Abstract
A colorimetric nucleic acid based test for label-free pathogen detection has been developed and used for the detection of the Zika virus. The test relies on nucleic acid sequence-based amplification (NASBA) of a viral RNA followed by interrogation of the amplicon by a cascade of deoxyribozymes constituting a visual split deoxyribozyme (vsDz) probe. The probe consists of a split phosphodiesterase deoxyribozyme, which forms its catalytic core upon binding to a specific amplicon fragment. The catalytically active complex recognizes and cleaves an inhibited peroxidase-like deoxyribozyme (PDz), thereby activating it. Active PDz catalyzes hydrogen peroxide-mediated oxidation of a colorless substrate into a colored product, thereby generating a visible signal. Viral RNA (106 copies/mL or higher) triggers intense color within 2 hr. The test selectively differentiates between Zika and closely related dengue and West Nile viruses. The reported technology combines isothermal amplification and visual detection and therefore represents a basis for the future development of a cost-efficient and instrument-free method for point-of-care nucleic acid analysis.
Keywords: Pathogen detection, isothermal amplification, deoxyribozyme cascade, G-quadruplex peroxidase, colorimetric test, NASBA
1. Introduction
Analytical tests with color change as a readout offer great convenience due to their ability to be read without instrumentation [1]. Among the most well-known colorimetric tests are pH strips, the Bradford assay for quantitative protein analysis, and the home pregnancy test. The Enzyme-Linked ImmunoSorbent Assay (ELISA) provides visual detection of human pathogens [2]. However, ELISA suffers from cross-reactivity, often provides only post-infection information when the pathogen is no longer present, and involves multistage protein-based manipulations requiring a skilled technician [2–5]. On the other hand, analysis of pathogenic nucleic acids allows for the detection of a current infection with ultimate sensitivity down to one molecule and the ability to resolve point mutations [6]. Conventional PCR-based nucleic acid amplification tests (NAATs), known also as molecular diagnostics, require a trained personal and sophisticated instrumentation [6]. As a result, both ELISA and molecular diagnostic tests are performed only in specialized diagnostic laboratories. Even though a number of nucleic acid-based tests for pathogen detection with a colorimetric output have been reported [7–16], molecular tests for “laboratory-free” diagnostics of bacterial or viral infections are not currently used in standard practice.
Unlike antibody-based approaches (e.g. ELISA), molecular diagnostic tests require the following stages: (i) isolation of a pathogenic DNA or RNA; (ii) nucleic acid amplification; and (iii) interrogation with a probe resulting in a signal output generation. In order to aim for point-of-care (POC) pathogen detection in resource-limited settings, the test should satisfy the ASSURED criteria of the World Health Organization: it should be Affordable, Sensitive, Specific, User-friendly, Rapid, Equipment-free, and Deliverable to those in need [17]. More general definitions stress a fast turnaround time from the sample collection to the results being available for a clinical decision to be made [18]. Therefore, all three stages should require minimal training for a person to perform the test, be rapid enough to complete within a typical doctor’s visit, and not rely on sophisticated or expensive equipment.
In this study, we developed a label-free test for colorimetric or visual pathogen detection. The test relies on isothermal amplification of a pathogen RNA combined with a deoxyribozyme cascade with color change as a readout. The test was designed for the detection of the Zika virus (ZIKV) as a model pathogen, which recently caused the outbreak in the Americas with severe complications [19]. Infection with ZIKV is predominantly asymptomatic, and any symptoms otherwise inferred are indistinguishable from other mosquito-borne infections. Therefore, reliable POC detection methods for the ZIKV are beneficial to ensure timely virus transmission surveillance and control over infected mosquito population.
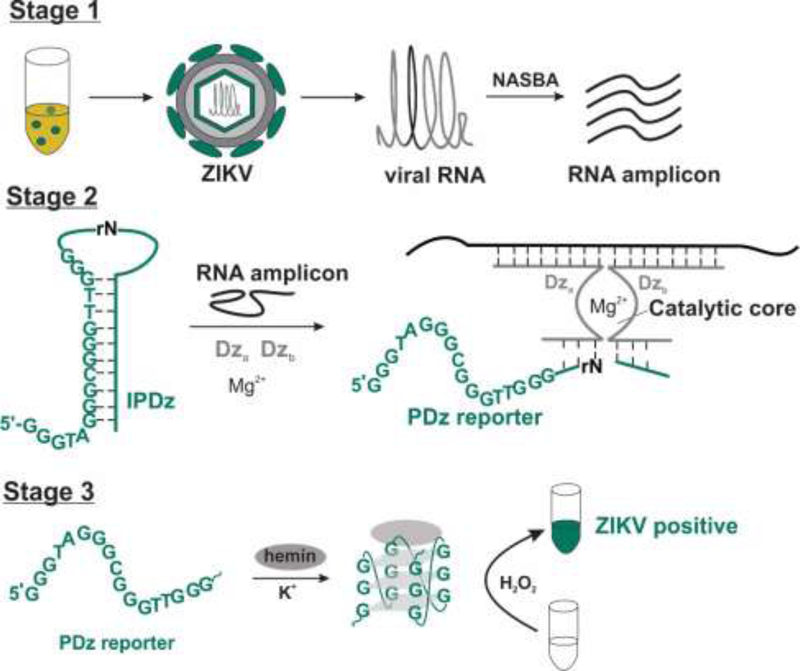
The test involves three stages (Scheme 1). First, a fragment of viral genome is amplified using the Nucleic Acid Sequence-Based Amplification (NASBA) technique, which has been suggested previously for amplification of bacterial and viral RNA [9,20–28]. This technique utilizes three enzymes – AMV reverse transcriptase, RNase H, and T7 RNA polymerase – to produce multiple copies of a single stranded RNA amplicon using viral RNA as a template (Scheme S1). RNA product being less stable than DNA offers an advantage of less incidence of sample cross-contamination when multiple samples are analyzed, which would result in less false-positive results. The NASBA amplicon then serves as a target in stage 2 to activate the first deoxyribozyme (Dz) of the cascade and cleave an RNA phosphodiester bond in the inhibited peroxidase-like deoxyribozyme (IPDz) to release a G-rich sequence (PDz reporter in Scheme 1). PDz reporter then folds into a G-quadruplex (G4) structure [29] and bind a hemin cofactor to form the second Dz - a catalytically active PDz-hemin complex, which catalyzes H2O2-mediated oxidation of a colorless peroxidase substrate, 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS), into a colored peroxidation product. Overall, after three stages, ZIKV RNA of 10 pg/mL (1.6×106 copies/mL) triggers intense green color with no cross-talk from closely related viruses. The test requires only a hot plate, a pipette, and reagents, allowing it to be performed by a user with minimal training.
