase N., Schmitz-Linneweber C., Barkan A. (2018). Unexpected functional versatility of the pentatricopeptide repeat proteins PGR3, PPR5 and PPR10. Nucleic Acids Res. 46: 10448–10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas M., Yu Q., Williams-Carrier R., Maliga P., Barkan A. (2019). Engineered PPR proteins as inducible switches to activate the expression of chloroplast transgenes. Nat. Plants 5: 505–511. [DOI] [PubMed] [Google Scholar]
- Ruwe H., Kupsch C., Teubner M., Schmitz-Linneweber C. (2011). The RNA-recognition motif in chloroplasts. J. Plant Physiol. 168: 1361–1371. [DOI] [PubMed] [Google Scholar]
- Scharff L.B., Childs L., Walther D., Bock R. (2011). Local absence of secondary structure permits translation of mRNAs that lack ribosome-binding sites. PLoS Genet. 7: e1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Williams-Carrier R., Barkan A. (2005). RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell 17: 2791–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schult K., Meierhoff K., Paradies S., Töller T., Wolff P., Westhoff P. (2007). The nuclear-encoded factor HCF173 is involved in the initiation of translation of the psbA mRNA in Arabidopsis thaliana. Plant Cell 19: 1329–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Wang X., Liu Y., Li Q., Yang Z., Yan N., Zou T., Yin P. (2015). Specific RNA recognition by designer pentatricopeptide repeat protein. Mol. Plant 8: 667–670. [DOI] [PubMed] [Google Scholar]
- Shen C., Zhang D., Guan Z., Liu Y., Yang Z., Yang Y., Wang X., Wang Q., Zhang Q., Fan S., Zou T., Yin P. (2016). Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat. Commun. 7: 11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiniello M., Knoener R.A., Steinbrink M.I., Yang B., Cesnik A.J., Buxton K.E., Scalf M., Jarrard D.F., Smith L.M. (2018). HyPR-MS for multiplexed discovery of MALAT1, NEAT1, and NORAD lncRNA protein interactomes. J. Proteome Res. 17: 3022–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Zerges W. (2015). Translational regulation in chloroplasts for development and homeostasis. Biochim. Biophys. Acta 1847: 809–820. [DOI] [PubMed] [Google Scholar]
- Wang M., Ogé L., Perez-Garcia M.D., Hamama L., Sakr S. (2018). The PUF protein family: Overview on PUF RNA targets, biological functions, and post transcriptional regulation. Int. J. Mol. Sci. 19: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cheong C.G., Hall T.M., Wang Z. (2009). Engineering splicing factors with designed specificities. Nat. Methods 6: 825–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins K.P., Kroeger T.S., Cooke A.M., Williams-Carrier R.E., Friso G., Belcher S.E., van Wijk K.J., Barkan A. (2007). A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell 19: 2606–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Carrier R., Brewster C., Belcher S.E., Rojas M., Chotewutmontri P., Ljungdahl S., Barkan A. (2019). The Arabidopsis pentatricopeptide repeat protein LPE1 and its maize ortholog are required for translation of the chloroplast psbJ RNA. Plant J. (in press). [DOI] [PubMed] [Google Scholar]
- Yagi Y., Nakamura T., Small I. (2014). The potential for manipulating RNA with pentatricopeptide repeat proteins. Plant J. 78: 772–782. [DOI] [PubMed] [Google Scholar]
- Yan J., Yao Y., Hong S., Yang Y., Shen C., Zhang Q., Zhang D., Zou T., Yin P. (2019). Delineation of pentatricopeptide repeat codes for target RNA prediction. Nucleic Acids Res. 47: 3728–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P., et al. (2013). Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504: 168–171. [DOI] [PubMed] [Google Scholar]
- Yoshimura H., Ozawa T. (2016). Monitoring of RNA dynamics in living cells using PUM-HD and fluorescent protein reconstitution technique. Methods Enzymol. 572: 65–85. [DOI] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.S., Niu Q.W., Chua N.H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wen Z., Washburn M.P., Florens L. (2010). Refinements to label free proteome quantitation: How to deal with peptides shared by multiple proteins. Anal. Chem. 82: 2272–2281. [DOI] [PubMed] [Google Scholar]
- Zhao Y.Y., Mao M.W., Zhang W.J., Wang J., Li H.T., Yang Y., Wang Z., Wu J.W. (2018). Expanding RNA binding specificity and affinity of engineered PUF domains. Nucleic Acids Res. 46: 4771–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]