Scheme 1.
Stages of the NASBA/vsDz test. Stage 1- RNA amplification; Stage 2 – recognition and activation of PDz; Stage 3 – color generation. Stages 2 and 3 show the components of the vsDz probe.
2. Material and Methods
2.1. Materials
All oligonucleotides were from Integrated DNA Technologies, Inc. (Coralville, IA). 2,2′-Azino-bis(3ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS), hemin, triton-X100 and HEPES were purchased from Sigma-Aldrich (St. Louis, MO). DMSO was from Fisher Scientific (Hampton, NH). Hydrogen peroxide solution (3%) was from VWR (Radnor, PA). Non-DEPC treated DNase/RNase-free water was from Boston Bio Products (Ashland, MA). GelRed® nucleic acid gel stain was from Biotium (Fremont, CA). All other reagents were of analytical grade.
2.2. Virus propagation and RNA isolation
The viruses listed in Table S1 were grown in Vero cells in DMEM media containing 10% fetal bovine serum. All ZIKV and DENV strains were obtained from World Reference Center for Emerging Viruses and Arboviruses. West Nile virus (NY99 strain) was kindly provided by Dr. Tesh at the University of Texas Medical Branch. Vero cells were obtained from ATCC, a large number of aliquots from early passages were frozen, and a new aliquot was thawed every two months. Only the cells within 4 passages are used to propagate viruses. Viral RNA was extracted using Trizol LS (Life Technologies) and quantified via absorbance at 260 nm using a NanoDrop. Additionally, to quantify viral RNA and authenticate viral identity, qPCR was performed using virus NS3-specific primers and a probe for ZIKV and WNV (Table S2), or the DENV qPCR kit (PCRmax). For this purpose, complementary DNA (cDNA) was synthesized from RNA, using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and random hexamers.
2.3. NASBA reaction
The amplification reaction was carried out using a NASBA liquid kit (Life Sciences Advanced Technologies, Inc.) according to the manufacturer’s protocol with a final volume of 15 μL. NASBA products were analyzed in 2% agarose gel and quantified using Qubit™ RNA HS assay. To convert the concentration into molar units, an average molecular weight of 340 g/mol for ribonucleotides was utilized to estimate the weight of the 147-nt long RNA sequence, yielding a molecular weight of 50,000 g/mol. A typical concentration of the amplified RNA obtained via the NASBA reaction with 50 pg/mL ZIKV RNA was calculated to be 6–10 μM.
2.4. vsDz probe reaction
Samples (30 μL) containing IPDz (1.0 µM), Dza (100 nM), Dzb, (100 nM) in the colorimetric buffer (50 mM HEPES-NaOH, pH 7.4, 50 mM MgCl2, 120 mM NaCl, 20 mM KCl, 0.03% Triton X-100, 1% DMSO) were mixed with either synthetic analyte T64 (0–100 nM, Table S2) or NASBA product (1.7–3.3%), incubated at 50 °C for 15–60 min, and briefly cooled to room temperature. Then, hemin (375 nM), ABTS (1 mM) and H2O2 (1 mM) were added to the samples. The stock solutions of hemin and ABTS were prepared daily in DMSO, and H2O2 was diluted daily with water. After 5–15 min incubation of the samples at room temperature, their absorbance at 420 nm was measured, and the tube images were captured using a smartphone camera. The absorbance data was measured with a NanoDrop spectrometer (Thermo Fisher) and processed in Microsoft Excel.
3. Results
3.1. Design of NASBA/vsDz test
As a target for the NASBA/vsDz test, a fragment of the envelope gene of ZIKV genomic RNA was selected based on the alignment data for genomes of ZIKV isolates circulating in the Americas. The fragment corresponded to nts 2622–2760 of ZIKV strain PRVABC-59 genome. The primers for NASBA reaction were designed according to previously reported guidelines [30]. The T7 promoter sequence was placed at the 5’-end of the forward primer to ensure the sequence of the amplified RNA product be the same as in the viral genome. The primer sequences are listed in Table S2. As a template for NASBA, RNA isolated from several strains of ZIKV Asian lineage were used (Table S1). In addition, MR766 strain of African lineage was tested. All ZIKV isolated were successfully amplified using the designed set of primers, as can be seen from gel electrophoresis analysis (Figure S1).
The amplified fragment of ZIKV genome was then tested with a vsDz probe. The probe represents a cascade of two Dz – a split phosphodiesterase based on the 10–23 Dz [31] and a peroxidase-like Dz (PDz) [29]. The split phosphodiesterase consisted of two Dz subunits – strands Dza and Dzb, each of which had a middle fragment comprising a half of 10–23 Dz catalytic core. In the absence of a complementary RNA analyte, the two halves were inactive. However, when both Dza and Dzb strands hybridized to the adjacent fragments of the amplified fragment of ZIKV RNA, the catalytic core of the 10–23 Dz was re-formed and capable of catalyzing the hydrolysis of a phosphodiester bond of a single ribonucleotide embedded in the deoxyribonucleotide sequence of IPDz substrate (Scheme 1, Stage 2). The 5’-terminal fragment IPDz substrate contained a G-rich PDz sequence sequestered in a stem-loop structure to render it catalytically inactive (Scheme 1, Stage 2, left). IPDz cleavage released PDz to enable its folding into a G4 structure that, in complex with a hemin cofactor, catalyzed the peroxidation of a colorless organic indicator (e.g. ABTS) into a colored product resulting in visual output of the test in response to the pathogen’s presence (Scheme 1, Stage 3).
3.2. Optimization of NASBA/vsDz test stages
3.2.1. NASBA reaction (Stage 1)
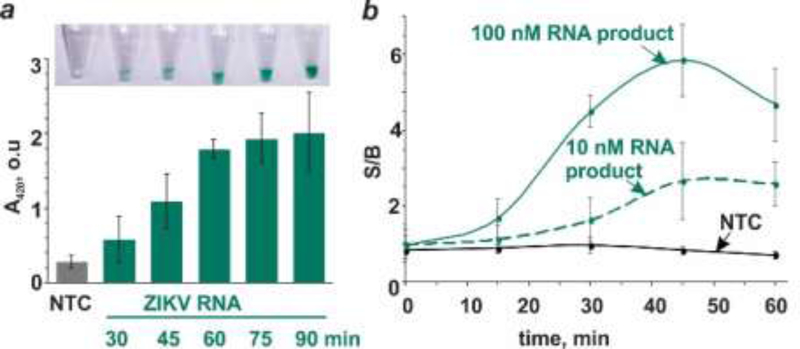
We optimized the duration of the NASBA reaction. Samples containing 50 pg/mL (8×106 copies/mL) ZIKV RNA were amplified for variable times, and then the amplified product was interrogated with a specific vsDz probe (Figure 1a). The color change was apparent with the RNA amplicon obtained by 30 min NASBA reaction (Figure 1a, inset), with greater color intensity than the no-target control (NTC) containing no RNA in the NASBA reaction. After 60 min of NASBA reaction, the color intensity was not significantly affected by longer amplification (Figure 1a). Adding more NASBA sample (3.3–10%) after shorter (30 or 45 min) amplification reaction did not result in high enough signal to reliably differentiate the presence of ZIKV RNA from its absence, especially by the naked eye (Figure S2). Therefore, for the subsequent experiments, stage 1 of the test was performed for 60 min.
Figure 1.
Time-dependence of NASBA/vsDz test stages. (a) Optimization of NASBA reaction time. NASBA reaction was allowed for the indicated time followed by the interrogation of the amplified RNA by the vsDz sensor. No-target control (NTC) containing no ZIKV RNA was amplified for 90 min and used as a control in the vsDz reaction. (b) Kinetics of IPDz cleavage. The RNA amplification product at either 10 or 100 nM concentration (estimated based on Qubit RNA HS assay) was incubated with IPDZ-11/2, Dza and Dzb for 0, 15, 30, 45 or 60 min. As a control, the NTC sample from panel (a) was used. The samples’ absorbance at 420 nm was normalized by the background absorbance of the blank samples containing only vsDz but not the RNA product or NTC. Signal-to-background ratios (S/B) were plotted over incubation time. The data are average values of three independent experiments with standard deviations as error bars.
3.2.2. ZIKV-dependent release of the signal reporter (Stage 2)
The product of NASBA is a single-stranded RNA, folded into a stable secondary structure (Figure S3A). In this study, we demonstrated that NASBA product can be used as a target for the vsDz interrogation without prior purification or annealing steps. In order to unwind the RNA secondary structure, we applied our original strategy that utilized a specially designed split deoxyribozyme probe [32–38]. In this design, two strands of the probe independently or semi-independently interrogate the abutting positions of the targeted RNA fragment (Scheme 1, stage 2, right). The catalytically active construct is formed only if both strands are simultaneously bound to the RNA analyte, i.e. if the analyte is fully complementary to the analyte binding arms of Dza and Dzb. One strand of the probe is designed to be longer and complementary to one of the single-stranded loop portions of the RNA analyte to enable efficient binding of the strand to the analyte and unwinding its secondary structure to assist binding of the second strand of the probe. We have tested four different designs for the vsDz probe targeting different fragments within the 147-nt product of NASBA reaction (Figure S3). Three vsDz probes demonstrated satisfactory performance (Figure S3b), and design 1 consisting of strands Dza and Dzb (Table S2) was selected for subsequent experiments.
Another important component of the vsDz probe is the stem-loop folded IPDz substrate (Scheme 1, stage 2, left), which contains the sequence of the color-generating signal reporter PDz, released upon IPDz cleavage. To ensure low background, the stem-loop structure of the intact IPDz should be stable enough at room temperature to prevent folding of its PDz fragment into a catalytically active G-quadruplex structure. At the same time, if the stem is too long, the two fragments of the cleaved IPDz can form a stable residual hybrid with each other, thus disfavoring the G4-peroxidase formation and inhibiting color generation. A too stable stem of IPDz could also interfere with its binding to Dza and Dzb, which would decrease the amount of PDz released and, correspondingly, the color intensity of the solution. Therefore, we optimized the stability of IPDz stem to ensure the highest turn-on ratio of vsDz sensor in the presence of its complementary target (Figure S4a). We tested six different IPDz designs, five of which folded into a hairpin with two wobble base-pairs and 8–12 Watson-Crick base-pairs. Another design (IPDZ-10/1) relied on different pattern of PDz sequestering. It was found that intact IPDz-8/2 and IPDz-9/2 are still capable of triggering the color change disregarding the presence of the target, with the color intensity being similar for IPDz alone or when Dza Dzb and the target were added (Figure S4, panels b and c). For IPDz-10/2 and IPDz-10/1, the target-induced color was more intense than in the absence of the target, but the background (IPDz only) was still too high due to the “leakage” of the stem. The best signal-to-background ratio was observed for IPDz-11/2 and IPDz-12/2. The former demonstrated lower background and, therefore, was selected for the NASBA/vsDz test.
Aiming at minimizing the time for the test, we studied the kinetics of IPDz cleavage (stage 2 of the test). The product of the NASBA reaction (10 nM or 100 nM, as estimated with Qubit RNA HS assay) was incubated in the presence of the vsDz probe for different time points followed by ABTS peroxidation reaction (Figure 1b). As a control, the NASBA NTC sample was used. The samples’ absorbance was normalized by the background absorbance (vsDz probe in the absence of the NASBA product). It can be seen that at high concentrations of the RNA analyte, the optimal incubation time is 45 min, since prolonged incubation resulted in a plateau of the signal and, at the same time, increase in the background, which decreased S/B over time (Figure 1b). At lower analyte concentrations, S/B plateaued after 45 min incubation as well. Therefore, stage 2 of the test was set to be 45 min.
3.2.3. Color generation (Stage 3)
This stage depends on the catalytic activity of a G4-peroxidase. The structure of a peroxidase substrate used as an indicator in the test will pre-determine the color, its intensity, as well as the duration of the color generation stage of the NASBA/vsDz test. To determine a suitable indicator for the test, we compared the PDz/hemin-catalyzed oxidation reaction of four peroxidase substrates – 3,3′-diaminobenzidine (DAB), 3,3′,5,5′-tetramethylbenzidine (TMB), 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), o-phenylenediamine (OPD) (Figure S5a) – in terms of color, time for color generation, and signal-to-background ratio. For both DAB and OPD, the rate of peroxidation reaction was relatively slow, with visible color change observed only 30–60 min after H2O2 addition (Figure S5, panels b and c). In addition, high background and low signal-to-background ratio (signal for PDz-containing versus PDz-free sample) was observed for OPD oxidation. Peroxidation of TMB in the presence of PDz rapidly generated royal blue reaction product, but the background was still too high (Figure S5d). Moreover, even at lower (100 nM) hemin concentration than for all other indicators an unstable product of TMB oxidation underwent further reaction to form a dark blue precipitate after 20 min, which made absorbance monitoring impossible (data not shown).
It should be noted that the blue-green color of the product of ABTS peroxidation, ABTS+•, can be observed within minutes upon addition of H2O2 to the sample containing ABTS, hemin and PDz (Figure S5e). Maximum absorbance at 420 nm was observed within 15–20 min, with about 9-to-10-fold absorbance increase over the background. Even though the color intensity gradually declined after 40 min due to low stability of ABTS+•, it was still visible for at least four hours (Figure S5e, inset).
3.3. Selectivity of NASBA/vsDz test
Molecular diagnostic tests, while powerful, can experience complications due to cross-talk with nonspecific, closely-related pathogens. For example, molecular diagnostics of ZIKV can cause false positive results in case of infection with Chikungunya virus and other flaviviruses, such as dengue virus (DENV) or West Nile virus (WNV), which are transmitted by the same mosquito species and have similar symptoms as ZIKV infection. Indeed, the homology between genomes of ZIKV and DENV is estimated to be 60–70% [39], which makes it highly probable for a ZIKV-specific probe to generate positive signal in response to DENV RNA, for example. At the same time, like all RNA-containing viruses, ZIKV genome is characterized by high mutation rate [40], which can give false negative results if a patient is infected with a viral isolate containing point-mutations in its genome when compared with the sequence of the original strain or isolate used for the probe design. For the diagnostic test to have low rate of false negatives and false positives, the signal should be observed in the presence of ideally all or at least most prevalent viral isolates, and, at the same time, not be triggered by the closely related viruses.
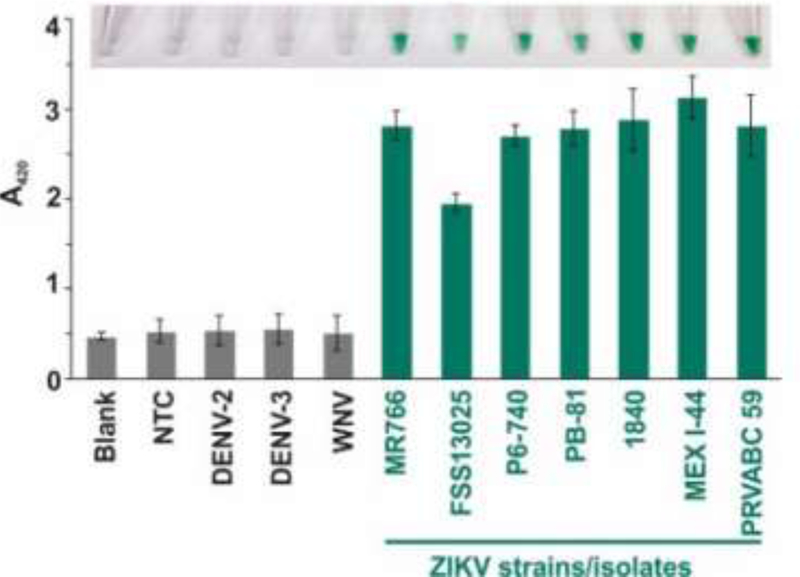
To test the selectivity of NASBA/vsDz test, we used RNA obtained from several isolates of ZIKV, as well as WNV and two subtypes of DENV (Table S1). All RNA samples underwent the NASBA reaction using the primers designed to amplify a fragment of the envelope gene, which is highly conserved among ZIKV isolates of the Asian genotype circulating in the Americas (Table S1). The products of NASBA reactions were then interrogated with the vsDz probe targeting the amplified fragment of the ZIKV genome. The intense color was observed only in the presence of ZIKV RNA, while no-target control or non-specific viral RNA triggered absorbance at the background level (blank sample, in the absence of RNA amplification product) (Figure 2). Remarkably, all tested ZIKV isolates responded positively to the test. All samples containing ZIKV amplicons turned green, while the samples containing NASBA mixtures obtained in the absence of viral RNA or presence of non-specific flaviviruses (DENV, WNV) stayed colorless (Figure 2, Inset), indicating a clear “yes/no” response. MR766, a strain representative of the African lineage, yielded positive signal despite thirteen mutations in the amplified sequence; only one such mutation was in the sensor-binding region yet, due to its location between the two binding arms, did not affect the formation of the Dz catalytic core (Figure S6).
Figure 2.
Selectivity of NASBA/vsDz test. Isolated RNA from different strains of ZIKV (indicated below the bar graph), DENV (type 2 and type 3) or WNV was used for NASBA reaction followed by vsDz reaction. The images of the sample tubes were taken and the absorbance at 420 nm was recorded 10–15 min after addition of H2O2. The data for three independent experiments were averaged, with standard deviations as error bars.
3.4. Detection limit of NASBA/vsDz test
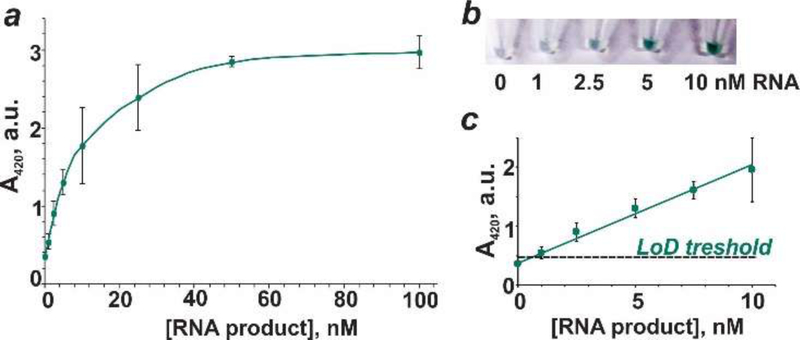
We determined the detection limit of NASBA product using vsDz probe. For this purpose, we used the product of NASBA reaction for ZIKV RNA (strain BR_SJRP1840) quantified with Qubit RNA HS assay. Different amounts of this stock solution were then mixed with the vsDz probe, and the absorbance of the samples was plotted as a function of RNA product concentration (Figures 3 and S7). The absorbance values increased with increased concentration of the amplified RNA and plateaued at ~40 nM RNA (Figure 3a). Visually, the analyte presence could be differentiated from its absence at as low as 2.5 nM amplicon concentration (Figure 3b). At the same time, the theoretical detection limit of ~1 nM was calculated based on the absorbance values using 3σ method, with the linear dynamic range to be 1–10 nM (Figure 3c).
Figure 3.
Absorbance for vsDz samples as a function of RNA concentration. (a) Serial dilutions of NASBA RNA product (0–100 nM) obtained using ZIKV RNA (strain BR_SJRP1840) were incubated with the vsDz probe, and the absorbance at 420 nm was measured upon addition of ABTS, hemin and H2O2. (b) Images of samples containing 0–10 nM RNA amplification product. (c) Linear dependence of absorbance at 420 nm on RNA concentration (0–10 nM). The data are average values of three independent experiments, with standard deviations as error bars.
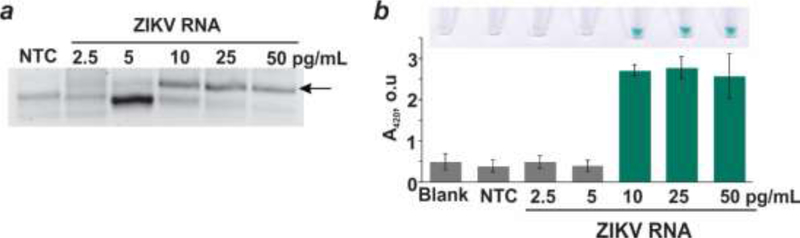
The concentration of ZIKV RNA in biological specimens (blood, urine, saliva) was reported to be from the femtomolar to subpicomolar range (~106-109 viral particles/mL) [41,42]. These values are beyond the capabilities of the vsDz probe for target amplification-free ZIKV detection. To circumvent this limitation, an isothermal NASBA reaction was employed for amplification of a targeted fragment of the viral genome. We determined the least amount of viral RNA that is required for the signal to be generated using the NASBA/vsDz test. Different amounts of ZIKV RNA (2.5–50 pg/mL) corresponding to viral count of (0.4–8)×106 copies/mL were used for NASBA reaction followed by vsDz test (Figure 4). When 10–50 pg/mL ZIKV RNA was used for NASBA reaction, the bands of similar intensity for the RNA product were observed, while very faint bands were seen in the case of 2.5 or 5 pg/mL viral RNA, which indicates inefficient amplification (Figure 4a). At the same time, only 1.7% or less of the NASBA reaction mixture with the starting amount of 10–50 pg/mL viral RNA was sufficient to trigger maximum absorbance values, with the intense green color to be clearly visible in these samples (Figure 4b). Therefore, based on the data obtained, it was concluded that at least 10 pg/mL (1.6×106 virions/mL) ZIKV RNA was needed for reliable and reproducible results of the NASBA/vsDz test. Thorough optimization of stage 1 of the test can potentially lower its detection limit.
Figure 4.
Detection limit of the NASBA/vsDz test. (a) Analysis of the products of NASBA reaction with different starting amount (2.5–50 pg/mL) of ZIKV RNA using 2% agarose gel electrophoresis. The bands corresponding to the amplification product are labeled with an arrow. NTC – no target control for NASBA reaction. (b) Absorbance of the samples containing the vsDz probe in the absence (blank) or presence of 1.7% NASBA reaction mixtures from panel (a). Inset. Images of the tubes with the correspondent samples.
4. Discussion
We proposed a test for viral pathogen detection with color change as a signal. The test consists of four steps: (0) pre-processing of the biological specimen (blood, serum etc.); (1) isothermal amplification of a highly conserved fragment of the pathogen’s genome, which eliminates the need of an expected thermocycler; (2) probing the amplified fragment with highly selective sDz sensors, which ensures accurate diagnosis of ZIKV even in the presence of closely related RNA from other flaviviruses; (3) signal generation due to the catalytic action of a G-quadruplex peroxidase release in response to the pathogen’s presence in the previous stage. The first stage of the test relies on isothermal amplification of viral RNA using the NASBA reaction. NASBA-based tests have been previously suggested for detection of viral infections [9,20,22–24,28]. For example, Pardee et al. reported a colorimetric detection of ZIKV RNA using isothermal RNA amplification combined with programmable toehold RNA switches and a CRISPR/Cas9-based module for ZIKV strain genotyping [9]. The color change depended on the activation of the lacZ gene by ZIKV RNA, which resulted in changing the color of the solution from yellow to purple. The test could detect clinically relevant concentrations of ZIKV RNA in 3 hr, but required the use of a multiprotein cell-free protein expression system for color generation. The NASBA/vsDz test developed in the present study required an enzyme mix only for the amplification stage, while the detection stage relied solely on protein-free components – catalytic DNA molecules were used instead.
Since the outbreaks of ZIKV infection in 2015–2016, several colorimetric tests for diagnostics of the infection have been reported [9,12,13,43–46]. The majority of the reported methods rely on reversetranscription loop-mediated amplification (RT-LAMP) of a fragment of ZIKV genome. This isothermal amplification technique is proven to be robust and efficient, but it produces a mixture of double-stranded DNA (dsDNA) product of different length. Such amplicons are difficult to interrogate with hybridization probes to enable genotype-specific detection, thus visual product detection relied on sequence-unspecific binding of a dye to dsDNA, or a pH indicator. In this case, any non-specific amplification would result in false-positives, since the selectivity of the tests depended solely on the primers (one-level recognition of the RNA target). Another isothermal method of nucleic acid amplification – recombinase polymerase amplification (RPA) [47] – has been suggested and is widely used in POC NAATs. Similar to LAMP, this methods uses DNA as a template, which imposes the need of an additional reverse transcription step for the conversion of viral RNA into a DNA template, and results in dsDNA as a product, unless asymmetric conditions are employed, which compromises the efficiency and increases time of the amplification stage.
The test reported here enabled detection of all tested ZIKV isolates including the isolates prevalent in the Americas. At the same time, RNA from closely related viruses (e.g. DENV and WNV) did not trigger the signal despite high homology between the viral genomes. High selectivity of the test relies on two-level recognition of viral RNA: (i) ZIKV-specific primers are used to amplify a fragment of viral genome; (ii) highly selective vsDz probe are designed to bind to and detect ZIKV-related nucleic acid targets. The high selectivity of split Dz probes has been previously demonstrated by distinguishing similar nucleic acids differing in as little as one nucleotide [32,34,38]. The discriminatory power of the probe is ensured by the split design. It allows for the shortening of the target-binding arm of one interrogating strand of the probe to ensure selective target recognition, while keeping the arm of another strand as long as needed for efficient target binding. The presence of mismatches in the hybrid between the target and the shorter probe strand compromises the hybrid formation, which prevents binding of both strands to the abutting positions of the target and, therefore, activation of the vsDz catalytic core. It is possible to uncouple the selectivity of the two test stages. This will allow for adapting the reported strategy for simultaneous detection of ZIKV and other flaviviruses (e.g. DENV). For example, the use of pan-flavivirus primers [48] would allow amplification of both ZIKV and DENV, while two vsDz probes, each targeting one virus, would produce color only in response to their fully complementary viral targets. In case of either ZIKV or DENV infection, the signal will be generated only for one of the probes. In the case of simultaneous ZIKV and DENV infection, which has been reported [49–52], both probes will exhibit high color intensity. A colorimetric test capable of detecting co-infection is beneficial, since such cases may require different treatment regimens and adjustment in the monitoring of viral spread.
In this proof-of-principle study, the total time for the NASBA/vsDz test was ~2 h: stage 1 – 60 min; stage 2 – 45 min, stage 3 – 15 min. At the same time, shortening of stages 1 and 2 to 45 and 30 min, respectively, can still result in the signal intense enough to differentiate between the absence and presence of the pathogen, especially at high viral load. Additional optimization experiments can improve the efficiency of NASBA reaction, thus shortening stage 1 and, potentially, improving the detection limit.
Currently, samples containing ~106 copies/mL viral RNA trigger maximum absorbance as a result of the NASBA/vsDa test. This is in agreement with the detection limits allowable by another NASBA-based test [9]. In addition, this correlates with viral RNA levels of 7×106–9×108 copies/mL in blood of symptomatic patients [19]. Even higher RNA levels were observed in urine or semen samples of ZIKV-positive donors [19,40,41]. At the same time, only 1.7% of the NASBA reaction mixture is used for the test with vsDz probe, which implies limitations on the starting material of viral RNA used in stage 1. Theoretically, if 50% of the NASBA reaction mixture is utilized for the subsequent stages of the test, the detection limit can be improved 50-fold. Unfortunately, in the current test format, the signal generation (stage 3 of the test) is inhibited by components of the NASBA buffer if more than 10% of the mixture is used (data not shown). Partial purification of the sample from the inhibitors of peroxidation reaction would contribute to better detection limit of the test.
Even though an ideal diagnostic test should not require any heating element and, therefore, should be efficient at ambient temperatures, NASBA/vsDz test required a thermostat for stage 1 (41oC) and for stage 2 (50 oC). Thermostats are, however, an inexpensive piece of equipment, which can be afforded by households or doctor’s offices. Despite that, minimization of heating is preferable in a test if it is to be used in rural areas. Eliminating stage 2 of the detection scheme would enable the test to be run at 41 oC, which can be provided by a simple battery-powered heating block. Alternatively, the use of a self-contained battery-powered device to perform the test is beneficial to implement this strategy in the laboratory-free settings.
In the developed test, the signal can be monitored with the naked eye with no instrumentation needed, which is advantageous for POC applications. The test serves to screen the patients for possible infection, and so requires only “yes/no” response: highly reproducible bright green color triggered in response to ZIKV infection can be reliably distinguished from a colorless solution when no infection occurs, even in hands of a first-time user. This test offers a promise to make diagnostics of viral infections more affordable for patients in terms of turnaround time, among other advantages.
4. Conclusions
We have reported on a strategy for visual pathogen detection based on a combination of isothermal amplification of a viral genome fragment using NASBA reaction and a color-generating split deoxyribozyme technology. We employed the strategy to detect ZIKV RNA. The use of pan-ZIKV primers for the NASBA reaction enabled amplification of highly conserved fragments of the viral genome, which open the perspective to use the same amplification conditions or even the same product of amplification in combination with strain- or lineage-specific vsDz probes for simultaneous detection of several ZIKV strains. The split approach enables efficient unwinding of a highly structured RNA amplification product and ensures high selectivity for target recognition. The advantages of the proposed NASBA/vsDz test include the following: (1) isothermal amplification of a targeted fragment of the viral genome; (2) color change as a signal, which can be detected by the naked eye without instrumentation; (3) highly selective target recognition since the vsDz probe can distinguish between closely related pathogens. It opens a possibility to differentiate closely related flaviviruses or even strains of ZIKV, which can be beneficial for case management and surveillance of virus distribution; (4) straightforward design of sDz sensor – minimal sequence adjustments and test optimization are required to adapt the platform for the detection of a new viral isolate or another virus. The reported strategy has a potential to advance diagnostics of viral infections, as well as improve monitoring and surveillance of the pathogen transmission.
Supplementary Material
Highlights.
A three-stage colorimetric test for the naked-eye detection of Zika virus.
Isothermal amplification combined with a label-free split deoxyribozyme probe.
Differentiation of Zika virus from closely related dengue and West Nile viruses.
Total test time of 2 hours without expensive instrumentation.
Detects clinically relevant concentrations of viral RNA.
Acknowledgments
This work was supported by the Florida Department of Health, Biomedical Research Program (grant 7ZK33), National Institute of Allergy and Infectious Diseases of the National Institutes of Health (grants R21AI123876 and R01AI110692).
Footnotes
Notes
The authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yoo SM; Lee SY Trends Biotechnol, 2016, 34, 7–19. [DOI] [PubMed] [Google Scholar]
- 2.Vashist SK; Luong JHT In Handbook of Immunoassay Technologies. Approaches, Performances, and Applications; Vashist SK and Luong JHT, Eds.; Academic Press: Cambridge, 2018; pp 97–127. [Google Scholar]
- 3.Rabe IB; Staples E; Villanueva J; Hummel KB; Johnson JA; Rose L; Hills S; Wasley A; Fischer M; Powers AM Morb. Mortal Wkly Rep, 2016, 65, 543–546. [DOI] [PubMed] [Google Scholar]
- 4.Felix AC; Souza NCS; Figueiredo WM; Costa AA; Inenami M; da Silva RMG; Levi JE; Pannuti CS; Romano CM J. Med Virol, 2017, 89, 1477–1479. [DOI] [PubMed] [Google Scholar]
- 5.Balmaseda A; Zambrana JV; Garcia N; Saborio S; Elizondo D; Mercado JC; Gonzalez K; Cerpas C; Nunez A; Corti D; Waggoner JJ; Kuan G; Burger-Calderon R; Harris EJ Clin. Microbiol 2018, 56, e01785–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mothershed EA; Whitney AM Clin. Chim. Acta, 2006, 363, 206–220. [DOI] [PubMed] [Google Scholar]
- 7.Pöhlmann C; Dieser I; Sprinzl M Analyst, 2014, 139, 1063–1071. [DOI] [PubMed] [Google Scholar]
- 8.Tian L; Sato T; Niwa K; Kawase M; Tanner AC; Takahashi N. Biomed. Res. Int, 2014, 2014, 180323–18332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pardee K; Gree AA; Takahashi MK; Braff D; Lambert G; Lee JW; Ferrante T; Ma D; Donghia N; Fan M; Daringer NM; Bosch I, Dudley DM; O’Connor DH; Gehrke L; Collins JJ Cell, 2016, 165, 1255–1266. [DOI] [PubMed] [Google Scholar]
- 10.Verma MS; Rogowski JL; Jones L; Gu FX Biotechnology Advances, 2015, 33, 666–680. [DOI] [PubMed] [Google Scholar]
- 11.Oh SJ; Park BH; Jung JH; Choi G; Lee DC; Kim DH; Seo TS Biosens. Bioelectron, 2016, 75, 293–300. [DOI] [PubMed] [Google Scholar]
- 12.Calvert AE; Biggerstaff BJ; Tanner NA; Lauterbach M; Lanciotti RS PLoS ONE, 2017, 12, e0185340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yaren O; Alto BW; Gangodkar PV; Ranade SR; Patil KN; Bradley KM; Yang Z; Phadke N; Benner SA BMC Infect. Dis, 2017, 17, 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy S; Mohd-Naim NF; Safavieh M; Ahmed MU ACS Sens, 2017, 2, 1713–1720. [DOI] [PubMed] [Google Scholar]
- 15.Batule BS; Kim SU; Mun H; Choi C; Shim W-B; Kim M-G J. Agric. Food Chem, 2018, 66, 3003–3008. [DOI] [PubMed] [Google Scholar]
- 16.James AS; Todd S; Pollak NM; Marsh GA; Macdonald J Virol. J, 2018, 15, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mabey D; Peeling RW; Ustianowski A; Perkins MD Nature Rev, 2004, 2, 231–240. [DOI] [PubMed] [Google Scholar]
- 18.Drain PK; Hyle EP; Noubary F; Freedberg KA; Wilson D; Bishai WR; Rodriguez W; Bassett IV Lancet Infect. Dis, 2014, 14, 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musso D; Gubler DJ Clin. Microbiol. Rev, 2016, 29, 487–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lunel F; Cresta P; Vitour D; Payan C; Dumont B; Frangeul L; Reboul D; Brault C; Piette JC; Huraux JM Hepatology, 1999, 29, 528–535. [DOI] [PubMed] [Google Scholar]
- 21.Mahony JB; Song X; Chong S; Faught M; Salonga T; Kapala JJ Clin. Microbiol, 2001, 39, 1429–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jean J; Blais B; Darveau A; Fliss I Appl. Environ. Microbiol, 2001, 67, 5593–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jean J; Blais B; Darveau A; Fliss IJ Virol. Methods, 2002, 105, 123–132. [DOI] [PubMed] [Google Scholar]
- 24.Greene SR; Moe C; Jaykus L-A; Cronin M; Grosso L; van Aarle PJ Virol. Methods, 2003, 108, 123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loens K; Ieven M; Ursi D; Beck T; Overdijk M; Sillekens P; Goossens HJ Clin. Microbiol, 2003, 41, 4448–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boulet GA; Micalessi IM; Horvath CAJ; Benoy IH; Depuydt CE; Bogers JJ J. Clin. Microbiol, 2010, 48, 2524–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mollasalehi H; Yazdanparast R Biosens. Bioelectron, 2013, 47, 231–236. [DOI] [PubMed] [Google Scholar]
- 28.Lu X; Shi X; Wu G; Wu T; Liu R; Wang Y Sci. Rep, 2017, 7, 44211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pavlov V; Xiao Y; Gill R; Dishon A; Kotler M; Willner I Anal. Chem, 2004, 76, 2152–2156. [DOI] [PubMed] [Google Scholar]
- 30.Deiman B; van Aarle P; Sillekens P Mol. Biotechnol, 2002, 20, 163–179. [DOI] [PubMed] [Google Scholar]
- 31.Santoro SW; Joyce GF Proc. Natl. Acad. Sci. USA, 1997, 94, 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolpashchikov DM Chembiochem, 2007, 8, 2039–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerasimova YV; Kolpashchikov DM Chem. Biol, 2010, 17, 104–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerasimova YV; Cornett E; Kolpashchikov DM Chembiochem, 2010, 11, 811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mokany E; Bone SM; Young PE; Doan TB; Todd AV J. Am. Chem. Soc, 2010, 132, 1051–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerasimova YV; Cornett EM; Edwards E; Su X; Rohde KH; Kolpashchikov DM Chembiochem, 2013, 14, 2087–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerasimova YV; Kolpashchikov DM Angew. Chem. Int. Ed. Engl, 2013, 52, 10586–10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerasimova YV; Yakovchuk P; Dedkova LM; Hecht SM; Kolpashchikov DM RNA, 2015, 21, 1834–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dey S; Roy P; Nandy A; Basak SC; Das S MOL2NET, 2017, 3, DOI: 10.3390/mol2net-0304966. [DOI] [Google Scholar]
- 40.Logan IS Zoological Research, 2016, 37; 110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gourinat AC; O’Connor O; Calvez E; Goarant C; Dupont-Rouzeyrol M Emerging infectious diseases, 2015, 21, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mansuy JM; Dutertre M; Mengelle C; Fourcade C; Marchu B; Delobel P; Izopet J; MartinBlondel G Lancet Infect. Dis, 2016, 16, 405. [DOI] [PubMed] [Google Scholar]
- 43.Song J; Mauk MG; Hackett BA; Cherry S; Bau HH; Liu C Anal. Chem, 2016, 88, 7289–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee D; Shin Y; Chung S; Hwang KS; Toon DS; Lee JH Anal. Chem, 2016, 88, 12272–12278. [DOI] [PubMed] [Google Scholar]
- 45.Kaarj K; Akarapipad P; Yoon J-Y Sci. Rep, 2018, 8, 12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang X; Loeb JC; Manzanas C; Lednicky JA; Fan ZH Angew. Chem. Int. Ed, 2018, 10.1002/anie.201809993 [DOI] [PubMed] [Google Scholar]
- 47.Lobato IM; O’Sullivan CK Trends Anal. Chem, 2018, 98, 19–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vina-Rodrigues A; Sachse K; Ziegler U; Chaintoutis SC; Keller M; Groschup MH; Eiden M Biomed. Res. Int, 2017, 2017, 4248756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cabral-Castro MJ; Cavalcanti MG; Peralta RH; Peralta JM J. Clin. Virol, 2016, 82, 108–111. [DOI] [PubMed] [Google Scholar]
- 50.Pessôa R; Patriota JV; de Souza LM; Felix AC; Mamede N; Sanabani SS Medicine, 2016, 95, e3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iovine NM; Lednicky J; Cherabuddi K; Crooke H; White SK; Loeb JC; Cella E; Ciccozzi M; Salemi M; Morris JG Jr. Clin. Infect. Dis, 2016, doi: 10.1093/cid/ciw667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carrillo-Hernandez MY; Ruiz-Saenz J; Villamizar LJ.; Gomez-Rangel SY; MartinezGutierrez M J. BMC Infect Dis, 2018, 18, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.